Abstract
Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV)-2 infection induces an exacerbated inflammation driven by innate immunity components. Dendritic cells (DCs) play a key role in the defense against viral infections, for instance plasmacytoid DCs (pDCs), have the capacity to produce vast amounts of interferon-alpha (IFN-α). In COVID-19 there is a deficit in DC numbers and IFN-α production, which has been associated with disease severity. In this work, we described that in addition to the DC deficiency, several DC activation and homing markers were altered in acute COVID-19 patients, which were associated with multiple inflammatory markers. Remarkably, previously hospitalized and nonhospitalized patients remained with decreased numbers of CD1c+ myeloid DCs and pDCs seven months after SARS-CoV-2 infection. Moreover, the expression of DC markers such as CD86 and CD4 were only restored in previously nonhospitalized patients, while no restoration of integrin β7 and indoleamine 2,3-dyoxigenase (IDO) levels were observed. These findings contribute to a better understanding of the immunological sequelae of COVID-19.
Keywords: SARS-CoV-2, COVID-19, Dendritic cell, Long-COVID
Subject terms: Viral infection, Dendritic cells
Introduction
Coronavirus disease 19 (COVID-19) is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and may progress with mild symptoms or asymptomatically in most of the individuals, while others experience an acute respiratory distress syndrome (ARDS) and poorer prognosis, including death [1]. Disease severity depends on the balance between host immune response, viral replication and tissue and organ damage. In severe COVID-19 there is a deregulation of this response, characterized by an hyperinflammation driven by innate immunity, characterized by very high levels of cytokines and pro-inflammatory biomarkers, also known as cytokine storm [2, 3].
One of the innate immune cell types that may play a pivotal role in the response against SARS-CoV-2 are the dendritic cells (DCs). There are two main DC types, conventional or myeloid DCs (mDCs) which include CD1c+, CD16+, and CD141+ mDC subsets, and plasmacytoid dendritic cells (pDCs). In general, DCs participate in antigen presentation, cytokine production, control of inflammatory responses, tolerance induction, immune cell recruitment, and viral dissemination. However, the role of these cells in response to acute SARS-CoV-2 infection and the recovery in convalescent subjects is not fully characterized. Some studies have shown a decrease of DC numbers in response to infection in peripheral blood [4] and also an association with disease severity [5]. This deficiency seems to be due to the migration of some DC subsets, such as CD1c, to the lung [6], and probably to other inflammatory foci. pDCs also seems to play a key role in COVID-19 [7]. pDCs are the main type I interferon (IFN-I) producers, with 1000-fold production compared to other immune cell types [8]. IFN-I is known to have an essential role in viral infections [9]. Significantly, pDCs depletion has been associated with poor COVID-19 prognosis [10]. Moreover, critical patients showed a highly impaired IFN-I response [7] associated with high viral load and aggravated inflammatory response [11].
The recovery of DC defects after COVID-19 could be crucial, since the normalization of the innate immune system after the acute insult would mean the system’s readiness to respond to new viral and bacterial challenges. However, the recovery of DC cell numbers and function after COVID-19 is unknown. This recovery is also important in the sense that a variable proportion of people who have overcome COVID-19 show clinical sequelae [12] which relation with innate immune defects needs to be clarified. Thus, the aim of the study was to analyze DC defects associated with SARS-CoV-2 infection, COVID-19 severity and whether these defects were restored after a median of 7 months after the resolution of the infection.
Results
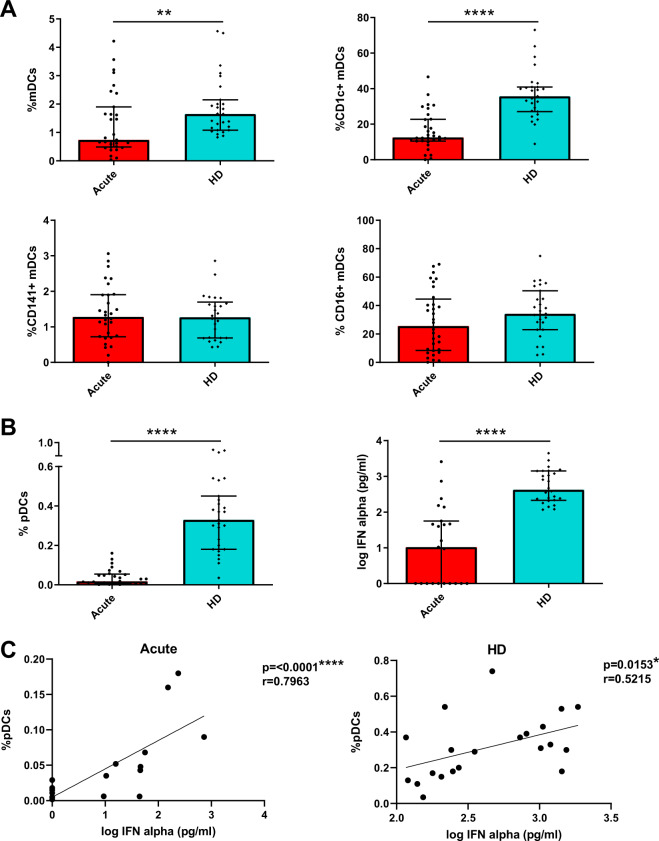
Patients with acute SARS-CoV-2 infection show a considerable decrease in DC percentages and TLR9-dependent IFN-α production
In order to investigate the effect of SARS-CoV-2 infection on the innate immune system, we first analyzed the percentages of total DCs and the different subsets in acute SARS-CoV-2 infected patients (COVID-19 patients) compared with age and sex matched healthy donors (HD). Specifically, we measured mDCs (CD123− CD11c+), including CD1c+, CD16+, and CD141+ mDC subsets, and pDCs (CD123+ CD11c−) (Supplementary Fig. 1a). Our results showed that acute COVID-19 patients exhibited a significant decrease in the percentages of total mDCs mainly due to CD1c+ mDCs decreased in comparison with HD. Meanwhile CD16+ and CD141+ mDCs remained at similar levels of HD (Fig. 1A). Remarkably, the percentage of pDCs in acute COVID-19 patients was considerably diminished with respect to HD (Fig. 1B left). Then, we calculated the ratio mDC/pDC in the different subjects, which was much lower in HD that in COVID-19 patients (Supplementary Fig. 2a). Additionally, based on previously published results [13], the following pDC subsets were analyzed: P1-pDC (CD86−PD-L1+), P2-pDC (CD86+PD-L1+) and P3-pDC (CD86+PD-L1−). Here, a lower percentage of P2- and P3-pDCs was observed in acute COVID-19 patients than in HD (Supplementary Fig. 2b). pDCs are known to be the main producers of IFN-α [8]. Therefore, to study their function in SARS-CoV-2 infection, we stimulated peripheral blood mononuclear cells (PBMCs) with CpG oligodeoxynucleotides class A (CpG)-A, a Toll-like receptor (TLR)-9 dependent stimulation, and we analyzed IFN-α production. We found that IFN-α production in acute COVID-19 was much lower than in HD (Fig. 1B right). To clarify if the decreased IFN-α production was due to a diminished percentage of pDCs, we performed a correlation analysis and we found that the IFN-α production was positively associated with the percentage of pDCs in both acute COVID-19 patients and HD (Fig. 1C). In conclusion, patients with acute SARS-CoV-2 infection exhibit a deficit in DC numbers and also decreased TLR9-dependent IFN-α production.
Fig. 1.
Patients with acute SARS-CoV-2 infection show a considerable decrease in DC percentages and TLR9-dependent IFN-α production. Bar graphs representing the percentage of total mDCs, CD1c+, CD141+, and CD16+ mDCs (A) and the percentage of pDCs and IFN-α production in response to CpG-A in acute SARS-CoV-2 infected patients (acute) and healthy donors (HD) (B). The median with the interquartile range is shown. Correlation between the percentage of pDCs and IFN-α production in acute patients and HD. Each dot represents an individual (C). *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Mann–Whitney U test was used for groups’ comparisons and Spearman test for nonparametric correlations
Acute SARS-CoV-2 infected patients show an altered pattern of DC activation markers
Afterwards, we analyzed the expression of DC activation markers in acute COVID-19 patients and HD. We measured the expression of homing receptors ((integrin-β7 (β7) and C-C chemokine receptor type 7 (CCR7)), co-stimulatory molecules (CD86 and CD4), and markers of immune tolerance and suppression ((Indoleamine 2,3-dioxygenase (IDO) and Programmed Death-ligand 1 (PD-L1), respectively) (Supplementary Fig. 1b). Most of the DC subpopulations, presented lower percentage of β7, specially total mDCs, CD1c+ mDCs, and pDCs and a higher percentage of CCR7+ DCs in acute COVID-19 patients compared with HD (Table 1). We also found lower percentage of CD86+ cells in acute patients in CD1c+ and CD16+ mDCs and pDCs. No differences were found in CD4+ DC levels (Table 1). Lastly, acute COVID-19 patients showed higher percentage of IDO+ cells within CD1c+ and CD16+ mDCs compared with HD, while a lower percentage PD-L1+ was seen within pDCs (Table 1). These results are indicative of alterations in different homing and activation patterns of DCs in response to SARS-CoV-2 infection.
Table 1.
Acute SARS-CoV-2 infected patients show an altered pattern of DC homing and activation markers
| Activation markers | Dendritic cells | Acute | HD | p |
|---|---|---|---|---|
| Beta7 | mDCs | 5.7 [3.2–11.1] | 22.1 [15.7–33.8] | <0.0001 |
| CD1c+ mDCs | 43.8 [26.2–62.2] | 62.0 [38.8–69.7] | 0.0340 | |
| CD16+ mDCs | 0.0 [0.0–0.0] | 0.03 [0.00–0.0] | 0.0851 | |
| CD141+ mDCs | 15.6 [9.9–28.2] | 25.9 [18.8–33.3] | 0.0547 | |
| pDCs | 2.2 [0.5–3.3] | 6.1 [3.2–10.4] | 0.0004 | |
| CCR7 | mDCs | 3.1 [1.4–21.7] | 0.9 [0.3–1.5] | <0.0001 |
| CD1c+ mDCs | 18.2 [6.4–94.3] | 4.5 [2.4–8.1] | <0.0001 | |
| CD16+ mDCs | 3.1 [0.3–14.4] | 0.3 [0.1–0.5] | 0.0015 | |
| CD141+ mDCs | 11.0 [3.1–18.5] | 1.9 [0.6–4.6] | 0.0005 | |
| pDCs | 0.9 [0.2–15.6] | 0.0 [0.0–0.0] | <0.0001 | |
| CD86 | mDCs | 64.9 [37.9–77.4] | 57.7 [50.4–65.2] | 0.4447 |
| CD1c+ mDCs | 5.5 [2.8–11.2] | 12.8 [6.7–17.4] | 0.0053 | |
| CD16+ mDCs | 95.8 [88.5–97.5] | 98.1 [96.7–98.7] | 0.0034 | |
| CD141+ mDCs | 7.6 [0.0–22.3] | 5.9 [2.6–13.6] | 0.9075 | |
| pDCs | 0.3 [0.0–0.9] | 1.4 [0.4–2.2] | 0.0024 | |
| CD4 | mDCs | 8.8 [2.5–24.1] | 5.6 [4.0–10.6] | 0.3229 |
| CD1c+ mDCs | 27.0 [7.7–61.5] | 23.0 [15.7–51.1] | 0.6457 | |
| CD16+ mDCs | 8.5 [2.2–16.5] | 10.7 [6.6–14.6] | 0.3194 | |
| CD141+ mDCs | 21.3 [10.6–51.1] | 40.0 [21.1–47.7] | 0.1109 | |
| pDCs | 57.3 [46.6–77.9] | 70.5 [59.0–87.9] | 0.0750 | |
| IDO | mDCs | 1.3 [0.8–3.1] | 2.0 [1.5–2.8] | 0.1469 |
| CD1c+ mDCs | 9.1 [2.4–23.2] | 2.2 [1.6–4.5] | 0.0039 | |
| CD16+ mDCs | 6.8 [0.1–27.2] | 0.4 [0.0–0.7] | 0.0004 | |
| CD141+ mDCs | 72.1 [56.9–85.7] | 69.2 [59.4–79.5] | 0.6460 | |
| pDCs | 0.0 [0.0–0.1] | 0.1 [0.0–0.3] | 0.2071 | |
| PD-L1 | mDCs | 17.4 [5.1–34.4] | 21.8 [10.1–41.3] | 0.2531 |
| CD1c+ mDCs | 0.9 [0.4–5.2] | 0.6 [0.2–1.7] | 0.1810 | |
| CD16+ mDCs | 28.6 [13.8–47.9] | 21.0 [6.1–41.0] | 0.2107 | |
| CD141+ mDCs | 0.0 [0.0–2.0] | 0.9 [0.0–2.7] | 0.2166 | |
| pDCs | 0.8 [0.2–2.1] | 5.6 [3.1–8.7] | <0.0001 |
Percentages of dendritic cells positive for activation markers in acute SARS-CoV-2 infected patients (Acute) and healthy donors (HD) are presented. The median with interquartile ranges [IQR] is shown. Significant differences are indicated in bold
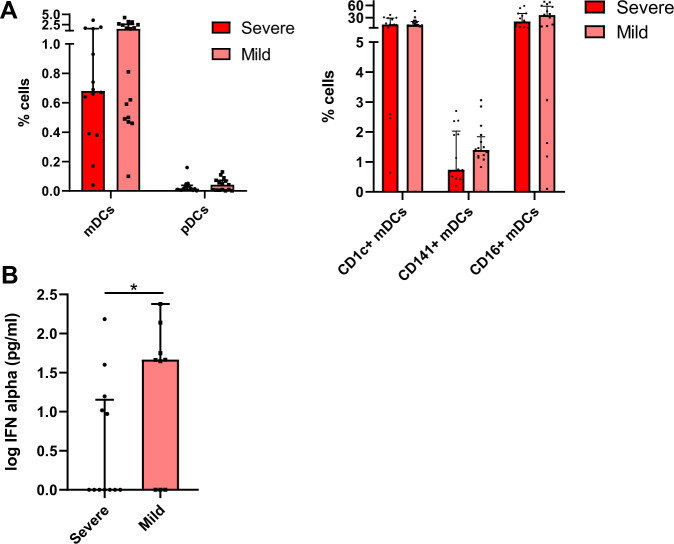
IFN-α production is associated with COVID-19 severity
The next step of this study was to investigate whether DC numbers and their function might be different in acute COVID-19 depending on disease severity. Therefore, we classified acute COVID-19 patients in two groups: severe ((high oxygen support requirement and Intensive Care Unit (ICU) admission or death)) and mild (low oxygen requirement and no ICU admission) (Supplementary Table 1). Our results did not show any significant difference in the percentage of mDCs and subpopulations and pDCs between severe and mild COVID-19 patients (Fig. 2A). However, we found increased levels of total CCR7+ mDCs and PD-L1+ CD141+ mDCs in severe patients (Supplementary Fig. 3). Importantly, we did find a considerable decrease in TLR9-dependent IFN-α production in severe subjects compared to mild patients (Fig. 2B). In summary, acute SARS-CoV-2 infected patients with severe symptoms exhibit a lower capacity to produce IFN-α than patients with mild symptoms.
Fig. 2.
IFN-α production is associated with COVID-19 severity. Bar graphs representing the percentage of pDCs and total mDCs, CD1c+, CD141+ and CD16+ mDCs subsets (A) and the percentage of IFN-α production in response to CpG-A (B) in acute severe and mild SARS-CoV-2 infected patients. The median with the interquartile range is shown and each dot represents an individual. *p < 0.05. Mann–Whitney U test was used for groups’ comparisons
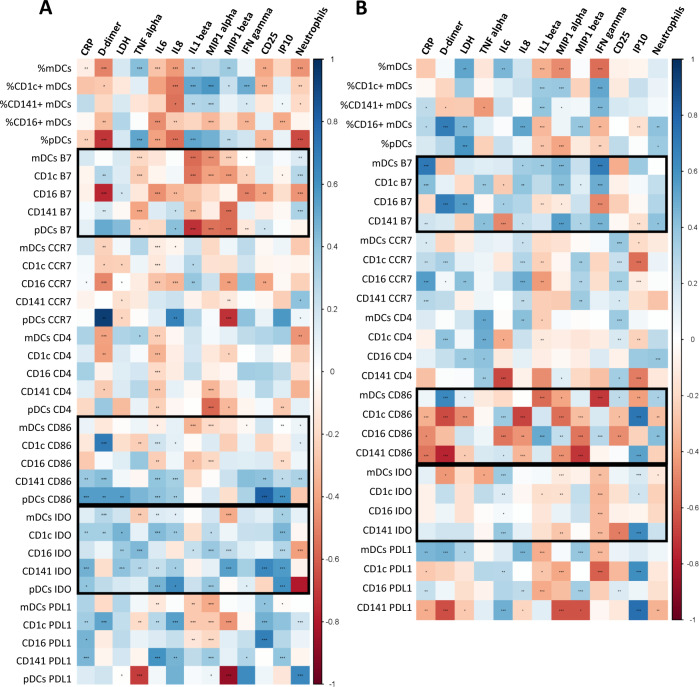
DC parameters are differentially associated to inflammation markers in mild and severe acute SARS-CoV-2 infected patients
DC numbers and activation markers were correlated to multiple inflammatory marker levels, including clinical biomarkers ((C-reactive protein (CRP), D-dimers and lactate dehydrogenase (LDH)), pro-inflammatory cytokines ((tumor necrosis factor (TNF)-α, interleukin (IL)-6, IL-8, IL1-β, macrophage inflammatory protein (MIP1)-α, MIP1-β, interferon inducible protein (IP)-10 and interferon (IFN)-γ) and soluble (sCD25)), and neutrophil numbers. These correlations were done in the overall group of patients during acute infection and also dividing in both severe and mild COVID-19. In the overall population, we observed correlations of dendritic cell subset levels with different pro-inflammatory cytokines and clinical biomarker levels (Supplementary Fig. 4). Interestingly, we observed a different correlation pattern in severe and mild patients and of note, more associations were found in mild patients (Fig. 3). On one hand, regarding COVID-19 patients with mild symptoms, the percentages of DC subpopulations were inversely correlated with D-dimers, IL-6, IL-8, sCD25 levels and neutrophil numbers, while they were positively correlated with TNF-α, IL-1β, MIP-1α, MIP1-β, and IFN-γ levels, with the exception of CD16+ mDCs that were negatively correlated with most of the inflammatory parameters. It is remarkable, that the percentage of pDCs showed a strong inverse correlation with D-dimer levels and neutrophil numbers. Focusing on DC homing and activation markers, regarding the expression of β7 in DCs, inverse associations prevailed, highlighting the strong correlations found in CD16+ β7+ mDCs with D-dimers and in β7+ pDCs with IL1-β. In contrast, the expression of CD86 and IDO in DCs was predominantly positively associated to several inflammatory markers, mainly in the case of CD141+ mDCs and pDCs (Fig. 3A). On the other hand, in severe COVID-19, many associations were lost (e.g., IDO expression) and others were opposite (e.g., CD86), comparing with mild patients. For instance, remarkably, the DC percentages and the expression of β7 and CD86, the associations found with inflammatory marker levels showed an opposite trend (Fig. 3B). Therefore, we conclude that DC levels and activation markers are associated to the inflammatory status of acute SARS-CoV-2 infected subjects, with a differential profile between patients with severe symptoms compared to those with mild symptoms.
Fig. 3.
DC parameters are differentially associated to inflammatory markers in mild and severe acute SARS-CoV-2 infected patients. Heatmap graphs representing correlations between the percentages of DC subpopulations and the percentages of DCs expressing activation and homing markers with inflammatory marker levels including CRP, D-dimer, LDH, TNF-α, IL-6, IL-8, IL1-β, MIP1-α, MIP1-β, IFN-γ, CD25, IP-10, and neutrophil numbers, in mild (A) and severe (B) SARS-CoV-2 infected patients. Blue color represents positive correlations and red color shows negative correlations. The intensity of the color indicates the R coefficient. The most relevant data are highlighted with black squares. *p < 0.05; **p < 0.01; ***p < 0.001. Spearman test was used for nonparametric correlations
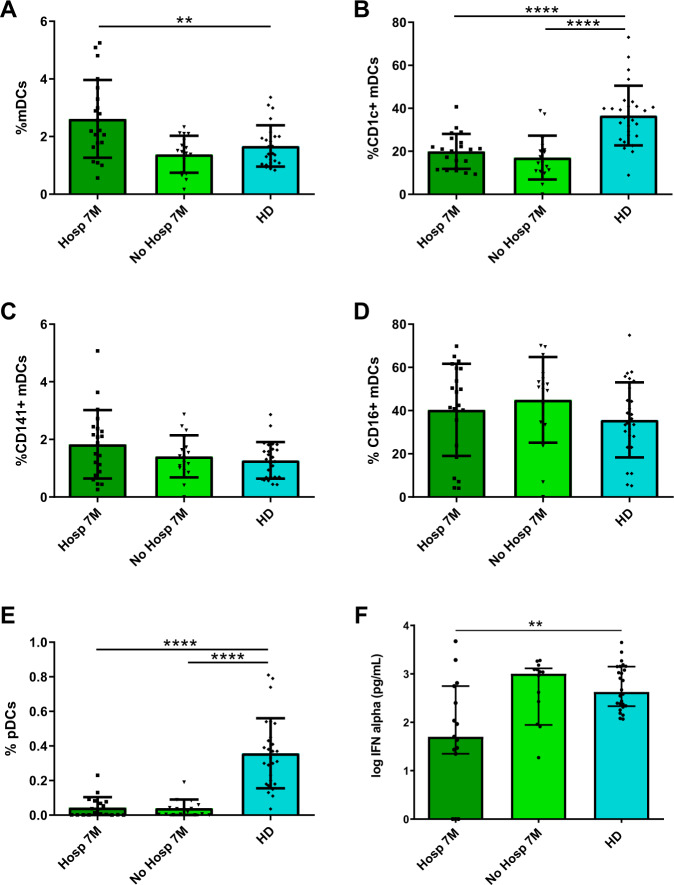
CD1c+ mDC and pDC levels and IFNα production are not normalized 7 months after SARS-CoV-2 infection
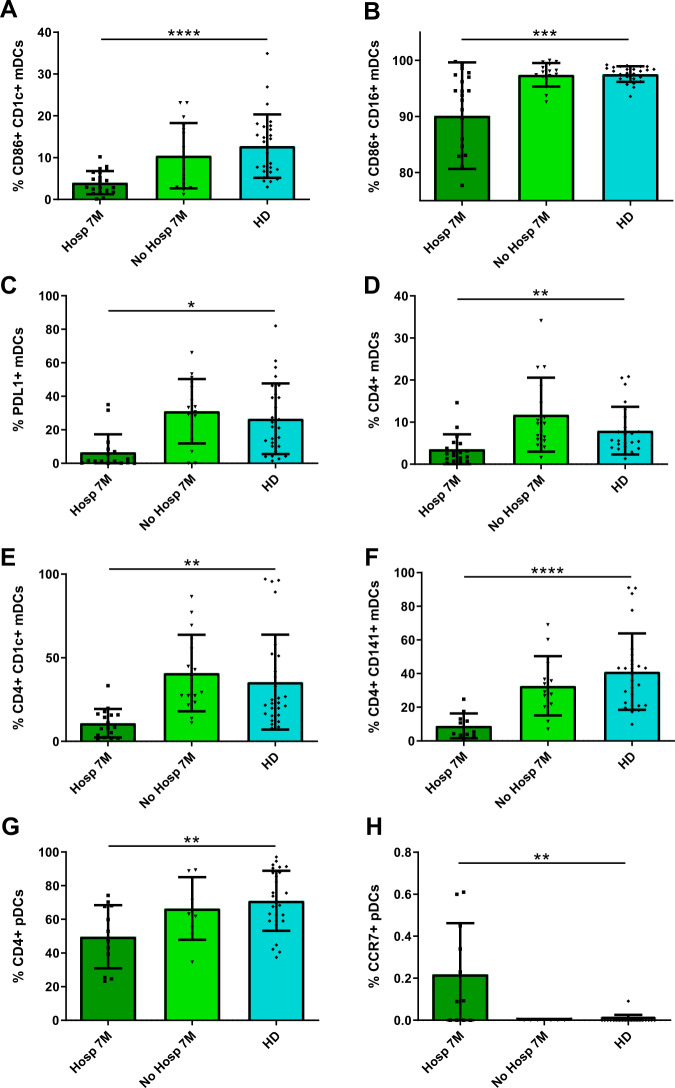
Apart from COVID-19 patients in acute phase, we also studied patients after 7 months of SARS-CoV-2 infection median 208 (interquartile range [IQR] [189–230]) days after symptoms’ onset, (Supplementary Table 1). Some of these patients were hospitalized during acute infection (Hosp 7M), while others were not (No Hosp 7M). We analyzed the percentages of DC subpopulations in these two groups and compared with HD’s levels. First, we observed a higher percentage of total mDCs on previously hospitalized patients compared with HD (Fig. 4A). Regarding mDC subpopulations, while the percentages of CD141+ and CD16+ were not altered, the percentage of CD1c+ mDCs remained lower in patients after 7 months compared with HD (Fig. 4B–D). Remarkably, the percentage of pDCs also persisted very low and was not restored 7 months after the infection in these both groups (Fig. 4E), confirmed by the mDC/pDC ratio (Supplementary Fig. 5a). Moreover, the percentage of P1-pDCs (CD86− PD-L1+) was only reduced in previously hospitalized patients comparing with HD, unlike P2− (CD86+PD-L1+) and P3-pDCs (CD86+PD-L1−), that were decreased in both hospitalized and nonhospitalized ones (Supplementary Fig. 5b). Next, to corroborate that our results were reproducible applying a paired analysis, we studied DC kinetic in a subgroup of subjects with available paired samples, analyzing the percentages of DC subpopulations in the acute phase, 6–8 months later and comparing them with HD. Even though the sample size was lower because of the sample availability, these results reproduced the analysis with unpaired samples (Supplementary Fig. 6).
Fig. 4.
CD1c + mDC and pDC levels and IFNα production are not normalized 7 months after SARS-CoV-2 infection. Bar graphs representing the percentage of total mDCs, CD1c+, CD141+, and CD16+ mDCs, pDCs (A–E) and the IFNα production (F) in previously hospitalized (Hosp 7M) or previously nonhospitalized (No Hosp 7M) patients 7 months after SARS-CoV-2 infection and in healthy donors (HD). The median with the interquartile range is shown and each dot represents an individual. **p < 0.01; ****p < 0.0001. Mann–Whitney U test was used for groups’ comparisons
When we measured the TLR9-dependent IFN-α production, we found that hospitalized patients 7 months after the infection showed a lower IFNα production than HD, unlike nonhospitalized patients, which display a similar production comparing with HD (Fig. 4F). Here, we conclude that the deficit of CD1c+ mDCs and pDCs is maintained 7 months after SARS-CoV-2 infection independently of whether the patients were or not previously hospitalized, and that IFNα production is not restored in previously hospitalized patients 7 months after infection.
Some DC activation markers are not normalized in previously hospitalized patients 7 months after SARS-CoV-2 infection
Afterwards, we measured the DC activation and homing markers in previously hospitalized and nonhospitalized patients 7 months after infection, and we compared them with the ones from HD. We observed that the expression of CD86 was lower in CD16+ and CD1c+ mDC subsets from hospitalized patients than in nonhospitalized ones and HD (Fig. 5A, B). Similar results were found in the expression of PD-L1 in total mDCs (Fig. 5C). Furthermore, hospitalized patients also showed lower levels of CD4 in total mDCs, CD1c+, and CD141+ mDCs and pDCs (Fig. 5D–G). In contrast, pDCs from hospitalized patients exhibited higher percentage of CCR7+ cells within pDCs compared with nonhospitalized ones and HD (Fig. 5H). In summary, these results show a recovery of some DC activation markers, mainly CD86 and CD4, only in previously nonhospitalized patients, while in more severe patients who required hospitalization, the defects in these markers persisted 7 months after infection.
Fig. 5.
Some DC activation markers are not normalized in previously hospitalized patients 7 months after SARS-CoV-2 infection. Bar graphs representing the percentage of DC subpopulations expressing CD86 (A, B), PD-L1 (C), CD4 (D–G), and CCR7 (H) in previously hospitalized (Hosp 7M) or previously nonhospitalized (No Hosp 7M) patients 7 months after SARS-CoV-2 infection and in healthy donors (HD). The median with the interquartile range is shown and each dot represents an individual. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Mann–Whitney U test was used for groups’ comparisons
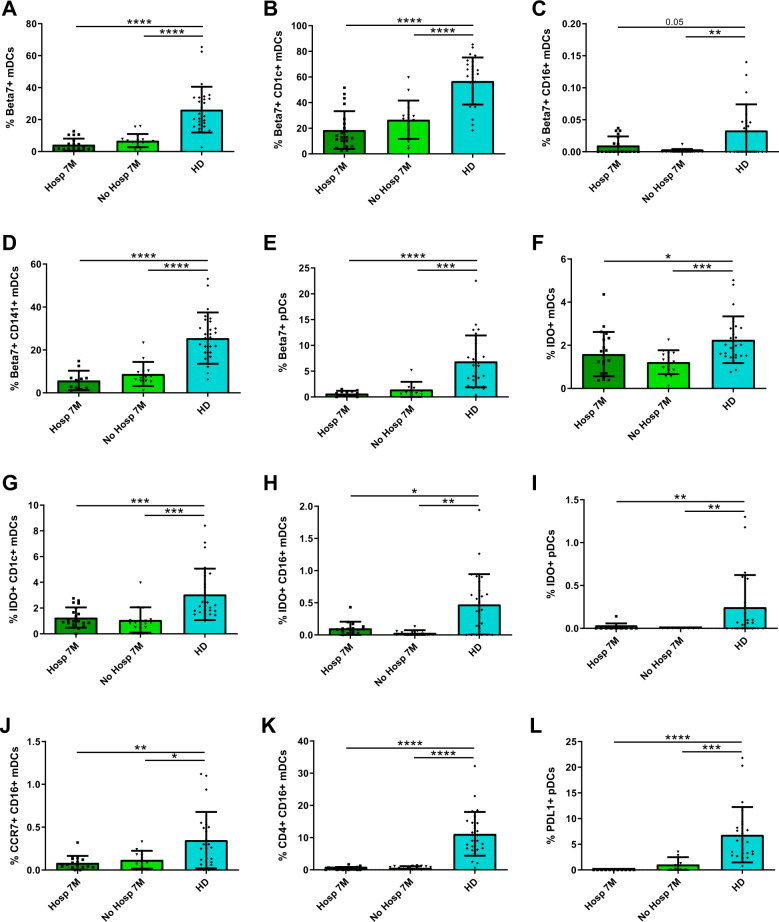
Some DC activation markers are not normalized neither in previously hospitalized nor in nonhospitalized patients 7 months after SARS-CoV-2 infection
Importantly, when we focused on the expression of other DC activation markers, we observed a lower percentage of β7+ cells in all mDCs and pDCs from both hospitalized and nonhospitalized patients after 7 months of infection compared to HD (Fig. 6A–E). The levels were also lower for IDO+ in total mDCs, CD1c+, and CD141+ mDCs and pDCs (Fig. 6F–I). Lastly, we also found that both hospitalized and nonhospitalized patients 7 months after infection showed lower percentages of CCR7+ and CD4+ cells within CD16+ mDCs and PD-L1+ cells within pDCs compared to HD (Fig. 6J–L). In conclusion, we demonstrated that the alterations in integrin β7 and IDO, associated with migration and tolerance, are not restored to normal levels neither in previously hospitalized nor in nonhospitalized patients 7 months after SARS-CoV-2 infection.
Fig. 6.
Some DC activation markers are not normalized neither in previously hospitalized nor in nonhospitalized patients 7 months after SARS-CoV-2 infection. Bar graphs representing the percentage of DC subpopulations expressing β7 (A–E), IDO (F–I), CCR7 (J), CD4 (K), and PD-L1 (L) in previously hospitalized (Hosp 7M) or previously nonhospitalized (No Hosp 7M) patients 7 months after SARS-CoV-2 infection and in healthy donors (HD). The median with the interquartile range is shown and each dot represents an individual. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. Mann–Whitney U test was used for groups’ comparisons
Discusion
The present study revealed that the deficits observed in CD1c+ mDCs and pDCs levels associated with altered homing and activation patterns in SARS-CoV-2 infected subjects in acute phase, were not restored beyond 7 months after infection. Importantly, this long-term defects related to DC migration and tolerogenesis (integrin β7 and IDO expression) were present independently of whether or not the patients were previously hospitalized. In addition, hospitalized patients showed additional deficiencies related with DC activation.
pDCs are known to have an important role in the first line of defense against viral replication, which mainly resides in their capacity to produce IFN-I via TLR-7/8 stimulation [14]. In this study, we first observed that acute SARS-CoV-2 infected patients displayed a dramatic decrease in pDC levels and a considerable reduction of IFN-α production. The strong direct correlation between pDC levels and IFN-α production suggested that this cell type was the main producer of this cytokine as it happens in other viral infections [15]. In fact, SARS-CoV-2 is known to induce pDC activation, accompanied by a high production of IFN-I and other cytokines, which is critically depended on IRAK4 and UNC93B [16]. The observed reduction of IFN-α is in accordance with previous studies in animal models of SARS-CoV-1 infection, which associated this deficit with lethal pneumonia [17] and is also consistent with recently published data following transcriptomic approaches [11] and intracellular cytokine staining after TLR stimulation in SARS-CoV-2 infection [7]. Importantly, the low IFN-α production was the main parameter associated with disease severity, in agreement with previous studies [7, 11], highlighting the potential use of this measurement as an early biomarker of disease progression. The mechanisms behind the attenuated IFN response have been related with viral antagonism of STAT1 (Signal transducer and activator of transcription 1) phosphorylation [18] and significantly, life-threatening ARDS in COVID-19 patients have been associated with neutralizing auto-antibodies against IFN-I [19, 20] and other inborn errors of IFN-I immunity [21]. Furthermore, single cell RNA sequencing of antigen-presenting cells revealed a lower expression of IFNAR1 and 2 in severe COVID-19 patients, suggesting a defect in IFN-α signaling, and also a downregulation of IFN-stimulated genes in both moderate and severe patients [22]. All these results support the essential role of IFN-I production in the first line of defense in COVID-19 for avoiding disease progression and point out to early immunotherapeutic strategies targeting this pathway. Remarkably, our results showed that, 7 months after SARS-CoV-2 infection, the IFNα production is not completely restored to normal levels, but only in previously hospitalized patients. This might be associated to the deficit in P1-pDCs found in hospitalized patients but not in nonhospitalized ones, being this pDC subset the main source of IFN-I [13]. Thus, our findings are indicative of a deficiency not only in pDC numbers but also in their function 7 months after SARS-CoV-2 infection in patients that were previously hospitalized.
Apart for IFN-I deficiency, one of the hallmarks of acute COVID-19 is the detection in plasma of heightened levels of soluble pro-inflammatory cytokines inducing a cytokine storm [23]. Here, we found multiple correlations between DC numbers and DC activation markers with inflammatory marker and cytokine levels in acute SARS-CoV-2 infected patients. It was remarkable that the lower percentage of DCs was associated to higher levels of IL-6 and higher neutrophil numbers. High levels of IL-6 in COVID-19 patients have been widely related to a poorer disease progression [10]. Moreover, neutrophils have been described as crucial drivers of hyperinflammation in COVID-19 [24]. It has to be also underlined, that the percentage of DCs expressing integrin β7 was inversely correlated to numerous inflammatory marker levels. These results suggest the hypothesis that not DC per se but DC migration to inflammatory sites may importantly contribute to the cytokine storm observed in SARS-CoV-2 infected patients. Our results also showed that patients with distinct level of disease severity displayed different associations of DC numbers and DC activation markers with inflammation. Therefore, DCs might be important contributors to the high inflammatory status characteristic of COVID-19 patients and this may dictate subsequent clinical progression.
The decreased numbers of total mDCs, CD1c+ mDCs and pDCs found in acute SARS-CoV-2 infected patients were in accordance with previous publications [5, 6]. This fact might be explained by different mechanisms, including apoptosis due to increased inflammatory mediators produced by abortive SARS-CoV-2 infection of myeloid cells [25]. Another nonexclusive explanation could be that DCs migrate from peripheral blood to tissues or inflammatory sites, such as CD1c+ mDCs preferential migration to the lungs in patients with severe COVID-19 [6]. These defects were accompanied by alterations mainly found in activation, migration and tolerogenic markers that importantly persisted 7 months after infection in previously hospitalized and also in nonhospitalized patients. Especially persistent in the total and DC subsets was the decreased expression of integrin β7. The expression of αEβ7 defines migration to antigen presentation sites within lymph nodes [26] and α4β7 on mDCs and pDCs is indicative of migration of these cells to gut [27]. Remarkably, SARS-CoV-2 has been shown to infect and productively replicate in human small intestinal organoids, increasing cytokine production and human angiotensin-converting enzyme 2 expression [28]. It has been also reported, that the disruption in gut barrier integrity contributes to COVID-19 severity [29]. Thus, the lower percentage of DCs expressing integrin β7 in peripheral blood might be a consequence of ongoing DC migration to the gut or other tissues or inflammatory sites up to 7 months after infection. In fact, necropsy studies in SARS-CoV-2 infected patients have shown mononuclear inflammatory infiltrates in different organs [30]. Also prominent was the deficit in IDO expressing DCs 7 months after infection. In contrast, IDO+ CD1c+ and CD16+ mDC levels in acute infection were dramatically increased compared to HD. This is in agreement with other acute respiratory infections such influenza [31] and respiratory syncytial virus [32] in which IDO expression is increased in order to counteract excessive inflammation as happen after acute SARS-CoV-2 infection. However, in this infection, the tissue damage in low respiratory tract is prominent [33] and may persist at the long-term what may cause the exhaustion of IDO producing DCs and/or migration of these cells to inflammatory focus even after 7 months after infection. Although these defects were present independently of whether or not the participants were previously at the hospital, hospitalized patients showed additional defects. These were, lower expression of the co-stimulatory molecule CD86, found in acute infection also by other authors [5, 7, 24] that persisted 7 months after infection together with lower levels of CD4+ DCs. Low levels of activation molecules, such as CD86 have been related with a possible impairment in T cell and DCs response to the virus. Specifically, we and others have found pDC hyporesponsiveness to HIV after CD4 downregulation in this cell type [15, 34]. On the contrary, CCR7+ pDCs remained at high levels even after 7 months after infection indicating again ongoing migration to lymph node or other inflammatory foci. In this line, the higher expression of other chemokine receptors such as CCR1, CCR3, and CCR5 has previously described in SARS-CoV-1 infected monocyte derived DCs [35].
It is unknown whether these defects in the DCs compartment will be reversible after longer follow up or specific therapies may be needed for the normalization of these defects. What is clear is that persisting symptoms and unexpected substantial organ dysfunction are observed in an increasing number of patients who have recovered from COVID-19 [12]. Actually, Huang et al. recently described that 7 months after illness onset, 76% of the SARS-CoV-2 infected patients reported at least one symptom that persisted, being fatigue or muscle weakness the most frequently reported symptoms [36]. In addition, many of those previously hospitalized patients presented residual chest imaging abnormalities, impaired pulmonary diffusion capacity and other extrapulmonary manifestations as a low estimated glomerular filtration rate [36]. The immune mechanisms that might be involved in the development of these persisting symptoms are still unknown. However, it would be expected that 7 months after SARS-CoV-2 infection there is still an inflammatory response due to persistent tissue damage or persistence presence of viral antigens in the absence of viral replication which may cause these deficits in DC. In fact, it has been reported that SARS-CoV-2 can persist in the intestines up to 7 months following symptoms resolution [37]. Thus, we postulate that the decrease in peripheral DCs numbers, along with the alterations in DC homing and activation markers 7 months after the infection might be indicative of DC migration to inflammatory sites which may be contributing to long-term symptoms, a phenomenon also known as long COVID.
One of the limitations of this study might be that, for a more precise identification of pDCs, CD2+, CD5+, and AXL+ cells should have been excluded [38]. Nevertheless, since these cell populations are barely represented within PBMCs, the showed results correspond mainly to pDCs, although some contamination with AS-DC cannot be excluded. The same happened with CD123+ mDCs, which were not included in our gating strategy, however, the levels of this subset was so low that did not change total mDC levels (data not shown). Moreover, a limitation of this work might be that all patients included in this study belong to the first wave of COVID-19 in Spain. It would have been interesting to have access to tissue samples, however due to safety issues at that moment of the pandemic it was not possible. At that time, different experimental treatments with very limited but transitory immunosuppressive effects were administered what may have affected the levels of immune parameters. However, the agreement of our observations with other data in the literature during acute infection and the persistence of these defects 7 months after infection minimized the potential bias of these treatments in our results.
In summary, we have demonstrated that SARS-CoV-2 infected patients showed a deficit in some DC subsets and alterations in DC homing and activation markers, which are not restored more than 7 months after the infection independently of previous hospitalization. Our results suggest that there is an ongoing inflammation which could be partially induced by DCs, these findings might contribute to a better understanding of the immunological sequelae of COVID-19.
Materials and methods
Study participants
Seventy one participants with confirmed detection of SARS-CoV-2 by reverse-transcription polymerase chain reaction (RT-PCR) were included. Out of these 71, 33 were hospitalized in acute phase of COVID-19 from March 25th to May 8th 2020, while 38 participants were recruited 7 months after being diagnosed with COVID-19, from September 9th to November 26th 2020. These participants came from the COVID-19 patients’ Cohort Virgen del Rocio University Hospital, Seville (Spain) and the COVID-19 Cohort IIS Galicia Sur (CohVID GS), Vigo (Spain). Twenty-seven healthy donors (HD), with cryopreserved pre-COVID-19 samples (May 12th to July 18th 2014) were included from the HD cohort, collection of samples of the Laboratory of HIV infection, Andalusian Health Public System Biobank, Seville (Spain) (C330024). Written or oral informed consent was obtained from all participants. The study was approved by the Ethics Committee of the Virgen del Rocio University Hospital (protocol code “pDCOVID”; internal code 0896-N-20). Hospitalized participants during the acute phase of infection were divided in Mild (n = 17) or Severe (n = 16), based on the highest grade of disease severity during course of hospitalization. Severe participants were those who required Intensive Care Unit admission, or having ≥6 points in the score on ordinal scale based on Beigel et al. [39] or death. Blood samples were collected at a median of 3 [interquartile range (IQR) 2–23] days after hospitalization and 14 [9–31] days after symptoms onset (Supplementary Table 1). The group of participants discharged after infection, included previously hospitalized (n = 21) and previously nonhospitalized subjects (n = 17). The samples from these participants were collected after a median of 201 [181–221] days after hospitalization and 208 [189–230] days after symptoms onset (Supplementary Table 1). COVID-19 participants in the different groups were age and sex matched with HDs’ group (Supplementary Table 1).
Cell and plasma isolation
PBMCs from healthy donors and participants were isolated from fresh blood samples using BD Vacutainer® CPT™ Mononuclear Cell Preparation Tubes (with Sodium Heparin, BD Cat# 362780) in a density gradient centrifugation at the same day of blood collection. Afterwards, PBMCs were cryopreserved in freezing medium (90% of fetal bovine serum (FBS) + 10% dimethyl sulfoxide) in liquid nitrogen until further use. Plasma samples were obtained using BD Vacutainer™ PET EDTA Tubes centrifugation, aliquoted and cryopreserved at −80 °C until further use.
Dendritic cell immunophenotyping
For DCs flow cytometry, PBMCs were centrifuged, pelleted and washed with Phosphate-buffered saline (PBS) and stained for 35 min at room temperature with LIVE/DEAD Fixable Aqua Dead Cell Stain (Life Technologies), BV421 CD86, BV650 CD11c, BV711 HLA-DR, BV786 CCR7 (CD197), FITC Lin-2 (CD3, CD14, CD19, CD20, CD56), BV605 CD16, PeCF594 PD-L1 (CD274), APC Integrin-β7 (BD Biosciences), PerCPCy5,5 CD4, APCCy7 CD1c, PeCy7 CD141 (BioLegend) and AF700 CD123 (R&D, San Diego, CA) antibodies. Then PBMCs were washed with Permeabilization Buffer 10X diluted 1:10 (eBioscience™), permeabilized by Fixation/Perm buffer (eBioscience™), and intracellularly stained with PE IDO (eBioscience, San Diego, CA, USA) antibody. DCs were gated based on Lin-2 HLA-DR expression. Each subset (mDCs and pDCS) was gated based on CD123 and CD11c expression. mDCs subsets were gated by using CD16, CD1c and CD141 staining, for gating strategy see Supplementary Fig. 1. Flow cytometry analyses were performed on an LRS Fortessa flow cytometer using FACS Diva software (BD Biosciences). Data were analyzed using the FlowJo software (Treestar, Ashland, OR). At least 1 × 106 events were acquired per sample.
Cell culture and IFN-α quantification
1 × 106 thawed PBMCs were incubated at 37 °C and 5% CO2 during 18 h in RPMI with 10% FBS without any stimuli or with 1 µM CpG-A (ODN 2216; InvivoGen). After incubation, cells were pelleted and the supernatants conserved for the subsequent quantification of IFN-α production at −80 °C. The amount of IFN-α in cell culture supernatants was assessed by an IFN-α multisubtype enzyme-linked immunosorbent assay kit (PBL Interferon Source Cat# 41105) according to the manufacturer’s instructions.
Cytokine quantification in plasma
Plasmas previously collected were used for the quantitative determination of cytokines. We used 3 different kits to quantify sCD25 by Human CD25/IL-2R alpha Quantikine ELISA Kit (R&D System, Cat# DR2A00), IP-10 by Human IP-10 ELISA Kit (CXCL10) (Abcam, Cat# ab173194) and IL-6, IL-8, IL-1β, TNF-α, IFN-γ, MIP-1α, and MIP-1β by MILLIPLEX MAP Human High Sensitivity T Cell Panel (Merck Cat# HSTCMAG-28SK) according to the manufacturer’s instructions.
Statistics
Statistical analyses were performed by using Statistical Package for the Social Sciences software (SPSS 25.0; SPSS, Inc., Chicago, IL) and R environment 4.0.3 (2020-10-10), using RStudio Version 1.3.959 as the work interface and GraphPad Prism, version 8.0 (GraphPad Software, Inc.). ROUT method was utilized to identify and discard outliers. Differences between conditions among different groups were analyzed by two-tailed Mann–Whitney U test. The Wilcoxon test was used to analyze paired samples. The Spearman test was used to analyze correlations between variables. All differences with a P value of <0.05 were considered statistically significant.
Supplementary information
Acknowledgements
This study would not have been possible without the collaboration of Virgen del Rocio University Hospital COVID Team and COHVID-GS and IISGS Biobank. All of the patients, nursing staff, and data managers who have taken part in this project. We would like to thank to Sara Bachiller for the critical reading of the paper. Members of COHVID-GS (Galicia Sur Health Research Institute): Alexandre Araujo, Jorge Julio Cabrera, Víctor del Campo, Manuel Crespo, Alberto Fernández, Beatriz Gil de Araujo, Carlos Gómez, Virginia Leiro, María Rebeca Longueira, Ana López-Domínguez, José Ramón Lorenzo, María Marcos, Alexandre Pérez, María Teresa Pérez, Lucia Patiño, Sonia Pérez, Silvia Pérez-Fernández, Eva Poveda, Cristina Ramos, Benito Regueiro, Cristina Retresas, Tania Rivera, Olga Souto, Isabel Taboada, Susana Teijeira, María Torres, Vanesa Val, Irene Viéitez. Members of the Virgen del Rocio University Hospital COVID Team: Jose Miguel Cisneros Herreros, Nuria Espinosa Aguilera, Cristina Roca Oporto, Julia Praena Segovia, José Molina, María Paniagua-García, Manuela Aguilar-Guisado, Almudena Aguilera, Clara Aguilera, Teresa Aldabo-Pallas, Verónica Alfaro-Lara, Cristina Amodeo, Javier Ampuero, Maribel Asensio, Bosco Barón-Franco, Lydia Barrera-Pulido, Rafael Bellido-Alba, Máximo Bernabeu-Wittel, Claudio Bueno, Candela Caballero-Eraso, Macarena Cabrera, Enrique Calderón, Jesús Carbajal-Guerrero, Manuela Cid-Cumplido, Juan Carlos Crespo, Yael Corcia-Palomo, Elisa Cordero, Juan Delgado, Alejandro Deniz, Reginal Dusseck-Brutus, Ana Escoresca Ortega, Fatima Espinosa, Michelle Espinoza, Carmen Ferrándiz-Millón, Marta Ferrer, Teresa Ferrer, Ignacio Gallego-Texeira, Rosa Gámez-Mancera, Emilio García, María Luisa Gascón-Castillo, Aurora González-Estrada, Demetrio González, Rocío GonzálezLeón, Carmen Grande-Cabrerizo, Sonia Gutiérrez, Carlos Hernández-Quiles, Concepción Herrera-Melero, Marta Herrero-Romero, Carmen Infante, Luis Jara, Carlos Jiménez-Juan, Silvia Jiménez-Jorge, Mercedes Jiménez-Sánchez, Julia LanserosTenllado, José María Lomas, Álvaro López, Carmina López, Isabel López, Luis F López-Cortés, Rafael Luque-Márquez, Daniel Macías-García, Luis Martín-Villén, Aurora Morillo, Dolores Nieto-Martín, Francisco Ortega, Amelia Peña-Rodríguez, Esther Pérez, Rafaela Ríos, Jesús F Rodríguez, María Jesús Rodríguez-Hernández, Santiago Rodríguez-Suárez, Ángel Rodríguez-Villodres, Nieves Romero-Rodríguez, Ricardo Ruiz, Zida Ruiz de Azua, Celia Salamanca, Sonia Sánchez, Javier Sánchez-Céspedez, Victor Manuel Sández-Montagut, Alejandro Suárez Benjumea, and Javier Toral. This work was supported by Consejeria de Transformacion Economica, Industria, Conocimiento y Universidades Junta de Andalucia (research Project CV20-85418), Consejeria de salud Junta de Andalucia (Research Contract RH-0037-2020 to JV) the Instituto de Salud Carlos III (CP19/00159 to AGV, FI17/00186 to MRJL, FI19/00083 to MCGC, CM20/00243 to APG, and COV20/00698 to support COHVID-GS) and the Red Temática de Investigación Cooperativa en SIDA (RD16/0025/0020 and RD16/0025/0026), which is included in the Acción Estratégica en Salud, Plan Nacional de Investigación Científica, Desarrollo e Innovación Tecnológica, 2008 to 2011 and 2013 to 2016, Instituto de Salud Carlos III, Fondos FEDER. ERM was supported by the Spanish Research Council (CSIC).
Author contributions
APG, JV, and MGC performed the experiments, analyzed and interpreted the data and participated in writing of the paper. AGV, MTR, ASG, and EMM participated in data collection, data analysis and interpretation and performed experiments, MRJL, MRIB participated in paper data interpretation, IRJ, CI, and JCC participated in data collection, CS, CRO, NE, AFV, and MC participated in data collection and paper interpretation. LFLC and EP participated in paper data analysis, patient and data collection, interpretation/ discussion of the results and coordination. ERM, participated in data analysis and interpretation, writing, conceived the idea and coordinate the project. APG, JV, and MCGC contributed equally to this work.
Competing interests
The authors declare no competing interests.
Footnotes
A list of authors and their affiliations appears at the end of the paper.
These authors contributed equally: Alberto Pérez-Gómez, Joana Vitallé, Carmen Gasca-Capote.
Contributor Information
Ezequiel Ruiz-Mateos, Email: eruizmateos-ibis@us.es.
the Virgen del Rocío Hospital COVID-19 Working Team:
José Miguel Cisneros, Sonsoles Salto-Alejandre, Judith Berastegui-Cabrera, Pedro Camacho-Martínez, Carmen Infante-Domínguez, Marta Carretero-Ledesma, Juan Carlos Crespo-Rivas, Eduardo Márquez, José Manuel Lomas, Claudio Bueno, Rosario Amaya, José Antonio Lepe, Jerónimo Pachón, Elisa Cordero, Javier Sánchez-Céspedes, Manuela Aguilar-Guisado, Almudena Aguilera, Clara Aguilera, Teresa Aldabo-Pallas, Verónica Alfaro-Lara, Cristina Amodeo, Javier Ampuero, María Dolores Avilés, Maribel Asensio, Bosco Barón-Franco, Lydia Barrera-Pulido, Rafael Bellido-Alba, Máximo Bernabeu-Wittel, Candela Caballero-Eraso, Macarena Cabrera, Enrique Calderón, Jesús Carbajal-Guerrero, Manuela Cid-Cumplido, Yael Corcia-Palomo, Juan Delgado, Antonio Domínguez-Petit, Alejandro Deniz, Reginal Dusseck-Brutus, Ana Escoresca-Ortega, Fátima Espinosa, Nuria Espinosa, Michelle Espinoza, Carmen Ferrándiz-Millón, Marta Ferrer, Teresa Ferrer, Ignacio Gallego-Texeira, Rosa Gámez-Mancera, Emilio García, Horacio García-Delgado, Manuel García-Gutiérrez, María Luisa Gascón-Castillo, Aurora González-Estrada, Demetrio González, Carmen Gómez-González, Rocío González-León, Carmen Grande-Cabrerizo, Sonia Gutiérrez, Carlos Hernández-Quiles, Inmaculada Concepción Herrera-Melero, Marta Herrero-Romero, Luis Jara, Carlos Jiménez-Juan, Silvia Jiménez-Jorge, Mercedes Jiménez-Sánchez, Julia Lanseros-Tenllado, Carmina López, Isabel López, Álvaro López-Barrios, Luis F. López-Cortés, Rafael Luque-Márquez, Daniel Macías-García, Guillermo Martín-Gutiérrez, Luis Martín-Villén, José Molina, Aurora Morillo, María Dolores Navarro-Amuedo, Dolores Nieto-Martín, Francisco Ortega, María Paniagua-García, Amelia Peña-Rodríguez, Esther Pérez, Manuel Poyato, Julia Praena-Segovia, Rafaela Ríos, Cristina Roca-Oporto, Jesús F. Rodríguez, María Jesús Rodríguez-Hernández, Santiago Rodríguez-Suárez, Ángel Rodríguez-Villodres, Nieves Romero-Rodríguez, Ricardo Ruiz, Zida Ruiz de Azua, Celia Salamanca, Sonia Sánchez, Víctor Manuel Sánchez-Montagut, César Sotomayor, Alejandro Suárez Benjumea, and Javier Toral
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-021-00728-2.
References
- 1.Guan W, Ni Z, Hu Y, Liang W, Ou C, He J, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–20. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhou Z, Ren L, Zhang L, Zhong J, Xiao Y, Jia Z, et al. Heightened innate immune responses in the respiratory tract of COVID-19 patients. Cell Host Microbe. 2020;27:883–90.e2. doi: 10.1016/j.chom.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chi Y, Ge Y, Wu B, Zhang W, Wu T, Wen T, et al. Serum cytokine and chemokine profile in relation to the severity of coronavirus disease 2019 in China. J Infect Dis. 2020;222:746–54. doi: 10.1093/infdis/jiaa363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kvedaraite E, Hertwig L, Sinha I, Ponzetta A, Hed Myrberg I, Lourda M, et al. Major alterations in the mononuclear phagocyte landscape associated with COVID-19 severity. Proc Natl Acad Sci USA. 2021;118:1–12. doi: 10.1073/pnas.2018587118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou R, To KKW, Wong YC, Liu L, Zhou B, Li X, et al. Acute SARS-CoV-2 infection impairs dendritic cell and T cell responses. Immunity. 2020;53:864–77.e5. doi: 10.1016/j.immuni.2020.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sánchez-Cerrillo I, Landete P, Aldave B, Sánchez-Alonso S, Sánchez-Azofra A, Marcos-Jiménez A, et al. COVID-19 severity associates with pulmonary redistribution of CD1c+ DCs and inflammatory transitional and nonclassical monocytes. J Clin Investig. 2020;130:6290–300. doi: 10.1172/JCI140335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arunachalam PS, Wimmers F, Mok CKP, Perera RAPM, Scott M, Hagan T, et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science. 2020;369:1210–20. [DOI] [PMC free article] [PubMed]
- 8.Swiecki M, Colonna M. Unraveling the functions of plasmacytoid dendritic cells during viral infections, autoimmunity, and tolerance. Immunol Rev. 2010;234:142–62. doi: 10.1111/j.0105-2896.2009.00881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muller U, Steinhoff U, Reis LF, Hemmi S, Pavlovic J, Zinkernagel RM, et al. Functional role of type I and type II interferons in antiviral defense. Science. 1994;264:1918 LP–21. doi: 10.1126/science.8009221. [DOI] [PubMed] [Google Scholar]
- 10.Laing AG, Lorenc A, del Molino del Barrio I, Das A, Fish M, Monin L, et al. A dynamic COVID-19 immune signature includes associations with poor prognosis. Nat Med. 2020;26:1623–35. doi: 10.1038/s41591-020-1038-6. [DOI] [PubMed] [Google Scholar]
- 11.Hadjadj J, Yatim N, Barnabei L, Corneau A, Boussier J, Smith N, et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science. 2020;369:718–24. [DOI] [PMC free article] [PubMed]
- 12.Carfì A, Bernabei R, Landi F. Group for the GAC-19 P-ACS. Persistent symptoms in patients after acute COVID-19. JAMA. 2020;324:603–5. doi: 10.1001/jama.2020.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alculumbre SG, Saint-André V, Di Domizio J, Vargas P, Sirven P, Bost P, et al. Diversification of human plasmacytoid predendritic cells in response to a single stimulus. Nat Immunol. 2018;19:63–75. doi: 10.1038/s41590-017-0012-z. [DOI] [PubMed] [Google Scholar]
- 14.Reizis B. Plasmacytoid dendritic cells: development, regulation, and function. Immunity. 2019;50:37–50. doi: 10.1016/j.immuni.2018.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Machmach K, Leal M, Gras C, Viciana P, Genebat M, Franco E, et al. Plasmacytoid dendritic cells reduce HIV Production in elite controllers. J Virol. 2012;86. 10.1128/JVI.07114-11. [DOI] [PMC free article] [PubMed]
- 16.Onodi F, Bonnet-Madin L, Meertens L, Karpf L, Poirot J, Zhang SY, et al. SARS-CoV-2 induces human plasmacytoid predendritic cell diversification via UNC93B and IRAK4. J Exp Med. 2021;218. 10.1084/JEM.20201387. [DOI] [PMC free article] [PubMed]
- 17.Channappanavar R, Fehr AR, Vijay R, Mack M, Zhao J, Meyerholz DK, et al. Dysregulated Type I interferon and inflammatory monocyte-macrophage responses cause lethal pneumonia in SARS-CoV-infected mice. Cell Host Microbe. 2016;19:181–93. doi: 10.1016/j.chom.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang D, Chu H, Hou Y, Chai Y, Shuai H, Lee ACY, et al. Attenuated interferon and proinflammatory response in SARS-CoV-2-infected human dendritic cells is associated with viral antagonism of STAT1 phosphorylation. J Infect Dis. 2020;222:734–45. doi: 10.1093/infdis/jiaa356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bastard P, Rosen LB, Zhang Q, Michailidis E, Hoffmann HH, Zhang Y, et al. Autoantibodies against type I IFNs in patients with life-threatening COVID-19. Science. 2020;370. 10.1126/science.abd4585. [DOI] [PMC free article] [PubMed]
- 20.Combes AJ, Courau T, Kuhn NF, Hu KH, Ray A, Chen WS, et al. Global absence and targeting of protective immune states in severe COVID-19. Nature. 2021. 10.1038/s41586-021-03234-7. [DOI] [PMC free article] [PubMed]
- 21.Zhang Q, Liu Z, Moncada-Velez M, Chen J, Ogishi M, Bigio B, et al. Inborn errors of type I IFN immunity in patients with life-threatening COVID-19. Science. 2020;370. 10.1126/science.abd4570. [DOI] [PMC free article] [PubMed]
- 22.Saichi M, Ladjemi MZ, Korniotis S, Rousseau C, Ait Hamou Z, Massenet-Regad L, et al. Single-cell RNA sequencing of blood antigen-presenting cells in severe COVID-19 reveals multi-process defects in antiviral immunity. Nat Cell Biol. 2021;23. 10.1038/s41556-021-00681-2. [DOI] [PubMed]
- 23.Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Investig. 2020;130:2620–9. doi: 10.1172/JCI137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parackova Z, Zentsova I, Bloomfield M, Vrabcova P, Smetanova J, Klocperk A, et al. Disharmonic Inflammatory Signatures in COVID-19: Augmented Neutrophils’ but Impaired Monocytes’ and Dendritic Cells’ Responsiveness. Cells. 2020;9:2206. [DOI] [PMC free article] [PubMed]
- 25.Zheng J, Wang Y, Li K, Meyerholz DK, Allamargot C, Perlman S. Severe acute respiratory syndrome coronavirus 2–induced immune activation and death of monocyte-derived human macrophages and dendritic cells. J Infect Dis. 2020;52242:1–11. doi: 10.1093/infdis/jiaa753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pribila JT, Itano AA, Mueller KL, Shimizu Y. The α 1 β 1 and α E β 7 integrins define a subset of dendritic cells in peripheral lymph nodes with unique adhesive and antigen uptake properties. J Immunol. 2004;172:282–91. doi: 10.4049/jimmunol.172.1.282. [DOI] [PubMed] [Google Scholar]
- 27.Clahsen T, Pabst O, Tenbrock K, Schippers A, Wagner N. Localization of dendritic cells in the gut epithelium requires MAdCAM-1. Clin Immunol. 2015;156:74–84. doi: 10.1016/j.clim.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 28.Lamers MM, Beumer J, van der Vaart J, Knoops K, Puschhof J, Breugem TI, et al. SARS-CoV-2 productively infects human gut enterocytes. Science. 2020;369:50–4. [DOI] [PMC free article] [PubMed]
- 29.Giron LB, Dweep H, Yin X, Wang H, Damra M, Goldman AR, et al. Plasma Markers of Disrupted Gut Permeability in Severe COVID-19 Patients. Front Immunol. 2021;12:686240. [DOI] [PMC free article] [PubMed]
- 30.Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420–2. doi: 10.1016/S2213-2600(20)30076-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fox JM, Sage LK, Poore S, Johnson S, Tompkins SM, Tripp RA. Drug analog inhibition of indoleamine 2,3-dioxygenase (IDO) activity modifies pattern recognition receptor expression and proinflammatory cytokine responses early during influenza virus infection. J Leukoc Biol. 2014;96:447–52. doi: 10.1189/jlb.3AB0114-046RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ajamian F, Wu Y, Ebeling C, Ilarraza R, Odemuyiwa SO, Moqbel R, et al. Respiratory syncytial virus induces indoleamine 2,3-dioxygenase activity: a potential novel role in the development of allergic disease. Clin Exp Allergy. 2015;45:644–59. doi: 10.1111/cea.12498. [DOI] [PubMed] [Google Scholar]
- 33.Broggi A, Ghosh S, Sposito B, Spreafico R, Balzarini F, Lo Cascio A, et al. Type III interferons disrupt the lung epithelial barrier upon viral recognition. Science. 2020;369:706–12. [DOI] [PMC free article] [PubMed]
- 34.Barblu L, MacHmach K, Gras C, Delfraissy J-F, Boufassa F, Leal M, et al. Plasmacytoid dendritic cells (pDCs) from HIV controllers produce interferon-and differentiate into functional killer pDCs under HIV activation. J Infect Dis. 2012;206. 10.1093/infdis/jis384. [DOI] [PubMed]
- 35.Law HKW, Cheung CY, Sia SF, Chan YO, Peiris JSM, Lau YL. Toll-like receptors, chemokine receptors and death receptor ligands responses in SARS coronavirus infected human monocyte derived dendritic cells. BMC Immunol. 2009;10:1–12. doi: 10.1186/1471-2172-10-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang C, Huang L, Wang Y, Li X, Ren L, Gu X, et al. 6-month consequences of COVID-19 in patients discharged from hospital: a cohort study. Lancet. 2021;397:220–32. doi: 10.1016/S0140-6736(20)32656-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tokuyama M, Ladinsky MS, Jha D, Cossarini F, Livanos AE, Reidy J, Tankelevich M, Martinez-Delgado G, Bjorkman PJ, Mehandru S. SARSCoV-2 persists in intestinal enterocytes up to 7 months after symptom resolution. Abstract 115. Conference on Retroviruses and Opportunistic Infections (CROI) 2021.
- 38.Villani AC, Satija R, Reynolds G, Sarkizova S, Shekhar K, Fletcher J, et al. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356:eaah4573. [DOI] [PMC free article] [PubMed]
- 39.Beigel JH, Tomashek KM, Dodd LE, Mehta AK, Zingman BS, Kalil AC, et al. Remdesivir for the treatment of Covid-19—final report. N Engl J Med. 2020;383:1813–26. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.