Abstract
Missing data is a common, difficult problem for network studies. Unfortunately, there are few clear guidelines about what a researcher should do when faced with incomplete information. We take up this problem in the third paper of a three-paper series on missing network data. Here, we compare the performance of different imputation methods across a wide range of circumstances characterized in terms of measures, networks and missing data types. We consider a number of imputation methods, going from simple imputation to more complex model- based approaches. Overall, we find that listwise deletion is almost always the worst option, while choosing the best strategy can be difficult, as it depends on the type of missing data, the type of network and the measure of interest. We end the paper by offering a set of practical outputs that researchers can use to identify the best imputation choice for their particular research setting.
Keywords: Missing data, Imputation, Network sampling, Network bias
Introduction
Network data are often incomplete, with nodes and edges missing from the network of interest. Missing data can create problems when analyzing network data because network measures are often defined with respect to a fully observed graph (Borgatti et al., 2006; Smith and Moody, 2013). For example, measures that depend on the paths between all actors (e.g., closeness centrality and betweenness centrality) tend to be particularly sensitive to missing data (Kossinets, 2006; Smith et al., 2017; Rosenblatt et al., 2020). A researcher faced with incomplete network data must decide what, if any, imputation should be employed to limit the biasing effect of missing data (Huisman, 2009; Žnidaršič et al., 2018). Unfortunately, there are few clear guidelines for making imputation decisions. Recent work has shown that imputation can reduce the bias resulting from missing data, but we are only beginning to understand the returns to imputation (e.g., Koskinen et al., 2010; Gile and Handock, 2017; de le Haye et al., 2017; Krause et al., 2018a,b, 2020). For instance, is it always best to impute network data, or can we sometimes get away with doing nothing? And more pressing, how should a researcher choose which imputation method is optimal? Does one approach offer a universally robust option, or does it depend on the circumstances of the study?
This paper is part of a three-paper series on missing network data, with the overall goal of offering practical advice and tools for network researchers faced with missing data (Smith and Moody, 2013; Smith et al., 2017). We focus on a common situation: where the network data is incomplete because a subset of actors has provided no information about their network ties (e.g., Galaskiewicz, 1991; Costenbader and Valente, 2003; Silk et al., 2018). For example, a network study of high school students may miss students because they were absent the day of the survey, or simply because they refused to participate in the study. Even automated data (e.g., based on Bluetooth proximity) may suffer from missing data problems (Wang et al., 2012).
Paper 1 and paper 2 of this series explored the effect of missing data on network measures across a wide range of networks and missing data scenarios. We find that bias can vary dramatically across settings, where the level of bias depends crucially on the measure of interest, the network being analyzed and the type of missing data (see also Frantz et al., 2009; Huisman, 2009; Martin and Niemeyer, 2019). For example, the same measure (e.g., Bonacich centrality) calculated on the same network can yield very different levels of bias, depending on if the missing nodes are central or peripheral actors (Smith et al., 2017).
In this paper, we focus on the question of imputation: how do different imputation methods fare under different research settings—in terms of measures, network features and missing data types.1 In this way, we combine the literature on missing data effects (where is missing data most problematic?) with the literature on imputation (what kinds of strategies can we use to impute network data?). We consider a range of imputation methods, from simple imputation to more complex model- based approaches (Žnidaršič et al., 2017; Krause et al., 2018a,b, 2020). Simple imputation approaches attempt to ‘rebuild’ the network as best as possible from the information found in the data itself (i.e., if i nominates j and j is missing we might impute a tie from j to i) (Huisman, 2009). Model-based approaches use sophisticated statistical models to probabilistically fill in the missing data (Koskinen et al., 2010; Wang et al., 2016). From the perspective of a researcher, it is crucial to know which approaches will work for their particular setting. An approach that is effective for one measure and/or network type may be ineffective for another. The effectiveness of the approach may even depend on the type of missing data.
Our ultimate goal is to offer a set of practical outputs for researchers trying to decide what imputation approach to implement. A researcher will be able to identify the case closest to their own (in terms of measures, missing data type, and network features) and then use our results to look up the optimal imputation choice. A researcher would, of course, have to balance the performance of the imputation method (in terms of lowering bias) with the difficulty of implementing it. Our results will make it easier for a researcher to perform such cost-benefit analyses, given the features of their research setting.
We begin the paper with a short background section on missing network data and imputation. We then describe the imputation methods of interest, as well as the networks, measures and sampling setup that will form the basis of the analysis. Our analysis follows past work in the literature. We begin by taking a complete network and simulating different missing data scenarios. We then impute the missing data (on the now incomplete network), recalculate the measures of interest, and compare the resulting value to the true value. We present results for three types of network measures: centrality, centralization and topology.
Theory
Our paper contributes to a growing literature on non-response treatments to missing network data (Wang et al., 2016; Žnidaršič et al., 2017; Krause et al., 2018a,b). We focus on a key form of missing data, actor non-response. We define a non-respondent as an actor that fails to offer any nomination information (i.e., no information on out-going ties). We assume that non-respondents are not completely missing, however, and can still be nominated by other actors.2 This is a common form of missing data, particularly in well-defined, bounded settings. For example, in a school, a student may be out sick the day of the survey but still be on the roster, so that other students could nominate them. In this way, we have observations of a non-respondent’s in-coming ties but not of their out-going ties.
The two most common approaches for inferring missing ties are simple imputation and model-based imputation. We discuss each in turn.
Simple imputation leverages information about the incoming ties to non-respondents to help reconstruct the network (Stork and Richards, 1992; Huisman, 2009). If node k is a non-respondent and is nominated by i and j (who are not missing), we begin by including the ties from i→k and j→k. Additional heuristics can then be applied to help fill in the network. For example, a researcher may assume reciprocal ties going out from the non-respondents to those who nominated them, imputing k→i and k→j. Assuming reciprocity, however, runs the risk of adding ties that do not really exist, while doing no further imputation may fail to add ties that do exist. Much of the methodological work on simple imputation has asked how well such methods work in practice. For example, Huisman (2009) compared an imputation strategy based on the observed network’s density to a preferential attachment strategy and a unit imputation strategy. He found that simple imputation generated stable estimates of reciprocity, mean degree, and inverse geodesic distance for undirected networks with a 40 % non-response rate or less, but preformed less well for directed networks.
Recent work has considered more complicated (non-model based) imputation methods; where the rules for adding ties are dependent on other properties of the graph, like the reciprocity rate or the indegree of a node’s nearest neighbors (Žnidaršič et al., 2017). For example, Žnidaršič et al. (2017) explored a range of actor non-response treatments, finding that imputing ties based on the incoming ties of ego’s k-nearest neighbors significantly reduces non-response bias in valued networks, but that the macro-structure of the network (e.g., core periphery networks, networks with cohesive subgroups, and hierarchical networks) significantly influences the effectiveness of such strategies (see also Žnidaršič et al., 2018).
Model-based imputation methods are an alternative approach for inferring missing data (Robins et al., 2004; Kolaczyk and Csárdi 2014´ ). As with simple imputation, model-based methods begin by leveraging the information provided by the incoming ties to non-respondents. Model-based approaches, however, go further by proposing a parametric model that derives the likelihood of the observed data as a marginalization of the complete-data likelihood over the possible states of the missing variable (in our case a given adjacency matrix) (Gile and Handcock, 2017). The advantage of this approach is that it allows the researcher to incorporate more information about the network’s nodes, dyads, and local structure when estimating the likelihood of a given tie, as well as information about the survey instrument such as number of alters each respondent could nominate (Wang et al., 2016). Similarly, a model-based approach makes it possible to impute ties between non-respondents, which is difficult with simple imputation approaches. Model-based imputation also has the advantage of considering multiple plausible states of the network to generate a summary measure that accounts for the increased variability of parameter estimates due to imputation (Huisman and Krause, 2017). In this way, a researcher is better able to take into account the uncertainties in the imputation process, consistent with best practices from the literature on multiple imputation (Allison, 2002).
For example, Wang et al., 2016 used an imputation method based on exponential random graph models (ERGM); they found, on average that, 73 % of the missing ties could be effectively imputed, although smaller, sparser networks were harder to fit. Krause et al., 2018a,b expanded on this approach by employing a Bayesian ERGM (see also Koskinen et al., 2013) to impute missing ties, finding that Bayesian models are particularly useful when the percentage of missing data approaches 50 % and the measure in question is sensitive to misspecification (such as with transitivity). The main disadvantage of model-based imputation is that it can be difficult to implement (given the need to specify and estimate a model). Model-based imputation may also introduce bias by over generalizing tendencies observed in information rich parts of the network to the entire network. Nevertheless, these methods are often warranted, particularly in longitudinal network analysis where missing data is likely due to the repeated nature of the sampling, and where biases present in the initial network will affect the estimates of all subsequent networks (Hipp et al., 2015; Krause et al., 2018a,b).
Imputation strategies thus have great potential to limit bias due to missing data. Nevertheless, the practical problem remains of how to choose an imputation approach in a given research setting, especially as an imputation method that works well in one setting may not work well in another (Hipp et al., 2015). Different factors such as the network type, the kind of missing data, and the measure of interest are likely to influence the performance of different imputation methods, as different conditions magnify (or hide) the relative weaknesses of each approach. For example, we might expect that estimates of transitivity for a directed core-periphery network are particularly vulnerable to approaches that add reciprocated ties, as this runs the risk of inflating the estimated number of closed triads. In this case, the researcher should avoid reciprocated imputation; but what imputation strategy should be used, and are there other cases where reciprocated imputation works well?
In short, different approaches are likely to work better/worse in different settings, and it is crucial for a researcher to understand the consequences of different imputation choices. With this is mind, our analysis will extend past work by considering different imputation options across a much wider range of network types, measures and missing data features than is typically considered. It is only by considering such a complex set of conditions that we can begin to offer practical advice to researchers, as we can say under what conditions a given imputation approach is most appropriate.
Data
We examine the efficacy of different imputation methods across twelve empirical networks, seven directed networks and five undirected networks. These networks vary widely in terms of network features and substantive contexts, although all networks are limited to under 1000 nodes. Medium to small networks are sensitive to missing data and are conducive to additional data collection efforts, making them particularly appropriate for the study of missing data and imputation methods (Gile and Handcock, 2017). All networks are binary. Binary network data are still commonly used, and it is important to understand how missing network information affects this baseline case. The networks are the same as in papers I and II of this study.3 They include: “data on elites (corporate interlocks: “Mizruchi Interlock” and “River City Elite”), young youth networks (“Gest 6th graders”, “Prosper s220”)4, adolescent and young adult networks (“Sorority Friendship”, “High School (p13 & p24)”, the Gagnon prison network (MacRae, 1960), science networks (the sociological abstracts collaboration graph, the Social Networks article co-citation graph, and the biotechnology exchange network) and epidemiological networks (Colorado Springs HIV risk network - Morris and Rothenberg, 2011)5 “ (quoted from Smith and Moody, 2013). See Fig. 1 for plots and summary statistics (Table 1).
Fig. 1.
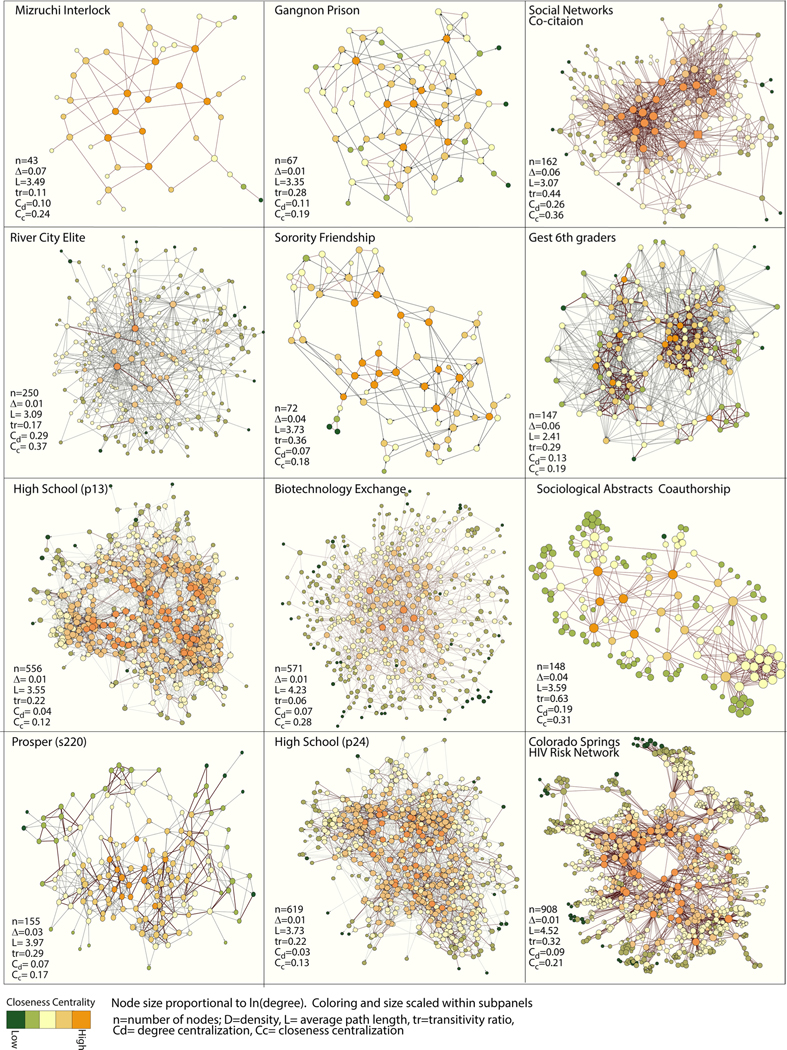
Networks Used for Sampling Simulations.
Table 1.
Sample network descriptive statistics.
| Inter-lock | Prison | Sorority | 6th Grade | Co-author | Prosper | Co-citation | Elite | HS 13 | Bio-tech | HS24 | HIV Risk | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Directed? | No | Yes | Yes | Yes | No | Yes | No | Yes | Yes | No | Yes | No |
| Centrality | ||||||||||||
| In – Degree | 3.02 | 2.72 | 2.89 | 8.86 | 6.16 | 3.83 | 9.32 | 2.39 | 6.06 | 3.85 | 5.71 | 6.05 |
| (1.93) | (2.02) | (1.75) | (5.26) | (5.98) | (2.69) | (10.62) | (7.59) | (4.42) | (4.99) | (3.96) | (8.12) | |
| Out – Degree | 3.02 | 2.72 | 2.89 | 8.86 | 6.16 | 3.83 | 9.32 | 2.39 | 6.06 | 3.85 | 5.71 | 6.05 |
| (1.93) | (1.48) | (1.85) | (4.67) | (5.98) | (2.36) | (10.62) | (1.63) | (2.90) | (4.99) | (2.99) | (8.12) | |
| Symmetric Degree | 3.02 | 5.43 | 5.78 | 17.7 | 6.16 | 7.65 | 9.32 | 4.78 | 12.1 | 3.85 | 11.43 | 6.05 |
| (1.93) | (2.73) | (2.88) | (7.73) | (5.98) | (3.85) | (10.62) | (7.83) | (6.04) | (4.99) | (5.84) | (8.12) | |
| Closeness | 0.36 | 0.18 | 0.15 | 0.35 | 0.32 | 0.18 | 0.38 | 0.03 | 0.22 | 0.26 | 0.2 | 0.25 |
| (0.08) | (0.08) | (0.09) | (0.12) | (0.06) | (0.09) | (0.09) | (0.02) | (0.05) | (0.04) | (0.06) | (0.04) | |
| Betweenness | 0.06 | 0.03 | 0.04 | 0.01 | 0.02 | 0.02 | 0.01 | 0 | 0.01 | 0.01 | 0.01 | 0 |
| (0.07) | (0.05) | (0.05) | (0.02) | (0.06) | (0.03) | (0.02) | (0) | (0.01) | (0.02) | (0.01) | (0.02) | |
| Bonacich Power | 0.82 | 0.86 | 0.86 | 0.9 | 0.58 | 0.82 | 0.63 | 0.64 | 0.83 | 0.6 | 0.83 | 0.57 |
| (0.58) | (0.52) | (0.51) | (0.43) | (0.82) | (0.57) | (0.78) | (0.77) | (0.55) | (0.8) | (0.56) | (0.82) | |
| Centralization | ||||||||||||
| In – Degree | 0.10 | 0.08 | 0.06 | 0.14 | 0.19 | 0.08 | 0.26 | 0.29 | 0.04 | 0.07 | 0.03 | 0.09 |
| Out – Degree | 0.10 | 0.08 | 0.06 | 0.15 | 0.19 | 0.02 | 0.26 | 0.03 | 0.01 | 0.07 | 0.01 | 0.09 |
| Symmetric Degree | 0.10 | 0.06 | 0.05 | 0.08 | 0.19 | 0.05 | 0.26 | 0.15 | 0.03 | 0.07 | 0.02 | 0.09 |
| Closeness | 0.27 | 0.12 | 0.17 | 0.16 | 0.35 | 0.11 | 0.48 | 0.06 | 0.08 | 0.36 | 0.08 | 0.28 |
| Betweenness | 0.20 | 0.17 | 0.16 | 0.06 | 0.37 | 0.16 | 0.12 | 0.01 | 0.05 | 0.24 | 0.03 | 0.17 |
| Bonacich Power | 0.20 | 0.18 | 0.14 | 0.13 | 0.26 | 0.16 | 0.22 | 0.41 | 0.13 | 0.29 | 0.12 | 0.23 |
| Topology | ||||||||||||
| Component Size | 43 | 67 | 72 | 147 | 148 | 155 | 162 | 250 | 556 | 571 | 619 | 908 |
| Bicomponent Size | 27 | 62 | 59 | 145 | 75 | 147 | 118 | 195 | 545 | 336 | 605 | 517 |
| Distance | 0.36 | 0.18 | 0.15 | 0.35 | 0.32 | 0.18 | 0.38 | 0.03 | 0.22 | 0.26 | 0.2 | 0.25 |
| Transitivity | 0.11 | 0.28 | 0.36 | 0.29 | 0.63 | 0.29 | 0.44 | 0.17 | 0.22 | 0.02 | 0.22 | 0.32 |
| TauRC | −0.91 | 2.35 | 4.65 | 17.75 | 17.65 | 7.22 | −27.36 | 163.6 | 23.66 | −91.61 | 22.91 | −104.22 |
Standard deviations are in parentheses.
Measures
Our analysis includes 16 different measures which we broadly divide into three classes: centrality, centralization and topology. By looking at a wide range of measures, we can better describe the conditions under which different imputation approaches offer the best choice.
Centrality
Our centrality measures include in-degree, total degree, Bonacich power centrality, closeness and betweenness. For the undirected networks, we only include a measure of degree (as total degree, out-degree, and in-degree are the same). Note that Bonacich power centrality is calculated on a symmetrized version of the network (for the directed networks). We calculate closeness centrality based on the inverse distance matrix, so that disconnected nodes have a value of 0 and directly connected nodes have a value of 1. We use the inverse distance matrix so that the summation does not include undefined values, a problem when pairs of people in the network cannot reach one another.
Centralization
Centralization measures the variation in the distribution of the given centrality measure, and is a graph-level statistic (whereas centrality captures an individual-level characteristic). We include centralization scores for each of our centrality measures. Our measure of centralization is a simple standard deviation of the individual centrality scores.
Topology
We include six topological measures. We include two global measures of connectivity, component size and bicomponent size. We measure component size as the proportion in the largest component. We first calculate the number of actors in the largest component, defined as the largest set of actors connected by at least one path. We then divide by the size of the network, defined as the number of nodes in the network being analyzed (i.e., the observed network after any nodes have been removed as part of the missing data treatment). A bicomponent is defined as a set of actors connected by at least two independent paths (Moody and White, 2003). As with component size, we divide the size of the largest bicomponent by the size of the network being analyzed, yielding the proportion in the largest bicomponent. Our third measure is distance, measured as the mean inverse distance between pairs of nodes (meaning that higher values actually indicate lower distances). We scale the value by the log of network size.6 Our fourth measure is transitivity, measured as the relative number of two-step paths that also have a direct path; more substantively, transitivity captures the tendency for a “friend of a friend to be a friend”. Our fifth measure is the tau statistic, a weighted summary statistic based on the triad distribution (Wasserman and Faust, 1994). The tau statistic captures the local processes that govern tie formation (like clustering and hierarchy). The tau statistic is a summation over the specified triads, conditioned on the dyad distribution in the network. Here, we use the ranked-cluster (RC) weighting scheme.7 Our last topological measure is based on blockmodeling the network (White et al., 1976). We begin by partitioning the full network into a set of equivalence blocks, where nodes with similar pattern of ties are placed together.8 We use the Rand statistic (Rand, 1971) to compare the partitioning found in the incomplete data to the partitioning observed in the full, true network. The unadjusted Rand statistic shows the proportion of pairs in one partition (the true partitioning) that are placed together in a second partition (the partitioning under missing data).
Missing data
Our study is focused on the efficacy of different imputation methods under different conditions of missing data. In order to explore different level of missing data), we identify a portion of the nodes as non-imputation approaches, it is first necessary to generate missing data respondents, individuals for whom we have no information on out- from the observed data in a controlled, systematic way. We follow a going edges. We construct the observed (incomplete) network by standard protocol when inducing missing data: for each network (and removing the out-going edges from the non-respondents. Once the appropriate edges are removed, we apply different imputation methods to the same network, with the same missing data. We repeat this process 1000 times for each level of missing data: 1 %, 2 %, 5 %, 10 %, 15 %, 20 %, 25 %, 30 %, 40 %, 50 %, 60 %, 70 %.9
We also consider different types of missing data, defined by which nodes are most likely to be non-respondents: central nodes, peripheral nodes, or nodes selected at random. Past work has shown that missing more central nodes generally yields larger bias (at least when the researcher takes no action to impute the missing cases) (Smith et al., 2017). Research contexts vary with respect to who is most likely to be a non-respondent. For example, central actors are less likely to provide nomination data when studying organizations (e.g., public officials in an elite network), where central actors are more likely to have scheduling conflicts and may be less willing to cooperate. Peripheral actors are more likely to be non-respondents in settings like schools, where actors who are not socially embedded in the network are more likely to be disengaged and thus less likely to take the survey. Network measures are typically robust to missing peripheral members of the network, but imputation procedures may struggle in such cases. We have very little information about nodes on the periphery (as few people nominate them), making it harder to impute where ties should be added for those actors. It is unclear how different imputation procedures will fare when periphery nodes are non-respondents (compared to more central nodes).
Formally, our missing data conditions are set based on the correlation between centrality and the probability of being a non-respondent. We have five correlation values: strong negative correlation (−.75), weak negative correlation (−.25), missing at random (0), weak positive correlation (.25) and strong positive correlation (.75). Cases with a negative correlation between centrality and missingness correspond to situations where those on the periphery are more likely to be non- respondents. A zero correlation corresponds to random selection of non-respondents, while a positive correlation means that more central actors are more likely to be non-respondents. We consider two definitions of central actors, one based on closeness and one based on in- degree. There are thus 9 different missing data types (−.75 closeness, −.75 in-degree, −.25 closeness, −.25 in-degree, 0, .25 closeness, .25 in- degree, 75 closeness, and .75 in-degree). We will not distinguish between the closeness and in-degree results here as they offer very similar findings, so we simply aggregate them in the final figures and tables.
Imputation methods
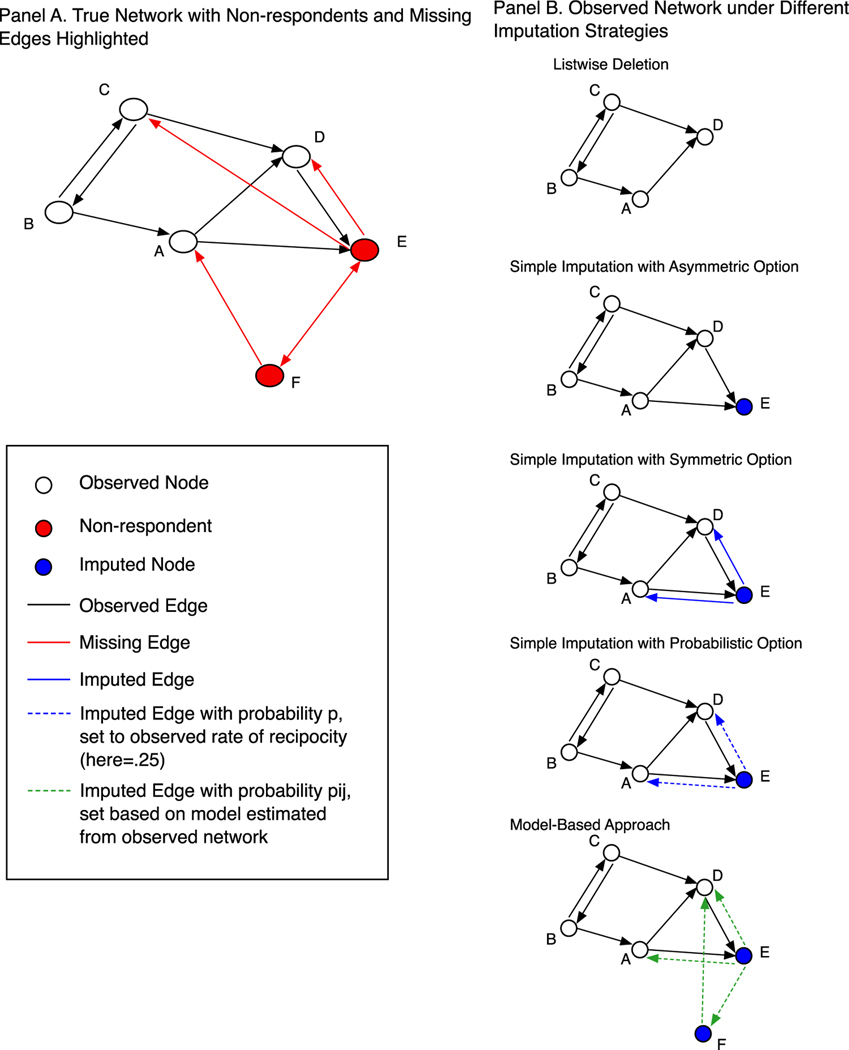
We consider the effectiveness of six different approaches for dealing with missing data. In each case, we first construct a network with missing data, where a subset of nodes is treated as non-respondents, with no out-going edge information. We then take the incomplete network and impute the missing edges using different imputation approaches. Fig. 2 presents the different imputation options in a simple network with 5 nodes. On the left-hand panel, we have the complete network with non-respondents and edges highlighted in red. Here, we see that nodes E and F are missing. All edges going from E or F to other actors (like E→D) will not be observed, including any edges between the non-respondents (see Handcock and Gile, 2010). A researcher will, however, typically have information on edges going to non-respondents (for example D→E), and we assume that this is the case for our analysis.
Fig. 2.
Demonstrating Imputation Strategies on Toy Network.
Given these conditions, we break out the imputation methods into three large classes: listwise deletion, simple imputation and model-based. The simple imputation methods include asymmetric, symmetric and probabilistic treatments; the model-based methods include simple and complex exponential random graph models. Thus, we consider six approaches: listwise deletion, asymmetric, symmetric, probabilistic, model-based simple and model-based complex.
Listwise deletion
The simplest option is to remove all nodes with incomplete information from the network. This is pictured in the top right-hand plot in Fig. 2. Here, non-respondents are not present in the network, while all incoming ties to non-respondents are not considered. This amounts to listwise deletion, where only those cases with full information, or those present at the time of data collection, are included in the network used for analysis. In this case, E and F and all ties going to and from E and F are missing. Note that this is the typical strategy for most network studies (Smith et al., 2017; Silk et al., 2015). Also, note that listwise deletion will yield a network that is smaller than the true network.
Simple imputation
The second class of imputation methods that we consider is simple imputation. Here, the researcher uses the tie information from the observed nodes to the non-respondents to help ‘fill in’, or reconstruct, the network (Huisman, 2009). The basic idea is that when an actor who is present in the survey nominates someone who is absent; that this is useful information that should not be thrown away, as is done in listwise deletion. For example, in Fig. 2, nodes A and D nominate E, who has missing data on all out-going ties. A simple imputation approach would begin by putting E back into the network with edges from A→E and D→E. This is demonstrated in the simple imputation plots in Panel B. Note that node F, who is also a non-respondent, is not put back into the network as none of the observed nodes nominated them. Once the missing nodes are put back, the researcher must decide on how to impute the missing edges from the non-respondents to the nodes who nominated them. No other ties are imputed. This means that all potential ties between non-respondents who were put back into the network are assumed not to exist, taking a value of 0 in the matrix.10 In our little example, we need to impute whether E sends ties back to A and D. Note that for undirected networks this choice is simple as all ties are reciprocated. The directed case is more difficult and we consider three different options.
Asymmetric imputation
First, a researcher could employ an asymmetric approach, where no ties from non-respondents are added to the nodes who nominated them (past work has also labeled this null tie imputation; Žnidaršič et al., 2017). In the second plot in Panel B, we see that there are ties from D to E and A to E but no ties from E, the non-respondent, back to D or A. This strategy privileges the observed data by removing from consideration ties from the non-respondents to the respondents who nominated them. The downside of this approach is that it assumes that an asymmetry exists between all non-respondents and respondents, an assumption that is unlikely to be true in many cases. It could, however, be useful for certain measures, especially those where adding an incorrect tie badly biases the results (like transitivity).
Symmetric imputation
The second option is symmetric imputation (also referred to as reconstruction in past work: Huisman, 2009; Žnidaršič et al., 2017). Here, a researcher always assumes that an edge from a non-respondent to a respondent is reciprocated. In our example, the edges from A and D to E (non-respondent) is returned, so we impute edges E→A and E→D. In this case, a researcher would get the E→D edge correct but would incorrectly add the E→A edge. A symmetric option is likely to work well when the reciprocity rate is high. It is also likely to work well in cases where the measure of interest does not rely heavily on the direction of the ties or in cases where missing edges are more consequential than adding incorrect edges.
Probabilistic imputation
The last simple imputation option is probabilistic imputation. Here, edges from non-respondents to the respondents who nominated them are imputed probabilistically, based on the rate of reciprocity in the observed network. A researcher first calculates the reciprocity rate as the proportion of ties that exist such that if i nominates j then j also nominates i. For this initial calculation, we only include dyads where both i and j are observed nodes (i.e., both respondents). For our example network in Fig. 2, the reciprocity rate in the observed, incomplete network is .25. A researcher would then take this rate and use it to impute the ties from non-respondents to respondents. Here, our researcher would basically flip a weighted coin, adding an edge with the probability set to .25. This would be done, in this case, for E→A and E→D. This process can itself be repeated a number of times (as there will be stochastic variation). Each iteration will yield a slightly different network, which can then be used in subsequent analysis. One could then summarize the results over the imputed networks. In our analysis, we repeat the imputation process over 100 networks, using the mean value (for the statistic of interest) over the 100 networks as the summary measure of interest. A probabilistic option falls somewhere between the asymmetric and symmetric options, in terms of adding or not adding edges. The probabilistic option is likely to be a fairly safe choice, although it may not always be the best option in every setting.
Overall, the simple imputation methods have the advantage of being very easy to implement while taking advantage of data from the survey itself. The disadvantage is that these methods miss any edge from non- respondents to other non-respondents (such as E↔F). It also systematically misses any asymmetric edge from a non-respondent to an observed node (such as E→C). Couple these built-in biases with the possibility of adding edges that are not really there, and it is unclear how far we can push simple imputation options, particularly given difficult combinations of measures and missing data types.
Model-based
The third class of imputation methods is model-based approaches. Here, a researcher takes the observed network, and estimates a statistical network model predicting the presence/absence of a tie between all ij pairs. The researcher then takes the underlying model and uses the model to predict, probabilistically, the ties that exist for nodes that are missing. For example, we know that networks tend to be homophilous (i. e., two actors who are similar are more likely to form a tie). A researcher can estimate the strength of this tendency (e.g., in terms of race or gender) and then use that information to help predict if a missing edge exists. We assume that the researcher has basic information on all actors, even the missing cases. Thus, an actor may have missing network data but basic demographic (or other) information about them may still be available. This may be acquired through administrative records, third hand reports or even from the ‘missing’ respondent, as they may begin the survey but not finish it.
A model-based imputation approach is depicted at the bottom of Panel B in Fig. 2. We assume that a researcher employing a model-based approach will begin by first performing a simple asymmetric imputation of the data. For example, in Fig. 2, Actor E is put back into the network and edges from A→E and D→E are added. The researcher will then take the remaining non-respondents, those who received no nominations from the observed nodes and put them back into the network. In our example actor F would be added to the network. The next step is to estimate a model predicting an edge between actors, only including the observed edges in the model.
We use exponential random graph models (ERGM) to impute the missing data. There are a number of possible options, but ERGM is a commonly used model and is quite flexible, making it an ideal choice. ERGMs are statistical models used to test hypotheses about network structure and formation (Hunter et al., 2008; Wasserman and Pattison, 1996). Formally, we define a network, Yij, over the set of nodes N, where Y is equal to 1 if a tie exists and 0 otherwise. Define y as the observed network. Y is then a random graph on N, where each possible tie, ij, is a random variable. ERGMs estimate the Pr(Y=y), where the “independent variables” are counts of local structural features in the network (Goodreau et al., 2009; Robins et al., 2007), such as number of ties and homophily. The model can be written as:
| (1) |
where g(y) is a vector of network statistics, θ is vector of parameters, and κ(θ) is a normalizing constant.
In this case, the model is estimated based on the incomplete network, where edges from non-respondents to other nodes are unobserved. Note that all missing edges are treated as NAs when estimating the model (i.e., missing instead of 0 s), and are thus not included in the estimation of the coefficients. The estimated coefficients may be biased, depending on the terms included and the type of missing data. It is, nonetheless, our best guess (given the data at hand) at what the underlying local tendencies are for the network. We can then take that estimated model and predict the missing edges. This amounts to simulating networks from the underlying model. Note that this includes missing edges from non-respondents to respondents (E→A) as well as edges between non-respondents (E and F). All observed edges (including edges from respondents to non-respondents) are held fixed and not allowed to vary as different networks are generated from the estimated model. In Fig. 2, we have plotted an example simulated network, with the missing edges colored green. This represents one possible imputation, or draw form the underlying model; another generated network would look slightly different (with different ‘green’ edges added to the network). A researcher could repeat this process a number of times, calculating the statistics of interest for each simulated network (with observed edges held fixed), summarizing over all of the calculated values.
The main benefit of a model-based approach is that we are able to recover nodes that are non-respondents and received no nominations from respondents (actor F in Fig. 2). We are also able to apply a better model to recover the edges between respondents and non-respondents (i.e., going beyond just reciprocity). Model-based approaches are thus likely to fare well when looking at measures that capture structural features at an aggregate level, like component size. The main drawback to a model- based approach is that the imputed network will deviate further from the raw data than with any of the simple imputation approaches. Measures that are sensitive to getting the specific edges correct, such as centrality measures, may be more difficult for a model-based approach.
We consider two versions of model-based imputation. One we denote as ‘simple’ and the other we denote as ‘complex’. Each has the same basic form but the simple model includes fewer terms, and is thus easier to estimate.
Simple model-based
The simple model-based approach includes three basic terms. First, we include a term for the base rate of tie formation in the network (edges). Second, we include terms for homophily on two attributes.11 We assume these attributes are known for both non-respondents and respondents. Third, for directed networks, we include a term capturing reciprocity, the count of the number of dyads where ij exists and ji exists. We then take the estimated model12, based on number of edges, reciprocity, homophily and simulate a set of networks from the underlying model. Only the missing edges are allowed to vary run to run, as the observed edges are held fixed.13
Within these simulations, we put constraints on the outdegree of each node, constraining the simulations so that each node has the same out- degree as the observed values (these values are imputed for the non-respondents).14 In this way, we ensure that out-degree in the simulated networks is consistent with the observed data, matching the respondents and matching our best guess for non-respondents. In the end, we calculate the statistics of interest for each generated network (we generate 100 networks each time)15 and take the mean over all networks as the measure of interest. All models are estimated in R using the ergm package (Handcock et al., 2019).
Complex model-based
The complex model-based approach is exactly the same as the simple approach expect that it includes a term to capture triadic processes. The simple model only includes terms at the node or dyadic level. For the complex model, we take the same model and add a GWESP (geometrically weighted edgewise shared partner) term to the model. GWESP is a weighted summation of the counts of how many shared partners each ij pair have (restricted to cases where i and j have a tie), capturing the tendency for groups (or local clusters) to emerge in the network. Adding GWESP allows us to capture tendencies towards transitivity and higher order closure. It also makes the model harder (and longer) to estimate.
In short, the model-based approach is more complicated than with simple imputation approaches. A researcher must make a number of difficult modeling choices (terms, constraints, etc.), and must then actually estimate and simulate from the specified model. Thus, one of the main questions of this study is about the relative payoffs and tradeoffs between simple imputation and model-based approaches. Simple imputation approaches are much easier to implement. But will they yield valid results? And if the model-based approach yields better estimates, is the improvement worth the added effort that a model-based approach requires? Thus, we want to identify the conditions (measure of interest, type of missing data, etc.) where one can ‘get away with’ the simpler network options, compared to the conditions where a more complicated model-based approach is necessary.
It is worth noting that our model-based imputation approaches employ particular forms, with particular terms and constraints included. It is possible that alternative specifications (i.e., using a Bayesian approach) would offer better results than that presented here (Krause et al., 2018a,b). Our results are, however, still instructive, as they represent the kinds of tradeoffs and models that a researcher in the field is likely to consider.
Presentation of results
Our results offer a presentation challenge. We have 12 networks, 5 missing data types, 12 missing data levels, 16 measures, and 6 imputation approaches. For each of these combinations, there are 1000 iterations (with different actors treated as non-respondents each time). The combinatorics make it difficult to present the results in a raw form. Our strategy is to calculate summary statistics across these iterations and then to use different plots and regression models as a means of summarizing the results.
For each scenario (network, measure, and missing data type), we begin by comparing the true values to that observed in the networks with missing data—where a subset of nodes are treated as non- respondents and different imputation strategies are used to deal with the missing data. We first calculate a bias score, capturing how far the observed value is from the true value. The observed value is the measure calculated on the imputed networks (i.e., the network we observe the imputation is performed). For the centrality scores, we calculate the centrality of the nodes in the complete network (no missing data), calculate it again using the networks with imputed data, and correlate these two vectors. The higher the correlation, the greater is the effectiveness of that imputation method for that missing data scenario. To make this a bias score, we calculate 1 minus the correlation between the true and observed centrality scores.
| (2) |
When we correlate the two vectors, we only include respondents, thus excluding non-respondents that we have brought back into the network through the imputation process. This makes the calculations consistent across different imputation strategies. It is also the likely choice that researchers would make in their own analysis (as the bias will be much higher for non-respondents). We have also performed an analysis where we keep all actors in the calculation (respondents and non-respondents). The imputation strategies fare much worse in this case, suggesting some of the costs of including the missing cases. The results for this additional analysis are presented in Table A13 in the Appendix.16
We use a standardized bias score for our graph level measures of centralization and topology. We define bias as:
| (3) |
A bias score captures how much the observed score (in the imputed networks) differs from the true value. The bias scores are relative to the size of the true value, making it easier to compare across networks and statistics. The bias scores can be negative (over-estimates) or positive (under-estimates). For simplicity, we take the absolute value of the bias scores, making them comparable in all analyses.
We begin each set of results with a bias ratio table. The bias ratio tables show the total improvement in the measure for each imputation type relative to listwise deletion. We first calculate the total bias that results from doing listwise deletion. We calculate the bias score (for the given measure) under listwise deletion, summing up the bias at each level of missing data to arrive at a total bias score. We then repeat this process for the same network and missing data, but now we assume that the data are imputed under different approaches. We take the total bias under each imputation approach and divide that by the total bias under listwise deletion, multiplying it by a 100 to arrive at a percent decrease in bias (or improvement in fit). Larger values are better, with negative values suggesting that the imputation method actually performed worse than listwise deletion.
For our second set of analyses, we take the bias scores and regress them on the level of missing data. We thus predict bias for each scenario as a function of percent missing nodes: . The estimated slope coefficient, β1, captures the expected increase in bias for a 10 % increase in number of non-respondents. The slope coefficients (β 1) are used as a summary measure, showing how quickly bias increases as the level of missing data goes up.
We then use a series of regression models to summarize the results, providing an overall picture of bias across all networks, measures, missing data types and imputation approaches. We run separate Hierarchical Linear Models (HLM) for each measure, using the bias slopes (β1) as the dependent variable (bias slopes are nested within networks). Larger coefficients mean that bias is predicted to be higher, as bias increases at a faster rate as missing data increases. Our main independent variable is the type of imputation, represented by a set of dummy variables: Asymmetric, Probabilistic, Symmetric, Model-based simple and Model-based complex. Listwise deletion serves as the reference category. We also include a variable for missing data type, ranging from −.75 (low degree nodes are more likely to be non-respondents) through .75 (high degree nodes are more likely to be non-respondents). The remaining variables capture network properties that may be correlated with higher levels of bias. We include predictors for network size (logged) and concentration (measured as the standard deviation of in- degree). We run separate models for the directed and undirected networks. We also include interactions between imputation type and the missing data type, as well as interactions between imputation type and network features. In this way, we can see the relative effectiveness of different imputation strategies under a variety of conditions. See Tables A2, A4, A6, A8, A10, and A12 in the Appendix.
We use the estimated regression models to produce a series of summary plots. We present two basic figures for each measure type (centrality, centralization, topology). The first figure focuses on the effectiveness of different imputation approaches across different measures and types of missing data. For each subplot, the x-axis is the level of missing data and the y-axis is the expected bias, under the scenario of interest. The results are based on a network with moderate features, one that is medium sized and moderately/weakly centralized. We have results for each measure and three different types of missing data (missing low centrality nodes, random missing data and missing high centrality nodes). Within each subplot there are 6 lines, one for each of the imputation approaches. The lines represent the predicted bias based on the regression model, using the regression coefficients for that model and setting the network features to the scenario of interest. Higher values in the plot indicate more bias and thus worse performing imputations. The second set of figures focuses on the effectiveness of imputation approaches across networks with different features. Here, we systemically vary the size and centralization of the networks, but hold the type of missing data fixed (only looking at random missing data). We include 4 combinations of size and centralization: large centralized, small centralized, large decentralized, small decentralized. The key question is how well different imputation methods fare for different measures under different conditions.
Results
Centrality
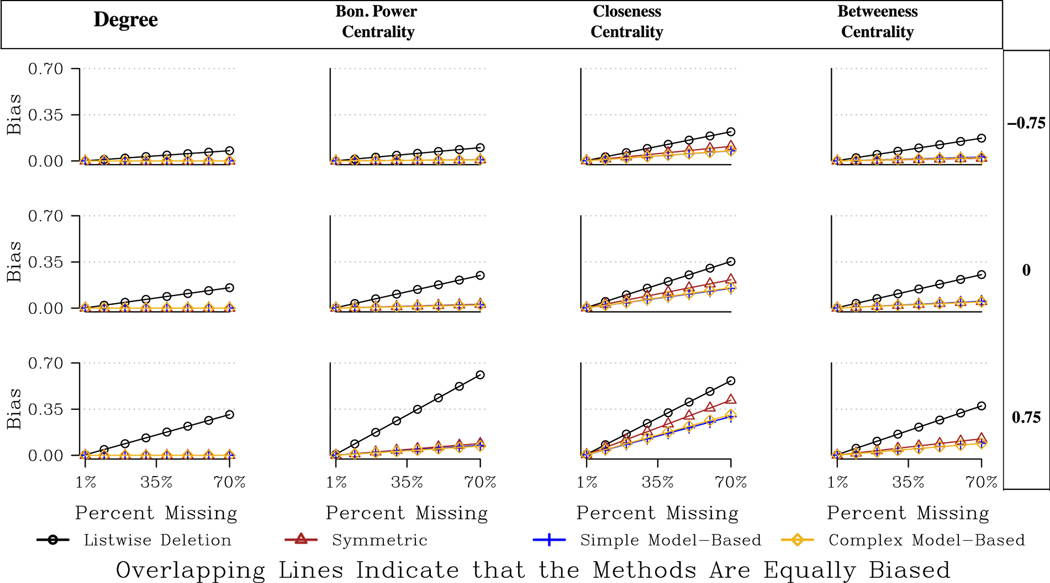
We begin our discussion of centrality by examining the undirected networks as they represent the simpler case. Table 2 captures a qualitative summary of the ‘best’ imputation approach under different conditions, while the main numerical results are presented in Fig. 3 and Tables A1 and A2. Table A1 is our bias ratio table, where each value in the table reports the decrease in total bias (over all levels of missing data and all runs) for that imputation approach compared to listwise deletion. Fig. 3 presents example predicted bias plots for one network for 3 different missing data types. The results are based on the HLM results presented in Table A2, predicting bias as a function of the missing data type and network characteristics.
Table 2.
Summary of Best Imputation Options for Centrality Measures.
| Measure | Directed or Undirected | Non-response Type | Size and Centralization | Best Imputation Option |
|---|---|---|---|---|
| Degree | Undirected | Any | Any | Any strategy except Listwise Deletion |
| Bon Power | Undirected | Central Nodes | Any | Model-Baseda |
| Betweenness | ||||
| Closeness | ||||
| Bon Power | Undirected | Less Central Nodes | Any | Any strategy except Listwise Deletion |
| Betweenness | ||||
| Closeness | ||||
| Indegree | Directed | Central Nodes | Any | Probabilistic |
| Indegree | Directed | Less Central Nodes | Any | Probabilistic, Listwise Deletion or Asymmetric |
| Total Degree | Directed | Any | Any | Asymmetric or Probabilistic |
| Bon Power | Directed | Any | Any | Probabilistic, Symmetric or Asymmetric |
| Closeness | Directed | Any | Any | Model-Based |
| Betweenness | Directed | Any | Any | Probabilistic |
Notes.
The model-based approach is assumed to correspond to complex or simple unless explicitly noted.
Fig. 3.
Predicted Bias for Centrality Measures for a Large, Undirected, Moderately Centralized Network.
The results for degree are very straightforward. We see that all imputation methods lower the bias to 0, or perfectly impute the missing data. The imputation methods add ties between i and j in cases where i nominates j and j is a non-respondent. There is no error possible here as the return tie (j to i) is assumed to be reciprocated, as the network is undirected. Note that this perfect correlation only holds if we restrict our attention to the degree of the respondents in the network (as there could be still unobserved edges between the missing cases that leads to bias).17
Bonacich, closeness and betweenness centrality offer a slightly more complicated story. In general, we find that the model-based imputation methods offer the best option, although the differences between the model-based and symmetric approaches are relatively small, especially for Bonacich centrality. For example, looking at closeness for the Biotech network, total bias decreases by 83 % using the model-based approaches and 78 % using the symmetric option, assuming we are missing high centrality nodes. The differences are even smaller when missing low centrality nodes, where the symmetric option is even sometimes preferred. More generally, we see that imputing (using any option) drastically reduces the bias compared with listwise deletion. Looking at betweenness with 40 % of the network missing and missing high degree nodes (assuming the network is medium-sized and decentralized), we would expect a bias of .21 under listwise deletion, .07 under the symmetric approach, and .052 under the simple and complex model-based approaches. The symmetric approach is particularly attractive here because it is so simple to implement, but still yields results that are close to the more onerous model-based approaches. See Fig. 3 for the full results.
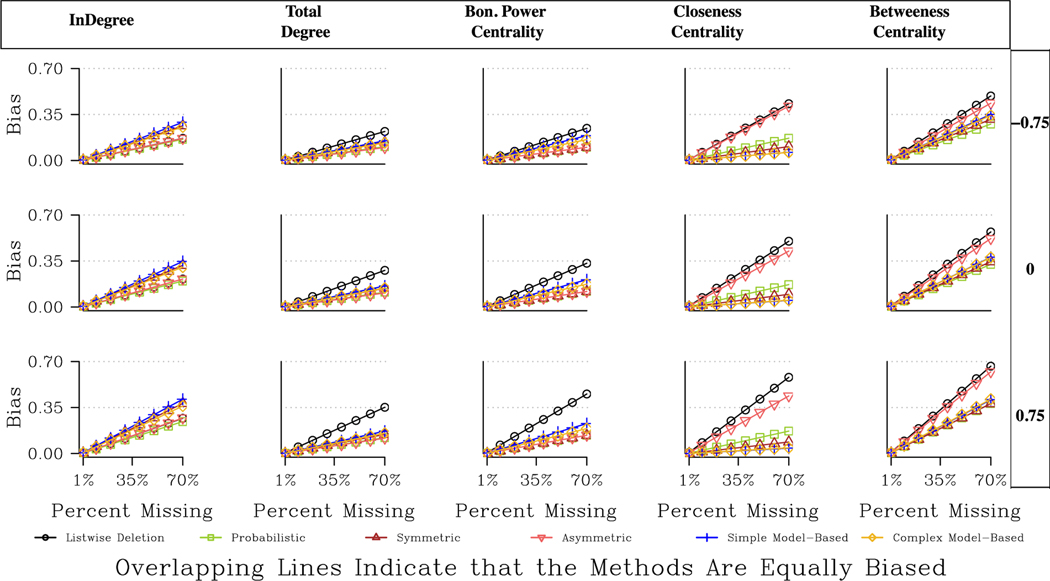
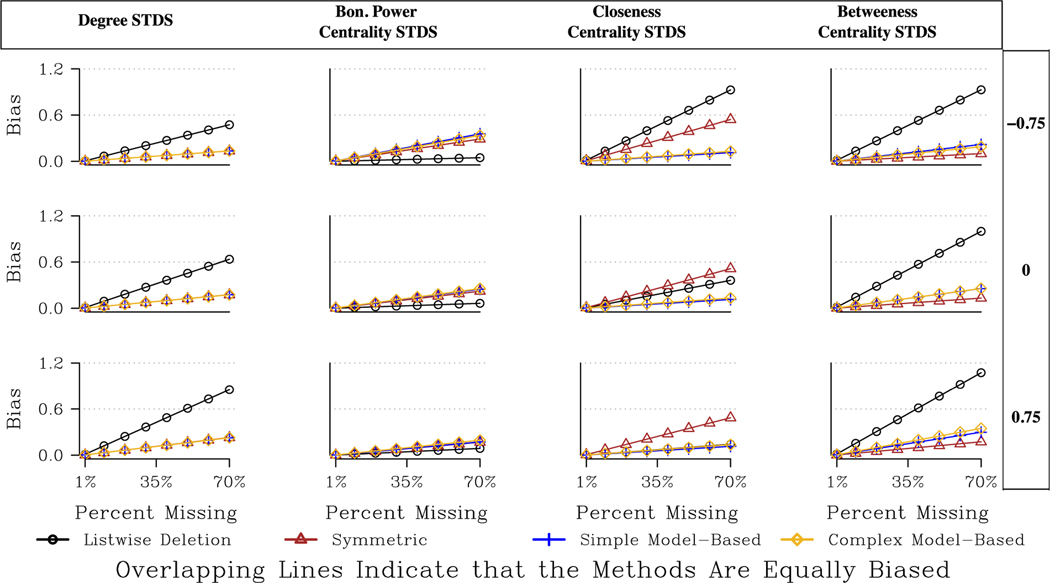
We now turn to the directed networks, where the results are more variable across measures and networks. The main results are presented in Tables A3 and A4 and Fig. 4. We will also refer to Table 2, capturing the best option for each scenario (in terms of measure, missing data type, etc.). Looking at indegree, we see that the probabilistic imputation method fares the best, although the real story is how badly imputation methods perform in general. The probabilistic approach is only marginally better than listwise deletion, while the symmetric and model-based approaches are actually worse than listwise deletion.
Fig. 4.
Predicted Bias for Centrality Measures for a Large, Directed, Moderately Centralized Network.
Looking at Table A3, the bias ratio table, we see negative numbers for the symmetric and model-based approaches, suggesting higher bias than listwise deletion. Indegree is difficult to impute because it is based on the specific number of nominations sent to each actor. An imputation method will add ties that are generally consistent with the existing data, but this does not mean that it will add the specific ties sent to a specific actor.
The imputation methods are more effective for total degree and Bonacich power centrality. With total degree, for example, the probabilistic and asymmetric approaches are almost always better than listwise deletion, and typically outperform the model-based and symmetric approaches. For example, in our large, moderately centralized network, the expected bias under 30 % missing data (with high degree nodes) is .049, .052, .062, .073, and .15 for the asymmetric, probabilistic, symmetric, model-based (simple) and listwise deletion approaches respectively. Fig. 4 also makes clear that the differences between listwise deletion and the imputation methods are highest when central nodes are more likely to be non-respondents. The returns to imputation are larger when central nodes are non-respondents because the bias under listwise deletion can be quite high (see Smith et al., 2017), while the imputation methods see only a slight increase in bias when missing central actors. The imputation methods tend to fare well when central nodes are non-respondents because there is so much information about central nodes (i.e. many people nominate them), making it easier to impute ties for those actors.
The exceptional case, as it often is, is the RC elite network, where imputation using the model-based or symmetric approaches are worse than listwise deletion, especially when missing less central nodes (the probabilistic and asymmetric approaches are similar to listwise deletion). The RC elite network is highly centralized, meaning if low centrality nodes are non-respondents, and we add reciprocated ties from the highly centralized node back to the peripheral nodes, and they do not exist, then we may greatly deviate from the true centralities in the network. There is, in that sense, the danger of over fitting/imputing what is essentially a hub and spoke structure.
Closeness offers a different story than the degree-based measures. All imputation methods are still better than listwise deletion, but here the model-based approaches fare considerably better. The model-based approaches are generally the best option, except for a few cases in the smaller networks (most noticeably the Sorority network where the probabilistic approach is best). For example, in the Proper network, the improvement using the model-based approach (simple or complex) is around 79 % compared to 61 % with probabilistic imputation or 67 % with symmetric imputation (missing high degree nodes).
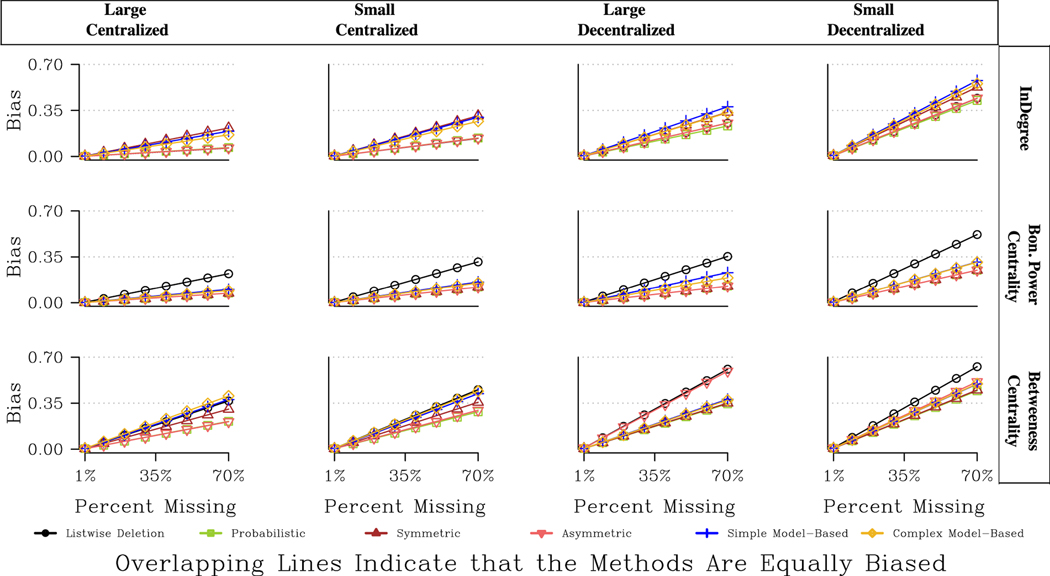
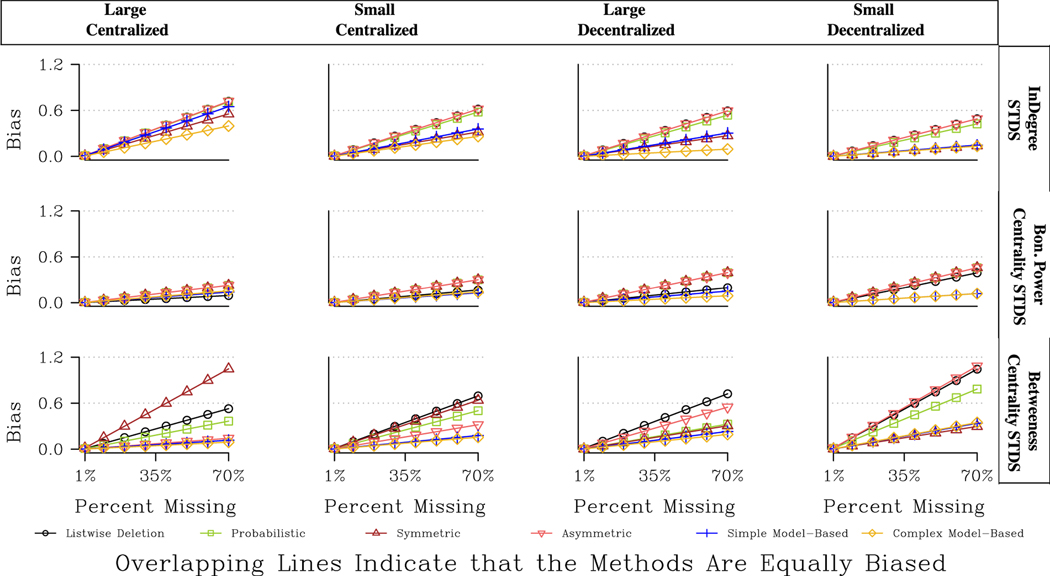
Fig. 5 offers a final set of comparisons, focusing on the performance of different imputation approaches in networks with different features. The figure presents the predicted bias for four different example networks, with varying combinations of size and centralization. The results are presented for directed networks with random non-response (also the case for Figs. 8 and 11) We focus on the results for betweenness centrality as the effect of centralization is so stark here. Overall, the probabilistic approach is consistently the best for betweenness, but otherwise the results are quite contingent. When the network is decentralized, the model-based and symmetric approaches perform adequately, offering better results than listwise deletion (although not as good as the probabilistic option). When the network is centralized, however, the model-based approach is the worst option, often little better than listwise deletion. We get similar results for closeness, where the model-based approaches are particularly preferred in large, decentralized networks (although the differences are less extreme). The symmetric and model- based approaches fare less well in centralized networks because both approaches tend to add more ties to the reconstructed network, potentially underestimating the centrality of the key actors while overestimating the centrality of the peripheral actors.
Fig. 5.
Predicted Bias for Centrality Measures for Four Network Types.
Fig. 8.
Predicted Bias for Centralization Measures for Four Network Types.
Fig. 11.
Predicted Bias for Topology Measures for Four Network Types.
Overall, for the directed networks, simple imputation strategies are preferred when estimating degree-based centrality measures, with probabilistic, asymmetric or sometimes even listwise deletion faring quite well. These imputation methods are less biased when estimating degree-based measures because they stick close to the actual data, and thus are better at recovering the specific number of alters. In contrast, with the path-based measures (particularly closeness), more complicated model-based approaches perform well, along with the probabilistic approach. For these path-based measures, the advantage of recovering more of the paths tends to outweigh the risk of (potentially) inflating the degree of any one node, as the model-based approaches are able to recover the pattern of ties. The probabilistic approach is unique in that it performs well in almost every case, a robust option when measuring centrality scores on directed networks.
Centralization
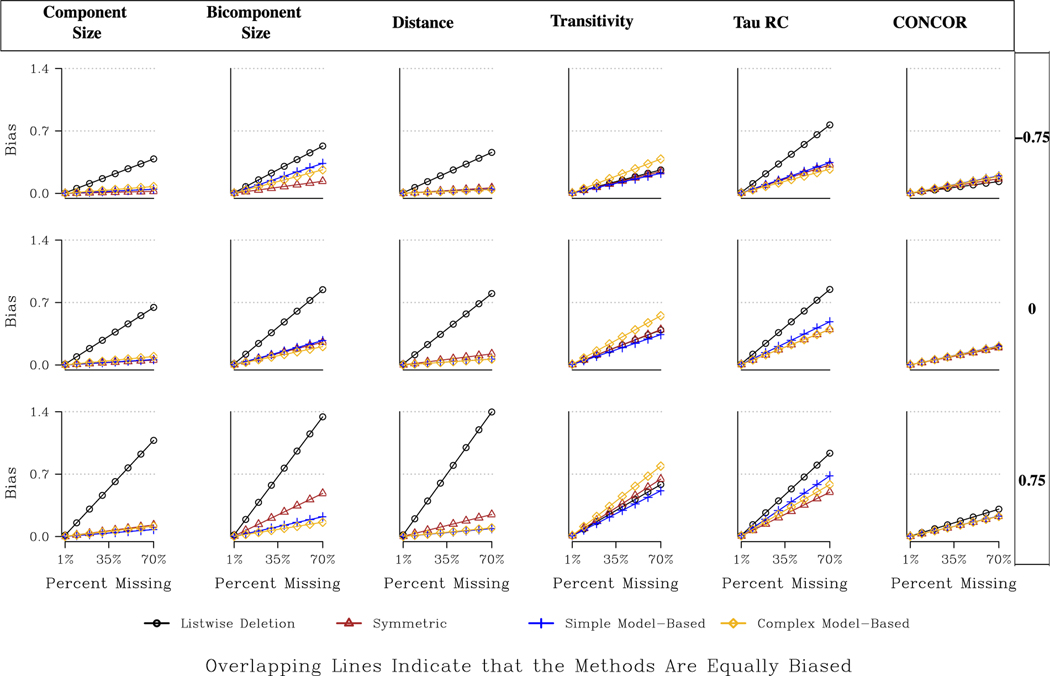
The centralization results are presented in Figs. 6 and 7, as well as Tables A5–A8. Table 3 offers a broad qualitative summary of the findings. We again start with a brief discussion of the undirected networks. Looking at degree centralization, all methods fare equally well in reducing bias and all outperform listwise deletion by a considerable margin (although some bias remains even after imputation). The returns to imputation are especially large when central nodes are more likely to be missing. For example, for an undirected, large, moderately centralized network with 40 % missing data, the predicted bias is .48 under listwise deletion and .13 after applying any of the imputation methods (assuming central nodes are more likely to be missing).
Fig. 6.
Predicted Bias for Centralization Measures for a Large, Undirected, Moderately Centralized Network.
Fig. 7.
Predicted Bias for Centralization Measures for a Large, Directed, Moderately Centralized Network.
Table 3.
Summary of Best Imputation Options for Centralization Measures.
| Measure | Directed or Undirected | Non-response Type | Size and Centralization | Best Imputation Option |
|---|---|---|---|---|
| Degree Std | Undirected | Any | Any | Any strategy except Listwise Deletion |
| Bon Power Std | Undirected | Any | Small | Model-Baseda |
| Bon Power Std | Undirected | Any | Medium/Large | Listwise Deletion |
| Closeness Std | Undirected | Any | Any | Model-Based |
| Betweenness Std | Undirected | Any | Any | Any strategy except Listwise Deletion |
| Indegree Std | Directed | Any | Any | Model-Based (complex) |
| Total Degree Std | Directed | Any | Any | Model-Based (complex) or Symmetric |
| Bon Power Std | Directed | Any | Any | Model-Based |
| Closeness Std | Directed | Central Nodes | Decentralized | Model-Based (complex) or Symmetric |
| Closeness Std | Directed | Less Central Nodes | Decentralized | Symmetric or Probabilistic |
| Closeness Std | Directed | Any | Centralized | Probabilistic or Listwise Deletion |
| Betweenness Std | Directed | Any | Decentralized | Model-Based or Symmetric |
| Betweenness Std | Directed | Any | Centralized | Model-Based or Probabilistic |
Notes.
The model-based approach is assumed to correspond to complex or simple unless explicitly noted.
The results are quite different for Bonacich Power centralization, as listwise deletion is actually a better option than any of the imputation methods for every network but the small Interlock network (see Table A5 and Fig. 6). In general, the bias for Bonacich Power centralization is quite low, even when high degree nodes are non-respondents and we use listwise deletion. Thus, any imputation method that adjusts the centrality of the respondents (while excluding the missing cases from the calculation) runs the risk of adding bias where little was present to begin with.
The closeness results show the clearest differentiation between imputation options, with the model-based approaches (simple or complex) being preferred over listwise deletion and the symmetric approach. This holds across all missing data types and networks. For example, for the Co-authorship network, the total decrease in bias is 85 % for the complex model (compared to listwise deletion) and 73 % under the symmetric option. The median bias for the Co-authorship network with 30 % missing data is less than .05 with the model-based approach (under random missing nodes); compare this to .88 bias under listwise deletion.
Overall, the undirected networks for the centralization measures yield a straightforward story. In every case besides Bonacich Power, it is better to impute than listwise deletion, and in most cases any of the imputation approaches will work, making simple imputation particularly attractive. Note that the returns to imputation are typically larger when more central nodes are non-respondents, as imputation tends to weaken the deleterious effects of missing central actors.
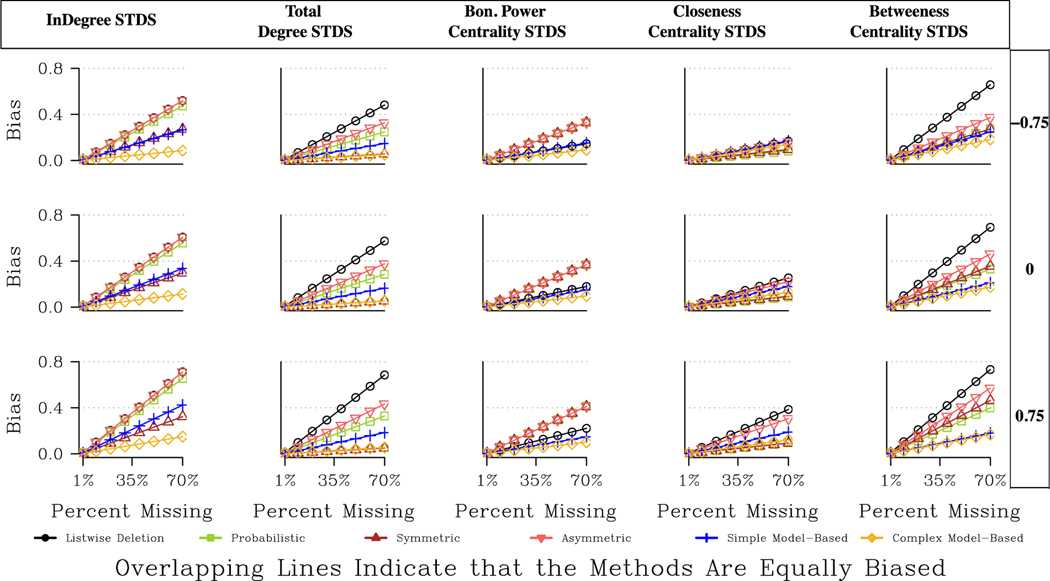
We now turn to the directed networks, presented in Fig. 7 and Table A7 (the bias ratio table). We start with indegree centralization. The results clearly point to the complex model-based approach offering the greatest reduction in bias, followed by the symmetric approach and the simple model-based approach. The asymmetric and probabilistic methods do not offer much (or any) improvement over listwise deletion. For example, looking at Fig. 7, the expected bias for a large, moderately centralized network (under random missing nodes) with 30 % missing data is about .05 for the complex-model imputation, .127 for symmetric imputation, and .26 for listwise deletion and asymmetric imputation. The results are similar for total degree centralization, although here the asymmetric and probabilistic approaches are somewhat better than listwise deletion. Additionally, the symmetric approach offers no worse estimates than the model-based approach, and is often the best option. For example, the decrease in total bias for the HS 24 network (missing low degree nodes) is 87 % for both the symmetric and the complex model-based approach. The decrease is 31 % for the asymmetric option and 48 % for the probabilistic option.
The Bonacich centralization results mirror the undirected results in many ways, with most imputation methods offering worse estimates than listwise deletion under conditions of missing data (note that the symmetric, asymmetric and probabilistic are all equivalent in this case). The exceptions are the model-based approaches, which consistently outperform listwise deletion for all directed networks, save for the RC elite network (where listwise deletion is preferred). Thus, while a symmetric imputation would be a good option for indegree or total degree centralization, this does not extend to the case of Bonacich power. Boncacich centrality depends on the degree of one’s neighbors, while the symmetric approach only imputes the degree of the missing nodes in a limited way, compared to the model-based approach. Symmetric imputation, thus, tends to overestimate the level of centralization in the network for Bonacich centrality (as the method potentially underestimates the degree of one’s neighbors).
Closeness and betweenness centralization offer more contingent, complicated stories. The best imputation approach for closeness centralization depends heavily on the features of the network and the kinds of nodes that are missing. When the network exhibits low to moderate centralization and central nodes are more likely to be non-respondents (bottom row in Fig. 7), the best option is either the model- based or the symmetric approach. On the other hand, when low centrality nodes are more likely to be non-respondents, the best options are the probabilistic or symmetric approaches. For example, consider the Prison network, which has low centralization. When low degree nodes are non-respondents, the median bias is .11, .12 and .15 under the probabilistic, symmetric and model-based approaches, assuming 40 % missing data. When high degree nodes are non-respondents, the analogous values are: .20 (probabilistic), .15 (symmetric), and .10 (model- based). In this case, the symmetric approach would be a robust choice, although not necessarily the best one in terms of lowering bias. Simple imputation works less well for the more centralized networks. Listwise deletion is often just as good (or even better) as the imputation methods, especially when the network is very centralized (such as the RC elite network) or the non-respondents are less central. See Table A7 for the full results.
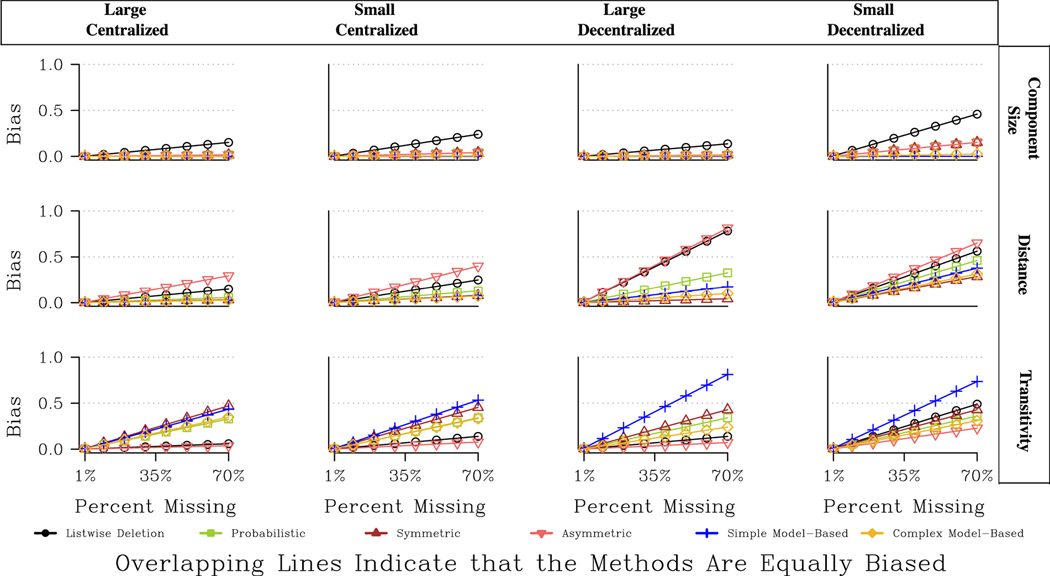
For betweenness, the results also depend heavily on the features of the network, and we can see this most clearly in Fig. 8. Fig. 8 presents the expected bias (based on the regression results presented in Table A8) for four different kinds of networks: large centralized, small centralized, large decentralized and small decentralized. The bottom row shows the results for betweenness. For the decentralized networks, the model- based and symmetric approaches are clearly preferred. For moderately centralized networks, the model-based approaches remain a good option. The model-based approaches fare quite poorly, however, for the RC elite network, the most centralized network. Here taking a model-based approach yields worse bias than listwise deletion, making the probabilistic and asymmetric approaches more appropriate. More striking, perhaps, is the poor performance of the symmetric option. In centralized networks, measures of betweenness centralization are sensitive to any imputation that changes the paths from the most central actors; thus, strategies that tend to add more ties (and thus paths between actors), like symmetric imputation, perform worse when the network is very centralized. Thus, the best option for decentralized networks is not an ideal choice for centralized ones.
In sum, the story is simple with undirected networks. All imputation methods are better than listwise deletion, with simple imputation methods being particularly attractive due to their ease of use. The case of directed networks is harder, as the ideal choice depends on the measure, the type of missing data and the type of network. In general, when there are contingent choices, the model-based approach performs comparatively well when the network is decentralized and/or the non-respondents have high degree.
Topology
We end the results section with a discussion of the topology measures, again starting with the undirected networks. The results are presented in Fig. 9 and Table A9. See Table 4 for an overall picture of the best imputation methods. Looking at Fig. 9, we can see that all imputation methods fare better than listwise deletion for component size, bicomponent size and distance, and that there are large returns to imputation. We also see that the ideal choice of imputation method depends on the type of missing data. When high centrality nodes are more likely to be non-respondents, the best choice is the model-based approach (simple or complex). For example, the expected bias for bicomponent size when the network is large, moderately centralized, and missing 30 % of the data (in Fig. 9) is .57 when listwise deletion is applied, .20 for the symmetric strategy, and .07 for the complex model- based strategy. The analogous values when low centrality nodes are non-respondents are: .23 (listwise deletion), .06 (symmetric), and .11 (model-based) suggesting that the symmetric approach is actually favored when missing less central nodes, although the model-based approach remains a good option.
Fig. 9.
Predicted Bias for Topology Measures for a Large, Undirected, Moderately Centralized Network.
Table 4.
Summary of Best Imputation Options for Topology Measures.
| Measure | Directed or Undirected | Non-response Type | Size and Centralization | Best Imputation Option |
|---|---|---|---|---|
| Component | Undirected | Central Nodes | Any | Model-baseda |
| Bicomponent | ||||
| Distance | ||||
| Component | Undirected | Less Central | Any | Symmetric |
| Bicomponent | ||||
| Distance | Undirected | Less Central | Any | Any strategy except Listwise Deletion |
| Transitivity | Undirected | Any | Decentralized | Model-based (simple) |
| Transitivity | Undirected | Any | Centralized | Listwise Deletion |
| Tau | Undirected | Any | Any | Model-based |
| CONCOR | Undirected | Central Nodes | Any | Any strategy except Listwise Deletion |
| CONCOR | Undirected | Less Central | Any | Symmetric |
| Component | Directed | Any | Any | Model-based |
| Bicomponent | ||||
| Distance | Directed | Central Nodes | Any | Model-based or Symmetric |
| Distance | Directed | Less Central | Any | Probabilistic or Symmetric |
| Transitivity | Directed | Any | Any | Asymmetric or Listwise Deletion |
| Tau | Directed | Any | Any | Probabilistic |
| CONCOR | Directed | Any | Any | Any strategy, including Listwise Deletion |
Notes.
The model-based approach is assumed to correspond to complex or simple unless explicitly noted.
The transitivity results show a different kind of pattern, where the best choice depends on the network being analyzed. When the network is decentralized, the simple model-based approach is the best option, followed by symmetric imputation (in most cases).18 The results are very different for the centralized networks, however. Here, all of the imputation methods perform poorly and listwise deletion is the most viable option. For example, looking at Table A9, we see negative values for the bias reduction in the HIV network and the Co-citation network (two highly centralized networks), meaning the imputation methods perform worse than listwise deletion. The imputation methods perform poorly in the centralized networks because transitivity tends to be unevenly distributed across the network (i.e., we may have lower levels with the most centralized actors), making imputation difficult for methods that do explicitly account for such variation.
Finally, looking at the CONCOR results, we see that the bias is quite low overall and the returns to imputation are small. For example, for the Co-authorship network, the median bias at 50 % missing is only .25 for listwise deletion and .20 for any of the imputation methods.
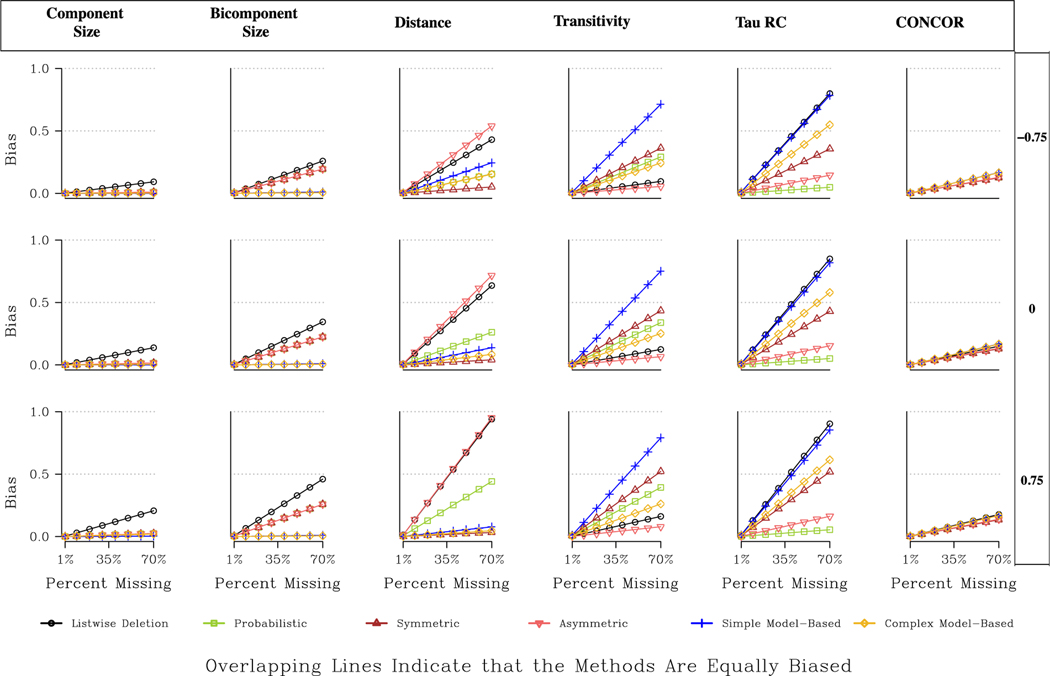
The results for the directed networks are presented in Fig. 10 and Table A11. Component and bicomponent size are similar to what we saw in the undirected case, but here the model-based approaches (simple or complex) are more uniformly the best option, followed by the simple imputation approaches.19 For example, for bicomponent size, the drop in total bias (compared with listwise deletion) under the model-based approach is 95 % for the Prosper network under random missing data; compare this to only 39 % using simple imputation. Or, looking at Fig. 10, the expected bias is almost 0 under the model-based approaches, outperforming the simpler options in all cases (but especially so when high degree nodes are more likely to be non-respondents).
Fig. 10.
Predicted Bias for Topology Measures for a Large, Directed, Moderately Centralized Network.
The distance results are much more sensitive to the type of missing data. For example, for the Prosper network under missing high degree nodes, the decrease in total bias is 81 % for the model-based approach, 73 % for the symmetric approach and 33 % for the probabilistic approach.20 When low centrality nodes are non-respondents, the model- based approach actually yields 7 % more bias than listwise deletion while the symmetric approach has a slight improvement over listwise deletion, with a 15 % decrease in total bias. The probabilistic approach offers the best option in this case, with a decrease of 35 %. The symmetric and probabilistic strategies are both relatively information light methods (assuming either full reciprocation or reciprocation based on the observed reciprocity rate), and thus tend to perform comparatively better when there is little information about the non-respondents, as is true when trying to impute ties for peripheral nodes. Note that the asymmetric option performs quite poorly in these cases, offering worse or similar estimates as listwise deletion.
The transitivity results are straightforward in the directed case. The only consistently viable option that performs better than listwise deletion is the asymmetric imputation approach. Thus, the worse option for distance is the best option for transitivity. For example, looking at Fig. 10, the expected bias for our directed, moderately centralized network is .03 under the asymmetric approach, .053 under listwise deletion, .11 under the model-based approach, .145 under the probabilistic approach, and .18 under the symmetric approach (assuming 30 % missing random data). These results indicate that imputation that attempts to go beyond the raw data alters the underlying transitivity estimate in a way that is worse than listwise deletion, which has low bias in itself. See Fig. 10 and Table A11 for tau statistic and CONCOR results.
Fig. 11 offers a different kind of comparison, presenting the predicted bias for four example networks with different combinations of size and centralization. The figure is limited to three measures, component size, distance and transitivity. Overall, looking at Fig. 11, the best imputation method does not strongly depend on the features of the network. The best choice for large decentralized networks tends to be the best choice for small centralized networks (making the choice easier from the point of view of the researcher). There are, however, differential returns to different methods, depending on the features of the network and the measure of interest. For example, for distance, the returns to symmetric imputation (in terms of how much is gained relative to listwise deletion) is highest in decentralized networks—especially large decentralized networks where the bias is high if listwise deletion is applied. For transitivity, we see that the asymmetric approach is consistently the best, but that the model-based and probabilistic approaches fare relatively better in the decentralized networks.
Overall, the topology results suggest that the best imputation choice depends on the type of missing data, the type of network and the measure of interest. For example, we see measures that capture large structural features, like component size or distance, are best imputed based on the model-based or symmetric options. Other measures that are more local, like transitivity, are harder to impute and often listwise deletion is the best option. The type of missing data also matters greatly here, as the model-based approaches tend to perform better when more central nodes are missing, with this being true in both the directed and undirected cases.
Conclusion
Missing data is a difficult problem faced by network researchers. Traditional measures assume a full census of a bounded population (Laumann et al., 1983; Wasserman and Faust, 1994). In practice, a full census is often difficult to come by, as nodes and/or edges may be missing, offering an incomplete picture of the full network structure. It is thus important to understand how much bias results from missing data and how successful different imputation methods are under different conditions. This can be difficult to gauge, however, as there are few general, practical guidelines on how to address missing network data. This paper takes up this problem directly, showing how different imputation methods fare across a range of circumstances, including different networks, missing data types and measures of interest. We also consider a number of different imputation methods, ranging from simple network imputation to more complicated model-based approaches. The hope is that our results will make it easier for a researcher to choose an imputation method, given the particular features of their study.
Overall, we find that doing listwise deletion is almost always the worst option. Which imputation method performs best, however, is quite contingent, depending on the type of missing data, the type of network and the measure of interest. In this way, it is an easy choice to impute, but a harder choice to decide which method to employ. For example, for degree-based measures of centrality (on directed networks), we see that very simple approaches, including the asymmetric option, fare well. On the other hand, path-based measures (like closeness and betweenness) tend to require more complicated options, either probabilistic imputation or a model-based approach. In a similar way, we find that model-based approaches are particularly effective for structural measures, like bicomponent size or distance, but fare less well when estimating more local measures, like transitivity. The results also suggest that the type of network and missing data affect the performance of the imputation methods (Žnidaršič et al., 2018; Krause et al., 2020). For example, the model-based approaches are comparatively more effective when the network is decentralized and non-respondents have high degree.
How to choose an imputation strategy
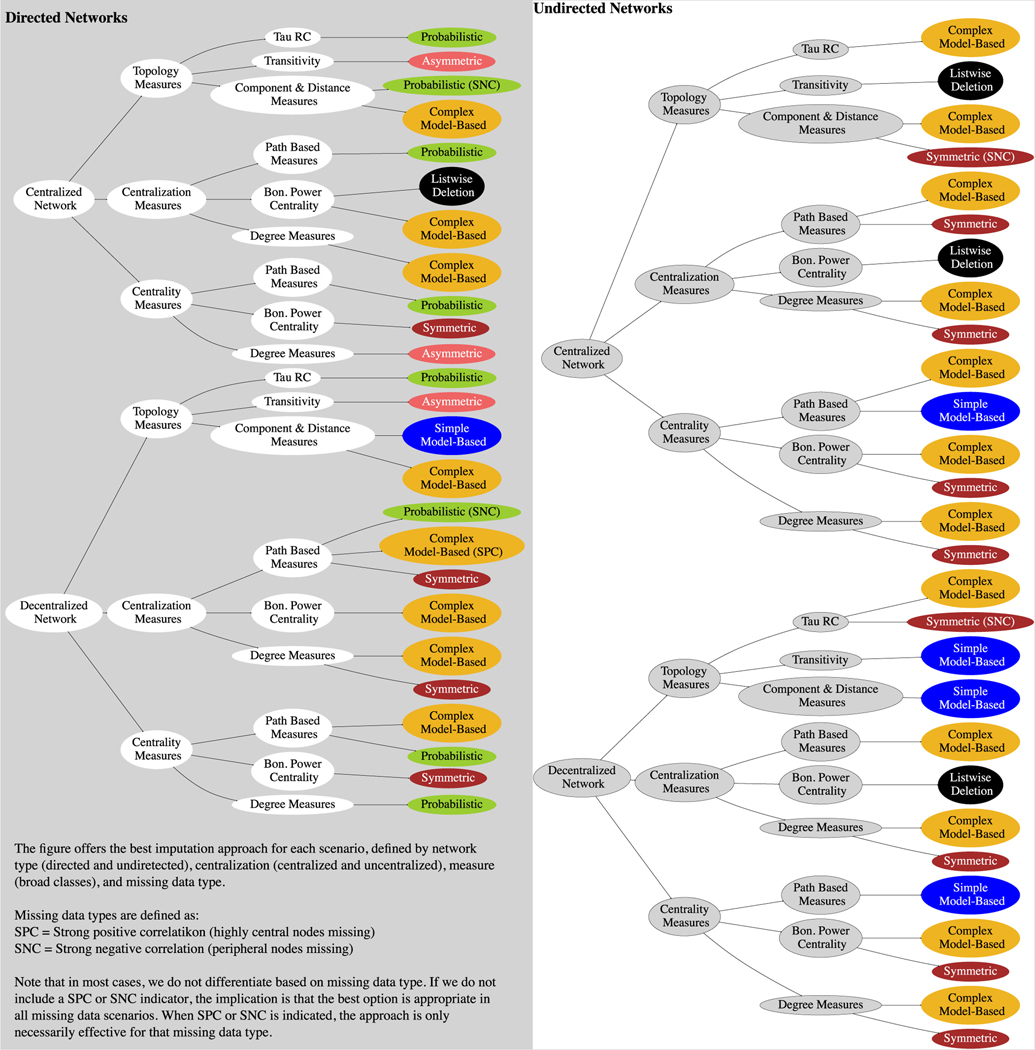
In short, different imputation methods are appropriate in different research settings, depending on the particular combination of missing data type, network and measure. Thus, a researcher must navigate a set of complex dependencies when making imputation decisions, especially since the size and type of network (e.g., large bipartite graphs) can make model-based imputation approaches intractable. To make this task easier, we have summarized our results in a simplified format in Fig. 12.
Fig. 12.
Summary Figure of Best Imputation Approach by Network Type, Measure, and Missing Data Condition.
The goal of Fig. 12 is to provide a user-friendly guide to imputation decisions. The figure is organized around a number of key factors, such as network type (directed/undirected; centralized/decentralized) and measure of interest.21 The figure is presented as a kind of branching structure, or decision tree, with the final branch showing the optimal choice of imputation method, given the features along the path. In selecting the optimal imputation strategy, we have tried to balance the best performing method with the difficulty of implementation. A researcher would simply follow the relevant path for their research setting and choose the best imputation method (noting that other imputation strategies may also offer reasonable results). For example, a researcher with a directed, decentralized network measuring betweenness or closeness centralization would do well using a symmetric imputation approach. The same researcher with a centralized network would do better using a probabilistic option. Such contingent decisions are hard to make without a guide, showing the clear utility of Fig. 12, as well as the more detailed summary tables presented earlier in the text.
It is important to note that Fig. 12 focuses on the optimal imputation method, but there are other factors that should be considered when making an imputation choice. First, a researcher must balance the performance of a method with the difficulty of implementing it. For example, a model-based approach can be difficult and expensive (time- wise) to implement. A researcher must already have a working knowledge of statistical network models, decide on the specific model to estimate, estimate the model, and so on. This is quite burdensome compared to the simple imputation options, which in many cases offer similar results. Thus, it is conceivable that a researcher would opt for simple imputation even when the model-based approach offers strictly lower bias. Of course, even with simple imputation options, a researcher must do a considerable amount of careful consideration, picking the right option for their particular scenario. In a similar way, a researcher may opt for a ‘safe’ choice that performs well across many settings, even if it is not necessarily the best (predicted) option in their particular case. Our results suggest that the probabilistic approach to network imputation is robust to different measures and networks, making it a good option overall. Finally, we note that a model-based approach may fare better under different specifications (e.g., Bayesian), as a better-specified model (i.e., one that captures the true tie formation processes) will offer improved estimates.
Limitations and considerations
Our analysis rests on a number of assumptions that must be considered when interpreting the results. For example, we assumed that the non-respondents are identifiable, so that respondents could still nominate them. This implicitly assumes a well-bounded, clearly defined population. Such conditions will not hold in all research settings, however, with important implications for the viability of different imputation strategies. Most clearly, the simple imputation approaches cannot be applied when missing nodes are unable to be identified (as there is no information to reconstruct the network). Model-based approaches could, in theory, still be used, as long as the size of the population was known, but their effectiveness will likely be reduced. There is no edge information on the missing nodes, making it difficult to predict how the missing nodes fit into the larger network. Listwise deletion could, of course, still be applied without complication.
Our analysis also assumes that the observed network is recorded without error. This assumption is unlikely to strictly hold in practice. Actors may forget to nominate people they should have (Brewer, 2000; Bell et al., 2007), while potentially nominating those they should not; for example, nominating an aspirational friend who is, in fact, not actually a friend (Almaatouq et al., 2016). Finally, there could be conflicting reports about the nature of the relationship (An and Schramski, 2015). Any measurement error in the observed data will end up being factored into the imputation process, and thus may lead to somewhat poorer results than reported in our own analysis. This may be particularly deleterious to the simple imputation approaches. If a researcher imputes a tie from j→i because i nominates j, then it is problematic if i→j does not really exist and is simply measurement error; as one imputes based on a false premise. Model-based approaches may fare better, as the model imputes probabilistically, based on general tendencies observed throughout the whole network, thus minimizing the effect of particular mistakes in the data.22 Ultimately, these are open questions deserving of more concerted work in the future.
Final thoughts
This set of papers began with a simple goal — to describe the consequences of missing data for typically used network measures (Smith and Moody, 2013). Overall, we have shown that the effect of missing data is highly contingent, depending on the circumstances of the study, as well as the actions of the researcher. Here, we emphasize the role of the researcher in reducing bias. Our results suggest that a researcher choosing an effective imputation method for their setting can greatly reduce the bias due to missing data, even in cases with difficult conditions (i.e., non-respondents tend to be central to the network). A study with over 50 % non-respondents can, potentially, still yield valid estimates. In the end, the hope is that our results can be used as a practical guide for researchers choosing an imputation strategy, and, more generally, dealing with the difficult problem of missing network data.
Acknowledgements
This work was supported by the National Institute of General Medical Sciences of the National Institutes of Health (Grant No. P20 GM130461) and the Rural Drug Addiction Research Center at the University of Nebraska-Lincoln.
We would like to thank Jake Fisher and Robin Gauthier for helpful comments on earlier versions of this article. We would like to thank the Prosper Peers project, Mark Mizruchi, Walter Powell, Lisa Keister, and Scott Gest for sharing network data files. The Prosper project is funded by NSF/HSD: 0624158, W. T. Grant Foundation 8316 & NIDA 1R01DA018225-01. The Colorado Springs HIV network was made available through: NIH R01 DA 12831 (PI Morris) Modeling HIV and STD in Drug User and Social Networks. This research uses data from Add Health, a program project designed by J. Richard Udry, Peter S. Bearman, and Kathleen Mullan Harris, and funded by a grant P01-HD31921 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development, with cooperative funding from 17 other agencies. Special acknowledgment is due Ronald R. Rindfuss and Barbara Entwisle for assistance in the original design. Persons interested in obtaining data files from Add Health should contact Add Health, Carolina Population Center, 123 W. Franklin Street, Chapel Hill, NC 27516-2524 (addhealth@unc.edu). No direct support was received from grant P01-HD31921 for this analysis.
Appendix A
Table A1.
Percent Decrease in Total Bias under Different Imputation Strategies: Centrality Measures for Undirected Networks.
| Measure | Imputation | Interlock |
Coauthor |
Co-citation |
Biotech |
HIV |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Correlation with Centrality |
Correlation with Centrality |
Correlation with Centrality |
Correlation with Centrality |
Correlation with Centrality |
||||||||||||
| −.75 | 0 | .75 | −.75 | 0 | .75 | −.75 | 0 | .75 | −.75 | 0 | .75 | −.75 | 0 | .75 | ||
| Degree | Symmetric | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Model-based Simple | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | |
| Model-based Complex | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | |
| Bon Power | Symmetric | 96 | 93 | 91 | 96 | 95 | 89 | 95 | 94 | 93 | 95 | 92 | 92 | 90 | 88 | 86 |
| Model-based Simple | 95 | 95 | 94 | 98 | 97 | 91 | 97 | 96 | 96 | 96 | 93 | 93 | 91 | 90 | 89 | |
| Model-based Complex | 93 | 94 | 94 | 98 | 96 | 91 | 98 | 97 | 97 | 96 | 94 | 93 | 93 | 92 | 92 | |
| Closeness | Symmetric | 91 | 85 | 80 | 89 | 82 | 75 | 92 | 90 | 86 | 85 | 83 | 78 | 88 | 86 | 82 |
| Model-based Simple | 93 | 91 | 87 | 90 | 86 | 80 | 94 | 94 | 92 | 91 | 89 | 84 | 91 | 90 | 85 | |
| Model-based Complex | 93 | 91 | 86 | 91 | 85 | 77 | 93 | 93 | 91 | 90 | 88 | 83 | 91 | 89 | 84 | |
| Betweenness | Symmetric | 86 | 79 | 67 | 86 | 78 | 56 | 83 | 74 | 49 | 90 | 88 | 79 | 87 | 82 | 62 |
| Model-based Simple | 87 | 86 | 77 | 74 | 81 | 68 | 79 | 74 | 60 | 88 | 88 | 84 | 65 | 75 | 70 | |
| Model-based Complex | 86 | 84 | 76 | 83 | 82 | 64 | 78 | 73 | 62 | 88 | 88 | 81 | 79 | 82 | 73 | |
Table A2.
Centrality Bias Slope Regressions: Undirected Networks.
| Variables | Model 1 Degree | Model 2 Bon. Power | Model 3 Closeness | Model 4 Betweeness |
|---|---|---|---|---|
| Intercept | −1.925*** | −2.572 | −2.834** | −1.802 |
| (0.46) | (1.7) | (0.94) | (1.33) | |
| Correlation with Centrality | 0.92*** | 1.199*** | 0.626*** | 0.517*** |
| (0.15) | (0.11) | (0.05) | (0.07) | |
| In-degree Std. Dev. | −0.166*** | −0.174 | −0.075 | −0.041 |
| (0.03) | (0.13) | (0.07) | (0.1) | |
| Log of Size | −0.166 | 0.016 | 0.035 | −0.206 |
| (0.09) | (0.35) | (0.19) | (0.27) | |
| Symmetric Imputation | −40.255*** | −3.407** | −2.313** | −0.397 |
| (0.6) | (1.06) | (0.89) | (0.63) | |
| Simple Model-Based Imputation | −39.492*** | −3.772*** | −2.771* | −1.214 |
| (1.22) | (1.1) | (1.21) | (0.87) | |
| Complex Model-Based Imputation | −39.492*** | −3.31** | −2.712* | −0.703 |
| (1.59) | (1.09) | (1.1) | (0.58) | |
| Correlation with Centrality* Symmetric Imputation | −0.871*** | 0.292 | 0.261*** | 0.68*** |
| (0.21) | (0.16) | (0.08) | (0.1) | |
| Correlation with Centrality* Simple Model-Based Imputation | −0.718*** | 0.225 | 0.267*** | 0.234* |
| (0.21) | (0.16) | (0.08) | (0.1) | |
| Correlation with Centrality* Complex Model-Based Imputation | −0.718*** | 0.232 | 0.291*** | 0.34*** |
| (0.21) | (0.16) | (0.08) | (0.1) | |
| In-degree Std. Dev.* Symmetric Imputation | 0.146** | −0.03 | −0.064 | 0.085 |
| (0.04) | (0.08) | (0.07) | (0.05) | |
| In-degree Std. Dev.* Simple Model-Based Imputation | 0.198* | −0.071 | −0.054 | 0.116 |
| (0.09) | (0.08) | (0.09) | (0.07) | |
| In-degree Std. Dev.* Complex Model-Based Imputation | 0.198 | −0.111 | −0.045 | 0.113* |
| (0.12) | (0.08) | (0.08) | (0.04) | |
| Log of Size* Symmetric Imputation | 0.211 | 0.224 | 0.337 | −0.257* |
| (0.12) | (0.22) | (0.18) | (0.13) | |
| Log of Size *Simple Model-Based Imputation | 0.029 | 0.294 | 0.345 | −0.15 |
| (0.25) | (0.23) | (0.25) | (0.18) | |
| Log of Size* Complex Model-Based Imputation | 0.029 | 0.241 | 0.333 | −0.242* |
| (0.33) | (0.22) | (0.23) | (0.12) | |
| N | 180 | 180 | 180 | 180 |
| Networks | 5 | 5 | 5 | 5 |
Note: The regression uses the betas slopes from each line as the dependent variable. The betas represent the expected drop in correlation (between the empirical and the observed) for a 10 % increase in the amount of missing data. Larger numbers mean larger bias with more missing data. The correlation with centrality takes four values: − .75, − .25, .25, and .75.
Table A3.
Percent Decrease in Total Bias under Different Imputation Strategies: Centrality Measures for Directed Networks.
| Measure | Imputation | Prison |
Sorority |
6th Grade |
Prosper |
RC Elite |
HS 13 |
HS 24 |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Correlation with Centrality |
Correlation with Centrality |
Correlation with Centrality |
Correlation with Centrality |
Correlation with Centrality |
Correlation with Centrality |
Correlation with Centrality |
||||||||||||||||
| −.75 | 0 | .75 | −.75 | 0 | .75 | −.75 | 0 | .75 | −.75 | 0 | .75 | −.75 | 0 | .75 | −.75 | 0 | .75 | −.75 | 0 | .75 | ||
| Indegree | Probabilistic | 4 | 5 | 6 | 6 | 14 | 20 | −6 | −2 | 2 | 2 | 3 | 3 | −1 | −1 | −1 | 6 | 7 | 9 | 10 | 12 | 15 |
| Symmetric | −31 | −25 | −27 | −24 | −7 | 5 | −103 | −92 | −71 | −54 | −51 | −50 | −226 | −238 | −277 | −42 | −41 | −41 | −29 | −25 | −21 | |
| Asymmetric | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Model-based Simple | −52 | −44 | −42 | −44 | −29 | −15 | −68 | −59 | −46 | −78 | −63 | −52 | −218 | −254 | −262 | −71 | −64 | −57 | −59 | −51 | −40 | |
| Model-based Complex | −43 | −37 | −34 | −35 | −20 | −9 | −63 | −52 | −37 | −65 | −51 | −41 | −168 | −190 | −210 | −50 | −44 | −34 | −42 | −33 | −22 | |
| Total Degree | Probabilistic | 60 | 63 | 68 | 69 | 74 | 77 | 52 | 55 | 61 | 61 | 63 | 67 | 0 | 6 | 47 | 57 | 60 | 67 | 63 | 66 | 71 |
| Symmetric | 51 | 56 | 63 | 64 | 71 | 76 | 34 | 40 | 52 | 49 | 53 | 60 | −120 | −108 | −15 | 44 | 48 | 57 | 53 | 58 | 66 | |
| Asymmetric | 60 | 62 | 67 | 67 | 71 | 74 | 57 | 58 | 63 | 62 | 63 | 67 | 1 | 7 | 48 | 59 | 62 | 67 | 64 | 66 | 71 | |
| Model-based Simple | 40 | 47 | 53 | 52 | 60 | 67 | 39 | 43 | 51 | 37 | 43 | 51 | −79 | −79 | −10 | 35 | 41 | 50 | 43 | 48 | 57 | |
| Model-based Complex | 43 | 48 | 55 | 55 | 62 | 68 | 43 | 47 | 55 | 41 | 47 | 55 | −45 | −44 | 7 | 42 | 47 | 57 | 48 | 53 | 62 | |
| Bon Power | Probabilistic | 55 | 57 | 62 | 56 | 60 | 64 | 57 | 58 | 60 | 57 | 57 | 61 | 66 | 73 | 87 | 63 | 66 | 73 | 64 | 67 | 70 |
| Symmetric | 55 | 57 | 62 | 56 | 60 | 64 | 57 | 58 | 60 | 57 | 57 | 61 | 66 | 73 | 87 | 63 | 66 | 73 | 64 | 67 | 70 | |
| Asymmetric | 55 | 57 | 62 | 56 | 60 | 64 | 57 | 58 | 60 | 57 | 57 | 61 | 66 | 73 | 87 | 63 | 66 | 73 | 64 | 67 | 70 | |
| Model-based Simple | 40 | 49 | 57 | 38 | 48 | 57 | 42 | 49 | 57 | 29 | 37 | 49 | 53 | 60 | 80 | 31 | 41 | 53 | 37 | 45 | 56 | |
| Model-based Complex | 39 | 49 | 57 | 36 | 47 | 56 | 47 | 53 | 61 | 32 | 40 | 52 | 57 | 63 | 80 | 43 | 51 | 63 | 44 | 52 | 62 | |
| Closeness | Probabilistic | 49 | 56 | 62 | 55 | 59 | 64 | 79 | 84 | 88 | 51 | 56 | 61 | 48 | 49 | 51 | 62 | 66 | 70 | 67 | 72 | 75 |
| Symmetric | 36 | 51 | 62 | 42 | 48 | 54 | 83 | 90 | 93 | 58 | 64 | 67 | −77 | −58 | −46 | 78 | 83 | 86 | 78 | 83 | 87 | |
| Asymmetric | 33 | 39 | 46 | 40 | 47 | 54 | 32 | 32 | 34 | 21 | 28 | 35 | 49 | 49 | 53 | 8 | 9 | 12 | 7 | 9 | 13 | |
| Model-based Simple | 49 | 65 | 73 | 33 | 52 | 61 | 87 | 94 | 95 | 66 | 76 | 80 | 0 | 15 | 18 | 79 | 85 | 88 | 84 | 89 | 91 | |
| Model-based Complex | 49 | 63 | 71 | 37 | 52 | 59 | 88 | 94 | 96 | 67 | 76 | 79 | 10 | 24 | 23 | 81 | 86 | 89 | 85 | 89 | 91 | |
| Betweenness | Probabilistic | 30 | 32 | 32 | 22 | 21 | 22 | 29 | 35 | 34 | 28 | 29 | 28 | 23 | 21 | 13 | 37 | 37 | 34 | 46 | 46 | 45 |
| Symmetric | 3 | 13 | 20 | 4 | 4 | 6 | −18 | −3 | 4 | 1 | 14 | 16 | −151 | −136 | −95 | 10 | 12 | 11 | 25 | 28 | 29 | |
| Asymmetric | 24 | 24 | 24 | 13 | 13 | 13 | 19 | 15 | 13 | 11 | 10 | 10 | 51 | 46 | 38 | 9 | 8 | 7 | 10 | 9 | 8 | |
| Model-based Simple | −22 | −3 | 9 | −8 | −2 | 3 | −4 | 6 | 8 | −10 | 6 | 11 | −145 | −144 | −110 | −6 | 0 | 1 | 10 | 15 | 19 | |
| Model-based Complex | −15 | 1 | 11 | −1 | 4 | 6 | 7 | 16 | 15 | −1 | 12 | 14 | −120 | −109 | −80 | 7 | 12 | 12 | 19 | 23 | 27 | |
Table A4.
Centrality Bias Slope Regressions: Directed Networks.
| Variables | Model 1 Indegree | Model 2 Total Degree | Model 3 Bon. Power | Model 4 Closeness | Model 5 Betweeness |
|---|---|---|---|---|---|
| Intercept | −2.598*** | −2.476*** | −2.198*** | −2.973* | −2.652*** |
| (0.7) | (0.6) | (0.23) | (1.37) | (0.19) | |
| Correlation with Centrality | 0.309*** | 0.31*** | 0.411*** | 0.197*** | 0.202*** |
| (0.04) | (0.03) | (0.03) | (0.05) | (0.02) | |
| In-degree Std. Dev. | −0.356*** | −0.273*** | −0.138*** | −0.058 | −0.114*** |
| (0.06) | (0.06) | (0.02) | (0.13) | (0.02) | |
| Log of Size | 0.107 | 0.073 | −0.037 | 0.094 | 0.103** |
| (0.15) | (0.13) | (0.05) | (0.29) | (0.04) | |
| Asymmetric Imputation | 0.019 | −0.845 | −0.286 | −2.184*** | −0.822 |
| (0.2) | (0.57) | (0.35) | (0.57) | (0.44) | |
| Simple Model-Based Imputation | 0.448 | −0.506 | −0.868* | 0.723 | 0.466 |
| (0.57) | (0.92) | (0.44) | (1.97) | (0.42) | |
| Complex Model-Based Imputation | 0.544 | −0.483 | −0.491 | 0.938 | 0.556 |
| (0.5) | (0.81) | (0.34) | (2.15) | (0.45) | |
| Probabilistic Imputation | 0.145 | −0.915 | −0.286 | −0.909 | 0.046 |
| (0.24) | (0.51) | (0.34) | (2.26) | (0.35) | |
| Symmetric Imputation | 0.544 | −0.811 | −0.286 | 0.358 | 0.262 |
| (0.42) | (0.97) | (0.32) | (1.5) | (0.32) | |
| Correlation with Centrality* Asymmetric Imputation | 0.01 | −0.196*** | −0.193*** | −0.158* | 0.035 |
| (0.06) | (0.04) | (0.04) | (0.07) | (0.03) | |
| Correlation with Centrality* Simple Model-Based Imputation | −0.075 | −0.224*** | −0.286*** | −0.475*** | −0.098** |
| (0.06) | (0.04) | (0.04) | (0.07) | (0.03) | |
| Correlation with Centrality* Complex Model-Based Imputation | −0.077 | −0.215*** | −0.292*** | −0.431*** | −0.077* |
| (0.06) | (0.04) | (0.04) | (0.07) | (0.03) | |
| Correlation with Centrality* Probabilistic Imputation | −0.033 | −0.219*** | −0.193*** | −0.192** | 0.009 |
| (0.06) | (0.04) | (0.04) | (0.07) | (0.03) | |
| Correlation with Centrality* Symmetric Imputation | −0.064 | −0.284*** | −0.193*** | −0.32*** | −0.074* |
| (0.06) | (0.04) | (0.04) | (0.07) | (0.03) | |
| In-degree Std. Dev.* Asymmetric Imputation | 0.003 | 0.207*** | −0.046 | −0.055 | −0.102* |
| (0.02) | (0.05) | (0.03) | (0.05) | (0.04) | |
| In-degree Std. Dev.*Simple Model-Based Imputation | 0.168** | 0.229** | −0.069 | 0.108 | 0.088* |
| (0.05) | (0.09) | (0.04) | (0.18) | (0.04) | |
| In-degree Std. Dev.*Complex Model-Based Imputation | 0.154*** | 0.205** | −0.064* | 0.109 | 0.105* |
| (0.05) | (0.07) | (0.03) | (0.2) | (0.04) | |
| In-degree Std. Dev.* Probabilistic Imputation | 0.029 | 0.21*** | −0.046 | −0.041 | −0.017 |
| (0.02) | (0.05) | (0.03) | (0.21) | (0.03) | |
| In-degree Std. Dev.* Symmetric Imputation | 0.218*** | 0.318*** | −0.046 | 0.059 | 0.059* |
| (0.04) | (0.09) | (0.03) | (0.14) | (0.03) | |
| Log of Size* Asymmetric Imputation | −0.005 | −0.163 | −0.087 | 0.358** | 0.187* |
| (0.04) | (0.12) | (0.08) | (0.12) | (0.09) | |
| Log of Size *Simple Model-Based Imputation | −0.111 | −0.169 | 0.111 | −0.553 | −0.2* |
| (0.12) | (0.2) | (0.09) | (0.42) | (0.09) | |
| Log of Size* Complex Model-Based Imputation | −0.138 | −0.171 | 0.019 | −0.584 | −0.227* |
| (0.11) | (0.17) | (0.07) | (0.46) | (0.1) | |
| Log of Size * Probabilistic Imputation | −0.055 | −0.145 | −0.087 | 0.002 | −0.085 |
| (0.05) | (0.11) | (0.07) | (0.48) | (0.07) | |
| Log of Size* Symmetric Imputation | −0.175 | −0.201 | −0.087 | −0.359 | −0.163* |
| (0.09) | (0.21) | (0.07) | (0.32) | (0.07) | |
| N | 378 | 378 | 378 | 378 | 378 |
| Networks | 7 | 7 | 7 | 7 | 7 |
Note: The regression uses the betas slopes from each line as the dependent variable. The betas represent the expected drop in correlation (between the empirical and the observed) for a 10 % increase in the amount of missing data. Larger numbers mean larger bias with more missing data. The correlation with centrality takes four values: −.75, −.25, .25, and .75.
Table A5.
Percent Decrease in Total Bias under Different Imputation Strategies: Centralization Measures for Undirected Networks.
| Measure | Imputation | Interlock |
Coauthor |
Cocitation |
Biotech |
HIV |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −.75 | 0 | .75 | −.75 | 0 | .75 | −.75 | 0 | .75 | −.75 | 0 | .75 | −.75 | 0 | .75 | ||
| Degree | Symmetric | 64 | 76 | 64 | 62 | 83 | 67 | 52 | 84 | 64 | 54 | 79 | 56 | 58 | 85 | 62 |
| Model-based Simple | 63 | 76 | 64 | 63 | 83 | 68 | 52 | 84 | 64 | 54 | 79 | 55 | 58 | 84 | 62 | |
| Model-based Complex | 63 | 76 | 64 | 63 | 83 | 68 | 52 | 84 | 64 | 54 | 79 | 55 | 58 | 84 | 62 | |
| Bon Power | Symmetric | −9 | 15 | 43 | −313 | −251 | −89 | −374 | −583 | −158 | −588 | −249 | −82 | −815 | −411 | −130 |
| Model-based Simple | 28 | 54 | 63 | −229 | −107 | −61 | −391 | −492 | −70 | −701 | −300 | −95 | −922 | −509 | −176 | |
| Model-based Complex | 25 | 54 | 61 | −175 | −76 | −131 | −308 | −226 | −100 | −689 | −292 | −134 | −855 | −413 | −127 | |
| Closeness | Symmetric | 78 | 66 | 61 | 82 | 73 | 65 | 79 | 80 | 77 | 80 | 77 | 76 | 86 | 83 | 84 |
| Model-based Simple | 86 | 82 | 77 | 89 | 86 | 78 | 87 | 87 | 78 | 93 | 93 | 91 | 96 | 95 | 91 | |
| Model-based Complex | 85 | 81 | 77 | 90 | 85 | 76 | 88 | 88 | 78 | 93 | 93 | 90 | 95 | 94 | 92 | |
| Betweenness | Symmetric | 81 | 74 | 62 | 80 | 83 | 76 | 76 | 64 | 44 | 78 | 78 | 67 | 83 | 89 | 86 |
| Model-based Simple | 78 | 79 | 76 | 69 | 58 | 50 | 88 | 85 | 77 | 66 | 72 | 69 | 88 | 81 | 69 | |
| Model-based Complex | 77 | 78 | 75 | 79 | 66 | 54 | 87 | 78 | 66 | 67 | 73 | 67 | 90 | 83 | 68 | |
Table A6.
Centralization Bias Slope Regressions: Undirected Networks.
| Variables | Model 1 Degree | Model 2 Bon. Power | Model 3 Closeness | Model 4 Betweeness |
|---|---|---|---|---|
| Intercept | −2.723*** | −2.284 | −2.387 | −1.912*** |
| (0.35) | (1.35) | (1.22) | (0.21) | |
| Correlation with Centrality | 0.39** | 0.423*** | −1.254*** | 0.097 |
| (0.13) | (0.1) | (0.06) | (0.07) | |
| In-degree Std. Dev. | 0.016 | −0.147 | 0.042 | −0.011 |
| (0.03) | (0.1) | (0.09) | (0.02) | |
| Log of Size | 0.038 | −0.265 | −0.123 | 0.003 |
| (0.07) | (0.27) | (0.25) | (0.04) | |
| Symmetric Imputation | −1.378* | −0.881 | −0.749 | 0.107 |
| (0.61) | (1.37) | (0.99) | (0.82) | |
| Simple Model-Based Imputation | −1.324* | −3.054** | 0.096 | −0.942 |
| (0.65) | (0.94) | (1.43) | (1.46) | |
| Complex Model-Based Imputation | −1.324 | −3.16** | −0.211 | −0.878 |
| (1.1) | (1.02) | (1.63) | (1.09) | |
| Correlation with Centrality* Symmetric Imputation | −0.036 | −0.784*** | 1.18*** | 0.271** |
| (0.18) | (0.15) | (0.09) | (0.1) | |
| Correlation with Centrality* Simple Model-Based Imputation | −0.044 | −0.909*** | 1.265*** | 0.111 |
| (0.18) | (0.15) | (0.09) | (0.1) | |
| Correlation with Centrality* Complex Model-Based Imputation | −0.044 | −0.788*** | 1.291*** | 0.311** |
| (0.18) | (0.15) | (0.09) | (0.1) | |
| In-degree Std. Dev.* Symmetric Imputation | −0.01 | 0.184 | −0.132 | 0.098 |
| (0.05) | (0.1) | (0.07) | (0.06) | |
| In-degree Std. Dev.* Simple Model-Based Imputation | −0.011 | 0.178* | −0.05 | −0.081 |
| (0.05) | (0.07) | (0.11) | (0.11) | |
| In-degree Std. Dev.* Complex Model-Based Imputation | −0.011 | 0.099 | −0.063 | −0.025 |
| (0.08) | (0.08) | (0.12) | (0.08) | |
| Log of Size* Symmetric Imputation | 0.022 | 0.189 | 0.277 | −0.413* |
| (0.13) | (0.28) | (0.2) | (0.17) | |
| Log of Size *Simple Model-Based Imputation | 0.014 | 0.555** | −0.158 | −0.003 |
| (0.13) | (0.19) | (0.29) | (0.3) | |
| Log of Size* Complex Model-Based Imputation | 0.014 | 0.638** | −0.075 | −0.056 |
| (0.22) | (0.21) | (0.33) | (0.22) | |
| N | 180 | 180 | 180 | 180 |
| Networks | 5 | 5 | 5 | 5 |
Note: The regression uses the beta slopes from each line as the dependent variable. The direction of the bias is ignored when calculating the regressions. The betas represent the expected increase in bias for a 10 % increase in the amount of missing data. Larger numbers mean larger bias with more missing data. The correlation with centrality takes four values: −.75, −.25, .25, and .75.
Table A7.
Percent Decrease in Total Bias under Different Imputation Strategies: Centralization Measures for Directed Networks.
| Measure | Imputation | Prison |
Sorority |
6th Grade |
Prosper |
RC Elite |
HS 13 |
HS 24 |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Correlation with Centrality |
Correlation with Centrality |
Correlation with Centrality |
Correlation with Centrality |
Correlation with Centrality |
Correlation with Centrality |
Correlation with Centrality |
||||||||||||||||
| −.75 | 0 | .75 | −.75 | 0 | .75 | −.75 | 0 | .75 | −.75 | 0 | .75 | −.75 | 0 | .75 | −.75 | 0 | .75 | −.75 | 0 | .75 | ||
| Indegree | Probabilistic | 4 | 5 | 5 | 14 | 15 | 16 | 7 | 7 | 8 | 6 | 6 | 5 | 0 | 0 | 0 | 6 | 6 | 7 | 9 | 10 | 10 |
| Symmetric | 43 | 44 | 44 | 58 | 66 | 70 | 49 | 50 | 55 | 51 | 50 | 51 | 6 | 6 | 8 | 36 | 40 | 44 | 46 | 51 | 54 | |
| Asymmetric | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | |
| Model-based Simple | 54 | 50 | 45 | 54 | 60 | 60 | 42 | 37 | 32 | 55 | 50 | 44 | 6 | 4 | 6 | 36 | 35 | 33 | 43 | 42 | 39 | |
| Model-based Complex | 56 | 62 | 58 | 36 | 62 | 68 | 77 | 70 | 61 | 66 | 73 | 65 | 27 | 20 | 17 | 72 | 67 | 60 | 79 | 74 | 66 | |
| Total Degree | Probabilistic | 43 | 42 | 39 | 60 | 66 | 67 | 60 | 61 | 63 | 58 | 59 | 57 | 7 | 7 | 8 | 39 | 43 | 46 | 48 | 52 | 54 |
| Symmetric | 52 | 66 | 72 | 40 | 58 | 66 | 78 | 79 | 80 | 70 | 77 | 81 | 20 | 19 | 22 | 76 | 82 | 87 | 87 | 92 | 93 | |
| Asymmetric | 25 | 25 | 23 | 38 | 42 | 42 | 43 | 45 | 48 | 38 | 40 | 40 | 7 | 6 | 8 | 26 | 30 | 33 | 31 | 35 | 37 | |
| Model-based Simple | 57 | 64 | 61 | 56 | 69 | 76 | 72 | 72 | 72 | 76 | 79 | 77 | 13 | 10 | 13 | 57 | 60 | 61 | 66 | 69 | 70 | |
| Model-based Complex | 45 | 61 | 67 | 33 | 61 | 71 | 80 | 82 | 82 | 62 | 76 | 82 | 33 | 26 | 23 | 83 | 83 | 80 | 87 | 89 | 85 | |
| Bon Power | Probabilistic | 1 | 10 | 19 | −13 | −7 | 1 | −86 | −79 | −62 | −58 | −43 | −25 | −213 | −192 | −203 | −74 | −69 | −55 | −82 | −82 | −63 |
| Symmetric | 1 | 10 | 19 | −13 | −7 | 1 | −86 | −79 | −62 | −58 | −43 | −25 | −213 | −192 | −203 | −74 | −69 | −55 | −82 | −82 | −63 | |
| Asymmetric | 1 | 10 | 19 | −13 | −7 | 1 | −86 | −79 | −62 | −58 | −43 | −25 | −213 | −192 | −203 | −74 | −69 | −55 | −82 | −82 | −63 | |
| Model-based Simple | 47 | 58 | 58 | 53 | 66 | 67 | 25 | 40 | 44 | 26 | 33 | 32 | −361 | −200 | −186 | 21 | 28 | 10 | 16 | 10 | −4 | |
| Model-based Complex | 46 | 57 | 59 | 56 | 64 | 66 | 40 | 56 | 52 | 35 | 46 | 39 | −391 | −227 | −233 | 45 | 51 | 30 | 46 | 46 | 25 | |
| Closeness | Probabilistic | 7 | 23 | 28 | 24 | 33 | 36 | −18 | 20 | 25 | 19 | 45 | 43 | −106 | −48 | 34 | −4 | 53 | 57 | −8 | 59 | 71 |
| Symmetric | −14 | 14 | 37 | 26 | 47 | 56 | −12 | 19 | −18 | 34 | 68 | 72 | −1824 | −2102 | −1381 | −19 | 12 | −3 | 5 | 48 | 35 | |
| Asymmetric | −3 | −8 | −4 | −13 | −5 | 1 | −12 | −3 | 15 | −11 | 2 | 6 | −80 | −26 | 44 | 3 | 3 | 5 | 5 | 15 | 17 | |
| Model-based Simple | −27 | 25 | 46 | 9 | 36 | 50 | −10 | 7 | −47 | 4 | 50 | 54 | −566 | −540 | −317 | −32 | −7 | −26 | −9 | 11 | −4 | |
| Model-based Complex | −33 | 28 | 52 | 23 | 53 | 63 | −12 | 16 | −25 | 15 | 70 | 75 | −264 | −223 | −127 | −36 | 35 | 17 | −9 | 52 | 33 | |
| Betweenness | Probabilistic | 46 | 40 | 36 | 48 | 42 | 39 | 35 | 30 | 25 | 62 | 56 | 50 | 35 | 30 | 27 | 65 | 58 | 51 | 50 | 36 | 25 |
| Symmetric | 72 | 72 | 68 | 71 | 69 | 64 | 7 | −2 | −16 | 67 | 64 | 62 | −771 | −675 | −556 | 43 | 33 | 19 | 45 | 30 | 14 | |
| Asymmetric | 15 | 11 | 8 | 10 | 7 | 5 | 58 | 44 | 25 | 26 | 16 | 11 | 24 | 19 | 16 | 35 | 12 | −20 | 40 | 7 | −33 | |
| Model-based Simple | 67 | 67 | 67 | 58 | 61 | 62 | 0 | −3 | −2 | 25 | 34 | 38 | −1231 | −1073 | −886 | 9 | 12 | 11 | −22 | −25 | −21 | |
| Model-based Complex | 69 | 68 | 66 | 63 | 65 | 63 | 34 | 32 | 27 | 42 | 50 | 53 | −939 | −724 | −526 | 42 | 43 | 39 | 21 | 18 | 19 | |
Table A8.
Centralization Bias Slope Regressions: Directed Networks.
| Variables | Model 1 Indegree | Model 2 Total Degree | Model 3 Bon. Power | Model 4 Closeness | Model 5 Betweeness |
|---|---|---|---|---|---|
| Intercept | −2.877*** | −3.012*** | −2.12*** | −3.022*** | −1.443 |
| (0.15) | (0.14) | (0.62) | (0.9) | (0.79) | |
| Correlation with Centrality | 0.21*** | 0.234*** | 0.28*** | 0.556*** | 0.072 |
| (0.03) | (0.04) | (0.05) | (0.1) | (0.04) | |
| In-degree Std. Dev. | 0.06*** | 0.074*** | −0.228*** | −0.278 | −0.103 |
| (0.01) | (0.01) | (0.06) | (0.15) | (0.13) | |
| Log of Size | 0.027 | 0.029 | −0.085 | 0.148 | −0.066 |
| (0.03) | (0.03) | (0.13) | (0.22) | (0.19) | |
| Asymmetric Imputation | 0 | −0.611 | −0.658* | 0.751 | −0.02 |
| (0.15) | (0.47) | (0.29) | (0.69) | (0.24) | |
| Simple Model-Based Imputation | −1.808*** | −1.489 | −2.423** | −0.973 | −1.321 |
| (0.42) | (0.92) | (0.82) | (0.81) | (1.2) | |
| Complex Model-Based Imputation | 0.346 | 1.953 | −1.173 | −0.567 | −0.969 |
| (1.16) | (1.03) | (1.1) | (0.97) | (1.06) | |
| Probabilistic Imputation | −0.176 | −1.426 | −0.658 | 1.792* | 0.844 |
| (0.28) | (0.82) | (0.42) | (0.91) | (0.79) | |
| Symmetric Imputation | −1.773 | 2.302* | −0.658 | 0.209 | −1.055 |
| (1.03) | (1.03) | (0.74) | (1.39) | (1.1) | |
| Correlation with Centrality* Asymmetric Imputation | 0 | −0.048 | −0.133 | −0.13 | 0.21*** |
| (0.04) | (0.06) | (0.07) | (0.14) | (0.05) | |
| Correlation with Centrality* Simple Model-Based Imputation | 0.101* | −0.085 | −0.302*** | −0.487*** | −0.283*** |
| (0.04) | (0.06) | (0.07) | (0.14) | (0.05) | |
| Correlation with Centrality* Complex Model-Based Imputation | 0.159*** | −0.19*** | −0.219** | −0.659*** | −0.134* |
| (0.04) | (0.06) | (0.07) | (0.14) | (0.05) | |
| Correlation with Centrality* Probabilistic Imputation | 0.005 | −0.047 | −0.133 | −0.46** | 0.177** |
| (0.04) | (0.06) | (0.07) | (0.14) | (0.05) | |
| Correlation with Centrality* Symmetric Imputation | −0.094* | −0.319*** | −0.133 | −0.572*** | 0.28*** |
| (0.04) | (0.06) | (0.07) | (0.14) | (0.05) | |
| In-degree Std. Dev.* Asymmetric Imputation | 0 | 0.054 | 0.094*** | −0.008 | −0.257*** |
| (0.01) | (0.04) | (0.03) | (0.11) | (0.04) | |
| In-degree Std. Dev.*Simple Model-Based Imputation | 0.173*** | 0.223** | 0.231** | 0.32* | −0.083 |
| (0.04) | (0.09) | (0.08) | (0.13) | (0.2) | |
| In-degree Std. Dev.*Complex Model-Based Imputation | 0.223* | 0.249** | 0.32** | 0.388* | −0.116 |
| (0.11) | (0.1) | (0.1) | (0.16) | (0.17) | |
| In-degree Std. Dev.* Probabilistic Imputation | 0.024 | 0.129 | 0.094* | 0.135 | 0.046 |
| (0.03) | (0.08) | (0.04) | (0.15) | (0.13) | |
| In-degree Std. Dev.*Symmetric Imputation | 0.159 | 0.308** | 0.094 | 0.384 | 0.376* |
| (0.1) | (0.1) | (0.07) | (0.23) | (0.18) | |
| Log of Size* Asymmetric Imputation | 0 | −0.008 | 0.153* | −0.135 | 0.119* |
| (0.03) | (0.1) | (0.06) | (0.17) | (0.06) | |
| Log of Size *Simple Model-Based Imputation | 0.07 | −0.119 | 0.193 | −0.126 | 0.079 |
| (0.09) | (0.2) | (0.18) | (0.19) | (0.29) | |
| Log of Size* Complex Model-Based Imputation | −0.477 | −0.86*** | −0.137 | −0.288 | 0.016 |
| (0.25) | (0.22) | (0.23) | (0.23) | (0.25) | |
| Log of Size * Probabilistic Imputation | −0.002 | 0.025 | 0.153 | −0.546* | −0.283 |
| (0.06) | (0.17) | (0.09) | (0.22) | (0.19) | |
| Log of Size*Symmetric Imputation | 0.055 | −0.958*** | 0.153 | −0.459 | −0.202 |
| (0.22) | (0.22) | (0.16) | (0.34) | (0.27) | |
| N | 378 | 378 | 378 | 324 | 324 |
| Networks | 7 | 7 | 7 | 7 | 7 |
Note: The regression uses the betas slopes from each line as the dependent variable. The betas represent the expected drop in correlation (between the empirical and the observed) for a 10 % increase in the amount of missing data. Larger numbers mean larger bias with more missing data. The correlation with centrality takes four values: −.75, −.25, .25, and .75.
Table A9.
Percent Decrease in Total Bias under Different Imputation Strategies: Topology Measures for Undirected Networks.
| Measure | Imputation | Interlock |
Co-author |
Co-citation |
Biotech |
HIV |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| −.75 | 0 | .75 | −.75 | 0 | .75 | −.75 | 0 | .75 | −.75 | 0 | .75 | −.75 | 0 | .75 | ||
| Component | Symmetric | 89 | 86 | 74 | 95 | 92 | 83 | 94 | 94 | 93 | 94 | 94 | 91 | 96 | 96 | 96 |
| Model-based Simple | 81 | 89 | 86 | 95 | 96 | 96 | 86 | 92 | 97 | 85 | 88 | 91 | 91 | 93 | 94 | |
| Model-based Complex | 79 | 88 | 85 | 86 | 90 | 91 | 75 | 87 | 93 | 84 | 87 | 90 | 82 | 87 | 90 | |
| Bicomponent | Symmetric | 68 | 67 | 55 | 81 | 82 | 73 | 76 | 85 | 82 | 77 | 79 | 77 | 87 | 89 | 85 |
| Model-based Simple | 41 | 79 | 80 | −100 | −1 | 42 | −28 | 49 | 83 | 63 | 80 | 92 | 17 | 50 | 74 | |
| Model-based Complex | 44 | 80 | 79 | −22 | 38 | 66 | 16 | 62 | 89 | 68 | 83 | 93 | 45 | 68 | 85 | |
| Distance | Symmetric | 30 | 72 | 59 | 70 | 83 | 68 | 14 | 81 | 68 | 92 | 90 | 78 | 89 | 91 | 81 |
| Model-based Simple | 38 | 80 | 70 | 67 | 92 | 88 | 18 | 84 | 76 | 93 | 93 | 84 | 83 | 97 | 91 | |
| Model-based Complex | 39 | 80 | 70 | 72 | 91 | 85 | 19 | 84 | 77 | 93 | 93 | 85 | 87 | 97 | 90 | |
| Transitivity | Symmetric | 75 | 70 | 72 | −8 | −31 | −51 | 30 | −101 | −58 | 74 | 47 | 33 | −140 | −92 | −45 |
| Model-based Simple | 76 | 79 | 82 | −42 | −56 | −64 | 5 | −123 | −55 | 79 | 61 | 48 | −217 | −159 | −97 | |
| Model-based Complex | 39 | 50 | 67 | 22 | 3 | −7 | 35 | −52 | −2 | 45 | 31 | 4 | −112 | −69 | −27 | |
| Tau | Symmetric | 39 | 53 | 20 | −27 | −16 | −12 | 57 | 49 | 63 | 86 | 68 | 46 | 72 | 82 | 64 |
| Model-based Simple | 40 | 42 | 28 | 13 | 7 | −3 | 79 | 72 | 74 | 71 | 60 | 48 | 88 | 82 | 66 | |
| Model-based Complex | 13 | 16 | 13 | 47 | 28 | 12 | 75 | 53 | 61 | 72 | 61 | 50 | 89 | 85 | 70 | |
| CONCOR | Symmetric | 44 | 42 | 46 | 50 | 49 | 52 | 54 | 42 | 38 | 39 | 36 | 40 | 47 | 43 | 44 |
| Model-based Simple | 21 | 30 | 42 | 22 | 31 | 44 | 38 | 30 | 33 | 30 | 31 | 38 | 37 | 37 | 41 | |
| Model-based Complex | 21 | 29 | 42 | 36 | 41 | 51 | 39 | 32 | 37 | 29 | 31 | 37 | 44 | 43 | 46 | |
Table A10.
Topology Bias Slope Regressions: Undirected Networks.
| Variables | Model 1 Component Size | Model 2 Bicomponent Size | Model 3 Distance | Model 4 Transitivity | Model 5 Tau RC | Model 6 CONCOR |
|---|---|---|---|---|---|---|
| Intercept | −2.752*** | −2.78*** | −3.262*** | −0.538 | −1.648 | −3.82*** |
| (0.44) | (0.45) | (0.42) | (1.68) | (0.84) | (0.24) | |
| Correlation with Centrality | 0.685*** | 0.619*** | 0.742*** | 0.53*** | 0.132 | 0.56*** |
| (0.07) | (0.06) | (0.13) | (0.05) | (0.08) | (0.02) | |
| In-degree Std. Dev. | −0.061 | −0.08* | −0.086** | −0.236 | −0.005 | −0.034 |
| (0.03) | (0.03) | (0.03) | (0.13) | (0.06) | (0.02) | |
| Log of Size | 0.106 | 0.168 | 0.24** | −0.184 | −0.07 | 0.07 |
| (0.09) | (0.09) | (0.08) | (0.34) | (0.17) | (0.05) | |
| Symmetric Imputation | −0.326 | 0.125 | 1.064* | −2.167 | 2.207* | −0.272 |
| (0.46) | (0.26) | (0.52) | (1.75) | (1.1) | (0.36) | |
| Simple Model-Based Imputation | −1.659 | −0.327 | 0.914 | −2.951 | 1.553 | −0.128 |
| (1.26) | (1.67) | (0.67) | (2.08) | (1.11) | (0.31) | |
| Complex Model-Based Imputation | −1.605*** | −0.077 | 1.052 | −1.784 | 2.683 | −0.145 |
| (0.42) | (1.53) | (1.15) | (1.53) | (1.38) | (0.24) | |
| Correlation with Centrality* Symmetric Imputation | 0.481*** | 0.231** | 0.179 | 0.117 | 0.161 | −0.341*** |
| (0.1) | (0.09) | (0.18) | (0.08) | (0.12) | (0.03) | |
| Correlation with Centrality* Simple Model-Based Imputation | −0.326** | −0.896*** | −0.332 | 0.02 | 0.322** | −0.458*** |
| (0.1) | (0.09) | (0.18) | (0.08) | (0.12) | (0.03) | |
| Correlation with Centrality* Complex Model-Based Imputation | −0.404*** | −0.951*** | −0.2 | −0.051 | 0.378** | −0.446*** |
| (0.1) | (0.09) | (0.18) | (0.08) | (0.12) | (0.03) | |
| In-degree Std. Dev.* Symmetric Imputation | −0.078* | −0.096*** | 0.045 | 0.227 | −0.001 | 0.02 |
| (0.03) | (0.02) | (0.04) | (0.13) | (0.08) | (0.03) | |
| In-degree Std. Dev.* Simple Model-Based Imputation | −0.078 | 0.085 | 0.067 | 0.308* | −0.126 | 0.021 |
| (0.09) | (0.13) | (0.05) | (0.16) | (0.08) | (0.02) | |
| In-degree Std. Dev.* Complex Model-Based Imputation | −0.026 | 0.07 | 0.062 | 0.1 | −0.066 | 0.018 |
| (0.03) | (0.12) | (0.09) | (0.11) | (0.1) | (0.02) | |
| Log of Size* Symmetric Imputation | −0.277** | −0.132* | −0.497*** | 0.164 | −0.465* | 0.021 |
| (0.09) | (0.05) | (0.11) | (0.36) | (0.22) | (0.07) | |
| Log of Size *Simple Model-Based Imputation | −0.051 | −0.193 | −0.59*** | 0.2 | −0.233 | 0.012 |
| (0.26) | (0.34) | (0.14) | (0.42) | (0.23) | (0.06) | |
| Log of Size* Complex Model-Based Imputation | −0.03 | −0.265 | −0.61** | 0.257 | −0.49 | 0.013 |
| (0.09) | (0.31) | (0.23) | (0.31) | (0.28) | (0.05) | |
| N | 180 | 180 | 180 | 180 | 180 | 180 |
| Networks | 5 | 5 | 5 | 5 | 5 | 5 |
Note: The regression uses the beta slopes from each line as the dependent variable. The direction of the bias is ignored when calculating the regressions. The betas represent the expected increase in bias for a 10 % increase in the amount of missing data. Larger numbers mean larger bias with more missing data. The correlation with centrality takes four values: -.75, -.25, .25, and .75.
Table A11.
Percent Decrease in Total Bias under Different Imputation Strategies: Topology Measures for Directed Networks.
| Measure | Imputation | Prison |
Sorority |
6th Grade |
Prosper |
RC Elite |
HS 13 |
HS 24 |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Correlation with Centrality |
Correlation with Centrality |
Correlation with Centrality |
Correlation with Centrality |
Correlation with Centrality |
Correlation with Centrality |
Correlation with Centrality |
||||||||||||||||
| −.75 | 0 | .75 | −.75 | 0 | .75 | −.75 | 0 | .75 | −.75 | 0 | .75 | −.75 | 0 | .75 | −.75 | 0 | .75 | −.75 | 0 | .75 | ||
| Component | Probabilistic | 69 | 69 | 68 | 61 | 62 | 59 | 77 | 76 | 77 | 73 | 76 | 76 | 85 | 86 | 91 | 82 | 82 | 86 | 80 | 81 | 84 |
| Symmetric | 69 | 69 | 68 | 61 | 62 | 59 | 77 | 76 | 77 | 73 | 76 | 76 | 85 | 86 | 91 | 82 | 82 | 86 | 80 | 81 | 84 | |
| Asymmetric | 69 | 69 | 68 | 61 | 62 | 59 | 77 | 76 | 77 | 73 | 76 | 76 | 85 | 86 | 91 | 82 | 82 | 86 | 80 | 81 | 84 | |
| Model-based Simple | 83 | 89 | 93 | 89 | 92 | 91 | 97 | 98 | 98 | 85 | 91 | 93 | 84 | 87 | 92 | 94 | 95 | 97 | 90 | 92 | 94 | |
| Model-based Complex | 79 | 86 | 91 | 84 | 88 | 87 | 90 | 89 | 92 | 81 | 88 | 91 | 78 | 81 | 89 | 86 | 88 | 92 | 85 | 88 | 91 | |
| Bicomponent | Probabilistic | 30 | 36 | 39 | 30 | 33 | 33 | −101 | −36 | 19 | 30 | 39 | 43 | 74 | 73 | 79 | −10 | 11 | 38 | −1 | 20 | 43 |
| Symmetric | 30 | 36 | 39 | 30 | 33 | 33 | −101 | −36 | 19 | 30 | 39 | 43 | 74 | 73 | 79 | −10 | 11 | 38 | −1 | 20 | 43 | |
| Asymmetric | 30 | 36 | 39 | 30 | 33 | 33 | −101 | −36 | 19 | 30 | 39 | 43 | 74 | 73 | 79 | −10 | 11 | 38 | −1 | 20 | 43 | |
| Model-based Simple | 86 | 88 | 89 | 74 | 84 | 87 | 47 | 69 | 84 | 93 | 95 | 95 | 66 | 77 | 84 | 85 | 90 | 94 | 90 | 94 | 96 | |
| Model-based Complex | 83 | 86 | 87 | 80 | 87 | 87 | 59 | 78 | 89 | 92 | 93 | 93 | 81 | 88 | 91 | 92 | 95 | 96 | 93 | 96 | 96 | |
| Distance | Probabilistic | 36 | 33 | 27 | 24 | 38 | 33 | 27 | 40 | 36 | 35 | 38 | 33 | 8 | 16 | 19 | 64 | 57 | 52 | 68 | 59 | 51 |
| Symmetric | −19 | 49 | 63 | −35 | 41 | 51 | −47 | 47 | 73 | 15 | 74 | 73 | −2616 | −2138 | −1463 | 55 | 75 | 82 | 59 | 81 | 85 | |
| Asymmetric | −22 | −13 | −8 | −26 | −15 | −9 | −42 | −83 | −40 | −22 | −11 | −7 | −3 | −6 | −3 | −23 | −16 | −10 | −23 | −14 | −9 | |
| Model-based Simple | −35 | 46 | 71 | −127 | 2 | 46 | −54 | 42 | 78 | −40 | 47 | 72 | −1075 | −662 | −392 | 32 | 58 | 76 | 22 | 56 | 76 | |
| Model-based Complex | −10 | 59 | 74 | −74 | 32 | 59 | −27 | 59 | 76 | −7 | 67 | 81 | −663 | −353 | −202 | 60 | 80 | 93 | 50 | 78 | 92 | |
| Transitivity | Probabilistic | 16 | 35 | 45 | 12 | 4 | 8 | −131 | −135 | −121 | −53 | −26 | −8 | 48 | 51 | 64 | −109 | −141 | −107 | −94 | −135 | −101 |
| Symmetric | 8 | 26 | 34 | 4 | −2 | 0 | −196 | −207 | −197 | −85 | −68 | −52 | −222 | −195 | −82 | −162 | −204 | −162 | −125 | −171 | −134 | |
| Asymmetric | 34 | 41 | 45 | 45 | 50 | 53 | 35 | 35 | 39 | 29 | 40 | 44 | 53 | 55 | 71 | 41 | 42 | 47 | 40 | 40 | 44 | |
| Model-based Simple | −82 | −48 | −20 | −142 | −112 | −74 | −379 | −365 | −301 | −282 | −235 | −171 | −190 | −147 | −46 | −538 | −583 | −396 | −490 | −534 | −355 | |
| Model-based Complex | 8 | 31 | 41 | 16 | 24 | 32 | −154 | −149 | −115 | −49 | −22 | 5 | 6 | 25 | 54 | −128 | −130 | −55 | −85 | −79 | −17 | |
| Tau | Probabilistic | 36 | 36 | 36 | 32 | 31 | 28 | 62 | 65 | 67 | 68 | 71 | 71 | 0 | 6 | 21 | 89 | 89 | 87 | 89 | 88 | 85 |
| Symmetric | −132 | −69 | −15 | 21 | 24 | 20 | −4 | 3 | 6 | −27 | 3 | 19 | −16 | −23 | −36 | 6 | 18 | 32 | 9 | 22 | 31 | |
| Asymmetric | 11 | 15 | 22 | −7 | 6 | 12 | 43 | 45 | 45 | 55 | 65 | 70 | 1 | 7 | 22 | 78 | 80 | 81 | 76 | 79 | 80 | |
| Model-based Simple | 15 | 20 | 24 | 8 | 10 | 6 | −16 | −15 | −12 | 0 | 7 | 10 | −19 | −11 | 6 | −10 | −5 | 4 | −15 | −8 | −3 | |
| Model-based Complex | 22 | 27 | 37 | 42 | 42 | 38 | 12 | 14 | 16 | 35 | 39 | 40 | −1 | 6 | 23 | 22 | 26 | 32 | 14 | 20 | 25 | |
| CONCOR | Probabilistic | 6 | 12 | 19 | 12 | 17 | 21 | 9 | 8 | 7 | 21 | 23 | 29 | 11 | 20 | 39 | 17 | 20 | 25 | 21 | 23 | 26 |
| Reciprocated | 7 | 12 | 19 | 10 | 16 | 21 | 8 | 7 | 6 | 22 | 24 | 29 | 4 | 14 | 32 | 17 | 20 | 25 | 22 | 24 | 27 | |
| Directed | 5 | 11 | 18 | 13 | 17 | 21 | 6 | 5 | 4 | 20 | 22 | 28 | 11 | 20 | 39 | 17 | 20 | 25 | 21 | 23 | 26 | |
| Model-based Simple | −13 | −3 | 8 | −9 | 2 | 11 | −7 | −6 | −3 | −4 | 2 | 11 | −15 | −3 | 21 | −5 | −1 | 6 | 3 | 7 | 14 | |
| Model-based Complex | −10 | 0 | 11 | −4 | 6 | 15 | −7 | −4 | −2 | −2 | 4 | 14 | −14 | −3 | 22 | 0 | 4 | 11 | 7 | 10 | 17 | |
Table A12.
Topology Bias Slope Regressions: Directed Networks.
| Variables | Model 1 Component Size | Model 2 Bicomponent Size | Model 3 Distance | Model 4 Transitivity | Model 5 Tau RC | Model 6 CONCOR |
|---|---|---|---|---|---|---|
| Intercept | −0.553 | −0.979 | −4.062*** | −0.957* | −2.251*** | −3.754*** |
| (3.77) | (1.45) | (0.37) | (0.46) | (0.11) | (0.41) | |
| Correlation with Centrality | 0.537 | 0.383*** | 0.521*** | 0.348*** | 0.081** | 0.224*** |
| (0.66) | (0.04) | (0.07) | (0.03) | (0.03) | (0.01) | |
| In-degree Std. Dev. | −0.089 | −0.084 | −0.333*** | −0.303*** | 0.014 | 0.045 |
| (0.35) | (0.13) | (0.06) | (0.08) | (0.01) | (0.04) | |
| Log of Size | −0.471 | −0.262 | 0.496*** | −0.274* | 0.013 | −0.046 |
| (0.81) | (0.31) | (0.09) | (0.11) | (0.02) | (0.09) | |
| Asymmetric Imputation | 0.696 | −0.141 | 0.71* | −0.84** | 1.884 | −0.117 |
| (7.31) | (0.97) | (0.35) | (0.26) | (1.37) | (0.33) | |
| Simple Model-Based Imputation | −14.491 | −0.026 | 1.411* | −1.834*** | −0.588 | −0.392*** |
| (41.66) | (1.17) | (0.68) | (0.39) | (0.62) | (0.11) | |
| Complex Model-Based Imputation | −3.342 | 1.705*** | 2.204** | −1.509** | −0.981* | −0.506*** |
| (7.28) | (0.44) | (0.69) | (0.48) | (0.49) | (0.14) | |
| Probabilistic Imputation | 0.696 | −0.141 | 0.925 | −1.934*** | 4.558*** | −0.212 |
| (4.26) | (1.01) | (0.72) | (0.48) | (1.1) | (0.3) | |
| Symmetric Imputation | 0.696 | −0.141 | 4.024*** | −1.784*** | −0.78 | −0.262 |
| (6.8) | (0.98) | (0.59) | (0.49) | (1.15) | (0.31) | |
| Correlation with Centrality* Asymmetric Imputation | 0.056 | −0.194** | −0.142 | −0.112* | −0.007 | −0.172*** |
| (0.93) | (0.06) | (0.11) | (0.05) | (0.04) | (0.02) | |
| Correlation with Centrality* Simple Model-Based Imputation | 2.348* | −0.443*** | −1.288*** | −0.28*** | −0.024 | −0.209*** |
| (0.93) | (0.06) | (0.11) | (0.05) | (0.04) | (0.02) | |
| Correlation with Centrality* Complex Model-Based Imputation | 1.109 | −0.346*** | −1.31*** | −0.301*** | −0.005 | −0.211*** |
| (0.93) | (0.06) | (0.11) | (0.05) | (0.04) | (0.02) | |
| Correlation with Centrality* Probabilistic Imputation | 0.056 | −0.194** | 0.176 | −0.149** | −0.005 | −0.169*** |
| (0.93) | (0.06) | (0.11) | (0.05) | (0.04) | (0.02) | |
| Correlation with Centrality* Symmetric Imputation | 0.056 | −0.194** | −0.82*** | −0.106* | 0.168*** | −0.176*** |
| (0.93) | (0.06) | (0.11) | (0.05) | (0.04) | (0.02) | |
| In-degree Std. Dev.* Asymmetric Imputation | −0.107 | −0.119 | 0.131* | 0.05 | 0.346** | −0.018 |
| (0.68) | (0.09) | (0.06) | (0.04) | (0.13) | (0.03) | |
| In-degree Std. Dev.*Simple Model-Based Imputation | −2.292 | 0.319** | −0.145 | 0.175** | 0.053 | −0.016 |
| (3.86) | (0.11) | (0.11) | (0.06) | (0.06) | (0.01) | |
| In-degree Std. Dev.*Complex Model-Based Imputation | −0.261 | 0.236*** | −0.056 | 0.361*** | 0.117* | −0.002 |
| (0.67) | (0.04) | (0.11) | (0.08) | (0.05) | (0.01) | |
| In-degree Std. Dev.* Probabilistic Imputation | −0.107 | −0.119 | −0.083 | 0.29*** | 0.423*** | −0.019 |
| (0.39) | (0.09) | (0.12) | (0.08) | (0.1) | (0.03) | |
| In-degree Std. Dev.* Symmetric Imputation | −0.107 | −0.119 | 0.098 | 0.323*** | 0.218* | −0.009 |
| (0.63) | (0.09) | (0.1) | (0.08) | (0.11) | (0.03) | |
| Log of Size* Asymmetric Imputation | −0.377 | 0.038 | −0.185* | −0.002 | −0.811** | 0.013 |
| (1.56) | (0.21) | (0.09) | (0.06) | (0.29) | (0.07) | |
| Log of Size *Simple Model-Based Imputation | 2.96 | −0.774** | −0.364* | 0.452*** | 0.05 | 0.091*** |
| (8.9) | (0.25) | (0.16) | (0.09) | (0.13) | (0.02) | |
| Log of Size* Complex Model-Based Imputation | 0.209 | −1.084*** | −0.629*** | 0.098 | 0.013 | 0.098*** |
| (1.56) | (0.09) | (0.17) | (0.12) | (0.1) | (0.03) | |
| Log of Size * Probabilistic Imputation | −0.377 | 0.038 | −0.229 | 0.262* | −1.461*** | 0.028 |
| (0.91) | (0.21) | (0.17) | (0.11) | (0.23) | (0.06) | |
| Log of Size* Symmetric Imputation | −0.377 | 0.038 | −1.14*** | 0.254* | −0.137 | 0.024 |
| (1.45) | (0.21) | (0.14) | (0.12) | (0.25) | (0.07) | |
| N | 378 | 378 | 324 | 324 | 378 | 378 |
| Networks | 7 | 7 | 7 | 7 | 7 | 7 |
Note: The regression uses the betas slopes from each line as the dependent variable. The betas represent the expected drop in correlation (between the empirical and the observed) for a 10 % increase in the amount of missing data. Larger numbers mean larger bias with more missing data. The correlation with centrality takes four values: − .75, − .25, .25, and .75.
Table A13.
Maximum percent missing to retain target correlation of .9 with true score.
| Network | Imputation Type | In-Degree |
Out-Degree |
Total Degree |
Bonacich Power |
Closeness |
Betweenness |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Non-respondents in Correlation |
Non-respondents in Correlation |
Non-respondents in Correlation |
Non-respondents in Correlation |
Non-respondents in Correlation |
Non-respondents in Correlation |
||||||||
| No | Yes | No | Yes | No | Yes | No | Yes | No | Yes | No | Yes | ||
| Undirected Networksa | Listwise Deletion | 55.8 | NAb | 55.8 | NA | 55.8 | NA | 47.6 | NA | 5 | NA | 30.2 | NA |
| Symmetric | 70c | 46.8 | 70 | 46.8 | 70 | 46.8 | 70 | 52.6 | 56.8 | 39.2 | 65.6 | 44.8 | |
| Model-Based Simple | 70 | 56.4 | 70 | 56.4 | 70 | 56.4 | 70 | 58.2 | 64.8 | 28.8 | 69.4 | 45.2 | |
| Model-Based Complex | 70 | 58.4 | 70 | 58.4 | 70 | 58.4 | 70 | 59.4 | 64.4 | 24.4 | 67.8 | 48.2 | |
| Directed Networksa | Listwise Deletion | 39 | NA | 25.6 | NA | 33.7 | NA | 26.3 | NA | 21.6 | NA | 15.4 | NA |
| Probabilistic | 40.9 | 39 | 70 | 9.6 | 58.6 | 24.4 | 51.1 | 23.9 | 42.3 | 9 | 20.4 | 11.1 | |
| Symmetric | 30.9 | 30.6 | 70 | 12.4 | 50.1 | 29.1 | 51.1 | 23.9 | 43.3 | 12.3 | 9.7 | 5.4 | |
| Asymmetric | 39 | 38.4 | 70 | 6.7 | 59.4 | 20.7 | 51.1 | 23.9 | 26.3 | 3.9 | 19.3 | 7.7 | |
| Model-Based Simple | 27.7 | 28.4 | 70 | 13.3 | 45.7 | 25.3 | 42 | 24 | 49.9 | 11.4 | 9.1 | 4.7 | |
| Model-Based Complex | 30 | 31.3 | 70 | 12.6 | 48.7 | 27.3 | 44 | 24.9 | 50.4 | 10.7 | 11.4 | 6.9 | |
The maximum percent missing was calculated based on a quadratic fit to the data.
Values represent the means (for the maximum percent missing to retain target correlation) taken over all directed or undirected networks.
Listwise deletion has no value for the case of non-respondents being kept in the correlation calculation.
Cases where percent missing is above 70, our observed maximum. In these cases, 70 is used to calculate overall means.
Footnotes
Note that the problem of imputation of missing data is distinct from the problem of network inference from independently sampled data, as the kind of approaches that are likely to be successful in each case are different (Handcock and Gile, 2010; Smith, 2012; McPherson and Smith, 2019).
This information was ignored in Part I and Part II of this study, which assumed that non-respondents were completely removed from the network when calculating summary statistics.
We thank the following authors for providing data for this study: Mark Mizruchi (Interlock network); Scott Gest (6th grade data); Lisa Keister (River City Elite); Walter Powell (Biotechnology exchange data).
The Prosper data were made available through the following grants: NSF/HSD:0624158, W.T. Grant Foundation 8316 &NIDA 1R01DA018225-01.
The Colorado Spring HIV network was made available through NIH R01 DA 12831 (PI Morris).
Different imputation strategies can yield different size networks to analyze, while component size, bicomponent size and distance are particularly sensitive to network size. We thus scale these measures, making it easier to interpret the results across imputation strategies.
We do not claim that a ranked cluster weighting scheme will offer the best fit for every network; we are only concerned if this summary measure of the triad distribution is measured better/worse across imputation strategies.
We utilize the simple CONCOR algorithm to place actors into equivalent blocks, setting the depth to 3 for all networks. We have also run analogous tests where the depth was allowed to vary across networks. Here, we determine the best fitting blockmodel on each network (without missing data), using that to set the depth when fitting the blockmodel on the networks with missing data. The results are very similar to what we see setting the depth to be constant and we only present those results here.
Note that for the small networks it is likely that some of the 1000 samples will be duplicates, with the same pattern of missing data. This means that the variability in the smaller networks might be biased downwards slightly, especially at lower sampling rates. Our analysis here focuses on the expected level of bias, rather than the variability, and such concerns are thus minimized.
This approach has the advantage of simplifying the analysis considerably, but also has the disadvantage of making pretty stringent, perhaps unrealistic, assumptions about the ties between missing actors. A researcher could alternatively opt for a more complicated approach. For example, one could assume that a ‘friend of a friend is a friend’, such that if i nominates j and i nominates k, then one would impute that j nominates k (with some probability), assuming that both j and k are missing. As a researcher adds increasingly more complicated imputation rules, however, the approach veers increasingly towards what a model-based approach is already doing and that is likely the better option; as one can include many terms, or rules, together in a single, systematic model.
The attributes themselves are based on constructed variables. The attributes are constructed to maintain a desired level of homophily (low and high) for that attribute. This was done as there is no common attribute across all networks to include in the analysis. In general, the attributes mimic the kinds of data a researcher is likely to have at their disposable when estimating the initial model.
The initial ERGM is estimated while constraining the max degree of each node to be below the observed outdegree (for the missing nodes this value is imputed).
We hold the observed edges fixed as we assume those values are known and thus do not need to be imputed. A more general view of a model-based approach could allow all ties to vary probabilistically based on the estimated model. A researcher would then calculate the network statistics of interest over a large set of possible networks, using the estimated features in the analysis of interest. The advantage of this approach is that captures the uncertainty in the underlying network. The disadvantage is that it requires the researcher to have a very good model of the network, otherwise the estimated features will not reflect the actual population.
For the non-missing nodes, we constrain out-degree to match the observed network perfectly (as we know all edges from i to other nodes). For missing nodes, we first predict what each missing node’s outdegree would have been, had they filled out the survey. We first estimate a model predicting outdegree based on indegree, restricted to the non-missing cases. Using this regression model as a basis for prediction, we then predict the outdegree of each missing case based on their indegree (the nominations from non-missing nodes to the missing node), adjusting for the fact that the indegree is itself biased (missing any nomination from other missing nodes).
Note that this is different than the sample of 1000 networks (for each level of missing data) that the entire analysis is run over.
Note that the main analysis does not include results for out-degree, but the additional results do. This is the case because out-degree will be recorded perfectly for those nodes that are respondents (as we know who they nominated), so no bias is possible there and the imputation results are not very informative. When we include non-respondents in the calculation, bias is once again possible for out-degree and the imputation results are worth reporting. See Table A13 for results that include out-degree.
We present additional results in the appendix. Table A13 shows the maximum level of missing data that a researcher could have and still maintain at least a .9 correlation with the true centrality values. Higher values suggest the strategy is more robust to missing data. We present these results for the case where non-respondents are not kept in the correlation calculation (as in the main analysis), and for the case where the non-respondents are kept in the correlation calculation. Overall, keeping the non-respondents in the calculation increases bias. This is particularly true for out-degree, which is perfectly recorded amongst respondents, while trying to impute the missing cases (where there is no information on out-going ties) can lead to considerable bias.
Note that the complex model-based approach performs poorly with transitivity, suggesting that the fitted model is predicting more transitive relations than found in the actual data.
Note that in this case the asymmetric probabilistic and symmetric options all offer the same levels of bias as the measures of interest are based on the symmetrized version of the (imputed) network.
Note that for the highly centralized RC elite network only the probabilistic option is better than listwise deletion, with the model-based approaches and symmetric options not performing well. Thus, the model-based approach and the symmetric option (which symmetrizes ties to the central actor) does not work so well when one or two actors dominate the network.
Note that we have collapsed some of the measures into broader classes (e.g. closeness and betweenness are placed together as path-based).
Fixed choice designs (where actors are restricted in the number of people they can nominate) offer similar kinds of problems, as edges that should exist in the network are not recorded (adams 2019). A researcher imputing under a fixed choice design could follow the basic model-based approach used here, but with the (potential) addition of setting all uncertain edges as missing. Thus, for any node that reached their maximum allowed outdegree, all other values in their row of the matrix would be set as missing, as we do not know if they would have nominated that person, had they had the opportunity.
References
- adams jimi, 2019. Gathering Social Network Data. SAGE Publications. Paul Allison, 2002. Missing Data. Sage Publications, Thousand Oaks, CA. [Google Scholar]
- Almaatouq Abdullah, Radaelli Laura, Pentland Alex, Shmueli Erez, 2016. Are you your friends’ friend? Poor perception of friendship ties limits the ability to promote behavioral change. PLoS One 11 (3), e0151588. 10.1371/journal.pone.0151588 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- An Weihua, Schramski Sam, 2015. Analysis of contested reports in exchange networks based on actors’ credibility. Soc. Networks 40, 25–33. [Google Scholar]
- Bell David C., Belli-McQueen Benedetta, Haider Ali, 2007. Partner naming and forgetting: recall of network members. Soc. Networks 29 (2), 279–299. 10.1016/j.socnet.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgatti Stephen P., Carley Kathleen M., Krackhardt David, 2006. On the robustness of centrality measures under conditions of imperfect data. Soc. Networks 28 (2), 124–136. [Google Scholar]
- Brewer Devon D., 2000. Forgetting in the recall-based elicitation of personal and social networks. Soc. Networks 22 (1), 29–43. [Google Scholar]
- Costenbader Elizabeth, Valente Thomas W., 2003. The stability of centrality measures when networks are sampled. Soc. Networks 25 (4), 283–307. [Google Scholar]
- de la Haye Kayla, Embree Joshua, Punkay Marc, Espelage Dorothy L., Tucker Joan S., Green Harold D. Jr., 2017. Analytic strategies for longitudinal networks with missing data. Soc. Networks 50, 17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz Terrill L., Cataldo Marcelo, Carley Kathleen M., 2009. Robustness of centrality measures under uncertainty: examining the role of network topology. Comput. Math. Organ. Theory 15 (4), 303–328. [Google Scholar]
- Galaskiewicz Joseph, 1991. Estimating point centrality using different network sampling techniques. Soc. Networks 13 (4), 347–386. [Google Scholar]
- Gile Krista J., Handcock Mark S., 2017. Analysis of networks with missing data with application to the national longitudinal study of adolescent health. J. R. Stat. Soc. Ser. C Appl. Stat 66 (3), 501–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodreau Steven M., Kitts James A., Morris Martina, 2009. Birds of a feather, or friend of a friend? Using exponential random graph models to investigate adolescent social networks. Demography 46 (1), 103–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handcock Mark S., Gile Krista J., 2010. Modeling social networks from sampled data. Ann. Appl. Stat 4 (1), 5–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handcock M, Hunter D, Butts C, Goodreau S, Krivitsky P, Morris M, 2019. _ergm: Fit, Simulate and Diagnose Exponential-Family Models for Networks_. The Statnet Project (<URL: https://statnet.org> ). R package version 3.10.4. <URL: https://CRAN.R-project.org/package=ergm>. [DOI] [PMC free article] [PubMed]
- Hipp John R., Wang Cheng, Butts Carter T., Jose Rupa, Lakon Cynthia M., 2015. Research note: the consequences of different methods for handling missing network data in stochastic actor based models. Soc. Networks 41, 56–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman Mark, 2009. Imputation of missing network data: some simple procedures. J. Soc. Struct 10 (1). [Google Scholar]
- Huisman Mark, Krause Robert W., 2017. Imputation of missing network data. In: Alhajj R, Rokne J. (Eds.), Encyclopedia of Social Network Analysis and Mining Springer, New York, pp. 1–10. [Google Scholar]
- Hunter David R., Handcock Mark S., Butts Carter T., Goodreau Steven M., Morris Martina, 2008. ergm: a package to fit, simulate and diagnose exponential- family models for networks. J. Stat. Softw 24 (3), 1–29 v24i03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczyk Eric D., Csardi Gabor, 2014. Statistical Analysis of Network Data with R.´ Springer. [Google Scholar]
- Koskinen Johan H., Robins Garry L., Pattison Philippa E., 2010. Analysing exponential random graph (p-star) models with missing data using Bayesian data augmentation. Stat. Methodol 7 (3), 366–384. [Google Scholar]
- Koskinen Johan H., Robins Garry L., Wang Peng, Pattison Philippa E., 2013. Bayesian analysis for partially observed network data, missing ties, attributes and actors. Soc. Networks 35 (4), 514–527. [Google Scholar]
- Kossinets Gueorgi, 2006. Effects of missing data in social networks. Soc. Networks 28 (3), 247–268. [Google Scholar]
- Krause Robert W., Huisman Mark, Snijders Tom A.B., 2018a. Multiple imputation for longitudinal network data. Ital. J. Appl. Stat 30, 33–58. [Google Scholar]
- Krause Robert W., Huisman Mark, Steglich Christian, Sniiders Tom A.B., 2018b. Missing network data a comparison of different imputation methods. In: The Ninth International Conference on Advances in Social Network Analysis and Mining (ASONAM). Barcelona, Spain: IEEE, pp. 159–163. [Google Scholar]
- Krause Robert W., Huisman Mark, Steglich Christian, Snijders Tom, 2020. Missing data in cross-sectional networks – an extensive comparison of missing data treatment methods. Soc. Networks 66 (July 2020), 99–112. [Google Scholar]
- Laumann Eward O., Marsden Peter V., Prensky Daivd, 1983. The boundary specification problem in network analysis. In: Burt RS, Minor MJ (Eds.), Applied Network Analysis: A Methodological Introduction. Sage Publications, Inc, Thousand Oaks, CA. [Google Scholar]
- MacRae Duncan, 1960. Direct factor analysis of sociometric data. Sociometry 23 (4), 360–371. [Google Scholar]
- Martin Christoph, Niemeyer Peter, 2019. Influence of measurement errors on networks: estimating the robustness of centrality measures. Network Sci. 7 (2), 180–195. [Google Scholar]
- McPherson Miller, Smith Jeffrey A., 2019. Network effects in blau space: imputing social context from survey data. Socius. 10.1177/2378023119868591 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moody James, White Douglass R., 2003. Structural cohesion and embeddedness: a hierarchical concept of social groups. Am. Sociol. Rev 68 (1), 103–127. [Google Scholar]
- Morris Martina, Rothenberg Richard, 2011. HIV Transmission Network Metastudy Project: an Archive of Data from Eight Network Studies, 1988–2001. Inter-university Consortium for Political and Social Research. [Google Scholar]
- Rand William M., 1971. Objective criteria for the evaluation of clustering methods. J. Am. Stat. Assoc 66 (336), 846–850. [Google Scholar]
- Robins Garry, Pattison Philippa, Woolcock Jodie, 2004. Missing data in networks: exponential random graph (P*) models for networks with non-respondents. Soc. Networks 26 (3), 257–283. [Google Scholar]
- Robins Garry, Pattison Pip, Kalish Yuval, Lusher Dean, 2007. An introduction to exponential random graph (P*) models for social networks. Soc. Networks 29 (2), 173–191. [Google Scholar]
- Rosenblatt Samuel F., Smith Jeffrey A., Robin Gauthier, G., Laurent Hébert-Dufresne, 2020. Immunization strategies in networks with missing data. PLoS Comput. Biol 16 (7), e1007897 10.1371/journal.pcbi.1007897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silk Matthew J., Jackson Andrew L., Croft Darren P., Colhoun Kendrew, Bearhop Stuart, 2015. The consequences of unidentifiable individuals for the analysis of an animal social network. Anim. Behav 104, 1–11. [Google Scholar]
- Silk Matthew J., 2018. The next steps in the study of missing individuals in networks: a comment on Smith et al. (2017). Soc. Networks 52, 37–41. [Google Scholar]
- Smith Jeffrey A., 2012. Macrostructure from microstructure: generating whole systems from ego networks. Sociol. Methodol 42 (1), 155–205. 10.1177/0081175012455628 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Jeffrey A., Moody James, 2013. Structural effects of network sampling coverage I: nodes missing at random. Soc. Networks 35 (4), 652–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Jeffrey A., Moody James, Morgan Jonathan H., 2017. Network sampling coverage II: the effect of non-random missing data on network measurement. Soc. Networks 48, 78–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stork Diana, Richards William D., 1992. Nonrespondents in communication network studies: problems and possibilities. Group Organ. Manag 17 (2), 193–209. [Google Scholar]
- Wang Dan J., Shi Xiaolin, McFarland Daniel A., Leskovec Jure, 2012. Measurement error in network data: a Re-classification. Soc. Networks 34 (4), 396–409. [Google Scholar]
- Wang Cheng, Butts Carter T., Hipp John R., Jose Rupa, Lakon Cynthia M., 2016. Multiple imputation for missing edge data: a predictive evaluation method with application to add health. Soc. Networks 45, 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman Stanley, Faust Katherine, 1994. Social Network Analysis: Methods and Applications, Vol. 8. Cambridge University Press. [Google Scholar]
- Wasserman Stanley, Pattison Philippa, 1996. Logit models and logistic regressions for social networks: I. An introduction to Markov graphs and p*. Psychometrika 61 (3), 401–425. [Google Scholar]
- White Harrison C., Boorman Scott A., Breiger Ronald L., 1976. Social structure from multiple networks. I. blockmodels of roles and positions. Am. J. Sociol 81 (4), 730–780. [Google Scholar]
- Žnidaršič Anja, Ferligoj Anuška, Doreian Patrick, 2017. Actor non-response in valued social networks: the impact of different non-response treatments on the stability of blockmodels. Soc. Networks 48, 46–56. [Google Scholar]
- Žnidaršič Anja, Ferligoj Anuška, Doreian Patrick, 2018. Stability of centrality measures in valued networks regarding different actor non-response treatments and macro- network structures. Network Sci. 6 (1), 1–33. [Google Scholar]