Abstract
Recent advances in lipopolysaccharide (LPS) biology have led to its use in drug discovery pipelines, including vaccine and vaccine adjuvant discovery. Desirable characteristics for LPS vaccine candidates include both the ability to produce a specific antibody titer in patients and a minimal host inflammatory response directed by the innate immune system. However, in-depth chemical characterization of most LPS extracts has not been performed; hence, biological activities of these extracts are unpredictable. Additionally, the most widely adopted workflow for LPS structure elucidation includes nonspecific chemical decomposition steps before analyses, making structures inferred and not necessarily biologically relevant. In this work, several different mass spectrometry workflows that have not been previously explored were employed to show proof-of-principle for top down LPS primary structure elucidation, specifically for a rough-type mutant (J5) E. coli-derived LPS component of a vaccine candidate. First, ion mobility filtered precursor ions were subjected to collision induced dissociation (CID) to define differences in native J5 LPS v. chemically detoxified J5 LPS (dLPS). Next, ultra-high mass resolving power, accurate mass spectrometry was employed for unequivocal precursor and product ion empirical formulae generation. Finally, MS3 analyses in an ion trap instrument showed that previous knowledge about dissociation of LPS components can be used to reconstruct and sequence LPS in a top down fashion. A structural rationale is also explained for differential inflammatory dose-response curves, in vitro, when HEK-Blue hTLR4 cells were administered increasing concentrations of native J5 LPS v. dLPS, which will be useful in future drug discovery efforts.
Graphical Abstract
Introduction
Lipopolysaccharide (LPS), also known as endotoxin, is a major component of the outer leaflet of most Gram-negative bacterial outer cell membranes [1, 2]. It is amphipathic, allowing it to interact with a wide range of ions and molecules to maintain cell membrane integrity [1], participate in cell-cell interactions [3–5], contribute to pathogenicity [6], and protect the bacterium from exogenous threats [7–10]. LPS is composed of three parts (listed in order from the membrane to the extracellular space): 1.) a lipophilic, multiply acylated diglucosamine membrane anchor that produces the canonical biologic activity of Gram-negative bacterial infection (lipid A), 2.) a non-repeating oligosaccharide core (core OS), and 3.) a polysaccharide, composed of repeating oligosaccharide units, that produces an immunodominant antigen responsible for the O serotype (O-antigen). LPS exists as a mixture of biosynthetic products that can be broadly classified into two groups: rough-type LPS (R-LPS) or lipooligosaccharide (LOS), and smooth-type LPS (S-LPS). S-LPS is a complete LPS molecule, comprising all three aforementioned parts, while R-LPS lacks the O-antigen portion, usually resulting in a loss of virulence. The biosynthesis pathway enzymes for making lipid A and core OS in Gram-negative bacteria are relatively well-conserved. However, due to differences in abundance or structure of LPS modifying enzymes, the resultant products from these syntheses can vary greatly, both in structure and function, even within one species [11]. Many non-stoichiometric substitutions of phosphate groups, sugars, amino acids, amines, and other R-groups are also observed in LPS extracts, making accurate structural analyses challenging.
The dramatic increase in antibiotic resistance has left clinicians with fewer options to treat Gram-negative bacterial infections. Vaccines have proven to be one of the most efficient strategies to prevent infectious disease-related mortality and morbidity [12]. Although some of the structural features of LPS resulting in specific illnesses like sepsis have been established (e.g. lipid A-TLR4 ligand-receptor binding induced cytokine storm), there is no FDA-approved drug or vaccine against Gram-negative bacteria-induced sepsis in large part because anti-lipid A antibodies have not shown much promise in clinical settings [13–16]. Lipid A three dimensional (3D) structure is usually quite flexible and varies greatly between species of Enterobacteriaceae; however, the 3D structure of core OS remains similar due to conservation of biosynthetic enzymes [1]. In the past, studies have drawn a correlation between survival after Gram-negative bacterial sepsis and measured levels of circulating anti-core endotoxin antibodies in patient sera [17, 18]. Consequently, one feasible strategy for preventing sepsis is modification of LPS to generate antibodies to conserved epitopes in the core OS without eliciting a strong innate immune response. Passive infusion of immune sera after immunization of human volunteers with a vaccine composed of a heat-killed mutant of E. coli O111 which lacked the O polysaccharide (J5, or Rc chemotype mutant) resulted in protection against Gram-negative bacterial sepsis. A subsequent vaccine was developed using the purified J5 R-LPS that was alkali-treated to reduce the lipid A-induced toxicity and make the vaccine less reactogenic. This detoxified J5 LPS (J5 dLPS) was non-covalently complexed with group B meningococcal outer membrane protein (OMP) to form a hydrophobic complex. The hydrophilic portion on the outer surface enhanced its solubility and delivery. When administered to rodents, it improved survival from polymicrobial sepsis [19–21] and was well tolerated in humans [22] where it elicited a 37–142-fold increase in anti-core LPS antibody titer post-vaccination [21]. Interestingly, anti-core LPS vaccines (e.g. J5 Bacterin®) have been successful in treating bovine mastitis and decreasing Gram-negative bacterial sepsis incidence in animals [23, 24]. Additional vaccines have been developed against core OS but have not progressed to clinical trial [18, 25].
The core glycolipid-carrier protein complex or liposome-associated preparations have been developed to facilitate vaccine delivery and immunization processes as lipid A in liposomes imparts significantly reduced toxicity [26] and liposome-encapsulated TLR4 ligands produce higher antibody response [27]. Since LPS is highly reactogenic and can impart severe toxicity, chemical modification of LPS vaccines, such as the J5 vaccine, to reduce lipid A toxicity while retaining core OS immunogenicity has been proposed [21, 24]. In addition, the potent pro-inflammatory activity of the lipid A portion of the LPS has been modified to safely provide adjuvant activity for many vaccines. In fact, a chemically modified LOS, monophosphoryl lipid A (MPL®), that has diminished reactogenicity but potent adjuvanticity [28, 29] is used as a vaccine adjuvant in GlaxoSmithKline’s hepatitis B vaccine Fendrix® and human papillomavirus (HPV) vaccine Cervarix®. Recently, rationally designed lipid A-based Toll-like receptor 4 (TLR4) ligands have been reported for vaccine adjuvant discovery and modulating the innate immune response [30, 31]. Given both the potential use of core OS as a vaccine and a modified lipid A as a vaccine adjuvant, LPS structure elucidation capability to support these efforts is critical.
In the past, LPS has been mostly analyzed after hydrolysis into its three distinct components by mass spectrometry (MS), nuclear magnetic resonance (NMR) spectroscopy, or most commonly, a combination of both techniques, and then reconstructed by inference to demonstrate “representative” LPS structures for a species or strain. The results from these experiments can be misleading for several reasons mentioned below; this list is not exhaustive. First – and most importantly – LPS extracts are always a mixture of very similar compounds, sometimes producing exactly isobaric ions, making exact data interpretation very difficult. This problem is compounded by relatively nonspecific methods like hydrolysis or solvolysis for dissociation. Mixture complexity inevitably increases when these methods are employed. Second, it is impossible, using this sub-component based structure definition approach, to infer which candidate LPS structures are actually present in the cell membrane and are consequently of biological relevance. Third, biological activities cannot be positively attributed to specific structures because the complete structures are unknown. Attribution of structure to activity, or structure-activity relationships (SAR), are also impossible to infer from mixtures of biologically active molecules unless the relative activities of each of the components, as well as any interactions they may have with one another, are known. Indeed, it has been shown that pure, chemically synthesized lipid As corresponding to molecular formulae found together in extracted E. coli lipid A mixtures cause markedly different cytokine responses in murine macrophages [32]. Therefore, even though the hypothesis has not been exhaustively tested, it is reasonable to work on the assumption that differences in activity between LPS extracts are cumulative, since any mixture of LPS molecules likely contains full agonists, partial agonists, and antagonists of TLR4.
Qureshi et al. showed that R-LPS could be purified based on the number of acyl chains present and analyzed offline by plasma desorption mass spectrometry in 1988 [33]. Recently, O’Brien et al. published a top down liquid chromatography-tandem MS approach to determining primary structural features of R-LPS from E. coli laboratory strains using collision induced dissociation (CID) and ultraviolet photodissociation (UVPD) [34]. Although it has been feasible to perform top down tandem MS experiments on R-LPS since the 1980’s, most researchers have opted for the divide-and-conquer approach of wet chemistry followed by MS analysis of the separate components.
With advances in MS instrument hardware, software, and electronics have come substantial increases in mass spectrometer capabilities since the heyday of plasma desorption. Modern instruments have become more sensitive, can analyze many more samples in the same amount of time, and possess greater mass resolving powers and mass accuracies than older mass spectrometers. These benefits allow operators to separate, detect, and identify many ions solely in the gas phase. This approach was applied to the following research, using several different MS instruments for confirmation of previous results and to add complementary data for strengthened structural conclusions. Since the primary chemical structure of the J5 vaccine’s LPS component has never been evaluated, and to investigate the hypothesis that the detoxified R-LPS vaccine from the J5 strain of E. coli derives its decreased inflammatory potential, while maintaining therapeutic value, from complete O-deacylation of the lipid A moiety, the following research was conducted:
Experimental
Materials
Purified (low protein and nucleic acid content) and lyophilized LPS from E. coli J5 strain was purchased from both List Biological Laboratories, Inc. (Campbell, CA) and Sigma Aldrich (St. Louis, MO). Note: LPS can be toxic if ingested or inhaled; proper personal protective equipment should be worn at all times. All solvents and water used throughout all experiments were Fisher (Waltham, MA) Optima LC/MS grade.
Detoxification of R-LPS and preparation of vaccine
To prepare a detoxified E. coli J5 LPS (J5 dLPS), purified LPS from List Biological Laboratories was re-suspended in water (4 mg mL−1) and an equal volume of 0.2 M NaOH solution was added slowly with gentle stirring, followed by heating in a water bath at 65 °C for 2 hours. The mixture was shaken every 5 minutes for the first hour and every 10 minutes thereafter. The solution was then neutralized with 1 M acetic acid, ethanol precipitated, and lyophilized to isolate the dry product. To prepare J5 dLPS/group B meningococcal outer membrane protein (OMP) complex vaccine, OMP extracted from phenol-killed bacteria was mixed with J5 dLPS as described elsewhere [19, 22]. Briefly, OMP was extracted from phenol-killed bacteria by Empigen BB (Huntsman Corporation, The Woodlands, TX; licensed to Sigma Aldrich for sale) detergent. Extracted OMP was mixed with J5 dLPS (in 0.9% NaCl) at a ratio of 1.2:1 (w/w) in Tris-EDTA buffer, pH 8.0 containing 0.1% Empigen BB. Extensive dialysis was achieved to remove detergent and the vaccine was further filter sterilized and stored at 5 °C.
Cell culture and cytokine reporter assay
HEK-Blue hTLR4 cells (Invitrogen, Waltham, MA) were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Gibco, Gaithersburg, MD) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Sigma Aldrich, St. Louis, MO), 100 IU mL−1 penicillin, 100 μg mL−1 streptomycin, 1 mM sodium pyruvate, and 200 mM L-glutamine in a humidified incubator at 37 °C, 5% CO2. For cell stimulation, lyophilized LPS was reconstituted in sterile, endotoxin-free water at a concentration of 1 mg mL−1. This stock was serially diluted in DMEM before addition to the cell culture for the stimulation experiment. Post-stimulation (16 h), supernatants were collected from HEK-Blue cells, and the production of SEAP reporter was detected using Quanti-Blue (Invitrogen) according to the manufacturer’s instructions. NF-κB reporter cell line stimulation data were plotted as the mean (± SD) from biological duplicates using GraphPad Prism 7.0 (La Jolla, CA).
Defining mixture composition differences between intact and detoxified LPS samples
Both intact J5 LPS and J5 dLPS samples from List Biological Laboratories, Inc. were dissolved in a solution, composed of 50% (v/v) 2-propanol and 50% water, and directly infused by syringe pump (5 μL min−1, at an estimated concentration of 20 μg mL−1) into the source of a Waters (Milford, MA) Synapt G2 HDMS quadrupole-ion mobility separation-orthogonal acceleration time of flight mass spectrometer, equipped with a 4 kDa quadrupole, and operated with negative polarity electrospray ionization (ESI) and in “Resolution” mode. Traveling wave ion mobility separation (TWIMS) was employed to isolate ions with similar size, shape, and charge and to consequently simplify mass spectra. TWIMS was also used for gas-phase separation after quadrupole precursor isolation and prior to tandem MS experiments. The ion mobility separations were all performed using N2 as buffer gas at a flow rate of 90 mL min−1, with a wave velocity of 650 m s−1 and a wave height of 40.0 V. The ESI source was operated with a capillary potential of 3.00 kV, source temperature of 100 °C, sampling cone at 40.0 V, source offset at 40.0 V, source gas (N2) flow at 0.0 mL min−1, desolvation temperature of 400 °C, cone gas flow at 25 L hr−1, desolvation gas flow at 400 L hr−1, and nebulizer gas pressure at 5.0 bar. Collision induced dissociation (CID) tandem MS experiments were performed using ultra-pure argon (Airgas, Radnor Township, PA) as collision gas, with manual collision energy ramping from 0 V to 100 V in increments of 10 V to produce comprehensive product ion spectra. All other instrument parameters are available upon request.
Ultra-high mass resolving power, high mass accuracy FT-ICR MS to determine empirical formulae and define J5 LPS primary structure
Intact E. coli J5 LPS from Sigma Aldrich (St. Louis, MO), dissolved in a solution of 50% 2-propanol and 50% water at an estimated total LPS concentration of 50 μg mL−1, was directly infused through a home-built nano-electrospray ionization (nESI) source at a flow rate of 1 μL min−1 into a hybrid linear ion trap – 21 Tesla Fourier transform-ion cyclotron resonance (FT-ICR) mass spectrometer, described in detail in a previous publication [35]. The ion spray was visually optimized for each experiment by adjusting capillary voltage and position while monitoring for constant signal as well as observation of a uniform electrospray plume. Multiple tandem MS experiments were performed, including trap CID, beam-type collisionally activated dissociation (beam CAD), and in-cell ultraviolet photodissociation (UVPD). Trap CID and beam CAD were performed with stepped collision energy. UVPD was carried out similarly to a previous publication [36] using a Coherent (Santa Clara, CA) Excistar XS ArF excimer laser operated at 193 nm wavelength and 522 μJ per pulse. The laser was previously aligned through a window in the rear of the ICR magnet housing on-axis with the ICR cell.
Multi-stage MS (MSn) to confirm structural inferences
Intact E. coli J5 LPS from both List Biological Laboratories, Inc. (Campbell, CA) and Sigma Aldrich (St. Louis, MO) were directly infused in a solution composed of 50% 2-propanol and 50% water at a flow rate of 2 μL min−1 into a home-built nESI source of a linear ion trap (linear trapping quadrupole; LTQ) mass spectrometer (Thermo Finnigan, San Jose, CA) with post-production ion funnel optics added for increased ion transmission efficiency. The mass spectrometer was operated in negative ionization mode with a capillary potential of 2.3 kV. Tandem MS experiments were performed in the ion trap with ultra-pure helium as collision gas. MS3 was carried out on product ions from LPS, corresponding to lipid A and core OS, at stepped normalized collision energies to evaluate its utility for top down sequencing.
Data analysis
Data acquired on the Synapt G2 HDMS instrument were initially processed in Driftscope version 2.7 and MassLynx version 4.1 software (Waters, Milford, MA). When necessary, data were converted to mzML format using the ProteoWizard msconvert utility. Automated peak picking was performed using mMass version 5.5 (www.mmass.org) [37]. Data acquired on the FT-ICR instrument were converted from magnitude mode to absorption mode and processed (including peak picking and absorption mode isotopic modeling) using AutoVectis (Spectroswiss Sàrl, Lausanne, Switzerland) [38–42]. For rapid visualization and spot checking of data, ICR mass spectra were saved as Thermo .raw files and manipulated in Thermo (San Jose, CA) Xcalibur version 3.0.63. Data acquired on the LTQ instrument were processed in Xcalibur version 3.0.63, and when necessary, were converted to mzML format for use in open source software suites. Peak picking for LTQ data was also performed using mMass version 5.5. Data were plotted using QtiPlot version 0.9.8.9 (www.qtiplot.com) and Figures were generated in Inkscape version 0.91 (https://inkscape.org) and when necessary, modified to an acceptable format using GIMP version 2.8.18 (www.gimp.org).
Results and Discussion
Cell culture and cytokine reporter assay
To test the pro-inflammatory capacity of various LPS described in this manuscript, LPS were incubated with the HEK-Blue hTLR4 reporter cell line from which NFκB activation can be directly measured (Fig. 1). Both E. coli strains tested had similar stimulation profiles with J5 LPS reaching maximum stimulation at a slightly higher concentration than the W3110 strain. The J5 dLPS reached a lower maximum signaling level at a lower concentration, indicating that it is less inflammatory but maintained a strong binding affinity to the activated signaling pathway. The J5 dLPS/GBOMP formulation reached a similar maximal signaling level as the J5 dLPS but activation could be titrated down at a much higher concentration. PHAD, an adjuvant molecule already in use, stimulated cells at a much lower level than all other molecules tested, only rising above baseline at the highest concentration tested. At low concentrations J5 dLPS/GBOMP is capable of maintaining NF-κB stimulation while PHAD, a known adjuvant molecule, is not. This novel method of detoxifying LPS could be used to create TLR4 agonists that are not toxic and still capable of stimulating the immune system to a desirable level for adjuvant use.
Fig. 1.
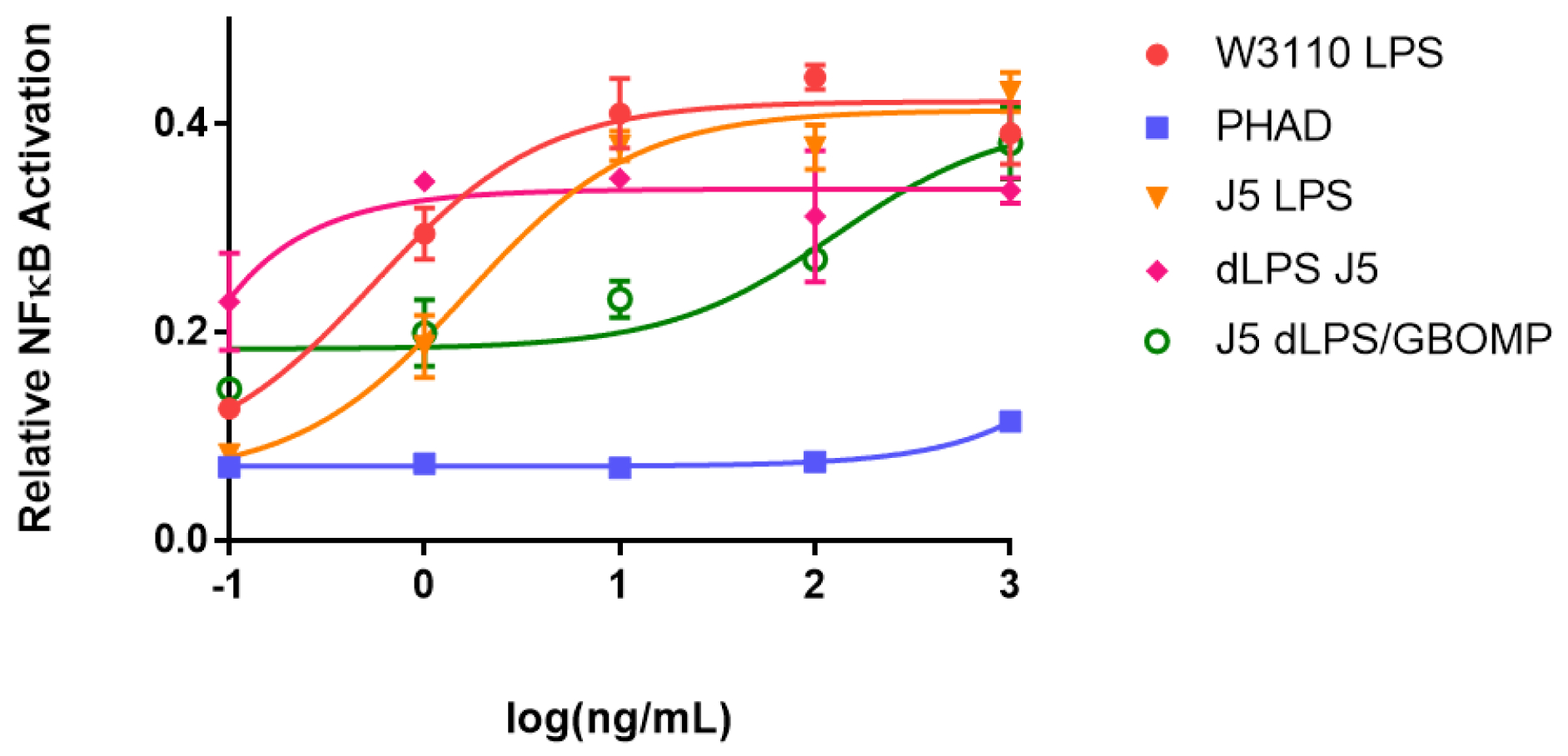
Agonists were cultured with HEK-Blue hTLR4 cells over a 5-log dose range from 0.1–1000 ng mL−1. W3110 E. coli LPS (red), J5 E. coli LPS (orange), J5 dLPS (pink), J5 dLPS/GBOMP (green), or PHAD (blue) were incubated for 16 hours. Then NF-κB activation was measured by quantification of SEAP in the supernatant. Mean ± SD of duplicate samples and an associated 4-parameter non-linear regression are shown
Defining mixture composition differences between intact and detoxified LPS samples
Direct infusion of J5 LPS and dLPS into the Synapt G2 HDMS produced many ions attributed to intact LPS and fragments thereof (Sup. Fig. 1). For the sake of simplicity, ions which could be easily associated with the “canonical” structures for E. coli LPS were isolated in the quadrupole, separated from isobars by ion mobility (Sup. Fig. 2), and subjected to CID in the “Transfer” region of the collision cell by ramping collision energy and averaging the resultant tandem mass spectra. The precursor ions used for direct comparison (m/z 1071 for LPS and m/z 789 for dLPS) happened to have the largest complete LPS peak amplitudes and represented [M-3H]3- ions.
In the single stage mass spectrum of dLPS, there were no detectable ions corresponding to the complete R-LPS structure. This was interpreted to mean that the detoxification chemistry had proceeded to completion. After dissociation of J5 LPS and dLPS, one difference in the tandem mass spectra was immediately obvious: Liberated O-linked fatty acids were not present in dLPS product ion spectra (Fig. 2). The observation of low m/z product ions, including deprotonated fatty acids and phosphorous-containing ions, is a particular advantage to using a beam-type mass spectrometer for LPS tandem MS experiments rather than any quadrupolar ion trap mass analyzer because the 1/3 cutoff rule, meaning product ions less than ~30% the m/z of the precursor ion are always unstable under trapping conditions, does not apply. These product ions can be directly diagnostic to structural changes in LPSs for which some structural information is already known. In aggregate, they can inform the analyst about which class of compound has been detected in an unknown.
Fig. 2.
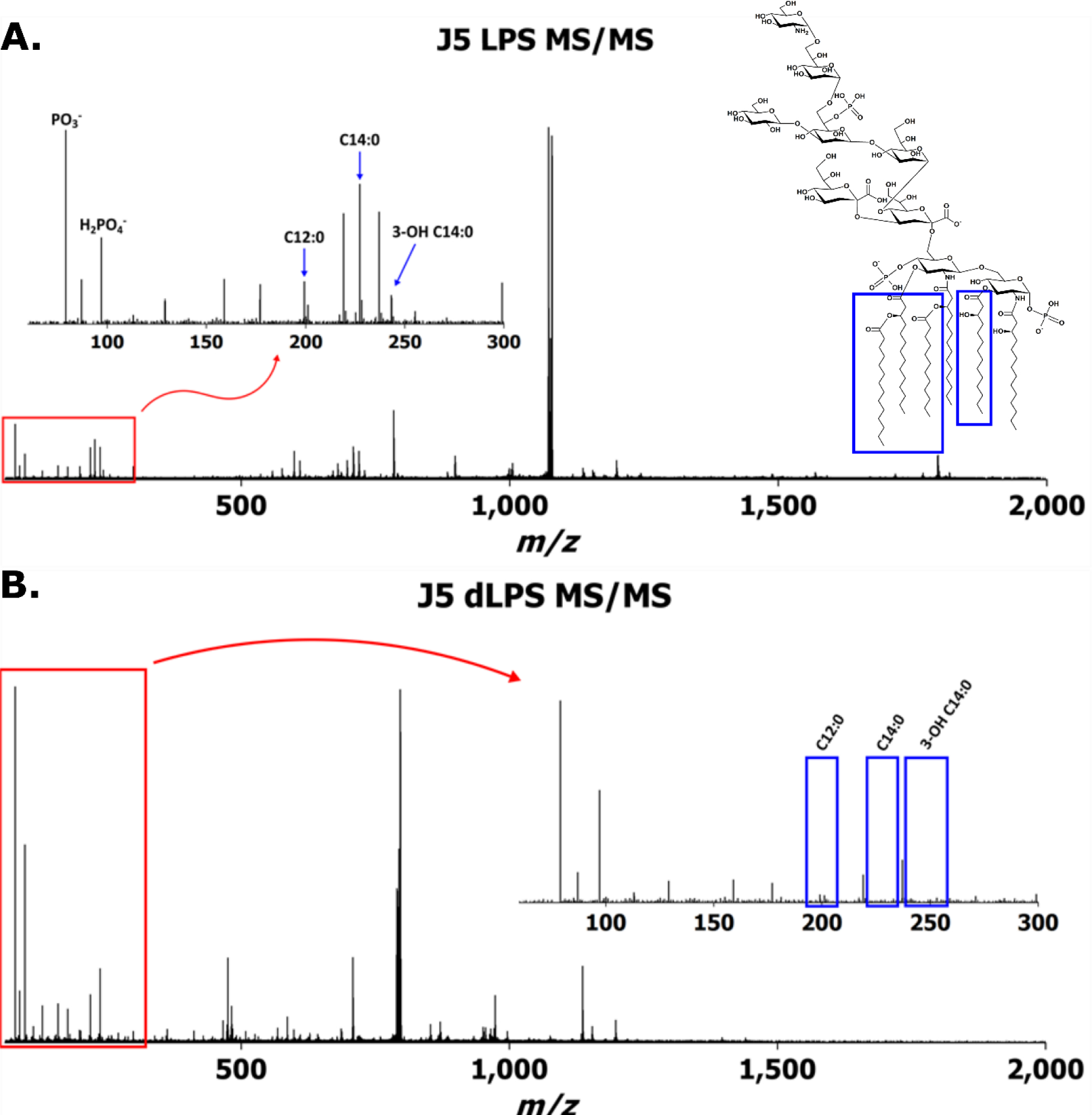
Averaged IMS-CID tandem mass spectra of (a) J5 LPS m/z 1071 and (b) J5 dLPS m/z 789 after collision energy ramping. Insets show deprotonated fatty acid product ions’ presence in (a) and absence in (b)
A total of 179 monoisotopic product ions were detected by the Synapt G2 HDMS above a signal to noise ratio (S:N) of 10:1 in the J5 LPS IMS-CID spectrum of m/z 1071; 221 were detected in the dLPS IMS-CID spectrum of m/z 789. Singly and doubly deprotonated product ions representing the full-length core OS (m/z 1418 and 708, respectively) were detected in both LPS and dLPS sample mass spectra, while the product ions representing the bis-phosphorylated, hexa-acylated E. coli lipid A (m/z 1796 and 897) were only present in the native LPS mass spectrum. Product ions assigned to the detoxified, di-acylated lipid A (m/z 951 and 475) were detected in the dLPS mass spectrum. These data further confirmed that the vaccine detoxification chemistry had proceeded as hypothesized.
It is important to emphasize that both the LPS and dLPS samples produced heterogeneous mass spectra, even after crude IMS filtering, presumably due to many different LPS structures in the samples. Other analyses of R-LPS by both ESI-MS and MALDI-MS have demonstrated similar results [34, 43]. This means that the activity data shown in the previous section are cumulative and cannot be attributed to a particular R-LPS structure. However, the alkaline hydrolysis that was performed produced an entirely different mass spectrum than the commercial product. Since the mass spectra are complex, quality control and quality assurance protocol development will be necessary to maximize the vaccine’s robustness. One example to illustrate this need can be seen in Sup Fig 1. Second and third envelopes of ions were observed in the dLPS spectrum corresponding to differences in the number of core OS sugars. It is unclear from this study whether these differences were part of the original LPS mixture or if they were a result of the sample processing.
Ultra-high mass resolving power, high mass accuracy FT-ICR MS to determine empirical formulae and define J5 LPS primary structure
In total, 252 unique, unambiguous monoisotopic masses were observed above a S:N of 10:1 in the Sigma J5 LPS sample with masses greater than the monoisotopic mass of KDO2-lipidIV A (J5 lipid A attached to two 2 – 4 linked 3-Deoxy-D-manno-oct-2-ulosonic acid residues through a 2 – 6’ glycosidic bond, [M] = 1844.971 Da), excluding redundant masses from additional charge states; 3-, 4-, and 5-ions were observed for intact R-LPS (Sup. Table 1). One hundred ninety-six of these masses were larger than the mass of the canonical E. coli, hexa-acylated lipid A attached to five core OS sugars. Interestingly, thirty-six monoisotopic masses observed were greater than the mass of the intact, canonical J5 E. coli R-LPS. Some of these ions corresponded to empirical formulae for E. coli R-LPS with known substitutions such as phosphoethanolamine and phosphate/pyrophosphate. A few were inferred to be cation adducts of multiply deprotonated ions. The vast majority, however, were unable to be inferred using accurate mass alone. In short, the R-LPS mixture was very heterogeneous, including ions not previously described in the literature for E. coli R-LPS. One example of the heterogeneity observed in only a 4 m/z window is shown in Fig. 3. In this window, at least eight different monoisotopic masses were observed, at a mass resolving power of ~300,000 FWHM, in absorption mode. There was also not a single dominant species in the mixture as has sometimes been reported.
Fig. 3.
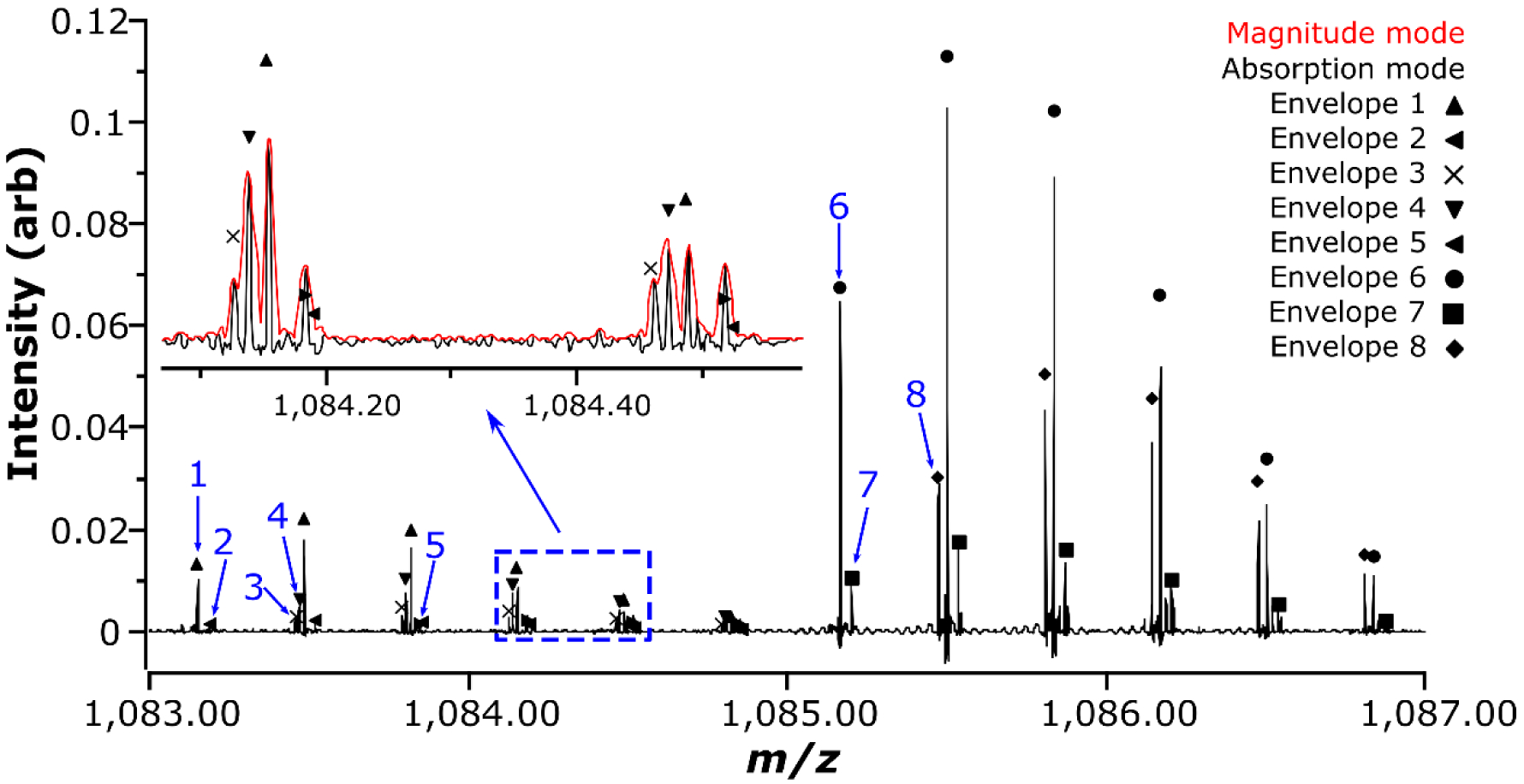
Zoomed negative mode FT-ICR mass spectrum (R ~ 300,000 FWHM, in absorption mode) after direct infusion of J5 LPS. Eight potential isotopic distribution envelopes can be identified in absorption mode in this 4 m/z window; these are denoted, at the m/z of their respective monoisotopic ions, with blue arrows. (inset) Magnified portion of the spectrum showing fine detail (including the magnitude mode and the proposed overlap between isotopologues from envelopes 2 and 5)
The highest intensity complete R-LPS monoisotopic mass in the J5 LPS spectrum, as in the previous experiment, was m/z 1071.1976. This mass corresponded to a similar structure to that reported in [44, 45] (Empirical formula: [C143H257N3O69P3]3-, Δ = 0.139 ppm). The only differences observed were an addition of a phosphate moiety on the second heptose and a terminal glucosamine (GlcN) rather than an N-acetylglucosamine (GlcNAc). An ion was also observed in the spectrum at m/z 1085.2009, corresponding to the previously published structure with an added phosphate group (Empirical formula: [C145H259N3O70P3]3-, Δ = −0.381 ppm). An isolation of m/z 1071 prior to CID experiments resulted in an error of 0.064 ppm, in magnitude mode, for a single 0.767 second transient.
Tandem MS experiments on m/z 1071 (3.5 m/z isolation) using trap CID, beam CAD, and UVPD all yielded feature-rich tandem mass spectra. Trap CID at normalized collision energy (NCE) of 30% yielded 201 monoisotopic product ions above a S:N of 4:1. Six pairs of product ions (twelve ions total) were found to be duplications due to multiple charge states. Beam CAD at 35 V yielded 152 product ions, with three pairs of duplicate identifications due to multiple charge states, using the same peak picking parameters. For many ions charge was not automatically identified, probably due to insufficient isotopic ion abundance. A wider precursor ion selection window and/or spectral averaging or summing would improve this statistic. Ninety-nine ions were detected in both trap CID and beam CAD experiments; either set of product ions was sufficient to confirm a hypothesized structure for the precursor ion (Fig. 4), when taking into account the cumulative effect of measuring every product ion at low ppb mass accuracy. These product ions corresponded to acyl chain cleavages, glycosidic bond cleavages, cross-ring cleavages, neutral losses of modifications, and combinations thereof. UVPD yielded ninety product ions, twenty-five of which were common to the CID and/or beam CAD experiments. This demonstrates, as in O’Brien et al.’s publication [34], a complementary set of product ions to those obtained through collisional activation. However, the efficiency of the UVPD process in this experiment was very low, so its value added would only be noticed with respect to very specific structural questions in this particular configuration and for this application (e.g. Are there hydroxylated fatty acyl chains in the lipid A moiety?). These types of questions are beyond the scope of this work.
Fig. 4.
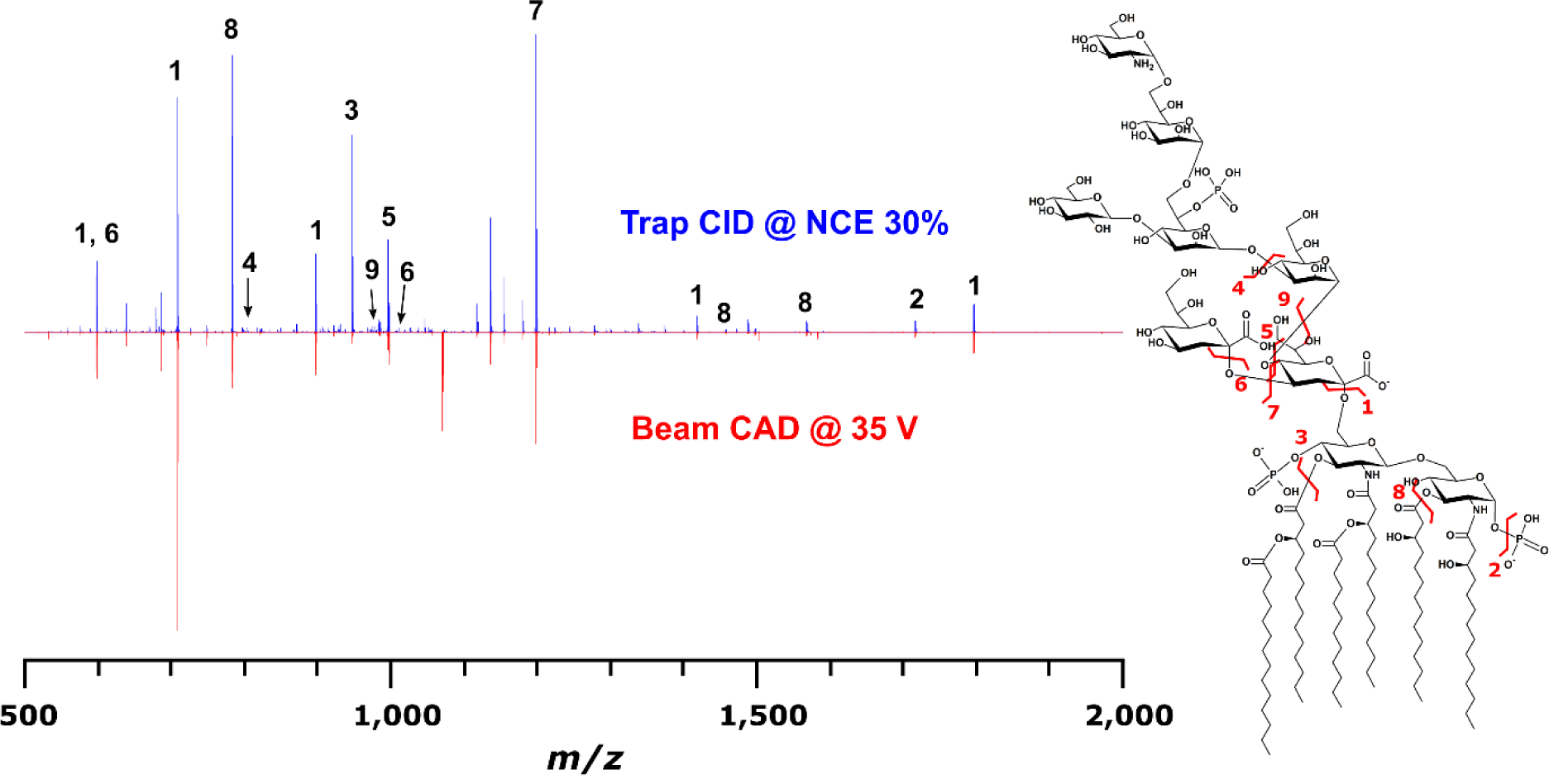
Comparison of trap CID (blue) and beam CAD (red) for the same precursor ion at m/z 1071. Ninety-nine monoisotopic product ions common to both experiments were observed, fifteen of which are annotated in the CID mass spectrum with corresponding bond cleavages in the structure on the right. All product ion m/z were measured with less than 100 ppb error
Multi-stage MS (MSn) to confirm structural inferences
After conducting multiple product ion scans on intact R-LPS precursor ions, it became apparent that abundant product ions were always formed which corresponded to both the lipid A and core OS moieties. Since there have been many published papers describing dissociation phenomena for both chemically isolated lipid A [46–51] and oligosaccharides [52–56], it seemed that the logical next step would be to dissociate these products to determine whether MS3 product ions would be formed according to these well-established rules. If so, top down sequencing data interpretation and deconvolution for R-LPS would become much less challenging, and could be performed with confidence on nominal mass accuracy, low resolving power ion trap instruments.
Fig. 5 shows an MS3 experiment for both J5 lipid A (m/z 1796) and core OS (m/z 1418) ions formed in the ion trap after dissociation from intact R-LPS (m/z = 1071). Characteristic tandem mass spectra for both lipid A and core OS were obtained, suggesting the feasibility of simplified top down sequencing for both moieties and attribution of these to a specific LPS precursor. These data demonstrate that gas phase decomposition chemistry proceeds similarly to the widely adopted approach of solution phase decomposition followed by analyses of the reaction products separately. The primary benefit of the gas phase decomposition approach is that both lipid A and core OS can be directly attributed to a R-LPS structure present in the sample precluding the need for inference when analyzed separately. For lipid A, product ions indicative of neutral losses of ester-linked fatty acyl chains and metaphosphoric acid were the most abundant features in the mass spectra. For core OS, B-and Y-ions, as described by Domon and Costello [52], corresponding to glycosidic bond cleavages were the most abundant product ions, with minor product ions corresponding to cross-ring cleavages. It is likely that any MS3-capable trapping instrument, as well as beam-type instruments outfitted with post-CID ion mobility separation capability and subsequent secondary CID, will be able to perform a similar experiment. As in our previous work with lipid A [57], stepped or ramped collision energy at the MS3 level was able to simulate subsequent levels of MSn, with qualitatively minimal ion losses, to provide more complete dissociation and primary structure coverage for lipid A and core OS (data not shown).
Fig. 5.
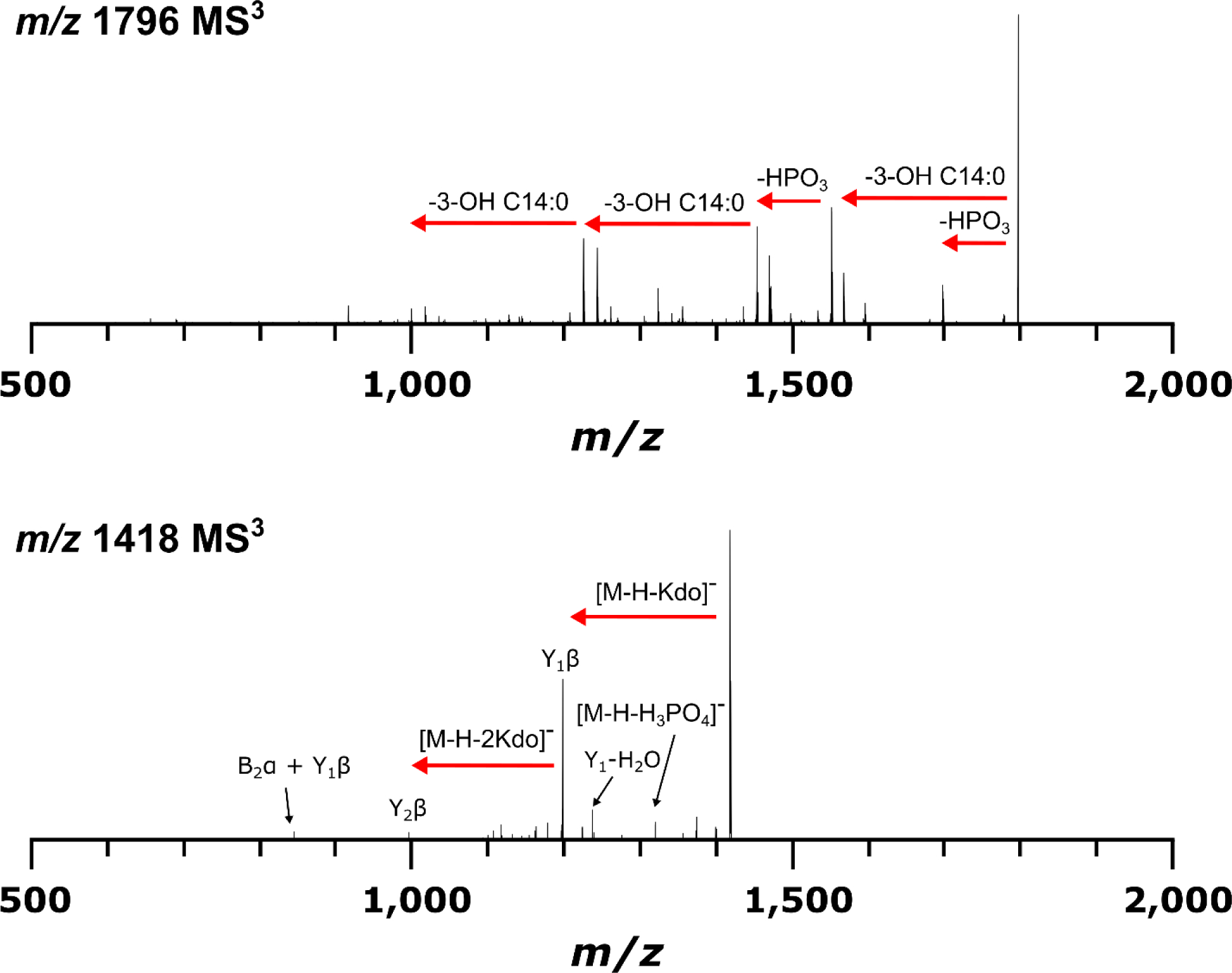
MS3 CID mass spectra from MS2 product ions representing J5 E. coli lipid A at m/z 1796 (top) and core OS at m/z 1418 (bottom). Similar dissociation phenomena were observed as in MS2 experiments for chemically isolated lipid A and oligosaccharides, indicating feasibility of LPS top down sequencing in this manner
Conclusions
As with all biologically active molecules, LPS activity is directly related to LPS structure. In this work, a chemically modified R-LPS vaccine candidate’s reduced innate immunogenicity was shown to be the result of O-deacylation of its lipid A moiety. Several different approaches were employed to more completely define structural features in commercially available, heterogeneous R-LPS mixtures. This study has shown that R-LPS can be analyzed on three different mass spectrometers with similar but complementary results. Success of LPS-based drugs in the clinic will be partially dependent on well-defined compositions of LPS extracts, if purification of single compounds or total chemical synthesis prove to be unfeasible, and batch-to-batch reproducibility of LPS production. This will inevitably lead to a better understanding of off-target effects and decrease probability of drug attrition. Reproducibility of immunological studies will also be improved through these efforts by improving quality control and quality assurance guidelines. In the past, this has been an onerous undertaking, but with improved data acquisition efficiency and the ability to acquire more complete data sets, the current rate limiting step is user-friendly, automated software development.
Supplementary Material
Acknowledgments
We would like to thank Melinda McFarland and Timothy Croley at the Center for Food Safety and Applied Nutrition, Food and Drug Administration for granting access to the Synapt G2 HDMS instrument for data acquisition. A portion of this work was performed at the National High Magnetic Field Laboratory, which is supported by National Science Foundation Cooperative Agreement No. DMR-1157490 and the State of Florida. M.M.K. is thankful to the American Association of Pharmaceutical Scientists (AAPS) foundation for a graduate student fellowship. This research was also supported in part by National Institutes of Health grant 5R01AI123820 (R.K.E. and D.R.G.).
References
- 1.Raetz CRH, Whitfield C: Lipopolysaccharide Endotoxins. Annu. Rev. Biochem 71, 635–700 (2002). doi: 10.1146/annurev.biochem.71.110601.135414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Galloway SM, Raetz CRH: A Mutant of Escherichia coli Defective in the First Step of Endotoxin Biosynthesis. J. Biol. Chem 265, 6394–6402 (1990) [PubMed] [Google Scholar]
- 3.Park BS, Song DH, Kim HM, Choi B-S, Lee H, Lee J-O: The structural basis of lipopolysaccharide recognition by the TLR4–MD-2 complex. Nature. 458, 1191–1195 (2009). doi: 10.1038/nature07830 [DOI] [PubMed] [Google Scholar]
- 4.Shi J, Zhao Y, Wang Y, Gao W, Ding J, Li P, Hu L, Shao F: Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. 514, 187–192 (2014). doi: 10.1038/nature13683 [DOI] [PubMed] [Google Scholar]
- 5.Yang J, Zhao Y, Shao F: Non-canonical activation of inflammatory caspases by cytosolic LPS in innate immunity. Curr. Opin. Immunol 32, 78–83 (2015). doi: 10.1016/j.coi.2015.01.007 [DOI] [PubMed] [Google Scholar]
- 6.Morrison DC: Bacterial Endotoxins and Pathogenesis. Rev. Infect. Dis 5, S733–S747 (1983). doi: 10.2307/4453209 [DOI] [PubMed] [Google Scholar]
- 7.Papo N, Shai Y: A molecular mechanism for lipopolysaccharide protection of gram-negative bacteria from antimicrobial peptides. J. Biol. Chem 280, 10378–10387 (2005). doi: 10.1074/jbc.M412865200 [DOI] [PubMed] [Google Scholar]
- 8.Rosenfeld Y, Shai Y: Lipopolysaccharide (Endotoxin)-host defense antibacterial peptides interactions: Role in bacterial resistance and prevention of sepsis. Biochim. Biophys. Acta 1758, 1513–1522 (2006). doi: 10.1016/j.bbamem.2006.05.017 [DOI] [PubMed] [Google Scholar]
- 9.Hornef MW, Wick MJ, Rhen M, Normark S: Bacterial strategies for overcoming host innate and adaptive immune responses. Nat Immunol. 3, 1033–1040 (2002). doi: 10.1038/ni1102-1033\rni1102-1033 [DOI] [PubMed] [Google Scholar]
- 10.Matsuura M: Structural modifications of bacterial lipopolysaccharide that facilitate gram-negative bacteria evasion of host innate immunity. Front. Immunol 4, 109 (2013). doi: 10.3389/fimmu.2013.00109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Scott AJ, Oyler BL, Goodlett DR, Ernst RK: Lipid A structural modifications in extreme conditions and identification of unique modifying enzymes to define the Toll-like receptor 4 structure-activity relationship. Biochim. Biophys. Acta 1862, 1439–1450 (2017). doi: 10.1016/j.bbalip.2017.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cross AS: Development of an anti-endotoxin vaccine for sepsis. Subcell. Biochem 53, 285–302 (2015). doi: 10.1007/978-90-481-9078-2_13 [DOI] [PubMed] [Google Scholar]
- 13.Morrison DC, Fujihara Y, Bogard WC, Lei MG, Daddona PE, Morrison DC, Fujihara Y, Bogard WC, Lei MG, Daddona PE: Monoclonal Anti-Lipid A IgM Antibodies HA-1A and E-5 Recognize Distinct Epitopes on Lipopolysaccharide and Lipid A. J. Infect. Dis 168, 1429–1435 (1993). doi: 10.1093/infdis/168.6.1429 [DOI] [PubMed] [Google Scholar]
- 14.Helmerhorst EJ, Maaskant JJ, Appelmelk BJ: Anti-lipid A monoclonal antibody Centoxin (HA-1A) binds to a wide variety of hydrophobic ligands. Infect. Immun 66, 870–873 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuhn HM: Cross-reactivity of monoclonal antibodies and sera directed against lipid A and lipopolysaccharides. Infection. 21, 179–186 (1993). doi: 10.1007/BF01710544 [DOI] [PubMed] [Google Scholar]
- 16.Kuhn HM, Brade L, Appelmelk BJ, Kusumoto S, Rietschel ET, Brade H: Characterization of the epitope specificity of murine monoclonal antibodies directed against lipid A. Infect. Immun 60, 2201–2210 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zinner SH, McCabe WR: Effects of IgM and IgG Antibody in Patients with Bacteremia Due to Gram-Negative Bacilli. J. Infect. Dis 133, 37–45 (1976). [DOI] [PubMed] [Google Scholar]
- 18.Pollack M, Huang AI, Prescott RK, Young LS, Hunter KW, Cruess DF, Tsai CM: Enhanced survival in Pseudomonas aeruginosa septicemia associated with high levels of circulating antibody to Escherichia coli endotoxin core. J. Clin. Invest 72, 1874–1881 (1983). doi: 10.1172/JCI111150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cross AS, Opal SM, Warren HS, Palardy JE, Glaser K, Parejo NA, Bhattacharjee AK: Active immunization with a detoxified Escherichia coli J5 lipopolysaccharide group B meningococcal outer membrane protein complex vaccine protects animals from experimental sepsis. J Infect Dis. 183, 1079–1086 (2001). doi: 10.1086/319297 [DOI] [PubMed] [Google Scholar]
- 20.Opal SM, Palardy JE, Chen WH, Parejo NA, Bhattacharjee AK, Cross AS: Active Immunization with a Detoxified Endotoxin Vaccine Protects against Lethal Polymicrobial Sepsis: Its Use with CpG Adjuvant and Potential Mechanisms. J. Infect. Dis 192, 2074–2080 (2005). doi: 10.1086/498167 [DOI] [PubMed] [Google Scholar]
- 21.Bhattacharjee AK, Opal SM, Taylor R, Naso R, Semenuk M, Zollinger WD, Moran EE, Young L, Hammack C, Sadoff JC, Cross AS: A noncovalent complex vaccine prepared with detoxified Escherichia coli J5 (Rc chemotype) lipopolysaccharide and Neisseria meningitidis Group B outer membrane protein produces protective antibodies against gram-negative bacteremia. J. Infect. Dis 173, 1157–1163 (1996) [DOI] [PubMed] [Google Scholar]
- 22.Cross AS, Opal SM, Palardy JE, Drabick JJ, Warren HS, Huber C, Cook P, Bhattacharjee AK: Phase I study of detoxified Escherichia coli J5 lipopolysaccharide (J5dLPS)/group B meningococcal outer membrane protein (OMP) complex vaccine in human subjects. Vaccine. 21, 4576–4587 (2003). doi: 10.1016/S0264-410X(03)00483-3 [DOI] [PubMed] [Google Scholar]
- 23.Cross AS, Karreman HJ, Zhang L, Rosenberg Z, Opal SM, Lees A: Immunization of cows with novel core glycolipid vaccine induces anti-endotoxin antibodies in bovine colostrum. Vaccine. 32, 6107–6114 (2014). doi: 10.1016/j.vaccine.2014.08.083 [DOI] [PubMed] [Google Scholar]
- 24.Cross AS: Anti-endotoxin vaccines: back to the future. Virulence. 5, 219–25 (2014). doi: 10.4161/viru.25965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nys M, Damas P, Joassin L, Lamy M: Sequential anti-core glycolipid immunoglobulin antibody activities in patients with and without septic shock and their relation to outcome. Ann. Surg 217, 300–306 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alving CR, Rao M: Lipid A and liposomes containing lipid A as antigens and adjuvants. Vaccine. 26, 3036–3045 (2008). doi: 10.1016/j.vaccine.2007.12.002 [DOI] [PubMed] [Google Scholar]
- 27.Richards RL, Rao M, Wassef NM, Glenn GM, Rothwell SW, Alving CR: Liposomes containing lipid a serve as an adjuvant for induction of antibody and cytotoxic T-cell responses against RTS,S malaria antigen. Infect. Immun 66, 2859–2865 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qureshi N, Takayama K, Ribi E: Purification and structural determination of nontoxic lipid A obtained from the lipopolysaccharide of Salmonella typhimurium. J. Biol. Chem 257, 11808–11815 (1982). [PubMed] [Google Scholar]
- 29.Casella CR, Mitchell TC: Putting endotoxin to work for us: Monophosphoryl lipid a as a safe and effective vaccine adjuvant. Cell Mol. Life Sci 65, 3231–3240 (2008). doi: 10.1007/s00018-008-8228-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gregg KA, Harberts E, Gardner FM, Pelletier MR, Cayatte C, Yu L, McCarthy MP, Marshall JD, Ernst RK: Rationally designed TLR4 ligands for vaccine adjuvant discovery. MBio. 8, e00492–17 (2017). doi: 10.1128/mBio.00492-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Needham BD, Carroll SM, Giles DK, Georgiou G, Whiteley M, Trent MS: Modulating the innate immune response by combinatorial engineering of endotoxin. Proc. Natl. Acad. Sci 110, 1464–1469 (2013). doi: 10.1073/pnas.1218080110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang Y Wolfert MA, Boons GJ, G.J.: Modulation of Innate Immune Responses with Synthetic Lipid A Derivatives. Jacs Artic. 5200–5216 (2007). doi: 10.1021/ja068922a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Qureshi N, Takayama K, Mascagni P, Honovich J, Wong R, Cotter RJ: Complete structural determination of lipopolysaccharide obtained from deep rough mutant of Escherichia coli. Purification by high performance liquid chromatography and direct analysis by plasma desorption mass spectrometry. J. Biol. Chem 263, 11971–11976 (1988). [PubMed] [Google Scholar]
- 34.O’Brien JP, Needham BD, Brown DB, Trent MS, Brodbelt JS: Top-down strategies for the structural elucidation of intact gram-negative bacterial endotoxins. Chem. Sci 5, 4291–4301 (2014). doi: 10.1039/C4SC01034E [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hendrickson CL, Quinn JP, Kaiser NK, Smith DF, Blakney GT, Chen T, Marshall AG, Weisbrod CR, Beu SC: 21 Tesla Fourier Transform Ion Cyclotron Resonance Mass Spectrometer: A National Resource for Ultrahigh Resolution Mass Analysis. J. Am. Soc. Mass Spectrom 26, 1626–1632 (2015). doi: 10.1007/s13361-015-1182-2 [DOI] [PubMed] [Google Scholar]
- 36.Shaw JB, Li W, Holden DD, Zhang Y, Griep-Raming J, Fellers RT, Early BP, Thomas PM, Kelleher NL, Brodbelt JS: Complete protein characterization using top-down mass spectrometry and ultraviolet photodissociation. J. Am. Chem. Soc 135, 12646–12651 (2013). doi: 10.1021/ja4029654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Strohalm M, Hassman M, Košata B, Kodíček M: mMass data miner: An open source alternative for mass spectrometric data analysis. Rapid Commun. Mass Spectrom 22, 905–908 (2008). doi: 10.1002/rcm.3444 [DOI] [PubMed] [Google Scholar]
- 38.Kilgour DPA, Hughes S, Kilgour SL, Mackay CL, Palmblad M, Tran BQ, Goo YA, Ernst RK, Clarke DJ, Goodlett DR: Autopiquer - a Robust and Reliable Peak Detection Algorithm for Mass Spectrometry. J. Am. Soc. Mass Spectrom 28, 253–262 (2017). doi: 10.1007/s13361-016-1549-z [DOI] [PubMed] [Google Scholar]
- 39.Kilgour DPA, Wills R, Qi Y, O’Connor PB: Autophaser: An algorithm for automated generation of absorption mode spectra for FT-ICR MS. Anal. Chem 85, 3903–3911 (2013). doi: 10.1021/ac303289c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kilgour D.P. a, Neal MJ, Soulby AJ, O’Connor PB: Improved optimization of the Fourier transform ion cyclotron resonance mass spectrometry phase correction function using a genetic algorithm. Rapid Commun. Mass Spectrom 27, 1977–1982 (2013). doi: 10.1002/rcm.6658 [DOI] [PubMed] [Google Scholar]
- 41.Kilgour DPA, Van Orden SL: Absorption mode Fourier transform mass spectrometry with no baseline correction using a novel asymmetric apodization function. Rapid Commun. Mass Spectrom 29, 1009–1018 (2015). doi: 10.1002/rcm.7190 [DOI] [PubMed] [Google Scholar]
- 42.Kilgour DPA, Van Orden SL, Tran BQ, Goo YA, Goodlett DR: Producing isotopic distribution models for fully apodized absorption mode FT-MS. Anal. Chem 87, 5797–5801 (2015). doi: 10.1021/acs.analchem.5b01032 [DOI] [PubMed] [Google Scholar]
- 43.Phillips NJ, John CM, Jarvis GA: Analysis of Bacterial Lipooligosaccharides by MALDI-TOF MS with Traveling Wave Ion Mobility. J. Am. Soc. Mass Spectrom 27, 1263–1276 (2016). doi: 10.1007/s13361-016-1383-3 [DOI] [PubMed] [Google Scholar]
- 44.Müller-Loennies S, Holst O, Brade H: Chemical structure of the core region of Escherichia coli J-5 lipopolysaccharide. Eur. J. Biochem 224, 751–760 (1994). doi: 10.1111/j.1432-1033.1994.t01-1-00751.x [DOI] [PubMed] [Google Scholar]
- 45.Holst O, Müller-Loennies S, Lindner B, Brade H: Chemical structure of the lipid A of Escherichia coli J-5. Eur. J. Biochem 214, 695–701 (1993) [DOI] [PubMed] [Google Scholar]
- 46.Qureshi N, Takayama K, Heller D, Fenselau C: Position of ester groups in the lipid A backbone of lipopolysaccharides obtained from Salmonella typhimurium. J. Biol. Chem 258, 12947–12951 (1983). [PubMed] [Google Scholar]
- 47.Chan S, Reinhold VN: Detailed structural characterization of lipid A: electrospray ionization coupled with tandem mass spectrometry. Anal. Biochem 218, 63–73 (1994). doi: 10.1006/abio.1994.1141 [DOI] [PubMed] [Google Scholar]
- 48.Kussak A, Weintraub A: Quadrupole ion-trap mass spectrometry to locate fatty acids on lipid A from Gram-negative bacteria. Anal. Biochem 307, 131–137 (2002). doi: 10.1016/S0003-2697(02)00004-0 [DOI] [PubMed] [Google Scholar]
- 49.Sforza S, Silipo A, Molinaro A, Marchelli R, Parrilli M, Lanzetta R: Determination of fatty acid positions in native lipid A by positive and negative electrospray ionization mass spectrometry. J. Mass Spectrom 39, 378–383 (2004). doi: 10.1002/jms.598 [DOI] [PubMed] [Google Scholar]
- 50.Boué SM, Cole RB: Confirmation of the structure of lipid A from Enterobacter agglomerans by electrospray ionization tandem mass spectrometry. J. Mass Spectrom 35, 361–368 (2000). doi: [DOI] [PubMed] [Google Scholar]
- 51.Shaffer SA, Harvey MD, Goodlett DR, Ernst RK: Structural Heterogeneity and Environmentally Regulated Remodeling of Francisella tularensis subspecies novicida Lipid A Characterized by Tandem Mass Spectrometry. J. Am. Soc. Mass Spectrom 18, 1080–1092 (2007). doi: 10.1016/j.jasms.2007.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Domon B, Costello CE: A systematic nomenclature for carbohydrate fragmentations in FAB-MS/MS spectra of glycoconjugates. Glycoconj. J 5, 397–409 (1988). doi: 10.1007/BF01049915 [DOI] [Google Scholar]
- 53.Zaia J: Mass spectrometry of oligosaccharides. Mass Spectrom. Rev 23, 161–227 (2004). doi: 10.1002/mas.10073 [DOI] [PubMed] [Google Scholar]
- 54.Li H, Bendiak B, Siems WF, Gang DR, Hill HH: Carbohydrate structure characterization by tandem ion mobility mass spectrometry (IMMS)2. Anal. Chem 85, 2760–2769 (2013). doi: 10.1021/ac303273z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kailemia MJ, Ruhaak LR, Lebrilla CB, Amster IJ: Oligosaccharide Analysis by Mass Spectrometry: A Review of Recent Developments. Anal. Chem 86, 196–212 (2014). doi: 10.1021/ac403969n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang Z, Linhardt RJ: Sequence Analysis of Native Oligosaccharides Using Negative ESI Tandem MS. Curr. Anal. Chem 5, 225–237 (2009). doi: 10.2174/157341109788680291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoon SH, Liang T, Schneider T, Oyler BL, Chandler CE, Ernst RK, Yen GS, Huang Y, Nilsson E, Goodlett DR: Rapid lipid a structure determination via surface acoustic wave nebulization and hierarchical tandem mass spectrometry algorithm. Rapid Commun. Mass Spectrom 30, 2555–2560 (2016). doi: 10.1002/rcm.7728 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


