Abstract
Transposable elements (TEs) are mobile DNA sequences that propagate within genomes. Through diverse invasion strategies, TEs have come to occupy a substantial fraction of nearly all eukaryotic genomes and they represent a major source of genetic variation and novelty. Here we review the defining features of each major group of eukaryotic TEs and explore their evolutionary origins and relationships. We discuss how the unique biology of different TEs influences their propagation and distribution within and across genomes. Environmental and genetic factors acting at the level of the host species further modulate the activity, diversification and fate of TEs, producing the dramatic variation in TE content observed in eukaryotes. We argue that cataloguing TE diversity and dissecting the idiosyncratic behaviour of individual elements is crucial to furthering our understanding of their impact on the biology of genomes and the evolution of species.
Keywords: Transposons, retrotransposons, transposition mechanisms, transposable element origins, genome evolution
INTRODUCTION
Transposable elements (TEs) are mobile DNA sequences capable of replicating themselves within genomes independently of the host cell DNA. They typically range in length from 100 to 10,000 base pairs, but are sometimes far larger (6). Along with viruses, TEs are the most intricate selfish genetic elements. They frequently encode proteins with multiple biochemical activities as well as complex noncoding regulatory sequences that promote their propagation.
The boundary between TEs and other invasive genetic elements such as viruses is fluid. Here we will define a TE as a genetic element capable of chromosomal and replicative mobilization in the germ line, thereby increasing in frequency through vertical inheritance. This definition incorporates non-autonomous elements such as short interspersed nuclear elements (SINEs) and miniature inverted-repeat TEs (MITEs). It also includes endogenous retroviruses (ERVs), but excludes endogenous elements that originate from viruses that do not typically integrate and further propagate in the host germline (47). Whilst the capacity for inheritance through the germ line is a defining feature of all TEs, it should be noted that horizontal transfer of TEs between species also occurs and is an important factor in their long-term success (60).
All eukaryotic genomes examined thus far, with a few notable exceptions (see below), are known to harbour TEs. Across most organisms, TE content correlates strongly with genome size, and in some species they constitute as much as 85% of the genome (158), with protein-coding regions little more than islands in a sea of TEs (44). However, the fraction of the genome occupied by TEs does not correlate with organismal complexity: both complex multicellular organisms such as conifers (118) and salamanders (117) as well as single-celled organisms such as Trichomonas vaginalis (22) and Anncaliia algerae (122) may contain substantial TE fractions. Thus, TEs are an omnipresent feature of eukaryotic genomes.
In the decades since Barbara McClintock’s far-seeing ideas on “controlling elements” (110), the profound effect that TEs have had on eukaryotic evolution has become clear. In everything from the size and structure of genomes, to the proteins they encode and the regulation of such, TEs play a critical role (1, 11, 14, 26, 28, 44, 49, 136). If we wish to understand how TEs have impacted the diversification and biology of species, we must therefore begin with an understanding of the diversity and biology of TEs themselves. In this review, we first provide an overview of the classification of eukaryotic TEs and a brief examination of their evolutionary origins and relationships. Next, we look at the variation of TE content across species, highlighting the extremes in abundance and diversity. We close with a discussion of the forces underlying such variation, focusing on the factors intrinsic to the TEs themselves.
CLASSIFICATION OF EUKARYOTIC TRANSPOSABLE ELEMENTS
The most fundamental division of eukaryotic TEs, introduced by David Finnegan in 1989 (51), distinguishes two major classes based on their transposition intermediates: class I – retrotransposons, and class II – DNA transposons. Class I elements replicate via an RNA intermediate, which is then reverse-transcribed back into a DNA copy and integrated into the genome. Because the original template element remains intact, retrotransposons are commonly referred to as “copy-and-paste” elements. In contrast, the majority of (but not all) class II elements mobilize through a “cut-and-paste” mechanism, in which the transposon itself is excised and moved to a new genomic location. Both classes can be subdivided further many times, first into subclasses (or orders (160)), which are primarily delineated according to their mechanisms of replication and/or chromosomal integration (Fig. 1), and then into superfamilies and families, which are more accurately characterised in terms of phylogenetic relationships (4, 36, 49, 160, 167).
Figure 1. Summary of replication mechanisms and transposition intermediates.
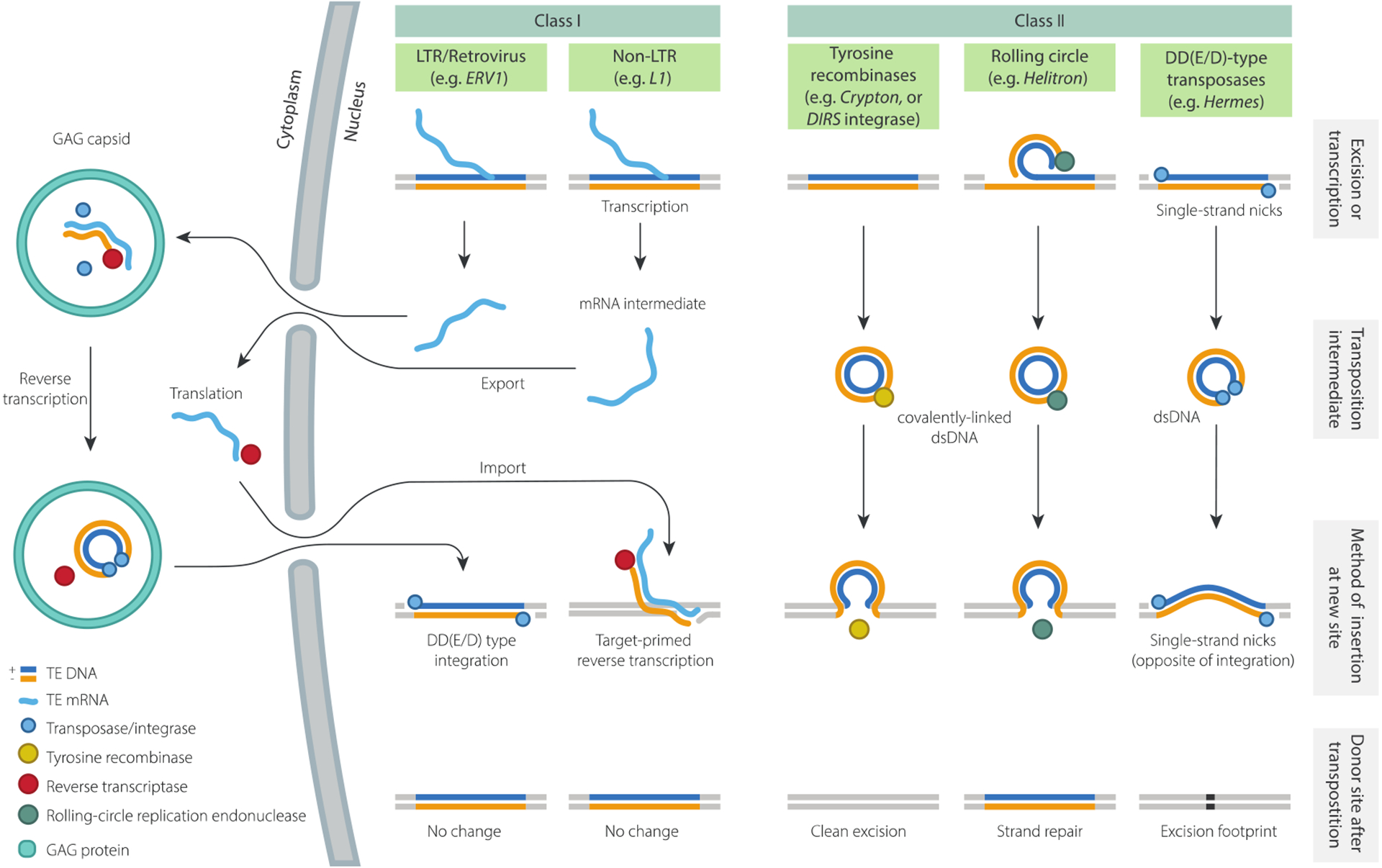
Proposed transposition intermediates and key replication steps for five TE subclasses. YR-retrotransposons and Maverick/Polintons are not shown, but the former are expected to transpose via the same intermediate as Class II YR-transposons (i.e. Cryptons). The mechanism of Mavericks/Polintons has not yet been studied, but based on the presence of protein-primed type B DNA polymerase (pPolB), they are expected to transpose by direct synthesis of a DNA copy (78). For comprehensive reviews on transposition mechanisms see (29, 68).
In practice, TE families are usually defined using the “80-80-80” rule, which specifies that insertions are members of the same family if they are longer than 80 base pairs, and share at least 80% sequence identity over 80% of their length (160). These families can then be represented by their majority-rule consensus sequence, as constructed from sequence alignments of multiple copies. In principle, the consensus sequence of a TE family represents an approximation of the ancestral TE that seeded the family (76, 142). This is particularly accurate if the family has expanded rapidly in a single burst of activity and each copy has evolved neutrally thereafter. There are many cases where these assumptions are violated however, and as such the 80-80-80 rule and corresponding consensus sequences do not always reflect the true phylogenetic structure of TE families. L1 elements in mammals, for example, produce distinctive ladder-like phylogenies which require more careful analyses to be defined as families or subfamilies (82, 142).
TEs can also be classified according to whether or not they are able to move autonomously. Autonomous elements are those that encode the enzymatic machinery necessary for their own transposition. Non-autonomous elements are typically noncoding but are still capable of mobilization in trans by hijacking the machinery produced by their autonomous counterparts. Families entirely composed of non-autonomous elements often emerge as parasites of other TEs. Some of these originate from deletion derivatives of autonomous elements, as is the case for most MITEs, which comprise only the terminal inverted repeats – and thus transposase binding sites – of ancestral, autonomous DNA transposons (50, 166). But others emerge ‘de novo’ from non-TE sequences. For instance, SINEs are usually derived from noncoding genes such as tRNAs, transcribed by RNA polymerase III (Pol III) and trans-mobilized by the machinery of Long Interspersed Nuclear Elements (LINEs) (32, 119). However, most SINEs are not merely retrogenes, but have acquired composite sequences promoting LINE parasitism and amplification (see below, and reviews (32, 119)).
Class I retrotransposons
Retrotransposons can be divided into three major subclasses according to their mechanism of replication and integration: (i) Long Terminal Repeat (LTR) elements (mobilized by an integrase); (ii) “target-primed” non-LTR elements and (iii) Tyrosine Recombinase (YR)-mobilized elements. Of these, non-LTR elements are the simplest structurally, and usually contain two open reading frames, ORF1 and ORF2. The function of ORF1 remains poorly understood and is dispensable or absent in some groups of non-LTR elements (it is not present in R2, for example (18)). When it is required, as in L1 elements, ORF1 proteins form an oligomeric product involved in recognition and transport of the template RNA to the nucleus (136). ORF2 encodes both endonuclease (EN) and reverse transcriptase (RT) activities, the latter of which is essential for target-primed reverse transcription (TPRT) (103, 113, 136). In L1, this process initiates with the formation of a single-stranded nick by EN, usually at a 5’-TT/AAAA-3’ site, followed by hybridisation of the host DNA with the 3’ end of the RNA template, reverse transcription and finally, integration of the newly synthesised cDNA strand (136) (Fig. 1). A hallmark of this process is that the reverse transcription step frequently terminates early, leading to 5’-truncation. Because non-LTR elements are expressed from an internal Pol II promoter located in their 5’ termini, such truncation generally prevents further propagation of the newly inserted copy (136).
The structures, coding capacity, and replication mechanisms of LTR elements are more complex and closely resemble those of retroviruses, to which they are evolutionarily related (36). Autonomous LTR elements contain a minimal set of two distinct genes (gag and pol), generally expressed as a single polycistronic RNA transcribed from a Pol II promoter located within the LTRs themselves. Both gag and pol encode polyproteins that are cleaved into multiple proteins by a pol-encoded protease (PR). Pol also encodes reverse transcriptase (RT), RNaseH and integrase (IN) activities. Reverse transcription uses a tRNA primer and occurs on a genomic RNA template encapsidated within a cytoplasmic viral-like particle assembled from gag-encoded proteins (for further details, see (161)). The cDNA product is bound by the IN protein, which mediates nuclear localization and integration into the host chromosome through a process similar to that of cut-and-paste transposases (29, 68). Indeed, the catalytic domain of IN belongs to the “DDE” nuclease family (see below).
The process of retroviral replication and integration is essentially the same as that of LTR elements, and the only substantive difference is linked to the acquisition of fusogenic env genes by retroviruses (36). Env genes are often lost, and consequently retroviruses that are active in the germline (e.g. Koala retrovirus (101)) frequently become endogenized (107). A classic example of this is IAP, of which there exists a single copy in the C57BL/6 mouse genome that is apparently still a functional retrovirus (135).
YR retrotransposons represent a third major subclass of class I elements, but they are relatively understudied (29). They are most similar to LTR elements in their genetic structure, but differ notably by encoding YR in place of IN. YR elements possess terminal repeat sequences, but the structure of these varies between the major superfamilies of YR retroelements: for example DIRS elements have inverted repeats, but these are non-identical, in contrast to true LTRs, whereas Ngaro, VIPER and TATE elements appear to have direct repeats laid out in a “split-repeat” pattern (61, 134). At present, the function of the terminal repeats and mode of replication of YR elements remains poorly characterized, but a proposed mechanism for DIRS involves reverse transcription of the mRNA template, circularization of the single-stranded cDNA copy (initiated by pairing of the terminal repeats), synthesis of the second cDNA strand and finally chromosomal integration mediated by YR (20, 126).
Finally, we must make a brief mention of Penelope elements. These curious TEs were first discovered as mutagenic agents in Drosophila virilis in 1997, but for some time remained the only known representatives of their class (43). Two features of Penelope-like elements stand out: firstly, the presence of pseudo-LTRs and secondly, a GIY-YIG (amino acid motif) endonuclease domain, which is not shared with any other retroelement subclasses (42). Based on their likely reliance on TPRT for transposition, they may be classified as non-LTR elements, but phylogenetic analyses of their RT domain suggest that they define a distinct monophyletic group. This group is equally distant from LTR and non-LTR elements and is most closely related to telomerase, implying that these elements diverged early in eukaryotic evolution (5, 42). Consequently, Penelope-like elements may be considered a separate subclass of retroelements (4).
Class II DNA transposons
At present, we know of four major groups of DNA transposons: (i) cut-and-paste elements mobilized by DDE transposases (named after the aspartic and glutamic acid catalytic residues) (33, 167), or (ii) by YR (called Cryptons) (88), (iii) rolling-circle elements (also known as Helitrons (77, 151)) and (iv) the most enigmatic – “self-synthesizing” transposons, known as Mavericks or Polintons (48, 78, 129). Of these, DDE transposons and Cryptons are the simplest, typically consisting of a single ORF encoding a recombinase flanked by short terminal inverted repeats (TIRs). As such, these elements resemble prokaryotic insertion sequences in their structure (141). Whilst Cryptons are relatively rare in eukaryotes (88), DDE transposons are the most diverse and widespread of all TEs, with at least 17 large superfamilies defined by phylogenetically distinct transposases (4, 9, 49, 167). In fact, the success of this subclass is such that the DDE recombinase is a contender for the oldest and most abundant gene on earth (7).
The precise mechanism of DDE transposition varies between superfamilies, but for all eukaryotic members thus far examined the process is initiated by nucleophilic attack of a water molecule in close proximity to the ends of each TIR, eventually resulting in direct excision and re-location of the transposon DNA (68). Whilst the process itself is non-replicative, these elements can still increase in copy number to form abundant families in the genome. One amplification strategy involves preferential transposition during host DNA synthesis from replicated to un-replicated sites, effectively causing the transposon to be replicated twice (58, 137, 144). Cut-and-paste transposons can also be duplicated when the double-strand break left at their excision site is repaired via homologous recombination. During this process, abortive repair, strand slippage and template switching commonly lead to the formation of internally deleted transposon copies (40, 70, 138). While these non-autonomous elements often lose their coding capacity, they may retain the binding site recognized by transposases from autonomous elements. These short elements often proliferate more effectively and to the expense of their autonomous counterparts, forming extensive families of MITEs (50, 114, 166).
Helitrons are abundant in many eukaryotic lineages, including in model organisms such as Drosophila melanogaster, Caenorhabditis elegans and Arabidopsis thaliana, but remained largely uncharacterized until the early 2000s (77, 151). This was in part because they are mostly represented by non-autonomous elements that lack TIRs and other features of canonical DNA transposons. The identification of the first autonomous Helitrons in various species – which code for a large Rep/Hel protein with a DNA helicase domain fused to a HUH nuclease domain related to that of bacterial rolling-circle transposons – led to the realisation that they must use a fundamentally different mobilization mechanism than that of cut-and-paste elements (77).
Significant insights into the Helitron transposition mechanism were recently gained through the study of Helraiser, an active autonomous element resurrected from inactive elements identified in bat genomes (63, 127). Functional studies of Helraiser suggest a “peel-and-paste” mechanism in which a covalently linked circular dsDNA intermediate is formed by peeling off the sense strand and (probably) synthesising the second strand as the circle rolls towards the 3’ end of the Helitron (62) (Fig. 1). However, whilst Helraiser transposes replicatively, genetic data from maize suggests that some Helitrons are able to directly excise rather than copy, indicating that there is still work to be done on the mechanisms of Helitron transposition (99).
Mavericks (or Polintons) are yet another poorly characterized class of DNA elements, which are exceptional for their size (15–20 kb) and complexity, consisting of up to twenty protein-coding genes flanked by long, 400–700 bp TIRs (48, 78, 129). These elements are widespread across eukaryotes, but they are generally present in low copy number (dozens per genome), with a few known exceptions such as in the protist Trichomonas vaginalis, where they have recently exploded to occupy one third of the genome (129). Maverick/Polintons share similarities to disparate groups of double-stranded DNA (dsDNA) viruses (78, 90, 129, 168). This includes a protein-primed family-B DNA polymerase (pPolB) most closely related to that of adenovirus, which suggests that they replicate via direct synthesis of a DNA copy (hence the proposed name “self-synthesizing transposons” (78)). They also encode a DDE nuclease most closely related to retroviral IN, which is consistent with the fact that they create 5- or 6-bp target site duplication upon chromosomal integration (48, 78, 129).
Many Maverick and Polinton elements are also predicted to encode double and single jelly-roll capsid-like proteins (90, 93). This observation, along with their close relationship to viruses and the Mavirus virophage, has led to the proposal that they may represent endogenous viruses or virophages (53, 93). Lending support to this idea is the recent discovery of abundant Polinton-like viral entities in freshwater lake habitats (12). The connection with virophages – satellite elements that parasitize much larger dsDNA viruses – is particularly intriguing, as it suggests that their integration and endogenization into the genome of eukaryotic organisms might confer protection against some of these giant viruses (52).
EVOLUTIONARY ORIGINS OF EUKARYOTIC TRANSPOSABLE ELEMENTS
When and how did the major groups of TEs described above originate, and how do they relate to each other? The best way to address these questions is through a phylogenomic framework, which integrates the taxonomic distribution of the elements with phylogenetic analyses of their shared core proteins (4, 160, 165, 167). This approach has gained power with the increasing diversity of host genome sequencing projects and the development of powerful tools to automate the annotation of TEs (2, 55, 120). However, it also has limitations. TE sequences tend to evolve rapidly, and even the most common and constrained TE protein domains (such as RT or the DDE catalytic region) can be difficult to align with confidence, especially when considering elements from different superfamilies (4). In addition, most TEs have undergone numerous horizontal transfers at different points in their history, even between distantly related taxa (e.g. between vertebrates and invertebrates (60)). Furthermore, entire TE lineages may be lost or go extinct during evolution (155). As a result, TE phylogenies often conflict with those of host species, making it difficult to trace the evolutionary history and origin of TEs.
These caveats aside, a number of important conclusions can be drawn from the observations gathered over the last few decades. First, all the major subclasses of elements (Fig. 2) are widely distributed across the eukaryotic tree, each being found in at least two of the nine or so currently defined “supergroups” (19). Second, phylogenetic topologies of the core TE proteins are consistent with the idea that each of these subclasses were already in existence early in eukaryotic evolution. Third, the evolution of TEs is highly modular, with recurrent gain and loss of proteins from a shared pool of conserved domains.
Figure 2. Structure and taxonomy of eukaryotic TEs.
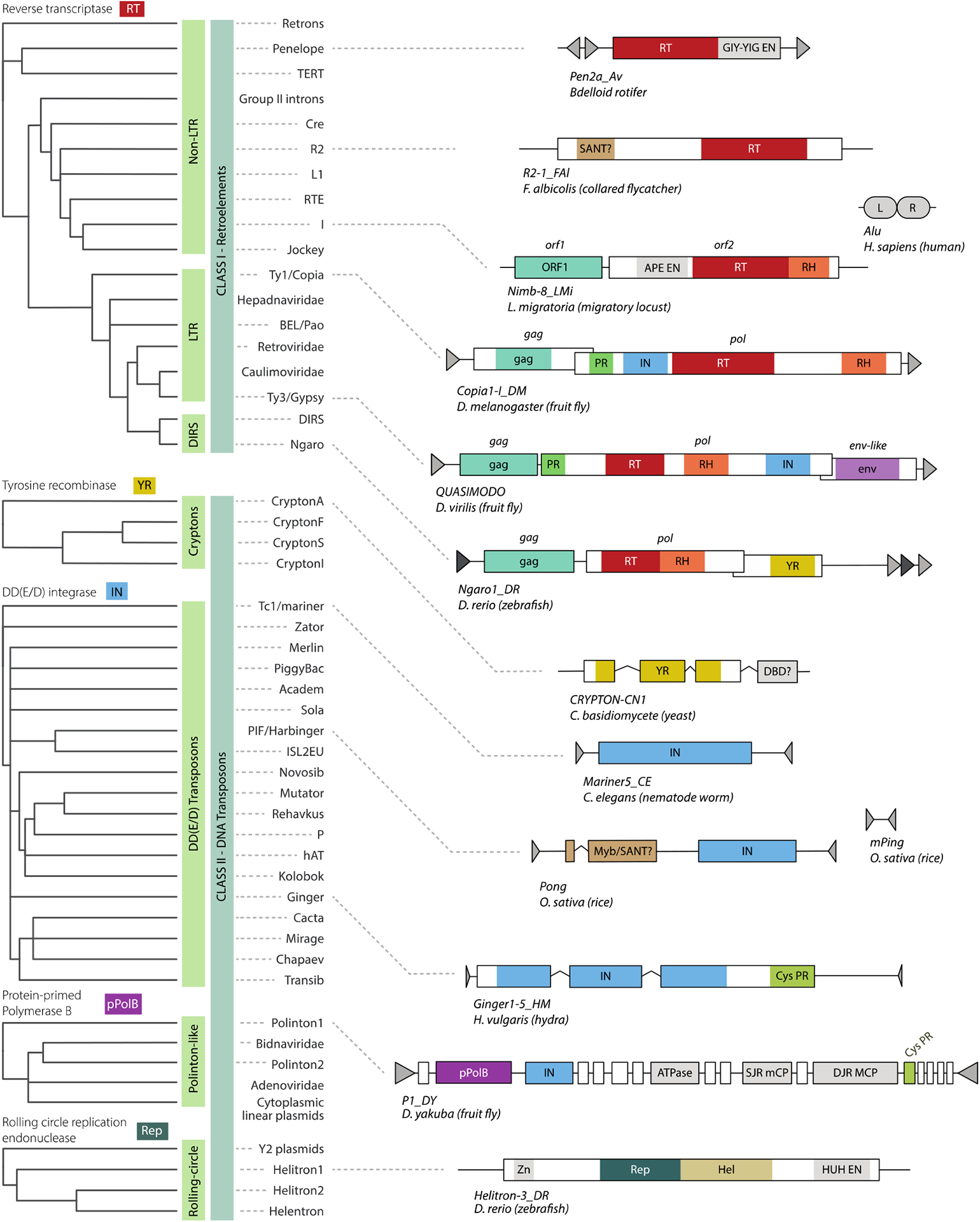
Left panel: unrooted cladograms showing putative relationships between the major TE superfamilies, based on phylogenies of core protein domains for five subclasses (4, 36, 67, 88, 95, 167). Right panel: genetic structures of representative elements from each subclass. Outlined boxes are open-reading frames, shaded regions are defining protein domains, kinked lines are introns, triangles are repeated sequences and rounded boxes (i.e. for Alu) are RNA elements. Domains with the same colours (excepting gray) indicate shared ancestry. Element lengths not to scale.
Deep evolutionary roots of TE proteins
Despite the bewildering diversity in the structure of different elements, the number of distinct protein families involved in replication and transposition is surprisingly small, comprising roughly five defining catalytic families (RT, DDE integrase, YR, Rep/Hel, and arguably pPolB) along with numerous accessory domains such as HUH endonuclease (Fig. 2). Remarkably, despite their seemingly disparate mechanisms, DDE integrases, HUH endonuclease and RT all share a deeply conserved structural fold termed the RNA recognition motif, which is thought to have played an important role in the transition from the primordial RNA world (for a recent review, see (94)). This fact, along with the widespread phylogenetic distribution of these proteins, indicates that the core enzymatic machinery of transposition – if not the TEs themselves – predates the emergence of eukaryotes.
At least six of the main DDE-transposase superfamilies can be phylogenetically clustered with well-defined prokaryotic IS transposases: Mutator with IS256, Tc1/mariner with IS630, PIF/Harbinger with IS5, Merlin with IS1016, piggyBac with IS1380, and Zator with ISAz013, suggesting that each of these DNA transposon superfamilies arose prior to the split of prokaryotes and eukaryotes (9, 49).
In contrast, none of the remaining eukaryotic TE subclasses have unambiguous prokaryotic homologs. While retroelements do occur in prokaryotes and phylogenies point to a direct affiliation between prokaryotic and eukaryotic RTs (notably between that of group II introns and non-LTR elements (24, 165, 171)), all extant eukaryotic retroelements are very distinct from their prokaryotic relatives.
In the case of rolling-circle replication elements, the HUH endonuclease involved in the transposition of Helitrons is also responsible for the mobilization of prokaryotic IS91 transposons, but it appears likely that prokaryotic and eukaryotic rolling-circle elements emerged independently from viruses or plasmids (67, 80). Similarly, although transposons mobilized by YR are common in prokaryotes, their enzymes are not directly related to those encoded by eukaryotic YR retrotransposons or class II Cryptons (61, 126, 134). Thus, most eukaryotic TE subclasses appear to have emerged shortly after the split of prokaryotes and eukaryotes.
Chimeric elements and modular evolution
While phylogenomic analyses reveal the deep relationships between the core transposition enzymes that define the major TE subclasses, they offer limited insight into the origin of individual families and superfamilies (Fig. 2). This is because TEs, together with other self-replicating elements like viruses and plasmids, form a densely connected evolutionary web characterized by frequent exchange of protein-coding units. These exchanges involve both the core domains essential for transposition, as well as accessory domains acquired from host genomes (4, 6, 90, 94) and they often blur the distinctions between TE classes and subclasses, and indeed between TEs and other invasive elements. For example, whilst Class I YR-retrotransposons cluster together with LTR elements and retroviruses based on phylogenies of their RT domains (Fig. 2), phylogenies based on YR show them closely related to Cryptons – Class II elements. Similarly, LTR retroelements, cut-and-paste DNA transposons and Mavericks/Polintons all use a DDE recombinase for chromosomal integration. The sharing of these enzymes points to chimerism and mosaic evolution as a major force in the emergence and diversification of TEs (4, 6, 90, 94, 97, 126).
LTR retrotransposons are a fascinating example of this process. These elements appear to have evolved a unique transposition mechanism that borrows components from non-LTR elements and cut-and-paste DDE transposons (109). Because both non-LTR and DDE elements appear to be evolutionarily older, LTR elements most likely arose by chimeric fusion between the two. One line of evidence supporting this scenario lies in the similarity between the RNase H domain (RH) of LTR and non-LTR elements. RH is another structural fold that arose near the origin of life, whose function is to degrade the RNA strand of DNA-RNA duplexes. A phylogenetic tree of RH domains from cellular genomes, LTRs and select non-LTR elements reveals that the LTR-derived RHs form a monophyletic group within the non-LTR clade (108). This tree is largely congruent with RT phylogenies, and by using host-derived RH sequences to root the tree, it can be inferred that non-LTR elements predate LTRs. The ‘ORF1’ protein encoded by several non-LTR elements also bear sequence, positional, and functional (RNA-binding and chaperone activities) similarities to the LTR Gag protein, although some of these features may be the result of convergent evolution (30, 83, 84, 132). While the acquisition of other attributes such as tRNA-priming or the LTRs themselves remain mysterious, the data currently point to a model where LTR elements emerged through a fusion of a non-LTR retrotransposon and DNA transposon.
SINEs also offer a compelling illustration of how highly successful TE families repeatedly emerge via chimeric assembly. Most SINEs are derived from Pol III-transcribed noncoding RNA such as tRNA, 7SL or 5S RNA, trans-mobilized by the machinery of LINEs (91). Whilst some SINE families consist of little more than fragments of Pol III sequence, many others have evolved complex mosaic structures which further enhance their transposition capacity. For instance, Alu elements arose early in primate evolution by a process involving the fusion of two monomeric 7SL-derived SINEs which emerged earlier in mammalian evolution (92). Since their appearance, Alus have spawned many subfamilies and new composite elements which not only outnumber their monomeric progenitors, but essentially all other TE families in primate genomes (11, 72). In the hominoid ancestor, a fusion between an Alu, a VNTR (Variable Number Tandem Repeat) and an LTR fragment gave rise to the SVA family (156). In the gibbon lineage, SVA in turn gave rise to another family of composite TEs called LAVA, which combines portions of L1, Alu, VNTR, and another Alu (21). Alu, SVA and LAVA are all non-autonomous elements mobilized by the L1 machinery, but SVA and LAVA have apparently acquired Pol II-driven promoters (65, 112, 131). Similarly tortuous stories of SINE diversification via fusion and accretion of additional sequences have been described in plants and other animals (91).
VARIABLE SUCCESS OF TRANSPOSABLE ELEMENTS ACROSS SPECIES
Half a century has passed since the realization that the TE content of genomes varies greatly between species (16, 17). The characterization of ever more genomes has continued to expose this variation, but we still have only partial clues to the factors influencing TE accumulation and diversification across species. Some genomes contain just a few, if any TE families, while others are bloated with a bewildering diversity (Fig. 3). Why? Answering this question is paramount to characterizing the impact of TEs on genome evolution.
Figure 3. Distribution of TEs across the eukaryote phylogeny.
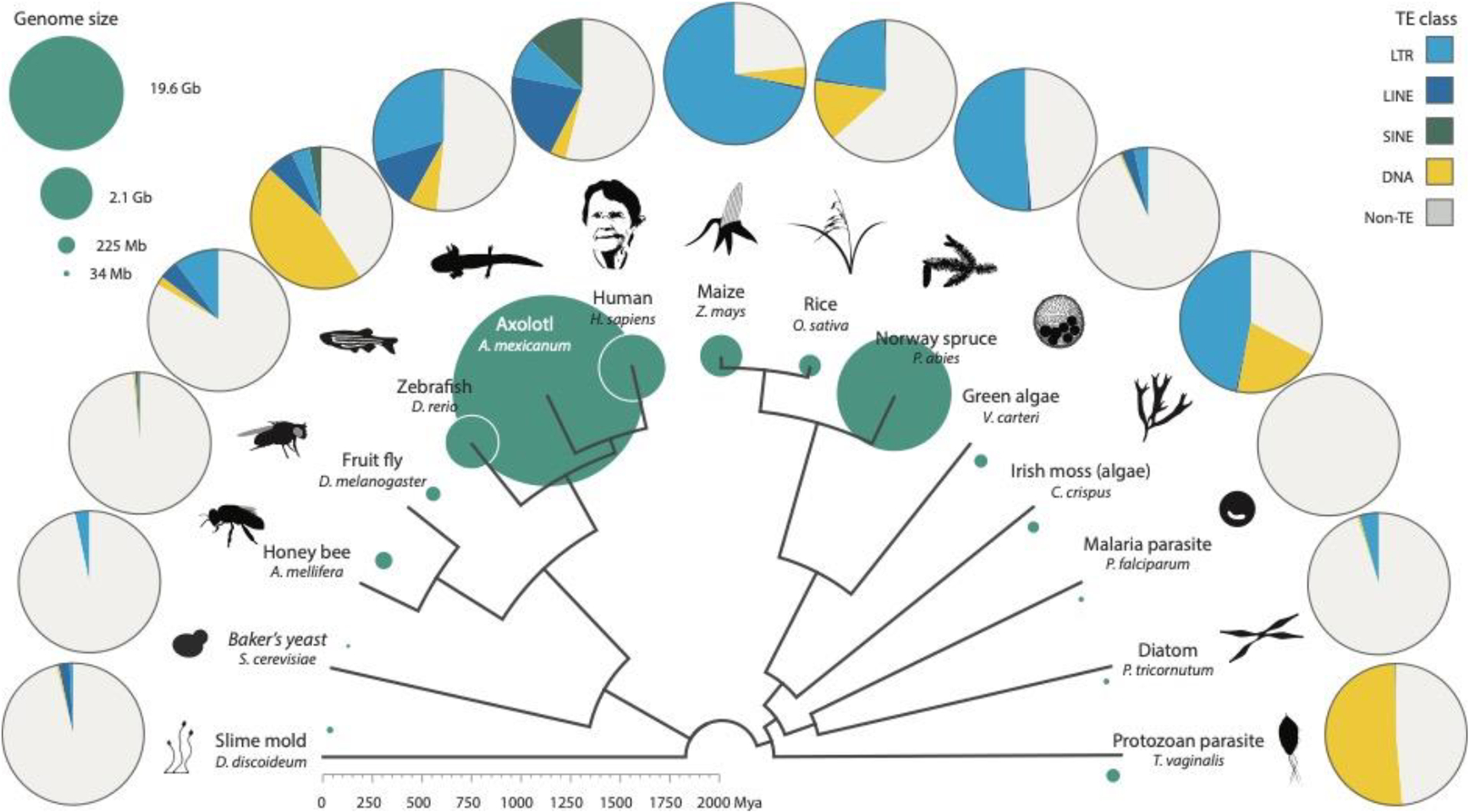
Reference genome size (sea green circles) varies dramatically across eukaryotes and is loosely correlated with transposable element content. Here, the honey bee TE content is likely an underestimate, as approximately 3% of the genome derives from unusual “large retrotransposon derivatives” (LARDs) (39). For ease of visualisation, DIRS elements have been included with LTRs and all Class II elements included under “DNA”. Data was acquired from genome RepeatMasker output files. Credit to Matt Crook for Volvox carteri silhouette and to Huang et al. for the figure inspiration (71).
Across broad phylogenetic scales, it has been proposed that the overall TE load of organisms is dictated by effective population size, or Ne (105). This is because the efficiency of selection in removing deleterious insertions decreases with Ne. This may explain, for instance, why TE insertions more frequently reach fixation in vertebrates than in fruit flies, which have relatively large Ne. But it cannot account for differences in TE abundance observed between species with comparable Ne, such as those within the same taxonomic order (79, 104, 123, 159) (Fig. 3). Similarly, it offers little explanation as to why the diversity of TEs should be so variable between species, or why certain TE types seem to be particularly successful in certain taxonomic groups. For instance, LTR elements are prevalent in flowering plants, while non-LTR elements dominate in mammals (116, 136) and DNA transposons prevail in zebrafish and Caenorhabditis nematodes (49). In the following section, we further illustrate such variation in TE abundance and diversity across species, before discussing some of the factors underlying this variation.
TE abundance and the relationship with genome size
Thus far, very few eukaryotic species appear to lack TEs altogether. The best-known exceptions are apicomplexan protists such as Plasmodium falciparum, Toxoplasma gondii, Encephalitozoon intestinalis and Theileria parva, which seem to have successfully purged TEs from their genomes (87). As a result, the latter two species now possess amongst the smallest known genomes of any eukaryotes (27, 64). It is probably not coincidental that all these species are single-celled, obligate intracellular parasites and are predominantly asexual except for brief periods in their lifecycle – a feature which has been predicted to reduce TE load (10). Although the dearth of TEs in apicomplexans may be related to their peculiar lifestyle and reduced genomes, several other parasitic unicellular eukaryotes do harbour diverse and active TE communities (22, 102, 122, 128). For instance, Anncaliia algerae is an obligately intracellular microsporidian parasite with a tiny genome of 23 Mb, but nonetheless, approximately 14% of its total DNA is TE-derived, originating from 240 different families, several of which appear to have been introduced by horizontal transfer (122).
At the other end of the size spectrum, many salamanders have undergone extreme genome expansions of as much as ~120 Gb since diverging from other amphibians, predominantly through the accumulation of LTR retroelements (37, 117, 149). Plant genomes too often grow very large through the rapid accumulation of LTR elements (1, 8, 150). Although this expansion is usually due to the combined effect of numerous families, in the absence of repression, individual TEs can have drastic effects on genome size: brown hydras diverged from the green hydra approximately 36 million years ago, but their genomes have roughly doubled in size in that time from ~300Mb to ~1 Gb, largely due to the activity of a single family of CR1 elements (162).
The rate of non-essential DNA removal is also a critical factor shaping TE content and genome size. Genomic gigantism in salamanders is associated with low deletion rates, whereas in rice and Arabidopsis transpositional gain of DNA appears to be buffered by high rates of deletion via ectopic recombination (31, 56, 106, 148, 149). This phenomena is also apparent in birds and mammals, and suggests an “accordion model” for genome size evolution, whereby bursts of TE activity promote the subsequent deletion of non-essential DNA via non-allelic recombination between copies of recently expanded TE families (79, 123).
Genomic TE diversity
In addition to variation in abundance, there are also differences in TE diversity between species. This can be measured at different levels, from the number of different classes or subclasses (Class I/II, LTR/DDE-type etc.) to the number of superfamilies, families and subfamilies. Family substructure can occur when, as with L1, arms-races develop between the TE and its host, leading to the independent expansion of multiple subfamilies (46, 73). Other elements such as Helitrons produce subfamilies due to the acquisition of gene fragments and other DNA (151).
Regardless of how you measure it, many eukaryotes harbour extraordinarily diverse TE repertoires. Zebrafish deserve a special mention here as both the most TE-abundant and -diverse vertebrate model organism currently in use, harbouring nearly 2000 distinct families with representatives from every subclass and almost every superfamily discussed in this review (Fig. 2, Fig. 3). Of these, DNA transposons are especially prevalent, with more than 1000 different families spanning a broad range of ages – this is unusual amongst fish and even closely related cyprinid species (59, 69, 139).
Large genomes might be assumed to be associated with wide TE diversity, but this is not necessarily true. Spruce pine, for example, is a gymnosperm conifer with a 20-Gb genome dominated by a relatively small number of very high-copy number LTR elements. Remarkably, the vast majority of insertions are estimated to be between 5 and 60 million years old, which stands in contrast to rice and maize (Fig. 3), where all insertions are less than 5 million years old (106, 118, 145). This indicates that whilst TE diversity is low in the spruce pine – as measured by the number of distinct families – elements that do establish in the genome are removed slowly. The opposite is true of most flowering plants, which tend to have smaller genomes but more diverse TE landscapes than gymnosperms; for reasons that are still unclear, across all land plants there is a negative correlation between genome size and TE diversity (38).
HOW THE BIOLOGY OF TEs AFFECTS THEIR SUCCESS
The fate of a TE family is dictated by three dynamic forces: (i) the rate of transposition, (ii) the rate of fixation of new TE insertions and (iii) the rate at which TE sequences are deleted or eroded. Each of these processes is influenced by a multitude of factors that fall into two broad categories: those intrinsic to the TE itself and those intrinsic to the host (genetics, development, physiology, ecology etc.). Both TE and host factors are in turn shaped by the environment, and the interplay between TE, host and environmental factors results in the dazzling variety of TE landscapes in eukaryotic genomes. In the following section, we will concentrate on the factors intrinsic to TEs that influence their survival and success within genomes.
TE insertion preference
A critical determinant of the fate of a TE is where it initially inserts in the genome. The most effective way to study insertion preference is through mapping of de novo insertions prior to the action of natural selection. Such studies have documented three general patterns: (i) TEs with apparently little insertional bias; (ii) TEs favouring insertion in genomic regions that minimize their deleterious effects; (iii) TEs targeting sites that likely facilitate their subsequent propagation. We will illustrate each with a few examples, but refer the readers to an excellent recent review for additional details on TE targeting (147).
Mechanistically, where a TE inserts is dictated by the nuclease that catalyses its chromosomal integration. Because all TE-encoded nucleases (endonucleases, integrases, transposases) have some degree of substrate specificity for particular DNA or chromatin attributes (e.g. sequence composition, nucleosome position), it follows that virtually all TEs show some level of insertion specificity. At the lowest level of specificity are TEs with nucleases that recognize highly degenerate or short sequence motifs, such as L1 elements (45). Indeed, L1 insertion profiles in human cells approach random distributions (54, 146).
Many TEs show much stronger insertion specificity, and a common theme involves targeting genomic sites where insertions are unlikely to disrupt cell function. A classic example includes several families of LINEs (e.g. R1, R2 etc.), which precisely target ribosomal RNA gene arrays (35). Such high copy-number genes offer an excellent niche for TEs because insertion in one or a few of the genes is unlikely to have immediate deleterious consequences, and TEs can be progressively purged out by recombination within the array. Precise targeting of these genes is achieved through highly sequence-specific endonucleases encoded by these elements. Remarkably different families have evolved different site preferences, which enables them to coexist within the same genome (89). The omnipresence of these elements across metazoans attest to the evolutionary stability of this strategy.
Targeting “safe havens” enables TEs to colonize compact genomes with little intergenic space. For example, all TEs in baker’s yeast are LTR elements that have evolved integration strategies to avoid genes (147). Ty1 and Ty3 preferentially insert upstream of Pol III-transcribed genes, where they usually do not disrupt gene expression. This is an evolutionary convergence because Ty1 and Ty3 belong to two very different superfamilies and achieve targeting via interaction of their IN with different Pol III subunits (15, 86). Ty5, which colonizes the genome of a closely related yeast, S. paradoxus, favors integration within silent chromatin primarily at telomeric regions through yet another molecular interaction between the Ty5 IN and Sir4p chromatin factor (164).
A wide variety of TEs are known to target the 5’ upstream region of genes, which attests to the evolutionary benefit for the TEs (and perhaps for the host). While insertion in this compartment may occasionally modulate the expression of adjacent genes, it alleviates the likelihood of disrupting coding sequences and places the newly inserted TE in a chromatin environment promoting further expression. Diverse DNA transposons have adopted this strategy, including the P-element in D. melanogaster (144), MuDR in maize (100), mPing in rice (115), and VANDAL21 in Arabidopsis (130). The fission yeast retrotransposon Tf1 also targets promoter regions, but with a preference for a subset of genes. Selectivity is achieved through Tf1 IN interaction with specific host transcription factors (96). Remarkably, Tf1 insertion around these genes can modulate their expression with adaptive effect under stressful environmental conditions (41).
Ty1/copia-like retrotransposons in Arabidopsis and possibly other plants have also evolved a mechanism to favour insertion into a subset of non-essential genes (130). This is achieved by targeting nucleosomes containing the histone variant H2A.Z, which are depleted within essential genes but enriched at a subset of environmentally responsive genes. Like Tf1 in fission yeast, this strategy suggests a non-random process of mutagenesis which could facilitate host adaptation in changing environmental conditions.
Another mitigating strategy is for TEs to target other TEs. Accordingly, several TE families have been found to be preferentially nested within other TE families (75, 98, 145). In most cases, it is difficult to distinguish between true targeting and the effects of differential retention of insertions due to selection. However in the case of the non-LTR element Tx1L in Xenopus laevis, which is almost exclusively found within Tx1D DNA transposons, targeting is achieved through the sequence-specificity of the Tx1L endonuclease (25). Consequently, the fate of Tx1L is dependent on the success of another TE – a form of hyper-parasitism.
Features affecting the long-term retention of TEs
All new TE insertions are subject to natural selection acting at the level of host fitness. The three major factors driving the deleterious effects of TE insertions are: disruption of gene expression, toxic effects of TE transcripts or protein products, and increased frequency of ectopic recombination between copies of the same TE family (13).
Current data point to ectopic recombination as the predominant factor affecting TE fixation in various species (13), albeit with some exceptions (140, 163). If correct, then longer TEs should be strongly selected against due to their increased likelihood of initiating recombination. This likely explains why LTR and LINE retroelements tend to accumulate in regions with low recombination rates (e.g. peri-centromeric heterochromatin), while shorter elements such as SINEs and MITEs accumulate in gene-rich regions, which are generally characterized by higher recombination rates (23, 34, 163). The relationship between TEs and recombination is a complex one, however, and is discussed in more depth elsewhere (81).
A second factor driving differential patterns of retention between TE types is their potential effect on gene expression. Since autonomous elements carry their own promoters and regulatory elements, they have a greater likelihood of disrupting expression of nearby genes upon insertion. In the human genome, L1 and LTR elements are significantly depleted within genes, and more severely so when considering insertions in the same orientation as the gene (111). Furthermore, older LTR insertions are also depleted in 5kb windows surrounding genes – an observation that is consistent with strong selection acting against the effect of LTR promoters on host gene expression. For intronic TEs in mouse and human genomes, there is a significant bias against insertion in “hazardous zones” near the exons, but the size of these regions differs between TE types. Highlighting the sensitivity of coding DNA, in humans most, if not all, disease-causing intronic TE insertions fall within these zones (170).
Horizontal transposon transfer
Sex provides the primary mechanism for the spread of TEs within populations, but horizontal transfer of TEs (HTT) is another important factor in their long-term success, and one which occurs regularly on evolutionary timescales (60). All major groups of TEs undergo HTT, but it is particularly common for some families. Notably, many DDE-type DNA transposons appear to pass between species with relative ease, whereas HTT events involving non-LTR retroelements are rare in comparison (125, 133, 169).
One possible explanation for this is that some DNA transposons have evolved mechanisms that reduce their dependence on specific host factors. For example, they encode either weak or no promoters: this makes them dependent on insertion near host regulatory elements for expression, but potentially reduces their dependence on specific transcription factors (60). This hypothesis has recently been tested using three elements from the Tc1/mariner superfamily, which share similar AT-rich, “blurry” promoters. These promoters drive reporter gene expression in cells derived from distantly related eukaryotes, in contrast to promoters isolated from an LTR retroelement and a hAT DNA transposon with more specific patterns of expression (121). Furthermore, the Tc1/mariner transposases are also known to be catalytically active in a wide range of organisms and even in cell-free assays. It is easy to envision how these properties could facilitate the spread of Tc1/mariner between widely diverged taxa (169).
The nature of transposition intermediates may also explain why some TEs can propagate horizontally more efficiently than others. DNA intermediates circularized and/or covalently bound by transposases/integrases are likely to be more stable than the ribonucleoprotein complexes mediating TPRT of non-LTR retroelements. The formation of cytoplasmic capsid- and (sometimes) envelope-coated viral-like particles by LTR elements may also facilitate their propagation between and outside of cells (85, 124, 154). Likewise, where these intermediates occur and traffic within cells will also affect their propensity to access potential vectors for HTT, such as viruses (133, 154). Elements with intermediates targeted (non-LTR) or even confined (DNA transposons) to the nucleus will be less likely to insert within viruses that replicate exclusively in the cytoplasm (e.g. poxviruses), but more likely to jump into those that are in the nucleus (e.g. herpesviruses, baculoviruses). If the former, for example, is a better vector for HTT, then it will favour the spread of these TEs. Thus, one can see how the intrinsic characteristics of TEs have a profound influence on their ability to propagate not just within but also between species.
Circumventing host defence systems
Numerous host-encoded systems control TE activity, the existence of which often manifests in signatures of genomic conflict: for example, as mentioned previously, L1 elements in placental mammals produce distinctive ladder-like phylogenetic trees (142, 143). These unusual trees are thought to arise from successive rounds of repression and mutational escape of elements from the defensive KRAB-zinc finger protein family, such that only one or two L1 subfamilies are active at any given time (46, 73).
This conflict is particularly intense in the germ cells, where dedicated silencing mechanisms such as the piRNA system exist (3, 28), and has led to inventive strategies by TEs to escape repression. One spectacular example is that of I-elements in D. melanogaster oogenesis (157). I-elements preferentially retrotranspose in the oocyte, but their RNA intermediates are exclusively produced in the nurse cells that surround the developing oocyte, before being trafficked into the mature oocyte via microtubule-mediated transport (153). Nurse cells are highly polyploid and outnumber the oocyte by fifteen to one, so this strategy allows I-element RNA to reach much higher concentrations than would be possible through expression in the germ cells alone, and simultaneously limits their exposure to piRNA silencing.
CONCLUSIONS
In the preceding pages, we have covered a minuscule fraction of the rich literature documenting the mechanisms by which TEs propagate, diversify and interact within their hosts. TEs exist in all domains of life, but their abundance and omnipresence in eukaryotes attest to their profound influence on genome architecture and organismal evolution. To take an anthropocentric example, we now understand that TEs account for the majority of cis-regulatory DNA in the human genome introduced during primate evolution (74, 152), and that TEs have given birth to numerous proteins coopted for mammalian physiology and development (14, 49, 57). Their movement, rearrangement and regulatory activities can also cause a plethora of diseases and exacerbate the effects of many more (14, 26, 66).
Despite their fundamental importance, however, the discovery of TEs did not immediately transform genome biology. The first six decades following McClintock’s initial breakthrough in maize were dominated by genetic and molecular characterization of a relatively small subset of active TEs and the myriad ways they cause mutant phenotypes in a few species, including model organisms, domesticated species and humans. Revolutionary advances in DNA sequencing since the early 2000s triggered a major shift in TE research to ‘genome-wide’ studies where virtually all TEs residing within any genome can be identified, compared and interrogated for their regulatory activities. While it was quickly realized that most TEs in any given species are inactive relics of past invasions, such genome-wide studies revealed with increasing breadth how TEs have fuelled genome evolution.
Today, TE research continues to be predominantly concerned with understanding their large-scale effects on genome architecture and function (14). But it is important not to lose sight of the fact that we can only interpret these effects when armed with an understanding of the mechanisms that promoted the propagation of the elements in the first place. Many of these insights have come from genetic, mechanistic and evolutionary studies of individual TEs. However, it seems this type of research has been in decline in recent years compared to genome-wide studies that attempt to discern broad patterns from an amalgam of diverse TEs lumped into a few groups.
Yet, as we have illustrated throughout this review, no two TE families look or behave exactly the same. Consequently, we can predict that the effects of TEs on their host genomes are as varied as the TEs themselves. It is therefore of paramount importance to continue cataloguing and organizing TE diversity in a wide range of species and Detailed studies of the molecular mechanisms and cellular activities of individual elements should also be encouraged, with priority given to TEs from widespread yet poorly characterized groups, such as Helitrons, Maverick/Polintons or YR elements. Inroads through the less travelled genomes are bound to uncover entirely new types of TEs and novel transposition strategies. While genomes are often dominated by defective and immobile elements, today’s technology offers the ability to revive these elements and reveal the idiosyncratic features that make each TE uniquely fascinating.
ACKNOWLEDGEMENTS
We would like to thank Ni-Chen (Sylvia) Chang, John A. Frank and Michelle C. Stitzer of the Feschotte Lab for feedback on figures. J.N.W. is supported by a postdoctoral fellowship from the Human Frontier Science Program Organisation (LT000017/2019-L). C.F is supported by supported by grants from the National Institutes of Health (R35GM122550, U01HG009391 and R01GM112972).
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any financial conflicts of interest affecting the objectivity of this review.
LITERATURE CITED
- 1.Ågren JA, Wright SI. 2011. Co-evolution between transposable elements and their hosts: A major factor in genome size evolution? Chromosom. Res 19(6):777–86 [DOI] [PubMed] [Google Scholar]
- 2.Amselem J, Cornut G, Choisne N, Alaux M, Alfama-Depauw F, et al. 2019. RepetDB: a unified resource for transposable element references. Mob. DNA 10:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aravin AA, Hannon GJ, Brennecke J. 2007. The Piwi-piRNA Pathway Provides an Adaptive Defense in the Transposon Arms Race. Science (80-. ). 318(5851):761–64 [DOI] [PubMed] [Google Scholar]
- 4.Arkhipova IR. 2017. Using bioinformatic and phylogenetic approaches to classify transposable elements and understand their complex evolutionary histories. Mob. DNA 8(1):19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arkhipova IR, Pyatkov KI, Meselson M, Evgen’ev MB. 2003. Retroelements containing introns in diverse invertebrate taxa. Nat. Genet 33(2):123–24 [DOI] [PubMed] [Google Scholar]
- 6.Arkhipova IR, Yushenova IA, Angert E. 2019. Giant Transposons in Eukaryotes: Is Bigger Better? Genome Biol. Evol 11(3):906–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aziz RK, Breitbart M, Edwards RA. 2010. Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res 38(13):4207–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baidouri M El, Panaud O. 2013. Comparative genomic paleontology across plant kingdom reveals the dynamics of TE-driven genome evolution. Genome Biol. Evol 5(5):954–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bao W, Jurka MG, Kapitonov VV, Jurka J. 2009. New superfamilies of eukaryotic DNA transposons and their internal divisions. Mol. Biol. Evol 26(5):983–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bast J, Jaron KS, Schuseil D, Roze D, Schwander T. 2019. Asexual reproduction reduces transposable element load in experimental yeast populations. Elife. 8: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Batzer MA, Deininger PL. 2002. Alu repeats and human genomic diversity. Nat. Rev. Genet 3(5):370–79 [DOI] [PubMed] [Google Scholar]
- 12.Bellas CM, Sommaruga R. 2019. Polinton-like viruses and virophages are widespread in aquatic ecosystems. bioRxiv. 2019.12.13.875310 [Google Scholar]
- 13.Bourgeois Y, Boissinot S. 2019. On the Population Dynamics of Junk: A Review on the Population Genomics of Transposable Elements. Genes (Basel). 10(6):419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bourque G, Burns KH, Gehring M, Gorbunova V, Seluanov A, et al. 2018. Ten things you should know about transposable elements. Genome Biol. 19(1):199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bridier-Nahmias A, Tchalikian-Cosson A, Baller JA, Menouni R, Fayol H, et al. 2015. An RNA polymerase III subunit determines sites of retrotransposon integration. Science (80-. ). 348(6234):585–88 [DOI] [PubMed] [Google Scholar]
- 16.Britten RJ, Davidson EH. 1969. Gene Regulation for Higher Cells: A Theory. Science (80-. ). 165(3891):349–57 [DOI] [PubMed] [Google Scholar]
- 17.Britten RJ, Kohne DE. 1968. Repeated Sequences in DNA. Science (80-. ). 161(3841):529–40 [DOI] [PubMed] [Google Scholar]
- 18.Burke WD, Calalang CC, Eickbush TH. 1987. The site-specific ribosomal insertion element type II of Bombyx mori (R2Bm) contains the coding sequence for a reverse transcriptase-like enzyme. Mol. Cell. Biol 7(6):2221–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burki F, Roger AJ, Brown MW, Simpson AGB. 2020. The New Tree of Eukaryotes. Trends Ecol. Evol 35(1):43–55 [DOI] [PubMed] [Google Scholar]
- 20.Cappello J 1985. Sequence of Dictyostelium DIRS-1: An apparent retrotransposon with inverted terminal repeats and an internal circle junction sequence. Cell. 43(1):105–15 [DOI] [PubMed] [Google Scholar]
- 21.Carbone L, Harris RA, Mootnick AR, Milosavljevic A, Martin DIK, et al. 2012. Centromere remodeling in Hoolock leuconedys (Hylobatidae) by a new transposable element unique to the gibbons. Genome Biol. Evol 4(7):648–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carlton JM, Hirt RP, Silva JC, Delcher AL, Schatz M, et al. 2007. Draft Genome Sequence of the Sexually Transmitted Pathogen Trichomonas vaginalis. Science (80-. ). 315(5809):207–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang C-H, Chavan A, Palladino J, Wei X, Martins NMC, et al. 2019. Islands of retroelements are major components of Drosophila centromeres. PLOS Biol. 17(5):e3000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang GS, Hong Y, Ko KD, Bhardwaj G, Holmes EC, et al. 2008. Phylogenetic profiles reveal evolutionary relationships within the “twilight zone” of sequence similarity. Proc. Natl. Acad. Sci 105(36):13474–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Christensen S, Pont-Kingdon G, Carroll D. 2000. Target Specificity of the Endonuclease from the Xenopus laevis Non-Long Terminal Repeat Retrotransposon, Tx1L. Mol. Cell. Biol 20(4):1219–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chuong EB, Elde NC, Feschotte C. 2017. Regulatory activities of transposable elements: from conflicts to benefits. Nat. Rev. Genet 18(2):71–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corradi N, Pombert J-F, Farinelli L, Didier ES, Keeling PJ. 2010. The complete sequence of the smallest known nuclear genome from the microsporidian Encephalitozoon intestinalis. Nat. Commun 1(1):77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cosby RL, Chang N-C, Feschotte C. 2019. Host-transposon interactions: conflict, cooperation, and cooption. Genes Dev. 33(17–18):1098–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Curcio MJ, Derbyshire KM. 2003. The outs and ins of transposition: from Mu to Kangaroo. Nat. Rev. Mol. Cell Biol 4(11):865–77 [DOI] [PubMed] [Google Scholar]
- 30.Dawson A, Hartswood E, Paterson T, Finnegan DJ. 1997. A LINE-like transposable element in Drosophila, the I factor, encodes a protein with properties similar to those of retroviral nucleocapsids. EMBO J. 16(14):4448–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Devos KM, Brown JKM, Bennetzen JL. 2002. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 12(7):1075–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dewannieux M, Heidmann T. 2005. LINEs, SINEs and processed pseudogenes: parasitic strategies for genome modeling. Cytogenet. Genome Res 110(1–4):35–48 [DOI] [PubMed] [Google Scholar]
- 33.Doak TG, Doerder FP, Jahn CL, Herrick G. 1994. A proposed superfamily of transposase genes: transposon-like elements in ciliated protozoa and a common “D35E” motif. Proc. Natl. Acad. Sci 91(3):942–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duret L, Marais G, Biémont C. 2000. Transposons but not retrotransposons are located preferentially in regions of high recombination rate in Caenorhabditis elegans. Genetics. 156(4):1661–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eickbush TH, Eickbush DG. 2015. Integration, Regulation, and Long-Term Stability of R2 Retrotransposons. Microbiol. Spectr 3(2): [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eickbush TH, Malik HS. 2002. Origins and Evolution of Retrotransposons. In Mobile DNA II, ed Craig NL, Craigie R, Gellert M, Lambowitz AM, pp. 1111–44. American Society of Microbiology [Google Scholar]
- 37.Elewa A, Wang H, Talavera-López C, Joven A, Brito G, et al. 2017. Reading and editing the Pleurodeles waltl genome reveals novel features of tetrapod regeneration. Nat. Commun 8(1):2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elliott TA, Gregory TR. 2015. Do larger genomes contain more diverse transposable elements? BMC Evol. Biol 15(1):69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elsik CG, Worley KC, Bennett AK, Beye M, Camara F, et al. 2014. Finding the missing honey bee genes: Lessons learned from a genome upgrade. BMC Genomics. 15(1):86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Engels WR, Johnson-Schlitz DM, Eggleston WB, Sved J. 1990. High-frequency P element loss in Drosophila is homolog dependent. Cell. 62(3):515–25 [DOI] [PubMed] [Google Scholar]
- 41.Esnault C, Lee M, Ham C, Levin HL. 2019. Transposable element insertions in fission yeast drive adaptation to environmental stress. Genome Res. 29(1):85–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Evgen’ev MB, Arkhipova IR. 2005. Penelope-like elements – a new class of retroelements: distribution, function and possible evolutionary significance. Cytogenet. Genome Res 110(1–4):510–21 [DOI] [PubMed] [Google Scholar]
- 43.Evgen’ev MB, Zelentsova H, Shostak N, Kozitsina M, Barskyi V, et al. 1997. Penelope, a new family of transposable elements and its possible role in hybrid dysgenesis in Drosophila virilis. Proc. Natl. Acad. Sci 94(1):196–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fedoroff NV. 2012. Transposable Elements, Epigenetics, and Genome Evolution. Science (80-. ). 338(6108):758–67 [DOI] [PubMed] [Google Scholar]
- 45.Feng Q, Moran JV, Kazazian HH, Boeke JD. 1996. Human L1 Retrotransposon Encodes a Conserved Endonuclease Required for Retrotransposition. Cell. 87(5):905–16 [DOI] [PubMed] [Google Scholar]
- 46.Fernandes JD, Haeussler M, Armstrong J, Tigyi K, Gu J, et al. 2018. KRAB Zinc Finger Proteins coordinate across evolutionary time scales to battle retroelements. bioRxiv [Google Scholar]
- 47.Feschotte C, Gilbert C. 2012. Endogenous viruses: insights into viral evolution and impact on host biology. Nat. Rev. Genet 13(4):283–96 [DOI] [PubMed] [Google Scholar]
- 48.Feschotte C, Pritham EJ. 2005. Non-mammalian c-integrases are encoded by giant transposable elements. Trends Genet. 21(10):551–52 [DOI] [PubMed] [Google Scholar]
- 49.Feschotte C, Pritham EJ. 2007. DNA Transposons and the Evolution of Eukaryotic Genomes. Annu. Rev. Genet 41(1):331–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feschotte C, Zhang X, Wessler SR. 2002. Miniature Inverted-Repeat Transposable Elements and Their Relationship to Established DNA Transposons. In Mobile DNA II, ed Craig N, Craigie R, Gellert M, Lambowitz A, pp. 1147–58. Washington, DC: American Society of Microbiology [Google Scholar]
- 51.Finnegan DJ. 1989. Eukaryotic transposable elements and genome evolution. Trends Genet. 5:103–7 [DOI] [PubMed] [Google Scholar]
- 52.Fischer MG, Hackl T. 2016. Host genome integration and giant virus-induced reactivation of the virophage mavirus. Nature. 540(7632):288–91 [DOI] [PubMed] [Google Scholar]
- 53.Fischer MG, Suttle CA. 2011. A Virophage at the Origin of Large DNA Transposons. Science (80-. ). 332(6026):231–34 [DOI] [PubMed] [Google Scholar]
- 54.Flasch DA, Macia Á, Sánchez L, Ljungman M, Heras SR, et al. 2019. Genome-wide de novo L1 Retrotransposition Connects Endonuclease Activity with Replication. Cell. 177(4):837–851.e28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Flynn JM, Hubley R, Goubert C, Rosen J, Clark AG, et al. 2020. RepeatModeler2: automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Frahry MB, Sun C, Chong RA, Mueller RL. 2015. Low Levels of LTR Retrotransposon Deletion by Ectopic Recombination in the Gigantic Genomes of Salamanders. J. Mol. Evol 80(2):120–29 [DOI] [PubMed] [Google Scholar]
- 57.Frank JA, Feschotte C. 2017. Co-option of endogenous viral sequences for host cell function. Curr. Opin. Virol 25:81–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fricker AD, Peters JE. 2014. Vulnerabilities on the Lagging-Strand Template: Opportunities for Mobile Elements. Annu. Rev. Genet 48(1):167–86 [DOI] [PubMed] [Google Scholar]
- 59.Gao B, Shen D, Xue S, Chen C, Cui H, Song C. 2016. The contribution of transposable elements to size variations between four teleost genomes. Mob. DNA 7(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gilbert C, Feschotte C. 2018. Horizontal acquisition of transposable elements and viral sequences: patterns and consequences. Curr. Opin. Genet. Dev 49(October 2017):15–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Goodwin TJD, Poulter RTM. 2004. A New Group of Tyrosine Recombinase-Encoding Retrotransposons. Mol. Biol. Evol 21(4):746–59 [DOI] [PubMed] [Google Scholar]
- 62.Grabundzija I, Hickman AB, Dyda F. 2018. Helraiser intermediates provide insight into the mechanism of eukaryotic replicative transposition. Nat. Commun 9(1): [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grabundzija I, Messing SA, Thomas J, Cosby RL, Bilic I, et al. 2016. A Helitron transposon reconstructed from bats reveals a novel mechanism of genome shuffling in eukaryotes. Nat. Commun 7(1):10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Guo X, Silva JC. 2008. Properties of non-coding DNA and identification of putative cis-regulatory elements in Theileria parva. BMC Genomics. 9(1):582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hancks DC, Goodier JL, Mandal PK, Cheung LE, Kazazian HH. 2011. Retrotransposition of marked SVA elements by human L1s in cultured cells. Hum. Mol. Genet 20(17):3386–3400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hancks DC, Kazazian HH. 2016. Roles for retrotransposon insertions in human disease. Mob. DNA 7(1):9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heringer P, Kuhn GCS. 2018. Exploring the remote ties between helitron transposases and other rolling circle replication proteins. Int. J. Mol. Sci 19(10): [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hickman AB, Dyda F. 2016. DNA Transposition at Work. Chem. Rev 116(20):12758–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, et al. 2013. The zebrafish reference genome sequence and its relationship to the human genome. Nature. 496(7446):498–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hsia AP, Schnable PS. 1996. DNA sequence analyses support the role of interrupted gap repair in the origin of internal deletions of the maize transposon, MuDR. Genetics. 142(2):603–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang CRL, Burns KH, Boeke JD. 2012. Active Transposition in Genomes. Annu. Rev. Genet 46(1):651–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ianc B, Ochis C, Persch R, Popescu O, Damert A. 2014. Hominoid Composite Non-LTR Retrotransposons—Variety, Assembly, Evolution, and Structural Determinants of Mobilization. Mol. Biol. Evol 31(11):2847–64 [DOI] [PubMed] [Google Scholar]
- 73.Jacobs FMJ, Greenberg D, Nguyen N, Haeussler M, Ewing AD, et al. 2014. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature. 516(7530):242–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jacques P-É, Jeyakani J, Bourque G. 2013. The Majority of Primate-Specific Regulatory Sequences Are Derived from Transposable Elements. PLoS Genet. 9(5):e1003504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jiang N, Wessler SR. 2001. Insertion Preference of Maize and Rice Miniature Inverted Repeat Transposable Elements as Revealed by the Analysis of Nested Elements. Plant Cell. 13(11):2553–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jurka J, Milosavljevic A. 1991. Reconstruction and analysis of human alu genes. J. Mol. Evol 32(2):105–21 [DOI] [PubMed] [Google Scholar]
- 77.Kapitonov VV., Jurka J. 2001. Rolling-circle transposons in eukaryotes. Proc. Natl. Acad. Sci 98(15):8714–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kapitonov VV., Jurka J. 2006. Self-synthesizing DNA transposons in eukaryotes. Proc. Natl. Acad. Sci 103(12):4540–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kapusta A, Suh A, Feschotte C. 2017. Dynamics of genome size evolution in birds and mammals. Proc. Natl. Acad. Sci 114(8):E1460–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kazlauskas D, Varsani A, Koonin EV., Krupovic M. 2019. Multiple origins of prokaryotic and eukaryotic single-stranded DNA viruses from bacterial and archaeal plasmids. Nat. Commun 10(1): [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kent TV, Uzunović J, Wright SI. 2017. Coevolution between transposable elements and recombination. Philos. Trans. R. Soc. B Biol. Sci 372(1736):20160458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Khan H, Smit A, Boissinot S. 2006. Molecular evolution and tempo of amplification of human LINE-1 retrotransposons since the origin of primates. Genome Res. 16(1):78–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Khazina E, Truffault V, Büttner R, Schmidt S, Coles M, Weichenrieder O. 2011. Trimeric structure and flexibility of the L1ORF1 protein in human L1 retrotransposition. Nat. Struct. Mol. Biol 18(9):1006–14 [DOI] [PubMed] [Google Scholar]
- 84.Khazina E, Weichenrieder O. 2009. Non-LTR retrotransposons encode noncanonical RRM domains in their first open reading frame. Proc. Natl. Acad. Sci. U. S. A 106(3):731–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim A, Terzian C, Santamaria P, Pelisson A, Purd’homme N, Bucheton A. 1994. Retroviruses in invertebrates: the gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc. Natl. Acad. Sci 91(4):1285–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kirchner J, Connolly C, Sandmeyer S. 1995. Requirement of RNA polymerase III transcription factors for in vitro position-specific integration of a retroviruslike element. Science (80-. ). 267(5203):1488–91 [DOI] [PubMed] [Google Scholar]
- 87.Kissinger JC, DeBarry J. 2011. Genome cartography: charting the apicomplexan genome. Trends Parasitol. 27(8):345–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kojima KK, Jurka J. 2011. Crypton transposons: Identification of new diverse families and ancient domestication events. Mob. DNA 2(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kojima KK, Seto Y, Fujiwara H. 2016. The wide distribution and change of target specificity of R2 non-LTR retrotransposons in animals. PLoS One. 11(9):e0163496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Koonin EV, Krupovic M. 2017. Polintons, virophages and transpovirons: a tangled web linking viruses, transposons and immunity. Curr. Opin. Virol 25:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kramerov DA, Vassetzky NS. 2011. Origin and evolution of SINEs in eukaryotic genomes. Heredity (Edinb). 107(6):487–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kriegs JO, Churakov G, Jurka J, Brosius J, Schmitz J. 2007. Evolutionary history of 7SL RNA-derived SINEs in Supraprimates. Trends Genet. 23(4):158–61 [DOI] [PubMed] [Google Scholar]
- 93.Krupovic M, Bamford DH, Koonin EV. 2014. Conservation of major and minor jelly-roll capsid proteins in Polinton (Maverick) transposons suggests that they are bona fide viruses. Biol. Direct 9(1):6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Krupovic M, Dolja VV., Koonin EV. 2019. Origin of viruses: primordial replicators recruiting capsids from hosts. Nat. Rev. Microbiol 17(7):449–58 [DOI] [PubMed] [Google Scholar]
- 95.Krupovic M, Koonin EV. 2015. Polintons: a hotbed of eukaryotic virus, transposon and plasmid evolution. Nat. Rev. Microbiol 13(2):105–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Leem Y-E, Ripmaster TL, Kelly FD, Ebina H, Heincelman ME, et al. 2008. Retrotransposon Tf1 Is Targeted to Pol II Promoters by Transcription Activators. Mol. Cell 30(1):98–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lerat E, Brunet F, Bazin C, Capy P. 1999. Is the evolution of transposable elements modular? Genetica. 107(1–3):15–25 [PubMed] [Google Scholar]
- 98.Levy A, Schwartz S, Ast G. 2009. Large-scale discovery of insertion hotspots and preferential integration sites of human transposed elements. Nucleic Acids Res. 38(5):1515–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Li Y, Dooner HK. 2009. Excision of Helitron transposons in maize. Genetics. 182(1):399–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu S, Yeh C-T, Ji T, Ying K, Wu H, et al. 2009. Mu transposon insertion sites and meiotic recombination events co-localize with epigenetic marks for open chromatin across the maize genome. PLoS Genet. 5(11):e1000733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Löber U, Hobbs M, Dayaram A, Tsangaras K, Jones K, et al. 2018. Degradation and remobilization of endogenous retroviruses by recombination during the earliest stages of a germ-line invasion. Proc. Natl. Acad. Sci 115(34):8609–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lorenzi H, Thiagarajan M, Haas B, Wortman J, Hall N, Caler E. 2008. Genome wide survey, discovery and evolution of repetitive elements in three Entamoeba species. BMC Genomics. 9(1):1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 72(4):595–605 [DOI] [PubMed] [Google Scholar]
- 104.Lynch M. 2011. Statistical Inference on the Mechanisms of Genome Evolution. PLoS Genet. 7(6):e1001389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lynch M, Conery JS. 2003. The Origins of Genome Complexity. Science (80-. ). 302(5649):1401–4 [DOI] [PubMed] [Google Scholar]
- 106.Ma J, Devos KM, Bennetzen JL. 2004. Analyses of LTR-Retrotransposon Structures Reveal Recent and Rapid Genomic DNA Loss in Rice. Genome Res. 14(5):860–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Magiorkinis G, Gifford RJ, Katzourakis A, De Ranter J, Belshaw R. 2012. Env-less endogenous retroviruses are genomic superspreaders. Proc. Natl. Acad. Sci 109(19):7385–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Malik HS. 2001. Phylogenetic Analysis of Ribonuclease H Domains Suggests a Late, Chimeric Origin of LTR Retrotransposable Elements and Retroviruses. Genome Res. 11(7):1187–97 [DOI] [PubMed] [Google Scholar]
- 109.Malik HS. 2005. Ribonuclease H evolution in retrotransposable elements. Cytogenet. Genome Res 110(1–4):392–401 [DOI] [PubMed] [Google Scholar]
- 110.McClintock B. 1948. Mutable loci in maize. In Carnegie Institution of Washington Yearbook. 47:155–69. Carnegie Institution of Washington, Cold Spring Harbor, New York [Google Scholar]
- 111.Medstrand P, Van De Lagemaat LN, Mager DL. 2002. Retroelement distributions in the human genome: Variations associated with age and proximity to genes. Genome Res. 12(10):1483–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Meyer TJ, Held U, Nevonen KA, Klawitter S, Pirzer T, et al. 2016. The Flow of the Gibbon LAVA Element Is Facilitated by the LINE-1 Retrotransposition Machinery. Genome Biol. Evol 8(10):3209–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH. 1996. High frequency retrotransposition in cultured mammalian cells. Cell. 87(5):917–27 [DOI] [PubMed] [Google Scholar]
- 114.Naito K, Cho E, Yang G, Campbell MA, Yano K, et al. 2006. Dramatic amplification of a rice transposable element during recent domestication. Proc. Natl. Acad. Sci 103(47):17620–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Naito K, Zhang F, Tsukiyama T, Saito H, Hancock CN, et al. 2009. Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature. 461(7267):1130–34 [DOI] [PubMed] [Google Scholar]
- 116.Neumann P, Novák P, Hoštáková N, Macas J. 2019. Systematic survey of plant LTR-retrotransposons elucidates phylogenetic relationships of their polyprotein domains and provides a reference for element classification. Mob. DNA 10(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nowoshilow S, Schloissnig S, Fei JF, Dahl A, Pang AWC, et al. 2018. The axolotl genome and the evolution of key tissue formation regulators. Nature. 554(7690):50–55 [DOI] [PubMed] [Google Scholar]
- 118.Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin YC, et al. 2013. The Norway spruce genome sequence and conifer genome evolution. Nature. 497(7451):579–84 [DOI] [PubMed] [Google Scholar]
- 119.Ohshima K, Okada N. 2005. SINEs and LINEs: symbionts of eukaryotic genomes with a common tail. Cytogenet. Genome Res 110(1–4):475–90 [DOI] [PubMed] [Google Scholar]
- 120.Ou S, Su W, Liao Y, Chougule K, Agda JRA, et al. 2019. Benchmarking transposable element annotation methods for creation of a streamlined, comprehensive pipeline. Genome Biol. 20(1):275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Palazzo A, Lorusso P, Miskey C, Walisko O, Gerbino A, et al. 2019. Transcriptionally promiscuous “blurry” promoters in Tc1/mariner transposons allow transcription in distantly related genomes. Mob. DNA 10(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Parisot N, Pelin A, Gasc C, Polonais V, Belkorchia A, et al. 2014. Microsporidian genomes harbor a diverse array of transposable elements that demonstrate an ancestry of horizontal exchange with metazoans. Genome Biol. Evol 6(9):2289–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pasquesi GIM, Adams RH, Card DC, Schield DR, Corbin AB, et al. 2018. Squamate reptiles challenge paradigms of genomic repeat element evolution set by birds and mammals. Nat. Commun 9(1):2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pastuzyn ED, Day CE, Kearns RB, Kyrke-Smith M, Taibi AV., et al. 2018. The Neuronal Gene Arc Encodes a Repurposed Retrotransposon Gag Protein that Mediates Intercellular RNA Transfer. Cell. 172(1–2):275–288.e18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Peccoud J, Loiseau V, Cordaux R, Gilbert C. 2017. Massive horizontal transfer of transposable elements in insects. Proc. Natl. Acad. Sci 114(18):4721–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Poulter RTM, Butler MI. 2015. Tyrosine Recombinase Retrotransposons and Transposons. In Mobile DNA III, ed Craig N, Chandler M, Gellert M, Lambowitz A, Rice P, Sandmeyer S, pp. 1271–91. American Society of Microbiology; [DOI] [PubMed] [Google Scholar]
- 127.Pritham EJ, Feschotte C. 2007. Massive amplification of rolling-circle transposons in the lineage of the bat Myotis lucifugus. Proc. Natl. Acad. Sci 104(6):1895–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Pritham EJ, Feschotte C, Wessler SR. 2005. Unexpected Diversity and Differential Success of DNA Transposons in Four Species of Entamoeba Protozoans. Mol. Biol. Evol 22(9):1751–63 [DOI] [PubMed] [Google Scholar]
- 129.Pritham EJ, Putliwala T, Feschotte C. 2007. Mavericks, a novel class of giant transposable elements widespread in eukaryotes and related to DNA viruses. Gene. 390(1–2):3–17 [DOI] [PubMed] [Google Scholar]
- 130.Quadrana L, Etcheverry M, Gilly A, Caillieux E, Madoui M-A, et al. 2019. Transposition favors the generation of large effect mutations that may facilitate rapid adaption. Nat. Commun 10(1):3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Raiz J, Damert A, Chira S, Held U, Klawitter S, et al. 2012. The non-autonomous retrotransposon SVA is trans -mobilized by the human LINE-1 protein machinery. Nucleic Acids Res. 40(4):1666–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Rashkova S, Athanasiadis A, Pardue M-L. 2003. Intracellular Targeting of Gag Proteins of the Drosophila Telomeric Retrotransposons. J. Virol 77(11):6376–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Reiss D, Mialdea G, Miele V, de Vienne DM, Peccoud J, et al. 2019. Global survey of mobile DNA horizontal transfer in arthropods reveals Lepidoptera as a prime hotspot. PLOS Genet. 15(2):e1007965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ribeiro YC, Robe LJ, Veluza DS, dos Santos CMB, Lopes ALK, et al. 2019. Study of VIPER and TATE in kinetoplastids and the evolution of tyrosine recombinase retrotransposons. Mob. DNA 10(1):34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ribet D, Harper F, Dupressoir A, Dewannieux M, Pierron G, Heidmann T. 2008. An infectious progenitor for the murine IAP retrotransposon: Emergence of an intracellular genetic parasite from an ancient retrovirus. Genome Res. 18(4):597–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Richardson S, Doucet A, Kopera H, Moldovan J, Garcia-Perez J, Moran J. 2015. The Influence of LINE-1 and SINE Retrotransposons on Mammalian Genomes. In Mobile DNA III, ed Craig N, Chandler M, Gellert M, Lambowitz A, Rice P, Sandmeyer S, pp. 1165–1208. American Society of Microbiology. 1st ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ros F, Kunze R. 2001. Regulation of activator/dissociation transposition by replication and DNA methylation. Genetics. 157(4):1723–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rubin E, Levy AA. 1997. Abortive gap repair: underlying mechanism for Ds element formation. Mol. Cell. Biol 17(11):6294–6302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shao F, Han M, Peng Z. 2019. Evolution and diversity of transposable elements in fish genomes. Sci. Rep 9(1):15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Shen JJ, Dushoff J, Bewick AJ, Chain FJJ, Evans BJ. 2013. Genomic Dynamics of Transposable Elements in the Western Clawed Frog (Silurana tropicalis). Genome Biol. Evol 5(5):998–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Siguier P, Gourbeyre E, Varani A, Ton-Hoang B, Chandler M. 2015. Everyman’s Guide to Bacterial Insertion Sequences. Microbiol. Spectr 3(2): [DOI] [PubMed] [Google Scholar]
- 142.Smit AFAA, Tóth G, Riggs AD, Jurka J, Toth G, et al. 1995. Ancestral, Mammalian-wide Subfamilies of LINE-1 Repetitive Sequences. J. Mol. Biol 246(3):401–17 [DOI] [PubMed] [Google Scholar]
- 143.Sookdeo A, Hepp CM, Boissinot S. 2018. Contrasted patterns of evolution of the LINE-1 retrotransposon in perissodactyls: The history of a LINE-1 extinction. Mob. DNA 9(1):12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Spradling AC, Bellen HJ, Hoskins RA. 2011. Drosophila P elements preferentially transpose to replication origins. Proc. Natl. Acad. Sci 108(38):15948–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Stitzer MC, Anderson SN, Springer NM, Ross-Ibarra J. 2019. The Genomic Ecosystem of Transposable Elements in Maize. bioRxiv [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sultana T, van Essen D, Siol O, Bailly-Bechet M, Philippe C, et al. 2019. The Landscape of L1 Retrotransposons in the Human Genome Is Shaped by Pre-insertion Sequence Biases and Post-insertion Selection. Mol. Cell 74(3):555–570.e7 [DOI] [PubMed] [Google Scholar]
- 147.Sultana T, Zamborlini A, Cristofari G, Lesage P. 2017. Integration site selection by retroviruses and transposable elements in eukaryotes. Nat. Rev. Genet 18(5):292–308 [DOI] [PubMed] [Google Scholar]
- 148.Sun C, López Arriaza JR, Mueller RL. 2012. Slow DNA Loss in the Gigantic Genomes of Salamanders. Genome Biol. Evol 4(12):1340–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sun C, Mueller RL. 2014. Hellbender genome sequences shed light on genomic expansion at the base of crown salamanders. Genome Biol. Evol 6(7):1818–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tenaillon MI, Hollister JD, Gaut BS. 2010. A triptych of the evolution of plant transposable elements. Trends Plant Sci. 15(8):471–78 [DOI] [PubMed] [Google Scholar]
- 151.Thomas J, Pritham EJ. 2015. Helitrons, the Eukaryotic Rolling-circle Transposable Elements. Microbiol. Spectr 3(4): [DOI] [PubMed] [Google Scholar]
- 152.Trizzino M, Park Y, Holsbach-Beltrame M, Aracena K, Mika K, et al. 2017. Transposable elements are the primary source of novelty in primate gene regulation. Genome Res. 27(10):1623–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Van De Bor V, Hartswood E, Jones C, Finnegan D, Davis I. 2005. gurken and the I Factor Retrotransposon RNAs Share Common Localization Signals and Machinery. Dev. Cell 9(1):51–62 [DOI] [PubMed] [Google Scholar]
- 154.Venner S, Miele V, Terzian C, Biémont C, Daubin V, et al. 2017. Ecological networks to unravel the routes to horizontal transposon transfers. PLOS Biol. 15(2):e2001536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wallau GL, Capy P, Loreto E, Le Rouzic A, Hua-Van A. 2016. VHICA, a New Method to Discriminate between Vertical and Horizontal Transposon Transfer: Application to the Mariner Family within Drosophila. Mol. Biol. Evol 33(4):1094–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wang H, Xing J, Grover D, Hedges DJ, Han K, et al. 2005. SVA Elements: A Hominid-specific Retroposon Family. J. Mol. Biol 354(4):994–1007 [DOI] [PubMed] [Google Scholar]
- 157.Wang L, Dou K, Moon S, Tan FJ, Zhang ZZ. 2018. Hijacking Oogenesis Enables Massive Propagation of LINE and Retroviral Transposons. Cell. 174(5):1082–1094.e12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wegrzyn JL, Lin BY, Zieve JJ, Dougherty WM, Martínez-García PJ, et al. 2013. Insights into the Loblolly Pine Genome: Characterization of BAC and Fosmid Sequences. PLoS One. 8(9):e72439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Whitney KD, Garland T. 2010. Did Genetic Drift Drive Increases in Genome Complexity? PLoS Genet. 6(8):e1001080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, et al. 2007. A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet 8(12):973–82 [DOI] [PubMed] [Google Scholar]
- 161.Wilhelm M, Wilhelm F-X. 2001. Reverse transcription of retroviruses and LTR retrotransposons. Cell. Mol. Life Sci 58(9):1246–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wong WY, Simakov O, Bridge DM, Cartwright P, Bellantuono AJ, et al. 2019. Expansion of a single transposable element family is associated with genome-size increase and radiation in the genus Hydra. Proc. Natl. Acad. Sci 116(46):22915–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wright SI, Agrawal N, Bureau TE. 2003. Effects of recombination rate and gene density on transposable element distributions in Arabidopsis thaliana. Genome Res. 13(8):1897–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Xie W, Gai X, Zhu Y, Zappulla DC, Sternglanz R, Voytas DF. 2001. Targeting of the Yeast Ty5 Retrotransposon to Silent Chromatin Is Mediated by Interactions between Integrase and Sir4p. Mol. Cell. Biol 21(19):6606–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Xiong Y, Eickbush TH. 1990. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 9(10):3353–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Yang G, Nagel DH, Feschotte C, Nathan Hancock C, Wessler SR. 2009. Tuned for transposition: Molecular determinants underlying the hyperactivity of a stowaway MITE. Science (80-. ). 325(5946):1391–94 [DOI] [PubMed] [Google Scholar]
- 167.Yuan Y-W, Wessler SR. 2011. The catalytic domain of all eukaryotic cut-and-paste transposase superfamilies. Proc. Natl. Acad. Sci 108(19):7884–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yutin N, Shevchenko S, Kapitonov V, Krupovic M, Koonin EV. 2015. A novel group of diverse Polinton-like viruses discovered by metagenome analysis. BMC Biol. 13(1):95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Zhang H-H, Peccoud J, Xu M-R-X, Zhang X-G, Gilbert C. 2020. Horizontal transfer and evolution of transposable elements in vertebrates. Nat. Commun 11(1):1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Zhang Y, Romanish MT, Mager DL. 2011. Distributions of Transposable Elements Reveal Hazardous Zones in Mammalian Introns. PLoS Comput. Biol 7(5):e1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zimmerly S, Semper C. 2015. Evolution of group II introns. Mob. DNA 6(1):7. [DOI] [PMC free article] [PubMed] [Google Scholar]


