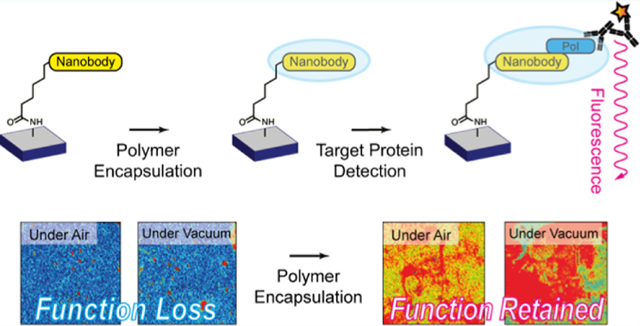
Abstract
To produce next-generation, shelf-stable biosensors for point-of-care diagnostics, a combination of rugged biomolecular recognition elements, efficient encapsulants, and innocuous deposition approaches is needed. Furthermore, to ensure that the sensitivity and specificity that are inherent to biological recognition elements are maintained in solid-state biosensing systems, site-specific immobilization chemistries must be invoked such that the function of the biomolecule remains unperturbed. In this work, we present a widely applicable strategy to develop robust solid-state biosensors using emergent nanobody (Nb) recognition elements coupled with a vapor-deposited polymer encapsulation layer. As compared to conventional immunoglobulin G antibodies, Nbs are smaller (12–15 kDa as opposed to ~150 kDa), have higher thermal stability and pH tolerance, boast greater ease of recombinant production, and are capable of binding antigens with high affinity and specificity. Photoinitiated chemical vapor deposition affords thin, protective polymer barrier layers over immobilized Nb arrays that allow for retention of Nb activity and specificity after both storage under ambient conditions and complete desiccation. Most importantly, we also demonstrate that vapor-deposited polymer encapsulation of Nb arrays enables specific detection of target proteins in complex heterogeneous samples, such as unpurified cell lysate, which is otherwise challenging to achieve with bare Nb arrays.
Keywords: photoinitiated chemical vapor deposition, nanobody, immobilization, biosensor, shelf life
Graphical Abstract
INTRODUCTION
Surface-immobilized proteins, antibodies, and other similar biomolecules form the basis of most known biosensors, biomolecular electronics, and nanodevices.1–5 Although a number of surface immobilization chemistries are reported, comparatively fewer strategies for maintaining the function and stability of these surface-immobilized biomolecules, especially upon long-term storage, are known.6 Particularly for biosensors, it is imperative to maintain constant reaction conditions and to suppress nonspecific binding interactions that could lead to decreased target diffusion to the sensing interface and result in significant signal loss.7,8 Careless choice of surface treatments can damage biomolecules involved in the sensing process and, eventually, significantly shorten the shelf-life and accuracy of the biosensors over time. Therefore, special emphasis has been placed on encapsulation techniques, as the resultant surface barrier can simultaneously suppress sensor fouling due to nonspecific binding and/or adhesion and produce a stable inner environment with optimal humidity and temperature for the biological sensing element.9,10
Typically, two kinds of polymer encapsulants are invoked for biosensors: hydrophilic polymers (or hydrogels) and zwitterionic polymers.11–14 These encapsulants are widely applied to form hydration layers at the sensor surface via either hydrogen bonds or ion solvation, respectively. Since the interaction force between water molecules or ions and the coating materials cannot be readily compensated by extra adsorption, this tightly bound water or ion layer becomes a physical, as well as an energetic, barrier against nonspecific adsorption of interferents.15 An emerging encapsulation strategy is to coat surfaces with polymers that present low surface energies, such as hyperbranched fluoropolymers, polypeptides, or silicone elastomers, and, thus, facilitate removal of adsorbed biomacromolecules by a limited shear force, like gentle rinsing.16,17
Layer-by-layer assemblies18,19 and in situ polymerized coatings6,20 also serve as encapsulants for intact biosensing systems. However, the non-conformal nature of these solution-processed coatings often hinders analyte diffusion to the underlying sensing element, which leads to inaccurate signal acquisition and increases signal integration times. Most critically, these methods require solvent usage, which is considered undesirable for reliably developing encapsulation layers on fragile biosensing elements, particularly proteins and antibodies that can degrade, denature, or adopt inactive conformations depending on solvent exposure and variations in pH or ionic strength.
To produce next-generation, shelf-stable biosensors for point-of-care diagnostics, a combination of rugged biomolecular recognition elements, efficient encapsulants, and innocuous deposition approaches is needed. Furthermore, to ensure that the sensitivity and specificity that are inherent to biological recognition elements are maintained in solid-state biosensing systems, site-specific immobilization chemistries must be invoked such that the function of the biomolecule remains unperturbed. In this work, we present a widely applicable strategy to develop robust solid-state biosensors using a uniquely stable nanobody (Nb) recognition element coupled with a vapor-deposited polymer encapsulation layer.
EXPERIMENTAL SECTION
Materials.
(3-Aminopropyl)trimethoxysilane (APTMS), 6-chlorohexanoic acid (CHA), N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC), dichloromethane (DCM), ethanol, 2-hydroxy-2-methylpropiophenone (HMPP), 2-hydroxyethyl acrylate (HEA), 1H,1H,2H,2H-perfluorooctyl acrylate (TFOA), and 1,1,1,3,3,3-hexafluoroisopropanol (HFIP) from MilliporeSigma and TCI America were used without purification.
Fabrication Protocols.
Clean Si wafers were immersed in APTMS 2% in an ethanol/water (90/10 v/v %) solution for 5 min. The treated wafers were washed with pure ethanol two times and further treated in an ultrasonic bath for 5 min to remove residual silanes on the surface. The washed substrates were blow-dried using compressed N2 gas. For the introduction of a chloroalkane group, 1.92 mg of EDC (0.01 mmol) and 1.50 mg of CHA (0.01 mmol) were added in 2 mL of DCM. Then, each substrate was immersed in the solution for 12 h. The grafted samples were further washed with pure DCM two times and then dried with N2 gas.
Protein Expression and Purification.
Halo-tagged Nb constructs were expressed in BL21(DE3) pLysS Escherichia coli cells in LB media supplemented with kanamycin (25 μg/mL) at 37 °C to OD600 ~ 1.0 and induced with 300 μM IPTG at 16 °C for 16 h. Cells were harvested by centrifugation at 5000g for 15 min. The resulting cell pellets were resuspended in lysis buffer (50 mM Tris-HCl pH 8.0, 300 mM NaCl, and 1 mM TCEP), lysed by sonication, and clarified by centrifugation at 30,000g for 30 min. The clarified lysate was incubated with Ni-NTA resin for 2 h at 4 °C, washed with binding buffer (lysis buffer plus 10 mM imidazole), and eluted into Ni-NTA elution buffer (lysis buffer plus 300 mM imidazole). Then, the eluate was buffer-exchanged into gel filtration buffer (50 mM Tris-HCl pH 8.0, 300 mM NaCl, and 1 mM DTT), concentrated, and run on a Superdex 75 (GE) gel filtration column in gel filtration buffer at 0.3 mL/min. Recombinant UCH37 was expressed and purified as previously described.21
Cell Lysate Generation.
Wild-type and UCH37 knockout (UCH37KO) HEK293 cells stably expressing RPN11-HTBH were grown, harvested, and lysed in Ptsm lysis buffer (40 mM HEPES pH7.4, 40 mM NaCl, 10 mM MgCl2, 2 mM ATP, 1 mM DTT, and 10% glycerol).21 The lysates were clarified at 20,000g for 20 min and the supernatant was collected and stored in −80 °C prior to use. The concentration of total cell lysate was determined by bicinchoninic acid assay.
Nb Immobilization.
The grafted wafers were first washed with phosphate-buffered saline (PBS) for 5 min at room temperature (RT). Halo-tagged control Nb and NbIII.15 were diluted to 5 μM in cold PBS and incubated with washed wafers at 4 °C overnight under rocking. Then, wafers were washed with PBS three times for 5 min to remove the unreacted Nb and dried under air.
Protein Target Treatment and Detection.
Prior to treating Nb-immobilized wafers with UCH37, the recombinant protein was diluted to 1 μM in cold PBS. When the cell lysate was used instead of the recombinant protein, it was diluted to 1 mg/mL in cold PBS. Wafers were incubated with the target protein at 4 °C overnight under rocking. Wafers were then washed with PBS three times for 5 min to remove the unbound material. To detect the presence of UCH37 on wafers, the samples were treated with a recombinant UCH37 antibody (Abcam, monoclonal Rabbit, 1:1000 dilution in TBS) at 4 °C overnight under rocking. The antibody-treated wafers were then washed three times with PBS and treated with goat anti-rabbit immunoglobulin G (IgG) fluorescent secondary antibody (Licor, 1:15,000 dilution in TBS) at RT for 30 min under rocking. The samples were again washed three times with PBS for 5 min, followed by brief sonication and dried under air. Fluorescent images were collected by scanning the wafer surface using a Licor Odyssey CLx fluorescent imager (700 channel, 21 μm).
Polymer Encapsulation.
A custom-built reactor (stainless-steel walls, 290 mm diameter, and 70 mm height) was used to polymerize HEA and TFOA directly on the surface of all samples using photoinitiated chemical vapor deposition (piCVD). A low-intensity UV lamp (UVP, UVLS-24 EL Series, 4 W, 254 nm) was turned on when the monomers were introduced into the reactor and the base pressure of the reactor had stabilized. During deposition, the base pressure of the reactor was kept at 300 mTorr, while the stage temperature was maintained at 20 °C with a recirculating water cooling system. Photoinitiator HMPP and monomer (HEA or TFOA) were stored in glass ampules wrapped with fiberglass heating tape and introduced into the reactor through articulated needle valves, as illustrated in Figure S1. HMPP was heated to 110 °C and flowed into the reactor first for 5 min, then the monomer, also heated to 110 °C, was introduced into the reactor, at which time polymerization and film deposition occurred. The deposition was allowed to proceed for 10 min to obtain an encapsulation layer. In the case of TFOA, 5 μL of HFIP was drop-cast onto the Nb-immobilized surface before deposition to promote TFOA wetting. After the deposition was completed, the samples were left on the substrate stage (maintained at 20 °C) for a further 30 min until the temperature of the reactants reaches RT. This post-annealing step allowed unreacted monomers to be pulled out of the film, which resulted in a porous coating on the substrate surface. The chemical structure and thickness of the piCVD film was determined by Fourier transform infrared (FTIR) spectroscopy (Bruker Alpha) and atomic force microscopy (AFM) performed on films grown on bare glass or silicon substrates; equivalent thickness and polymer structure were assumed for encapsulation layers grown on Nb arrays.
Surface Characterization.
A Jupiter atomic force microscope (Asylum Research, USA) was used to characterize the surface morphology of the samples. AFM images were obtained in the tapping mode using PPP-NCHR probes (Nanosensors, Switzerland). The thickness of the samples was analyzed by spectroscopic ellipsometry (SE) using an RC2 Ellipsometer (JA Woollam, USA). At each measurement, at least three different points of the surface were taken and averaged.
RESULTS AND DISCUSSION
Nb Immobilization.
Single-variable domain (VHH) antibodies from camelids (referred to as nanobodies) have attracted great attention as biosensors.22–25 Similar to conventional IgG antibodies, nanobodies are capable of binding antigens with high affinity and specificity. What set nanobodies apart from IgGs are their smaller size (12–15 kDa as opposed to ~150 kDa), high thermal stability, pH tolerance, and ease of recombinant production. Due to these properties, we decided to build a robust, encapsulated sensing platform based on nanobodies. The Nb (NbIII.15) chosen for these studies was derived from a synthetic yeast surface display library and engineered to bind with high affinity and specificity to the human protein UCHL5/UCH37 (LHC unpublished results).
Surface immobilization of proteins typically entails physical adsorption or covalent chemistry involving the canonical amino acid side chains, both of which provide little specificity in terms of surface orientation.26 Thus, to avoid surface heterogeneity, we immobilized NbIII.15 on silane-treated silicon surface using the HaloTag system.27 According to isothermal titration calorimetry, the HaloTagged variant of NbIII.15 binds UCH37 with a Kd of 60 ± 20 nM and can be lyophilized without impairing function (Figure S2). As a control, a Nb unable to bind UCH37 (NbCtrl) was also immobilized using the same approach. The entire fabrication process is illustrated in Figure 1a. At each step of the experiment, the thickness and the surface morphology of the samples were monitored by SE (Table S1) and AFM measurements (Figures 1b–f and S3), respectively.
Figure 1.
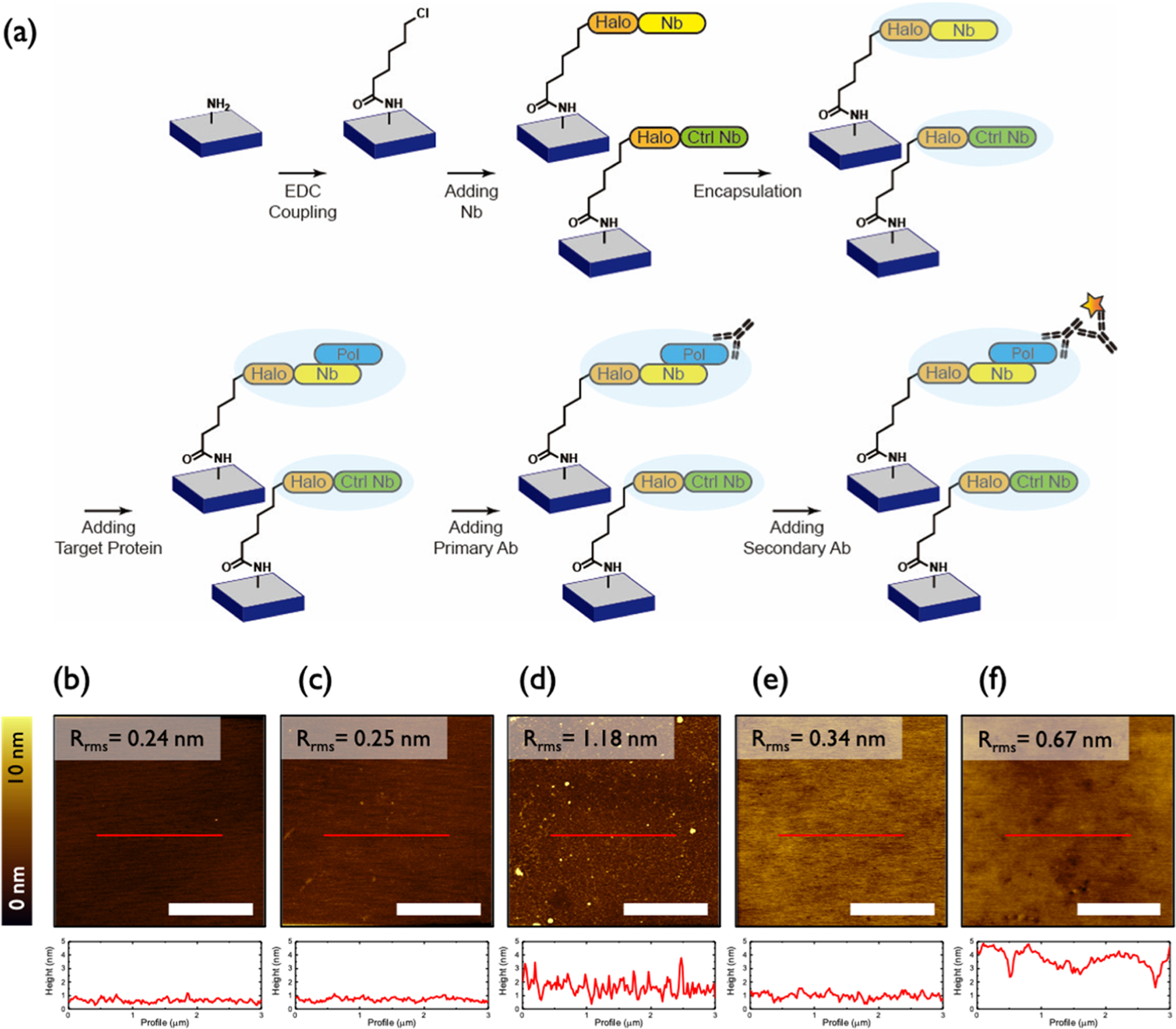
(a) Schematic illustration of the sensor fabrication process. (b–f) AFM topography images (top) and height profiles (bottom) of sequentially (b) ATPMS-treated, (c) CHA-treated, (d) Nb-treated, (e) pTFOA-encapsulated, and (f) target protein-/antibody-exposed samples. All scale bars are 2 μm.
Silanization with APTMS was readily achieved on silicon substrates. Within 5 min, a smooth, amine-terminated surface was isolated, as shown Figure 1b. EDC coupling with CHA provided the chloroalkane moiety, necessary for covalent attachment to HaloTag. AFM topography images (Figure 1b,c) and the corresponding root-mean-square roughness (Rrms) data show that smooth surfaces were obtained even after EDC coupling with CHA. HaloTag-fused nanobodies, NbIII.15 and NbCtrl, were then immobilized by covalent attachment to the chloroalkane linker. As summarized in Table S1, the thickness of the samples increased as the sensing layers were articulated. The resulting Nb-decorated surfaces exhibit an increase in Rrms of 0.9 nm and are uniform according to AFM topography (Figure 1d).
Sensing and Binding Specificity of Immobilized Nb Arrays.
To determine whether NbIII.15 retains function once immobilized, we monitored surface binding by fluorescence and SE. Fluorescence imaging reveals that recombinant UCH37 binds the surface decorated with NbIII.15 but not the surface decorated with NbCtrl (Figure 2a). These images also confirm that fluorescent antibodies can be sequentially bound to the Nb-decorated surface, further demonstrating that surface-immobilized nanobodies can provide a suitable sensing platform in the solid state.
Figure 2.
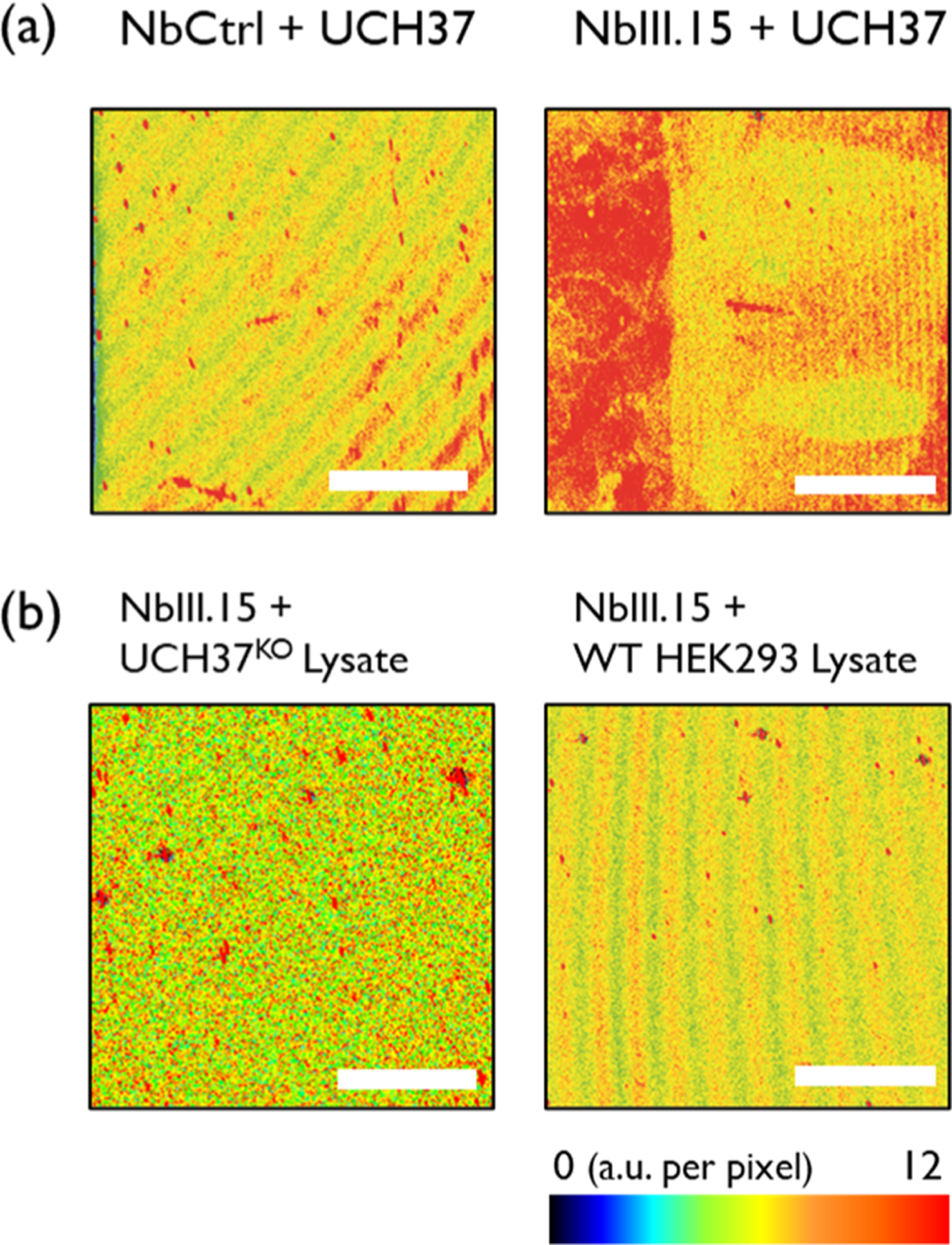
Fluorescence images of (a) purified UCH37-treated samples on NbCtrl (left) and on the NbIII.15 arrays (right). (b) 1 mg/mL of UCH37KO lysate-treated (left) and 1 mg/mL of HEK293 lysate-treated (right) samples on the NbIII.15 arrays. The estimated concentration of UCH37 in 1 mg/mL of cell lysate is 53 pM.21 Scale bars are 2 mm.
The fluorescence microscopy images were also reinforced by SE; the thickness increases from 3.0 to 4.4 nm upon adding recombinant UCH37 to immobilized NbIII.15, but no change in thickness is observed when UCH37 is added to immobilized NbCtrl (Table S1). This confirms that the Ctrl Nb has no binding affinity to the protein targets. In addition, the Rrms of the surface increases from 0.95 to 1.46 nm (Figure S3c,d), indicating that Nb has a stronger affinity toward target proteins.
While the results with purified recombinant UCH37 suggest that surface-bound NbIII.15 functions as predicted, a more stringent test requires the presentation of UCH37 in a complex, heterogeneous mixture such as cell lysate. Thus, lysates derived from human embryonic kidney (HEK293) cells and UCH37-deficient HEK293 (UCH37KO) cells were added to surfaces coated with either NbIII.15 or NbCtrl and analyzed by fluorescence microscopy. The images in Figure 2 show that, in contrast to the purified system, little binding occurs.
Encapsulation.
We postulated that the lack of binding to the protein of interest in a complex, heterogeneous environment was a result of sensor fouling due to nonspecific binding and/or interferent adhesion to the sensor surface. Thus, we decided to apply a mild, solvent-free vapor deposition process to create a polymer barrier layer on the Nb-immobilized surface.
CVD is a solvent-free process in which vaporized precursors chemically react in the gas phase to generate a thin, solid film on a target surface.28,29 CVD is also singularly able to produce films that are otherwise inaccessible via low-temperature solution casting, such as fluoropolymer films and highly crosslinked hydrophilic networks. Among several CVD methods to create polymer films, photoinitiated CVD (piCVD) is an especially mild iteration that does not require high-energy sources, high process temperatures, or reactive initiators.30,31 Previous studies have demonstrated that specific protein-resistant hydrogels made of poly(2-hydroxyethyl methacrylate) can be synthesized via piCVD on optical sensors without sacrificing sensitivity and accuracy.30 However, direct deposition of antifouling layers on surface-immobilized proteins and subsequent use of these sensing surfaces to screen for target analytes in complicated mixtures, such as cell lysate, have not been investigated.
We pursued piCVD of two polymer encapsulants: a biocompatible hydrogel, poly(2-hydroxyethyl acrylate) (pHEA), and a fluoropolymer, poly(1H,1H,2H,2H-perfluorooctyl acrylate) (pTFOA). Although the use of a super-hydrophobic barrier layer for biosensors is counterintuitive, we posited that a microstructured or porous fluoropolymer encapsulant could mitigate undesired surface adhesion and fouling in complex sample fluids, such as cell lysates. Further, we hypothesized that the high intrinsic binding affinity of nanobodies to proteins of interest would drive target binding and signal generation, even if mass transport to the underlying Nb recognition elements was attenuated by the pTFOA barrier layer.
piCVD afforded uniform, nanometer-thick polymer coatings on the Nb-decorated silicon surfaces. To reduce the time over which the Nb arrays are subjected to vacuum conditions and UV light (which may cause photodegradation), we used a stepwise deposition algorithm to enact piCVD. Photoexcited carbonyl species are known to decompose into radicals from readily accessible triplet excited states, which can, in turn, initiate a free-radical polymerization. In our piCVD process, HMPP, the photoinitiator, was first introduced to the chamber, followed by the monomer, HEA or TFOA, to affect polymerization. Under UV radiation of low intensity, HMPP decomposes and initializes the polymerization over the excited-state lifetime of a carbonyl triplet state (ca. 100 ns). As illustrated in Figure S4, the FTIR spectra of HEA and pHEA showed a broad peak at 3200–3600 cm−1, corresponding to the hydroxyl group peak, and a sharp peak at 1720 cm−1, corresponding to the carbonyl stretching. Significantly, pHEA did not exhibit a peak located at 1637 cm−1, where the stretching of an unsaturated C=C double bond of HEA monomer should appear, thus confirming that the starting acrylate monomer was polymerized through the piCVD process. Likewise, the FTIR spectra of TFOA and pTFOA also showed that TFOA was polymerized with HMPP using piCVD. In addition, residual monomers could have also been removed from the films during the post-deposition vacuum annealing step.
Water contact angle measurements were conducted (Figure S5) to confirm the hydrophilic and hydrophobic nature of pHEA and pTFOA films, respectively. AFM topography images revealed that uniform coatings were obtained over the entire Nb-decorated surface (images for a pTFOA barrier layer are shown in Figures 1e and S6), without apparent defects. Moreover, the starting surface morphology of Nb-decorated surfaces was not noticeably altered after polymer coating, tentatively suggesting that the dry piCVD process did not perturb the Nb arrays.
Sensing and Binding Specificity of Encapsulated Arrays.
Next, we confirmed that the Nb arrays retained their function after being encapsulated using piCVD. After encapsulation with pHEA, the Nb-immobilized sample still displayed very high fluorescence signals, while the Ctrl Nb did not yield a notable fluorescence signal (Figure 3a), indicating that the mild piCVD process does not interfere with the function or specificity of the Nb arrays. A similar behavior was also observed for Nb arrays encapsulated with pTFOA (Figure 3c), confirming that this fluoropolymer barrier layer does not inhibit mass transport of target proteins. We posit that the pTFOA films created via piCVD enables the diffusion of target proteins to the underlying Nb due to its discontinuous, wrinkled morphology, as shown in Figure S7, which originates from small gaps between monomer nucleation sites during the piCVD process.
Figure 3.
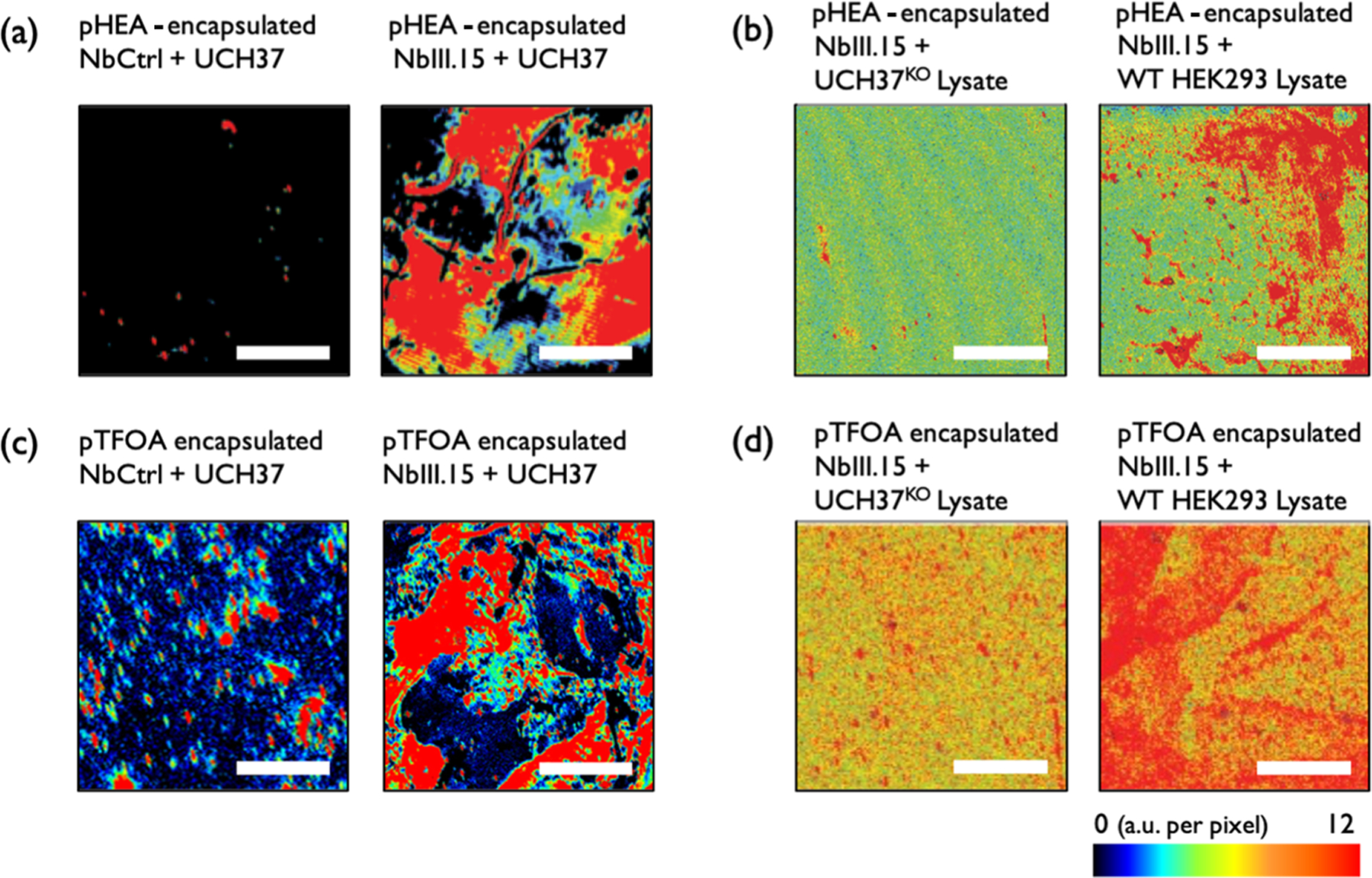
Fluorescence images of (a) purified UCH37-treated samples on pHEA-encapsulated NbCtrl (left) and on pHEA-encapsulated NbIII.15 arrays (right). (b) 1 mg/mL of UCH37KO lysate-treated (left) and 1 mg/mL HEK293 lysate-treated (right) samples on pHEA-encapsulated NbIII.15 arrays. (c) Purified UCH37-treated samples on pTFOA-encapsulated NbCtrl (left) and on pHEA-encapsulated NbIII.15 arrays (right). (d) 1 mg/mL of UCH37KO lysate-treated (left) and 1 mg/mL HEK293 lysate-treated (right) samples on pTFOA-encapsulated NbIII.15 arrays. The estimated concentration of UCH37 in 1 mg/mL of cell lysate is 53 pM.21 Scale bars are 2 mm.
The utility of the polymer barrier layers became evident when the Nb arrays were presented with cell lysates. As opposed to the bare Nb arrays, which did not demonstrate target binding in these complex, heterogeneous matrices, pHEA- and pTFOA-encapsulated Nb arrays readily displayed selective target binding with a limit of detection close to 10 pM based on an estimated concentration of UCH37 in cell lysates (Figures 3b,d and S8). The fluorescence obtained from lysate-treated pHEA/Nb demonstrated that pHEA encapsulation was able to reduce inference while maintaining target mass transfer toward the Nb platform (Figure 3b). The effect of the encapsulation layer becomes stronger with pTFOA. As shown in Figure 3d, the lysate-treated pTFOA/Nb sample shows higher fluorescence signals compared to other lysate-treated samples in Figure 3b. This demonstrates that the pTFOA film most effectively mitigates undesired nonspecific binding without hampering the activity of the underlying Nb.
Stability of Arrays after Storage and Desiccation.
The stability of the Nb arrays was investigated under harsh environments in order to probe the efficacy of vapor-deposited polymer encapsulants in protecting biological recognition elements. We first investigated the stability of bare, unencapsulated Nb arrays by placing them under vacuum or storing them in air overnight at RT. As shown in Figure 4a,b, the bare Nb arrays became inactive after both desiccation and storing in air overnight. This highlights the need for a protective barrier. In contrast, both pHEA- and pTFOA-encapsulated Nb arrays retained their target binding activity after being exposed to air and vacuum (desiccation) storage conditions for more than 12 h. Based on the low deviation in fluorescence signals (Figure 4c–f), and AFM morphologies (Figure S9) observed for air- and vacuum-treated samples, we infer that the morphology and surface chemistry of these vapor-deposited encapsulation layers were not notably altered in these harsh storage conditions. Moreover, the stronger fluorescence signal observed for pTFOA-encapsulated samples indicated that a larger number of target binding events occurred in these samples compared to the pHEA-encapsulated samples. A strong fluorescence signal can even be observed after storing the array for 4 days at RT (Figure S10). Therefore, we conclude that pTFOA is a strong candidate for biosensor encapsulation.
Figure 4.
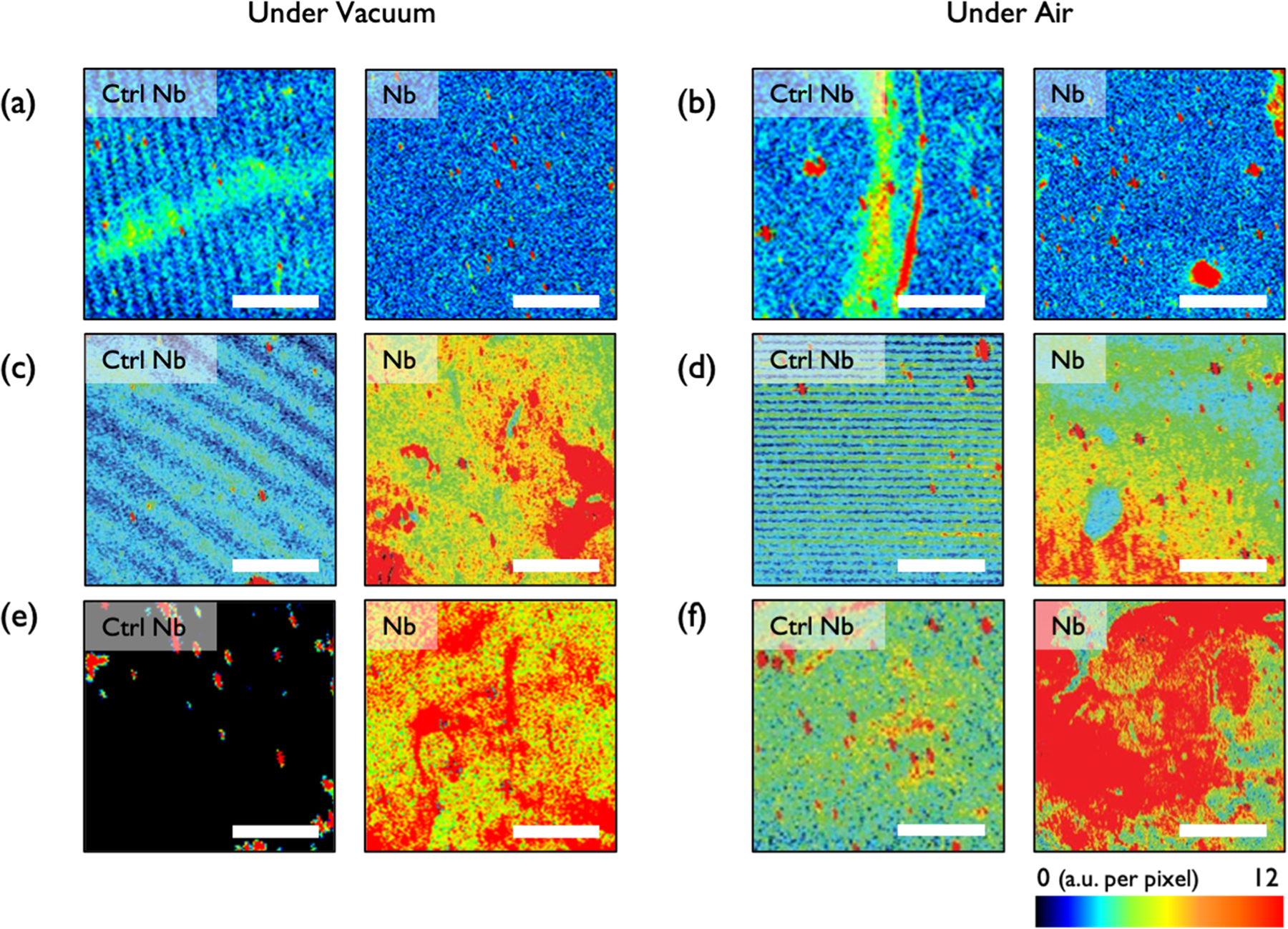
Fluorescence images of 1 mg/mL HEK293 lysate-treated samples on NbCtrl (left) and Nb arrays (right). (a,c,e) Samples kept under vacuum and (b,d,f) under desiccation: (a,b) samples without encapsulation, (c,d) pHEA-encapsulated samples, and (e,f) pTFOA-encapsulated samples. The estimated concentration of UCH37 in 1 mg/mL of cell lysate is 53 pM.21 Scale bars are 2 mm.
CONCLUSIONS
Here, we demonstrate that vapor-deposited polymer encapsulation of immobilized Nb arrays enables specific detection of target proteins in complex heterogeneous samples. The Nb chosen for this study was based on the availability, but the site-specific, HaloTag immobilization approach and subsequent encapsulation are generalizable to any Nb and the corresponding target protein of interest. The key advance is the use of piCVD to create a polymer matrix that both protects the integrity of the Nb array and allows for target protein diffusion. Future studies will focus on implementing this strategy to rapidly sense the presence of pathogens.
Supplementary Material
ACKNOWLEDGMENTS
This material was based on the work partially supported by the National Science Foundation under CBET 1706633 and the National Institutes of Health under R01GM110543.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsapm.1c00140.
AFM images and SE data (PDF)
Complete contact information is available at: https://pubs.acs.org/10.1021/acsapm.1c00140
The authors declare no competing financial interest.
REFERENCES
- (1).Perumal V; Hashim U Advances in biosensors: Principle, architecture and applications. J. Appl. Biomed 2014, 12, 1–15. [Google Scholar]
- (2).Limberis L; Magda JJ; Stewart RJ Polarized Alignment and Surface Immobilization of Microtubules for Kinesin-Powered Nanodevices. Nano Lett. 2001, 1, 277–280. [Google Scholar]
- (3).Skottrup PD; Nicolaisen M; Justesen AF Towards on-site pathogen detection using antibody-based sensors. Biosens. Bioelectron 2008, 24, 339–348. [DOI] [PubMed] [Google Scholar]
- (4).Steen Redeker E; Ta DT; Cortens D; Billen B; Guedens W; Adriaensens P Protein Engineering For Directed Immobilization. Bioconjugate Chem. 2013, 24, 1761–1777. [DOI] [PubMed] [Google Scholar]
- (5).Yoon J; Shin M; Lee T; Choi J-W Highly sensitive biosensors based on biomolecules and functional nanomaterials depending on the types of nanomaterials: A perspective review. Materials 2020, 13, 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Yang Q; Wu BC; Eles JR; Vazquez AL; Kozai TDY; Cui XT Zwitterionic Polymer Coating Suppresses Microglial Encapsulation to Neural Implants In Vitro and In Vivo. Adv. Biosyst 2020, 4, 1900287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Lichtenberg JY; Ling Y; Kim S Non-specific adsorption reduction methods in biosensing. Sensors 2019, 19, 2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Contreras-Naranjo J; Aguilar O Suppressing non-specific binding of proteins onto electrode surfaces in the development of electrochemical immunosensors. Biosensors 2019, 9, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Campuzano S; Pedrero M; Yáñez-Sedeño P; Pingarrón J Antifouling (bio) materials for electrochemical (bio) sensing. Int. J. Mol. Sci 2019, 20, 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Jiang C; Wang G; Hein R; Liu N; Luo X; Davis JJ Antifouling strategies for selective in vitro and in vivo sensing. Chem. Rev 2020, 120, 3852–3889. [DOI] [PubMed] [Google Scholar]
- (11).Zhang L; Cao Z; Bai T; Carr L; Ella-Menye J-R; Irvin C; Ratner BD; Jiang S Zwitterionic hydrogels implanted in mice resist the foreign-body reaction. Nat. Biotechnol 2013, 31, 553–556. [DOI] [PubMed] [Google Scholar]
- (12).Xu J; Lee H Anti-Biofouling Strategies for Long-Term Continuous Use of Implantable Biosensors. Chemosensors 2020, 8, 66. [Google Scholar]
- (13).Erfani A; Seaberg J; Aichele CP; Ramsey JD Interactions between Biomolecules and Zwitterionic Moieties: A Review. Biomacromolecules 2020, 21, 2557–2573. [DOI] [PubMed] [Google Scholar]
- (14).Vaisocherová H; Brynda E; Homola J Functionalizable low-fouling coatings for label-free biosensing in complex biological media: advances and applications. Anal. Bioanal. Chem 2015, 407, 3927–3953. [DOI] [PubMed] [Google Scholar]
- (15).Paschke S; Lienkamp K Polyzwitterions: from surface properties and bioactivity profiles to biomedical applications. ACS Appl. Polym. Mater 2020, 2, 129–151. [Google Scholar]
- (16).Damodaran VB; Murthy NS Bio-inspired strategies for designing antifouling biomaterials. Biomater. Res 2016, 20, 18–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Li Z; Guo Z Bioinspired surfaces with wettability for antifouling application. Nanoscale 2019, 11, 22636–22663. [DOI] [PubMed] [Google Scholar]
- (18).Liu T; Wang Y; Zhong W; Li B; Mequanint K; Luo G; Xing M Biomedical Applications of Layer-by-Layer Self-Assembly for Cell Encapsulation: Current Status and Future Perspectives. Adv. Healthcare Mater 2019, 8, 1800939. [DOI] [PubMed] [Google Scholar]
- (19).Sakr OS; Borchard G Encapsulation of enzymes in Layer-by-Layer (LbL) structures: latest advances and applications. Biomacromolecules 2013, 14, 2117–2135. [DOI] [PubMed] [Google Scholar]
- (20).Wang J; Hui N Zwitterionic poly(carboxybetaine) functionalized conducting polymer polyaniline nanowires for the electrochemical detection of carcinoembryonic antigen in undiluted blood serum. Bioelectrochemistry 2019, 125, 90–96. [DOI] [PubMed] [Google Scholar]
- (21).Deol KK; Crowe SO; Du J; Bisbee HA; Guenette RG; Strieter ER Proteasome-Bound UCH37/UCHL5 Debranches Ubiquitin Chains to Promote Degradation. Mol. Cell 2020, 80, 796–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Nevoltris D; Lombard B; Dupuis E; Mathis G; Chames P; Baty D Conformational Nanobodies Reveal Tethered Epidermal Growth Factor Receptor Involved in EGFR/ErbB2 Predimers. ACS Nano 2015, 9, 1388–1399. [DOI] [PubMed] [Google Scholar]
- (23).Buchfellner A; Yurlova L; Nüske S; Scholz AM; Bogner J; Ruf B; Zolghadr K; Drexler SE; Drexler GA; Girst S; Greubel C; Reindl J; Siebenwirth C; Romer T; Friedl AA; Rothbauer U A new nanobody-based biosensor to study endogenous PARP1 in vitro and in live human cells. PLoS One 2016, 11, No. e0151041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Irannejad R; Tomshine JC; Tomshine JR; Chevalier M; Mahoney JP; Steyaert J; Rasmussen SGF; Sunahara RK; El-Samad H; Huang B; von Zastrow M Conformational biosensors reveal GPCR signalling from endosomes. Nature 2013, 495, 534–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).De Meyer T; Muyldermans S; Depicker A Nanobody-based products as research and diagnostic tools. Trends Biotechnol. 2014, 32, 263–270. [DOI] [PubMed] [Google Scholar]
- (26).Anderson GP; Liu JL; Shriver-Lake LC; Zabetakis D; Sugiharto VA; Chen H-W; Lee C-R; Defang GN; Wu S-JL; Venkateswaran N; Goldman ER Oriented Immobilization of Single-Domain Antibodies Using SpyTag/SpyCatcher Yields Improved Limits of Detection. Anal. Chem 2019, 91, 9424–9429. [DOI] [PubMed] [Google Scholar]
- (27).Los GV; Encell LP; McDougall MG; Hartzell DD; Karassina N; Zimprich C; Wood MG; Learish R; Ohana RF; Urh M; Simpson D; Mendez J; Zimmerman K; Otto P; Vidugiris G; Zhu J; Darzins A; Klaubert DH; Bulleit RF; Wood KV HaloTag: a novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol 2008, 3, 373–382. [DOI] [PubMed] [Google Scholar]
- (28).Bilger D; Homayounfar SZ; Andrew TL A critical review of reactive vapor deposition for conjugated polymer synthesis. J. Mater. Chem. C 2019, 7, 7159–7174. [Google Scholar]
- (29).Yagüe JL; Gleason KK Enhanced cross-linked density by annealing on fluorinated polymers synthesized via initiated chemical vapor deposition to prevent surface reconstruction. Macromolecules 2013, 46, 6548–6554. [Google Scholar]
- (30).Baxamusa SH; Montero L; Dubach JM; Clark HA; Borros S; Gleason KK Protection of sensors for biological applications by photoinitiated chemical vapor deposition of hydrogel thin films. Biomacromolecules 2008, 9, 2857–2862. [DOI] [PubMed] [Google Scholar]
- (31).Baxamusa SH; Gleason KK Random copolymer films with molecular-scale compositional heterogeneities that interfere with protein adsorption. Adv. Funct. Mater 2009, 19, 3489–3496. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


