Abstract
Sensitive, reliable and cost-effective detection of pathogens has wide ranging applications in clinical diagnostics and therapeutics, water and food safety, environmental monitoring, biosafety and epidemiology. Nucleic acid amplification tests (NAATs) such as PCR and isothermal amplification methods provide excellent analytical performance and significantly faster turnaround times than conventional culture-based methods. However, the inherent cost and complexity of NAATs limit their application in resource-limited settings and the developing world. To help address this urgent need, we have developed a sensitive method for nucleic acid analysis based on padlock probe rolling circle amplification (PLRCA), nuclease protection (NP) and lateral flow detection (LFA), referred to as PLAN-LFA, that can be used in resource-limited settings. The assay involves solution-phase hybridization of a padlock probe to target, sequence-specific ligation of the probe to form a circular template that undergoes isothermal rolling circle amplification in the presence of a polymerase and a labeled probe DNA. The RCA product is a long, linear concatenated single-stranded DNA that contains binding sites for the labeled probe. The sample is then exposed to a nuclease which selectively cleaves single-stranded DNA, the double-stranded labeled probe is protected from nuclease digestion and detected in a lateral flow immunoassay format to provide a visual, colorimetric readout of results. We have developed specific assays targeting beta-lactamase resistance gene for monitoring of antimicrobial resistance and Severe Acute Respiratory Syndrome-Coronavirus-2 (SARS-CoV-2, the novel coronavirus discovered in 2019) using the PLAN-LFA platform. The assay provides a limit of detection of 1.1 pM target DNA (or 1.3×106 copies/reaction). We also demonstrate the versatility and robustness of the method by performing analysis on DNA and RNA targets, and perform analysis in complex sample matrices like saliva, plant tissue extract and bacterial culture without any sample pretreatment steps.
1. Introduction
Accurate, reliable and efficient pathogen detection methods are critical in lowering the global burden of communicable diseases.1, 2 Conventional culture-based methods of pathogen detection have long turnaround times.2 Increasingly nucleic acid amplification tests (NAATs) such as polymerase chain reaction (PCR) and isothermal amplification techniques have been employed for sensitive, sequence-specific pathogen detection. NAATs amplify the amount of target nucleic acids in the sample enabling detection using fluorescent or colorimetric methods. PCR, the most utilized NAAT, involves three main steps, i.e., denaturation, annealing and extension performed at three different temperatures requiring a thermocycler for precise temperature control and cycling.3 PCR is also prone to inhibition from compounds present in unprocessed biological, environmental and food samples, requiring significant sample matrix-specific pre-processing steps.4–7 Isothermal amplification techniques such as recombinase polymerase amplification (RPA), template-mediated amplification (TMA), helicase-dependent amplification (HAD), loop-mediated isothermal amplification (LAMP) and rolling circle amplification (RCA) perform nucleic acid amplification at a fixed lower temperature, therefore do not require a thermocycler.3 Furthermore, isothermal techniques can be more robust and resilient than PCR to unprocessed samples, though they still require significant infrastructure, equipment and highly trained personnel to perform.8, 9 Inexpensive, portable, accurate and reliable nucleic acid sensor platforms that can be used by minimally trained personnel are needed to lower the global burden of communicable diseases and improve global health.3, 10
Rolling circle amplification offers a simple and versatile method for nucleic acid amplification frequently employed in biosensing, sequencing, and cloning applications.11, 12 RCA uses a small primer sequence to amplify a circular DNA template in the presence of specialty polymerases (like phi29 or Bst DNA polymerase) and deoxynucleotides (dNTPs).13 RCA can also be used to detect a short strand of nucleic acid using a circular primer in the presence of dNTPs and DNA polymerase. The DNA polymerases used in the RCA reactions possess both polymerase and strand displacement activity, and therefore the RCA product is a long, concatenated single stranded DNA with 100s to 1000s of tandem repeats.14–17 The RCA product has a high molecular weight (100–1000× of the primers) and can be detected using gel electrophoresis or fluorescence or colorimetric methods.11–13 Lateral flow assays offer a simple and cost-effective method for the visualization of results.18 RCA has also been combined with lateral flow assays for equipment-free detection.11, 13, 19, 20 Another popular variation of RCA is padlock rolling circle amplification, wherein, a linear DNA probe is specifically ligated in the presence of the target sequence forming a primed-circular template that can undergo rolling circle amplification in the presence of DNA polymerase.21–27 We previously reported a nucleic acid sensor platform based on nuclease protection and lateral flow detection.28 Nuclease protection or hybridization protection assay is a simple technique wherein post-denaturation and annealing of a labeled probe, the sample is exposed to a single-strand specific nuclease. The nuclease cleaves single-stranded DNA while perfectly hybridized dsDNA is protected from nuclease digestion. These protected probes can be detected using a lateral flow immunoassay format.28 This assay lacked the sensitivity needed for clinical diagnosis of communicable diseases, though the platform offered a simple, and inexpensive method for nucleic acid analysis.28
To improve the sensitivity of the assay we have developed a padlock rolling circle amplification, nuclease protection and lateral flow detection (PLAN-LFA). Combining padlock rolling circle amplification with our paper-based nuclease protection assay gave a 1000-fold improvement in the limit of detection of the assay.28 The PLAN-LFA assay provides 1.1 pM target DNA limit of detection in 3 h of analysis time and can be used to develop point-of-need nucleic acid sensor platforms. PLAN-LFA can be used for the detection of pathogens (human, animal or plant pathogens) and infectious disease diagnosis, to monitor the spread of antimicrobial resistance, and monitoring of nucleic acid biomarkers like miRNA. We have developed specific assays for beta-lactamase resistance gene detection to enable environmental monitoring of antimicrobial resistance, and for the detection of SARS-CoV-2.
The primer design for padlock rolling circle amplification is simpler than other isothermal amplification methods like LAMP.12 The assay can be performed in a variety of sample matrices, such as serum, urine, saliva, plant extract, without the need for sample pretreatment making it an attractive platform for development of point-of-need nucleic acid sensors.
2. Materials and Methods:
All DNA sequences used in the development of the PLAN-LFA assay shown in Table 1 were purchased from Integrated DNA Technologies Inc. (Coralville, IA, USA). T4 DNA ligase, adenosine triphosphate (ATP), P1 nuclease (from P. citrinum), Phi29 DNA polymerase and deoxynucleotides were purchased from New England Biolabs, Inc. (Ipswich, MA, USA). The supplied buffers were used for each of the ligation, amplification and nuclease protection steps of the assay. Nitrocellulose (FF120HP) and Whatman Grade 4 filter paper were purchased from GE Lifesciences (Pittsburgh, PA, USA). Streptavidin conjugated to Horseradish Peroxidase (Strep-HRP) was purchased from R&D Systems, Inc. (Minneapolis, MN). Pierce™ 1-Step Ultra TMB-Blotting Solution was purchased from Thermo Scientific (Waltham, MA). Anti-digoxigenin monoclonal antibody was purchased from Abcam, Inc (Cambridge, MA). StabilGuard® Immunoassay Stabilizer (BSA-free) was purchased from Surmodics, Inc. (Eden Prairie, MN). Trehalose dihydrate was purchased from EMD Millipore (Burlington, MA). Glycerol was purchased from Mallinkrodt (Staines-upon-Thames, United Kingdom).
Table 1:
Oligonucleotide padlock probe and primers used for PLAN-LFA
| Sequence Name | DNA sequence (5’ – 3’) |
|---|---|
| Padlock Probe (Beta-lactamase resistance) | 5’Phosphate/CAACGATCAAGGCGAGAGATTTAGGTGACACTATAGCGCCCCTATAGTGAGTCGTATTAGCGTTCAGCTCCGGTTCC |
| Beta-lactamase resistance target | 5’/CATGTAACTCGCCTTGATCGTTGGGAACCGGAGCTGAATGAAGCCATAC/3’ |
| Nuclease Protection Probe 1 | 5’biotin/GATTTAGGTGACACTATAG/3’Digoxigenin |
| Padlock probe (SARS-CoV-2) | /5’Phosphate/GGGCAAATTGTAAAAAAAAAAAAAAAAAAAAAAAAAAAAAACTGAAGCGCTGG |
| SARS-CoV-2 Target | 5’/TTACAAACATTGGCCGCAAATTGCACAATTTGCCCCCAGCGCTTCAGCGTTCTTCGGAATGTCGCGC/3’ |
| Nuclease Protection Probe 2 | /5’Biotin/AAAAAAAAAAAAAAAAAAAA/3’Digoxigenin |
2.1. PLAN-LFA Assay
In a 200 mL centrifuge tube, 2 mL of 10X ligation buffer, 5 mL of 100 nM Padlock Probe, 2 mL of varying concentration of target DNA and 11 mL of nuclease-free water was added. The sample was denatured at 95°C for 2 min and hybridization was performed at 37°C for 10 min. After hybridization, 2 mL of 10 mM adenosine triphosphate (ATP) and 20U of T4 DNA ligase were added to the reaction. Ligation was performed at 37°C for 30 min. After ligation, 3 mL of 10X Phi29 buffer, 0.5 mL of 10 mM dNTP mix, 0.5 mL of 10 mg/mL bovine serum albumin (BSA) and 20U of Phi29 DNA polymerase were added to the reaction. Rolling circle amplification was performed at 30°C for 2 h. After amplification 2 mL of 100 nM nuclease protection probe was added to the reaction, and the sample was denatured at 95°C for 2 min and annealed at 37°C for 10 min. Finally, 4 mL of 10X P1 nuclease buffer, and 20U of P1 nuclease were added to the reaction and the reaction was incubated at 37°C for 20 min. Then, 10 mL of the reaction were pipetted on to the lateral flow strip inlet, followed by wash buffer (100 mM Tris buffer saline + 0.05% Tween20), 10 mL of 1:200 diluted strep-HRP enzyme, 10 mL of wash buffer, and 20 mL of 1-step Ultra TMB blotting solution. After the development of the colorimetric signal, the lateral flow strips were scanned using an Epson Perfection V600 scanner. ImageJ (open-source image processing software, National Institute of Health) was used to analyze the images and quantitate the colorimetric signals.
2.2. Assay Feasibility
The PLAN-LFA assay was run in the presence of all the necessary components, and without either the padlock probe or target DNA or T4 DNA ligase or the Phi29 DNA polymerase to show that the assay does not produce non-specific signal in the absence of one of the constituents. 5 mL of the undigested samples were run on a 1% agarose gel in duplicates to analyze the results of the amplification reaction. Gel electrophoresis was run in 0.5X Tris Acetate EDTA buffer, at 120V for 30 minutes. Nuclease digested and undigested samples were also run on the lateral flow strips along with a negative control (no Target DNA).
2.3. Analytical Performance
Different concentrations (100 nM, 1 nM, 10 pM, 100 fM, 1 fM) of target DNA solutions were prepared in DI water along with negative controls (no target and non-target (random) DNA of equal length as the target). 2 mL of each of these solutions were added to the PLAN-LFA reactions. Assay limit of detection was calculated using mean blank + 3.3×standard deviation (). All samples were tested in triplicate. The lateral flow strips were scanned after completion of the test. The color intensity of the developed signal was quantified using ImageJ.
2.4. Matrix Effects
Human saliva was collected from a healthy volunteer into a sterile tube and spiked with the target oligonucleotide. PLAN-LFA were run with positive (10 pM SARS-CoV-2 target) and negative (no target) samples in undiluted human saliva as per the procedure outlined above. Target and no target samples prepared in 1X PBS were used as controls for the experiment. The colored signal obtained in the paired saliva and buffer positive and negative samples were compared to determine the effect of sample type on assay performance. To prepare cacao plant extract, five-0.5 cm cacao leaf punches were added to 1 mL PBS and heated at 98°C for 5 min. The extract was cooled to room temperature, aliquoted and stored at –20°C until use. Positive (10 pM beta-lactamase resistance target oligo) and negative (no target) samples were prepared in leaf extract and buffer; and tested using the assay protocol described above. The color signal obtained from leaf extract and buffer samples was compared to determine the effect of plant extract on assay performance.
2.5. RNA detection using PLAN-LFA
The beta-lactamase padlock probe is designed to bind a sequence in the beta-lactamase resistance gene, and therefore also binds the mRNA expressed by ampicillin resistant cells in the presence of the antibiotics. To demonstrate that the PLAN-LFA assay can be used to detect a specific target RNA, we performed the assay on ampicillin-resistant cells and chloramphenicol resistant cells grown in the presence of ampicillin and chloramphenicol respectively. Briefly, E. coli (XL10-Gold) cells were transformed with pUC19 plasmid DNA (carries ampicillin resistance gene) by heat shocking the cells at 42°C. 20 μL of transformed cells were added to 5 mL of LB broth along with 5 μL of 50 mg/mL Ampicillin. The cells were grown overnight in a shaker incubator set to 37°C. E. coli (BL-21 (DE3) pLysS) cells with chloramphenicol resistance grown overnight in the presence of chloramphenicol was used as a negative control. Buffer samples spiked with ssDNA target oligo and no target controls were used as controls for the PLAN-LFA assay performance. Chloramphenicol resistant culture was also spiked with ssDNA target oligo was used to control for sample matrix effects arising from the presence of bacterial culture. All samples were tested in triplicates. No changes were made to the PLAN-LFA assay protocol for mRNA detection. To further support our claim that the PLAN-LFA assay can be utilized for RNA targets, we used the SARS-CoV-2 padlock probe to analyze SARS-CoV-2 genomic RNA. SARS-CoV-2 virus (USA-WA1/20, BEI Resources NR52281) was grown in Vero cells and RNA concentrations were determined via ddPCR using the Bio-Rad 2019-nCoV CDC ddPCR Triplex Probe Assay. SARS-CoV-2 RNA stock concentrations were determined to be 2.6×1010 copies/mL. Different concentrations of the SARS-CoV-2 RNA along with appropriate controls were tested using the PLAN-LFA assay protocol. The mean intensity of blank + 3.3 was used to calculate the limit of detection of the assay.
3. Results and Discussion
Beta-lactamase resistance genes and SARS-CoV-2 were used as model sequences to demonstrate PLAN-LFA assay feasibility and characterize assay performance. Padlock probe was designed such that the 5’ and 3’ ends of the probe are complementary to the target sequence; these ends of the probe are referred to as hybridization arms. The middle region of the padlock probe is referred to as the padlock probe backbone is designed such that the RCA product that is generated contains a region complementary to a biotin-and-digoxigenin-labeled nuclease protection probe, referred to as nuclease protection probe or NPA probe. A second padlock probe was designed based on a poly-adenine backbone sequence and targeting the novel coronavirus discovered in 2019 (SARS-CoV-2).29 Spacer sequences were added between the padlock probe hybridization arms and padlock probe backbone regions to minimize probe secondary structure formation, which is known to interfere with efficient ligation and rolling circle amplification.30, 31
3.1. Assay Feasibility
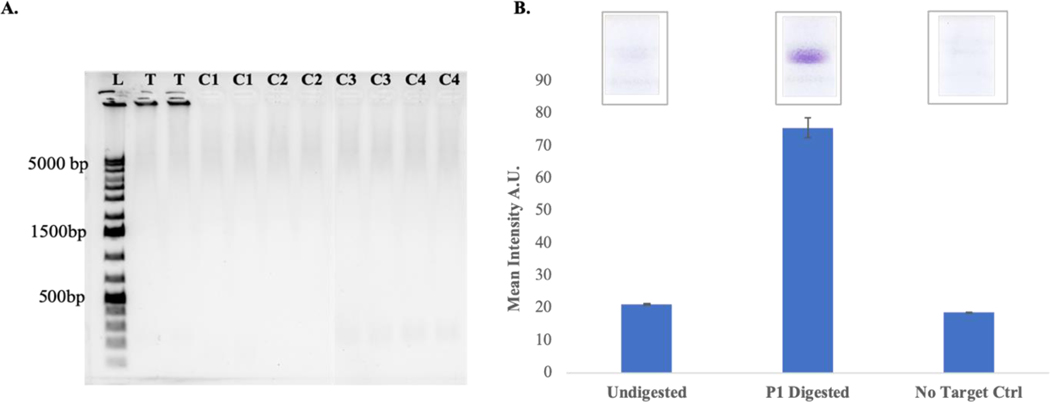
After determining the melting temperature and appropriate annealing temperature for the padlock probe and target sequences, assay feasibility was determined by performing the PLAN-LFA reactions in the presence of all assay components. Control reactions were run in the absence of one of the assay components. The samples were tested on a 1% agarose gel (0.5X Tris Acetate EDTA running buffer, 120 V for 35 min) in duplicate wells (Figure 2A). The RCA product is a large, single-stranded concatenated DNA which tends to remain in the well of the gel. RCA product is only observed in the Test condition (T) which includes all assay components. No RCA product was observed in the absence of the padlock probe (C1) or target DNA oligonucleotide (C2) or T4 DNA ligase (C3) or Phi29 DNA polymerase (C4). Nuclease protection probe was added to each of the target (T) and no target control conditions followed by P1 nuclease digestion of the samples. These samples were tested on lateral flow strips as per previously published protocol.28
Figure 2. PLAN-LFA Assay Feasibility.
A. Gel electrophoresis data showing large molecular weight RCA product is generated in the presence of all assay components (T) which remains in the well of the gel. No product is observed in the absence of the padlock probe (C1) or Target oligo (C2) or T4 DNA ligase (C3) or Phi29 DNA polymerase (C4). B. P1 nuclease digested and undigested samples were run on lateral flow strips (n =3).
The lateral flow strips (Figure 2B, upper) show that strong signal is produced when the samples are P1 nuclease digested while the undigested sample or no target control samples do not produce a signal. Given that the RCA product is a large, concatenated DNA, it is likely that the nuclease protection probe bound to its target site on the RCA product is unable to flow through the pores of the nitrocellulose membrane or the digoxigenin label on the nuclease protection probe is unavailable to the anti-digoxigenin antibody on the detection region of the strip. Upon P1 nuclease digestion, the protected NPA probe-hybrid becomes available for binding on the detection region. No signal is observed in the absence of the target DNA oligonucleotide indicating that the assay is specific to the target sequence.
3.2. Analytical Performance
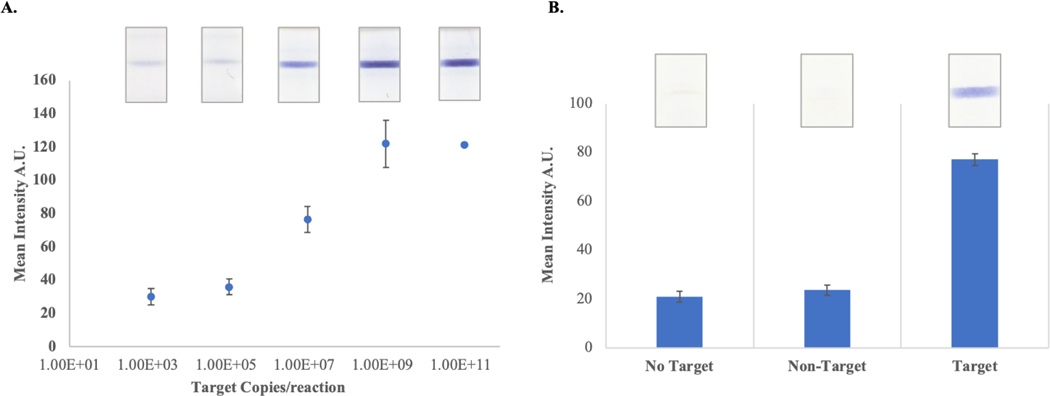
After showing that the PLAN-LFA was feasible, we studied the analytical performance of the assay. Sensitivity analysis was performed by running the assay in the presence of different amounts of target oligo (Figure 3A). Assay specificity was studied by performing reactions in the presence and absence of the target oligo and in the presence of a non-target oligonucleotide of similar length as the target.
Figure 3. PLAN-LFA Assay Performance.
A. Assay sensitivity was studied in the presence of increasing amount of target oligonucleotides. The color response observed for each sample is shown in the images above each data point. (n=3 per sample) B. Specificity analysis was performed by running the assay in the absence of target oligo and in the presence of non-target and target oligonucleotides. (n=3 per sample)
The limit of detection of the assay was 1.1 pM (2.2×10−18 moles or 1.3×106 copies) of target oligo, which is 20- to 40-fold improvement over other reported methods using linear rolling circle amplification and lateral flow detection.32 The assay also provides excellent specificity as no target and non-target conditions do not produce a signal, while strong colorimetric response is observed in the presence of target oligo (Figure 3B). These results indicate that the assay provides reasonable assay performance and can be applied for the detection of pathogenic nucleic acids in a simple, cost-effective assay that can be used by minimally trained personnel as it only requires a few reagent additions and visual colorimetric detection.
3.3. Sample Matrix Effects
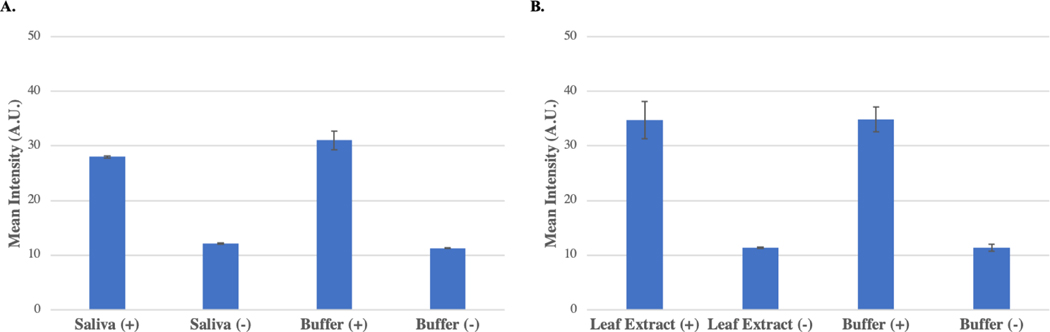
PCR can often produce false-negative results due to PCR inhibition caused by complex biomolecules present in samples. For example, polyphenolic compounds present in plant tissues are known inhibitors of PCR, and therefore detection of plant pathogens by PCR often requires laborious and costly sample pretreatment steps.33 To understand the effect of sample matrices on the performance of the PLAN-LFA assay, we performed analysis in saliva and cacao leaf extract. SARS-CoV-2 positive (10 pM) and negative samples were prepared by spiking saliva collected from a healthy individual, while positive beta-lactam resistance (10 pM) and negative samples were prepared by spiking cacao healthy leaf extract.
The color intensity of the positive samples was compared to the positive sample in buffer to determine if PLAN-LFA was affected by the presence of the complex sample matrix. Figure 4A shows that the assay performance in saliva was nearly identical to the performance in 1X PBS buffer samples with less than 10% decrease in signal intensity compared to the buffer control. Figure 4B shows that the assay performance was unaffected by the presence of cacao leaf extract. The negative samples were free of any background signal indicating that the presence of these complex sample matrices does not affect produce a false-positive response in the PLAN-LFA assay.
Figure 4. Sample Matrix Effects:
A. Positive (10pM) and negative samples prepared in saliva and buffer were tested using the PLAN-LFA assay protocol (n = 3 per sample). B. Positive (10 pM) and negative samples prepared in cacao leaf extract and buffer were tested using the PLAN-LFA assay protocol (n=3 per sample).
3.4. RNA Detection using PLAN-LFA
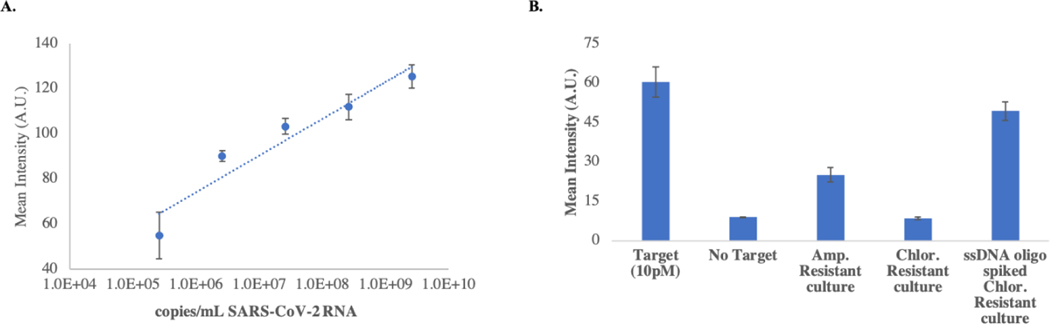
To demonstrate the versatility of the PLAN-LFA assay, we performed analysis on SARS-CoV-2 genomic RNA using the SARS-CoV-2 padlock probe. T4 DNA ligase can ligate the DNA padlock probe when in a DNA-RNA hybrid, and therefore no changes to the protocol or components were necessary to detect RNA targets. Different concentrations of purified SARS-CoV-2 RNA were prepared in water and tested using the protocol described in the methods section. A limit of detection of approximately 1.78×105 copies/mL of SARS-CoV-2 RNA was obtained using the mean intensity of the blank + 3.3. We also tested bacterial cultures of ampicillin resistant cells and chloramphenicol resistant cells were tested using the beta-lactam padlock probe. Figure 5 shows the mean intensity obtained from the lateral flow strips run with different samples. The ampicillin resistant cells produced a positive signal while chloramphenicol resistant culture did not produce a positive signal. The ssDNA oligo spiked (10 pM) in chloramphenicol resistant culture gave a strong color response indicating that the PLAN-LFA assay is unaffected by the presence of bacterial culture.
Figure 5. RNA detection using PLAN-LFA:
A. Dose-response with purified SARS-CoV-2 RNA. Using blank + 3.3 a limit of detection of approximately 1.78×105 copies/mL was obtained. B. PLAN-LFA performed on bacterial cultures of E. coli transformed with ampicillin resistance (test condition) and chloramphenicol resistance (negative control). ssDNA oligo target positive (10 pM) and negative samples were used to control for assay performance. The ssDNA oligo spiked in chloramphenicol resistant culture was used to control for sample matrix effects.
Though the signal intensity obtained with the ampicillin resistant culture was fainter than expected, the clear difference between positive and negative cultures indicates that the PLAN-LFA assay can be used to detect RNA of interest. It should also be noted that no changes were made to the assay protocol to perform RNA detection. Assay performance for RNA detection can be improved by altering the assay protocol specifically for RNA targets by optimizing denaturation and hybridization conditions, along with the use of other ligases (T2 and T4 RNA ligase) that may improve ligation efficiency for RNA targets.25, 34
4. Conclusions
We describe a nucleic acid sensor platform based on padlock probe rolling circle amplification, nuclease protection and lateral flow detection. The assay provides a visual colorimetric readout of results with the appearance of a test line if the sequence of interest is present in the sample. Semi-quantitation of the copy number is possible by performing image analysis on a photograph or scan of the lateral flow test strip further improving the utility of the test. The assay provides picomolar sensitivity (1.3×106 copies/reaction) and can be used to detect both DNA and RNA targets with excellent specificity. The assay does not require a cDNA synthesis step for RNA detection which is a significant advantage over LAMP and PCR where a reverse transcription step is necessary. We demonstrated assay feasibility by developing specific assays for SARS-CoV-2 and beta-lactamase resistance genes. While the assay limit of detection is too high for COVID-19 diagnostics, we believe the assay has value as a screening tool in nucleic acid sensing applications where higher pathogen loads would be expected for positive samples. The PLAN-LFA assay can be completed within 3 hours, making it an attractive platform for point-of-need nucleic acid sensing applications. We also demonstrate assay robustness by performing nucleic acid analysis in complex sample matrices like saliva, plant extract and bacterial culture without the need for any sample pretreatment steps. The probe design is simplified in PLAN-LFA compared to other isothermal amplification methods such as LAMP, as the padlock probe hybridization arms are complementary to the target sequence. Alternative targets can be assayed by simply changing the hybridization arm sequences, while a common padlock probe backbone region such as the poly-adenine sequence can be used to design probes. The assay sensitivity, turnaround time and ease-of-use would need to be further improved to make it more applicable for point-of-need nucleic acid sensing applications. Using exponential mode RCA or hyperbranched RCA can significantly improve both assay sensitivity and turnaround times.35 We are presently working on an exponential mode PLAN-LFA assay to further improve the assay sensitivity and turnaround times.
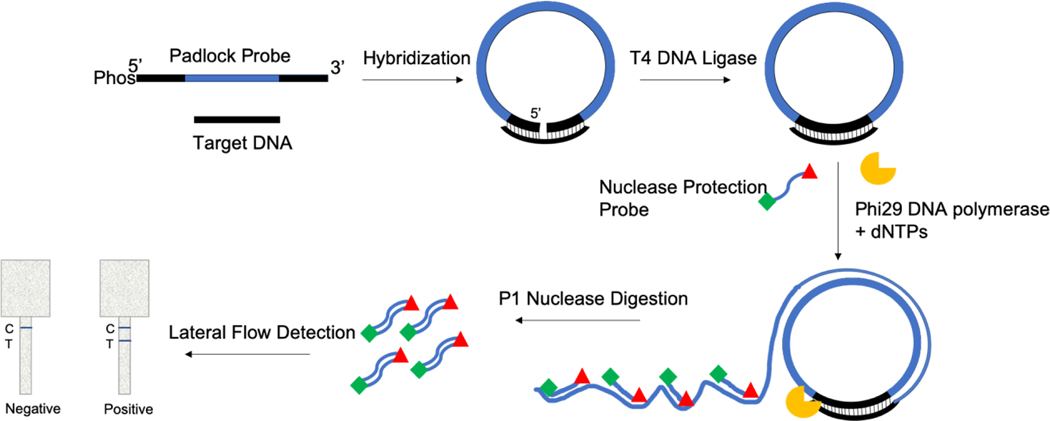
Figure 1. Padlock Rolling Circle Amplification Nuclease Protection Lateral Flow Assay (PLAN-LFA) Schematic.
Padlock probe hybridization with target DNA oligo enables sequence specific ligation of the 5’ and 3’ end of the padlock probe by T4 DNA ligase. The target-primed circular template undergoes rolling circle amplification in the presence of Phi29 DNA polymerase. The resulting ssDNA product binds the labeled nuclease protection probe. Upon P1 nuclease digestion, the protected labeled NPA probes are detected in an immunoassay format.
Acknowledgements
The authors acknowledge financial support for this project from the National Institutes of Health (R33ES024719 / R01AI132668).
Footnotes
Conflict of Interest
The authors have no conflicts to declare.
References
- 1.Mabey D, Peeling RW, Ustianowski A and Perkins MD, Nature Reviews Microbiology, 2004, 2, 231. [DOI] [PubMed] [Google Scholar]
- 2.Peeling R and Mabey D, Clinical microbiology and infection, 2010, 16, 1062–1069. [DOI] [PubMed] [Google Scholar]
- 3.Abou Tayoun AN, Burchard PR, Malik I, Scherer A and Tsongalis GJ, American journal of clinical pathology, 2014, 141, 17–24. [DOI] [PubMed] [Google Scholar]
- 4.Schrader C, Schielke A, Ellerbroek L and Johne R, Journal of applied microbiology, 2012, 113, 1014–1026. [DOI] [PubMed] [Google Scholar]
- 5.Rådström P, Knutsson R, Wolffs P, Lövenklev M and Löfström C, Molecular biotechnology, 2004, 26, 133–146. [DOI] [PubMed] [Google Scholar]
- 6.Jiang J, Alderisio KA, Singh A and Xiao L, Applied and environmental microbiology, 2005, 71, 1135–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rossen L, Nørskov P, Holmstrøm K and Rasmussen OF, International journal of food microbiology, 1992, 17, 37–45. [DOI] [PubMed] [Google Scholar]
- 8.Francois P, Tangomo M, Hibbs J, Bonetti E-J, Boehme CC, Notomi T, Perkins MD and Schrenzel J, FEMS Immunology & Medical Microbiology, 2011, 62, 41–48. [DOI] [PubMed] [Google Scholar]
- 9.Kaneko H, Kawana T, Fukushima E and Suzutani T, Journal of biochemical and biophysical methods, 2007, 70, 499–501. [DOI] [PubMed] [Google Scholar]
- 10.Drain PK, Hyle EP, Noubary F, Freedberg KA, Wilson D, Bishai WR, Rodriguez W and Bassett IV, The Lancet infectious diseases, 2014, 14, 239–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ali MM, Li F, Zhang Z, Zhang K, Kang D-K, Ankrum JA, Le XC and Zhao W, Chemical Society Reviews, 2014, 43, 3324–3341. [DOI] [PubMed] [Google Scholar]
- 12.Demidov VV, Expert review of molecular diagnostics, 2002, 2, 542–548. [DOI] [PubMed] [Google Scholar]
- 13.Zhao W, Ali MM, Brook MA and Li Y, Angewandte Chemie International Edition, 2008, 47, 6330–6337. [DOI] [PubMed] [Google Scholar]
- 14.Dean FB, Nelson JR, Giesler TL and Lasken RS, Genome research, 2001, 11, 1095–1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johne R, Müller H, Rector A, Van Ranst M and Stevens H, Trends in microbiology, 2009, 17, 205–211. [DOI] [PubMed] [Google Scholar]
- 16.Lagunavicius A, Merkiene E, Kiveryte Z, Savaneviciute A, Zimbaite-Ruskuliene V, Radzvilavicius T and Janulaitis A, Rna, 2009, 15, 765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li X-Y, Du Y-C, Zhang Y-P and Kong D-M, Scientific reports, 2017, 7, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koczula KM and Gallotta A, Essays in biochemistry, 2016, 60, 111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng Y, Zhang X, Li Z, Jiao X, Wang Y and Zhang Y, Angewandte Chemie International Edition, 2009, 48, 3268–3272. [DOI] [PubMed] [Google Scholar]
- 20.Haible D, Kober S and Jeske H, Journal of virological methods, 2006, 135, 9–16. [DOI] [PubMed] [Google Scholar]
- 21.Baner J, Nilsson M, Mendel-Hartvig M and Landegren U, Nucleic acids research, 1998, 26, 5073–5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He Y, Yang X, Yuan R and Chai Y, Analytical chemistry, 2017, 89, 2866–2872. [DOI] [PubMed] [Google Scholar]
- 23.Jonstrup SP, Koch J and Kjems J, Rna, 2006, 12, 1747–1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Larsson C, Koch J, Nygren A, Janssen G, Raap AK, Landegren U and Nilsson M, Nature methods, 2004, 1, 227–232. [DOI] [PubMed] [Google Scholar]
- 25.Liu H, Li L, Duan L, Wang X, Xie Y, Tong L, Wang Q and Tang B, Analytical chemistry, 2013, 85, 7941–7947. [DOI] [PubMed] [Google Scholar]
- 26.Sato K, Ishii R, Sasaki N, Sato K and Nilsson M, Analytical biochemistry, 2013, 437, 43–45. [DOI] [PubMed] [Google Scholar]
- 27.Thomas DC, Nardone GA and Randall SK, Archives of pathology & laboratory medicine, 1999, 123, 1170–1176. [DOI] [PubMed] [Google Scholar]
- 28.Noviana E, Jain S, Hofstetter J, Geiss BJ, Dandy DS and Henry CS, Analytical and Bioanalytical Chemistry, 2020, 1–11. [DOI] [PubMed] [Google Scholar]
- 29.Park KW, Lee CY, Batule BS, Park KS and Park HG, RSC advances, 2018, 8, 1958–1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohsen MG and Kool ET, Accounts of chemical research, 2016, 49, 2540–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Connor L and Glynn B, Fungal Diagnostics: Methods and Protocols, Springer, 2013. [Google Scholar]
- 32.Yao M, Lv X, Deng Y and Rasheed M, Analytica Chimica Acta, 2019, 1055, 115–125. [DOI] [PubMed] [Google Scholar]
- 33.Singh R, Nie X, Singh M, Coffin R and Duplessis P, Journal of Virological Methods, 2002, 99, 123–131. [DOI] [PubMed] [Google Scholar]
- 34.Zhuang J, Lai W, Chen G and Tang D, Chemical communications, 2014, 50, 2935–2938. [DOI] [PubMed] [Google Scholar]
- 35.Kaocharoen S, Wang B, Tsui KM, Trilles L, Kong F and Meyer W, Electrophoresis, 2008, 29, 3183–3191. [DOI] [PubMed] [Google Scholar]