Abstract
Background:
Accumulated evidence shows that DNA repair gene X-ray repair cross complementing group 1 (XRCC1) may determine individual susceptibility to head and neck cancer (HNC) as a major DNA repair gene. However, the results from previous studies have been conflictive and inconsistent. In order to more accurately estimate and integrate the association between XRCC1 Arg399Gln polymorphism and HNC risk, we conducted a meta-analysis including 14586 subjects.
Methods:
In this meta-analysis, literatures were collected up until September 15, 2020 through multifarious retrieval strategies by searching through electronic databases of PubMed, Cochrane Library, EMBASE, Medline, Web of Science and CNKI. The association between the XRCC1 Arg399Gln polymorphism and HNC was analyzed through calculating summary odds ratios (OR) and 95% confidence intervals (CI).
Results:
Thirty-one studies consisting of 6025 cases and 8561 controls were identified and analyzed. No significant association between XRCC1 Arg399Gln polymorphisms and HNC risk was found under the allelic (OR = 0.94, 95% CI: 0.82-1.07, P = 0.35), homozygous (OR = 0.99, 95% CI: 0.81-1.21, P = 0.91), heterozygous (OR = 1.01, 95% CI: 0.90-1.13, P = 0.91), dominant (OR = 1.05, 95% CI: 0.85-1.29, P = 0.67) or recessive (OR = 0.93, 95% CI: 0.80-1.08, P = 0.35) genetic models in the overall comparison. In addition, subgroup analyses according to tumor site also displayed no significant association between XRCC1 Arg399Gln polymorphisms and HNC risk. However, subgroup analyses based on ethnicity indicated that HNC risk was significantly related to Arg399Gln genetic heterozygous model (OR = 1.21, 95%CI: 1.04-1.42, P = 0.02) and dominant model (OR = 1.27, 95%CI: 1.02-1.60, P = 0.04) in Caucasians populations.
Conclusion:
The results from this meta-analysis suggest that the XRCC1 Arg399Gln variants (Arg/Gln and Arg/Arg+Arg/Gln) may contribute to high HNC risk among Caucasians. Further well-designed studies and larger sample sizes are needed to validate our findings.
Keywords: XRCC1, Arg399gln, polymorphism, head and neck cancer
Introduction
Head and neck cancer (HNC) is the sixth most common cancer, with a 5-year survival rate less than 50%, and includes cancers of the oral cavity, pharynx and larynx. 1 Risk factors for HNC include smoking, alcohol abuse, and high-risk human papilloma virus (HPV) infection, and the most common histologic classification is squamous cell carcinoma. 2 In addition, many studies in recent years have shown that family history, gene polymorphism and other genetic factors play an important role in the occurrence and development of HNC. 3
Recent evidence indicates that DNA damage caused by ultraviolet light, ionizing radiation or environmental chemicals is probably the most important factor in initiating human cancers. 4 DNA damage stimulates the cell to begin the DNA repair process. DNA repair systems are key to maintain genomic integrity and play crucial roles in preventing mutations. X-ray Repair cross complementing Group 1 (XRCC1) is a common DNA repair gene that mainly engages in DNA base excision repair (BER), nucleotide excision repair (NER) and chain fracture repair, 5 Studies have shown that single nucleotide polymorphisms (SNP) in the coding region or population can affect DNA repair ability and is closely related to genetic susceptibility of many tumors including HNC. 5 The most common SNPs leading to amino acid substitution are at exons 6 (Arg194Trp) and 9 (Arg280His) and 10 (Arg399Gln). Interestingly, these amino acid changes may affect protein-protein interactions between XRCC1 and other BER proteins, which in turn may alter DNA repair capabilities. 6
Previous studies have concentrated on the SNP gene of XRCC1 Arg399Gln, which has been shown to be correlated with the risk of several cancers, including HNC. 7,8 In addition, a number of scholars integrated previous research reports and conducted relevant meta-analysis, and finally they came to conflictive and inconsistent conclusions. 9 -11 Wang et al suggest that Arg399Gln variants of XRCC1 are able to contribute to head and neck squamous cell carcinoma risk among Caucasians and to the risk of larynx squamous cell carcinoma. 10 However, XRCC1 Arg399Gln polymorphisms are probably not associated with the increased risk of HNC in Wu’s study. 9 In view of these conflicting results, we conducted an updated meta-analysis to derive reasonable conclusions about the relationship between XRCC1 Arg399Gln polymorphism and HNC risk. Subgroup analyses according to ethnicity, tumor site were performed respectively, which probably provide more comprehensive evidence for the correlation of XRCC1 Arg399Gln polymorphisms with HNC risk.
Materials and Methods
Search Strategy
This meta-analysis was performed according to the PRISMA statement. 12 A comprehensive and systematic literature search was performed up until September 15, 2020 via reasonable retrieval strategies. Two authors worked for completely searching in electronic databases including PubMed, Cochrane Library, EMBASE, Medline, Web of Science and China National Knowledge Internet (CNKI). The following combinations of search terms were used for literature search: “head and neck,” “oral,” “oropharyngeal,” “laryngeal,” “pharyngeal,” “cancer,” “tumor,” “carcinoma,” “x-ray repair cross complementing group 1,” “XRCC1,” “Arg399Gln” and “polymorphism.” We have registered this meta-analysis with INPLASY (https://inplasy.com/), and our registration number is INPLASY202150104.
Inclusion and Exclusion Criteria
The criteria for inclusion of literature in the study were as follows: (1) The study should evaluate the association between XRCC1 Arg399Gln polymorphisms and HNC risk. (2) The studies were published in English. (3) Case-control studies or cohort studies. (4) The studies described sufficient genotype frequencies, which could estimate odds ratios (ORs) and 95% confidence intervals (CIs).
The criteria for exclusion were as follows: (1) Insufficient information about the frequency or quantity of genotypes; (2) duplicate publications; (3) non-human studies, letters, case reports, meta-analysis and review articles.
Data Extraction
The data was collected according to the standard protocol and collated by 2 authors. Information extracted from each study included the first author’s name, year of publication, country, tumor site, ethnicity and origin of the case and control, characteristics of the sample population, and genotype number of the case and control.
Statistical Analysis
The hardy-Weinberg balance (HWE) test in the control group using the goodness-of-fit test (Chi-square test or Fisher exact test) was performed to assess the genetic balance of each study. P > 0.05 indicated no significant imbalance. In order to avoid the inclusion of unknown heterogeneity, subsequent analysis excluded studies that the genotype distribution of XRCC1 gene polymorphism was inconsistent with HWE. Review Manager (RevMan) 5.3 software and STATA 14 software were used to combine odds ratio and 95%CI for this meta-analysis. Publication bias was assessed using Begg’s funnel plot visual inspection or Egger’s inspection in meta-analysis. The heterogeneity of results was estimated by Q test and I 2 statistics. The fixed-effects model and the random effects model were respectively selected for data analysis when I 2 < 50% and I 2 > 50%.
Results
Study Characteristics
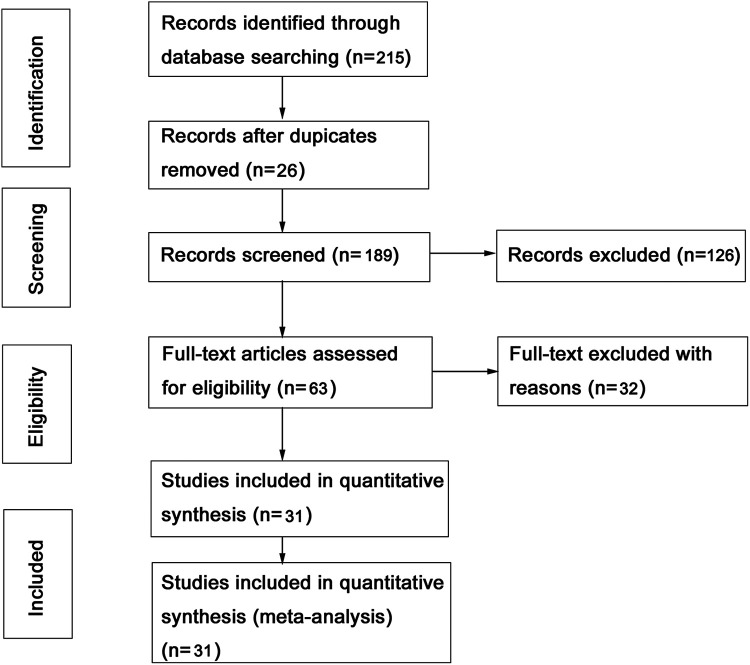
Detailed search strategies were performed to select all eligible articles and the results were summarized in Figure 1. There were a total of 215 potentially relevant studies identified and screened. After screening and reading the full text, 30 publications including 14586 subjects were eventually identified in this meta-analysis for further analysis. 5,13 -41 The main characteristics of each included study were summarized in Table 1. The eligible studies were published from 1999 to 2020, and the most of samples were Caucasians from the Europe or the United States. Unfortunately, there were 3 studies whose P values were less than 0.05 by HWE test, suggesting that the genetic equilibrium might be out of balance, so they were excluded from the subsequent analysis. Subsequently, 9 studies including Asians were respectively from China, India, Thailand and Korea. Among the eligible studies, the tumor types mainly involved head and neck cancer (18 studies), oral (7 studies) and larynx (5 studies). The association of the XRCC1 Arg399Gln polymorphisms with the risk of HNC was summarized in Table 1.
Figure 1.
Flow diagram of the literatures selection procedure in this meta-analysis.
Table 1.
The Main Characteristics of the Eligible Literatures Included in the Meta-Analysis.
| No. of case | No. of control | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| First author | Year | Control source | Country | Ethnicity | Tumor site | N | AA | AG | GG | N | AA | AG | GG | HWE |
| Ammar | 2020 | Hospital | Jordan | Asian | Head & neck | 99 | 67 | 30 | 2 | 89 | 43 | 43 | 3 | 0.047 |
| Applebaum | 2009 | Healthy | USA | Mixed | Head & neck | 483 | 192 | 229 | 62 | 547 | 232 | 246 | 69 | 0.762 |
| Bogela | 2011 | Hospital | China | Asian | Larynx | 58 | 32 | 22 | 4 | 116 | 61 | 48 | 7 | 0.542 |
| Csejtei | 2009 | Healthy | Hungary | Caucasians | Head & neck | 108 | 50 | 47 | 11 | 102 | 53 | 41 | 8 | 0.985 |
| Demokan | 2005 | Healthy | Turkey | Caucasians | Oral | 95 | 42 | 41 | 12 | 98 | 39 | 46 | 13 | 0.922 |
| Dos | 2013 | Healthy | Brazil | Brazilian | Oral | 150 | 64 | 62 | 24 | 150 | 62 | 54 | 34 | 0.002 |
| Gajecka | 2005 | Healthy | Poland | Caucasians | Larynx | 293 | 106 | 153 | 34 | 319 | 124 | 145 | 50 | 0.484 |
| Gugatschka | 2011 | Healthy | Austria | Caucasians | Head & neck | 168 | 70 | 74 | 24 | 463 | 204 | 198 | 61 | 0.24 |
| Hakan | 2017 | Hospital | Turkey | Mixed | Oral | 111 | 44 | 22 | 45 | 148 | 133 | 15 | 0 | 0.516 |
| Harth | 2008 | Healthy | Germany | Caucasians | Head & neck | 310 | 114 | 166 | 30 | 300 | 143 | 121 | 36 | 0.189 |
| He | 2010 | Hospital | China | Asian | Larynx | 72 | 22 | 38 | 12 | 72 | 43 | 22 | 7 | 0.116 |
| Jelonek | 2010 | Healthy | Poland | Caucasians | Head & neck | 104 | 47 | 50 | 7 | 110 | 35 | 62 | 13 | 0.068 |
| Kietthubthew | 2006 | Hospital | Thailand | Asian | Oral | 106 | 55 | 45 | 6 | 164 | 67 | 74 | 23 | 0.724 |
| Kostrzewska-Poczekai | 2013 | Healthy | Poland | Caucasians | Head & neck | 290 | 110 | 154 | 26 | 158 | 50 | 81 | 27 | 0.55 |
| Kowalski | 2009 | Hospital | Poland | Caucasians | Head & neck | 92 | 37 | 44 | 11 | 124 | 49 | 53 | 13 | 0.253 |
| Krupa | 2011 | Hospital | Poland | Caucasians | Larynx | 253 | 93 | 111 | 49 | 253 | 105 | 113 | 35 | 0.238 |
| Kumar | 2012 | Healthy | India | Asian | Head & neck | 278 | 128 | 124 | 26 | 278 | 98 | 144 | 36 | 0.132 |
| Li | 2007 | Hospital | USA | Caucasians | Head & neck | 830 | 335 | 374 | 121 | 854 | 360 | 285 | 109 | 0.577 |
| Majumder | 2005 | Hospital | India | Asian | Oral | 310 | 135 | 143 | 32 | 348 | 158 | 163 | 27 | 0.088 |
| Majumder | 2007 | Healthy | India | Asian | Oral | 309 | 134 | 143 | 32 | 385 | 170 | 179 | 36 | 0.255 |
| Matullo | 2006 | Healthy | Italy | Caucasians | Head & neck | 82 | 34 | 38 | 10 | 1094 | 484 | 482 | 128 | 0.632 |
| Olshan | 2002 | Hospital | USA | Caucasians | Head & neck | 98 | 45 | 50 | 3 | 161 | 62 | 82 | 17 | 0.183 |
| Pelin | 2015 | Hospital | Turkey | Caucasians | Head & neck | 55 | 21 | 27 | 7 | 69 | 22 | 35 | 12 | 0.763 |
| Ramachandran | 2006 | Healthy | India | Asian | Oral | 110 | 46 | 48 | 16 | 110 | 73 | 33 | 4 | 0.91 |
| Rim | 2014 | Hospital | Tunisia | Caucasians | Head & neck | 169 | 12 | 78 | 79 | 261 | 14 | 165 | 82 | 0.001 |
| Rydzanicz | 2005 | Healthy | Poland | Caucasians | Head & neck | 182 | 63 | 98 | 21 | 143 | 59 | 63 | 21 | 0.535 |
| Sturgis | 1999 | Hospital | USA | Mixed | Head & neck | 203 | 94 | 77 | 32 | 424 | 181 | 197 | 46 | 0.483 |
| Tae | 2004 | Hospital | Korea | Asian | Head & neck | 129 | 69 | 51 | 9 | 157 | 86 | 64 | 7 | 0.25 |
| Varzim | 2003 | Healthy | Portugal | Caucasians | Larynx | 88 | 37 | 40 | 11 | 178 | 80 | 80 | 18 | 0.759 |
| Yuan | 2012 | Healthy | China | Asian | Head & neck | 390 | 221 | 146 | 23 | 886 | 481 | 339 | 66 | 0.558 |
Quantitative Data Synthesis
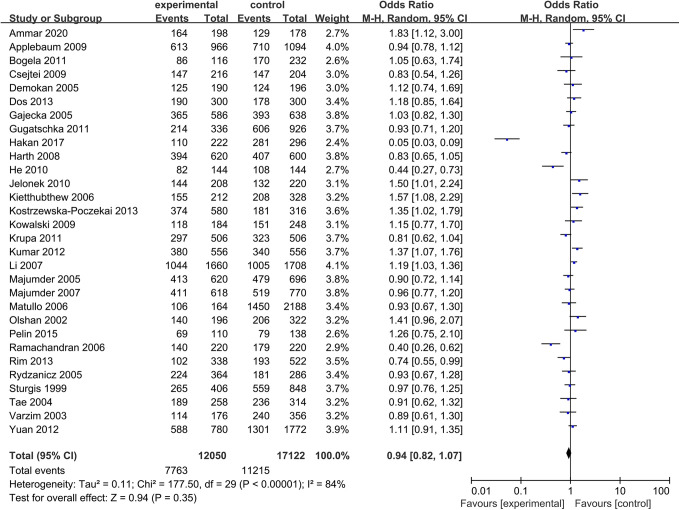
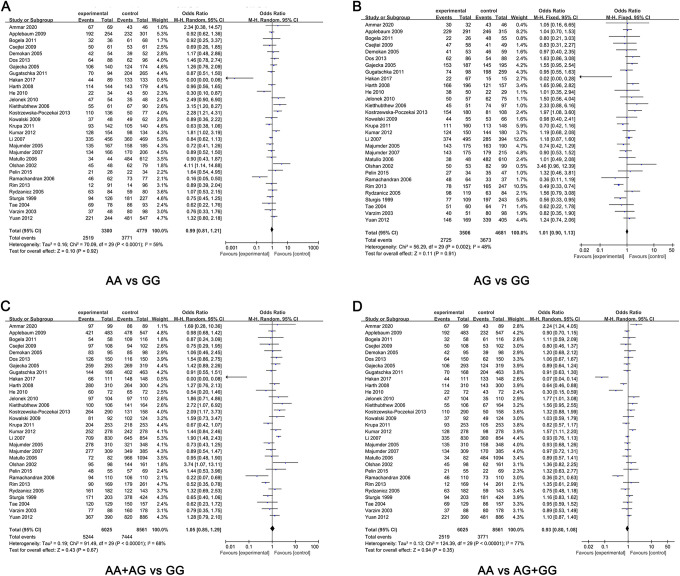
The ORs and heterogeneity tests of XRCC1 Arg399Gln polymorphisms related to HNC risk were summarized in Table 2. The pooled results indicated that no significant associations were found between XRCC1 Arg399Gln polymorphisms and HNC risk (Figure 2). Except heterozygous model, the rest of genetic models used the random-effect model in the subsequent meta analysis according to the heterogeneity analysis. There was no significant connection between XRCC1 Arg399Gln and HNC risk in any genetic model of total populations (Figure 3). The results were as follows, allelic (OR = 0.94, 95% CI: 0.82-1.07, P = 0.35), homozygous (OR = 0.99, 95% CI: 0.81-1.21, P = 0.91), heterozygous (OR = 1.01, 95% CI: 0.90-1.13, P = 0.91), dominant (OR = 1.05, 95% CI: 0.85-1.29, P = 0.67) and recessive (OR = 0.93, 95% CI: 0.80-1.08, P = 0.35). To further optimize our analysis results, we excluded 3 studies that the genotype distribution of XRCC1 gene polymorphism was inconsistent with HWE. However, there were also no significant associations in any genetic models, such as allelic model (OR = 0.92, 95%CI: 0.80-1.06, P = 0.25), heterozygous model (OR = 1.05, 95%CI: 0.93-1.18, P = 0.41), homozygous model (OR = 0.97, 95%CI: 0.78-1.20, P = 0.76), dominant model (OR = 1.06, 95%CI: 0.86-1.31, P = 0.57), recessive model (OR = 0.89, 95%CI: 0.76-1.05, P = 0.16).
Table 2.
Stratified Analyses of the Association of the XRCC1 Arg399Gln Polymorphisms With HNC Risk.
| Comparisons | No. of studies | Test of association | Analysis model | Test of heterogeneity | ||||
|---|---|---|---|---|---|---|---|---|
| OR | 95%CI | P-valuea | χ2 | P-value | I2 (%) | |||
| XRCC1 gene Arg399Gln polymorphism in total populations | ||||||||
| Allelic (A versus G) | 30 | 0.94 | 0.82-1.07 | 0.35 | Random | 177.50 | 0.001 | 84% |
| Heterozygous (AG versus GG) | 30 | 1.01 | 0.90-1.13 | 0.91 | Fixed | 56.29 | 0.002 | 48% |
| Homozygous (AA versus GG) | 30 | 0.99 | 0.81-1.21 | 0.92 | Random | 70.09 | 0.001 | 59% |
| Dominant (AA + AG versus GG) | 30 | 1.05 | 0.85-1.29 | 0.67 | Random | 91.49 | 0.001 | 68% |
| Recessive (AA versus AG + GG) | 30 | 0.93 | 0.80-1.08 | 0.35 | Random | 124.39 | 0.001 | 78% |
| XRCC1 gene Arg399Gln polymorphism in HWE | ||||||||
| Allelic (A versus G) | 27 | 0.92 | 0.80-1.06 | 0.25 | Random | 166.54 | 0.001 | 84% |
| Heterozygous (AG versus GG) | 27 | 1.05 | 0.93-1.18 | 0.41 | Fixed | 41.34 | 0.03 | 37% |
| Homozygous (AA versus GG) | 27 | 0.97 | 0.78-1.20 | 0.76 | Random | 67.57 | 0.001 | 63% |
| Dominant (AA + AG versus GG) | 27 | 1.06 | 0.86-1.31 | 0.57 | Random | 75.83 | 0.001 | 66% |
| Recessive (AA versus AG + GG) | 27 | 0.89 | 0.76-1.05 | 0.16 | Random | 115.14 | 0.001 | 77% |
| XRCC1 gene Arg399Gln polymorphism in Asian populations | ||||||||
| Allelic (A versus G) | 9 | 0.92 | 0.73-1.16 | 0.48 | Random | 41.10 | 0.001 | 81% |
| Heterozygous (AG versus GG) | 9 | 0.98 | 0.78-1.24 | 0.88 | Fixed | 8.9 | 0.35 | 10% |
| Homozygous (AA versus GG) | 9 | 0.87 | 0.54-1.39 | 0.55 | Random | 27.82 | 0.001 | 71% |
| Dominant (AA + AG versus GG) | 9 | 0.92 | 0.64-1.32 | 0.65 | Random | 17.87 | 0.02 | 55% |
| Recessive (AA versus AG + GG) | 9 | 0.91 | 0.69-1.21 | 0.53 | Random | 35.91 | 0.001 | 78% |
| XRCC1 gene Arg399Gln polymorphism in Caucasians populations | ||||||||
| Allelic (A versus G) | 15 | 1.04 | 0.94-1.15 | 0.66 | Fixed | 24.30 | 0.04 | 42% |
| Heterozygous (AG versus GG) | 15 | 1.21 | 1.04-1.42 | 0.02 | Fixed | 15.40 | 0.35 | 9% |
| Homozygous (AA versus GG) | 15 | 1.01 | 0.86-1.18 | 0.91 | Fixed | 21.72 | 0.08 | 36% |
| Dominant (AA + AG versus GG) | 15 | 1.27 | 1.02-1.60 | 0.04 | Random | 27.92 | 0.01 | 50% |
| Recessive (AA versus AG + GG) | 15 | 0.93 | 0.84-1.03 | 0.17 | Fixed | 18.70 | 0.18 | 25% |
| XRCC1 gene Arg399Gln polymorphism in Oral tumor populations | ||||||||
| Allelic (A versus G) | 6 | 0.58 | 0.30-1.12 | 0.11 | Random | 113.46 | 0.001 | 96% |
| Heterozygous (AG versus GG) | 6 | 0.77 | 0.42-1.42 | 0.40 | Random | 14.48 | 0.01 | 65% |
| Homozygous (AA versus GG) | 6 | 0.55 | 0.21-1.46 | 0.23 | Random | 36.84 | 0.001 | 86% |
| Dominant (AA + AG versus GG) | 6 | 0.61 | 0.26-1.43 | 0.26 | Random | 30.67 | 0.001 | 84% |
| Recessive (AA versus AG + GG) | 6 | 0.70 | 0.38-1.29 | 0.25 | Random | 62.09 | 0.001 | 92% |
| XRCC1 gene Arg399Gln polymorphism in Larynx tumor populations | ||||||||
| Allelic (A versus G) | 5 | 0.83 | 0.65-1.07 | 0.15 | Random | 9.99 | 0.04 | 60% |
| Heterozygous (AG versus GG) | 5 | 1.01 | 0.74-1.36 | 0.97 | Fixed | 5.27 | 0.26 | 24% |
| Homozygous (AA versus GG) | 5 | 0.80 | 0.59-1.08 | 0.15 | Fixed | 7.20 | 0.13 | 44% |
| Dominant (AA + AG versus GG) | 5 | 0.90 | 0.68-1.19 | 0.47 | Fixed | 6.25 | 0.18 | 36% |
| Recessive (AA versus AG + GG) | 5 | 0.77 | 0.56-1.07 | 0.13 | Random | 9.59 | 0.05 | 58% |
aThe bold value means P < 0.05.
Figure 2.
Forest plots of the included literatures evaluating the association between XRCC1 Arg399Gln polymorphisms with HNC risk. Arg vs Gln.
Figure 3.
Forest plots of the included literatures evaluating the correlation between XRCC1 Arg399Gln variants with HNC risk. (A) AA vs GG; (B) AG vs GG; (C) AA + AG vs GG; (D) AA vs AG + GG.
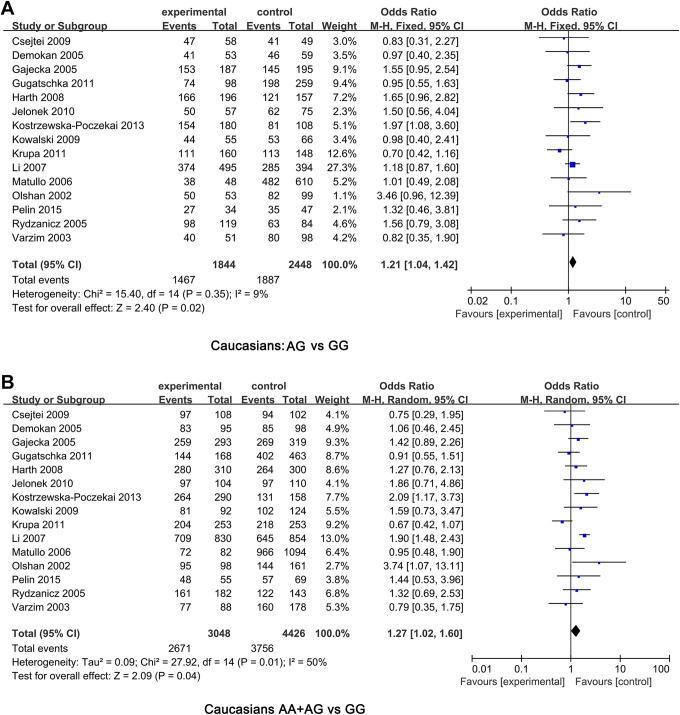
In the overall comparison, we found that there was no significant association between XRCC1 Arg399Gln polymorphisms and HNC risk. So the subgroup analyses respectively based on ethnicity and tumor site were performed to further refine the analysis association between XRCC1 Arg399Gln polymorphisms and HNC risk. The results suggested that XRCC1 Arg399Gln were significantly related to HNC risk in heterozygous model (OR = 1.21, 95%CI: 1.04-1.42, P = 0.02) and dominant model (OR = 1.27, 95%CI:1.02-1.60, P = 0.04) in Caucasians populations (Figure 4). However, no association was shown in heterozygous and recessive models. In addition, the association between XRCC1 gene Arg399Gln polymorphism and HNC risk seemed to be more likely to occur among Caucasians populations than among Asians populations. Unfortunately, based on tumor site, we found that all models showed no significant association between XRCC1 Arg399Gln polymorphism and oral tumor. Meanwhile, no significant association between larynx tumor and XRCC1 Arg399Gln under different genetic models. The detailed results were shown in Table 2. Subsequently, we analyzed the subgroups of oropharyngeal cancer. As we know, numbers of Asians have a habit of chewing betel nut, which is a risk factor for oral cancer. Therefore, subgroup analysis of studies involving Asians suggested that oral cancer and Arg399Gln polymorphism were lack of associations (Table 3).
Figure 4.
The subgroup analyses respectively based on Caucasians populations were performed to further refine the analysis association between XRCC1 Arg399Gln polymorphisms and HNC risk. (A) AG vs GG; (B) AA + AG vs GG.
Table 3.
Subgroup Analyses of the Association of the XRCC1 Arg399Gln Polymorphisms in Asians With Oral Tumors.
| Comparisons | No. of studies | Test of association | Analysis model | Test of heterogeneity | ||||
|---|---|---|---|---|---|---|---|---|
| OR | 95%CI | P-value | χ2 | P-value | I2 (%) | |||
| XRCC1 gene Arg399Gln polymorphism in Asians with oral tumor | ||||||||
| Allelic (A versus G) | 4 | 0.87 | 0.59-1.31 | 0.51 | Random | 21.74 | 0.001 | 86% |
| Heterozygous (AG versus GG) | 4 | 0.89 | 0.52-1.52 | 0.66 | Random | 6.38 | 0.09 | 53% |
| Homozygous (AA versus GG) | 4 | 0.79 | 0.34-1.83 | 0.58 | Random | 15.54 | 0.001 | 81% |
| Dominant (AA + AG versus GG) | 4 | 0.83 | 0.41-1.68 | 0.60 | Random | 11.72 | 0.008 | 74% |
| Recessive (AA versus AG + GG) | 4 | 0.87 | 0.55-1.35 | 0.53 | Random | 15.54 | 0.001 | 81% |
Sensitivity Analysis and Publication Bias
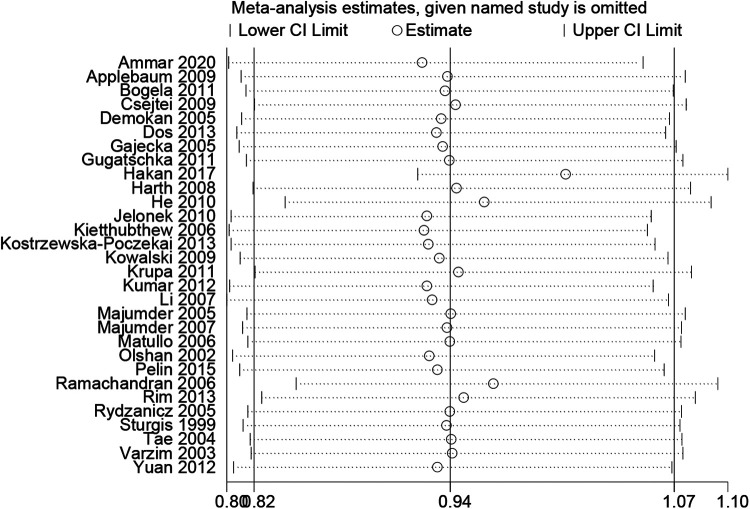
Sensitivity analysis was performed to assess the robustness of the results of the meta-analysis. We found that the study of Hakan seemed to influence the merged results, however, the lower CI limit did not cross the middle line and the circle of estimate did not beyond the upper CI limit, indicating Hakan’s study had less influences on merged results. The final results indicated that there was no substantial change in merged ORs, suggesting that no single study significantly influenced the outcome of the merged results (Figure 5).
Figure 5.
Sensitivity analysis for pooled results in this meta-analysis.
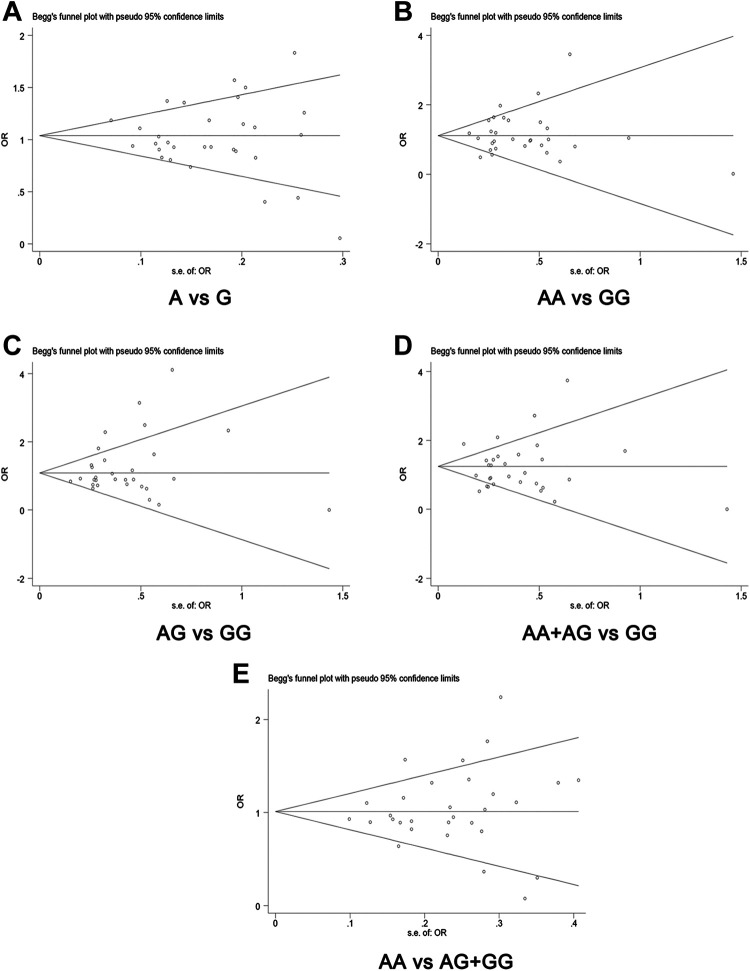
To assess publication bias statistically, the Begg’s funnel plot and the Egger’s regression method was performed to analyze the bias. The results showed that statistical evidence of publication bias did not exist (Table 4). Funnel plot analysis also did not show any strong publication bias, since visual inspection funnel plot did not show an asymmetric comparison model, indicating that the results were indeed feasible (Figure 6).
Table 4.
Results of Publication Bias by Egger’s and Begg’s Test for the Arg399Gln Polymorphism With HNC Risk.
| A vs G | GA vs GG | AA vs GG | AA + GA vs GG | AA vs GA + GG | ||
|---|---|---|---|---|---|---|
| Begg’s test | z | 0.21 | 0.29 | 1.36 | 1.07 | 1.03 |
| P-value | 0.830 | 0.775 | 0.175 | 0.284 | 0.301 | |
| Egger’s test | t | −0.55 | 0.68 | 1.55 | −0.58 | 0.70 |
| P-value | 0.586 | 0.499 | 0.132 | 0.565 | 0.489 |
Figure 6.
Publication bias for pooled results in this meta-analysis. (A) A vs G; (B) AA vs GG; (C) AG vs GG; (D) AA + AG vs GG; (E) AA vs AG + G.
Discussion
XRCC1, an important component of basilectomy repair system, plays a key role in protecting cells from DNA damage and maintaining genomic integrity. In recent years, many studies have shown that common genetic polymorphisms in XRCC1 gene are significant correlated with development and procession of cancer, such as colorectal carcinoma, 42 lung cancer, 43 breast cancer 44 and HNC. Since 1999, accumulated evidence has identified specific associations between XRCC1 polymorphisms and an increased risk of HNC. A meta-analysis study has shown that XRCC1 Arg399Gln SNP is a high risk factor for lymphocytic leukemia in Asian children. 45 Among them, XRCC1 Arg399Gln, as the most common type of polymorphisms, has been widely studied. However, different studies often draw inconsistent conclusions, which might not effectively explain its correlation with the increased risk of HNC.
Emerging evidence has shown XRCC1 Arg399Gln polymorphism is associated with the risk of HNC, and the frequency of Arg allele is significantly higher in the HNC patients than normal peoples, suggesting that allele may act as a genetic biomarker for HNC. 5 Meanwhile, Hakan Avci’s study indicated that Gln/Gln genotype of XRCC1 Arg399Gln polymorphism and Gln allele were high risk factor for oral squamous cell carcinoma. 20 In addition, another study also suggested that 399Gln allele increased over 3-fold the risk of local disease relapse for irradiated oral and oropharyngeal patients, indicating this polymorphism was related to poor prognosis. 7 However, Arg399Gln polymorphisms were defined as invalid and had no significant correlation with the development of HNC. 34 Collectively, the function and association of Arg399Gln polymorphisms with the risk of HNC were contradictory and inconsistent. So we conducted this meta-analysis to pool the latest results and try to uncover strong evidence for the association between Arg399Gln polymorphisms and HNC susceptibility.
In this meta-analysis, the results indicated that the interaction of HNC and Arg399Gln variant genotypes displayed no statistical significance in all genetic models with a overall analysis. This conclusion is same to Wei Wu’ meta-analysis in 2014 and Yadong Wang’ meta-analysis in 2013. 9,10 Further, in the subgroup analyses based on ethnicity and tumor site, interestingly, we found that there were significant associations between HNC susceptibility and Arg399Gln with heterozygous (AG vs GG) and dominant (AA+AG vs GG) models in Caucasians but not among Asian populations. The results suggested that the HNC susceptibility of different ethnicities was a key factor for Arg399Gln polymorphisms. In Yadong Wang’ meta-analysis, their results also supported that polymorphism of Arg399Gln was associated with ethnicity. Meanwhile, the heterozygous model showed a positive correlation to HNC risk, which was consistent with our results. Moreover, they also found that subgroup analysis in tumor site displayed a significant association between larynx squamous cell carcinoma and Gln/Gln genetic model. However, the results of subgroup based on oral or larynx tumor both indicated little associations between them. Their findings are inconsistent with this meta-analysis study. Subsequently, considering that some Asian people have the habit of eating betel nut, which is a high risk factor for oral fibrosis or oral cancer, 46 we further performed a subgroup analysis involved in XRCC1 Arg399Gln polymorphisms and the risk in Asians with oral tumor. Unfortunately, the results turned out they did not correlate. Collectively, we found new results compared to previous meta-analysis studies.
Although we perform a comprehensive and updated analysis, our study have a number of limitations. First, our positive results mainly concentrated on Caucasians populations, while we only added 2 eligible study involved Caucasians compared with the meta-analysis of Wei et al 2014. Then, Arg399Gln polymorphisms is not only related to hereditary susceptibility, but also related to environmental factors. This study did not analyze environmental factors such as smoking. Third, biological factor is also the key factor for HNC, for example, HPV-positive HNC maybe different with HPV-negative HNC. However, the included studies did not distinguish the HPV-positive and HPV-negative HNC so that we were not able to analyze subgroup about HPV-relevant HNC. These are deficiencies and limitations for this meta-analysis
Conclusion
In this meta-analysis, the XRCC1 Arg399Gln variants (Arg/Gln and Arg/Arg+Arg/Gln) may contribute to HNC risk among Caucasians. Further studies with a larger sample are needed to confirm these findings.
Acknowledgments
Thanks for the English editing for this manuscript by Dr. Yang.
Authors’ Note: This research does not involve animal experiments and clinical trials involving humans, so ethics is not suitable for this study.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Shitong Xia, MD  https://orcid.org/0000-0001-7835-5037
https://orcid.org/0000-0001-7835-5037
References
- 1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. [DOI] [PubMed] [Google Scholar]
- 2. Wei T, Choi S, Buehler D, Anderson RA, Lambert PF. A PI3K/AKT scaffolding protein, IQ motif-containing GTPase associating protein 1 (IQGAP1), promotes head and neck carcinogenesis. Clin Cancer Res. 2020;26(1):301–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hiyama T, Yoshihara M, Tanaka S, Chayama K. Genetic polymorphisms and head and neck cancer risk (review). Int J Oncol. 2008;32(5):945–973. [PubMed] [Google Scholar]
- 4. Hopkins J, Cescon DW, Tse D, et al. Genetic polymorphisms and head and neck cancer outcomes: a review. Cancer Epidemiol Biomarkers Prev. 2008;17(3):490–499. [DOI] [PubMed] [Google Scholar]
- 5. Sobiahe A, Hijazi E, Al-Ameer HJ, et al. Arg399Gln XRCC1 polymorphism and risk of squamous cell carcinoma of the head and neck in Jordanian patients. Asian Pac J Cancer Prev. 2020;21(3):663–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shen MR, Jones IM, Mohrenweiser H. Nonconservative amino acid substitution variants exist at polymorphic frequency in DNA repair genes in healthy humans. Cancer Res. 1998;58(4):604–608. [PubMed] [Google Scholar]
- 7. Stur E, Agostini LP, Garcia FM, et al. Prognostic significance of head and neck squamous cell carcinoma repair gene polymorphism. Genet Mol Res. 2015;14(4):12446–12454. [DOI] [PubMed] [Google Scholar]
- 8. Azad AK, Bairati I, Samson E, et al. Validation of genetic sequence variants as prognostic factors in early-stage head and neck squamous cell cancer survival. Clin Cancer Res. 2012;18(1):196–206. [DOI] [PubMed] [Google Scholar]
- 9. Wu W, Liu L, Yin Z, Guan P, Li X, Zhou B. Association of X-ray repair cross-complementing group 1 Arg194Trp, Arg399Gln and Arg280His polymorphisms with head and neck cancer susceptibility: a meta-analysis. PLoS One. 2014;9(1):e86798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang Y, Chu X, Meng X, Zou F. Association of X-ray repair cross complementing group 1 Arg399Gln polymorphisms with the risk of squamous cell carcinoma of the head and neck: evidence from an updated meta-analysis. PLoS One. 2013;8(10):e77898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gal TJ, Huang WY, Chen C, Hayes RB, Schwartz SM. DNA repair gene polymorphisms and risk of second primary neoplasms and mortality in oral cancer patients. Laryngoscope. 2005;115(12):2221–2231. [DOI] [PubMed] [Google Scholar]
- 12. Moher D Liberati A Tetzlaff J Altman DG;. PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Ann Intern Med. 2009;151(4):264–269, W64. [DOI] [PubMed] [Google Scholar]
- 13. Applebaum KM, McClean MD, Nelson HH, Marsit CJ, Christensen BC, Kelsey KT. Smoking modifies the relationship between XRCC1 haplotypes and HPV16-negative head and neck squamous cell carcinoma. Int J Cancer. 2009;124(11):2690–2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ayiheng Q, Bogela A. Study on laryngeal cancer related on polymorphism of the Arg399Gln of XRCC1 DNA repair gene in different nationalities in Xinjiang [in Chinese]. Lin Chung Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2013;27(17):948–951, 954. [PubMed] [Google Scholar]
- 15. Demokan S, Demir D, Suoglu Y, Kiyak E, Akar U, Dalay N. Polymorphisms of the XRCC1 DNA repair gene in head and neck cancer. Pathol Oncol Res. 2005;11(1):22–25. [DOI] [PubMed] [Google Scholar]
- 16. Csejtei A, Tibold A, Koltai K, et al. Association between XRCC1 polymorphisms and head and neck cancer in a Hungarian population. Anticancer Res. 2009;29(10):4169–4173. [PubMed] [Google Scholar]
- 17. Dos RM, Losi-Guembarovski R, de Souza FRE, et al. Allelic variants of XRCC1 and XRCC3 repair genes and susceptibility of oral cancer in Brazilian patients. J Oral Pathol Med. 2013;42(2):180–185. [DOI] [PubMed] [Google Scholar]
- 18. Gajecka M, Rydzanicz M, Jaskula-Sztul R, et al. Reduced DNA repair capacity in laryngeal cancer subjects. A comparison of phenotypic and genotypic results. Adv Otorhinolaryngol. 2005;62:25–37. [DOI] [PubMed] [Google Scholar]
- 19. Gugatschka M, Dehchamani D, Wascher TC, Friedrich G, Renner W. DNA repair gene ERCC2 polymorphisms and risk of squamous cell carcinoma of the head and neck. Exp Mol Pathol. 2011;91(1):331–334. [DOI] [PubMed] [Google Scholar]
- 20. Avci H, Ergen A, Bireller ES, Ertugrul B, Cakmakoglu B. A strong relationship between oral squamous cell carcinoma and DNA repair genes. Biochem Genet. 2017;55(5-6):378–386. [DOI] [PubMed] [Google Scholar]
- 21. Harth V, Schafer M, Abel J, et al. Head and neck squamous-cell cancer and its association with polymorphic enzymes of xenobiotic metabolism and repair. J Toxicol Environ Health A. 2008;71(13-14):887–897. [DOI] [PubMed] [Google Scholar]
- 22. Tian H, Yang Y, Sheng HE. Association of the XRCC1 and XPD polymorphism with the risk of laryngeal carcinoma. Chi J Rehabilitat. 2010;25(1):9–12. [Google Scholar]
- 23. Jelonek K, Gdowicz-Klosok A, Pietrowska M, et al. Association between single-nucleotide polymorphisms of selected genes involved in the response to DNA damage and risk of colon, head and neck, and breast cancers in a polish population. J Appl Genet. 2010;51(3):343–352. [DOI] [PubMed] [Google Scholar]
- 24. Kietthubthew S, Sriplung H, Au WW, Ishida T. Polymorphism in DNA repair genes and oral squamous cell carcinoma in Thailand. Int J Hyg Environ Health. 2006;209(1):21–29. [DOI] [PubMed] [Google Scholar]
- 25. Kostrzewska-Poczekaj M, Gawecki W, Illmer J, et al. Polymorphisms of DNA repair genes and risk of squamous cell carcinoma of the head and neck in young adults. Eur Arch Otorhinolaryngol. 2013;270(1):271–276. [DOI] [PubMed] [Google Scholar]
- 26. Kowalski M, Przybylowska K, Rusin P, et al. Genetic polymorphisms in DNA base excision repair gene XRCC1 and the risk of squamous cell carcinoma of the head and neck. J Exp Clin Cancer Res. 2009;28(1):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Krupa R, Kasznicki J, Gajecka M, et al. Polymorphisms of the DNA repair genes XRCC1 and ERCC4 are not associated with smoking- and drinking-dependent larynx cancer in a polish population. Exp Oncol. 2011;33(1):55–56. [PubMed] [Google Scholar]
- 28. Kumar A, Pant MC, Singh HS, Khandelwal S. Associated risk of XRCC1 and XPD cross talk and life style factors in progression of head and neck cancer in north Indian population. Mutat Res. 2012;729(1-2):24–34. [DOI] [PubMed] [Google Scholar]
- 29. Li C, Hu Z, Lu J, et al. Genetic polymorphisms in DNA base-excision repair genes ADPRT, XRCC1, and APE1 and the risk of squamous cell carcinoma of the head and neck. Cancer. 2007;110(4):867–875. [DOI] [PubMed] [Google Scholar]
- 30. Majumder M, Sikdar N, Paul RR, Roy B. Increased risk of oral leukoplakia and cancer among mixed tobacco users carrying XRCC1 variant haplotypes and cancer among smokers carrying two risk genotypes: one on each of two loci, GSTM3 and XRCC1 (codon 280). Cancer Epidemiol Biomarkers Prev. 2005;14(9):2106–2112. [DOI] [PubMed] [Google Scholar]
- 31. Majumder M, Sikdar N, Ghosh S, Roy B. Polymorphisms at XPD and XRCC1 DNA repair loci and increased risk of oral leukoplakia and cancer among NAT2 slow acetylators. Int J Cancer. 2007;120(10):2148–2156. [DOI] [PubMed] [Google Scholar]
- 32. Matullo G, Palli D, Peluso M, et al. XRCC1, XRCC3, XPD gene polymorphisms, smoking and (32)P-DNA adducts in a sample of healthy subjects. Carcinogenesis. 2001;22(9):1437–1445. [DOI] [PubMed] [Google Scholar]
- 33. Olshan AF, Watson MA, Weissler MC, Bell DA. XRCC1 polymorphisms and head and neck cancer. Cancer Lett. 2002;178(2):181–186. [DOI] [PubMed] [Google Scholar]
- 34. Mutlu P, Mutlu M, Yalcin S, et al. Detection of XRCC1 gene polymorphisms in Turkish head and neck squamous cell carcinoma patients: a comparative analysis with different populations. J BUON. 2015;20(2):540–547. [PubMed] [Google Scholar]
- 35. Ramachandran S, Ramadas K, Hariharan R, Kumar RR, Radhakrishna Pillai M. Single nucleotide polymorphisms of DNA repair genes XRCC1 and XPD and its molecular mapping in Indian oral cancer. Oral Oncol. 2006;42(4):350–362. [DOI] [PubMed] [Google Scholar]
- 36. Khlifi R, Kallel I, Hammami B, Hamza-Chaffai A, Rebai A. DNA repair gene polymorphisms and risk of head and neck cancer in the Tunisian population. J Oral Pathol Med. 2014;43(3):217–224. [DOI] [PubMed] [Google Scholar]
- 37. Rydzanicz M, Wierzbicka M, Gajecka M, Szyfter W, Szyfter K. The impact of genetic factors on the incidence of multiple primary tumors (MPT) of the head and neck. Cancer Lett. 2005;224(2):263–278. [DOI] [PubMed] [Google Scholar]
- 38. Sturgis EM, Castillo EJ, Li L, et al. Polymorphisms of DNA repair gene XRCC1 in squamous cell carcinoma of the head and neck. Carcinogenesis. 1999;20(11):2125–2129. [DOI] [PubMed] [Google Scholar]
- 39. Tae K, Lee HS, Park BJ, et al. Association of DNA repair gene XRCC1 polymorphisms with head and neck cancer in Korean population. Int J Cancer. 2004;111(5):805–808. [DOI] [PubMed] [Google Scholar]
- 40. Varzim G, Monteiro E, Silva RA, Fernandes J, Lopes C. CYP1A1 and XRCC1 gene polymorphisms in SCC of the larynx. Eur J Cancer Prev. 2003;12(6):495–499. [DOI] [PubMed] [Google Scholar]
- 41. Yuan H, Li H, Ma H, et al. Genetic polymorphisms in key DNA repair genes and risk of head and neck cancer in a Chinese population. Exp Ther Med. 2012;3(4):719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Puerta-Garcia E, Urbano-Perez D, Carrasco-Campos MI, et al. Effect of DPYD, MTHFR, ABCB1, XRCC1, ERCC1 and GSTP1 on chemotherapy related toxicity in colorectal carcinoma. Surg Oncol. 2020;35:388–398. [DOI] [PubMed] [Google Scholar]
- 43. Xie X, Lin SH, Welsh JW, et al. Radiation-induced lymphopenia during chemoradiation therapy for non-small cell lung cancer is linked with age, lung V5, and XRCC1 rs25487 genotypes in lymphocytes. Radiother Oncol. 2021;154:187–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Howlader NR, Rahman MM, Hossain MA, et al. Genetic polymorphisms in DNA repair genes XRCC1 and 3 are associated with increased risk of breast cancer in Bangladeshi population. Breast Cancer Res Treat. 2020;182(3):739–750. [DOI] [PubMed] [Google Scholar]
- 45. Wang F, Zhao Q, He HR, et al. The association between XRCC1 Arg399Gln polymorphism and risk of leukemia in different populations: a meta-analysis of case-control studies. Onco Targets Ther. 2015;8:3277–3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nagao T, Warnakulasuriya S. Screening for oral cancer: future prospects, research and policy development for Asia. Oral Oncol. 2020;105:104632. [DOI] [PubMed] [Google Scholar]