Abstract
The systematic classification of the cells that compose a tissue or an organ is key to understanding how these cells cooperate and interact as a functional unit. Our capacity to detect features that define cell identity has evolved from morphological and chemical analyses, through the use of predefined genetic markers, to unbiased transcriptomic and epigenetic profiling. The innovative technology of single-cell RNA sequencing (scRNA-seq) enables transcriptional profiling of thousands of individual cells. Since its development, scRNA-seq has been extensively applied to numerous organs and tissues in a wide range of animal models and human samples, thereby providing a plethora of fundamental biological insights into their development, homeostasis, and pathology. In this review, we present the findings of 3 recent studies that employed scRNA-seq to unravel the complexity of cellular composition in mammalian teeth. These findings offer an unprecedented catalogue of cell types in the mouse incisor, which is a convenient model system for studying continuous tooth growth. These studies identified novel cell types in the tooth epithelium and mesenchyme, as well as new markers for known cell types. Computational analyses of the data also uncovered the lineage and dynamics of cell states during ameloblast and odontoblast differentiation during both normal homeostasis and injury repair. The transcriptional differences between the mouse incisor and mouse and human molars uncover species-specific as well as shared features in tooth cell composition. Here, we highlight these findings and discuss important similarities and differences between these studies. We also discuss potential future applications of scRNA-seq in dental research and dentistry. Together, these studies demonstrate how the rapidly evolving technology of scRNA-seq can advance the study of tooth development and function and provide putative targets for regenerative approaches.
Keywords: single-cell analysis, sequence analysis, cell differentiation, tooth components, regeneration, stem cells
Introduction
While tooth function is essential for proper feeding and digestion, this organ has also become an important model system for developmental and regeneration studies. Indeed, many years of research have provided a wealth of information regarding the initiation, patterning, and morphogenesis of teeth (Beadles 1893; Tucker and Sharpe 2004). These studies have also uncovered genetic mechanisms and signaling pathways underlying various pathological conditions (Miletich and Sharpe 2003; Fleischmannova et al. 2008). However, these analyses have focused mainly on regulation at the tissue or cell population level. Consequently, we still know very little about the development of the different cell types of the tooth, the genetic programs that regulate them, and how these different cell types function together.
Mouse teeth are the most commonly used in vivo model to study tooth development and regeneration. This is due to their high availability, their structural and molecular similarity to human teeth, and the accessibility of genetic and imaging tools. Mice have a reduced dental formula with a single incisor and 3 molar teeth per dental quadrant, and they are monophyodont, producing only a single set of teeth. In mice, molars and incisors undergo a similar developmental process (Tucker and Sharpe 2004). However, a striking difference between the 2 tooth types is that the mouse incisor, like that of every Glires species, continues to grow and to replace lost tissue throughout life (Fig. 1). This is achieved by the sustained presence of stem cells, which are capable of giving rise to the various dental cell lineages, including enamel-forming ameloblasts and dentin-forming odontoblasts. In contrast, mouse molars are similar to human teeth and cease to grow after the completion of root development, at which stage their ectoderm-derived ameloblasts terminate their life cycle. These teeth also differ in the structure and function of their periodontium, the unique tissue that anchors them to the jaw.
Figure 1.
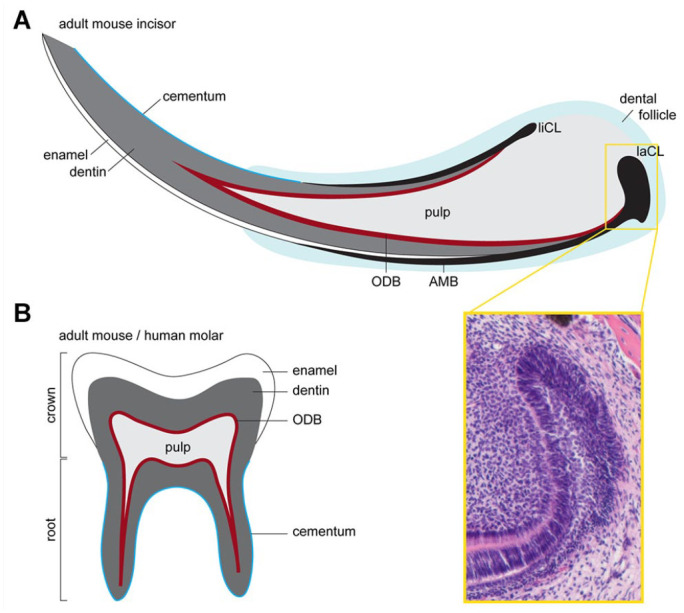
Mouse tooth structure. (A) Schematics of the continuously gowning mouse incisor. This growth is made possible by the presence of epithelial and mesenchymal stem cells residing at the labial cervical loop (laCL) and the dental pulp, respectively. The epithelial stem cells give rise to enamel secreting ameloblasts (AMBs), while mesenchymal progenitors give rise to odontoblasts (ODBs), which secrete dentin, and also to the cementum-producing cementoblasts and periodontal ligaments. The yellow box shows a hematoxylin and eosin staining of the laCL. Ameloblasts are formed on the labial side, whereas the lingual cervical loop (liCL) is less developed and does not normally produce AMBs. (B) Schematic of the adult mouse molar, which shares many morphological features with adult human teeth. The main difference between mouse and human molars to mouse incisors is that the former have a finite growth phase. Once the molar crown is formed, the progenitors in the epithelial cervical loop are gradually lost, and roots, which anchor the molar to the jawbone, are formed. Pulp also fills the molar and is lined by a layer of ODBs. A layer of enamel covers the dentin at the tooth crown, while the roots are covered with cementum.
Understanding the cellular heterogeneity of the mouse incisor and how it arises in comparison to nongrowing teeth is therefore fundamental not only for developmental biology but also for stem cell–based tooth regeneration and bioengineering. The challenge remains to find strategies to regenerate epithelial tissues in human teeth. This is because these tissues disappear upon completion of tooth morphogenesis, with the exception of the epithelial rests of Malassez, remnants of the Hertwig’s epithelial root sheath that are thought to act as stem cells in periodontal regeneration (Tsunematsu et al. 2016). Efforts to develop dental regenerative strategies have been also limited by our incomplete understanding of how and where stem cells are generated and maintained. To expand this understanding, we must first acquire a comprehensive picture of the expression profiles of the various cell types in different teeth.
The recent development of single-cell RNA sequencing (scRNA-seq) technology allows an unbiased transcriptional profiling of thousands of individual cells in many organs and conditions. To illustrate the prospects of scRNA-seq to advance dental research and to create a framework for future studies in this field, we present the findings of 3 recent studies (Sharir et al. 2019; Chiba et al. 2020; Krivanek et al. 2020). These studies used scRNA-seq to uncover a unique catalogue of dental cell types, including newly identified types, and their hierarchical relationships.
A Methodological Comparison of the Reviewed Studies
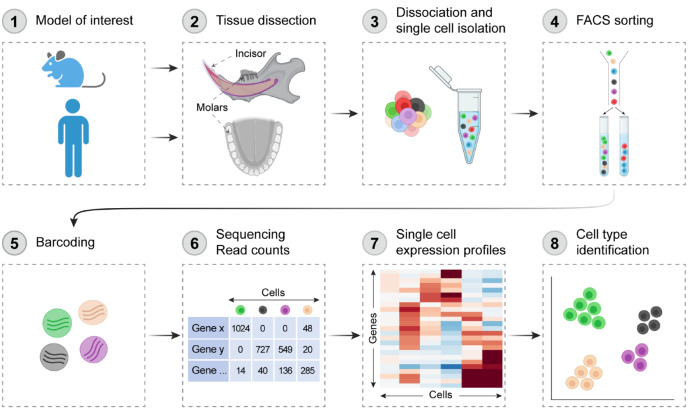
The 3 studies applied well-established scRNA-seq protocols, which include the typical steps outlined in Figure 2. However, the research design and computational analysis of the scRNA-seq data differed in several important aspects. Sharir et al. (2019) and Chiba et al. (2020) sequenced only the mouse incisor epithelium, whereas Krivanek et al. (2020) also sequenced the incisor mesenchyme, the mouse first molar, and human wisdom tooth. Regarding methodology, different approaches for cell clustering and data visualization were employed, as summarized in Table 1. Data set size, clustering methodology, and user-defined parameters have profound effects on the number of identified cell types and hence on the characteristics of each cell type and downstream analyses (Krzak et al. 2019). Thus, these differences should be considered when interpreting and comparing the findings of these studies.
Figure 2.
Workflow of single-cell RNA sequencing analysis of mouse and human teeth. Teeth are excised/isolated (1, 2), and dental tissues are dissociated into single-cell suspensions (3). Cell of interest may be sorted by a fluorescent activated cell sorter (FACS) (4) or directly examined for live cells. Cells are then individually barcoded (5) before complementary DNA libraries are prepared and sequenced. The resulting files undergo rounds of quality control and filtering (6). Cells can then be grouped based on the similarity of their transcriptomes through unsupervised clustering (7), and markers are used to identify cell types in the data set (8). Created with BioRender.com.
Table 1.
Summary of the Main Similarities and Differences in Study Design, Sample Preparation, Single-Cell RNA Sequencing Strategies, and Bioinformatic Analysis Pipelines between the 3 Studies.
| Study | Sharir et al. (2019) | Krivanek et al. (2020) | Chiba et al. (2020) |
|---|---|---|---|
| Species | Mouse | Mouse a | Mouse |
| Strain | C57BL/6N | C57BL/6N; Sox2-RFP | Krt14-RFP |
| Age | 8 wk | 2–4 mo | 7 d |
| Sex | Males | Males and females | Not stated |
| Number of individuals | 5 | 39 | 7 |
| Tooth | Incisor | Incisor a | Incisor |
| Region | Proximal region | Entire tooth | Entire tooth |
| Dissociation enzyme | Collagenase P | Collagenase P | Dispase II |
| Strategy to reduce the impact of cellular stress | FACS with live/dead stain + mitochondrial gene expression regression | Rapid FACS sorting of cells onto plates | No FACS sorting + mitochondrial gene expression regression |
| Number of analyzed cells | 3,173 | 2,889 | 6,260 b |
| scRNA-seq strategy | 10× Chromium | Smart-seq2 | 10× Chromium |
| Clustering method | Spectral clustering of a K-nearest neighbor graph | Hierarchical clustering using Ward method and Pearson correlation distance (PAGODA) | Hierarchical clustering based on Euclidean distance and complete linkage (Seurat) |
| Visualization | SPRING (Weinreb et al. 2018) | t-SNE | t-SNE |
FACS, Fluorescence Activated Cell Sorter; scRNA-seq, single-cell RNA sequencing; t-SNE, t-distributed stochastic neighbor embedding.
Krivanek et al. (2020) also sequenced the incisor mesenchyme, the mouse first molar, and human wisdom tooth.
Including the mesenchyme and immune cells.
Using scRNA-seq to Study the Mouse Incisor Epithelium
Historically, 4 main cell types have been described in the tooth epithelial cervical loop based on their microscopic appearances and spatial locations (Thesleff 2003; Gulabivala and Ng 2014). These include cells at the inner or outer enamel epithelium (IEE or OEE, respectively), star-shaped cells of the stellate reticulum (SR) layer, and cells within a layer between the ameloblasts and the SR known as the stratum intermedium (SI) (Fig. 3A). IEE cells differentiate into enamel-producing ameloblasts. However, despite extensive research on mouse incisor development and homeostasis, the lineage relations and the function of the nonameloblast cell types remain less understood (Liu et al. 2016).
Figure 3.
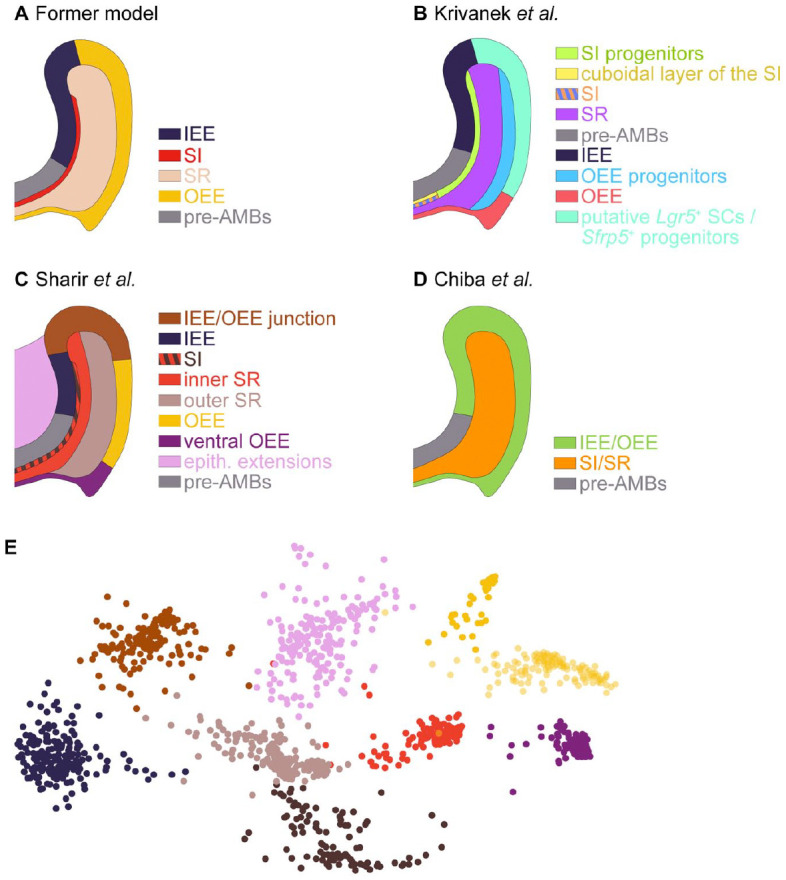
Single-cell RNA sequencing (scRNA-seq) identifies cell types in the mouse incisor epithelium. (A) The former model, which was based on microscopic appearances, posits the existence of 4 cell types in the mouse incisor epithelium, in addition to the ameloblast lineage. Results from Krivanek et al. (2020) (B) and Sharir et al. (2019) (C) indicate that the nonameloblast incisor epithelium is divided into more than the 4 traditional cell types, underscoring the cellular heterogeneity of this tissue. (D) By contrast, Chiba et al. (2020) found a more homogeneous configuration of only 2 cell types: one in the inner and outer enamel epithelium (IEE and OEE, respectively) and another in the stratum intermedium (SI) and stellate reticulum (SR). The schemes are inferred from the transcriptional signatures and marker validations presented in each study. (E) Example of scRNA-seq data from the nonameloblast epithelium. The Spring plot shows the identification of 9 color-coded clusters. The expression of several markers was validated by in situ hybridization. Modified from Sharir et al. (2019).
Using scRNA-seq, Sharir et al. (2019) and Krivanek et al. (2020) have revealed that the mouse incisor epithelium is more heterogeneous than previously thought, containing 8 (Krivanek et al. 2020) or 9 (Sharir et al. 2019) nonameloblast cell subpopulations (Fig. 3). The unique transcriptional signature of each subpopulation included both specific markers and markers that were shared between several subpopulations. By contrast, the nonameloblast epithelium in Chiba et al. (2020) appeared more homogeneous, as the IEE/OEE and SR/SI are each considered a single cell type (Fig. 3D). The small number of cell types reported by Chiba et al. (2020) can be due to the early age of the sampled mice, a stage at which cell types are possibly less demarcated, or, more likely, due to variations in the computational analysis tools and parameters used. For example, the relatively low-resolution value used to cluster the large number of cells in this data set (0.2 compared to the standard 0.4 to 1.2 range recommended when using the FindCluster function in the Seurat package) (Butler et al. 2018). This resolution value does not directly affect the biological identity of the clusters but rather their transcriptional definition. Nevertheless, the combination of this low value and the large number of cells in the data set are likely to cause underrepresentation of cellular heterogeneity.
Once cells are clustered, different methods have been used to assign them to specific cell types, either based on existing markers or using an unbiased approach. Chiba et al. (2020) relied on previously established markers, such as Sfrp5 and Notch1, to annotate the OEE and the SI, respectively. Krivanek et al. (2020) used a combination of established as well as novel markers. For example, they relied on the canonical dental epithelial stem cell markers Sox2, Lrig1, and Bmi1 to designate a presumed stem cell population in the OEE or to enrich for the presumptive epithelial stem cells using a Sox2 GFP reporter mouse. Concurrently, they used newly identified markers such as Cygb and Rhcg to tag a cell population in the SI. In contrast, identification of the various cell groups by Sharir et al. (2019) was independent of any preexisting markers. The results gained through this approach question the existing models of the incisor epithelium composition, as discussed below.
Many of the enriched genes identified by the 3 studies are novel markers, which had not been recognized to mark the nonameloblast epithelium before. While there were differences between studies, of note is the independent identification of keratin 15 (Krt15) in the OEE and Cldn10 in the SI by all 3 groups. Predictions regarding cell type definition were confirmed by in situ imaging, which is an essential step to validate the biological relevance of sequencing data. The spatial distribution of different cell types was examined by either immunohistochemistry (Chiba et al. 2020; Krivanek et al. 2020) or single-molecule RNA hybridization (Sharir et al. 2019; Krivanek et al. 2020). Interrogating the newly identified genes may illuminate the function of each cell type in the nonameloblast epithelium. However, because many of these genes are expressed in more than 1 cell population, we need to develop new genetic tools, such as Cre drivers, to probe their function in the intended population (Buckingham and Meilhac 2011).
A better understanding of the different cell types identified by scRNA-seq also allowed scientists to revisit previous models of stem cell–fueled incisor renewal. Past studies of the mouse incisor epithelium, which were based on work done in hair follicles, posited that quiescent stem cells are located in the proximal portion of the OEE or SR (Harada et al. 1999; Seidel et al. 2010; Juuri et al. 2012). This model suggests that these quiescent stem cells give rise to short-lived transit-amplifying cell in the IEE, which in turn differentiate into preameloblasts and then ameloblasts as the cells move toward the distal end of the incisor and lay down the enamel matrix. However, these earlier studies relied on a few candidate markers and a presumed cellular hierarchy. scRNA-seq offers the opportunity to question this dogma using unbiased information on the spatial organization and lineage relationships of all cell types in the ever-growing rodent incisors. For example, Sharir et al. (2019) concluded that the expression of many of the previously reported stem cell markers, such as Sox2 and Bmi1, was not exclusive to any cell subpopulation. This was confirmed by single-molecule RNA hybridization. Furthermore, integrating sequencing data with results obtained by quantitative kinetics tools, such as label-dilution assays and quantitative 3-dimensional analysis of cell proliferation, indicated that epithelial stem cells in the tooth are in fact not quiescent. This repositions the dental renewal process in line with what is now known about other self-renewing tissues (Clevers and Watt 2018). In this new model, a pool of actively cycling progenitors in the IEE fuels adult renewal of the incisor epithelium. RNA velocity, which uses the abundance of spliced and unspliced messenger RNAs (mRNAs) in single cells to predict their future state (La Manno et al. 2018), was used by Sharir et al. (2019) and Chiba et al. (2020) to independently verify the predicted origin of new cell production in the IEE and the direction of cell flow from the IEE to the rest of the incisor epithelium.
While lineage relationships and cell trajectories can be imputed from transcriptomic data, the gold standard for determining long-term cell dynamics and fate is by genetic lineage tracing (Blanpain and Simons 2013). This technique involves irreversible genetic labeling of specific cell populations (pulse) and then following their progeny at later time points (chase). The lines used for in vivo genetic lineage tracing are listed in Appendix 1. Using this tool, Krivanek et al. (2020) suggested that Fos+/Sox2− OEE cells represent a group of intermediate progenitors, possibly downstream of more long-lasting stem cells. Lineage tracing of the Notch1+ SI cell by Sharir et al. (2019) indicated that at least some of the SR and OEE cells are derived from SI cells and not vice versa, as previously suggested (Harada et al. 2006; Juuri et al. 2012).
Together, these analyses provide a wealth of information on the cellular identity and dynamic nature of different cell types within the mouse incisor epithelium and expand the knowledge base regarding the identity, location, and function of dental epithelial progenitors in this tissue.
Amelogenesis at a Single-Cell Level
During amelogenesis, the process of enamel formation, epithelial-derived preameloblasts undergo a highly regulated differentiation sequence resulting in the formation of a partially mineralized extracellular matrix (Reith 1970). This enamel matrix is then further mineralized to produce the hardest tissue in the body, which covers the crown of the tooth. The life cycle of ameloblasts involves a series of morphological changes and functional adaptations, which ultimately ensure proper enamel formation (Warshawsky and Smith 1974). Amelogenesis is traditionally divided into 4 stages: the presecretory, secretory, transition, and maturation stages (Bartlett 2013). However, this somewhat simplistic view does not fully reflect the genetic and biochemical progression of amelogenesis (Smith 1998). For example, ameloblasts gradually transition between stages, and secretion of enamel matrix proteins and hydroxyapatite is not restricted to the secretory stage.
Information on the genetic program that controls the progression of ameloblasts through these stages has been obtained mainly through candidate gene approaches, in which ameloblasts from patients and animal models exhibiting enamel malformations were analyzed. While this reverse genetic approach may identify specific mutations in individuals, it falls short of revealing the full genetic program involved in coordinating amelogenesis. Over the past decade, a few efforts have been made for unbiased profiling of the transcriptome of amelogenesis using microarrays of rodent incisors and human embryonic tooth buds (Lacruz et al. 2012; Yin et al. 2014; Hu et al. 2015; Seidel et al. 2017). While these investigations revealed hundreds of genes that are expressed by ameloblasts at various differentiation stages, they still represent an average expression of collections of cells at different phases. The detailed description of the sequence of transcriptomic changes during amelogenesis that is provided by scRNA-seq allows for a better mechanistic understanding of this process.
Due to the continuous eruption of the mouse incisor, the successive stages of ameloblasts are present adjacent to each other in the adult tooth. Therefore, the various cell states can be viewed as a continuous lineage that displays a gradient of spatial variability. In scRNA-seq, this spatial information is lost during cell preparation and measurement. Hence, to recover the sequence of transcriptional events during amelogenesis, transcriptomes of individual cells derived from all ameloblast differentiation stages were aligned along a single trajectory, a process also known as pseudotime analysis (Trapnell et al. 2014), which reconstructs the differentiation trajectory according to expression similarity (Fig. 4). As amelogenesis involves a single lineage trajectory, the task of ordering cells throughout the process is relatively straightforward.
Figure 4.
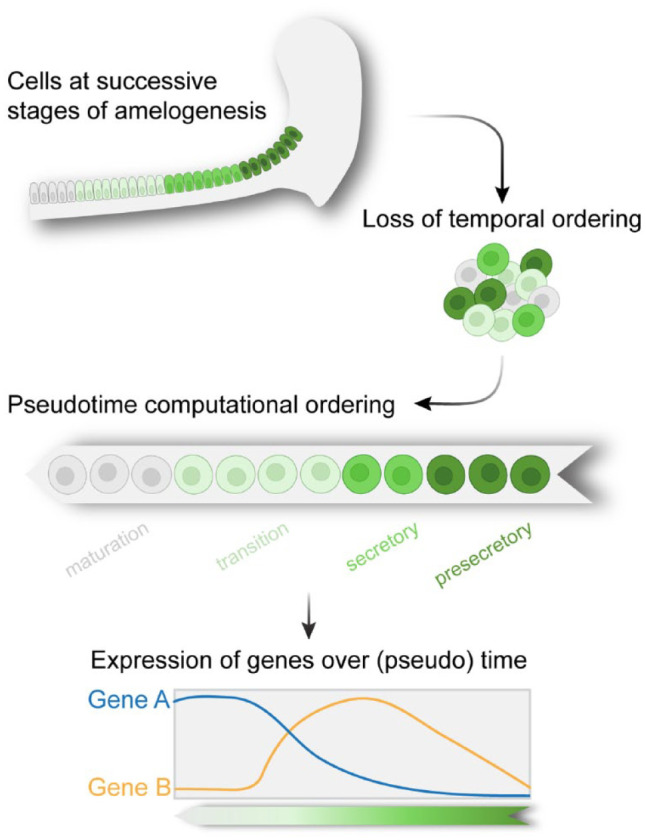
Single-cell RNA sequencing (scRNA-seq) analysis shows the sequence of transcriptional events during amelogenesis. The spatial identity of ameloblasts at different stages of differentiation is lost during single-cell preparations. Pseudotime algorithm reconstructs the spatiotemporal dynamics of the cells and can be used to identify genes that are differentially expressed over (pseudo)time. Created with BioRender.com.
The 3 studies manually defined the source (origin) and sink (endpoint) for the reconstructed lineage trajectory. This prelabeling, as well as the algorithm used to order the cells, differed between the studies, which may explain some differences in the observations. For example, Krivanek et al. (2020) selected the SR as the source of preameloblasts, whereas the other 2 studies defined the IEE as the source of trajectory. Sharir et al. (2019) centered their investigation on the proximal region of the incisor and, hence, their terminal cell identity was a secretory ameloblast. By contrast, the other 2 studies considered the entire process of amelogenesis, and their lineage terminated in postmaturation stage ameloblasts.
The predicted trajectories uncovered the detailed changes in expression of known markers of amelogenesis. In addition, it identified hundreds of genes that displayed variability along the ameloblast differentiation trajectory, many of which represent novel markers of ameloblasts at different differentiation stages. Several of these genes displayed similar trends of expression in pseudotime, being downregulated early, upregulated late, or transiently upregulated. It will be interesting to investigate if these groups of genes also share biological functions or regulators. For example, Krivanek et al. (2020) found a group of cells that appeared during the maturation stage, which expressed several genes encoding ion channels, including Piezo2, Trpm2, Trpm3, and Trpm6. These genes were previously associated with mechanotransduction, the process by which cells convert a mechanical stimulus into chemical signaling (Chen et al. 2004). While the number of these cells did not change upon reducing the occlusal load by trimming the tooth end, further investigation is required to determine the specific role of these genes in ameloblasts.
The Mouse Incisor Mesenchyme
Mesenchymal populations make up most of the mass of the tooth tissue. They include dentin-producing odontoblasts, the pulp, and the dental follicle, also known as periodontium (Schroeder 1986). Both dental pulp and periodontium are gelatinous-like connective tissue with blood supply and innervation and are home to many resident cell types of mesenchymal and ectomesenchymal origin (Goldberg and Smith 2004). Mammalian dental mesenchymal tissues possess a limited capacity to regenerate dentin, cementum, and pulp cells (Gronthos et al. 2000); thus, they play an important role in tooth homeostasis and repair. Many attempts to isolate cells from human dental pulp for regeneration applications have been reported (Miura et al. 2003; Monteiro and Yelick 2019).
The mouse incisor mesenchyme is composed of several cell types, including connective tissue, blood vessels, and nerves. Yet, this cellular heterogeneity had not been considered at the single-cell resolution level. Studies of the incisor mesenchyme found that quiescent cells are located in a region between the labial and lingual epithelial cervical loops. Genetic labeling using several putative stem cell markers, including Gli1, Thy1, Igfbp5, and Lrig1, showed that labeled cells colocalize with the quiescent region of the mesenchyme and give rise to differentiated odontoblasts, pulp cells, and/or dental follicle cells, further supporting this region as the putative stem cell niche (Balic and Mina 2010; Seidel et al. 2017; An et al. 2018). Progeny of glial cells and pericytes have also been suggested as precursors for dental mesenchymal stem cells (Kaukua et al. 2014; Zhao et al. 2014). Recent studies indicated that the various mesenchymal lineages are maintained by different pools of progenitors. For example, a gene coexpression analysis suggested that the outer region of the mesenchyme contains Lrig1-expressing stem cells that contribute solely to periodontium cells (Seidel et al. 2017), whereas stem cells associated with nearby nerves are biased toward the odontoblast lineage (Walker et al. 2019). The diversity of mesenchymal progenitors was also indicated by another study, which employed scRNA-seq to sequence parathyroid hormone–related protein (PTHrP)–expressing cells in the developing molar mesenchyme (Takahashi et al. 2019).
To build on these earlier analyses, scRNA-seq experiments by Krivanek et al. (2020) identified a high level of heterogeneity in the mesenchyme. The anatomical locations of several of the transcriptionally distinct cell types were verified using both known and newly identified markers. Transcriptional trajectory modeling predicted the existence of an active pool of progenitor cells that give rise to 3 main mesenchymal branches—namely, odontoblasts and distal and apical pulp cells—which form a continuum of transient cell states. The apical region contains 2 subpopulations: active progenitor-like cells and upstream stromal-like cells. The existence of subpopulations within the mesenchymal stem cells and the segregation of the apical region into quiescent and active cell populations were also noticed by another recent scRNA-seq study, which found a niche-like subpopulation of Runx2+ cells within the Gli1+ mesenchymal stem cells (Chen et al. 2020).
Analysis of sequencing data also classified Foxd1, expressed by mesenchymal cells located near the epithelial labial cervical loop (laCL), as a novel marker of mesenchymal stem cells. Using lineage tracing, Krivanek et al. (2020) further showed that Foxd1+ cells are specified at the apical region toward either the odontoblast or the pulp lineage.
In agreement with earlier models of odontoblast differentiation (Garant et al. 1968; Ruch et al. 1995; Vidovic et al. 2017), pseudotime analysis revealed at least 4 transcriptional stage transitions, from odontoblast progenitors to preodontoblast, then to early odontoblasts and, finally, to late odontoblasts. Each step was characterized by distinct transcriptomic signatures and transcription factors, which were validated using single-molecule in situ hybridization, providing a useful resource for future investigations of odontogenesis.
Using scRNA-seq to Understand Tooth Repair Process
During homeostasis, to guarantee organ integrity and function, the number of cells generated by resident stem cells must equal the number of differentiated cells that are lost due to physiological wear (Simons and Clevers 2011). By contrast, injury increases the demand for differentiated cells, and in response, stem cells are redirected from a tissue maintenance program toward tissue repair.
To examine how the incisor epithelium transcriptome is affected by injury, Sharir et al. (2019) compared scRNA-seq data between regenerating and uninjured teeth. Proliferating progenitor cells were targeted by treating mice with the chemotherapeutic drug 5-fluorouracil (5-FU), a pyrimidine analogue that causes cell cycle arrest and apoptosis (Longley et al. 2003). To identify cell states that emerge or expand during regeneration, scRNA-seq was conducted 3 d after injury, when the recovery process peaked. A classification-based approach, where each injured cell is given the label of its transcriptionally closest counterpart in the control condition, showed that no new cell states appeared during the repair process. The most striking change was the increase in cell proliferation. Moreover, the increase in the fraction of cycling cells also included cell types that are normally noncycling. Such an increase in the proliferation rate of progenitors, together with recruitment of additional ad hoc progenitors, is a common cellular mechanism in stem cell–based tissue repair (Wabik and Jones 2015). This mechanism often involves a single population of progenitors, which switches from the homeostatic to the regenerative behavior in response to injury (Doupé et al. 2012). However, it is still possible that a pool of dormant incisor stem cells is activated following a more severe injury. For example, Lgr5+ cells at the OEE can be triggered during recovery from ablation of Sox2+ cells (Sanz-Navarro et al. 2018). Furthermore, Sharir et al. (2019) probed the recovery at a single time point, while a cell state may become detectable at a different repair phase, similar to observations in other organs (Plasschaert et al. 2018).
To explore the biological significance of the transcriptional changes in the recovering incisor, differentially expressed genes (DEGs) in each pair of corresponding clusters were computed. While gene ontology (GO) and gene set enrichment analysis (GSEA) did not pinpoint any pathway enrichment, several Wnt antagonists, such as Sfrp5, WISE (Sostdc1), and Wisp1, were enriched during the repair process. Uncovering the role of Wnt signaling in the repair process is an important future direction because Wnt antagonists have been previously implicated in tooth development and enamel regeneration (Ahn et al. 2010; Juuri et al. 2012). Of note, Notch1-expressing cells of the SI have a key function in recovery. The role of SI cells is discussed in detail in Sharir et al. (2019).
Overall, scRNA-seq analysis of both homeostatic and injured incisor epithelia uncovered cellular behaviors as well as composition and transcriptomic changes that enable this tissue to regenerate. Understanding these plastic modifications during endogenous enamel regeneration can be used to develop strategies for stem cell–based tooth renewal.
Comparison of scRNA-seq Analyses of the Mouse Incisor versus the Mouse Molar and Human Teeth
Uncovering species-specific as well as shared features in tooth composition and structure can affirm the use of mouse as a model system for dental studies. Comparing the transcriptional differences between ever-growing and nongrowing adult teeth can also elucidate the transcriptional program that maintains and enables the tooth self-renewal capacity. The study by Krivanek et al. (2020) highlights the differences in cell subtypes and gene expression between the mouse incisor and molar, as well as between mouse and human growing and mature molars.
As expected, the adult mouse molar entirely lacked epithelial cell types, as most of them disappear during tooth eruption (Lungová et al. 2011). However, surprisingly, it also lacked all the stem-like cell types at the incisor apical pulp, such that all the molar mesenchymal pulp cells resembled a single incisor distal pulp cell type. Consistent with the reduced ability of the mouse molar pulp to self-renew, these distal pulp-like cells expressed more genes associated with terminally differentiated cells and low levels of genes that are expressed in the incisor apical pulp. Although both human and mouse adult molars have terminated their growth phase, the human molar mesenchyme was more transcriptionally diverse than that of the mouse. It contained several subpopulations that resembled the incisor mesenchyme, including remnants of a stem cell–like population to be further explored. Interestingly, the human molar exhibited a distinct subpopulation in the periodontal layer that does not appear to have a clear parallel in the mouse dentition. Continued investigation of this unique cell type is urged.
That wisdom teeth continue to grow until later in life enabled Krivanek et al. (2020) to examine whether there are differences between apical and distal pulp cells of mature and growing human molars, as observed in the mouse incisor. Comparing the human molar subpopulations to the apical-distal components of the mouse incisor pulp showed that the pulp of nongrowing human tooth was biased toward the distal pulp with a transcriptional state of more mature cells. In contrast, the pulp of a human growing tooth exhibited a transcriptome that is more apical-like. Of special interest is a group of cells uniquely present in the growing human papilla, which did not share transcriptional similarities with mature nongrowing papilla populations; however, they displayed similarities to the incisor apical pulp in both transcriptomic signatures and anatomic location. This analysis also revealed hundreds of differentially expressed genes, which provide targets for future study aimed at elucidating the evolutionary divergence of the genetic program of ever-growing teeth.
These results led the authors to postulate that in both human and mouse, nongrowing teeth are characterized by a default distal pulp-like state, while an apical-like state is a signature of growing tissue.
Conclusions, Challenges, and Future Directions of scRNA-seq in Dental Research
The reviewed studies represent important first applications of scRNA-seq in dental research. The ability to comprehensively define cell types and states, uncover their functions, and reconstruct their hierarchal dynamics and relationships is changing the way we explore and understand tooth biology. It provides better tools for developmental studies, improved diagnostic tools for dental pathologies, and potential molecular targets for regenerative approaches. In addition to the typical technical and computational difficulties of scRNA-seq experiments, sequencing of dental samples poses unique challenges. Both human and rodent nongrowing teeth contain a limited number of live cells, relative to tissues such as skin or gut, and these cells are difficult to dissociate. This requires pooling cells from a large number of individuals, which can complicate study design, analyses, and data interpretation. Another challenge is that compared to other organs, teeth lack comprehensive references to annotate the various cell types. We anticipate an ever-increasing application of scRNA-seq in dental research. It will be exciting to see how scRNA-seq analyses are integrated with additional multiomics methods to profile epigenetic marks, DNA alterations, protein expression, and metabolite states of tooth cells (Stuart and Satija 2019). Assessing transcriptional signatures while preserving the spatial context, for example, by using 10× Genomics spatial transcriptomics technology, STARmap (Wang et al. 2018), or Slide-seq (Stickels et al. 2020) protocols, is another exciting innovation that is likely to be applied soon to single-cell analysis of dental tissues. Together, these multimodal profiling strategies will provide a more complete understanding of the cellular and molecular mechanisms regulating the development, function, and repair of teeth.
Author Contributions
R. Fresia, P. Marangoni, T. Burstyn-Cohen, A. Sharir, contributed to conception and design, drafted and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, sj-pdf-1-jdr-10.1177_00220345211001848 for From Bite to Byte: Dental Structures Resolved at a Single-Cell Resolution by R. Fresia, P. Marangoni, T. Burstyn-Cohen and A. Sharir in Journal of Dental Research
Acknowledgments
We apologize to our colleagues if their original contributions are missed in the references owing to space limitations. We thank J. Hu, R. Žilionis, and O. D. Klein for helpful feedback and discussions and N. Konstantin for expert editorial assistance.
Footnotes
A supplemental appendix to this article is available online.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: T. Burstyn-Cohen is supported by Israel Science Foundation Grant 655/18.
References
- Ahn Y, Sanderson BW, Klein OD, Krumlauf R. 2010. Inhibition of Wnt signaling by Wise (Sostdc1) and negative feedback from Shh controls tooth number and patterning. Development. 137(19):3221–3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An Z, Sabalic M, Bloomquist RF, Fowler TE, Streelman T, Sharpe PT. 2018. A quiescent cell population replenishes mesenchymal stem cells to drive accelerated growth in mouse incisors. Nat Commun. 9(1):378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balic A, Mina M. 2010. Characterization of progenitor cells in pulps of murine incisors. J Dent Res. 89(11):1287–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett JD. 2013. Dental enamel development: proteinases and their enamel matrix substrates. ISRN Dent. 2013:684607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beadles EP. 1893. Development of the human tooth. Am J Dent Sci. 27(8):351–355. [PMC free article] [PubMed] [Google Scholar]
- Blanpain C, Simons BD. 2013. Unravelling stem cell dynamics by lineage tracing. Nat Rev Mol Cell Biol. 14(8):489–502. [DOI] [PubMed] [Google Scholar]
- Buckingham ME, Meilhac SM. 2011. Tracing cells for tracking cell lineage and clonal behavior. Dev Cell. 21(3):394–409. [DOI] [PubMed] [Google Scholar]
- Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 36(5):411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CS, Tan J, Tien J. 2004. Mechanotransduction at cell-matrix and cell-cell contacts. Annu Rev Biomed Eng. 6(1):275–302. [DOI] [PubMed] [Google Scholar]
- Chen S, Jing J, Yuan Y, Feng J, Han X, Wen Q, Ho T-V, Lee C, Chai Y. 2020. Runx2+ niche cells maintain incisor mesenchymal tissue homeostasis through IGF signaling. Cell Rep. 32(6):108007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba Y, Saito K, Martin D, Boger ET, Rhodes C, Yoshizaki K, Nakamura T, Yamada A, Morell RJ, Yamada Y, et al. 2020. Single-cell RNA-sequencing from mouse incisor reveals dental epithelial cell-type specific genes. Front Cell Dev Biol. 8:841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clevers H, Watt FM. 2018. Defining adult stem cells by function, not by phenotype. Annu Rev Biochem. 87:1015–1027. [DOI] [PubMed] [Google Scholar]
- Doupé DP, Alcolea MP, Roshan A, Zhang G, Klein AM, Simons BD, Jones PH. 2012. A single progenitor population switches behavior to maintain and repair esophageal epithelium. Science. 337(6098):1091–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmannova J, Matalova E, Tucker AS, Sharpe PT. 2008. Mouse models of tooth abnormalities. Eur J Oral Sci. 116(1):1–10. [DOI] [PubMed] [Google Scholar]
- Garant PR, Szabo G, Nalbandian J. 1968. The fine structure of the mouse odontoblast. Arch Oral Biol. 13(8):857–876. [DOI] [PubMed] [Google Scholar]
- Goldberg M, Smith AJ. 2004. Cells and extracellular matrix of dentin and pulp: a biological basis for repair and tissue engineering. Crit Rev Oral Biol Med. 15(1):13–27. [DOI] [PubMed] [Google Scholar]
- Gronthos S, Mankani M, Brahim J, Robey PG, Shi S. 2000. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc Natl Acad Sci U S A. 97(25):13625–13630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulabivala K, Ng Y-L. 2014. Tooth organogenesis, morphology and physiology. In: Gulabivala K, Ng Y-L, editors. Endodontics. 4th ed. St. Louis (MO): Mosby. p. 2–32. [Google Scholar]
- Harada H, Ichimori Y, Yokohama-Tamaki T, Ohshima H, Kawano S, Katsube K, Wakisaka S. 2006. Stratum intermedium lineage diverges from ameloblast lineage via Notch signaling. Biochem Biophys Res Commun. 340(2):611–616. [DOI] [PubMed] [Google Scholar]
- Harada H, Kettunen P, Jung H-S, Mustonen T, Wang YA, Thesleff I. 1999. Localization of putative stem cells in dental epithelium and their association with Notch and FGF signaling. J Cell Biol. 147(1):105–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Parker J, Wright JT. 2015. Towards unraveling the human tooth transcriptome: the dentome. PLoS One. 10(4):e0124801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juuri E, Saito K, Ahtiainen L, Seidel K, Tummers M, Hochedlinger K, Klein OD, Thesleff I, Michon F. 2012. Sox2+ stem cells contribute to all epithelial lineages of the tooth via Sfrp5+ progenitors. Dev Cell. 23(2):317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaukua N, Shahidi MK, Konstantinidou C, Dyachuk V, Kaucka M, Furlan A, An Z, Wang L, Hultman I, Ahrlund-Richter L, et al. 2014. Glial origin of mesenchymal stem cells in a tooth model system. Nature. 513(7519):551–554. [DOI] [PubMed] [Google Scholar]
- Krivanek J, Soldatov RA, Kastriti ME, Chontorotzea T, Herdina AN, Petersen J, Szarowska B, Landova M, Matejova VK, Holla LI, et al. 2020. Dental cell type atlas reveals stem and differentiated cell types in mouse and human teeth. Nat Commun. 11(1):4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzak M, Raykov Y, Boukouvalas A, Cutillo L, Angelini C. 2019. Benchmark and parameter sensitivity analysis of single-cell RNA sequencing clustering methods. Front Genet. 10:1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A, et al. 2018. RNA velocity of single cells. Nature. 560(7719):494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacruz RS, Smith CE, Bringas P, Jr, Chen Y-B, Smith SM, Snead ML, Kurtz I, Hacia JG, Hubbard MJ, Paine ML. 2012. Identification of novel candidate genes involved in mineralization of dental enamel by genome-wide transcript profiling. J Cell Physiol. 227(5):2264–2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Yan X, Pandya M, Luan X, Diekwisch TGH. 2016. Daughters of the enamel organ: development, fate, and function of the stratum intermedium, stellate reticulum, and outer enamel epithelium. Stem Cells Dev. 25(20):1580–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longley DB, Harkin DP, Johnston PG. 2003. 5-Fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer. 3(5):330–338. [DOI] [PubMed] [Google Scholar]
- Lungová V, Radlanski RJ, Tucker AS, Renz H, Míšek I, Matalová E. 2011. Tooth-bone morphogenesis during postnatal stages of mouse first molar development. J Anat. 218(6):699–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miletich I, Sharpe PT. 2003. Normal and abnormal dental development. Hum Mol Genet. 12(Suppl 1):R69–R73. [DOI] [PubMed] [Google Scholar]
- Miura M, Gronthos S, Zhao M, Lu B, Fisher LW, Robey PG, Shi S. 2003. SHED: stem cells from human exfoliated deciduous teeth. Proc Natl Acad Sci U S A. 100(10):5807–5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro N, Yelick PC. 2019. Dental tissue engineering. In: Atala A, Lanza R, Mikos AG, Nerem R, editors. Principles of regenerative medicine. 3rd ed. Boston (MA): Academic Press. p. 907–921. [Google Scholar]
- Plasschaert LW, Žilionis R, Choo-Wing R, Savova V, Knehr J, Roma G, Klein AM, Jaffe AB. 2018. A single cell atlas of the tracheal epithelium reveals the CFTR-rich pulmonary ionocyte. Nature. 560(7718):377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reith EJ. 1970. The stages of amelogenesis as observed in molar teeth of young rats. J Ultrastruct Res. 30(1):111–151. [DOI] [PubMed] [Google Scholar]
- Ruch JV, Lesot H, Bègue-Kirn C. 1995. Odontoblast differentiation. Int J Dev Biol. 39(1):51–68. [PubMed] [Google Scholar]
- Sanz-Navarro M, Seidel K, Sun Z, Bertonnier-Brouty L, Amendt BA, Klein OD, Michon F. 2018. Plasticity within the niche ensures the maintenance of a Sox2+ stem cell population in the mouse incisor. Development. 145(1):dev155929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder HE. 1986. Periodontium, a developmental and functional unit. In: Schroeder HE, editor. The periodontium: handbook of microscopic anatomy. Berlin (Germany): Springer. p. 12–22. [Google Scholar]
- Seidel K, Ahn CP, Lyons D, Nee A, Ting K, Brownell I, Cao T, Carano RAD, Curran T, Schober M, et al. 2010. Hedgehog signaling regulates the generation of ameloblast progenitors in the continuously growing mouse incisor. Development. 137(22):3753–3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidel K, Marangoni P, Tang C, Houshmand B, Du W, Maas RL, Murray S, Oldham MC, Klein OD. 2017. Resolving stem and progenitor cells in the adult mouse incisor through gene co-expression analysis. Elife. 6:e24712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharir A, Marangoni P, Zilionis R, Wan M, Wald T, Hu JK, Kawaguchi K, Castillo-Azofeifa D, Epstein L, Harrington K, et al. 2019. A large pool of actively cycling progenitors orchestrates self-renewal and injury repair of an ectodermal appendage. Nat Cell Biol. 21(9):1102–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons BD, Clevers H. 2011. Strategies for homeostatic stem cell self-renewal in adult tissues. Cell. 145(6):851–862. [DOI] [PubMed] [Google Scholar]
- Smith CE. 1998. Cellular and chemical events during enamel maturation. Crit Rev Oral Biol Med. 9(2):128–161. [DOI] [PubMed] [Google Scholar]
- Stickels RR, Murray E, Kumar P, Li J, Marshall JL, Di Bella DJ, Arlotta P, Macosko EZ, Chen F. 2020. Highly sensitive spatial transcriptomics at near-cellular resolution with Slide-seqV2. Nat Biotechnol [epub ahead of print 7 Dec 2020]. doi: 10.1038/s41587-020-0739-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T, Satija R. 2019. Integrative single-cell analysis. Nat Rev Genet. 20(5):257–272. [DOI] [PubMed] [Google Scholar]
- Takahashi A, Nagata M, Gupta A, Matsushita Y, Yamaguchi T, Mizuhashi K, Maki K, Ruellas AC, Cevidanes LS, Kronenberg HM, et al. 2019. Autocrine regulation of mesenchymal progenitor cell fates orchestrates tooth eruption. Proc Natl Acad Sci U S A. 116(2):575–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thesleff I. 2003. Epithelial-mesenchymal signalling regulating tooth morphogenesis. J Cell Sci. 116(Pt 9):1647–1648. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, Pokharel P, Li S, Morse M, Lennon NJ, Livak KJ, Mikkelsen TS, Rinn JL. 2014. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 32(4):381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsunematsu T, Fujiwara N, Yoshida M, Takayama Y, Kujiraoka S, Qi G, Kitagawa M, Kondo T, Yamada A, Arakaki R, et al. 2016. Human odontogenic epithelial cells derived from epithelial rests of Malassez possess stem cell properties. Lab Invest. 96(10):1063–1075. [DOI] [PubMed] [Google Scholar]
- Tucker A, Sharpe P. 2004. The cutting-edge of mammalian development: how the embryo makes teeth. Nat Rev Genet. 5(7):499–508. [DOI] [PubMed] [Google Scholar]
- Vidovic I, Banerjee A, Fatahi R, Matthews BG, Dyment NA, Kalajzic I, Mina M. 2017. αSMA-expressing perivascular cells represent dental pulp progenitors in vivo. J Dent Res. 96(3):323–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wabik A, Jones PH. 2015. Switching roles: the functional plasticity of adult tissue stem cells. EMBO J. 34(9):1164–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JV, Zhuang H, Singer D, Illsley CS, Kok WL, Sivaraj KK, Gao Y, Bolton C, Liu Y, Zhao M, et al. 2019. Transit amplifying cells coordinate mouse incisor mesenchymal stem cell activation. Nat Commun. 10(1):3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Allen WE, Wright MA, Sylwestrak EL, Samusik N, Vesuna S, Evans K, Liu C, Ramakrishnan C, Liu J, et al. 2018. Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science. 361(6400):eaat5691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warshawsky H, Smith CE. 1974. Morphological classification of rat incisor ameloblasts. Anat Rec. 179(4):423–446. [DOI] [PubMed] [Google Scholar]
- Weinreb C, Wolock S, Klein AM. 2018. SPRING: a Kinetic interface for visualizing high dimensional single-cell expression data. Bioinformatics. 34(7):1246:1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin K, Hacia JG, Zhong Z, Paine ML. 2014. Genome-wide analysis of miRNA and mRNA transcriptomes during amelogenesis. BMC Genomics. 15(1):998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Feng J, Seidel K, Shi S, Klein O, Sharpe P, Chai Y. 2014. Secretion of Shh by a neurovascular bundle niche supports mesenchymal stem cell homeostasis in the adult mouse incisor. Cell Stem Cell. 14(2):160–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-jdr-10.1177_00220345211001848 for From Bite to Byte: Dental Structures Resolved at a Single-Cell Resolution by R. Fresia, P. Marangoni, T. Burstyn-Cohen and A. Sharir in Journal of Dental Research