Abstract
RNA interference (RNAi) is a cellular process involved in the silencing of genes, which makes RNAi important for observing and understanding the function of specific gene products. Short interfering RNA (siRNA) pathway is a RNAi pathway, where exogenous double stranded RNA is introduced to the cell and cleaved by an endoribonuclease, Dicer, to form siRNA, which interacts with a protein complex to scan mRNAs to bind to its complementary sequence. The binding of the siRNA to its complementary mRNA, the mRNA is cleaved and degraded by the cell, significantly reducing the levels of the target protein product. The discovery of this mechanism made it a powerful tool to use as a technique for therapeutics, agricultural biology, and cellular and molecular biology.
Keywords: RNAi, siRNA, Gene silencing, Gene knockdown, Cell cycle regulation, p53
1. Introduction
RNA interference (RNAi) is a biological process where double-stranded RNAs inhibit the expression of its target genes. It was first defined as posttranscriptional gene silencing (PTGS) when the overexpression of transgenes showed silencing of itself and the endogenous genes with sequence similarities. The mechanism behind PTGS was later shown to involve double-stranded RNA that significantly downregulated gene expression [1, 2]. These double stranded RNAs are cleaved by and endoribonuclease, Dicer, where the resulting short interfering RNA (siRNA), of about 25 nucleotide in length [3, 4], bind to the RISC complex and scans for its complementary mRNA sequence. The mRNA complementary to the siRNA is cleaved and degraded by the cell [5]. The elucidation of this pathway became a useful tool for cell biology, therapeutics, and agricultural biology [5, 6].
In cell biology, double-stranded RNA can be injected into mammalian cell cultures to knock down the expression of its target sequence mRNA and thereby significantly decrease the levels of protein product in the cell [4]. The knockdown of p53 can be used to help understand its role in cell cycle regulation. In response to stress, p53, a transcription factor, regulates the expression of genes involved in various cellular processes [7-9]. Some of these p53 responses include apoptosis, cell cycle arrest, and senescence, which are three of the most studied p53 regulated processes [7]. Since p53 is involved in preventing the progression of cell proliferation, elucidating the role of p53 is important in understanding cell cycle regulation, which is usually impaired in disease models such as tumorigenesis [8, 10] and neurodegenerative diseases [11]. In addition to p53, other players of the cell cycle can be knocked down to further shed light on the regulation of cell cycle and to develop therapeutics for cancer and neurodegenerative diseases.
2. Materials
On-TARGET plus siRNA or gene of interest (20 μM).
Allstars Negative Control siRNA (20 μM).
Oligofectamine™ Transfection Reagent.
Opti-MEM™∣Reduced Serum Medium.
DMEM.
DMEM with 20% FBS: For 50 ml solution, 40 ml DMEM and 10 ml FBS.
3. Methods
All steps must be done under the tissue culture hood (see Note 1).
For one 100 mm dish (or 2 × 60 mm dishes) → 0.1 μM of siRNA per 100 mm dish (see Notes 2-3):
Make siRNA mix in Opti-MEM solution in falcon tubes. Per siRNA: 15 μl of siRNA (20 μM), 540 μl of Opti-MEM. Be sure to have one mix with negative control siRNA, so there must be at least two mixes made (see Notes 4-5).
Make Oligofectamine mix in Opti-MEM solution: 12 μl of Oligofectamine, 33 μl of Opti-MEM. Multiply the volumes by the number of mixes made in step 1.
Let the two mixtures sit in room temperature for 15 min.
Take the Oligofectamine mix (per 1 × 100 mm dish volume = 45 μl) and pipette mix into the siRNA mix.
Let the mix sit in room temperature for 15 min.
While the siRNA+Oligofectamine mix is sitting in room temperature for 15 min, wash the cells three times with DMEM without FBS (or 2× PBS and last time with DMEM without FBS).
Add DMEM (without FBS) into the siRNA–Oligofectamine mix so that 3 ml of the mixture is added to each 100 mm dish (for 1 × 100 mm dish, add 2.380 ml DMEM to the 620 μl siRNA+Oligofectamine mix).
Add 3 ml of the final mix (siRNA + Oligofectamine + Opti-MEM + DMEM) to each 100 mm dish (or 1.5 ml of mix to each 60 mm dish). This brings the final siRNA concentration per dish to 0.1 μM.
Incubate cells at 37 °C for 5 h.
Add 5 ml of DMEM with 20% FBS to each 100 mm dish (or 2.5 ml to each 60 mm dish).
Incubate at 37 °C for 24 h for siRNA knockdown. After 24 h the cells will be ready for collection or further experimental procedures (see Note 6).
Fig. 1.
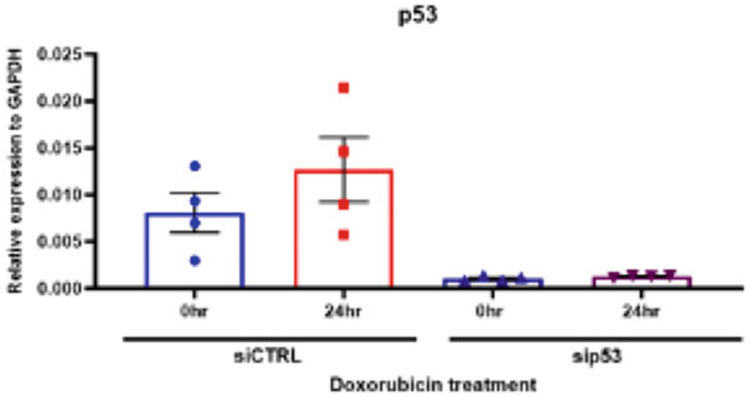
Quantitative RT-PCR of p53 after knockdown and doxorubicin treatment in U2OS cells. U2OS cells were transfected with p53 siRNA or negative control siRNA and treated with 0.1 μM doxorubicin for 24 h. RNA levels determined by RT-PCR
Fig. 2.
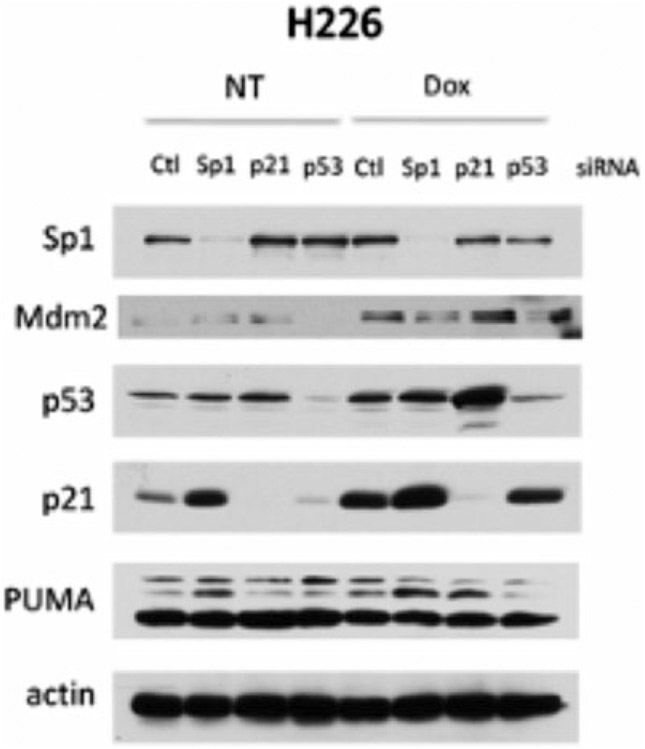
Western blot of Sp1, p21, and p53 after knockdown and doxorubicin treatment. H226 cells were treated with each respective siRNA or negative control siRNA and treated with 0.1 μM doxorubicin for 24 h. Protein levels determined by Western blot
Footnotes
Be sure to have all calculations ready before the start of the knockdown.
Split cells 24 h before they are treated with siRNA.
Cells should be at about 50–75% confluency for the transfection.
Pipette mix the solutions and do not vortex. Or gently flip tubes up and down to mix the solutions.
For multiple gene knockdowns, be sure to keep the total siRNA concentration at 0.1 μM per dish. Divide the total volume of siRNA needed by the number of genes that need to be knocked down for the volume of siRNA needed per gene.
To confirm knockdown, use RT-PCR and Western blot or immunocytochemistry. RT-PCR will check if the target mRNA is still present (Fig. 1), but this does not confirm if the protein levels has decreased. Western blot (Fig. 2) and immunocytochemistry will check if the target protein product is present.
References
- 1.Fire A et al. (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391(6669):806–811 [DOI] [PubMed] [Google Scholar]
- 2.Fire A et al. (1991) Production of antisense RNA leads to effective and specific inhibition of gene expression in C. elegans muscle. Development 113(2):503–514 [DOI] [PubMed] [Google Scholar]
- 3.Hamilton AJ, Baulcombe DC (1999) A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286(5441):950–952 [DOI] [PubMed] [Google Scholar]
- 4.Elbashir SM et al. (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411(6836):494–498 [DOI] [PubMed] [Google Scholar]
- 5.Dana H et al. (2017) Molecular mechanisms and biological functions of siRNA. Int J Biomed Sci 13(2):48–57 [PMC free article] [PubMed] [Google Scholar]
- 6.Joga MR et al. (2016) RNAi efficiency, systemic properties, and novel delivery methods for Pest insect control: what we know so far. Front Physiol 7:553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vousden KH, Prives C (2009) Blinded by the light: the growing complexity of p53. Cell 137(3):413–431 [DOI] [PubMed] [Google Scholar]
- 8.Chen J (2016) The cell-cycle arrest and apoptotic functions of p53 in tumor initiation and progression. Cold Spring Harb Perspect Med 6(3):a026104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sullivan KD et al. (2018) Mechanisms of transcriptional regulation by p53. Cell Death Differ 25(1):133–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bieging KT, Mello SS, Attardi LD (2014) Unravelling mechanisms of p53-mediated tumour suppression. Nat Rev Cancer 14(5):359–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang JR et al. (2012) Role of p53 in neurodegenerative diseases. Neurodegener Dis 9(2):68–80 [DOI] [PMC free article] [PubMed] [Google Scholar]


