Abstract
Human carboxylesterases (CESs) are serine hydrolases that are responsible for the phase I metabolism of an assortment of ester, amide, thioester, carbonate, and carbamate containing drugs. CES activity is known to be influenced by a variety of factors including single nucleotide polymorphisms, alternative splicing, and drug–drug interactions. These different factors contribute to interindividual variability of CES activity which has been demonstrated to influence clinical outcomes among people treated with CES-substrate therapeutics. Detailed exploration of the factors that influence CES activity is emerging as an important area of research. The use of fluorescent probes with live cell imaging techniques can selectively visualize the real-time activity of CESs and have the potential to be useful tools to help reveal the impacts of CES activity variations on human health. This review summarizes the properties of the five known human CESs including factors reported to or that could potentially influence their activity before discussing the design aspects and use considerations of CES fluorescent probes in general in addition to highlighting several well-characterized probes.
In this review, drug metabolizing human carboxylesterases and fluorescent probes capable of studying their activity in live cells are discussed.
Introduction
Human carboxylesterases (CESs, EC 3.1.1.1) are serine hydrolases that belong to the α/β-hydrolase superfamily and are responsible for the hydrolysis of a variety of endogenous and xenobiotic esters, amides, thioesters, carbonates, and carbamates (Fig. 1).1–8 As these functional groups are commonly used in drug design, in part to mask polar groups in prodrug strategies, CESs play a key role in the phase I metabolism of many drugs. However, there are multiple factors that result in interindividual variability of CES activity. This variation is particular problematic in the design and use of prodrugs as it is normally assumed that there is little difference in the enzymatic activity that unmasks the active form person to person.2,5 Inconstancy in CES activity among individuals has been demonstrated to cause different clinical outcomes,5,6,9 presaging the need for a complete understanding of CES activity in humans. Fluorescent probes that are capable of monitoring the activity of specific CESs could help address this need. The aim of this review is to summarize the factors that are known or have the potential to modulate CES activity as well as describe the merits, design aspects, and use considerations of fluorescent probes that can be deployed to study CES activity in live cells. A special emphasis is placed on well-characterized probes that are capable of preferentially reporting on specific CES enzymes.
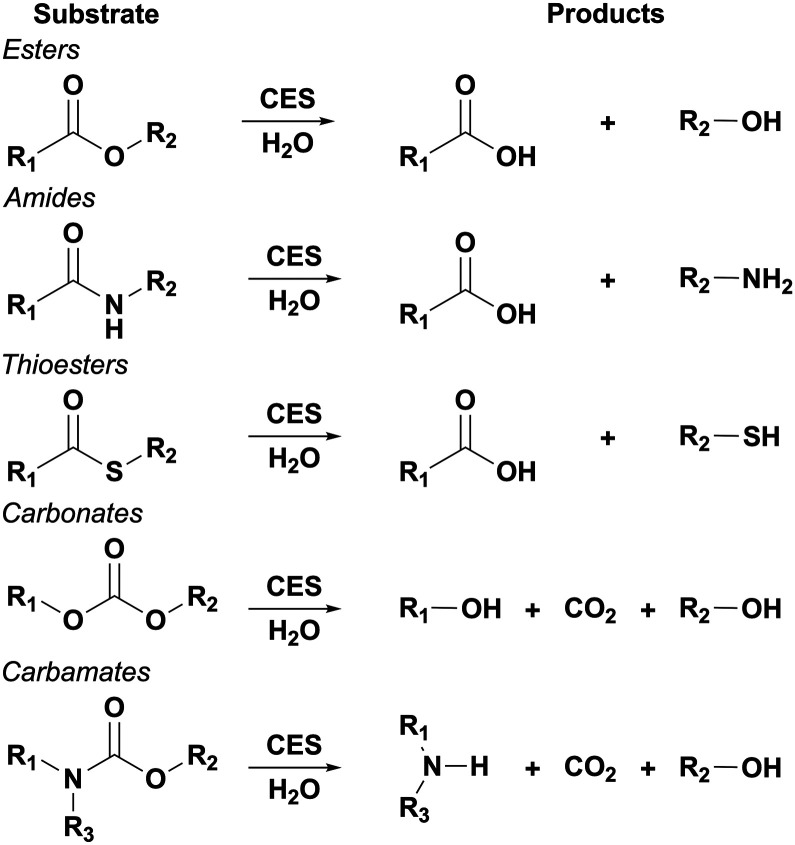
Fig. 1. Known CES substrates and products after hydrolysis. Hydrolysis of carbonates and carbamates initially produce a short lived R1OCOOH or R2OCOOH and R1R3NCOOH species, respectively, that undergoes rapid rearrangement into the shown products.
Human carboxylesterases
Five human carboxylesterases (CES1, CES2, CES3, CES4A, and CES5A) have been reported to date.3,10–13 CES1 and CES2 are the most well studied in humans with CES3 gaining more attention recently. Human CESs share ca. 40% sequence homology, despite this homology they are known to have different substrate preferences and distribution.4,12,13,38,46 A summary of the characteristics of these human carboxylesterases can be found in Table 1.
Summary of human CES properties.
| Namea | Isoforms | Human body localizationb | Subcellular localizationb | Substrate specificity | Known drug substrates |
|---|---|---|---|---|---|
| CES1 | 4 | Liver > lungs ≫ heart, spleen, stomach, testis6,36–38 | Endoplasmic reticulum | Large acyl moiety; small alcohol/amine/thiol group | Benazepril,14 candesartan cilexetil,15 capecitabine,16 ciclesonide,17 cilazapril,14 clopidogrel,18 cocaine,19 dabigatran,20 delapril,14 enalapril,21 flumazenil,22 fosinopril,23 heroin,24 imidapril,14 mepridine,25 methylphenidate,26 moexipril,23 mycophenolate mofetil,27 perindopril,23 oseltamivir,28 oxybutynin,29 quinapril,14 ramipril,21 rufinamide,30 sacubitril,31 sofosbuvir,32 telotristat etiprate,33 tenofovir alafenamide,34 trandolapril35 |
| CES2 | 6 | Colon, intestine > liver, kidneys, heart, brain, testis6,36–40 | Endoplasmic reticulum | Small acyl moiety; large alcohol/amine/thiol group | Aspirin,18 capecitabine,16 cocaine,19 dabigatran,20 gemcitabine,41 heroin,24 irinotecan (CPT-11),42 methylprednisolone 21-hemisuccinate,43 mycophenolate mofetil,27 tenofovir disoproxil44 |
| CES3 | Up to 8 | Liver, colon, intestine, trachea, placenta13,45 | Potentially endoplasmic reticulum or Golgi bodies | Unknown | Poor for irinotecan (CPT-11)45 |
| CES4A | 10 | Brain, lungs, skinc 46 | Unknown, potentially secreted | Unknown | Unknown |
| CES5A | 4 | Brain, kidneys, lungs, testis11,12 | Unknown, potentially secreted | Unknown | Unknown |
Nomenclature as recommended by Holmes et al.10
Based on canonical CES sequence, different isoforms and other sequence variants may be localized differently.
Reported to be in the melanocytes of skin.
CES1
CES1 is located to the endoplasmic reticulum by an N-terminal hydrophobic signal and C-terminal HIEL retention sequence.4,12,13,38,47 In the human body, CES1 is found primarily in the liver and to a lesser degree in the lungs, heart, spleen, stomach, and testis (Table 1).6,36–38 Several crystal structures of CES1 have been reported with different ligands.48–51 These structures reveal that substrates bind in a gorge that has a binding pocket on either side of the catalytic serine. These pockets have different properties with one being considered small and rigid with the other being large and flexible.50 This helps to explain the known substrate preference of CES1 for molecules that have a small alcohol (or amine or thiol) and a large acyl group.37,38,50 The alcohol or alcohol analog portion will bind in the small pocket and the large pocket is flexible to accommodate the binding of the large acyl moiety. Additionally, these crystal structures and atomic force microscopy (AFM) studies demonstrate that CES1 exists in an equilibrium between monomeric, trimeric, and hexameric forms that interact with each other at a Z-shaped dimer interface (Z-site).50,51 The ratio of each species present has been found to be substrate-dependent with the trimeric form being the most active. Furthermore, the Z-site likely serves as an allosteric regulation site that controls oligomerization.49–51
CES2
CES2, similar to CES1, is localized to the endoplasmic reticulum by an N-terminal hydrophobic signal and a C-terminal retrieval sequence of HTEL.4,12,13,38,47 The localization of CES2 in the human body is different from CES1 with CES2 being highly expressed in the colon and intestine but lower levels of expression in the liver, kidney, heart, brain and testis.6,36–40 CES2 is known to prefer substrates that have a large alcohol (or amine or thiol) and a small acyl group.37,38,52 A crystal structure of CES2 has not been reported, however, homology models based on the crystal structures of CES1 have been used to help explain differences in the substrate preferences of CES1 and CES2.52–54 These homology models suggest that the absence of a large loop near the active site of CES2 enlarges the substrate binding gorge which may allow for accommodation of the larger alcohol groups.52 Additionally, these models suggest that CES2's region analogous to CES1's Z-site differs in the amino acid sequence which may explain why CES2 has only been shown to exist in a monomeric state.13,38,52
CES3
CES3 is less studied compared to CES1 and CES2. CES3 also has an N-terminal hydrophobic signal and a C-terminal QEDL sequence that may localize it to the endoplasmic reticulum.11,45 While QEDL is not a classically recognized endoplasmic reticulum retrieval signal, a bioinformatic study has identified it as one of the most common KDEL-like C-terminal sequences in proteins known to be localized to the endoplasmic reticulum.55 Conversely, studies with fluorescent proteins tagged with the QEDL sequence suggest this signal results in localization to the Golgi bodies.56 In the human body, it has been found in the liver, colon, intestine, trachea, and placenta (Table 1).13,45 Like CES2, no crystal structure has been reported for CES3, but homology models based on the crystal structures of CES1 have been generated.13,38 These homology models indicate that CES3 has a similar structure to CES2, likely existing in a monomeric state with a more solvent exposed active site. This suggests CES3 may have a substrate scope closer to CES2 than CES1.13
CES4A and CES5A
CES4A (also known as CES6 and CES8)10,12,46 and CES5A (also known as cauxin and CES7)10–12 have not been studied in significant detail. Both CES4A and CES5A lack a known endoplasmic reticulum retrieval signal suggesting they may be secreted from cells. Expression of CES4A is known to be in the brain, lungs, and skin (melanocytes),46 while CES5A is known to be expressed in the brain, kidneys, lungs, and testis (Table 1).11,12 This difference in localization compared to other CESs has led to the suggestion that CES4A and CES5A play a distinctly different role in xenobiotic and lipid metabolism, though this remains to be established experimentally.11,46 In the absence of crystal structures for CES4A and CES5A, analysis of the sequence and models have been performed. These models suggest CES4A may be oligomeric while CES5A is likely monomeric.11,12,46 The major differences of CES4A and CES5A's structure compared to CES1 involve an alpha helix near the site where the acyl product is released. The exact influence of these structural differences on substrate scope has not been hypothesized or experimentally determined.
CES activity in drug metabolism
The two main carboxylesterases, CES1 and CES2, have been reported to be responsible for the majority of the hydrolysis of xenobiotic esters in humans. Thus, CES1 and CES2 have been found to activate or deactivate many therapeutics across different drug types and classes including antiplatelets/anticoagulants, angiotensin-receptor blockers (ARBs), angiotensin-converting enzyme (ACE) inhibitors, antivirals, antihyperlipidemics, immunosuppressants, chemotherapeutics, and antispasmodics (Table 1).5,7 Generally, CES1 prefers substrates that have a small alcohol and large acyl group, while CES2 prefers substrates with a large alcohol and small acyl group (vide supra). This substrate preference is likely predominantly determined by the shape of the active sites as described earlier.37,57 Substrate preference differences may also be controlled by differences in the catalytic activity of CESs. CES1 is known to catalyze transesterification of carboxylic acids in the presence of large hydrophobic alcohols.57 CES2's transesterification activity is significantly lower in comparison. This could help explain the lower apparent hydrolysis rate by CES1 towards known CES2 substrates which have larger alcohol moieties. CES3's substrate specificity has not been studied in detail.38 CES3 was found to hydrolyze CPT-11, a known CES2 substrate, less efficiently than CES2 in one of the few reports on CES3's substrate scope.45
Drug–drug interactions influencing CES activity
Given the prevalence of CES substrates across many drug classes, drug–drug interactions (DDIs) influencing CES activity are possible.5–7 There are several reports of DDIs for CESs in vitro.5,28,58–60 Animal and clinical DDI studies on CES1 have also been carried out.6,7,61–63 One of the most studied examples is the effect of ethanol on cocaine metabolism by CES1.6,7,19,64 Normally, CES1 catalyzes the hydrolysis of cocaine's methyl ester to form benzoylecgonine which can be excreted. In the presence of ethanol, however, CES1 will catalyze the transesterification of cocaine to form cocaethylene, a more toxic and longer-lived metabolite.19,64–66 The susceptibility of CES-metabolized drugs to DDIs has been highlighted in the US Food and Drug Administration's most recent guidance on in vitro drug interaction studies; where it is now specifically stated that investigational drugs should be evaluated for CES metabolism and checked as a source for potential DDIs.67
Variability in CES sequences and effect on activity
In addition to DDIs influencing CES-mediated drug metabolism, CES activity is known to be different among individuals.5,38 One source stems from interindividual variability of the transcripts that express CES (Fig. 2). CES1 genes have a particularly high degree of variability. In contrast to other human CESs, the copy number of CES1 genes is known to vary.5,13 Copy number variations (CNVs) can change the sequence of the gene creating paralogs with different expression levels and activity.13,68 Evidence of this occurring for CES1, however, is conflicting with some studies showing an effect of multiple copy numbers increasing substrate metabolism while others show no effect.5 Additional CES genetic dissimilarities among individuals can arise from single-nucleotide polymorphisms (SNPs) in CES1, CES2, and CES3 genes.5,7,9,13,38 Some non-synonymous SNPs of CES1 and CES2 have been found to have decreased or no carboxylesterase activity towards known substrates.5,9,38
Fig. 2. Factors influencing diversity of CES activity resulting in interindividual variability of CES-mediated drug metabolism.
Another source of variability in CES activity comes from alternative RNA splicing and alternative translation start sites.11,13,46 All human CES genes are known to produce multiple isoforms through these processes (Table 1). Since they are typically regulated, different cell types depending on cell state and conditions may express variable amounts of the possible CES isoforms.69 This has the potential to result in significant context specific changes to the sequence of a CES which could produce protein products with different activity and subcellular localization. Combined with the fact that several of the CES isoforms studied have little to no carboxylesterase activity, this can create heterogeneity of CES activity among individuals.11,13,46 Two of the four known CES1 isoforms have been found to have significantly decreased activity towards a few CES1 drug substrates when expressed in live cells.70 CES2 has six isoforms known with at least two of the isoforms having no carboxylase activity.13,71 Up to eight CES3 isoforms have been discovered, but no studies have reported on their activity.13,72 Likewise, carboxylase activity of the reported ten isoforms of CES4A and the four isoforms of CES5A activities are also unknown.11,46
Glycosylation state influence on CES activity
Glycosylation is a major regulator of protein stability, folding, and localization.73–75 All human CESs have been reported to be glycoproteins.11,13,46,66,72,76 As the glycosylation state of a protein is regulated and can be modulated in different diseases, this could also contribute to the variability of CES activity.73,75N-Glycosylation for CES1 at N79 has been shown to contribute to enzyme stability and increase catalytic activity while the effects of glycosylation on other CESs have yet to be determined.50,76 CES3 also contains one glycosylation site at N105 while CES2 contains two glycosylation sites at N111 and N276.13,72 CES4A and CES5A have more potential glycosylation sites with CES4A having up to three sites (N214, N276, and N388)46 and CES5A having up to four (N281, N363, N511, and N522).11 The additional glycosylation sites for CES4A and CES5A may help increase their stability and activity if they are secreted into body fluids as has been proposed.11,46
Other factors modulating CES activity
Other factors can also influence CES activity. CES1 and CES2 expression in the liver has been shown to increase as humans age.5,6,77,78 This difference in expression levels has been demonstrated to influence hydrolysis of known CES1 and CES2 substrates in ex vivo liver samples.77,79 CES2 has been found to not have different levels of expression between males and females when corrected for age.6,80 While some clinical studies have suggested that CES1 activity may be higher in females than males, later ex vivo liver sample studies have not supported this.5–7 Certain transcription factors and disease states have also been implicated in changing CES expression.6,81,82 Interleukin 6 (IL-6) has been shown to decrease CES1 and CES2 expression and reduce hydrolysis of CES substrate drugs in cell cultures.81 IL-6 plays a key role in inflammation and is involved in the response to infection and injury as well as being implicated in chronic inflammatory diseases like arthritis and colitis.83,84 Therefore, CES activity could also be altered depending on disease state.
As a whole, DDIs, variability in CES sequences, glycosylation, age, sex, and disease state all can contribute to interindividual variability in the pharmacokinetic and pharmacodynamic properties of CES substrate drugs (Fig. 2). Most of the focus has been on the genetic variability of CES1 where studies have indicated certain SNPs result in different clinical outcomes upon treatment with CES1 substrate drugs.5,6,9 However, it is likely that these other factors could also influence clinical outcomes.
Approaches to study CES activity
A clear understanding of all the factors that contribute to interindividual variability of CES could lead to a more personalized approach in treatments and better clinical outcomes. Recent biotechnological advances that have led to the cost reduction of various techniques that allow for rapid determination of DNA, mRNA, and protein levels have been used to study CESs.85 These methods, however, do not provide information on the activity levels of CESs. Studying the activity of CESs is more complicated. In vitro studies with purified enzymes, microsomes, or cell lysates, are limited by their nature of not being a complete system. In contrast, model organism studies could be complicated by the differences between human CESs and the organism's CESs.86 For example, mice have 20 different CES genes which make assigning orthologs difficult.10,12 Additionally, substrate specificity can vary among orthologous CESs.12 This can add complexity to studying CESs in organisms and the results obtained could potentially not correspond to the properties of human CESs. Thus, cultures of live human cells have been utilized as they are a more complete system that express human CESs.
Typical methods to study CES activity in live cells
There are only a few methods to study CES activity in live cells. Chromatographic methods often paired with mass spectrometry can be utilized to study both endogenous or overexpressed CES.5,70,87 This method uses drugs that are known to be processed by specific CES enzymes to compare the hydrolysis activity of CES forms to those considered normal. This approach has utilized clopidogrel,70,87,88 enalapril,23,70,87 and sacubitril31,70,87 to study CES1 SNPs and isoforms produced by alternative RNA splicing.
Another approach for studying CES2 activity in live cells utilizes CPT-11, a known CES2 substrate.42,89 In this method, the relative activity of CES2 isoforms are determined by monitoring cell viability after treatment with CPT-11. CES2 hydrolyses CPT-11 to produce SN-38, a toxic topoisomerase I inhibitor, which results in cell death. Therefore, the active CES2 forms will generate SN-38 reducing cell viability.
Fluorescent probes to study CES activity in live cells
Fluorescent probes paired with fluorescence microscopy have several advantages over the current methods typically used to study CES activity in live cells. This approach is minimally invasive, requires little sample preparation, and can be adapted into high-throughput assays.90–97 Additionally, fluorescence microscopy can be carried out quantitatively and with spatiotemporal resolution which allows for the additional information of where, when, and how much enzymatic activity is occurring in live cells and tissues.91,93,98 Two reviews of fluorescent probes reported to be able to monitor CES activity have recently been published.85,99 Here, we focus on the general design aspects and considerations that impact the use of these probes to study CES enzymes highlighting several probes that researchers may find particularly useful.
Specificity of fluorescent probes for CESs
Understanding the specificity of a particular fluorescent probe is necessary for its proper use.100–103 A potential probe should be evaluated for specificity among the known CESs as well as interference from other esterases that have similar activity as CESs.3,104–106 Acetylcholinesterase (AChE) catalyzes the hydrolysis of acetylcholine and is predominately in the brain, muscles, and erythrocytes.3,107 AChE is relatively rigid in structure with a narrow active site gorge that has a potential-gradient that may help “pull” substrates towards the anionic choline binding pocket.107,108 This limits the substrate scope of AChE.105,107,109 AChE is known to be inhibited by Irinotecan (CPT-11), a known CES2 substrate, suggesting that similar CES2 substrates have the potential to interact with AChE.110 Butyrylcholinesterase (BChE) has a larger acyl binding pocket compared to AChE, resulting in a larger substrate scope.105 It is known to hydrolyze a variety of substrates that overlap with CES2 including CPT-11, aspirin, and cocaine.3,105,106 Interference from BChE is also more likely to occur due to its localization in similar tissues including the liver and is also known to be in human plasma.105,106 Arylacetamide deacetylase (AADAC) could also be a source of interference as it is localized to the ER in the liver and digestive tract in humans and prefers small acyl moieties similar to CES2.3,104 It has been suggested, however, that AADAC prefers smaller acyl groups than CES2.104 Biphenyl hydrolase-like protein (BPHL; also known as valacyclovir hydrolase or valacyclovirase) is predominately found in the liver and kidneys.114 Their preference for α-amino acids as the acyl group results in less of a concern of interference with CES probes.115,116 Paraoxonases (PONs) are known to hydrolyze cyclic esters, carbonates, and thioesters.111 They are not serine hydrolases like the other enzymes discussed here, instead the catalytic activity of PONs are believed to occur through general base catalysis to activate water utilizing an active site calcium and an aspartic acid or histidine R-group.112,113 PONs are expressed in liver and are secreted into the plasma.3,111,113 Their dependence on calcium likely limits their activity to the plasma where calcium concentrations are high.3 Similarly, human serum albumin (HSA) is secreted from the liver into the plasma. HSA has a relatively low esterase activity compared to the other esterases, but the large quantities present in the plasma could result in meaningful hydrolysis of probes.105,117 Careful consideration of the substrate specificities of these esterases when designing CES specific probes could help limit interference.
In vitro characterization with purified enzymes can be utilized to explore the specificity of probes for a particular CES and to determine if any other esterase could also hydrolyze the probe. Results from this approach should be considered carefully as in vitro hydrolysis of a probe by one of these interfering enzymes may not be relevant in all cases. For example, interference from secreted enzymes like PONs and HSA as well as potentially CES4A and CES5A would be limited in cell cultures as the culture media containing any secreted enzymes could be removed before adding a probe. Likewise, a probe that has apparent high specificity in cell culture could lose that specificity when applied to whole organisms due to the activity of these secreted enzymes.
Additionally, the specificity of probes can also be characterized in cells using inhibitors. Troglitazone20,118,119 and 3-O-(β-carboxypropionyl)-ursolic acid (UKA)120 have been used in live cells to inhibit CES1, while loperamide is commonly used for inhibiting CES2.16,97,118,119 It is important to note that these inhibitors have been found to preferentially inhibit either CES1 or CES2, but have not been tested against CES3, CES4A, or CES5A. Therefore, genetic knockdown/knockout of specific CES genes may provide better information on the specificity of a fluorescent probe. While genetic methods may provide more precision in targeting the activity of a particular CES, the longer timeline of these experiments can allow for compensatory mechanisms to balance for the loss of a CES.121,122 This has been demonstrated in THP-1 cells where knocking down CES1 results in an increase in CES3 expression.123 This may make a probe appear to be less sensitive to the loss of a particular CES activity but in reality it is responding to the expression increase of another CES. Thus, both small molecule inhibitors and genetic manipulations should be used to obtain a more complete understanding of a probe's susceptibility to hydrolysis by a specific CES.
Choice of cell line is also important when characterizing the specificity of a CES fluorescent probe as well as during the use of these probes. Different human cell lines will have different expression levels of each CES and potentially interfering esterases.40,97,124 A probe could appear to be specific for a CES if the cell line employed has low levels of expression of the off target esterase. This could also lead to erroneous results if a probe is used in a different cell line without fully recharacterizing the probe's specificity. In contrast, clever use of cell lines with certain CES and other esterase expression profiles could be exploited to study specific CESs with probes that have suboptimal selectivity.
General design of CES fluorescent probes
Reported CES fluorescent probes follow a typical probe design strategy where a molecule's fluorescence properties are controlled by a pendent CES substrate.91,125–129 Upon this substrate moiety being processed by a CES, the probe undergoes a change in its fluorescent properties which can be monitored by fluorescence microscope. There are three general design classifications of fluorescent probes: turn-on, turn-off, and ratiometric (Fig. 3).125,130–132 Probes of these design types have various advantages and disadvantages.
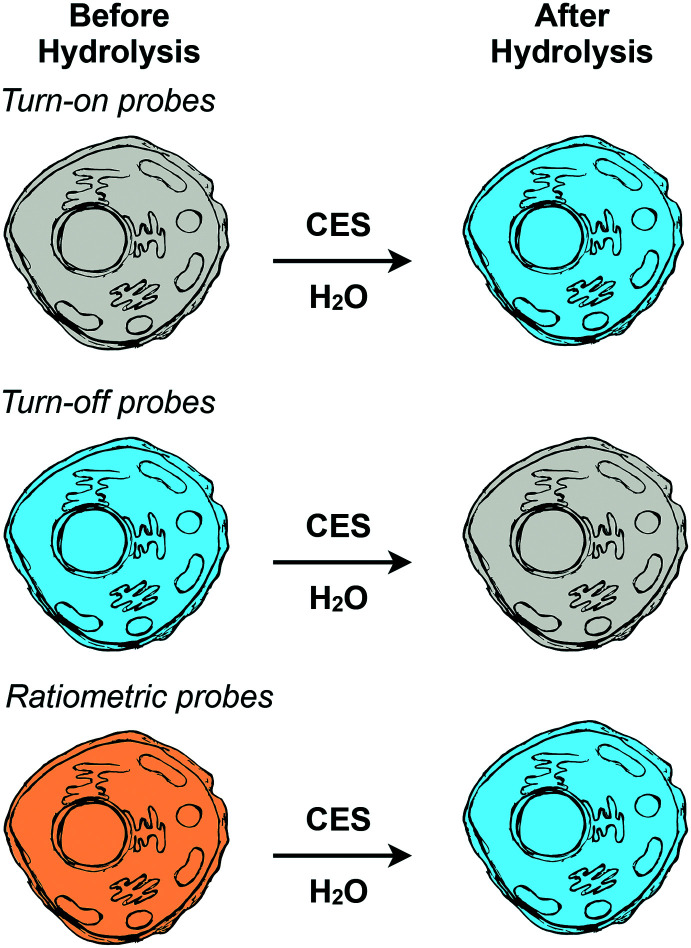
Fig. 3. Design approaches for making fluorescent probes. Turn-on probes initially have little to no fluorescence (grey) and become fluorescent (blue) after hydrolysis. Turn-off probes are fluorescent (blue) before hydrolysis and decrease in fluorescence after hydrolysis (grey). Ratiometric probes are fluorescent in both states with different fluorescence properties before (orange) and after (blue) hydrolysis.
Turn-on and turn-off probes depend on measuring changes in fluorescence intensity.125,130,131 Turn-on probes have little to no fluorescence until their substrate moiety is hydrolyzed by a CES. After hydrolysis, these molecules have a significant increase in fluorescence intensity. Turn-off probes work in the opposite manner. Initially, before being acted upon by CESs, they are fluorescent and after hydrolysis they “turn off” resulting in a form of the probe with little to no fluorescence. Both of these design approaches are conceptually straightforward to use experimentally where after treatment of cells with the probe the fluorescence is imaged. The simplicity of this probe design does have some disadvantages as the change in fluorescence intensity is dependent on the concentration of the probe in the cell.130,131 This means that uptake differences between cells can introduce error in measurements of CES activity.
Ratiometric probes are fluorescent both before and after hydrolysis by CES but exhibit different excitation and/or emission wavelengths depending on the hydrolysis state of the substrate moiety.125,130,131 Experimentally, this requires taking images at two different excitation/emission pairings (channels) and using image analysis software to create new images that represent the ratio of the hydrolyzed probe to unhydrolyzed probe for each pixel. While slightly more complex, this allows for the correction of the uptake differences between cells enabling higher sensitivity measurements of CES activity.130–134
Fluorescent probes for CESs
There are many fluorescent probes reported that claim to be specific for CESs, but they have not been fully demonstrated to be specific for CESs in general or for a particular CES enzyme in live cells (Table S1†). The use of high-quality well-characterized probes and understanding their limitations is important to be able to produce impactful results that accurately reflect the role of CES enzymes in the process that is being evaluated.100–102 In the next sections we have highlighted several probes that are particularly well-characterized for their activity towards CES1 or CES2. This, however, is not to completely dismiss any probes that we do not highlight as these compounds may be useful in certain contexts and if the user understands their limitations.
CES1 specific fluorescent probes
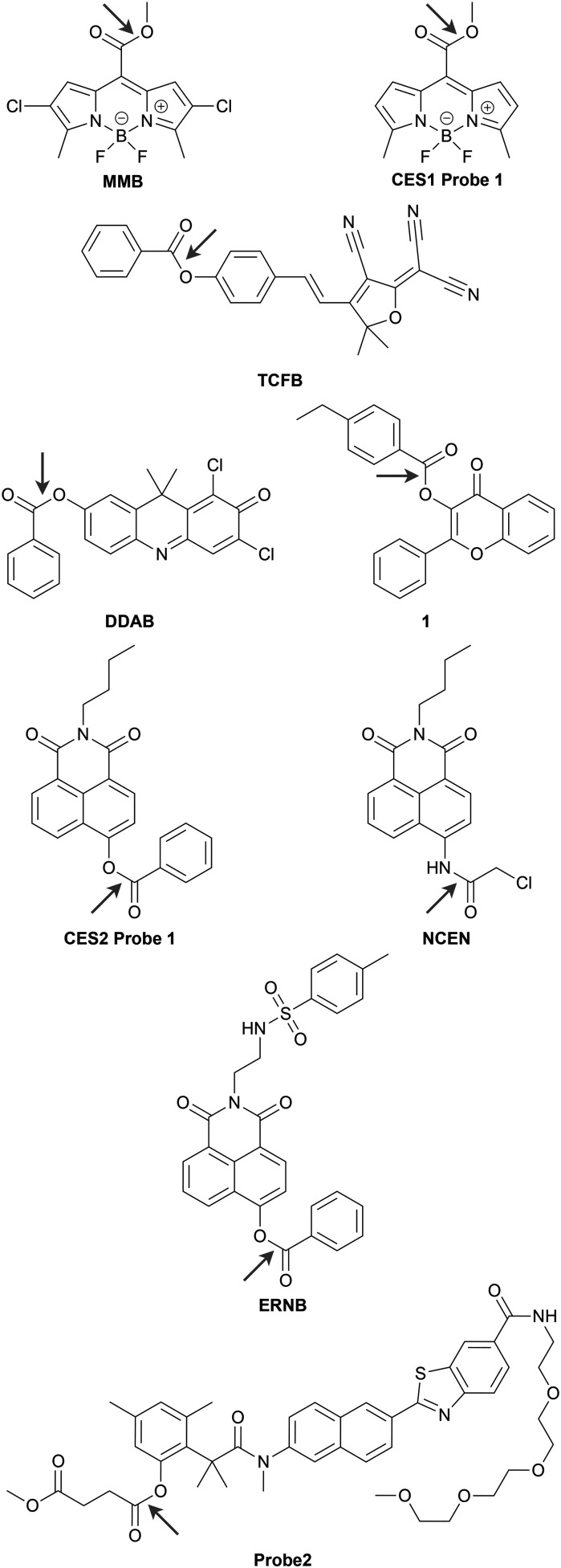
There are only few fluorescent probes that are characterized well enough to be considered specific for CES1. The best characterized is MMB from the Guangbo Ge, Jingnan Cui, and co-workers (Chart 1).54 MMB is a turn-on probe based on a BODIPY fluorophore framework with the fluorescence controlled by CES1 hydrolysis of a pendant methyl ester. The probe was characterized in vitro using purified enzymes and human liver microsomes which are enriched in CES1. The Caco-2 cell line was used to validate the probe's ability to report on endogenous CES1 in live cells using CES1 siRNA. The probe's red-shifted fluorescence properties (λex = 530 nm, λem = 600 nm) also allowed for imaging CES1 ortholog activity in ex vivo mouse organs and in live zebrafish. This group earlier in the same year also reported “Probe 1”, a turn-on probe based on a similar scaffold BODIPY scaffold, to be specific for CES1 (CES1 probe 1 in Chart 1).135 This probe, however, is not as fully characterized as MMB with the specificity for CES1 being determined using inhibitors with purified recombinant CES1, human liver microsomes, and A549 cell lysates as well as UKA in live HepG2 cells. While likely to be specific for CES1 based on the similarity to MMB, the specificity of probe 1 towards CES1 should be completely validated using inhibitors and genetic knockdown/knockout in live cells. It is important to emphasize that not having a fully characterized specificity does not completely abolish the utility of probe 1. It was demonstrated that probe 1 can respond to CES1 activity in live cells and can be used to assay for CES1 inhibitors.
Chart 1. Well-characterized fluorescent probes for monitoring CES1 (MMB and CES1 probe 1) and CES2 (TCFB, DDAB, CES2 probe 1, NCEN, ERNB, and probe2) activity in living cells with fluorescence microscopy. Arrow indicates the bond hydrolysed by CES1 or CES2.
CES2 specific fluorescent probes
There are more probes that are reported to be specific for CES2 that are well-characterized. This is likely due to the substrate preference of CES2 allows for these probes to use a wider variety of fluorophore scaffolds that are sensitive to the acylation state of a heteroatom like oxygen or nitrogen.91,93,125,126 Guang-Bo Ge, Jing-Nan Cui, Ling Yang, and co-workers have several benzoate and benzoate-derivative CES2 substrate-based probes that are well-characterized utilizing turn-on136–138 and ratiometric134 fluorophore scaffolds (Chart 1). The same group has also reported a two-photon ratiometric probe, NCEN,133 based on the hydrolysis of a chloroacetyl amide by CES2 (Chart 1). Xiaochi Ma and Tony D. James and co-workers created a ratiometric ER targeted CES2 probe, ERNB, by replacing the butyl amine group of CES2 probe 1 with an ethylenediamine linked p-toluenesulfonamide (Chart 1).139
All of these probes have been characterized in vitro with purified enzymes to be specific for CES2 and in live cells using loperamide to inhibit CES2. Some of the probes were also able to be used with different biological samples depending on their fluorescence properties. DDAB's fluorescence in the near-IR region (λex = 630 nm, λem = 700 nm) allowed for imaging orthologous mouse CES2 activity in ex vivo organs of mice and in live nude mice in addition to being able to monitor CES2 activity in SKOV-3 and HL-7702 cells (Chart 1).137 ERNB's ratiometric fluorescence was used to image mouse CES2 ortholog activity in ex vivo liver slices and was utilized to determine that mouse CES2 activity is decreased under ER stress after acetaminophen (APAP)-induced acute mouse liver injury.139 Similarly, NCEN's two-photon fluorescence properties permitted ratiometric measurements of mouse CES2 ortholog activity in ex vivo mouse liver slices.133 Additionally, NCEN was adapted for use in a fluorescence microscopy-based high-content analysis (HCA) assay in a later report.97 This allowed for screening several natural products for their CES2 inhibitory activity in HepG2 cells.
Kyeong Sook Choi, Hwan Myung Kim, and co-workers have also reported a CES2 selective probe using a different acyl substrate for CES2 (Chart 1).140 Their ratiometric probe, “Probe2”, utilizes a succinate ester as the CES2 substrate based on a known CES2 inhibitor, 18β-glycyrrhetinic acid derivative (GAD). This probe shows preference, but not complete selectivity, for CES2 over CES1 and BChE in vitro. After validation of specificity for CES2 in live RKO cells using loperamide, probe 2 was used to quantitatively measure endogenous CES2 activity in several breast cell lines which allowed for the authors to develop a method using probe 2 to predict the responsiveness of cells to CPT-11, a CES2 substrate drug. It is important to point out that all the CES2 probes discussed here were not evaluated by genetic knockdown/knockout of CES2 in live cells to ensure the response is indeed coming from CES2 and not from off-target inhibition of another CES or esterase.
Future directions
To date, there are two probes for CES1 and seven probes for CES2 that have enough specificity to allow for their use to study CES activity in live cells (Chart 1). There is little diversity in the fluorophores utilized for CES1 due to the limited number of well-characterized CES1 probes. Current probes are limited to the BODIPY scaffold. The development of CES1 probes that use different molecules for their fluorescent cores would help create a color palette of fluorescent probes for monitoring CES activity when paired with existing CES2 probes. This would enable multicolor fluorescence imaging129 to monitor multiple CESs at the same time.
Additionally, even the most well-characterized probes that are highlighted here have not been directly tested if they could be activated by CES3, CES4A, or CES5A. This could lead to error in studies that use these fluorescent probes given the potential of these other CES enzymes to have similar substrate scopes to CES1 and CES2. Thus, future efforts should also include characterization of fluorescent probes activity towards CES3, CES4A, and CES5A as well as work towards the development of probes that are able to specifically report on the activity of these enzymes.
Another area of future focus could also be studying the activity of CESs in different cellular compartments. While CES1 and CES2 are annotated and known to be in the endoplasmic reticulum due to their targeting sequences, CES3's subcellular localization is less well defined due to having a noncanonical endoplasmic reticulum retrieval signal and both CES4A and CES5A lack this signal. A few lysozyme-targeted probes for CESs have been reported,141–143 however, their specificity for CESs are poorly characterized. Currently, known human CESs have not been suggested to be lysosomal. Future exploration using well-characterized targeted probes may lead to the discovery of new CESs, reveal the subcellular localization of specific CES forms, and/or annotate new enzymes with CES-like activity.
Finally, adaptation of more probes for use in HCA assays, similar to the assay developed for CES2 using NCEN,97 would enable rapid analysis of CES activity. These HCA assays would have a particularly high utility in determining potential DDI interactions involving specific CES forms. Overall, having a variety of fluorescent CES probes with different properties would allow researchers to choose the one that is best suited for use in the system of their interest.
Conclusions
CES enzymes are relatively understudied despite their key role in the metabolism of drugs across several classes. The variation of CES activity caused by a variety of factors has the potential to and have been demonstrated in some cases to influence clinical outcomes. Fluorescent probes, when properly characterized and used in the right context, are capable of studying CES activity in live cells and can rapidly provide information on activity under different conditions. Overall, we believe fluorescent probes capable of specifically reporting the activity of CESs will help expand the understanding of the effects of interindividual variation of CES activity in drug metabolism leading personalized treatments resulting in better clinical outcomes.
Author contributions
A. S., M. G., and M. W. B. were responsible for writing and editing.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This work was supported by Eastern Illinois University (EIU) and EIU Department of Chemistry and Biochemistry. A. S. thanks the EIU Department of Chemistry and Biochemistry and EIU Graduate School for their support through a graduate assistantship.
Biographies
Biography
Anchal Singh.

Anchal Singh born in Durg, India obtained her B.S. in Biotechnology in 2015 from Sam Higginbottom University of Agricultural Technology and Sciences in Allahabad, India. Her interest in biotechnology was cemented during her undergraduate studies when completing an undergraduate research thesis on genetic alterations in wound healing of diabetes mellitus patients under the guidance of Prof. Kiran Singh and Kanhaiya Kumar. In 2019, she moved to the United States to pursue a M.S. in Biochemistry at Eastern Illinois University working with Michael W. Beck as part of an interdisciplinary research team to complete her thesis studies on human carboxylesterase enzymes.
Biography
Mingze Gao.

Mingze Gao, born in Xianyang, China, started his undergraduate education at North University of China before transferring to Eastern Illinois University where he is currently pursuing a B.S. in Biological Sciences. He began his research career in 2019 studying transcription factors that are involved in cell migration and outgrowth with Gary Bulla. In 2020, he joined Michael W. Beck's group to study human carboxylesterase enzymes. His research interests revolve around projects that have eventual impacts to human health.
Biography
Michael W. Beck.

Michael W. Beck earned his B.S. in Chemistry (Biochemistry Concentration) at Tennessee Technological University in 2011 where he started his research career by performing undergraduate research with Edward Lisic. He then obtained his Ph.D. in Chemistry (2015) at the University of Michigan with Mi Hee Lim. After completing his postdoctoral studies with Bryan Dickinson at the University of Chicago, he joined the Department of Chemistry and Biochemistry at Eastern Illinois University as an assistant professor in 2019. His research group focuses on designing chemical tools to study biological problems in live cells while training the next generation of scientists.
Electronic supplementary information (ESI) available. See DOI: 10.1039/d1md00073j
References
- Testa B. and Mayer J. M., in Hydrolysis in Drug and Prodrug Metabolism, Verlag Helvetica Chimica Acta, Zürich, 2006, pp. 1–9 [Google Scholar]
- Jornada D. dos Santos Fernandes G. Chiba D. de Melo T. dos Santos J. Chung M. Molecules. 2015;21:42. doi: 10.3390/molecules21010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukami T. Yokoi T. Drug Metab. Pharmacokinet. 2012;27:466. doi: 10.2133/dmpk.dmpk-12-rv-042. [DOI] [PubMed] [Google Scholar]
- Satoh T. Hosokawa M. Annu. Rev. Pharmacol. Toxicol. 1998;38:257. doi: 10.1146/annurev.pharmtox.38.1.257. [DOI] [PubMed] [Google Scholar]
- Her L. Zhu H.-J. Drug Metab. Dispos. 2020;48:230. doi: 10.1124/dmd.119.089680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di L. Curr. Drug Metab. 2019;20:91. doi: 10.2174/1389200219666180821094502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey Laizure S. Herring V. Hu Z. Witbrodt K. Parker R. B. Pharmacotherapy. 2013;33:210. doi: 10.1002/phar.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa B. and Mayer J. M., in Hydrolysis in Drug and Prodrug Metabolism, Verlag Helvetica Chimica Acta, Zürich, 2006, pp. 419–534 [Google Scholar]
- Merali Z. Ross S. Paré G. Drug Metab. Drug Interact. 2014;29:143. doi: 10.1515/dmdi-2014-0009. [DOI] [PubMed] [Google Scholar]
- Holmes R. S. Wright M. W. Laulederkind S. J. F. Cox L. A. Hosokawa M. Imai T. Ishibashi S. Lehner R. Miyazaki M. Perkins E. J. Potter P. M. Redinbo M. R. Robert J. Satoh T. Yamashita T. Yan B. Yokoi T. Zechner R. Maltais L. J. Mamm. Genome. 2010;21:427. doi: 10.1007/s00335-010-9284-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes R. S. Cox L. A. VandeBerg J. L. Comp. Biochem. Physiol., Part D: Genomics Proteomics. 2008;3:195. doi: 10.1016/j.cbd.2008.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian J. Nelson R. Lehner R. Protein Cell. 2018;9:178. doi: 10.1007/s13238-017-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanghani S. P. Sanghani P. C. Schiel M. A. Bosron W. F. Protein Pept. Lett. 2009;16:1207. doi: 10.2174/092986609789071324. [DOI] [PubMed] [Google Scholar]
- Takai S. Matsuda A. Usami Y. Adachi T. Sugiyama T. Katagiri Y. Tatematsu M. Hirano K. Biol. Pharm. Bull. 1997;20:869. doi: 10.1248/bpb.20.869. [DOI] [PubMed] [Google Scholar]
- Ishizuka T. Yoshigae Y. Murayama N. Izumi T. Drug Metab. Dispos. 2013;41:1888. doi: 10.1124/dmd.113.053595. [DOI] [PubMed] [Google Scholar]
- Quinney S. K. Sanghani S. P. Davis W. I. Hurley T. D. Sun Z. Murry D. J. Bosron W. F. J. Pharmacol. Exp. Ther. 2005;313:1011. doi: 10.1124/jpet.104.081265. [DOI] [PubMed] [Google Scholar]
- Mutch E. Nave R. McCracken N. Zech K. Williams F. M. Biochem. Pharmacol. 2007;73:1657. doi: 10.1016/j.bcp.2007.01.031. [DOI] [PubMed] [Google Scholar]
- Tang M. Mukundan M. Yang J. Charpentier N. LeCluyse E. L. Black C. Yang D. Shi D. Yan B. J. Pharmacol. Exp. Ther. 2006;319:1467. doi: 10.1124/jpet.106.110577. [DOI] [PubMed] [Google Scholar]
- Laizure S. C. Mandrell T. Gades N. M. Parker R. B. Drug Metab. Dispos. 2003;31:16. doi: 10.1124/dmd.31.1.16. [DOI] [PubMed] [Google Scholar]
- Fukami T. Takahashi S. Nakagawa N. Maruichi T. Nakajima M. Yokoi T. Drug Metab. Dispos. 2010;38:2173. doi: 10.1124/dmd.110.034454. [DOI] [PubMed] [Google Scholar]
- Thomsen R. Rasmussen H. B. Linnet K. The INDICES Consortium Drug Metab. Dispos. 2014;42:126. doi: 10.1124/dmd.113.053512. [DOI] [PubMed] [Google Scholar]
- Kleingeist B. Böcker R. Geisslinger G. Brugger R. J. Pharm. Pharm. Sci. 1998;1:38. [PubMed] [Google Scholar]
- Wang X. Wang G. Shi J. Aa J. Comas R. Liang Y. Zhu H.-J. Pharmacogenomics J. 2016;16:220. doi: 10.1038/tpj.2015.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamendulis L. M. Brzezinski M. R. Pindel E. V. Bosron W. F. Dean R. A. J. Pharmacol. Exp. Ther. 1996;279:713. [PubMed] [Google Scholar]
- Zhang J. Burnell J. C. Dumaual N. Bosron W. F. J. Pharmacol. Exp. Ther. 1999;290:314. [PubMed] [Google Scholar]
- Sun Z. Murry D. J. Sanghani S. P. Davis W. I. Kedishvili N. Y. Zou Q. Hurley T. D. Bosron W. F. J. Pharmacol. Exp. Ther. 2004;310:469. doi: 10.1124/jpet.104.067116. [DOI] [PubMed] [Google Scholar]
- Fujiyama N. Miura M. Kato S. Sone T. Isobe M. Satoh S. Drug Metab. Dispos. 2010;38:2210. doi: 10.1124/dmd.110.034249. [DOI] [PubMed] [Google Scholar]
- Shi D. Yang J. Yang D. LeCluyse E. L. Black C. You L. Akhlaghi F. Yan B. J. Pharmacol. Exp. Ther. 2006;319:1477. doi: 10.1124/jpet.106.111807. [DOI] [PubMed] [Google Scholar]
- Sato Y. Miyashita A. Iwatsubo T. Usui T. Drug Metab. Dispos. 2012;40:902. doi: 10.1124/dmd.111.043208. [DOI] [PubMed] [Google Scholar]
- Williams E. T. Eric Carlson J. George Lai W. Nancy Wong Y. Yoshimura T. Critchley D. J. Narurkar M. Drug Metab. Lett. 2011;5:280. doi: 10.2174/187231211798472511. [DOI] [PubMed] [Google Scholar]
- Shi J. Wang X. Nguyen J. Wu A. H. Bleske B. E. Zhu H.-J. Drug Metab. Dispos. 2016;44:554. doi: 10.1124/dmd.115.068536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami E. Tolstykh T. Bao H. Niu C. Micolochick Steuer H. M. Bao D. Chang W. Espiritu C. Bansal S. Lam A. M. Otto M. J. Sofia M. J. Furman P. A. J. Biol. Chem. 2010;285:34337. doi: 10.1074/jbc.M110.161802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center for Drug Evaluation and Research, Xermelo, 2009
- Birkus G. Bam R. A. Willkom M. Frey C. R. Tsai L. Stray K. M. Yant S. R. Cihlar T. Antimicrob. Agents Chemother. 2016;60:316. doi: 10.1128/AAC.01834-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H.-J. Appel D. I. Johnson J. A. Chavin K. D. Markowitz J. S. Biochem. Pharmacol. 2009;77:1266. doi: 10.1016/j.bcp.2008.12.017. [DOI] [PubMed] [Google Scholar]
- Satoh T. Taylor P. Bosron W. F. Sanghani S. P. Hosokawa M. La Du B. N. Drug Metab. Dispos. 2002;30:488. doi: 10.1124/dmd.30.5.488. [DOI] [PubMed] [Google Scholar]
- Imai T. Drug Metab. Pharmacokinet. 2006;21:173. doi: 10.2133/dmpk.21.173. [DOI] [PubMed] [Google Scholar]
- Wang D. Zou L. Jin Q. Hou J. Ge G. Yang L. Acta Pharm. Sin. B. 2018;8:699. doi: 10.1016/j.apsb.2018.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G. Zhang W. Ma M. K. McLeod H. L. Clin. Cancer Res. 2002;8:2605. [PubMed] [Google Scholar]
- Imai T. Imoto M. Sakamoto H. Hashimoto M. Drug Metab. Dispos. 2005;33:1185. doi: 10.1124/dmd.105.004226. [DOI] [PubMed] [Google Scholar]
- Wickremsinhe E. Bao J. Smith R. Burton R. Dow S. Perkins E. Pharmaceutics. 2013;5:261. doi: 10.3390/pharmaceutics5020261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humerickhouse R. Lohrbach K. Li L. Bosron W. F. Dolan M. E. Cancer Res. 2000;60:1189. [PubMed] [Google Scholar]
- Furihata T. Hosokawa M. Fujii A. Derbel M. Satoh T. Chiba K. Biochem. Pharmacol. 2005;69:1287. doi: 10.1016/j.bcp.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Shen Y. Yan B. J. Hepatol. 2017;66:660. doi: 10.1016/j.jhep.2016.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanghani S. P. Quinney S. K. Fredenburg T. B. Davis W. I. Murry D. J. Bosron W. F. Drug Metab. Dispos. 2004;32:505. doi: 10.1124/dmd.32.5.505. [DOI] [PubMed] [Google Scholar]
- Holmes R. S. Cox L. A. VandeBerg J. L. Comp. Biochem. Physiol., Part D: Genomics Proteomics. 2009;4:209. doi: 10.1016/j.cbd.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbi M. Beaufay H. J. Biol. Chem. 1991;266:20498. [PubMed] [Google Scholar]
- Fleming C. D. Edwards C. C. Kirby S. D. Maxwell D. M. Potter P. M. Cerasoli D. M. Redinbo M. R. Biochemistry. 2007;46:5063. doi: 10.1021/bi700246n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming C. D. Bencharit S. Edwards C. C. Hyatt J. L. Tsurkan L. Bai F. Fraga C. Morton C. L. Howard-Williams E. L. Potter P. M. Redinbo M. R. J. Mol. Biol. 2005;352:165. doi: 10.1016/j.jmb.2005.07.016. [DOI] [PubMed] [Google Scholar]
- Bencharit S. Morton C. L. Xue Y. Potter P. M. Redinbo M. R. Nat. Struct. Biol. 2003;10:349. doi: 10.1038/nsb919. [DOI] [PubMed] [Google Scholar]
- Bencharit S. Edwards C. C. Morton C. L. Howard-Williams E. L. Kuhn P. Potter P. M. Redinbo M. R. J. Mol. Biol. 2006;363:201. doi: 10.1016/j.jmb.2006.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vistoli G. Pedretti A. Mazzolari A. Testa B. J. Comput.-Aided Mol. Des. 2010;24:771. doi: 10.1007/s10822-010-9373-1. [DOI] [PubMed] [Google Scholar]
- Yu Y. Kong R. Cao H. Yin Z. Liu J. Nan X. Phan A. T. Ding T. Zhao H. Wong S. T. C. Oncotarget. 2018;9:27958. doi: 10.18632/oncotarget.24563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z. Ding L. Li K. Song Y. Dou T. Hou J. Tian X. Feng L. Ge G. Cui J. Anal. Chem. 2019;91:5638. doi: 10.1021/acs.analchem.8b05417. [DOI] [PubMed] [Google Scholar]
- Scott M. Lu G. Hallett M. Thomas D. Y. Bioinformatics. 2004;20:937. doi: 10.1093/bioinformatics/bth010. [DOI] [PubMed] [Google Scholar]
- Raykhel I. Alanen H. Salo K. Jurvansuu J. Van D. N. Latva-Ranta M. Ruddock L. J. Cell Biol. 2007;179:1193. doi: 10.1083/jcb.200705180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T. Taketani M. Shii M. Hosokawa M. Chiba K. Drug Metab. Dispos. 2006;34:1734. doi: 10.1124/dmd.106.009381. [DOI] [PubMed] [Google Scholar]
- Rhoades J. A. Peterson Y. K. Zhu H.-J. Appel D. I. Peloquin C. A. Markowitz J. S. Pharm. Res. 2012;29:972. doi: 10.1007/s11095-011-0637-9. [DOI] [PubMed] [Google Scholar]
- Qian Y. Wang X. Markowitz J. S. Drug Metab. Dispos. 2019;47:465. doi: 10.1124/dmd.118.086074. [DOI] [PubMed] [Google Scholar]
- Shen Y. Eades W. Yan B. Fundam. Clin. Pharmacol. 2021;35(2):432. doi: 10.1111/fcp.12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R. B. Hu Z.-Y. Meibohm B. Laizure S. C. Clin. Pharmacokinet. 2015;54:627. doi: 10.1007/s40262-014-0226-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick K. S. Straughn A. B. Minhinnett R. R. Yeatts S. D. Herrin A. E. DeVane C. L. Malcolm R. Janis G. C. Markowitz J. S. Clin. Pharmacol. Ther. 2007;81:346. doi: 10.1038/sj.clpt.6100082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge P.-X. Jiang L.-P. Tai T. Zhu T. Ji J.-Z. Li Y.-F. Mi Q.-Y. Xie H.-G. Life Sci. 2021:119268. doi: 10.1016/j.lfs.2021.119268. [DOI] [PubMed] [Google Scholar]
- Harris D. S. Everhart E. T. Mendelson J. Jones R. T. Drug Alcohol Depend. 2003;72:169. doi: 10.1016/s0376-8716(03)00200-x. [DOI] [PubMed] [Google Scholar]
- Andrews P. J. Addict. Dis. 1997;16:75. doi: 10.1300/J069v16n03_08. [DOI] [PubMed] [Google Scholar]
- Bencharit S. Morton C. L. Hyatt J. L. Kuhn P. Danks M. K. Potter P. M. Redinbo M. R. Chem. Biol. 2003;10:341. doi: 10.1016/s1074-5521(03)00071-1. [DOI] [PubMed] [Google Scholar]
- Food and Drug Administration and Center for Drug Evaluation and Research, In Vitro Drug Interaction Studies — Cytochrome P450 Enzyme- and Transporter-Mediated Drug Interactions Guidance for Industry, 2020
- Iskow R. C. Gokcumen O. Lee C. Trends Genet. 2012;28:245. doi: 10.1016/j.tig.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen T. W. Graveley B. R. Nature. 2010;463:457. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Shi J. Zhu H.-J. Proteomics. 2019;19:1800288. doi: 10.1002/pmic.201800288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiel M. A. Green S. Davis W. I. Sanghani P. C. Bosron W. F. Sanghani S. P. J. Pharmacol. Exp. Ther. 2007;323:94. doi: 10.1124/jpet.107.127027. [DOI] [PubMed] [Google Scholar]
- Holmes R. S. Cox L. A. VandeBerg J. L. Genetica. 2010;138:695. doi: 10.1007/s10709-010-9438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reily C. Stewart T. J. Renfrow M. B. Novak J. Nat. Rev. Nephrol. 2019;15:346. doi: 10.1038/s41581-019-0129-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieberich E. Adv. Neurobiol. 2014;9:47. doi: 10.1007/978-1-4939-1154-7_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo K. Marth J. D. Cell. 2006;126:855. doi: 10.1016/j.cell.2006.08.019. [DOI] [PubMed] [Google Scholar]
- Kroetz D. L. Gonzalez F. J. McBride O. W. Biochemistry. 1993;32:11606. doi: 10.1021/bi00094a018. [DOI] [PubMed] [Google Scholar]
- Yang D. Pearce R. E. Wang X. Gaedigk R. Wan Y.-J. Y. Yan B. Biochem. Pharmacol. 2009;77:238. doi: 10.1016/j.bcp.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H.-J. Appel D. I. Jiang Y. Markowitz J. S. Drug Metab. Dispos. 2009;37:1819. doi: 10.1124/dmd.109.028209. [DOI] [PubMed] [Google Scholar]
- Shi D. Yang D. Prinssen E. P. Davies B. E. Yan B. J. Infect. Dis. 2011;203:937. doi: 10.1093/infdis/jiq145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines R. N. Simpson P. M. McCarver D. G. Drug Metab. Dispos. 2016;44:959. doi: 10.1124/dmd.115.068957. [DOI] [PubMed] [Google Scholar]
- Yang J. Shi D. Yang D. Song X. Yan B. Mol. Pharmacol. 2007;72:686. doi: 10.1124/mol.107.036889. [DOI] [PubMed] [Google Scholar]
- Shen Y. Shi Z. Yan B. Nucl. Recept. Res. 2019;6:101435. [Google Scholar]
- Tanaka T. Narazaki M. Kishimoto T. Cold Spring Harbor Perspect. Biol. 2014;6:a016295. doi: 10.1101/cshperspect.a016295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabay C. Arthritis Res. Ther. 2006;8(Suppl 2):S3. doi: 10.1186/ar1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan L. Ren X. Yang J. Liu D. Zhang C. Bioorg. Chem. 2020;94:103388. doi: 10.1016/j.bioorg.2019.103388. [DOI] [PubMed] [Google Scholar]
- Hunter P. EMBO Rep. 2008;9:717. doi: 10.1038/embor.2008.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. Rida N. Shi J. Wu A. H. Bleske B. E. Zhu H.-J. Drug Metab. Dispos. 2017;45:1149. doi: 10.1124/dmd.117.077669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H.-J. Wang X. Gawronski B. E. Brinda B. J. Angiolillo D. J. Markowitz J. S. J. Pharmacol. Exp. Ther. 2013;344:665. doi: 10.1124/jpet.112.201640. [DOI] [PubMed] [Google Scholar]
- Bellott R. Le Morvan V. Charasson V. Laurand A. Colotte M. Zanger U. M. Klein K. Smith D. Bonnet J. Robert J. Cancer Chemother. Pharmacol. 2008;61:481. doi: 10.1007/s00280-007-0493-9. [DOI] [PubMed] [Google Scholar]
- Ptaszek M., in Progress in Molecular Biology and Translational Science, Elsevier Inc., 1st edn, 2013, vol. 113, pp. 59–108 [DOI] [PubMed] [Google Scholar]
- Chyan W. Raines R. T. ACS Chem. Biol. 2018;13:1810. doi: 10.1021/acschembio.8b00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M. Kwok S. J. J. Yun S. H. Physiology. 2015;30:40. doi: 10.1152/physiol.00019.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y. Finney N. S. RSC Adv. 2018;8:29051. doi: 10.1039/c8ra02297f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettinger A. and Wittmann T., in Methods in Cell Biology, Elsevier Inc., 1st edn, 2014, vol. 123, pp. 77–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollman R. Stuurman N. J. Cell Sci. 2007;120:3715. doi: 10.1242/jcs.013623. [DOI] [PubMed] [Google Scholar]
- Baruch A. Jeffery D. A. Bogyo M. Trends Cell Biol. 2004;14:29. doi: 10.1016/j.tcb.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Xue L. Qian X. Jin Q. Zhu Y. Wang X. Wang D. Ge G. Yang L. Anal. Bioanal. Chem. 2020;412:2645. doi: 10.1007/s00216-020-02494-y. [DOI] [PubMed] [Google Scholar]
- Pécot T. Zengzhen L. Boulanger J. Salamero J. Kervrann C. eLife. 2018;7:1. doi: 10.7554/eLife.32311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J. Hou Y. Wu J. Shen B. ChemistrySelect. 2020;5:11185. [Google Scholar]
- Arrowsmith C. H. Audia J. E. Austin C. Baell J. Bennett J. Blagg J. Bountra C. Brennan P. E. Brown P. J. Bunnage M. E. Buser-Doepner C. Campbell R. M. Carter A. J. Cohen P. Copeland R. A. Cravatt B. Dahlin J. L. Dhanak D. Edwards A. M. Frye S. V. Gray N. Grimshaw C. E. Hepworth D. Howe T. Huber K. V. M. Jin J. Knapp S. Kotz J. D. Kruger R. G. Lowe D. Mader M. M. Marsden B. Mueller-Fahrnow A. Müller S. O'Hagan R. C. Overington J. P. Owen D. R. Rosenberg S. H. Roth B. Ross R. Schapira M. Schreiber S. L. Shoichet B. Sundström M. Superti-Furga G. Taunton J. Toledo-Sherman L. Walpole C. Walters M. A. Willson T. M. Workman P. Young R. N. Zuercher W. J. Nat. Chem. Biol. 2015;11:536. doi: 10.1038/nchembio.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blagg J. Workman P. Cancer Cell. 2017;32:9. doi: 10.1016/j.ccell.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman P. Collins I. Chem. Biol. 2010;17:561. doi: 10.1016/j.chembiol.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner S. H. Reinhardt C. J. Chan J. Angew. Chem., Int. Ed. 2021;60:5000. doi: 10.1002/anie.202003687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukami T. Kariya M. Kurokawa T. Iida A. Nakajima M. Eur. J. Pharm. Sci. 2015;78:47. doi: 10.1016/j.ejps.2015.07.006. [DOI] [PubMed] [Google Scholar]
- Liederer B. M. Borchardt R. T. J. Pharm. Sci. 2006;95:1177. doi: 10.1002/jps.20542. [DOI] [PubMed] [Google Scholar]
- Li B. Sedlacek M. Manoharan I. Boopathy R. Duysen E. G. Masson P. Lockridge O. Biochem. Pharmacol. 2005;70:1673. doi: 10.1016/j.bcp.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Dvir H. Silman I. Harel M. Rosenberry T. L. Sussman J. L. Chem.-Biol. Interact. 2010;187:10. doi: 10.1016/j.cbi.2010.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kua J. Zhang Y. McCammon J. A. J. Am. Chem. Soc. 2002;124:8260. doi: 10.1021/ja020429l. [DOI] [PubMed] [Google Scholar]
- Harel M. Schalk I. Ehret-Sabatier L. Bouet F. Goeldner M. Hirth C. Axelsen P. H. Silman I. Sussman J. L. Proc. Natl. Acad. Sci. U. S. A. 1993;90:9031. doi: 10.1073/pnas.90.19.9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds H. M. Rivory L. P. Mol. Pharmacol. 1999;56:1346. doi: 10.1124/mol.56.6.1346. [DOI] [PubMed] [Google Scholar]
- Kim D. S. Marsillach J. Furlong C. E. Jarvik G. P. Pharmacogenomics. 2013;14:1495. doi: 10.2217/pgs.13.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaha-Nelson D. Krüger D. M. Szeler K. Ben-David M. Kamerlin S. C. L. J. Am. Chem. Soc. 2017;139:1155. doi: 10.1021/jacs.6b10801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shunmoogam N. Naidoo P. Chilton R. Vasc. Health Risk Manage. 2018;14:137. doi: 10.2147/VHRM.S165173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente X. S. López-Otín C. J. Biol. Chem. 1995;270:12926. doi: 10.1074/jbc.270.21.12926. [DOI] [PubMed] [Google Scholar]
- Lai L. Xu Z. Zhou J. Lee K.-D. Amidon G. L. J. Biol. Chem. 2008;283:9318. doi: 10.1074/jbc.M709530200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I. Song X. Vig B. S. Mittal S. Shin H.-C. Lorenzi P. J. Amidon G. L. Mol. Pharmaceutics. 2004;1:117. doi: 10.1021/mp0499757. [DOI] [PubMed] [Google Scholar]
- Sakurai Y. Ma S. F. Watanabe H. Yamaotsu N. Hirono S. Kurono Y. Kragh-Hansen U. Otagiri M. Pharm. Res. 2004;21:285. doi: 10.1023/b:pham.0000016241.84630.06. [DOI] [PubMed] [Google Scholar]
- Feng Z. Wang H. Zhou R. Li J. Xu B. J. Am. Chem. Soc. 2017;139:3950. doi: 10.1021/jacs.7b00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L.-W. Jin Q. Wang D.-D. Qian Q.-K. Hao D.-C. Ge G.-B. Yang L. Curr. Med. Chem. 2017;25:1627. doi: 10.2174/0929867325666171204155558. [DOI] [PubMed] [Google Scholar]
- Zou L.-W. Dou T.-Y. Wang P. Lei W. Weng Z.-M. Hou J. Wang D.-D. Fan Y.-M. Zhang W.-D. Ge G.-B. Yang L. Front. Pharmacol. 2017;8:1. doi: 10.3389/fphar.2017.00435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florian S. Hümmer S. Catarinella M. Mayer T. U. HFSP J. 2007;1:104. doi: 10.2976/1.2752600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Brolosy M. A. Stainier D. Y. R. PLoS Genet. 2017;13:1. doi: 10.1371/journal.pgen.1006780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B. Bie J. Wang J. Marqueen S. A. Ghosh S. Am. J. Physiol. 2012;303:C427. doi: 10.1152/ajpcell.00103.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt S. E. Durland-Busbice S. Shepard R. L. Heinz-Taheny K. Iversen P. W. Dantzig A. H. Clin. Cancer Res. 2013;19:1159. doi: 10.1158/1078-0432.CCR-12-1184. [DOI] [PubMed] [Google Scholar]
- Lavis L. D. Raines R. T. ACS Chem. Biol. 2008;3:142–155. doi: 10.1021/cb700248m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavis L. D. Raines R. T. ACS Chem. Biol. 2014;9:855. doi: 10.1021/cb500078u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. Dodani S. C. Chang C. J. Nat. Chem. 2012;4:973. doi: 10.1038/nchem.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu T. Urano Y. Anal. Sci. 2015;31:257. doi: 10.2116/analsci.31.257. [DOI] [PubMed] [Google Scholar]
- Kobayashi H. Ogawa M. Alford R. Choyke P. L. Urano Y. Chem. Rev. 2010;110:2620. doi: 10.1021/cr900263j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demchenko A. P., Klymchenko A. S., Pivovarenko V. G. and Ercelen S., in Fluorescence Spectroscopy, Imaging and Probes, ed. R. Kraayenhof, A. J. W. G. Visser and H. Gerritsen, Springer Berlin Heidelberg, 2002, pp. 101–110 [Google Scholar]
- Yuan L. Lin W. Zheng K. Zhu S. Acc. Chem. Res. 2013;46:1462. doi: 10.1021/ar300273v. [DOI] [PubMed] [Google Scholar]
- Lee M. H. Kim J. S. Sessler J. L. Chem. Soc. Rev. 2015;44:4185. doi: 10.1039/c4cs00280f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q. Feng L. Wang D.-D. Dai Z.-R. Wang P. Zou L.-W. Liu Z.-H. Wang J.-Y. Yu Y. Ge G.-B. Cui J.-N. Yang L. ACS Appl. Mater. Interfaces. 2015;7:28474. doi: 10.1021/acsami.5b09573. [DOI] [PubMed] [Google Scholar]
- Liu Z.-M. Feng L. Hou J. Lv X. Ning J. Ge G.-B. Wang K.-W. Cui J.-N. Yang L. Sens. Actuators, B. 2014;205:151. [Google Scholar]
- Ding L. Tian Z. Hou J. Dou T. Jin Q. Wang D. Zou L. Zhu Y. Song Y. Cui J. Ge G. Chin. Chem. Lett. 2019;30:558. [Google Scholar]
- Feng L. Liu Z.-M. Xu L. Lv X. Ning J. Hou J. Ge G.-B. Cui J.-N. Yang L. Chem. Commun. 2014;50:14519. doi: 10.1039/c4cc06642a. [DOI] [PubMed] [Google Scholar]
- Jin Q. Feng L. Wang D.-D. Wu J.-J. Hou J. Dai Z.-R. Sun S.-G. Wang J.-Y. Ge G.-B. Cui J.-N. Yang L. Biosens. Bioelectron. 2016;83:193. doi: 10.1016/j.bios.2016.04.075. [DOI] [PubMed] [Google Scholar]
- Feng L. Liu Z.-M. Hou J. Lv X. Ning J. Ge G.-B. Cui J.-N. Yang L. Biosens. Bioelectron. 2015;65:9. doi: 10.1016/j.bios.2014.10.002. [DOI] [PubMed] [Google Scholar]
- Tian X. Yan F. Zheng J. Cui X. Feng L. Li S. Jin L. James T. D. Ma X. Anal. Chem. 2019;91:15840. doi: 10.1021/acs.analchem.9b04189. [DOI] [PubMed] [Google Scholar]
- Park S. J. Kim Y. J. Kang J. S. Kim I. Y. Choi K. S. Kim H. M. Anal. Chem. 2018;90:9465. doi: 10.1021/acs.analchem.8b02101. [DOI] [PubMed] [Google Scholar]
- Zhou H. Tang J. Zhang J. Chen B. Kan J. Zhang W. Zhou J. Ma H. J. Mater. Chem. B. 2019;7:2989. [Google Scholar]
- Ma C. Wu J. Sun W. Hou Y. Zhong G. Gao R. Shen B. Huang H. Sens. Actuators, B. 2020;325:128798. [Google Scholar]
- Gao M. Hu Q. Feng G. Tang B. Z. Liu B. J. Mater. Chem. B. 2014;2:3438. doi: 10.1039/c4tb00345d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.