Abstract
An in-depth understanding of lung cancer biology and mechanisms of tumor progression has facilitated significant advances in the treatment of lung cancer. There remains a pressing need for the development of innovative approaches to detect and intercept lung cancer at its earliest stage of development. Recent advances in genomics, computational biology and innovative technologies offer unique opportunities to identify the immune landscape in the tumor microenvironment associated with early-stage lung carcinogenesis, and provide further insight in the mechanism of lung cancer evolution. This review will highlight the concept of immunoediting and focus on recent studies assessing immune changes and biomarkers in pulmonary premalignancy and early-stage NSCLC. A pro-tumor immune response hallmarked by an increase in checkpoint inhibition and inhibitory immune cells and a simultaneous reduction in anti-tumor immune response have been correlated with tumor progression. The potential systemic biomarkers associated with early lung cancer will be highlighted along with current clinical efforts for lung cancer interception. Research focusing on the development of novel strategies for cancer interception prior to the progression to advanced stages will potentially lead to a paradigm shift in the treatment of lung cancer and have a major impact on clinical outcomes.
Keywords: BIOMARKERS/Cancer risk biomarkers, CANCER PREVENTION/Early detection, CANCER INTERCEPTION/Cancer surveillance and screening, IMMUNOLOGY/Immune responses to cancer, TUMOR MICROENVIRONMENT/Immune cells and the microenvironment
Introduction
An in-depth understanding of lung cancer biology and mechanisms of tumor progression has facilitated significant advances in the treatment of lung cancer [1]. For example, tyrosine kinase inhibitor (TKI) targeted therapies and immune checkpoint blockade (ICB) immunotherapies have resulted in durable responses and survival benefits in subsets of non-small cell lung cancer (NSCLC) patients. The lung cancer survival rate, however, remains low, particularly for metastatic disease. There is a pressing need for the development of innovative approaches to detect and intercept lung cancer at its earliest point. Recent advances in genomics, computational biology and new technologies offer unique opportunities to identify the earliest cellular and molecular events associated with lung carcinogenesis as well as the immune landscape in the tumor microenvironment (TME). This will afford the development of novel strategies for cancer interception prior to the progression to advanced stages [2, 3]. In this review, we will highlight the concept of immunoediting and focus on recent studies assessing immune changes in the context of lung cancer pathogenesis and early detection.
Materials and Methods
A comprehensive literature search was conducted in PubMed and Early Detection Research Network (EDRN) Public Portal to identify lung cancer and immunity markers research. Search terms were combined with “lung cancer”, “immunity markers”, “immune changes”, “biomarkers” and “interception”. These previous publications were summarized in this review to highlight immunological changes and biomarkers during development of lung cancer.
Results
1. History of Immunosurveillance and Cancer Immunoediting
Among the earliest demonstrations of the capacity of the immune system to affect tumor development were those formulated by Paul Ehrlich in 1909. He found that the injection of weakened cancer cells generated tumor immunity in mice and concluded that the host defense system can keep emerging tumors at bay [4]. This revolutionary concept was galvanized by Lewis Thomas in 1959 who postulated that the immune system has the capacity to protect against the development of cancer [5]. These early notions laid the foundation for Burnet’s concept of immunosurveillance. He wrote: “it is an evolutionary necessity that there should be some mechanism for eliminating or inactivating such potentially dangerous mutant cells and it is postulated that this mechanism is of immunological character.” [6] Studies from Old and Boyse at that time supported this hypothesis through the identification of murine tumor antigens [7].
These studies shaped the early concepts of immunosurveillance but they were limited by scientific approaches to fully demonstrate that the immune system played a role in cancer pathogenesis. It was not until the early 2000s with advances in knockout mice and genetic models that the immunosurveillance concept was experimentally confirmed. For example, immunogenic tumor cells expressing dominant negative Interferon Gamma (IFN-γ) receptors were found to have enhanced growth in vivo [8]. Other studies using perforin knockout mice and eventually recombination activating gene 1 (RAG-1) or RAG-2 knockout mice solidified the concept that the immune system, specifically lymphocytes, protected the host against chemically induced tumors and spontaneously developing epithelial tumors [9]. The studies by Smyth et al. and Dunn et al. shed further light on the mechanisms of immunosurveillance [10–13]. Subsequent studies not only suggested the involvement of the immune system in protecting humans, but also demonstrated the emergence of tumors with reduced immunogenicity.
These studies established immunosurveillance while simultaneously raising a new question: if the immune system has the capability to prevent and eradicate neoplastic cells, why does cancer still develop? This question, along with the discovery of tumors with reduced immunogenicity, led to the concept of cancer immunoediting. Cancer immunoediting highlights a three-phase model of tumor growth: elimination, equilibrium and escape [11]. Elimination embodies the classical concept of cancer immunosurveillance in which the immune system eradicates neoplastic disease. Equilibrium defines a state of continued immune elimination with incomplete tumor destruction. Finally, escape is the outgrowth of tumor cells that have successfully evaded the immune response of the previous phase.
2. Premalignancy, Lung Cancer and Early Detection
Lung cancer is the leading cause of cancer death worldwide, accounting for 25% of all cancer deaths. In 2017, there were 2.2 million incident cases and 1.9 million deaths. The Global Burden of Disease Collaboration has reported that from 2007 to 2017, lung cancer cases have increased by 37% worldwide [14]. Approximately 85% lung cancer patients have NSCLC, which are composed of two major subtypes, lung squamous cell carcinoma (LUSC) and lung adenocarcinoma (LUAD) [1]. Lung cancer is often diagnosed at advanced stages leading to a poor prognosis. The 5-year survival rate is approximately 17%. Multiple efforts are now being directed at diagnosing lung cancer at earlier stages. The National Lung Screening Trial (NSLT) [15, 16] and the Nederlands-Leuvens Longkanker Screenings Onderzoek, (NELSON) trial [17] both used low-dose computed tomography (LDCT) to screen high-risk populations to reduce lung cancer mortality. These ground-breaking studies demonstrate the importance and impact of early detection. Studies are now focused on improving the performance characteristics of LDCT scans via radiomics and machine learning as well as using additional non-invasively obtained biomarkers to complement imaging based assessments. An abnormal LDCT scan could result from benign, premalignant or invasive disease. Understanding the cellular and molecular determinants of pulmonary premalignancy progression to invasive cancer will facilitate detection and interception of lung cancer at its earliest stages.
With the recognition that a better understanding of early stages of lung cancer pathogenesis can improve patient outcomes through early diagnosis and interception, the National Cancer Institute (NCI) launched multiple initiatives to direct the research effort to lung cancer early detection. The Early Detection Research Network (EDRN) was initiated by the NCI to bring together multiple institutions to identify biomarkers for clinical applications for early cancer detection. Another effort facilitated by the NCI is the PreCancer Atlas (PCA) initiative, which aims to utilize a comprehensive multi-omic strategy to establish detailed molecular and cellular characteristics of premalignant lesions and their evolution to invasive cancer [18–20]. Technological advances in autofluorescence bronchoscopy (AFB), multispectral imaging and laser capture microdissection (LCM) have allowed for better characterization of these lesions. Coupled with enhanced analysis tools in multiplex immunofluorescence (MIF), mass cytometry by time-of-flight (CyTOF), genome-wide omic approaches and single cell sequencing, studies are assessing premalignancy and the immune microenvironment to better understand the molecular and cellular determinants of lung cancer progression. Below, we highlight selected key studies that exemplify research aimed at defining immunity markers in pulmonary premalignancy and early-stage lung cancer.
3. Immune Changes and Biomarkers in Pulmonary Premalignancy and Early-Stage Lung Cancer
3.1. Immune Changes in Premalignancy and Early-Stage NSCLC (Figure 1)
Figure 1. Immune Changes during Lung Cancer Progression.
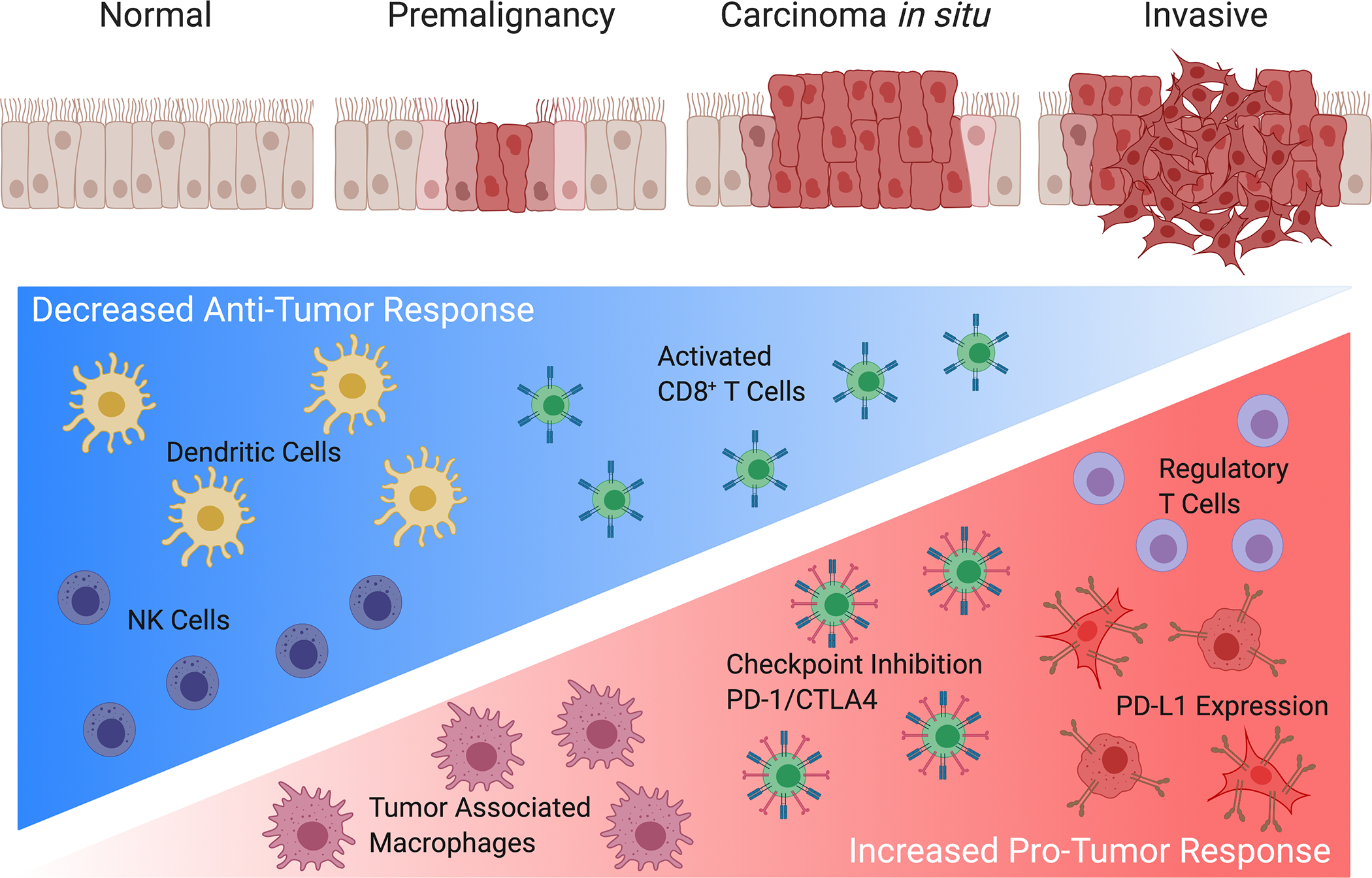
Graphical presentation showing progression from normal epithelium to premalignant cells advancing to invasive malignancy. Lung tumorigenesis is accompanied by a decrease in anti-tumor responses with the hallmark of decreased dendritic cells, activated natural killer (NK) cells and granzyme B secreting activated CD8+ T cells. In addition, lung cancer progression is coupled with an increase in tumor associated macrophages, a prevalence of regulatory CD4+FOXP3+ T cells, an upregulation of PD-L1 expression in tumor and myeloid populations, and transition from activated T cells to an exhausted state identified by the expression of checkpoint inhibition markers, such as PD-1 and CTLA4.
3.1.1. Changes in LUAD Precursor Atypical Adenomatous Hyperplasia (AAH)
To survey the premalignant lesions, Sivakumar et al. examined the early mutation events and gene expression changes in atypical adenomatous hyperplasia (AAH), the precursor of LUAD [21]. Deep targeted DNA and RNA sequencing of AAH and LUAD along with matched normal lung tissues was performed. The study revealed various patterns of expression profiles and variants of BRAF (encoding B-Raf, a serine-threonine kinase), EGFR (encoding epidermal growth factor receptor), and KRAS (encoding K-Ras, a GTPase) genes. Gene set enrichment analysis showed elevated immune cell trafficking and WNT/β-catenin signaling along with an inhibition of both the type 1 T helper (Th1) antitumor inflammatory response and transforming growth factor beta 1 (TGFB1) signaling in AAH as compared to normal lung. The overall immune marker profiling showed a shift from the antitumor response defined by Th1-derived IFN-γ signaling to a dominance of pro-tumorigenic type 2 T helper (Th2) immune pathways. Gene sets of suppressed anti-tumor genes included IL12A, as well as chemokines (CCL3, CCL4 and TLR4) and apoptosis-inducing protease granzyme B (GZMB). An increase in pro-tumor gene sets, including CCR2 and CTLA4, was also evident in AAH relative to normal lung. CCL2/CCR2 signaling has been shown to enhance tumor progression, and were found to be overexpressed in multiple tumor tissues [22]. Compared to AAH lesions, increased CTLA4 expression was noted in LUAD, suggesting an immune suppressive pathway favoring invasive disease. Decreased expression of TNFRSF9 was observed in BRAF-mutant AAHs. This gene encodes CD137, known to regulate the activation of T cells to enhance anti-tumor immune responses [23]. With the recent success and rapid advances in immunotherapy, identification of key immune regulatory genes and signaling pathways will afford the opportunities to develop novel biomarkers and immune-based strategies for early intervention.
3.1.2. Mutational Landscape and the Associated Immune Contexture in AAH
The mutational landscape and the associated immune contexture in the LUAD continuum were interrogated by Krysan et al. who performed whole exome sequencing (WES) of resection biospecimens from 41 lung cancer patients, which included laser captured microdissection of 89 AAH lesions, 15 adenocarcinomas in situ (AIS), 55 invasive LUAD and their adjacent normal lung tissues [24]. The authors designated the somatic mutations detected in both lung adenomatous premalignant lesions and the associated tumors as progression-associated mutations (PAMs). Putative neoantigens from these mutations correlated with the infiltration of CD4+ and CD8+ T cells and the upregulation of PD-L1, suggesting adaptive immunity and possible recognition of neoepitopes occurring in pulmonary premalignancy. The percentage of putative progression-associated neoantigens significantly correlated with the percentage of CD8+ T cells infiltrating AAH lesions, whereas the overall neoantigen load in AAH was associated with CD4+ T cell infiltration and PD-L1 expression. Analysis of the immune-related gene expression in the Cancer Genome Atlas (TCGA) LUAD dataset revealed that patients with higher immune-related gene expression had better survival compared to patients with the lowest expression of immune genes. This difference was most prominent in stage I patients, suggesting that modulation of the immune-related pathways, especially at the early stages of LUAD, may have a significant impact on patient outcomes.
These results are consistent with the findings of Angelova et al. who studied the evolution of colorectal cancer metastasis [25]. The authors demonstrated that clonal evolution or the selection of certain tumor cell types that progress was defined by the immune contexture. They further demonstrated that clones expressing neoantigens can be immunoedited out while progressing clones tend to be immune-privileged despite the presence of tumor-infiltrating lymphocytes. This suggests that immunoediting may occur in some premalignant lesions leading to regression, while other lesions escape immunosurveillance and progress to invasive disease. Longitudinal studies are required to validate this hypothesis.
3.1.3. Immune Landscape in Early-Stage LUAD
To help identify potential immunotherapy strategies for patients with early lung cancer, Lavin et al. evaluated the immune cell landscapes in 28 early stage LUAD patients by utilizing a designed barcoding method in combination with single cell analysis to characterize the infiltrating immune cells in the primary tumor, non-involved lung and blood in to search for tumor driven immune changes [26]. Specifically, the authors evaluated the immune cell landscapes in different compartments of stage I adenocarcinoma utilizing CyTOF combined with single cell transcriptomics, multiplex IHC and cytokine profiling. The study revealed an increased number of immune cells accumulated in the tumor tissue compared to normal lung with all the major immune lineages present but with T lymphocytes and mononuclear phagocytes being prevalent. Though both phagocytes and granulocytes were equally represented in both normal and tumor tissue, T and B lymphocytes were more abundant in the TME compared to normal lung. Regulatory T cell (Tregs) were significantly increased in the tumors and expressed high levels of CLTA4, CD39, ICOS and 4–1BB suggesting inhibitory functions. T cells were clearly distinguishable from those that resided in normal lung based on higher expression of FOXP3, CTLA4 and PD-1, suggesting the immune exhaustion. In contrast, there were fewer CD8+ T cells in the tumor compared to normal lung and blood. Functionally, tumor-infiltrating CD8+ T cells expressed significantly less GZMB and IFN-γ than their normal lung counterparts. PD-1 was primarily expressed on a small subset of CD4+ and CD8+ T cells in the tumor, while increased TCR clonality was found in the tumor-infiltrating CD8+ PD1+ T cells but not in normal lung, suggestive of tumor-specific T cell expansion. Natural killer (NK) cells were significantly reduced in all LUAD patients examined, and these NK cells had high CXCR3 levels coupled with reduced cytolytic activity (GZMB and IFN-γ). Tumor lesions with reduced MHC I expression had the highest numbers of tumor infiltrating NK cells suggesting an NK-driven immunoediting process that could circumvent the paucity of tumor MHC class I.
To further categorize the tumor-infiltrating myeloid cells, this study focused on macrophages, monocytes and dendritic cells (DCs). Two predominant monocyte subsets, CD16+ and CD14+ were identified. CD16+ monocytes were reduced at the tumor site, which correlated with reduced NK cells. In contrast, an increase in intratumoral CD14+ monocytes producing high levels of IL-8 was evident. CX3CL1 was the only cytokine that increased in tumor tissue compared to normal lung and correlated strongly with tumor infiltrating CD14+ monocytes. In the DC subset, there were reduced intratumoral CD141+ DCs localized adjacent to T cells thus, limiting the opportunity for T cell activation and clonal expansion. CD103+ DCs, recently demonstrated as the murine counterpart of CD141+ DCs, are critical in the cross presentation and priming of CD8+ T cells [27].
The authors also found increased tumor associated macrophages (TAMs) with a distinct transcriptomic signature. These TAMs had higher levels of the immunomodulatory transcription factor PPARγ, higher CD64, CD16 and CD11c, and lower levels of CD86 and CD206 when compared to normal lung macrophages. PD-L1 expression was the highest in macrophages and mast cells in both the tumor and normal lung as compared to other cell types. Tumor macrophages produced significantly more IL-6 than normal lung macrophages. A significant survival disadvantage was observed in patients with a high ratio of TAMs to normal lung macrophages. These results support an immunosuppressive role of TAMs in early lung adenocarcinoma lesions.
3.1.4. Changes in LUSC Precursor Bronchial Premalignant Lesions
In LUSC, high-grade persistent or progressive dysplasia is a marker of increased lung cancer risk. These lesions may regress, persist, or progress to invasive disease. The determinants of progression or regression and the underlying molecular mechanisms of the differential outcomes are not fully defined.
To characterize premalignant lesions (PMLs) that have the highest risk of progressing to LUSC, Beane et al. utilized mRNA sequencing (mRNA-Seq) to profile cells derived from bronchial brushings and endobronchial biopsies from patients undergoing longitudinal lung cancer screening by AFB [3]. Based on the transcriptional signatures, these PMLs were divided into four distinct molecular subtypes: proliferative, inflammatory, secretory and normal-like. Among the dysregulated immune genes of the proliferative PML, the authors found decreased expression of genes involved in IFN signaling and antigen processing/presentation pathways, including human leukocyte antigen (HLA) class I genes and Beta-2 microglobulin (B2M). Consistent with this finding, CD68+/CD163+ macrophages with an M2 anti-inflammatory phenotype and CD8+ T cells were also decreased in the proliferative PMLs. These lesions also contained greater numbers of CD4+ T cells. Further analysis will be required to determine whether these are T regulatory cells that promote an immunosuppressive environment. An increase in M2 CD163+ macrophages coupled with increased expression of IFNγ signaling genes was associated with a regressive phenotype of proliferative PMLs and potentially better outcomes. In contrast, both inflammatory and secretory PML subtypes showed increase in genes involved in inflammation and lymphocyte/leukocyte regulation, of which Interleukin-1 beta (IL-1β) has been suggested as a target for lung cancer interception. These data suggest that a better understanding of progression-associated immune changes in PMLs may lead to strategies for immunoprevention in the context of lung cancer interception. These potential biomarkers can be measured either directly at the lesion site or nearby surrogate tissue such as bronchial airway epithelium. Further studies will be required to understand the determinants of impaired immunosurveillance in progressive lesions.
3.1.5. The Evolution of Immune Changes through Progression of LUSC
To understand the evolution of immune changes through various stages of LUSC, Mascaux et al. performed gene expression profiling and divided the progression of LUSC into four distinct molecular steps, including bronchial mucosa with normal histology and hyperplasia, metaplasia and low-grade lesions, carcinoma in situ and high-grade lesions and finally invasive LUSC [28]. A module of 150 co-expressed immune-related genes was identified that had increasing expression as the lesions progressed from low to high grade. To analyze the trajectory of the immune response, the authors delineated gene expression patterns of activated T cells, neutrophils, M1 macrophages and myeloid cells, and found that infiltration of all of these cell types was highest in the high-grade lesions prior to tumor invasion. These data support immune sensing at the early stages of tumorigenesis, during which immune activation or escape occur before invasive LUSC.
The authors also assessed changes in the activation states of T cells, macrophages, B cells and DCs following progression. As the lesion progressed into high-grade hyperplasia, these cells demonstrated a shift from a resting to a more activated phenotype, and from naïve to memory phenotypes. Lesions within the same patient can have different immune compositions at different developmental stages. The authors performed functional analysis to validate this finding, which revealed altered immune functions with tumor progression that coincided with the altered immune gene signatures. Negative regulation of the immune system and antigen presentation were implicated in all developmental stages, while genes involved in immune suppression, including CD274, TIGIT, CTLA4, IL10 and IL6, were highly expressed in high grade dysplasia as compared to normal tissue. Stimulatory molecules such as TNFRSF9, TNFRSF18, ICOS, CD80, CD86, CD70, and TNFRSF25 showed increased expression in high-grade dysplasia and, to a greater extent, at the invasive stage. The enhanced expression of multiple immune checkpoints (IDO1, PD-L1, CTLA4, TIGIT and TIM3) in high-grade lesions was confirmed by IHC.
Seven-plex multiplex immunofluorescence staining was utilized to reveal the spatial relationships of various immune cell subtypes and the microenvironment architecture, which supported the findings by gene expression data. For example, the immune cell densities of CD4+ and CD8+ T cells, myeloid cells, neutrophils and macrophages increased in high-grade pre-invasive lesions. An increase in PD-L1 was observed through progressive stages with the highest levels found in LUSC suggesting immune exhaustion associated with progression. As tumorigenesis progressed, second-order spatial relationships, indicated a greater separation between cytokeratin-labeled epithelial cells and CD3+ T cells suggesting segregation as the tumor progressed. These results confirmed that dynamic immune changes occur before tumor invasion and highlighted the need for identifying immune markers for early lung cancer detection as well as immunotherapy-based strategies for early prevention.
3.2. Biomarkers for Early Detection of Lung Cancer (Figure 2)
Figure 2. Potential Systemic Biomarkers for Early Detection of Lung Cancer.
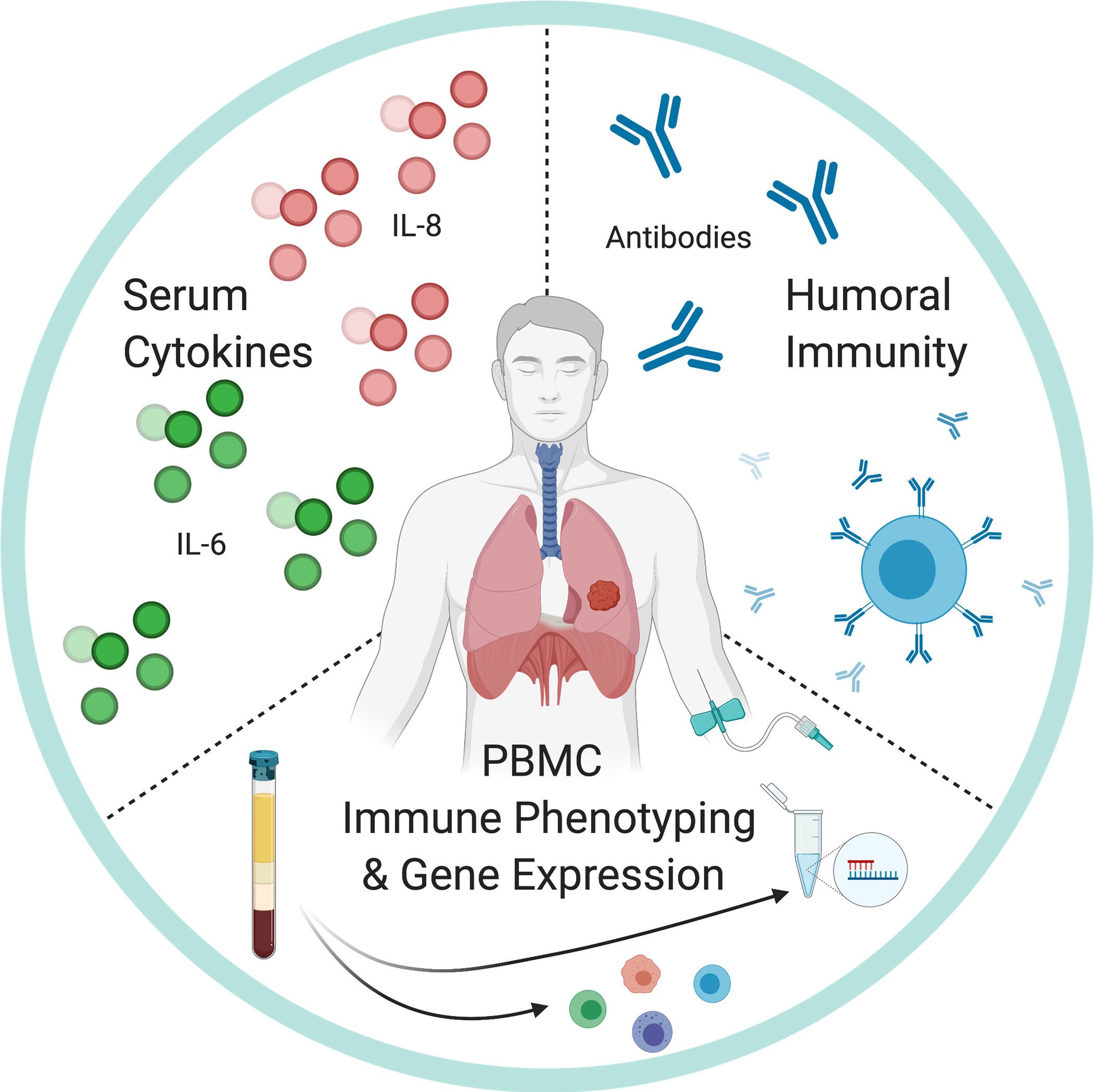
Pursuits in early detection include 1) the upregulation of systemic chemokines, such as IL-6 and IL-8, 2) the identification of autoantibodies specific to potential cancer associated antigens, which is indicative of humoral immunity, and 3) global changes in gene expression of whole blood and peripheral blood mononuclear cells (PBMCs).
3.2.1. Cytokines as Systemic Biomarkers
Inflammation has been shown to impact both cancer initiation, progression and metastasis. In an effort to determine if cytokine signatures in noncancerous lung tissue could predict the metastatic capability of adjacent lung adenocarcinoma, Seike et al. developed an 11-gene Cytokine Lung Adenocarcinoma Survival Signature (CLASS-11) that was able to identify stage I LUAD patients with poor prognosis, where the 5-yr survival for this population is approximately 61% [29]. One of the genes in this signature was IL-6, which has previously been identified as a prognostic marker of lung cancer [30, 31].
To further investigate the cytokine signature associated with early lung cancer, Enewold et al. determined the serum levels of ten circulating cytokines, namely IL-1β, IL-4, IL-5, IL-6, IL-8, IL-10, IL-12, granulocyte macrophage colony-stimulating factor (GM-CSF), IFN-γ, and tumor necrosis factor-alpha (TNF-α), in 353 NSCLC cases from a case-control study [32]. This confirmed that higher levels of IL-6 in the serum (≥ 4.0 pg/mL) correlated significantly with poorer survival in both African Americans and Caucasians. Increased IL-10 and IL-12 were associated with poorer survival only in African Americans, while higher TNF-α levels showed a trend in Caucasians. In a follow-up study using the National Cancer Institute (NCI-MD) and the Prostate, Lung, Colorectal, and Ovarian (PLCO) Cancer Screening Trial, Pine et al. investigated the associations between IL-6, IL-8 or C-reactive protein (CRP), a systemic inflammation biomarker and lung cancer [33]. Elevated serum levels of IL-6 and IL-8 were found to be associated with lung cancer in the NCI-MD study and predicted lung cancer risk in the PLCO study. The authors showed that higher levels of IL-6 predicted lung cancer within two years, while IL-8 was associated with lung cancer risk several years prior to diagnosis. IL-8 in combination with CRP levels was found to be a better predictor than either one alone. Additional studies have sought to develop peripheral blood inflammatory markers as tools for therapeutic decision making for patients with early stage lung cancer [34] [35].
3.2.2. Humoral Immunity as Systemic Biomarkers
In addition to cytokines, humoral immunity-based biomarkers have been studied for early detection of NSCLC. One method, based on the hypothesis that tumor neoantigens lead to a humoral response, is the autoantibody profiling in serum. This approach can potentially allow for enhanced sensitivity by taking advantage of the inherent biological amplification provided by autoantibodies to tumor proteins.
Early studies explored antibody targets combined with proteomic approaches to identify patient antibodies to detect early lung cancer. Patient sera were used to probe the A549 (LUAD cell line) proteins or tumor tissues on a Polyacrylamide gel electrophoresis (PAGE) gels [36, 37]. Later studies utilized lung cancer phage display or protein microarrays to help identify potential antibody targets that can be identified to predict the presence of lung cancer [38, 39]. It is now recognized that development of accurate humoral immunity-based screens required more specific information about potential lung cancer targets.
Assessment of known tumor associated antigens (TAAs) as targets to autoantibodies underwent validation utilizing enzyme-linked immunosorbant assay (ELISA). With the heterogeneity of antigen expression, a panel of assays for autoantibodies to various TAA targets as opposed to a single antigen was hypothesized to provide better sensitivity in detection of lung cancer. These targets included p53, NY-ESO-1, cancer associated antigen (CAGE), GBU4–5, SOX2 MAGE A4 and HuD (n-ELAV) [40, 41]. Massion et al. utilized this approach, in conjunction with LDCT to assess the risk for malignancy in the assessment of indeterminate pulmonary nodules [42]. Results showed that nodules between 4 to 20mm with a positive autoantibody test result had a twofold greater risk for development of lung cancer. The combination of LDCT with this ELISA (EarlyCDT®-Lung) test allowed the reclassification of lower-risk false-negative scans to true positives. This autoantibody test can potentially be a biomarker for physicians to predict the probability of malignancy in relatively small nodules leading to early detection of lung cancer. With continued genome wide sequencing of tumor tissues and pulmonary nodules, further refinement of humoral immunity-based biomarkers can be developed as alternative means for tumor antigen identification and early cancer diagnosis. In combination with cytokine markers, humoral immunity markers could help identify an immune response to lung cancer with improved sensitivity.
3.2.3. Gene Expression Changes of Circulating Peripheral Blood Mononuclear Cells (PBMCs) and Whole Blood
To identify systemic immune biomarkers for early detection of lung cancer, Kossenkov et al. assessed mRNA and miRNA expression profiles of PBMC before and after tumor resection utilizing Illumina arrays [43, 44]. The presence of tumor led to increased expression of 67% of the PBMC genes prior to resection and allowed for the distinction of benign from malignant nodules.
With potential limitations in PBMC collection and the reliability of microarrays for clinical use, the investigators evaluated the use of whole blood RNA expression as a potential biomarker for early detection of lung cancer [45]. They utilized RNA-stabilizing PAXgene tubes in combination with Illumina microarrays to screen 821 samples from five clinical sites to develop a pulmonary nodule classifier (PNC). This PNC with a biomarker of 559 gene probes was later refined on a NanoString nCounter platform to generate a NanoString PNC (nNPC) with only 41 genes while maintaining the accuracy of the prediction. The nNPC algorithm was tested on various data sets and accurately identified smaller benign nodules and larger malignant cancers. There was a minor decrement in sensitivity in indeterminate pulmonary nodules in the 6–20mm range. These 41 genes did not include the PBMC changes shown previously, though those genes were in the top 100 ranked probes. Additional data is being accumulated and analyzed to further asses the utility of a prognostic biomarker incorporating PBMC and whole blood gene expression.
4. Cancer Interception
These studies demonstrate that early detection of lung cancer in premalignancy is feasible and informative. Knowledge of lung cancer-associated biomarkers not only provides in-depth understanding of the pathogenesis of the disease, but also affords the opportunities for lung cancer early detection and interception. The concept of cancer interception introduced by Blackburn implies that in addition to preventative measures and cancer risk reduction, such as tobacco cessation, pharmacological and therapeutic approaches can be considered to prevent, delay or reverse carcinogenic progression to invasive disease in high risk patients [2]. Emerging techniques to help understand immune changes in cancer premalignancy may reveal potential targets for cancer interception at the earliest and most effective stages. In the following section we review selected recent studies targeting inflammation or modulating immune responses for lung cancer interception.
4.1. Canakinumab
A recent trial that targeted inflammatory pathways to prevent myocardial infarction additionally suggested the potential for lung cancer interception. Canakinumab, a humanized monoclonal antibody targeting IL-1β was utilized in the Canakinumab Anti-inflammatory Thrombosis Outcomes Study (CANTOS) trial [46] in which a significant decrease in lung cancer incidence (61%) and mortality following interleukin-1β inhibition was observed. This was a randomized double blind, placebo controlled trial that examined more than 10,000 patients assigned to three dosage groups of 50mg, 150mg and 300mg or placebo. Patients who were subsequently diagnosed with lung cancer had higher concentrations of C-reactive protein (hsCRP) and IL-6. During the three to seven year follow-up, patients who had been treated with canakinumab had a dose-dependent reduction in hsCRP and IL-6 levels. All patient groups treated with canakinumab had lower lung cancer mortality compared to the placebo group, and this was the most pronounced in the 300 mg canakinumab group. The participants that received 150 and 300mg canakinumab had a significantly lower incidence of lung cancer. These findings suggest that the anti-inflammatory effects of canakinumab may lead to the observed clinical benefits. Given the established roles of IL-1β in facilitating tumor growth, invasiveness and metastasis, the reduction of lung cancer incidence and mortality by canakinumab is likely due to either a prevention of PML progression to tumor or to effective treatment of early stage lung cancer. Additional clinical trials are currently assessing the anti-tumor effects of canakinumab as a single agent or in combination with chemotherapy or immunotherapy (NCT03447769, NCT03631199).
4.2. MUC1 Vaccine
Mucin 1 (MUC1) is a transmembrane glycoprotein aberrantly expressed in a variety of cancers, including NSCLC. It is the first reported human cancer antigen targeted by cytotoxic T cells [47]. MUC1 is ranked as the second most promising antigen among 75 selected cancer-associated antigens, largely based on its therapeutic benefit and immunogenicity [48]. In vivo delivering MUC1 peptide, injecting autologous DCs loaded with MUC1 or fusing chimeric antigen receptors T cells (CAR-T) targeting MUC1, all elicit adaptive immune response and demonstrate therapeutic benefits in preclinical and clinical studies [49]. In a randomized, double-blind and placebo-controlled phase II trial, a modified vaccinia Ankara virus vector expressing MUC1 and interleukin-2 (IL-2) in combination with chemotherapy significantly improved progression-free survival compared to chemotherapy alone [50]. The potential of MUC1 vaccine in cancer prevention is currently being explored in a phase I trial where its immunogenicity is evaluated in current and former smokers at high risk of developing lung cancer (NCT03300817).
4.3. Immunotherapy for Cancer Prevention and Early-Stage Lung Cancer
Recent studies revealed immune cell infiltration and activation in lung premalignant lesions, which correlated with immunogenic somatic mutations and upregulation of immune checkpoints such as PD-L1 [24, 28], suggesting that ICB may be a viable preventative strategy for lung cancer interception. An ongoing clinical trial is to evaluate pembrolizumab in treating patients with high-risk pulmonary nodules (NCT03634241).
ICB therapy has also been explored in early stage NSCLC patients in either adjuvant or neoadjuvant settings. Results from the PACIFIC trial, which studied adjuvant anti-PD-L1 therapy, durvalumab, compared with placebo in unresectable stage III patients treated with concurrent chemoradiation, demonstrated a significant increase in PFS [51]. Durvalumab is now approved as consolidation therapy following concurrent chemoradiation in unresectable stage III NSCLC patients. Currently, several large phase III trials are evaluating the use of adjuvant PD-1/PD-L1 blockade after surgical resection of NSCLC [52]. Initial results from a phase II study testing neoadjuvant atezolizumab in resectable NSCLC revealed that preoperative treatment of atezolizumab is well tolerated, and the initial major pathologic response (MPR) rate is approximately 21%, while longer assessment is pending [53] [NCT02927301]. A recent phase II trial showed that Atezolizumab plus carboplatin and nab-paclitaxel given as a neoadjuvant regimen, achieved 56% major pathological response rate with no compromise to surgical resection and manageable treatment-related toxic effects [54] [NCT02716038]. Given the potential synergy between chemoradiation and ICB therapies, several ongoing trials are evaluating the combination of neoadjuvant chemo or radiation therapy with PD-1/PD-L1 blockade in early stage NSCLC with promising initial and interim results [52]. These studies highlight the potential of ICB as an effective therapeutic approach for lung cancer interception at early stages.
Discussion
Although recent advances in TKI and ICB therapies have revolutionized the treatment of lung cancer, durable responses are limited to only a subset of patients, and the overall survival rate for metastatic disease remains low. Therefore, innovative strategies to detect and intercept lung cancer at the earliest points of disease progression will have a major impact on patient care and clinical outcomes.
Among the most challenging problems in the field of precancer investigation is the availability of sufficient premalignant tissues collected spatially and temporally to allow for precise determination of genomic, epigenomic, transcriptomic and immune changes associated with tumor progression. Advances in obtaining precancerous tissues, and the possibility of serial biopsies of the same lesions longitudinally will enable studies that fully define the determinants of progression to invasive disease. Studies that focus on the molecular profiling of premalignant lung tissues and their associated immune microenvironment, as well as the course of immune recognition and adaptive responses across the spectrum of disease will provide further insight in the mechanisms of lung cancer evolution and progression. This will identify actionable and personalized targets for lung cancer early interception.
Acknowledgments
We thank Lauren Winter for her administrative assistance. We thank Dr. Linh M. Tran for helpful discussions.
Financial Support:
Grant support: UC Tobacco-Related Disease Research Program (TRDRP) T30DT0963 (Raymond J. Lim), DOD W81XWH-16-1-0194 (Kostyantyn Krysan), UC Tobacco-Related Disease Research Program (TRDRP) 27IR-0036 (Kostyantyn Krysan), DOD W81XWH-17-1-0399 (Steven M. Dubinett), Stand Up To Cancer-LUNGevity-American Lung Association Lung Cancer Interception Dream Team Translational Cancer Research Grant (Grant Number: SU2C-AACR-DT23-17). Stand Up To Cancer is a division of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the scientific partner of SU2C (Steven M. Dubinett), NCI HTAN (PCA) 1U2CCA233238-01 (Steven M. Dubinett), NIH/NCI Molecular Characterization Laboratory 5U01CA196408-04 (Steven M. Dubinett), NIH/NCI EDRN (Steven M. Dubinett) 1U01CA214182, NIH/NCATS—UCLA Clinical and Translational Science Institute UL1TR001881 (Steven M. Dubinett), VA Merit Review 1I01CX000345-01 (Steven M. Dubinett).
Disclosures:
S.M. Dubinett reports receiving a research grant from Johnson & Johnson and is on scientific advisory boards for LungLifeAI, Early Dx, Inc, the Johnson & Johnson Lung Cancer Initiative, and T-Cure Biosciences, Inc.
References
- 1.Herbst RS, Morgensztern D, and Boshoff C, The biology and management of non-small cell lung cancer. Nature, 2018. 553(7689): p. 446–454. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn EH, Cancer interception. Cancer Prev Res (Phila), 2011. 4(6): p. 787–92. [DOI] [PubMed] [Google Scholar]
- 3.Beane J, et al. , Detecting the Presence and Progression of Premalignant Lung Lesions via Airway Gene Expression. Clin Cancer Res, 2017. 23(17): p. 5091–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ehrlich P, Ueber den jetzigen Stand der Karzinomforschung. Ned Tijdschr Geneeskd, 1909. 5: p. 273–290. [Google Scholar]
- 5.Lawrence HS, Cellular and humoral aspects of the hypersensitive states; a symposium held at the New York Academy of Medicine. Symposia of the Section on Microbiology, the New York Academy of Medicine,. 1959, New York,: P.B. Hoeber. xii, 667 p. [Google Scholar]
- 6.Burnet FM, The concept of immunological surveillance. Prog Exp Tumor Res, 1970. 13: p. 1–27. [DOI] [PubMed] [Google Scholar]
- 7.Old LJ and Boyse EA, Antigens of tumors and leukemias induced by viruses. Fed Proc, 1965. 24(5): p. 1009–17. [PubMed] [Google Scholar]
- 8.Dighe AS, et al. , Enhanced in vivo growth and resistance to rejection of tumor cells expressing dominant negative IFN gamma receptors. Immunity, 1994. 1(6): p. 447–56. [DOI] [PubMed] [Google Scholar]
- 9.Shinkai Y, et al. , RAG-2-deficient mice lack mature lymphocytes owing to inability to initiate V(D)J rearrangement. Cell, 1992. 68(5): p. 855–67. [DOI] [PubMed] [Google Scholar]
- 10.Dunn GP, et al. , Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol, 2002. 3(11): p. 991–8. [DOI] [PubMed] [Google Scholar]
- 11.Dunn GP, Old LJ, and Schreiber RD, The three Es of cancer immunoediting. Annu Rev Immunol, 2004. 22: p. 329–60. [DOI] [PubMed] [Google Scholar]
- 12.Dunn GP, Old LJ, and Schreiber RD, The immunobiology of cancer immunosurveillance and immunoediting. Immunity, 2004. 21(2): p. 137–48. [DOI] [PubMed] [Google Scholar]
- 13.Smyth MJ, Godfrey DI, and Trapani JA, A fresh look at tumor immunosurveillance and immunotherapy. Nat Immunol, 2001. 2(4): p. 293–9. [DOI] [PubMed] [Google Scholar]
- 14.Global Burden of Disease Cancer C, et al. , Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-Years for 29 Cancer Groups, 1990 to 2017: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.National Lung Screening Trial Research T, et al. , Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med, 2011. 365(5): p. 395–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.National Lung Screening Trial Research T, et al. , Results of initial low-dose computed tomographic screening for lung cancer. N Engl J Med, 2013. 368(21): p. 1980–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Koning HJ, et al. , Reduced Lung-Cancer Mortality with Volume CT Screening in a Randomized Trial. N Engl J Med, 2020. 382(6): p. 503–513. [DOI] [PubMed] [Google Scholar]
- 18.Campbell JD, et al. , The Case for a Pre-Cancer Genome Atlas (PCGA). Cancer Prev Res (Phila), 2016. 9(2): p. 119–24. [DOI] [PubMed] [Google Scholar]
- 19.Srivastava S, et al. , The Making of a PreCancer Atlas: Promises, Challenges, and Opportunities. Trends Cancer, 2018. 4(8): p. 523–536. [DOI] [PubMed] [Google Scholar]
- 20.Srivastava S, et al. , The PreCancer Atlas (PCA). Trends Cancer, 2018. 4(8): p. 513–514. [DOI] [PubMed] [Google Scholar]
- 21.Sivakumar S, et al. , Genomic Landscape of Atypical Adenomatous Hyperplasia Reveals Divergent Modes to Lung Adenocarcinoma. Cancer Res, 2017. 77(22): p. 6119–6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lim SY, et al. , Targeting the CCL2-CCR2 signaling axis in cancer metastasis. Oncotarget, 2016. 7(19): p. 28697–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yonezawa A, et al. , Boosting Cancer Immunotherapy with Anti-CD137 Antibody Therapy. Clin Cancer Res, 2015. 21(14): p. 3113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krysan K, et al. , The Immune Contexture Associates with the Genomic Landscape in Lung Adenomatous Premalignancy. Cancer Res, 2019. 79(19): p. 5022–5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Angelova M, et al. , Evolution of Metastases in Space and Time under Immune Selection. Cell, 2018. 175(3): p. 751–765 e16. [DOI] [PubMed] [Google Scholar]
- 26.Lavin Y, et al. , Innate Immune Landscape in Early Lung Adenocarcinoma by Paired Single-Cell Analyses. Cell, 2017. 169(4): p. 750–765 e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roberts EW, et al. , Critical Role for CD103(+)/CD141(+) Dendritic Cells Bearing CCR7 for Tumor Antigen Trafficking and Priming of T Cell Immunity in Melanoma. Cancer Cell, 2016. 30(2): p. 324–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mascaux C, et al. , Immune evasion before tumour invasion in early lung squamous carcinogenesis. Nature, 2019. 571(7766): p. 570–575. [DOI] [PubMed] [Google Scholar]
- 29.Seike M, et al. , Use of a cytokine gene expression signature in lung adenocarcinoma and the surrounding tissue as a prognostic classifier. J Natl Cancer Inst, 2007. 99(16): p. 1257–69. [DOI] [PubMed] [Google Scholar]
- 30.De Vita F, et al. , Serum levels of interleukin-6 as a prognostic factor in advanced non-small cell lung cancer. Oncol Rep, 1998. 5(3): p. 649–52. [PubMed] [Google Scholar]
- 31.Yanagawa H, et al. , Serum levels of interleukin 6 in patients with lung cancer. Br J Cancer, 1995. 71(5): p. 1095–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Enewold L, et al. , Serum concentrations of cytokines and lung cancer survival in African Americans and Caucasians. Cancer Epidemiol Biomarkers Prev, 2009. 18(1): p. 215–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pine SR, et al. , Increased levels of circulating interleukin 6, interleukin 8, C-reactive protein, and risk of lung cancer. J Natl Cancer Inst, 2011. 103(14): p. 1112–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryan BM, et al. , A combined prognostic serum interleukin-8 and interleukin-6 classifier for stage 1 lung cancer in the prostate, lung, colorectal, and ovarian cancer screening trial. J Thorac Oncol, 2014. 9(10): p. 1494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pine SR, et al. , Differential Serum Cytokine Levels and Risk of Lung Cancer Between African and European Americans. Cancer Epidemiol Biomarkers Prev, 2016. 25(3): p. 488–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brichory FM, et al. , An immune response manifested by the common occurrence of annexins I and II autoantibodies and high circulating levels of IL-6 in lung cancer. Proc Natl Acad Sci U S A, 2001. 98(17): p. 9824–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brichory F, et al. , Proteomics-based identification of protein gene product 9.5 as a tumor antigen that induces a humoral immune response in lung cancer. Cancer Res, 2001. 61(21): p. 7908–12. [PubMed] [Google Scholar]
- 38.Chen G, et al. , Autoantibody profiles reveal ubiquilin 1 as a humoral immune response target in lung adenocarcinoma. Cancer Res, 2007. 67(7): p. 3461–7. [DOI] [PubMed] [Google Scholar]
- 39.Madoz-Gurpide J, et al. , Integral protein microarrays for the identification of lung cancer antigens in sera that induce a humoral immune response. Mol Cell Proteomics, 2008. 7(2): p. 268–81. [DOI] [PubMed] [Google Scholar]
- 40.Murray A, et al. , Technical validation of an autoantibody test for lung cancer. Ann Oncol, 2010. 21(8): p. 1687–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chapman CJ, et al. , EarlyCDT(R)-Lung test: improved clinical utility through additional autoantibody assays. Tumour Biol, 2012. 33(5): p. 1319–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Massion PP, et al. , Autoantibody Signature Enhances the Positive Predictive Power of Computed Tomography and Nodule-Based Risk Models for Detection of Lung Cancer. J Thorac Oncol, 2017. 12(3): p. 578–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Showe MK, et al. , Gene expression profiles in peripheral blood mononuclear cells can distinguish patients with non-small cell lung cancer from patients with nonmalignant lung disease. Cancer Res, 2009. 69(24): p. 9202–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kossenkov AV, et al. , Resection of non-small cell lung cancers reverses tumor-induced gene expression changes in the peripheral immune system. Clin Cancer Res, 2011. 17(18): p. 5867–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kossenkov AV, et al. , A Gene Expression Classifier from Whole Blood Distinguishes Benign from Malignant Lung Nodules Detected by Low-Dose CT. Cancer Res, 2019. 79(1): p. 263–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ridker PM, et al. , Effect of interleukin-1beta inhibition with canakinumab on incident lung cancer in patients with atherosclerosis: exploratory results from a randomised, double-blind, placebo-controlled trial. Lancet, 2017. 390(10105): p. 1833–1842. [DOI] [PubMed] [Google Scholar]
- 47.Barnd DL, et al. , Specific, major histocompatibility complex-unrestricted recognition of tumor-associated mucins by human cytotoxic T cells. Proc Natl Acad Sci U S A, 1989. 86(18): p. 7159–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheever MA, et al. , The prioritization of cancer antigens: a national cancer institute pilot project for the acceleration of translational research. Clin Cancer Res, 2009. 15(17): p. 5323–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Taylor-Papadimitriou J, et al. , Latest developments in MUC1 immunotherapy. Biochem Soc Trans, 2018. 46(3): p. 659–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quoix E, et al. , TG4010 immunotherapy and first-line chemotherapy for advanced non-small-cell lung cancer (TIME): results from the phase 2b part of a randomised, double-blind, placebo-controlled, phase 2b/3 trial. Lancet Oncol, 2016. 17(2): p. 212–223. [DOI] [PubMed] [Google Scholar]
- 51.Antonia SJ, et al. , Durvalumab after Chemoradiotherapy in Stage III Non-Small-Cell Lung Cancer. N Engl J Med, 2017. 377(20): p. 1919–1929. [DOI] [PubMed] [Google Scholar]
- 52.Rosner S, Reuss JE, and Forde PM, PD-1 Blockade in Early-Stage Lung Cancer. Annu Rev Med, 2019. 70: p. 425–435. [DOI] [PubMed] [Google Scholar]
- 53.Rusch VW, et al. , Neoadjuvant atezolizumab in resectable non-small cell lung cancer (NSCLC): Initial results from a multicenter study (LCMC3). Journal of Clinical Oncology, 2018. 36(15_suppl): p. 8541–8541. [Google Scholar]
- 54.Shu CA, et al. , Neoadjuvant atezolizumab and chemotherapy in patients with resectable non-small-cell lung cancer: an open-label, multicentre, single-arm, phase 2 trial. Lancet Oncol, 2020. 21(6): p. 786–795. [DOI] [PubMed] [Google Scholar]


