Abstract
Background
Whole genome sequencing (WGS) is increasingly used for Mycobacterium tuberculosis (Mtb) research. Countries with the highest tuberculosis (TB) burden face important challenges to integrate WGS into surveillance and research.
Methods
We assessed the global status of Mtb WGS and developed a 3-week training course coupled with long-term mentoring and WGS infrastructure building. Training focused on genome sequencing, bioinformatics and development of a locally relevant WGS research project. The aim of the long-term mentoring was to support trainees in project implementation and funding acquisition. The focus of WGS infrastructure building was on the DNA extraction process and bioinformatics.
Findings
Compared to their TB burden, Asia and Africa are grossly underrepresented in Mtb WGS research. Challenges faced resulted in adaptations to the training, mentoring and infrastructure building. Out-of-date laptop hardware and operating systems were overcome by using online tools and a Galaxy WGS analysis pipeline. A case studies approach created a safe atmosphere for students to formulate and defend opinions. Because quality DNA extraction is paramount for WGS, a biosafety level 3 and general laboratory skill training session were added, use of commercial DNA extraction kits was introduced and a 2-week training in a highly equipped laboratory was combined with a 1-week training in the local setting.
Interpretation
By developing and sharing the components of and experiences with a sequencing and bioinformatics training program, we hope to stimulate capacity building programs for Mtb WGS and empower high-burden countries to play an important role in WGS-based TB surveillance and research.
Keywords: Mycobacterium tuberculosis, whole genome sequencing, bioinformatics, Africa, capacity building
Introduction
Tuberculosis (TB) remains a major public health problem with an estimated 10 million new cases annually [1, 2]. Two decades ago, the first complete genome of Mycobacterium tuberculosis (Mtb) was described [3]. Even though bioinformatics and genomics are relatively new biomedical disciplines, they have already made important contributions to the health of patients and populations by mapping TB transmission dynamics and predicting the comprehensive drug resistance profile in individual patients [4]. By unraveling the complete DNA sequence of the Mtb genome, whole genome sequencing (WGS) is also increasingly used in basic science research aimed at understanding the evolution and pathogenicity of Mtb. WGS has thus become an important tool in TB research and surveillance [5].
In Europe and the USA, WGS of Mtb is increasingly being used in routine care settings for species identification, determination of drug resistance profiles and to complement epidemiological source investigation [6, 7]. For example, in 2017, Public Health England introduced WGS in the National Health Service for diagnosis of TB, detection of drug resistance and typing of Mtb strains at the population level [8]. Similarly, the New York State Department of Health’s Wadsworth Centre and the Dutch National Institute for Public Health and the Environment implemented WGS for routine drug resistance profiling [9, 10].
In the past decade, important progress has been made in the standardization of the technical approach to WGS, and its cost has dropped dramatically [5]. Consequently, incorporation of WGS into TB research has now become a realistic option for high TB burden countries. Scientists at reference laboratories and universities in high TB burden countries are thus well positioned to play an important role in WGS-based surveillance and research. Unfortunately, most countries face important challenges when implementing WGS in a research setting and even greater challenges in the context of a clinical setting.
Bringing the scientific and technological advances of genomics to resource-poor countries in a way that is relevant to the local health priorities poses a major challenge [11, 12]. Establishing WGS facilities requires investment in human, laboratory and computational infrastructure [5]. Laboratory and biosafety equipment is needed for Mtb culture, DNA extraction, library preparation and sequencing. In addition to the initial investment in equipment, WGS requires continuous funding to cover the purchase of reagents and instrument maintenance and insurance. Computational infrastructure has to be upgraded to ensure that it is powerful enough to store, transfer and analyse the vast amounts of genomic data generated by WGS. With regard to human resources, WGS research requires a multidisciplinary team with knowledge of relevant laboratory skills, biology (of Mtb) and bioinformatics, and preferably also knowledge of computer sciences, genetics, epidemiology and medicine.
Many countries with a high burden of TB suffer from limited capacities in education and human skills development [13]. Currently, the interdisciplinary field of bioinformatics is still in its infancy in most high TB burden countries, and the number of institutes that offer formal bioinformatics degrees are too few to meet the demand. As a consequence, many universities and reference laboratories in high TB burden countries lack bioinformaticians with experience in Mtb research. Organizing effective training programs to advance the genome sequencing and bioinformatics skills of current academic researchers and staff of reference laboratories will thus be critical to facilitate the integration of WGS into Mtb research and surveillance activities in developing countries [14, 15].
In this article, we present an overview of the global status of Mtb WGS research, outline the development of a training program and highlight the main challenges faced during the first two trainings.
Methods
Global status of Mtb WGS research
To explore the status of Mtb WGS research capacity at the global level, we performed a PubMed search on 3 March 2020, to identify all published manuscripts on WGS of Mtb using the following search terms (‘whole genome sequencing’ AND tuberculosis). We extracted data from 466 eligible articles. We assessed the number of publications over time by geographic location of the samples included in the analysis and by region of affiliation of the first author. To assess for possible imbalance between regional participation in Mtb WGS research and the burden of TB, we compared the location of data collection, location of affiliation of first author and the burden of TB in 2018 (most recent data reported by WHO [1]) between four regions: Europe, North America and Oceania (predominantly Australia and New Zealand); Asia; Africa and South and Central America.
Development of the Mtb WGS and bioinformatics short course
Instead of organizing a ‘fly in, teach and leave’ workshop, which often fails to have durable impact [16], we aimed to build a critical mass of junior researchers and scientists who, upon completion of the training course, would integrate their newly acquired skills into their TB research and/or surveillance work. The short course focused on acquisition of both theoretical and practical skills related to genome sequencing of Mtb, bioinformatics analysis of Mtb WGS data and development of a research project. The short course was complemented with exposure to relevant research using WGS of Mtb and long-term mentoring of trainees. To have maximal impact, the training was intended for a group of 10–15 scholars from academic institutions (preferably employed at lecturer or assistant professor level, at the start of their career) and reference laboratories (preferably holders of a master’s degree).
In addition to human capacity building, the program also aimed to build infrastructure in bioinformatics and sequencing by creating functional bioinformatics and genome sequencing units that use standard operating procedures. WGS has infrastructure requirements for sample preparation, DNA extraction, library preparation, sequencing and data analysis. Because sequencing instruments become outdated very rapidly, we opted to focus on high-quality DNA extraction and data analysis steps. Following DNA extraction, the Mtb DNA can then be shipped for library preparation and sequencing. Outsourcing the library preparation and sequencing steps can be highly cost-effective and is an approach that is also employed by many sequencing research groups in high-income, low TB burden countries.
To test and refine the training program, we built upon an existing collaboration with Jimma University in Ethiopia. Ethiopia is one of the 30 high TB burden countries with an estimated 165 000 new TB cases and 1600 new cases of rifampicin-resistant TB in 2018 [1]. The Tuberculosis Omics ResearCH (TORCH) consortium acquired funding from VLIR-UOS (Flemish Interuniversities Council–—University Development Co-operation) to develop a bioinformatics and sequencing training program through an academic collaboration between Jimma University in Ethiopia, Stellenbosch University in South Africa and the University of Antwerp and the Institute of Tropical Medicine in Belgium.
Results
Global status of Mtb WGS research reporting on original WGS-based TB research
After reviewing title, abstract and full text, 466 articles were eligible for inclusion, with the first manuscript being published in 2009. The number of articles published increased gradually over time and reached a peak of 98 articles in 2018 (Figure 1). Of the 444 manuscripts published on Mtb WGS in the past decade (2009–2019, 2020 excluded), the data collection and performance of research took place in Europe, North America or Australia/New Zealand in 57% (n = 254), Asia in 25% (n = 111), Africa in 11% (n = 51) and South and Central America in 6% (n = 28) of studies. For the African region, most (61%) studies took place in South Africa (31 of the 51 manuscripts).
Figure 1.
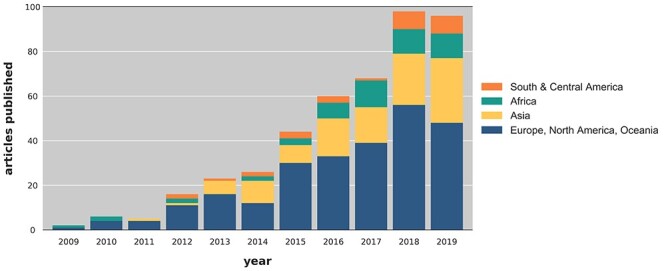
Number of published articles on WGS of Mtb in peer reviewed journals by year and geographic region.
Next, we investigated the region of the affiliation of the first author. Of the 51 manuscripts with study location in Africa, the first author was from an African institution in 33 (65%) of the manuscripts (23 of the 31 papers from South Africa and 10 of 20 manuscripts from other African countries). Of the 28 manuscripts with study location in South or Central America, the first author was from a South or Central American institute in 23 (82%) of the manuscripts. Of the 111 manuscripts with study location in Asia, the first author was from an Asian institute in 104 (94%) of the manuscripts. Lastly, of the manuscripts with study location in Europe, North America or Oceania, all (100%) first authors were affiliated with an institute in one of these regions. In total, the first author was affiliated with Europe, North America or Oceania in 284 (64%) of the manuscripts, while 254 (57%) of the manuscripts had a study location in these regions.
The distribution of the region where the WGS of Mtb data were collected differed from the relative burden of TB (Figure 2). In 2018, 11% of published articles on WGS of Mtb originated from Africa, while the continent accounted for 26% of the global TB burden. Likewise, 23% of articles originated from Asia, while accounting for 69% of the global TB burden. Eight percent of articles originated from South or Central America, while accounting for 3% of the global TB burden. The greatest imbalance occurred in Europe, North America and Oceania, as 57% of articles originated from there even though these regions combined accounted for only 3% of the global TB burden [1].
Figure 2.
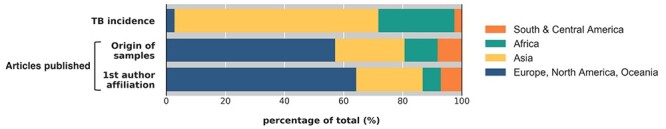
Regional comparison of relative burden of TB (proportion of global TB incidence by region), origin of samples included in research articles on WGS of Mtb published in peer reviewed journals by continent in 2018, and region of first author of such publications.
Initial program goals and development: Mtb WGS, bioinformatics and research proposal writing
The Mtb WGS laboratory training started with a theoretical overview of a typical WGS experiment, the underlying mechanics and applications of different WGS technologies. The training mainly focused on Illumina technology and only briefly touched upon PacBio and Oxford Nanopore Technologies platforms as these are currently less commonly used. During the interactive practical sessions, the trainees received instruction on the sample preparation steps of a sequencing experiment. DNA extraction was performed under supervision using a cetyltrimethylammonium bromide (CTAB) method. M. smegmatis solid cultures were used so that inactivation of the samples could be demonstrated outside of biosafety level 3 (BSL-3) laboratory conditions. Other practical sessions included template DNA quality control, library preparation and clean-up, library quality control and starting a sequencing run on an Illumina MiSeq instrument.
Bioinformatics training included a hands-on session on computing in UNIX operating systems followed by a theoretical session on WGS reference-mapping approaches and a tutorial on variant calling. To demonstrate a standard bioinformatics analysis of Mtb WGS data, we selected the MTBseq pipeline because this is a modular, easy to install, publicly available pipeline consisting of open source software implemented in Perl. MTBseq can be invoked by a single Linux command, is customizable, expandable and can be used without an Internet connection [17]. Next, the WGS reference-mapping training was taught over 2 days. First, a theoretical explanation was given of the process with extensive examples to highlight the advantages and disadvantages for the three primary tasks of strain identification, drug resistance profiling and transmission studies. Hands-on sessions were split into online tools and UNIX tools. PhyResSE and TBProfiler were used for the former, indicating that each has a command line equivalent that trainees can use when more comfortable with UNIX [18, 19]. A Galaxy interface (allowing for UNIX tools to be used through a graphical user interface) was provided for a more lightweight WGS analysis pipeline involving the tools snippy and tb_variant_filter [20–22]. This approach allowed the trainees to see the benefits of WGS without making the UNIX learning curve too steep.
To increase the likelihood that trainees would implement their newly acquired skills after completion of the short course, we included the development of a research proposal by each of the trainees as a component of the course. Trainees were given an overview of past and on-going WGS of Mtb research by the instructors to provide examples of Mtb WGS research. In addition, other opportunities for exposure to relevant research were created through attendance of fora where young researchers present research in the area of bioinformatics, medical informatics and TB research (Biomina lunch talk at the University of Antwerp or annual ‘Acid Fast Club’ at the University of Stellenbosch). For the program, trainees were asked to identify a topic of interest for Mtb WGS research. During an interactive group discussion lead by senior TB researchers (epidemiologist, molecular biologist and bioinformatician), the relevance of WGS for the research question, the relevance of the research question for the local setting and the novelty and feasibility were discussed. A few days later, a similar second session took place, so trainees could present their revised or refined research idea based on the feedback received. Half a day was scheduled to spend in the library to perform a literature review and build the rationale for the proposed question and place their idea in context of published studies. At the end of the 2nd week, students presented the rationale for and expected impact of their research idea. In the 3rd week, research ideas were discussed in an interactive manner to demonstrate how one translates a research idea into specific aims and hypotheses, chooses the optimal study design, study setting and study population to address the specific aim, and how to generate a sample size calculation. After each session, students were asked to apply the information to their own research idea and presented their work at the next session. Finally, issues relating to ethics, risks and risk management of the research projects were highlighted and discussed.
To support the transition of trainees into independent researchers, we aimed to mentor each trainee to support the completion of the research proposal developed during the training. To incentivize this process, each trainee who generated a high-quality research proposal was promised 2100 euro in research funds. We also aimed to assist trainees with the identification of additional freely available software relevant to their research project and to help identify external funding opportunities. We also aimed to establish a network for sustained dialogue among participants, including peer review of research proposals and journal clubs.
Refining the training course based on experiences and trainees’ feedback
In June 2018 and July 2019, 15 individuals from southwest, central and northwest Ethiopia participated in one of the two trainings. During the first two short courses, we faced multiple challenges. Based on the experiences gained and trainees’ feedback on the first training, the program was modified for the 2nd year. After the second training, further adjustments were made to generate the final training program. The main challenges and the solutions implemented to overcome these are discussed below and summarized in Table 1. The complete final program is provided in Table 2.
Table 1.
Challenges and implemented solutions experienced in capacity building for Mtb WGS and bioinformatics in Ethiopia
| Topic | Challenge | Solution |
|---|---|---|
| Identification of junior scientists | Obtaining a gender balance | Lower requirements for female trainees (e.g. BA degree instead of MA) |
| Brain–drain after recruitment process | Complement competitive selection process with targeted selection by university, lower requirements to BA | |
| Bioinformatics training | Computing infrastructure is lacking | Develop WGS pipelines that can run on laptops and expand online tool usage |
| Laboratory skills for DNA extraction | Poor adherence to BSL-3 good laboratory practice | Add a session on good BSL-3 practice |
| Poor basic laboratory skills | Add hands-on session on pipetting skill training | |
| Difficult transition from supervised DNA extraction training to performing DNA extraction in local laboratory setting | Perform 3rd week training in local laboratory and add independent hands-on trainings to the supervised sessions | |
| Equipment for DNA extraction | Lack of reagents and safety equipment (fume hood) for CTAB DNA extraction method in some laboratories | Switch to use of commercially available DNA extraction kits |
| Lack of spectrophotometer to assess quality and quantity of extracted DNA in most laboratories | Purchase spectrophotometer | |
| Identification of research ideas and create research proposal | Communication barrier: limited experience with the use of interactive teaching methods | Awareness of this culture clash, use of experienced instructors, use of case studies and explicit creation of a safe atmosphere where opinions can be voiced |
| Long-term mentoring and creation of trainee network | Poor Internet infrastructure and Internet outages during civil unrest hampered communication and the ability to hold monthly conference and organize e-journal clubs | Use of Slack to create chat rooms by topic and facilitate communication between trainees and between trainees and instructors |
| Transition towards independent scientist | Limited funding available to support WGS-based research projects | Promote use of existing samples for the first research project |
Table 2.
Training program for 3-week short course training on WGS of Mtb bioinformatics for academic and reference laboratory scientists from high burden low-income countries
| Day | Area | Topic | Format | Rationale |
|---|---|---|---|---|
| 1 | General | Introduction to training | Formal presentation | Introduce trainees and instructors give overview of training program |
| General | Overview of WGS research performed in TORCH consortium | Formal presentation | Give overview of WGS research performed by instructors | |
| Genome sequencing | WGS theory | Formal presentation | Teach the theoretical basis underlying WGS | |
| General | Basic BSL-3 and pipetting skills | Hands-on laboratory training | To ensure everyone implements good BSL-3 and basic laboratory practices | |
| 2 | General | Mtb culture in BSL-3 | Hands-on laboratory training | To implement good biosafety and culture practices |
| Genome sequencing | DNA extraction from liquid clinical Mtb culture: heat killing, pelleting and enzymatic lysis steps | Hands-on laboratory training under supervision | Acquire practical skills for DNA extraction | |
| 3 | Genome sequencing | DNA extraction from liquid culture using optimized protocol with Qiagen kit | Hands-on laboratory training under supervision | Acquire practical skills for DNA extraction |
| Genome sequencing | DNA extraction from solid culture: heat killing, pelleting and enzymatic lysis steps | Hands-on laboratory training under supervision | Important to acquire skills to extract DNA from both liquid and solid culture | |
| Genome sequencing | Spectrophotometer to measure DNA quantity and purity | Hands-on laboratory training under supervision | Validation of DNA extraction by measuring DNA quality and quantity (when fluorometer is not available) | |
| Genome sequencing | DNA library preparation | Formal presentation | Presentation on library preparation with emphasis on outsourcing | |
| 4 | Genome sequencing | DNA extraction from solid culture using optimized protocol with Qiagen kit | Hands-on laboratory training under supervision | Important to acquire skills to extract DNA from both liquid and solid culture |
| Genome sequencing | Spectrophotometer to measure DNA quantity and purity | Hands-on laboratory training under supervision | Validation of DNA extraction by measuring DNA quality (spectrophotometer) and quantity (fluorometer) if fluorometer is available | |
| 5 | Research proposal development | Evaluation of research ideas suggested by trainees: relevance for WGS, relevance for Ethiopia, novelty, feasibility | Interactive group discussion led by senior TB researchers | After a week of exposure to WGS, trainees have first opportunity to develop a research idea |
| General | Local research conference | Conference | Expose trainees to presentations of relevant research projects at different stages of completion | |
| 6 | Research proposal development | Evaluation of updated research ideas suggested by trainees: relevance for WGS, relevance for Ethiopia, novelty, feasibility | Interactive group discussion led by senior TB researchers | Based on feedback in first session, trainees present their updated research idea |
| Bioinformatics | UNIX tutorial—using Linux for windows | Hands-on computer training | Acquire basic Linux skills for bioinformatic analysis | |
| 7 | Bioinformatics | Galaxy WGS pipeline—Snippy | Hands-on computer training | Introduce user-friendly bioinformatics platform for basic WGS analysis |
| Bioinformatics | PhyResSE and TBProfiler | Hands-on computer training | Use of freely available online bioinformatics pipeline resources | |
| 8 | Bioinformatics | Phylogeny and phylodynamics of Mtb | Formal presentation | Teach the theoretical basis underlying WGS transmission studies |
| General | Findings of Mtb transmission studies | Formal presentation | Expose trainees to the field of transmission research | |
| Research proposal development | Library time | Literature review | Self-study to update research idea | |
| 9 | Bioinformatics | Data analysis | Hands-on computer training | Wrap up bioinformatics sessions with Q&A |
| Genome sequencing | Library preparation demonstration | Supervised laboratory training | Acquire practical skills to understand where library preparation can go wrong | |
| Genome sequencing | DNA shipping | Formal presentation | Present optimal way to package DNA sample for shipping | |
| 10 | Research proposal development | Rationale, novelty and impact of research idea | Interactive group discussion led by senior TB researchers | Based on literature review, trainees present updated research idea, its rationale, novelty and impact |
| General | Social event | Promote group coherence and social interaction with instructors | ||
| 11 | Genome sequencing | DNA extraction from liquid and solid clinical Mtb culture: heat killing, pelleting and enzymatic lysis steps | Independent hands-on laboratory training | Apply DNA extraction skills in local setting in an independent manner |
| Research proposal development | Develop specific aims for research idea | Interactive group discussion led by senior TB researchers | Teach how a research idea is translated into specific aims using one of the proposed research ideas as case study | |
| Research proposal development | Develop specific aims for research idea | Homework assignment | Apply newly acquired skills to own research idea | |
| 12 | Genome sequencing | DNA extraction from liquid and solid culture using optimized protocol with Qiagen kit | Independent hands-on laboratory training | Apply DNA extraction skills in local setting in an independent manner |
| Genome sequencing | Spectrophotometer to measure DNA quantity and purity | Independent hands-on laboratory training | Apply DNA extraction skills in local setting in an independent manner (if infrastructure available) | |
| 13 | Research proposal development | Develop specific aims for research idea | Interactive group discussion led by senior TB researchers | Group discussion of specific aims developed by each of the trainees |
| Research proposal development | Select appropriate study design, study setting and study population for research idea | Interactive group discussion led by senior TB researchers | Teach how to select the appropriate study design, study setting and study population using one of the proposed research ideas as case study | |
| Research proposal development | Select appropriate study design, study setting and study population for research idea | Homework assignment | Apply newly acquired skills to own research idea | |
| 14 | Research proposal development | Presentation of homework. Formulate hypothesis and generate sample size |
Interactive group discussion led by senior TB researchers | Teach how a specific aim translates into a hypothesis and how to generate a sample size using one of the proposed research ideas as case study |
| Research proposal development | Formulate hypothesis and define assumptions for sample size calculations | Homework assignment | Apply newly acquired skills to own research idea | |
| 15 | Research proposal development | Presentation of homework. Select appropriate study procedures, reflect on ethical issues, risk and risk management strategies |
Interactive group discussion led by senior TB researchers | Teach how to translate a specific aim into study procedures using one of the proposed research ideas as case study. Identify key ethical issues and risks in the different studies proposed by trainees |
| General | Certificate ceremony and social event | Promote group coherence and wrap up training |
Trainees were selected based on their motivation and active involvement in TB research, TB diagnostics or TB surveillance activities. During the selection process, the number of female applicants was low. After the selection process, several trainees dropped out before the start of the training due to a change in employment or emigration. To overcome these challenges, we lowered the requirement from holder of a master’s degree to holder of a relevant bachelor’s degree and complemented open recruitment with hand-picked, targeted recruitment paying special attention to the recruitment of female scientists.
We experienced many challenges with training on the laboratory aspects of Mtb WGS. Even though we recruited trainees from academic and reference laboratories, we observed during the first hands-on training session that the level of experience with and adherence to good BSL-3 practices and general pipetting skill training were not optimal for all trainees. We therefore added a session on BSL-3 practices and a general pipetting skill training session. A follow-up visit to Ethiopia after the first training highlighted greater than expected challenges with DNA extraction. We therefore decided to focus even more practical laboratory training on the DNA extraction process. In the second training, the steps of library preparation and a sequencing run on an Illumina instrument were limited to a theoretical introduction. On-site follow-up of the trainees also demonstrated that laboratory reagents, such as lysozyme, proteinase K and sodium acetate required for the standard CTAB DNA extraction method were not readily available and that a fume hood (needed for safe handling of chloroform and isoamyl alcohol) was not present in all laboratories. We therefore switched to a user-friendly commercial DNEasy Ultraclean Microbial DNA extraction kit (Qiagen, Hilden, Germany), with an added overnight enzymatic lysing step (lysozyme 10 mg/ml) to maximize the amount of Mtb DNA extracted (supplementary data: supplementary file 1). This kit provides a simple and standardized approach suitable for low-income countries with varied levels of laboratory infrastructure. In the second training, trainees used the kit to perform DNA extraction on both solid and liquid bacterial cultures, to highlight the differences between these two culturing methods as starting material. Additionally, we switched to cultures of Mtb instead of M. smegmatis to more accurately represent a real Mtb experiment and simultaneously emphasize good BSL-3 practices. Finally, the validation of the DNA extraction process proved challenging as a functioning spectrophotometer was not available. Funding had to be freed to purchase a new spectrophotometer. Finally, because of the many challenges with the transition of supervised demonstration of DNA extraction and WGS techniques in a well-equipped laboratory during the training to implementation in the home laboratory, we changed from a 3-week training in a well-resourced setting (Antwerp University) to a 2-week training in a well-resourced laboratory (Stellenbosch University) complemented with a 1-week hands-on training in the trainees’ local setting (Jimma University) so that trainees could individually apply their acquired skills in their home laboratory.
With regard to bioinformatics infrastructure, it quickly became clear that the trainees’ personal computers did not meet the hardware specifications required. The computer hardware needed to run the standard bioinformatics pipelines for strain typing, drug resistance profiling and transmission studies was lacking. Furthermore, most existing bioinformatics pipelines are not configured to run on computers with low memory, making even basic analyses impossible. For example, the minimum 8GB RAM required to run MTBseq was not available on many of the trainees’ personal laptops and many had out-of-date operating systems. During the first training, we resolved this by using a local server to run analysis pipelines in a Linux virtual machine. In the second training, we used online tools and a Galaxy implementation of a WGS pipeline running on the Ilifu cloud (https://www.ilifu.ac.za) to circumvent this. Another hurdle was that, for many trainees, the jump from inexperienced computer user to UNIX user for interpreting the WGS outputs was too large. Therefore, for the second training course, the UNIX tutorial was expanded to give more time for trainees to grasp the importance and usefulness of this platform. The switch to online tools proved more intuitive, allowed them to see the benefit of this approach and encouraged them to spend time learning such skills.
During the research proposal development session, it became clear that trainees were not familiar with the interactive teaching methods that are considered integral to higher education in many high-income countries. The lack of experience with peer review, critical thinking, freedom of expression and mentoring in Ethiopian trainees resulted initially in poor interaction during the sessions. Awareness of differences in educational styles and use of experienced instructors allowed the program to still strive towards maximal interaction between the instructor and the trainees and between trainees. In addition, case studies were used to promote collective analyses of a case in order to identify challenges and proposed solutions and created an atmosphere where students felt safe to formulate and defend their opinions. Once this was established, interesting research plans were developed, indicating that trainees were highly capable of being independent researchers once given the correct guidance.
During the long-term follow-up of the 15 trainees, we were faced with several challenges that we either did not expect or underestimated the impact of. One student dropped out during the training and two within a few months after completion of the training. The organization of monthly conference calls and journal e-clubs failed due to poor Internet access and disruption of the Internet access during civil unrest in Ethiopia. In the 2nd year, we switched to the use of Slack, a chat-based system that provides private message boards organized by topic and direct messaging between users. The initial budget of 2100 euro available per trainee proved to be too small for a prospective pilot studies and was raised to make a meaningful project possible. To increase the development of research projects, the use of existing samples was actively promoted in the second training session. Within 1 year of training, nine trainees submitted a fundable proposal. One student successfully obtained additional external funding.
Discussion
We found a gradually increasing number of WGS of Mtb research papers in the past decade but a striking imbalance between the geographic origin of the research and the burden of TB. Specifically, WGS of Mtb research in Europe and North America is hugely overrepresented while the African and Asian regions are grossly underrepresented. Furthermore, ownership of research was unjust, especially for research in the African region. This is similar to findings of a review of genomics research in Africa published between 2004 and 2013, where less than half (47%) of first authors were affiliated with an African institution [23]. This highlights insufficient progress despite the existence of multiple training initiatives, including the African Society for Bioinformatics and Computational Biology founded in 2004 and the H3ABioNet pan-African bioinformatics network founded in 2012 [24–27]. This may be due to the challenges of online courses, including a relatively high drop-out rate [27, 28], or because these courses focus on bioinformatics without hands-on training for the laboratory component of pathogen genomics research.
To achieve equity and maximum impact of the genomics revolution for TB, it is essential that scientists from high TB burden countries lead WGS research of Mtb activities [29]. Local capacity needs to be built in both the laboratory and bioinformatics aspects of Mtb WGS as many institutions continue to suffer from an insufficient number of experienced personnel that can perform, supervise and train others in bioinformatics and sequencing research [25, 26]. To address this dearth of expertise, we built a WGS of Mtb training program on five pillars: combine short course training with long-term mentoring, include both theoretical training and hands-on laboratory training, focus on DNA extraction as library preparation and sequencing can be centralized or outsourced, guide trainees on the development of locally relevant WGS research proposals and use bioinformatics tools that require low computer resources but achieve accurate results. We faced many challenges during the first two trainings. We experienced that trainees were often underprepared for basic bioinformatics instruction and we struggled to maintain long-term commitment from the trainees after the trainings. Many challenges could be overcome by tweaking the program, including a 1-week in-country training, and switching to a commercial DNA extraction kit. Nevertheless, additional resources will be needed to complement training programs by providing secure access to remote computing services, developing light-weight pipelines that can run in resource poor settings, investment in WGS research in high TB burden settings by both national and international agencies and ensuring that trainees are kept up-to-date as the field and tools change rapidly. In the future, one could explore the use of video training to reduce the cost of on-site training for the laboratory component of Mtb WGS. In addition to training on the standard Illumina sequencing technology, other platforms such as the Oxford Nanopore Technologies MinION platform could be included in the training. This technology holds great potential for low-income countries given its portability, ease of use and minimal instrumental investment cost. This WGS technology also has a library preparation method that can be performed in a decentralized laboratory, which allows for the complete WGS workflow to be carried out in the local setting. However, for Mtb, MinION sequencing and sample processing is still in its infancy and would need to be further developed before becoming core to WGS training instead of Illumina technologies [30].
Conclusion
There continues to be an important underrepresentation of scientists from high TB burden countries in Mtb WGS research, especially from the African and Asian continents. While infrastructural issues can in part be overcome by outsourcing the library preparation and sequencing steps, development of expertise in extraction of quality Mtb DNA and acquisition of bioinformatics skills for in-country analysis of WGS data continues to pose great challenges. By developing and sharing the components of a 3-week course on WGS and bioinformatics for TB research, we hope to stimulate the development of such programs and empower scientists from high TB burden countries to play an important role in WGS-based TB surveillance and TB research.
Key Points
Africa and Asia are grossly underrepresented in Mycobacterium tuberculosis WGS research, compared to their tuberculosis burden.
We developed a 3-week training course on WGS of tuberculosis, combined with long-term mentoring and infrastructure building.
We faced several challenges, resulting in iterative adaptations to the training program, mentoring and infrastructure building.
We present our optimized training concept and framework, which could stimulate other capacity building initiatives.
Supplementary Material
Acknowledgments
We thank all the trainees that participated and completed our training programme and provided valuable feedback: Dr. Mulualem Tadesse Jano (Jimma University, Jimma), Muluwork Getahun Worku (Ethiopian Public Health Institute, Addis Ababa), Shewki Moga Siraj (Ethiopian Public Health Institute, Addis Ababa), Adane Worku Shana (Aklilu Lemma Institute of Pathobiology, Addis Ababa), Dr. Solomon Ali Mohammed (Jimma University, Jimma), Abyot Meaza Dasho (Ethiopian Public Health Institute, Addis Ababa), Agumas Shibabaw Tiruye (University of Gondar, Gondar), Getu Balay Adugna (Jimma University, Jimma), Melashu Balew Shiferaw (Amhara Public Health Institute, Bahir Dar), Olifan Zewdie Abil (Wollega University, Nekemte), Setegn Eshetie Kebede (University of Gondar, Gondar), Wubet Birhan Yigzaw (University of Gondar, Gondar), Yihun Mulugeta Alemu (Bahir Dar University, Bahir Dar), Zegeye Bonsa Ayano (Jimma University, Jimma), Mebrat Ejo Kitata (University of Gondar, Gondar). We thank members of the VLIR-UOS Jimma Network University Cooperation for logistic support. We are grateful to the ILIFU research cloud for providing resources to run a Galaxy server and virtual compute cluster during the second training. We like to thank Sabine Chapelle of the Laboratory of Medical Microbiology, University of Antwerp, for her assistance with the training.
Emmanuel Rivière is a PhD candidate at the University of Antwerp. He is interested in conducting systems biology research on tuberculosis.
Tim H. Heupink is a post-doctoral researcher at the University of Antwerp. He is interested in real-time evolutionary dynamics of Mycobacterium tuberculosis in individuals and populations.
Nabila Ismail is a post-doctoral researcher at the Stellenbosch University. She is interested in the evolution of drug resistance in Mycobacterium tuberculosis, especially to new and repurposed drugs.
Anzaan Dippenaar is a post-doctoral researcher at the University of Antwerp. She is interested in using next-generation sequencing approaches to investigate microevolution in the Mycobacterium tuberculosis complex.
Charlene Clarke is a PhD candidate at the Stellenbosch University. She is interested in conducting systems biology research on tuberculosis.
Gemeda Abebe is a professor at the Jimma University and unit director of the Mycobacteriology Research Center in Jimma, Ethiopia. He is interested in conducting systems biology research on tuberculosis.
Peter Van Heusden is a Senior Systems Developer at the South African National Bioinformatics Institute (SANBI). He is interested in pathogen bioinformatics and works to improve accessibility of bioinformatics tools.
Rob Warren is a professor at the Stellenbosch University and unit director of the SAMRC Centre for Tuberculosis Research. He is interested in conducting systems biology research on tuberculosis.
Conor J. Meehan is a lecturer in molecular microbiology at the University of Bradford. He is interested in evolution and epidemiology of microbes, primarily pathogenic mycobacteria.
Annelies Van Rie is a professor at the University of Antwerp and unit director of the Tuberculosis Omics Research Consortium. She is interested in conducting clinical and translational research on tuberculosis.
Contributor Information
Emmanuel Rivière, University of Antwerp.
Tim H Heupink, University of Antwerp.
Nabila Ismail, Stellenbosch University.
Anzaan Dippenaar, University of Antwerp.
Charlene Clarke, Stellenbosch University.
Gemeda Abebe, Mycobacteriology Research Center in Jimma, Ethiopia.
van Peter Heusden, South African National Bioinformatics Institute (SANBI).
Rob Warren, SAMRC Centre for Tuberculosis Research.
Conor J Meehan, University of Bradford.
Annelies Van Rie, Tuberculosis Omics Research Consortium.
Funding
Vlaamse Interuniversitaire Raad-secretariaat voor universitaire ontwikkelingssamenwerking (ET2018JOI008A10); the Research Foundation Flanders under FWO Odysseus (grant G0F8316N); the South African Research Chairs Initiative of the Department of Science and Technology and National Research Foundation of South Africa (64751); the South African Medical Research Council.
Conflict of Interest
The authors declare that there is no conflict of interest regarding the publication of this article.
References
- 1. World Health Organization . Global Tuberculosis Report. 2019. https://apps.who.int/iris/bitstream/handle/10665/329368/9789241565714-eng.pdf?ua=1.
- 2. Dookie N, Rambaran S, Padayatchi N, et al. Evolution of drug resistance in Mycobacterium tuberculosis: a review on the molecular determinants of resistance and implications for personalized care. J Antimicrob Chemother 2018;73(5):1138–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cole ST, Brosch R, Parkhill J, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 1998;393(6685):537–44. [DOI] [PubMed] [Google Scholar]
- 4. Satta G, Lipman M, Smith GP, et al. Mycobacterium tuberculosis and whole-genome sequencing: how close are we to unleashing its full potential? Clin Microbiol Infect 2018;24(6):604–9. [DOI] [PubMed] [Google Scholar]
- 5. Meehan CJ, Goig GA, Kohl TA, et al. Whole genome sequencing of Mycobacterium tuberculosis: current standards and open issues. Nat Rev Microbiol 2019;17(9):533–45. [DOI] [PubMed] [Google Scholar]
- 6. Werf MJ, Kodmon C. Whole-genome sequencing as tool for investigating international tuberculosis outbreaks: a systematic review. Front Public Health 2019;7:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zakham F, Laurent S, Esteves Carreira AL, et al. Whole-genome sequencing for rapid, reliable and routine investigation of Mycobacterium tuberculosis transmission in local communities. Microbes Infect 2019;31:100582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Public Health England . England World Leaders in the Use of Whole Genome Sequencing to Diagnose TB. 2017. https://www.gov.uk/government/news/england-world-leaders-in-the-use-of-whole-genome-sequencing-to-diagnose-tb.
- 9. New York State Department of Health . Wadsworth Center Chosen as a Pilot Site for Mycobacterium tuberculosis complex (MTBC) Whole Genome Sequencing (WGS). 2016. https://www.wadsworth.org/news/wadsworth-center-chosen-as-a-pilot-site-for-mycobacterium-tuberculosis-complex-mtbc-whole-genome-sequencing-wgs.
- 10. Rijksinstituut voor volksgezondheid en mileu . 2019. https://www.rivm.nl/tuberculose/tuberculose-referentielaboratorium.
- 11. Smith AC, Mugabe J, Singer PA, et al. “Harnessing genomics to improve health in Africa”—an executive course to support genomics policy. Health Res Policy Syst 2005;3(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tekola-Ayele F, Rotimi CN. Translational genomics in low- and middle-income countries: opportunities and challenges. Public Health Genomics 2015;18(4):242–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. United Nations . Sustaining Human Progress: Reducing Vulnerabilities and Building Resilience. Human Development Report, 2014.
- 14. Helmy M, Awad M, Mosa KA. Limited resources of genome sequencing in developing countries: challenges and solutions. Appl Transl Genom 2016;9:15–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Karikari TK, Quansah E, Mohamed WM. Widening participation would be key in enhancing bioinformatics and genomics research in Africa. Appl Transl Genom. 2015;6:35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feldon DF, Jeong S, Peugh J, et al. Null effects of boot camps and short-format training for PhD students in life sciences. Proc Natl Acad Sci USA 2017;114(37):9854–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kohl TA, Utpatel C, Schleusener V, et al. MTBseq: a comprehensive pipeline for whole genome sequence analysis of Mycobacterium tuberculosis complex isolates. PeerJ 2018;6:e5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Feuerriegel S, Schleusener V, Beckert P, et al. PhyResSE: a web tool delineating Mycobacterium tuberculosis antibiotic resistance and lineage from whole-genome sequencing data. J Clin Microbiol 2015;53(6):1908–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Coll F, McNerney R, Preston MD, et al. Rapid determination of anti-tuberculosis drug resistance from whole-genome sequences. Genome Med 2015;7(1):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Afgan E, Baker D, Batut B, et al. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update. Nucleic Acids Res 2018;46(W1):W537–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Seemann T. Rapid haploid variant calling and core genome alignment snippy. 2018. GitHub repository. https://github.com/tseemann/snippy.
- 22. Van Heusden P. tb_variant_filter. 2019. GitHub repository. https://github.com/COMBAT-TB/tb_variant_filter.
- 23. Adedokun BO, Olopade CO, Olopade OI. Building local capacity for genomics research in Africa: recommendations from analysis of publications in Sub-Saharan Africa from 2004 to 2013. Glob Health Action 2016;9:31026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Karikari TK, Quansah E, Mohamed WM. Developing expertise in bioinformatics for biomedical research in Africa. Appl Transl Genom 2015;6:31–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mulder NJ, Adebiyi E, Alami R, et al. H3ABioNet, a sustainable pan-African bioinformatics network for human heredity and health in Africa. Genome Res 2016;26(2):271–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tastan Bishop O, Adebiyi EF, Alzohairy AM, et al. Bioinformatics education—perspectives and challenges out of Africa. Brief Bioinform 2015;16(2):355–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gurwitz KT, Aron S, Panji S, et al. Designing a course model for distance-based online bioinformatics training in Africa: the H3ABioNet experience. PLoS Comput Biol 2017;13(10):e1005715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ahmed AE, Awadallah AA, Tagelsir M, et al. Delivering blended bioinformatics training in resource-limited settings: a case study on the university of Khartoum H3ABioNet node. Brief Bioinform 2020;21(2):719–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bah SY, Morang'a CM, Kengne-Ouafo JA, et al. Highlights on the application of genomics and bioinformatics in the fight against infectious diseases: challenges and opportunities in Africa. Front Genet 2018;9:575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cabibbe AM, Spitaleri A, Battaglia S, et al. Application of targeted next generation sequencing assay on a portable sequencing platform for culture-free detection of drug resistant tuberculosis from clinical samples. J Clin Microbiol 2020. https://jcm.asm.org/content/jcm/early/2020/07/20/JCM.00632-20.full.pdf. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


