Abstract
Alcohol is among the most widely used psychoactive substances worldwide. Ethanol metabolites such as acetate, thought to be primarily the result of ethanol breakdown by hepatic aldehyde dehydrogenase 2 (ALDH2), contribute to alcohol’s behavioural effects and alcoholism. Here, we show that ALDH2 is expressed in astrocytes in the mouse cerebellum and that ethanol metabolism by astrocytic ALDH2 mediates behavioural effects associated with ethanol intoxication. We show that ALDH2 is expressed in astrocytes in specific brain regions and that astrocytic, but not hepatocytic, ALDH2 is required to produce ethanol-derived acetate in the mouse cerebellum. Cerebellar astrocytic ALDH2 mediates low-dose ethanol-induced elevation of GABA levels, enhancement of tonic inhibition and impairment of balance and coordination skills. Thus, astrocytic ALDH2 controls the production, cellular and behavioural effects of alcohol metabolites in a brain-region-specific manner. Our data indicate that astrocytic ALDH2 is an important, but previously under-recognized, target in the brain to alter alcohol pharmacokinetics and potentially treat alcohol use disorder.
Alcohol is one commonly misused psychoactive substance causing notable pathology across the medical spectrum. Unlike other psychoactive chemicals such as opioids, nicotine and cannabinoids, ethanol does not bind to one specific target or receptor in the brain1,2. The metabolites of ethanol can produce psychoactive effects for longer durations than ethanol in the human body2,3. Human alcohol metabolism is controlled by genetic variations in the enzymes that break down alcohol. The most notable one is a genetic deficiency in ALDH2, the key enzyme for the conversion of acetaldehyde to acetate. This deficiency, which occurs in nearly half of the Asian population, causes people to experience elevated levels of blood acetaldehyde and a flushing reaction even with low doses of alcohol4,5. The liver is the primary organ for alcohol detoxification, although the overall picture of the central mechanism that underlies alcohol metabolism remains unclear. Most of the ethanol is oxidized by alcohol dehydrogenase (ADH) and ALDH2 in the liver6. However, the signal levels of both enzymes in the brain are very low6. Although catalase and CYP2E1 may oxidize ethanol in the brain7, the sensitivity of these enzymes to break down ethanol is limited especially at low ethanol exposure. ALDH1a1 is proposed to metabolize acetaldehyde; however, its role in alcohol metabolism in the brain has not been fully characterized8,9. In addition, understanding of the cell-type-specific distribution of ALDH2 in the brain is not clear.
At the behavioural level, much of the research on alcohol intermediate metabolites has focused on acetaldehyde, whose pattern of effects is similar to that of ethanol10. Until recently, acetate had been considered a harmless alcohol by-product, and brain acetate is thought to derive largely from liver alcohol metabolism11. There is strong evidence suggesting that acetate is primarily utilized by the astrocytes in the brain12. Acetate is also found to contribute to ethanol-induced acute intoxication and alcohol preference in mice13–15. Several neuroimaging studies have demonstrated that the uptake of acetate is significantly increased in the cerebellum and other brain areas after ethanol consumption in people with alcohol use disorder14,16. The cerebellum is a primary brain region involved in alcohol metabolism and alcohol motor impairment. However, recent studies provide strong evidence for cerebellar contributions to non-motor functions such as cognition, language, emotions and social behaviours17–19. The cerebellum also plays a role in many clinical neurological conditions, including attention deficit hyperactivity disorder, schizophrenia, bipolar disorder, major depressive disorder and anxiety disorders20–22. Relative to the most frequently investigated brain regions, for example, the amygdala and basal ganglia, the cerebellum has been largely overlooked regarding its impact on neurological mechanisms underlying alcohol use disorder.
Nevertheless, it remains unclear whether there is a central target that mediates ethanol-derived acetate production and acetate-induced behaviours. Specifically, we have only limited knowledge about the possible role of brain ALDH2 in alcohol metabolism and alcohol intoxication. One major barrier to our understanding of brain ALDH2 is a lack of specific approaches to distinguish between central and peripheral ALDH2-mediated effects. To address these questions, we implemented in vitro and in vivo techniques to explore the cell-type-specific distribution of ALDH2 in the cerebellum and the neurological mechanism by which central ALDH2 regulates alcohol metabolism and intoxication. We observed that ALDH2 was expressed in the cerebellum and astrocytes in humans and mice. Astrocytic ALDH2 mediated the production of the ethanol metabolite acetate and ethanol-induced cellular and behavioural effects in a brain-region-specific manner. These findings suggest that astrocytic ALDH2 is an important target that mediates alcohol metabolism and action in the brain.
Results
Astrocyte-specific expression of ALDH2 in cerebellum.
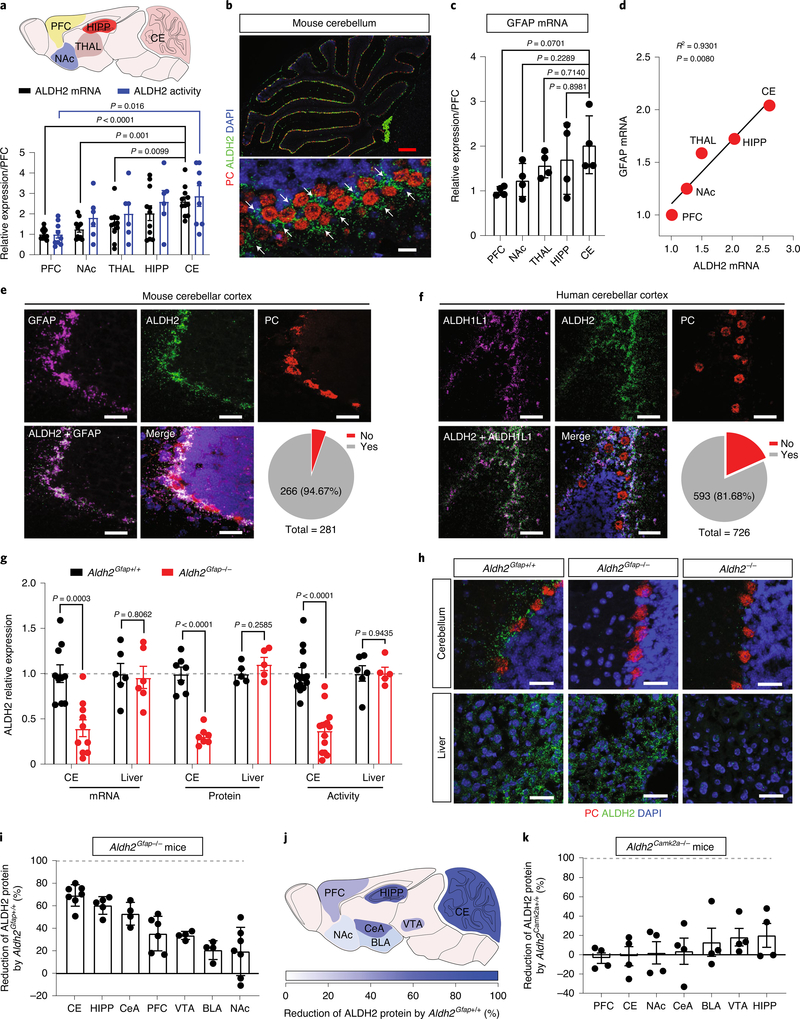
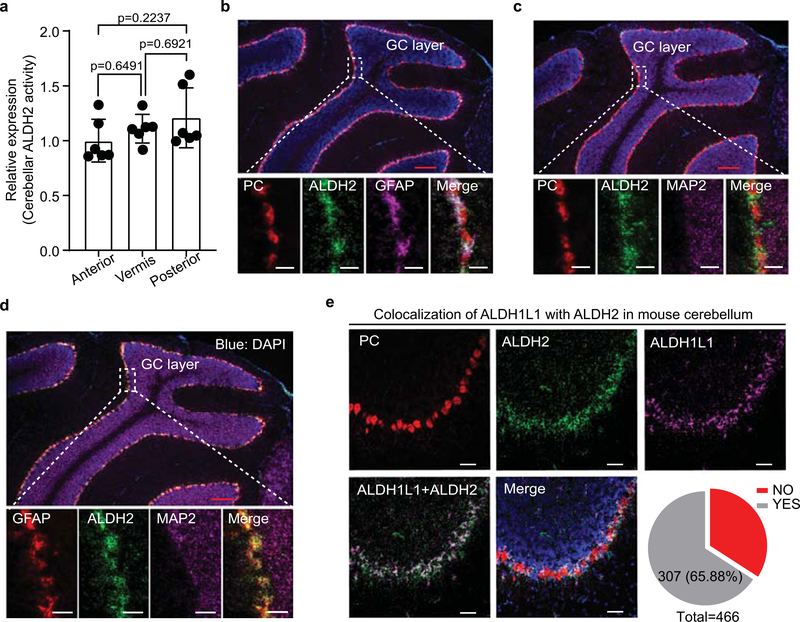
We first examined ALDH2 mRNA expression in different brain areas by using quantitative PCR (Fig. 1a). Among five tested regions, the expression level of ALDH2 mRNA was most abundant in the cerebellum and least abundant in the prefrontal cortex (PFC). A similar pattern was observed in a subsequent assay of ALDH2 enzymatic activity. Posterior cerebellum displayed the highest level of ALDH2 enzymatic activity among three different cerebellar subregions (Extended Data Fig. 1a). The picture of cell-type-specific distribution of brain ALDH2 remains unclear because a specific ALDH2 antibody for immunostaining of tissue slices is not available. To address this issue, we conducted RNA in situ hybridization using the RNAscope technique. A strong signal of ALDH2 was detected at low magnification in all lobes of mouse cerebellar sagittal slices (Fig. 1b). The ALDH2 signal was detected in the Purkinje cell layers identified by the signal of the gene encoding Purkinje cell protein 2 or L7 (Pcp2), a specific marker for Purkinje cells. However, further analysis at high magnification revealed that ALDH2 was not colocalized with either Purkinje cells or MAP2, a neuronal marker in cerebellar slices (Fig. 1b and Extended Data Fig. 1b,c). Instead, ALDH2 mRNA was primarily expressed in glial fibrillary acidic protein (GFAP)-positive cells (Extended Data Fig. 1d). There was a strong correlation between the levels of GFAP and ALDH2 mRNAs in five brain regions tested (Fig. 1c,d). We next quantified the colocalization of ALDH2 mRNA with two astrocyte biomarkers, GFAP and ALDH1L1, in cerebellar cortex. The majority of GFAP-positive cells (94.67%) and ALDH1L1-positive cells (65.88%) were colocalized with ALDH2-positive cells, whereas no colocalization was observed between Pcp2/L7 and ALDH2 (Fig. 1e and Extended Data Fig. 1e). Next, we asked if this astrocytic-specific distribution of ALDH2 is replicable in human tissues. Cerebellar tissues from four individuals were examined, and a strong ALDH2 signal was detected in cerebellar slices from two of the individuals. Around 81% of ALDH1L1-positive cells were found to express ALDH2 signal in human cerebellar cortex slices (Fig. 1f).
Fig. 1 |. Astrocytic ALDH2 expression in the cerebellum.
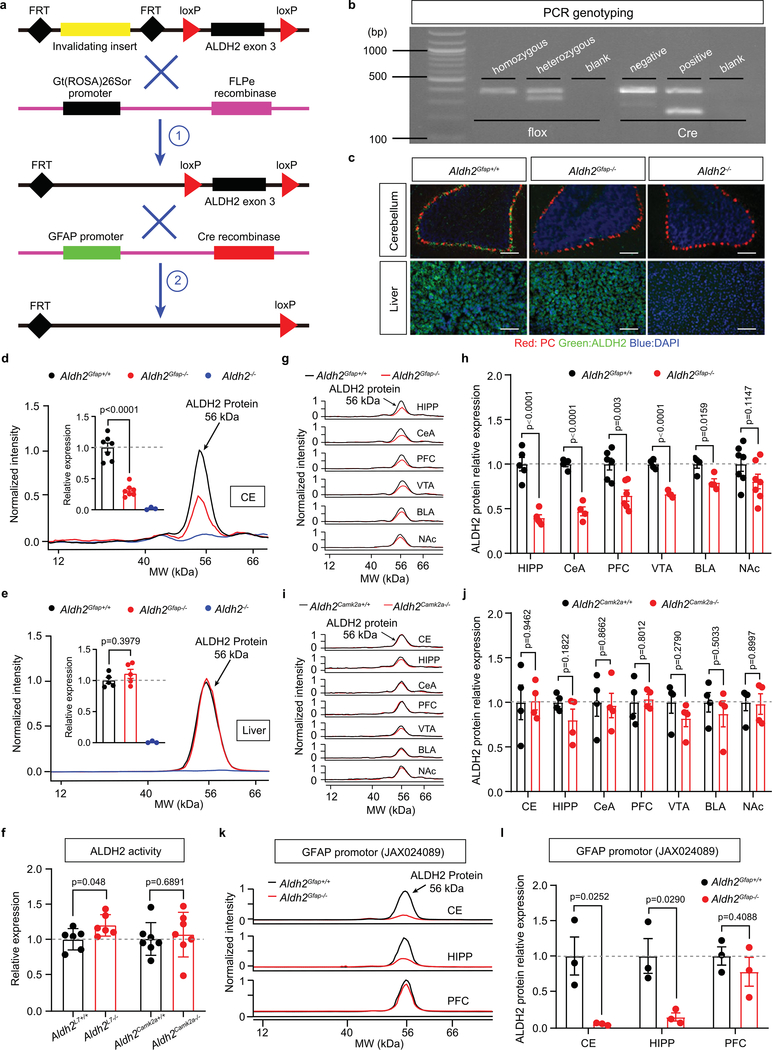
a, Upper: schematic diagram of sample collection from different brain regions. Lower: quantitative analysis of ALDH2 mRNA. Bar graphs represent the relative expression of ALDH2 mRNA (black) and ALDH2 enzymatic activity (blue). Data were plotted and normalized to values of the PFC (ALDH2 mRNA: n = 11 mice per group; ALDH2 enzymatic activity: n = 9, 6, 6, 6 and 9 mice per region, respectively). b, Microscopy imaging of mRNA signals in slices from GFAP–GFP mice. GFAP mRNA was detected using an EGFP probe (indicated by white arrows). Scale bars: red, 200 μm; white, 30 μm. c, Quantitative analysis of GFAP mRNA by RT–PCR. Data were normalized to values of PFC (n = 4 mice per group). d, Correlational analysis of the expression levels of ALDH2 and GFAP mRNA in various brain areas (P = 0.008 by linear regression). e,f, Confocal microscopy images showing the colocalization of ALDH2 mRNA with GFAP mRNA in mouse cerebellar slices (e) and ALDH2 mRNA with ALDH1L1 mRNA in human cerebellar slices (f). The grey area represents the colocalization between ALDH2 and GFAP or ALDH1L1. The experiment was repeated four times for each biologically independent mouse or human brain sample, with similar results obtained (n = 3 mice). More detailed information is provided in Supplementary Tables 1 and 2. Yes: colocalization; No: no colocalization; white scale bars, 50 μm. g, Bars represent a brain-specific reduction of ALDH2 mRNA, protein and enzymatic activity in Aldh2Gfap−/− mice. Data were normalized to values of Aldh2Gfap+/+ mice (ALDH2 mRNA: n = 10, 10, 6 and 6 mice, respectively; ALDH2 protein: n = 7, 7, 5 and 5 mice, respectively; ALDH2 enzymatic activity: n = 14, 14, 6 and 5 mice, respectively). h, Confocal microscopy imaging of cerebellar and liver ALDH2 in slices from Aldh2Gfap−/− and Aldh2−/− mice. Each experiment was repeated four times with similar results obtained. White scale bars, 50 μm. i, Measurement of brain ALDH2 proteins by using an automated size-based capillary western blot system in Aldh2Gfap−/− and Aldh2Gfap+/+ mice. Each bar represents the ratio (percentage change) of ALDH2 proteins in Aldh2Gfap−/− versus Aldh2Gfap+/+ mice (n = 7, 5, 4, 6, 4, 4 and 7 mice, respectively). j, Schematic of the relative deficiency of ALDH2 proteins in different brain regions from astrocytic ALDH2-deficient mice. k, Measurement of ALDH2 proteins in different brain areas in Aldh2Camk2a−/− and Aldh2Camk2a+/+ mice (n = 4 mice per group). All data are expressed as the mean ± s.e.m. Analysis was performed using an unpaired two-tailed Student’s t-test (g, i and k) or one-way analysis of variance (ANOVA) followed by Tukey’s test (a and c). CE, cerebellum; HIPP, hippocampus; CeA, central nucleus of the amygdala; VTA, ventral tegmental area; BLA, basolateral amygdala; NAc, nucleus accumbens; THAL, thalamus.
The impact of the alcohol metabolic pathway in the brain on human behaviour is poorly understood, in large part because approaches are lacking to distinguish between central and peripheral ALDH2-mediated effects. To address this issue, we generated cell-type-specific ALDH2-deficient mice by crossing three Cre mouse lines individually with Aldh2 flox mice23. Each of these Cre lines targets astrocytes, or two subsets of neurons (L7/Purkinje cells) and calmodulin-dependent protein kinase II alpha (Camk2a; Extended Data Fig. 2a,b). Significant reductions of cerebellar ALDH2 mRNA, protein and enzymatic activity levels were observed in Aldh2Gfap−/− mice (Fig. 1g,h and Extended Data Fig. 2c,d). No change was observed in liver ALDH2 mRNA, protein and enzymatic activity in Aldh2Gfap−/− mice (Fig. 1g,h and Extended Data Fig. 2c,e). No difference was observed in cerebellar ALDH2 enzymatic activity between Aldh2L7−/− mice, Aldh2Camk2a−/− mice and their wild-type littermates (Extended Data Fig. 2f). Aldh2−/− mice displayed a universal depletion of ALDH2 mRNAs and proteins from both brain and liver (Fig. 1h and Extended Data Fig. 2c–e). We next examined the levels of ALDH2 protein expression in selected brain areas that are relevant to ethanol-induced behavioural effects in Aldh2Gfap−/− mice. The reduction of ALDH2 protein was brain-region specific in Aldh2Gfap−/− mice. The magnitude of ALDH2 protein reduction in Aldh2Gfap−/− mice was calculated as a percentage change relative to their littermates, Aldh2Gfap+/+ mice. Consistent with our findings in PCR with reverse transcription (RT–PCR) and enzymatic assays, the largest reduction of ALDH2 protein levels was found in the cerebellum (Fig. 1i,j and Extended Data Fig. 2d,g,h). The rank order of the reduction of ALDH2 was: cerebellum > hippocampus > central nucleus of the amygdala > ventral tegmental area > basolateral amygdala > PFC > nucleus accumbens (Fig. 1i,j). No significant difference was observed across all seven brain areas between Aldh2Camk2a−/− mice and their wild-type littermates (Fig. 1k and Extended Data Fig. 2i,j). To carefully document cell-specific manipulation by Cre/lox site-specific recombination because of unwanted events24, we generated the additional line of Aldh2 conditional knockout mice using a Cre mouse line driven by a different GFAP promoter). We observed a significant reduction in ALDH2 protein in both cerebellar and hippocampal regions but not in PFC (Extended Data Fig. 2k,l). This expression pattern is similar to our observation above in Aldh2Gfap−/− mice (GFAP-Cre). These findings suggest a predominant expression of ALDH2 in the astrocytes of some brain areas.
Astrocytic ALDH2 mediates ethanol-induced elevation in cerebellar acetate levels.
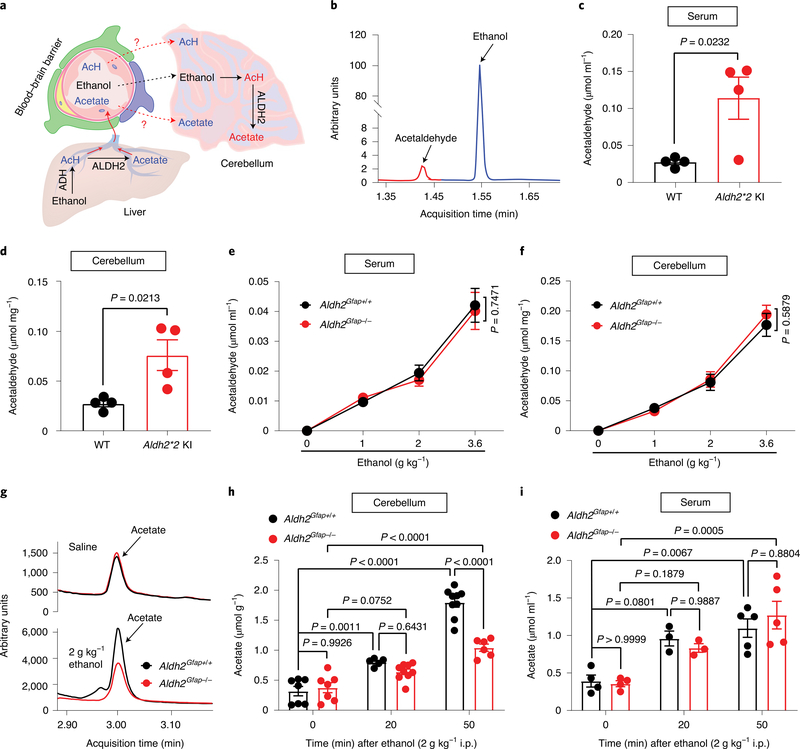
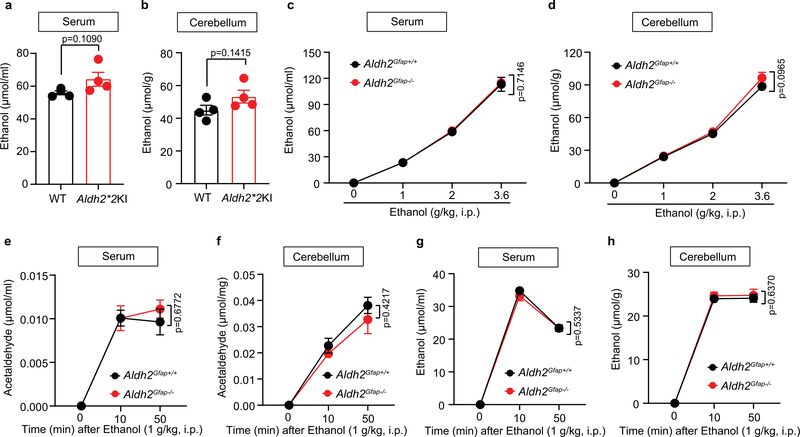
The possibility that ethanol is metabolized inside the brain has been one of the major questions in alcohol research (Fig. 2a). However, previous studies have been hindered by a lack of specific tools in vivo. The data presented above suggest that Aldh2Gfap−/− mice are a valuable animal model to distinguish between central and peripheral ALDH2-mediated effects on alcohol metabolism. We first examined serum and cerebellar ethanol and acetaldehyde levels in mice with global ALDH2 deficiency (Aldh2*2 knock-in (KI)) by using gas chromatography–mass spectrometry (GC–MS; Fig. 2b)25. Aldh2*2 KI mice carry a dominant point mutation in Aldh2 that causes ALDH2 deficiency and blood acetaldehyde elevation after ethanol intake in humans5. Indeed, we observed that Aldh2*2 KI mice exhibited a significant increase in blood and brain (cerebellum) acetaldehyde concentrations 50 min after systemic ethanol administration (2 g kg−1; Fig. 2c,d). Global ALDH2 deficiency did not significantly affect blood and brain ethanol contents (Extended Data Fig. 3a,b). Astrocytic ALDH2 deficiency did not significantly alter ethanol and acetaldehyde contents in either serum or brain tissues following an intraperitoneal (i.p.) injection of low-dose ethanol (Fig. 2e,f and Extended Data Fig. 3c–h). In contrast, astrocytic ALDH2 deficiency significantly reduced the elevation of cerebellar acetate derived from ethanol metabolism (Fig. 2g,h). This reduction was specific to the cerebellum because astrocytic ALDH2 deficiency did not significantly alter serum acetate content (Fig. 2i).
Fig. 2 |. Astrocytic ALDH2 selectively regulates ethanol metabolite acetate in cerebellar tissues.
a, Schematic of metabolic pathways from ethanol to acetate in the liver and brain. AcH, acetaldehyde. b, Representative peaks for ethanol and acetaldehyde from GC–MS in serum. c,d, Bar graphs of acetaldehyde concentrations in serum (c) and cerebellum (d) from Aldh2*2 KI and wild-type mice 50 min after ethanol administration (2 g kg−1, i.p.; n = 4 mice per group). e,f, Graphs showing acetaldehyde concentrations in the serum and cerebellum 50 min after ethanol administration in Aldh2Gfap−/− and Aldh2Gfap+/+ mice (0 g kg−1 ethanol: n = 4 mice per group; 1 g kg−1 ethanol: n = 11 mice per group in serum, n = 7 mice (Aldh2Gfap+/+) and 8 mice (Aldh2Gfap−/−) in cerebellum; 2 g kg−1 ethanol: n = 5 mice per group; 3.6 g kg−1 ethanol: n = 5 mice per group). g, Tracing records of acetate peaks detected by GC–MS measurement of cerebellar tissues isolated from Aldh2Gfap−/− and Aldh2Gfap+/+ mice. h,i, Bar graphs showing acetate concentrations in the cerebellum and serum 20 and 50 min after ethanol administration (2 g kg−1, i.p.) in Aldh2Gfap−/− and Aldh2Gfap+/+ mice (serum: n = 4 mice (0 min), n = 3 mice (20 min) and n = 5 mice (50 min); cerebellum: n = 7 mice (0 min), n = 5 and 9 from Aldh2Gfap−/− and Aldh2Gfap+/+ mice, respectively (20 min), n = 9 and 6 from Aldh2Gfap−/− and Aldh2Gfap+/+ mice, respectively (50 min)). All data are expressed as the mean ± s.e.m. Groups were compared by an unpaired two-tailed Student’s t-test (c and d) or two-way ANOVA followed by Tukey’s test (e, f, h and i).
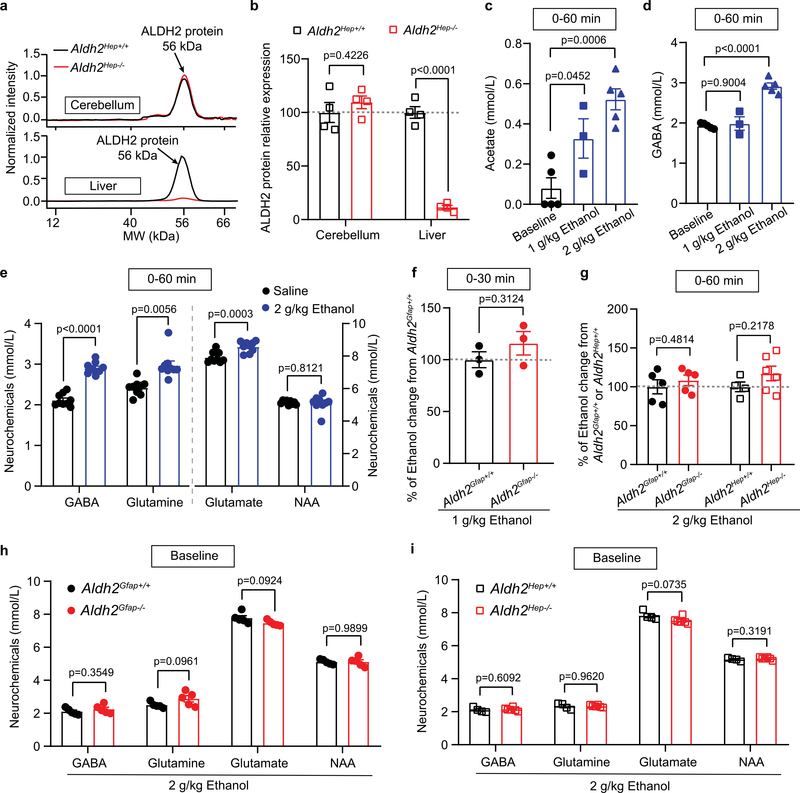
Astrocytic ALDH2 mediates ethanol-induced elevation in cerebellar GABA levels.
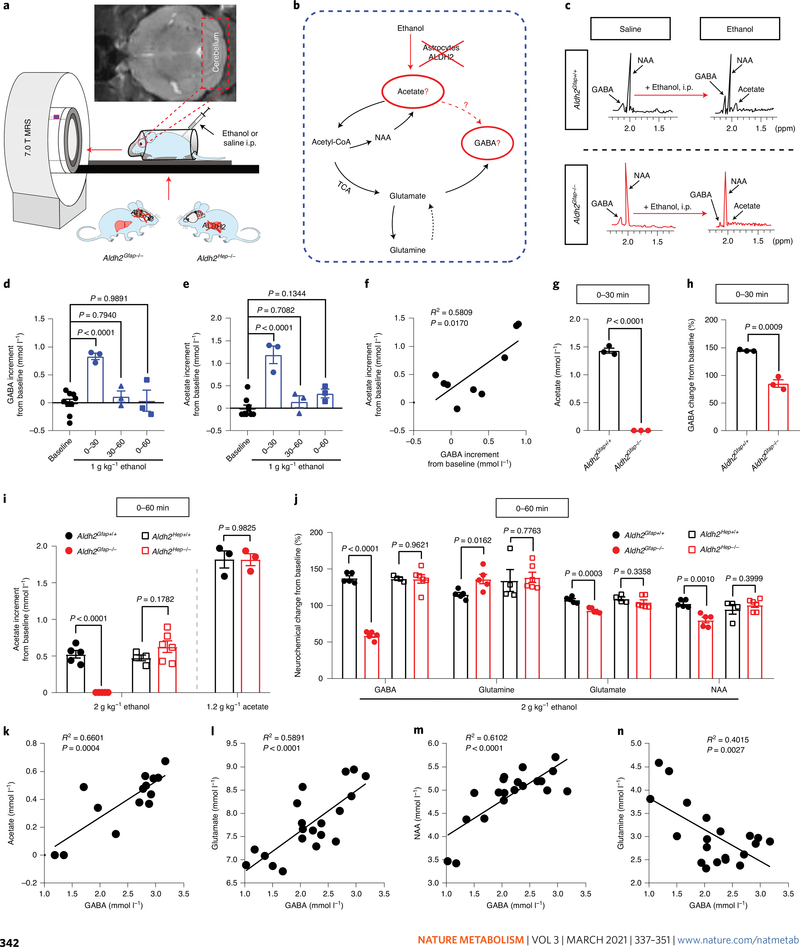
In vivo quantitative magnetic resonance spectroscopy (MRS) has been used to study brain alcohol metabolites and alcohol-induced neurochemical changes in animals and humans21,26. In cell-type ALDH2-deficient mice, we next explored the cell-type-specific mechanism that mediates alcohol metabolism and action in the brain (Fig. 3a–c). A recent study from our laboratory has shown that hepatocytic ALDH2 deficiency caused elevated levels of blood acetaldehyde after ethanol administration27. While hepatocytic ALDH2 deficiency reduced liver ALDH2 protein level by 90%, no change was observed in ALDH2 protein levels of cerebellar tissues between hepatocytic ALDH2-deficient mice (Aldh2Hep−/−) and their wild-type littermates (Aldh2Hep+/+; Extended Data Fig. 4a,b). Together, these astrocytic ALDH2-deficient and hepatocytic ALDH2-deficient mice provide us with valuable in vivo approaches to differentiate between central and peripheral effects of ALDH2 activity. We first tested GABA contents by collecting and analysing the data every 30 min after administration of 1 g kg−1 ethanol. Cerebellar GABA was significantly elevated within the first 30-min interval but not the second interval after systemic ethanol (Fig. 3d). Cerebellar acetate rose with a pattern similar to that of GABA (Fig. 3e). There was a strong correlation between the concentrations of GABA and acetate in the cerebellum (Fig. 3f). Injection of ethanol (i.p.) increased the levels of cerebellar acetate and GABA in a concentration-dependent manner in mice (Extended Data Fig. 4c,d). In line with previous studies26,28, systemic ethanol also elevated the levels of glutamine and glutamate in mice (Extended Data Fig. 4e). Astrocytic ALDH2 deficiency prevented acetate elevation induced by systemic ethanol (1–2 g kg−1, i.p.) but not by systemic acetate (Fig. 3g,i). Hepatocytic ALDH2 deficiency did not significantly affect ethanol-induced elevation in cerebellar acetate. Neither astrocytic nor hepatocytic ALDH2 deficiency significantly altered ethanol contents in the cerebellum (Extended Data Fig. 4f,g). In contrast, astrocytic ALDH2 deficiency reduced GABA contents to a level that was even significantly lower than baseline after administration of ethanol at 1–2 g kg−1 i.p (Fig. 3h,j). Astrocytic ALDH2 deficiency reduced cerebellar glutamate and N-acetylaspartate (NAA) contents, but to a lesser extent than GABA. On the other hand, astrocytic ALDH2 deficiency increased the level of cerebellar glutamine. Hepatocytic ALDH2 deficiency did not significantly alter cerebellar neurochemicals after ethanol administration (Fig. 3j). Neither astrocytic nor hepatocytic ALDH2 deficiency significantly altered the basal levels of cerebellar neurochemical contents (Extended Data Fig. 4h,i). To further explore the interrelationships between cerebellar acetate and neurochemicals, we conducted a correlational analysis based on the data presented above. The level of GABA content was positively correlated with that of acetate, glutamate and NAA (Fig. 3k–m). Conversely, there was an inverse correlation between GABA and glutamine (Fig. 3n).
Fig. 3 |. Astrocytic ALDH2 mediates ethanol-induced elevation in cerebellar acetate and GABA levels.
a, Schematic of in vivo quantitative MRS measurement of mouse brain ethanol metabolites. b, Schematic of ethanol–acetate–GABA metabolic pathways. c, Representative spectrometry peaks of cerebellar acetate and GABA before and after i.p. ethanol administration in Aldh2Gfap+/+ mice (upper, black) and Aldh2Gfap−/− mice (lower, red). d, Summary statistics of cerebellar GABA detected by MRS at the different time periods (min) after 1 g kg−1 ethanol administration in Aldh2Gfap+/+ mice (n = 3 mice per group). e, Summary statistics of cerebellar acetate detected by MRS at the different time periods (min) after 1 g kg−1 ethanol administration in Aldh2Gfap+/+ mice (n = 3 mice per group). f, Correlation of cerebellar GABA and acetate after systemic injection of ethanol (P = 0.017 by linear regression). g,h, Summary statistics of GABA and acetate contents by MRS from 0–30 min after systemic injection of 1 g kg−1 ethanol in Aldh2Gfap−/− and Aldh2Gfap+/+ mice. Data were calculated as the percentage change in GABA (before versus after ethanol) in the cerebellum (n = 3 mice per group). i, Summary statistics of cerebellar acetate by MRS from 0–60 min after systemic injection of 2 g kg−1 ethanol or 1.2 g kg−1 acetate in astrocytic ALDH2-deficient mice and in hepatocytic ALDH2-deficient mice (n = 5, 5, 4, 6, 3 and 3 mice, respectively). j, Neurochemical changes after 2 g kg−1 ethanol administration in astrocytic ALDH2-deficient mice (n = 5 mice per group), hepatocytic ALDH2-deficient mice (n = 4 mice in Aldh2Hep+/+ and 6 mice in Aldh2Hep−/−) and their wild-type littermates. Data were calculated as the percentage change in neurochemicals (before versus after ethanol) in the cerebellum. k–n, Correlation analysis of cerebellar GABA with acetate, glutamine, NAA and glutamate detected by MRS (P value was calculated by linear regression). All data are expressed as the mean ± s.e.m. Analysis was performed using an unpaired two-tailed Student’s t-test (g–j) or one-way ANOVA followed by Tukey’s test (d and e).
Astrocytic ALDH2 mediates ethanol enhancement of GABA tonic inhibition.
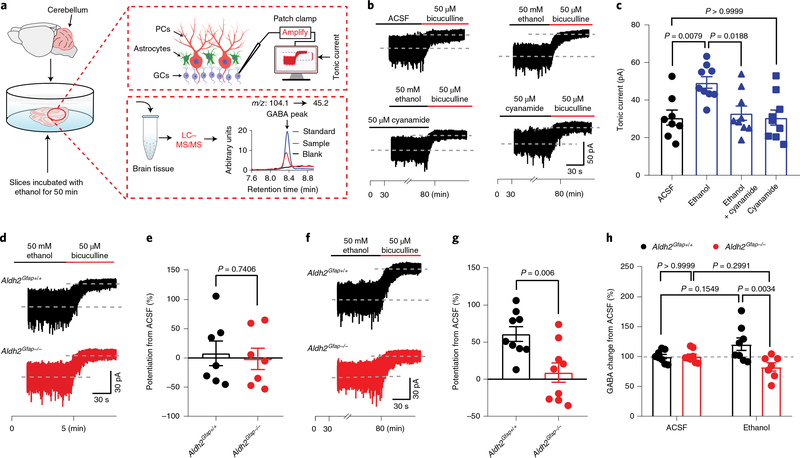
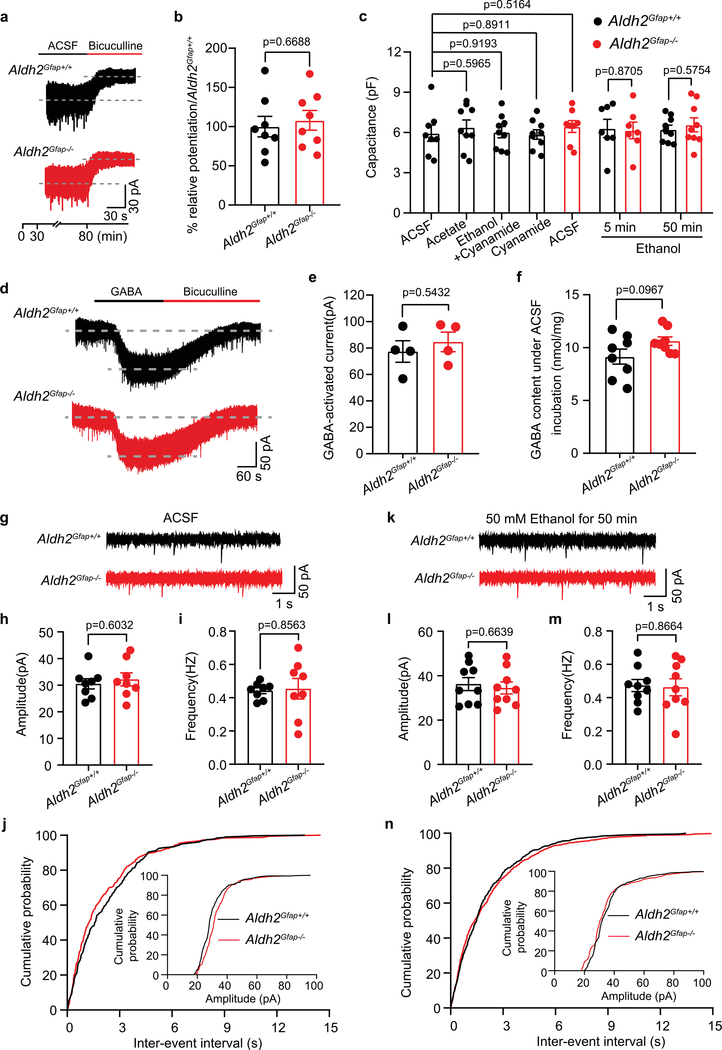
Low-dose ethanol enhances GABAergic tonic currents of cerebellar neurons29,30. Emerging evidence has suggested that astrocytic GABA synthesis and release mediates GABA tonic inhibition in the granule cells (GCs) from the granule layers of the cerebellar cortex31,32. To explore the mechanism underlying astrocytic ALDH2 control of the ethanol–GABA interaction, we next tested the effect of ethanol on GABA tonic inhibition, GABAergic inhibitory postsynaptic currents (IPSCs) and GABA synthesis in cerebellar slices. We conducted patch-clamp recordings of GCs of cerebellar slices (Fig. 4a). We subsequently measured GABA content in these cerebellar slices by using liquid chromatography–mass spectrometry (LC–MS/MS)33. Ethanol has been shown to enhance GABA-activated tonic currents in GCs the cerebellum and hippocampus34,35. GABAA receptor-mediated tonic currents were indicated by reduction of the amplitude of inward currents after 50 μM bicuculline (Fig. 4b and Extended Data Fig. 5a). The average amplitudes of GABA-activated tonic currents did not significantly differ in slices between wild-type and mutant mice (Extended Data Fig. 5b). The continuous incubation of cerebellar slices with ethanol for 50 min potentiated the amplitude of GABA-activated tonic currents by 49% (Fig. 4b,c). This pretreatment was required for ethanol potentiation because a simultaneous application of ethanol did not significantly alter GABA-activated tonic currents (Fig. 4d,e). Pretreatment with an ALDH inhibitor, cyanamide, inhibited the ethanol-induced potentiating effect on GABA tonic currents (Fig. 4c). Astrocytic ALDH2 deficiency also significantly inhibited ethanol potentiation of GABA tonic currents (Fig. 4f,g). This suggests that ethanol potentiation of GABA tonic current is ALDH2 dependent. Ethanol pretreatment did not significantly alter cell membrane capacitance, whole-cell currents (Extended Data Fig. 5c–e) and the frequency and amplitude of GABAergic miniature IPSCs (mIPSCs) in cerebellar slices (Extended Data Fig. 5g–n). Finally, we directly measured GABA content in cerebellar slices with and without ethanol treatment by using LC–MS/MS. Ethanol treatment slightly but nonsignificantly increased GABA content in cerebellar slices from Aldh2Gfap+/+ mice (Fig. 4h). On the other hand, astrocytic ALDH2 deficiency reduced GABA content to a level significantly lower than the GABA content collected from Aldh2Gfap+/+ mice. There was no significant difference in the basal levels of cerebellar GABA between Aldh2Gfap+/+ and Aldh2Gfap−/− mice (Extended Data Fig. 5f). These observations suggest that astrocytic ALDH2 is essential for the control of ethanol-induced GABA tonic inhibition in cerebellar slices.
Fig. 4 |. Astrocytic ALDH2 mediates ethanol enhancement of GABA tonic inhibition.
a, Schematics of electrophysiological patch-clamp recording of GCs and the measurement of GABA by LC–MS/MS in cerebellar slices. b, Representative trace records of GABA tonic currents induced by bicuculline (50 μM) in a cerebellar slice and ethanol enhancement of GABA tonic currents in slices with and without cyanamide (50 μM) pretreatment. c, Summary statistics of the effect of cyanamide on the ethanol-induced potentiation of GABA tonic currents (n = 8, 9, 9 and 9 cells, respectively; 6 mice). d,e, Trace records and summary statistics of the effect of incubation with 50 mM ethanol for 5 min on GABA tonic currents in GCs of cerebellar slices from Aldh2Gfap+/+ and Aldh2Gfap−/− mice (n = 7 cells per group, 6 mice). f,g, Trace records and summary statistics of the effect of incubation with 50 mM ethanol for 50 min on GABA tonic currents in GCs of cerebellar slices from Aldh2Gfap+/+ and Aldh2Gfap−/− mice (n = 9 cells per group, 6 mice). h, Summary statistics of GABA content in cerebellar slices measured by LC–MS/MS (n = 8, 8, 8 and 7 mice, respectively). All data are expressed as the mean ± s.e.m. Analysis was performed using an unpaired two-tailed Student’s t-test (e and g), one-way ANOVA followed by Tukey’s test (c) or two-way ANOVA followed by Tukey’s test (h).
Acetate promotes GABA synthesis and GABA tonic inhibition.
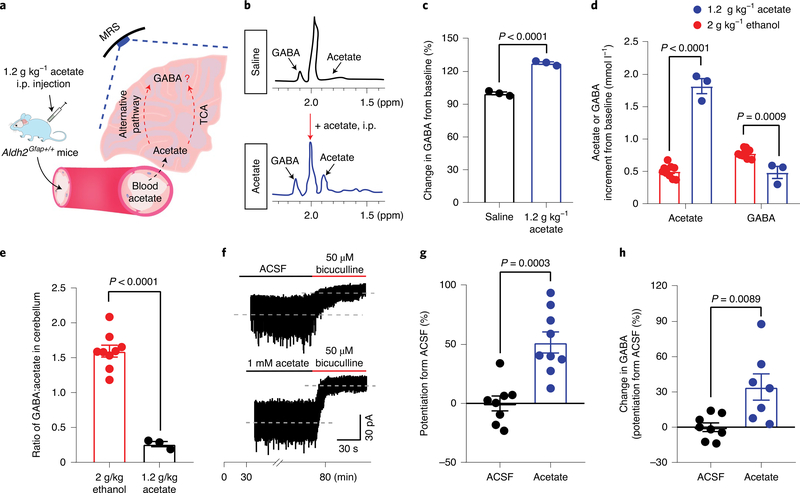
We next examined if acetate can directly promote GABA synthesis in vivo and GABA tonic inhibition in vitro (Fig. 5a). Cerebellar acetate rose after systemic injection of acetate (Fig. 5b). Astrocytic ALDH2 deficiency did not significantly alter cerebellar acetate content derived from systemic acetate (Fig. 3i). Systemic acetate significantly increased cerebellar GABA content in mice (Fig. 5c). While cerebellar acetate content derived from systemic acetate was fourfold higher than that derived from systemic ethanol, the level of GABA content derived from systemic acetate was significantly lower than that of GABA content derived from systemic ethanol (Fig. 5d). The ratio of GABA to acetate after ethanol was eightfold higher than that after acetate (Fig. 5e). This suggests that acetate from central but not peripheral ethanol metabolism primarily promotes GABA synthesis in the brain. Whether acetate can potentiate GABA tonic inhibition has not been reported. We next tested the effect of acetate on GABA tonic current in the cerebellar slices. The slices were preincubated with 1 mM acetate for 50 min before application of bicuculline. Acetate significantly enhanced GABA tonic currents (Fig. 5f,g). Moreover, acetate pretreatment increased the GABA concentration of these slices as detected by LC–MS (Fig. 5h). Collectively, these observations favour the idea that acetate mediates ethanol-induced GABA synthesis and GABA tonic inhibition.
Fig. 5 |. Acetate enhancement of GABA synthesis and GABA tonic inhibition in cerebellum.
a, Schematic of brain GABA synthesis after systemic acetate administration. b, Representative MRS peaks of acetate and GABA after systemic acetate (1.2 g kg−1, i.p., blue) and saline (black) in mice. c, Summary statistics of GABA content from baseline after systemic administration of either saline or acetate. Data were calculated as the percentage change in GABA (before versus after acetate) in the cerebellum (n = 3 mice per group). d, Acetate or GABA increment from baseline after systemic ethanol or acetate administration in the cerebellum (1.2 g kg−1 acetate: n = 3 mice; 2 g kg−1 ethanol: n = 9 mice) e, The ratio of GABA:acetate after systemic ethanol or acetate administration in the cerebellum of mice (1.2 g kg−1 acetate: n = 3 mice; 2 g kg−1 ethanol: n = 9 mice). f,g Trace records and summary statistics of GABA tonic currents recorded from GCs of cerebellar slices without and with acetate pretreatment (n = 8 and 9 cells, respectively; 6 mice). h, Summary statistics of GABA content in cerebellar slices after acetate pretreatment as measured by LC–MS/MS (n = 8 and 7 mice, respectively). All data are the mean ± s.e.m. Groups were compared by an unpaired two-tailed Student’s t-test (c–e, g and h).
ALDH2 mediates ethanol enhancement of GABA synthesis at the single-astrocyte level.
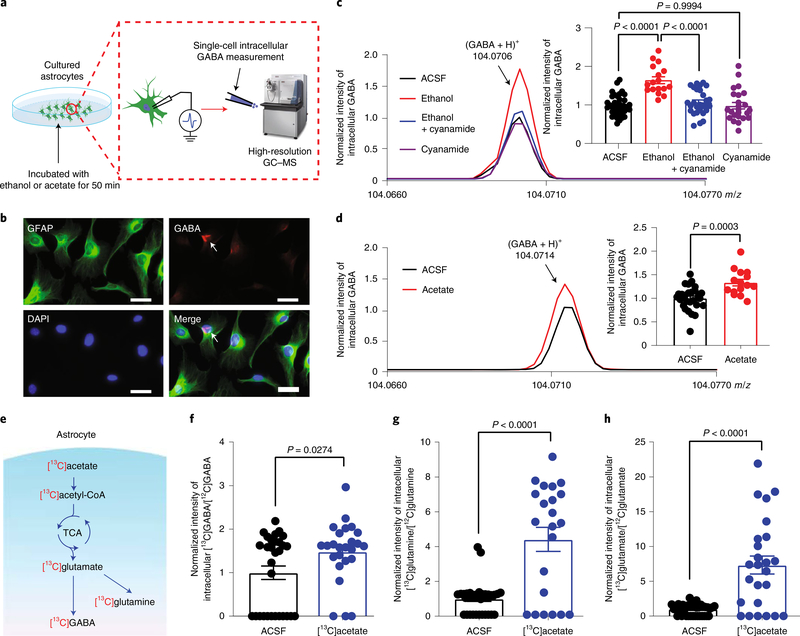
To test if astrocytic ALDH2 regulation of acetate and GABA synthesis occurs in single astrocytes, we used a high-resolution GC–MS/MS technique to measure GABA contents at the single-cell level (Fig. 6a)36. Nearly all cultured cells were positive for GFAP after 2 weeks of culture (Fig. 6b). Some of these cells exhibited GABA antibody fluorescence. Single-cell desorption electrospray ionization–mass spectrometry detected intracellular GABA content in almost all astrocytes (Fig. 6c,d). Pretreatment of cultured astrocytes with ethanol (50 mM) significantly increased the GABA signal by 57% (Fig. 6c). A similar result was observed in astrocytes pretreated with 1 mM of acetate (Fig. 6d). To test whether such ethanol-induced enhancement of GABA is ALDH2 dependent, we preincubated the astrocytes with cyanamide (50 μM). Pretreatment with cyanamide alone did not significantly alter astrocytic GABA content; however, cyanamide completely abolished the ethanol-induced enhancement of GABA content (Fig. 6c). These observations suggest that ethanol promotes GABA synthesis via an ALDH2-dependent mechanism at the single-astrocyte level. The precise pathways for ethanol-induced GABA synthesis are unclear. 13C-labelled acetate can be converted to 13C-labelled glutamate and GABA via the tricarboxylic acid (TCA) cycle in vitro and in vivo37–39. Whether this metabolic mechanism occurs in single astrocytes has not been reported. To address this question, we treated cultured astrocytes with stable isotope-labelled [13C]acetate (1 mM; Fig. 6e). The signals of all three labelled amino acids, 13C-labelled GABA, glutamate and glutamine, were substantially increased in cells incubated with [13C] acetate (Fig. 6f–h).
Fig. 6 |. Single-astrocyte analysis of the effects of ethanol and acetate on GABA synthesis.
a, Schematic of high-resolution GC–MS to quantitatively measure intracellular GABA content at the single-astrocyte level. b, Microscopic images of GFAP and GABA immunostaining in cultured astrocytes from the cerebellar cortex. White scale bars, 50 μm. c, Representative GC–MS spectra and normalized intensities of GABA signal in the presence of 50 mM ethanol with and without pretreatment with cyanamide (n = 37, 17, 28 and 24 cells, respectively). d, Representative GC–MS spectra and normalized intensities of GABA signal in the presence of 1 mM acetate (n = 27 and 15 cells, respectively). e, Hypothetical diagram of ethanol, acetate and GABA synthesis in astrocytes. f, Relative intensity of 13C-labelled GABA in astrocytes after [13C]acetate incubation (n = 39 and 38 cells, respectively). g, Relative intensity of 13C-labelled glutamine in astrocytes after [13C]acetate incubation (n = 38 and 22 cells, respectively). h, Relative intensity of 13C-labelled glutamate in astrocytes after [13C]acetate incubation (n = 30 and 28 cells, respectively). All data are the mean ± s.e.m. Analysis was performed using an unpaired two-tailed Student’s t-test (d and f–h) or one-way ANOVA followed by Tukey’s test (c).
Cerebellar astrocytic ALDH2 mediates ethanol-induced motor impairment via a GABAergic mechanism.
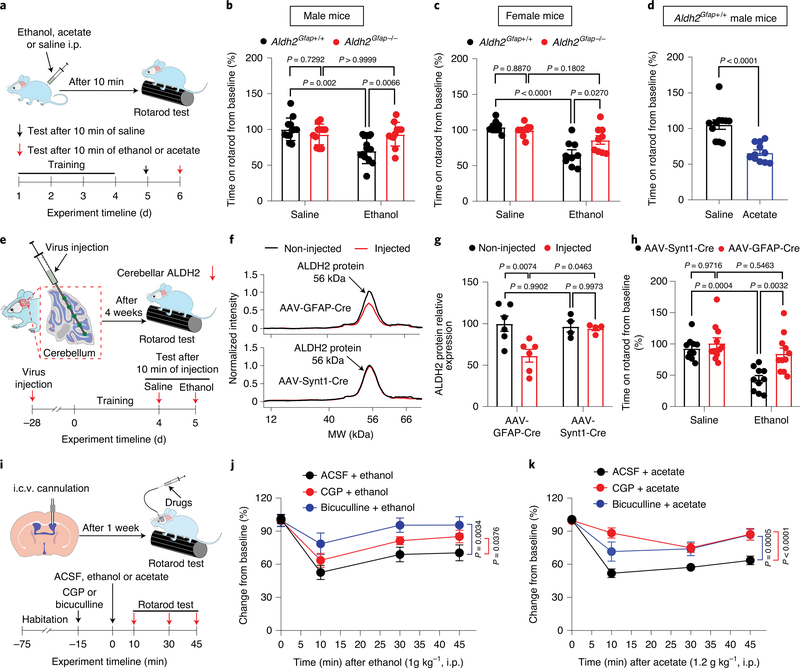
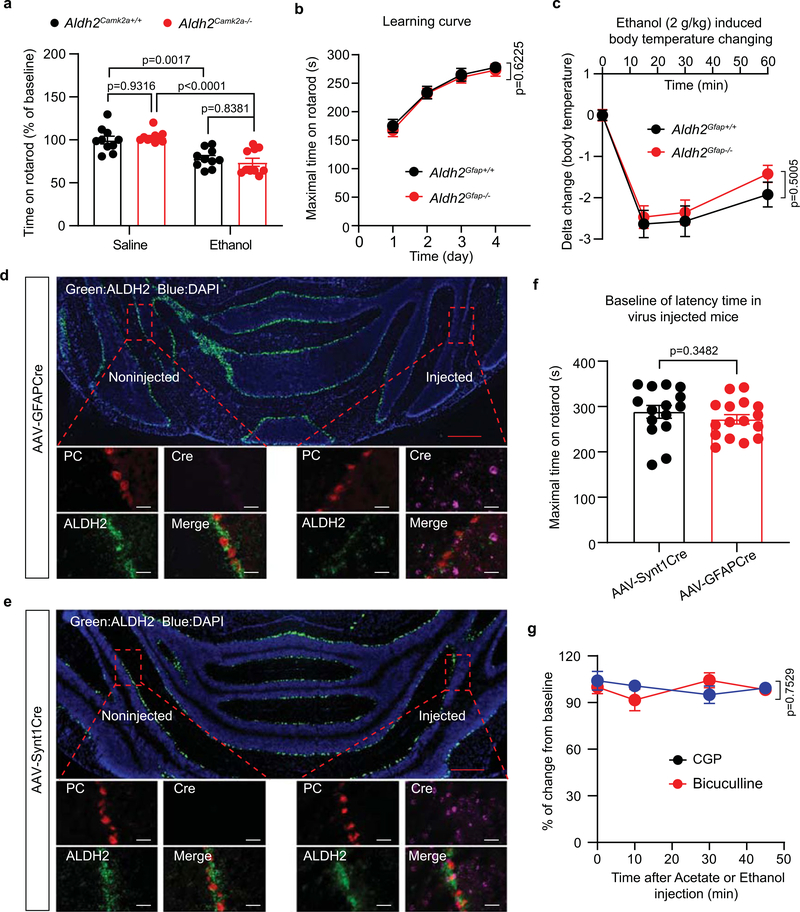
Next, we investigated whether astrocytic ALDH2 contributes to ethanol-induced impairment of balance and coordination skills (Fig. 7a). Previous studies suggested that ethanol at a dose of 1 g kg−1 i.p. is ideal to assess the genetic susceptibility to ethanol intoxication in mice40,41. As we have shown, systemic administration of such a low dose of ethanol produced a binge level of blood ethanol (33 mM; Extended Data Fig. 3g). Consistent with this, ethanol significantly impaired accelerating rotarod performance in wild-type mice (Fig. 7b). However, such ethanol-induced motor impairment was not observed in Aldh2Gfap−/− mice. In contrast, Aldh2Camk2a−/− mice did not alter ethanol-induced motor impairment (Extended Data Fig. 6a). No significant difference in baseline rotarod performance was found between Aldh2Gfap+/+ and Aldh2Gfap−/− mice during training for 4 consecutive days (Extended Data Fig. 6b). We also examined the sensitivity of astrocytic Aldh2−/− female mice to ethanol-induced incoordination (Fig. 7c). We observed that astrocytic Aldh2−/− female mice reduced ethanol-induced incoordination in a similar manner to their male counterparts. In line with findings from a previous study12, systemic acetate also produced motor impairment in mice (Fig. 7d). Interestingly, astrocytic ALDH2 deficiency did not significantly alter ethanol-induced hypothermia (Extended Data Fig. 6c), indicating that astrocytic ALDH2 selectively contributes to some but not all ethanol-induced in vivo consequences. To explore whether astrocytic ALDH2 mediates ethanol-induced discoordination in a brain-region-specific manner (cerebellum), we microinjected an adeno-associated virus bearing Cre recombinase under the control of the GFAP promoter (AAV5-GFAP-Cre) into the entire right hemisphere of the cerebellum in Aldh2 flox mice (Fig. 7e). AAV9-Synt1-Cre virus, which targets neuronal cells, was also injected into the same brain area in a separate group of mice. We first used the RNAscope technique to examine the expression of Cre in cerebellar slices. The Cre signal was detected in both areas that received injections of AAV5-GFAP-Cre and AAV9-Synt1-Cre but not in non-injected areas (Extended Data Fig. 6d,e). The ALDH2 signal only decreased in the AAV5-GFAP-Cre-injected hemisphere (Extended Data Fig. 6d,e). Consistent with this, the level of ALDH2 protein significantly decreased in the brain region that was injected with AAV5-GFAP-Cre but not in the brain region that was injected with AAV9-synt1-Cre (Fig. 7f,g). These findings suggest that AAV5-GFAP-Cre selectively reduces cerebellar ALDH2 expression in astrocytes but not in neurons. There was no significance in the baseline of rotarod performance in virus-injected mice (Extended Data Fig. 6f). The mice injected with AAV5-GFAP-Cre but not AAV9-Synt1-Cre were significantly less sensitive to ethanol-induced motor impairment in the rotarod test (Fig. 7h). To test whether ethanol-induced motor impairment is mediated by a GABAergic-dependent mechanism, we delivered a GABAA receptor antagonist, bicuculline, or a GABAB receptor antagonist, CGP55845 (CGP), via intracerebroventricular (i.c.v.) infusion in mice (Fig. 7i). Neither bicuculline nor CGP (200 ng, i.c.v.) altered baseline rotarod performance (Extended Data Fig. 6g). Consistent with previous studies4,42, bicuculline and CGP reduced the levels of ethanol-induced discoordination in the rotarod test (Fig. 7j). We also observed that both GABAA and GABAB receptor antagonists attenuated the acetate-induced impairment of motor performance in the rotarod test (Fig. 7k). These findings support the hypothesis that acetate mediates low-dose ethanol-induced intoxication via GABAergic signalling.
Fig. 7 |. Astrocytic ALDH2 mediates ethanol-induced discoordination.
a, Schematic of rotarod test and experimental procedure. b,c, Bar graphs showing the percentage changes from baseline in the latency to fall from the accelerating rotarod 10 min after i.p. administration of ethanol (1 g kg−1) in Aldh2Gfap−/− and Aldh2Gfap+/+ male and female mice (male: n = 12, 11, 12 and 11 mice, respectively; female: n = 9 mice per group). d, Bar graphs showing the percentage changes from baseline in the latency to fall from the accelerating rotarod 10 min after i.p. administration of acetate (1.2 g kg−1) in male mice (n = 11 mice per group). e, Schematic of AAV microinjection in the cerebellar hemisphere and experimental procedure. f, Representative electropherogram traces from the automated size-based capillary western blot of ALDH2 protein in the AAV-injected (red) and non-injected (black) hemispheres. g, Summary statistics of brain ALDH2 proteins without and with AAV Cre injection (n = 4 mice for AAV9-Synt1-Cre; n = 6 mice for AAV5-GFAP-Cre). h, Bar graphs showing the latency to fall from the rotarod 10 min after i.p. administration of ethanol (1 g kg−1) in mice with and without intracerebellar AAV injection (n = 10, 11, 10 and 11 mice, respectively). i, Schematic of i.c.v. cannulation and microinjection of GABAA receptor antagonist (bicuculline) or GABAB receptor antagonist (CGP). j, Time courses showing central administration of GABAA or GABAB receptor antagonist inhibited ethanol-induced motor impairment (n = 7 mice for CGP + ethanol and bicuculline + ethanol; n = 8 mice for artificial cerebrospinal fluid (ACSF) + ethanol). k, Time courses showing central administration of GABAA or GABAB receptor antagonist inhibited acetate-induced motor impairment (n = 7 mice for bicuculline + acetate; n = 14 mice for ACSF + acetate; n = 6 mice for CGP + acetate). All data are the mean ± s.e.m. Analysis was performed using an unpaired two-tailed Student’s t-test (d), two-way ANOVA followed by Tukey’s test (b, c, g and h) or two-way repeated-measures ANOVA (j and k).
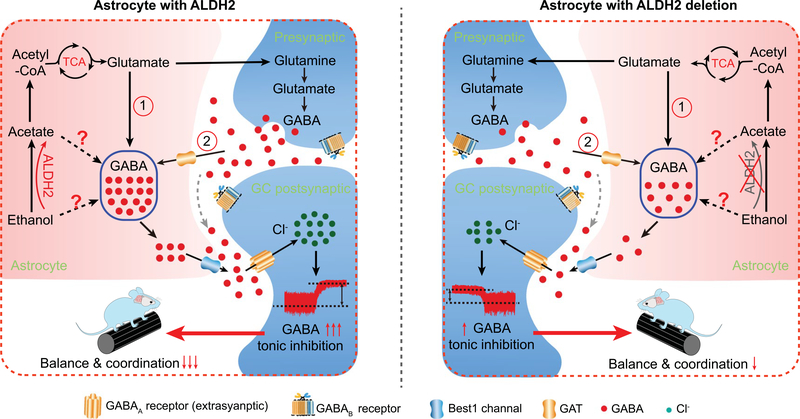
Thus, astrocytic ALDH2 mediates the conversion of ethanol–acetate–GABA signalling. The precise molecular details for this conversion are unclear, but it could involve multiple pathways as illustrated by a working model (Extended Data Fig. 7). The pathways may include (1) an astrocytic TCA pathway and (2) a neuron-dependent pathway from acetate–glutamate to GABA via the TCA cycle. ALDH2 also regulates ethanol and GABA synthesis through other pathways. For instance, ALDH2 is the key enzyme in the diamine oxidase pathway that synthesizes GABA from putrescine in astrocytes and neurons8,31.
Discussion
The present results reveal ALDH2 control of alcohol metabolism in a brain-region-specific and cell-type-specific mechanism. Astrocytic ALDH2 mediated the production of acetate in the cerebellum after low-dose ethanol administration. Acetate from the central ethanol metabolic pathway mediated ethanol-induced motor impairment by promoting GABA synthesis and GABA tonic inhibition in the cerebellum. The astrocytic ALDH2-dependent mechanism also contributes to ethanol-induced GABA synthesis and motor impairment.
The possibility of brain alcohol metabolism has been a controversial topic within the field of alcohol research for several decades10,43. There is a long-standing idea that brain acetate derives largely from liver alcohol metabolism11,44. Indeed, acetate can be transported through the blood–brain barrier with a high capacity12. Our data presented here directly challenge this idea and suggest that the central but not the peripheral alcohol metabolic pathway produces the alcohol metabolite acetate. Data from both ex vivo and in vivo experiments suggest an astrocytic ALDH2 control of acetate production from ethanol metabolism in the brain. Consistent with this idea, there was no change in cerebellar acetate content in hepatocytic ALDH2-deficient mice. This ALDH2 deficiency did not alter the level of blood ethanol27. Astrocytic ALDH2 deficiency inhibited ethanol-induced enhancement of GABA tonic inhibition and GABA content. As a result, such astrocytic ALDH2 deficiency reduced ethanol-induced behavioural changes. We also observed that enhancement of cerebellar GABA content was largely dependent on the central but not the peripheral metabolic pathway that breaks ethanol down into acetate. For instance, although systemic administration of acetate resulted in a higher level of cerebellar acetate, the extent of the increase in GABA by systemic acetate was much smaller than that of systemic ethanol. In line with our observation, there is strong evidence suggesting a rate-limiting step for glial metabolism of acetate after its entry into the brain45. It is worth mentioning that we cannot exclude the possibility that astrocytic ALDH2 deficiency may alter the processes of GABA degradation and uptake in addition to inhibiting the acetate metabolic pathway.
It has long been suggested that penetration of peripherally circulating acetaldehyde past the blood–brain barrier is very low43,46. This scenario is particularly true when exposure to ethanol is low due to the high ALDH2 activity at the blood–brain barrier43,47. We failed to detect any difference in brain acetaldehyde content in astrocytic and hepatocytic ALDH2-deficient mice as compared to their wild-type littermates even after low-dose ethanol. While astrocytic ALDH2 did not alter blood acetaldehyde, hepatocytic ALDH2 deficiency substantially increased blood acetaldehyde levels27. One possibility to explain this observation is that acetaldehyde is dynamically buffered between central and peripheral pools. We only evaluated the metabolic mechanism for low-dose alcohol (1–2 g kg−1) in this study. It is possible that liver ALDH2 regulates the contents of brain alcohol metabolites such as acetaldehyde and acetate after exposure to high-dose alcohol.
Our results demonstrate that astrocytic ALDH2 mediated ethanol-induced behaviours in a brain-region-specific manner. For instance, the mice with cerebellar astrocytic ALDH2 deficiency were less sensitive to ethanol-induced discoordination. This also suggests that alcohol metabolites such as acetate can induce behavioural changes in a brain-region-specific manner. Until recently, we had limited knowledge about the cell-type-specific and brain-region-specific mechanisms underlying alcohol-metabolite-induced behavioural effects. The cell-type-specific ALDH2-deficient mice are a valuable resource for exploring the in vivo impacts of alcohol metabolites in discrete brain areas. In addition to the cerebellum, several other brain areas were found to express astrocytic ALDH2. They include the hippocampus, central nucleus of the amygdala and ventral tegmental area in Aldh2Gfap−/− mice. These brain regions are well known for their involvement in a variety of ethanol’s actions on the brain and behaviours3. ALDH2 is also expressed in various neurons and possibly in other types of glial cells apart from astrocytes8. A combination of in vivo MRS with brain-region, cell-type-specific ALDH2-deficient mice should allow us to uncover the mechanism of action of alcohol metabolites in alcohol use disorder.
One unexpected finding is that cerebellar GABA content was decreased below baseline in astrocytic ALDH2-deficient mice after systemic ethanol. The precise mechanism underlying this GABA reduction is not clear. In general, these transgenic mice have not shown any developmental or behavioural changes. No difference was observed in all neurochemicals at the baseline between wild-type and mutant mice as revealed by MRS experiments. Thus, the reduction of GABA was selectively caused by systemic ethanol in astrocytic ALDH2-deficient mice. In this regard, we cannot exclude the possibility that this GABA reduction was caused by elevated ethanol or acetaldehyde in some subpopulations of cells (glial cells and neurons). In this case, we were unable to detect the changes of alcohol metabolites because of technical limitations in our MRS experiment. Further studies are needed to investigate the reason for this observation and its potential clinical significance.
Our study suggests that acetate is a key ethanol metabolite that mediates GABA synthesis through multiple ALDH2-dependent pathways. It is very likely that acetate-induced GABA synthesis is mainly mediated though the neuron–glia TCA cycle. We also provide evidence indicating that ethanol and acetate can promote GABA synthesis in a single astrocyte, supporting the idea that astrocytes can directly synthesize GABA in the brain31,48,49. The regulation of GABA synthesis by astrocytic ALDH2 may have an impact on many common neurological diseases other than ethanol use disorder. ALDH2 deficiency has been found to contribute to aging50, anxiety51, nociceptive state52 and addiction mechanisms9,53. Additionally, astrocytic GABA synthesis and release has been linked with Alzheimer’s disease pathology54–56.
Our data show that cerebellar astrocytic ALDH2 critically regulates ethanol metabolism, GABA synthesis and ethanol-induced motor impairment. This is consistent with a previous study showing that the cerebellum was one brain region with high acetate uptake following ethanol consumption in humans14. Recent studies have suggested that the cerebellum is the most underestimated brain area critical for major brain functions and neurological diseases18,19,57,58. Its role in ethanol-induced behavioural effects has also been overlooked.
Although the Aldh2Gfap−/− mouse model is valuable for studying the in vivo consequence of astrocytic ALDH2 in the brain, this mouse line has some limitations. For example, Cre activity in this particular mouse line is mainly detected in the cerebellum and hippocampus in adulthood (http://www.informatics.jax.org/allele/MGI:3522215?romRibbon=open). In this regard, the tissue-specific depletion of ALDH2 in Aldh2Gfap−/− mice may not truly represent the actual distribution of astrocytic ALDH2 in normal mice. We cannot fully exclude the potential role of neuronal ALDH2 in the actions of ethanol, given that ALDH2 inhibitors selectively inhibit dopamine release in the midbrain53. However, it is unlikely that the depletion of brain ALDH2 observed in Aldh2Gfap−/− mice is contaminated with neuronal stem cells59. First, we examined two Aldh2 conditional knockout mouse lines driven by different GFAP promoters. We observed a similar pattern of ALDH2 depletion in several brain regions from these mice. Second, a significant and similar magnitude of reduction in ALDH2 protein was also observed in cerebellar cortex previously injected with GFAP-Cre virus but not with AAV9-Synt1-Cre. Additionally, no significant reduction of ALDH2 protein levels was observed in all tested brain regions in Aldh2Camk2a−/− mice. One weakness of MRS is that this technique has a limited sensitivity in detecting low levels of metabolites. In this study, we conducted both ex vivo (GC–MS and LC–MS/MS) and in vivo (MRS) assays to measure brain acetate and GABA content. Overall results from these experiments are consistent in supporting our main hypothesis that astrocytic ALDH2 mediates ethanol metabolism and neurochemical changes in the brain. Finally, it is worth mentioning that subcellular localization of ALDH2 may differ between humans and rodents. While mouse ALDH2 is predominantly located in the mitochondrial subcellular fraction, there is evidence that human ALDH2 is distributed in both cytosolic and mitochondrial pools9,60–62. These differences in alcohol metabolic pathways may potentially affect the translation of our findings to human studies.
In summary, these findings suggest that central but not peripheral ALDH2 mediates the production of acetate from alcohol metabolism through an astrocyte-dependent mechanism. Astrocytic ALDH2 regulates both ethanol- and acetate-induced cellular and behavioural effects on the brain via distinct GABA synthesis pathways. Astrocytic ALDH2 is an important target not only for alcohol use disorders but also for other neurological diseases.
Methods
Animals.
Mice had free access to food and water unless otherwise specified and were housed under a 12-h light/dark cycle. Unless otherwise indicated, male mice between 10–20 weeks of age were used. Aldh2-knockout (Aldh2−/−) mice on a C57BL/6N background were described previously63. Aldh2*2 KI mice on a C57BL/6N background were described previously27. The Aldh2 flox mice were generated by activating the Aldh2 gene in Aldh2tm1a(EUCOMM)Wtsi mice, by crossing of Aldh2tm1a(EUCOMM)Wtsi mice (kindly provided by K. J. Patel)23 with homozygous FLPcR mice (Jackson Laboratory), which express flippase in the germline cells. Tissue-specific Aldh2-knockout mice were generated via crossing Aldh2 flox mice with several strains of Cre transgenic mice from the Jackson Laboratory, including B6.Cg-Tg (Gfap-cre)73.12Mvs/J (stock no. 012886), B6.Cg-Tg (Gfap-cre)77.6Mvs/2J (stock no. 024098), B6.Cg-Tg (Pcp2-cre)3555jdhu/j (stock no. 010536; also known as L7), B6.Cg-Tg (Camk2a-cre)T29–1Stl/J (stock no. 005359) and B6.Cg-Tg (Alb-cre)21Mgn/J (stock no. 003574). The newly generated astrocyte- (Aldh2Gfap−/−), Purkinje cell- (Aldh2L7−/−) and neuron-specific (Aldh2Camk2a−/−) Aldh2−/− mice were then used in this study. Animal behavioural tests were carried out in a double-blinded manner. All procedures used in this study were approved by the National Institute on Alcohol Abuse and Alcoholism (NIAAA) Animal Use and Animal Care Committee and performed in accordance with the National Institutes of Health (NIH) guidelines for care and use of laboratory animals.
Cultured astrocytes from the cerebellar cortex.
Cerebellar cortex tissues were collected from C57BL/6J mouse pups (postnatal days 0–3). Briefly, the cerebellar cortex was removed and transferred to a 2.5-cm dish in cold Hank’s buffer on ice. Cerebellar tissue was minced gently with sterile scissors and transferred to 15-ml tubes. The tissue was sequentially triturated with a 5-ml pipette and a fire-polished Pasteur pipette after incubation with 5 ml of 1× trypsin for 10 min at 37 °C. The suspension of dissociated cells was transferred to a fresh tube in 10 ml of medium. The cell suspension was passed through a 40-μm strainer and centrifuged at 1,200 r.p.m. for 5 min. The cells were resuspended and seeded in 12.5-cm2 flask. The primary cells were cultured in DMEM supplemented with 10% FBS at 37 °C and 5% CO2. The medium was replaced with fresh medium every 2 d. The astrocytes were seeded onto glass coverslips and could be used for experiments after 2–3 generations.
Conditional Aldh2 knockdown in cerebellum induced by AAV Cre injection.
Female Aldh2 flox (Aldh2Gfap+/+) mice aged 3–6 months were anaesthetized with isoflurane and placed into a stereotaxic frame. AAV5-GFAP-Cre (105550-AAV5, Addgene) and AAV9-Synt1-Cre (105553-AAV9, Addgene) lentiviral vectors were diluted to a concentration of 1012 DNA copies per ml, and they were loaded into a micro-dispenser and injected unilaterally into the right cerebellar hemisphere of Aldh2 flox mice. To infect the vermis and hemispheres of the cerebellar cortex (lobes V–VIII), the stereotaxic coordinates of the injection were set up as follows: AP, −4.5 mm; ML, −1.5 mm; DV, −4.5 mm; with an angle of 60°. The pipette was stopped every 0.5 mm for virus delivery during pull back. The virus was injected into tissues slowly at 500 nl at each stop for 5 min. The mice were tested with rotarod performance 4 weeks after virus injection.
Intraventricular cannulation and microinjection.
C57BL/6J male mice aged 3 months were anaesthetized by isoflurane and placed into stereotaxic frames. After the surface of the skull was exposed, a 1-mm burr hole was drilled (AP, −0.5 mm; lateral, −1.0 mm from bregma) on the right side of the animal. A 26-gauge guide cannula of 2.0 mm in length under pedestal (C315G-SPC, Plastics One) was gently inserted into the brain and held in place with dental acrylic bonded to two stainless steel screws anchored to the skull. The dummy and injector cannulae were used to seal the top of the guide cannula and to infuse testing chemicals. Mice with normal activity and no occlusion of the cannula were used for rotarod testing after 7 d.
RNAscope assay.
Human cerebellar tissues from four individuals (IDs: 836, 845, 852 and 913) were provided by the Human Brain Collection Core at the National Institute of Mental Health. Adult mice were deeply anaesthetized by isoflurane. The brain and liver were quickly removed and frozen on dry ice after euthanasia. The fresh-frozen tissue sections (10–12 μm) were cut using a cryostat (CM3050S, Leica Biosystem) and mounted on positively charged microscopic glass slides (Thermo Fisher Scientific). All probes were designed and provided by Advanced Cell Diagnostics. These probes include mouse (Mm) Mm-ALDH2-C3 (507121-C3) targeting 634–1640 of NM_009656.4, Cre-C2 (312281-C2) targeting 1058–2032 of KC845567.1, Mm-MAP2-C2 (ref. 431151) targeting 1265–2363 of NM_001039934.1, Mm-GFAP-C2 RNA (ref. 313211-C2) targeting 2–1761 of NM_001131020.1, Mm-ALDH1L1-C2, RNA (405891-C2) targeting 1256–2112 of NM_027406.1, EGFP-C1 (400281-C1) targeting 628–1352 of U55763.1, Mm-PCP2-C1 targeting 2–302 of NM_001129804.1, human (Hs) Hs-PcP2-C1 targeting 2–692 of NM_174895.2, Hs-ALDH2-C3 targeting 439–1485 of NM_000690.3-C3, and Hs-ALDH1L1-C2 (ref. 438881-C3) targeting 1999–2982 of NM_001270364.1. The experimental procedures are provided from the manufacturer’s instructions for the RNAscope Fluorescent Multiplex V2 assay (Advanced Cell Diagnostics). Briefly, the slides were immersed in 4% paraformaldehyde and incubated at 4 °C for 60 min, washed twice in PBS and then fixed by 50%, 70% and 100% ethanol in turn at room temperature (RT) for 5 min. Subsequently, the slides were treated with hydrogen peroxide for 10 min and digested by protease IV for 20 min at RT. The slides were washed with PBS three to five times, incubated with probes and placed in an incubation oven (HybEZ II, ACD) at 40 °C for 2 h. The slides were washed twice with wash buffer at RT and then hybridized with FLv2 Amp 1, Amp 2 or Amp 3 and labelled with FLv2 HRP C1, C2 or C3 signal; all slides were blocked by FLv2 HRP blocker followed by DAPI to stain the nucleus. Stained slides were cover-slipped with fluorescence mounting medium (Prolong Gold Antifade Reagent, P36930; Thermo Fisher Scientific) and scanned using a Zeiss LSM880 fluorescence or confocal microscope.
Automated size-based capillary western blotting.
Tissue samples were taken from various mouse brain areas as indicated in the main text for measurement of ALDH2 protein. Western blot analysis was performed using an automated size-based capillary system (Sally Instrument) from ProteinSimple. Briefly, mouse brain was removed after euthanasia and immersed in ice-cold RNAlater Stabilization Solution (AM7021, Thermo Fisher Scientific). The 100-μm brain slices were prepared using a DSK Zero 1N microslicer (Ted Pella). The tissue of each specific nucleus was punched and collected via a disposable biopsy punch with a diameter of 1 mm (Integra Miltex, 33–31AA-P/25). The protein was extracted using RIPA buffer containing phosphatase inhibitor coktail (ThermoFisher Scientific). Total protein concentrations were measured using a Pierce BCA Protein Assay Kit. The lysate was then diluted using 0.1× sample buffer at a final concentration of 0.3 mg ml−1 for chemiluminescence. ALDH2 protein expression was quantified with an automated size-based capillary electroresis system (ProteinSimple). The procedures were set as follows: separation time of 25 min with separation voltage of 375 V followed by antibody diluent for 5 min, primary antibody time of 60 min and secondary antibody time of 30 min. The primary antibodies and their dilutions included goat anti-ALDH2 polyclonal (1:20; Santa Cruz Biotechnology) and mouse anti-actin (1:100; Cell signaling technology). The horseradish-peroxidase-conjugated secondary antibodies included donkey anti-goat (1:100; Santa Cruz Biotechnology) and anti-mouse (Protein simple). The chemiluminescence signals were automatically converted to electropherograms in Compass software (ProteinSimple, version 3.0). ALDH2 protein expression was quantified by integrating the electropherogram peak conrresponding to the ALDH2 signal.
Quantitative PCR with reverse transcription.
Total RNA was extracted from the cerebellum, hippocampus, PFC, thalamus, striatum and liver of Aldh2Gfap+/+ and Aldh2Gfap−/− mice for relative quantitative RT–PCR analysis. A total of 1,000 ng RNA was reverse transcribed to cDNA and amplified using the QuantiTeck Reverse Transcription kit (QIAGEN). The primers for mouse 18S (4331182/Hs01375212_g1), Aldh2 (4331182/Mm00477469_m1) and Gfap (4331182/Mm01253033_m1) genes were designed and synthesized by Thermo Fisher Scientific. The PCR analysis was performed with an ABI Prism 9700HT system (Foster City) according to the manufacturer’s instructions. The PCR cycling conditions were set as follows: 95 °C for 20 s followed by 40 cycles of denaturation at 95 °C for 3 s and annealing/extension for 30 s at 60 °C. In each experimental run, triplicate-distilled water served as the negative control. All amplification reactions were performed in triplicate for each sample. Threshold cycles (Ct) were automatically calculated by the PCR system and relative gene expression levels were calculated as 2−ΔΔCT. The relative expression of ALDH2 mRNA in different brain regions of Aldh2Gfap+/+ mice was normalized to values of the PFC and the reduction of ALDH2 mRNA in cerebellum or liver of Aldh2Gfap−/− mice was normalized to values of Aldh2Gfap+/+ mice.
ALDH2 enzyme activity.
Brain tissues were freshly isolated and homogenized, and ALDH2 enzyme activity was measured using the mitochondrial ALDH2 enzyme activity kit (ab115348, Abcam) following the manufacturer’s instructions. The prepared sample aliquots of 100 μl were added to the provided microplate, covered and incubated for 3 h at RT. Each sample well was washed twice and mixed with 200 μl of cocktail solution (including coupler, reagent dye, 1 mM NAD+ and 25 mM acetaldehyde). The colour development with elapsed time at 450 nm was recorded in duplicates for 120 min. The ALDH2 activity was normalized to that of the PFC, and the value in the PFC was assigned as 1.
Immunofluorescence.
The cultured astrocyte slides were immersed in ice-cold 4% paraformaldehyde for 40 min. The slides were rinsed in PBS three times and incubated for 1 h at RT with blocking solution (0.3% Triton-X and 2% normal serum in 0.1 M PBS). Slides were incubated overnight in a mixture of the following primary antibodies with blocking solution at 4 °C on a shaker: rabbit anti-GABA (1:500 dilution; Abcam, Ab8891) and goat anti-GFAP (1:250 dilution; Santa Cruz, sc6171). After washing three times in PBS, sections were incubated with corresponding secondary antibodies: Alexa Fluor 488–conjugated donkey anti-rabbit (1:1,000 dilution; Thermo Fisher, A21206), Alexa Fluor 568–conjugated donkey anti-goat (1:1,000 dilution; Thermo Fisher, A11057) for 2 h at RT. The slides were mounted with an anti-fade mounting medium (Fluoromount G) before short exposure to DAPI. The stained images were examined and recorded with a fluorescence microscope (Zeiss, LSM 700).
Single-cell mass spectrometry (GC–MS/MS).
To test whether ethanol or acetate could raise the level of GABA in the astrocyte, ethanol and acetate were added into culture medium. The cultured astrocytes were incubated with 50 mM ethanol, 1 mM acetate or [13C]acetate for 50 min at 37 °C and 5% CO2, and then transferred onto the recording chambers with ACSF. The astrocytes were randomly chosen for the subsequent electrophysiological recording and mass spectrometry analysis as described in our recent work36,64. We approached the astrocytes using a micromanipulator (MP-285, Sutter) and patched the neuron with borosilicate glass pipettes filled with pipette solution (185 mM NH4HCO3 and 80 mM NH4Cl) by applying negative pressure. The astrocytes were clamped at −60 mV after the patched cell membrane was broken by rapidly applying negative pressure. After the electrophysiological data were recorded, the cytoplasmic chemical constituents were obtained from the assayed astrocytes by applying negative pressure. Only astrocytes with tightly held seals (>1 GΩ) and nonruptured membranes were selected for analysis, avoiding the dilution of intracellular fluid by ACSF. Once enough fluid was withdrawn from the cell, the patch pipette was quickly removed from the slice and then assayed via mass spectrometry. An AC voltage with an amplitude of 3 kV at ∼200 Hz was applied outside the spray capillary micropipette. The tip of the spray micropipette was kept ∼5 mm away from the orifice of the mass spectrometer. High-resolution mass measurements were analysed using a Thermo Exactive Plus mass spectrometer (Thermo Fisher Scientific). The main experimental parameters were: capillary temperature, 275 °C; S-lens radio-frequency level, 50%; mass resolution, 70,000; maximum injection time, 10 ms; microscan, 1. Other mass spectrometry experiments were conducted with LTQ Velos Pro Mass spectrometry instrument (Thermo Fisher Scientific). Mass spectrometry parameters were: capillary temperature, 275 °C, S-lens radio-frequency level, 42%; maximum injection time, 300 ms; microscan, 1. Chemical structure was confirmed by tandem mass spectrometry using LTQ Velos Pro Mass spectrometry instrument based on collision-induced dissociation (CID) with helium as background gas at 28% energy. Commercial electrospray ionization (ESI) source was removed before our experiments. Positive-ion mode was used in our experiments.
In vivo magnetic resonance spectroscopy measurement of ethanol metabolites.
In vivo MRS experiments were performed on a Bruker BioSpec 70/30USR Advance III 7T horizontal bore MR scanner (Bruker Biospin MRI) as described previously65,66. All mice were weighed, anaesthetized and placed in a prone position on a specially designed cradle and inserted into the MR scanner for a baseline measurement. The animals were under 1–2% isoflurane anesthesia at all times during the experiment. After the baseline measurement, mice were injected with ethanol (1–2 g kg−1, i.p.) or acetate (1.2 g kg−1 i.p.). The spectra were acquired at an interval of 0–30 min, 30–60 min or 0–60 min after systemic administration of ethanol or saline. A three-slice (transaxial, midsagittal and coronal) scout image using a fast, low-angle shot sequence was obtained to centre the mouse’s brain in the imaging field of view. A fast shimming procedure (FASTMAP) was used to improve the B0 homogeneity in the region of interest. Both proton density- and T2-weighted images were obtained for anatomical reference using a two-dimensional rapid acquisition with relaxation enhancement sequence covering the entire brain in the coronal plane (repetition time (TR), echo time (TE) eff1 and TE eff2 were 3,500, 19.4 and 56.8 ms, respectively; slice thickness was 0.5 mm; in-plane resolution was 100 × 100 μm2). For proton MRS, adjustments of all first- and second-order shims over the voxel of interest were accomplished using FASTMAP. The high quality of the MR spectrum was controlled by this procedure. A short echo-time point-resolved spectroscopy pulse sequence (TR and TE were 2,500 and 10 ms, respectively; number of averages was 300) was used for MRS data acquisition from the cerebellum and hippocampus. The unsuppressed water signal from each of the prescribed voxels was obtained to serve as a reference for determining metabolite concentrations.
Quantification of the MRS was based on frequency domain analysis using a ‘linear combination of model spectra’ (LCModel). In vivo spectra were analysed by a superposition of a simulated basis set provided by the LCModel software (version 6.3–0G; S. Provencher). The reference for determining metabolite concentration was the water signal, which was acquired from the same voxel. The metabolic profile was measured with the same parameters as those for biochemical data acquisition except the number of averages was set at 8. The results were normalized by LCModel to the chemical concentrations and expressed as millimolar. Cramér-Rao lower bounds as reported from the LCModel analysis were used for assessing the reliability of the major metabolites. The simulated spectra of alcohol metabolites and neurochemicals were included in the LCModel basis set. Only chemicals with Cramér-Rao lower bounds not greater than 25% were considered in each measurement.
Cerebellar slice preparation.
Aldh2Gfap+/+ and Aldh2Gfap−/− mice were used for cerebellar slice preparation. Animals were deeply anaesthetized with isoflurane. After decapitation, the brain was quickly excised from the skull and submerged in an ice-cold cutting solution that contained: 194 mM sucrose, 30 mM NaCl, 26 mM NaHCO3, 1.2 mM NaH2PO4, 4.5 mM KCl, 1 mM MgCl2 and 10 mM glucose (pH 7.4). The whole solution was oxygenated with 95% O2 and 5% CO2. After trimming the cerebellum from the brain, 250-μm parasagittal slices were cut using a vibratome (VT 1200S, Leica). The slides were transferred to ACSF oxygenated with 95% O2 and 5% CO2 and containing 126 mM NaCl, 24 mM NaHCO3, 1 mM NaH2PO4, 2.5 mM KCl, 2.5 mM CaCl2, 2 mM MgCl2, 10 mM glucose, 6 mM sucrose, 5 μM AP-5 (dl-2-amino-5-phosphonopentanoic acid), 5 μM DNQX (6,7-dinitroquinoxaline-2,3-dione) and 0.4 μM tetrodotoxin (pH 7.4), with an osmolality of 300 mOsm kg−1 at 32 °C for 30 min and cooled to RT for at least 30 min before incubation with 50 mM ethanol and 1 mM acetate for 5 min or 50 min before recording or GABA measurement.
Whole-cell patch-clamp recordings.
The protocol used in this experiment was described previously29,30. Cerebellar slices were transferred to a recording chamber that was continuously perfused with ASCF solution at RT. The slice chamber was mounted on the stage of an upright Olympus microscope and viewed with an ×40, 0.7-NA water immersion objective with infrared differential interference contrast optics. Whole-cell recordings were made under visual guidance from cerebellar GCs located in lobules 2–5 using pipettes (5–8 MΩ) filled with internal solution, and the holding potential was −60 mV. The internal solution used to measure tonic current contained 135 mM CsCl, 4 mM NaCl, 0.5 mM CaCl2, 10 mM HEPES, 5 mM EGTA, 2 mM Mg-ATP, 0.5 mM Na2-GTP, 5 mM QX-314, and pH was adjusted to 7.2 with CsOH (275–285 mOsmol). GABA-mediated tonic current was estimated by blocking with 50 μM bicuculline. Electrical signals were recorded and amplified with a MultiClamp 700B amplifier (Molecular Devices, AXON), digitized at 10 kHz and filtered at 2 kHz with a Digidata 1500A (Molecular Devices, AXON) using pCLAMP 10.3 software. Tonic current and mIPSCs were analysed by Clampfit and Mini analysis software (6.0.7, Synaptosoft). For the analysis of tonic GABAergic currents, we selected the gap-free mode of a 30-min recording. The tonic current was defined as the difference of the mean values between 2 to 3 min before and 3 min after administration of bicuculline. The percentage potentiation of tonic current mediated by ethanol or acetate was calculated from baseline. For the analysis of mIPSCs, we selected the epoch in each event before adding bicuculline. mIPSCs were distinguished from the background noise based on their fast rise times and slower decay times.
Measurement of cerebellar cortex GABA content by LC–MS/MS.
GABA was measured in the brain according to analytical conditions described previously with slight modifications33. Briefly, after the brain slices were incubated with 50 mM ethanol, 1 mM acetate or the same volume of ACSF for 50 min, the slices were harvested and weighed. The tissue was homogenized in 300 μl of ice-cold 0.1 N perchloric acid (PCA) containing 2% EDTA and 1% ethanol using a Precellys tissue homogenizer. Homogenized samples were vortexed and centrifuged at 10,000g for 10 min, and 10 μl hydrolysate was diluted 100-fold by the addition of 990 μl of 0.1 N PCA. LC–MS/MS analyses were conducted on an Agilent 6470 triple quadrupole mass spectrometer coupled to an Agilent 1260 LC system. GABA was separated using an Intrada Amino Acid column (50 mm × 3 mm column, 3 μm; Imtakt) at 40 °C. Mobile phase A consisted of 9/75/16/0.3 acetonitrile/tetrahydrofuran/25 mM ammonium formate/formic acid (vol/vol/vol/vol). Mobile phase B contained 20/80 acetonitrile/100 mM ammonium formate (vol/vol). Gradient elution (600 μl min−1) was initiated and held at 0% B for 3 min, followed by a linear increase to 17% B by 6.5 min. A step was increased to 100% B and held until 10 min, followed by a linear decrease to 0% B by 11 min and held until 13 min. The mass spectrometer was set for Agilent jet stream ionization source and operated in positive-ion mode. The source parameters were capillary voltage, 3,500 V; gas temperature, 300 °C; sheath gas temperature, 25 °C, sheath gas flow, 10 l min−1, gas flow, 5 l min−1; nitrogen was used as the nebulizing gas. CID was conducted using nitrogen. GABA level was analysed by multiple-reaction monitoring. The molecular ion and fragments for GABA were measured as follows: m/z 104.1 → 87.1 and 104.1 → 45.2 (CID energy of 8 V and 24 V, respectively). Levels of GABA were determined against a standard curve, using GABA as standard (Sigma). Values are expressed as nmol mg−1 wet tissue. The percentage change from baseline in GABA content induced by ethanol or acetate was calculated. Independent unpaired t-tests were used to test for significant differences between Aldh2Gfap+/+ and Aldh2Gfap−/− mice.
Measurement of serum and brain tissue ethanol and acetaldehyde by GC–MS.
Aldh2Gfap+/+, Aldh2Gfap−/−, Aldh2 KI and wild-type littermates were used for ethanol and acetaldehyde measurement in the cerebellum and blood. All mice were randomly assigned to different groups, weighed, individually housed and acclimatized for at least 1 h before the test. Aldh2Gfap+/+ and Aldh2Gfap−/− mice were injected with ethanol (1, 2 and 3.6 g kg−1) or an equal volume of saline intraperitoneally. Mice were anaesthetized with isoflurane and decapitated to retrieve the whole blood and the cerebellar and hippocampal tissue 10 or 50 min after systemic ethanol or saline. The GC/MS method was modified from work by Sun et al.25. All procedures were conducted at 0–4 °C. In brief, 50 μl of serum or about 30 mg of brain tissue was mixed with 5 μmol of 2H6-ethanol (internal standard for ethanol) and 0.04 μmol of 2H4-acetaldehyde (internal standard for acetaldehyde) before adding 200 μl of 0.6 N PCA into each sample. Plasma samples were centrifuged at 1,780g × 15 min at 2 °C after vortexing for 30 s. Brain samples were homogenized and then centrifuged at 13,200g × 15 min at 2 °C. The supernatant of each sample was quantitatively transferred into a 20-ml headspace vial and capped immediately. Headspace vials were then loaded onto the 111-vial tray of a headspace sampler coupled to a GC–MS instrument (Agilent Technologies). The concentrations of ethanol and acetaldehyde in plasma and brain were calculated by comparing the integrated areas of ethanol and acetaldehyde peaks on the gas chromatograms with those of internal standards added in each sample.
Measurement of serum and brain tissue acetate by GC–MS.
Procedures for the determination of sera and cerebellum acetate by GC–MS were described previously67. Acetate was analysed as its tertiary butyl dimethyl silyl ether derivative using GC–MS in the electron impact mode and quantified using [13C3]acetate. Aldh2Gfap+/+ and Aldh2Gfap−/− littermates were randomly assigned to different groups, weighed, individually housed and acclimatized for at least 1 h before the test. Aldh2Gfap+/+ and Aldh2Gfap−/− mice were injected with ethanol (2 g kg−1) or an equal volume of saline intraperitoneally. The mice were anaesthetized with isoflurane and decapitated to retrieve the whole blood and the cerebellar tissue 20 or 50 min after i.p. ethanol or saline.
For PCA extraction of sera acetate, 50 μl of a 3.6% PCA solution and 5 μl (32 μM solution) of a sodium [13C3]acetate (Sigma-Aldrich) was added to 50 μl of frozen sera in 100-μl polypropylene conical snap-capped tubes. Samples were vortexed for 1 min and neutralized with 5 μl KHCO3 (3 M), according to previously described methods. For PCA extraction of cerebellar acetate, approximately 30 mg of frozen brain tissue was added to 100 μl of a 3.6% PCA solution and 10 μl (32 μM solution) of sodium [13C3]acetate in 2-ml polypropylene screw-capped tubes with 10–15 small glass beads. Samples were shaken on a Mini Bead Beater (Biopsec Products) for 30 s (2×) until samples were homogeneous. Samples were placed on ice until centrifugation at 4 °C in a Sorvall benchtop centrifuge at a speed of 10.7g for 2 min. Eighty microliters of the upper layer was pipetted into clean tubes and neutralized with 9 μl KHCO3 (3 M).
For sample derivatization, 10 μl of the sample extracts was evaporated under a stream of nitrogen to dryness in 1.5-ml sylinized screw-capped vials. Samples were immediately reacted with 5 μl of the silylating reagent, N-methyl-N-(tert-butylmethylsilyl) trifluoroacetamide with 1% tert-butyldimethylchlorosilane reagent and bis-trimethylfluoro methylsilyl (Pierce Chemical) in 15 μl of acetonitrile and heated to 60 °C for 5 min. For GC–MS determinations, samples were analysed on an Agilent 5973 quadrupole GC–MS instrument, and 1 μl of the sample solution was injected onto a 250 × 30 μm2 capillary DB-1 (Agilent) column in split mode (100:1) using helium as the carrier gas. The injector temperature was set at 250 °C and the transfer line at 280 °C. The gas chromatography oven temperature was programmed from 40 to 325 °C at 15 °C per min. The mass spectrometer was operated in the electron impact mode (70 eV) and the quadrupole mass analyser scanned for ions that corresponded to a loss of 15 mass units (−CH3) from the molecular ion of the acetate derivative (m/z 159) and its corresponding 13C-labelled internal standard (m/z 162) using selected ion monitoring mode. The m/z 162:159 ratio was used to quantify the quantity of acetate in each sample.
Rotarod test.
The protocol was based on a previous study with a modification68. A computer-interfaced rotarod accelerating from 4 to 40 rotations per min over 300 s was used (ENV-575M, MED Associates). Mice were trained using five trials per day with a 30-min intertrial interval for 4 consecutive days. Each trial ended when the mouse fell off the rotarod or after 300 s had elapsed. The time that each mouse maintained its balance on the rotating rod was measured as the latency to fall. Values for latency to fall were averaged results for five trials every day. Mice that failed to stand on the rotarod longer than 250 s were disqualified after 4 d of training. On testing days, each mouse was placed on the rotarod to test and to record the latency to fall 10 min after i.p. drug injection. Mice with i.c.v. cannulas were injected with ACSF or drugs 15 min before i.p. saline or drugs.
Statistical analysis.
Data are expressed as the mean ± s.e.m. and analysed with GraphPad Prism version 7.0 for Windows. An unpaired two-tailed Student’s t-test was used for comparison between two groups, and one-way ANOVA or two-way ANOVA followed by Tukey’s test was used for multiple comparisons of more than two groups, as indicated in specific figures. Repeated-measures ANOVA was performed to analyse the ethanol consumption and the values of rotarod performance at different time points. P < 0.05 was considered statistically significant.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All data from these studies are contained within this manuscript or are available from the corresponding author upon reasonable request. Source data are provided with this paper.
Extended Data
Extended Data Fig. 1 |. Astrocyte-specific distribution of ALDH2 in the cerebellum.
a, Bar graphs of ALDH2 activity in cerebellar anterior, vermis and posterior regions. (n=6 mice/group). Data are presented as means±s.e.m. Analysis was performed using one-way analysis of variance (ANOVA) followed by Tukey’s test. b, Epifluorescence images of ALDH2 (green), GFAP (violet) and L7/PC (red) gene in a cerebellar sagittal slice. Red scale bar, 200 μm; White scale bars, 50 μm. c, Epifluorescence images of ALDH2 (green), MAP2 (violet) and L7/PC (red) gene. No colocalization across these genes. Red scale bar, 200 μm; White scale bars, 50 μm. d, Images showing a colocalization of ALDH2 (green) with GFAP (red) but not MAP2 (violet) gene in cerebellum. Red scale bar, 200 μm; White scale bars, 50 μm. For b, c and d, Experiments were repeated four times for each biologically independent mouse, with similar results obtained. e, Quantification of cluster colocalization between ALDH2 and ALDH1L1 in cerebellum. The experiment was repeated four times for each biologically independent mouse, with similar results obtained (n=3 mice). Quantitative data was provided in Supplementary Table 2. YES: colocalization; NO: no colocalization; White scale bars, 50 μm.
Extended Data Fig. 2 |. ALDH2 expression in different brain regions of Aldh2Gfap−/− mice.
a, the schematic diagram shows the generation of astrocytic specific Aldh2 deficient mice. b, A representative image of PCR genotyping result. The image of agar gel migration shows flox (homozygous: one band, 365 bp; heterozygous: two bands, 365 and 300 bp) and Cre (negative: one band, 350 bp; positive: two bands, 350 and 200 bp) gene. c, RNAscope images of ALDH2 signals in cerebellum and liver slices from conditional and global Aldh2−/− mice. Noted that ALDH2 was selectively depleted from microdomains surrounding PC signal in cerebellar slice from Aldh2Gfap−/− mice. Liver ALDH2 mRNA was intact in Aldh2Gfap−/− mice, whereas liver ALDH2 signal was abolished in Aldh2−/− mice. Each experiment was repeated four times with similar results obtained. White scale bars, 100 μm. d, e, Representative peaks from electrophoresis densitometry of cerebellar (n=7, 7 and 3 mice, respectively) and liver (n=5, 5 and 3 mice, respectively) ALDH2 proteins from Aldh2Gfap+/+ (black line or rectangle), Aldh2Gfap−/− and Aldh2−/− mice. f, Summary statistics of cerebellar ALDH2 enzymatic activity detected in Aldh2L7−/− and Aldh2Camk2a−/− mice (n=6, 6, 7 and 7 mice, respectively). g, Representative electropherogram traces of ALDH2 protein samples collected from different brain areas of Aldh2Gfap+/+ and Aldh2Gfap−/− mice. h, Summary statistics of brain ALDH2 protein detected in difference brain areas from Aldh2Gfap+/+ (n=5, 4, 6, 4, 4 and 7 mice, respectively) and Aldh2Gfap−/− (n=5, 4, 6, 4, 4 and 7 mice, respectively) mice. i, Representative electropherogram traces of protein samples collected from different brain areas from Aldh2Camk2a+/+ and Aldh2Camk2a−/− mice. j, Summary statistics of brain ALDH2 protein detected in difference brain areas from Aldh2Camk2a+/+ and Aldh2Camk2a−/− mice (n=4 mice/group). k, l, Representative electropherogram traces and summary statistics of brain ALDH2 protein detected in different brain areas from astrocytic ALDH2 deficient mice generated by crossing Aldh2 flox with GFAPCre (Jax, 024098), Aldh2Gfap−/− mice and their wildtype littermates, Aldh2Gfap+/+ (n=3 mice/group). Data are presented as means ±s.e.m. Analysis was performed using unpaired two tailed student’s t test (h, j, f, l) or one-way analysis of variance (ANOVA) followed by Tukey’s test (d, e). PFC, Prefrontal cortex; CE, Cerebellum; HIPP, Hippocampus; CeA, Central nucleus of the amygdala; VTA, Ventral tegmental area; BLA, Basolateral amygdala; NAc, Nucleus accumbent.
Extended Data Fig. 3 |. Astrocytic Aldh2 deficiency does not alter serum and brain (cerebellar cortex) ethanol and acetaldehyde contents.
a, b, Serum and cerebellum ethanol concentrations measured 50 min after ethanol (2 g/kg, i.p.) in WT and Aldh2*2KI mice (n=4 mice/group). c, d, Graphs showing ethanol concentrations in the serum and cerebellum 50 min after ethanol in Aldh2Gfap+/+ and Aldh2Gfap−/− mice (0 g/kg ethanol: n=4 mice/group; 1 g/kg ethanol: n=12 and 13 from Aldh2Gfap+/+ and Aldh2Gfap−/− mice respectively (serum), n=7 and 8 from Aldh2Gfap+/+ and Aldh2Gfap−/− mice respectively (cerebellum); 2 g/kg ethanol: n=5 mice/group; 3.6 g/kg ethanol: n=5 mice/group). e, f, Serum and cerebellum acetaldehyde contents measured after ethanol (1 g/kg, i.p.) in Aldh2Gfap+/+ and Aldh2Gfap−/− mice (0 min: n=4 mice/group; 10 min: n=9 mice/group (serum), n=4 mice/group (cerebellum); 50 min: n=11 mice/group (serum), n=7 and 8 from Aldh2Gfap+/+ and Aldh2Gfap−/− mice respectively (cerebellum)). g, h, Serum and cerebellum ethanol contents measured after ethanol (1 g/kg, i.p.) in Aldh2Gfap+/+ and Aldh2Gfap−/− mice (0 min: n=4 mice/group; 10 min: n=9 mice/group (serum), n=4 mice/group (cerebellum); 50 min: n=12 and 13 from Aldh2Gfap+/+ and Aldh2Gfap−/− mice respectively (serum), n=7 and 8 from Aldh2Gfap+/+ and Aldh2Gfap−/− mice respectively (cerebellum)). Data are presented as mean±s.e.m. Analysis was performed using unpaired two tailed student’s t test (a, b) or two-way analysis of variance (ANOVA) (c-h).
Extended Data Fig. 4 |. In vivo MRS quantitative measurement of brain ethanol metabolites and neurochemicals.
a, b, Representative peaks and Summary statistics of brain ALDH2 protein in cerebellum and liver from Aldh2Hep+/+ and Aldh2Hep−/−mice (n=4 mice/group).c, d, In vivo MRS measurement of cerebellar acetate and GABA contents after systemic ethanol (1–2 g/kg, i.p.) in Aldh2Gfap+/+ mice (n=5, 3 and 5 mice, respectively). e, Ethanol enhancement of cerebellar GABA, Glutamine, Glutamate and NAA in Aldh2 flox mice (n=9 mice/group). f, g, Statistic summaries of cerebellar ethanol by MRS after systemic administration of ethanol (1–2 g/kg, i.p.) in astrocytic ALDH2 deficient mice (f: n=3 mice/group; g: n=5 mice/group), hepatocytic ALDH2 deficient mice (Aldh2Hep+/+: n=4 mice and Aldh2Hep−/−: n=6 mice) and their wild type littermates mice. g, Bar graphs showing cerebellar ethanol concentrations after ethanol (2 g/kg, i.p.) by in vivo MRS measurement in Aldh2Hep+/+ (n=4) and Aldh2Hep−/− (n=4) mice. h, Bar graph showing the basal levels of GABA, Glutamine, Glutamate and NAA in cerebellum in Aldh2Gfap+/+ and Aldh2Gfap−/− mice (n=5 mice/group). i, Bar graph showing the basal levels of GABA, Glutamine, Glutamate and NAA in cerebellum in Aldh2Hep+/+ (n=4) and Aldh2Hep−/− (n=6) mice. Data are presented as mean±s.e.m. Groups were compared by unpaired two tailed student’s t test (b, e-i), or one-way analysis of variance (ANOVA) followed by Tukey’s test (c, d).
Extended Data Fig. 5 |. Ethanol does not significantly alter GABAergic mIPSCs and GABA-activated current in GCs.
a, b, Trace records and summary statistics of tonic current recorded using whole cell recording in cerebellar GCs from Aldh2Gfap+/+ and Aldh2Gfap−/− mice (ACSF: n=8 cells, 6 mice). c, Summary statistics of the cell membrane capacitance with and without ethanol and acetate (n=8, 8, 7, 7, 7, 8, 9, 9, 9 and 10 cells, respectively). d, e, Left, Trace records of GABA (20 μM) activated currents in cerebellar GCs from Aldh2Gfap+/+ and Aldh2Gfap−/− mice. Right, Summary of the average amplitudes of GABA-activated currents (n=4 cells/group, 4 mice). f, Statistic summary of the average levels of cerebellar GABA contents measured by LC-MS/MS in Aldh2Gfap+/+ and Aldh2Gfap−/− mice (n=8 mice/group). g-j, Trace records and summary statistics of mIPSCs in GCs from Aldh2Gfap+/+ and Aldh2Gfap−/− mice (n=8 cells/group). k-n, The effect of ethanol on GABAergic IPSCs (n=9 cells/group, 6 mice). Data are presented as mean±s.e.m. Groups were compared by one-way analysis of variance (ANOVA) (c), or unpaired two tailed student’s t test (b, c, e, f, h, i, l, m).
Extended Data Fig. 6 |. Characterization of unilateral conditional deletion of cerebellar astrocytic ALDH2.
a, Bar graph showing the rotarod performance of Aldh2Camk2a+/+ and Aldh2Camk2a−/− mice (n=10 mice/group) after 10 min 1 g/kg ethanol i.p. injection. b, Time course of the basal levels of the rotarod performance in consecutive trails in Aldh2Gfap+/+ (n=9) and Aldh2Gfap−/− (n=10) mice. c, Time courses of core-body temperature after ethanol (2 g/kg, i.p.) in Aldh2Gfap+/+ (n=9) and Aldh2Gfap−/− (n=13) mice. d,e, Florescent microscopy images of ALDH2 signal (green) in cerebellar slices from Aldh2 flox mice previously injected with AAV-GFAPCre and AAV-Synt1Cre virus (4 weeks after injection). Experiments were repeated four times for each biologically independent mouse, with similar results obtained (n=3 mice). Red scale bar, 500 μm; White scale bars, 50 μm. f, Bar graph showing the baseline levels of rotarod performance in cerebellar virus injected mice (n=15 mice for AAV-Synt1Cre; n=16 mice for AAV-GFAPCre). g, Time courses of the levels of rotarod performance after i.c.v. injection of CGP (200 ng, n=7 mice) or Bicuculline (200 ng, n=5 mice). Data are presented as mean±s.e.m. Analysis was performed using unpaired two tailed student’s t test (f) or two-way analysis of variance (ANOVA) followed by Turkey’s test (a), or two-way repeated-measures analysis of variance (ANOVA) (b,c,g).
Extended Data Fig. 7 |. A hypothetical mechanism for astrocytic ALDH2 control of alcohol metabolism and action in the brain.
Simplified diagram of ALDH2-dependent pathways that mediate ethanol-induced elevation in cerebellar GABA levels and discoordination.
Supplementary Material
Acknowledgements
We thank F. Langevin and K. J. Patel (University of Cambridge) for Aldh2 flox mice. We especially thank M. Chen, X. Sun and X. Li (Department of Diagnostic Radiology and Nuclear Medicine, University of Maryland) for their technical support for in vivo MRS experiments and data analysis. We thank A. Salinas, G. Luo, T. Ren, A. Guillot and H.-J. White (NIAAA, NIH) for technical assistance and comments on the manuscript. We thank the Human Brain Collection Core at the National Institute of Mental Health for providing human cerebellar tissues. This work was supported by grants 1UL1TR003098 (to Q. Cao) from the University of Maryland, Baltimore, Institute for Clinical & Translational Research (ICTR) and the National Center for Advancing Translational Sciences (NCATS) Clinical Translational Science Award (CTSA), K08AA024895-01A1 to (Q. Cao) from the National Institue on Alcohol Abuse and Alcoholism (NIAAA), National Institutes of Health (NIH), chairman seed grant award (to Q. Cao) of the Department of Diagnostic Radiology and Nuclear Medicine, University of Maryland, Baltimore and 81801938 (to S.J.) from the National Natural Science Foundation of China.
Footnotes
Competing interests
The authors declare no competing interests.
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s42255-021-00357-z.
Supplementary information The online version contains supplementary material available at https://doi.org/10.1038/s42255-021-00357-z.
Peer review information Primary Handling Editors: Pooja Jha; Isabella Samuelson. Nature Metabolism thanks Juan Bolaños and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Reprints and permissions information is available at www.nature.com/reprints.
This is a U.S. government work and not under copyright protection in the U.S.; foreign copyright protection may apply 2021
References
- 1.Volkow ND, Koob G & Baler R Biomarkers in substance use disorders. ACS Chem. Neurosci 6, 522–525 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Cui C & Koob GF Titrating tipsy targets: the neurobiology of low-dose alcohol. Trends Pharmacol. Sci 38, 556–568 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abrahao KP, Salinas AG & Lovinger DM Alcohol and the brain: neuronal molecular targets, synapses and circuits. Neuron 96, 1223–1238 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McLaughlin PJ, Chuck TL, Arizzi-LaFrance MN, Salamone JD & Correa M Central versus peripheral administration of ethanol, acetaldehyde and acetate in rats: effects on lever pressing and response initiation. Pharmacol. Biochem. Behav 89, 304–313 (2008). [DOI] [PubMed] [Google Scholar]
- 5.Eng MY LS & Wall TL ALDH2, ADH1B and ADH1C genotypes in Asians: a literature review. Alcohol Res. Health 30, 22–27 (2007). [PMC free article] [PubMed] [Google Scholar]
- 6.Edenberg HJ The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res. Health 30, 5–13 (2007). [PMC free article] [PubMed] [Google Scholar]
- 7.Aragon CM, Rogan F & Amit Z Ethanol metabolism in rat brain homogenates by a catalase-H2O2 system. Biochem. Pharmacol 44, 93–98 (1992). [DOI] [PubMed] [Google Scholar]
- 8.Kim JI et al. Aldehyde dehydrogenase 1a1 mediates a GABA synthesis pathway in midbrain dopaminergic neurons. Science 350, 102–106 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen CH, Ferreira JC, Gross ER & Mochly-Rosen D Targeting aldehyde dehydrogenase 2: new therapeutic opportunities. Physiol. Rev 94, 1–34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deitrich R, Zimatkin S & Pronko S Oxidation of ethanol in the brain and its consequences. Alcohol Res. Health 29, 266–273 (2006). [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang L et al. Increased brain uptake and oxidation of acetate in heavy drinkers. J. Clin. Invest 123, 1605–1614 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deelchand DK, Shestov AA, Koski DM, Ugurbil K & Henry PG Acetate transport and utilization in the rat brain. J. Neurochem 109, 46–54 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carmichael FJ et al. Central nervous system effects of acetate: contribution to the central effects of ethanol. J. Pharmacol. Exp. Ther 259, 403–408 (1991). [PubMed] [Google Scholar]
- 14.Volkow ND et al. Acute alcohol intoxication decreases glucose metabolism but increases acetate uptake in the human brain. Neuroimage 64, 277–283 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mews P et al. Alcohol metabolism contributes to brain histone acetylation. Nature 574, 717–721 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nuutinen H, Lindros K, Hekali P & Salaspuro M Elevated blood acetate as indicator of fast ethanol elimination in chronic alcoholics. Alcohol 2, 623–626 (1985). [DOI] [PubMed] [Google Scholar]
- 17.Wagner MJ, Kim TH, Savall J, Schnitzer MJ & Luo L Cerebellar granule cells encode the expectation of reward. Nature 544, 96–100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carta I, Chen CH, Schott AL, Dorizan S & Khodakhah K Cerebellar modulation of the reward circuitry and social behaviour. Science 10.1126/science.aav0581 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen CH, Fremont R, Arteaga-Bracho EE & Khodakhah K Short latency cerebellar modulation of the basal ganglia. Nat. Neurosci 17, 1767–1775 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sathyanesan A et al. Emerging connections between cerebellar development, behaviour and complex brain disorders. Nat. Rev. Neurosci 20, 298–313 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lackey EP, Heck DH & Sillitoe RV Recent advances in understanding the mechanisms of cerebellar granule cell development and function and their contribution to behavior. F1000Res 10.12688/f1000research.15021.1 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reeber SL, Otis TS & Sillitoe RV New roles for the cerebellum in health and disease. Front Syst. Neurosci 7, 83 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langevin F, Crossan GP, Rosado IV, Arends MJ & Patel KJ Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature 475, 53–58 (2011). [DOI] [PubMed] [Google Scholar]
- 24.Song AJ & Palmiter RD Detecting and avoiding problems when using the Cre-lox system. Trends Genet. 34, 333–340 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun Q, Zhang W, Zhong W, Sun X & Zhou Z Dietary fisetin supplementation protects against alcohol-induced liver injury in mice. Alcohol Clin. Exp. Res 40, 2076–2084 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xiang Y & Shen J In vivo detection of intermediate metabolic products of [1-13C]ethanol in the brain using 13C magnetic resonance spectroscopy. NMR Biomed. 24, 1054–1062 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guillot A et al. Targeting liver aldehyde dehydrogenase 2 prevents heavy but not moderate alcohol drinking. Proc. Natl Acad. Sci. USA 116, 25974–25981 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang WT, Lee P, Hui D, Michaelis EK & Choi IY Effects of ethanol exposure on the neurochemical profile of a transgenic mouse model with enhanced glutamate release using in vivo 1H MRS. Neurochem. Res 44, 133–146 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carta M, Mameli M & Valenzuela CF Alcohol enhances GABAergic transmission to cerebellar granule cells via an increase in Golgi cell excitability. J. Neurosci 24, 3746–3751 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Diaz MR & Valenzuela CF Sensitivity of GABAergic tonic currents to acute ethanol in cerebellar granule neurons is not age- or delta subunit-dependent in developing rats. Alcohol Clin. Exp. Res 40, 83–92 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee S et al. Channel-mediated tonic GABA release from glia. Science 330, 790–796 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Woo J et al. Control of motor coordination by astrocytic tonic GABA release through modulation of excitation/inhibition balance in cerebellum. Proc. Natl Acad. Sci. USA 115, 5004–5009 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cinar R et al. Hybrid inhibitor of peripheral cannabinoid-1 receptors and inducible nitric oxide synthase mitigates liver fibrosis. JCI Insight 10.1172/jci.insight.87336 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang J et al. Chronic intermittent ethanol-induced switch of ethanol actions from extrasynaptic to synaptic hippocampal GABAA receptors. J. Neurosci 26, 1749–1758 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valenzuela CF & Jotty K Mini-review: effects of ethanol on GABAA receptor-mediated neurotransmission in the cerebellar cortex–recent advances. Cerebellum 14, 438–446 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Zhu H et al. Moderate UV exposure enhances learning and memory by promoting a novel glutamate biosynthetic pathway in the brain. Cell 173, 1716–1727 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Sonnewald U et al. NMR spectroscopic studies of [13C]acetate and [13C] glucose metabolism in neocortical astrocytes: evidence for mitochondrial heterogeneity. Dev. Neurosci 15, 351–358 (1993). [DOI] [PubMed] [Google Scholar]
- 38.Sonnewald U et al. Direct demonstration by [13C]NMR spectroscopy that glutamine from astrocytes is a precursor for GABA synthesis in neurons. Neurochem. Int 22, 19–29 (1993). [DOI] [PubMed] [Google Scholar]
- 39.Tiwari V, Veeraiah P, Subramaniam V & Patel AB Differential effects of ethanol on regional glutamatergic and GABAergic neurotransmitter pathways in mouse brain. J. Neurochem 128, 628–640 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Rustay NR et al. Sensitivity and tolerance to ethanol-induced incoordination and hypothermia in HAFT and LAFT mice. Pharmacol. Biochem. Behav 70, 167–174 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Rustay NR, Wahlsten D & Crabbe JC Assessment of genetic susceptibility to ethanol intoxication in mice. Proc. Natl Acad. Sci. USA 100, 2917–2922 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saeed Dar M Co-modulation of acute ethanol-induced motor impairment by mouse cerebellar adenosinergic A1 and GABAA receptor systems. Brain Res. Bull 71, 287–295 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Hipolito L, Sanchez MJ, Polache A & Granero L Brain metabolism of ethanol and alcoholism: an update. Curr. Drug Metab 8, 716–727 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Wilson DF & Matschinsky FM Ethanol metabolism: The good, the bad, and the ugly. Med. Hypotheses 140, 109638 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Deelchand DK, Nelson C, Shestov AA, Ugurbil K & Henry PG Simultaneous measurement of neuronal and glial metabolism in rat brain in vivo using co-infusion of [1,6-13C2]glucose and [1,2-13C2]acetate. J. Magn. Reson 196, 157–163 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zimatkin SM, Pronko SP, Vasiliou V, Gonzalez FJ & Deitrich RA Enzymatic mechanisms of ethanol oxidation in the brain. Alcohol Clin. Exp. Res 30, 1500–1505 (2006). [DOI] [PubMed] [Google Scholar]
- 47.Isse T, Matsuno K, Oyama T, Kitagawa K & Kawamoto T Aldehyde dehydrogenase 2 gene targeting mouse lacking enzyme activity shows high acetaldehyde level in blood, brain and liver after ethanol gavages. Alcohol Clin. Exp. Res 29, 1959–1964 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Martinez-Rodriguez R et al. Synaptic and non-synaptic immunolocalization of GABA and glutamate acid decarboxylase in cerebellar cortex of rat. Cell Mol. Biol 39, 115–123 (1993). [PubMed] [Google Scholar]
- 49.Le Meur K, Mendizabal-Zubiaga J, Grandes P & Audinat E GABA release by hippocampal astrocytes. Front Comput Neurosci 6, 59 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Joshi AU et al. Aldehyde dehydrogenase 2 activity and aldehydic load contribute to neuroinflammation and Alzheimer’s disease related pathology. Acta Neuropathol. Commun 7, 190 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Overstreet DH, Knapp DJ, Breese GR & Diamond I A selective ALDH-2 inhibitor reduces anxiety in rats. Pharmacol. Biochem. Behav 94, 255–261 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zambelli VO et al. Aldehyde dehydrogenase 2 regulates nociception in rodent models of acute inflammatory pain. Sci. Transl. Med 6, 251ra118 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yao L et al. Inhibition of aldehyde dehydrogenase 2 suppresses cocaine seeking by generating THP, a cocaine use-dependent inhibitor of dopamine synthesis. Nat. Med 16, 1024–1028 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mitew S, Kirkcaldie MT, Dickson TC & Vickers JC Altered synapses and gliotransmission in Alzheimer’s disease and AD model mice. Neurobiol. Aging 34, 2341–2351 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Jo S et al. GABA from reactive astrocytes impairs memory in mouse models of Alzheimer’s disease. Nat. Med 20, 886–896 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu Z, Guo Z, Gearing M & Chen G Tonic inhibition in dentate gyrus impairs long-term potentiation and memory in an Alzheimer’s disease model. Nat. Commun 5, 4159 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Marek S et al. Spatial and temporal organization of the individual human cerebellum. Neuron 100, 977–993 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin CY, Chen CH, Tom SE & Kuo SH, Alzheimer’s Disease Neuroimaging Initiative. Cerebellar volume is associated with cognitive decline in mild cognitive impairment: results from ADNI. Cerebellum 19, 217–225 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Garcia AD, Doan NB, Imura T, Bush TG & Sofroniew MV GFAP-expressing progenitors are the principal source of constitutive neurogenesis in adult mouse forebrain. Nat. Neurosci 7, 1233–1241 (2004). [DOI] [PubMed] [Google Scholar]
- 60.Maeda M, Hasumura Y & Takeuchi J Localization of cytoplasmic and mitochondrial aldehyde dehydrogenase isozymes in human liver. Lab. Invest 59, 75–81 (1988). [PubMed] [Google Scholar]
- 61.Kitagawa K et al. Aldehyde dehydrogenase 2 associates with oxidation of methoxyacetaldehyde; in vitro analysis with liver subcellular fraction derived from human and Aldh2 gene targeting mouse. FEBS Lett. 476, 306–311 (2000). [DOI] [PubMed] [Google Scholar]
- 62.Koivisto T, Carr LG, Li TK & Eriksson CJ Mitochondrial aldehyde dehydrogenase polymorphism in AA and ANA rats: lack of genotype and phenotype line differences. Pharmacol. Biochem. Behav 45, 215–220 (1993). [DOI] [PubMed] [Google Scholar]
- 63.Gao Y et al. Alcohol inhibits T cell glucose metabolism and hepatitis in ALDH2-deficient mice and humans: roles of acetaldehyde and glucocorticoids. Gut 68, 1311–1322 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu H et al. Single-neuron identification of chemical constituents, physiological changes and metabolism using mass spectrometry. Proc. Natl Acad. Sci. USA 114, 2586–2591 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ende G Proton magnetic resonance spectroscopy: relevance of glutamate and GABA to neuropsychology. Neuropsychol. Rev 25, 315–325 (2015). [DOI] [PubMed] [Google Scholar]
- 66.Xu S et al. Decreased taurine and creatine in the thalamus may relate to behavioral impairments in ethanol-fed mice: a pilot study of proton magnetic resonance spectroscopy. Mol Imaging 17, 1536012117749051 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pawlosky RJ et al. Alterations in brain glucose utilization accompanying elevations in blood ethanol and acetate concentrations in the rat. Alcohol Clin. Exp. Res 34, 375–381 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xiong W et al. Cannabinoids suppress inflammatory and neuropathic pain by targeting α3 glycine receptors. J. Exp. Med 209, 1121–1134 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data from these studies are contained within this manuscript or are available from the corresponding author upon reasonable request. Source data are provided with this paper.