Abstract
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), is a highly transmissible virus causing the ongoing global pandemic, COVID-19. Evidence suggests that viral and host microRNAs play pivotal roles in progression of such infections. The decisive impact of viral miRNAs and their putative targets in modulating the transcriptomic profile of its host, however remains unexplored. We hypothesized that the SARS-CoV-2 derived miRNAs can potentially play a contributory role in its pathogenicity and aid in its survival. A series of computational tools predicted 34 SARS-CoV-2 encoded miRNAs and their putative targets in the host. Immune and apoptotic pathways were identified as most enriched pathways. Further investigation using a dataset of SARS-CoV-2 infected cells (available from public repository- GSE150392) revealed that 46 genes related to immune and apoptosis-related functions were deregulated. Of these 46 genes, 42 genes were identified to be significantly up-regulated and 4 genes were down-regulated. In silico analysis revealed all of the these significantly down-regulated genes to be putative targets of 9 out of 34 of our predicted viral miRNAs. Overall, 123 out of 324 genes that are differentially regulated in SARS-CoV2 infected cells, and also identified as putative targets of viral miRNAs, were found to be significantly down-regulated. KEGG pathway analysis using these genes revealed p53 signaling as the most enriched pathway – a pathway that is known to influence immune responses. This study thus provides the theoretical foundation for the underlying molecular mechanisms involved in progression of viral pathogenesis.
Keywords: SARS-CoV-2, miRNA, COVID-19 pathogenesis, Immune system, Apoptosis, Functional enrichment
1. Introduction
In late 2019, an unidentified strain of human novel beta coronavirus, later termed as SARS-CoV-2, emerged in Wuhan, Hubei province, China which led to a sudden formidable outbreak of the novel coronavirus disease (COVID-19). The disease is generally characterized by dry cough, fever and shortness of breath, although varied presentations ranging from asymptomatic cases to severely affected individuals with respiratory and cardiovascular complications is not uncommon. SARS-CoV-2 is an enveloped, positive single-stranded RNA virus of the Coronaviridae family with a genome spanning approximately 30 Kb in length and encoding 16 non-structural and four structural proteins typical of Coronavirus; Spike(S), Nucleocapsid(N), Membrane(M) and Envelope(E) [1]. It is a highly aggressive viral strain with moderate to low mortality rate (2–3%) compared to other related human coronaviruses [2]. As of March 29, 2021, a total of 126,697,603 confirmed cases with 2,776,175 deaths globally were reported by the World Health Organization [3]. As a rapidly evolving pandemic with frequent emergence of novel variants, COVID-19 has posed a serious threat to global health and world economy, peddling lives of millions in distress. Infection with this potentially lethal virus can lead to Acute Respiratory Distress Syndrome (ARDS), and the resulting lung pathogenesis, and associated severe respiratory complications may lead to death. Furthermore, reports have suggested that subclinical cases with mild or no symptoms could act as reservoirs, spreading the virus unknowingly across the population cohort and thus making the situation even worse. Evidences have suggested that covert cases pose a serious threat in compliance of public health mitigation strategies, creating great difficulties in combating the disease [4]. Although 2019-nCoV patients show resemblance in symptoms to the MERS-CoV (Middle East respiratory syndrome-CoV) and SARS-CoV (Severe acute respiratory syndrome-CoV) infections, pathophysiology of 2019-nCoV infection and exact molecular mechanism behind the unusually high virulence and infectivity of SARS-CoV-2 is not clearly understood yet. Early studies have indicated towards an emerging role of immune cells in disease progression. It has been shown that a sudden storm of pro-inflammatory cytokines and chemokines might lead to inflammation-induced lung failure and ARDS which are the main cause of death in this disease [5]. Some early evidences also link cellular apoptosis to the COVID-19 severity [6]. Nevertheless, the exuberant mechanism of viral invasion, host adaptation, transmissibility and pathogenesis are remained largely unknown, attracting prime attention of the researchers to explore.
MicroRNAs are small endogenous non-coding RNA molecules of approximately 22 nucleotides (NTs) in length. They are capable of regulating gene expression of a variety of biological processes including apoptosis, cellular division, growth and development via post-transcriptional mechanism, either by mRNA degradation or translational repression [7]. They are known to influence different cell signaling pathways and thereby get associated with various pathophysiological conditions [8]. miRNAs regulate a wide variety of protein-coding genes in plants, animals and some viruses. Similar to plants and animals, some viruses are also reported to express viral miRNAs that are associated with host-pathogen interplay, controlling apoptosis and modulating host immune systems, which could ensure viral survival within infected cells, and propel viral pathogenesis [9,10]. There are several instances where viral miRNAs target host genes and play a decisive role in prolonging viral longevity within the infected cells. For example, miR-BART-5 regulates the pro-apoptotic gene PUMA in Epstein-Barr virus(EPV) [11]. The first viral miRNAs were discovered in Epstein-Barr Virus(EPV) in 2004 [12], and since then more than 20 viruses(both DNA and RNA viruses), have been reported to express miRNAs which can target host as well as viral genes [11]. To date, more than 300 viral miRNAs have already been reported across different genera, contributing immense insights to the viral infection panorama and host defense machinery. In 2017, L Morales et al. reported the presence of small RNAs encoded by SARS-CoV which are linked to inflammation and lung pathogenesis [13]. However, role of viral miRNAs in 2019-nCoV disease progression and pathogenicity has remained largely unexplored and needs critical attention.
With the rapid progress in bioinformatics and availability of computational tools, we have achieved a great feat in proteomics as well as transcriptomic studies. Recently, in silico methods have proven to be indispensable in deciphering pathophysiology of COVID-19 disease. Recent evidences from both experimental and in silico studies have suggested that miRNAs of viral origin might play an immensely important role in progression of the disease by targeting host genes of various signaling pathways to evade host immune system. Thus, identifying viral miRNAs and their respective target genes within host genome is crucially important for understanding molecular mechanism and development of miRNA-targeted therapeutic strategies. In order to survive within host cells, the virus needs to replicate inside the host cells. Therefore, it needs to forge a way which could prolong the sustenance of infected cell, impeding host defense mechanisms and cellular apoptotic pathways. In this study, we have explored the intricate regulatory mechanism of SARS-CoV-2 encoded miRNAs that evade host defense mechanism and apoptotic machinery through down-regulation of important immune system-related and apoptotic genes to facilitate invasion and sustenance of infected host cells, thus ensuring its own survival and disease progression.
2. Methods and materials
2.1. Extraction of pre-miRNA signatures in SARS-CoV-2 genome
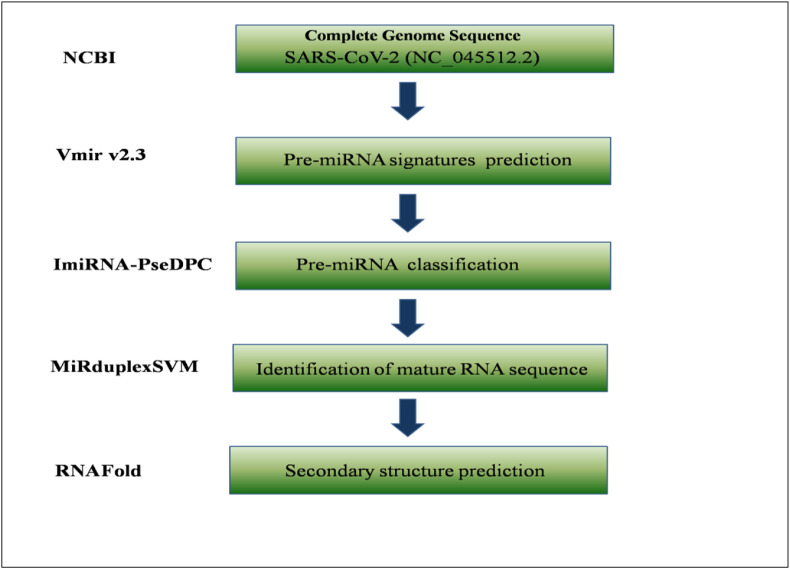
In this study, we have downloaded the complete genome sequence of SARS-CoV-2, Wuhan 1(GenBank accession no. NC_045512.2), as submitted on 17 January 2020, from National Centre for Biotechnology Information (NCBI). The VMir, a virus-encoded miRNA prediction programme (v2.3) [14], was used to predict pre-miRNA signatures in viral genome. The program screened for all possible pre-miRNA hairpin structures and predicted all potential candidates within viral genome. Stringent filters (minimum hairpin size of 60 nucleotides, maximum hairpin size of 120 nucleotides, minimum hairpin score of 160, and minimum window count of 20) were used to reduce ambiguity and predict miRNA precursors accurately.
The Minimum Free Energy (MFE) of the filtered pre-miRNAs was calculated by using RNAFold programme of Vienna package [15]. Pre-miRNA hairpins with MFE > −20 kCal/mol were excluded from our study due to poor stability. ImiRNA-PseDPC webserver [16] was used to discriminate the real pre-miRNA precursors from false ones(pseudo hairpins).Mature sequences of selected hairpins were predicted using MiRduplexSVM [17]based on sequential and structural information of miRNA precursors. Finally, RNAFold webserver [15] was used to predict the secondary structures of miRNA precursors by using default parameters (Fig. 1 ).
Fig. 1.
Schematic representation of SARS-CoV-2 encoded miRNA prediction.
2.2. Conserved miRNAs prediction among different coronavirus genera
To identify the evolutionary conserved pre-miRNA precursors and mature miRNA sequences across different genera, we utilized the same pipeline of miRNA predictions in other coronaviruses. The complete genome sequence of Human coronavirus NL63 (HCoV-NL63), SARS-CoV and MERS with GenBank accession no.NC_005831.1, NC_004718.3 and NC_019843.3 respectively were retrieved from NCBI database. The predicted hairpins were compared against different genera of the family Coronaviridae.
2.3. Target predictions of viral miRNA
In order to identify putative gene targets of viral miRNAs which may provide selective advantage to the virus when inside the host cells, target predictions of mature miRNAs were performed using Diana (MR-microT software) [18] and miRDB (custom target prediction) [19]tools. For the custom prediction, miRDB uses the MirTarget algorithm which undertakes the target prediction process. MirTarget algorithm generates a probability score between 0 and 100 for the miRNA-target pairs. The pairs with a score of ≥50 delivered as the output were then selected for further study. In case of Diana (MR-microT software), author′s recommendations were maintained to filter out the targets with scores less than 50. The overlapping gene transcripts between two prediction tools were extracted as reliable gene targets of mature viral miRNA to avoid false positives.
2.4. Differential gene expression analysis and functional annotation
In order to obtain the differentially expressed genes (DEGs) that are highly deregulated during viral infection, we performed differential gene expression analysis. RNA-seq data for mock and infected human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) 72 h post infection were downloaded from https://www.ncbi.nlm.nih.gov/GEO (Accession no. GSE150392) [20] and analyzed with DESeq2 [21]. Low expressed genes were discarded. False discovery rate (FDR) < 0.05, adjusted pvalue< 0.05 and minimum score 1 for absolute log2fold change was kept according to author's recommendation to obtain differentially expressed genes (DEGs).The lists of DEGs were then annotated in terms of their biological functions, cellular components and molecular functions using Gene Ontology Consortium (GO) of Database for Annotation, Visualization and Integrated Discovery (DAVID) [22]. Functional enrichment of signaling pathways was also evaluated using Kyoto Encyclopedia of Genes and Genomes (KEGG) of DAVID tool. The list of DEGs was matched with putative targets of vmiRNA in host genome to determine DEG targets predicted in the host genome. The overlapping DEGs were identified and functionally characterized using DAVID.
We searched for “apoptosis” and “immune functions” related genes in KEGG database and extracted the genes related to apoptotic and immune functions along with their expression values in RNA-seq data. The selected differentially expressed immune and apoptosis-related genes were retrieved for heatmap construction using R packages. Viral miRNA with overlapping target genes were identified to establish regulatory relationship of viral miRNA with host genes which could give virus selective advantage and helps to pervade host defense system, thus ensuring survival. Cytoscape v 3.8.2 [23] was used to construct miRNA-mRNA-KEGG pathway networks to visualize the molecular dynamics of different genes regulated by predicted viral miRNAs.
2.5. Construction of miRNA-miRNA functional synergistic network (MFSN)
To uncover the functional synergistic relationships between viral miRNAs and their putative gene targets, we constructed the miRNA-miRNA functional synergistic network (MFSN) by using sequence-based target binding information. The miRNA synergism was computed based on: (1) significant overlap of target genes between each miRNA-miRNA pairs using hypergeometric distribution test(p < 0.05) (2) overlap of significantly enriched target genes between each miRNA-miRNA pairs in Gene Ontology(GO) terms of DAVID functional annotation tool. In order to ascertain the role of synergistic regulatory network in progression of SARS-CoV-2 pathogenesis, we focused on studying the relationship between viral miRNAs and down-regulated host genes. The down-regulated genes with log fold change two or more and their corresponding viral miRNAs were selected to construct the miRNA synergistic network. R scripts used to make the synergistic networks are provided as supplementary data (Supplementary file 1).
2.6. Hybridization between important host gene transcripts and mature SARS-CoV-2 encoded miRNA
In order to ascertain the direct interactions between SARS-CoV-2 encoded mature miRNAs and their putative gene transcript targets, we have employed the RNAhybrid tool [24]. RNAhybrid is a miRNA target prediction tool which could be utilized to find pairing energy or minimum free energy (MFE) of RNA hybridization. Here, we have retrieved 3′UTR sequence of significantly down-regulated genes from UTRdb [25] and hybridized with their corresponding mature miRNAs. We set −15 kcal/mol pairing energy as cut off value for effective hybridization.
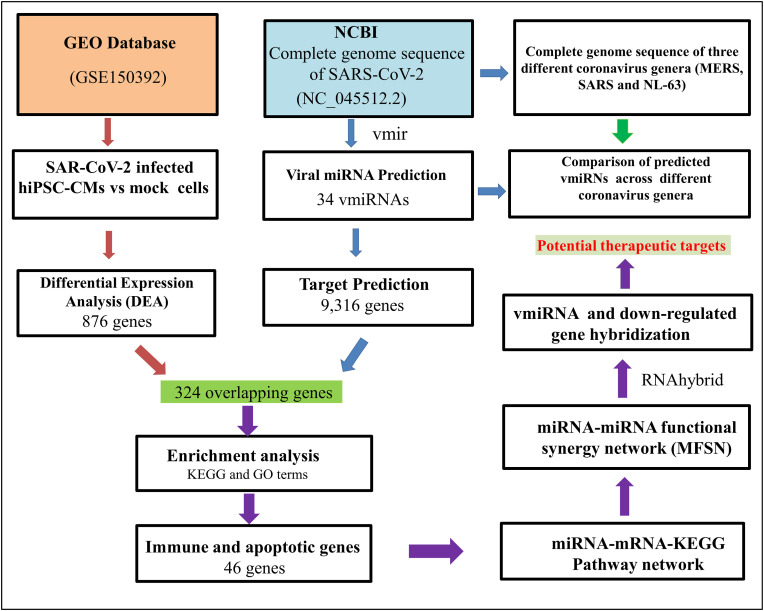
The overall workflow for studying the SARS-CoV-2 encoded miRNAs in viral pathogenesis is schematically represented in Fig. 2 .
Fig. 2.
Flowchart depicting the methodological workflow for studying the SARS-CoV-2 encoded miRNAs in COVID-19 pathogenesis.
2.7. Statistical analysis
In this study, we have used R v3.6.3 (https://www.r-project.org/) and Bioconductor (https://www.bioconductor.org/) to perform differential expression analysis (DEA), enrichment analysis and visualization. DESeq2 package of Bioconductor was used for differential analysis. Wald test (DESeq2 package) was used for hypothesis testing in differential expression analysis. R scripts of the analysis are provided as supplementary material (Supplementary file 1).
3. Results
3.1. Computational prediction of SARS-CoV-2-encoded pre-miRNAs
In order to obtain all possible miRNA precursors encoded by SARS-CoV-2, we applied computational method using VMir analyzer, an ab initio program which could precisely predict miRNA-like hairpins in 2019-nCoV genome. Initially, a total of 1114 miRNA precursors were predicted which ultimately reduced to 48 (26 directly and 22 reversely oriented) after applying the filters specific for viral pre-miRNAs prediction. Next, the pre-miRNAs were classified into real or pseudo by means of iMiRNA-PseDPC approach [16]. Forty-one out of 48 pre-miRNAs were classified as real which were further analyzed using RNAfold software to calculate MFE and thus predict the secondary structure of hairpin loops. A total of 34 pre-miRNAs were obtained after removing candidates with MFE > -20kCal/mol. Finally, mature sequence and position respective to SARS-CoV-2 genome were obtained by using MiRduplexSVM algorithm (see Table 1 ).
Table 1.
Predicted hairpin and mature sequences within SARS-CoV-2 genome. (Position relative to SARS-CoV-2 genome; MFE: minimum free energy in kcal/mol; Sequences are 5’ mature sequence).
|
Name |
Classification | Position | Score | MFE | 5′ mature Sequence |
|---|---|---|---|---|---|
| 1. MD20 | Real | 2107–2213 | 180.1 | −30.30 | UAUGAAAAACUCAAACCCGUCC |
| 2. MD22 | Real | 2284–2403 | 123.7 | −34.50 | UAAAUAAAUUUUUGGCUUUGUG |
| 3. MD52 | Real | 5317–5398 | 135 | −23.00 | AAGUUUAAUCCACCUGCUCUAC |
| 4. MD64 | Real | 6134–6224 | 136.3 | −23.30 | UGACUUAAAUGGUGAUGUGGUG |
| 5. MD83 | Real | 7961–8026 | 167.1 | −25.50 | CAACCUAUACUGUUACUAGAUC |
| 6. MD85 | Real | 8144–8221 | 170.2 | −23.60 | GACAAUGUCUUAUCUACUUUU |
| 7. MD135 | Real | 11234–11334 | 211.4 | −36.50 | UGCGUAUUAUGACAUGGUUGGA |
| 8. MD198 | Real | 17386–17448 | 138.5 | −21.90 | UUGAGUGUUGUCAAUGCCAGAUU |
| 9. MD210 | Real | 18702–18785 | 159.2 | −21.10 | UGUUGGCAUCAUUCUAUUGGAU |
| 10. MD221 | Real | 19425–19492 | 164.8 | −29.10 | UUAUGUACCACUAAAGUCUGC |
| 11. MD229 | Real | 20002–20081 | 159.3 | −21.50 | AAGUAGACUUAUUUAGAAAUGCC |
| 12. MD241 | Real | 21131–21240 | 204.7 | −38.30 | GGAGGUUCCGUGGCUAUAAAGA |
| 13. MD279 | Real | 24532–24607 | 130.1 | −20.10 | UGAUAGGUUGAUCACAGGCA |
| 14. MD283 | Real | 24799–24860 | 140.1 | −20.50 | UGCCAUUUGUCAUGAUGGAAAAGC |
| 15. MD288 | Real | 25217–25276 | 126.9 | −21.50 | GGUUUUAUAGCUGGCUUGAU |
| 16. MD306 | Real | 26743–26861 | 188.9 | −39.10 | AUUGGAUCACCGGUGGAAUUGC |
| 17. MD308 | Real | 26995–27113 | 169.1 | −36.20 | UGCUACAUCACGAACGCUUUC |
| 18. MD312 | Real | 27666–27769 | 205.6 | −24.70 | UUUUUCUUAUUGUUGCGGCAAUA |
| 19. MR3 | Real | 396–485 | 126.3 | −24.10 | AACACAUAGGGCUGUUCAAGUU |
| 20. MR8 | Real | 756–823 | 144.5 | −24.20 | CGAGUGUAUGCCCCUCCGUUAAG |
| 21. MR47 | Real | 4506–4577 | 133.8 | −20.20 | GUGUGUUGAUAAGUGACGCUAC |
| 22. MR63 | Real | 6126–6231 | 191.8 | −22.90 | UUCUUAAAAGAGGGUGUGUAGUG |
| 23. MR101 | Real | 9806–9908 | 127 | −22.90 | UAAAGAGCUAAGUAUCUAUUAU |
| 24. MR103 | Real | 10010–10091 | 132.7 | −23.20 | AUGCCAUUUUUCUAAAACCAC |
| 25. MR155 | Real | 14973–15081 | 183.9 | −27.30 | UAUAGUAGGGAUGACAUUACGU |
| 26. MR162 | Real | 15799–15905 | 127.3 | −22.60 | UGUUGAGAGCAAAAUUCAUGA |
| 27. MR165 | Real | 16284–16395 | 150.4 | −30.50 | AGACUAAUUUAUGUGAUGUUGAU |
| 28. MR186 | Real | 18089–18189 | 152.7 | −33.00 | AUGUCAACACAUAAACCUUCAG |
| 29. MR231 | Real | 22614–22679 | 143.9 | −23.90 | AUGCGGAAUUAUAUAGGACAGAA |
| 30. MR252 | Real | 24680–24190 | 151.8 | −29.10 | UCAUUUCAUCUGUGAGCAAAGG |
| 31. MR274 | Real | 26059–26138 | 174.1 | −25.00 | GACAUGUUCUUCAGGCUCAUCA |
| 32. MR285 | Real | 27102–27180 | 142.8 | −24.80 | AUUGUCACUGCUACUGGAAUGGU |
| 33. MR299 | Real | 28925–29007 | 145.5 | −23.10 | UGUUGUUGGCCUUUACCAGACA |
| 34. MR304 | Real | 29532–29625 | 184.6 | −28.40 | CACAAGAGUAGACUAUAUAUCG |
3.2. Common pre-miRNAs identified across different coronavirus genera
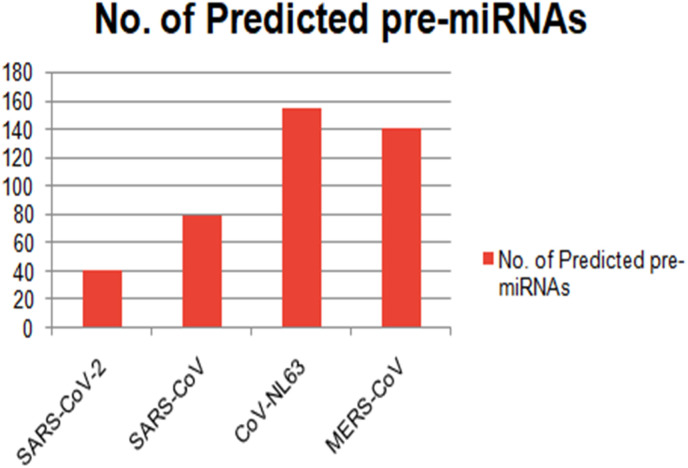
Viral miRNAs seldom show conservation across different virus genera. SARS-CoV-2, as a RNA virus, shows high degree of genetic heterogeneity. However, as a member of Coronaviridae family, it shares high degree of genetic homology with MERS-CoV, SARS-CoV and CoV-NL63 [26]. Therefore, we intended to identify the common vmiRNAs that were present in these four related coronavirus genomes. We performed same prediction analysis with other three viruses too (MERS, SARS and NL-63), resulting in a total of 154 for CoV-NL63, 78 for SARS-CoV and 139 for MERS-CoV pre-miRNA hairpins prediction. We have identified 44 pre-miRNAs that were common among three genera (MERS, SARS and NL-63) (Fig. 3 ). Interestingly, SARS-CoV-2 shared only two miRNAs (MD20 and MD22) with other genera (Supplementary file 2).
Fig. 3.
Showing comparison study of predicted miRNAs of different coronaviruses.
3.3. Identification of potential target genes for the predicted SARS-CoV-2 encoded miRNAs
The interactions between viral miRNAs and host genes are extremely important to comprehend complex molecular dynamics of viral infection and pathogenicity. Identification of viral miRNA-target genes interactions through experimental methods is costly and time-consuming. In silico methods have provided useful alternatives, leading to accurate predictions of target sites based on complete or partial binding with miRNAs. Researchers have developed several target prediction tools to investigate like TargetScan [27], PicTar [28], miRanda [29], miRnalyze [30], etc. For this study we used two independent prediction tools, miRDB [19] and Diana [18] to obtain robust gene targets. Candidate genes, predicted by both tools independently were extracted for further analysis. A total of 9,316 combined unique gene targets of 34 vmiRNAs were predicted by both tools (Supplementary file 3).
3.4. Gene expression changes upon SARS-CoV-2 infection
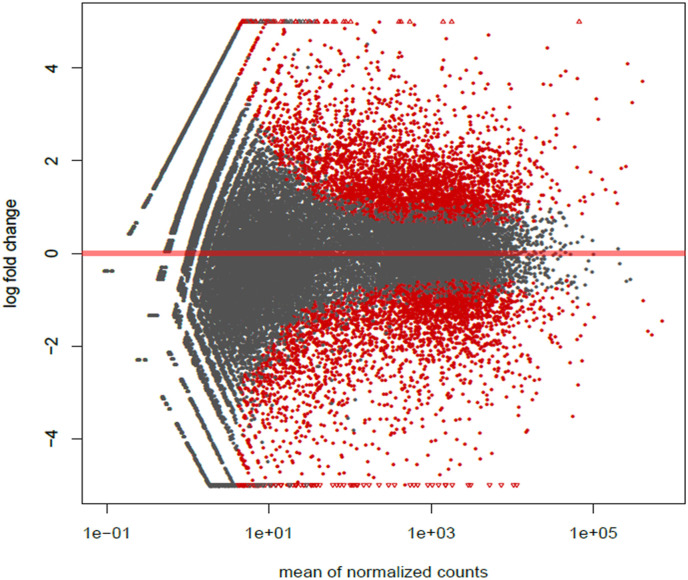
It is well-documented that viral miRNA can regulate gene expression upon infection [31]. To ascertain steady-state expression level of predicted mRNA targets upon SARS-CoV-2 infection, we have performed differential expression analysis (DEA) of bulk RNA-seq data, obtained from mock and infected hiPSC-CMs with GEO Accession no. GSE150392 (Fig. 4 ).
Fig. 4.
MA plot showing differentially expressed genes (DEGs) in GSE150392. Genes with significant difference (adjusted p-value <0.05) are shown in red. Wald test were used for hypothesis testing (DESeq2 package).
The number of differentially expressed genes was 876 for SARS-CoV2 infected vs mock infected hiPSC-CMs (Supplementary file 4). We compared targets predicted by prediction tools with these DEGs. A total of 324 genes were identified to be common, suggesting paramount importance of viral miRNA in determining transcriptomic profiles after infection (Supplementary file 5). Among them, 123 genes were identified to be significantly(|logFC|>2 and adj P < 0.05) down-regulated.
3.5. Functional enrichment analysis to ascertain biological roles and pathways regulated by predicted viral miRNAs
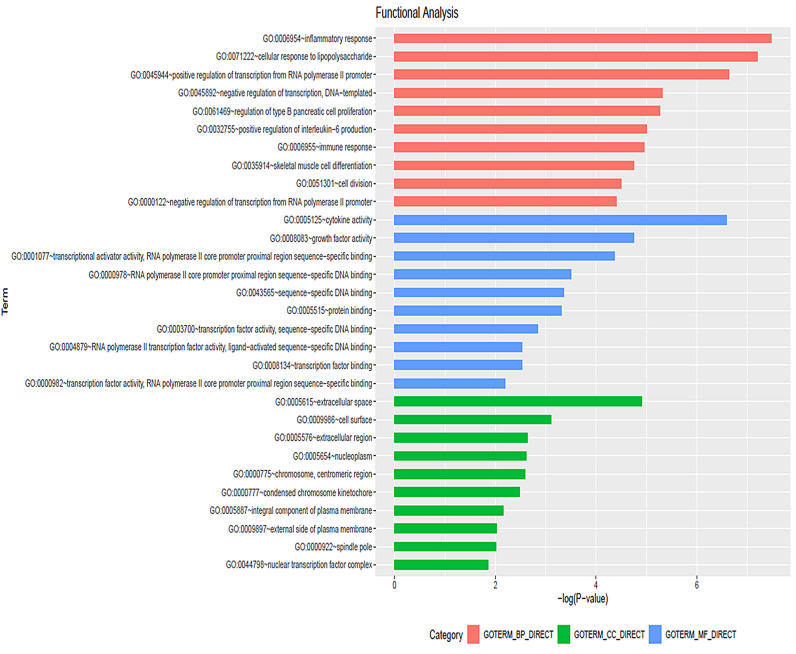
To identify specific biological roles and pathways associated with common DEGs targeted by predicted viral miRNAs, we conducted functional enrichment analysis using the Database for Annotation, Visualization and Integrated Discovery (DAVID) tool [22]. Results revealed that the most enriched Biological Process (BP) was inflammatory response (GO: 0006954), the most enriched Cellular Components (CC) was extracellular space (GO: 0005615) and the most enriched Molecular Function (MF) was cytokine activity (GO: 0005125) (Fig. 5 ).
Fig. 5.
Gene Ontology (GO) analysis of differentially expressed common target genes using DAVID. Top 10 of significantly enriched (P < 0.05) components in terms of Biological process (Red), Cellular component (Green) and Molecular function (Blue) are shown. All the adjusted values were negative log transformed.
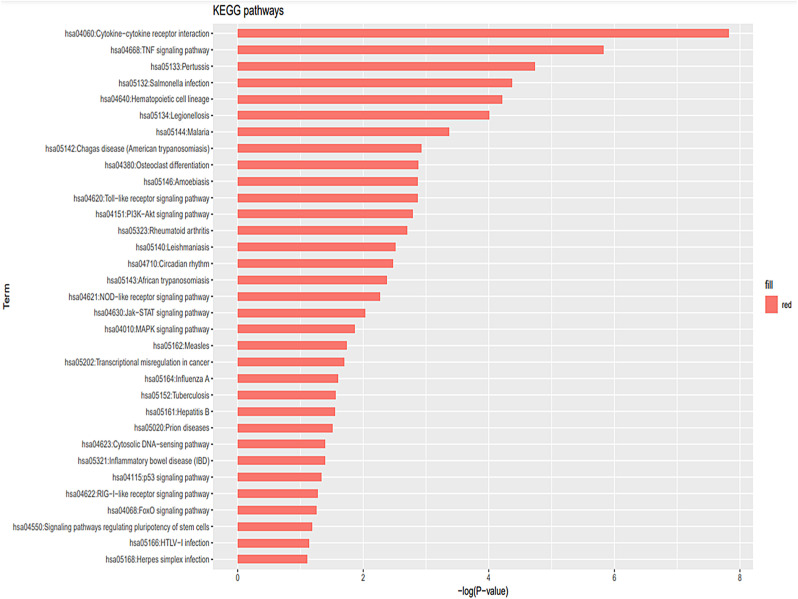
KEGG pathway analysis revealed significantly associated signaling pathways upon infection. Obtained results showed that majority of the retrieved pathways are associated with apoptotic and immune response with “hsa04060: cytokine-cytokine receptor interaction” being the most significantly enriched (adjP<0.05) pathway which is followed by TNF signaling pathway (hsa04668) and Pertussis (hsa05133) (Fig. 6 ). Further, KEGG pathway analysis using only the down-regulated genes revealed that hsa04115: p53 signaling pathway as the most enriched pathway in infected human host.
Fig. 6.
Results obtained from enriched pathway analysis using KEGG of DAVID tool are shown. All significant values are negative log transformed.
3.6. Viral miRNA-targeted genes related to immune response and apoptosis provide insights on aggressive virulence and prolonged sustenance of SARS-CoV-2 within host cells
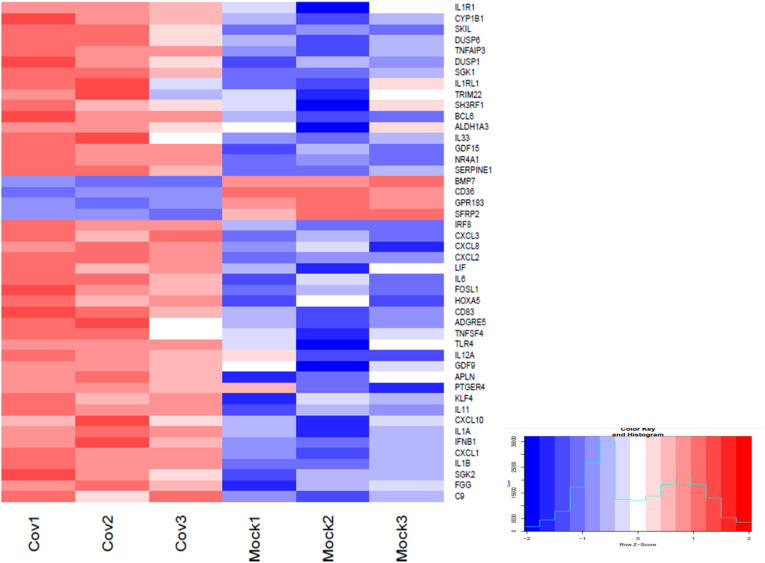
As viral miRNAs evade host defense mechanisms and prolong its own survival by deregulating important immune and apoptotic genes, we searched for “immune response” and “apoptosis” related genes in KEGG database to better understand the progression of viral infection and high virulence. We obtained list of 119(46 unique genes) genes targeted by SARS-CoV-2 encoded miRNAs (Fig. 7 ) (Supplementary file 6). Among them, 42 genes were found to be up-regulated, and rest, 4 genes were down-regulated. We have identified these host genes to be the targets of 26 predicted viral miRNAs. These genes and their corresponding miRNAs were further utilized to predict miRNA-mRNA-KEGG pathway network to delineate the molecular interrelations involved in viral pathogenesis.
Fig. 7.
Expression heatmap of differentially expressed immune and apoptotic genes in GSE150392 [20] which are potentially targeted by predicted SARS-CoV-2 miRNAs. Heatmap was generated using R v3.6.3. Red= up-regulated genes, Blue= down-regulated genes.
3.7. Construction of miRNA-mRNA-KEGG pathway network
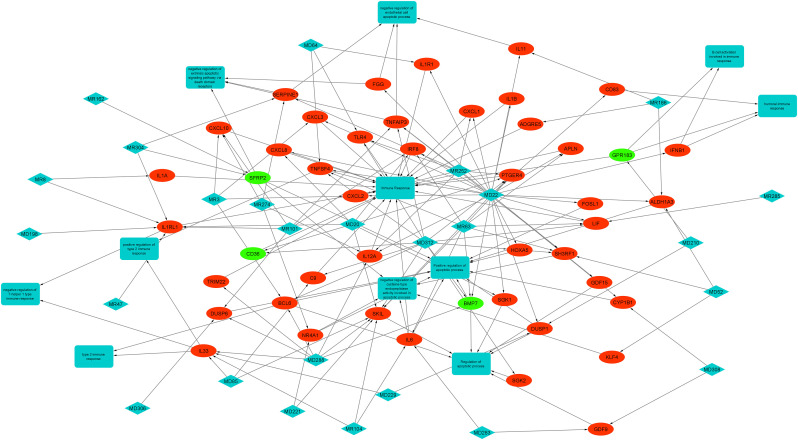
To comprehensively understand the interactions among different viral miRNAs and host genes involved in dynamics of host-pathogen interplay and thus, promote pathogenesis, we constructed the miRNA-mRNA-KEGG pathway network axis using yfiles layout and edge routing algorithm in Cytoscape v 3.8.2 [23] (Fig. 8 ).
Fig. 8.
The interactions among viral miRNAs, host mRNAs and different KEGG pathways (related to immune and apoptotic functions) are shown. The diamond, elliptical and rectangular shapes denote viral miRNAs, host immune and apoptosis-related genes and KEGG pathways, respectively. Red = Up-regulation; Green = Down-regulation. The arrows show interactions.
In miRNA-mRNA-KEGG pathway network, 26 predicted viral miRNAs and 46 genes related to immune and apoptotic functions were identified to be interlinked. In addition, 11 KEGG immune and apoptotic pathways were found to be part of the network. The network consists of 83 nodes and 184 edges. All of these interactions together formed an intricate network which might promote pathogenesis in human host. To elucidate the molecular interactions involved during pathogenesis, we selected significantly down-regulated genes (|logFC|>2 and adj P < 0.05) and viral miRNAs which target them.
3.8. Identification of miRNA pairs that synergistically down-regulate important host genes related to immune and apoptotic functions
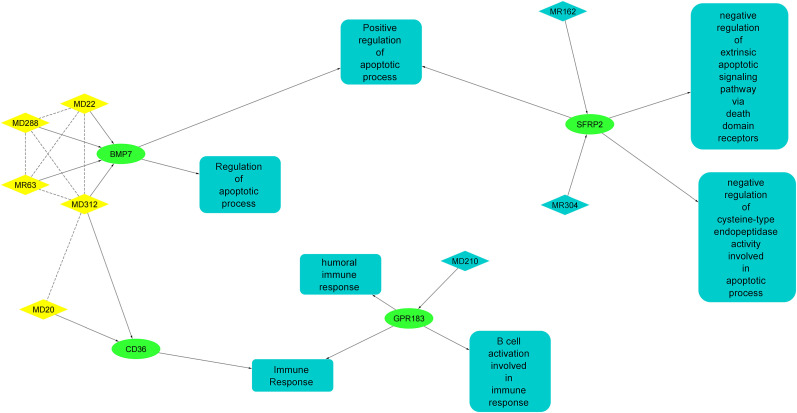
In order to obtain intricate network of viral miRNAs synergism that promote viral pathogenesis, we have constructed the miRNA-miRNA functional synergistic networks (MFSN) based on miRNA pairs regulating at least one functional module. The MFSN is the objective representation of all the synergistic actions of miRNAs and their putative targets. The synergistic regulatory network between viral miRNAs and down-regulated host target genes are shown in Fig. 9 . The predicted network comprised of 19 nodes and 25 edges. We identified 5 viral miRNAs synergistically down-regulated 2 important host genes (BMP7 and CD36) related to immune and apoptotic functions.
Fig. 9.
miRNA-miRNA functional synergistic network (MFSN). The figure is showing synergism among different viral miRNAs and their co-regulating functional networks. Non-directed dashed lines represent miRNA synergism and direct lines represent miRNA regulation. Yellow color = viral miRNA synergy; Green color = down-regulated genes related to immune and apoptotic functions.
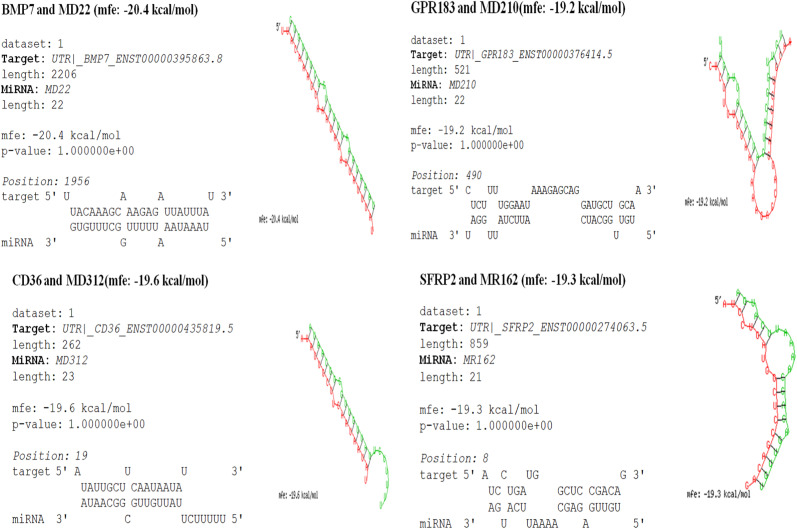
3.9. Effective hybridization of significantly down-regulated genes and their corresponding SARS-CoV-2 miRNAs
In order to comprehend stability between viral miRNA and significantly down-regulated putative target genes, we have calculated the pairing energy or minimum free energy of binding using RNAhybrid. We have observed effective partial binding of all of the 4 down-regulated gene transcripts with moderately negative pairing energy or MFE. Results with best pairing are shown in Fig. 10 .
Fig. 10.
Effective hybridization between 4 down-regulated genes (BMP7, GPR183, CD36 and SFRP2) and their corresponding SARS-CoV-2 mature miRNAs using RNAhybrid tool.
4. Discussion
The presence of numerous viral-encoded miRNAs has been reported to provide protection to virus against host immunity during infection. Viral-encoded miRNAs can mitigate cellular defense mechanism by down-regulating pivotal immune and apoptotic genes to sustain favorable intercellular milieu [32]. In 2017, Morales et al. confirmed the role of three small viral RNAs(svRNA) encoded by SARS-CoV genome, contributing in disease progression and lung pathology [13]. SARS-CoV-2 shares considerable amount of genetic homology with SARS-CoV, suggesting the possibilities of occurrence of similar svRNAs in SARS-CoV-2 genome too. Although extensive studies have already been carried out on SARS-CoV-2 pathogenesis, only few studies have been reported to enunciate the role of viral miRNA-mediated molecular regulation involved viral pathogenesis. In recent years, miRNAs have been reported to be important regulatory molecules which are involved in various biological processes and pathophysiological conditions [33]. Therefore, studying viral miRNA could provide invaluable opportunities to gain insights regarding detailed molecular regulation involved in host-pathogen interactions which could be utilized to develop novel therapeutic interventions [34]. Previously, it has been reported that host miRNAs could target various viral genes as an antiviral response [35]. Interestingly, research has also suggested that viral miRNAs target host immune genes to evade host immune response and thus, promote viral replication within host cells [36]. Another study has revealed that viral miRNA can down-regulate apoptosis-related host genes to ensure sustenance within infected cells [37]. Therefore, we hypothesized that SARS-CoV-2 encoded miRNAs might achieve a selective advantage and protection against anti-viral response by down-regulating host immune and apoptosis-related genes. With this aim, we have used a viral miRNA prediction pipeline similar to previously applied workflow for miRNA prediction in Zika virus genome [38] to predict miRNAs encoded in SARS-CoV-2 genome to elucidate underlying molecular mechanism involved in pathophysiology and progression of COVID-19 disease. Stringent filters were used for viral pre-miRNA prediction. These filters are based on statistical analysis of nearly 6500 mammalian and viral pre-miRNA structures deposited in miRNA registry [39] and popularly used in SARS-CoV-2 encoded pre-miRNA prediction [40]. By performing series of bioinformatics analyses, we have predicted 34 viral miRNAs in SARS-CoV-2 genome which could effectively target human genes to promote viral pathogenesis and survival within host. To get insights on how these miRNAs potentially regulate the transcriptomic profile of host cells upon infection, we have carried out differential expression analysis (DEA) of mock vs infected hiPSC-CM cells, using a GEO dataset with accession no. GSE150392, [20].Computational investigations have predicted deregulated genes in host cells as putative targets of our predicted viral miRNAs. In the same study, Sharma et al.(2020) have reported that human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) can be utilized as efficient in vitro platform to elucidate mechanism of viral invasion and pathogenesis [20]. Therefore, we have analyzed RNA-seq data obtained from the same study to gain insights about possible underlying molecular mechanism of the SARS-CoV-2 infection.
Functional enrichment analysis revealed several physiological processes which are significantly enriched during infection. We have found that various immune cell-mediated pathways such as Toll-like receptor signaling pathway, MAPK signaling pathway, NOD-like receptor signaling pathway and apoptotic pathways were mainly enriched following infection. Toll-like receptor signaling pathway plays a pivotal role in immune modulation and recognition of viral particles which leads to the secretion of pro-inflammatory cytokines such as type 1 interferon, IL-6, tumor necrosis factor-α, etc. in SARS-CoV-2 infection [41].Similar studies have reported possible role of MAPK signaling pathway and NOD-like receptor signaling pathway in the SARS-CoV-2 infected cells [42]. Another crucial pathway in viral infection panorama is apoptotic pathway. Apoptosis is a complex cellular process which is characterized by regulated self-demolition of cells in response to aging or infection [43]. It can prevent viral pathogenesis by regulated destruction of infected cells, and thus, maintains cellular homeostasis. Functional enrichment analysis of only the down-regulated genes revealed p53 signaling pathway as the most enriched pathway in infected cells. Previous studies have reported the association of Tumor suppressor p53 genes with the innate immune response [44]. The p53 gene is also reported to regulate apoptosis [45].Here, we have identified enrichment of immune response and apoptotic pathways and their corresponding target genes which were found to be deregulated by SARS-CoV-2 encoded miRNAs. This study provides strong indications that predicted SARS-CoV-2 miRNAs potentially target important immune and apoptotic genes to confer survival against host immunity. Notably, 46 differentially expressed immune and apoptosis-related genes that were predicted as targets of 26 vmiRNAs, were identified to be significantly deregulated in the SARS-CoV2 infected hiPSC-CM cells, suggesting a pivotal role of viral miRNAs in disease progression and pathogenicity. The miRNA-mRNA-KEGG pathway networks revealed 4 down-regulated genes (BMP7, CD36, GPR183 and SFRP2) that are targeted by 9 vmiRNAs. RNAhybrid analysis revealed the stable interactions of all of these down-regulated genes with their corresponding viral miRNAs. SARS-CoV-2 miRNAs are found to be hybridized with the 3’ UTR of these gene transcripts with partial complementarity which suggests possible translational repression. These interactions possibly could provide important insights regarding the escape mechanism of SARS-CoV-2 from immune elimination in human host. Previous reports have suggested that multiple miRNAs may act synergistically to regulate target genes [46]. A study reported that viral miRNAs act in synergy with host miRNAs to escape immune elimination [47]. To best of our knowledge, no previous study has reported the involvement of viral miRNAs synergy in progression of SARS-CoV-2 infection. In this study, we were also interested to explore the relationships between viral miRNA clusters and COVID-19 pathogenesis. Five viral miRNAs were predicted to act in synergy to regulate 2 genes (BMP7 and CD36) out of 4 significantly down-regulated genes related to immune and apoptotic functions. Previous studies reported that BMP-7 is associated with pathophysiology of several diseases [48]. Recent evidence suggested that antagonists of BMP signaling pathways may play critical role in the treatment of COVID-19 induced multi-organ failure [49]. Another study reported that CD-36 expression is reduced during viral infection [50]. A recent study finds the possible links between the CD-36 and SARS-CoV-2 pathogenesis [51].In a similar study, Lee et al., 2020, have reported the role of GPR183 in regulation of severe inflammatory response in COVID-19 patients [52]. These evidences strengthen our hypothesis regarding the selective targeting of host immune and apoptotic pathways by SARS-CoV-2 encoded miRNAs in order to attain protective immunity and thereby ensure its own survival.
5. Conclusion
In summary, this in silico study envisages the possible role of viral-encoded miRNA in modulating the underlying molecular dynamics involved in host-pathogen interactions which could provide insights into viral miRNA-mediated regulatory network involved in promotion of SARS-CoV-2 pathogenicity. Interestingly, systemic analysis revealed that immune and apoptosis-related genes are among the top putative targets of SARS-CoV-2 encoded miRNAs. By silencing these genes, viral miRNAs could potentially ensure continued survival of infected cell, and thus, promote disease progression and viral pathogenesis. This study therefore paves the way for more in-depth experimental research to confirm the findings. Identification of miRNA-miRNA synergistic regulatory networks and their putative target genes such as, BMP7, CD36, SFRP2 , etc., if confirmed experimentally, can be utilized to develop novel diagnostic and treatment strategies against ongoing COVID-19 pandemic.
Declaration of competing interest
None.
Acknowledgements
The authors are thankful to Indian Institute of Technology Kharagpur for providing the infrastructural support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.compbiomed.2021.104669.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Interactome S.-H., Guzzi P.H., Mercatelli D., Ceraolo C., Giorgi F.M. vol. 4. December 2019. p. 2020. (Master Regulator Analysis of the). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Heymann D.L. A novel coronavirus outbreak of global health concern. 2020;395:15–18. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO . Vol. 48. World Health Organization; 2021. Weekly Operational Update on COVID-19; pp. 1–10. no. [Google Scholar]
- 4.Stringari L.L., et al. Covert cases of severe acute respiratory syndrome coronavirus 2: an obscure but present danger in regions endemic for Dengue and Chikungunya viruses. PLoS One. 2021;16(1):1–21. doi: 10.1371/journal.pone.0244937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang C., et al. 2020. Articles Clinical features of patients infected with 2019 novel coronavirus in Wuhan , China, pp. 497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cizmecioglu A., Prof A. “Apoptosis ‐ induced T ‐ cell lymphopenia is related to COVID ‐ 19 severity. 2021;2:1–8. doi: 10.1002/jmv.26742. September 2020. [DOI] [PubMed] [Google Scholar]
- 7.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 8.S. S. Das, P. Saha, and N. Chakravorty, “miRwayDB: a database for experimentally validated microRNA-pathway associations in pathophysiological conditions.,” Database., vol. 2018, Jan. 2018. [DOI] [PMC free article] [PubMed]
- 9.Bruscella P., Bottini S., Baudesson C., Pawlotsky J.-M., Feray C., Trabucchi M. Viruses and miRNAs: more friends than foes. Front. Microbiol. 2017;8:824. doi: 10.3389/fmicb.2017.00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kincaid R.P., Sullivan C.S. vol. 8. 2012. (“Virus-Encoded microRNAs : an Overview and a Look to the Future). 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jr J.W.C., Trgovcich J., Hannenhalli S. Widespread evidence of viral miRNAs targeting host pathways. 2013;14(Suppl 2):1–9. doi: 10.1186/1471-2105-14-S2-S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pfeffer S., et al. Identification of virus-encoded microRNAs. Science. Apr. 2004;304(5671):734–736. doi: 10.1126/science.1096781. [DOI] [PubMed] [Google Scholar]
- 13.Morales L., Oliveros J.C., Fernandez-Delgado R., tenOever B.R., Enjuanes L., Sola I. SARS-CoV-Encoded small RNAs contribute to infection-associated lung pathology. Cell Host Microbe. Mar. 2017;21(3):344–355. doi: 10.1016/j.chom.2017.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grundhoff A., Sullivan C.S., Ganem D.O.N. 2006. A Combined Computational and Microarray-Based Approach Identifies Novel microRNAs Encoded by Human Gamma-Herpesviruses; pp. 733–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruber A.R., Lorenz R., Bernhart S.H., Neuböck R., Hofacker I.L. The Vienna RNA websuite. Nucleic Acids Res. Jul. 2008;36:W70–W74. doi: 10.1093/nar/gkn188. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu B., Fang L., Liu F., Wang X., Chou K.-C., iMiRNA-PseDPC “. microRNA precursor identification with a pseudo distance-pair composition approach. J. Biomol. Struct. Dyn. 2016;34(1):223–235. doi: 10.1080/07391102.2015.1014422. [DOI] [PubMed] [Google Scholar]
- 17.Karathanasis N., Tsamardinos I., Poirazi P. MiRduplexSVM: a high-performing MiRNA-duplex prediction and evaluation methodology. PLoS One. May 2015;10(5) doi: 10.1371/journal.pone.0126151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paraskevopoulou M., et al. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. May 2013;41 doi: 10.1093/nar/gkt393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y., Wang X. “miRDB : an online database for prediction of functional microRNA targets. 2020;48:127–131. doi: 10.1093/nar/gkz757. August 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma A., et al. Report human iPSC-derived cardiomyocytes are susceptible to SARS-CoV-2 infection ll human iPSC-derived cardiomyocytes are susceptible to SARS-CoV-2 infection. Cell Rep. Med. 2020;1(4):100052. doi: 10.1016/j.xcrm.2020.100052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Love M.I., Huber W., Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2; pp. 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dennis G., et al. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4(9):R60. [PubMed] [Google Scholar]
- 23.Shannon P., et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. Nov. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krüger J., Rehmsmeier M. RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res. 2006;34(2):W451–W454. doi: 10.1093/nar/gkl243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pesole G., Liuni S., Grillo G., Saccone C., UTRdb “. A specialized database of 5′- and 3′-untranslated regions of eukaryotic mRNAs. Nucleic Acids Res. 1998;26(1):192–195. doi: 10.1093/nar/26.1.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petrosillo N., Viceconte G., Ergonul O., Ippolito G., Petersen E. COVID-19, SARS and MERS: are they closely related? Clin. Microbiol. Infect. 2020;26(6):729–734. doi: 10.1016/j.cmi.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agarwal V., Bell G.W., Nam J.-W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. Elife. Aug. 2015;4 doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krek A., et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37(5):495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 29.Enright A.J., John B., Gaul U., Tuschl T., Sander C., Marks D.S. MicroRNA targets in Drosophila. Genome Biol. 2003;5(1):R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das S.S., James M., Paul S., Chakravorty N. miRnalyze: an interactive database linking tool to unlock intuitive microRNA regulation of cell signaling pathways. Database. 2017;2017(1):1–9. doi: 10.1093/database/bax015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mishra R., Kumar A., Ingle H., Kumar H. The interplay between viral-derived miRNAs and host immunity during infection. Front. Immunol. 2020;10:3079. doi: 10.3389/fimmu.2019.03079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mishra R., Kumar A., Ingle H., Kumar H. The interplay between viral-derived miRNAs and host immunity during infection. Front. Immunol. 2019;10:3079. doi: 10.3389/fimmu.2019.03079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gebert L.F.R., MacRae I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019;20(1):21–37. doi: 10.1038/s41580-018-0045-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gordon D.E., et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583(7816):459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Girardi E., López P., Pfeffer S. On the importance of host MicroRNAs during viral infection. Front. Genet. 2018;9:439. doi: 10.3389/fgene.2018.00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Skalsky R.L., Cullen B.R. Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 2010;64:123–141. doi: 10.1146/annurev.micro.112408.134243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Islam M.S., Islam A.B.M.M.K. Viral miRNAs confer survival in host cells by targeting apoptosis related host genes. Inf. Med. 2021;22:100501. [Google Scholar]
- 38.Cristina J., Echeverría N., Gambaro F., Fajardo A. 2016. Genome-wide Prediction of microRNAs in Zika Virus Genomes Reveals Possible Interactions with Human Genes Involved in the Nervous System Development. [Google Scholar]
- 39.Grundhoff A. In: In Antiviral RNAi: Concepts, Methods, and Applications. van Rij R.P., editor. Humana Press; Totowa, NJ: 2011. Computational prediction of viral miRNAs; pp. 143–152. [Google Scholar]
- 40.Verma S., Dwivedy A., Kumar N., Biswal B.K. ” bioRxiv; 2020. Computational Prediction of SARS-CoV-2 Encoded miRNAs and Their Putative Host Targets. [Google Scholar]
- 41.Khanmohammadi S., Rezaei N. Role of Toll-like receptors in the pathogenesis of COVID-19. J. Med. Virol. 2021;93(5):2735–2739. doi: 10.1002/jmv.26826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wehbe Z., Hammoud S., Soudani N., Zaraket H., El-Yazbi A., Eid A.H. Molecular insights into SARS COV-2 interaction with cardiovascular disease: role of RAAS and MAPK signaling. Front. Pharmacol. 2020;11:836. doi: 10.3389/fphar.2020.00836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hengartner M.O. The biochemistry of apoptosis. Nature. 2000;407(6805):770–776. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- 44.Menendez D., Shatz M., Resnick M.A. Interactions between the tumor suppressor p53 and immune responses. Curr. Opin. Oncol. Jan. 2013;25(1):85–92. doi: 10.1097/CCO.0b013e32835b6386. [DOI] [PubMed] [Google Scholar]
- 45.Fridman J.S., Lowe S.W. Control of apoptosis by p53. Oncogene. 2003;22(56):9030–9040. doi: 10.1038/sj.onc.1207116. [DOI] [PubMed] [Google Scholar]
- 46.Chen X., et al. MicroRNAs tend to synergistically control expression of genes encoding extensively-expressed proteins in humans. PeerJ. 2017;5 doi: 10.7717/peerj.3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nachmani D., Lankry D., Wolf D.G., Mandelboim O. The human cytomegalovirus microRNA miR-UL112 acts synergistically with a cellular microRNA to escape immune elimination. Nat. Immunol. Sep. 2010;11(9):806–813. doi: 10.1038/ni.1916. [DOI] [PubMed] [Google Scholar]
- 48.Aluganti Narasimhulu C., Singla D.K. The role of bone morphogenetic protein 7 (BMP-7) in inflammation in heart diseases. Cells. Jan. 2020;9(2) doi: 10.3390/cells9020280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carlson F.R., Bosukonda D., Keck P.C., Carlson W.D. Multiorgan damage in patients with COVID-19: is the TGF-β/BMP pathway the missing link? JACC Basic Transl. Sci. 2020;5(11):1145–1148. doi: 10.1016/j.jacbts.2020.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cooper G., et al. 2015. Viral Infection of Macrophages Reduces CD36 Expression: Role of IFNβ. [Google Scholar]
- 51.Pérez M.M., et al. ” medRxiv; 2021. Cholinergic and Lipid Mediators Crosstalk in Covid-19 and the Impact of Glucocorticoid Therapy. 2021.01.07.20248970. [Google Scholar]
- 52.Lee H., Im H.-J., Na K.J., Choi H. 2020. Discovery of Potential Imaging and Therapeutic Targets for Severe Inflammation in COVID-19 Patients. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.