Abstract
The past 20 years have witnessed enormous progress in synthetic biology in the development of engineered cells for diverse applications, including biomanufacturing, materials fabrication, and potential therapeutics and diagnostics. However, it still remains a major challenge to maintain long-term performance of synthetic gene circuits, due to the emergence of mutants that lose circuit function. Here, we highlight major vulnerabilities of synthetic gene circuits resulting in circuit failure and mutant escape. We also discuss engineering strategies to enhance long-term circuit stability and performance. These approaches can be divided into two strategies: the suppression of the emergence of mutants and the suppression of their relative fitness if mutants do emerge. We anticipate that mechanistic understanding of the modes of circuit failure will facilitate future efforts to design evolutionarily robust synthetic biology-inspired applications.
Keywords: gene circuits, metabolic burden, design principles
1. Introduction
A major objective in synthetic biology is to program cells in a predictable manner using synthetic gene circuits. These circuits can serve as well-defined models to gain insights into the operation of their natural counterparts [1]. Programmed genetic devices also have demonstrated their potential utility in diverse applications, including biosensors [2,3], biomanufacturing [4–7], living therapeutics [8], and diagnostics [9].
Despite recent advancements, the predictable programming of cells is still limited by several fundamental challenges. For example, a target circuit function may be corrupted by cell-cell variability due to the stochastic nature of intracellular dynamics [10]. Moreover, due to an insufficient mechanistic understanding of the circuit components or the host cells, the circuit function can be inadvertently impacted by unanticipated host-circuit interactions [11–13].
Another challenge is to maintain the long-term operation of a circuit. Upon circuit activation, the expression of circuit elements requires the host to allocate intracellular resources, which results in a metabolic burden on the host [14]. Additionally, either by design or as a side effect, a circuit can encode components that are toxic to the host cell [15,16]. A mutant can have a fitness advantage by losing the circuit or by inactivating the expression of burdensome or toxic components. If so, the mutant can eventually take over a population, leading to the corruption or loss of the circuit function in the entire population. Thus, the success of many synthetic biology applications hinges on our ability to predictably engineering biological components that can maintain their genetic stability against evolutionary pressure to sustain their programmed function over time.
In this review, we identify sources of synthetic circuit failure and highlight advances in engineering strategies to circumvent the evolutionary forces that lead to genetic circuit instability. While diverse in application contexts and specific implementations, current approaches reveal two complementary engineering strategies: (1) suppressing mutant emergence, i.e., reducing the evolutionary potential to lower the chances of mutant emergence; and (2) suppressing the relative fitness of mutants, i.e., targeting genetic variants by restricting their fitness to inhibit them from taking over the population.
2. Modes of circuit failure
The link between the burden of a circuit and its failure has been experimentally demonstrated in several systems [7,14]. Gorochowski et al. observed a stress-response-like transcriptomic profile in the host cell upon the introduction and activation of a synthetic gene circuit. This global response in turn reduced the host fitness and the circuit stability [17]. Using inducible expression of the green fluorescent protein (GFP) as a model, Sleight et al. demonstrated that the half-life of a circuit decreased exponentially with target gene expression level [18]. In a different context, Ceroni et al. also showed that reducing circuit burden improved circuit stability [14]. For a circuit that incurs toxicity, it has been shown that its stability can be enhanced by reducing the toxicity [7].
The emergence of a mutant and its interplay with the cells carrying the functional circuit can be illustrated by a simple model (Figure 1A, Eqs 1&2). In this model, the wildtype population (W) represents the population carrying a functional synthetic circuit. Loss of the circuit or inactivation of the circuit’s function gives rise to a mutant cell population (M). We describe the rate of failure as η(W), which can be a continuous or discrete function. W and M reproduce with specific growth rates μW and μM, respectively. They are eliminated at death rates δW and δM, respectively.
Figure 1. Modes of circuit failure.
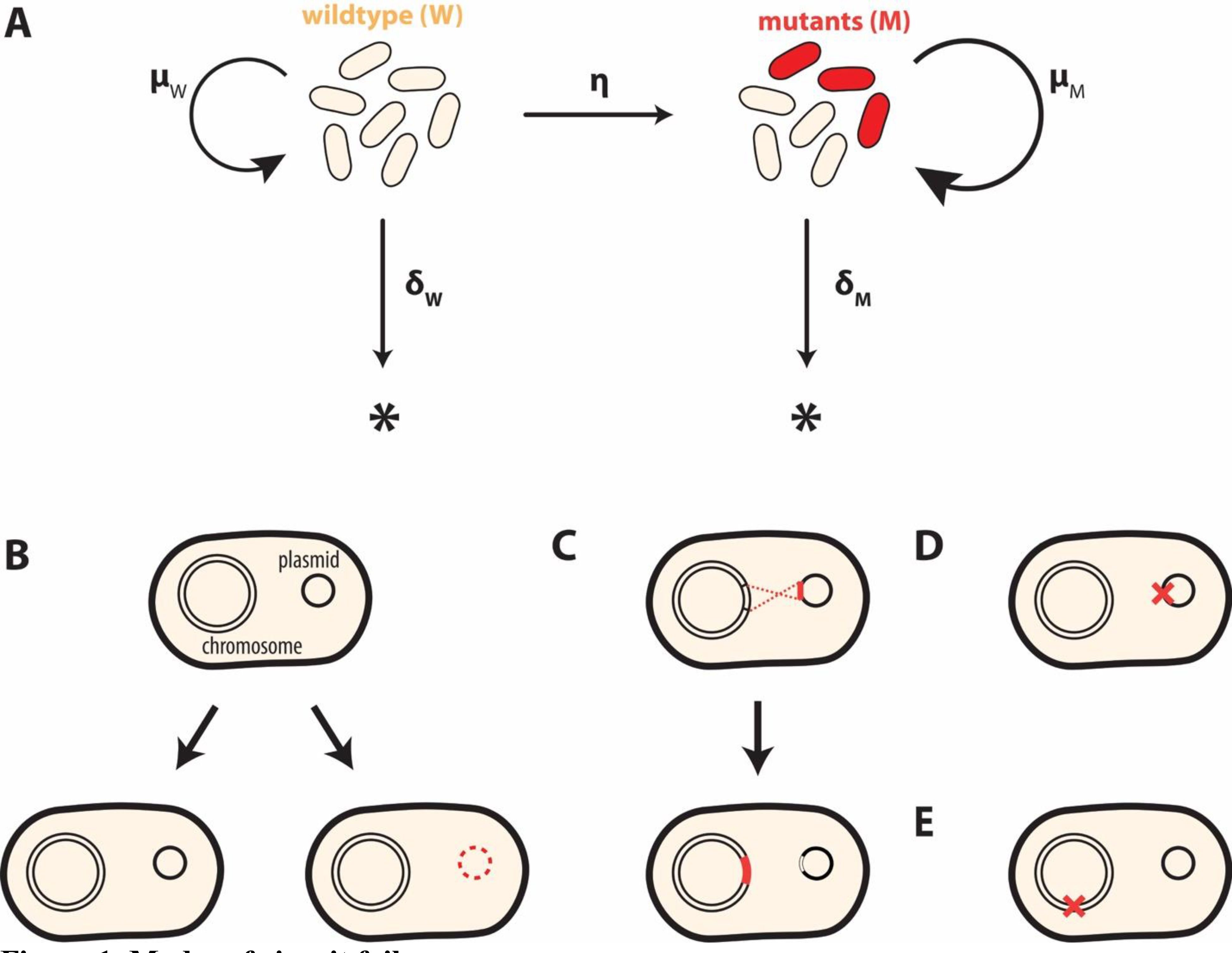
(A) Dynamics of circuit failure. The wildtype population (W) can lose the programmed function at a rate η(W), giving rise to a mutant (M). Each population grows at a specific growth rate (μi) and is eliminated at death rate (δi), where i = W or M. The stability of the circuit in the overall population depends on the magnitudes of the kinetic parameters, which are determined by modes of circuit failure and intervention strategies.
(B) Segregation error. If the designed construct is implemented on the plasmid, segregation error can lead to a subpopulation of cells not carrying the circuit.
(C) DNA recombination. Recombination can occur between different regions of chromosome or plasmids, leading to physical disruption of a circuit.
(D, E) Spontaneous mutations. These mutations can disrupt a circuit element or a host element required for circuit operation.
| (1) |
| (2) |
In general, the growth rates and the death rates are a function of the circuit function and the growth environment. For example, an increase in circuit burden is reflected by a decrease in μW; an increasing toxicity of a circuit component corresponds to an increasing δW. Once it emerges, the relative fitness advantage of the mutant is reflected by . The mutant will eventually dominate if α > 1. This modeling framework is generally applicable for different systems and different modes of mutant generation. Moreover, while we focus on genetic mutants here, the same conceptual framework is applicable to analysis of circuit functional stability involving non-genetic phenotypic bifurcation [13,19].
Depending on its implementation and operation condition, a circuit can fail in multiple ways. If the circuit is encoded on a plasmid, segregation error can result in the loss of the plasmid in daughter cells during cell division (Figure 1B) [20]. This plasmid instability is a major challenge in industrial bioproduction, where imposing an antibiotic selection pressure is impractical due to the associated cost [20]. If some of the circuit elements, such as promoters and terminators, contain repeated sequences, the circuit is prone to recombination-mediated deletion (Figure 1C) [21–23]. Transposable elements can disrupt the circuit function directly, if they are inserted into one or more circuit elements (Figure 1D) [24], or indirectly, if they disrupt host functions required for circuit operation (Figure 1E). These host functions include those involved in plasmid maintenance or gene expression regulation [16,18,21,25]. Finally, spontaneous point mutations or small indels can also result in a partial or complete circuit inactivation. This alleviates the host organism of the costly heterologous metabolic burden if the deleterious mutations occur in the engineered device (Figure 1D) [18,26] or the host chromosome (Figure 1E) [21,27].
3. Strategies to enhance circuit stability
The simple model (Figure 1A, Eqs 1&2) reveals two intervention strategies for enhancing circuit stability. One is to delay or prevent the emergence of mutants (i.e., decreasing η); the other is to minimize the relative fitness benefit of mutants (i.e., reducing α). This concept is reflected in recent efforts to increase stability that spans over diverse application contexts and implementations.
3.1. Suppressing the emergence of circuit mutants
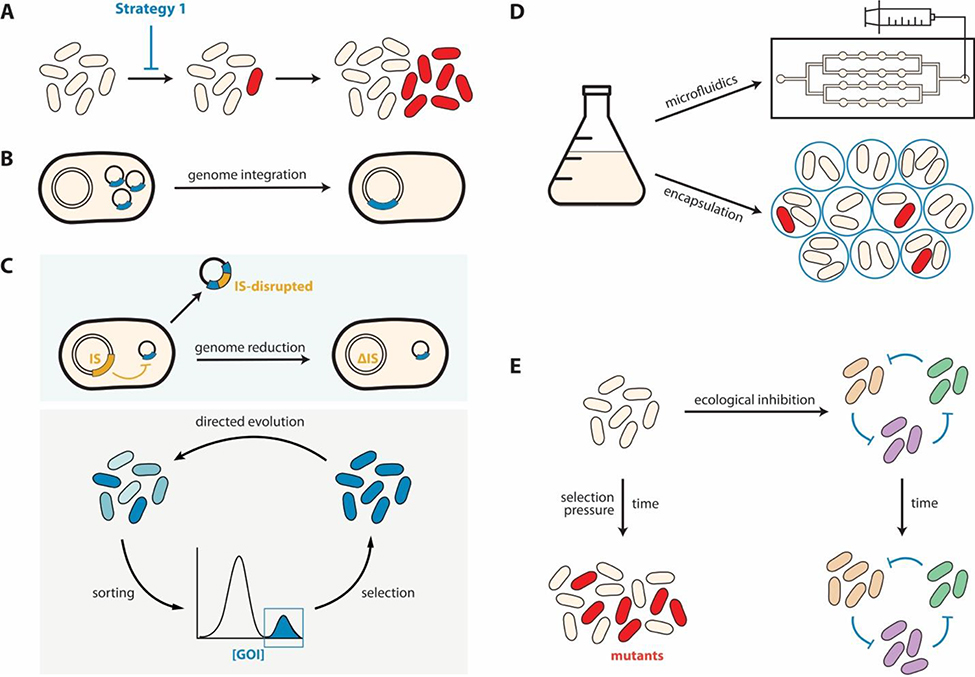
Several strategies have been used to delay or prevent the emergence of mutants, aiming to reduce the failure rate η (Figure 2A, Eqs. 1&2). These approaches include implementing the circuit on the host genome, reducing the host background mutation rate, reducing the population size, and by exploiting external ecological intervention (Figure 2).
Figure 2. Delaying or preventing the emergence of mutants.
(A) Schematic illustration.
(B) Genome integration. Integration into the genome avoids circuit loss due to plasmid segregation error. It can also reduce the advantage of a mutant if it appears (see Strategy 2).
(C) Reducing the host mutation rate. This can be achieved by (Top) removing transposable elements and IS elements and by (Bottom) optimizing the host for a specific circuit by directed evolution.
(D) Reducing the local population size. Enclosing the cells into smaller culture compartments, such as microfluidics, microwells, or microcapsules, reduces the probability by which a mutant can emerge in a local population.
(E) External ecological intervention. Before a mutant emerges, one can eliminate the current population with another carrying the same circuit function. Cycling through different populations allows prevention of mutants while maintaining a common circuit function.
Genomic integration of a circuit precludes segregation error-mediated circuit loss (Figure 1B). This strategy has been shown to enhance the long-term stability of a number of circuits [7,28,29]. In metabolic engineering, heterologous genes are frequently integrated in host chromosome to enhance genetic stability and enable antibiotic-free large-scale fermentation [6,7]. Optimizing the integration location can further enhance the circuit integrity. Recent studies have also suggested that using mobilizable plasmids may compensate for plasmid loss in a population. In particular, if a plasmid can transfer at a sufficiently fast rate, it can be maintained in the absence of positive selection [30].
An alternative approach is to optimize the host to reduce its mutation rate. Since a major source of mutations that cause loss of circuit function is transposable elements or insertion sequences (IS), removing these elements has been shown to reduce the background mutation rate in E. coli (Figure 2C) [24,25,31–33]. Several studies have demonstrated enhanced stability of circuits by implementing in different reduced-genome strains [7,16] or by directly targeting IS [34]. For instance, Chan et al. demonstrated a drastic improvement of the stability of a toxin-mediated biocontainment system: the IS-mediated circuit failure rate was curtailed by 103–105 fold when the circuit was implemented in a reduced-genome E. coli strain [16]. Similarly, the deletion of genomic islands and mobile DNA elements in Pseudomonas putida KT2440 [35] and Acinetobacter baylyi ADP1 genomes [25,33] significantly increased genetic reliability.
The host can also be optimized to be more tolerant to a specific heterologous system via directed evolution. In particular, engineered cells are repeatedly selected for subpopulations that produce target proteins at a high level while maintaining a fast growth rate (Figure 2C) [28,36]. This approach has identified E. coli with reduced mutation rates. In a study that chose fluorescence protein expression level as the target phenotype, the optimized E. coli cells adopted mutations in DNA polymerase I and RNase E, manifesting plasmid mutation rates that are 6- to 30-fold lower compared to the parental strain [36].
Reducing the population size can suppress the emergence of mutants in an engineered cell population (Figure 2D) [5,7,37]. For the same per cell mutation rate, the probability of having at least one mutant emerging increases with the population size [5,7,37]. Rugbjerg et al. used modeling and experiments to demonstrate that the final population in an industrial-scale fermenter was composed of 96% non-producer cells, but only 3% in a lab-scale culture [7]. To this end, minimized culture, such as microfluidic devices [37] or microencapsulation [5], can help preventing escape mutants as compared to large batch culture. The distribution of an engineered population into spatially-segregated compartments provides an additional benefit: when a mutant emerges, it is confined in a local community and it will not overtake the entire population [5].
Finally, ecological interactions can be exploited to reduce the probability of mutants arising. For example, Liao et al. demonstrated this notion by extending a population controller [15] into three engineered populations (R, P, and S) forming a rock-paper-scissors logic (Figure 2E) [38]. Each population carries a circuit to program autonomous population control. In addition, each can eliminate another in a mixture: P kills R, S kills P, and R kills S. By itself, each population is prone to being taken over by a mutant that has lost the population-control function. Before this happens, however, another population can be introduced to eliminate the previous population. Cycling through the three populations at the right pace can thus periodically reboot the common circuit function (population control), while suppressing emergence of escaping mutants [38].
3.2. Reducing the relative advantage of circuit mutants
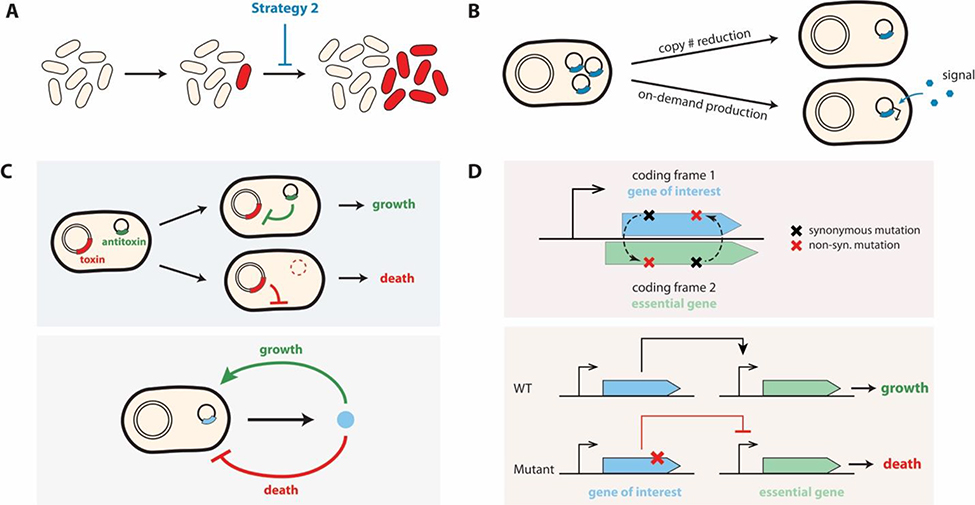
In general, the emergence of an escaping mutant is inevitable given enough time or a sufficiently large population. The second strategy to prevent a mutant’s dominance is to reduce its advantage relative to that of the wild type (Figure 3A), corresponding to reducing α (Figure 1E, Eqs 1&2). This reduction can be achieved by reducing the burden (increasing μW) or toxicity (reducing δW) of circuit activation, reducing the benefit of losing circuit function (reducing μM), or penalizing the mutant (increasing δM).
Figure 3. Reducing the growth advantage of a mutants.
(A) Schematic illustration.
(B) Reducing circuit-mediated metabolic burden, by reducing circuit copy number and on-demand circuit activation.
(C) Targeted killing of mutants. (Top) The toxin-antitoxin system induces cell death in the plasmid-free subpopulation that emerges due to plasmid segregation error. Toxin-producing host strain can survive only if carrying the plasmid encoding antitoxin. Once the cell loses the plasmid, toxin kills the host. (Bottom) A paradoxical circuit design, in which the same signal induces both cell growth and death, results in a biphasic effect of overall growth that depends on the signal intensity. In low-signal environments, cells with mutations that reduce their sensitivity to this signal are inherently selected against in this system, disfavoring the survival of such mutants.
(D) Direct coupling of circuit activation to survival. (Top) A gene of interest (GOI) and a survival gene is entangled by using an overlapping sequence; disruption of the GOI also disrupts the survival gene. (Bottom) Expression of a survival gene depends on the expression of the GOI; disruption of the latter would eliminate expression of the survival gene.
Tuning the synthetic gene expression level is one method to reduce the burden [14] (Figure 3B). Studies avoided overloading the host metabolism by reducing unnecessary transcriptional [39] and translational activities [14] or by reducing circuit copy number, which can be achieved by using low-copy plasmids (Figure 3B) [40] or chromosome integration (Figure 2B) [27,41]. Genome-integrated circuits can impose less burden compared to the plasmid-encoded counterpart, even if their copy numbers are similar [39,41]. This notion is related to the observations that genome-integrated genes often have lower basal expression level [29,41].
However, reducing the circuit copy number can be a double-edged sword. Although a low copy number reduces the chance of burden-driven circuit failure, a high copy number may better conserve the circuit function when failure occurs [8,26,29]. If the device is duplicated in multiple independent integration sites on the host chromosome, the host cell has to break all copies to lose the circuit function completely [7,8,28,29]. To this end, Caliando and Voigt increased the copy numbers of arabinose repressor and cas genes to inhibit their complete loss-of-function over several months[29].
Another method of avoiding an excess of synthetic circuit-derived stress is to implement an inducible or dynamic control [2,3,8,26,29]. Such on-demand production restricts the burdensome and toxic protein production to be active only when needed. Riglar et al. genomically integrated their engineered construct, which was activated in the presence of tetrathionate, a signal for inflammation in the gut [42]. Their circuit remained genetically stable for 6 months under both a continuous induction and infection-inducible inflammation in two different murine models. Ceroni et al. developed a burden-driven gene expression regulation system, where the target protein production rate is negatively regulated by the increasing host burden via dCas9-mediated feedback regulation [43]. Their design ensures that the host does not experience excessive metabolic stress caused by circuit activation.
The toxin-antitoxin (TA) system is a common mechanism found in nature that augments plasmid prevalence. Because the antitoxin degrades more rapidly than the toxin, the plasmid-free daughter cells are targeted via post-segregational killing (Figure 3B) [44]. Several studies have exploited the TA mechanism to enhance the long-term stability of their engineered plasmids [2,3,9,20]. For example, Danino et al. applied a two-pronged approach to maintaining the stability of programmable probiotics for cancer detection in urine in mice [9]. To prevent segregation error, their construct consists of a hok/sok TA system, as well as actin-like protein (ALP). ALP produces cytoskeletal structures to promote the bipolar distribution of plasmids during cell division, reducing the probability of segregation errors [9]. Combining the TA system with bacteriocins can improve the plasmid stabilization. The hok/sok system was used in conjugation with microcin-V, a bacteriocin, that is secreted into the environment providing population level control by killing plasmid-free cells [20]. The efficacy of this approach was demonstrated in murine tumor xenograft model to demonstrate plasmid stability in an in vivo application [20].
Gallagher et al. used an EcoRI endonuclease-EcoRI methylase pair to implement an analogous safeguard mechanism, where EcoRI methylase protects the host from the nuclease-mediated cell death only in the presence of exogenously supplemented synthetic signal [26]. This approach, when layered with riboregulation of essential genes, reduced the frequency of escape mutants to <1.3 ×10−12 over 110 generations as compared to ~10−6 for all of the initial ribo-essential strains. In another study, a different type of endonuclease, Cas9, was used to selectively kill mutants carrying known genomic single nucleotide polymorphism mutations using complementary guide-RNAs [45].
Biphasic regulation of a target gene can enhance circuit stability when implemented with a paradoxical regulatory motif. In paradoxical regulation, an input signal promotes both growth and death. Once genetic variants lose stable control of intracellular homeostasis, they are penalized with a significant growth defect [46]. In human tissues, a transcriptional factor c-Myc paradoxically induces apoptosis as part of the intrinsic mutant prevention strategy for tumor suppression [46]. Ma et al. recently applied this motif to a population control circuit in a mammalian system, in which the toxin was induced by a quorum-sensing molecule, auxin. They demonstrated that the paradoxical control suppressed mutants that lost the engineered auxin sensing module, thus enhancing circuit stability (Y Ma et al., bioRxiv doi: 10.1101/2020.09.02.278564).
A target gene encoding a burdensome or toxic protein can also be entangled with a survival gene by using an overlapping sequence. As such, loss or interruption of the target gene can cause death by simultaneously breaking the survival gene (Figure 3D). The role of genetic entanglement in enhancing genetic fidelity has been recognized in a number of natural systems [47,48]. Blazejewski et al. recently demonstrated the use of this strategy to enhance genetic stability of engineered systems [48]. The investigators used computational methods to design de novo overlapping sequences to entangle their gene of interest with an essential gene. Their design restricts evolution of the target gene, as even a synonymous mutation can detrimentally influence the host fitness [48]. This entanglement can also be applied to regulatory elements. Yang et al. constructed a bidirectional promoter, in which the forward and reverse promoters are overlapped. They demonstrated a four- to ten-fold increase in the evolutionary half-life of their target gene when an essential gene and the target gene were expressed from either side of this bidirectional promoter [23].
The exogeneous gene expression can also be coupled with host fitness by locating the circuit in the same orientation between host essential genes. Consequently, the integrated genes are less prone to complete loss via recombination, which leads to cell death [8]. However, this is not universally true, as the host transcription and circuit performance can interfere with each other depending on the genome location, thus reducing circuit integrity and host fitness [39]. Another approach is to express an essential gene under a promoter that is inducible by the target synthetic protein (Figure 3D). This “synthetic addiction” strategy penalizes escape mutants lacking functional target protein [4].
Synthetic auxotrophs are frequently used as a method of biocontainment. The host essential genes are recoded to incorporate non-standard amino acids (NSAA). These strains are hindered from escaping the programmed containment, as losing the engineered features will result in death in the absence of the environmentally supplemented NSAAs [27,49].
4. Conclusion
“The optimist invents the aeroplane, the pessimist the parachute.”
— George Bernard Shaw
In the backdrop of the extensive efforts to generate new circuit functions and expand the scope of their applications, there have been limited endeavors to develop strategies for enhanced circuit stability. Yet, being able to ensure long-term operation of a circuit has implications both for practical applications (as noted above) and for basic understanding of biology. For instance, many microbes undergo autonomous lysis at high densities [50], which has inspired the engineering of a population controller and its variants. That this phenomenon persists over evolutionary time scales begs the question of how nature is able to maintain the genetic stability of the underlying circuit. Analysis of corresponding engineered circuits can provide insights into the understanding their natural system.
Examples presented here reveal a set of design patterns that are rooted in modulating the evolutionary dynamics between the functional and mutant populations. In many cases, however, while the design choices are appropriate, quantitative analysis of the efficacy of circuit stabilization by each chosen strategy is generally lacking. In certain cases, tradeoff exists such that a design can either enhance or reduce the stability of a circuit. Filling this gap through systematic modeling and experiments represents a major frontier in synthetic biology. We anticipate that these efforts will benefit from drawing inspiration for molecular implementations from a myriad of natural systems and from theories and conceptual frameworks rooted in ecology and evolutionary biology.
Acknowledgements
We thank Ryan Tsoi and Helena Ma for commenting on the manuscript.
Funding
Research in our lab related to the topic of this article is in part supported by the National Science Foundation, the Office of Naval Research, the Air Force Office of Scientific Research, the National Institutes of Health, and the David and Lucile Packard Foundation.
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bashor CJ, Collins JJ: Understanding Biological Regulation Through Synthetic Biology. Annu Rev Biophys 2018, 47:399–423. [DOI] [PubMed] [Google Scholar]
- 2.Stirling F, Naydich A, Bramante J, Barocio R, Certo M, Wellington H, Redfield E, O’Keefe S, Gao S, Cusolito A, et al. : Synthetic Cassettes for pH-Mediated Sensing, Counting, and Containment. Cell Rep 2020, 30:3139–3148 e3134.• Enhanced biocontainment efficacy using a 2-factor containment system combining pH- and temperature-sensitive kill switches, demonstrating advances in synthetic biology allowing for higher-order circuits. The authors improved stability by balancing toxin-antitoxin expression levels using degenerate bases in the promoter RNA polymerase and RBSs of the toxin and antitoxin.
- 3.Stirling F, Bitzan L, O’Keefe S, Redfield E, Oliver JWK, Way J, Silver PA: Rational Design of Evolutionarily Stable Microbial Kill Switches. Mol Cell 2017, 68:686–697 e683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rugbjerg P, Sarup-Lytzen K, Nagy M, Sommer MOA: Synthetic addiction extends the productive life time of engineered Escherichia coli populations. Proc Natl Acad Sci U S A 2018, 115:2347–2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dai Z, Lee AJ, Roberts S, Sysoeva TA, Huang S, Dzuricky M, Yang X, Zhang X, Liu Z, Chilkoti A, et al. : Versatile biomanufacturing through stimulus-responsive cell-material feedback. Nat Chem Biol 2019, 15:1017–1024.• Encapsulated an engineered ‘microbial swarmbot’ that underwent a partial lysis at a high density to release the protein products of interest in response to the local chemical environment. The population size was reduced by discrete spatial compartments, which reduced the prabability of mutant emergence and prevented mutants from outcompeting the wildtype population if mutants did emerge.
- 6.Rugbjerg P, Sommer MOA: Overcoming genetic heterogeneity in industrial fermentations. Nat Biotechnol 2019, 37:869–876. [DOI] [PubMed] [Google Scholar]
- 7.Rugbjerg P, Myling-Petersen N, Porse A, Sarup-Lytzen K, Sommer MOA: Diverse genetic error modes constrain large-scale bio-based production. Nat Commun 2018, 9:787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Isabella VM, Ha BN, Castillo MJ, Lubkowicz DJ, Rowe SE, Millet YA, Anderson CL, Li N, Fisher AB, West KA, et al. : Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nat Biotechnol 2018, 36:857–864. [DOI] [PubMed] [Google Scholar]
- 9.Danino T, Prindle A, Kwong GA, Skalak M, Li H, Allen K, Hasty J, Bhatia SN: Programmable probiotics for detection of cancer in urine. Sci Transl Med 2015, 7:289ra284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eldar A, Elowitz MB: Functional roles for noise in genetic circuits. Nature 2010, 467:167–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorochowski TE, Chelysheva I, Eriksen M, Nair P, Pedersen S, Ignatova Z: Absolute quantification of translational regulation and burden using combined sequencing approaches. Mol Syst Biol 2019, 15:e8719.• Simultaneously quantified 779 translation initiation and 750 translation termination rates using RNA-seq and ribo-seq in absolute measurement units. The authors utilized the technology to assess and analyze the synthetic circuit-mediated burden on the host cells.
- 12.Tan C, Marguet P, You L: Emergent bistability by a growth-modulating positive feedback circuit. Nat Chem Biol 2009, 5:842–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu F, Menn DJ, Wang X: Quorum-sensing crosstalk-driven synthetic circuits: from unimodality to trimodality. Chem Biol 2014, 21:1629–1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ceroni F, Algar R, Stan GB, Ellis T: Quantifying cellular capacity identifies gene expression designs with reduced burden. Nat Methods 2015, 12:415–418. [DOI] [PubMed] [Google Scholar]
- 15.You L, Cox RS 3rd, Weiss R, Arnold FH: Programmed population control by cell-cell communication and regulated killing. Nature 2004, 428:868–871. [DOI] [PubMed] [Google Scholar]
- 16.Chan CT, Lee JW, Cameron DE, Bashor CJ, Collins JJ: ‘Deadman’ and ‘Passcode’ microbial kill switches for bacterial containment. Nat Chem Biol 2016, 12:82–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorochowski TE, Espah Borujeni A, Park Y, Nielsen AA, Zhang J, Der BS, Gordon DB, Voigt CA: Genetic circuit characterization and debugging using RNA-seq. Mol Syst Biol 2017, 13:952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sleight SC, Bartley BA, Lieviant JA, Sauro HM: Designing and engineering evolutionary robust genetic circuits. J Biol Eng 2010, 4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xiao Y, Bowen CH, Liu D, Zhang F: Exploiting nongenetic cell-to-cell variation for enhanced biosynthesis. Nat Chem Biol 2016, 12:339–344. [DOI] [PubMed] [Google Scholar]
- 20.Fedorec AJH, Ozdemir T, Doshi A, Ho YK, Rosa L, Rutter J, Velazquez O, Pinheiro VB, Danino T, Barnes CP: Two New Plasmid Post-segregational Killing Mechanisms for the Implementation of Synthetic Gene Networks in Escherichia coli. iScience 2019, 14:323–334.• Multiplexed the axe/txe TA and microcin-V bacteriocin systems to reduce segregation error-mediated plasmid instability in the probiotic E. coli strain Nissle 1917. The authors quantified the enhanced plasmid stability in an in vivo mouse cancer model using their multiplexed plasmid stabilization system and Bayesian statistical modeling methods.
- 21.Fernandez-Rodriguez J, Yang L, Gorochowski TE, Gordon DB, Voigt CA: Memory and Combinatorial Logic Based on DNA Inversions: Dynamics and Evolutionary Stability. ACS Synth Biol 2015, 4:1361–1372. [DOI] [PubMed] [Google Scholar]
- 22.Sleight SC, Sauro HM: Randomized BioBrick assembly: a novel DNA assembly method for randomizing and optimizing genetic circuits and metabolic pathways. ACS Synth Biol 2013, 2:506–518. [DOI] [PubMed] [Google Scholar]
- 23.Yang S, Sleight SC, Sauro HM: Rationally designed bidirectional promoter improves the evolutionary stability of synthetic genetic circuits. Nucleic Acids Res 2013, 41:e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Umenhoffer K, Feher T, Baliko G, Ayaydin F, Posfai J, Blattner FR, Posfai G: Reduced evolvability of Escherichia coli MDS42, an IS-less cellular chassis for molecular and synthetic biology applications. Microb Cell Fact 2010, 9:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geng P, Leonard SP, Mishler DM, Barrick JE: Synthetic Genome Defenses against Selfish DNA Elements Stabilize Engineered Bacteria against Evolutionary Failure. ACS Synth Biol 2019, 8:521–531. [DOI] [PubMed] [Google Scholar]
- 26.Gallagher RR, Patel JR, Interiano AL, Rovner AJ, Isaacs FJ: Multilayered genetic safeguards limit growth of microorganisms to defined environments. Nucleic Acids Res 2015, 43:1945–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandell DJ, Lajoie MJ, Mee MT, Takeuchi R, Kuznetsov G, Norville JE, Gregg CJ, Stoddard BL, Church GM: Biocontainment of genetically modified organisms by synthetic protein design. Nature 2015, 518:55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santos CN, Regitsky DD, Yoshikuni Y: Implementation of stable and complex biological systems through recombinase-assisted genome engineering. Nat Commun 2013, 4:2503. [DOI] [PubMed] [Google Scholar]
- 29.Caliando BJ, Voigt CA: Targeted DNA degradation using a CRISPR device stably carried in the host genome. Nat Commun 2015, 6:6989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lopatkin AJ, Meredith HR, Srimani JK, Pfeiffer C, Durrett R, You L: Persistence and reversal of plasmid-mediated antibiotic resistance. Nat Commun 2017, 8:1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Posfai G, Plunkett G 3rd, Feher T, Frisch D, Keil GM, Umenhoffer K, Kolisnychenko V, Stahl B, Sharma SS, de Arruda M, et al. : Emergent properties of reduced-genome Escherichia coli. Science 2006, 312:1044–1046. [DOI] [PubMed] [Google Scholar]
- 32.Ostrov N, Landon M, Guell M, Kuznetsov G, Teramoto J, Cervantes N, Zhou M, Singh K, Napolitano MG, Moosburner M, et al. : Design, synthesis, and testing toward a 57-codon genome. Science 2016, 353:819–822. [DOI] [PubMed] [Google Scholar]
- 33.Suarez GA, Renda BA, Dasgupta A, Barrick JE: Reduced Mutation Rate and Increased Transformability of Transposon-Free Acinetobacter baylyi ADP1-ISx. Appl Environ Microbiol 2017, 83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nyerges A, Balint B, Cseklye J, Nagy I, Pal C, Feher T: CRISPR-interference-based modulation of mobile genetic elements in bacteria. Synth Biol (Oxf) 2019, 4:ysz008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liang P, Zhang Y, Xu B, Zhao Y, Liu X, Gao W, Ma T, Yang C, Wang S, Liu R: Deletion of genomic islands in the Pseudomonas putida KT2440 genome can create an optimal chassis for synthetic biology applications. Microb Cell Fact 2020, 19:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Deatherage DE, Leon D, Rodriguez AE, Omar SK, Barrick JE: Directed evolution of Escherichia coli with lower-than-natural plasmid mutation rates. Nucleic Acids Res 2018, 46:9236–9250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balagadde FK, You L, Hansen CL, Arnold FH, Quake SR: Long-term monitoring of bacteria undergoing programmed population control in a microchemostat. Science 2005, 309:137–140. [DOI] [PubMed] [Google Scholar]
- 38.Liao MJ, Din MO, Tsimring L, Hasty J: Rock-paper-scissors: Engineered population dynamics increase genetic stability. Science 2019, 365:1045–1049.•• Expanded on the use of population control circuits and manipulated inhibitory ecological interactions among the engineered subpopualations to safeguard against the evolution of mutants.
- 39.Park Y, Espah Borujeni A, Gorochowski TE, Shin J, Voigt CA: Precision design of stable genetic circuits carried in highly-insulated E. coli genomic landing pads. Mol Syst Biol 2020, 16:e9584.•• Identified E. coli genomic locations in which integrated synthetic circuits are highly expressive but do not adversely affect host endogenous gene expression. By flanking the genome-integrated circuit by double terminators, the authors reduced cirucit-derived burden and enhanced its long-term stability.
- 40.Sleight SC, Sauro HM: Visualization of evolutionary stability dynamics and competitive fitness of Escherichia coli engineered with randomized multigene circuits. ACS Synth Biol 2013, 2:519–528. [DOI] [PubMed] [Google Scholar]
- 41.Lee JW, Gyorgy A, Cameron DE, Pyenson N, Choi KR, Way JC, Silver PA, Del Vecchio D, Collins JJ: Creating Single-Copy Genetic Circuits. Mol Cell 2016, 63:329–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Riglar DT, Giessen TW, Baym M, Kerns SJ, Niederhuber MJ, Bronson RT, Kotula JW, Gerber GK, Way JC, Silver PA: Engineered bacteria can function in the mammalian gut long-term as live diagnostics of inflammation. Nat Biotechnol 2017, 35:653–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ceroni F, Boo A, Furini S, Gorochowski TE, Borkowski O, Ladak YN, Awan AR, Gilbert C, Stan GB, Ellis T: Burden-driven feedback control of gene expression. Nat Methods 2018, 15:387–393. [DOI] [PubMed] [Google Scholar]
- 44.Hernandez-Arriaga AM, Chan WT, Espinosa M, Diaz-Orejas R: Conditional Activation of Toxin-Antitoxin Systems: Postsegregational Killing and Beyond. Microbiol Spectr 2014, 2. [DOI] [PubMed] [Google Scholar]
- 45.Chavez A, Pruitt BW, Tuttle M, Shapiro RS, Cecchi RJ, Winston J, Turczyk BM, Tung M, Collins JJ, Church GM: Precise Cas9 targeting enables genomic mutation prevention. Proc Natl Acad Sci U S A 2018, 115:3669–3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karin O, Alon U: Biphasic response as a mechanism against mutant takeover in tissue homeostasis circuits. Mol Syst Biol 2017, 13:933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Barrell BG, Air GM, Hutchison CA 3rd: Overlapping genes in bacteriophage phiX174. Nature 1976, 264:34–41. [DOI] [PubMed] [Google Scholar]
- 48.Blazejewski T, Ho HI, Wang HH: Synthetic sequence entanglement augments stability and containment of genetic information in cells. Science 2019, 365:595–598.•• Computationally designed de novo overlapping sequences to entangle the target gene with an essential gene and restricted the mutation accumulation within the designed genetic elements.
- 49.Rovner AJ, Haimovich AD, Katz SR, Li Z, Grome MW, Gassaway BM, Amiram M, Patel JR, Gallagher RR, Rinehart J, et al. : Recoded organisms engineered to depend on synthetic amino acids. Nature 2015, 518:89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lewis K: Programmed death in bacteria. Microbiol Mol Biol Rev 2000, 64:503–514. [DOI] [PMC free article] [PubMed] [Google Scholar]