Abstract
Alzheimer’s disease (AD) is a progressive incurable neurodegenerative disease and a major health problem in aging population. We show that the combined use of Laser-Induced Breakdown Spectroscopy (LIBS) and machine learning applied for the analysis of micro-drops of plasma samples of AD and healthy controls (HC) yields robust classification. Following the acquisition of LIBS spectra of 67 plasma samples from a cohort of 31 AD patients and 36 healthy controls (HC), we successfully diagnose late-onset AD (> 65 years old), with a total classification accuracy of 80%, a specificity of 75% and a sensitivity of 85%.
1. Introduction
It is estimated that, by the year 2050, the number of Alzheimer’s disease (AD) cases in the US will exceed 20 million and the related healthcare costs will reach $ 1 trillion. AD, a neurodegenerative disease characterized by progressive and irreversible cognitive loss, is currently incurable. An efficient management of symptoms, as well as the success of clinical trials of drug candidates, relies on early diagnosis and clear discrimination of AD from other dementias [1,2]. Three biomarkers, collectively identified as ATN, were defined for AD, i.e. amyloid deposit (A), Tau pathology (T), and neurodegeneration (N) [3]. The ATN biomarkers can be detected in living individuals, but since the required procedures are either costly (e.g. brain imaging) or invasive (e.g. lumbar puncture for cerebrospinal fluid harvesting), diagnoses still largely rely on neurological and cognitive symptoms. This approach has two main pitfalls: 1) AD pathophysiological processes may start long before symptoms appear; and 2) symptoms themselves do not allow discriminating AD from other dementias due to their low specificity for AD. Therefore, there is a strong need for non-invasive cost-effective tests for AD early diagnosis.
Laser-Induced Breakdown Spectroscopy (LIBS) is the optical emission spectroscopy of Laser-Induced Plasmas (LIP), i.e. highly ionized gases that can be produced by the interaction of intense laser pulses and matter. LIPs are dynamic systems, that emit ultraviolet, visible and infrared radiation during their persistence time. This emission constitutes the LIBS signal, and, since the elemental composition of LIPs is the same as the irradiated target, assuming the ablation is stoichiometric, LIBS spectra can be used for qualitative and quantitative analysis of the target itself. Some of the most useful features of LIBS as an analytical technique include: simultaneous analysis of all elements present in the target, irrespective of their atomic mass; possibility of analyzing samples of virtually any chemical nature and state of aggregation; limited or absent sample preparation; immediate response; relatively low cost; and possibility of in situ and stand-off analyses. For this reason, LIBS lends itself to a large number of analytical applications, in a variety of diverse fields [4–10]. Biomedical applications of LIBS are currently in a phase of great vitality, as recently reviewed in [11]. Recently, our group proposed a minimally invasive approach to medical diagnosis, i.e. LIBS liquid biopsy. Instead of tissues, this method employs easily harvested biological fluids, such as blood, urine or saliva, that are deposited and dried on solid substrates prior to laser irradiation. Our previous studies on Epithelial Ovarian Cancer (EOC) [12] and melanoma [13] in mice demonstrated that classification accuracies up to 97% could be achieved. High classification accuracy (up to 100%) was also obtained by other authors, with serum and whole blood from lymphoma and multiple myeloma patients [14,15]. LIBS liquid biopsy makes it intrinsically versatile and applicable to problems of different nature, such as, in the biomedical field, the diagnosis of different diseases. The use of machine learning, moreover, can provide several advantages such as automated procedures; speed of analysis; high classification accuracy; generation of spectral libraries containing a large number of samples [16–18].
In this paper, we propose the combined use of LIBS and machine learning for the diagnosis of AD using biomedical fluids. Using an elemental analysis technique for fundamental studies and early diagnosis of AD is justified by the increasing attention that the role of metals in AD onset and progress has been receiving in the past three decades [19–22]. Despite these efforts, a clear correlation between the blood levels of various metals and AD is yet to be established [20]. In this work, we analyzed micro-drops of plasma from a cohort of AD patients and HC and focused on distinguishing the two classes with a data analysis approach based on the use of LIBS difference spectra.
2. Experimental setup
The LIBS experimental set used for this work is standard and described by several research groups [4–7]. The laser source used to generate the LIP is a 7-ns (ns) Nd: YAG laser (Surelite II, Continuum) operating at 1064 nm, focused on the sample with an air-spaced doublet lens with focal length of 30 mm. The samples were loaded onto a motorized and computer-controlled x-y-z translation stage located within a chamber (SciTrace, AtomTrace). The plasma emission was collected at an angle of 45° with respect to the laser beam by means of a 50 μm core-diameter optical fiber. This fiber was coupled to the Echelle spectrograph (Andor Technology, ME 5000) and a thermoelectrically cooled iStar Intensified Charge Coupled Device (ICCD) camera (Andor Technology, DH734–18F-03). The time parameters were: 1 μs starting delay after the laser pulse; 5 μs pulse width. The focused laser spot diameter was about 100 μm, the repetition rate was ½ Hz, and the laser energy was 130 ± 2 mJ. All measurements were carried out in air at atmospheric pressure. As described in our previous work on cancer diagnosis with LIBS liquid biopsy [12,13], we deposited the plasma samples on a solid target prior to laser irradiation. In this case, we deposited 5 μl of individual plasma specimen on the unpolished side of pure Si wafers, previously rinsed in 2-propanol, and dried the samples for 10 min with a Tungsten infrared lamp. To make up for possible inhomogeneity in the liquid distribution on the substrate, we acquired 100 single shot spectra for each sample. We ensured that each spectrum came from a fresh spot on the surface of the dried plasma drop by displacing the substrate with respect to the incoming laser beam with a computer-programmable target holder. After each laser shot, the substrate was displaced 200 mm from the previous position. As previously described [12,13], prior to data analysis, for each sample we removed all the spectra having total emission intensity lying beyond one standard deviation around the mean total emission intensity. Since the number of outliers was not the same for each sample, the resulting number of selected spectra was also not the same, after the filtering procedure.
Finally, we note that the subjects in this study were recruited from the Bedford VA Hospital Dementia Care Unit. The protocol was approved by the Bedford VA Hospital Institutional Review Board and written informed consent for each participant was obtained before initiation of the study and blood collection.
3. Results and discussion
3.1. Using difference spectrum method
Fig. 1 shows an example of LIBS spectrum of plasma from an AD patient a) and of an HC subject b), deposited and dried on a Si substrate. This figure shows that there is no clearly distinguishable difference between the two classes as the spectra are virtually identical, therefore LIBS spectra as such do not appear to provide an obvious discrimination between AD and HC samples.
Fig. 1.
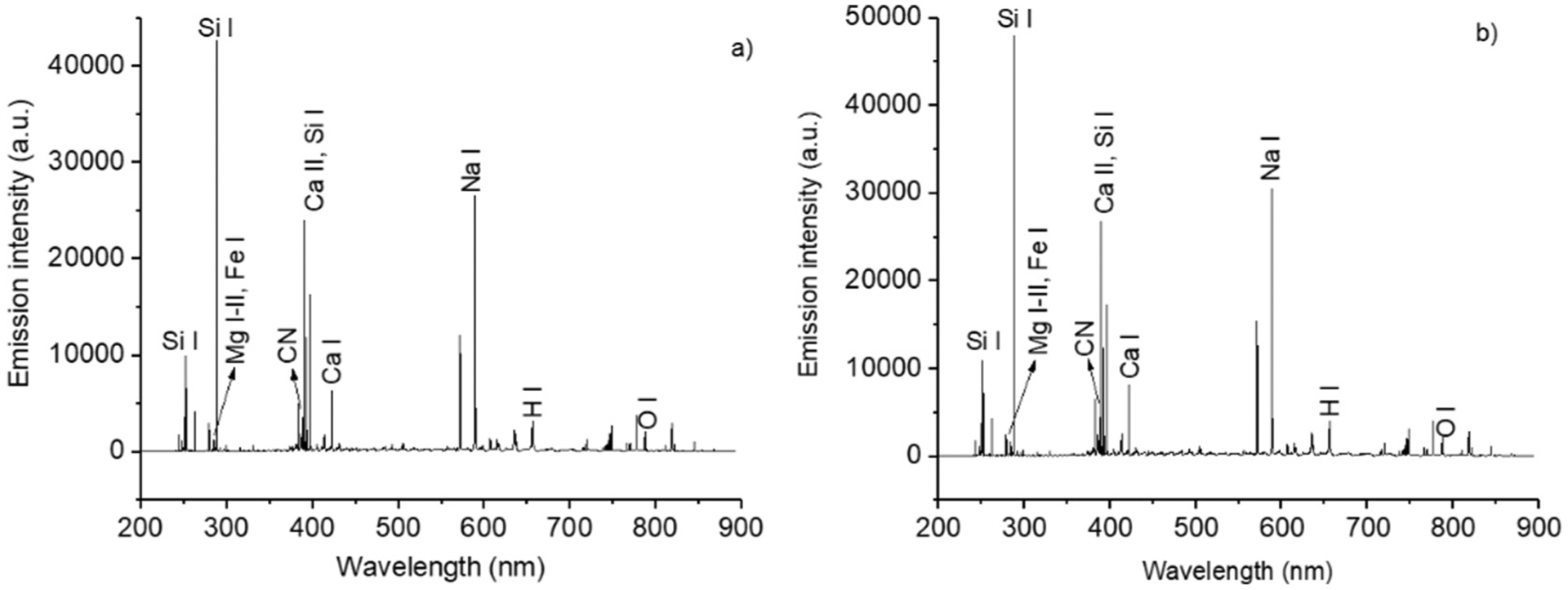
a)–b): LIBS spectra of the blood plasma of an AD patient deposited on a Si substrate. The species responsible for the emission of some of the main peaks are identified in the two spectra.
To seek to differentiate the AD and HC classes of samples, we generated difference spectra as a preprocessing step to enhance spectral differences between AD and HC samples. A group of 23 samples (11 AD and 12 HC) was selected randomly from our AD and HC pool of subjects as our training set and used to obtain the mean AD and mean HC spectra. After normalizing the spectra over the total emission intensity, we subtracted the mean HC spectrum from that of AD. This yielded a difference spectrum that displays positive and negative peaks (Fig. 2) and is representative of the differences between AD and HC samples.
Fig. 2.
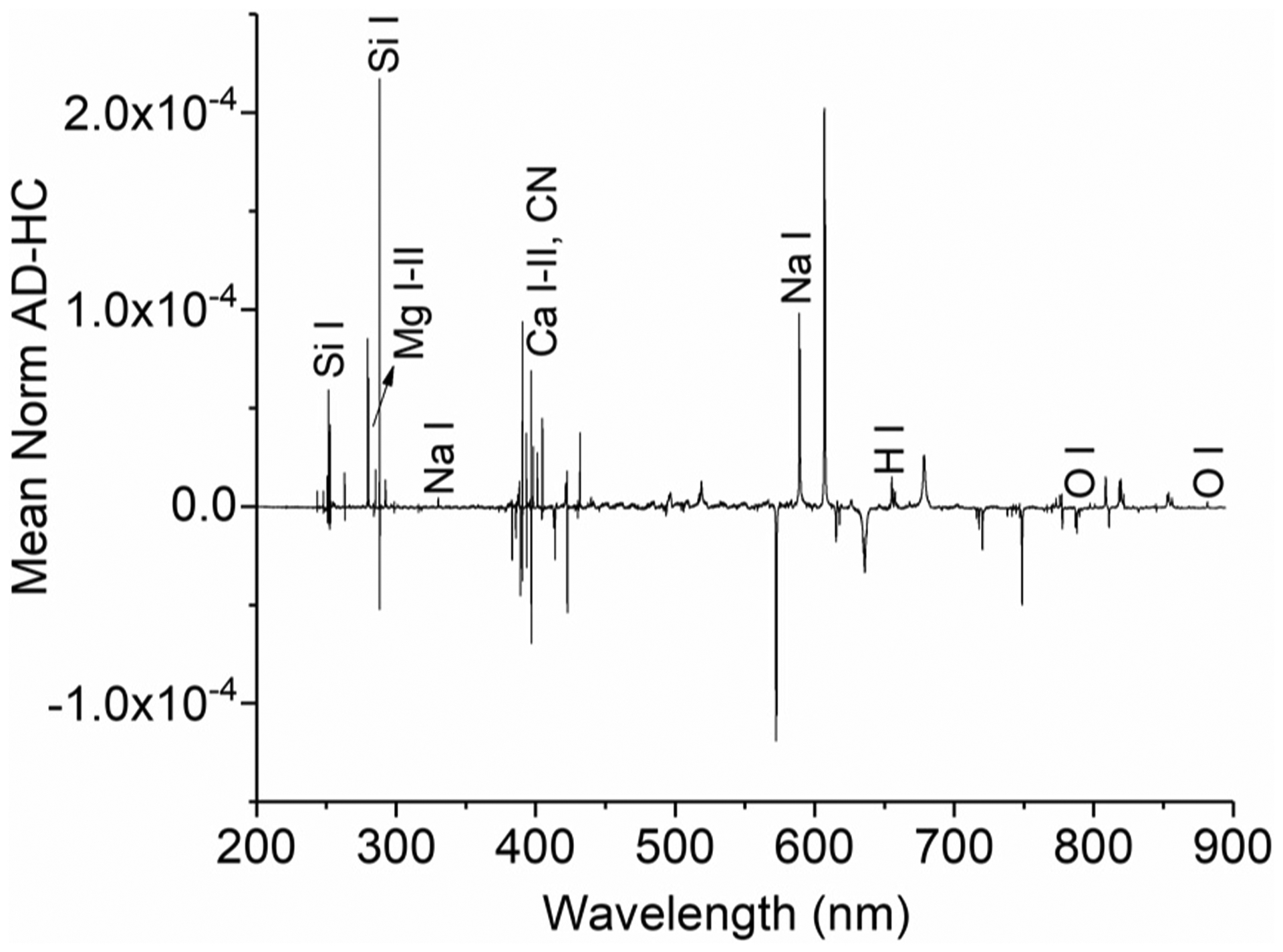
Model difference spectrum, showing typical net positive and negative peaks that were used for the diagnosis of unknown samples. Some of the difference peaks are assigned in the spectrum.
The transitions appearing as positive or negative peaks (Fig. 2) indicate an enrichment (positive) or a depletion (negative) of the given emitter in AD samples with respect to HC ones. Therefore, the difference spectrum was used as a model template to diagnose 66 testing spectra. These corresponded to 44 different patients. Samples from 10 of these patients were acquired more than once, and their spectra treated independently as belonging to different patients. For each unknown sample, we computed a difference spectrum, using a similar procedure to that described above.
We determined a mean HC spectrum and subtracted it from each of the unknown spectra. This provided a difference spectrum for each unknown, that we compared with the model. Since the latter was obtained as AD – HC, we assumed that if transitions in the unknown difference spectra had the same polarity as in the model, they should be classified as AD, and if the opposite polarity, as HC. Fig. 3 illustrates the difference spectra for one AD sample a) and one HC sample, b) in the spectral region 570–575 nm. Fig. 3 a)–b) shows an unambiguous difference between the AD (same polarity as in the reference, a) and HC samples (opposite polarity). This observation was used to obtain a diagnosis, i.e., each of the net transitions appearing as negative or positive peaks in the model difference spectrum was checked in the unknown ones and labeled “AD” or “HC”. Each sample was then diagnosed as either AD or HC, based on the number of AD or HC labels it received.
Fig. 3.
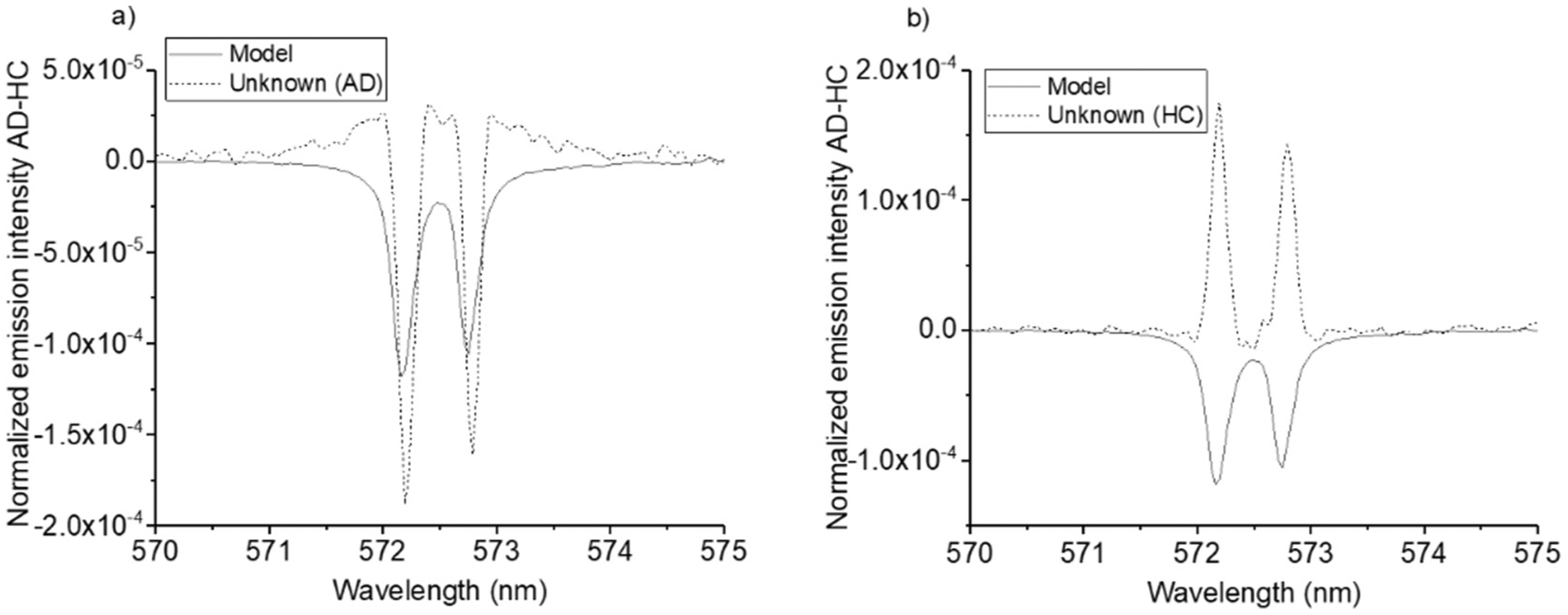
a)–b): Difference spectra of two samples, treated as unknowns in the difference spectrum diagnostic test, plotted together with the model spectrum difference. The sample in a) is an AD case, the sample in b) is an HC. The transition at 572.14 nm is Na II emission line while the one at 572.71 nm cannot be identified.
It should be noted that a slight shift of the wavelengths in the mean AD and mean HC spectra can cause an asymmetry in the difference peak. In this study, we did not use these; only non-asymmetric emission lines (reported in Table 1) were used to carry out the diagnostic test.
Table 1.
spectral features appearing in the difference spectra and examples of the application of the difference spectrum “majority vote” to two samples, treated as unknowns, i.e. patient 22 (clinical diagnosis: AD) and patient 104 (clinical diagnosis: HC). For patient 22, the number of AD and HC labels was, respectively, 17 and 14. The sample was therefore classified as AD. For patient 104, the number of AD and HC labels was, respectively, 8 and 14, and the sample was therefore classified as HC. It was not possible to assign some emission lines because of the lack of relevant information in the atomic spectra databases [23,24]. A description of the process by which assignation of these transitions was performed is provided in Appendix 1.
| Emitter | Wavelength (nm) | Polarity in model | Patient 22 | Label | Patient 104 | Label |
|---|---|---|---|---|---|---|
| Mg II | 279.5 | + | + | AD | + | AD |
| Mg II | 280.27 | + | + | AD | + | AD |
| Mg I | 285.21 | + | + | AD | + | AD |
| − | 292.35 | + | − | HC | − | HC |
| Na I | 330.24 | + | + | AD | + | AD |
| Ca II | 373.69 | − | − | AD | absent | |
| − | 383.19 | − | absent | + | HC | |
| CN | 387.08 | + | − | HC | − | HC |
| CN | 388.29 | + | − | H | − | HC |
| − | 389.16 | − | − | AD | + | HC |
| − | 398.25 | + | − | HC | − | HC |
| − | 401.27 | + | − | HC | absent | |
| − | 414.01 | − | − | AD | + | HC |
| CN | 421.52 | + | − | HC | − | HC |
| − | 431.72 | + | − | HC | absent | |
| − | 572.15 | − | + | HC | + | HC |
| − | 572.71 | − | + | HC | + | HC |
| − | 606.93 | + | + | AD | − | HC |
| − | 607.48 | + | + | AD | − | HC |
| − | 617.74 | − | − | AD | + | HC |
| − | 738.13 | − | + | HC | absent | |
| N I | 744.33 | − | − | AD | absent | |
| − | 787.95 | − | + | HC | + | HC |
| Na I | 818.37 | + | + | AD | + | AD |
| N I | 818.86 | + | + | AD | + | AD |
| Na I | 819.48 | + | + | AD | + | AD |
| N I | 821.63 | + | + | AD | + | AD |
| − | 852.57 | + | + | AD | absent | |
| − | 853.79 | + | + | AD | absent | |
| − | 856.03 | + | − | HC | absent | |
| − | 881.48 | + | − | HC | absent |
We found that not all transitions were visible in the difference spectra of all samples (Table 1). This stems from the fact that some transitions have very similar intensity to that of the mean HC spectrum, resulting in their absence in the difference spectrum. Therefore, due to unavoidable individual differences between the patients, the number of detectable peaks in difference spectra was not always the same. In addition, we observed that not all transitions unequivocally identified patients as either an AD case or a control, and a mix of different labels was the most common occurrence. In such cases, the diagnosis was obtained based on the majority of labels (in this case, AD for patient 22 and HC for patient 104, but we estimated the frequency of such mixed situations, in which the number of AD/HC label may be very similar or even identical. To do this, we introduced a parameter, that we named index of confidence, IC:
| (1) |
The possible values of this index range between +1 (all top transitions visible in the spectrum difference and providing an AD label) and − 1 (all top transitions visible in the spectrum difference and providing an HC label.) IC = 0 corresponds to an equal number of AD and HC labels, which would make it impossible to obtain a diagnosis only based on the difference spectrum method.
In this work, we did not encounter a situation where IC is equal to zero. As this may occur and to reduce its impact, carrying at least 3 experimental replicas for each patient is advisable. An alternative approach specific to LIBS could be based on performing the difference spectrum test with each single-shot spectrum, rather than with the average spectrum of each patient. However, such an approach is relatively time consuming. We determined that the frequency of cases with similar numbers of AD and HC labels was 6%, using a threshold values IC < 0.1 for AD and IC > −0.1 for HC. These values, with 31 selected spectral features, corresponded to diagnoses obtained with a difference between the number of AD and HC labels less than 3.
The transitions listed in Table 1 were used to diagnose 66 blood plasma samples. It is important to underline that no single spectral transition provided a clear-cut distinction between AD and HC. We were able to discriminate between the two classes only by considering all the spectral transitions and the resulting relative numbers of AD vs HC labels. It is also worth mentioning that the samples used to generate the reference difference spectrum were randomly chosen and were selected only with the goal of having a congruous and similar number of AD and HC specimens. We used our 23 training samples (11 AD and 12 HC) and applied the resulting model for subsequent analysis of remaining samples.
The cohort of donors analyzed consisted of a sample of patients 50 to 97 years old. We divided this sample set into two separate age groups, one for individuals older than 65 (HC and late-onset AD), and one for individuals younger than 65. The older group had 48 samples and contained a similar number of AD cases and HC (respectively, 26 and 22), while the younger group contained 18 samples, of whom only 2 AD cases and 16 HC samples. Table 2 shows the results, obtained using the same model, of the two separate tests, expressed in terms of total classification accuracy, specificity and sensitivity. These are defined as such:
| (2) |
| (3) |
| (4) |
where TP = true positive (samples correctly classified as AD), FN = false negative (AD samples wrongly classified as HC), TN = true negative (samples correctly classified as HC), FP = false positive (samples HC samples wrongly classified as AD.)
Table 2.
Results of the difference spectrum diagnostic test for the two age groups, i.e. 65 years old and younger, and older than 65.
| ≤ 65 years old (2 AD, 16 HC) | > 65 years old (26 AD, 22 HC) | |
|---|---|---|
| Specificity | 47% | 60% |
| Sensitivity | 50% | 68% |
| Total classification accuracy | 50% | 65% |
These data indicate that, while in the younger age group we were unable to distinguish AD and HC, in contrast 31 out of 48 patients were correctly identified in the older age group. This age “gap” in the results can be explained by the fact that the number of AD patients in the younger age group available for this study is very small. As a result, it is not possible to draw any conclusion regarding the impact of the age of the patients on our diagnostics method. For the remainder of our investigation, we therefore focused our investigation on the older age group for which we had more patients and an almost equal number of AD/HC subjects.
3.2. Using machine learning approach
Several studies have demonstrated the potential of LIBS to diagnose a variety of diseases, based on the different emission intensity of key elements (e.g. Ca and Mg enrichment in biopsied colorectal and breast cancer tissues [11] (and references therein.) Moreover, as previously observed [13], multivariate statistical approaches can provide high classification performance, even when a direct comparison between the emission intensity of individual spectral transitions does not yield a distinction between healthy and diseased samples. In this work, the difference spectrum method provided a moderate differentiation between plasma samples from AD and HC subjects in the older age group.
To improve the classification and investigate the development of an automated minimally invasive method for AD diagnosis, we explored the use of advanced statistical methods. Due to its simplicity and the fact that it does not require a parameter for optimization, we selected Quadratic Discriminant Analysis (QDA) implemented in scikit-learn [25,26]. This supervised learning technique provides the possibility to account for non-linear decision boundaries between classes. In addition, in QDA, each class is modeled with its own covariance matrix [23]. We note that we tested other statistical methods such as linear discriminant analysis (LDA) and partial least square analysis (PLS) using different type of input data (i.e. raw LIBS spectra vs. spectra resulting from the difference method and by selecting only those features that are either positive or negative in the difference spectra. We found that QDA with difference spectra as input data and manual feature selection provided the best classification results.
We used a leave-one-out cross-validation approach, that is, we divided the patients set into a training subset (all patients but one) and a testing subset (the one left-out-patient.) The training set was used to develop a prediction model, which was then used to provide the label of the one left-out patient. This procedure was repeated as many times as we had patients (N), by swapping the subsets, so that eventually each patient was used both in the training set (N – 1 times) and as a testing sample (once.) As shown in Fig. 4, the results for classification accuracy, sensitivity and specificity are 80%, 85% and 75% respectively.
Fig. 4.
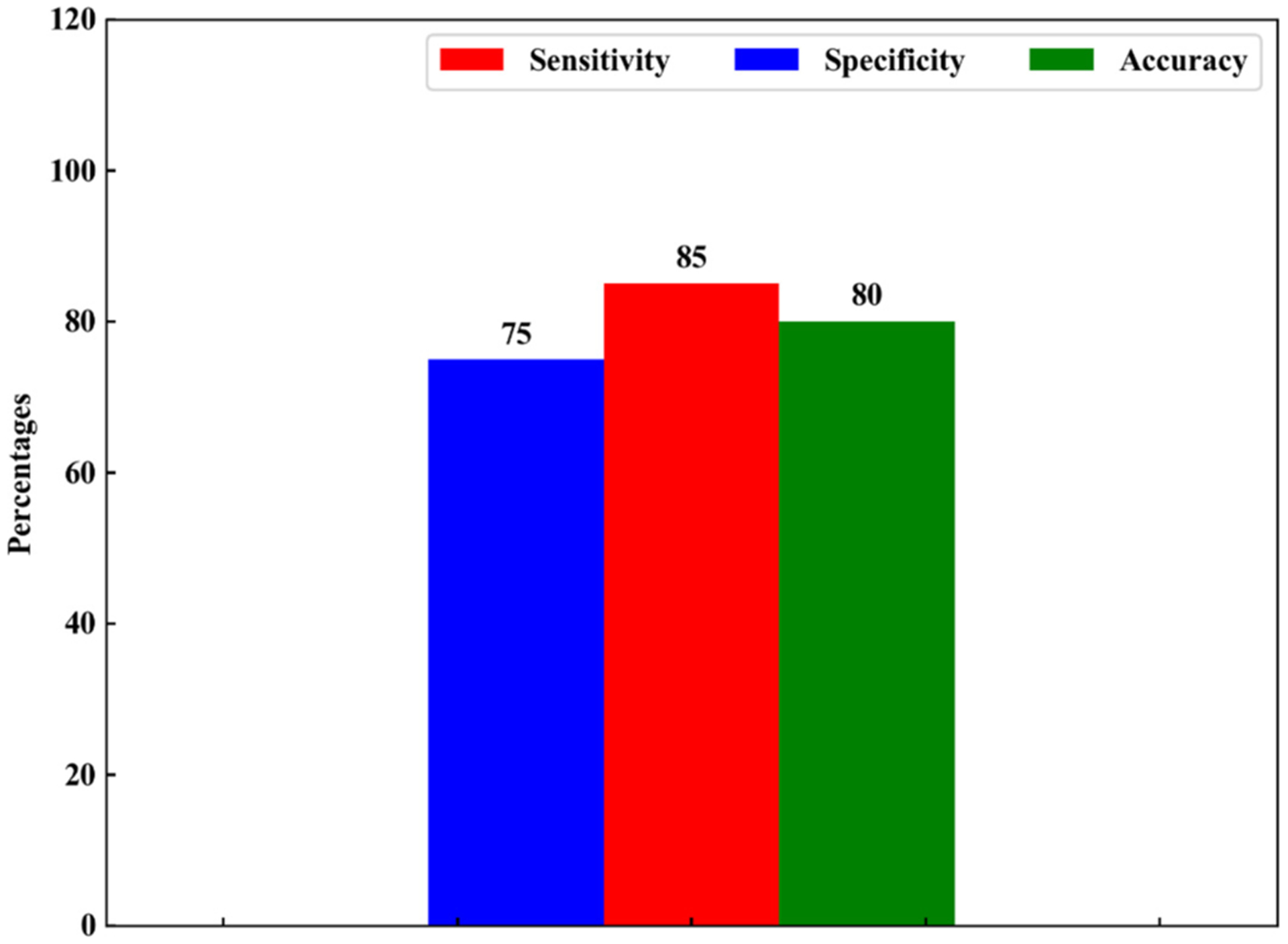
classification results obtained with QDA with manual feature selection (i.e. by using only the spectral features appearing as positive or negative peaks in the difference spectra.)
For all the tests we conducted, sensitivity was higher than specificity, which may indicate that our experimental approach is more appropriate to detect the presence of AD than its absence. This may be rationalized by hypothesizing the existence of a specific spectral profile, possibly of one or more elements, for AD patients, that can be captured by LIBS, either with the difference spectrum method or with statistical approaches. Nonetheless, controls are a group of individuals that do not share this common feature, while at the same time presenting all the individual variability associated to any group of people, which can ultimately contribute to make their identification more difficult.
4. Conclusions
We performed a feasibility study of LIBS liquid biopsy for the diagnosis of AD, using micro-drops of plasma from AD patients and HC. We developed a data analysis method based on the use of difference spectra as input data for one supervised machine learning algorithm (QDA), which provided classification accuracy, sensitivity and specificity, respectively, of 80%, 85% and 75%. These results were obtained by using manually selected features from the difference spectra (i.e. all the features that appeared as positive or negative peaks in difference spectra). These results open the way to further investigate and pursue the use of LIBS as a fast and minimally invasive methodology for the diagnosis of AD and possibly better understand its progression. This study contributes to the growing literature focused on investigating the potential of the combined use of LIBS and multivariate statistical approaches to analyze rapidly and relatively simply a large number of biomedical samples for the diagnosis of asymptomatic diseases.
Acknowledgments
This study was partially supported by the Kerith Foundation and by the award IO1 BX003527 from the Biomedical Laboratory Research and Development Service of the Veterans Affairs Office of Research and Development (WX), RF1AG063913 from National Institute of Aging (WX), and the Cure Alzheimer’s Fund (WX). One of us (NM) thanks Dr. Sean Wang for his support for this work. We thank the anonymous reviewer for useful comments and sggestions.
This work is dedicated to the memory of Mr. Trari M’hamed, Professor of Physics at the Lycée Abane Ramdane in Algiers, Algeria who dedicated his life to teaching physics and chemistry and the memory of Mr. Nicola Gaudiuso for his love of science and his battle against Alzheimer’s disease.
Appendix A. Appendix 1
The spectral assignation of the transitions reported in Table 1, and which are included in our classification analysis, was performed using information available in the widely employed databases, i.e. NIST and Kurucz [23,24]. In addition, we performed a series of cross-checks:
First, we determined the correlation coefficient between the intensity of each of the transitions appearing in the LIBS spectra: transitions from the same emitter (both in the same ionization stage and in different ionization stages) are typically correlated, which can be used to tentatively identify the emitter even when the databases do not list any transition at the given wavelength. However, the correlation analysis should be considered with caution, because we observed high correlation between different elements (e.g. Mg I 285.21 nm is highly correlated with Na I 589.00 nm and 589.58 nm; H 656.27 nm is highly correlated with O I triplet at 777 nm and with N I transitions at 744.33 nm and 742.36 nm). This suggests that not only the chemical identity of the emitter, but also its origin and sample-specific considerations contribute to the correlation;
Second, we considered LIBS plasmas to be in Local Thermodynamic Equilibrium (LTE) and therefore, based on Boltzmann distribution, high-energy transitions should be less populated than low-energy ones. Therefore, if low-energy transitions of a given element were missing from the spectrum, we ruled out that some of the other transitions we observed in our spectra could be assigned to higher-energy transitions of that same element.
Third, we included other spectroscopic parameters, i.e. Einstein coefficient of spontaneous emission, Aul, and relative intensity. With this regard, we ruled out the possibility of assigning a given peak to a transition with lower Aul if other transitions of the same element with higher Aul were not visible in the spectra. We used relative intensity in a similar way, always taking into account that the latter is only a qualitative indication of the expected intensity of emission peaks.
Based on these considerations, we were unable to assign with absolute certainty some of the transitions present in the spectra. Below, we report the tentative assignations for the unassigned transitions reported in Table 1. This is done by relaxing one or more of the constraints described above. For the sake of completeness, for each transition we provide the possible caveats associated with the assignations thus obtained. In the following, we will refer to the lower and upper level involved in the transition as, respectively, El and Eu.
292.35 nm: this transition does not correlate with any of the other transitions appearing in the spectrum. It may be tentatively assigned it to a high-energy transition of Na II (292.35 nm, Aul 1.41e +07 s−1, El 265,689.62 cm−1, Eu 299,885.37 cm−1). Nonetheless, several Na II transitions between the similar energy levels and higher Aul are not visible in the spectra (e.g. 292.10 nm, 293.77 nm.)
383.16 nm: this transition does not correlate with any of the other transitions appearing in the spectrum. It may be tentatively assigned to a high-energy transition of C II (383.17 nm), though another C II transition at 383.57 nm, with virtually identical spectroscopic parameters and energy levels, is absent from the spectra.
389.16 nm: this transition correlates with only one transition, at 404.81 nm, that we did not use for our analysis. The transition at 404.81 nm may be tentatively assigned to a high-energy transition of O II (404.82 nm, El 231,427.970 cm−1 - Eu 256,123.231 cm−1, no available data for Aul). Other O II transitions between lower energy levels and with higher Aul are not visible in the spectra, e.g. 397.33 nm. The transition at 389.16 nm may be tentatively assigned to a high-energy transition of Mg I (389.19 nm, El 57,833.40 cm−1, Eu 83,520.47 cm−1). Note that a similar Mg I transition at 389.56 nm is absent from the spectra.
398.25 nm: this transition correlates with one CN band at 421.62 nm, one Na I transition at 808.34 nm and one transition at 775.73 nm (the latter, which we did not use for our analysis, may not be assigned to any element, as the only transitions listed in databases at this wavelength are: Ar II 775.70 nm, Nb I 775.73 nm and Rb I 775.76 nm). The 398.25 nm transition may be tentatively assigned to one Ti I transition (398.18 nm, El 0 - Eu 25,102.875 cm−1, Aul 4.42e+07 s−1, though other resonance Ti lines with higher Aul are absent from the spectra, e.g. 264.11 nm and 294.20 nm) or one high-energy O II transition (398.27 nm, El 189,068.514 cm−1 - Eu 214,169.920 cm−1, Aul 4.16e+07 s−1, though other O II transitions involving similar energy levels and having higher Aul are not visible in the spectra, e.g. 397.33 nm).
401.27 nm: this transition does not correlate with any of the other transitions appearing in the spectra. It might be tentatively assigned to a high-energy Fe II transition (401.27 nm, El 88,614.523 cm−1 - Eu 113,528.091 cm−1, no available Aul data). Nonetheless, ground-state and low-energy Fe II transitions with high Aul and relative intensity (respectively in the order of 108 s−1 and 105 a.u.) are absent from the spectra.
431.72 nm: this transition does not correlate with any of the other transitions appearing in the spectrum. Three possible candidates for its assignation are high-energy transitions from the following species: O II (431.71 nm, El 185,235.281 cm−1 – Eu 208,392.258 cm−1, Aul 3.68e+07 s −1), C II (431.73 nm, El 186,466.02 cm−1 – Eu 209,622.32 cm−1, Aul 5.89e+07 s−1), Ca I (431.71 nm, El 47,843.760 cm−1 - Eu 71,001.000 cm−1, Aul 1.082e+06 s−1).
572.15–572.75 nm: these transitions most likely belong to the same element, based on the observed behavior in spectra and difference spectra, as well as on the fact that they are mutually highly correlated. They are also correlated with three unknown transitions at 741.40, 786.90, 787.94 nm, none of which was included in our analysis. The transition at 572.15 nm may be tentatively assigned to a Na II transition (572.14 nm) for which no data about Einstein coefficient, energy levels or terms is available in either database. No attribution whatsoever could be attempted for the transition at 572.71 nm. In [27], one of the few papers in the LIBS literature about biological samples that features detailed assignation of individual transitions, these peaks were observed and left unassigned.
606.93–607.48 nm: these transitions most likely belong to the same element, based on the observed behavior in spectra and to the fact that they are mutually highly correlated. They are also correlated with one unknown transition at 771.12 nm, that we did not include in our analysis. No attribution could be attempted for either transition.
617.74 nm: this transition does not correlate with any of the other transitions appearing in the spectrum. It might be tentatively assigned to Br I (El 75,697.05 cm−1 - Eu 91,880.64 cm−1, no data for Aul available). Nonetheless, the main Br I peak (827.24 nm, Aul 3.5e +07, El 63,436.45 cm−1 - Eu 75,521.50 cm−1) is less intense that the 617.74 nm transition.
738.13 nm: this transition correlates with two unknown transitions at 741.40 nm and 786.90 nm, neither of which we used for our analysis. While the transition at 741.40 nm might be tentatively assigned to Cl I (El 71,958.363 cm−1 – Eu 85,442.430 cm−1, Aul 4.7e +06 s−1), the main Cl I lines are either barely visible (837.6 nm, El 71,958.363 cm−1 – Eu 83,894.037 cm−1, Aul 2.8e+07 s−1; 857.53 nm, El 72,827.038 cm−1 – Eu 84,485.309 cm−1, Aul 1.2e +07 s−1) or absent (833.33 nm, El 72,488.568 cm−1 - Eu 84,485.309 cm−1, Aul 1.6e+07 s−1). The 741.40 nm transition was also observed in [1] and left unassigned. The transition at 786.90 nm might be tentatively assigned to a high-energy Fe I transition (El 40,842.154 cm−1 – Eu 53,545.833 cm−1, Aul 105 s−1), but main Fe I lines with El = 0 and Aul in the order of 108 s−1 are not visible in the spectra (e.g. 252.28 nm and 271.90 nm). The transition at 738.13 nm may be tentatively assigned to a high-energy O II transition (El 232,745.981 cm−1 - Eu 246,291.822, no Aul data available, though other O II transitions involving lower energy levels and having Aul in the order of 107 s−1 are not visible in the spectra, as mentioned earlier) or to a high-energy, low Aul Fe I transition (El 43,163.323 cm−1 – Eu 56,707.280 cm−1, Aul 5.049e +05 s−1, but main Fe I lines with El = 0 and Aul in the order of 108 s−1 are not visible in the spectra, e.g. 252.28 nm and 271.90 nm).
787.95 nm: this transition correlated with the following unknown transitions: 572.15, 572.71 nm; 741.40 nm; 786.90 (the latter was not used in our analysis,) whose assignation has been discussed above. The transition at 787.95 nm may be tentatively assigned to a high-energy, low-Aul Fe I transition (787.97 nm, El 40,594.432 cm−1 – Eu 53,281.689 cm−1, Aul 2.185e+05 s−1, but main Fe I lines with El = 0 and Aul in the order of 108 s−1 are not visible in the spectra, e.g. 252.28 nm and 271.90 nm).
852.57 nm: this transition does not correlate with any of the other transitions appearing in the spectra. It may be tentatively assigned to a Ca I transition (852.57 nm, El 35,730.454 cm−1 - Eu 47,456.452 cm−1, Aul 1.920e+06 s−1). Transitions between lower energy levels and having Aul in the order of 108 s−1, nonetheless, are not visible in the spectra (e.g. 300.086 nm, 551.30 nm).
853.79 nm: this transition does not correlate with any other transition appearing in the spectra. It may be tentatively assigned to a Ca I transition (853.69 nm, El 36,547.688 cm−1 - Eu 48,258.300 cm−1, Aul 1.397e+05 s−1). Transitions between lower energy levels and having Aul in the order of 108 s−1, nonetheless, are not visible in the spectra (e.g. 300.086 nm, 551.30 nm).
856.03 nm: this transition correlates with several others, most of which we did not use for our analysis. These are: 678.35 nm (tentative assignation: Fe I 678.33 nm); 773.03 nm (tentative assignation: Ca I 772.97); 775.3 nm (Mg I 775.33 nm); 808.63 nm (Cl I 808.56 nm); 856.63 nm (tentative assignation: N II 856.68 nm); 878.59 nm (no assignation possible); 881.49 nm (see below). For the tentative assignation of all these transitions, the aforementioned caveats should be kept in mind. The 856.03 nm transition may be tentatively assigned to Fe I 855.97 nm (El 41,178.409 cm−1 – Eu 52,857.800 cm−1, Aul 1.261e+06 s−1) but main Fe I lines with El = 0 and Aul in the order of 108 s−1 are not visible in the spectra, e.g. 252.28 nm and 271.90 nm.
881.48 nm: this transition correlates with several others, most of which we did not use for our analysis: 421.61 (CN), 518.91 nm (Ca I 518.89 nm), 678.35 nm (tentative assignation: Fe I 678.33 nm); 742.49 nm (N I 742.44 nm); 773.03 nm (tentative assignation: Ca I 772.97 nm); 775.73 nm (tentative assignation: Mg I 775.33 nm); 808.34 nm (tentative assignation: C I 808.38 nm); 808.63 nm (Cl I 808.56 nm); 856.03 nm (see above); 856.63 nm (tentative assignation: N II 856.68 nm); 878.59 nm (no assignation possible). The 881.49 nm transition may be tentatively assigned to a high-energy, low-Aul Ca I transition (881.40 nm, El 47,456.452 cm−1 - Eu 58,798.920 cm−1, Aul 2.082e+05 s−1) or to a very high-energy Fe I transition (881.45 nm, El 98,114.577 cm−1 – Eu 109,456.381 cm−1, Aul 3.0e+07 s−1.) The aforementioned caveats about stronger Ca I and Fe I transitions being absent from the spectra should be kept in mind for what concerns these tentative assignations.
Footnotes
Declaration of Competing Interest
The authors declare no conflicts of interest.
The views expressed in this article are those of the authors and do not represent the views of the US Department of Veterans Affairs or the US Government.
References
- [1].Huang C, Isidoro C, Raman spectrometric detection methods for early and non-invasive diagnosis of Alzheimer’s disease, J. Alzheimers Dis 57 (2017) 1145–1156. [DOI] [PubMed] [Google Scholar]
- [2].Olsson B, Lautner R, Andreasson U, Öhrfelt A, Portelius E, Bjerke M, Hölttä M, er Rosén C, Olsson C, Strobel G, Wu E, Dakin K, Petzold M, Blennow K, Zetterberg H, CSF and blood biomarkers for the diagnosis of Alzheimer’s disease: a systematic review and meta-analysis, Lancet Neurol. 15 (2016) 673–684. [DOI] [PubMed] [Google Scholar]
- [3].Jack CR Jr. , Bennett DA, Blennow K, Carrillo MC, Feldman HH, Frisoni GB, Hampel H, Jagust WJ, Johnson KA, Knopman DS, Petersen RC, Scheltens P, Sperling RA, Dubois B, A/T/N: an unbiased descriptive classification scheme for Alzheimer disease biomarkers, Neurology 87 (2016) 539–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Radziemski LJ, Cremers DA (Eds.), Laser-induced Plasma and Applications, Marcel Dekker, New York, NY, USA, 1989. [Google Scholar]
- [5].Radziemski LJ, Cremers DA (Eds.), Handbook of Laser-Induced Breakdown Spectroscopy, Second edition, John Wiley & Sons, Ltd, 2013. [Google Scholar]
- [6].Miziolek AW, Palleschi V, Schechter I (Eds.), Laser Induced Breakdown Spectroscopy (LIBS): Fundamentals and Applications, Cambridge University press, 2006. [Google Scholar]
- [7].Musazzi S, Perrini U (Eds.), Laser-Induced Breakdown Spectroscopy. Theory and Applications, 182 Springer Series in Optical Sciences, 2014. [Google Scholar]
- [8].Markushin Y, Sivakumar P, Connolly D, Melikechi N, Tag-femtosecond laser-induced breakdown spectroscopy for the sensitive detection of cancer antigen 125 in blood plasma, Anal. Bioanal. Chem 407 (2015) 1849–1855. [DOI] [PubMed] [Google Scholar]
- [9].Sivakumar P, Fernández-Bravo A, Taleh L, Biddle JF, Melikechi N, Detection and classification of live and dead escherichia coli by laser-induced breakdown spectroscopy, Astrobiology 15 (2015) 144–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Pokrajac D, Lazarevic A, Kecman V, Marcano A, Markushin Y, Vance T, Reljin N, McDaniel S, Melikechi N, Automatic classification of laser-induced breakdown spectroscopy (LIBS) data of protein biomarker solutions, Appl. Spectrosc 68 (2015) 1067–1075. [DOI] [PubMed] [Google Scholar]
- [11].Gaudiuso R, Melikechi N, Abdel-Salam ZA, Harith MA, Palleschi V, Motto-Ros V, Busser B, Laser-induced breakdown spectroscopy for human and animal health: a review, Spectrochim. Acta B 152 (2019) 123–148. [Google Scholar]
- [12].Melikechi N, Markushin Y, Connolly DC, Lasue J, Ewusi-Annan E, Makrogiannis S, Age-specific discrimination of blood plasma samples of healthy and ovarian cancer prone mice using laser-induced breakdown spectroscopy, Spectrochim. Acta B 123 (2016) 33–41. [Google Scholar]
- [13].Gaudiuso R, Ewusi-Annan E, Melikechi N, Sun X, Liu B, Campesato LF, Merghoub T, Using LIBS to diagnose melanoma in biomedical fluids deposited on solid substrates: limits of direct spectral analysis and capability of machine learning, Spectrochim. Acta B 146 (2018) 106–114. [Google Scholar]
- [14].Chen X, Li X, Yu X, Chen D, Liu A, Diagnosis of human malignancies using laser-induced breakdown spectroscopy in combination with chemometric methods, Spectrochim. Acta B 139 (2018) 63–69. [Google Scholar]
- [15].Chen X, Li X, Yang S, Yu X, Liu A, Discrimination of lymphoma using laser induced breakdown spectroscopy conducted on whole blood samples, Biomed. Opt. Express 9 (2018) 1057–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Torrione P, Collins LM, Morton KD Jr., Multivariate analysis, chemometrics, and machine learning in laser spectroscopy, in: Baudelet M (Ed.), Laser Spectroscopy for Sensing, Woodhead Publishing Series in Electronic and Optical Materials, 2014Number 43. [Google Scholar]
- [17].El Haddad J, Canioni L, Bousquet B, Good practices in LIBS analysis: review and advices, Spectrochim. Acta B 101 (2014) 171–182. [Google Scholar]
- [18].Pořízka P, Klus J, Képeš E, Prochazka D, Hahn DW, Kaiser J, On the utilization of principal component analysis in laser-induced breakdown spectroscopy data analysis, a review, Spectrochim. Acta B 148 (2018) 65–82. [Google Scholar]
- [19].Bush AI, Pettingell WH, Multhaup G, d Paradis M, Vonsattel JP, Gusella JF, Beyreuther K, Masters CL, Tanzi RE, Rapid induction of Alzheimer a beta amyloid formation by zinc, Science 265 (1994) 1464–1467. [DOI] [PubMed] [Google Scholar]
- [20].Cicero CE, Mostile G, Vasta R, Rapisarda V, Signorelli SS, Ferrante M, Zappia M, Nicoletti A, Metals and neurodegenerative diseases. A systematic review, Environ. Res 159 (2017) 82–94. [DOI] [PubMed] [Google Scholar]
- [21].Adlard PA, Bush AI, Metals and Alzheimer’s disease: how far have we come in the clinic? J. Alzheimers Dis 62 (2018) 1369–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bush AI, The metal theory of Alzheimer’s disease, J. Alzheimers Dis 30 (2012) 1–5. [DOI] [PubMed] [Google Scholar]
- [23].NIST Atomic Spectra Database, https://www.nist.gov/pml/atomic-spectra-database (latest access: April 7, 2020.).
- [24].Kurucz Atomic Spectra Database, https://www.cfa.harvard.edu/amp/ampdata/kurucz23/sekur.html (latest access: April 7, 2020.).
- [25].Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay É, Scikit-learn: machine learning in Python, J. Mach. Learn. Res 12 (2011) 2825–2830. [Google Scholar]
- [26].Hastie T, Tibshirani R, Friedman J, The Elements of Statistical Learning, Springer, 2008. [Google Scholar]
- [27].Baudelet M, Guyon L, Yu J, Wolf J-P, Amodeo T, Fréjafon E, Laloi P, Spectral signature of native CN bonds for bacterium detection and identification using femtosecond laser-induced breakdown spectroscopy, Appl. Phys. Lett 88 (2006) 063901. [Google Scholar]


