Abstract
The novel coronavirus SARS-CoV-2 has been affecting the world, causing severe pneumonia and acute respiratory syndrome, leading people to death. Therefore, the search for anti-SARS-CoV-2 compounds is pivotal for public health. Natural products may present sources of bioactive compounds; among them, flavonoids are known in literature for their antiviral activity. Siparuna species are used in Brazilian folk medicine for the treatment of colds and flu. This work describes the isolation of 3,3′,4′-tri-O-methyl-quercetin, 3,7,3′,4′-tetra-O-methyl-quercetin (retusin), and 3,7-di-O-methyl-kaempferol (kumatakenin) from the dichloromethane extract of leaves of Siparuna cristata (Poepp. & Endl.) A.DC., Siparunaceae, using high-speed countercurrent chromatography in addition to the investigation of their inhibitory effect against SARS-CoV-2 viral replication. Retusin and kumatakenin inhibited SARS-CoV-2 replication in Vero E6 and Calu-3 cells, with a selective index greater than lopinavir/ritonavir and chloroquine, used as control. Flavonoids and their derivatives may stand for target compounds to be tested in future clinical trials to enrich the drug arsenal against coronavirus infections.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s43450-021-00162-5.
Keywords: Countercurrent chromatography, Siparunaceae, Mass spectrometry, O-Methyl flavonoids, Coronavirus
Introduction
Coronaviruses (CoVs), Coronaviridae family, are subdivided into the Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus genera, which are etiologic agents causing several acute and chronic respiratory, enteric, and central nervous system diseases (Chinsembu, 2020; Mani et al. 2020). The betacoronoviruses SARS-CoV and MERS-CoV are the etiologic agents of the severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) that occurred from 2002 and 2012. In 2019, the new coronavirus SARS-CoV-2 (Betacoronavirus) appeared in Wuhan, China, causing the worldwide pandemic of COVID-19 and public health concerns (Mani et al. 2020). Due to the highly complex pathophysiology of SARS-CoV-2 infection (Elizalde-González 2020), involving not only the activation of the immune and hematologic systems but also the involvement and impairment of different organs and potential for systemic complications, the term multiple organ dysfunction syndrome, MODS-CoV-2 was proposed (Maisch, 2020; Robba et al. 2020). As of May 2021, there are over 160 million positive cases reported in 220 affected countries and regions, with death numbers surpassing the figure of 3 million (WHO 2021). In Brazil, the deaths from the disease have so far surpassed the number of 500,000 (JHU CSSE COVID-19 Data 2021). Due to this worldwide scenario, the search for vaccines, medicines, monoclonal antibodies, interferon therapies, peptides, and natural medicines has been developed for fighting the new coronavirus. The non-structural proteins 3-chymotrypsin-like protease (3CLpro), papain-like protease (PLpro), and RNA-dependent RNA polymerase (RdRp) and the structural protein spike (S) protein, present in the SARS-CoV-2 genome, have been research targets for drug interventions against this new virus (Mani et al. 2020). Proteases are important therapeutic targets due their crucial activity in the replicative cycle of the virus. Both main protease (Mpro) and PLpro act by cleaving the pp1a and pp1ab polyproteins that are translated from the viral genome shortly after SARS-CoV-2 enters the host cell. Together, they give rise to sixteen functional non-structural proteins. PLpro does proteolytic cleavage from nsp1 to nsp3, while nsp4 to nsp16 are excised by Mpro (Abdul et al., 2021). Earlier studies showed SARS-CoV 3CLpro and SARS-CoV PLpro have been considered potential targets for the design and development of antiviral drugs. Several in silico simulations suggested the possibility that flavonoids can affect key factors responsible for the virus viability replication. In the course of 2020 and 2021, works have been published dealing with the screening of natural flavonoids as a promising class of SARS-CoV-2 inhibitors (Komolafe et al. 2021), blocking its entry or replication (El-Mordy et al. 2020; Jo et al. 2020; Cherrak et al. 2020; Russo et al. 2020; Mouffouk et al. 2021; Pandey et al. 2021).
In silico screening studies with different plant species from traditional Chinese medicines (TCMs) showed that flavonoids such as baicalin, epigallocatechin gallate, herbacetin, isobavaschalcone, kaempferol derivatives, luteolin, myricetin, quercetin 3-β-d-glucoside, rhoifolin, and scutellarein have been described as potential inhibitors of SARS-CoV-2 PLpro and 3CLpro (Chinsembu, 2020; Mani et al. 2020). Flavonoids commonly present in propolis samples have also been highlighted as promising agents that could attenuate SARS-CoV-2 infection and its consequences (Berretta et al. 2020). A recent work described three flavonoids that were found to efficiently block the enzymatic activity of SARS-CoV 3CLpro, among them is pectolinarin, a dimethylated flavone glycoside (Jo et al. 2020). Furthermore, flavonoids have demonstrated an efficient modulation potential against the SARS-COV-2–induced inflammatory storm and counteracting lung inflammation (Liskova et al. 2021; Santana et al., 2021).
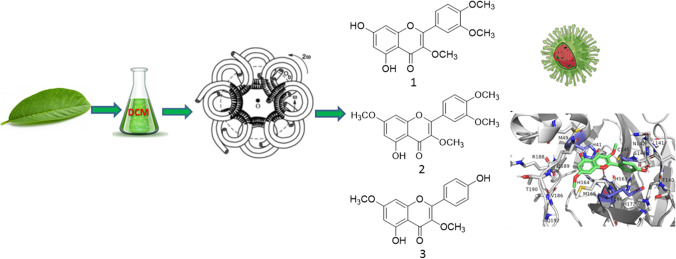
According to “Diagnosis and Treatment Program for Corona Virus Disease 2019 (COVID-19)” in China, treatment with traditional medicine is recommended, which has achieved good clinical effects (Ren et al. 2020). In the same way, a recent work (Cock and Vuuren 2020) revealed the potential of South African medicinal plants used to treat viral respiratory diseases in screening studies against the SARS-CoV-2 virus. Brazil has a long tradition of medicinal plant use, and in many regions of the country where medical care units are scarce or inexistent, traditional medicinal therapies are the only option which is being used to overcome COVID-19. Plants of the genus Siparuna are commonly used in Brazilian folk medicine in the treatment of colds, fever, headache, and rheumatism, as well as in rituals (Leitão et al. 1999). A syrup prepared with Siparuna apiosyce, a species from the first Brazilian Pharmacopeia, has long been commercialized in Brazil for the treatment of colds and flu with the name of “limão-bravo.” During the investigation on the chemistry of Siparuna species for the management of colds and fever, we came across a methylated flavonoid-rich extract with anti-influenza activity (Leal et al. 2021) from the Amazonian species Siparuna cristata (Poepp. & Endl.) A.DC., Siparunaceae. In this paper, metabolic profiling by HPLC, isolation by high-speed countercurrent chromatography (HSCCC), structure elucidation, and molecular docking studies of the methylated flavonoids 1–3 from S. cristata were performed to investigate their in vitro inhibitory effect against SARS-CoV-2 and to evaluate their in silico inhibitory effect against 3CLpro and PLpro SARS-CoV-2 proteases.
Materials and Methods
Plant Material
Leaves of Siparuna cristata (Poepp. & Endl.) A.DC., Siparunaceae, were collected at Reserva Ducke, Manaus, in August 2015. A voucher specimen is deposited at Instituto Nacional de Pesquisas da Amazônia (INPA) herbarium (Manaus, AM) under the registration INPA 269,731. This work was authorized by the Directing Council of Genetic Heritage (Conselho de Gestão do Patrimônio Genético, CGEN) by the authorization A3C04CB. The leaves were dried in a ventilated oven (Marconi, model lMA037) and ground in a Wiley-type mill (Marconi, model MA340, serial 9,304,176). The powdered material of leaves (1325.72 g) was exhaustively extracted by percolation with ethanol 96° GL, filtrated, and evaporated under reduced pressure. Then, the ethanol extract (151 g, 11.4% yield dry weight) was sequentially partitioned in a separatory funnel between water–methanol 7:3 (v/v) and hexane (23.72 g), dichloromethane (20.11 g), ethyl acetate (25.46 g), and butanol (35.43 g) in this order. The solvents were removed by rotary evaporation.
Fractionation by Countercurrent Chromatography
Part of the CH2Cl2 (DCM) extract (600 mg) from leaves of S. cristata was submitted to HSCCC fractionation using a Quattro HTPrep apparatus equipped with two bobbins containing two polytetrafluoroethylene multilayer coils each (26 ml, 1.0 mm i.d., + 224 ml, 3.2 mm i.d., and 95 ml, 2.0 mm i.d., + 98 ml, 2.0 mm i.d.). The 98-ml coil was used, and the solvent system chosen was hexane–ethyl acetate–methanol–water 1:1:1:1 (v/v). The upper organic layer served as a stationary phase (reversed-phase elution mode), and the aqueous phase was the mobile phase at a flow rate of 2 ml/min. The sample was dissolved in 5 ml of the solvent system (1:1, v/v), and the solution was introduced in the coil through a manual sample injection valve using a 5-ml sample loop. Thirty fractions of 4 ml were collected during elution with a rotation of 860 rpm followed by extrusion of another 30 fractions of 4 ml. Fractions were analyzed by TLC and grouped into 13 subfractions: Fr-1 (54.4 mg), Fr-2 (40.6 mg), Fr-3 (47.6 mg), Fr-4 (43.5 mg), Fr-5 (40.2 mg), Fr-6 (64.2 mg), Fr-7 (45.5 mg, 3,3′,4′-tri-O-methyl-quercetin, 1), Fr-8 (53.1 mg), Fr-9 (45.2 mg), Fr-10 (49.3 mg, 3,7-di-O-methyl-kaempferol or kumatakenin, 3), Fr-11 (16 mg), Fr-12 (53.2 mg), and Fr-13 (47.2 mg). Fraction Fr-11 (16 mg) was further fractionated by HSCCC under the same conditions as above, to afford Fr-11A (7 mg, tetra-O-methyl-quercetin or retusin, 2) and Fr-11B (9 mg, 3,7-di-O-methyl-kaempferol or kumatakenin, 3) (Fig. S1).
3,3′,4′-Tri-O-methyl-quercetin (1): UV–Vis λ/nm (λmax. 253, 355); 1H and 13C NMR data (Figs. S20 and S27), see Table S1. Positive DI-APCI-MS/MS m/z 345.2 [M + H]+ (calcd for C18H17O7+, 345.1), which was identified by comparison with previously described data (Awad et al. 2018).
3,7,3′,4′-Tetra-O-methyl-quercetin (retusin) (2): UV–Vis λ/nm (λmax. 250, 350); 1H and 13C NMR data (Figs. S41 and S44). Positive DI-APCI-MS/MS m/z 359.3 [M + H]+ (calcd for C19H19O7+, 359.1), which was identified by comparison with previously described data (Silva et al. 2009).
3,7-Di-O-methyl-kaempferol (kumatakenin) (3): UV–Vis λ/nm (λmax. 265, 345); 1H and 13C NMR data (Figs. S30 and S37), see Table S2. Positive DI-APCI-MS/MS m/z 315.2 [M + H]+ (calcd for C17H15O6+, 315.1), which was identified by comparison with previously described data (Silva et al. 2009).
Analysis by HPLC–DAD
HPLC–DAD (280 nm) analyses were performed using an Agilent 1260 Infinity Series with a Poroshell 120 EC-C18 column (2.1 × 100 mm i.d.; 2.7 µm particle size; Agilent) at 30 °C. Gradient conditions were as follows: solvent A = water–0.01% formic acid, solvent B = acetonitrile, B = 60% in t = 40 min, and B = 100% in t = 45 min.
Analysis by APCI-MS/MS
The MS analyses were performed using LCQ Fleet (Thermo Fisher Scientific, Waltham, MA, USA) through direct infusion of the diluted samples in MeOH:H2O (9:1) containing 0.1% formic acid as a modifier for positive ionization mode in a flow rate of 0.1 ml/min. The mass spectrometer, equipped with APCI font and ion trap analyzer, was operated in positive mode. High-purity nitrogen (N2) was used as sheath gas (10 arbitrary units) and auxiliary gas (5 arbitrary units). High-purity helium (He) was used as collision gas. Mass spectrometry parameters used were a source voltage of 6.0 kV, a capillary voltage of 10 V, a tube lens voltage of − 13 V, a capillary temperature of 400 °C, and an APCI vaporizer temperature of 450 °C. Full-scan data acquisition (mass range: m/z 50–1000). The normalized collision energy of the collision-induced dissociation (CID) cell was set at 35 eV. The spectra were processed using the Xcalibur software, version 2.2 SP1.
Identification of Isolated Compounds
1H, 13C, APT, HMBC, and HSQC NMR data were acquired in deuterated dimethyl sulfoxide (DMSO-d6) and chloroform-d (CDCl3) at 25 °C (Varian VNMRS 500 MHz spectrometer). UV-1240 ultraviolet spectrometer (Shimadzu, Japan) uses MeOH and anhydrous sodium acetate (NaOAc) P.A. as displacement reagent.
Inhibition of SARS-CoV-2 Replication
Vero E6 (African green monkey kidney) and Calu-3 (human lung adenocarcinoma) cells were infected with SARS-CoV-2 isolate (GISAID EPI ISL #414,045) in multiplicity of infection (MOI) 0.01 and 0.1, respectively. After 1 h, the supernatants were harvested, and the cells were incubated with 1–3 at log and semi-log concentrations (from 10 to 0.01 µM) or DCM extract from leaves of Siparuna cristata (from 250 to 31.25 µg/ml). Lopinavir/ritonavir (LPV/RTV) in combination and chloroquine (CLQ) were used as control. LPV/RTV was prepared in the proportion of 3:1 as the pharmaceutical pills were composed of 300 mg LPV and 100 mg RTV (Fintelman-Rodrigues et al. 2020). The concentrations of LPV/RTV showed in the present study were based on LPV concentration. After 24 h of infection in Vero E6 cells or 48 h of infection in Calu-3 cells, the supernatants were titrated by plaque-forming units (PFU/ml).
For PFU assay, monolayers of Vero E6 (105 cell/well) in 24-well plates were infected with 300 µl of supernatant dilutions (10−3, 10−4, or 10−5). After 1 h at 37 °C in 5% CO2, the medium was replaced by 500 µl of carboxymethylcellulose solution (DMEM-HG 10 × , 1.8% carboxymethylcellulose and 2% fetal bovine serum). After 72 h of infection, the cytopathic effect (CPE) was analyzed on an optical microscope and 500 µl of 10% formalin was added to fix the cells. After 3 h, this solution was harvested, and plaques were colored by 0.4% bromophenol blue and PFU was counted. All procedures related to virus culture were handled at biosafety level 3 (BSL3) multiuser facility, according to WHO guidelines (WHO 2020).
For cytotoxicity analysis, monolayers of 104 Vero E6 cells and 105 Calu-3 cells in 96-well plates were treated for 72 h with semi-log dilutions (from 6000 to 50 µM) of all compounds tested or DCM extract (200 µg/ml). Then, 5 mg/ml of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, Sigma) in 1 × PBS was added to the cells, according to the manufacturer’s instructions. After 4 h at 37 °C, 10% SDS was added. After incubating for 2 h at 37 °C, the plates were read in a spectrophotometer at 570 nm. The 50% cytotoxic concentration (CC50) was calculated by a non-linear regression analysis of the dose–response curves. All the compounds were resuspended in 100% dimethyl sulfoxide (DMSO) for the in vitro tests. In the assays, the DMSO final concentrations were equal or lower than 1% (v/v) diluted in Dulbecco’s modified Eagle’s medium (DMEM, Gibco), not affecting the growth of the cells.
In SilicoAnalyses
Preparation of Receptors and Ligands
The selected crystal structures for 3CLpro (PDBid 6XQT) and for PLpro (PDBid 7JRN) were obtained from the Protein Data Bank. Through the Pymol and UCSF Chimera software programs, all ligands and identical chains present in the molecules were removed (De Lano 2002; Pettersen et al. 2004). The ligand files were processed by the PDB2PQR server (http://server.poissonboltzmann.org/pdb2pqr) (Dolinsky et al. 2007) with AMBER force field in order to assess the pKa prediction of the ionizable protein residuals at pH 7.4. For 3CLpro, the selection of the probable 3D-structure protonation state was performed through the pdb2gmx module from the computational package GROMACS with the AMBER99SB-ILDN protein field force, nucleic AMBER94 (Abrahan et al. 2015). The conversion of the structure and the bind to the pdbqt format was performed with AutoDock tools (Morris et al. 2009), while the PLpro PDB2PQR output was converted by using UCSF Chimera.
Molecular Docking
Through the Zone function, which is available at the Chimera software (Pettersen et al. 2004), the far residuals were selected until 5 Å from the selected bind is achieved. With this selected perimeter, it was developed with the grid center x = − 11, center y = 1, and center z = 45, and size x = 32, size y = 35, and size z = 33 for 3CLpro, and center x = 13, center y = − 9, and center z = 30, and size x = 30, size y = 30, and size z = 30 for PLpro. The protein redocking was performed with the AutoDock Vina software (Trott et al. 2009). Six different dockings were performed for both receptors using the following substances: lopinavir, ritonavir, chloroquine, 3,3′,4′-tri-O-methyl-quercetin (1), 3,7,3′,4′-tetra-O-methyl-quercetin (2), and 3,7-di-O-methyl-kaempferol (3). The results were reranked based on the distances in Å from hydrogen bonds between the His41, Cys145, and Glu166 residues for 3CLpro, and from the one created with residue Tyr268, described as the main bonding interaction for PLpro.
Results and Discussion
Isolation and Characterization of Flavonoids
To isolate the flavonoids (1–3) from S. cristata, the DCM extract was fractionated by HSCCC. DCM extracts normally contain compounds of medium polarity, and therefore, the solvent system hexane–ethyl acetate–methanol–water (HEMWat) was used because of the versatility and range of polarity of this solvent system family (Costa and Leitão 2010; Costa et al. 2013). In general, the selection of HEMWat ratios should start with 1:1:1:1 (v/v) and then be adjusted with the proper polarity for K near 1 for the target compounds (Costa and Leitão 2010). In the present case, these ratios were appropriate for the fractionation of the DCM extract of leaves of S. cristata. HPLC–DAD analysis of the resulting 13 fractions (Figs. S2 to S7) revealed the presence of the flavonoids in fractions Fr-7 to Fr-12 (Tables S1–S3), which annotation results and data obtained with HPLC–DAD and DI-APCI ( +)-MS/MS analyses are summarized in Table S3. A detailed description of fraction annotations can be found in the Supplementary Material.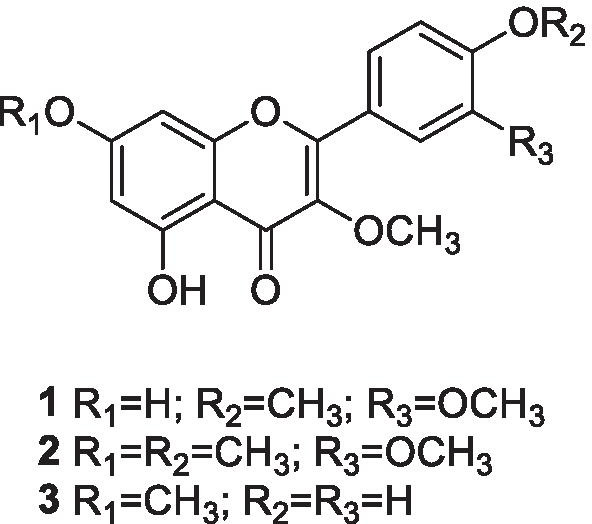
Cell Toxicity and Inhibition of SARS-CoV-2 Replication
Flavonoids 1–3 and DCM extract were evaluated against SARS-CoV-2 replication and cell viability (Table 1). The combination of LPV/RTV and CLQ was used as a positive control to inhibit viral protease and replication. Retusin (2), kumatakenin (3), and the DCM extract were able to inhibit SARS-CoV-2 replication, while the flavonoid 3,3′,4′-tri-O-methyl-quercetin (1) did not exert any inhibition, in both Vero E6 and Calu-3 cells. In Vero E6 and Calu-3 cells, retusin (2) was active at the lowest concentrations tested and displayed a lower EC50 than the tested controls (LPV/RTV and CLQ), with a selective index (SI) which is 1257 and 7 times greater than LPV/RTV and CLQ, respectively, in Vero E6 cells. In Calu-3, retusin (2) showed an inhibitory effect 417 times greater than LPV/RTV. CLQ did not have an inhibitory effect, as shown in previous studies (Hoffmann et al. 2020). The three analyzed flavonoids have high CC50 values and are less toxic than the compounds used as control in both cell lineages analyzed (Table 1).
Table 1.
CC50, EC50, and SI values for 3,3′,4′-tri-O-methyl-quercetin, 1; 3,7,3′,4′-tetra-O-methyl-quercetin (retusin), 2; 3,7-di-O-methyl-kaempferol (kumatakenin) 3; LPV/RTV; and CLQ
| Vero E6 | Calu-3 | |||||
|---|---|---|---|---|---|---|
| CC50 (µM) | EC50 (µM) | SI | CC50 (µM) | EC50 (µM) | SI | |
| 3,3′,4′-Tri-O-methyl-quercetin (1) | 3000 ± 150 | NA | NA | 3500 ± 130 | NA | NA |
| 3,7-Di-O-methyl-kaempferol (kumatakenin) (3) | 2000 ± 230 | 10 ± 0.7 | 200 | 2080 ± 135 | 0.3 ± 0.02 | 6933 |
| 3,7,3′,4′-Tetra-O-methyl-quercetin (retusin) (2) | 4575 ± 300 | 0.4 ± 0.05 | 11,438 | 5000 ± 200 | 0.6 ± 0.06 | 8333 |
| LPV/RTV | 91 ± 3 | 10 ± 3 | 9.1 | 100 ± 3 | 5 ± 0.5 | 20 |
| CLQ | 1664 ± 75 | 1 ± 0.15 | 1664 | 500 ± 50 | NA | NA |
| Siparuna cristata dichloromethane crude extract | > 200 µg/ml | < 31.25 µg/ml | > 6.4 | > 200 µg/ml | < 31.25 µg/ml | > 6.4 |
CC50, the concentration required to reduce normal, non-infected cell viability by 50%. The values represent the mean of duplicate samples from three independent experiments. EC50, the concentration required to reduced inhibition of viral infection-induced cytopathogenicity by 50%. The values represent the mean of duplicate samples from three independent experiments
SI selective index determined by the ratio between CC50 and EC50, LPV/RTV the combination of lopinavir/ritonavir, CLQ chloroquine, NA not applicable
The inhibitory activity of 3CLpro and PLpro SARS-CoV-2 proteases by flavonoids was recently described through attaching to their active site and inactivating them (Tutunchi et al. 2020). Quercetin reduced the infectivity of human and bovine coronaviruses, showing activities against SARS-CoV and MERS-CoV (Russo et al. 2020; Solnier et al. 2020). Tetramethyl derivatives of quercetin have shown to display antiviral and cytotoxic activities; e.g., retusin (2), isolated from rhizomes of Kaempferia parviflora Wall. ex Baker, Zingiberaceae, showed an inhibitory effect on the feline foamy virus (FFV) (Lee et al. 2017). Also, 5,7,3′,4′-tetra-O-methyl-quercetin, isolated from Sambucus nigra L., Adoxaceae, inhibited influenza A (H1N1) infection in vitro (Roschek Jr. et al. 2009). Studies show inhibition of neuraminidase activity for kumatakenin (3) (Ryu et al. 2010). Recently, pectolinarin (5,7-dihydroxy-4′,6-dimethoxyflavone-7-rutinoside) demonstrated inhibitory activity by efficiently blocking the enzymatic activity of SARS-CoV-2 3CL-Pro (Jo et al. 2020). Therefore, the study with flavonoids is interesting from the point of view of viral proteases.
Molecular Docking with 3CLpro
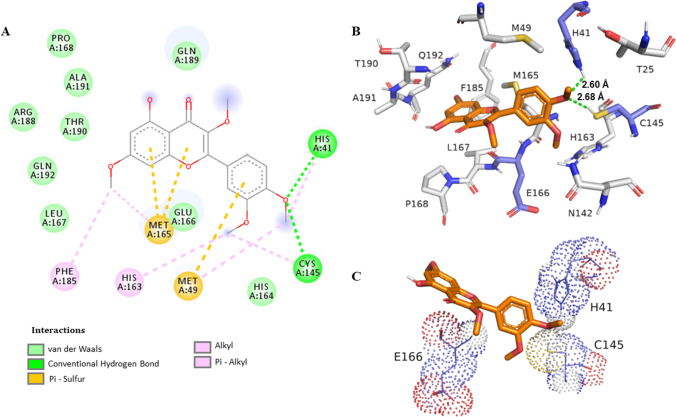
Previous studies have shown hydrophobic π-π stacking interactions with His41 residue via molecular docking (Xu et al. 2020). Therefore, His41 was the choice to analyze the binding distance from the ligand, due the importance of this residue on the enzyme activity. Also, flavonoids have demonstrated a wide range of binding affinity to SARS-CoV-2 3CLpro due to their hydrophobic aromatic rings and hydrophilic hydroxyl groups (Jo et al. 2020). The first bind mode generated on the redocking step presented a bind energy of − 10.4 kcal/mol, and the root mean square deviation (RMSD) is equal to 0.97 Å (Fig. S51) between the pose and the crystal original bind (PDBid 6XQT) calculated with the Open Babel software (Table S4) (Murray-Rust et al. 2004). The simulations revealed the best interactions with their respective binding energy value: thus, the value of − 6.5 kcal/mol was obtained for 1 (Fig. S53), − 7.2 kcal/mol for 3 (Fig. S54), and − 6.3 kcal/mol for 2, and finally, the following values were obtained for the control drugs: − 6.4 kcal/mol for CLQ, − 5.1 kcal/mol (Fig. S52) for LPV, and − 8.1 kcal/mol for RTV. Figure 1 illustrates the most frequent and stable retusin (2)–3CLpro protease complex where it was possible to identify a hydrogen-bonding interaction of the OCH3 at C-4′ with the His41 residue with a distance of 2.6 Å. Table 2 summarizes the best binding energies, as well as the distances between the His41, Cys145, and Glu166 residues from the receptor protein 3CLpro (PDBid: 6XQT).
Fig. 1.
Interaction of SARS-CoV-2 3CLpro protease residues with retusin (2). A Map of the interaction of residues. B Protease 3CLpro (PDBid: 6XQT) in gray and residues within a radius of proximity equal to 5 Å of the ligand, represented by sticks. The ligand is in orange, and the catalytic dyad residues His41 and Cys145 and the residue Glu166 are in lavender. C The representation shows the interaction of these residues with the ligand
Table 2.
Energy docking values for 3CLpro and PLpro hydrogen-bonding interaction with compounds 1–3
| Compound | 3CLpro | PLpro | ||||||
|---|---|---|---|---|---|---|---|---|
| Affinity for the best distance mode (kcal/mol) | Mode | Distance His41 (Å) |
Distance Cys145 (Å) |
Distance Glu166 (Å) |
Affinity for the best distance mode (kcal/mol) | Mode | Distance Tyr268 (Å) |
|
| 1 | − 6.5 | 9 | – | 2.77 | – | − 7.1 | 1 | 2.2 |
| 2 | − 6.3 | 6 | 2.60 | 2.68 | – | − 5.5 | 7 | 1.9 |
| 3 | − 7.2 | 3 | – | 3.01 | 2.19 | − 5.7 | 7 | 2.1 |
| LPVa | − 5.1 | 9 | 2.92 | – | – | − 5.7 | 9 | 1.8 |
| RTVb | − 8.1 | 11 | 2.41 | 2.68 | 2.42 | − 6.9 | 2 | 1.6 |
| CLQc | − 6.4 | 1 | – | – | 2.11 | − 5.9 | 8 | 1.1 |
aPositive control: lopinavir
bPositive control: ritonavir
cPositive control: chloroquine
Molecular Docking with PLpro
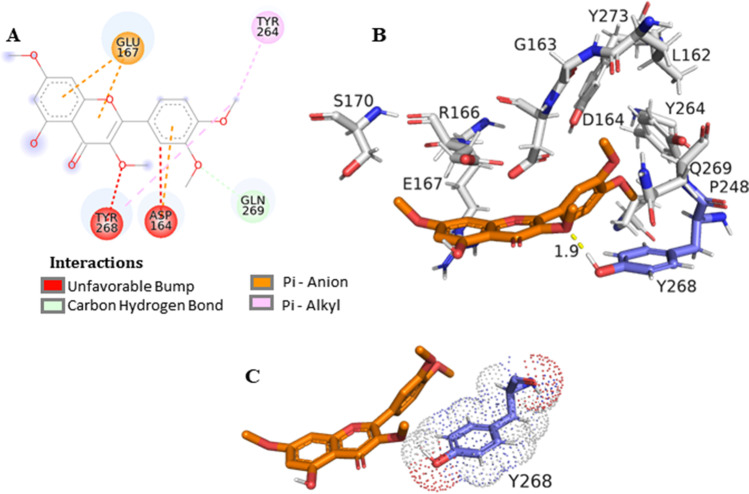
The first bind mode generated on the redocking step presented a bind energy of − 9.6 kcal/mol and the RMSD of 0.45356 Å (Fig. S55) between the pose and the crystal original bind (PDBid: 7JRN) calculated with the Open Babel software (Table S4) (Murray-Rust et al. 2004). Molecular docking simulations afforded the following results based on affinity energy and distances for ligands and the Tyr268 residue (Table 2): RTV − 6.9 kcal/mol, distance 1.6 Å; LPV − 5.7 kcal/mol, distance 1.8 Å; CLQ − 5.9 kcal/mol, distance 1.1 Å (Fig. S56); 3,3′,4′-tri-O-methyl-quercetin (1) − 7.2 kcal/mol, distance 2.2 Å (Fig. S57); retusin (2) − 5.5 kcal/mol, distance 1.9 Å; and kumatakenin (3) − 5.7 kcal/mol, distance 2.1 Å (Fig. S58). Based on these results, the interaction between PLpro and the ligand retusin (2) may be favorable due the proximity of the OCH3 at C-3 from the Tyr268 residue (Fig. 2). Docking simulations have shown the SARS-CoV-2 PLpro BL2 loop having significant flexibility in ligand-free proteins. Residues Asn267, Gln269, and most importantly, Tyr268 account for most of this motion, making Tyr268 the key residue to calculate the distance in Å between the enzyme and inhibitor (Bosken et al. 2020). Flavonoids were previously described as potential inhibitors, by assuming that the hydrophobic flavonoids have shown higher affinity to SARS-CoV PLpro than to other proteases, which might be due to certain structural differences in the protein sequences (Solnier et al. 2020).
Fig. 2.
Interaction of SARS-CoV-2 PLpro residues with retusin (2). A Map of the interaction of residues. B Protease PLpro (PDBid:7JRN) in gray and residues within a radius of proximity equal to 5 Å of the ligand, represented by sticks. The ligand is in orange, and the residue Tyr268 is in lavender. C This representation shows the interaction of this residue with the ligand
Conclusions
In this work, we describe the anti-SARS-CoV-2 potential of a Brazilian medicinal plant traditionally used to treat respiratory infections such as colds and flu with a long history of commercialization for this purpose. Two isolated flavonoids inhibited SARS-CoV-2 viral replication with higher efficiency and lower cytotoxicity than lopinavir/ritonavir and chloroquine treatment. Among the isolated flavonoids, tetra-O-methyl-quercetin is being reported for the first time in the genus as well as the inhibitory potential of free O-methyl-flavonoids against SARS-CoV-2. The in silico results demonstrated the potential interaction between flavonoids and key residues of COVID-19 3CLpro as well as PLpro, in a similar way to that of the screened potential inhibitors against COVID-19. Retusin (3,7,3′,4′-tetra-O-methyl-quercetin) demonstrated the best results in the assays and in the molecular docking, based on the hydrogen-bonding distance between selected amino acid residues of the catalytic site from 3CLpro and PLpro SARS-CoV-2 proteases and all tested flavonoids. This study highlights the possible application of methylated flavonoids, for example retusin, as an antiviral or adjuvant therapy in the treatment of COVID-19.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We are indebted to the Laboratório Multiusuário de Análises por RMN at the Federal University of Rio de Janeiro for the access to NMR facilities, and to the Central Analítica, Departamento de Produtos Naturais e Alimentos for the MS analyses. Thanks are due to Dr. Carmen Beatriz Wagner Giacoia Gripp for the assessments related to BSL3 facility.
Author Contribution
Collecting and identifying plant material: PHAN; HSCCC experiments: CML, RS, and PHAN; interpretation of the spectrometric data: CML and SCM; supervision of the chromatographic and MS analyses: FNC, RCS, SGL, and GGL; biological assays: TSG, MESM, ART, NFR, MMS, and MDM; molecular docking: CFARC, MEAE, and MLS; drafting the manuscript: all authors; critical revision of the manuscript: GGL, SGL, RCS, MDM, and MLS. All the authors have read the final manuscript and approved its submission.
Funding
Financial support was provided by the Conselho Nacional de Desenvolvimento Científico e Tecnológico, Fundação de Amparo à Pesquisa do Estado do Rio de Janeiro, and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior.
Declarations
Competing Interests
The authors declare no competing interests.
Footnotes
This article is part of a Special Issue to celebrate the 35th anniversary of the Brazilian Journal of Pharmacognosy
Contributor Information
Suzana Guimarães Leitão, Email: sgleitao@pharma.ufrj.br.
Gilda Guimarães Leitão, Email: ggleitao@ippn.ufrj.br.
References
- Abdul S, Banerjee S, Ghosh K, Gayen S, Jha T. Protease targeted COVID-19 drug discovery and its challenges: insight into viral main protease (Mpro) and papain-like protease (PLpro) inhibitors. Bioorgan Med Chem. 2021;29:115860. doi: 10.1016/j.bmc.2020.115860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awad HM, Abd-Alla HI, Ibrahim MA, El-Sawy ER, Abdalla MM. Flavones from heavenly blue as modulators of Alzheimer’s amyloid-beta peptide (Aβ) production. Med Chem Res. 2018;27:768–776. doi: 10.1007/s00044-017-2100-x. [DOI] [Google Scholar]
- Berretta AA, Silveira MAD, Capcha JMC, De Jong D. Propolis and its potential against SARS-CoV-2 infection mechanisms and COVID-19 disease. Biomed Pharmacother. 2020;131:110622. doi: 10.1016/j.biopha.2020.110622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosken YK, Cholko T, Lou YC, Wu KP, Chang CA. Insights into dynamics of inhibitor and ubiquitin-like protein binding in SARS-CoV-2 papain-like protease. Front Mol Biosci. 2020;7:174. doi: 10.3389/fmolb.2020.00174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrak SA, Merzouk H, Mokhtari-Soulimane N. Potential bioactive glycosylated flavonoids as SARS-CoV-2 main protease inhibitors: a molecular docking and simulation studies. PLoS ONE. 2020;15:e0240653. doi: 10.1371/journal.pone.0240653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinsembu KC. Coronaviruses and nature’s pharmacy for the relief of coronavirus disease 2019. Rev Bras Farmacogn. 2020;30:603–621. doi: 10.1007/s43450-020-00104-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cock IE, Vuuren SFV. The traditional use of southern African medicinal plants in the treatment of viral respiratory diseases: a review of the ethnobotany and scientific evaluations. J Ethnopharmacol. 2020;262:113194. doi: 10.1016/j.jep.2020.113194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa FN, Leitão GG. Strategies of solvent system selection for the isolation of flavonoids by countercurrent chromatography. J Sep Sci. 2010;33:336–347. doi: 10.1002/jssc.200900632. [DOI] [PubMed] [Google Scholar]
- Costa FN, Garrard I, Da Silva AJ, Leitão GG. Changes in the mobile phase composition on a stepwise counter-current chromatography elution for the isolation of flavonoids from Siparuna glycycarpa. J Sep Sci. 2013;36:2253–2259. doi: 10.1002/jssc.201201054. [DOI] [PubMed] [Google Scholar]
- Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 2007;35(Suppl. 2):522–525. doi: 10.1093/nar/gkm276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elizalde-González JJ. SARS-CoV-2 and COVID-19. A pandemic review. Med Crit. 2020;3:53–67. doi: 10.35366/93281. [DOI] [Google Scholar]
- El-Mordy FMA, El-Hamouly MM, Ibrahim MT, El-Rheem GA, Aly OM, El-Kader AMA, Youssif KA, Abdelmohsen UR. Inhibition of SARS-CoV-2 main protease by phenolic compounds from Manilkara hexandra (Roxb.) Dubard assisted by metabolite profiling and in silico virtual screening. RSC Adv. 2020;10:32148–32155. doi: 10.1039/d0ra05679k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fintelman-Rodrigues N, Sacramento SQ, Lima CR, Da Silva FS, Ferreira AC, Mattos M, De Freitas CS, Soares VC, Dias SSG, Temerozo JR, Miranda MD, Matos AR, Bozza FA, Carels N, Alves CR, Siqueira MM, Bozza PT, Souza TM. Atazanavir, alone or in combination with ritonavir, inhibits SARS-CoV-2 replication and proinflammatory cytokine production. Antimicrob Agents Chemother. 2020;64:e00825–e920. doi: 10.1128/AAC.00825-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann M, Mosbauer K, Hofmann-Winkler H, Kaul A, Kleine-Weber H, Kruger N, Gassen NC, Muller MA, Drosten C, Pohlmann S. Chloroquine does not inhibit infection of human lung cells with SARS-CoV-2. Nature. 2020;585:588–590. doi: 10.1038/s41586-020-2575-3. [DOI] [PubMed] [Google Scholar]
- Jo S, Kim S, Kim DY, Kim MS, Shin DH. Flavonoids with inhibitory activity against SARS-CoV-2 3CLpro. J Enzyme Inhib Med Chem. 2020;35:1539–1544. doi: 10.1080/14756366.2020.1801672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komolafe K, Komolafe TR, Fatoki TH, Akinmoladun AC, Brai BIC, Olaleye MT, Akindahunsi AA. Coronavirus disease 2019 and herbal therapy: pertinent issues relating to toxicity and standardization of phytopharmaceuticals. Rev Bras Farmacogn. 2021;31:142–161. doi: 10.1007/s43450-021-00132-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal CM, Simas RC, Miranda M, Campos MF, Gomes BA, Siqueira MM, Do Vale G, De Almeida CVG, Leitão SG, Leitão GG. Amazonian Siparuna extracts as potential anti-influenza agents: metabolic fingerprinting. J Ethnopharmacol. 2021;270:113788. doi: 10.1016/j.jep.2021.113788. [DOI] [PubMed] [Google Scholar]
- Lee GE, Kim J, Shin CG. Antiviral activities of hydroxylated flavones on feline foamy viral proliferation. Appl Biol Chem. 2017;60:419–425. doi: 10.1007/s13765-017-0294-8. [DOI] [Google Scholar]
- Leitão GG, Simas NK, Soares SS, De Brito AP, Claros BM, Brito TB, DelleMonache F. Chemistry and pharmacology of Monimiaceae: a special focus on Siparuna and Mollinedia. J Ethnopharmacol. 1999;65:87–102. doi: 10.1016/s0378-8741(98)00233-5. [DOI] [PubMed] [Google Scholar]
- Leitão GG, Soares SSV, Brito M, DelleMonache F, De Barros T. Kaempferol glycosides from Siparuna apiosyce. Phytochemistry. 2000;55:679–682. doi: 10.1016/S0031-9422(00)00222-3. [DOI] [PubMed] [Google Scholar]
- Leitão GG, El-Adji SS, De Melo WAL, Leitão SG, Brown L. Separation of free and glycosylated flavonoids from Siparuna guianensis by gradient and isocratic CCC. J Liq Chromatogr R T. 2005;28:2041–2051. doi: 10.1081/JLC-200063669. [DOI] [Google Scholar]
- Liskova A, Samec M, Koklesova L, Samuel SM, Zhai K, Al-Ishaq RK, Abotaleb M, Nosal V, Kajo K, Ashrafizadeh M, Zarrabi A, Brockmueller A, Shakibaei M, Sabaka P, Mozos I, Ullrich D, Prosecky R, La Rocca G, Caprnda M, Busselberg D, Rodrigo L, Kruzliak P, Kubatka P. Flavonoids against the SARS-CoV-2 induced inflammatory storm. Biomed Pharmacother. 2021;138:111430. doi: 10.1016/j.biopha.2021.111430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisch B. SARS-CoV-2 as potential cause of cardiac inflammation and heart failure. Is it the virus, hyperinflammation, or MODS? Herz. 2020;22:321–322. doi: 10.1007/s00059-020-04925-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani JS, Johnson JB, Steel JC, Broszczak DA, Neilsen PM, Walsh KB, Naiker M. Natural product-derived phytochemicals as potential agents against coronaviruses: a review. Virus Res. 2020;284:197989. doi: 10.1016/j.virusres.2020.197989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GM, Ruth H, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. Software news and updates AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouffouk C, Mouffouk S, Mouffouk S, Hambaba L, Haba H. Flavonols as potential antiviral drugs targeting SARS-CoV-2 proteases (3CLpro and PLpro), spike protein, RNA-dependent RNA polymerase (RdRp) and angiotensin-converting enzyme II receptor (ACE2) Eur J Pharmacol. 2021;891:173759. doi: 10.1016/j.ejphar.2020.173759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray-Rust P, Rzepa HS, Williamson MJ, Willighagen EL. Chemical markup, XML, and the world wide web. 5. Applications of chemical metadata in RSS aggregators. J Chem Inform Comput Sci. 2004;44:462–469. doi: 10.1021/ci034244p. [DOI] [PubMed] [Google Scholar]
- Pandey P, Khan F, Rana AK, Srivastava Y, Jha SK, Jha NK. A drug repurposing approach towards elucidating the potential of flavonoids as COVID-19 spike protein inhibitors. Biointerface Res Appl Chem. 2021;11:8482–8501. doi: 10.33263/BRIAC111.84828501. [DOI] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera. A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Ren X, Shao XX, Li XX, Jia XH, Song T, Zhou WY, Wang P, Li Y, Wang XL, Cui QH, Qiu PJ, Zhao YG, Li XB, Zhang FC, Li ZY, Zhong Y, Wang ZG, Fu XJ. Identifying potential treatments of COVID-19 from traditional Chinese medicine (TCM) by using a data-driven approach. J Ethnopharmacol. 2020;258:112932. doi: 10.1016/j.jep.2020.112932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robba C, Battaglini D, Pelosi P, Rocco PRM. Multiple organ dysfunction in SARS-CoV-2: MODS-CoV-2. Expert Rev Respir Med. 2020;9:865–868. doi: 10.1080/17476348.2020.1778470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschek B, Jr, Fink RC, McMichael MD, Li D, Alberte RS. Elderberry flavonoids bind to and prevent H1N1 infection in vitro. Phytochemistry. 2009;70:1255–1261. doi: 10.1016/j.phytochem.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Russo M, Moccia S, Spagnuolo C, Tedesco I, Russo GL. Roles of flavonoids against coronavirus infection. Chem-Biol Interact. 2020;328:109211. doi: 10.1016/j.cbi.2020.109211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu YB, Kim JH, Park SJ, Chang JS, Rho MC, Bae KH, Park KH, Lee WS. Inhibition of neuraminidase activity by polyphenol compounds isolated from the roots of Glycyrrhiza uralensis. Bioorg Med Chem Lett. 2010;20:971–974. doi: 10.1016/j.bmcl.2009.12.106. [DOI] [PubMed] [Google Scholar]
- Santana FPR, Thevenard F, Gomes KS, Taguchi L, Câmara NOS, Stilhano RS, Ureshino RP, Prado CM, Lago JHG (2021) New perspectives on natural flavonoids on COVID-19-induced lung injuries. Phytoter Res. 10.1002/ptr.7131 [DOI] [PMC free article] [PubMed]
- Solnier J, Fladerer JP (2020) Flavonoids: a complementary approach to conventional therapy of COVID-19?. Phytochem Rev. 10.1007/s11101-020-09720-6 [DOI] [PMC free article] [PubMed]
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2009;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutunchi H, Naeini F, Ostadrahimi A, Hosseinzadeh-Attar MJ. Naringenin, a flavanone with antiviral and anti-inflammatory effects: a promising treatment strategy against COVID-19. Phytother Res. 2020;34:3137–3147. doi: 10.1002/ptr.6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Yang L, Zhang X, Zhang Q, Yang Z, Liu Y, Wei S, Liu W. Discovery of potential flavonoid inhibitors against COVID-19 3CL proteinase based on virtual screening strategy. Front Mol Biosci. 2020;7:556481. doi: 10.3389/fmolb.2020.556481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.