Abstract
Triggers and biological processes controlling male or female gonadal differentiation vary in vertebrates, with sex determination (SD) governed by environmental factors or simple to complex genetic mechanisms that evolved repeatedly and independently in various groups. Here, we review sex evolution across major clades of vertebrates with information on SD, sexual development and reproductive modes. We offer an up-to-date review of divergence times, species diversity, genomic resources, genome size, occurrence and nature of polyploids, SD systems, sex chromosomes, SD genes, dosage compensation and sex-biased gene expression. Advances in sequencing technologies now enable us to study the evolution of SD at broader evolutionary scales, and we now hope to pursue a sexomics integrative research initiative across vertebrates. The vertebrate sexome comprises interdisciplinary and integrated information on sexual differentiation, development and reproduction at all biological levels, from genomes, transcriptomes and proteomes, to the organs involved in sexual and sex-specific processes, including gonads, secondary sex organs and those with transcriptional sex-bias. The sexome also includes ontogenetic and behavioural aspects of sexual differentiation, including malfunction and impairment of SD, sexual differentiation and fertility. Starting from data generated by high-throughput approaches, we encourage others to contribute expertise to building understanding of the sexomes of many key vertebrate species.
This article is part of the theme issue ‘Challenging the paradigm in sex chromosome evolution: empirical and theoretical insights with a focus on vertebrates (Part I)’.
Keywords: evolution, genomics, reproduction, vertebrates, sex chromosomes, sex determination
1. Introduction
(a) . Towards an integrative understanding of vertebrate sexual differentiation, development and sex determination
In gonochoristic (for this and other terms see Glossary) vertebrates, the genetic and cellular biological processes determining whether an undifferentiated gonad develops towards male or female exhibit great diversity [1,2]. Sex determination (SD) in vertebrates ranges from environmental SD (ESD) to simple or complex genetic systems (genotypic SD (GSD)) that have evolved repeatedly and independently [3–6]. Great plasticity of the developmental processes determining gonads and their initiation during embryogenesis contrasts with the evolutionary conservation of pathways that regulate development of most other tissues and organs [3,7]. In poikilothermic vertebrates, much of the epigenetics and genetics of SD, sex differentiation and sexual development remains poorly understood, and knowledge in homeotherms is mostly restricted to a few models such as humans, mice and chickens [7]. For fishes and amphibians, a diversity of master SD genes defining sex chromosomes was early postulated [8], with some downstream components of the SD networks appearing conserved. Fascinatingly, recent work has illustrated that the molecular control and regulation of SD factors and gonadal differentiation can substantially differ even among closely related groups with indistinguishable gonadal development at the morphological, histological and cellular levels [3,7,9,10].
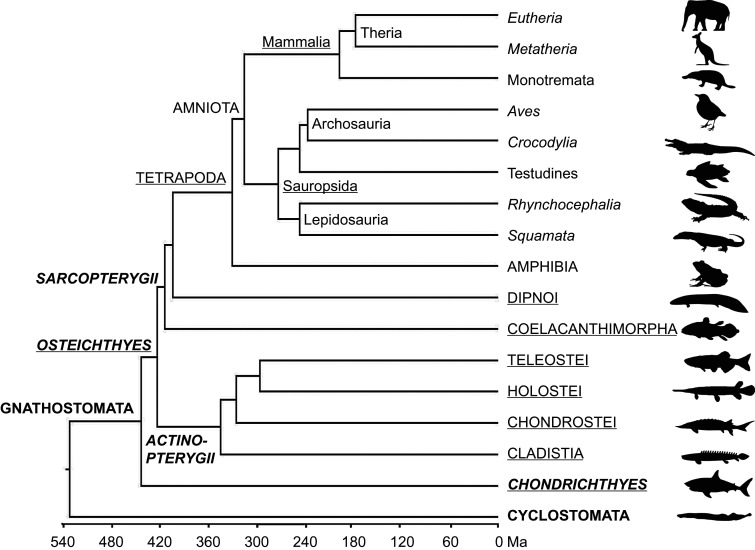
An interesting heterogeneity exists in the evolution of SD in that some clades exhibit very ancient conservation of sex chromosomes (e.g. birds, therian mammals and many reptile lineages, figure 1), whereas others show frequent evolutionary turnovers with variation even between related clades or even species, such as in many amphibians and fishes, and some reptilian lineages [11]. Highly diverse sex chromosomes may derive from frequent turnovers of SD genes [12,13], suggesting that new SD systems may evolve de novo and independently. Deep homology of some sex chromosome systems across disparate taxa suggest that gene content may predispose certain linkage groups to become sex chromosomes [4,14–16], however, so far with relatively weak support in amniotes [17]. Numerous theoretical concepts and models about transitions among SD systems, degeneration and turnover of sex chromosomes [18–23] often remain to be empirically tested in vertebrates. To understand the diversity of SD and sexual development, a deeper and broader knowledge in multiple species from major phylogenetic lineages is necessary. This may have far-reaching consequences also for other fields, owing to likely coevolution of SD, reproductive modes and life history, which are up to now poorly studied, especially in poikilothermic vertebrates [24–26], although these aspects are very relevant for theoretical and empirical studies of sex ratio ecology and evolution [27].
Figure 1.
Phylogenetic tree of major clades of vertebrates. Divergence times in millions of years ago (Ma) according to sources provided in the text; typesetting indicates cladistic hierarchies as also used in the text and in table 1.
Here we present an overview of the current knowledge about SD and the genomic resources available for each vertebrate group, as an overture towards a more comprehensive understanding of vertebrate sex evolution. We review the available whole-genome information in all major clades across the vertebrate tree of life, in relation to knowledge about SD, sexual development and reproductive modes, and available genomic resources. We provide an up-to-date overview on divergence times, species numbers, available genomes, genome size, occurrence and nature of polyploids, SD systems, sex chromosomes, SD genes, dosage compensation and sex-biased gene expression.
Despite the fast-developing sequencing technologies allowing genome assemblies of many vertebrates, we consider high-quality genomes only as a starting point that should be complemented by and synthesized with additional information types in order to comprehensively understand sex evolution. We then pledge for an integrative sexomics research initiative, which uses high-throughput approaches (e.g. RADSex, PoolSex, RNASex, epigenomics) that would integrate the growing numbers of vertebrate species with an available genome assembly to better understand the evolution of genetic SD and differentiation in vertebrates. This sexomics approach could be a starting point for a more in-depth characterization of the ‘complete’ sexome of representative species that would require physiological, cell-biological, behavioural information and beyond to better understand sexual reproduction across lineages.
2. Overview of current knowledge about sex evolution across the vertebrate phylogeny
(a) . Vertebrate sister groups: CEPHALOCHORDATA (LANCELETS) and TUNICATA (TUNICATES)
Extant fish-like lancelets (also called amphioxi; [28,29] are considered the sister group to tunicates and vertebrates (e.g. [30]). Lancelets are gonochorists, but little is known about their SD. Recent genetic evidence suggests a female-heterogametic (ZZ/ZW) GSD system [31]. The karyotype of lancelets is considered to resemble that of ancestral vertebrates [32]. According to traditional models, the early vertebrate ancestors experienced two successive rounds of whole-genome duplications (assigned as 1R, 2R) between approximately 500 and 450 Ma [33,34]. However, Simakov et al. [29] suggested three duplication events—the first before the diversification of extant chordates, the second in the ancestor of lampreys, and the third in the ancestor of jawed vertebrates.
Tunicates, the putative sister group of vertebrates, possess a wide array of reproductive systems. Sedentary ascidians are mostly sequential hermaphrodites, but some produce sperms and eggs simultaneously with incompatible cell-surface proteins, preventing self-fertilization [35]. Colonial species reproduce asexually by budding. Appendicularian tunicates are mostly sequential hermaphrodites [36] but the pelagic tunicate Oikopleura dioica has an XX/XY genetic sex-determining system with possible dosage compensation [37]. Pyrosomes are hermaphroditic as well, reproducing both asexually and sexually with internal fertilization. Thaliaceans (salps) have complex life cycles, obligatorily alternating between sexual and asexual reproduction, allowing rapid population growth while preserving genetic variability [38]. The oozooid develops from a zygote produced by budding, resulting in a chain of individuals that contains an ovary and a testis. The eggs are fertilized internally and the embryo is brooded by the ‘mother’. The life cycle of doliolids is the most complex, again including asexual reproduction with a sequential hermaphroditic phase (for overview: [39]).
With respect to the chordate ancestor, extant lancelets and tunicates may have a derived sexual development, life cycles and SD systems, which evolved during the hundreds of millions of years of divergence from the vertebrate lineage. Nevertheless, as the closest living vertebrate outgroups, they might provide important insights into the deep evolutionary history of sex-related traits and SD genes in vertebrates.
(b) . VERTEBRATA (VERTEBRATES)
CYCLOSTOMATA (JAWLESS FISHES)
The branch of jawless vertebrates with its approximately 120 living species branched off approximately 540 Ma from the lineage leading to all other vertebrates during the Cambrian (figure 1). Agnatha comprises the extant clades of hagfishes (Myxini) and lampreys (Petromyzontiformes). Four lamprey genomes are available [40–42], and an assembly from hagfish (Myxinidae) is available. Hagfish genome size (c-value: 2.4–4.5 Gb; [43]) exceeds that of lampreys (1.4–2.4 Gb). Hagfishes and lampreys are oviparous (table 1). SD and sexual development of hagfishes is poorly understood. Some species appear as protogynous hermaphrodites [44], but no functional simultaneous hermaphrodites have been documented [45,55], other species are gonochoristic [56]. It is possible that SD in lampreys is epigenetic/environmental [57]. However, the critical sex differentiation period is unexplored, and the evidence for ESD in lampreys remains equivocal since GSD as a possible alternative has been proposed recently [58]. The sea lamprey genome contains several hundred genes that are eliminated from somatic cells during early development [41]. Other lampreys and hagfish likewise undergo genome elimination [46], but it remains unknown whether genome elimination plays a role in sexual development.
Table 1.
Overview on available genome assemblies of vertebrates ([42], as December 2020) contrasted with generally known information on sexual phenotype, sex determination mode (SD mode) in gonochorists, system of genotypic sex determination (GSD), reproductive mode, reproduction and master sex determination genes (in GSD species). (Total numbers of species and families were derived from the NCBI taxonomy database and may contain higher than expected numbers owing to the presence of extinct species/families. Herein: x, trait is present; NRSF, trait ‘not reported so far’ despite the availability of at least some data on this topic; no data, to our knowledge, this topic has not been studied/examined; ?, questionable/equivocal evidence.)
| vertebrate genome assemblies at NCBI as of December 2020: |
sexual phenotype |
SD mode in gonochorists |
system of GSD |
reproductive mode |
reproduction |
master SD gene | comments | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| families | species | gonochorism | hermaphroditism | ESD | GSD | male heterogamety | female heterogamety | multilocus sex det. | oviparous | viviparous | bisexual | parthenogenesis, facultative | parthenogenesis, obligate | gynogenesis | hybridogenesis | |||
| CYCLOSTOMATA | 2 (of 3) | 5 (of 546) | x | ? | no data | no data | no data | no data | no data | x | x | x | no data | NRSF | no data | no data | no data | hermaphroditism in hagfish questionable [44,45]; genome elimination in both lampreys and hagfishes [46] |
| GNATHOSTOMATA | 487 (of 1061) | 1646 (of 78 773) | ||||||||||||||||
| CHONDRICHTHYES | 8 (of 56) | 10 (of 1736) | x | NRSF | no data | x | x | ? | no data | x | x | x | x | NRSF | NRSF | NRSF | no data | female heterogamety in a stingray questionable; cases of hermaphroditism report rather on intersexuality (rudimentary hermaphroditism) [47] |
| OSTEICHTHYES | 479 (of 1005) | 1636 (of 77039) | ||||||||||||||||
| ACTINOPTERYGII | 137 (of 498) | 631 (of 38854) | ||||||||||||||||
| CLADISTIA | 1 (of 1) | 1 (of 16) | x | no data | no data | no data | no data | no data | no data | x | NRSF | x | no data | no data | no data | no data | no data | |
| CHONDROSTEI | 1 (of 2) | 2 (of 62) | x | ? | NRSF | x | ? | x | NRSF | x | NRSF | x | NRSF | NRSF | NRSF | NRSF | no data | questionable (male or female heterogamety) SD reported in Polyodon [48,49] |
| HOLOSTEI | 1 (of 2) | 1 (of 14) | x | no data | no data | no data | no data | no data | no data | x | NRSF | x | no data | no data | no data | no data | no data | |
| TELEOSTEI | 134 (of 493) | 627 (of 38659) | x | x | x | x | x | x | x | x | x | x | NRSF | NRSF | x | x | multiple | |
| SARCOPTERYGII | 342 (of 507) | 1005 (of 38 185) | ||||||||||||||||
| COELACANTHIMORPHA | 1 (of 1) | 1 (of 2) | x | no data | no data | no data | no data | no data | no data | NRSF | x | x | no data | no data | no data | no data | no data | |
| DIPNOI | 0 (of 3) | 0 (of 17) | x | no data | no data | no data | no data | no data | no data | x | NRSF | x | no data | no data | no data | no data | no data | |
| TETRAPODA | 341 (of 503) | 1004 (of 38 168) | ||||||||||||||||
| AMPHIBIA | 13 (of 71) | 19 (of 10 180) | x | ? | ? | x | x | x | x | x | x | x | NRSF | NRSF | x (see comment) | x | Dmrt1?, Dm-W (some Xenopus), other suggested | Ambystoma salamanders reproduce by kleptogenesis [50], previously reported as ‘gynogenesis’ [51] |
| AMNIOTA | 328 (of 432) | 985 (of 27 990) | x | x | x | x | x | x | x | x | x | |||||||
| Sauropsida | 206 (of 274) | 564 (of 19 731) | ||||||||||||||||
| Archosauria | 180 (of 197) | 506 (of 10 173) | ||||||||||||||||
| Aves | 177 (of 194) | 502 (of 10 137) | x | NRSF | NRSF | x | NRSF | x | NRSF | x | NRSF | x | ? | NRSF | NRSF | NRSF | Dmrt1 | see main text on parthenogenetic development in domestic birds and [52,53] |
| Crocodylia | 3 (of 3) | 4 (of 36) | x | NRSF | x | NRSF | NRSF | M | NRSF | x | NRSF | x | NRSF | NRSF | NRSF | NRSF | NRSF | |
| Testudines | 13 (of 14) | 22 (of 366) | x | NRSF | x | x | x | x | NRSF | x | NRSF | x | NRSF | NRSF | NRSF | ? | no in ESD/unknown in GSD | see text on Platemys and [54] |
| Lepidosauria | 13 (of 61) | 36 (of 9110) | ||||||||||||||||
| Squamata | 12 (of 60) | 35 (of 9109) | x | NRSF | x | x | x | x | NRSF | x | x | x | x | x | NRSF | NRSF | no in ESD/unknown in GSD | |
| Rhynchocephalia/Sphenodontia | 1 (of 1) | 1 (of 1) | x | NRSF | x | NRSF | NRSF | NRSF | NRSF | x | NRSF | x | NRSF | NRSF | NRSF | NRSF | NRSF | |
| Mammalia | 122 (of 158) | 421 (of 8259) | ||||||||||||||||
| Monotremata | 2 (of 2) | 2 (of 7) | x | NRSF | NRSF | x | x | NRSF | NRSF | x | NRSF | x | NRSF | NRSF | NRSF | NRSF | Amh? | |
| Theria | 120 (156) | 419 (of 8247) | ||||||||||||||||
| Metatheria | 8 (of 19) | 8 (of 422) | x | NRSF | NRSF | x | x | NRSF | NRSF | NRSF | x | x | NRSF | NRSF | NRSF | NRSF | Sry | |
| Eutheria | 112 (of 137) | 411 (of 7825) | x | NRSF | NRSF | x | x | NRSF | x | NRSF | x | x | NRSF | NRSF | NRSF | NRSF | Sry | |
GNATHOSTOMATA (JAWED VERTEBRATES) CHONDRICHTHYES (CARTILAGINOUS FISHES)
With approximately 1200 extant species, cartilaginous fishes comprise the sister group to all other living jawed vertebrates, with elasmobranchs (sharks, rays and relatives) and holocephalans (chimaeras) sharing an Ordovician common ancestor with Osteichthyes approximately 450 Ma [59]. The genomes of only a few species have been characterized, hindered by large genome sizes (2.6–16.6 Gb; [43]). Currently, six sharks [60], two skates [61] and two chimaera genomes [62] have been assembled (recent overview: table 1). The modes of reproduction are very diverse, including yolk-sac viviparity, histotrophy (nutrition of an embryo by uterine secretions), oophagy and placental reproduction [45,63]. Several studies report cases of occasional (facultative) parthenogenetic reproduction giving rise to all-female offspring [64]. Intersexes (often reported as hermaphrodites) were reported in more than 30 elasmobranch species. They frequently showed improper development or maturation rendering one or both sexes nonfunctional [47]. Nevertheless, no functional hermaphroditism was described in this group. SD in Chondrichthyes appears to be largely genotypic with cytogenetic data suggesting XX/XY sex chromosomes in the few studied species of sharks [65] and rays [66], or possibly other forms of male heterogamety in freshwater stingrays (Potamotrygon; [67,68]). A ZZ/ZW system was tentatively reported in the stingray Hypanus americana [65]. We can conclude that there is currently more information about the evolution of the male genitalia, the claspers [69], than on genetic or possible environmental triggers of SD.
OSTEICHTHYES (BONY FISHES)
ACTINOPTERYGII (RAY-FINNED FISHES)
With some more than 31 000 species, the ray-finned fishes are a very diverse vertebrate class, largely comprising extant Teleostei and few non-teleosts: Cladistia (bichirs), Chondrostei (sturgeons and paddlefish, and Holostei (bowfins and gars). The ancestor of Teleostei underwent another round of whole-genome duplication (traditionally called ‘3R WGD’, or ‘teleost-specific WGD’; [33,70]).
CLADISTIA (BICHIRS)
These ray-finned fishes diverged more than 350 Ma in the Devonian from other actinopterygians [71]. Cladistia comprise 13 Polypterus and a single Erpetoichthys species, with large genomes (4.6 Gb to possibly 7.00 Gb) in Polypterus, and a similar genome size (4.4 Gb) in Erpetoichthys [43]. A BAC-library of the Senegal bichir, Polypterus senegalus, was prepared [72]. For the reedfish, Erpetoichthys calabaricus, a chromosome-scale genome assembly is available (table 1). Bichirs are egg-laying and share holoblastic embryonic cleavage with sturgeons. Heteromorphic sex chromosomes have not been found in bichirs [73–75]. Given their phylogenetic position, information on SD and development might provide important insights into the ancestral condition of Actinopterygii.
CHONDROSTEI (STURGEONS and PADDLEFISH)
Sturgeons and paddlefish comprise 27 living species [76] that diverged 330 Ma [77] from the ancestor of the Teleostei and Holostei [33,70]. After their divergence from the other ray-finned fish lineages, sturgeons and paddlefish experienced several polyploidization events, yielding extant species karyotypes from basal approximately 120 up to as many as about 380 chromosomes [78] (and even more in single individuals), and moderate to large genome sizes from 1.4 to 4.4 Gb [43]. Several projects are underway to assemble high-quality sturgeon genomes [77], and a paddlefish genome has been recently published [79]. In contrast to other polyploid fishes, sturgeon genomes maintain a high proportion of ohnologues, i.e. they exhibit a slow deduplication process and loss of several homeologous chromosomes (segmental rediploidization), posing major challenges for genome assembly [77,80]. Chondrostei have exclusively oviparous reproduction [76] and share holoblastic cleavage with most amphibians but not teleosts [81]. Sturgeons do not possess cytologically differentiated sex chromosomes [77,82]. The sex ratio of offspring from experimental gynogenesis yielded contradictory results suggesting either male (XX/XY; [48]) or female heterogamety (ZZ/ZW; [49]) in a paddlefish (Polyodon spathula) and a female-heterogametic (ZZ/ZW) SD system in sturgeon [83], yet a sex-linked marker was not found for decades [77]. Using chromosome-scale assemblies and pool-sequencing, an approximately 16 kb female-specific sequence from sterlet (Acipenser ruthenus) was detected by Kuhl et al. [84]. A polymerase chain reaction-genotyping test, yielding female-specific products in six sturgeon species, spanning the entire phylogeny with the most divergent extant lineages (Acipenser sturio, Acipenser oxyrinchus versus Acipenser ruthenus, Huso huso), stemming from an ancient tetraploidization. Similar results were obtained in two octoploid species (Acipenser gueldenstaedtii, Acipenser baerii). Phylogenetic conservation during 180 Myr of sturgeon evolution and across at least one polyploidization event revealed the oldest known vertebrate system with undifferentiated sex chromosomes, based presumably on a ZZ/ZW-mode of sex determination [84].
HOLOSTEI (BOWFINS and GARS)
A single species of extant bowfin (Amia calva) [85] from North America and closely related gars (Lepisosteiformes), occurring in North and Central America plus the Caribbean, with seven living species [86] represent the sister taxon of teleosts. These two lineages diverged in the Early Permian (approx. 300 Ma; [71]), before the teleost-specific WGD. Eased by reasonable genome sizes in bowfin (1.0–1.3 Gb) and gars (1.0–1.3 Gb), a gar [87], and most recently the bowfin genome [88] have been assembled. Gars and bowfins are oviparous [89,90] and show holoblastic embryonal cleavage. No information on the SD in Holostei is available and no sex-specific genome regions have been identified so far [88].
TELEOSTEI (TELEOSTS)
The rise of teleosts, which comprise approximately 31 000 species [91]) and thus make up over 99% of all ray-finned fishes (Actinopterygii), was accompanied by the teleost-specific WGD (traditionally assigned as ‘3R WGD’) in their common ancestor approximately 300 Ma [34,71]. Some lineages, e.g. salmonids and carps, independently experienced yet additional WGD events. Teleosts evolved meroblastic embryonal cleavage [92]. To date several hundreds of teleost genomes have been assembled (table 1). Owing to advanced deduplication and diploidization of genomes and relatively small to large genome sizes (0.4–5.3 Gb; with most genomes less than 2.0 Gb; [43], whole-genome sequencing (WGS) of teleosts shows great progress among vertebrates. Teleosts feature the largest diversity of reproductive modes [93]. All-female sperm-dependent parthenogenetic (gynogenetic) or hybridogenetic species of hybrid origin [94,95], and even sequential (protandrous, protogynous or serial, i.e. bidirectional) hermaphroditism [96], in some cases involving socially controlled sex change [97,98] and simultaneous hermaphroditism exist [99,100], the latter including the only self-fertilizing vertebrate [101]. Sexual development of teleosts is also very plastic [102], and sex reversal can be easily induced by hormonal and sometimes by environmental triggers or treatments [103], rendering them susceptible to endocrine-disruptive pollution [104]. Data in zebrafish and medaka indicate that germ cell number can drive SD [105,106].
Teleosts show the widest variety of sex-determining mechanisms among vertebrates [107]. This includes gonochorism with ESD and GSD (as well as its environmental modulation), GSD ranging from homomorphic to heteromorphic female (ZZ/ZW) or male heterogametic (XX/XY) systems, plus polygenic SD [108,109] or multiple sex chromosomes [110], with different systems evolved in closely related species. Pure temperature-dependent sex determination (TSD, e.g. [111]) appears to occur in teleosts relatively rarely [107]. In teleosts, the largest number of master SD genes in vertebrates has been characterized [112]. Teleost master SD genes evolved from well-known members of the sexual development regulatory network (the ‘usual suspects’, [3]), stemming in some cases from transcription factors (Dmrt1, Sox3), Tgf-beta signalling pathway members (Amh, Amhr2, Gsdf, Gdf6), or exceptionally from an immune gene in salmonids (Irf9; [113]), triggering male gonadogenesis through an unknown mechanism. The non-recombining region of young teleost sex chromosomes may be remarkably small, e.g. 300 kb in Atlantic herring (Clupea harengus; [114]), some sex chromosomes may even freely recombine, and rarely, the X and Y may differ by just a single nucleotide polymorphism, as reported in the Japanese pufferfish, Takifugu rubripes [115]. In teleosts, the research on rewiring of SD- and sex differentiation gene networks is the most advanced [3]. Compared to the huge teleost biodiversity, the discovery of novel SD genes and systems can be expected from WGS of additional species. Many teleosts, among them sequential or simultaneous hermaphrodites and recent polyploids with specific reproductive modes, such as gynogenesis or hybridogenesis [116], still lack the characterization of their genomes, SD systems and SD genes as well as interactions of allospecific sex chromosomes in taxa of hybrid origin.
SARCOPTERYGII (LOBE-FINNED FISHES)
Coelacanths and lungfishes are the only living sarcopterygian fishes [117] that all trace back to a divergence in Silurian times, i.e. more than 420 Ma [71]; all other extant sarcopterygians comprise tetrapods.
COELACANTHIMORPHA (COELACANTHS)
There are two coelacanth species from southeastern Africa and Sulawesi [118]. Coelacanths are ovoviviparous [119,120]. A coelacanth 2.86 Gb genome of Latimeria chalumnae has been assembled [121]. Over 50 genes involved in sex differentiation and gametogenesis were sequenced in L. chalumnae and Latimeria menadoensis, but no master SD genes have been characterized [122,123]. This situation may not change, given the secretive deep-sea lifestyle of these species and their conservation status (CITES).
DIPNOI (LUNGFISHES)
The six living known species of lungfish occur in Africa, South America and Australia. As their closest living relatives, lungfishes are in a uniquely informative phylogenetic position to infer the ancestral condition of tetrapods [124]. Lungfishes are oviparous [125] and show a pattern similar to holoblastic cleavage [92,125]. While coelacanths have moderate vertebrate genome sizes (2.6 Gb; [121]), lungfish genomes range among the largest in vertebrates (49–60 Gb; [43]). Despite their huge size, the assemblies without information about SD systems from the Australian lungfish (Neoceratodus forsteri) and the African lungfish (Protopterus annectens) have recently been obtained [126,127]. In P. annectens, more than 50 genes related to sex differentiation and gametogenesis have been characterized [123]. Master SD genes have not been identified in lungfishes. The availability of captive breeding in some lungfish species might ease elucidation of SD.
TETRAPODA (TETRAPODS)
AMPHIBIA (AMPHIBIANS)
Soon after their Devonian divergence (335 Ma; [128]) from Amniota, the amphibian lineage to Gymnophiona (caecilians) branched off from that of Anura (frogs and toads) and Urodela (= Caudata: newts and salamanders), while the latter two clades (Anura, Caudata) diverged in the Early Permian (300 Ma, [129]). Many amphibian families are deeply (100–150 Ma) diverged [130,131], with recent evidence that 88% of anurans (Hyloidea, Microhylidae, Natatanura) underwent a rapid Cretaceous–Palaeogene boundary diversification [132]. Gymnophiona also exhibit deep divergences, raising expectations for major genomic evolutionary differences [133]. Cleavage in most frog and salamander embryos is radially symmetrical and holoblastic. The limited knowledge on caecilians, however, suggests meroblastic cleavage in this group [134].
Although there are more species of amphibians (over 8260; [135]) than mammals (6485; [136,137], to date only 19 amphibian genomes of various quality have been assembled, including 15 out of 7291 Anura, 1 out of 760 of Urodela (Caudata) and 3 out of 213 Gymnophiona [42,135]. Even fewer assemblies have reached chromosome-scale quality. The so far slow progress in amphibian genomics is mostly caused by large genome sizes, reaching from 3.9 to 9.8 Gb in Gymnophiona, 1.9–13.1 Gb in Anura, and huge 16.6–78.2 Gb in Urodela [138], and by the large proportions of repetitive sequences. The ongoing dawn of amphibian genomics will be much enlightened by long-read and three-dimensional technologies [139], with many amphibian families still awaiting their first WGS.
Anurans evolved a great diversity of reproductive modes, with terrestrial eggs and exotrophic aquatic larvae, preceding the frequent and repeated evolutionary rise of direct development (terrestrial eggs, no tadpoles), while non-feeding (endotrophic) larvae never led to direct developers [140]. Newts and salamanders exhibit aquatic larvae (rarely involving exceptional or even obligate neoteny, i.e. larval reproduction), as well as terrestrial eggs, and ovo-viviparity with birth of larvae or fully metamorphosed offspring, rarely boosting development by intrauterine cannibalism [141]. Gymnophiona are oviparous or viviparous [142,143], including rare direct developers [144].
While true parthenogenesis most likely did not evolve in amphibians (table 1), hybridogenetic systems, including male- or female-biased and probably GSD-governed population systems occur in anurans [145] as well as kleptogenesis (previously called ‘gynogenesis’; [51]) in salamanders [50], where all-female hybrids of five ploidy levels acquire full or partial genomes from allospecific males and ‘purge’ genomes from deleterious alleles. Recent auto- and allo- (i.e. hybrid origin) polyploids, presenting in amphibians the highest frequency of all vertebrates, are known from several families of anurans and salamanders [146] but are so far unknown in Gymnophiona. Occasional reports on natural sex change in adult anurans (e.g. [147] in Hyperolius viridiflavus) require further examination.
About 96% of the amphibians exhibit undifferentiated sex chromosomes [148,149]. All studied amphibians show GSD and either male (XY/XX) or female (ZZ/ZW) heterogamety [150–152], in addition a putative case of a female W0/00 male SD system [153] and several cases of multiple sex chromosomes [154] have been reported, which form a ring during meiosis in the smoky jungle frog, Leptodactylus pentadactylus [155]. While the vast majority of amphibians exhibit homomorphic XX/XY or ZZ/ZW sex chromosome systems, there are several prominent examples of cytogenetically differentiated sex chromosomes [156,157], and for the African bullfrog, Pyxicephalus adspersus, a draft genome is pre-published [158], from which potential upregulation of the heterogametic W-chromosome and/or repression in the homogametic Z might inform about dosage compensation. In the cytologically indistinguishable sex chromosomes of the western clawed frog, Xenopus tropicalis, male-biased expression of sex-linked transcripts is suspected to be owing to degeneration of the non-recombining portion of the W-chromosome, coupled with incomplete or absent dosage compensation [159]. Cases of sex chromosome-autosome translocations have been shown by cytogenetics [160]. A balanced lethal system in newts (Triturus) may have evolved from a vestigial sex chromosome pair [161,162]. Sex chromosomes of most newts and salamanders are homomorphic [157,163], and the observation of balanced sex ratios from clutches is interpreted as indication for GSD but has remained without genetic evidence [164]. Whole-genome approaches in multiple individuals identified the homomorphic sex chromosome of axolotl (Ambystoma mexicanum), and a putative approximately 300 kb SD region on the W-chromosome [164,165]. Genomic approaches recently also suggested sex-linked loci in ancient clades of giant salamanders (family Cryptobranchidae; [166,167]). Transcriptomic approaches try to circumvent limitations of huge urodelean genome sizes to address sexual developmental aspects [168,169]. Evidence for heteromorphic sex chromosomes exists for at least one species of Gymnophiona [170].
Homomorphic sex chromosomes in amphibians may be caused by high turnover rates [171], where autosomes evolve into new sex chromosomes [8], as documented in ranid frogs [15] and pipid frogs [10]. Another hypothesis to explain homomorphy is occasional X-Y recombination (‘fountain-of-youth’-model; [9]), assuming recombination arrest in males to be controlled by maleness (i.e. by the sexual phenotype rather than the sex chromosomal genotype). Thus, Y chromosomes may recombine, for example, in sex-reversed XY-females, preventing long-term Y degeneration, supported by data from tree frogs [172], true frogs [173] and Palaearctic green toads [174]. Generally, sex reversal in early developmental stages owing to environmental cues is possible, making semi-aquatic amphibians, like fishes, vulnerable to pollution of aquatic ecosystems with endocrine-disruptive compounds [104,175].
Early studies on SD involved experimental sex reversal [176,177], cytogenetics and crossing experiments [148,149]. In-depth molecular studies on amphibian SD stem mostly from clawed frogs (Xenopus), where LG7 is sex-linked in diploid X. tropicalis [178,179] and coexisting X, Y and W-chromosomes are suggested [154,159] but no master SD gene is known [180]. The only well-characterized anuran master SD gene is a Dmrt1-paralogue, the W-linked Dm-w of Xenopus laevis [151,181], present in some closely related Xenopus species but not in the entire pipid radiation [10,182]. Dm-w arose after (and perhaps in response to) tetraploidization [182–184] and may initially not have governed sexual differentiation. Dmrt1 itself is considered a candidate master SD gene in some hylid frogs [185,186], bufonid toads [174] and common frogs (Rana temporaria; [173]), and is also sex-linked in several other ranids [15]. The male versus female-determining molecular mechanisms suggest that parallel amino acid substitutions contributed to the establishment of Dmrt1Y (medaka fish) and Dm-w (Xenopus) as SD genes [187]. A well-studied ranid frog system is that of the Japanese wrinkled frog, Glandirana rugosa, with five genetic lineages: the west Japan, east Japan and XY-groups possess XX/XY systems; the ZW- and Neo-ZW groups ZZ/ZW SD systems [188]. In all lineages, the genes androgen receptor (Ar), splicing factor 1 (Sf-1) and Sry-box transcription factor 3 (Sox3) are located on the Z and W or X and Y chromosomes [189,190]. In most amphibians, the characterization of diploid and polyploid SD systems, evolution by hybridization and introgression and generally the characterization of SD systems, sex chromosomes and their evolution remain unknown from a genomic perspective.
AMNIOTA (AMNIOTES) Sauropsida (sauropsids, reptiles and birds)
Lepidosauria (lepidosaurs)
Rhynchocephalia/Sphenodontia (tuatara) The only extant species in the reptilian order Rhynchocephalia (Sphenodontia), diverged approximately 250 Ma from their sister taxon Squamata (lizards and snakes), is the tuatara (Sphenodon punctatus), endemic to New Zealand. The 5 Gb tuatara genome has been recently reported [191], including a list of sex developmental genes [192]. The oviparous tuataras exhibit a unique form of TSD, with females produced below, and males above 22°C [193]. Tuataras possess no sex chromosomes with neither population genomic resources nor global CG-methylation patterns revealing sex specificity [191,193]. Orthologues of genes acting antagonistically in masculinizing (e.g. Sf1, Sox9) or feminizing (e.g. Rspo1, Wnt4) networks promoting testicular or ovarian development, have been identified, as were genes implicated in TSD (e.g. Cirbp; [7]). This example shows that WGS alone can be insufficient to understand SD, particularly TSD. However, a high-quality genome is an important resource for the evaluation of embryonic transcriptomes or proteomes, which are critical data sources for characterization of genes related to sexual development.
Squamata (squamates, lizards and snakes) Squamates, comprising currently more than 11 000 species [194], diverged approximately 250 Ma from Rhynchocephalia [195], while lepidosaurs diverged 277 Ma from archosaurs and turtles [191]. To date, more than 35 genomes have been assembled (table 1). Genome studies are eased by moderate genome sizes, ranging from 1.3 to 3.7 Gb [196]. Squamate reptiles are oviparous or viviparous; ‘ovo-viviparity’ may be difficult to distinguish from viviparity [197]. They mostly exhibit gonochorism, and very rarely true parthenogenesis (females give birth to genetically identical—‘clonal’—daughters; [94,198]). In several clades of lizards and in a blind snake, diploid or triploid all-female obligate parthenogenetic complexes, mostly of hybrid origin, are known [199,200] or arose in the laboratory [201]. Interestingly, natural polyploid reptiles appear only fertile as triploids [202]. Squamates exhibit GSD with male (XX/XY) or female (ZZ/ZW) heterogamety, having undifferentiated or often differentiated, heteromorphic sex chromosomes, or ESD, mostly in the form of TSD [6,18,203–205]. ESD seems relatively rare, currently estimated to occur in roughly 5% of non-avian reptile species [206]. Multiple neo-sex chromosomes evolved via sex chromosome-autosome fusions more frequently in iguanas with male heterogamety than in snakes with female heterogamety [207], which agrees with similar apparent patterns in other vertebrates [110,207,208]. Neither simultaneous nor sequential hermaphrodite species are known in reptiles [209]. Facultative parthenogenesis is well documented in many snake and lizard lineages, with all-female progeny under male heterogamety but all-male progeny under female heterogamety with degenerated W-chromosomes [210]. Facultative parthenogenesis yielding genetically variable offspring of both sexes was discovered in a xantusiid lizard [211]. Five squamate clades (iguanas, lacertid lizards, varanids, skinks and caenophidian snakes) covering approximately 60% of extant squamates show evolutionary conserved sex chromosomes [206,212–216], while other lineages, particularly Acrodonta (agamid lizards and chameleons), boas and pythons, and geckos exhibit more variable SD [18,205,217–219]. In two snake families and the Komodo dragon (Varanus komodoensis) with female heterogamety, substantial W-chromosome degeneration and the absence of global Z-chromosome dosage compensation has been shown, dosage balance is largely lacking in Z-specific genes in these species [215,220,221]. By contrast, X-linked genes are twofold upregulated in males and thus fully dosage-compensated in Anolis carolinensis [222,223], a species with a 160 Myr-old sex chromosome system [212]. However, a lack of dosage balance under male heterogamety was found in Basiliscus vittatus and Lialis burtonis [224–226]. Rates of evolution in Z-linked genes were demonstrated to be increased, relative to their autosomal homologues in snakes, supporting the fast-Z effect [220]. Nevertheless, many questions remain regarding SD, dosage compensation and evolutionary rates of sex-linked loci, including the reasons for differences in the variability of SD among squamate lineages.
Testudines (turtles)
Despite their derived anatomy, turtles, containing 361 extant species [194], are related to the bird-crocodilian (Archosaurian) lineage, from which they split between the Upper Permian and Triassic, approximately 270–250 Ma [227], or earlier, in the Carboniferous, 320 Ma [228]. Twenty-two species have draft genomes assembled [42], specifically 18 Cryptodira and four Pleurodira (table 1). Turtles exhibit highly homologous and similarly sized genomes as crocodiles and some birds [229], ranging from 2 to 2.9 Gb [43]. Turtles are exclusively oviparous [197]. They comprise ESD (TSD) or GSD species, the latter with either ZZ/ZW or XX/XY systems [204,230–233]. While ESD is possibly ancestral to turtles and has been found in most studied species, GSD evolved independently at least five times and stayed notably stable in trionychids (ZZ/ZW) and probably also in chelids for many millions of years [233–235], although in chelids their XX/XY sex chromosomes display considerable morphological evolution, including a Y-to-autosome fusion [236]. No global dosage compensation was found in the female-heterogametic trionychid Apalone ferox [237], yet, dosage compensation varying by tissue, age, and temperature is suggested in Apalone spinifera [238]. Preliminary analyses of few sex-linked genes hint to fast-Z and slow-X effects in turtles [239,240]. Despite efforts to elucidate the molecular basis of GSD in turtles by searching for reptilian homologues of genes [232] involved in sexual development of mammals [241] and birds [242], no master SD gene has been identified yet [204]. However, Sf1 (a testis development gene) is translocated to the ZW-chromosomes in Apalone and remains a candidate [243]. Natural polyploids are found in Platemys platycephala, specifically triploids, diploid–triploid mosaic and triploid–tetraploid mosaicism [54]. Transcriptomic analyses in turtles with ESD targeted the network of gonadal development [244–246], including its epigenetic regulation [246,247]. In early embryos of Trachemys scripta, the histone H3 lysine 27 (H3K27) demethylase Kdm6b has temperature-dependent sexually dimorphic expression. Knockdown of Kdm6b at 26°C (all-male offspring) triggers male-to-female sex reversal in more than 80% of embryos. Kdm6b directly promotes transcription of Dmrt1 by eliminating the trimethylation of H3K27 near its promoter. Additionally, overexpression of Dmrt1 was sufficient to rescue the sex reversal induced by disruption of Kdm6b [248]. Recent research revealed that temperature-mediated influx of calcium at 31°C drives phosphorylation of Stat3, which represses transcription of Kdm6b [249]. Still, many research questions on the genomics and molecular mechanisms of SD remain unanswered.
Archosauria (archosaurs)
Crocodilia (crocodiles) Crocodiles, containing only 24 extant species [194] diverged from birds more than 240 Ma [250,251], whereas forms, morphologically similar to the living crocodilians (Alligatoridae, Crocodylidae, Gavialidae), first appear in the fossil record 80–90 Ma [252]. With moderately large genome sizes (2.3–2.9 Gb; [251]), four genomes (Alligator mississippiensis, Alligator sinensis, Crocodylus porosus, Gavialis gangeticus) have been sequenced [251,253]. All crocodiles are oviparous [254]. Crocodiles have no sex chromosomes [255], and sexual differentiation is determined during development by a temperature-sensing mechanism with a poorly understood molecular basis. Earlier gene expression studies [256,257] have more recently been extended using gonadal RNAseq and revealed 41 differentially expressed/spliced genes at a male-producing temperature, including Wnt1, Kdm6b, C/EBP [258] and Jumonji chromatin modifiers [259]. In the Chinese alligator, orthologues of male-determining genes show an increasing or steady expression during gonadogenesis under the male-inducing but a decreasing expression pattern under the female-inducing temperature [260].
Aves (birds) Birds contain more than 10 000 extant species [261]. They shared the last common ancestor with the sister taxon of crocodiles earlier than 240 Ma [251,252]. Eased by high synteny [262] and compact genome sizes (0.9–2.1 Gb; [43]), over 502 [42] of bird genome assemblies have been published [263] and more are in preparation (table 1). Birds share homologous female-heterogametic sex chromosomes, i.e. a ZZ/ZW system [264]. No candidate for a female (W-specific) SD gene has been identified [265,266] and current knowledge strongly suggests that SD in birds is based on copy-number (i.e. dosage) variation of the Z-linked master SD gene Dmrt1 with a key role in testis development, which is missing on the W [267]. The gene Dmrt1 resides in the oldest evolutionary stratum of the Z-chromosome [268], shared by palaeognath and neognath birds [269–271]. A recent study using a CRISPR-Cas9 based mono-allelic targeting approach with sterile surrogate chicken hosts supports this hypothesis [272]. Such a chromosomally male (ZZ) chicken with a single functional copy of Dmrt1 developed ovaries with typical female markers and exhibited follicular development. Interestingly, these animals were indistinguishable in external appearance from wild-type adult males, supporting that the development of male secondary sexual characters is driven by cell-autonomous sex identity and independent of gonadal hormones [272,273]. The rarity of Z0 and ZZW individuals in birds may suggest that these genotypes are often lethal or infertile [274], and that a locus on the W might control dosage compensation of some Z-linked genes [275,276]. Lethality of polyploid bird embryos may be owing to a general disruption of development. Mortality was high among ZZZ individuals, which developed as males [277]. In a study of 4182 chicken embryos, haploids (1.4%), triploids (0.8%, 9 ZZZ, 7 ZZW, 15 ZWW) and tetraploids (0.1%, 1 ZZZZ, 1 ZZWW) were found, none of which survived to hatching ([278]; discussed in: [279]). ZZ-eggs can be sex-reversed to female by oestrogen-exposure during the critical period of gonad formation [7]. Gynandromorphs with male versus female bilateral morphology can arise from double fertilization of a binucleate egg and this bilaterally distinctive chromosomal constitution of cells governs perception of the hormone environment [7]. Facultative parthenogenesis in birds mostly leads to early embryonic mortality, but hatchlings or even adults (all males) were reported in turkey and chicken [52,53]. Multiple neo-sex chromosomes have been found only extremely rarely in birds [280]; however, extended Z and W chromosomes, formed by addition of autosomal material to both Z and W chromosomes, evolved within songbirds in the Sylvioidea superfamily [281–283] and in Eopsaltria australis [284].
Genomics of avian sex chromosomes is well studied and revealed great interspecies diversity of pseudoautosomal regions (PAR) and Z/W differentiation, from relatively modest degradation in some palaeognath species to extreme degradation in most modern birds [285,286]. The PAR is short in many neognaths, and even without genes in chicken [287]. Similar to the surviving genes on the mammalian Y chromosomes, the retained genes on the bird W chromosomes are enriched for housekeeping or putative dosage-sensitive genes with stronger selective constraints than the lost ones, and are conserved between distantly related lineages of birds [287,288]. Shared or lineage-specific recombination suppression produced ‘evolutionary strata’, i.e. punctuated sequence divergence owing to stepwise suppression of recombination between Z and W [268]. These strata evolved by a complex process of W- and Z-linked inversions, the latter comprising 25 in total across avian lineages [270].
All studied birds exhibit incomplete ZZ/ZW dosage compensation [289], which seems gene-specific and partial [290]. Moderation of expression levels partially balances out the otherwise twofold difference [291,292], presumably because not all genes are equally sensitive to dosage differences. For many genes, this twofold expression difference does not appear to be associated with severe fitness costs. In addition, other bird genes have evolved sex-biased expression [285,293]. Likewise, in palaeognath birds, sex chromosome genomics recently revealed incomplete dosage compensation, confirmed large (more than 100 Myr-old) PARs, where genes in some species, however, evolve faster than autosomal ones [294]. Like other sex chromosomes, those of birds accumulate transposable elements in the non-recombining regions of the W [295]. On the W, Peona et al. [296] revealed enrichment of endogenous retroviruses, which can be expressed and may retrotranspose, inducing genome-wide female-biased mutation rates. Furthermore, probably all songbirds have a germline-restricted chromosome (GRC) and thus undergo a form of partial genome elimination [40,41]. First cytogenetically described in zebra finch, Taeniopygia guttata [297], GRC is absent in somatic cells but present in one copy in male germline cells (but eliminated during spermatogenesis) and two copies in female germline cells (reviewed in [298,299]). Recent genomic, transcriptomic and comparative cytogenetic work suggests that the GRC is enriched in genes [300–302]. The zebra finch GRC contains more than 115 paralogues to single-copy genes on 18 autosomes and the Z is enriched in genes involved in female gonadal development. These genes are transcribed in testes and ovaries [301]. Although the exact function of GRC is currently unclear, the GRC resembles an XX/X0 system, albeit one limited to the germline on top of a ZZ/ZW system in germline and soma. Another level of complexity for understanding the songbird sexome arises from the proposed maternal inheritance of the GRC (but see [303]), implying that it is co-inherited with the W and the mitochondrial genome.
Mammalia (mammals)
Monotremata (monotremes) With five extant species [137], this order includes the sole representatives of the subclass Prototheria, which diverged 200 Ma from viviparous mammals (Theria; [304]), represented by Ornithorhynchidae with a single species (platypus) and the 50 Ma diverged Tachyglossidae (echidnas) with four species. Platypus and echidna genomes are among the smallest in mammals (2.7–2.8 Gb; [43]). Monotremes display a fascinating mixture of derived mammalian and primitive amniote morphological and physiological features shared with sauropsids (reptiles including birds), and have a unique reproductive system that combines egg-laying with lactation. Likewise, the platypus genome exhibits a combination of derived and plesiomorphic characters [305]. The echidna genome has just become available [306]. The monotreme karyotypes have been controversial for almost half a century (cf. [307]) but turned out to contain multiple sex chromosomes, which probably arose from sequential rearrangements between ancient sex chromosomes and several autosomes. During gametogenesis, meiotic chains form that comprise 10 sex chromosomes (five Xs and five Ys) in male platypus and 9 (five Xs and four Ys) in male echidnas [307,308]. This monotreme sex chromosome system evolved independently of the sex chromosomes of viviparous mammals approximately 175 Ma [304,309]. The mammalian master SD gene, Sry, is absent from the genome, while the putative avian SD gene, Dmrt1, is located on the chromosome X5, in two copies in females and one in males, i.e. the opposite situation from birds [310]. The most promising master SD candidate is Amh (AmhY), which is known to have a fundamental role in SD of fishes, and is carried by the Y5 chromosome that corresponds to the oldest of the evolutionary strata of the monotreme sex chromosomes [304,311]. The recent improvement of a male platypus genome revealed seven strata, distributed across the five Xs, which sequentially suppressed recombination with their homologous Ys, five of which are shared with echidna [306]. This work also provided insights into the origin and evolution of the 10 platypus sex chromosomes. Sequence homology was found between the chromosome Y5, where AmhY is located, and the chromosome X1, suggesting that the 10 platypus sex chromosomes ancestrally formed a ring, rather than a chain. In contrast to autosomes, there are extensive interchromosomal contacts between the extant platypus sex chromosome pairs. Unusually frequent interchromosomal contacts were also found between the autosomal regions in humans homologous to the platypus sex chromosomes, suggesting that reciprocal translocations leading to the evolution of the multiple platypus sex chromosomes were facilitated by spatial proximity of these chromosomes that pre-existed in the mammalian ancestor. Monotreme dosage compensation of X-linked genes occurs on a gene-by-gene basis [312], rather than through chromosome-wide silencing, as in eutherians and marsupials [313,314].
Theria (viviparous mammals)
Metatheria (marsupials) Marsupials diverged approximately 180 Ma from Eutheria (placentals) [304] and contain 385 extant species [136,137], inhabiting Australasia and the Americas. Marsupials exhibit moderate genome sizes of approximately 3.9 Gb [43,315]. To date, eight genomes have been sequenced [42] and genomic evolution has recently been reviewed [316]. Marsupials differ from eutherian mammals in many features of reproduction and development, e.g. extraembryonic tissues have undergone remarkable modifications to accommodate reduced egg size and quantity of yolk/deutoplasm versus increasing emphasis on viviparity and placentation [317]. While all marsupials show male heterogamety (XX/XY), the X of marsupials vary substantially in size, morphology and banding patterns, even between species with an ancestral-like 2n = 14 karyotype [318]. The marsupial X shares complete homology with two-thirds of the eutherian X, the remaining third is autosomal in marsupials and corresponds to an early addition on the eutherian lineage. The marsupial X, therefore, represents the ancestral therian X [285]. Translocations or fusions between autosomes and sex chromosomes have been observed in several marsupials [207,319]. While marsupials usually inactivate the paternal X chromosome in the female soma by a marsupial-specific non-coding RNA (RSX: RNA on silent X; [320]), dosage compensation often remains incomplete, contrasting to random but tightly controlled eutherian X inactivation [321]. Marsupial dosage compensation is associated with specific epigenetic modifications [322]. Cytogenetics in some bandicoots (family Peramelidae) revealed somatic elimination of one X in females and the Y in males at different ontogenetic stages, resulting in sex chromosome mosaics in various tissues [323]. The marsupial Y is much smaller than the eutherian Y; marsupial X and Y do not share a PAR, and thus cannot form a synaptonemal complex or recombine during the first meiotic division, but a special structure, the dense plate, maintains sex chromosome association to ensure proper segregation [319,324,325]. Marsupial Y chromosomes share the master male SD gene, Sry, with placental Y chromosomes [316,326].
Eutheria (placentals) Placental mammals diverged 180 Ma from marsupials [304] and with 6992 species comprise the vast majority of living mammals [136,137]. Genome size varies between approximately 2.7 Gb in Laurasiatheria, approximately 3.3 Gb in Supraprimates/Euarchontoglires, approximately 4.4 Gb in Xenarthra and approximately 5.3 Gb in Afrotheria [315], with the largest mammalian genome (approx. 7.7 Gb) being that of a rodent from South America, Tympanoctomys barrerae [327]. Placental genome assemblies are available from 411 species [42] (table 1). Presumably owing to sex-specific methylation [328] and/or other aspects of development [279], no polyploid mammals are viable and reports on natural polyploids have been disproved [327,329]. Eutherian sex chromosomes evolved from a pair of autosomes in the therian lineage around 180 Ma, they are nowadays highly differentiated in both size and gene content owing to the arrest of recombination causing the degeneration of the Y [285,330]. The eutherian X chromosome carries more than 1000 genes, whereas the Y contains only a few protein-coding genes [304]. The degree of heteromorphism and PAR of eutherian sex chromosomes can differ dramatically, e.g. humans exhibit two PARs with the larger of about 2.5 Mb, whereas the house mouse PAR is only 0.5 Mb, and other species have even lost their PAR [331,332]. Lineages with multiple neo-sex chromosomes (X1X2X1X2/X1X2Y or XX/XY1Y2) have independently evolved by fusion with an autosome at least 20 times [207]. In some rare cases, a translocation of an autosome to both sex chromosomes has restored a large segment of homology between X and Y, creating a neo-PAR, as found in the African pygmy mouse (Mus minutoides), where it appears to show signs of early stages of sex chromosome differentiation [333]. Eutherians randomly inactivate one of two Xs in female somatic cells by a non-coding RNA (Xist: X-inactive specific transcript; [334]). Active and inactive X chromosomes localize to different subnuclear positions with distinct chromosomal architectures and epigenetic signatures, reflecting their activity state [335]. The eutherian Y exhibits strata that stopped recombining at well-dated time points [304] and carries the master SD gene (Sry). This testis development initiating transcription factor is homologous to the X-linked Sox3 [7,336]. The gene regulatory network of male and female SD- and developmental pathways are best-studied in laboratory house mice [7]. While for 30 years Sry has been thought to comprise a single exon, a cryptic second exon, essential for male SD in mice has just been identified [337]. Although eutherian XX/XY sex determination is extremely conserved, a few rodent species evolved unusual, derived sex chromosome systems [338]. For example, spiny rats, Tokudaia osimensis, and mole voles, Ellobius lutescens, have lost their Y chromosomes including Sry [339,340], and the gene etv is hypothesized to activate Sox9 [7]. On the other hand, fertile females with a Y chromosome are known in some rodents (e.g. Akodon azarae). The situation is probably best explored in the African pygmy mouse (Mus minutoides), with a sex reversal mutation on a mutant X (called X*) and only XY individuals presenting phenotypic males, while genotypic XX, XX* and X*Y mice are females [341]. Genotypic XX females in moles (Talpa occidentalis) develop ovotestes instead of ovaries and exhibit a masculinized phenotype (musculature, external genitalia, aggressiveness). The testicular part of the ovotestes lacks fertile germ cells but contains typical male androgen-producing cells. Recently, it was uncovered that the increased androgen synthesis in female moles is caused by a tandem triplication of a region containing Cyp17A1, a gene controlling androgen synthesis, and an intrachromosomal inversion involving the pro-testicular growth factor gene Fgf9, heterochronically expressed in the ovotestes [342]. Adult mammals cannot perform sex reversal but genetic perturbations can destabilize the commitment to Sertoli and granulosa cell fate in adult life [7], showing that adult mammalian testes or ovaries require repression of the alternative state [343,344]. Gene expression in eutherians across 12 tissues (human, macaque, mouse, rat, dog) revealed hundreds of genes with conserved sex-biased expression but showed that it has arisen recently and is thus not shared between most mammals [345]. XX-genotypes have been experimentally shown to increase lifespan in mice [346].
3. Beyond whole vertebrate genomes: a pledge for ‘sexomics’
There are several ongoing initiatives to sequence many of the 71 000 vertebrate genomes [347–350]. In context to future research on vertebrate SD and differentiation, we hereby suggest that future sequencing efforts target species with missing information on their SD system, the sex chromosomes or special developmental and/or reproductive modes of interest. As an overview, we have prepared table 1, a summary of the electronic supplementary material, table S1, which summarizes currently (December 2020) available whole-genome information in the context of knowledge on sex evolution from [42]. This is where we are now and we think that sequencing technology and bioinformatics will make it increasingly easier to obtain high-quality genomes from non-model species.
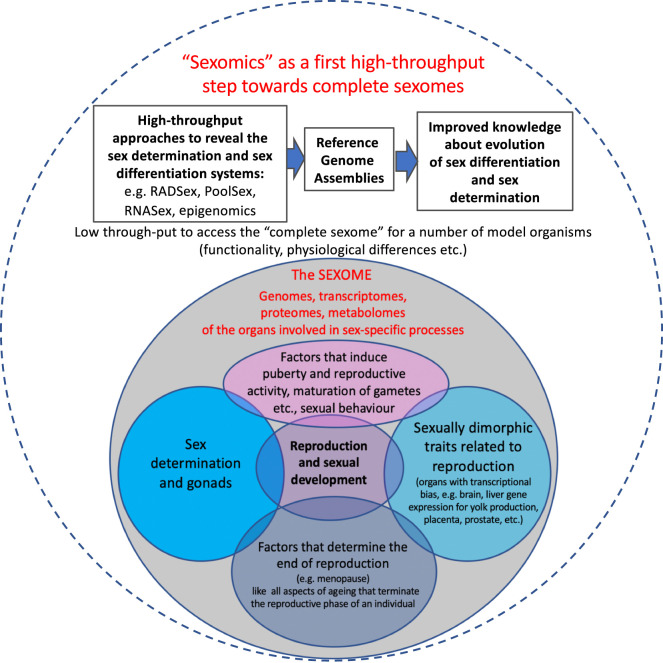
An obvious priority for the sexome (figure 2) to be examined by sexomics is the sequencing and assembly of sex chromosomes in taxa possessing them. Assembling the sex-limited sex chromosome, the Y or W, has been historically difficult owing to the accumulation of repetitive elements and palindromic sequences on the Y and W [287,351]. Many early genome assembly projects chose to sequence the homogametic sex (XX or ZZ individuals) to avoid problems with assembling sex chromosomes and prevent mis-assembly [352,353]. The advent of long-read sequencing, e.g. PacBio and Oxford Nanopore, has made assembly of the hemizygous sex chromosome (Y or W) feasible, and many genome assembly consortia are now using the heterogametic sex as the reference assembly [354,355]. However, sexomics is more than just including sex chromosomes in genome assemblies.
Figure 2.
Scheme on sexomics as a first high-throughput step to improve our understanding of the complete sexome of vertebrates.
While genome sequencing per se will undoubtedly present a driving force towards our understanding of vertebrate sex, we wish to point out that genome sequencing is only a starting point to comprehensively understand SD and sex evolution. For integrative research from here and far beyond, we propose to introduce the terms sexome (and sexomics; figure 2). As the sexome, we consider the information about an individual regarding its sexual differentiation, development and reproduction on all levels of biological organization. This includes the genomic and epigenetic information, the transcriptomes of the organs involved in these sex-specific processes. These organs comprise the gonads, secondary sex organs and characters, organs with a transcriptional sex-bias (e.g. brain, liver gene expression for yolk production, placenta, prostate), the respective proteomes as well as information about environmental factors that induce puberty and reproductive activity, maturation of gametes, etc., sexual behaviour, and finally the factors that determine fertility and the end of or transitions in reproduction (e.g. menopause). It should also include information on malfunction and impairment (e.g. teratology and endocrine disruption).
While we will not be able to cover this universe, we first focus on the analyses of genomes, transcriptomes and proteomes and how they influence the whole picture. We also would like to encourage others (neurobiologists, ethologists, ecologists) to contribute their expertise to complete the sexome (figure 2) of as many species as possible.
Like other ‘-omics’ terms, sexomics describes a special feature of an organism, and the sexomics idea is a term to gather all relevant ‘-omics’ approaches, applicable in high-throughput mode. We argue that the sexome in the first place is a comprehensive description, which comprises all aspects of sexual development and is an archive of data that characterizes a complex phenotype, specific to the reproductive mode of an organism (e.g. female, male, hermaphrodite). Information about the sexome feeds into the classical disciplines (see above) and should be considered at the level of ‘comparative sexomics' as a tool for improving the approaches to a better understanding of molecular and phenotypic evolution, population dynamics, ecology and more. We are convinced that only such comparative approaches across the phylogeny as well as information on intraspecific and intra-population variation, and its regulation will lead to substantial scientific progress. We are sure that this holds particularly true for the sexome.
Elucidating the evolution of sex chromosomes and SD in non-model vertebrates primarily addresses fundamental research questions [2,11], including turnovers of SD systems [356], speciation [357], hybridization [358] and evolutionary development [359]. Likewise, based on similarities and differences in SD and sexual differentiation in non-vertebrates, such as insects and other arthropods, genomic and molecular links between these major taxonomic groups (e.g. the role of the Dmrt gene family [360]) may allow us to consider sexomics-like approaches in other organismal groups.
Beyond basic research, we emphasize that integrative sexomics research in vertebrates will also be of high relevance for many fields of applied research. For example, a major, still poorly understood and complex threat for aquatic and semi-terrestrial vertebrates is endocrine disruption [361]. Missing knowledge on the developmental biology of sex and the genetics of SD remains a major obstacle to study endocrine-disruptive effects in many non-model fishes and amphibians [175,362], and such applications would improve bioindication in freshwater bodies to sense threats for humans. A second field, with relevance to better protect wild fish populations, is sustainable aquaculture with an increasing demand to control sex ratios or to foster mono-sex production [363]. We hope that our review, which does not claim to be complete, will provide a stimulus for upcoming and future research.
Acknowledgement
We thank Jana Thomayerová and Sylvia Kanzler for technical assistance in preparing the manuscript.
Contributor Information
Matthias Stöck, Email: matthias.stoeck@igb-berlin.de.
Lukáš Kratochvíl, Email: lukas.kratochvil@natur.cuni.cz.
Data accessibility
This article has no additional data.
Authors' contributions
M.St. and L.K. drafted the paper to which first M.Sc. and Y.G., and then all co-authors contributed.
Competing interests
We declare we have no competing interests.
Funding
M.St. and M.Sc. were in part supported by COFASP/ERANET (STURGEoNOMICS) by the German Federal Ministry of Food and Agriculture through the Federal Office for Agriculture and Food (grant nos. 2816ERA04G, and 2816ERA05G); M.Sc. was also funded by the German Research Foundation (DFG), grant nos. SCHA408/14-1 und 15-1; H.K. was funded by the German Research Foundation (DFG), grant no. KU 3596/1-1 (project no. 324050651); L.K. and M.R. were supported by the Czech Science Foundation (project no. 17-22604S); B.J.E. was supported by the Natural Sciences and Engineering Research Council of Canada (RGPIN-2017-05770); A.S. was supported by the Swedish Research Council Vetenskapsrådet (grant nos. 2016-05139547 and 2020-04436); N.V. was supported by a grant from the National Science Foundation (grant no. IOS-1555999); B.C. was supported by a grant from the National Science Foundation (grant no. IOS-1256675); Q.Z. is supported by a European Research Council Starting Grant (grant no. 677696).
Glossary
- Autosomes:
all chromosomes of the nucleus, which are not sex chromosomes (or B chromosomes).
- Budding:
a type of asexual reproduction, where the new organism emerges gradually as a bud from the cellular membrane or a body part of the parent.
- Deduplication:
the process by which duplicated genes revert to single-copy genes owing to loss or silencing of one copy.
- Diploidization:
the process by which a polyploid genome turns functionally and structurally (e.g. by gene expression changes, chromosome loss or divergence between homologous chromosomes in autopolyploids) into a diploid state.
- Dosage balance:
(also ‘male-to-female expression balance’, or ‘parity in the expression between sexes’): the molecular mechanisms that equalize the expression of X-/Z-linked genes missing copies on the Y/W chromosomes between sexes, regardless of whether the ancestral expression levels are restored.
- Dosage compensation:
the molecular mechanisms that restore the expression of X-/Z-linked genes with missing copies on the Y/W chromosome equalizing the expression of these genes between sexes, according to some definitions, to the ancestral expression levels.
- Environmental sex determination:
sex is determined by environmental factors, most commonly temperature (TSD) during a sensitive embryonic stage of gonadal development, in species without sex-specific sequence differences in their genome.
- Epigenetic modification:
reversible changes that modify the genetic material and regulate expression without affecting the DNA sequence (e.g. DNA methylation, histone modification).
- Evolutionary stratum:
a region of the sex chromosomes, which ceased recombination in a single step and thus evolutionary period (plural: ‘evolutionary strata’).
- Exotrophic larvae:
larvae feeding on external resources (including maternal trophic eggs) (opposite term ‘endotrophic’).
- Facultative parthenogenesis:
occasional parthenogenetic reproduction in a species that typically reproduces sexually.
- Fast(er)-X/Z effect:
the accelerated evolutionary rate of X- or Z-linked sequences relative to that of autosomal loci.
- Female heterogamety:
species with sex determination controlled by a ZZ/ZW sex chromosome system (and similar derived systems, e.g. with multiple sex chromosomes).
- Gametogenesis:
the developmental process to produce (usually sex-specific and usually haploid) cells specialized for reproduction (gametes), commonly referred to as ovum/egg (in females) and sperm (in males).
- Gametologues:
homologous genes shared by sex chromosomes (e.g. between X and Y chromosomes) in their non-recombining parts.
- Genome elimination:
regulated loss of genomic regions during development of an organism.
- Genotypic sex determination (GSD):
sex is determined by a sex-specific genomic region (at least a single-nucleotide polymorphism), most commonly by a sex-specific combination of chromosomes (i.e. sex chromosomes).
- Germline-restricted chromosomes (GRCs):
type of partial genome elimination where one or more entire chromosomes, the GRCs, are lost during germline-soma differentiation.
- Gonochorism:
having just one of at least two distinct sexes in any one individual organism throughout its lifetime.
- Gynogenesis:
reproductive mode, requiring the activation of embryogenesis by sperm without contribution of paternal DNA (often also called ‘sperm-dependent parthenogenesis' or sometimes ‘pseudogamy’).
- Hemizygous genes:
genes present in a single copy in an otherwise diploid cell, typically Y- and W-specific genes, and likewise X-specific genes in males (i.e. genes present on the X and missing on the Y chromosome or X-linked genes in X0-males of XX/X0 systems), and Z-specific genes in females.
- Hermaphroditism:
developing both male and female gametes during the life cycle of an organism.
- Heterogametic sex:
the sex that produces two types of gametes that each contain one of two different types of sex chromosomes, e.g. XY-males or ZW-females.
- Heteromorphic sex chromosomes:
sex chromosomes that are morphologically distinguishable when viewed with a light microscope (opposite term: ‘homomorphic’).
- Holoblastic (total) embryonal cleavage:
the zygote and blastomeres are completely divided during the cleavage.
- Homeologues:
orthologous genes derived from different lineages or species, combined by hybridization in the same diploid or polyploid genome.
- Homeologous chromosomes:
homologous chromosomes derived from different lineages or species, combined in the same genome of hybrid origin (diploid or polyploid).
- Homeothermic:
having stable, usually physiologically maintained, internal temperature.
- Homogametic sex:
the sex that produces gametes that all contain the same type of sex chromosomes, e.g. XX females or ZZ males.
- Homomorphic sex chromosomes:
sex chromosomes that are morphologically indistinguishable in size and shape (opposite term: ‘heteromorphic’).
- Hybridogenesis:
reproductive mode with selective (usually clonal: ‘hemiclonal’, i.e. ‘half clonal’) transmission of one of the two parental genomes of hybrids to their offspring; more complex inheritance patterns may occur in hybrid polyploid organisms, sometimes collectively termed ‘hybridogenesis’ (or ‘meroclonal’, i.e. ‘partly clonal’, inheritance).
- Introgression:
the transfer/moving of genetic/genomic material from one population or species into the gene pool of another, by hybridization.
- Kleptogenesis:
reproductive mode of a hybrid unisexual species, requiring sperm from a related, often parental, species to trigger the embryonic development; the sperm can either be eliminated (see Gynogenesis) or its genome can be partially or completely incorporated.
- Male heterogamety:
species with sex determination controlled by an XX/XY sex chromosome system (and similar derived systems, e.g. with multiple sex chromosomes).
- Menopause:
post-reproductive life period after the end of female reproduction; known from few cetaceans and hominins.
- Meroblastic (partial) embryonal cleavage:
the cleavage furrows do not completely divide the fertilized egg (usually in eggs with large amounts of yolk).
- Mosaicism:
the presence of more than one population of somatic or germline cells with different genotypes or ploidies within an individual.
- Neo-sex chromosome:
derived sex chromosome, formed by fusion of the ancestral sex chromosome with an autosome.
- Neoteny:
reproduction while retaining juvenile characteristics, e.g. in amphibians (urodela) without full metamorphosis.
- Obligate parthenogenesis:
offspring are produced exclusively by parthenogenesis.
- Ohnologues:
paralogous genes originated from whole-genome duplications, in vertebrate research usually understood as diversified paralogues, evolved by ancient whole-genome duplications.
- Orthologues:
homologous genes originated from a single common ancestor, now present in different genomes (usually separated by a speciation event).
- Oviparity:
the egg is expelled and the embryo largely develops and hatches outside the body of the mother.
- Ovo-viviparity:
the egg develops until hatching within the mother, where the embryo is feeding exclusively on nutrients pre-deposited in the egg.
- Ovotestis:
a gonad with both ovarian and testicular tissues (irrespective of their functionality).
- Palindromic sequence:
complementary short DNA or RNA sequence motifs, arranged in close proximity but with opposite orientation; they can potentially form secondary structures, such as hairpins.
- Parthenogenesis:
reproductive mode by which offspring (or at least an embryo) is produced from an egg without genetic contribution from sperm.
- Paralogues:
homologous genes in the same genome, originated by duplication (local gene duplication, whole-genome duplication).
- Poikilothermic:
having variable, usually environmentally dependent, internal temperature.
- Ploidy:
the number of the complete sets of chromosomes in a eukaryotic cell (i.e. one set = haploidy, two sets = diploidy).
- Polygenic sex determination:
sex is controlled by multiple genes (also called ‘multilocus sex determination’).
- Polyploidy:
more than two complete sets of chromosomes occur in a eukaryotic cell.
- Protandrous hermaphroditism:
producing male gametes in the first stage of the life cycle, and female gametes in a later stage of an organism.
- Protogynous hermaphroditism:
producing female gametes in the first stage of the life cycle, and male gametes in a later stage of an organism.
- Pseudoautosomal region:
recombining part of sex chromosomes.
- Recombination:
the exchange of genetic material between homologous chromosomes, occurring during meiosis (most frequent and regularly) or mitosis in eukaryotes.
- Sequential hermaphroditism:
producing female and male gametes at different periods of the life cycle of an organism.
- Sex determination:
the developmental process deciding the sex of the individual.
- Sex-determining locus:
a locus determining the sex of an individual, in vertebrates triggering the differentiation of the initially bipotential gonad either towards testis or ovary.
- Sex chromosomes:
chromosomes that carry a sex-determining locus (or loci) and segregate in a sex-specific manner.
- Sex chromosome differentiation:
the process leading to changes in content and structure between the homologous X and Y (or Z and W) sex chromosomes, involving one or more of the following events: accumulation of sexually antagonistic alleles, loss of functional genes, recombination arrest, chromosomal rearrangements, heterochromatinization and/or accumulation of repetitive elements.
- Sex chromosome turnover:
evolutionary switch from one sex chromosome system to another, e.g. by the emergence of new master SD genes on new chromosomes or translocation of the sex-determining locus to another chromosome.
- Sex reversal:
the change of sex during the development of an organism, evident by a mismatch between gonadal phenotype (phenotypic sex) and sex-specific genotype (genotypic sex).
- Simultaneous hermaphroditism:
producing both female and male gametes at the same time in one organism.
- Synaptonemal complex:
a protein structure that connects paired homologous chromosomes during the meiotic prophase in eukaryotes.
- Transcription factor:
protein that regulates the transcription of genes.
- Transposable element:
genetic elements capable of mobilizing via copy-and-paste (retrotransposons, including endogenous retroviruses) or cut-and-paste mechanisms (most DNA transposons).
- Viviparity:
the egg develops exclusively within the mother, where the embryo is feeding on nutrients, regularly provided by the mother.
References
- 1.Matson CK, Zarkower D. 2012. Sex and the singular DM domain: insights into sexual regulation, evolution and plasticity. Nat. Rev. Genet. 13, 163-174. ( 10.1038/nrg3161) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bachtrog DKirkpatrick M, Mank JE, Mcdaniel SF, Pires JC, Rice W, Valenzuela N. 2014. Sex determination: why so many ways of doing it? PLoS Biol. 12, e1001899. ( 10.1016/j.tig.2011.05.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Herpin A, Schartl M. 2015. Plasticity of gene-regulatory networks controlling sex determination: of masters, slaves, usual suspects, newcomers, and usurpators. EMBO Rep. 16, 1260-1274. ( 10.15252/embr.201540667) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pan Q, Anderson J, Bertho S, Herpin A, Wilson C, Postlethwait JH, Schartl M, Guiguen Y. 2016. Vertebrate sex-determining genes play musical chairs. C. R. Biol. 339, 258-262. ( 10.1016/j.crvi.2016.05.010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson Pokorná M, Kratochvíl L. 2016. What was the ancestral sex-determining mechanism in amniote vertebrates? Biol. Rev. 91, 1-12. ( 10.1111/brv.12156) [DOI] [PubMed] [Google Scholar]
- 6.Straková B, Rovatsos M, Kubička L, Kratochvíl L. 2020. Evolution of sex determination in amniotes: Did stress and sequential hermaphroditism produce environmental determination? BioEssays 42, e2000050. ( 10.1002/bies.202000050) [DOI] [PubMed] [Google Scholar]
- 7.Capel B. 2017. Vertebrate sex determination: evolutionary plasticity of a fundamental switch. Nat. Rev. Genet. 18, 675-689. ( 10.1038/nrg.2017.60) [DOI] [PubMed] [Google Scholar]
- 8.Schartl M. 2004. Sex chromosome evolution in non-mammalian vertebrates. Curr. Opin. Genet. Dev. 14, 634-641. ( 10.1016/j.gde.2004.09.005) [DOI] [PubMed] [Google Scholar]
- 9.Perrin N. 2009. Sex reversal: a fountain of youth for sex chromosomes? Evolution 63, 3043-3049. ( 10.1111/j.1558-5646.2009.00837.x) [DOI] [PubMed] [Google Scholar]
- 10.Cauret CMS, Gansauge M-T, Tupper AS, Furman BLS, Knytl M, Song XY, Greenbaum E, Meyer M, Evans BJ. 2020. Developmental systems drift and the drivers of sex chromosome evolution. Mol. Biol. Evol. 37, 799-810. ( 10.1093/molbev/msz268) [DOI] [PubMed] [Google Scholar]
- 11.Beukeboom LW, Perrin N. 2014. The evolution of sex determination. Oxford, UK: Oxford University Press. [Google Scholar]
- 12.van Doorn, GS, Kirkpatrick M. 2007. Turnover of sex chromosomes induced by sexual conflict. Nature 449, 909-912. ( 10.1038/nature06178) [DOI] [PubMed] [Google Scholar]
- 13.Ross JA, Urton JR, Boland J, Shapiro MD, Peichel CL. 2009. Turnover of sex chromosomes in the stickleback fishes (Gasterosteidae). PLoS Genet. 5, e1000391. ( 10.1371/journal.pgen.1000391) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.O'Meally D, Ezaz T, Georges A, Sarre SD, Graves JAM. 2012. Are some chromosomes particularly good at sex? Insights from amniotes. Chromosome Res. 20, 7-19. ( 10.1007/s10577-011-9266-8) [DOI] [PubMed] [Google Scholar]
- 15.Jeffries DL, et al. 2018. A rapid rate of sex-chromosome turnover and nonrandom transitions in true frogs. Nat. Comm. 9, 4088. ( 10.1038/s41467-018-06517-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Montiel EE, Badenhorst D, Lee LS, Literman R, Trifonov V, Valenzuela N. 2016. Cytogenetic insights into the evolution of chromosomes and sex determination reveal striking homology of turtle sex chromosomes to amphibian autosomes. Cytogenet. Genome Res. 148, 292-304. ( 10.1159/000447478) [DOI] [PubMed] [Google Scholar]
- 17.Kratochvíl L, Gamble T, Rovatsos M. 2021. Sex chromosome evolution among amniotes: is the origin of sex chromosomes non-random? Phil. Trans. R. Soc. B 376, 20200108. ( 10.1098/rstb.2020.0108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pokorná M, Kratochvíl L. 2009. Phylogeny of sex-determining mechanisms in squamate reptiles: are sex chromosomes an evolutionary trap? Zool. J. Linn. Soc. 156, 168-183. ( 10.1111/j.1096-3642.2008.00481.x) [DOI] [Google Scholar]
- 19.Leonard J.L. 2018. The evolution of sexual systems in animals. In Transitions between sexual systems (ed. Leonard J), pp. 1-58. Cham, Switzerland: Springer. [Google Scholar]
- 20.Ponnikas S, Sigeman H, Abbott JK, Hansson B. 2018. Why do sex chromosomes stop recombining? Trends Genet. 34, 492-503. ( 10.1016/j.tig.2018.04.001) [DOI] [PubMed] [Google Scholar]
- 21.Meisel RP. 2020. Evolution of sex determination and sex chromosomes: a novel alternative paradigm. BioEssays 42, 1900212. ( 10.1002/bies.20200152) [DOI] [PubMed] [Google Scholar]
- 22.Lenormand T, Fyon F, Sun E, Roze D. 2020. Sex chromosome degeneration by regulatory evolution. Curr. Biol. 30, 3001– 3006. ( 10.1016/j.cub.2020.05.052) [DOI] [PubMed] [Google Scholar]
- 23.Charlesworth B, Charlesworth D. 2020. Evolution: a new idea about the degeneration of Y and W chromosomes. Curr. Biol. 30, R871-R896. ( 10.1016/j.cub.2020.06.008) [DOI] [PubMed] [Google Scholar]
- 24.Charnov EL, Bull J. 1977. When is sex environmentally determined? Nature 266, 828-830. ( 10.1038/266828a0). [DOI] [PubMed] [Google Scholar]
- 25.Warner D, Shine R. 2008. The adaptive significance of temperature-dependent sex determination in a reptile. Nature 451, 566-568. ( 10.1038/nature06519). [DOI] [PubMed] [Google Scholar]
- 26.Organ CL, Janes DE, Meade A, Pagel M. 2009. Genotypic sex determination enabled adaptive radiations of extinct marine reptiles. Nature 461, 389-392 ( 10.1038/nature08350) [DOI] [PubMed] [Google Scholar]
- 27.Geffroy B, Douhard M. 2019. The adaptive sex in stressful environments. Trends Ecol. Evol. 34, 628-640 ( 10.1016/j.tree.2019.02.012) [DOI] [PubMed] [Google Scholar]
- 28.Putnam N, et al. 2008. The amphioxus genome and the evolution of the chordate karyotype. Nature 453, 1064-1071. ( 10.1038/nature06967) [DOI] [PubMed] [Google Scholar]
- 29.Simakov O, et al. 2020. Deeply conserved synteny resolves early events in vertebrate evolution. Nat. Ecol. Evol. 4, 820-830 ( 10.1038/s41559-020-1156-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Satoh N, Rokhsar D, Nishikawa T.. 2014. Chordate evolution and the three-phylum system. Proc. R. Soc. B 281, 20141729. ( 10.1098/rspb.2014.1729) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi C, Wu X, Su L, Shang C, Li X, Wang Y, Li G. 2020. A ZZ/ZW sex chromosome system in cephalochordate amphioxus. Genetics 214, 617-622. ( 10.1534/genetics.120.303051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Howell WM, Boschung HT. 1971. Chromosomes of the lancelet, Branchiostoma floridae (order amphioxi). Experientia 27, 1495-1496. ( 10.1007/BF02154315). [DOI] [PubMed] [Google Scholar]
- 33.Meyer A, Van de Peer Y.. 2005. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD). BioEssays 27, 937-945. ( 10.1002/bies.20293) [DOI] [PubMed] [Google Scholar]
- 34.Sacerdot C, Louis A, Bon C, Berthelot C, Crollius HR. 2018. Chromosome evolution at the origin of the ancestral vertebrate genome. Genome Biol. 19, 166. ( 10.1186/s13059-018-1559-1) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sawada H, Shirae-Kurabayashi M.. 2020. Chapter 9. Self- and nonself-recognition of gametes in Ascidians. In Reproduction in aquatic animals (eds Yoshida M, Asturiano J), pp. 179-192. Singapore: Springer. [Google Scholar]
- 36.Holland LZ, Gorsky G, Fenaux R. 1988. Fertilization in Oikopleura dioica (Tunicata, Appendicularia): acrosome reaction, cortical reaction and sperm-egg fusion. Zoomorphology 108, 229-243. ( 10.1007/BF00312223). [DOI] [Google Scholar]
- 37.Navratilova P, Danks GB, Long A, Butcher S, Manak JR, Thompson EM. 2017. Sex-specific chromatin landscapes in an ultra-compact chordate genome. Epigenetics Chromatin 10, 3. ( 10.1186/s13072-016-0110-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henschke N, Everett JD, Anthony J, Richardson AJ, Suthers IM. 2016. Rethinking the role of salps in the ocean. Trends Ecol. Evol. 31, 720-733. ( 10.1016/j.tree.2016.06.007) [DOI] [PubMed] [Google Scholar]
- 39.Holland LZ. 2016. Tunicates. Curr. Biol. 26, R146-R152. ( 10.1016/j.cub.2015.12.024) [DOI] [PubMed] [Google Scholar]
- 40.Smith JJ, et al. 2013. Sequencing of the sea lamprey (Petromyzon marinus) genome provides insights into vertebrate evolution. Nat. Genet. 45, 415-421. ( 10.1038/ng.2568) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith JJ, et al. 2018. The sea lamprey germline genome provides insights into programmed genome rearrangement and vertebrate evolution. Nat. Genet. 50, 270-277 ( 10.1038/s41588-017-0036-1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.NCBI. 2020. www.ncbi.nlm.nih.gov (accessed December 2020).
- 43.Gregory TR. 2015. Animal genome size database. See http://www.genomesize.com. [Google Scholar]
- 44.Gorbman A. 1990. Sex differentiation in the hagfish Eptatretus stouti. Gen. Comp. Endocrinol. 77, 309-323. ( 10.1016/0016-6480(90)90315-D) [DOI] [PubMed] [Google Scholar]
- 45.Adolfi MC, Nakajima RT, N'Nobrega RH, Schartl M. 2019. Intersex, hermaphroditism, and gonadal plasticity in vertebrates: evolution of the Müllerian duct and Amh/Amhr2 signaling. Annu. Rev. Anim. Biosci. 7, 7.1-7.24. ( 10.1146/annurev-animal-020518-114955) [DOI] [PubMed] [Google Scholar]
- 46.Smith JJ, Timoshevskiy VA, Saraceno C. 2021. Programmed DNA elimination in vertebrates. Annu. Rev. Anim. Biosci. 9, 173-201. ( 10.1146/annurev-animal-061220-023220) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hendon JM, Koester DM, Hoffmayer ER, Driggers WB III, Cicia AM. 2013. Occurrence of an intersexual blacktip shark in the northern Gulf of Mexico, with notes on the standardization of classifications for this condition in elasmobranchs. Mar. Coast. Fish. 5, 174-180. ( 10.1080/19425120.2013.799618) [DOI] [Google Scholar]
- 48.Mims SD, Shelton WL, Linhart O, Wang C. 1997. Induced meiotic gynogenesis of paddlefish Polyodon spathula. J. World Aquacult. Soc. 28, 334-343. ( 10.1111/j.1749-7345.1997.tb00280.x) [DOI] [Google Scholar]
- 49.Shelton WL, Mims SD. 2012. Evidence for female heterogametic sex determination in paddlefish Polyodon spathula based on gynogenesis. Aquaculture 356–357, 116-118. ( 10.1016/j.aquaculture.2012.05.029) [DOI] [Google Scholar]
- 50.Bogart JP. 2019. Unisexual salamanders in the genus Ambystoma. Herpetologica 75, 259-267 ( 10.1655/Herpetologica-D-19-00043.1) [DOI] [Google Scholar]
- 51.Macgregor HC, Uzzell TM. 1964. Gynogenesis in salamanders related to Ambystoma jeffersonianum. Science 143, 1043-1045. ( 10.1126/science.143.3610.1043) [DOI] [PubMed] [Google Scholar]
- 52.Olsen MW. 1975. Avian parthenogenesis. Agricultural Research Service USDA, ARS-NE 65, 1-82. [Google Scholar]
- 53.Ramachandran R, McDaniel CD. 2018. Parthenogenesis in birds: a review. Reproduction 155, R245-R257. ( 10.1530/REP-17-0728) [DOI] [PubMed] [Google Scholar]
- 54.Bickham JW, Hanks BG, Hale DW, Martin JE. 1993. Ploidy diversity and the production of balanced gametes in male twist-necked turtles (Platemys platycephala). Copeia 1993, 723 ( 10.2307/1447233) [DOI] [Google Scholar]
- 55.Jørgensen JM, Lomholt JP, Weber RE, Malte H. 1998. The biology of hagfishes. London, UK: Chapman & Hall. [Google Scholar]
- 56.Powell ML, Kavanaugh SI, Sower SA. 2005. Current knowledge of hagfish reproduction: implications for fisheries management. Integr. Comp. Biol. 45: 158-165. ( 10.1093/icb/45.1.158) [DOI] [PubMed] [Google Scholar]
- 57.Johnson NS, Swink WD, Brenden TO.. 2017. Field study suggests that sex determination in sea lamprey is directly influenced by larval growth rate. Proc. R. Soc. B 284, 20170262. ( 10.1098/rspb.2017.0262) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Docker MF, Beamish FWH, Yasmin T, Bryan MB, Khan A.. 2019. Chapter 1: the lamprey gonad. In Lampreys: biology, conservation and control (ed. Docker MF), pp. 1-186. Dordrecht, The Netherlands: Springer. [Google Scholar]
- 59.Irisarri I, et al. 2017. Phylotranscriptomic consolidation of the jawed vertebrate timetree. Nat. Ecol. Evol. 1, 1370-1378. ( 10.1038/s41559-017-0240-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hara Y, et al. 2018. Shark genomes provide insights into elasmobranch evolution and the origin of vertebrates. Nat. Ecol. Evol. 2, 1761-1771. ( 10.1038/s41559-018-0673-5) [DOI] [PubMed] [Google Scholar]
- 61.King BL, Gillis JA, Carlisle HR, Dahn RD. 2011. A natural deletion of the HoxC cluster in elasmobranch fishes. Science 334, 1517. ( 10.1126/science.1210912) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Venkatesh B, et al. 2014. Elephant shark genome provides unique insights into gnathostome evolution. Nature 505, 174-179. ( 10.1038/nature12826) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Musick JA, Ellis J. 2005. Reproductive evolution of chondrichthyans. In Reproductive biology and phylogeny of chondrichthyans (ed. Hamlett WC), pp. 45-79. Enfield, NH: Scientific Publication. [Google Scholar]
- 64.Dudgeon CL, Coulton L, Bone R, Ovenden JR, Thomas S. 2017. Switch from sexual to parthenogenetic reproduction in a zebra shark. Sci. Rep. 7, 40537. ( 10.1038/srep40537) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maddock MB, Schwartz FJ. 1996. Elasmobranch cytogenetics: methods and sex chromosomes. Bull. Mar. Sci. 58, 147-155. [Google Scholar]
- 66.Donahue WHA. 1974. A karyotypic study of three species of rajiformes (Chondrichthyes, Pisces). Can. J. Genet. Cytol. 16, 203-211. ( 10.1139/g74-020) [DOI] [PubMed] [Google Scholar]
- 67.da Cruz VP, Shimabukuro-Dias CK, Oliveira C, Fausto Foresti F.. 2011. Karyotype description and evidence of multiple sex chromosome system X1X1X2X2/X1X2Y in Potamotrygon aff. motoro and P. falkneri (Chondrichthyes: Potamotrygonidae) in the upper Paraná River basin, Brazil. Neotrop. Ichthyol. 9, 201-208. ( 10.1590/S1679-62252011000100020) [DOI] [Google Scholar]
- 68.Valentim FCS, Porto JIR, Bertollo LAC, Gross MC, Feldberg E. 2013. XX/X0, a rare sex chromosome system in Potamotrygon freshwater stingray from the Amazon Basin, Brazil. Genetica 141, 381-387. ( 10.1007/s10709-013-9737-2) [DOI] [PubMed] [Google Scholar]
- 69.O'Shaughnessy KL, Dahn RD, Cohn MJ. 2015. Molecular development of chondrichthyan claspers and the evolution of copulatory organs. Nat. Comm. 6, 6698. ( 10.1038/ncomms7698) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Braasch I, Postlethwait JH.. 2012. Polyploidy in fish and the teleost genome duplication. In Polyploidy and genome evolution (eds Soltis P, Soltis D), pp. 341-383. Berlin, Germany: Springer. [Google Scholar]
- 71.Hughes LC, et al. 2018. Comprehensive phylogeny of ray-finned fishes (Actinopterygii) based on transcriptomic and genomic data. Proc. Natl Acad. Sci. USA 24, 6249-6254. ( 10.1073/pnas.1719358115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Raincrow JD, Dewar K, Stocsits C, Prohaska SJ, Amemiya CT, Stadler PF, Chiu CH. 2011. Hox clusters of the bichir (Actinopterygii, Polypterus senegalus) highlight unique patterns of sequence evolution in gnathostome phylogeny. J. Exp. Zool. (Mol. Dev. Evol.) 316, 451-464. ( 10.1002/jez.b.21420) [DOI] [PubMed] [Google Scholar]
- 73.Morescalchi MA, Stingoa V, Capriglione T. 2011. Cytogenetic analysis in Polypterus ornatipinnis (Actinopterygii, Cladistia, Polypteridae) and 5S rDNA. Mar. Genomics 4, 25-31. ( 10.1016/j.margen.2010.12.002) [DOI] [PubMed] [Google Scholar]
- 74.Morescalchi MA, Liguori I, Rocco L, Archimandritis A, Stingo V. 2008. Karyotypic characterization and genomic organization of the 5S rDNA in Polypterus senegalus (Osteichthyes, Polypteridae). Genetica 132, 179-186. ( 10.1007/s10709-007-9160-7) [DOI] [PubMed] [Google Scholar]
- 75.Morescalchi MA, Liguori I, Rocco L, Stingo V. 2007. Karyotypic characterization and genomic organization of the 5S rDNA in Erpetoichthys calabaricus (Osteichthyes, Polypteridae). Genetica 131, 209-216. ( 10.1007/s10709-006-9119-0) [DOI] [PubMed] [Google Scholar]
- 76.Hochleithner M, Gessner J. 2001. The sturgeons and paddlefishes of the world — biology and aquaculture. Aquatech. Publ. 106, 81-82. [Google Scholar]
- 77.Du K, et al. 2020. The sterlet sturgeon genome sequence and the mechanisms of segmental rediploidization. Nat. Ecol. Evol. 4, 841-852. ( 10.1038/s41559-020-1166-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Havelka M, Hulák M, Bailie D, Prodöhl P, Flajšhans M. 2013. Extensive genome duplications in sturgeons: new evidence from microsatellite data. J. Appl. Ichthyol. 29, 704-708. ( 10.1111/jai.12224) [DOI] [Google Scholar]
- 79.Cheng P, et al. 2020. The American paddlefish genome provides novel insights into chromosomal evolution and bone mineralization in early vertebrates. Mol. Biol. Evol. 38, 1595-1607. ( 10.1093/molbev/msaa326) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Romanenko SA, et al. 2019. Segmental paleotetraploidy revealed in sterlet (Acipenser ruthenus) genome by chromosome painting. Mol. Cytogenet. 8, 90. ( 10.1186/s13039-015-0194-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Saito T, Pšenička M, Goto R, Adachi S, Inoue K, Arai K, Yamaha E. 2014. The origin and migration of primordial germ cells in sturgeons. PLoS ONE 9, e86861. ( 10.1371/journal.pone.0086861) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Keyvanshokooh S, Gharaei A. 2010. A review of sex determination and searches for sex-specific markers in sturgeon. Aquacult. Res. 41, e1-e7. ( 10.1111/j.1365-2109.2009.02463.x) [DOI] [Google Scholar]
- 83.Fopp-Bayat D, Kolman R, Woznicki P. 2007. Induction of meiotic gynogenesis in sterlet (Acipenser ruthenus) using UV-irradiated bester sperm. Aquaculture 264, 54-58. ( 10.1016/j.aquaculture.2006.12.006) [DOI] [Google Scholar]
- 84.Kuhl H, et al. 2021. A 180 Myr-old female-specific genome region in sturgeon reveals the oldest known vertebrate sex determining system with undifferentiated sex chromosomes. Phil. Trans. R. Soc. B 376, 20200089. ( 10.1098/rstb.2020.0089) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Majtánová Z, Symonová R, Arias-Rodriguez L, Sallan L, Ráb P. 2017. ‘Holostei versus halecostomi’ problem: insight from cytogenetics of ancient nonteleost actinopterygian fish, bowfin Amia calva. J. Exp. Zool. (Mol. Dev. Evol.) 328B, 620-628. ( 10.1002/jez.b.22720) [DOI] [PubMed] [Google Scholar]
- 86.Nelson JS, Grande TC, Wilson MVH. 2016. Fishes of the world, 5th edn. New York, NY: John Wiley and Sons. [Google Scholar]
- 87.Braasch I, et al. 2016. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat. Genet. 48, 427-437. ( 10.1038/ng.3526) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thompson A, et al. 2020. The genome of the bowfin (Amia calva) illuminates the developmental evolution of ray-finned fish. Preprint. ( 10.21203/rs.3.rs-92055/v1) [DOI] [Google Scholar]
- 89.Herrington SJ, Hettiger KN, Heist EJ, Keeney DB. 2008. Hybridization between longnose and alligator gars in captivity, with comments on possible gar hybridization in nature. Transact. Am. Fish. Soc. 137, 158-164. ( 10.1577/T07-044.1) [DOI] [Google Scholar]
- 90.McGinn Daniels KL. 1993. Reproductive biology of the bowfin, Amia calva Linnaeus, from the green bottom wildlife management area, Cabell County, West Virginia. Theses, Dissertations Capstones 309. (https://mds.marshall.edu/etd/309) [Google Scholar]
- 91.Froese R, Pauly D (eds). 2019. FishBase World Wide Web electronic publication. See www.fishbase.org.
- 92.Hasley A, Chavez S, Danilchik M, Wühr M, Pelegri F.. 2017. Vertebrate embryonic cleavage pattern determination. In Vertebrate development. Advances in experimental medicine and biology, vol. 953 (eds Pelegri F, Danilchik M, Sutherland A), pp. 117-171. Cham, Switzerland: Springer. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Smith C, Wootton RJ. 2016. The remarkable diversity of teleost fishes. Fish Fish. 17, 1208-1215. ( 10.1111/faf.12116) [DOI] [Google Scholar]
- 94.Avise JC. 2008. Clonality. The genetics, ecology and evolution of sexual abstinence in vertebrate animals, pp. 1-237, New York, NY: Oxford University Press. [Google Scholar]
- 95.Warren W, et al. 2018. Clonal polymorphism and high heterozygosity in the celibate genome of the Amazon molly. Nat. Ecol. Evol. 2, 669-679. ( 10.1038/s41559-018-0473-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Avise JC, Mank JE. 2009. Evolutionary perspectives on hermaphroditism in fishes. Sex. Dev. 3, 152-163. ( 10.1159/000223079) [DOI] [PubMed] [Google Scholar]
- 97.Fricke H, Fricke S. 1977. Monogamy and sex change by aggressive dominance in coral reef fish. Nature 266, 830-832. ( 10.1038/266830a0) [DOI] [PubMed] [Google Scholar]
- 98.Todd EV, et al. 2019. Stress, novel sex genes, and epigenetic reprogramming orchestrate socially controlled sex change. Sci. Adv. 5, eaaw7006. ( 10.1126/sciadv.aaw7006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Erisman BE, Petersen CW, Hastings PA, Warner RR. 2013. Phylogenetic perspectives on the evolution of functional hermaphroditism in teleost fishes. Integ. Comp. Biol. 53, 736-754. ( 10.1093/icb/ict077) [DOI] [PubMed] [Google Scholar]
- 100.Hart MK, Kratter AW, Crowley PH. 2016. Partner fidelity and reciprocal investments in the mating system of a simultaneous hermaphrodite. Behav. Ecol. 27, 1471-1479. ( 10.1093/beheco/arw065) [DOI] [Google Scholar]
- 101.Kanamori A, Sugita Y, Yuasa Y, Suzuki T, Kawamura K. 2016. A Genetic map for the only self-fertilizing vertebrate. G3: Genes Genom. Genet. 4, 1095-1106. ( 10.1534/g3.115.022699) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liu H, Todd E, Lokman M, Laamm MS, Godwin JR, Gemmell NJ. 2017. Sexual plasticity: a fishy tale. Mol. Reprod. Dev. 84, 171-194. ( 10.1002/mrd.22691) [DOI] [PubMed] [Google Scholar]
- 103.Baroiller JF, D'Cotta H. 2016. The reversible sex of gonochoristic fish: insights and consequences. Sex. Dev. 10, 242-266. ( 10.1159/000452362) [DOI] [PubMed] [Google Scholar]
- 104.Martyniuk CJ, Feswick A, Munkittrick KR, Dreier DA, Denslow ND. 2020. Twenty years of transcriptomics, 17alpha-ethinylestradiol, and fish. Gen. Comp. Endocrinol. 286, 113325. ( 10.1016/j.ygcen.2019.113325) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nishimura T, Tanaka M. 2016. The mechanism of germline sex determination in vertebrates. Biol. Reprod. 95, 30. ( 10.1095/biolreprod.115.138271) [DOI] [PubMed] [Google Scholar]
- 106.Nakamura S, Watakabe I, Nishimura T, Picard J-Y, Toyoda A, Taniguchi Y, Di Clemente N, Tanaka M. 2012. Hyperproliferation of mitotically active germ cells due to defective anti-Müllerian hormone signaling mediates sex reversal in medaka. Development 139, 2283-2287. ( 10.1242/dev.076307) [DOI] [PubMed] [Google Scholar]
- 107.Penman DJ, Piferrer F. 2008. Fish gonadogenesis. Part I: genetic and environmental mechanisms of sex determination. Rev. Fisher. Sci. 16(Suppl. 1), 16-34. ( 10.1080/10641260802324610) [DOI] [Google Scholar]
- 108.Ser JR, Roberts RB, Kocher T. 2010. Multiple interacting loci control sex determination in lake Malawi cichlids. Evolution 64, 486-501. ( 10.1111/j.1558-5646.2009.00871.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Moore EC, Roberts RB. 2013. Polygenic sex determination. Curr. Biol. 23, R510-R513. ( 10.1016/j.cub.2013.04.004) [DOI] [PubMed] [Google Scholar]
- 110.Sember A, Nguyen P, Perez MF, Altmanová M, Ráb P, Cioffi M de B. 2021. Multiple sex chromosomes in teleost fishes from a cytogenetic perspective: state of the art and future challenges. Phil. Trans. R. Soc. B 376, 20200098. ( 10.1098/rstb.2020.0098) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yamamoto Y, Zhang Y, Sarida M, Hattori RS, Strüssmann CA. 2014. Coexistence of genotypic and temperature-dependent sex determination in pejerrey Odontesthes bonariensis. PLoS ONE 9, e102574. ( 10.1371/journal.pone.0102574) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Guiguen Y, Fostier A, Herpin A. 2019. Sex determination and differentiation in fish. In Sex control in aquaculture. vol. 1 (eds Wang H-P, Pifferrer F, Chen S-L, Shen Z-G), pp. 35-63. Hoboken, NJ: John Wiley & Sons. [Google Scholar]
- 113.Yano A, et al. 2012. An immune-related gene evolved into the master sex-determining gene in rainbow trout, Oncorhynchus mykiss. Curr. Biol. 22, 1423-1428. ( 10.1016/j.cub.2012.05.045) [DOI] [PubMed] [Google Scholar]
- 114.Rafati N, et al. 2020. Reconstruction of the birth of a male sex chromosome present in Atlantic herring. Proc. Natl Acad. Sci. USA 117, 24 359-24 368. ( 10.1073/pnas.2009925117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kamiya T, et al. 2012. A trans-species missense SNP in Amhr2 is associated with sex determination in the tiger pufferfish, Takifugu rubripes (fugu). PLoS Genet. 8, e1002798. ( 10.1371/journal.pgen.1002798) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lamatsch DK, Stöck M.. 2009. Sperm-dependent parthenogenesis and hybridogenesis in teleost fishes. Chapter 19. In Lost sex – the evolutionary biology of parthenogenesis (eds Schoen I, Martens K, van Dijk P), pp. 399-432. Berlin, Germany: Springer. [Google Scholar]
- 117.Biscotti M, Gerdol M, Canapa A, Forconi M, Olmo E, Pallavicini A, Barucca M, Schartl M. 2016. The lungfish transcriptome: a glimpse into molecular evolution events at the transition from water to land. Sci. Rep. 6, 21571. ( 10.1038/srep21571) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pouyaud L, Wirjoatmodjo S, Rachmatika I, Tjakrawidjaja A, Hadiaty R, Hadie W. 1999. Une nouvelle espèce de coelacanthe. Preuves génétiques et morphologiques. C. R. Acad. Sci. 322, 261-267. ( 10.1016/S0764-4469(99)80061-4) [DOI] [PubMed] [Google Scholar]
- 119.Smith CL, Rand CS, Schaeffer B, Atz JW. 1975. Latimeria, the living coelacanth, is ovoviviparous. Science 190, 1105-1106. ( 10.1126/science.190.4219.1105) [DOI] [Google Scholar]
- 120.Wourms JP, Atz JW, Stribling MD.. 1991. Viviparity and the maternal-embryonic relationship in the coelacanth Latimeria chalumnae. In The biology of Latimeria chalumnae and evolution of coelacanths. Developments in environmental biology of fishes, vol. 12 (eds Musick JA, Bruton MN, Balon EK), pp. 225-248. Dordrecht, The Netherlands: Springer. [Google Scholar]
- 121.Amemiya CT, et al. 2013. The African coelacanth genome provides insights into tetrapod evolution. Nature 496, 311-316. ( 10.1038/nature12027) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Forconi M, et al. 2013. Characterization of sex determination and sex differentiation genes in Latimeria. PLoS ONE 8, e56006. ( 10.1371/journal.pone.0056006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Biscotti MA, Adolfi MC, Barucca M, Forconi M, Pallavicini A, Gerdol M, Canapa A, Costantini M. 2018. A comparative view on sex differentiation and gametogenesis genes in lungfish and coelacanths. Genome Biol. Evol. 10, 1430-1444. ( 10.1093/gbe/evy101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Brinkmann H, Venkatesh B, Brenner S, Meyer A. 2004. Nuclear protein-coding genes support lungfish and not the coelacanth as the closest living relatives of land vertebrates. Proc. Natl Acad. Sci. USA 101, 4900-4905. ( 10.1073/pnas.0400609101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kershaw F, Joss GH, Joss JMP. 2009. Early development in sarcopterygian fishes. In Development of non-teleost fishes (eds Kunz YW, Luer CA, Kapoor BG), pp. 275-289. Enfield, NJ: Scientific Publication. [Google Scholar]
- 126.Meyer A, et al. 2021. Giant lungfish genome elucidates the conquest of land by vertebrates. Nature 590, 284-299. ( 10.1038/s41586-021-03198-8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wang K, et al. 2021. African lungfish genome sheds light on the vertebrate water-to-land transition. Cell 184, 1362-1376. ( 10.1016/j.cell.2021.01.047) [DOI] [PubMed] [Google Scholar]
- 128.Betancur RR, et al. 2013. The tree of life and a new classification of bony fishes. PLoS Curr. Tree Life 1. ( 10.1371/currents.tol.53ba26640df0ccaee75bb165c8c26288) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pyron A. 2011. Divergence time estimation using fossils as terminal taxa and the origins of Lissamphibia. Syst. Biol. 60, 4466-4481. ( 10.1093/sysbio/syr047) [DOI] [PubMed] [Google Scholar]
- 130.Bossuyt F, Roelants K. 2009. Anura. In The timetree of life (eds Hedges SB, Kumar S), pp. 357-364. New York, NY: Oxford University Press. [Google Scholar]
- 131.Pyron RA, Wiens JJ. 2011. A large-scale phylogeny of Amphibia including over 2800 species, and a revised classification of extant frogs, salamanders, and caecilians. Mol. Phylogenet. Evol. 61, 543-583. ( 10.1016/j.ympev.2011.06.012) [DOI] [PubMed] [Google Scholar]
- 132.Feng Y-J, Blackburn DC, Liang D, Hillis DM, Wake DB, Cannatella DC, Zhang P. 2017. Phylogenomics reveals rapid, simultaneous diversification of three major clades of Gondwanan frogs at the Cretaceous–Paleogene boundary. Proc. Natl Acad. Sci. USA 114, E5864-E5870. ( 10.1073/pnas.1704632114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.San Mauro D, Gower DJ, Müller H, Loader SP, Zardoya R, Nussbaum RA, Wilkinson M. 2014. Life-history evolution and mitogenomic phylogeny of caecilian amphibians. Mol. Phylogenet. Evol. 73, 177-189. ( 10.1016/j.ympev.2014.01.009) [DOI] [PubMed] [Google Scholar]
- 134.Dünker N, Wake MH, Olson WM. 2000. Embryonic and larval development in the caecilian Ichthyophis kohtaoensis (Amphibia, Gymnophiona) a staging table. J. Morph. 243, 3-34. () [DOI] [PubMed] [Google Scholar]
- 135.AmphibiaWeb. See www.amphibiaweb.org (accessed December 2020).
- 136.Burgin CJ, Colella JP, Kahn PL, Upham NS. 2018. How many species of mammals are there? J. Mammal. 99, 1-11. ( 10.1093/jmammal/gyz052) [DOI] [Google Scholar]
- 137.Mammal Diversity Database. 2020. Mammal Diversity Database (Version 1.2) (Dataset). Zenodo. Accessed December 2020. ( 10.5281/zenodo.4139818) [DOI]
- 138.Liedtke HC, Gower DJ, Wilkinson M, Gomez-Mestre I. 2018. Macroevolutionary shift in the size of amphibian genomes and the role of life history and climate. Nat. Ecol. Evol. 2, 1792-1799. ( 10.1038/s41559-018-0674-4) [DOI] [PubMed] [Google Scholar]
- 139.Li J, Yu H, Wang W, Fu C, Zhang W, Han F, Wu H. 2019. Genomic and transcriptomic insights into molecular basis of sexually dimorphic nuptial spines in Leptobrachium leishanense. Nat. Comm. 10, 5551. ( 10.1038/s41467-019-13531-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gomez-Mestre I, Pyron RA, Wiens JJ. 2011. Phylogenetic analyses reveal unexpected patterns in the evolution of reproductive modes in frogs. Evolution 66, 3687-3700. ( 10.1111/j.1558-5646.2012.01715.x) [DOI] [PubMed] [Google Scholar]
- 141.Buckley D, Alcobendas M, Garcia-Paris M, Wake MH. 2007. Heterochrony, cannibalism, and the evolution of viviparity in Salamandra salamandra. Evol. Dev. 9, 105-115. ( 10.1111/j.1525-142X.2006.00141.x) [DOI] [PubMed] [Google Scholar]
- 142.Wake MH. 1977. The reproductive biology of caecilians. An evolutionary perspective. In The reproductive biology of amphibians (eds Taylor DH, Guttman SI), pp. 73-100. Oxford, OH: Miami University. [Google Scholar]
- 143.Exbrayat J-M. 2009. Oogenesis and female reproductive system in Gymnophiona. In Reproduction in amphibians (ed. Ogielska M), pp. 305-342. Enfield, NH: Scientific Publication. [Google Scholar]
- 144.Wake MH. 1989. Phylogenesis of direct development and viviparity in vertebrates. In Complex organismal functions: integration and evolution in vertebrates (eds Wake D, Roth G), pp. 235-250. New York, NY: John Wiley & Sons Ltd. [Google Scholar]
- 145.Hoffman A, et al. 2015. Genetic diversity and distribution patterns of diploid and polyploid hybrid water frog populations (Pelophylax esculentus complex) across Europe. Mol. Ecol. 24, 4371-4391. ( 10.1111/mec.13325) [DOI] [PubMed] [Google Scholar]
- 146.Schmid M, Evans BJ, Bogart JP. 2015. Polyploidy in amphibia. Cytogenet. Genome Res. 145, 315-330. ( 10.1159/000431388) [DOI] [PubMed] [Google Scholar]
- 147.Grafe TU, Linsenmair KE. 1989. Protogynous sex change in the reed frog (Hyperolius viridiflavus). Copeia 1989, 1024-1029. ( 10.2307/1445989) [DOI] [Google Scholar]
- 148.Schmid M, Nanda I, Steinlein C, Kausch K, Epplen JT, Haaf T. 1991. Sex-determining mechanisms and sex chromosomes in amphibia. In Amphibian cytogenetics and evolution (eds Green DM, Sessions SK), pp. 393-430. San Diego, CA: Academic Press. [Google Scholar]
- 149.Eggert C. 2004. Sex determination: the amphibian models. Reprod. Nutr. Dev. 44, 539-549. ( 10.1051/rnd:2004062) [DOI] [PubMed] [Google Scholar]
- 150.Hillis DM, Green DM. 1990. Evolutionary changes of heterogametic sex in the phylogenetic history of amphibians. J. Evol. Biol. 3, 49-64. ( 10.1046/j.1420-9101.1990.3010049.x) [DOI] [Google Scholar]
- 151.Ito M. 2018. Sex determination and differentiation in frogs. Reproductive and developmental strategies. Diversity and commonality in animals (eds Kobayashi K, Kitano T, Iwao Y, Kondo M), pp. 349-366. Tokyo, Japan: Springer. [Google Scholar]
- 152.Nakamura M. 2010. The mechanism of sex determination in vertebrates: are sex steroids the key-factor? J. Exp. Zool . 313A, 381-398. ( 10.1002/jez.616) [DOI] [PubMed] [Google Scholar]
- 153.Green DM. 1988. Cytogenetics of the endemic New Zealand frog, Leiopelma hochstetteri: extraordinary supernumerary chromosome variation and a unique sex-chromosome system. Chromosoma 97, 55-77. ( 10.1007/BF00331795) [DOI] [PubMed] [Google Scholar]
- 154.Roco AS, Olmstead AW, Degitz SJ, Amano T, Zimmerman LB, Bullejos M. 2015. Coexistence of Y, W, and Z sex chromosomes in Xenopus tropicalis. Proc. Natl Acad. Sci. USA 112, E4752-E4761. ( 10.1073/pnas.1505291112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Gazoni T, Haddad CFB, Narimatsu H, Cabral-de-Mello DC, Lyra ML, Parise-Maltempi PP. 2018. More sex chromosomes than autosomes in the Amazonian frog Leptodactylus pentadactylus. Chromosoma 127, 269-278. ( 10.1007/s00412-018-0663-z) [DOI] [PubMed] [Google Scholar]
- 156.Schmid M, Steinlein C. 2001. Sex chromosomes, sex-linked genes, and sex determination in the vertebrate class Amphibia. In Genes and mechanisms in vertebrate sex determination (eds Scherer G, Schmid M), pp. 143-176. Basel, Switzerland: Birkhäuser Verlag, [DOI] [PubMed] [Google Scholar]
- 157.Sessions SK, Bizjak Malib L, Green DM, Trifonov V, Ferguson-Smith M. 2016. Evidence for sex chromosome turnover in proteid salamanders. Cytogenet. Genome Res. 148, 305-313. ( 10.1159/000446882) [DOI] [PubMed] [Google Scholar]
- 158.Denton RD, Kudra RS, Malcom JW, Du Preez L, Malone JH.. 2018. The African bullfrog (Pyxicephalus adspersus) genome unites the two ancestral ingredients for making vertebrate sex chromosomes. BioRxiv ( 10.1101/329847) [DOI]
- 159.Furman BLS, Cauret CMS, Knytl M, Song X-Y, Premachandra T, Ofori-Boateng C, Jordan DC, Horb ME, Evans BJ. 2020. A frog with three sex chromosomes that co-mingle together in nature: Xenopus tropicalis has a degenerate W and a Y that evolved from a Z chromosome. PLoS Genet. 16, e1009121. ( 10.1371/journal.pgen.1009121) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Schmid M, Steinlein C. 2018. Chromosome banding in Amphibia. XXXVII. Y-Autosome translocations in Anura. Cytogenet. Genome Res. 154, 153-180. ( 10.1159/000487907) [DOI] [PubMed] [Google Scholar]
- 161.Wallace H. 1994. The balanced lethal system of crested newts. Heredity 73, 41-46. ( 10.1038/hdy.1994.96) [DOI] [Google Scholar]
- 162.Sessions SK, Macgregor HC, Schmid M, Haaf T. 1988. Cytology, embryology and evolution of the developmental arrest syndrome in newts of the genus Triturus (Caudata: Salamandridae). J. Exp. Zool. 248, 321-334. ( 10.1002/jez.1402480311) [DOI] [PubMed] [Google Scholar]
- 163.Green DM, Sessions SK (eds). 1991. Amphibian cytogenetics and evolution. San Diego, CA: Academic Press. [Google Scholar]
- 164.Keinath, MC, Timoshevskaya N, Timoshevskiy VA, Voss R, Smith JJ. 2018. Miniscule differences between sex chromosomes in the giant genome of a salamander. Sci. Rep. 8, 17882. ( 10.1038/s41598-018-36209-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Smith JJ, Voss SR. 2009. Amphibian sex determination: segregation and linkage analysis using members of the tiger salamander species complex (Ambystoma mexicanum and A. t. tigrinum). Heredity 102, 542– 548. ( 10.1038/hdy.2009.15). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Hu Q, Chang C, Wang Q, Tian H, Qiao Z, Wang L, Meng Y, Xu C, Xiao H. 2019. Genome-wide RAD sequencing to identify a sex-specific marker in Chinese giant salamander Andrias davidianus . BMC Genomics 20, 415. ( 10.1186/s12864-019-5771-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Hime PM, Briggler JT, Reece JS, Weisrock DW. 2019. Genomic data reveal conserved female heterogamety in giant salamanders with gigantic nuclear genomes. G3: Genes Genom. Genet. 9, 3467-3476. ( 10.1534/g3.119.400556) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Stöck M, et al. 2019. Shedding light on a secretive Tertiary urodelean relict: Hynobiid salamanders (Paradactylodon persicus s.l.) from Iran, illuminated by phylogeographic, developmental, and transcriptomic data. Genes 10, 306 ( 10.3390/genes10040306) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Biscotti MA, Carducci F, Barucca M, Gerdol M, Pallavicini A, Schartl M, Canapa A, Adolfi MC. 2020. The transcriptome of the newt Cynops orientalis provides new insights into evolution and function of sexual gene networks in sarcopterygians. Sci. Rep. 10, 5445. ( 10.1038/s41598-020-62408-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Venu G, Venkatachalaiah G. 2005. Karyology of two species of caecilians (Caeciliidae: Gymnophiona): evolution through tandem fusion and sex chromosome dimorphism. Caryologia 58, 140-151. ( 10.1080/00087114.2005.10589444) [DOI] [Google Scholar]
- 171.Evans BJ, Pyron RA, Wiens JJ. 2012. Polyploidization and sex chromosome evolution in amphibians. In Polyploidy and genome evolution (eds Soltis PS, Soltis DE), pp. 385-410. Berlin, Germany: Springer. [Google Scholar]
- 172.Stöck M, et al. 2011. Ever-young sex chromosomes in European tree frogs. PLoS Biol. 9, e1001062. ( 10.1371/journal.pbio.1001062) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Rodrigues N, Studer T, Dufresnes C, Perrin N. 2018. Sex-chromosome recombination in common frogs brings water to the fountain-of-youth. Mol. Biol. Evol. 35, 942-948. ( 10.1093/molbev/msy057) [DOI] [PubMed] [Google Scholar]
- 174.Stöck M, Savary R, Betto-Colliard C, Biollay S, Jourdan-Pineau H, Perrin N. 2013. Low rates of X-Y recombination, not turnovers, account for homomorphic sex chromosomes in several diploid species of Palearctic green toads (Bufo viridis subgroup). J. Evol. Biol. 3, 674-682. [DOI] [PubMed] [Google Scholar]
- 175.Tamschick S, Rozenblut-Kościsty B, Ogielska M, Lehmann A, Lymberakis P, Hoffmann F, Lutz I, Kloas W, Stöck M. 2016. Sex reversal assessments reveal different vulnerability to endocrine disruption between deeply diverged anuran lineages. Sci. Rep. 6, 23825. ( 10.1038/srep23825) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ponse K. 1941. La proportion sexuelle dans la descendance issue des œufs produits par l'organe de Bidder des crapauds femelles. Rev. Suisse Zool. 48, 541-544. [Google Scholar]
- 177.Ponse K. 1949. La différentiation du sexe et l'intersexualité chez les vértebrés. Lausanne, Switzerland: F. Rouge. [Google Scholar]
- 178.Olmstead AW, Lindberg-Livingston A, Degitz SJ. 2010. Genotyping sex in the amphibian, Xenopus (Silurana) tropicalis, for endocrine disruptor bioassays. Aquatic Toxicol. 98, 60-66. ( 10.1016/j.aquatox.2010.01.012) [DOI] [PubMed] [Google Scholar]
- 179.Bewick AJ, Anderson DW, Evans BJ. 2013. A large pseudoautosomal region on the sex chromosomes of the frog Silurana tropicalis. Genome Biol. Evol. 5, 1087-1098. ( 10.1093/gbe/evt073) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Mitros T, et al. 2019. A chromosome-scale genome assembly and dense genetic map for Xenopus tropicalis. Dev. Biol. 452, 8-20. ( 10.1016/j.ydbio.2019.03.015) [DOI] [PubMed] [Google Scholar]
- 181.Yoshimoto S, et al. 2008. A W-linked DM-domain gene, DM-W, participates in primary ovary development in Xenopus laevis. Proc. Natl Acad. Sci. USA 105, 2469-2474. ( 10.1073/pnas.0712244105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Bewick AJ, Anderson DW, Evans BJ. 2011. Evolution of the closely related, sex-related genes dm-w and Dmrt1 in African clawed frogs (Xenopus). Evolution 65, 698-712. ( 10.1111/j.1558-5646.2010.01163.x) [DOI] [PubMed] [Google Scholar]
- 183.Furman BLS, Evans BJ. 2016. Sequential turnovers of sex chromosomes in African clawed frogs (Xenopus) suggest some genomic regions are good at sex determination. G3: Genes Genom. Genet. 6, 3625-3633. ( 10.1534/g3.116.033423) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Mawaribuchi S, et al. 2017. Sex chromosome differentiation and the W- and Z-specific loci in Xenopus laevis. Dev. Biol. 426, 393-400. ( 10.1016/j.ydbio.2016.06.015) [DOI] [PubMed] [Google Scholar]
- 185.Brelsford A, et al. 2013. Homologous sex chromosomes in three deeply divergent anuran species. Evolution 67, 2434-2440. ( 10.1111/evo.12151) [DOI] [PubMed] [Google Scholar]
- 186.Brelsford A, Dufresnes C, Perrin N. 2016. Trans-species variation in Dmrt1 is associated with sex determination in four European tree-frog species. Evolution 70, 840-847. ( 10.1111/evo.12891) [DOI] [PubMed] [Google Scholar]
- 187.Ogita Y, et al. 2020. Parallel evolution of two dmrt1-derived genes, dmy and dm-W, for vertebrate sex determination. iScience 23, 100757. ( 10.1016/j.isci.2019.100757) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Miura I. 2008. An evolutionary witness: the frog Rana rugosa underwent change of heterogametic sex from XY male to ZW female. Sex. Dev. 1, 323-331. ( 10.1159/000111764) [DOI] [PubMed] [Google Scholar]
- 189.Uno Y, Nishida C, Oshima Y, Yokoyama S, Miura I. 2008. Comparative chromosome mapping of sex-linked genes and identification of sex chromosomal rearrangements in the Japanese wrinkled frog (Rana rugosa, Ranidae) with ZW and XY sex chromosome systems. Chromosome Res. 16, 637-647. ( 10.1007/s10577-008-1217-7) [DOI] [PubMed] [Google Scholar]
- 190.Miura I. 2017. Sex determination and sex chromosomes in Amphibia. Sex. Dev. 11, 298-306. ( 10.1159/000485270) [DOI] [PubMed] [Google Scholar]
- 191.Gemmell NJ, et al. 2020. The tuatara genome: insights into vertebrate evolution from the sole survivor of an ancient reptilian order. Nature 584, 403-409. ( 10.1038/s41586-020-2561-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Janes DE, et al. 2014. Molecular evolution of Dmrt1 accompanies change of sex-determining mechanisms in reptilia. Biol. Lett. 10, 20140809. ( 10.1098/rsbl.2014.0809) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Cree A, Thompson MB, Daugherty CH. 1995. Tuatara sex determination. Nature 375, 543. ( 10.1038/375543a0)7791870 [DOI] [Google Scholar]
- 194.Uetz P (ed.) 2020. The Reptile Database. See http://www.reptile-database.org (accessed December 2020). [Google Scholar]
- 195.Zheng Y, Wiens JJ. 2016. Combining phylogenomic and supermatrix approaches, and a time-calibrated phylogeny for squamate reptiles (lizards and snakes) based on 52 genes and 4162 species. Mol. Phylogenet. Evol. 94, 537-547. ( 10.1016/j.ympev.2015.10.009) [DOI] [PubMed] [Google Scholar]
- 196.Organ CL, Moreno GR, Edwards SV. 2008. Three tiers of genome evolution in reptiles. Integr. Comp. Biol. 48, 494-504. ( 10.1093/icb/icn046) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Shine R. 1983. Reptilian reproductive modes: the oviparity-viviparity continuum. Herpetologica 39, 1-8. [Google Scholar]
- 198.Darevsky IS. 1958. Natural parthenogenesis in certain subspecies of rock lizards (Lacerta saxicola Eversmann). Dokl. Akad. Nauk SSSR Biol. Sci. 122, 730-732. [Google Scholar]
- 199.Kearney M, Fujita MK, Ridenour J. 2009. Lost sex in the reptiles: constraints and correlations. In Lost sex: the evolutionary biology of parthenogenesis (eds Schön I, Martens K, van Dijk P), pp. 447-474. Dordrecht, The Netherlands: Springer. [Google Scholar]
- 200.Fujita MK, Moritz C. 2009. Origin and evolution of parthenogenetic genomes in lizards: current state and future directions. Cytogenet. Genome Res. 127, 261-272. ( 10.1159/000295177) [DOI] [PubMed] [Google Scholar]
- 201.Lutes AA, Baumann DP, Neaves WB, Baumann P.. 2011. Laboratory synthesis of an independently reproducing vertebrate species. Proc. Natl Acad. Sci. USA 108, 9910-9915. ( 10.1073/pnas.1102811108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Moritz C, Bi K.. 2011. Spontaneous speciation by ploidy elevation: laboratory synthesis of a new clonal vertebrate. Proc. Natl Acad. Sci. USA 108, 9733-9734. ( 10.1073/pnas.1106455108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Ezaz T, Sarre SD, O'Meally D, Graves JAM, Georges A. 2009. Sex chromosome evolution in lizards: independent origins and rapid transitions. Cytogenet. Genome Res. 127, 249-260. ( 10.1159/000300507) [DOI] [PubMed] [Google Scholar]
- 204.Valenzuela N. 2018. Causes and consequences of evolutionary transitions in the level of phenotypic plasticity of reptilian sex determination. In Transitions between sexual systems, (ed. Leonard J), pp. 345-363. Cham, Switzerland: Springer. [Google Scholar]
- 205.Gamble T, Coryell J, Ezaz T, Lynch J, Scantlebury DP, Zarkower D. 2015. Restriction site-associated DNA sequencing (RAD-seq) reveals an extraordinary number of transitions among gecko sex-determining systems. Mol. Biol. Evol. 32, 1296-1309. ( 10.1093/molbev/msv023) [DOI] [PubMed] [Google Scholar]
- 206.Kostmann A, Kratochvíl L, Rovatsos M.. 2021. Poorly differentiated XX/XY sex chromosomes are widely shared across skink radiation. Proc. R. Soc. B 288, 20202139. ( 10.1098/rspb.2020.2139) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Pokorná M, Altmanová M, Kratochvíl L. 2014. Multiple sex chromosomes in the light of female meiotic drive in amniote vertebrate. Chromosome Res. 22, 35-44. ( 10.1007/s10577-014-9403-2) [DOI] [PubMed] [Google Scholar]
- 208.Pennell MW, Kirkpatrick M, Otto SP, Vamosi JC, Peichel CL, Valenzuela N, Kitano J. 2015. Y Fuse? Sex chromosome fusions in fishes and reptiles. PLoS Genet. 11, e1005237. ( 10.1371/journal.pgen.1005237) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Leonard JL. 2013. Williams' paradox and the role of phenotypic plasticity in sexual systems. Integr. Comp. Biol. 53, 671-688. ( 10.1093/icb/ict088) [DOI] [PubMed] [Google Scholar]
- 210.Booth W, Schuett GW. 2016. The emerging phylogenetic pattern of parthenogenesis in snakes. Biol. J. Linn. Soc. Lond. 118, 172-186. ( 10.1111/bij.12744) [DOI] [Google Scholar]
- 211.Kratochvíl L, Vukić J, Červenka J, Kubička L, Johnson Pokorná M, Kukačková D, Rovatsos M, Piálek L. 2020. Mixed-sex offspring produced via cryptic parthenogenesis in a lizard. Mol. Ecol. 9, 4118-4127 ( 10.1111/mec.15617) [DOI] [PubMed] [Google Scholar]
- 212.Rovatsos M, Pokorná M, Altmanová M, Kratochvíl L. 2014. Cretaceous park of sex determination: sex chromosomes are conserved across iguanas. Biol. Lett. 10, 20131093. ( 10.1098/rsbl.2013.1093) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Rovatsos M, Vukić J, Altmanová M, Johnson Pokorná M, Moravec J, Kratochvíl L. 2016. Conservation of sex chromosomes in lacertid lizards. Mol. Ecol. 25, 3120-3126. [DOI] [PubMed] [Google Scholar]
- 214.Augstenová B, Johnson Pokorná M, Altmanová M, Frynta D, Rovatsos M, Kratochvíl L. et al. 2018. ZW, XY, and yet ZW: sex chromosome evolution in snakes even more complicated. Evolution 72, 1701-1707. ( 10.1111/evo.13543) [DOI] [PubMed] [Google Scholar]
- 215.Rovatsos M, Rehák I, Velenský P, Kratochvíl L. 2019. Shared ancient sex chromosomes in varanids, beaded lizards, and alligator lizards. Mol. Biol. Evol. 36, 1113-1120. ( 10.1093/molbev/msz024) [DOI] [PubMed] [Google Scholar]
- 216.Iannucci A, et al. 2019. Conserved sex chromosomes and karyotype evolution in monitor lizards (Varanidae). Heredity 123, 215-227. ( 10.1038/s41437-018-0179-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Nielsen SV, Banks JL, Diaz RE Jr, Trainor PA, Gamble T. 2018. Dynamic sex chromosomes in Old World chameleons (Squamata: Chamaeleonidae). J. Evol. Biol. 31, 484-490. ( 10.1111/jeb.13242) [DOI] [PubMed] [Google Scholar]
- 218.Gamble T. 2010. A review of sex determining mechanisms in geckos (Gekkota: Squamata). Sex. Dev. 4, 88-103. ( 10.1159/000289578) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Gamble T, Castoe TA, Nielsen SV, Banks JL, Card DC, Schield DR, Schuett GW, Booth W. 2017. The discovery of XY sex chromosomes in a Boa and Python. Curr. Biol. 27, 2148-2153. ( 10.1016/j.cub.2017.06.010) [DOI] [PubMed] [Google Scholar]
- 220.Vicoso B, Emerson JJ, Zektser Y, Mahajan S, Bachtrog D. 2013. Comparative sex chromosome genomics in snakes: differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biol. 11, e1001643. ( 10.1371/journal.pbio.1001643) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Lind AL, et al. 2019. Genome of the Komodo dragon reveals adaptations in the cardiovascular and chemosensory systems of monitor lizards. Nat. Ecol. Evol. 3, 1241-1252. ( 10.1038/s41559-019-0945-8) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Rupp SM, Webster TH, Olney KC, Hutchins ED, Kusumi K, Wilson Sayres MA. 2017. Evolution of dosage compensation in Anolis carolinensis, a reptile with XX/XY chromosomal sex determination. Genome Biol. Evol. 9, 231-240. ( 10.1093/gbe/evw263) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Marin R, et al. 2017. Convergent origination of a Drosophila-like dosage compensation mechanism in a reptile lineage. Genome Res. 27, 1974-1987. ( 10.1101/gr.223727.117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Acosta A, et al. 2019. Corytophanids replaced the pleurodont XY system with a new pair of XY chromosomes. Genome Biol. Evol. 9, 666-2677. ( 10.1093/gbe/evz196) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Rovatsos M, Gamble T, Nielsen SV, Georges A, Ezaz T, Kratochvíl L. 2021. Do male and female heterogamety really differ in expression regulation? Lack of global dosage balance in pygopodid geckos. Phil. Trans. R. Soc. B 376, 20200102. ( 10.1098/rstb.2020.0102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Nielsen SV, Guzmán-Méndez IA, Gamble T, Blumer M, Pinto BJ, Kratochvíl L, Rovatsos M. 2015. Escaping the evolutionary trap? Sex chromosome turnover in basilisks and related lizards (Corytophanidae: Squamata). Biol. Lett. 15, 20190498. ( 10.1098/rsbl.2019.0498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Wang Z, et al. 2013. The draft genomes of softshell turtle and green sea turtle yield insights into the development and evolution of the turtle-specific body plan. Nat. Gen. 45, 701-708. ( 10.1038/ng.2615) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Shaffer B, McCartney-Melstad E, Near TJ, Mountac GG, Spinks PQ. 2017. Phylogenomic analyses of 539 highly informative loci dates a fully resolved time tree for the major clades of living turtles (Testudines). Mol. Phylogenet. Evol. 115, 7-15. ( 10.1016/j.ympev.2017.07.006) [DOI] [PubMed] [Google Scholar]
- 229.Kasai F, O'Brien PCM, Ferguson-Smith MA. 2012. Reassessment of genome size in turtle and crocodile based on chromosome measurement by flow karyotyping: close similarity to chicken. Biol. Lett. 8, 631-635. ( 10.1098/rsbl.2012.0141) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Kawagoshi T, Nishida C, Matsuda Y. 2012. The origin and differentiation process of X and Y chromosomes of the black marsh turtle (Siebenrockiella crassicollis, Geoemydidae, Testudines). Chromosome Res. 20, 95-110. ( 10.1007/s10577-011-9267-7) [DOI] [PubMed] [Google Scholar]
- 231.Kawagoshi T, Uno Y, Matsubara K, Matsuda Y, Nishida C. 2009. The ZW micro-sex chromosomes of the Chinese soft-shelled turtle (Pelodiscus sinensis, Trionychidae, Testudines) have the same origin as chicken chromosome 15. Cytogenet. Genome Res. 125, 125-131. ( 10.1159/000227837) [DOI] [PubMed] [Google Scholar]
- 232.Badenhorst D, Stanyon R, Engstrom T, Valenzuela N. 2013. A ZZ/ZW microchromosome system in the spiny softshell turtle, Apalone spinifera, reveals an intriguing sex chromosome conservation in Trionychidae. Chromosome Res. 21, 137-147. ( 10.1007/s10577-013-9343-2) [DOI] [PubMed] [Google Scholar]
- 233.Bista B, Valenzuela N. 2020. Turtle insights into the evolution of the reptilian karyotype and the genomic architecture of sex determination. Genes 11, 416 ( 10.3390/genes11040416) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Mazzoleni S, et al. 2020. Sex is determined by XX/XY sex chromosomes in Australasian side-necked turtles (Testudines: Chelidae). Sci. Rep. 10, 4276. ( 10.1038/s41598-020-61116-w) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Rovatsos M, Praschag P, Fritz U, Kratochvil L. 2017. Stable Cretaceous sex chromosomes enable molecular sexing in softshell turtles (Testudines: Trionychidae). Sci. Rep. 7, 42150. ( 10.1038/srep42150) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Lee LS, Montiel EE, Valenzuela N. 2019. Discovery of putative XX/XY male heterogamety in Emydura subglobosa turtles exposes a novel trajectory of sex chromosome evolution in Emydura. Cytogenet. Genome Res. 158, 160-169. ( 10.1159/000501891) [DOI] [PubMed] [Google Scholar]
- 237.Rovatsos M, Kratochvil L. 2021. Evolution of dosage compensation does not depend on genomic background. Mol. Ecol. 30, 1836-1845. ( 10.1101/2020.08.14.251801) [DOI] [PubMed] [Google Scholar]
- 238.Bista B, Wu Z, Literman R, Valenzuela N. 2021. Thermosensitive sex chromosome dosage compensation in ZZ/ZW softshell turtles, Apalone spinifera. Phil. Trans. R. Soc. B 376, 20200101. ( 10.1098/rstb.2015.2020.0101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Radhakrishnan S, Valenzuela N. 2017. Chromosomal context affects the molecular evolution of sex-linked genes and their autosomal counterparts in turtles and other vertebrates. J. Hered. 108, 720-730. [DOI] [PubMed] [Google Scholar]
- 240.Literman R, Burret A, Bista B, Valenzuela N. 2018. Putative independent evolutionary reversals from genotypic to temperature-dependent sex determination are associated with accelerated evolution of sex-determining genes in turtles. J. Mol. Evol. 86, 11-26. [DOI] [PubMed] [Google Scholar]
- 241.Eggers S, Ohnesorg T, Sinclair A. 2014. Genetic regulation of mammalian gonad development. Nat. Rev. Endocrinol. 10, 673-683. ( 10.1038/nrendo.2014.163) [DOI] [PubMed] [Google Scholar]
- 242.Smith CA. 2010. Sex determination in birds: a review. Emu 110, 364-377. [Google Scholar]
- 243.Lee LS, Montiel Jiménez EE, Navarro-Domínguez BM, Valenzuela N. 2019. Chromosomal rearrangements during turtle evolution altered the synteny of genes involved in vertebrate sex determination. Cytogenet. Genome Res. 157, 77-88. ( 10.1159/000497302). [DOI] [PubMed] [Google Scholar]
- 244.Czerwinski M, Natarajan A, Barske L, Looger LL, Capel B. 2016. A timecourse analysis of systemic and gonadal effects of temperature on sexual development of the red-eared slider turtle Trachemys scripta elegans. Dev. Biol. 420, 166-177. ( 10.1016/j.ydbio.2016.09.018) [DOI] [PubMed] [Google Scholar]
- 245.Radhakrishnan S, Literman R, Neuwald J, Severin A, Valenzuela N. 2017. Transcriptomic responses to environmental temperature by turtles with temperature-dependent and genotypic sex determination assessed by RNAseq inform the genetic architecture of embryonic gonadal development. PLoS ONE 12, e0172044. ( 10.1371/journal.pone.0172044) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Radhakrishnan S, Literman R, Neuwald JL, Valenzuela N. 2018. Thermal response of epigenetic genes informs turtle sex determination with and without sex chromosomes. Sex. Dev. 12, 308-319. ( 10.1159/000492188) [DOI] [PubMed] [Google Scholar]
- 247.Radhakrishnan S, Literman R, Mizoguchi BA, Valenzuela N. 2017. MeDIPseq and nCpG analyses illuminate sexually dimorphic methylation of gonadal development genes with high historic methylation in turtle hatchlings with temperature-dependent sex determination. Epigenetics Chromatin 10, 1-16. ( 10.1186/s13072-017-0136-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Ge C, Ye J, Weber C, Sun W, Zhang H, Zhou Y, Cai C, Qian G, Capel B. 2018. The histone demethylase Kdm6b regulates temperature-dependent sex determination in a turtle species. Science 360, 645-648. ( 10.1126/science.aap8328) [DOI] [PubMed] [Google Scholar]
- 249.Weber C, Capel B. 2021. Sex determination without sex chromosomes. Phil. Trans. R. Soc. B 376, 20200109. ( 10.1098/rstb.2020.0109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Chiari Y, Cahais V, Galtier N, Delsuc F. 2012. Phylogenomic analyses support the position of turtles as the sister group of birds and crocodiles (Archosauria). BMC Biol. 10, 1-14. ( 10.1186/1741-7007-10-65) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Green RE, Braun EL, Armstrong J, Earl D, Nguyen N. 2014. Three crocodilian genomes reveal ancestral patterns of evolution among archosaurs. Science 346, 1254449. ( 10.1126/science.1254449) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Brochu CA. 2003. Phylogenetic approaches toward crocodylian history. Annu. Rev. Earth Planet. Sci. 31, 357-397. ( 10.1146/annurev.earth.31.100901.141308) [DOI] [Google Scholar]
- 253.Wan Q-H, et al. 2013. Genome analysis and signature discovery for diving and sensory properties of the endangered Chinese alligator. Cell Res. 23, 1091-1105. ( 10.1038/cr.2013.104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Lewis JL, FitzSimmons NN, Jamerlan ML, Buchan JC, Grigg GC. 2013. Mating systems and multiple paternity in the estuarine crocodile (Crocodylus porosus). J. Herpetol. 47, 24-33. ( 10.1670/10-303) [DOI] [Google Scholar]
- 255.González EJ, Martínez-López M, Morales-Garduza MA, García-Morales R, Charruau P, Gallardo-Cruz JA. 2019. The sex-determination pattern in crocodilians: a systematic review of three decades of research. J. Anim. Ecol. 88, 1417-1427. ( 10.1111/1365-2656.13037) [DOI] [PubMed] [Google Scholar]
- 256.Smith CA, Joss JMP. 1994. Steroidogenic enzyme activity and ovarian differentiation in the saltwater crocodile, Crocodylus porosus. Gen. Comp. Endocrinol. 93, 232-245. ( 10.1006/gcen.1994.1027) [DOI] [PubMed] [Google Scholar]
- 257.Smith CA, Elf PK, Lang JW, Joss JMP. 1995. Aromatase enzyme activity during gonadal sex differentiation in alligator embryos. Differentiation 58, 281-290. ( 10.1046/j.1432-0436.1995.5840281.x) [DOI] [Google Scholar]
- 258.Yatsu R, et al. 2016. RNA-seq analysis of the gonadal transcriptome during Alligator mississippiensis temperature-dependent sex determination and differentiation. BMC Genomics 17, 1. ( 10.1186/s12864-016-2396-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Deveson IW, Holleley CE, Blackburn J, Graves JAM, Mattick JS, Waters PD, Georges A. 2017. Differential intron retention in Jumonji chromatin modifier genes is implicated in reptile temperature-dependent sex determination. Sci. Adv. 3, e1700731. ( 10.1126/sciadv.1700731) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Lin JQ, Zhou Q, Yang HQ, Fang LM, Tang K-Y, Sun L, Wan Q-H, Fang S-G. 2018. Molecular mechanism of temperature-dependent sex determination and differentiation in Chinese alligator revealed by developmental transcriptome profiling. Sci. Bull. 63, 209-212. ( 10.1016/j.scib.2018.01.004) [DOI] [PubMed] [Google Scholar]
- 261.AVIBASE. 2020. See https://avibase.bsc-eoc.org/avibase.jsp?lang=EN.
- 262.Zhang G. 2018. The bird's-eye view on chromosome evolution. Genome Biol. 19, 201. ( 10.1186/s13059-018-1585-z) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Feng S, et al. 2020. Dense sampling of bird diversity increases power of comparative genomics. Nature 587, 252-257. ( 10.1038/s41586-020-2873-9) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Fridolfsson AK, et al. 1998. Evolution of the avian sex chromosomes from an ancestral pair of autosomes. Proc. Natl Acad. Sci. USA 95, 8147-8152. ( 10.1073/pnas.95.14.8147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Hirst CE, Major AT, Ayes KL, Brown RJ, Mariette M, Sackton TB, Smith CA. 2017. Sex reversal and comparative data undermine the W chromosome and support Z-linked Dmrt1 as the regulator of gonadal sex differentiation in birds. Endocrinology 158, 2970-2987. ( 10.1210/en.2017-00316) [DOI] [PubMed] [Google Scholar]
- 266.Estermann MA, Williams S, Hirst CE, Roly, ZY, Serralbo O, Adhikari D, Powell D, Major AT, Smith CA. 2020. Insights into gonadal sex differentiation provided by single-cell transcriptomics in the chicken embryo. Cell Rep. 31, 107491. ( 10.1016/j.celrep.2020.03.055) [DOI] [PubMed] [Google Scholar]
- 267.Smith CA, Roeszler KN, Ohnesorg T, Cummins DM, Farlie FG, Doran TJ, Sinclair AH. 2009. The avian Z-linked gene Dmrt1 is required for male sex determination in the chicken. Nature 461, 267-271. ( 10.1038/nature08298) [DOI] [PubMed] [Google Scholar]
- 268.Zhou Q, Zhang J, Bachtrog D, An N, Huang Q, Jarvis ED, Gilbert MTP, Zhang G. 2014. Complex evolutionary trajectories of sex chromosomes across bird taxa. Science 346, 1246338. ( 10.1126/science.1246338) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Shetty S, Kirby P, Zarkower D, Graves JAM. 2002. Dmrt1 in a ratite bird: evidence for a role in sex determination and discovery of a putative regulatory element. Cytogenet. Genome Res. 99, 245-251. ( 10.1159/000071600) [DOI] [PubMed] [Google Scholar]
- 270.Yazdi HP, Ellegren H. 2019. A genetic map of ostrich Z chromosome and the role of inversions in avian sex chromosome evolution. Genome Biol. Evol. 10, 2049-2060. ( 10.1093/gbe/evy163) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Vicoso B, Kaiser VB, Bachtrog D. 2013. Sex-biased gene expression at homomorphic sex chromosomes in emus and its implication for sex chromosome evolution. Proc. Natl Acad. Sci. USA 110, 6453-6458. ( 10.1073/pnas.1217027110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Ioannidis J, et al. 2020. Primary sex determination in birds depends on DMRT1 dosage, but gonadal sex does not determine secondary sexual characteristics. Proc. Natl Acad. Sci. USA 118, e2020909118. ( 10.1073/pnas.2020909118) [DOI] [Google Scholar]
- 273.Zhao D, McBride D, Nandi S, McQueen HA, McGrew MJ. 2010. Somatic sex identity is cell autonomous in the chicken. Nature 464, 237-242. ( 10.1038/nature08852) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Lin M, Thorne MH, Martin IC, Sheldon BL, Jones RC. 1995. Development of the gonads in the triploid (ZZW and ZZZ) fowl, Gallus domesticus, and comparison with normal diploid males (ZZ) and females (ZW). Reprod. Fertility Dev. 7, 1185-1197. ( 10.1071/RD9951185) [DOI] [PubMed] [Google Scholar]
- 275.Graves JAM. 2003. Sex and death in birds: a model of dosage compensation that predicts lethality of sex chromosome an-euploids. Cytogenet. Genome Res. 101, 278-282. ( 10.1159/000074349) [DOI] [PubMed] [Google Scholar]
- 276.Kuroiwa A. 2017. Sex-determining mechanism in avians. In Avian reproduction. Advances in experimental medicine and biology, vol. 1001 (ed. Sasanami T), pp. 19-31. Singapore: Springer. [DOI] [PubMed] [Google Scholar]
- 277.Clinton M. 1998. Sex determination and gonadal development: a bird's eye view. J. Exp. Zool. 281, 457-465. [DOI] [PubMed] [Google Scholar]
- 278.Bloom SE. 1972. Chromosome abnormalities in chicken (Gallus domesticus) embryos: types, frequencies and phenotypic effects. Chromosoma 37, 309-326. ( 10.1007/bf00319873) [DOI] [PubMed] [Google Scholar]
- 279.Otto SP, Whitton J. 2000. Polyploidy: incidence and evolution. Annu. Rev. Genet. 34, 401-437. ( 10.1146/annurev.genet.34.1.401) [DOI] [PubMed] [Google Scholar]
- 280.Gunski RJ, Cañedo AD, Garnero ADV, Ledesma MA, Coria N, Montalti D, Degrandi TM. 2017. Multiple sex chromosome system in penguins (Pygoscelis, Spheniscidae). Comp. Cytogenet. 11, 541-552. ( 10.3897/CompCytogen.v11i3.13795) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Pala I, Naurin S, Stervander M, Hasselquist D, Bensch S, Hansson B. 2012. Evidence of a neo-sex chromosome in birds. Heredity 108, 264-272. ( 10.1038/hdy.2011.70) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Pala I, Hasselquist D, Bensch S, Hansson B. 2012. Patterns of molecular evolution of an avian neo-sex chromosome. Mol. Biol. Evol. 12, 3741-3754. ( 10.1093/molbev/mss177) [DOI] [PubMed] [Google Scholar]
- 283.Sigeman H, Ponnikas S, Chauhan P, Dierickx E, Brooke MD, Hansson B. 2019. Repeated sex chromosome evolution in vertebrates supported by expanded avian sex chromosomes. Proc. R. Soc. B 286, 20192051. ( 10.1098/rspb.2019.2051) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Gan HM, Falk S, Morales HE, Austin CM, Sunnucks P, Pavlova A. 2019. Genomic evidence of neo-sex chromosomes in the eastern yellow robin. GigaScience 8, giz111. ( 10.1093/gigascience/giz111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Graves JAM. 2016. Evolution of vertebrate sex chromosomes and dosage compensation. Nat. Rev. Gen. 17, 33-46. ( 10.1038/nrg.2015.2) [DOI] [PubMed] [Google Scholar]
- 286.Yazdi HP, Silva WTAF, Suh A. 2020. Why do some sex chromosomes degenerate more slowly than others? The odd case of ratite sex chromosomes. Genes 11, 1153. ( 10.3390/genes11101153) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Bellott D, et al. 2017. Avian W and mammalian Y chromosomes convergently retained dosage-sensitive regulators. Nat. Genet. 49, 387-394. ( 10.1038/ng.3778) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Xu L, Zhou Q. 2020. The female-specific W chromosomes of birds have conserved gene contents but are not feminized. Genes 11, 1126. ( 10.3390/genes11101126) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Uebbing S, Künstner A, Makinen H, Ellegren H. 2013. Transcriptome sequencing reveals the character of incomplete dosage compensation across multiple tissues in flycatchers. Genome Biol. Evol. 5, 1555-1566. ( 10.1093/gbe/evt114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Itoh Y, et al. 2007. Dosage compensation is less effective in birds than in mammals. J. Biol. 6, 2. ( 10.1186/jbiol53) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Irwin DE. 2018. Sex chromosomes and speciation in birds and other ZW systems. Mol. Ecol. 27, 3831-3851. ( 10.1111/mec.14537) [DOI] [PubMed] [Google Scholar]
- 292.Warnefors M, Mossinger K, Halbert J, Studer T, VandeBerg JL. 2017. Sex-biased microRNA expression in mammals and birds reveals underlying regulatory mechanisms and a role in dosage compensation. Genome Res. 27, 1961-1973. ( 10.1101/gr.225391.117) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Graves JAM. 2014. Avian sex, sex chromosomes, and dosage compensation in the age of genomics. Chromosome Res. 22, 45-57. ( 10.1007/s10577-014-9409-9) [DOI] [PubMed] [Google Scholar]
- 294.Xu L, Wa Sin SY, Grayson P, Edwards SV, Sackton TB. 2019. Evolutionary dynamics of sex chromosomes of paleognathous birds. Genome Biol. Evol. 11, 2376-2390. ( 10.1093/gbe/evz154) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Kapusta A, Suh A. 2016. Evolution of bird genomes - a transposon's-eye view. Annu. NY Acad. Sci. 1389, 164-185. ( 10.1111/nyas.13295) [DOI] [PubMed] [Google Scholar]
- 296.Peona V, et al. 2021. The avian W chromosome is a refugium for endogenous retroviruses with likely effects on female-biased mutational load and genetic incompatibilities. Phil. Trans. R. Soc. B 376, 20200186. ( 10.1098/rstb.2020.0186) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Pigozzi MI, Solari AJ. 1998. Germ cell restriction and regular transmission of an accessory chromosome that mimics a sex body in the zebra finch, Taeniopygia guttata. Chromosome Res. 6, 105-113 ( 10.1023/A:1009234912307) [DOI] [PubMed] [Google Scholar]
- 298.Itoh Y, Kampf K, Pigozzi MI, Arnold AP. 2009. Molecular cloning and characterization of the germline-restricted chromosome sequence in the zebra finch. Chromosoma 118, 527-536. ( 10.1007/s00412-009-0216-6) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Malinovskaya LP, et al. 2020. Germline-restricted chromosome (GRC) in the sand martin and the pale martin (Hirundinidae, Aves): synapsis, recombination and copy number variation. Sci. Rep. 10, 1058. ( 10.1038/s41598-020-58032-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Biederman MK, Nelson MM, Asalone KC, Pedersen AL, Saldanha CJ, Bracht JR. 2018. Discovery of the first germline-restricted gene by subtractive transcriptomic analysis in the zebra finch, Taeniopygia guttata. Curr. Biol. 28, 1620-1627. ( 10.1016/j.cub.2018.03.067) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Kinsella CM, et al. 2019. Programmed DNA elimination of germline development genes in songbirds. Nat. Comm. 10, 5468. ( 10.1038/s41467-019-13427-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Torgasheva AA, et al. 2019. Germline-restricted chromosome (GRC) is widespread among songbirds. Proc. Natl Acad. Sci. USA 116, 11 845-11 850. ( 10.1073/pnas.1817373116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Pei Y, et al. 2021. Occasional paternal inheritance of the germline-restricted chromosome in songbirds. BioRxiv. See https://www.biorxiv.org/content/10.1101/2021.01.28.428604v1. ( 10.1101/2021.01.28.428604) [DOI]
- 304.Cortez D, Marin R, Toledo-Flores D, Froidevaux L, Liechti A, Waters PD, Grützner F, Kaessmann H. 2014. Origins and functional evolution of Y chromosomes across mammals. Nature 508, 488-493. ( 10.1038/nature13151) [DOI] [PubMed] [Google Scholar]
- 305.Warren WC, et al. 2008. Genome analysis of the platypus reveals unique signatures of evolution. Nature 453, 175-183. ( 10.1038/nature06936) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Zhou Y, et al. 2021. Platypus and echidna genomes reveal mammalian biology and evolution. Nature 592, 756-762. ( 10.1038/s41586-020-03039-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Rens W, et al. 2007. The multiple sex chromosomes of platypus and echidna are not completely identical and several share homology with the avian Z. Genome Biol. 8, R243. ( 10.1186/gb-2007-8-11-r243) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Grützner F, Rens W, Tsend-Ayush E, El-Mogharbel N, O'Brien PCM, Jones RC, Ferguson-Smith MA, Graves JAM. 2004. In the platypus a meiotic chain of ten sex chromosomes shares genes with the bird Z and mammal X chromosomes. Nature 432, 913-917. ( 10.1038/nature03021) [DOI] [PubMed] [Google Scholar]
- 309.Veyrunes F, et al. 2008. Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res. 18, 965-973. ( 10.1101/gr.7101908) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.El-Mogharbel N, Wakefield M, Deakin JE, Tsend-Ayush E, Grützner F, Alsop A, Ezaz T, Marshall Graves JA. 2007. Dmrt gene cluster analysis in the platypus: new insights into genomic organization and regulatory regions. Genomics 89, 10-21. ( 10.1016/j.ygeno.2006.07.017) [DOI] [PubMed] [Google Scholar]
- 311.Deakin JE. 2017. Implications of monotreme and marsupial chromosome evolution on sex determination and differentiation. Gen. Comp. Endocrinol. 244, 130-138. ( 10.1016/j.ygcen.2015.09.029) [DOI] [PubMed] [Google Scholar]
- 312.Deakin JE, Hore TA, Koina E, Graves JAM. 2008. The status of dosage compensation in the multiple X chromosomes of the platypus. PLoS Genet. 4, e1000140. ( 10.1371/journal.pgen.1000140) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Julien P, Brawand D, Soumillon M, Necsulea A, Liechti A, Schütz F, Daish T, Grützner F, Kaessmann H. 2012. Mechanisms and evolutionary patterns of mammalian and avian dosage compensation. PLoS Biol. 10, e1001328. ( 10.1371/journal.pbio.1001328) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Whitworth DJ, Pask AJ. 2016. The X factor: X chromosome dosage compensation in the evolutionarily divergent monotremes and marsupials. Semin. Cell Dev. Biol. 56, 117-121. ( 10.1016/j.semcdb.2016.01.006) [DOI] [PubMed] [Google Scholar]
- 315.Redia CA, Capanna E. 2012. Genome size evolution: sizing mammalian genomes. Cytogenet. Genome Res. 137, 97-112. ( 10.1159/000338820) [DOI] [PubMed] [Google Scholar]
- 316.Deakin JE, O'Neill RJ. 2020. Evolution of marsupial genomes. Annu. Rev. Anim. Biosci. 8, 25-45. ( 10.1146/annurev-animal-021419-083555) [DOI] [PubMed] [Google Scholar]
- 317.Frankenberg S. 2018. Pre-gastrula development of non-eutherian mammals. Curr. Top. Dev. Biol. 128, 237-265. ( 10.1016/bs.ctdb.2017.10.013) [DOI] [PubMed] [Google Scholar]
- 318.Hayman D. 1989. Marsupial cytogenetics. Aust. J. Zool. 37, 331-349. ( 10.1071/ZO9890331) [DOI] [Google Scholar]
- 319.Deakin JE. 2018. Chromosome evolution in marsupials. Genes 9, 72. ( 10.3390/genes9020072) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Grant J, et al. 2012. Rsx is a metatherian RNA with Xist-like properties in X-chromosome inactivation. Nature 487, 254-258. ( 10.1038/nature11171) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 321.Deakin JE. 2013. Marsupial X chromosome inactivation: past, present and future. Aust. J. Zool. 61, 13-23. [Google Scholar]
- 322.Rens W, Wallduck MS, Lovell FL, Ferguson-Smith MA, Ferguson-Smith AC. 2010. Epigenetic modifications on X chromosomes in marsupial and monotreme mammals and implications for evolution of dosage compensation. Proc. Natl Acad. Sci. USA 107, 17 657-17 662. ( 10.1073/pnas.0910322107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 323.Johnson PG, Watson CM, Adams M, Paull DJ. 2002. Sex chromosome elimination, X chromosome inactivation and reactivation in the southern brown bandicoot Isoodon obesulus (Marsupialia: Peramelidae). Cytogenet. Genome Res. 99, 119-124. ( 10.1159/000071583) [DOI] [PubMed] [Google Scholar]
- 324.Sharp P. 1982. Sex chromosome pairing during male meiosis in marsupials. Chromosoma 86, 27-47. ( 10.1007/BF00330728) [DOI] [PubMed] [Google Scholar]
- 325.Page J, Berrios S, Parra MT, Viera A, Suja JA, Prieto I, Barbero JL, Rufas JS, Fernández-Donoso R. 2005. The program of sex chromosome pairing in meiosis is highly conserved across marsupial species: implications for sex chromosome evolution. Genetics 170, 793-799. ( 10.1534/genetics.104.039073) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Foster JW, et al. 1992. Evolution of sex determination and the Y chromosome: SRY-related sequences in marsupials. Nature 359, 531-533. ( 10.1038/359531a0) [DOI] [PubMed] [Google Scholar]
- 327.Evans BJ, Upham NS, Golding GB, Ojeda RA, Ojeda AA. 2017. Evolution of the largest mammalian genome. Genome Biol. Evol. 9, 1711-1724. ( 10.1093/gbe/evx113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Georgiades P, Watkins M, Burton GJ, Ferguson-Smith AC. 2001. Roles for genomic imprinting and the zygotic genome in placental development. Proc. Natl Acad. Sci. USA 98, 4522-4527. ( 10.1073/pnas.081540898) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Svartman MT, Stone G, Stanyon R. 2005. Molecular cytogenetics discards polyploidy in mammals. Genomics 85, 425-430. ( 10.1016/j.ygeno.2004.12.004) [DOI] [PubMed] [Google Scholar]
- 330.Bachtrog D. 2013. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat. Rev. Genet. 14, 113-124. ( 10.1038/nrg3366) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.De la Fuente R, Parra MT, Viera A, Calvente A, Gomez R, Suja J, Rufas JS, Page J. 2007. Meiotic pairing and segregation of achiasmate sex chromosomes in eutherian mammals: the role of SYCP3 protein. PLoS Genet. 3, e198. ( 10.1371/journal.pgen.0030198) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Tasman Daish T, Grützner F. 2019. Evolution and meiotic organization of heteromorphic sex chromosomes. Curr. Topics Dev. Biol. 134, 1-48. ( 10.1016/bs.ctdb.2019.009) [DOI] [PubMed] [Google Scholar]
- 333.Gil-Fernandez G-F, et al. 2020. Meiosis reveals the early steps in the evolution of a neo-XY sex chromosome pair in the African pygmy mouse Mus minutoides. PLoS Genet. 16, e1008959. ( 10.1371/journal.pgen.1008959) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Wutz A, Rasmussen TP, Jaenisch R. 2002. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat. Genet. 30, 167-174. ( 10.1038/ng820) [DOI] [PubMed] [Google Scholar]
- 335.Jégu T, Aeby E, Lee JT. 2017. The X chromosome in space. Nat. Rev. Genet. 18, 377-389. [DOI] [PubMed] [Google Scholar]
- 336.Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R. 1991. Male development of chromosomally female mice transgenic for Sry. Nature 351, 117-121. [DOI] [PubMed] [Google Scholar]
- 337.Miyawaki S, Kuroki S, Maeda R, Okashita N, Koopman P, Tichibana M. 2020. The mouse Sry locus harbors a cryptic exon that is essential for male sex determination. Science 370, 121-124. [DOI] [PubMed] [Google Scholar]
- 338.Parma P, Veyrunes F, Pailhoux E. 2016. Sex reversal in non-human placental mammals. Sex. Dev. 10, 326-344. ( 10.1159/000448361) [DOI] [PubMed] [Google Scholar]
- 339.Kuroiwa A, Ishiguchi Y, Yamada F, Shintaro A, Matsuda Y. 2010. The process of a Y-loss event in an XO/XO mammal, the Ryukyu spiny rat. Chromosoma 119, 519-526. [DOI] [PubMed] [Google Scholar]
- 340.Just W, Baumstark A, Süss A, Graphodatsky A, Rens W, Schäfer N, Bakloushinskaya I, Hameister H, Vogel W. 2007. Ellobius lutescens: sex determination and sex chromosome. Sex. Dev. 1, 211-221. ( 10.1159/000104771) [DOI] [PubMed] [Google Scholar]
- 341.Veyrunes F, Chevret P, Catalan J, Castiglia R, Watson J, Dobigny G, Robinson TJ, Britton-Davidian J. 2010. A novel sex determination system in a close relative of the house mouse. Proc. R. Soc. B 277, 1049-1056. ( 10.1098/rspb.2009.1925) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Real FM, et al. 2020. The mole genome reveals regulatory rearrangements associated with adaptive intersexuality. Science 370, 208-214. ( 10.1126/science.aaz2582) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Uhlenhaut NH, et al. 2009. Somatic sex reprogramming of adult ovaries to testes by foxl2 ablation. Cell 139, 1130-1142. [DOI] [PubMed] [Google Scholar]
- 344.Matson CK, Murphy MW, Sarver AL, Griswold MD, Bardwell VJ, Zarkower D. 2011. Dmrt1 prevents female reprogramming in the postnatal mammalian testis. Nature 476, 101-104. ( 10.1038/nature10239) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Naqvi S, Godfrey AK, Hughes, JF, Goodheart ML, Mitchell RN, Page DC. 2019. Conservation, acquisition, and functional impact of sex-biased gene expression in mammals. Science 365, eaaw7317. ( 10.1126/science.aaw7317) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Davis EJ, Lobach I, Dubal DB. 2019: Female XX sex chromosomes increase survival and extend lifespan in aging mice. Aging Cell 18, e12871. ( 10.1111/acel.12871) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.VGP. 2020. See https://vertebrategenomesproject.org.
- 348.EBG. 2020. See https://www.earthbiogenome.org/.
- 349.Fish1 k. 2020. See https://db.cngb.org/fisht1k.
- 350.Birds10 K. 2020. See https://b10k.genomics.cn.
- 351.Skaletsky H, et al. 2003. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423, 825-837. ( 10.1038/nature01722). [DOI] [PubMed] [Google Scholar]
- 352.Carvalho AB, Clark AG. 2013. Efficient identification of Y chromosome sequences in the human and Drosophila genomes. Genome Res. 23, 1894-1907. ( 10.1101/gr.156034.113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Clark AG. 2014. The vital Y chromosome. Nature 508, 463-465. ( 10.1038/508463a) [DOI] [PubMed] [Google Scholar]
- 354.Rhie A, et al. 2021. Towards complete and error-free genome assemblies of all vertebrate species. Nature 592, 737-746. ( 10.1038/s41586-021-03451-0) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Koepfli K-P, Paten B, Antunes A, Belov K, Bustamante Cet al. 2015. The genome 10 K project: a way forward. Ann. Rev. Anim. Biosci. 3, 57-111. ( 10.1146/annurev-animal-090414-014900) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Pennell MW, Mank JE, Peichel CL. 2018. Transitions in sex determination and sex chromosomes across vertebrate species. Mol. Ecol. 2, 3950-3963. ( 10.1111/mec.14540) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Payseur BA, Presgraves DC, Filatov DA. 2018. Sex chromosomes and speciation. Mol. Ecol. 27, 3745-3748. ( 10.1111/mec.14828) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Runemark A, Eroukhmanoff F, Nava-Bolaños A, Hermansen JS, Meier JI. 2018. Hybridization, sex-specific genomic architecture and local adaptation. Phil. Trans. R. Soc. B 373, 20170419. ( 10.1098/rstb.2017.0419) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Capel B (ed). 2019. Sex determination in vertebrates. Current topics in developmental biology, vol. 104, pp. 1-376. New York, NY: Academic Press. [Google Scholar]
- 360.Kopp A. 2012. Dmrt genes in the development and evolution of sexual dimorphism. Trends Genet. 28, 175-184. ( 10.1016/j.tig.2012.02.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Darbre PD. 2019. The history of endocrine-disrupting chemicals. Curr. Opin. Endocrinol. Metab. Res. 7, 26-33. ( 10.1016/j.coemr.2019.06.007) [DOI] [Google Scholar]
- 362.Kloas W, et al. 2009. Endocrine disruption in aquatic vertebrates. Ann. NY Acad. Sci. 1163, 187-200. ( 10.1111/j.1749-6632.2009.04453.x) [DOI] [PubMed] [Google Scholar]
- 363.Wang H, Piferrer F, Chen S (eds). 2019. Sex control in aquaculture, vol. 1. New York, NY: John Wiley & Sons. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This article has no additional data.