Abstract
Background
Pseudomonas aeruginosa is a notorious opportunistic pathogen causing various types of biofilm-related infections. Biofilm formation is a unique microbial strategy that allows P. aeruginosa to survive adverse conditions such as antibiotic treatment and human immune clearance.
Results
In this study, we experimentally evolved P. aeruginosa PAO1 biofilms for cyclic treatment in the presence of high dose of imipenem, and enriched hyperbiofilm mutants within six cycles in two independent lineages. The competition assay showed that the evolved hyperbiofilm mutants can outcompete the ancestral strain within biofilms but not in planktonic cultures. Whole-genome sequencing analysis revealed the hyperbiofilm phenotype is caused by point mutations in rpoS gene in all independently evolved mutants and the same mutation was found in P. aeruginosa clinical isolates. We further showed that mutation in rpoS gene increased the intracellular c-di-GMP level by turning on the expression of the diguanylate cyclases. Mutation in rpoS increased pyocyanin production and virulence in hyperbiofilm variants.
Conclusion
Here, our study revealed that antibiotic treatment of biofilm-related P. aeruginosa infections might induce a hyperbiofilm phenotype via rpoS mutation, which might partially explain antimicrobial treatment failure of many P. aeruginosa biofilm-related infections.
Supplementary Information
The online version contains supplementary material available at 10.1186/s13578-021-00655-9.
Keywords: Experimental biofilm evolution, Pseudomonas aeruginosa, Sigma factor RpoS, Biofilms, Cyclic-di-GMP, Virulence
Introduction
Microbial cells undergo rapid evolution under stress to adapt to the environment in nature [1] or inside the host [2]. Certain key mutations on genome can greatly help bacterial populations to gain a competitive advantage in adverse environment [2]. To identify these adaptive traits of microbes, experimental evolution experiments are usually conducted to mimic these diverse environmental conditions for accelerating the emergence of well-fitted variants [3]. Employment of next-generation sequencing approaches will facilitate the identification of the mutations of experimental adapted bacterial variants and elucidation of the underlying molecular mechanism of the evolved traits [4]. So far, adaptive experimental evolution has been applied to reveal the molecular basis of drug resistance [5], persistence [6], biofilm formation [7] and etc. An important question for experimental evolution is the relevance of laboratory observation to the evolution in natural conditions. For this point, studies have indicated the mutation derived phenotypes in laboratory evolution could be found in clinical isolates [8–11].
Bacterial pathogens can form biofilms on both biotic and abiotic surfaces, causing many hospital-acquired and recurrent infections. Biofilms are densely-packed microbial cells embedded in self-secreted hydrated matrix consisting of polysaccharides, proteins, extracellular DNA and lipids [12]. Previous studies have shown that the biofilm-grown bacteria have distinct phenotypes from planktonic cultures, including gene expression [13, 14] and increased antibiotic resistance [15]. The tolerance to antimicrobial agents by biofilm cells can be increased up to 1000-fold compared to planktonic cells [16]. Investigating evolution traits of biofilm cells against antibiotics might provide knowledge about bacterial adaptation during chronic infections.
P. aeruginosa is a notorious opportunistic pathogen, which causes a variety of infections, including wounds, urinary tract and respiratory tract infections, and is the leading cause of morbidity and mortality for people suffering from cystic fibrosis (CF) [17]. Infections caused by P. aeruginosa can be very difficult to treat due to its intrinsic resistance to a variety of antibiotics and tends to form biofilms at the sites of infection [18, 19]. As an important nosocomial pathogen, P. aeruginosa biofilms have been found on various surfaces of indwelling medical devices, including urinary catheters, bone plates, ventricular assist device drivelines and pacemakers [20]. Biofilms provide P. aeruginosa an enormous advantage in clinical infections by protecting biofilm cells from the immune clearance [21] and tolerance to antimicrobial agents [22, 23]. Persisters are subpopulation of isogenic bacteria that tolerance to antibiotics [24] and the persister cells were dormant in biofilms, which significantly contributes to P. aeruginosa biofilm recalcitrance after the cessation of antibiotic therapy [25]. Considering the issue of drug tolerance and recalcitrance of P. aeruginosa biofilm-related infections, novel anti-biofilm therapeutics are urgently needed.
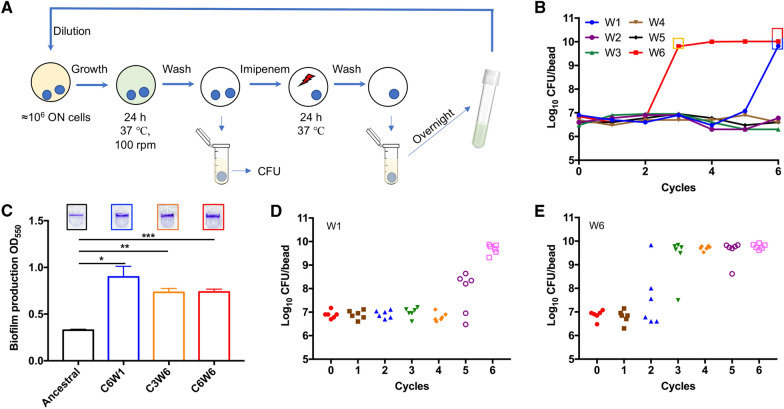
The long-term use of antibiotics in the treatment of P. aeruginosa infections in cystic fibrosis patients is well-known to drive emergence of diversified drug-resistant variants. Clinical isolates of P. aeruginosa have shown distinct biofilm formation capacity and many of them were strong biofilm producers [26]. However, current adaptive experimental evolution studies of P. aeruginosa are mainly focusing on planktonic cultures [10, 27, 28]. Carbapenems, including Imipenem, were considered as the last resort of drugs for the treatment of multi drug resistant P. aeruginosa infections. The emergence of resistance to carbapenems limits its use for treatment [29]. In this study, we established an experimental evolution model to investigate the evolution traits of imipenem-treated biofilms of P. aeruginosa, and the set-up of this model is presented in Fig. 1A.
Fig. 1.
Experimental biofilm evolution of P. aeruginosa under antibiotic stress. A The setup of experimental biofilm evolution of P. aeruginosa. The biofilm of six independent lineages of the P. aeruginosa PAO1 were grown on 5 mm glass bead. After 24 h cultivation, one bead was vortex and sonicated for CFU counts, another bead was transferred to a 24 well microplate and treated with 160 μg/mL imipenem. After 24 h treatment, the surviving cells were grown overnight in fresh medium and start another cycle. B Evolution of biofilm bacteria exposed to imipenem resulted in a rapid increase in biofilm bacteria CFU on bead. C Crystal violet (CV) staining of biofilms formed by ancestral and hyperbiofilm variant strains on PVC plate. Data are presented as the mean ± s.d. of five biological replicates. Significance was determined using a Student’s t test: *P < 0.05, **P < 0.01 and ***P < 0.001. D and E The time frame of emergence of hyperbiofilm variants in linage W1 (D) and W6 (E). The biofilm formation by the different colonies was displayed with CFU of biofilm cells on 5 mm glass bead
We show here that cyclic exposure of P. aeruginosa biofilms to high concentration of imipenem led to emergence of variants with a hyperbiofilm phenotype. The competition assay showed that the evolved hyperbiofilm variants can outcompete ancestral strain within biofilms but not in planktonic cultures. Genome sequencing analysis revealed that the hyperbiofilm phenotype is caused by single-point mutations in the sigma factor RpoS. Importantly, the mutations on rpoS identified in the in vitro experimental biofilm model also occurred in P. aeruginosa clinic isolates with a hyperbiofilm phenotype at a substantial rate. Overall, our data show that under imipenem treatment, mutations in rpoS could be selected in P. aeruginosa and subsequently lead to enhanced biofilm formation.
Results
Experimental biofilm evolution selects for P. aeruginosa hyperbiofilm mutants
In order to examine the evolutionary traits of P. aeruginosa biofilms under antibiotic stress condition, we exposed biofilms of P. aeruginosa PAO1 to different concentrations of imipenem (40, 80 and 160 μg/mL), which is 10, 20 and 40 times of the minimum inhibitory concentration (MIC), in a cyclic manner (Fig. 1). Imipenem, a widely used last resort antibiotic, has been chosen as the selective pressure for experimental biofilm evolution owing to its’ commonly prescribed for treatment of P. aeruginosa infections [30].
The biofilms of six independent lineages initiated from a common ancestor PAO1 strain were formed on the surface of 5 mm glass beads [31] and treated with different concentrations of imipenem for 24 h. Survivor cells on beads were quantitated by CFU counts. Biofilm survivors were collected and reinoculated in fresh LB medium for the 2nd cycle (Fig. 1A). At the first cycle, the CFU counts on bead of each lineage were between 6.30 and 6.90 log10. After 6 cycles, no hyperbiofilm variant was observed in control group or 10 × and 20 × MIC (Additional file 1: Figure S1A–C) imipenem treated groups, and two lineages treated by 40 × MIC of imipenem accumulated hyperbiofilm variants (Additional file 1: Figure S1D). The CFU counts within lineage W1 and W6 biofilms on bead reached 9.82 and 10.01 log10 in 40 × MIC imipenem treated group (Fig. 1B). Next, the biofilm formation capacity of the ancestral, C6W1, C3W6 and C6W6 (C refer to the cycle number, W refer to the linage number) population were further confirmed by the crystal violet (CV) biofilm assay. Similarly, the CV method revealed that biofilms formed by C6W1, C3W6 and C6W6 population were between 2- and threefold higher than the ancestral population (Fig. 1C). In order to track when the hyperbiofilm variants have emerged within lineage W1 and W6 population, we picked 6 colonies form each cycle in random and measured the CFU of biofilms on bead. We found that the hyperbiofilm variants of lineage W1 and W6 appeared since cycle 5 and cycle 2 and enriched at cycle 6 (Fig. 1D) and cycle 3 (Fig. 1E), respectively. These results indicate that the hyperbiofilm variants only accumulated upon higher concentration imipenem treatment, rather than lower concentration of imipenem. The different appearance time of hyperbiofilm variants in the two independent linages is possibly due to the variation of evolutionary rate [32]. More lineages might accumulate hyperbiofilm variants in 40 × MIC imipenem treated group if we increase the treatment cycles.
Point mutations in rpoS lead to hyperbiofilm phenotype of P. aeruginosa
In order to elucidate the genetic mechanisms underlying the hyperbiofilm phenotype, we sequenced the C6W1 and C6W6 population, and choose C6W5 population as the negative control. Through comparative genomic analysis, we identified only SNPs in one gene, PA3622 (encodes sigma factor RpoS) (Table 1), which was mutated in C6W1 and C6W6 population, and in contrast, no non-synonymous SNP was identified in C6W5 population when compared to the ancestral strain. The sigma factor RpoS is well known as a master regulator that controls the expression of genes involved in stress response and virulence factors production in P. aeruginosa [33, 34]. A previous study used transcript profiling and found that 772 genes were regulated by RpoS in stationary phase and it affects expression of more than 40% quorum-sensing controlled genes [35].
Table 1.
Whole genome sequencing data for evolved P. aeruginosa populations
| Strains | Region | Mutation | Mutation effect | Gene | Annotation |
|---|---|---|---|---|---|
| C6W1C | 794674..794675 | AC to GT | D89G | PA0727 | Pf replication initiator protein |
| 4058162 | G to A | P251L | rpoS | RNA polymerase sigma factor | |
| C3W6F | 794674..794675 | AC to GT | G89G | PA0727 | Pf replication initiator protein |
| 4058118 | G to A | Q266Stop | rpoS | RNA polymerase sigma factor | |
| C6W6F | 4058118 | G to A | Q266Stop | rpoS | RNA polymerase sigma factor |
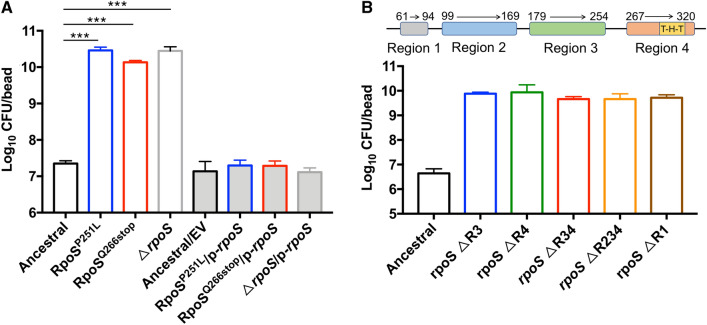
Targeted resequencing of rpoS in C6W1C, C3W6F and C6W6F (colonies isolated from C6W1, C3W6 and C6W6 population) identified the nonsynonymous mutations in RpoS of C6W1C (P251L), C3W6F (Q266stop) and C6W6F (Q266stop). To further confirm the causality of rpoS mutations for the hyperbiofilm phenotype, we constructed a de novo mutant allele with a SNP on the ancestor PAO1 genome to yielded RpoSP251L and RpoSQ266stop mutant strains. We found that single point mutation in rpoS could produce the hyperbiofilm phenotype (Fig. 2A). We also tested the biofilm formation capacity of ΔrpoS strain and confirmed that knockout rpoS in P. aeruginosa PAO1 indeed increased the biofilm formation (Fig. 3A). Complementing the mutation strains with wild type rpoS reverted the hyperbiofilm phenotype to the wild-type level (Fig. 2A).
Fig. 2.
Hyperbiofilm phenotype is caused by rpoS mutation. rpoS point mutation, full deletion (A) and regions deletion strains (B) were increased the biofilm formation. The biofilm formation by the indicated strains was displayed with CFU of biofilm cells on 5 mm glass bead. Data are presented as the mean ± s.d. of four biological replicates. Significance was determined using a Student’s t test: ***P < 0.001. EV represents the empty vector pHERD20T in this assay
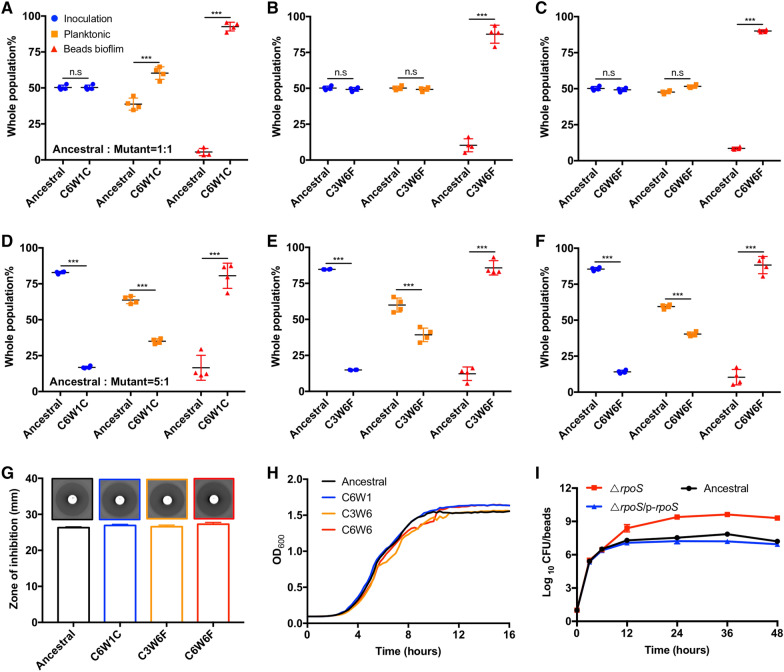
Fig. 3.
Hyperbiofilm variants prevail in biofilm competitions. A–F The competition of ancestor and hyperbiofilm variants in planktonic cultures and biofilms when inoculated at the same ratio (A–C) or ancestral: hyperbiofilm mutant = 5:1 (D–F). G Disc diffusion antibiotic sensitivity testing, H growth curves measurement, and I biofilm growth curve was measured. Data are presented as the mean ± s.d. of four biological replicates. Significance was determined using a Student’s t test: n.s indicates no significant difference (P ≥ 0.05); ***P < 0.001
Protein domain analysis showed that RpoS consists of 4 regions, and region 4 contains a DNA binding domain. P251L located on the end of region 3, and knockout this region increased the biofilm formation. Q266stop mutation leads to RpoS lacking region 4, and knockout region 4 has the same phenotype of Q266stop mutation. Moreover, we have constructed region 1, region 3–4 and region 2–4 deletion strains, all of those region deletion mutants produced the hyperbiofilm phenotype (Fig. 2B). Together, these results provide a solid evidence that the SNPs identified are necessary and sufficient to cause hyperbiofilm phenotype in P. aeruginosa. Moreover, P251L and Q266stop mutations are very likely lead to the inactivation of RpoS.
Hyperbiofilm variants outcompete the ancestral strain during biofilm competitions
The convergent emergency of P. aeruginosa hyperbiofilm variants from independent lineages suggested a competitive advantage for these variants over the ancestor. We have previously showed that P. aeruginosa cells did not share its EPS with its neighboring cells [36] and thus we hypothesized that the evolved hyperbiofilm variants only gain advantage to the ancestor strain during biofilm mode of growth. We then tested the competition of isolated hyperbiofilm variants with the ancestral strain in both planktonic cultures and biofilms. The fluorescently tagged hyperbiofilm variants (tagged with mCherry) were mixed with ancestral strain (tagged with GFP) in different ratio and inoculated into the bead-containing 24 well microplates. After 24 h cultivation, the planktonic and biofilm cells were analyzed using flow cytometry. We found that, the proportion of planktonic cells of C6W1C was slightly higher than ancestral strain when inoculated at the same ratio (Fig. 3A), while no difference was found between C3W6F and C6W6F with ancestral strain when inoculated at the same ratio (Fig. 3B, C). Interestingly, the competitions in biofilms confirmed that the hyperbiofilm variants have a significant and predominant selective advantage against the ancestral strain (Fig. 3A–C). Next, we increased the inoculation ratio of ancestor and hyperbiofilm variants to 5:1, the proportion of ancestor within biofilm was still much less than hyperbiofilm variants (Fig. 3D–F). These results indicated that the hyperbiofilm variants increased their fitness against the ancestor strain only in biofilms and this phenotype is not related to the change of growth rate, drug resistance and tolerance. Moreover, the competition advantage of hyperbiofilm variants is growth model specific and did not occur in planktonic culture.
Previous studies showed that acquired mutations conferring beneficial traits such as antibiotic resistance will dominate the populations when exposing biofilm bacteria to high concentrations of antibiotic [37]. Therefore, the enrichment of the hyperbiofilm rpoS mutation variants could have been achieved due to their enhanced resistance to imipenem. We found, however, that the MIC of imipenem for colonies isolated from the evolved lines (C6W1C, C3W6F and C6W6F) was indistinguishable from that for their ancestor (Fig. 3G). One of the most straightforward ways to gain a competitive advantage is increasing the growth rate. To test this point, we measured the growth rates of the evolved hyperbiofilm variants and the ancestor in LB medium and found there is no significant difference between the hyperbiofilm variants and ancestor (Fig. 3H). Next, we measured the biofilm growth curve of PAO1, ΔrpoS and the complementation strain ΔrpoS/p-rpoS and confirmed that ΔrpoS mutant formed more biofilms than the PAO1 wild type at all stages of biofilm formation (Fig. 3I). These results indicated that it is not the resistance level and planktonic growth rate select ΔrpoS mutant.
Mutations in rpoS lead to an elevated intracellular c-di-GMP levels
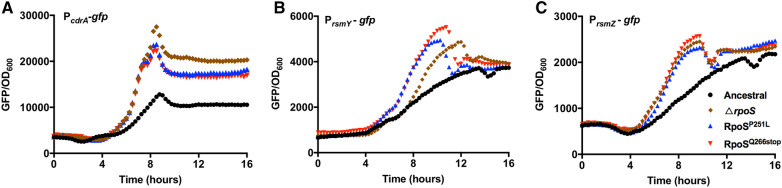
Quorum-sensing (QS) [38] and c-di-GMP [39] have been well documented to play important roles in regulating P. aeruginosa biofilm formation. To assess whether quorum-sensing and c-di-GMP levels were changed in the ΔrpoS variants, we introduced the quorum-sensing and c-di-GMP reporter systems [40–43] into the variants isolated from biofilm evolution experiments and ancestral strain to determine the relative levels of the corresponding signaling pathways. We found that the fluorescent signal of PlasB-gfp and PrhlA-gfp in ancestral strain were higher than that of hyperbiofilm variants (Additional file 1: Figure S2A and B), while there was no difference in fluorescent signal of PpqsA-gfp between the ancestral and hyperbiofilm variants (Additional file 1: Figure S2C). For the fluorescent signal of PcdrA-gfp, the hyperbiofilm variants showed twofold higher in expression level than the ancestral strain, indicating that the hyperbiofilm variants might have elevated intracellular c-di-GMP levels (Figure S2D). We further showed that the PcdrA-gfp expression level were increased in RpoSP251L, RpoSQ266stop and ΔrpoS strain compared to the PAO1 wild-type strain (Fig. 4A). The second messenger c-di-GMP is a key regulator of P. aeruginosa biofilm formation, which is synthesized from two GTP molecules by diguanylate cyclases (DGC) and is degraded into 5ʹ-phosphoguanylyl-(3ʹ–5ʹ) guanosine (pGpG) and/or GMP by phosphodiesterases (PDE) [39]. Till now, 43 DGC and PDE proteins have been identified in P. aeruginosa [44].
Fig. 4.
Expression of PcdrA-gfp, PrsmY-gfp and PrsmZ-gfp reporter fusions in rpoS mutants and PAO1 wild-type strain. Relative fluorescence intensity (reflected as GFP/OD600) was measured in representative strains containing the PcdrA-gfp (A), rsmY (B) and rsmZ (C) reporter fusions. Data are presented as the mean ± s.d. of five biological replicates
To further investigate the regulatory roles of rpoS gene on c-di-GMP signaling pathway, we performed transcriptomic analysis of PAO1, RpoSP251L and RpoSQ266stop strains using RNA-sequencing. Samples were collected after 8.5 h culture owing to the PcdrA-gfp fluorescent intensity (Additional file 1: Figure S3) between mutants and wild type PAO1 strain have the biggest difference at this time point. We found that, 15 DGC and PDE encoding genes were upregulated by at least twofold in both RpoSP251L and RpoSQ266stop strains compared to PAO1 (Table 2). This result indicated that the c-di-GMP metabolism in rpoS mutant strains were more active than PAO1.
Table 2.
The expression of GGDEF, EAL, HD-GYP proteins in rpoS mutation and ancestral strains
| ORF | Name | Domain signature | Activity* | RpoSP251L vs Ancestral FC | p value | RpoSQ266stop vs Ancestral FC | p value |
|---|---|---|---|---|---|---|---|
| PA0169 | siaD | GGEEF | DGC | − 2.17 | 3.39E-10 | − 2.21 | 2.09E-10 |
| PA2572 | YN-GYP | – | 7.12 | 5.02E-202 | 19.45 | 1.16E-234 | |
| PA0847 | GGDEF | DGC | 2.94 | 7.01E-42 | 3.09 | 4.48E-43 | |
| PA1107 | roeA | GGEEF | DGC | 2.16 | 3.15E-18 | 2.52 | 3.39E-23 |
| PA1120 | yfiN, | GGDEF | DGC | 2.27 | 1.85E-19 | 2.85 | 2.19E-27 |
| PA3343 | GGEEF | DGC | – | – | 2.09 | 1.07E-29 | |
| PA3702 | wspR | GGEEF | DGC | 2.17 | 5.79E-25 | 2.55 | 3.33E-34 |
| PA4929 | nicD | GGDEF | DGC | 5.4 | 1.61E-174 | 8.85 | 2.60E-236 |
| PA0290 | GGEEF | ND | – | – | 2 | 3.72E-17 | |
| PA0575 | rmcA | GGDEF, EAL | ND | 3.5 | 4.78E-92 | 4.63 | 3.54E-125 |
| PA1181 | GGDEF, ELL | ND | 3.18 | 3.48E-79 | 3.56 | 3.54E-125 | |
| PA2771 | GGEEF | ND | 3.4 | 2.17E-60 | 4.25 | 4.65E-74 | |
| PA0861 | rbdA | GGDEF, ELL | PDE | 4.19 | 2.22E-140 | 6.93 | 7.11E-230 |
| PA2072 | GGDEF, EAL | PDE | 7.55 | 8.41E-222 | 14.62 | 6.89E-293 | |
| PA3311 | nbdA | AGDEF, EAL | PDE | 7.65 | 6.06E-252 | 13.56 | 1.66E-279 |
| PA3825 | EVL | PDE | 3 | 9.51E-46 | 3.45 | 9.00E-52 | |
| PA4108 | HD-GYP | PDE | 3.42 | 2.03E-80 | 3.9 | 2.38E-92 | |
| PA4781 | HD-GYP | PDE | 8.8 | 2.99E-306 | 14.18 | 2.78E-130 |
*ND not determined, – not active, FC fold change
RpoS regulates the expression of small regulatory RNAs rsmY and rsmZ in Legionella pneumophila [45]. Moreover, rsmY/Z participate in the regulation of c-di-GMP production in P. aeruginosa, the c-di-GMP levels were strongly reduced in the rsmY/Z double deletion mutant [46]. Our transcriptomic analysis showed that the expression of rsmY and rsmZ was increased 3.84 and 5.04-fold in RpoSP251L compared to the PAO1 wild-type, respectively. Next, we measured the expression of rsmY/Z in PAO1, RpoSP251L, RpoSQ266stop and ΔrpoS strains using reporter fusions [13]. We found the rsmY/Z expressions were increased in RpoSP251L, RpoSQ266stop and ΔrpoS (Fig. 4B, C), which is consistent with the increased level of c-di-GMP of these mutants. These results showed that the mutation of rpoS has led to the increase in rsmY/Z expression and intracellular c-di-GMP content in P. aeruginosa.
rpoS mutation associated hyperbiofilm phenotype in clinical isolates
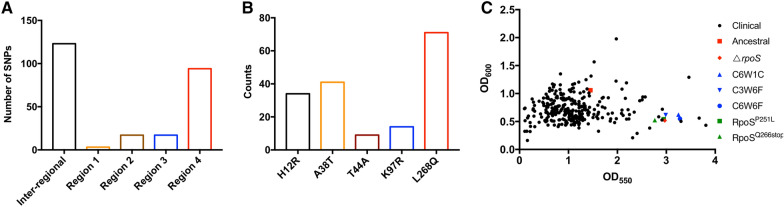
Our experimental biofilm evolution data has revealed that single-nucleotide mutations in rpoS confer P. aeruginosa hyperbiofilm phenotype and produce a pronounced competitive advantage within the biofilm microenvironment. In order to analyze the preference of rpoS mutation, we downloaded 4000 sequences of rpoS from the pseudomonas genome database (www.pseudomonas.com). Through comparative analysis, we have identified 241 non-synonymous mutations (6.03% of total sequence), 8 insertion or deletion mutations (0.2% of total sequence) and 5 stop coding mutations (0.13% of total sequence) compared to the PAO1 wild-type strain. Among those mutations, 123 mutations were in the sequence between annotated regions (inter-region) of RpoS and 131 mutations were within 4 regions (Fig. 5A). Moreover, we have identified 2 sequences harbored RpoSP251L mutation. We also analyzed the top 5 mutation sites among the 4000 sequences, and found L268Q was the top one with 71 sequences (Fig. 5B).
Fig. 5.
rpoS mutations were existed in P. aeruginosa clinical isolates. The distribution of non-synonymous mutation A and the top 5 mutations B on RpoS of P. aeruginosa. 4000 sequences of rpoS were download form pseudomonas genome database, and the non-synonymous mutations were analyzed by CLC Genomics Workbench. C Biofilm formation ability assessment of clinical isolates. A total of 288 clinical isolates from the first affiliated hospital of Guangxi Medical University (shown in black) were analyzed for rpoS mutations and biofilm assays on 96 well PVC plates
Since imipenem has been used for clinical treatment of P. aeruginosa infections, we wondered whether rpoS mutation caused hyperbiofilm strains exist in clinical isolates. Therefore, we examined the biofilm formation capacity of 288 clinical P. aeruginosa isolates obtained from the patients with culture confirmed P. aeruginosa infections (Additional file 1: Table S1). Through quantitative analysis of biofilm formation by measuring crystal violet staining at OD550 nm and total bacterial growth at OD600 nm to exclude growth variation, we identified 29 hyperbiofilm isolates (10.07% of total isolates) in this collection (Fig. 5C). Next, we target sequenced rpoS of the 29 hyperbiofilm isolates and confirmed that #16 isolate harbored non-synonymous mutation in rpoS. Interestingly, #16 isolate, which is isolated from the peritoneal drainage fluid, has the same mutation RpoSP251L as our experimental evolved variant C6W6F.
The evolved rpoS variants are hypervirulent against host cells
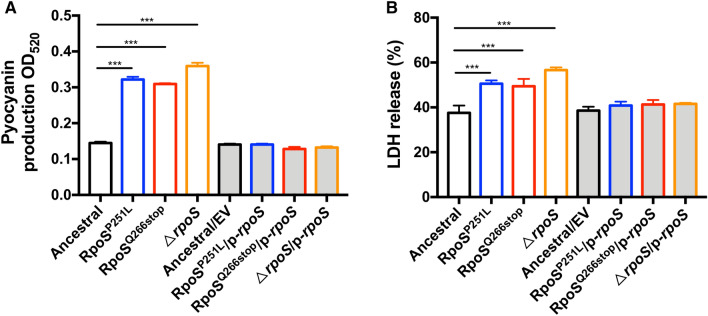
Pyocyanin is one of the major virulence factors of P. aeruginosa, which causes oxidative stress to the host cells and induces apoptosis in neutrophils and inhibits phagocytosis of macrophages [47, 48]. Previous studies showed that the pyocyanin production was increased in a rpoS-deletion mutant [34]. To test the impact of rpoS point mutations of the biofilm evolved variants on pyocyanin production, we compared the production of pyocyanin by P. aeruginosa PAO1 strain, RpoSP251L, RpoSQ266stop and ΔrpoS. As we expected, similar to the ΔrpoS mutant, the P. aeruginosa RpoSP251L and RpoSQ266stop produced higher amounts of pyocyanin than the wild-type PAO1 strain (Fig. 6A). This result suggests that point mutations accumulated in the rpoS gene in P. aeruginosa have similar effect as rpoS gene deletion on its physiology.
Fig. 6.
Pyocyanin production and virulence are increased in rpoS mutants. The production of pyocyanin (A) and cytotoxicity effect against macrophage cells (B) of P. aeruginosa PAO1 wild-type, RpoSP251L, RpoSQ266stop and ΔrpoS. Data are presented as the mean ± s.d. of four biological replicates. Significance was determined using a Student’s t test: *P < 0.05, **P < 0.01, ***P < 0.001
Next, we further assessed the impact of evolved rpoS point mutations on virulence using the macrophage cytotoxicity model [49]. The RAW264.7 macrophages were infected with P. aeruginosa PAO1, RpoSP251L, RpoSQ266stop and ΔrpoS, and the release of cytosolic lactate dehydrogenase (LDH) was determined. We found that macrophages infected with RpoSP251L, RpoSQ266stop and ΔrpoS released more LDH compared to P. aeruginosa PAO1 after 4 h infection (Fig. 6B). Altogether, these results suggest that mutations in rpoS can enhance the virulence in P. aeruginosa.
Discussion
Biofilms represent the predominant lifestyle for most microorganisms in nature. Understanding how microorganisms evolve in biofilms can reveal novel insights of adaptive evolution, especially under stress conditions. For example, small colony variants are enriched in P. aeruginosa biofilms after exposure to sodium dodecyl sulfate [50]. The P. aeruginosa hyperbiofilm forming variants are often observed from patients who are suffering chronic infections such as CF [51]. We have previously using a planktonic experimental evolution model to demonstrate that oxidative stress drives the evolution P. aeruginosa hyperbiofilm variants carrying point mutations in the wspF gene, which lead to increase in intracellular c-di-GMP content and exopolysaccharide synthesis [27]. Here, we performed biofilm experimental evolution to examine the adaptive evolution of P. aeruginosa biofilms under the treatment of imipenem (160 μg/mL), which is a case can be encountered in clinical settings. The keratitis infection caused by P. aeruginosa usually form corneal biofilms [52]. In a case of the treatment of bacterial keratitis in patients, topical imipenem (50 mg/mL) has been selected as monotherapy for corneal infection [53]. In another study, 1–5 mg/mL imipenem has been used for bacterial keratitis treatment via topical administration [54]. Moreover, we showed that the hyperbiofilm variants could be accumulated in biofilms after cycle antibiotic treatment and these variants were able to outcompete the ancestor strain shortly after appearance. Genome sequencing analysis revealed that the adapted P. aeruginosa hyperbiofilm variants in different linages shared single-point mutations in the same gene, which encodes the sigma factor RpoS.
The rpoS gene has been previously well characterized in P. aeruginosa for its regulatory role on quorum sensing and virulence. A DNA microarray-based transcriptomic study showed that the expression of rpoS in P. aeruginosa biofilm cells was downregulated compared to the planktonic cells, inactivation of rpoS in P. aeruginosa PAO1 increased biofilm formation in flow-cell reactor [14]. In our study, we demonstrated that rpoS point mutations have similar impact to the rpoS deletion on biofilm formation in P. aeruginosa. Function domain analysis indicates that RpoS contain 4 regions, RpoS as a global regulator, the DNA binding domain was located on region 4. Proline residues are restricts to the first four positions of an α-helix [55], which plays a special role in the stable of protein structure. RpoSP251L and RpoSQ266stop on P. aeruginosa genome have shown the same phenotype of hyperbiofilm, pyocyanin production and virulence as ΔrpoS strain, which means the 251proline to leucine mutation in rpoS might results loss of the function on regulation. Moreover, we showed that mutation in rpoS increased the production of c-di-GMP and pyocyanin, both of those two molecules were play a very important role in biofilm formation [39] and virulence [56] in P. aeruginosa.
It is generally accepted that bacterial cells employ biofilm mode of growth during chronic infections with reduced level of virulence. For example, the alginate-over producing P. aeruginosa ΔmucA mutant and the high c-di-GMP containing ΔwspF mutant both produce less amounts of virulence factors compared to the wild-type strains [57, 58]. However, here we showed that rpoS mutation enable enhancement of both biofilm formation and production of virulence factors such as pyocyanin. The reactive oxygen species generated by redox cycling of pyocyanin kills host and pathogen cells, resulting in extracellular DNA (eDNA) release [59, 60]. eDNA has been shown plays a central role in biofilm formation by increasing biofilm integrity and strength [61–63]. Not limited to the P. aeruginosa species, rpoS mutants were also reported to contribute to population heterogeneity in Escherichia coli O157:H7 strains [64].
Biofilm formation as a response to ecological competition [65], PAO1 wild-type and rpoS mutants live with identical niches and also have identical needs within biofilm, thus they will compete for precisely the same resources. Our observation that rpoS mutants can outcompete the PAO1 within biofilms could be explained by the competitive exclusion principle [66, 67]. The P. aeruginosa small colony variants (e.g., with wspF mutations) are also well known being evolved in P. aeruginosa biofilms, which have even higher biofilm formation capacity than the rpoS mutants. However, these small colony variants often have a lower planktonic growth rate than the P. aeruginosa wild-type strain, and thus can easily be outcompeted by the wild-type in the planktonic phase of growth [27]. Instead, our study showed that the P. aeruginosa rpoS mutants are able to outcompete the wild-type PAO1 strain within biofilm while not compromising its planktonic growth rate. MIC measurement experiments showed no antimicrobial resistance difference between the rpoS mutants and PAO1, suggesting a form of antimicrobial tolerance of biofilm cells. Antimicrobial tolerance is one of the most important features of microbial biofilms, which does not involve drug resistance mutations [68]. The main mechanism of antimicrobial tolerance of biofilms is the reduction of antibiotic penetration [69]. Our study here showed that rapid selection of the hyperbiofilm variants carrying the rpoS mutations represents a novel antimicrobial tolerance mechanism of biofilms.
Conclusions
Our study showed that imipenem treatment drives rapid evolution of P. aeruginosa rpoS deficient mutants within biofilms. We provided evidence that rpoS mutation not only increase P. aeruginosa virulence, but also enhance its intracellular c-di-GMP content. Importantly, the major obstacle for treatment of P. aeruginosa infection in clinical is the formation of biofilms. As a sigma factor, RpoS controls a wide range of genes under stationary phase of growth, which shared many characters with biofilm mode of growth, such as lack of nutrients and accumulation of waste products. Further studies should be carried to examining the regulatory roles of RpoS on P. aeruginosa physiology and virulence factors under biofilm mode of growth. This study raises the possibility that some clinical P. aeruginosa strains with rpoS mutations could have a selective advantage during imipenem administration, which might have an impact on the antibiotic therapy against P. aeruginosa biofilm-associated infections.
Methods
For details see Additional file 1.
Biofilm experimental evolution
The experimental evolution of P. aeruginosa PAO1 biofilm was carried out on glass beads formed biofilm [31]. Two autoclaved 5 mm glass beads (Merck KGaA, Darmstadt, Germany) were placed into each well of a 24-well microtiter plate (Jet Biofil, Guangzhou, China). A LB overnight culture of P. aeruginosa was diluted in LB to approx. 1 × 106 bacteria per mL and dispensed into the bead-containing 24 well microplate (1 mL per well). The microplate was then placed in a moisture box and incubated at 37 °C for 24 h at 100 rpm on an orbital shaker. After 24 h, the liquid culture was removed, and beads were washed by 0.9% NaCl for twice to remove loosely attached bacteria. Then one bead was transferred into a 2 mL microcentrifuge tube containing 1 mL 0.9% NaCl, subjected to 6 × 10 s vortex and sonicated in an ultrasonic bath (Worldvicon, Shenzhen, China) at 40 kHz for 5 min. Bacterial suspensions were subsequently serially diluted in 0.9% NaCl before being drop-plated onto lysogeny broth agar plates (Difco). After 24 h of incubation at 37 °C, the residual biofilm was quantified as CFU/bead. Another bead was transferring to a 24 well microplate contain 1 mL LB with 160 μg/mL imipenem. The microplate was then placed in a moisture box and incubated at 37 °C for 24 h without shaking. After 24 h treatment, this bead was washed by 0.9% NaCl for twice and transfer into 2 mL microcentrifuge tube containing 1 mL LB, after vortex and sonicated. 20 μL of bacterial suspensions were subsequently serially diluted in 0.9% NaCl before being drop-plated onto lysogeny broth agar plates (Difco), the rest bacterial suspensions were cultured at 37 °C for 24 h at 200 rpm. After 24 h of cultivation, 100 μL P. aeruginosa was diluted in LB to approx. 1 × 106 bacteria per mL and start a new cycle. The rest culture was glycerol stocked at − 80 °C. The CFU/bead increased over 100-fold compared to the ancestral strain was defined as hyperbiofilm phenotype variants.
Biofilm competition assay
The biofilm competition assay was carried out on glass beads formed biofilm. The ancestor strain PAO1 and mutants were tagged with gfp and mcherry at the attB site to generate the strain PAO1 attB::gfp and mutant attB::mcherry as previously described [70]. Overnight cultures were adjusted OD600 to 1.0, cells were mixed 1:1 or 1:5 and confirmed by flow cytometer analysis. The mixed bacteria were diluted in LB to approx.1 × 106 of per mL and dispensed into the bead-containing 24 well microplate (1 mL per well). The microplate was then placed in a moisture box and incubated at 37 °C for 24 h at 100 rpm on an orbital shaker. After 24 h treatment, the cells in planktonic and biofilm were analyzed by Beckman Cytoflex S flow cytometer. All samples were assayed with lasers emitting at 488 nm for GFP or 561 nm for mCherry. Fluorescence was collected by 530/30 nm bandpass filter for GFP and 615/20 nm bandpass filter for mCherry. In each run, we measured 100,000 events. Sterile PBS, wild-type (WT) P. aeruginosa PAO1, mCherry positive and GFP-positive cells were used to gate the cell populations on flow-cytometry diagrams.
DNA extraction, sequencing, and SNP analysis
Genomic DNA of the ancestor and evolved bacterial populations were extracted form glycerol stocked by AxyPerp Bacterial Genomic DNA Miniprep Kit (Corning) and sequenced by Illumina NovaSeq platform. Illumina genomic reads of the isolates were analyzed by CLC Genomics Workbench 20 (Qiagen) using Resequencing analysis module with default parameters for single nucleotide polymorphism (SNP) with P. aeruginosa PAO1 as reference genome.
RNA extraction, sequencing, and transcriptomic analysis
Samples were collected at the peak of PcdrA-gfp fluorescence intensity, RNA extraction was performed using the miRNeasy kit (Qiagen) according to the manufacturer’s instructions. RNA samples were submitted to Guangdong Magigene Biotechnology Co., Ltd. (Guangzhou, China) for ribosomal RNA depletion and sequencing. RNA samples were sequenced on an Illumina Hiseq Xten platform and 150 bp paired-end reads were generated.
The quality of raw sequence data was assessed using FastQC (Babraham Bioinformatics). Adaptor sequences were removed by adaptor trimming function in CLC. RNA sequence analysis was done using “RNA-seq analysis’ module in CLC genomics Workbench 20 (CLC Bio, Aarhus, Denmark) using P. aeruginosa PAO1 reference genome downloaded from NCBI database. Adaptor sequences were removed by adaptor trimming function in CLC. Differential gene expression was analyzed using DESeq2 package in R software.
Statistical analysis
Data are presented as mean ± standard deviation (SD). All other comparisons were made using a one-way analysis of variance (ANOVA) with Student’s t test. Analyses were performed using GraphPad Prism v.7 (GraphPad Software). Statistical significance was determined using a P value of < 0.05.
Supplementary Information
Additional file 1. Supplementary methods, figures and tables for this manuscript.
Acknowledgements
Not applicable.
Abbreviations
- CF
Cystic fibrosis
- CFU
Colony forming units
- MIC
Minimum inhibitory concentration
- CV
Crystal violet
- QS
Quorum-sensing
- DGC
Diguanylate cyclases
- PDE
Phosphodiesterases
- LDH
Lactate dehydrogenase
Authors’ contributions
XD and YL designed methods and experiments; XD carried out the laboratory experiments, analyzed the data and interpreted the results; YP performed DNA manipulation and cytotoxicity assay; ZC and YL co-designed the DNA sequencing experiments and worked on associated data collection and its interpretation; YML and YZ performed motility and transcriptional fusions reporter assay; KW and ML collected the clinical strains and performed high biofilm screening; XD, LZ and LY wrote the paper; all authors have contributed to, seen and approved the manuscript. All authors read and approved the final manuscript.
Funding
This research was supported by the Shenzhen Key Laboratory of Gene Regulation and Systems Biology (ZDSYS20200811144002008), the Shenzhen Overseas High-level Talent Team (KQTD20200909113758004), Guangdong Natural Science Foundation for Distinguished Young Scholar (2020B1515020003), the Guangdong Natural Science Foundation (2019A1515110640, 2020A1515010316), National Natural Science Foundation of China (81760024), the Guangzhou Municipal Science and Technology Bureau (201607020044).
Availability of data and materials
The DNA and RNA sequence data that support the findings of this study have been deposited in the NCBI Sequence Read Archive (SRA) with the accession number: PRJNA678555. The materials that support the findings of this study are available from the corresponding author upon reasonable request.
Declarations
Ethics approval and consent to participate
Working with the P. aeruginosa clinical isolates is approved by the Ethics Committee of the first affiliated hospital of Guangxi Medical University [2017(KY-E-080)].
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Lenski RE. What is adaptation by natural selection? Perspectives of an experimental microbiologist. PLoS Genet. 2017;13:1006668. doi: 10.1371/journal.pgen.1006668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Elena SF, Lenski RE. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat Rev Genet. 2003;4:457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- 3.Lenski RE. Experimental evolution and the dynamics of adaptation and genome evolution in microbial populations. ISME J. 2017;11:2181–2194. doi: 10.1038/ismej.2017.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duan X, Fu Y, Yang L. Molecular and systems biology approaches for analyzing drug-tolerant bacterial persister cells. In Sustainable Agriculture Reviews 46. Springer. 2020, pp 109–128.
- 5.Santos-Lopez A, Marshall CW, Snyder DJ, Cooper VS. Evolutionary pathways to antibiotic resistance are dependent upon environmental structure and bacterial lifestyle. Elife. 2016;8:e47612. doi: 10.7554/eLife.47612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van den Bergh B, Michiels JE, Wenseleers T, Windels E, Vanden Boer P, Kestemont D, De Meester L, Verstrepen KJ, Verstraeten N, Michiels J. Frequency of antibiotic application drives rapid evolutionary adaptation of Escherichia coli persistence. Nat Microbiol. 2016;1:1–7. doi: 10.1038/nmicrobiol.2016.20. [DOI] [PubMed] [Google Scholar]
- 7.Steenackers HP, Parijs I, Foster KR, Vanderleyden J. Experimental evolution in biofilm populations. FEMS Microbiol Rev. 2016;40:373–397. doi: 10.1093/femsre/fuw002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Penterman J, Nguyen D, Anderson E, Staudinger BJ, Greenberg EP, Lam JS, Singh PK. Rapid Evolution of culture-impaired bacteria during adaptation to biofilm growth. Cell Rep. 2014;6:293–300. doi: 10.1016/j.celrep.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Savage VJ, Chopra I, O'Neill AJ. Population diversification in Staphylococcus aureus biofilms may promote dissemination and persistence. Plos One. 2013;8:e62513. doi: 10.1371/journal.pone.0062513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jorth VJ, McLean I, Ratjen AJ, Secor AJ, Bautista AJ, Ravishankar AJ, Rezayat A, Garudathri J, Harrison JJ, Harwood RA. Evolved aztreonam resistance is multifactorial and can produce hypervirulence in Pseudomonas aeruginosa. MBio. 2017;8:e00517-17. doi: 10.1128/mBio.00517-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Traverse CC, Mayo-Smith LM, Poltak SR, Cooper VS. Tangled bank of experimentally evolved Burkholderia biofilms reflects selection during chronic infections. Proc Natl Acad Sci USA. 2013;110:E250–E259. doi: 10.1073/pnas.1207025110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8:623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 13.Chua SL, Liu Y, Yam JK, Chen Y, Vejborg RM, Tan BG, Kjelleberg S, Tolker-Nielsen T, Givskov M, Yang L. Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyles. Nat Commun. 2014;5:4462. doi: 10.1038/ncomms5462. [DOI] [PubMed] [Google Scholar]
- 14.Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S, Greenberg EP. Gene expression in Pseudomonas aeruginosa biofilms. Nature. 2001;413:860–864. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- 15.Olsen I. Biofilm-specific antibiotic tolerance and resistance. Eur J Clin Microbiol Infect Dis. 2015;34:877–886. doi: 10.1007/s10096-015-2323-z. [DOI] [PubMed] [Google Scholar]
- 16.Brooun A, Liu S, Lewis K. A dose-response study of antibiotic resistance in Pseudomonas aeruginosa biofilms. Antimicrob Agents Chemother. 2000;44:640–646. doi: 10.1128/AAC.44.3.640-646.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malhotra S, Hayes D, Wozniak DJ. Cystic fibrosis and Pseudomonas aeruginosa: the host-microbe interface. Clin Microbiol Rev. 2019;32:e00138–18. doi: 10.1128/CMR.00138-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mulcahy LR, Isabella VM, Lewis K. Pseudomonas aeruginosa biofilms in disease. Microb Ecol. 2014;68:1–12. doi: 10.1007/s00248-013-0297-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lebeaux D, Ghigo J-M, Beloin C. Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol Mol Biol Rev. 2014;78:510–543. doi: 10.1128/MMBR.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donlan RM. Biofilms and device-associated infections. Emerg Infect Dis. 2001;7:277–281. doi: 10.3201/eid0702.010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jensen PO, Givskov M, Bjarnsholt T, Moser C. The immune system vs. Pseudomonas aeruginosa biofilms. FEMS Immunol Med Microbiol. 2010;59:292–305. doi: 10.1111/j.1574-695X.2010.00706.x. [DOI] [PubMed] [Google Scholar]
- 22.Arciola CR, Campoccia D, Montanaro L. Implant infections: adhesion, biofilm formation and immune evasion. Nat Rev Microbiol. 2018;16:397–409. doi: 10.1038/s41579-018-0019-y. [DOI] [PubMed] [Google Scholar]
- 23.Hall CW, Mah TF. Molecular mechanisms of biofilm-based antibiotic resistance and tolerance in pathogenic bacteria. FEMS Microbiol Rev. 2017;41:276–301. doi: 10.1093/femsre/fux010. [DOI] [PubMed] [Google Scholar]
- 24.Lewis K. Persister cells. Ann Rev Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 25.Lewis K. Persister cells, dormancy and infectious disease. Nat Rev Microbiol. 2007;5:48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 26.Abdelraheem WM, Abdelkader AE, Mohamed ES, Mohammed MS. Detection of biofilm formation and assessment of biofilm genes expression in different Pseudomonas aeruginosa clinical isolates. Meta Gene. 2020;23:100646. doi: 10.1016/j.mgene.2020.100646. [DOI] [Google Scholar]
- 27.Chua SL, Ding Y, Liu Y, Cai Z, Zhou J, Swarup S, Drautz-Moses DI, Schuster SC, Kjelleberg S, Givskov M. Reactive oxygen species drive evolution of pro-biofilm variants in pathogens by modulating cyclic-di-GMP levels. Open Biol. 2016;6:160162. doi: 10.1098/rsob.160162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jørgensen KM, Wassermann T, Jensen PØ, Hengzuang W, Molin S, Høiby N, Ciofu O. Sublethal ciprofloxacin treatment leads to rapid development of high-level ciprofloxacin resistance during long-term experimental evolution of Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2013;57:4215–4221. doi: 10.1128/AAC.00493-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pragasam AK, Raghanivedha M, Anandan S, Veeraraghavan B. Characterization of Pseudomonas aeruginosa with discrepant carbapenem susceptibility profile. Ann Clin Microbiol Antimicrob. 2016;15:1–4. doi: 10.1186/s12941-016-0127-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winston DJ, McGrattan M, Busuttil R. Imipenem therapy of Pseudomonas aeruginosa and other serious bacterial infections. Antimicrob Agents Chemother. 1984;26:673–677. doi: 10.1128/AAC.26.5.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Konrat K, Schwebke I, Laue M, Dittmann C, Levin K, Andrich R, Arvand M, Schaudinn C. The Bead assay for biofilms: a quick, easy and robust method for testing disinfectants. Plos One. 2016;11:e0157663. doi: 10.1371/journal.pone.0157663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brookfield JFY. Experimental evolution: the rate of adaptive evolution. Curr Biol. 2010;20:R23–R25. doi: 10.1016/j.cub.2009.11.044. [DOI] [PubMed] [Google Scholar]
- 33.Jorgensen F, Bally M, Chapon-Herve V, Michel G, Lazdunski A, Williams P, Stewart G. RpoS-dependent stress tolerance in Pseudomonas aeruginosa. Microbiology (Reading) 1999;145(Pt 4):835–844. doi: 10.1099/13500872-145-4-835. [DOI] [PubMed] [Google Scholar]
- 34.Suh SJ, Silo-Suh L, Woods DE, Hassett DJ, West SE, Ohman DE. Effect of rpoS mutation on the stress response and expression of virulence factors in Pseudomonas aeruginosa. J Bacteriol. 1999;181:3890–3897. doi: 10.1128/JB.181.13.3890-3897.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schuster M, Hawkins AC, Harwood CS, Greenberg EP. The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol Microbiol. 2004;51:973–985. doi: 10.1046/j.1365-2958.2003.03886.x. [DOI] [PubMed] [Google Scholar]
- 36.Chua SL, Sivakumar K, Rybtke M, Yuan M, Andersen JB, Nielsen TE, Givskov M, Tolker-Nielsen T, Cao B, Kjelleberg S. C-di-GMP regulates Pseudomonas aeruginosa stress response to tellurite during both planktonic and biofilm modes of growth. Sci Rep. 2015;5:10052. doi: 10.1038/srep10052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahmed MN, Abdelsamad A, Wassermann T, Porse A, Becker J, Sommer MOA, Hoiby N, Ciofu O. The evolutionary trajectories of P. aeruginosa in biofilm and planktonic growth modes exposed to ciprofloxacin: beyond selection of antibiotic resistance. NPJ Biofilms Microbiomes. 2020;6:28. doi: 10.1038/s41522-020-00138-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Kievit TR. Quorum sensing in Pseudomonas aeruginosa biofilms. Environ Microbiol. 2009;11:279–288. doi: 10.1111/j.1462-2920.2008.01792.x. [DOI] [PubMed] [Google Scholar]
- 39.Ha D-G, O’Toole GA. c‐di‐GMP and its effects on biofilm formation and dispersion: a Pseudomonas aeruginosa review. Microbial Biofilms. 2015;3:301–317. doi: 10.1128/9781555817466.ch15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hentzer M, Riedel K, Rasmussen TB, Heydorn A, Andersen JB, Parsek MR, Rice SA, Eberl L, Molin S, Hoiby N, et al. Inhibition of quorum sensing in Pseudomonas aeruginosa biofilm bacteria by a halogenated furanone compound. Microbiology. 2002;148:87–102. doi: 10.1099/00221287-148-1-87. [DOI] [PubMed] [Google Scholar]
- 41.Yang L, Barken KB, Skindersoe ME, Christensen AB, Givskov M, Tolker-Nielsen T. Effects of iron on DNA release and biofilm development by Pseudomonas aeruginosa. Microbiology. 2007;153:1318–1328. doi: 10.1099/mic.0.2006/004911-0. [DOI] [PubMed] [Google Scholar]
- 42.Yang L, Rybtke MT, Jakobsen TH, Hentzer M, Bjarnsholt T, Givskov M, Tolker-Nielsen T. Computer-aided identification of recognized drugs as Pseudomonas aeruginosa quorum-sensing inhibitors. Antimicrob Agents Chemother. 2009;53:2432–2443. doi: 10.1128/AAC.01283-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rybtke MT, Borlee BR, Murakami K, Irie Y, Hentzer M, Nielsen TE, Givskov M, Parsek MR, Tolker-Nielsen T. Fluorescence-based reporter for gauging cyclic di-GMP levels in Pseudomonas aeruginosa. Appl Environ Microbiol. 2012;78:5060–5069. doi: 10.1128/AEM.00414-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Valentini M, Filloux A. Biofilms and cyclic di-GMP (c-di-GMP) signaling: lessons from Pseudomonas aeruginosa and other bacteria. J Biol Chem. 2016;291:12547–12555. doi: 10.1074/jbc.R115.711507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rasis M, Segal G. The LetA-RsmYZ-CsrA regulatory cascade, together with RpoS and PmrA, post-transcriptionally regulates stationary phase activation of Legionella pneumophila Icm/Dot effectors. Mol Microbiol. 2009;72:995–1010. doi: 10.1111/j.1365-2958.2009.06705.x. [DOI] [PubMed] [Google Scholar]
- 46.Frangipani E, Visaggio D, Heeb S, Kaever V, Camara M, Visca P, Imperi F. The Gac/Rsm and cyclic-di-GMP signalling networks coordinately regulate iron uptake in Pseudomonas aeruginosa. Environ Microbiol. 2014;16:676–688. doi: 10.1111/1462-2920.12164. [DOI] [PubMed] [Google Scholar]
- 47.Lau GW, Ran HM, Kong FS, Hassett DJ, Mavrodi D. Pseudomonas aeruginosa pyocyanin is critical for lung infection in mice. Infect Immun. 2004;72:4275–4278. doi: 10.1128/IAI.72.7.4275-4278.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lau GW, Hassett DJ, Ran HM, Kong FS. The role of pyocyanin in Pseudomonas aeruginosa infection. Trends Mol Med. 2004;10:599–606. doi: 10.1016/j.molmed.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 49.Garai P, Berry L, Moussouni M, Bleves S, Blanc-Potard A-B. Killing from the inside: intracellular role of T3SS in the fate of Pseudomonas aeruginosa within macrophages revealed by mgtC and oprF mutants. PLoS Pathog. 2019;15:1007812. doi: 10.1371/journal.ppat.1007812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee KWK, Yam JKH, Mukherjee M, Periasamy S, Steinberg PD, Kjelleberg S, Rice SA. Interspecific diversity reduces and functionally substitutes for intraspecific variation in biofilm communities. ISME J. 2016;10:846–857. doi: 10.1038/ismej.2015.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Starkey M, Hickman JH, Ma L, Zhang N, De Long S, Hinz A, Palacios S, Manoil C, Kirisits MJ, Starner TD. Pseudomonas aeruginosa rugose small-colony variants have adaptations that likely promote persistence in the cystic fibrosis lung. J Bacteriol. 2009;191:3492–3503. doi: 10.1128/JB.00119-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Saraswathi P, Beuerman RW. Corneal biofilms: from planktonic to microcolony formation in an experimental keratitis infection with Pseudomonas aeruginosa. Ocul Surf. 2015;13:331–345. doi: 10.1016/j.jtos.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 53.Turkyilmaz K, Kurt A, Dilek AR, Sekeryapan B, Erturk A. A case of suture-related bacterial keratitis and its treatment with topical imipenem. J Ocular Biol, Dis, Inf. 2011;4:141–144. doi: 10.1007/s12177-012-9081-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Egrilmez S, Yildirim-Theveny Ş. Treatment-resistant bacterial keratitis: challenges and solutions. Clin Ophthalmol (Auckland, NZ) 2020;14:287. doi: 10.2147/OPTH.S181997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Woolfson DN, Williams DH. The influence of proline residues on alpha-helical structure. FEBS Lett. 1990;277:185–188. doi: 10.1016/0014-5793(90)80839-B. [DOI] [PubMed] [Google Scholar]
- 56.Jayaseelan S, Ramaswamy D, Dharmaraj S. Pyocyanin: production, applications, challenges and new insights. World J Microbiol Biotechnol. 2014;30:1159–1168. doi: 10.1007/s11274-013-1552-5. [DOI] [PubMed] [Google Scholar]
- 57.Silo-Suh L, Suh S-J, Sokol PA, Ohman DE. A simple alfalfa seedling infection model for Pseudomonas aeruginosa strains associated with cystic fibrosis shows AlgT (sigma-22) and RhlR contribute to pathogenesis. Proc Natl Acad Sci. 2002;99:15699–15704. doi: 10.1073/pnas.242343999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chung I-Y, Choi KB, Heo Y-J, Cho Y-H. Effect of PEL exopolysaccharide on the wspF mutant phenotypes in Pseudomonas aeruginosa PA14. J Microbiol Biotechnol. 2008;18:1227–1234. [PubMed] [Google Scholar]
- 59.Rada B, Jendrysik MA, Pang L, Hayes CP, Yoo D-g, Park JJ, Moskowitz SM, Malech HL, Leto TL. Pyocyanin-enhanced neutrophil extracellular trap formation requires the NADPH oxidase. PLoS ONE. 2013;8:54205. doi: 10.1371/journal.pone.0054205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Das T, Manefield M. Pyocyanin promotes extracellular DNA release in Pseudomonas aeruginosa. PLoS ONE. 2012;7:46718. doi: 10.1371/journal.pone.0046718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487–1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 62.Dominiak DM, Nielsen JL, Nielsen PH. Extracellular DNA is abundant and important for microcolony strength in mixed microbial biofilms. Environ Microbiol. 2011;13:710–721. doi: 10.1111/j.1462-2920.2010.02375.x. [DOI] [PubMed] [Google Scholar]
- 63.Saunders SH, Edmund C, Yates MD, Otero FJ, Trammell SA, Stemp ED, Barton JK, Tender LM, Newman DK. Extracellular DNA promotes efficient extracellular electron transfer by pyocyanin in Pseudomonas aeruginosa biofilms. Cell. 2020;182:919–932. doi: 10.1016/j.cell.2020.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Carter MQ, Louie JW, Huynh S, Parker CT. Natural rpoS mutations contribute to population heterogeneity in Escherichia coli O157: H7 strains linked to the 2006 US spinach-associated outbreak. Food Microbiol. 2014;44:108–118. doi: 10.1016/j.fm.2014.05.021. [DOI] [PubMed] [Google Scholar]
- 65.Oliveira NM, Martinez-Garcia E, Xavier J, Durham WM, Kolter R, Kim W, Foster KR. Biofilm formation as a response to ecological competition. PLoS Biol. 2015;13:1002191. doi: 10.1371/journal.pbio.1002191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Anderson MS, Garcia EC, Cotter PA. Kind discrimination and competitive exclusion mediated by contact-dependent growth inhibition systems shape biofilm community structure. PLoS Pathog. 2014;10:1004076. doi: 10.1371/journal.ppat.1004076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nadell CD, Drescher K, Foster KR. Spatial structure, cooperation and competition in biofilms. Nat Rev Microbiol. 2016;14:589–600. doi: 10.1038/nrmicro.2016.84. [DOI] [PubMed] [Google Scholar]
- 68.Lewis K. Persister cells. Annu Rev Microbiol. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- 69.Alipour M, Suntres ZE, Omri A. Importance of DNase and alginate lyase for enhancing free and liposome encapsulated aminoglycoside activity against Pseudomonas aeruginosa. J Antimicrob Chemother. 2009;64:317–325. doi: 10.1093/jac/dkp165. [DOI] [PubMed] [Google Scholar]
- 70.Bao Y, Lies DP, Fu H, Roberts GP. An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene. 1991;109:167–168. doi: 10.1016/0378-1119(91)90604-A. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Supplementary methods, figures and tables for this manuscript.
Data Availability Statement
The DNA and RNA sequence data that support the findings of this study have been deposited in the NCBI Sequence Read Archive (SRA) with the accession number: PRJNA678555. The materials that support the findings of this study are available from the corresponding author upon reasonable request.