Abstract
Lipids play a critical role in cell membrane integrity, signaling, and energy storage. However, in-depth structural characterization of lipids is still challenging and not routinely possible in lipidomics experiments. Techniques such as collision-induced dissociation (CID) tandem mass spectrometry (MS/MS), ion mobility (IM) spectrometry, and ultrahigh-performance liquid chromatography are not yet capable of fully characterizing double-bond and sn-chain position of lipids in a high-throughput manner. Herein, we report on the ability to structurally characterize lipids using large-area triboelectric nanogenerators (TENG) coupled with time-aligned parallel (TAP) fragmentation IM-MS analysis. Gas-phase lipid epoxidation during TENG ionization, coupled to mobility-resolved MS3 via TAP IM-MS, enabled the acquisition of detailed information on the presence and position of lipid C═C double bonds, the fatty acyl sn-chain position and composition, and the cis/trans geometrical C═C isomerism. The proposed methodology proved useful for the shotgun lipidomics analysis of lipid extracts from biological samples, enabling the detailed annotation of numerous lipid isobars.
Graphical Abstract
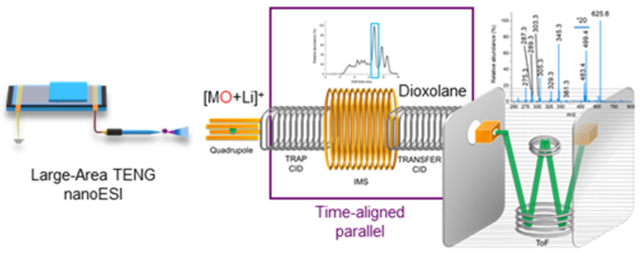
INTRODUCTION
Glycerophospholipids (GPs) play multiple physiological roles, including the preservation of cell membrane structure, energy storage, and cellular signaling.1–3 Lipid imbalances have been correlated to disease pathophysiology, including chronic inflammation, allergy, autoimmunity, and cancer, among others.4,5 Characterization of lipids only at their basic compositional level, however, is insufficient to robustly establish their biological role. Therefore, tools to more comprehensively annotate lipids are greatly needed.
The fields of mass spectrometry (MS) and lipidomics have seen tremendous growth in the last decade.6,7 Technical improvements in MS instrumentation now enable high-throughput lipid analysis via shotgun or chromatographic approaches.8–12 Combination of high-resolution accurate mass measurements with tandem MS experiments readily reveals a multitude of GP structural features, such as lipid class (e.g., phosphatidylcholine or PC), number of carbon atoms and double bonds (e.g. PC(36:1)), and fatty acyl (FA) chain composition (e.g., PC 18:1_18:0), leading to better, but still incomplete lipid annotations. Mass spectral overlaps and inconclusive fragmentation data hamper chain sn-position assignment, C═C bond location pinpointing, and C═C bond stereochemistry elucidation.13,14
The quintessential separation technique for lipid analysis is liquid chromatography (LC) coupled to tandem MS.15,16 Its specificity can be leveraged for resolving lipid regioisomers, but it typically requires long separation times or specialized stationary phases.17,18 Shotgun lipidomics approaches enable higher-throughput analysis, but may suffer from bias towards high abundance species due to ionization suppression.19 Ion mobility spectrometry coupled to MS (IM-MS) has enabled lipid separations by class and subclass20 and, under certain experimental conditions, may also allow the resolution of lipid isobars.21 When direct infusion (DI) analysis and IM-MS are combined, rapid lipidomic experiments can be carried out in a matter of seconds.22
A number of creative MS approaches have been proposed for C═C bond location pinpointing and/or chain sn-position assignment. Gross et al. were one of the first to show that unique fragments enabling C═C bond location pinpointing can be produced by collision-induced dissociation (CID).23 Later, Hsu et al. demonstrated how lithiated lipid adducts and low-energy CID could distinguish various C═C bond locations, and even chain sn-position.24,25 Although innovative, those methods produced complex tandem mass spectra, making lipid characterization less straightforward. In more recent times, a number of new approaches to C═C bond location pinpointing have been reported. A subset of these involve gas-phase reactions through ozone-induced fragmentation (OzID),26 ultraviolet photodissociation (UVPD),27 charge transfer dissociation (CTD),28 radical-directed dissociation (RDD),29 and electron impact excitation of ions from organics (EIEIO).30 Other approaches to C═C bond location pinpointing leverage derivatization and/or ion source electrochemistry. These include Paternó-Büchi (PB) chemistry,31 ozonolysis,32 plasma-induced oxidation,33 meta-chloroperoxybenzoic acid epoxidation,34 and nanoelectrospray-chloride electrochemistry.35 Most of these methodologies have been mainly used to locate C═C bonds, with OzID,36 UVPD,27 and PB37 chemistry used also for sn-chain position assignment. Downsides of some of these approaches include the need for highly customized MS instrumentation, specific reagents, or long acquisition times, limiting their widespread adoption.
In recent work, our team reported on the formation of transient corona discharges that enable C═C bond location pinpointing in unsaturated fatty acids using large-area triboelectric nanogenerator (TENG) nanoelectrospray (nanoESI) MS.38 The unique control over the number of charges supplied to the ion source in TENG MS makes on-demand, reproducible analyte gas-phase oxidation possible. Following mass selection of the epoxidized precursor ions, the diagnostic fragment ions formed by CID are separated by 16 Da, with their specific m/z values unveiling the specific double-bond position(s). Here, we report on a shotgun lipidomics strategy that harnesses TENG gas-phase epoxidation of unsaturated GP, followed by fragmentation of the corresponding cationized or anionized species by IM-MS using time-aligned parallel (TAP) fragmentation.39 This approach does not require specific reagents or sample pretreatment, nor it requires any sort of complex instrument modifications. This method was applied to various model analytes and GP from a complex egg phospholipid extract, enabling in-depth structural lipid annotation.
EXPERIMENTAL SECTION
Materials.
All GP chemical standards and the egg PC extract were purchased from Avanti Polar Lipids (Alabaster, Alabama). Optima LC-MS-grade acetonitrile (ACN), acetone, and methanol (MeOH) were acquired from Fisher Chemical (Suwanee, GA). Ultrapure 18.2 MΩ·cm deionized water (Barnstead Nanopure Diamond, Van Nuys, CA) was used to prepare all solutions. Ammonium acetate (NH4OAc) was purchased from Fisher Chemical (Suwanee, GA), lithium acetate (LiOAc), sodium acetate (NaOAc), and hydrogen peroxide were purchased from Sigma-Aldrich (Saint Louis, MO).
TENG Analysis.
Experiments were carried out using a large-area sliding freestanding TENG that powered a nanoESI ion source identical to that described previously.40 The TENG sliding electrode was made of Nylon (12 × 12 cm), whereas the two stationary electrodes were made of fluorinated ethylene propylene (FEP, 24 × 12 cm). NanoESI glass emitter tips (Econo 12, New Objective, i.d. 0.69 mm, o.d. 1.2 mm, part no. ECONO 12N) were employed in all cases. The nanoESI emitters were mounted on an x, y, z manual linear stage (Thorlabs, Newton, NJ) to control their position with respect to the mass spectrometer inlet. Unless specified otherwise, the emitter tip was held 8–10 mm away from the inlet. A 0.25 mm diameter platinum wire was inserted into the emitters to provide electric contact with the sample solution. The device and operational principle of large-area TENG have been previously described.41 A linear motor (LinMot USA, Inc.) was used to operate the movable TENG slider electrode in all experiments (0.25 Hz). The TENG device was operated asynchronously with the mass spectrometer used for detection, but synchronization is also possible.38
Ion Mobility–Mass Spectrometry TAP Analysis.
All experiments were conducted on a Synapt G2-S from Waters (Wilmslow, U.K.). The instrument was operated in resolution mode (ToF resolving power, 20 000). The inlet settings were as follows: cone voltage, 0 V; source offset, 0 V; source temperature, 120 °C. Two different sets of parameters were used for TAP analysis:
Positive-ion mode: LM resolution, 10; trap collision energy, 35 eV; transfer collision energy, 50 eV; trap gas flow, 3 mL min−1; helium gas flow, 180 mL min−1; IMS gas flow (nitrogen), 90 mL min−1; trap T-wave, 311 ms−1; IM T-wave, 650 m s−1; transfer T-wave, 175 m s−1.
Negative-ion mode: LM resolution, 10; trap collision energy, 50 eV; transfer collision energy, 30 eV; trap gas flow, 3 mL min−1; helium gas flow, 180 mL min−1; IM gas flow (nitrogen), 90 mL min−1; trap T-wave, 311 ms−1; IM T-wave, 650 m s−1; transfer T-wave, 175 m s−1.
ATD and mass spectra were extracted using MassLynx 4.1, and the data were analyzed and plotted in OriginPro 9.1.
RESULTS AND DISCUSSION
TENG-TAP IM-MS Analysis of Model PC Analytes.
A schematic of the TENG ion source coupled to the IM-MS system and the various steps involved in TAP MS/MS lipid analysis are shown in Figure 1. To optimize this experiment, various electrospray conditions were investigated with the goal of maximizing epoxidation yields for unsaturated GP while minimizing side products. Various solvent systems and additives were evaluated using 1-oleoyl-2-hydroxy-sn-glycero-3-phosphocholine (lysoPC(18:1(9Z))) as the test analyte (Figure S1). Negative-ion mode results indicated that the solvent combination that maximized the yield of [MO + OAc]− while minimizing the yields of [M-CH3 + OAc]−, [MO-2H + OAc]−, and [M+2O + OAc]− was acetone/water/methanol (75:12.5:12.5) with 25 mM NH4OAc added (we note the neutral lipid as “M” and “MO” for the oxidized lipid; see additional discussion in Figure S1). Acetone photoexcitation has been previously reported as being a key step in the formation of oxetane ions during Paternò-Büchi reactions, enabling the detection of two diagnostic fragments ions that localize double-bond positions following tandem MS experiments.42 Here, lipid C═C bond oxidation occurs in the gas phase and is promoted by radical reactions within the corona discharge-generated plasma. Although oxetanes or its corresponding diagnostic fragments were not detected in our experiments, acetone still seems to play a key role, likely by boosting the population of radical species in the gas phase. Future experiments will likely be necessary to unveil the exact role of acetone in large-area TENG nanoESI lipid oxidation. Further discussion is provided in Figure S1.
Figure 1.
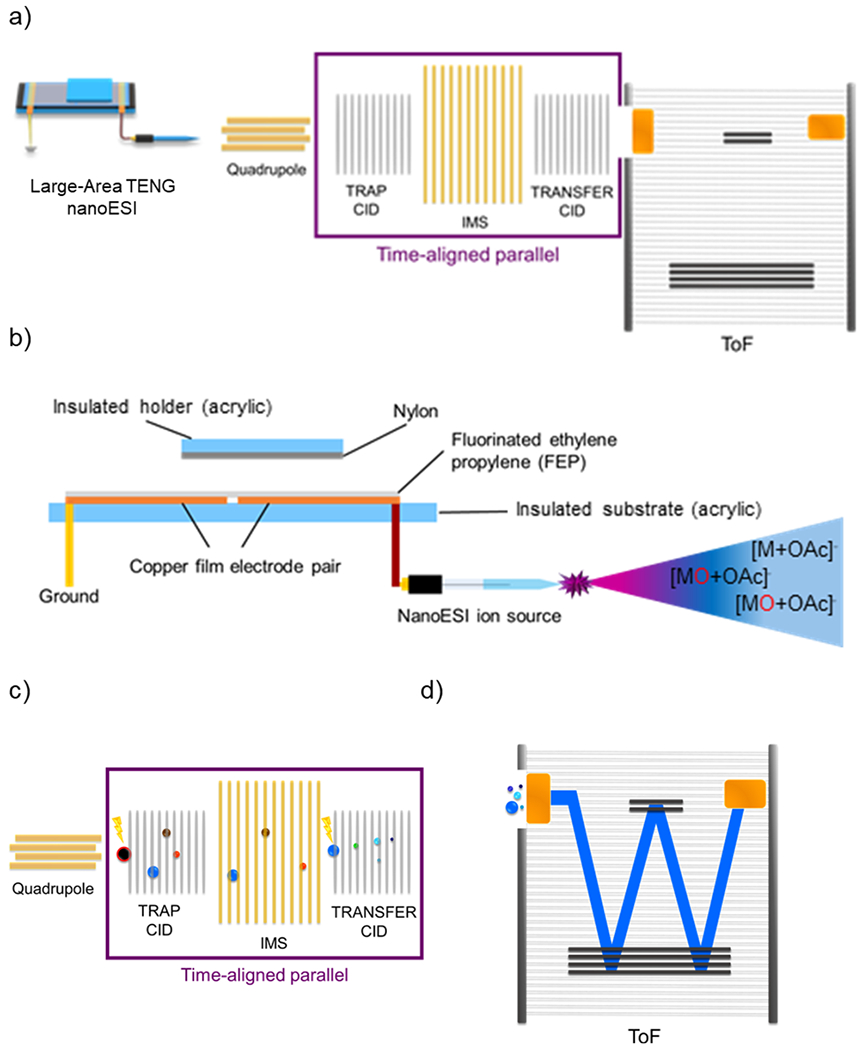
(a) Schematic representation of the TENG TAP fragmentation IM-MS instrumentation used in the present work. (b) Schematic of the large-area TENG ion source. Displacement of the TENG sliding electrode generates a transient corona discharge accompanied by electrospray, promoting unsaturated lipid epoxidation. (c) Schematic of the most relevant events in TAP fragmentation IM-MS experiments, enabling mobility-resolved MS3 analysis. (d) Ion packages separated by ion mobility and fragmented in the trap collision cell are monitored by time-of-flight (ToF) MS.
Solvent optimization experiments in the positive-ion mode used the same acetone/water/methanol solvent system as in the negative-ion mode. A Li salt was chosen for cationization, following reports by Hsu et al. that CID on Li-cationized lipid precursor ions enable the determination of sn1/sn2 regioisomerism.25 The main variable optimized was the concentration of LiOAc added to the acetone/methanol/water solvent system. Results showed that a 5 mM LiOAc concentration was sufficient to produce optimum yields of [MO + Li]+ ions and was therefore selected for all further experiments. Concentrations higher than 5 mM resulted in notable signal suppression and were avoided. Overall, epoxidation yields in negative-ion mode were 30% higher than in positive-ion mode, suggesting that high sensitivity measurements would be better pursued in the former.
Once electrospray conditions were optimized, negative-ion mode TENG TAP IM-MS analysis of the model lipid 1,2-dioleyl-sn-glycero-3-phosphocholine (PC(18:1(9Z)/18:1-(9Z))) was undertaken. As shown in Figure 2a, TENG MS led to the detection of the [M + OAc]− species at m/z 844.6, with the corresponding [MO + OAc]− species at m/z 860.6. The epoxidation yield observed was 5.5% of the precursor ion for a single TENG pulse consuming ~3 nL of sample in 2 s. When DC-nanoESI was attempted for analysis of this model analyte at a voltage of 1.5 kV, epoxidation was not observed, as no corona discharge was generated (Figure S2).
Figure 2.
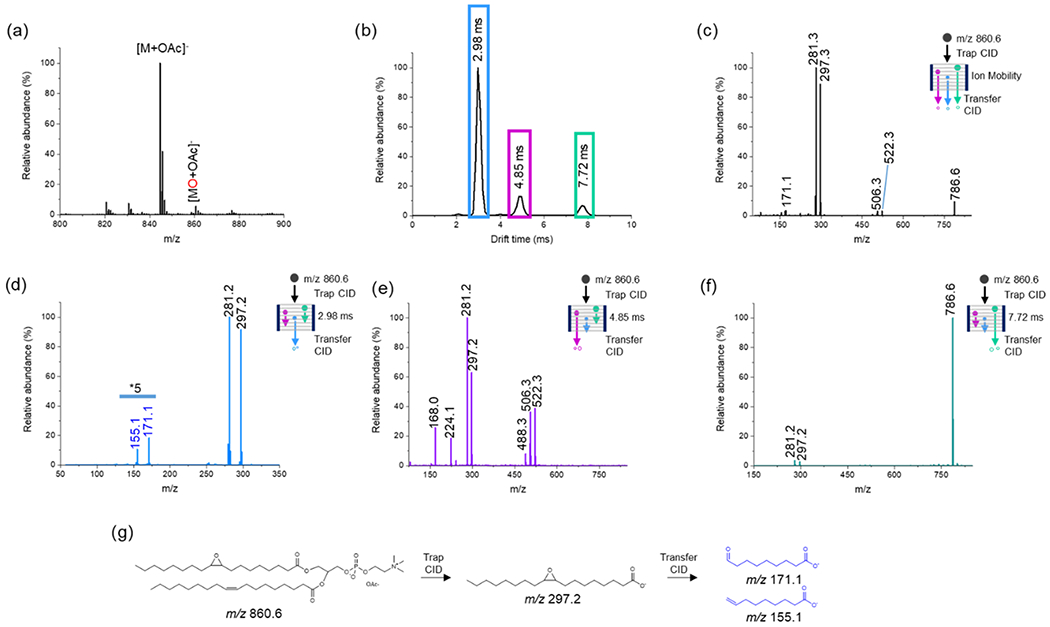
Negative-ion mode analysis of 20 μM PC(18:1(9Z)/18:1(9Z)) in acetone/water/methanol (75:12.5:12.5) with 25 mM NH4OAC added. (a) TENG-MS analysis showing the [M + OAc]− epoxidation product. (b) IM total ion ATD following selection of the [MO + OAc]− ion (m/z 860.6) in the quadrupole analyzer and trap CID. The blue, purple, and green boxes depict the various individual (or groups) of product ions produced by trap CID; their TAP product ion spectra are shown in (d–f). (c) Averaged TAP mass spectrum for the full arrival time distribution (ATD) shown in (b). (d) TAP transfer CID spectrum of the group of fragment ions outlined with a blue box in (b). The fragment ions at m/z 171.1 and 155.1 originating from m/z 297.2 indicate the Δ9 double-bond position. (e) TAP transfer CID spectrum of the group of fragment ions outlined with the purple box in (b). The fragment ion at m/z 522.3 corresponds to the loss of the sn1 or sn2 acyl chain as a ketene (RCH═C═O) and loss of CH3 from the head group with the remaining acyl chain oxidized. The fragment ion at m/z 506.3 corresponds to the loss of the sn1 or sn2 acyl chain as a ketene (RCH═C═O) and loss of CH3 from the head group with the remaining acyl chain unoxidized. The fragment ion at m/z 488.3 corresponds to the neutral loss of an RCOOH from sn1 or sn2 with concurrent loss of CH3. (f) TAP transfer CID spectrum of the fragment ion at 7.72 ms fragment with m/z 786.6, [MO + OAc-74]− (green box). The m/z 281.2 and 297.2 fragment ions correspond to the unoxidized and oxidized fatty acyl chains. (g) Fragmentation pathway for the formation of the diagnostic fragments at m/z 171.1 and 155.1.
TAP analysis utilizes both the trap and transfer collision cells to achieve a mobility-resolved pseudo-MS3 experiment (Figure 1c). To study the behavior of model analytes in TENG TAP IM-MS, the PC(18:1(9Z)/18:1(9Z)) epoxidized ion at m/z 860.6 was first isolated in the quadrupole mass analyzer and fragmented via CID in the trap cell (Figure 1c). To observe the fragmentation patterns of the evaluated lipids, different transfer collision cell energies were evaluated for PC(18:1(9Z)/18:1(9Z)) in both negative-and positive-ion modes (Figure S3). The main groups of ions resolved in the IM stage are shown in Figure 2b, highlighted with blue, purple, and green boxes. CID of these ion groups in the transfer cell yielded second-generation fragment ions (Figure 2c). The prominent m/z 297.2 fragment ion was assigned to the epoxidized [C18H33O3]− 18:1 fatty acyl chain, whereas the m/z 281.2 fragment was assigned to the nonepoxidized equivalent.
Fragmentation of the ion group outlined with the blue box in Figure 2b led to information regarding double-bond position (Figure 2d). The fragment ions at m/z 155.1 and 171.1 originated from the primary fragment at m/z 297.2. These diagnostic ions pointed at the double-bond position in the ninth carbon, as expected. The fragment-ion group within the 4.85 ms IM region (purple box in Figure 2b) yielded numerous fragments (Figure 2e). Interestingly, this group of ions was actually composed of several unresolved species with slightly different ion mobilities. These ions informed correctly about the expected acyl chain composition, but no information regarding sn1/sn2-chain position was obtained. A detailed analysis of the origin of these product ions is given in Figure S4. The TAP mass spectrum corresponding to the 7.72 ms IM region (Figure 2f) was not very informative, yielding only low abundance ions at m/z 281.2 and 297.2 originating from the nonepoxidated and epoxidated 18:1 acyl chain fragments. The m/z 786.6 ion resulted from the demethylation and acetate loss (CH3+ + CH3COO−, −74 Da) from the precursor ion. A simplified reaction scheme for the m/z 860.6 TAP fragmentation process is provided in Figure 2g.
Positive-ion-mode TENG TAP IM-MS experiments were conducted to establish how results compared to negative-ion mode. Because under atmospheric pressure conditions the air breakdown voltage for corona discharges is higher in positive mode than in negative mode, the observed unsaturated lipid epoxidation yields (~3.8%) were lower (Figure 3a). However, these lower yields were offset by the gain in structural information when proper cationization was implemented. Although sodiated adducts have been successfully used to yield structural information with Paternò–Büchi or UV photodissociation approaches,27,37 those adducts, and the corresponding epoxidated species, produced rather uninformative spectra by TENG TAP IM-MS (Figure S5a,b). Typically, protonated PC lipids are selected for analysis and quantitation. Castro-Perez et al., for example, have shown that TAP experiments of lipid protonated ions could provide fatty acyl and double-bond position pinpointing information.39 However, at the lower concentrations evaluated here (20 μM) compared to previous work (100 μMS), sensitivity was insufficient to detect any diagnostic ions. As an alternative to protonated or sodiated ions, lithium cationization has been used for CID tandem MS,25 prompting us to further investigate this approach. Results showed that abundant diagnostic fragments were produced from [MO + Li]+ ions (Figure S5c) with as little as one TENG pulse (60 fmol, Figure S6a). Similar results were obtained in negative-ion mode (Figure S6b). As expected, averaging of more than one pulse improved signal-to-noise (SNR) ratios in all cases.
Figure 3.
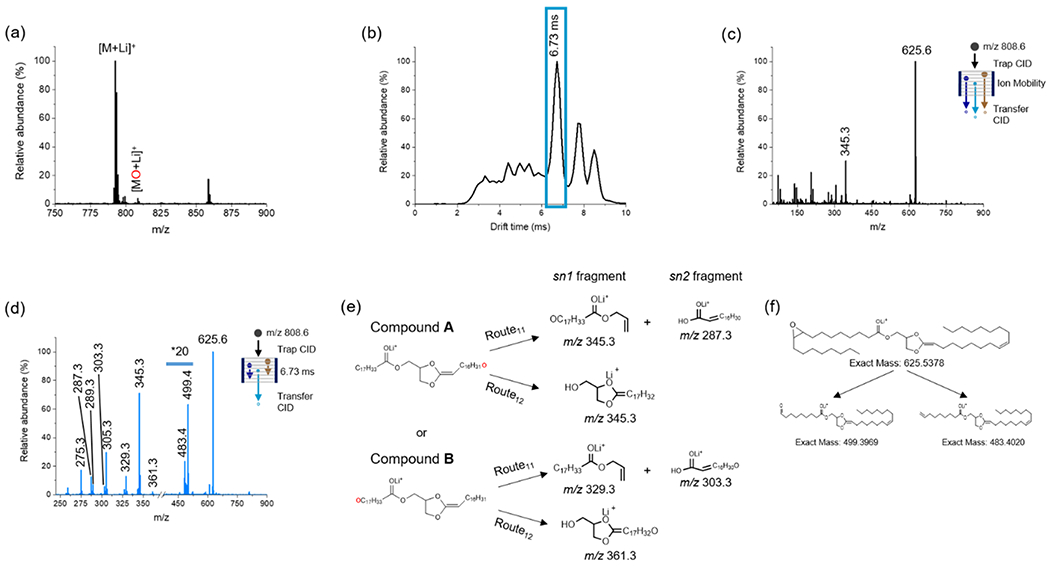
Positive-ion mode analysis of 20 μM PC(18:1(9Z)/18:1(9Z)) in acetone/water/methanol (75:12.5:12.5) with 5 mM LiOAc added. (a) TENG-MS analysis showing both the [M + Li]+ and epoxidated [MO + Li]+ions. (b) Total ion ATD following selection and trap CID of the [MO + Li]+ ion (m/z 808.6). The blue box represents the region corresponding to the dioxolane fragment ion at m/z 625.6. (c) Averaged TAP mass spectrum across the full ATD. (d) TAP transfer CID spectrum of fragment ions outlined with the blue box in (b). (e) Fragmentation pathways for the two possible dioxolane ions derived from [MO + Li]+. Generic pathways for lipids with other acyl chain lengths are shown in Figure S7. (f) Proposed structures for diagnostic fragments species detected at m/z 499.4 and 483.4 indicating the Δ9 double-bond position.
The arrival time distribution (ATD) observed in positive-ion mode was much more complex than in negative-ion mode (Figure 3b), with a maximum at 6.73 ms corresponding to the fragment ion at m/z 625.6. Averaging of this ATD showed a TAP mass spectrum with salient ions at m/z 345.3 and 625.6 (Figure 3c). The m/z 625.6 ion was assigned to the N(CH3)3 + C2H5PO4 neutral loss from the PC head group, yielding [MO + Li-183]+. This species contains a five-membered dioxolane ring that has been reported to form through two possible routes.43 These can involve five-member (Route 1, Figure S7) or six-member (Route 2, Figure S7) rings. Our own results suggested that the m/z 625.6 ion was formed through Route 1, as fragments were observed at m/z 287.3, 303.3, 329.3 345.3, and 361.3 (Figure 3d), matching Route11 and Route12 (Figure 3e), and in full agreement with the literature.27,44
Further fragmentation of the [MO + Li-183]+ ion yielded fragments at m/z 483.4 and 499.4 that successfully informed on the C═C double bond at the Δ9 position. These two ions, separated by 16 Da, correspond to aldehyde and ethylene fragments (Figure 3f). Testing of an alternate PC(36:2) positional isomer with a Δ6 cis double bond (Figure S8) showed the formation of two different diagnostic fragment ions at m/z 457.4 and 441.1, consistent with the expected fragmentation for the Δ6 position. These results suggest that TENG TAP IM-MS provides a robust means of determining double-bond position, avoiding higher stages of MS analysis (MS4,5),45 or the need for monitoring complex allylic or vinillic fragment ion series.39
The major ion at m/z 345.3 observed in Figure 3c,d is an example of a regioisomer-specific species that allows sn1/sn2 chain position elucidation. Its likely origin is from species A, the dioxolane fragment ion with the epoxidized double bond in the sn1 chain, through Route11 and not Route12 (Figure 3e). This fragmentation route proceeds by loss of the sn2 18:1 chain as an α,β-unsaturated fatty acid (C15H30CH═CHCO2H). The complementary fragment ion produced by Route11, [C15H30CH═CHCO2H + Li]+ at m/z 287.3, is produced by localization of the Li+ cation on the opposite side of the cleavage site (Figure 3e). The presence of a lower-abundance ion at m/z 329.3 and two minor ions at m/z 361.3 and 303.3 suggested the formation of a less abundant sn2 epoxidation product B. Fragmentation of the [M + Li]+ ion at m/z 792.6 yielded three fragments at m/z 287.3, 329.3, and 345.3 (Figure S9), confirming the composition of the fatty acid chains as 18:1/18:1. Analysis of two different PC lipids with uneven acyl chains, PC(16:0/18:0) and PC(18:0/16:0), yielded the expected diagnostic fragments, enabling the correct assignment of sn1/sn2 chain position, as discussed in Figure S10.
TENG/TAP Analysis of Other Glycerophospholipid Subclasses.
To examine further applicability of TENG TAP IM-MS beyond PC lipids, experiments with phosphatidic acids (PA), phosphatidylethanolamines (PE), and phosphatidylserines (PS) were conducted. As shown in Figure 4, comprehensive structural analysis was possible for these subclasses, with negative-ion mode providing double-bond position information with higher sensitivity (Figure 4a–c) and positive-ion mode enabling both C═C bond location pinpointing and sn-position elucidation (Figure 4d).
Figure 4.
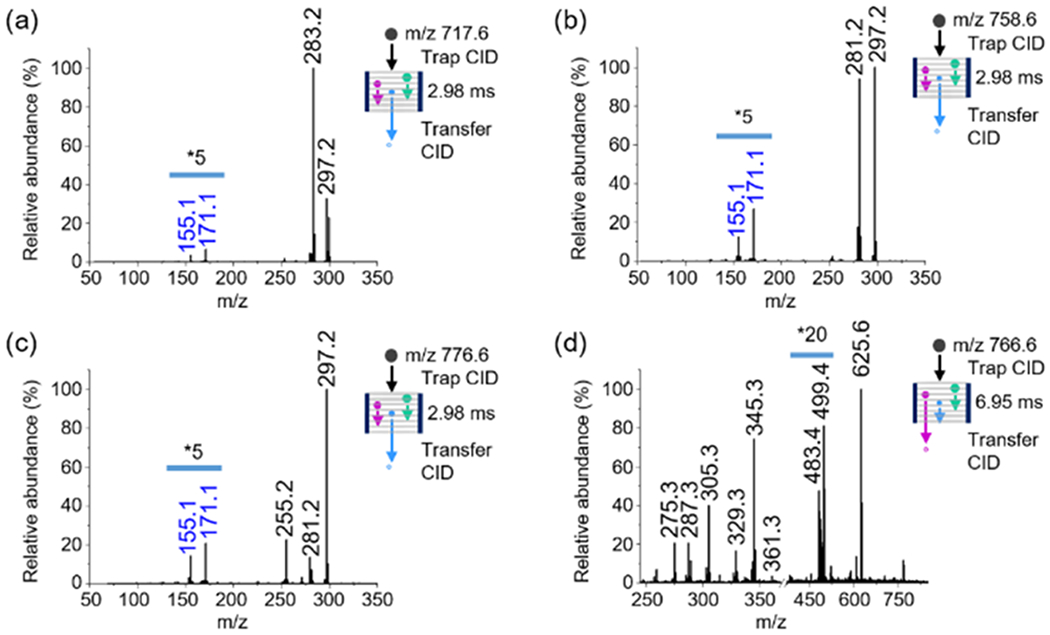
TENG TAP IM-MS analysis of different GP. (a) PA(18:0/18:1(9Z)) in negative-ion mode, (b) PE(18:1(9E)/18:1(9E)) in negative-ion mode, (c) PS(16:0/18:1(9Z)) in negative-ion mode, and (d) PE(18:1(9E)/18:1(9E)) in positive-ion mode. Negative-ion mode experiments used 25 mM NH4OAc as an additive. Positive-ion mode experiments used 5 mM LiOAc as an additive.
Negative-ion mode TENG TAP IM-MS experiments for PA, PE, and PS with Δ9 C═C bonds showed the same diagnostic fragments as for PC(18:1(9Z)/18:1(9Z)), with a maximum in the ATD at 2.98 ms similar to the results shown in Figure 2b. Information on fatty acyl chain stereochemistry was also readily obtained for these alternative lipid subclasses (Figure S11).
The anionic nature of PA and PS lipids meant that positive-ion mode experiments for these species were not successful. Other lipid subclasses such as PE, however, produced satisfactory results. Figure 4d shows the TAP spectrum for the PE(18:1/18:1) [MO + Li]+ ion. The 18:1 fatty acyl chains produced diagnostic ions identical to the ones in Figure 3d (m/z 361.3, 345.3, 329.3 287.3). A five-member ring ion similar to that for PC lipids but with a shift in the maximum of the ATD towards longer times (6.95 ms) was observed for PE(18:1(9-E)/18:1(9E)) due to the trans orientation of the C═C bond. The two fragment ions at m/z 499.4 and 483.4 were assigned to the Δ9 C═C bond position. Overall, these results confirmed the feasibility of characterizing different GP subclasses using the proposed methodology.
Lipid Geometrical Isomerism.
Differences in the overall shape and therefore IM collision cross section values of isobaric lipid pairs have been previously reported.46–48 To evaluate the ability of TENG TAP IM-MS to differentiate geometrical isomers, cis and trans PC(18:1(9)/18:1(9)) were analyzed in both positive- and negative-ion modes. In positive-ion mode, the [MO + Li]+ precursor ion (m/z 808.6) did not show significant ATD differences between both geometric isomers (Figure 5a). However, differences were more clearly observed by monitoring the ATD of the double-bond position diagnostic fragments (Figure 5b). For these species, the different orientations of the remaining fatty acyl chain in the fragment ions were sufficient to partially resolve Δ9 cis from Δ9 trans isomers. These results are in agreement with previous reports,49 where the cis isomer displayed shorter arrival times than the trans isomer. In the case of negative-ion-mode experiments, IM data for the [MO + OAc-74]− ion also showed a slightly more compact structure for the cis isomer (Figure 5c). However, the epoxidized fatty acyl chain ion [R-CO2-H]−, key for C═C bond location pinpointing, did not show significant ATD differences (Figure 5d). We envision that TENG TAP IM-MS experiments using higher-resolving-power IM instrumentation, such as the new cyclic IM spectrometers,50,51 would offer the required figures of merit for furthering this type of studies.
Figure 5.
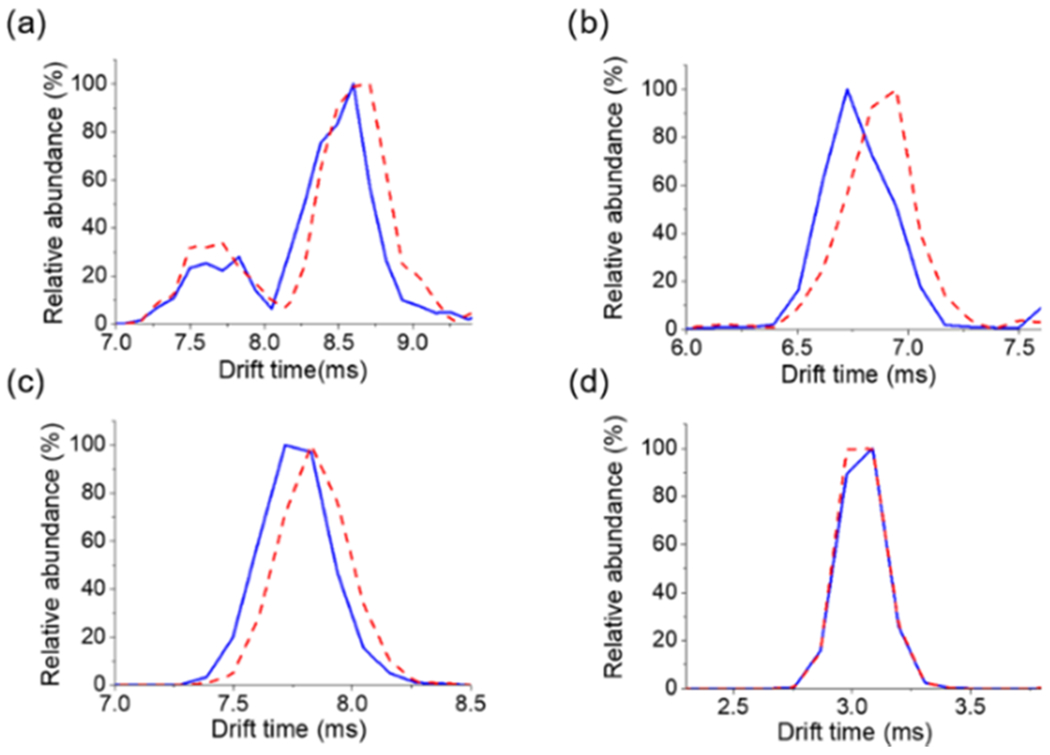
ATD obtained from TENG TAP IM-MS experiments of PC(18:1(9Z)/18:1(9Z)) (blue solid trace) or PC(18:1(9E)/18:1(9E)) (red dashed trace). (a) Positive-ion mode [MO + Li]+ precursor ion at m/z 808.6. (b) Positive-ion mode fragment ion at m/z 499.4 originating from the m/z 625.6 ion (see Figure 2f). (c) Negative-ion mode [MO + OAc-74]− at m/z 786.6. (d) Fragment ion at m/z 297.2 (see Figure 2g). Negative-ion mode experiments used 25 mM NH4OAc as an additive. Positive-ion mode experiments used 5 mM LiOAc as the additive.
TENG IM-MS of Complex Lipid Extracts.
The applicability of TENG TAP IM-MS to real-life samples was tested using a complex chicken egg PC extract (Figure 6). In these experiments, 40 TENG pulses (240 nL) were averaged to enable the detection of lower abundance species, resulting in multiple epoxidation products of unsaturated lipids being detected and structurally characterized. Figures S12–S18 describe results for a number of species of interest, with Tables S1 and S2 listing all precursor and fragment ions detected in both ionization modes.
Figure 6.
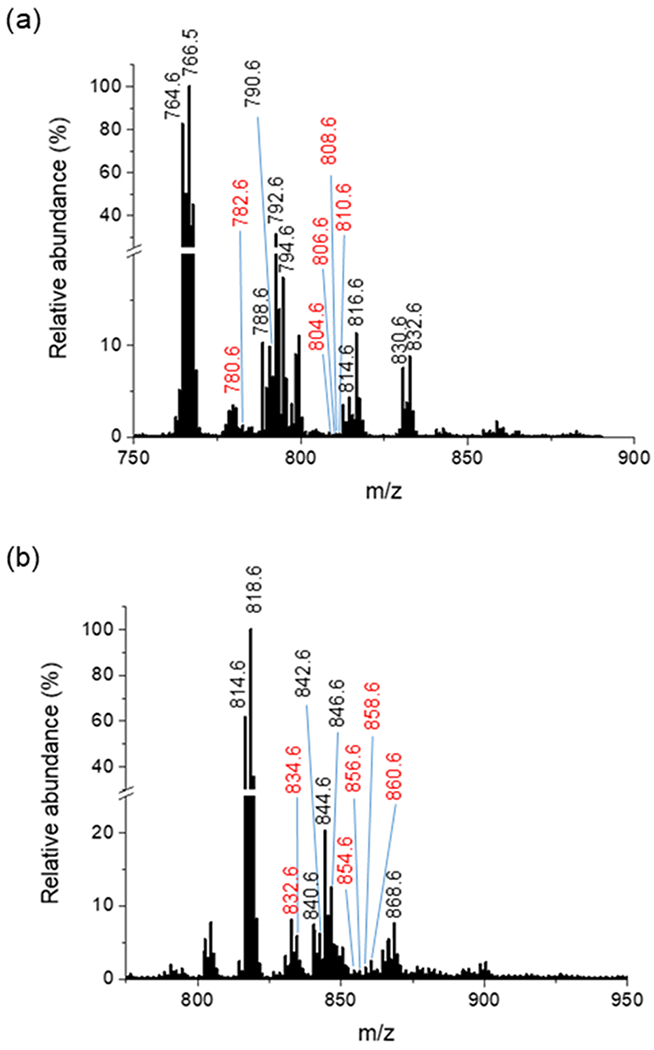
TENG IM-MS analysis of an egg PC extract in (a) positive-ion mode and (b) negative-ion mode. Ions labeled in red indicate some of the epoxidated lipid species that were further identified by TAP experiments.
As an example of the in-depth structural information that these experiments yielded, we discuss results for PC(34:2). In positive-ion mode, TAP IM-MS of the [MO + Li]+ ion at m/z 780.6 showed an ATD (Figure S12a) with features similar to those observed for PC(18:1(9E)/18:1(9E)) (see Figure 3b). For this lipid, however, the arrival time maximum for the dioxolane ion at m/z 597.5 was at 6.5 ms. The 5.80–7.20 ms ATD region yielded numerous diagnostic fragments that were used to assign structural features (Figure S12b). Isobaric lipids corresponding to identical precursor ions were distinguished by differences in the ion mobilities of their fragments. Three different arrival times (6.28, 6.55, and 6.95 ms) were linked to three unique product ions: m/z 317.3 for PC(16:1/18:1 + O), m/z 303.3 for PC(16:0/18:2 + O), and m/z 331.3 for the 14Da loss of PC(36:1) (Figure S12c).
The PC(16:1/18:1 + O) ion showed sn-position diagnostic ions at m/z 303.3, 317.3, and 361.3 (Table S1b). Its nonepoxidized counterpart showed species at m/z 301.3, 287.3, and 345.3, corresponding to 16:1 and 18:1 sn1 and sn2 chains (Table S1a). The 16:1 and 18:1 epoxidized chains produced diagnostic ion pairs at m/z 499.4/483.4 and 471.4/455.4, respectively, indicating C═C bonds at the Δ9 position for both chains (Figure S12d and Table S1b).
Two other PC(34:2) isobars, PC(16:0/18:2) and PC(18:2/16:0), were detected in the 6.30–6.90 ms mobility region when analyzing the chicken egg PC extract in positive-ion mode. Figure S12e shows a set of ions at m/z 261.3, 319.3, and 343.3 that were assigned to PC(18:2/16:0), and a second set at m/z 301.3, 303.3, and 359.3 corresponding to PC(16:0/18:2) (Table S1b). The region between m/z 450 and 525 showed fragment ion pairs for two double bonds, one Δ9 (m/z 473.4/457.4) and another one Δ12 (m/z 513.4/497.4) in the 18:2 acyl chain.
Positive-ion mode TAP analysis of PC(34:2) also revealed the presence of an interferent species, likely produced by in-source demethylation of PC(36:1), isobaric with the PC(34:2) product ions studied. As shown in Figure S12f, an atypical dioxolane product ion at m/z 611.5 was detected, likely originating from PC(36:1) with an 18:0 fatty acyl chain in the sn1 position, as indicated by the fragment ion at m/z 331.3.
TENG TAP IM-MS negative-ion mode analysis of PC(34:2) in the chicken egg PC extract also enabled distinguishing various isobars while pinpointing C═C bond location and assigning acyl chain composition. (Figure S13). The ATD showed three major features (Figure S13a). In the main ATD feature with a maximum at 2.98 ms, three different IM peaks of interest were detected. These corresponded to the 16:1 + O (m/z 269.2), 18:2 + O (m/z 295.2), and 20:1 (m/z 309.2) fatty acyl chains (Figure S13b). These chains originated from PC(16:1_18:1), PC(16:0_18:2), and PC(14:1_20:1). The total ion TAP mass spectrum (Figure S13c) showed multiple fragment ions in the sn-chain composition region (m/z 410–540). The product ions at m/z 476.3, 494.3, 504.3, and 522.3 were assigned to PC(16:1_18:1) (Table S2b). The ions at m/z 462.3, 480.3, 502.3, and 520.3 corresponded to the PC(16:0_18:2) isomer (Table S2b). A third PC(34:2) isomer, PC(14:1_20:1), was also detected, but its structure assignment was much less confident. For this lipid, an ion at m/z 309.2 ([FA 20:1-H]−) that agreed with the proposed structure was observed, but the [FA 14:1-H]− and [FA 14:1 + O-H]− acyl chain ions at m/z 224.2 and 240.2, respectively, were not (Figure S13d). As shown in Figure S13e,f, multiple double-bond diagnostic fragment ions were observed. Product ions at m/z 171.1 and 155 pinpointed the C═C bond at the Δ9 position, and m/z 195.1 at the Δ12 carbon. Table S2 summarizes all diagnostic fragment ions that enabled C═C bond location pinpointing and/or chain sn composition for the species identified in negative-ion mode analysis.
CONCLUSIONS
The results presented here demonstrate the capabilities of TENG TAP IM-MS for shotgun lipidomics with more in-depth structural information. The combination of gas-phase oxidation in the TENG ion source with ion-mobility separations and sequential ion activation enabled detailed lipid structural analysis. Various degrees of structural information were obtained depending on the chosen ion-generation mode. The higher epoxidation efficiency observed in negative-ion mode increased sensitivity, but the structural information was limited to double-bond location pinpointing and chain composition. Although less-intense corona discharges were generated in positive-ion mode, a careful study of TAP fragments of lithiated species enabled both C═C bond location pinpointing and chain sn-position assignment. The inclusion of the ion mobility dimension also helped us to distinguish geometrical lipid isomers (cis vs trans.) Overall, the proposed approach offers a rapid and simple platform for lipid structural annotation that is easily adapted to other IM-MS systems without the need for specific reagents for double-bond location pinpointing (i.e., ozone, meta-chloroperoxybenzoic acid, 2-acetylpyridine, etc.). Future research will involve incorporating a liquid chromatography dimension to minimize signal suppression and adding quantitative capabilities to the proposed approach. To achieve this goal, new solvent systems may need to be evaluated. Acetone has been shown as an LC-compatible solvent for online PB reactions to annotate lipid double-bond position and could also be employed with TENG.52 Moreover, the TENG operation frequency and instrument duty cycle should be adapted to LC operation. The TENG pulse frequency of 0.25 Hz used in this work could be boosted to LC-compatible values by changing the TENG design to that of a rotating device,53 for example.
Supplementary Material
ACKNOWLEDGMENTS
This work was partially supported by NSF and the NASA Astrobiology Program under the NSF Center for Chemical Evolution, CHE-1504217. F.M.F. also acknowledges support by NIH 1R01CA218664-01, 1U2CES0 30167-01, 1U24DK112341, and the CMaT NSF Research Center (EEC-1648035).
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.0c05145.
Solvent optimization for TENG-TAP-IM-MS analysis, comparison between DC-nanoESI and TENG nanoESI GP analysis, ion mobility discrimination of key product ions informing on acyl chain composition, influence of cation selection for TAP IM-MS analysis, effect of the number of TENG pulses, fragmentation pathway for lithium-adducted phosphocholines, TENG TAP IM-MS of PC(18:1(6Z)/18:1(6Z)), TENG TAP IM-MS of [M + Li]+ species, TENGTAP-IM-MS of PC(34:0) in positive-ion mode, mass spectra resulting from integration of different ATD regions for several glycerophospholipids, various TENG TAP IM-MS spectra for egg PC lipids in both positive- and negative-ion modes, and tables with precursor and fragment-ion information for species detected during analysis of egg PC extracts (PDF)
Stand-alone spreadsheet with all spectral data displayed in Figures 2–6 (XLSX)
Complete contact information is available at: https://pubs.acs.org/10.1021/acs.analchem.0c05145
The authors declare no competing financial interest.
Contributor Information
Marcos Bouza, School of Chemistry and Biochemistry, Georgia Institute of Technology, Atlanta, Georgia 30332, United States; NSF/NASA Center for Chemical Evolution, Atlanta, Georgia 30332, United States.
Yafeng Li, School of Chemistry and Biochemistry, Georgia Institute of Technology, Atlanta, Georgia 30332, United States.
Aurelia C. Wang, School of Materials Science and Engineering, Georgia Institute of Technology, Atlanta, Georgia 30332, United States
Zhong Lin Wang, School of Materials Science and Engineering, Georgia Institute of Technology, Atlanta, Georgia 30332, United States; Beijing Institute of Nanoenergy and Nanosystems, Chinese Academy of Sciences, Beijing 101400, China.
Facundo M. Fernández, School of Chemistry and Biochemistry, Georgia Institute of Technology, Atlanta, Georgia 30332, United States; NSF/NASA Center for Chemical Evolution, Atlanta, Georgia 30332, United States.
REFERENCES
- (1).Piomelli D; Astarita G; Rapaka R Nat. Rev. Neurosci 2007, 8, 743–754. [DOI] [PubMed] [Google Scholar]
- (2).Barrera NP; Zhou M; Robinson CV Trends Cell Biol. 2013, 23, 185–192. [DOI] [PubMed] [Google Scholar]
- (3).Saliba A-E; Vonkova I; Gavin A-C Nat. Rev. Mol. Cell Biol 2015, 16, 753–761. [DOI] [PubMed] [Google Scholar]
- (4).Wymann MP; Schneiter R Nat. Rev. Mol. Cell Biol 2008, 9, 162–176. [DOI] [PubMed] [Google Scholar]
- (5).Santos CR; Schulze A FEBS J. 2012, 279, 2610–2623. [DOI] [PubMed] [Google Scholar]
- (6).Ivanova PT; Milne SB; Myers DS; Brown HA Curr. Opin. Chem. Biol 2009, 13, 526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Blanksby SJ; Mitchell TW Annu. Rev. Anal. Chem 2010, 3, 433–465. [DOI] [PubMed] [Google Scholar]
- (8).Han X; Yang K; Gross RW Mass Spectrom. Rev. 2012, 31, 134–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Ryan E; Reid GE Acc. Chem. Res 2016, 49, 1596–1604. [DOI] [PubMed] [Google Scholar]
- (10).Murphy RC; Axelsen PH Mass Spectrom. Rev 2011,30, 579–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Nygren H; Seppänen-Laakso T; Castillo S; Hyötyläinen T; Orešič M Metabolic Profiling: Methods and Protocols; Metz TO, Ed.; Humana Press: Totowa, NJ, 2011; pp 247–257. [DOI] [PubMed] [Google Scholar]
- (12).Zhao Y-Y; Wu S-P; Liu S; Zhang Y; Lin R-C Chem.-Biol. Interact 2014, 220, 181–192. [DOI] [PubMed] [Google Scholar]
- (13).Mitchell TW; Pham H; Thomas MC; Blanksby SJ J. Chromatogr. B 2009, 877, 2722–2735. [DOI] [PubMed] [Google Scholar]
- (14).Hancock SE; Poad BLJ; Batarseh A; Abbott SK; Mitchell TW Anal. Biochem 2017, 524, 45–55. [DOI] [PubMed] [Google Scholar]
- (15).Lin J-T; McKeon TA; Woodruff CL; Singleton JA J. Chromatogr. A 1998, 824, 169–174. [Google Scholar]
- (16).Damen CWN; Isaac G; Langridge J; Hankemeier T; Vreeken RJ J. Lipid Res 2014, 55, No. 1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Nakanishi H; Iida Y; Shimizu T; Taguchi R J. Biochem 2010, 147, 245–256. [DOI] [PubMed] [Google Scholar]
- (18).Kozlowski RL; Campbell JL; Mitchell TW; Blanksby SJ Anal. Bioanal. Chem 2015, 407, 5053–5064. [DOI] [PubMed] [Google Scholar]
- (19).Han X; Gross RW Mass Spectrom. Rev 2005, 24, 367–412. [DOI] [PubMed] [Google Scholar]
- (20).Leaptrot KL; May JC; Dodds JN; McLean JA Nat. Commun 2019, 10, No. 985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Jeanne Dit Fouque K; Ramirez CE; Lewis RL; Koelmel JP; Garrett TJ; Yost RA; Fernandez-Lima F Anal. Chem 2019, 91, 5021–5027. [DOI] [PubMed] [Google Scholar]
- (22).Keating JE; Glish GL Anal. Chem 2018, 90, 9117–9124. [DOI] [PubMed] [Google Scholar]
- (23).Tomer KB; Crow FW; Gross ML J. Am. Chem. Soc 1983, 105, 5487–5488. [Google Scholar]
- (24).Hsu F-F; Turk J J. Am. Soc. Mass Spectrom 2008, 19, 1673–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Hsu F-F; Turk J J. Chromatogr. B 2009, 877, 2673–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Thomas MC; Mitchell TW; Harman DG; Deeley JM; Nealon JR; Blanksby SJ Anal. Chem 2008, 80, 303–311. [DOI] [PubMed] [Google Scholar]
- (27).Williams PE; Klein DR; Greer SM; Brodbelt JS J. Am. Chem. Soc 2017, 139, 15681–15690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Li P; Jackson GP J. Mass Spectrom 2017, 52, 271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Pham HT; Ly T; Trevitt AJ; Mitchell TW; Blanksby SJ Anal. Chem 2012, 84, 7525–7532. [DOI] [PubMed] [Google Scholar]
- (30).Campbell JL; Baba T Anal. Chem 2015, 87, 5837–5845. [DOI] [PubMed] [Google Scholar]
- (31).Ma X; Xia Y Angew. Chem. Int. Ed 2014, 53, 2592–2596. [DOI] [PubMed] [Google Scholar]
- (32).Thomas MC; Mitchell TW; Blanksby SJ J. Am. Chem. Soc 2006, 128, 58–59. [DOI] [PubMed] [Google Scholar]
- (33).Cao W; Ma X; Li Z; Zhou X; Ouyang Z Anal. Chem 2018, 90, 10286–10292. [DOI] [PubMed] [Google Scholar]
- (34).Feng Y; Chen B; Yu Q; Li L Anal. Chem. 2019, 91, 1791–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Tang S; Cheng H; Yan X Angew. Chem., Int. Ed 2020, 59, 209–214. [DOI] [PubMed] [Google Scholar]
- (36).Poad BLJ; Pham HT; Thomas MC; Nealon JR; Campbell JL; Mitchell TW; Blanksby SJ J. Am. Soc. Mass Spectrom. 2010, 21, 1989–1999. [DOI] [PubMed] [Google Scholar]
- (37).Cao W; Cheng S; Yang J; Feng J; Zhang W; Li Z; Chen Q; Xia Y; Ouyang Z; Ma X Nat. Commun 2020, 11, No. 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Bouza M; Li Y; Wu C; Guo H; Wang ZL; Fernández FM J. Am. Soc. Mass Spectrom 2020, 31, 727–734. [DOI] [PubMed] [Google Scholar]
- (39).Castro-Perez J; Roddy TP; Nibbering NMM; Shah V; McLaren DG; Previs S; Attygalle AB; Herath K; Chen Z; Wang S-P; Mitnaul L; Hubbard BK; Vreeken RJ; Johns DG; Hankemeier T J. Am. Soc. Mass Spectrom 2011, 22, 1552–1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Li Y; Bouza M; Wu C; Guo H; Huang D; Doron G; Temenoff JS; Stecenko AA; Wang ZL; Fernández FM Nat. Commun 2020, 11, No. 5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Li A; Zi Y; Guo H; Wang ZL; Fernández FM Nat. Nanotechnol 2017, 12, 481–487. [DOI] [PubMed] [Google Scholar]
- (42).Ma X; Chong L; Tian R; Shi R; Hu TY; Ouyang Z; Xia Y Proc. Natl. Acad. Sci. U.S.A 2016, 113, 2573–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Hsu F-F; Turk J; Thukkani AK; Messner MC; Wildsmith KR; Ford DA J. Mass Spectrom 2003, 38, 752–763. [DOI] [PubMed] [Google Scholar]
- (44).Hsu F-F; Turk J J. Am. Soc. Mass Spectrom 2003,14, 352–363. [DOI] [PubMed] [Google Scholar]
- (45).Hsu F-F; Turk J J. Am. Soc. Mass Spectrom 2008, 19, 1681–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Kyle JE; Zhang X; Weitz KK; Monroe ME; Ibrahim YM; Moo RJ; Cha J; Sun X; Lovelace ES; Wagoner J; Polyak SJ; Metz TO; Dey SK; Smith RD; Burnum-Johnson KE; Baker ES Analyst 2016, 141, 1649–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Xie X; Xia Y Anal. Chem 2019, 91, 7173–7180. [DOI] [PubMed] [Google Scholar]
- (48).Harris RA; May JC; Stinson CA; Xia Y; McLean JA Anal. Chem 2018, 90, 1915–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Poad BLJ; Zheng X; Mitchell TW; Smith RD; Baker ES; Blanksby SJ Anal. Chem 2018, 90, 1292–1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Giles K; Ujma J; Wildgoose J; Pringle S; Richardson K; Langridge D; Green M Anal. Chem 2019, 91, 8564–8573. [DOI] [PubMed] [Google Scholar]
- (51).McKenna KR; Li L; Baker AG; Ujma J; Krishnamurthy R; Liotta CL; Fernández FM Analyst 2019, 144, 7220–7226. [DOI] [PubMed] [Google Scholar]
- (52).Franklin ET; Xia Y Analyst 2020, 145, 6532–6540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Zhu G; Chen J; Zhang T; Jing Q; Wang ZL Nat. Commun 2014, 5, No. 3426. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


