Summary
Bone stroma contributes to the regulation of osteogenesis and hematopoiesis but also to fracture healing and disease processes. Mesenchymal stromal cells from bone (BMSCs) represent a heterogenous mixture of different subpopulations with distinct molecular and functional properties. The lineage relationship between BMSC subsets and their regulation by intrinsic and extrinsic factors are not well understood. Here, we show with mouse genetics, ex vivo cell differentiation assays, and transcriptional profiling that BMSCs from metaphysis (mpMSCs) and diaphysis (dpMSCs) are fundamentally distinct. Fate-tracking experiments and single-cell RNA sequencing indicate that bone-forming osteoblast lineage cells and dpMSCs, including leptin receptor-positive (LepR+) reticular cells in bone marrow, emerge from mpMSCs in the postnatal metaphysis. Finally, we show that BMSC fate is controlled by platelet-derived growth factor receptor β (PDGFRβ) signaling and the transcription factor Jun-B. The sum of our findings improves our understanding of BMSC development, lineage relationships, and differentiation.
Keywords: bone mesenchymal stromal cells, metaphysis, diaphysis, osteoblast lineage cells, adipocytes, mouse genetics, single-cell RNA sequencing
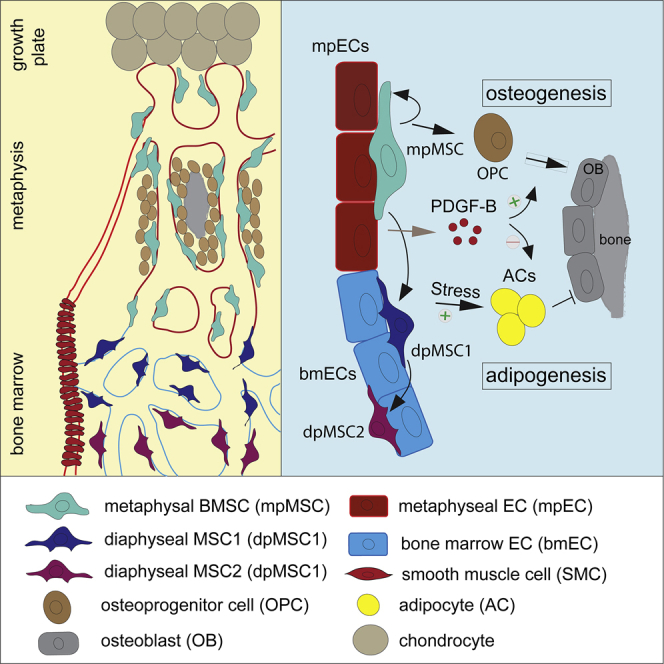
Graphical abstract
Highlights
-
•
Mesenchymal stromal cells from metaphysis and diaphysis have distinct properties
-
•
Metaphyseal mpMSCs include cells with multi-lineage differentiation potential
-
•
mpMSCs give rise to osteoprogenitors and reticular cells in marrow during development
-
•
Microenvironmental cues and cell-autonomous transcription factors control BMSC fate
Sivaraj et al. characterize the heterogeneity of bone mesenchymal stromal cells (BMSCs) during skeletal development. A subpopulation of metaphyseal MSCs (mpMSCs) has self-renewing and multi-lineage differentiation potential to generate bone cells and LepR+ marrow stromal cells. BMSCs fate determination is controlled extrinsically by PDGF-B and intrinsically by Jun-B.
Introduction
Developmental growth of the skeletal system involves extensive communication between different stromal cell types, including mesenchymal stem/progenitor cells, committed osteoblast lineage cells, mature osteocytes, chondrocytes, vascular cells, and bone-degrading osteoclasts (Bianco et al., 2008, 2014; Cook and Genever, 2013; Olsen et al., 2000). Some of these interactions persist after the completion of developmental bone formation and remain relevant in the context of lifelong hematopoiesis, homeostasis of the skeletal system, fracture healing, or osteoporotic bone loss (Gerber and Ferrara, 2000; Maes and Clemens, 2014; Sacchetti et al., 2007; Sivaraj and Adams, 2016; Zelzer and Olsen, 2005). BMSCs are a heterogenous mixture of different cell subpopulations, which are often associated with progenitor properties, colony-forming capacity ex vivo, and the potential to generate bone, cartilage, fat, fibroblasts, and vascular support cells (i.e., pericytes and vascular smooth muscle cells) (Bianco et al., 2008; Nombela-Arrieta et al., 2011; Ortinau et al., 2019; Zhou et al., 2014). Some key features are not shared by all BMSCs and, instead, are probably confined to comparably rare mesenchymal stem and progenitor cells (MSPCs) or skeletal stem cells (SSCs), which are characterized by self-renewal capacity and multipotency, i.e., the ability to give rise to multiple differentiated cell types in a clonal fashion (Bianco et al., 2008; Nombela-Arrieta et al., 2011; Ono et al., 2019; Sacchetti et al., 2007; Uccelli et al., 2008). SSCs exist within the growth plate or periosteum in early long-bone development but have been also been found in adult periosteum and bone marrow (BM) (Chan et al., 2015, 2018; Debnath et al., 2018; Mizuhashi et al., 2018; Ono et al., 2019; Ortinau et al., 2019). Another important BMSC subpopulation is CXCL12-abundant reticular (CAR) cells, which are similar or identical to leptin receptor-positive (LepR+) cells, are tightly associated with the sinusoidal BM vasculature, and function as critical regulators of hematopoiesis (Ding et al., 2012; Sugiyama et al., 2006). LepR+ cells can also give rise to osteoblast lineage cells and have been implicated in bone formation (Ding et al., 2012; Zhou et al., 2014). LepR+ BMSCs include cells that express markers of stem cells and neurons, namely Thy (CD90) and Nestin (Isern et al., 2014; Méndez-Ferrer et al., 2010; Worthley et al., 2015). Expression of the markers Gremlin 1 and CD105 labels a population of reticular SSCs, which can give rise to chondrocytes, osteoblasts, and BM perivascular stromal cells (Worthley et al., 2015). SSCs/MSPCs were also reported to express the receptor kinase PDGFRβ, which, in response to activation by platelet-derived growth factor B (PDGF-B), promotes bone formation and regeneration (Böhm et al., 2019; Xie et al., 2014).
Recently, single-cell RNA sequencing (scRNA-seq) has provided further insight into the heterogeneity of BMSCs in healthy adults and challenged mice (Addo et al., 2019; Baccin et al., 2020; Baryawno et al., 2019; Tikhonova et al., 2019; Wolock et al., 2019; Zhong et al., 2020). Although these studies have led to the identification of numerous BMSC subsets, the spatial distribution, lineage specification, and functional roles of many of these populations remain to be determined. In addition, little is known about BMSC heterogeneity in the context of bone growth and development. Here, we provide insight into the properties of BMSCs in developing long bone with a combination of bulk and scRNA-seq, ex vivo colony formation assays, genetic fate tracking in postnatal mice, and gene inactivation experiments. The sum of our work indicates that metaphyseal BMSCs (i.e., mpMSCs) possess growth and multipotential differentiation potential that is not shared by diaphyseal dpMSCs from the BM cavity. PDGF-B functions as a BMSC niche factor, which promotes osteogenic differentiation and represses adipogenesis. Finally, we show that loss of the AP-1 family transcription factor Jun-B impairs the osteogenic differentiation capacity of BMSCs and thereby enhances adipogenesis during skeletal development.
Results
Distribution and features of BMSCs in long bone
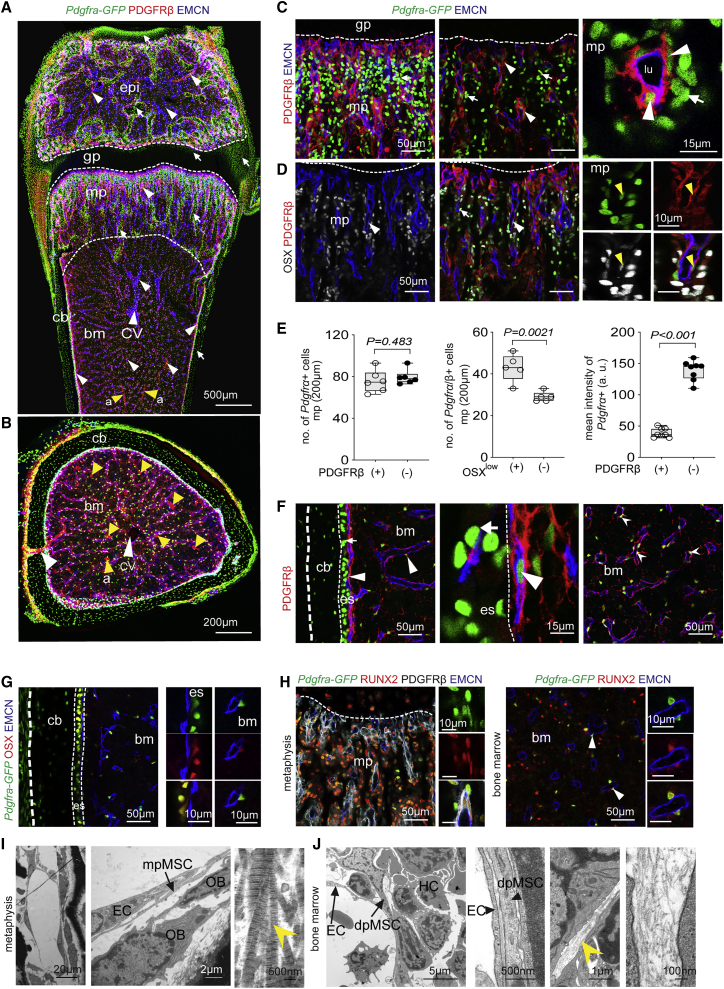
To study the spatial distribution and morphology of mesenchymal stromal cells during postnatal bone growth at single-cell resolution, we analyzed the femurs of 3-week-old Pdgfra-H2BGFP (Pdgfra-GFP) knock-in reporter mice, which express nuclear green fluorescent protein (GFP) in PDGFRα+ cells (Hamilton et al., 2003) by immunostaining. In the metaphysis (mp), the region mediating the growth of long bone, Pdgfra-GFP labels cells in trabecular bone (tb) and compact bone (cb) but also a subset of growth-plate chondrocytes. Although Pdgfra-GFP+ cells lack PDGFRβ immunosignal in cb and cartilage, double-positive cells (PDGFRα+β+) are located around blood vessels in mp, BM, and epiphysis (Figures 1A, 1B, S1A, and S1B). These perivascular, PDGFRβ+ BMSCs express comparably low levels of Pdgfra-GFP both in mp and diaphysis (Figures 1C and S1B). Co-immunostaining for osterix (OSX) shows low levels of the nuclear transcription factor in a fraction of vessel-associated PDGFRα+β+ cells, whereas high OSX signal identifies PDGFRα+β− osteoprogenitor cells (OPCs) in the mp and endosteum lining the inner surface of cb (Figures 1D, 1E, S1C, and S1D). Differentiated bone cells in compact and trabecular bone are also PDGFRα+β− but lack OSX expression (Figures 1F, 1G, S1E, and S1F). Using a genetic reporter for OSX expression, Sp7-mCherry, in combination with Pdgfra-GFP, we confirm the presence of double-positive cells in trabecular and cb but also in epiphysis (Figures S1G and S1H). Although OSX is low or absent in vessel-associated PDGFRα+β+ BMSCs in the mp, those cells express RUNX2, a transcription factor marking immature osteoblast-lineage cells (Figures 1G and 1H). Larger arteries in the BM are covered by PDGFRα+β+ cells, which represent vascular smooth muscle cells (SMCs) and show expression of α-smooth muscle actin (α-SMA) and the proteoglycan Cspg4/NG2 (Figures S1I and S1J). In contrast, reticular cells associated with sinusoidal vessels in the BM are PDGFRα+β+ but lack appreciable NG2 and α-SMA expression and extend long matrix fibers containing collagen IIIA1 and IV (Figures S2A and S2B). Ultrastructural analysis confirms that mesenchymal stromal cells in BM are associated with thin reticular fibers, whereas thick collagen bundles are abundant in the mp (Figures 1I and 1J). The analysis of Ki67, a marker of proliferating cells, reveals further differences among PDGFRα+β+ cell populations, which show much higher Ki67 labeling in the mp than in the diaphysis (Figure S1K).
Figure 1.
Characterization and heterogeneity of BMSCs
(A and B) Tile-scan confocal images of 3-week-old Pdgfra-GFP (green) femur in a longitudinal section (A) and transverse section through diaphysis (B), immunostained for PDGFRβ (red) and the EC marker EMCN (blue). Epiphysis (epi), metaphysis (mp), growth plate (gp), cortical bone (cb), bone marrow (bm), central vein (cv; white arrowheads), and arteries (a; yellow arrowheads) are indicated. Dashed lines illustrate borders of growth plate and metaphysis. White arrowheads indicate Pdgfra-GFP+ PDGFRβ+ double-positive cells in metaphysis, bone marrow, growth plate, and epiphysis, whereas white arrows point at Pdgfra-GFP+ PDGFRβ− cells, including osteocytes in cortical and trabecular bone as well as chondrocytes in the growth plate and articular cartilage (top).
(C) Representative maximum intensity projections and optical z stack single plane (center) of metaphyseal region showing Pdgfra-GFP (green, arrow) expression in PDGFRβ (red) perivascular cells (arrowheads). (Right) Higher magnification of vessel with lumen (lu) lined by EMCN+ (blue) ECs.
(D) Confocal image showing Osterix+ (OSX, gray) osteoprogenitor cells together with Pdgfra-GFP (green) and PDGFRβ (red) staining in metaphysis. OSX+ and Pdgfra-GFP double-positive cells are marked by arrowheads, whereas cells with only Pdgfra-GFP+ signal are shown by arrows. ECs, EMCN+ (blue). High-magnification image on the right highlights Pdgfra-GFP+ PDGFRβ+ OSX+ triple-positive cells (yellow arrowhead).
(E) Quantification of Pdgfra-GFP+ PDGFRβ+ double-positive cells and Pdgfra-GFP+ PDGFRβ− cells in metaphysis. Pdgfra-GFP expression (mean intensity) is higher in single-positive cells compared with Pdgfra+β+ cells. (n = 5–8; data are presented as means ± SEM, p values, two-tailed unpaired t test).
(F and G) High-magnification confocal images of Pdgfra-GFP (green) femoral diaphysis and marrow immunostained for PDGFRβ (red) and EMCN (blue). In endosteum (eo) next to cortical bone (cb) and marrow, PDGFRβ+ cells co-express Pdgfra-GFP, whereas Pdgfra-GFP+ cells in periosteum lack PDGFRβ (F) but express OSX+ (red) (G).
(H) Representative image of perivascular (arrowheads) Pdgfra-GFP+ PDGFRβ+ BMSCs in metaphysis and BM co-expressing Runt-related transcription factor 2 (RUNX2) (red). ECs, EMCN (blue).
(I and J) Electron micrographs of 3-week-old wild-type metaphysis (I) and marrow (J). Arrows highlight ECs, perivascular MSCs (PVC), hematopoietic cells (HC), and osteoblasts (OB). Collagen bundle in metaphysis and thin fiber structures in BM (yellow arrowheads) are indicated in high-magnification images.
The data above are consistent with substantial heterogeneity among BMSCs and indicate fundamental differences between the diaphyseal (dpMSC) and metaphyseal (mpMSC) BMSC subpopulations. Furthermore, expression of Hairy/enhancer-of-split related with YRPW motif protein 1 (Hey1), a Notch-pathway component, which we detected with a Hey1-GFP reporter allele, prominently labels mpMSCs but is absent in dpMSCs (Figures 2A and S2C). To gain insight into the molecular signatures of PDGFRα+β+Hey1+ mpMSCs and PDGFRα+β+Hey1− dpMSCs, we used flow cytometry to sort those cell populations from 3-week-old Pdgfra-GFP+ or Hey1-GFP+ reporter mice (see Method details) (Figure S2D) for bulk RNA-seq. Principle component analysis (PCA) of the resulting RNA-seq data shows low variation within each sample group but major differences (PC1, 56%) between mpMSCs and dpMSCs (Figures 2B and S2E): 2,044 transcripts are upregulated and 1,686 downregulated in mpMSCs compared with dpMSCs (Figures 2C and S2F). Numerous BMSC markers and, as gene ontology analysis reveals, genes associated with vascular and skeletal development are enriched in mpMSCs relative to dpMSCs (Figures 2D, S2G, and S2H). When mpMSCs and dpMSCs are isolated and cultured in vitro, the former population has significantly greater potential to generate colonies and can readily form spheroid-like structures consisting of PDGFRα+β+ cells (Figures 2E, 2F, and S2I). Furthermore, mpMSCs can be efficiently differentiated to osteogenic, adipogenic, and chondrogenic lineage cells in vitro (Figures 2G–2J). These data indicate that mpMSCs have multipotent properties, whereas dpMSCs showed only limited growth in vitro under the same conditions, precluding ex vivo cell differentiation assays.
Figure 2.
Properties of metaphyseal BMSCs
(A) Hey1-GFP (green) expression in 3-week-old femur is predominantly found in metaphysis. GFP co-localizes with PDGFRβ (red, left) and PDGFRa (red, right) in the metaphysis (arrowheads) but also labels arterial ECs (arrow) in BM. ECs, EMCN+ (blue).
(B) Principal component analysis (PCA) of RNA sequencing data shows 56% variance (PC1) between sorted mpMSCs and dpMSCs and 17% variance (PC2) within sample groups.
(C) Differentially regulated genes in MA plots of mpMSCs relative to dpMSCs. The x axis represents mean normalized counts, and the y axis shows the log2-fold change between the two populations. Differentially regulated genes are represented by red-colored points (false-detection rate [FDR]-adjusted p < 0.01 and absolute log2-fold change < 1). Data points outside of the range of the y axis are represented as triangles.
(D) Heatmap of differentially expressed BMSC marker genes in mpMSCs and BM-derived dpMSCs.
(E) Quantification of colony-forming units (CFU) in mpMSCs compared with dpMSCs. (n = 6; data are presented as mean ± SEM, p values, two-tailed unpaired t test).
(F) Confocal image of spheroid formed by Pdgfra-GFP+ PDGFRβ+ mpMSCs after 15–20 days in culture. Inset marks spheroid shown at high magnification on the right.
(G and H) Osteogenic cells derived from mpMSCs in culture express the osteoblast lineage markers osteopontin (OPN, blue) and osteocalcin (OCN, red) (G). Upregulation of osteogenic markers in differentiated cells relative to mpMSCs (H) (n = 4–8; data are presented as mean ± SEM, p values, two-tailed unpaired t test).
(I) Adipogenic differentiation of mpMSCs in culture indicated by perilipin immunostaining (left) and significantly increased transcription of Adipoq, Cfd, and Pparg relative to control (n = 4; data are presented as mean ± SEM, p values, two-tailed unpaired t test).
(J) Chondrogenic differentiation of cultured mpMSCs indicated by Aggrecan immunostaining (left) and significantly increased transcription of Acan and Col11a2 relative to control (n = 4; data are presented as mean ± SEM, p values, two-tailed unpaired t test).
See also Figure S2
BMSC expansion and lineage-specific differentiation
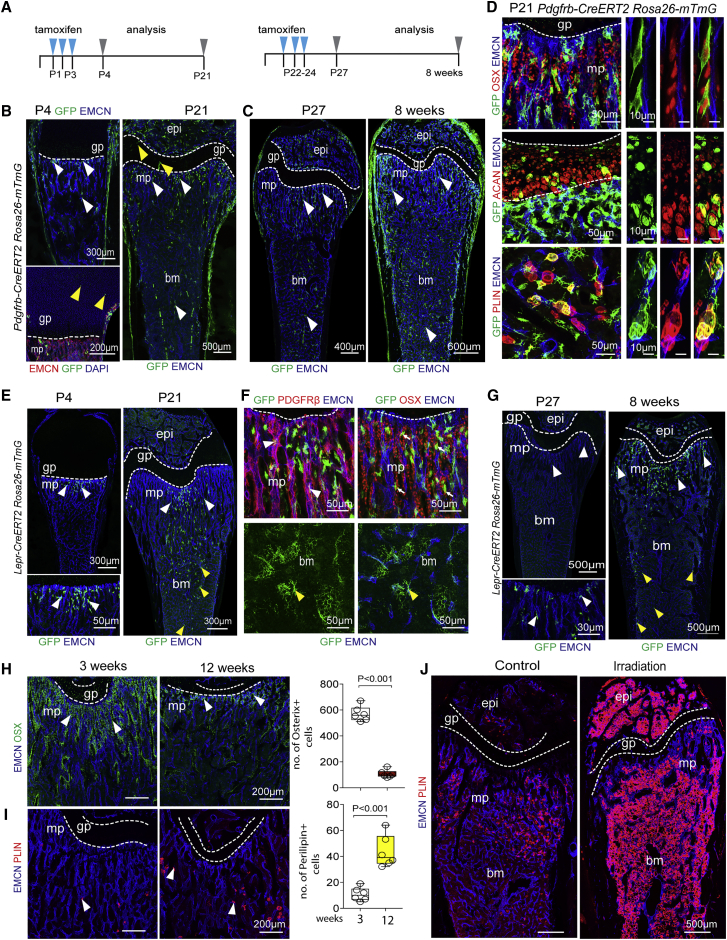
Next, we used genetic approaches to study BMSC expansion and lineage-specific differentiation in bone development in vivo. BMSC populations were genetically labeled with a range of tamoxifen-inducible Cre alleles, namely Pdgfrb-CreERT2 (Walker et al., 2019), Lepr-CreERT2 (see Method details), Gli1-CreERT2 (Ahn and Joyner, 2004), and Myh11-CreERT2 (Wirth et al., 2008), in combination with the Rosa26-mTmG Cre reporter (Muzumdar et al., 2007). For fate tracking analysis, pups received tamoxifen from P1–P3 and were analyzed at P4 and P21. In another series of experiments, tamoxifen was given at P22–P24, followed by analysis at P27 and 8 weeks (Figure 3A). At P4, Pdgfrb-CreERT2-labeled GFP+ cells represent predominantly mpMSCs in the mp, cells in the periosteum covering the outer bone surface, and isolated growth-plate chondrocytes (Figure 3B). These freshly labeled GFP+ cells show the expected PDGFRβ immunosignal but lack staining for OSX, the cartilage-specific proteoglycan core protein aggrecan or the adipocyte marker perilipin (Figure S3A). By P21, GFP+ cells have substantially expanded throughout the femur and include very different cell subpopulations, including OSX+ osteoprogenitors, aggrecan+ chondrocytes, perilipin+ adipocytes, and highly branched reticular cells in BM (Figures 3B, 3D, and S3B). Similarly, after tamoxifen administration to juvenile mice (P22–P24), femurs showed strong expansion of GFP+ cells in mp and BM at 8 weeks relative to P27, whereas chondroprogenitor cells in the growth plate were no longer labeled by Pdgfrb-CreERT2 (Figure 3C). Next, we generated tamoxifen-inducible Lepr-CreERT2 transgenic mice (see Method details) to target LepR+ cells, which have been previously associated with bone formation (Zhou et al., 2014) (Figure S3C). After tamoxifen administration from P1–P3, Lepr-CreERT2-labeled GFP+ cells represented predominantly mpMSCs in the mp at P4 but show substantial expansion into the BM and differentiation into OPCs and reticular cells by P21 (Figures 3E, 3F, and S3D). Likewise, the late tamoxifen administration regime (P22–P24) labels mpMSCs in the mp at P27, which is followed by expansion of GFP+ cells into the BM cavity in the following 5 weeks (Figure 3G). Like Pdgfrb-CreERT2 and Lepr-Cre-ERT2, Gli1-Cre-ERT2, which has been previously been used to mark osteogenic progenitors (Shi et al., 2017), targets mpMSCs at P21, which later differentiated into OSX+ OPCs or expanded into the BM cavity (Figures S3E and S3F). All three mouse lines tested above consistently indicated that mpMSCs from the mp included progenitors, which proliferate substantially and give rise to other cell types in bone and marrow. By contrast, genetic labeling and tracking of PDGFRα+β+ SMCs covering arteries with Myh11-CreERT2 did not result in expansion or fate changes of GFP+ cells, indicating that this cell population lacks appreciable progenitor capacity (Figure S3G). Thus, PDGFRα+β+ subpopulations have distinct properties and differentiation potential under physiological conditions. Vascular SMCs are firmly restricted to the coverage of arteries, whereas vessel-associated mpMSCs undergo substantial expansion and give rise to OPCs and reticular cells in BM. This developmental hierarchy is directly supported by genetic fate tracking with tamoxifen-inducible Pdgfrb-CreERT2, Lepr-CreERT2, and Gli1-CreERT2 mice. Constitutive Lepr-Cre (DeFalco et al., 2001) mice label both mpMSCs and dpMSCs in the postnatal femur (Figures S4A–S4E), which is, even in the absence of temporally controlled cell labeling, consistent with the emergence of dpMSCs from mpMSCs presented above.
Figure 3.
mpMSC expansion and differentiation in postnatal bone in vivo
(A) Scheme showing CreERT2-mediated recombination of R26-mT/mG Cre reporter for lineage tracing. Tamoxifen was administered at P1–P3 for analysis at P4 and P21 or at P22–P24 for analysis at P27 and 8 weeks, respectively.
(B and C) Tile scan confocal image of Pdgfrb-CreERT2 R26-mT/mG femurs. Mice received tamoxifen at P1–P3 or P22–P24 for analysis at P4 and P21 (B) or at P27 and 8 weeks (C), respectively. Yellow arrowheads mark GFP+ chondrocytes in growth plate, white arrowheads GFP+ cells in metaphysis, and bone marrow.
(D) Representative confocal images of 3-week-old Pdgfrb-CreERT2 R26-mT/mG femur showing differentiation of GFP+ (green) cells into OSX+ osteoprogenitors (top), ACAN+ chondrocytes (center), and PLIN+ adipocytes (bottom).
(E–G) Tile scan confocal image of tamoxifen-treated Lepr-CreERT2 R26-mT/mG mice analyzed at P4 and P21 (E). Higher magnification shows that GFP+ cells co-express the BMSC marker PDGFRβ (red, white arrowheads) and osterix (OSX, small arrows). Yellow arrowheads mark GFP+ cells in BM, which represent reticular cells; see bottom in (F) for higher magnification. Analysis of Lepr-CreERT2 R26-mT/mG lineage tracing at P27 and 8 weeks uncovers expansion of GFP+ cells from the metaphysis (white arrowheads) into BM (yellow arrowheads) (G).
(H and I) Representative confocal image of young (3 weeks) and adult (12 weeks) wild-type femur and graphs showing reduction of OSX+ osteoprogenitors (H) and increase of PLIN+ adipocytes (arrowheads) (I) with increased age (n = 6; data are presented as mean ± SEM, p values, two-tailed unpaired t test).
(J) Tile-scan confocal images showing strong increase of PLIN+ (red) adipocytes after irradiation.
See also Figures S3 and S4
In adult (12-week-old) postnatal long bone, the number of OSX+ osteoprogenitors is significantly reduced relative to juvenile (3-week-old) samples (Figure 3H) (Kusumbe et al., 2014). Another age-related change is the increased appearance of adipocytes in the adult transition zone, i.e., the region at the interface of mp and BM, compared with juvenile bone (Figure 3I). In response to gamma-irradiation (9 Gy), adult long bones become filled with Perilipin+ adipocytes both in mp and BM cavity (Figures 3J and S4F). Genetic lineage tracing indicates that these adipocytes are derived from Pdgfrb-CreERT2-labeled (GFP+) stromal cells or their progeny (Figure S4G).
Overall, these data show that PDGFRα+β+ mpMSCs can differentiate into multiple lineages, including dpMSCs in BM, during bone development. In the adult organism, mpMSC-derived BMSCs retain plasticity and can undergo adipogenic specification in response to stress.
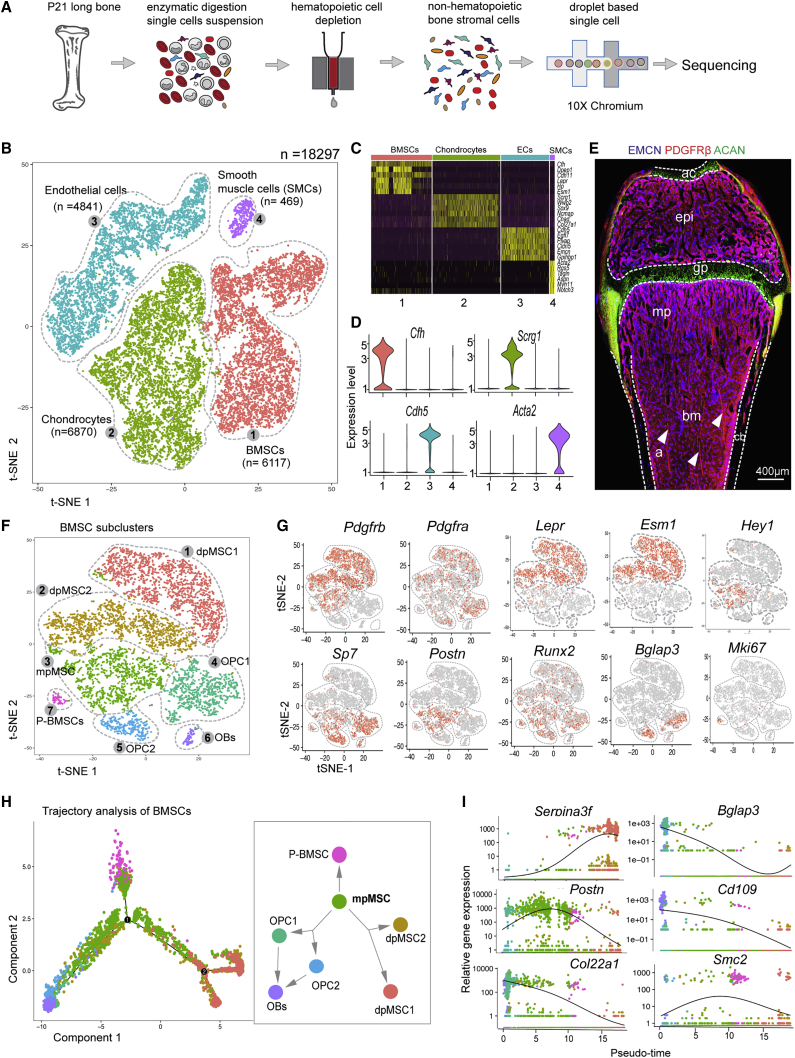
Transcriptional landscape and heterogeneity of BMSCs at single-cell resolution
To identify cellular heterogeneity and molecular signatures of non-hematopoietic cells in bone, we used microbeads containing lineage cocktail, CD45, and CD117 (c-Kit) antibodies (see Method details) to deplete hematopoietic stem/progenitor and lineage-committed cells and thereby enrich BMSCs from 3-week-old long bone (Figure 4A). Next, we performed droplet-based scRNA-seq using the 10× Genomics chromium system. We obtained a total of 18,297 cells by combining two experiments and, after trimming down doublets and residual hematopoietic cells, using bioinformatics (Figure S5A). Clustering and t-distributed stochastic neighbor-embedding (t-SNE) identified four cell clusters, as visualized in a color-coded t-SNE plot and cluster-frequency graph (Figures 4B and S5B). Cluster-specific marker genes were identified by analysis of pairwise differential gene expression of each cluster against all other clusters. The top six highly expressed genes for each cluster are displayed in a heatmap (Figure 4C), and specific markers for each individual cluster are shown in violin plots (Figure 4D). Cell types were identified based on the expression of a known marker combined with new marker genes, such as Pdgfrb and Cfh, encoding complement component factor H, for BMSCs. Chondrocytes express Acan (encoding aggrecan) and Scrg1 (scrapie responsive gene 1/stimulator of chondrogenesis 1). Endothelial cells express Cdh5 (encoding the cell adhesion molecule VE-cadherin) and Emcn (a mucin-like sialoglycoprotein), whereas transcripts for Pdgfrb, Myh11 (encoding SMC-specific myosin heavy chain), and Acta2 (α-SMA) are found in vascular smooth muscle cells (Figure S5C). Immunostaining of sections from 3-week-old femur for endomucin, PDGFRβ, and aggrecan followed by tile-scan confocal imaging reveals the anatomical location of these major cell populations (Figures 4E and S5D). Consistent with previous studies, our scRNA-seq approach confirmed that the expression of crucial niche factors for HSCs, namely Kitl (encoding stem cell factor; SCF) and Cxcl12 (SDF-1), is found in Lepr+ BMSCs, specifically dpMSCs (Figures S5E). Kitl transcripts are also highly enriched in Lepr+ dpMSCs/reticular cells, whereas Cxcl12 is more broadly expressed and also found in chondrocytes, SMCs, and some ECs.
Figure 4.
Transcriptional landscape and heterogeneity of BMSCs at single-cell resolution
(A) Schematic overview of bone sample preparation for scRNA-seq.
(B–D) t-SNE plot with color-coded subclusters. Cell numbers are indicated for each cluster (B). Heatmap of the top-six most-significantly expressed genes in all clusters (C) and violin plots of individual cluster markers (D).
(E) Tile-scan confocal image of 3-week-old femur stained for EMCN (blue) PDGFRβ+ (red) mark and ACAN (green, chondrocytes). Arrowheads indicate PDGFRβ+ smooth muscle cells covering arteries.
(F and G) t-SNE plot with color-coded BMSC subclusters (F). Expression of mesenchymal (Pdgfrb, Pdgfra, Lepr, and Esm1), osteogenic (Hey1, Sp7, Postn, Runx2, and Bglap3), and proliferative (Mki67) marker genes in BMSC subclusters (G).
(H and I) Monocle trajectory analysis of BMSC lineage differentiation. Clusters are colored according to their identity (H) and relative expression of key genes is displayed in pseudo-time (I).
See also Figure S5
Sub-clustering of BMSCs with Seurat (Butler et al., 2018) identified seven subpopulations with distinct gene expression and substantial molecular heterogeneity (Figures 4F and S5F): BMSC subclusters 1 and 2 represent Lepr+ Esm1+ diaphyseal BMSCs (dpMSC1 and dpMSC2), which are distinguished by expression of Serpina3f (serine protease inhibitor A3F) and Gdpd2 (glycerophosphoinositol inositolphosphodiesterase). Subcluster 3 defines metaphyseal mpMSCs characterized by transcription of the integrin ligand periostin (Postn) and the Notch pathway gene Hey1. BMSC sub-clusters 4 and 5 are osteoprogenitor cells (OPC1 and OPC2) expressing transcripts for the transcription factors OSX (Sp7) and Runx2, osteocalcin-related protein (Bglap3), and the alpha 1 chain of collagen XXII (Col22a1). Subcluster 6, expressing the surface protein CD109 and carbonic anhydrase 3 (Car3), represents differentiated osteoblasts (OBs). Proliferating BMSCs (P-BMSCs) in subcluster 7 express Ki-67 (Mki67) and DNA topoisomerase II alpha (Top2a) (Figures 4G and S5G–S5I). We confirmed the identity of selected clusters with a combination of reporter strains and immunostaining of bone sections. Expression of a Hey1-GFP reporter allele marks PDGFRβ+ mpMSCs and OSX+ cells in 3-week-old mp but also PDGFRβ+ vascular smooth muscle cells (Figures S5J–S5L). Labeling of cells by Lepr-Cre-controlled expression of tdTomato fluorescent protein in combination with ESM1 or PDGFRβ immunostaining shows that dpMSC1 and dpMSC2 are associated with the sinusoidal vasculature in BM, whereas tdTomato+ cells in the mp are largely devoid of ESM1 and PDGFRβ immunosignals (Figure S5M). Monocle trajectory analysis of BMSC clusters places mpMSCs in the center, with three direction paths toward P-BMSCs, dpMSC1 and dpMSC2, and OPC1 and OPC2 cells, respectively (Figure 4H). Relative gene expression in pseudo-time further argues for a close relationship between mpMSCs and osteoprogenitors/osteoblasts but also dpMSCs/reticular cells (Figure 4I). Together, these data indicate that the PDGFRβ+ mpMSC population in bone contains progenitors with the potential to generate major BMSC subsets.
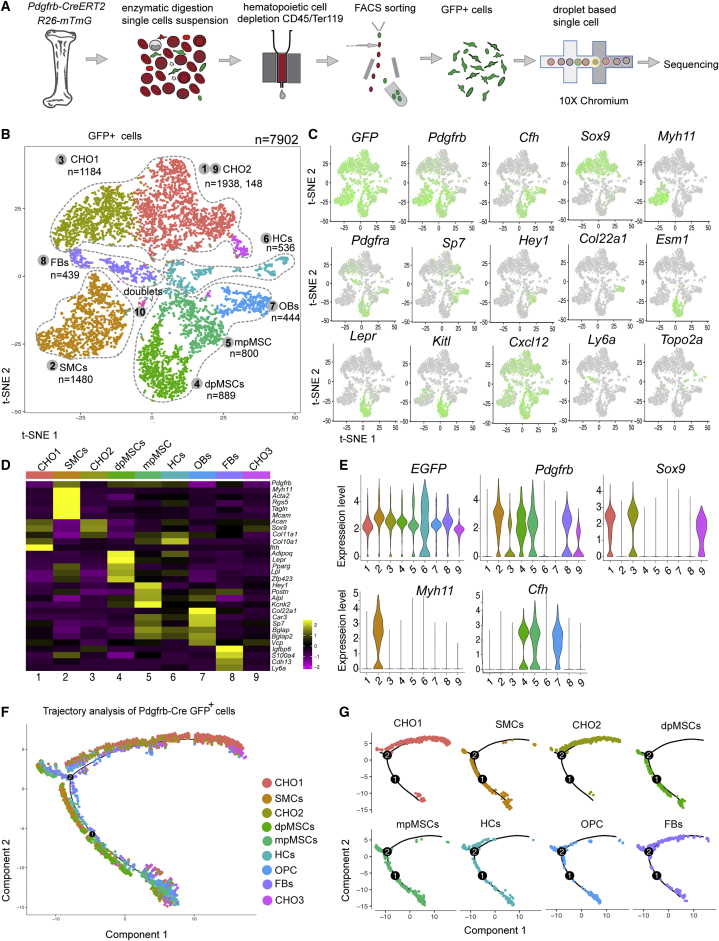
Single-cell analysis of PDGFRβ+ MSC lineage differentiation
To gain further insight into BMSC differentiation in vivo, Pdgfrb-CreERT2 Rosa26-mTmG (Pdgfrb-Cre R26-mT/mG) double-transgenic mice were treated with tamoxifen at P1–P3 and euthanized at P21 for the isolation of GFP+ cells from long bone, followed by scRNA-seq analysis (Figures 5A and S6A). This approach yielded 7,902 cells belonging to three major cell populations, namely BMSCs (clusters 4, 5, and 7), chondrocytes (clusters 1, 3, and 9), and SMCs (cluster 2), and two minor cell populations, namely fibroblasts (FBs, cluster 8, Ly6a+, and Cdh13+) and hematopoietic cells (HCs, cluster 9, Ctse+, and Tuba4a+) (Figures 5B and S6B). Pdgfrb-Cre R26-mT/mG-controlled expression of GFP transcripts was detectable in all five cell populations, which also expressed the expected cell type and subpopulation-specific marker genes (Figures 5C–5E, S6B, and S6C). In addition, our data contain a minor population of Cdh5+ ECs, which presumably represents a contamination because of cell doublets or fragments (Figure S6C).
Figure 5.
Single-cell analysis of PDGFRβ+ bone cells
(A) Experimental strategy for scRNA-seq analysis of Pdgfrb-CreERT2-labeled (GFP+) cells.
(B and C) t-SNE plot with color-coded cell clusters in scRNA-seq analysis of GFP+ cells. Clusters represent chondrocytes (CHO1, CHO2, and CHO3), smooth muscle cells (SMCs), mpMSCs and dpMSCs, a small population of GFP+ hematopoietic cells (HCs), osteoblastic cells (OBs), and fibroblasts (FBs) (B). Feature plots showing GFP transcripts in all clusters and expression of cell-type-specific marker genes (green) (C).
(D and E) Heatmap displaying expression of selected cell-type-specific mark genes (D). Violin plots showing expression of selected markers in cell subclusters (E).
(F and G) Monocle trajectory analysis of color-coded Pdgfrb-CreERT2-labeled clusters (F). Differentiation pathway of each cell cluster (G).
See also Figure S6
Trajectory analysis of Pdgfrb-Cre R26-mT/mG cells and relative gene expression in pseudo-time further support the genetic lineage tracing results and argue that Pdgfrb+ BMSCs can undergo osteogenic, chondrogenic, adipogenic, and smooth muscle cell differentiation (Figures 5F, 5G, and S6D). Similarly, culture of GFP+ BMSCs isolated from Pdgfrb-Cre R26-mT/mG bone confirms the results obtained with sorted mpMSCs (Figures 2F–2J) and supports PDGFRβ+ (GFP+) cells possessing osteogenic, adipogenic, and chondrogenic differentiation potential in vitro (Figures S6E and S6F).
Function of PDGFRβ+ BMSCs in bone formation
To determine the function of PDGFRβ+ BMSCs in bone formation, we ablated this cell population with a genetic strategy using a combination of Pdgfrb-CreERT2 and R26-DTA mice (Wu et al., 2006), enabling tamoxifen-inducible, Cre-controlled expression of diphtheria toxin (R26-DTAiΔMSC). Double-transgenic pups received tamoxifen at P6–P8 and were analyzed at P21 (Figure S7A). DTA-mediated ablation of PDGFRβ+ cells leads to decreased femur length (Figure S7B) and profound alterations in the mp. The region containing columnar CD31hi Emcnhi (type H) vessels, which has previously been shown to couple angiogenesis and osteogenesis (Kusumbe et al., 2014), as well as the number of OSX+ osteoprogenitors and deposition of the matrix protein osteopontin, are strongly reduced in R26-DTAiΔMSC femur compared with Cre-negative littermate control samples (Figures S7C and S7D). Microtomography (MicroCT) analysis unveils significant reduction of R26-DTAiΔMSC trabecular bone volume and cb thickness (Figures S7E and S7F).
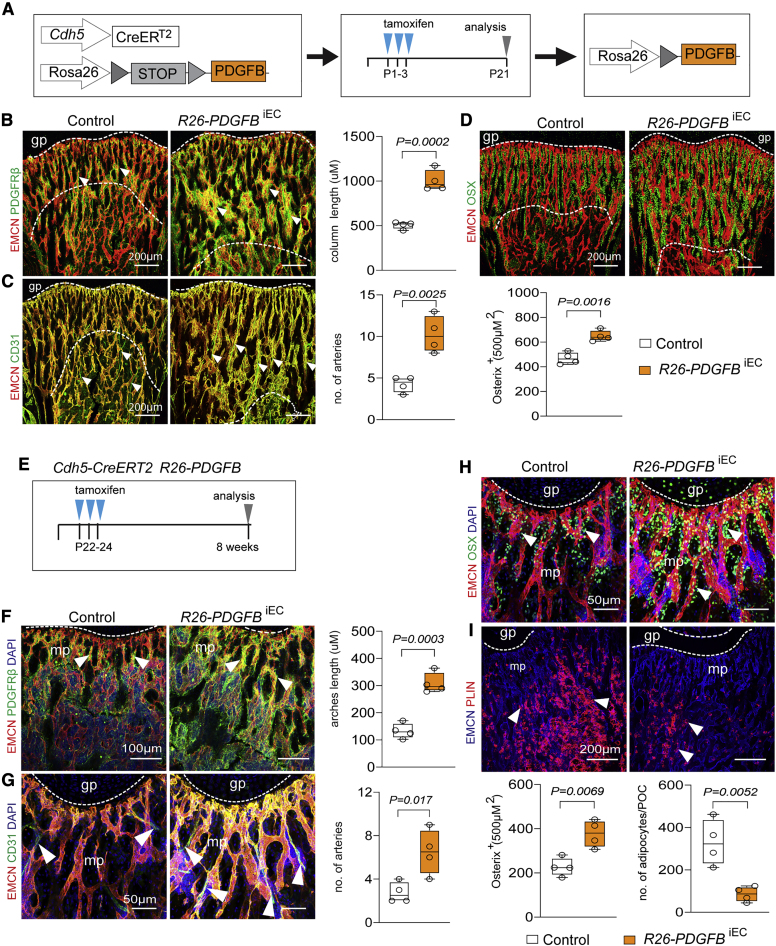
As previous reports have shown that PDGFRβ signaling promotes osteogenesis and triggers the activation of skeletal stem and progenitor cells during bone repair (Böhm et al., 2019; Xie et al., 2014), we used an inducible genetic approach in mice to overexpress human PDGF-B, specifically in ECs (see Method details; Figure 6A). In the resulting PDGF-B gain-of-function mice, the area of the femoral mp, the length of columnar type H vessels, the abundance of PDGFRβ+ mpMSCs and OSX+ osteoprogenitors, and the number of CD31+ EMCN− arteries are increased (Figures 6B–6D). Together, these results indicate that signaling through PDGF-B and PDGFRβ controls key aspects of both osteogenesis and angiogenesis in postnatal bone.
Figure 6.
Effect of endothelial-cell-derived PDGF-B on BMSC differentiation in bone
(A) Scheme of tamoxifen-inducible PDGFB overexpression in ECs with Cdh5-Cre-ERT2 mice.
(B) Representative confocal image of 3-week-old control and R26-PDGFBiEC femoral EMCN+ (red) blood vessels associated with PDGFRβ+ (green) BMSCs (arrowheads). Length of metaphyseal vessel columns (indicated by dashed lines) is increased in R26-PDGFBiEC bone (n = 4; data are presented as mean ± SEM, p values, two-tailed unpaired t test).
(C) Expansion of EMCNhigh (red) CD31high (green) vessels (indicated by dashed lines) and increase of arterioles (arrowheads) in R26-PDGFBiEC bone (n = 4; data are presented as mean ± SEM, p values, two-tailed unpaired t test).
(D) Confocal image and quantitation showing increase of OSX+ (green) cells in PDGFBiEC bone (n = 4; data are presented as mean ± SEM, p values, two-tailed unpaired t test).
(E) Tamoxifen-inducible overexpression of human PDGF-B (PDGFB) in ECs.
(F and G) Representative confocal images and quantitative analysis of 8-week-old R26-PDGFBiEC femur showing increase in PDGFRβ+ mpMSCs (F), arrowheads) and arterioles (G), arrowheads) relative to control (n = 4; data are presented as mean ± SEM, p values, two-tailed unpaired t test).
(H and I) Representative confocal images showing increased OSX+ cells (H) and decrease of PLIN+ adipocytes (arrowheads) (I) in 8-week-old R26-PDGFBiEC femur relative to control (n = 4; data are presented as mean ± SEM, p values, two-tailed unpaired t test).
See also Figures S7 and S8
Extrinsic and intrinsic signals control BMSC lineage-specific differentiation
Previous work has shown that CAR cells associated with BM sinusoidal vessels show adipocyte-like gene expression, whereas cells that are enriched for osteo-lineage-related transcripts preferentially localize to arterioles or non-vascular areas (Baccin et al., 2020; Baryawno et al., 2019; Zhong et al., 2020). In our single-cell analysis, dpMSCs also show pronounced expression of adipocyte markers under baseline conditions (Figure S7H). Genetic cell tracking and scRNA-seq results support BMSC specification being dynamically regulated, leading to adipogenic differentiation after irradiation (Figures S4F, S4G, S7G, S7H, and S8A). Cre-controlled overexpression of human PDGF-B in murine ECs in vivo (R26-PDGFBiEC), on the other hand, boosts the abundance of PDGFRβ+ mpMSCs and dpMSCs. The number of osteogenic (OSX+) cells is also increased, relative to littermate controls, whereas the number of perilipin+ adipocytes in is significantly reduced in adult R26-PDGFBiEC mice (Figures 6E–6I, S8B, and S8C). The changes in postnatal R26-PDGFBiEC gain-of-function mutants are accompanied by alterations in the long bone vasculature, namely, increases in the length of vessel columns in the mp and the number of arteries (Figures 6F and 6G). Taken together, these results show that PDGF-B boosts the number of BMSCs and promotes osteogenic differentiation, whereas adipocyte formation is inhibited.
The AP-1 family transcription factor Jun-B (encoded by Junb) is a key regulator of adipogenic and osteogenic differentiation (Rauch et al., 2019). Junb is highly expressed in mpMSCs (Figure 7A). Immunostaining of bone sections shows that Jun-B protein is predominantly found in the mp, where the transcription factor labels the nuclei of Pdgfra-GFP+, PDGFRβ+, and Hey1-GFP+ mpMSCs (Figures 7B, 7C, S8D, and S8E). To investigate the function of Jun-B in those cells, animals generated by interbreeding of mice carrying a loxP-flanked Junb allele (Hess et al., 2003) and the Pdgfrb-CreERT2 transgenic line were treated with tamoxifen from P1–P3 and analyzed at P21 (Figure S8F). Inactivation of Junb in PDGFRβ+ cells results in the profound reduction of the area covered by metaphyseal type H vessel columns and of PDGFRα+ and PDGFRβ+ cells relative to the control (Figure 7D). The number of OSX+ OPCs, type H vessel columns and arteries are reduced in Junb mutant bone, whereas perilipin+ adipocytes are increased, relative to control littermates (Figures 7E, 7F, and S8GS8I). Furthermore, long bone longitudinal growth and weight are also reduced in JunbiΔMSC-mutant bone compared with that of controls (Figures 7G, S8J, and S8K). These data indicate that EC-derived PDGF-B acts as a critical niche factor for BMSCs and promotes osteogenic differentiation of these cells. The AP-1 family transcription factor Jun-B controls BMSC fate and represses adipogenic differentiation during bone development.
Figure 7.
Function of Jun-B in PDGFRβ+ BMSCs in vivo
(A) Junb transcripts in mpMSCs relative to dpMSCs (n = 3–4; Fpkm, fragments per kilobase million; data are presented as mean ± SEM, p values, two-tailed unpaired t test).
(B and C) 3-week-old Pdgfra-GFP (green) femur immunostained for Jun-B (red) and PDGFRβ (blue) (B). Higher magnification confirms Jun-B expression in Pdgfrb-GFP+ PDGFRβ+ mpMSCs (arrowheads) (C).
(D) Maximum-intensity projection shows the reduction of PDGFRβ+ (top) and PDGFRα+ (bottom) cells (arrowheads) associated with metaphyseal vessel columns (indicated by dashed lines) near the growth plate in 3-week-old JunbiΔMSC bone.
(E) Decrease of OSX+ (green, arrowhead) cells in 3-week-old JunbiΔMSC metaphysis (n = 5; data are presented as mean ± SEM, p values, two-tailed unpaired t test).
(F) Representative confocal images and quantitation showing increase of PLIN+ adipocytes in JunbiΔMSC epiphysis and metaphysis relative to control (n = 5; data are presented as mean ± SEM, p values, two-tailed unpaired t test).
(G) Image of control and JunbiΔMSC mutant femurs. Femur length is significantly reduced after inactivation of Junb. (n = 6; data are presented as mean ± SEM, p values, two-tailed unpaired t test).
See also Figures S7 and S8
Discussion
There is increasing evidence that BMSCs represent a heterogenous mixture of different cell populations with distinct molecular and functional properties (Baccin et al., 2020; Baryawno et al., 2019; Tikhonova et al., 2019). This issue is further complicated by the plasticity of BMSCs, which can enable the acquisition of different terminal fates depending on external signals and stress factors (Ambrosi et al., 2017; Cook and Genever, 2013; Tencerova and Kassem, 2016). Given these complications, it has remained unclear whether BMSC subsets are derived from several different or a single (common) stem/progenitor population. Our genetic fate mapping approach with tamoxifen-inducible Pdgfrb-CreERT2, Lepr-CreERT2, and Gli1-CreERT2 lines argues that the mp harbors multipotent cells, which provide several different stromal cell populations during development. Shortly after tamoxifen administration, recombined BMSCs are predominantly confined to the mp and represent vessel-associated PDGFRα+β+ mpMSCs. Furthermore, we provide evidence that proliferation, colony-forming capacity, and multipotency in vivo and ex vivo are primarily associated with PDGFRα+β+ Hey1+ mpMSCs. This is also supported by the trajectory analysis of our scRNA-seq data, which shows that proliferating BMSCs emerge from mpMSCs but not from dpMSCs/reticular cells in postnatal BM. Moreover, trajectory analysis and genetic fate tracking indicate that mpMSCs have the capacity to generate osteoblast lineage cells as well as dpMSCs.
Recently, scRNA-seq approaches were used to investigate the composition of adult BM stroma (Baccin et al., 2020; Baryawno et al., 2019; Tikhonova et al., 2019; Zhong et al., 2020). Given their critical role in hematopoietic stem cell maintenance and function, vessel-associated reticular (CAR) cells in the BM were a focus of these studies. LepR+ CAR cells express high levels of the chemokine CXCL12 and stem cell factor (SCF), which are both important regulators of hematopoietic stem and progenitor cells (HSPCs). In addition, LepR+ BMSCs contain transcripts for adipogenic genes, which is consistent with their ability to give rise to adipocytes in response to myeloablative treatment and other challenges (Baryawno et al., 2019; Tikhonova et al., 2019). This population of LepR+ BMSCs might be similar to adipo-CAR cells identified by Baccin et al. (2020) and adipogenic lineage precursors (MALPs) reported by Zhong et al. (2020). Based on the expression of Lepr, HSPC niche factors and adipogenic genes, these cells also appear to be equivalent to the dpMSCs described in our study. We, therefore, propose that dpMSCs/reticular cells are derived from mpMSCs under physiological conditions. It is feasible, however, that mpMSCs and dpMSCs retain plasticity and might be interconvertible in response to tissue damage, during regeneration or in the context of aging and disease. It also needs to be noted that clonal analysis by in vivo transplantation would give more-direct and -conclusive evidence for the multipotent nature of mpMSCs and their differentiation potential.
Several previous publications have made use of genetic approaches and fluorescence-activated cell sorting (FACS) isolation strategies to study the nature and spatial distribution of MSPC/SSCs. This has, for example, established that lineage-restricted stem and progenitor cells give rise to bone, cartilage, and stromal tissue in a clonal fashion preferentially around the femoral growth plate but do not generate hematopoietic, adipose, or muscle cells under normal conditions (Chan et al., 2015). Arguing further for the enrichment of MSPC/SSCs in the mp, similar to what we have observed in our study, cells labeled with a tamoxifen-inducible Col2–CreER (Col2a1-CreERT2) allele continued to generate osteoblasts and stromal cells in the mp and epiphysis in addition to chondrocytes in the growth plate and articular cartilage for more than a year. Because the same approach did not mark BMSCs in the middle of the diaphysis, it was proposed that those cells might be derived from a separate population of LepR+ precursor cells (Nakamura et al., 2006; Zhou et al., 2014). That notion is not, however, fully consistent with our new data indicating that Lepr-CreERT2-labeled cells emerge initially in the mp before they populate the postnatal BM. Adding further complexity, several studies have also identified the periosteum as a source of SSCs with high bone regenerative capacity (Debnath et al., 2018; Duchamp de Lageneste et al., 2018). Thus, future studies will have to compare the molecular profiles and functional properties of putative MSPC/SSCs from different bone regions, including the mp, diaphysis, and periosteum.
In addition to insight on the localization and maintenance of MSPC/SSCs, it is also important to understand the regulators controlling the differentiation and lineage specification of those cells. The analysis of global knockout mice has previously established that the transcription factor Jun-B is necessary for stromal cell proliferation and differentiation into bone-forming cells (Hess et al., 2003; Kenner et al., 2004). Indeed, we also found that cell-type-specific JunbiΔMSC mutants, generated with the Pdgfrb-CreERT2 line, have strongly impaired bone growth and a significant reduction of OSX+ OPCs, whereas the number of perilipin+ adipocytes is increased. These results indicate that Jun-B controls the behavior of PDGFRβ+ BMSCs, presumably mpMSCs, which indirectly also affects the bone vasculature. These results highlight the cell-autonomous function of Jun-B in BMSCs but also the extensive crosstalk between vessel-associated cells and the adjacent endothelium. Blood vessels are known to be an important source of PDGF-B, which promotes the proliferation and recruitment of PDGFRβ+ pericytes and vascular smooth muscle cells (Hellström et al., 1999). In bone, PDGFRβ controls the behavior of skeletal stem and progenitor cells during regeneration (Böhm et al., 2019; Doherty et al., 2019). In agreement with these results, we show that overexpression of PDGF-B enhances the number of OSX+ OPCs, suppresses the emergence perilipin+ adipocytes, and promotes type-H vessel and artery formation in the bone vasculature. Together, these data establish that BMSC fate is controlled by a combination of cell-autonomous and extrinsic signals, with implications for vascular growth and, thereby, the communication between different cell types in bone. Given the great relevance of BMSCs for biotechnology and the development or therapeutic approaches aiming at improved fracture healing, future work might well gain important insights from our analysis of postnatal bone BMSCs. Some findings might also be relevant in the context of osteoporosis and other diseases characterized by impaired bone formation (Aghebati-Maleki et al., 2019; Hu et al., 2018).
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rat monoclonal anti-Endomucin (V.7C7) | Santa Cruz | Cat# sc-65495; RRID: AB_2100037 |
| Rabbit polyclonal anti-Acan | Millipore | Cat#AB1031; RRID: AB_90460 |
| Goat anti-Pdgfrb | R & D | Cat# AF1042; RRID: AB_2162633 |
| Rabbit polyclonal anti-Osterix/sp7 | Abcamabcam | Cat# ab22552 RRID: AB_2194492 |
| Rabbit monoclonal anti-Runx2 | Abcam | Cat# ab22552; RRID: AB_2713945 |
| Mouse monoclonal anti-alpha-smooth muscle actin (Cy3 - conjugated) | Sigma-Aldrich | Cat#C6198; RRID: AB_476856 |
| Rabbit polyclonal anti-Ki67 | Abcam | Cat#ab15580; RRID: AB_443209 |
| Chicken polyclonal anti-GFP | Abcam | Cat#ab13970; RRID: AB_300798 |
| Goat anti-Pdgfra | R&D | Cat#AF1062; RRID: AB_2236897 |
| Goat polyclonal anti-CD31 | R&D | Cat#AF3628; RRID: AB_2161028 |
| Rabbit monoclonal anti-Perilipin | Cell signaling | Cat#9349; RRID: AB_10829911 |
| Goat polyclonal anti-Esm1 | R&D | Cat# AF1810; RRID: AB_2246741 |
| Goat polyclonal anti-Osteopontin | R&D | Cat# AF808; RRID: AB_2194992 |
| Rabbit polyclonal anti-Osteocalcin | Lifespan Bioscience | Cat#LS-C17044; RRID: AB_798874 |
| Rabbit polyclonal anti-Collagen IV | AbD Serotec | Cat# 2150-1470; RRID: AB_2082660 |
| Rabbit polyclonal anti-Collagen IIIA1 | Abcam | Cat# ab7778; RRID: AB_306066 |
| Rabbit monoclonal anti-JunB | Cell signaling | Cat#3753; RRID: AB_2130002 |
| Anti rat Alexa Fluor 488 | ThermoFisher Scientific | Cat#A21208 RRID: AB_141709 |
| Anti rat Alexa Fluor 594 | ThermoFisher Scientific | Cat#A21209 RRID: AB_2535795 |
| Anti goat Alexa Fluor 546 | ThermoFisher Scientific | Cat#A11056 RRID: AB_142628 |
| Anti goat Alexa Fluor 647 | ThermoFisher Scientific | Cat#A21447 RRID: AB_141844 |
| Anti rabbit Alexa Fluor 647 | ThermoFisher Scientific | Cat#A31573 RRID: AB_2536183 |
| Anti rabbit Alexa Fluor 488 | ThermoFisher Scientific | Cat#A21206 RRID: AB_141708 |
| Anti chicken Alexa Fluor 488 | Jackson Laboratories | Cat#703-545-155; RRID: AB_2340375 |
| Chemicals, peptides, and recombinant proteins | ||
| Tamoxifen | Sigma | Cat#T5648 |
| 4’,6’-diamidino-2-phenylindole (DAPI) | Sigma | Cat#D9542 |
| Kolliphor EL | Sigma | Cat#C5135 |
| Critical commercial assays | ||
| lineage cell depletion kit | MACS | Cat#130-090-858 |
| CD45 MicroBeads | MACS | Cat#130-052-301 |
| CD117 MicroBeads | MACS | Cat#130-091-224 |
| RNeasy Plus Mini Kit | QIAGEN | Cat# 74134 |
| iScript cDNA Synthesis kit | BIO-RAD | Cat# 1708890 |
| TaqMan Gene Expression Master Mix | ThermoFisher Scientific | Cat#4369016 |
| PowerUp SYBR Green Master Mix | ThermoFisher Scientific | Cat#A25742 |
| Quantification-RNA Qubit Fluorometric quantitation | ThermoFisher Scientific | Cat#Q32851 |
| High sensitive DNA kit | Agilent Bioanalyzer | Cat#5067-4626 |
| Chromium Single cell 3′ Reagent Kit v2 | 10X genomics | Cat#PN-120237 |
| NextSeq 500/550 High Output Kit V2 (150 cycles) | Illumina | Cat#20024907 |
| NextSeq 500/550 Mid Output Kit V2 | llumina | Cat#20024904 |
| Deposited data | ||
| Bulk-RNA seq data mpMSC and dpMSC | This paper | GSE156636:GSE156634 |
| single cell-RNA seq data BMSCs | This paper | GSE156636:GSE156635 |
| single cell-RNA seq data -Pdgfrb-Cre GFP+ cells | This paper | GSE156636:GSE156635 |
| Experimental models: Cell lines | ||
| Primary mouse bone MSC | This paper | N/A |
| Experimental models: Organisms/strains | ||
| C57/BL6 | Janvier Labs | N/A |
| Gt(Rosa26) ACTB-tdTomato-EGFP | The Jackson Laboratory | JAX: 007676 |
| Pdgfra-H2B-GFP | The Jackson Laboratory | JAX:007669 |
| Hey1GFP | Gensat | |
| Cspg4-DsRed | The Jackson Laboratory | JAX:008241 |
| Sp7 (Osterix)-mCherry | The Jackson Laboratory | JAX:024850 |
| Pdgfrb(BAC) CreERT2 | Laboratory of Ralf H.Adams | (Kato et al., 2018) |
| Myh11(BAC)Cre ERT2 | The Jackson Laboratory | JAX:019079 |
| Rosa26-hPDGFB-GOF | Laboratory of Christer Betsholtz | (Armulik et al., 2010) |
| Cdh5(PAC) Cre-ERT2 | Laboratory of Ralf H.Adams | (Sivaraj et al., 2020) |
| Leprtm2(cre)Rck/J | The Jackson Laboratory | JAX:008320 |
| Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J | The Jackson Laboratory | JAX:007908 |
| Lepr(BAC) Cre-ERT2 | This paper | |
| Rosa26-DTA | The Jackson Laboratory | JAX:010527 |
| Gli1-CreERT2 | The Jackson Laboratory | JAX:007913 |
| Junbflox/flox | The Jackson Laboratory | JAX:012369 |
| Oligonucleotides | ||
| Eukaryotic 18S rRNA | ThermoFisher Scientific | Cat#4319413E |
| Pdgfrb (Mm00435546_m1) | ThermoFisher Scientific | Cat#4331182 |
| Pdgfra (Mm00440701_m1) | ThermoFisher Scientific | Cat# 4331182 |
| Sp7 (Mm04209856_m1) | ThermoFisher Scientific | Cat# 4331182 |
| Alpl (Mm00475834_m1) | ThermoFisher Scientific | Cat# 4331182 |
| Spp1(Mm00436767_m1) | ThermoFisher Scientific | Cat# 4331182 |
| Bglap (Mm03413826_m1) | ThermoFisher Scientific | Cat#4331182 |
| Runx2 (Mm00501584_m1) | ThermoFisher Scientific | Cat# 4331182 |
| Adipoq (Mm00456425_m1) | ThermoFisher Scientific | Cat# 4331182 |
| Cfd (Mm01143935_m1) | ThermoFisher Scientific | Cat#4331182 |
| Pparg (Mm00440940_m1) | ThermoFisher Scientific | Cat# 4331182 |
| Acan (Mm00545794_m1) | ThermoFisher Scientific | Cat#4331182 |
| Col11a2 (Mm00487046_m1) | ThermoFisher Scientific | Cat# 4331182 |
| Software and algorithms | ||
| Volocity 6.3 (image analysis) | PerkinElmer | N/A |
| FACS software, FlowJo | FLOWJO, LLC | N/A |
| GraphPad Prism 7 | GraphPad Software, inc | N/A |
| Adobe illustrator | Adobe | N/A |
| Adobe Photoshop | Adobe | N/A |
Resource availability
Lead contact
The lead contact, Ralf H. Adams (ralf.adams@mpi-muenster.mpg.de), should be contacted for requests regarding resources, data, and reagents used in the study.
Materials availability
Mice generated in this study can be requested from the lead contact.
Data and code availability
The NGS datasets (bulk-RNA and single cell -RNA sequencing) used in this article are available at GEO (https://www.ncbi.nlm.nih.gov/geo/) with the accession number GEO: GSE156636.
Experimental models and subject details
Animal models
C57BL/6J male mice were used for all analysis of wild-type bone. Pdgfra-GFP heterozygous knock-in, Sp7-mcherry and Cspg4-DsRed reporter mice were used for MSC characterization. Pdgfra-GFP and Sp7-mcherry mice were crossed to generate double transgenic reporter mice. Hey1-GFP (Tg(Hey1-EGFP)ID40Gsat) (http://www.gensat.org) heterozygotes were used for analysis of mpMSCs from bone. For the characterization of MSC differentiation and lineage tracing, Pdgfrb-CreERT2 (Chen et al., 2016), Myh11-CreERT2 (Wirth et al., 2008), Gli1-CreERT2 (Ahn and Joyner, 2004), Lepr-Cre (DeFalco et al., 2001) and Lepr-CreERT2 (see below) transgenic mice were interbred with Gt(Rosa26) ACTB-tdTomato-EGFP reporter mice (Muzumdar et al., 2007) or Gt(Rosa26)–dTomato (Madisen et al., 2010) reporter mice, respectively. Double transgenic offspring (R26-DTAiΔMSC) generated by interbreeding of Pdgfrb-CreERT2 and R26-iDTA (Wu et al., 2006) mice was used for tamoxifen-inducible cell ablation. To overexpress PDGF-B in endothelial cells, we interbred R26-hPDGFB (Armulik et al., 2010) and Cdh5-CreERT2 mice (Sivaraj et al., 2020). Junb floxed mice (Kenner et al., 2004) were bred to Pdgfrb-CreERT2 transgenic animals for conditional inactivation of Junb in MSCs. Cre positive (JunbiΔMSC or R26-DTAiΔMSC) mutants were compared to Cre negative control littermates.
For the generation of Lepr-CreERT2 transgenic mice, a cDNA encoding tamoxifen-inducible Cre recombinase (CreERT2) followed by a polyadenylation signal sequence and an FRT-flanked ampicillin resistance cassette was introduced by recombineering into the start codon of Lepr in the BAC clone RP23-135C17 (BACPAC Resources Center, Children’s Hospital Oakland Research Institute, Oakland, USA). After Flp-mediated excision of the ampicillin resistance cassette in bacteria, the resulting constructs were validated by PCR analysis and used for pronuclear injection into fertilized mouse oocytes. Founders, identified by PCR genotyping, were bred to Cre-mediated reporter transgenic Rosa26-mT/mG mice.
For inducible Cre-mediated recombination, pups received daily intraperitoneal injections (IP) of 50 μg or 100 μg of tamoxifen from postnatal day 1 (P1) to P3 or from P22 to P24. Tamoxifen stocks were prepared by dissolving 20mg in 500 μL of ethanol and vortex for 10mins before an equal volume of Kolliphor EL was added. 1mg aliquots were stored at −20°C and dissolved in the required volume of PBS prior to injection (Sivaraj et al., 2020).
Age or developmental stage of the mice used are indicated in the text and figures. All husbandry and experiments involving animals were performed in compliance with the relevant laws and institutional guidelines, following protocols previously approved by local animal ethics committees and conducted with permission granted by the Landesamt für Natur, Umwelt und Verbraucherschutz (LANUV) of North Rhine-Westfalia to the Max Planck Institute of Molecular Biomedicine. Our research fully complies with national and international standards for animal husbandry, welfare, and monitoring as well as the 3Rs (replacement, reduction, and refinement).
BMSC isolation and culture
Femur and tibia were harvested from 3-week-old mice, surrounding muscle tissue was cleaned off and the epiphysis removed. The metaphysis was separated with a scalpel to obtain metaphyseal BMSCs (mpMSCs). Next, bone marrow was gently removed from the bone shaft into suspension medium (1% FBS, 1mM EDTA in PBS without Ca2+/Mg2+) to obtain BM-derived BMSCs (dpMSCs). Metaphyseal fragments were cut into small pieces and crushed using mortar and pestle. Crushed samples were digested using 1 mL of Collagenase type II and I enzymes (2mg/ml) for 20mins at 37°C under gentle agitation. Digested metaphysis and BM samples were transferred through 70 μm strainers into 50ml tubes to obtain single–cell suspensions, which were resuspended in suspension medium, centrifuged at 300 g for 5mins, and washed 2 to 3 times. Cell pellets were resuspended in the respective volume of suspension medium. Next, we used lineage cell depletion kit (Milteyi Biotec, cat#130-090-858) to deplete hematopoietic cells. BMSCs were isolated from Pdgfra-GFP or Hey1-GFP reporter mice by using FACS. Sorted cells were collected in MesenCult MSC medium (STEMCELL™ Technology. Cat#05501,05502). BMSCs were cultured in MesenCult BMSC medium, maintained with 5% CO2 in a water-jacketed incubator at 37°C. Half of the medium was changed weekly.
Method details
Bone sample preparation and immunofluorescence staining
Mice were euthanized and long bones (femur and tibia) were harvested and fixed immediately in ice-cold 2% paraformaldehyde (PFA) for 6 to 8 hours under gentle agitation. Bones were decalcified in 0.5M EDTA for 16 to 24 hours (for samples from 3-week-old animals) or 2-3 days (adult samples) at 4°C under agitation, which was followed by overnight incubation in sucrose solution (20% sucrose, 2%PVP) and immersion in bone embedding (8% gelatine, 20% sucrose, 2% PVP). Samples were stored overnight at −80°C. 60-100 μm-thick cryosections were prepared for immunofluorescence staining (Kusumbe et al., 2015).
Bone immunostaining was done as described previously (Kusumbe et al., 2015; Sivaraj et al., 2020). Bone sections were washed in ice-cold PBS and permeabilized with ice-cold 0.3% Triton X-100 in PBS for 10 mins at room temperature (RT). Samples were incubated in blocking solution (5% heat-inactivated donkey serum in 0.3% Triton X-100) for 30mins at RT. Primary antibodies (see Key resources table) were diluted in 5% donkey serum and PBS and incubated overnight at 4°C. Next, slides were washed 3 to 5 times in PBS in 5-10min intervals. Species-specific Alexa Fluor secondary antibodies (ThermoFisher) diluted in PBS were added and incubated for 2-3 hours at RT.
Electron microscopy (EM)
Femurs of 3 weeks old mouse were removed and cut directly longitudinally into half in fixative, containing 2% glutaraldehyde, 2% paraformaldehyde, 20mM CaCl2, 20mM MgCl2 in 0,1M cacodylatbuffer, pH 7.4. The samples were fixed for 3 hours at room temperature. For post-fixation, specimen was incubated in 1% OsmO4, containing 1.5% KFeCN in 0.1 M cacodylate buffer, pH 7.4, followed by stepwise dehydration, uranyl-en-bloc staining and epon embedding. Ultrathin sections were cut from the area of interest and brought on formvar-filmed single-slot grids. Sections were counterstained with lead citrate and analyzed at a transmission electron microscope at 80kV (Tecnai-12-biotwin, Thermo-Fisher, Eindhoven, Netherlands). Representative images were taken with a 2K-CCD camera (Veleta, EMSIS, Muenster, Germany).
MSC CFU-F and differentiation assay
For colony formation assays, mpMSCs or dpMSCs were freshly isolated from Hey1-GFP or Pdgfra-GFP reporter mice. Single cell suspensions (1000 cells per cm2) were plated and incubated at 37°C under 5% hypoxia conditions for 14 days with MesenCult MSC medium. Half of the medium was changed after a week. 14 days later, cells were fixed, stained, and number of colonies counted.
To analyze osteogenic, adipogenic, and chondrogenic differentiation, mpMSCs were cultured in 6 well plates with MesenCult medium. At 50 – 70% confluency the culture medium was replaced with differentiation medium (R&D system, base medium cat#CCM007, osteogenic supplement cat#CCM009 and adipogenic cat#CCM011; Chondrogenic differentiation medium cat#CCM005 and supplement cat#CCM006). After 2 – 3 days the medium was replaced once and after 15 days cells were analyzed for differentiation by immunostaining and quantitative PCR.
Quantitative PCR (qPCR)
Total RNA was isolated from BMSCs or BMSC-derived cells using RNA Plus mini kit (QIAGEN, Cat#74134) according to the manufacturer’s instructions. RNA was reverse-transcribed using the iScript cDNA synthesis kit (Bio-Rad, Cat#1708890). Quantitative PCR was carried out using gene specific Taqman probes (see Key resources table) using C1000 Touch Thermal cycler (BIORAD).
Fluorescence activated cell sorting (FACS)
Single cell suspensions were prepared from 3-weeks-old Hey1-GFP, Pdgfra-H2B-GFP, PdgfrbCre Rosa26mT/mG reporter mice femur and tibia as described above. DAPI was added and the incubation continued for 10 mins on ice. Next, samples were washed 2-3 times with blocking solution and resuspended using ice-cold PBS. Cell sorting was performed on a FACS Aria II cell sorter (BD Bioscience). Dead cells and debris were excluded by FSC, SSC and DAPI positive signal. Sorted GFP were collected in blocking buffer for scRNA-seq or RTL buffer for bulk RNA-seq or MSC cell culture medium.
Bulk-RNA sequencing and data analysis
Metaphyseal and diaphyseal BMSCs were sorted from three weeks old Hey1-GFP metaphysis and Pdgfra-EGFP BM using a flow cytometer. RNA was isolated using the RNeasy Plus Micro Kit (QIAGEN) according to the manufacturer’s instructions. RNA quality was checked using a 2100 BioAnalyzer (Agilent). 10ng of RNA were used for preparation of sequencing libraries with SMARTer Ultra Low input RNA Kit-V3 (Clontech laboratories) and followed by Nextera XT DANN Library Prep Kit (Illumina) according to the manufacturer’s instructions. Libraries were validated using a BioAnalyzer, quantified by qPCR and Qubit Fluorometric Quantitation (Thermo Fisher). Libraries were diluted to a final concentration of 1.6pM for sequencing. The NextSeq 500/550 Mid Output Kit v2.5 (Illumina) was used for sequencing with a NextSeq 500 (Illumina).
Bulk-RNA sequencing data analysis was performed as described previously (Sivaraj et al., 2020) with some modifications. The quality assessment of raw sequence data was performed using FastQC (Version: FastQC 0.11.3). Paired-end sequence reads were mapped to the mm10 mouse genome assembly (GRCm38) using TopHat-2 (Version: tophat-2.0.13). The mouse genome was downloaded from the iGenome portal. HTSeq was used to count the aligned reads on a per gene basis (Version: HTSeq-0.6.1).
The count data were normalized using the Variance Stabilizing Transformation (VST) function from the DESeq2 package. Principal Component Analysis (PCA) was performed on transformed read counts using the variable genes to assess the overall similarity between the samples. Differential gene expression analysis between control and mutants were performed using DESeq2. Differentially expressed genes were selected using an FRD-adjusted p value cut-off < 0.01 and an absolute log2 fold change > 0.5. Gene symbols were annotated using biomaRt (BioConductor version 3.1). Gene ontology analysis and cellular signaling pathways were performed with PANTHER online tool (http://www.pantherdb.org/).
Bone marrow stromal cell preparation for single cell RNA sequencing
3-week-old (n = 2) male mice were used for non-hematopoietic single cell analysis. Femur and tibia were harvested, cleaned from attached surrounding tissue and collected in ice cold sterile PBS (without Ca2+/Mg2+). Next, bones were cut into small pieces and crushed using mortar and pestle. Samples were digested using 1 mL of collagenase type II and I enzymes (2mg/ml) for 20mins at 37°C under gentle agitation. Digested samples were transferred to 70 μm strainers in 50ml tubes to obtain a single–cell suspension, which was resuspended in blocking solution (1% BSA, 1mM EDTA in PBS without Ca2+/Mg2+), centrifuged at 300 g for 5mins, washed 2 to 3 times with ice-cold blocking solution, and filtered through 50 μm strainers. Pellets were resuspended in the equivalent volume of blocking solution. To enrich bone marrow stromal cells, hematopoietic cells were removed using a lineage cell depletion kit (MACS, cat#130-090-858) following the manufacturer’s instructions. Next, lineage negative cells were depleted by CD45 and CD117 using microbeads (MACS, cat. no. 130-052-301 and 130-091-224) from lineage negative (lin-) bone cells. Bone marrow stromal cells were examined by microscopy and resuspended at 106 cells/ml in 0.05% BSA in PBS.
For Pdgfrb-CreERT2 Rosa26-mT/mG reporter mice (n = 4), bone single cell suspension was prepared and lineage-positive cells were depleted as described above. GFP positive cells were sorted by FACS Aria II (BD Bioscience), collected in blocking buffer and used for scRNA sequencing.
Bone marrow stromal single cells or Pdgfrb-CreERT2 GFP+ sorted cells were prepared for microfluidic droplets-based scRNA-seq. Single cells were encapsulated into emulsion droplets using Chromium controller (10X Genomics). The scRNA seq libraries were prepared using the chromium single cells 3′ regent kit (V2) (10X genomics, cat#PN-120237) according to the manufacturer’s protocol. scRNA-seq libraries were evaluated and quantified by Agilent Bioanalyzer using High sensitivity DNA kit (cat#5067-4626) and Qubit (ThermoFisher, car#Q32851). Individual libraries were diluted to 4nM and pooled for sequencing. Pooled libraries were sequenced by High Output kit (150 cycle) (Illumina cat#20024907) with Next500/550 sequencer (Illumina).
Single cell RNA sequencing (scRNA-seq) analysis
Sequencing data were processed with UMI-tools (Smith et al., 2017) (version 1.0.0), aligned to the mouse reference genome (mm10) with STAR (Dobin et al., 2013) (version 2.7.1a), and quantified with Subread featureCounts (Liao et al., 2014) (version 1.6.4). Data normalization, cell clustering and visualization were performed using Seurat (Butler et al., 2018) (version 2.3.4, version 3.1.1). For initial quality control of the extracted gene-cell matrices, we filtered cells with parameters low.threshold = 1,000, high.threshold = 6,000 for number of genes per cell (nGene), high.threshold = 0.3 for percentage of mitochondrial genes (percent.mito) and genes with parameter min.cell = 3. Filtered matrices were normalized by LogNormalize method with scale factor = 10,000. Variable genes were found with parameters of mean.function = ExpMean, dispersion.function = LogVMR, x.low.cutoff = 0.0125, x.high.cutoff = 3 and y.cutoff = 0.5, trimmed for the genes related to cell cycle (GO:0007049) and then used for principal component analysis. Statistically significant principal components were determined by JackStraw method and the first 12 principle components were used for non-linear dimensional reduction (tSNE), canonical correlation analysis (CCA) and clustering analysis with resolution = 0.4. Monocle (Trapnell et al., 2014) (version 2.10.1) was used for pseudotime trajectory analysis. We imported Seurat objects to Monocle R package and then performed dimensionality reduction with DDRTree method with parameters max_components = 2 and norm_method = ”log.” Cell cycle phases were classified by cyclone function of scran (Lun et al., 2016) (version 1.14.5). Detailed scripts and parameters used for the study are available from the authors upon reasonable request.
Micro computed tomography (μCT) analysis
Femurs were fixed overnight in 4% PFA at 4°C and analyzed using μCT. Measurements were carried out by Scanco Medical AG, Switzerland. The chosen voxel size was 6 μm in all three spatial dimensions. For each sample, 248 slices were evaluated with the following specifications: covering a total of 1.488 mm, X-ray voltage was 70 kVp, Intensity 114 μA, Integration Time 300 ms, Frame Averaging 4.
Quantification and statistical analysis
Immunofluorescence stained bone sections were imaged with a Leica SP8 and ZEISS LSM 880 confocal microscope. Images were analyzed, quantified, and processed using Volocity (Perkin Elmer), Adobe CS6 Photoshop and Illustrator software.
Statistical analysis was performed using Graphpad Prism software or the R statistical environment (http://www.r-project.org). All data are presented as mean ± s.e.m. unless indicated otherwise. Unpaired two tailed Student’s t tests were used to determine statistical significance. p < 0.05 was considered significant unless stated otherwise. Sample number was chosen based on experience from previous experiments. Reproducibility was ensured by several independent experiments. No animals were excluded from analysis.
Acknowledgments
The authors thank Silke Schröder and Karin Mildner for excellent technical assistance and M. Stehling for cell sorting. Funding was provided by the Max Planck Society (R.H.A. and M.P.), the Leducq Foundation (R.H.A. and M.P.), and the European Research Council (R.H.A.; AdG 786672 PROVEC).
Author contributions
Conceptualization, writing – original draft, writing – review and editing, K.K.S. and R.H.A; investigation, methodology, K.K.S., B.D., D.Z., S.A., and M.P.; data curation, formal analysis, K.K.S. and H.-W.J.; resources, supervision, funding acquisition, project administration, R.H.A.
Declaration of interests
R.H.A. is investigator on patent EP 2 860 243 A1 (Reprogramming bone endothelial cells for bone angiogenesis and osteogenesis).
Published: July 13, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.109352.
Supplemental information
References
- Addo R.K., Heinrich F., Heinz G.A., Schulz D., Sercan-Alp Ö., Lehmann K., Tran C.L., Bardua M., Matz M., Löhning M. Single-cell transcriptomes of murine bone marrow stromal cells reveal niche-associated heterogeneity. Eur. J. Immunol. 2019;49:1372–1379. doi: 10.1002/eji.201848053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghebati-Maleki L., Dolati S., Zandi R., Fotouhi A., Ahmadi M., Aghebati A., Nouri M., Kazem Shakouri S., Yousefi M. Prospect of mesenchymal stem cells in therapy of osteoporosis: A review. J. Cell. Physiol. 2019;234:8570–8578. doi: 10.1002/jcp.27833. [DOI] [PubMed] [Google Scholar]
- Ahn S., Joyner A.L. Dynamic changes in the response of cells to positive hedgehog signaling during mouse limb patterning. Cell. 2004;118:505–516. doi: 10.1016/j.cell.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Ambrosi T.H., Scialdone A., Graja A., Gohlke S., Jank A.M., Bocian C., Woelk L., Fan H., Logan D.W., Schürmann A. Adipocyte accumulation in the bone marrow during obesity and aging impairs stem cell-based hematopoietic and bone regeneration. Cell Stem Cell. 2017;20:771–784.e6. doi: 10.1016/j.stem.2017.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armulik A., Genové G., Mäe M., Nisancioglu M.H., Wallgard E., Niaudet C., He L., Norlin J., Lindblom P., Strittmatter K. Pericytes regulate the blood-brain barrier. Nature. 2010;468:557–561. doi: 10.1038/nature09522. [DOI] [PubMed] [Google Scholar]
- Baccin C., Al-Sabah J., Velten L., Helbling P.M., Grünschläger F., Hernández-Malmierca P., Nombela-Arrieta C., Steinmetz L.M., Trumpp A., Haas S. Combined single-cell and spatial transcriptomics reveal the molecular, cellular and spatial bone marrow niche organization. Nat. Cell Biol. 2020;22:38–48. doi: 10.1038/s41556-019-0439-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baryawno N., Przybylski D., Kowalczyk M.S., Kfoury Y., Severe N., Gustafsson K., Kokkaliaris K.D., Mercier F., Tabaka M., Hofree M. A cellular taxonomy of the bone marrow stroma in homeostasis and leukemia. Cell. 2019;177:1915–1932.e1916. doi: 10.1016/j.cell.2019.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianco P. “Mesenchymal” stem cells. Annu. Rev. Cell Dev. Biol. 2014;30:677–704. doi: 10.1146/annurev-cellbio-100913-013132. [DOI] [PubMed] [Google Scholar]
- Bianco P., Robey P.G., Simmons P.J. Mesenchymal stem cells: revisiting history, concepts, and assays. Cell Stem Cell. 2008;2:313–319. doi: 10.1016/j.stem.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böhm A.M., Dirckx N., Tower R.J., Peredo N., Vanuytven S., Theunis K., Nefyodova E., Cardoen R., Lindner V., Voet T. Activation of skeletal stem and progenitor cells for bone regeneration is driven by PDGFRβ signaling. Dev. Cell. 2019;51:236–254.e12. doi: 10.1016/j.devcel.2019.08.013. [DOI] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.K., Seo E.Y., Chen J.Y., Lo D., McArdle A., Sinha R., Tevlin R., Seita J., Vincent-Tompkins J., Wearda T. Identification and specification of the mouse skeletal stem cell. Cell. 2015;160:285–298. doi: 10.1016/j.cell.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C.K.F., Gulati G.S., Sinha R., Tompkins J.V., Lopez M., Carter A.C., Ransom R.C., Reinisch A., Wearda T., Murphy M. Identification of the human skeletal stem cell. Cell. 2018;175:43–56.e21. doi: 10.1016/j.cell.2018.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Zhang H., Liu Y., Adams S., Eilken H., Stehling M., Corada M., Dejana E., Zhou B., Adams R.H. Endothelial cells are progenitors of cardiac pericytes and vascular smooth muscle cells. Nat. Commun. 2016;7:12422. doi: 10.1038/ncomms12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook D., Genever P. Regulation of mesenchymal stem cell differentiation. Adv. Exp. Med. Biol. 2013;786:213–229. doi: 10.1007/978-94-007-6621-1_12. [DOI] [PubMed] [Google Scholar]
- Debnath S., Yallowitz A.R., McCormick J., Lalani S., Zhang T., Xu R., Li N., Liu Y., Yang Y.S., Eiseman M. Discovery of a periosteal stem cell mediating intramembranous bone formation. Nature. 2018;562:133–139. doi: 10.1038/s41586-018-0554-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFalco J., Tomishima M., Liu H., Zhao C., Cai X., Marth J.D., Enquist L., Friedman J.M. Virus-assisted mapping of neural inputs to a feeding center in the hypothalamus. Science. 2001;291:2608–2613. doi: 10.1126/science.1056602. [DOI] [PubMed] [Google Scholar]
- Ding L., Saunders T.L., Enikolopov G., Morrison S.J. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481:457–462. doi: 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty L., Yu J., Wang X., Hankenson K.D., Kalajzic I., Sanjay A. A PDGFRβ-PI3K signaling axis mediates periosteal cell activation during fracture healing. PLoS ONE. 2019;14:e0223846. doi: 10.1371/journal.pone.0223846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchamp de Lageneste O., Julien A., Abou-Khalil R., Frangi G., Carvalho C., Cagnard N., Cordier C., Conway S.J., Colnot C. Periosteum contains skeletal stem cells with high bone regenerative potential controlled by Periostin. Nat. Commun. 2018;9:773. doi: 10.1038/s41467-018-03124-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber H.P., Ferrara N. Angiogenesis and bone growth. Trends Cardiovasc. Med. 2000;10:223–228. doi: 10.1016/s1050-1738(00)00074-8. [DOI] [PubMed] [Google Scholar]
- Hamilton T.G., Klinghoffer R.A., Corrin P.D., Soriano P. Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Mol. Cell. Biol. 2003;23:4013–4025. doi: 10.1128/MCB.23.11.4013-4025.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellström M., Kalén M., Lindahl P., Abramsson A., Betsholtz C. Role of PDGF-B and PDGFR-beta in recruitment of vascular smooth muscle cells and pericytes during embryonic blood vessel formation in the mouse. Development. 1999;126:3047–3055. doi: 10.1242/dev.126.14.3047. [DOI] [PubMed] [Google Scholar]
- Hess J., Hartenstein B., Teurich S., Schmidt D., Schorpp-Kistner M., Angel P. Defective endochondral ossification in mice with strongly compromised expression of JunB. J. Cell Sci. 2003;116:4587–4596. doi: 10.1242/jcs.00772. [DOI] [PubMed] [Google Scholar]
- Hu L., Yin C., Zhao F., Ali A., Ma J., Qian A. Mesenchymal stem cells: cell fate decision to osteoblast or adipocyte and application in osteoporosis treatment. Int. J. Mol. Sci. 2018;19:360. doi: 10.3390/ijms19020360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isern J., García-García A., Martín A.M., Arranz L., Martín-Pérez D., Torroja C., Sánchez-Cabo F., Méndez-Ferrer S. The neural crest is a source of mesenchymal stem cells with specialized hematopoietic stem cell niche function. eLife. 2014;3:e03696. doi: 10.7554/eLife.03696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K., Diéguez-Hurtado R., Park D.Y., Hong S.P., Kato-Azuma S., Adams S., Stehling M., Trappmann B., Wrana J.L., Koh G.Y., Adams R.H. Pulmonary pericytes regulate lung morphogenesis. Nat. Commun. 2018;9:2448. doi: 10.1038/s41467-018-04913-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenner L., Hoebertz A., Beil F.T., Keon N., Karreth F., Eferl R., Scheuch H., Szremska A., Amling M., Schorpp-Kistner M. Mice lacking JunB are osteopenic due to cell-autonomous osteoblast and osteoclast defects. J. Cell Biol. 2004;164:613–623. doi: 10.1083/jcb.200308155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumbe A.P., Ramasamy S.K., Adams R.H. Coupling of angiogenesis and osteogenesis by a specific vessel subtype in bone. Nature. 2014;507:323–328. doi: 10.1038/nature13145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusumbe A.P., Ramasamy S.K., Starsichova A., Adams R.H. Sample preparation for high-resolution 3D confocal imaging of mouse skeletal tissue. Nat. Protoc. 2015;10:1904–1914. doi: 10.1038/nprot.2015.125. [DOI] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Lun A.T., McCarthy D.J., Marioni J.C. A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res. 2016;5:2122. doi: 10.12688/f1000research.9501.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes C., Clemens T.L. Angiogenic-osteogenic coupling: the endothelial perspective. Bonekey Rep. 2014;3:578. doi: 10.1038/bonekey.2014.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Méndez-Ferrer S., Michurina T.V., Ferraro F., Mazloom A.R., Macarthur B.D., Lira S.A., Scadden D.T., Ma’ayan A., Enikolopov G.N., Frenette P.S. Mesenchymal and haematopoietic stem cells form a unique bone marrow niche. Nature. 2010;466:829–834. doi: 10.1038/nature09262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuhashi K., Ono W., Matsushita Y., Sakagami N., Takahashi A., Saunders T.L., Nagasawa T., Kronenberg H.M., Ono N. Resting zone of the growth plate houses a unique class of skeletal stem cells. Nature. 2018;563:254–258. doi: 10.1038/s41586-018-0662-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar M.D., Tasic B., Miyamichi K., Li L., Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Nakamura E., Nguyen M.T., Mackem S. Kinetics of tamoxifen-regulated Cre activity in mice using a cartilage-specific CreER(T) to assay temporal activity windows along the proximodistal limb skeleton. Dev. Dyn. 2006;235:2603–2612. doi: 10.1002/dvdy.20892. [DOI] [PubMed] [Google Scholar]
- Nombela-Arrieta C., Ritz J., Silberstein L.E. The elusive nature and function of mesenchymal stem cells. Nat. Rev. Mol. Cell Biol. 2011;12:126–131. doi: 10.1038/nrm3049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen B.R., Reginato A.M., Wang W. Bone development. Annu. Rev. Cell Dev. Biol. 2000;16:191–220. doi: 10.1146/annurev.cellbio.16.1.191. [DOI] [PubMed] [Google Scholar]
- Ono N., Balani D.H., Kronenberg H.M. Stem and progenitor cells in skeletal development. Curr. Top. Dev. Biol. 2019;133:1–24. doi: 10.1016/bs.ctdb.2019.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortinau L.C., Wang H., Lei K., Deveza L., Jeong Y., Hara Y., Grafe I., Rosenfeld S.B., Lee D., Lee B. Identification of Functionally Distinct Mx1+αSMA+ Periosteal Skeletal Stem Cells. Cell Stem Cell. 2019;25:784–796.e5. doi: 10.1016/j.stem.2019.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauch A., Haakonsson A.K., Madsen J.G.S., Larsen M., Forss I., Madsen M.R., Van Hauwaert E.L., Wiwie C., Jespersen N.Z., Tencerova M. Osteogenesis depends on commissioning of a network of stem cell transcription factors that act as repressors of adipogenesis. Nat. Genet. 2019;51:716–727. doi: 10.1038/s41588-019-0359-1. [DOI] [PubMed] [Google Scholar]
- Sacchetti B., Funari A., Michienzi S., Di Cesare S., Piersanti S., Saggio I., Tagliafico E., Ferrari S., Robey P.G., Riminucci M., Bianco P. Self-renewing osteoprogenitors in bone marrow sinusoids can organize a hematopoietic microenvironment. Cell. 2007;131:324–336. doi: 10.1016/j.cell.2007.08.025. [DOI] [PubMed] [Google Scholar]
- Shi Y., He G., Lee W.C., McKenzie J.A., Silva M.J., Long F. Gli1 identifies osteogenic progenitors for bone formation and fracture repair. Nat. Commun. 2017;8:2043. doi: 10.1038/s41467-017-02171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivaraj K.K., Adams R.H. Blood vessel formation and function in bone. Development. 2016;143:2706–2715. doi: 10.1242/dev.136861. [DOI] [PubMed] [Google Scholar]
- Sivaraj K.K., Dharmalingam B., Mohanakrishnan V., Jeong H.W., Kato K., Schröder S., Adams S., Koh G.Y., Adams R.H. YAP1 and TAZ negatively control bone angiogenesis by limiting hypoxia-inducible factor signaling in endothelial cells. eLife. 2020;9:e50770. doi: 10.7554/eLife.50770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T., Heger A., Sudbery I. UMI-tools: modeling sequencing errors in unique molecular identifiers to improve quantification accuracy. Genome Res. 2017;27:491–499. doi: 10.1101/gr.209601.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama T., Kohara H., Noda M., Nagasawa T. Maintenance of the hematopoietic stem cell pool by CXCL12-CXCR4 chemokine signaling in bone marrow stromal cell niches. Immunity. 2006;25:977–988. doi: 10.1016/j.immuni.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Tencerova M., Kassem M. The bone marrow-derived stromal cells: commitment and regulation of adipogenesis. Front. Endocrinol. (Lausanne) 2016;7:127. doi: 10.3389/fendo.2016.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tikhonova A.N., Dolgalev I., Hu H., Sivaraj K.K., Hoxha E., Cuesta-Dominguez A., Pinho S., Akhmetzyanova I., Gao J., Witkowski M. The bone marrow microenvironment at single-cell resolution. Nature. 2019;569:222–228. doi: 10.1038/s41586-019-1104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uccelli A., Moretta L., Pistoia V. Mesenchymal stem cells in health and disease. Nat. Rev. Immunol. 2008;8:726–736. doi: 10.1038/nri2395. [DOI] [PubMed] [Google Scholar]
- Walker J.V., Zhuang H., Singer D., Illsley C.S., Kok W.L., Sivaraj K.K., Gao Y., Bolton C., Liu Y., Zhao M. Transit amplifying cells coordinate mouse incisor mesenchymal stem cell activation. Nat. Commun. 2019;10:3596. doi: 10.1038/s41467-019-11611-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth A., Benyó Z., Lukasova M., Leutgeb B., Wettschureck N., Gorbey S., Orsy P., Horváth B., Maser-Gluth C., Greiner E. G12-G13-LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nat. Med. 2008;14:64–68. doi: 10.1038/nm1666. [DOI] [PubMed] [Google Scholar]
- Wolock S.L., Krishnan I., Tenen D.E., Matkins V., Camacho V., Patel S., Agarwal P., Bhatia R., Tenen D.G., Klein A.M., Welner R.S. Mapping distinct bone marrow niche populations and their differentiation paths. Cell Rep. 2019;28:302–311.e5. doi: 10.1016/j.celrep.2019.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthley D.L., Churchill M., Compton J.T., Tailor Y., Rao M., Si Y., Levin D., Schwartz M.G., Uygur A., Hayakawa Y. Gremlin 1 identifies a skeletal stem cell with bone, cartilage, and reticular stromal potential. Cell. 2015;160:269–284. doi: 10.1016/j.cell.2014.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S., Wu Y., Capecchi M.R. Motoneurons and oligodendrocytes are sequentially generated from neural stem cells but do not appear to share common lineage-restricted progenitors in vivo. Development. 2006;133:581–590. doi: 10.1242/dev.02236. [DOI] [PubMed] [Google Scholar]
- Xie H., Cui Z., Wang L., Xia Z., Hu Y., Xian L., Li C., Xie L., Crane J., Wan M. PDGF-BB secreted by preosteoclasts induces angiogenesis during coupling with osteogenesis. Nat. Med. 2014;20:1270–1278. doi: 10.1038/nm.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelzer E., Olsen B.R. Multiple roles of vascular endothelial growth factor (VEGF) in skeletal development, growth, and repair. Curr. Top. Dev. Biol. 2005;65:169–187. doi: 10.1016/S0070-2153(04)65006-X. [DOI] [PubMed] [Google Scholar]
- Zhong L., Yao L., Tower R.J., Wei Y., Miao Z., Park J., Shrestha R., Wang L., Yu W., Holdreith N. Single cell transcriptomics identifies a unique adipose lineage cell population that regulates bone marrow environment. eLife. 2020;9:e54695. doi: 10.7554/eLife.54695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B.O., Yue R., Murphy M.M., Peyer J.G., Morrison S.J. Leptin-receptor-expressing mesenchymal stromal cells represent the main source of bone formed by adult bone marrow. Cell Stem Cell. 2014;15:154–168. doi: 10.1016/j.stem.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The NGS datasets (bulk-RNA and single cell -RNA sequencing) used in this article are available at GEO (https://www.ncbi.nlm.nih.gov/geo/) with the accession number GEO: GSE156636.