Abstract
Purpose
Transcriptome changes can be expected in survivors after lethal irradiation. We aimed to characterize these in males and females and after different cytokine treatments 60 days after irradiation.
Material and methods
Male and female rhesus macaques (n = 142) received a whole-body exposure with 700 cGy, from which 60 animals survived. Peripheral whole blood was drawn pre-exposure and before sacrificing the surviving animals after 60 days.
Results
We evaluated gene expression in a three-phase study design. Phase I was a whole-genome screening (NGS) for mRNAs using five pre- and post-exposure RNA samples from both sexes (n = 20). Differential gene expression (DGE) was calculated between samples of survivors and pre-exposure samples (reference), separately for males and females. 1,243 up- and down-regulated genes were identified with 30–50% more deregulated genes in females. 37 candidate mRNAs were chosen for qRT-PCR validation in phase II using the remaining samples (n = 117). Altogether 17 genes showed (borderline) significant (t-test) DGE in groups of untreated or treated animals. Nine genes (CD248, EDAR, FAM19A5, GAL3ST4, GCNT4, HBG2/1, LRRN1, NOG, SYT14) remained with significant changes and were detected in at least 50% of samples per group. Panther analysis revealed an overlap between both sexes, related to the WNT signaling pathway, cell adhesion and immunological functions. For phase III, we validated the nine genes with candidate genes (n = 32) from an earlier conducted study on male baboons. Altogether 14 out of 41 genes showed a concordantly DGE across both species in a bilateral comparison.
Conclusions
Sixty days after radiation exposure, we identified (1) sex and cytokine treatment independent transcriptional changes, (2) females with almost twice as much deregulated genes appeared more radio-responsive than males, (3) Panther analysis revealed an association with immunological processes and WNT pathway for both sexes.
Introduction
Gene expression changes caused by high dose irradiation have been investigated intensively over the last decades [1]. Radiation-induced long-term effects are known [2] and for example, stable chromosomal aberrations changes after high dose irradiation are used for retrospective dosimetry [3]. Effects of high dose irradiation include cancer as well as chronic non-cancer diseases [4–6] affecting e.g. the cardiovascular [7] and central nervous system [8, 9]. As almost two-third of all cancer patients undergo partial or whole body radiotherapy [10], the overall number of people with radiation-associated long-term effects is high making the topic pivotal [11]. Delayed effects of acute radiation exposure (DEARE) are other known health effects occurring after high radiation exposures. They are associated with degenerative and inflammatory conditions affecting multiple organs [12] and induce cerebrovascular injury [13] or lung injury [14]. Additionally, high dose irradiation with potentially lethal dose levels might occur [15] during large-scale radiological events like nuclear power plant disasters, the use of radiological weapons by terrorists, or during military conflicts. DEARE or long term effects have to be expected in survivors of these scenarios [6, 16].
In irradiated healthy individuals so far little is known about long-term changes in the transcriptome/post-transcriptome [17–19]. Recently, we identified persistent gene expression changes in the peripheral blood of sub-lethally irradiated baboons up to about three months after exposure and discussed their impact on long-term health effects [19]. A persistent alteration of the immune response 30 days after high dose partial body irradiation was postulated by another group [18], but overall, the molecular mechanisms and the entanglement with known long-term effects are not well characterized [1].
We measured gene expression in pre- and post-exposure (60 days) peripheral blood samples of irradiated 142 male and female rhesus macaques. Altogether 60 animals survived a severe and potentially lethal exposure to 700 cGy for 60 days. We hypothesized that high dose irradiation leads to sex-specific changes in the transcriptome 60 days after irradiation and wanted to examine whether previous results of our group performed in baboons could be validated in another species and at least at one time point. In collaboration with Neumedicines Inc., we assessed blood samples obtained from rhesus macaque that were involved in a study to determine the efficacy of IL-12 or/and G-CSF in the treatment of the hematopoietic acute radiation syndrome. The study was designed under the FDA Animal Rule. We performed a whole-genome screening and identified protein-coding mRNA genes in survivors separately for males and females. These mRNAs were then validated using the gold standard qRT-PCR methodology for gene expression analysis. Animals were either untreated (vehicle) or treated using recombinant human cytokines (IL-12), or IL-12 and G-CSF combined. As an additional step of validation, we did an inter-species comparison of candidate genes examined 60 days after irradiation with earlier generated results of a recent baboon study where we checked for long-term gene expression changes utilizing similar techniques and covering several months after irradiation.
Materials and methods
Animals and irradiation
Rhesus macaques were obtained from the Yongfu County Xingui Wild Animals Raising, China. At the beginning of the treatment, animal age ranged from 2 years, 5 months to 3 years, 10 months, and all animals weighed between 2.8 and 5.7 kg. Total body irradiation (TBI) comprised 700 cGy (60 cGy/min from a Theratron 1000 Co60 source [Best Theratronics; Ottawa, Ontario, Canada]). Details regarding animal characteristics as well as TBI procedure and other design details are described elsewhere [20]. Non-human primates (NHP) were randomized (separated by sex; stratified by weight) and either untreated (vehicle) or treated by subcutaneously administering either recombinant human IL12 (175 ng/kg), G-CSF (10 mg/kg), or a combination (for more details see [20]). Clinical signs were recorded twice daily and animals were euthanized at the occurrence of predefined criteria [20]. On day 60, all surviving animals (n = 60) were euthanized. Details of clinical assessments, supportive care, and euthanasia criteria are specified at [20]. Animals were housed individually in stainless steel cages equipped with an automatic watering system. Males and females were housed in separate rooms. The animal room environment was controlled to maintain a temperature of 21 ± 3°C; a relative humidity of 50 ± 20%; a light dark cycle of 12 hours light/12 hours dark, except during procedures; and 10–15 air changes per hour. Temperature and relative humidity were monitored continuously except during animal transportation and inside the radiation facility where only temperature was recorded every 5 minutes. A standard certified commercial chow (Harlan Teklad Certified Hi-Fiber Primate Diet #7195C) was provided to the animals twice daily except during designated procedures. Treats or fresh fruits/vegetables or juice (except orange or grapefruit juice) was provided as part of the animal enrichment program. If animals became appetiteless during the study, the standard diet was supplemented at the discretion of the Study Director or Clinical Veterinarian. Specifically, nutritional support (in the form of crushed cookies with banana, fruit and/or vegetable buffets, juice and/or Ensure®) was provided when body weight decreases by >15% of baseline or when an animal presented mouth lesion which hindered its food intake and was continued as long as the body weight remained >15% lower than baseline, and the animal was not eating. Municipal tap water (which has been exposed to ultraviolet light and purified by reverse osmosis) was provided to the animals ad libitum, except during procedures, via an automatic watering system or water bottles when considered necessary. Regarding symptomatic palliative care, buprenorphine was given for pain; bupivacaine (0.25%) was administered topically for the management of mouth ulcers; Pepto-Bismol was given for the management of diarrhea; topical hydrotherapy and/or iodine 1% was administered to wounds. Snacks or supplements (Rhesus Liquid Diet [BIO-SERV; Frenchtown, New Jersey], EnsureVR [Abbott Laboratories, Abbott Park, IL], vegetables, juices, or crushed cookies with banana) were given for anorexia. Animals in unrelievable pain or distress were euthanized based on the clinical judgment of the Clinical Veterinarians who was blinded to experimental treatments. Euthanasia endpoints were: Respiratory distress; Anorexia/Decreased appetite (Complete anorexia for 3 days with deteriorating conditions based on the clinical examination; animal not taking nutritive supplement offered orally); weight Loss, more than 20% over a 3-day period; decreased level of activity; acute, gross blood loss; status epilepticus; clinical appearance associated with abnormal vital signs. All procedures involving animals were conducted in compliance with the Good Laboratory Practice (GLP; 21 CFR Part 58) and performed at CiToxLAB North America (Montreal, Quebec, Canada) after approval by the Institutional Animal Care and Use Committee (IACUC) of CiToxLAB North America, Inc.
RNA extraction and quality control
Whole blood samples (2.5 ml) were processed following the PAXgene Blood RNA System (BD Diagnostics, PreAnalytiX GmbH, Hombrechtikon, Switzerland). In brief, blood was drawn into a PAXgene Blood RNA tube at CiToxLAB. The tube was gently inverted (10 times), stored at room temperature overnight then at -20°. After all samples were collected, the PaxGene tubes were sent to Germany for further processing. After thawing, washing and centrifugation, cells in the supernatant were lysed (Proteinase K) followed by the addition of Lysis/Binding Solution taken from the mirVana Kit (Life Technologies, Darmstadt, Germany). With the mirVana kit, total RNA, including small RNA species, was isolated by combining a Phenol-Chloroform RNA precipitation with further processing using a silica membrane. After several washing procedures, DNA residuals became digested on the membrane (RNAse free DNAse Set, Qiagen, Hilden, Germany). RNA was eluted in a collection tube and frozen at -20°C. Quality and quantity of isolated total RNA were measured spectrophotometrically (NanoDrop, PeqLab Biotechnology, Erlangen, Germany). RNA integrity was assessed by the 2100 Agilent Bioanalyser (Life Science Group, Penzberg, Germany) and DNA contamination was controlled by conventional PCR using an actin primer. We used only RNA specimens with a ratio of A260/A280 ≥ 2.0 (Nanodrop) and RNA integrity number (RIN) ≥ 7.2 for RNA seq NGS (IMGM Laboratories, Martinsried, Germany) or RIN ≥ 7.3 for qRT-PCR analyses or no signs of degradation in agarose gel electrophoresis.
Phase I screening
Whole-genome screening for differentially expressed genes (protein-coding mRNAs) was performed on 20 RNA samples (four groups comprised pre- and post-exposure samples originating from five male and five female rhesus macaques, n = 4x5) among the untreated arm (Fig 1).
Fig 1. Flow diagram of included samples, three-phase study design, gene expression results, bioinformatics and inter-species validation between rhesus macaques and baboons.
The overview shows the samples used in untreated (phase I) as well as untreated and treated rhesus macaques (phase II). Comparisons are shown separately for males and females in the lower panel for phase I and phase II.
RNA Seq library preparation was based on the TruSeq® Stranded mRNA HT Library Prep Kit (Illumina, San Diego, CA) following the manufacturer’s recommendations (Illumina Inc. TruSeq® Stranded mRNA Sample Preparation Guide—Catalog # RS-122-9004DOC—Part # 15031047 Rev. E. 2013). In short, RNA was fragmented followed by cDNA synthesis, 3’ adenylation and adaptor ligation comprising indices for multiplex sequencing. After library quantification using the Qubit® dsDNA HS Assay Kit (Thermo Scientific, Waltham, MA) and assessment of length distribution using an Agilent Bioanalyzer system (Agilent 2100 Bioanalyzer, Agilent, Santa Clara, CA) equimolar pooling into a final sequencing library was performed. Before sequencing adapter dimers were removed using AMPure® XP Beads (Beckman Coulter, Brea, CA) followed by additional quality control using library quantification and electrophoresis as described above. Subsequently, single-end sequencing (1x75bp) was performed on the Illumina NextSeq 500 sequencing platform (Illumina, San Diego, CA).
For in-depth analysis of differential gene expression, we used the CLC Genomics Workbench (CLC Bio-Qiagen, Aarhus, Denmark; version 9.0.1). After the import of read data into the analysis tool, sequence reads were mapped against the Macaca mulatta genome (Mm1_8.0.1, NCBI). The incorporated edgeR Bioconductor package was used for the identification of DGEs [21]. In this study, only those gene transcripts that had a call “present” in at least 60% of RNA specimens were included in the analysis of gene expression and only genes with FDR corrected p-values ≤ 0.05 and with fold changes >2/<-2 among compared groups were considered to represent a candidate gene for validation in phase II. We considered multiple comparisons in the bioinformatic approach and in the validation part (phase II) of our study where the number of hypotheses tested in parallel was reduced from about 32,141 transcripts (phase I) to 37 mRNAs (see below).
Phase II: Validation of phase I candidate genes via qRT-PCR
We independently validated the mRNA candidate genes from phase I (screening with NGS) using the remaining RNA samples (n = 117, Fig 1) and employing qRT-PCR, the gold standard in quantitative gene expression analysis. These samples comprised 79 pre-exposure samples taken from survivor and non-survivor as well as 38 post-exposure samples taken from survivor only (Fig 1). This strategy increased the sample size available for statistical analysis. We used a custom low density array (LDA, high throughput qRT-PCR platform) for quantification of our 37 candidate genes employing TaqMan chemistry (S1 Table). A 1 μg RNA aliquot of each RNA sample was reverse transcribed using a two-step PCR protocol (High Capacity Kit). 400 μl cDNA (1 μg RNA equivalent) was mixed with 400 μl 2 x RT-PCR master mix and pipetted into the fill ports of the LDA. Cards were centrifuged twice (1,200 rpm, 1 min, Multifuge3S-R, Heraeus, Germany), sealed, and transferred into the 7900 qRT-PCR instrument. The qRT-PCR was processed following the qRT-PCR protocol for 384-well LDA format.
For the LDA Ct-values (threshold cycles) we used the median mRNA expression on each LDA for normalization purposes, because this proved to be the more robust and the slightly more precise method compared to a normalization approach using a housekeeping mRNA species provided on the LDA.
All technical procedures for qRT-PCR were performed in accordance with standard operating procedures implemented in our laboratory in 2008 when the Bundeswehr Institute of Radiobiology became certified according to DIN EN ISO 9001/2008. All chemicals for qRT-PCR using TaqMan chemistry were provided by Life Technologies, Darmstadt, Germany.
Phase III: Inter-species validation
The results from phase II were compared to whole-genome screening results from our earlier conducted male baboon study [19] (S2 Table). Altogether 32 candidate genes from the baboon screening data (Microarray) were compared to rhesus macaques data (NGS). Vice versa, nine candidate genes, that were positively validated in rhesus macaques (NGS) from the current study were examined in an already generated baboon whole genome (microarray) screening data file [19]. Only male individuals were included, as the baboon study did not involve females. The pre-treatment samples served as reference. Inter-species validation was defined as a fold change (FC) ≥ |1.5| in the comparison group. This less robust FC was chosen to adjust for methodological differences inherent to both studies and to avoid false negative results.
Bioinformatics
For differentially expressed genes of Phase I we performed gene set enrichment analysis using PANTHER pathway software (http://www.pantherdb.org/, version 15, released 2020-02-14 [22]. PANTHER groups genes with similar biological functions based on their annotation (reference list was the current homo sapiens GO database). The p-values were corrected for multiple testing employing the False Discovery Rate algorithm (Table 1).
Table 1. Gene ontology (Panther) classification of differentially expressed genes.
| up-regulated genes | |||||||
| Classification | male | female | |||||
| fold enrichment | p-value* | fold enrichment | p-value* | ||||
| GO biological process complete | |||||||
| anatomical structure regression | 36.13 | 4.14E-02 | |||||
| gland morphogenesis | 9.67 | 5.57E-03 | |||||
| positive regulation of T cell activation | 7.08 | 1.83E-03 | |||||
| positive regulation of leukocyte cell-cell adhesion | 6.48 | 3.00E-03 | |||||
| regulation of leukocyte cell-cell adhesion | 4.99 | 1.61E-02 | |||||
| regulation of cell-cell adhesion | 4.38 | 1.06E-02 | |||||
| regulation of cell adhesion | 3.33 | 1.28E-02 | |||||
| positive regulation of cell-cell adhesion | 6.7 | 5.11E-04 | |||||
| positive regulation of cell adhesion | 4.44 | 5.10E-03 | |||||
| positive regulation of lymphocyte activation | 4.71 | 2.33E-02 | |||||
| positive regulation of leukocyte activation | 4.24 | 4.26E-02 | |||||
| positive regulation of cell activation | 4.13 | 4.91E-02 | |||||
| regulation of T cell activation | 5.36 | 4.69E-03 | |||||
| T cell differentiation | 7.02 | 1.06E-02 | |||||
| T cell activation | 6.2 | 4.07E-03 | |||||
| immune system development | 3.34 | 1.28E-0 | |||||
| translation | 4.2 | 5.58E-04 | |||||
| peptide biosynthetic process | 4.1 | 4.74E-04 | |||||
| peptide metabolic process | 3.56 | 1.18E-03 | |||||
| cellular amide metabolic process | 2.91 | 4.79E-03 | |||||
| amide biosynthetic process | 3.8 | 5.03E-04 | |||||
| cell-cell signaling | 2.97 | 2.52E-02 | |||||
| T cell costimulation | 18.99 | 4.63E-02 | |||||
| positive regulation of T cell activation | 5.03 | 4.60E-02 | |||||
| positive regulation of cell-cell adhesion | 4.84 | 1.51E-02 | |||||
| regulation of ossification | 5.58 | 6.83E-03 | |||||
| GO molecular function complete | |||||||
| coreceptor activity | 18.73 | 4.09E-02 | |||||
| structural constituent of ribosome | 7.31 | 2.47E-06 | |||||
| structural molecule activity | 4.35 | 1.11E-05 | |||||
| cytokine receptor activity | 7.28 | 3.12E-02 | |||||
| cytokine binding | 6.61 | 2.30E-02 | |||||
| GO cellular component complete | |||||||
| protein complex involved in cell adhesion | 13.6 | 5.71E-03 | 12.91 | 2.46E-03 | |||
| cytosolic large ribosomal subunit | 10.12 | 8.92E-05 | |||||
| cytosolic ribosome | 8.09 | 1.51E-05 | |||||
| ribosome | 5.9 | 3.17E-06 | |||||
| large ribosomal subunit | 6.27 | 2.67E-03 | |||||
| ribosomal subunit | 6.21 | 2.20E-05 | |||||
| tight junction | 7.44 | 1.91E-02 | |||||
| external side of plasma membrane | 5.55 | 1.65E-05 | |||||
| cell surface | 3.02 | 5.06E-03 | |||||
| side of membrane | 4.54 | 1.62E-05 | |||||
| plasma membrane signaling receptor complex | 5.35 | 1.61E-02 | 5.29 | 4.69E-03 | |||
| receptor complex | 3.7 | 7.13E-03 | |||||
| external side of plasma membrane | 3.57 | 1.39E-02 | |||||
| side of membrane | 3.4 | 2.21E-03 | |||||
| PANTHER Pathways | |||||||
| Gonadotropin-releasing hormone receptor pathway | 3.36 | 6.01E-02 | |||||
| Wnt signaling pathway | 3.69 | 2.48E-02 | 3.16 | 3.02E-02 | |||
| PANTHER GO-Slim Molecular Function | |||||||
| cytokine receptor activity | 7.55 | 3.82E-02 | |||||
| cytokine binding | 6.93 | 4.41E-02 | |||||
| PANTHER GO-Slim Biological Process | |||||||
| positive regulation of cytosolic calcium ion concentration | 7.17 | 1.12E-02 | |||||
| regulation of cytosolic calcium ion concentration | 6.02 | 2.23E-02 | |||||
| Wnt signaling pathway | 6.02 | 4.95E-02 | 6.12 | 1.04E-02 | |||
| cell-cell signaling by wnt | 5.96 | 4.38E-02 | 6.06 | 9.58E-03 | |||
| cell surface receptor signaling pathway involved in cell-cell signaling | 6.3 | 2.21E-02 | 6.23 | 3.92E-03 | |||
| cell surface receptor signaling pathway | 3.19 | 6.92E-04 | |||||
| transmembrane receptor protein serine/threonine kinase signaling pathway | 6.39 | 2.20E-02 | |||||
| enzyme linked receptor protein signaling pathway | 4.1 | 1.45E-02 | |||||
| cellular response to growth factor stimulus | 6.2 | 1.32E-02 | |||||
| response to growth factor | 6.2 | 1.44E-02 | |||||
| negative regulation of signal transduction | 4.47 | 1.59E-02 | |||||
| negative regulation of signaling | 4.29 | 1.29E-02 | |||||
| PANTHER GO-Slim Cellular Component | |||||||
| external side of plasma membrane | 6.18 | 1.70E-03 | |||||
| side of membrane | 4.75 | 3.88E-03 | |||||
| cell surface | 3.91 | 2.28E-02 | |||||
| leaflet of membrane bilayer | 4.75 | 2.58E-03 | |||||
| dendritic spine | 11.11 | 6.26E-02 | |||||
| neuron spine | 11.11 | 5.36E-02 | |||||
| down-regulated genes | |||||||
| Classification | male | female | |||||
| fold enrichment | p-value* | fold enrichment | p-value* | ||||
| GO biological process complete | |||||||
| DNA-dependent DNA replication | 10.86 | 5.95E-03 | |||||
| DNA replication | 11.21 | 1.53E-04 | |||||
| cell division | 9.07 | 4.87E-04 | |||||
| chromosome segregation | 6.45 | 1.36E-02 | |||||
| mitotic cell cycle process | 5.85 | 4.04E-04 | |||||
| mitotic cell cycle | 5.09 | 1.42E-03 | |||||
| cell cycle | 4.23 | 1.88E-04 | |||||
| cell cycle process | 4.7 | 7.27E-05 | |||||
| cellular response to cytokine stimulus | 4.05 | 2.05E-02 | |||||
| immune response | 3.26 | 2.98E-02 | |||||
| response to molecule of fungal origin | |||||||
| response to external biotic stimulus | 5.59 | 8.06E-22 | |||||
| response to biotic stimulus | 5.54 | 9.15E-22 | |||||
| pentose-phosphate shunt, non-oxidative branch | 46.08 | 1.14E-02 | |||||
| pentose-phosphate shunt | 34.13 | 3.29E-04 | |||||
| glucose 6-phosphate metabolic process | 19.4 | 3.85E-04 | |||||
| NADPH regeneration | 30.72 | 4.57E-04 | |||||
| NADP metabolic process | 18.43 | 4.70E-04 | |||||
| type I interferon signaling pathway | 36.86 | 2.65E-05 | |||||
| cellular response to type I interferon | 39.1 | 2.01E-06 | |||||
| response to type I interferon | 30.72 | 7.97E-07 | |||||
| innate immune response | 6.94 | 2.61E-14 | |||||
| defense response to other organism | 6.54 | 1.73E-19 | |||||
| defense response | 5.43 | 3.96E-20 | |||||
| immune response | 4.6 | 3.69E-15 | |||||
| immune system process | 3.93 | 2.01E-20 | |||||
| cytokine-mediated signaling pathway | 3.89 | 3.11E-03 | |||||
| interferon-gamma-mediated signaling pathway | 36.86 | 1.64E-02 | |||||
| cytidine to uridine editing | 30.72 | 2.17E-02 | |||||
| response to peptidoglycan | 26.33 | 2.93E-02 | |||||
| regulation of ribonuclease activity | 26.33 | 2.92E-02 | |||||
| detection of other organism | 23.63 | 9.83E-04 | |||||
| detection of external biotic stimulus | 21.94 | 1.26E-03 | |||||
| positive regulation of cytokine production involved in inflammatory response | 21.94 | 1.24E-03 | |||||
| regulation of cytokine production involved in inflammatory response | 11.89 | 2.82E-03 | |||||
| negative regulation of viral genome replication | 21.59 | 4.67E-10 | |||||
| regulation of viral genome replication | 14.01 | 3.33E-08 | |||||
| negative regulation of viral life cycle | 16.3 | 7.79E-09 | |||||
| response to interferon-alpha | 20.48 | 4.65E-02 | |||||
| cytoplasmic pattern recognition receptor signaling pathway | 20.48 | 4.63E-02 | |||||
| negative regulation of interleukin-10 production | 18.9 | 1.12E-02 | |||||
| defense response to protozoan | 18.9 | 4.10E-16 | |||||
| defense response to virus | 17.55 | 4.10E-16 | |||||
| positive regulation of interleukin-6 production | 14.18 | 1.55E-05 | |||||
| positive regulation of interleukin-8 production | 12.93 | 2.91E-02 | |||||
| positive regulation of cytokine biosynthetic process | 12.8 | 8.02E-03 | |||||
| regulation of toll-like receptor signaling pathway | 12.71 | 2.11E-03 | |||||
| lipopolysaccharide-mediated signaling pathway | 11.81 | 1.08E-02 | |||||
| toll-like receptor signaling pathway | 12.71 | 9.38E-04 | |||||
| activation of MAPKK activity | 10.97 | 1.35E-02 | |||||
| positive regulation of tumor necrosis factor production | 9.83 | 5.88E-04 | |||||
| regulation of erythrocyte differentiation | 9.6 | 2.05E-02 | |||||
| positive regulation of chemokine production | 9.03 | 2.52E-02 | |||||
| regulation of defense response to virus | 8.19 | 1.30E-02 | |||||
| granulocyte chemotaxis | 7.06 | 2.05E-02 | |||||
| positive regulation of NF-kappaB transcription factor activity | |||||||
| GO molecular function complete | |||||||
| single-stranded DNA helicase activity | 28.58 | 4.55E-02 | |||||
| DNA helicase activity | 12.58 | 1.87E-02 | |||||
| DNA-dependent ATPase activity | 9.31 | 1.83E-02 | |||||
| ATPase activity | 4.75 | 5.42E-03 | |||||
| immune receptor activity | 8.51 | 2.54E-02 | |||||
| cytokine binding | 7.85 | 3.11E-02 | |||||
| microtubule binding | 6.03 | 9.52E-03 | |||||
| 2’-5’-oligoadenylate synthetase activity | 46.08 | 2.81E-02 | |||||
| pattern recognition receptor activity | 20.48 | 2.48E-02 | |||||
| double-stranded RNA binding | 8.11 | 1.55E-02 | |||||
| GO cellular component complete | |||||||
| replication fork protection complex | 44.46 | 1.83E-02 | |||||
| Midbody | 7.69 | 2.74E-02 | |||||
| Spindle | 5.83 | 8.50E-03 | |||||
| external side of plasma membrane | 5.49 | 8.90E-03 | |||||
| PANTHER Pathways | |||||||
| Pentose phosphate pathway | 24.57 | 9.35E-03 | |||||
| Toll receptor signaling pathway | 5.76 | 4.89E-02 | |||||
| PANTHER GO-Slim Molecular Function | |||||||
| cytokine receptor activity | 9.95 | 1.06E-01 | |||||
| cytokine binding | 9.14 | 5.16E-02 | |||||
| PANTHER GO-Slim Biological Process | |||||||
| cell motility | 4.61 | 4.35E-02 | |||||
| localization of cell | 4.61 | 5.80E-02 | |||||
| organelle fission | 4.45 | 5.31E-02 | |||||
| cell cycle | 3.81 | 1.03E-01 | |||||
| defense response to virus | 24.57 | 6.97E-10 | |||||
| negative regulation of cell-cell adhesion | 14.46 | 3.79E-02 | |||||
| regulation of T cell proliferation | 12.93 | 5.16E-02 | |||||
| T cell proliferation | 12.93 | 4.89E-02 | |||||
| regulation of multi-organism process | 9.64 | 6.40E-04 | |||||
| RNA phosphodiester bond hydrolysis | 8.3 | 4.80E-02 | |||||
| PANTHER Protein Class | |||||||
| Apolipoprotein | 21.34 | 1.18E-02 | |||||
| basic leucine zipper transcription factor | 13.03 | 5.87E-04 | |||||
Using the differentially expressed genes (DEG) from irradiated male and female rhesus macaque, a classification of overrepresented and underrepresented genes coding, e.g., biological processes or protein classes, was conducted using the bioinformatic tool PANTHER (http://www.pantherdb.org; version 15) which comprises Gene Ontology (GO) annotations directly imported from the GO database. Based on the comparison of observed vs expected numbers of upregulated or downregulated genes (reference database was Macaca mulatta) a fold enrichment in the number of genes annotated to this process and a corresponding p value (False Discovery Rate (FDR) corrected) was calculated. Numbers highlighted in grey refer to processes that were similar between male and female rhesus macaque.
Statistical analysis
Quantitative gene expression data of the 37 candidate genes from phase II were examined using descriptive statistics (n, mean, standard deviation) and groups were compared employing the t-test. We assessed the assumptions of normality and equal variance and if required utilized either the pooled (equal variance) or the Satterthwaite variant of the t-test (unequal variance). Unconditional logistic regression analysis was performed for each of the independent variables (genes) of interest separately (univariate) on the binary outcome variable where the probability modeled was gene expression changes measured in the 60-day post-exposure sample relative to the pre-exposure sample used as the reference. Using logistic regression, we determined the area under a receiver-operator characteristic (ROC) curve providing a reasonable indication of overall diagnostic accuracy. ROC areas of 1.0 indicate complete agreement between the predictive model and the binary group comparison and thus a clear distinction between a pre- and a post-exposure sample. All calculations were performed using SAS (release 9.4, Cary NC, USA).
Results
Material available for the three-phase study design
For this study, Neumedicines Inc entrusted peripheral blood samples from 142 rhesus macaque comprising 72 samples from males and 70 from females (Fig 1, upper panel). Based on RNA quality and quantity (availability) criteria altogether 137 RNA samples from 113 rhesus macaque were eligible for this analysis (Fig 1).
During the screening approach (phase I), we assessed 20 whole-genome mRNA sequencing (RNA seq) for 20 blood samples, randomly selected from the group of untreated NHP. Samples were collected before (pre-exposure) and 60 days after irradiation (post-exposure) from surviving animals (Fig 1, overview upper panel and sample overview lower panel). Five RNA samples were utilized for each, the pre- and post-exposure groups as well as males and females, summing up to 20 NGS examinations (n = 4x5). All samples originated from different animals, increasing inter-individual variance and decreasing false positive results occurring by chance.
For the validation of mRNAs, we used the remaining 117 blood samples comprising another 55 and 62 RNA samples from untreated and treated NHP, respectively (Fig 1, sample overview, phase II panel and lower panel). In phase II, we performed the validation separately for males and females, in groups of untreated (vehicle), treated (IL-12 and IL-12 + G-CSF combined) and merged untreated and treated NHP, thereby increasing the sample size examined.
Phase I: RNA isolation and screening results
From 2.5 ml whole blood we isolated 10.6 μg (stdev 5.1 μg) total RNA on average before irradiation. RNA integrity (RIN) with a mean value of 8.0 (stdev 2.3) suggested high-quality RNA sufficient for running both methods.
The quality parameter for a successful sequencing run expressed as the percentage of Q30 bases (translates into a 99.9% accuracy) is supposed to be > 80% and was on average 90.6% in our study. The average number of PF reads (passed filter reads) was 21.6 x 106 (stdev 3.8 x 106) and ranged between 17.3–32.5 x 106 reads/run. In total, 87–91% of the reads mapped to known gene regions (exons and introns).
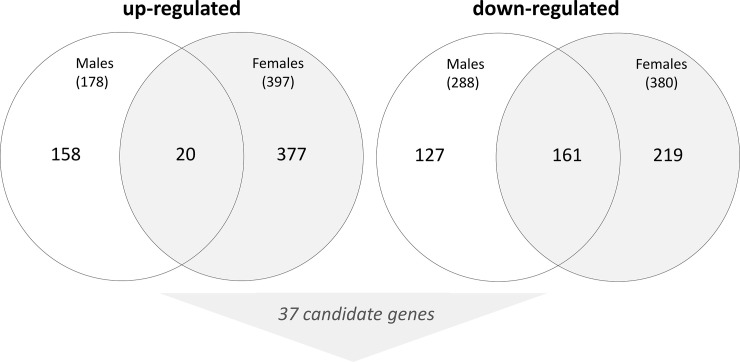
Based on our filter (Fold change ≥ |2|, p<0.05) we identified 1,243 DEG (668 up-regulated, 575 down-regulated, Fig 1) with 161 and 20 of them being similarly up-regulated and down-regulated in both sexes (Fig 2). Bioinformatic analysis regarding the significant enrichment of genes to certain gene ontology (GO) classifications revealed a small overlap between both sexes, namely related to the WNT signaling pathway (males: fold enrichment, 6.02, p-value 0.049; females: fold enrichment 6.12, p-value 0.01) and cell adhesion (males: fold enrichment, 13.6, p-value 0.006; females: fold enrichment 12.91, p-value 0.0025; Table 1). Genes coding for immunological functions (e.g. T-cell costimulation or leukocyte cell-cell stimulation) were differently enriched in both sexes. The number of deregulated genes was about 30–50% higher in females relative to males and about 2-time more genes appeared up-regulated relative to the down-regulated genes in both sexes (Fig 2). Using the fold-difference and the p-value, we selected 37 most promising candidate mRNAs for validation at phase II employing qRT-PCR.
Fig 2. Venn diagram of up- and down-regulated genes that were differentially expressed by pre- and postexposure samples in male and female rhesus macaques.
An overlapping number of genes being similarly up- and down-regulated in both sexes are presented by the numbers in the overlapping circles. The number of genes differentially expressed in males or females only is shown outside the overlapping circle. Numbers in parenthesis represent the total number of differentially expressed genes in males and females. Based on the fold-change, the p-value and the preference to genes differentially expressed in both sexes, 37 candidate genes were selected and put forward in phase II for validation purposes.
Phase II: Independent methodological validation of NGS using qRT-PCR
In a first step, where we examined for genes expressed in 117 RNA samples, we identified 20 out of 37 candidate genes eligible for analysis (S1 Fig). Thirteen of these mRNAs independently examined in 117 samples using qRT-PCR appeared up-regulated (HBG2/1) or down-regulated similarly to the other 20 RNA samples used to employ the NGS whole-genome screening (S1 Fig). False-positive results (control values detected with qRT-PCR, while NGS data indicated an up- or down-regulation) were identified in six mRNAs and an inverse regulation with both methods could be detected in one case.
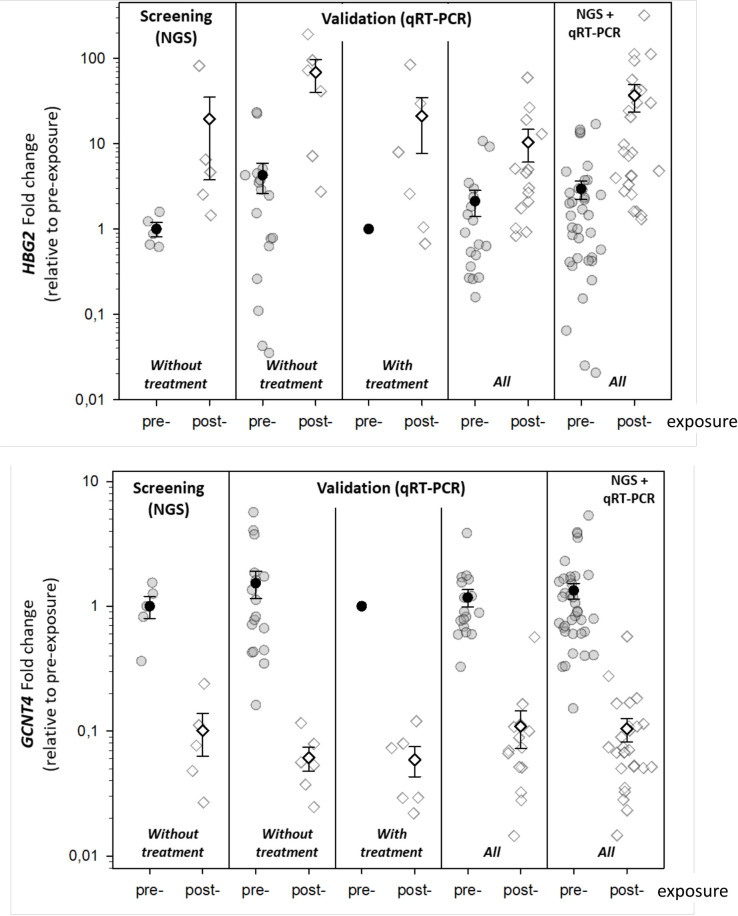
From these 13 validated candidate mRNA species appeared expressed and showed significant or borderline significant DGE in post-exposure relative to pre-exposure samples in separate analysis considering sexes and treatment groups. In a second analysis step, we considered significant changes and a minimal number of 50% expressed samples per group. Based on that, nine most promising validated genes were identified, namely CD248, EDAR, FAM19A5, GAL3ST4, GCNT4, HBG2, LRRN1, NOG and SYT14 (Table 2). All nine genes revealed the same direction in DGE in the post-exposure samples relative to the pre-exposure sample irrespective of the examined group of untreated or cytokine treated NHP or males and females (Table 2). This is graphically visualized for HBG2/1 (up-regulated) and GCNT4 (down-regulated) using the NGS screening data and the corresponding qRT-PCR data which reveal the results separately for untreated, cytokine treated and merged untreated and treated groups (Fig 3). The last plot in Fig 3 combines NGS and all qRT-PCR data.
Table 2. Overview of nine genes showing significantly different DGE in post-exposure relative to pre-exposure samples.
| Males | Females | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| treatment | mRNA species | n (pre-exposure) | n (60 days after irr.) | mean Ct-value (pre-exposure) | mean Ct-value (60 days after irr.) | Fold-change | p-value | ROC | n (pre-exposure) | n (60 days after irr.) | mean Ct-value (pre-exposure) | mean Ct-value (60 days after irr.) | Fold-change | p-value | ROC |
| without treatment | CD248 | 14 | 1 | 15.3 | 17.8 | 0.2 | 1.0 | 22 | 2 | 15.5 | 17.7 | 0.2 | 0.08 | 0.91 | |
| with treatment, IL-12 | 1 | 3 | 15.1 | 17.7 | 0.2 | 1.0 | 5 | 1 | 14.8 | 17.7 | 0.1 | 1.0 | |||
| with treatment, IL-12+G-CSF | 18 | 3 | 15.0 | 17.8 | 0.1 | 0.002 | 1.0 | 9 | 2 | 14.4 | 16.0 | 0.3 | 0.027 | 0.94 | |
| all | 33 | 7 | 15.1 | 17.8 | 0.2 | <0.0001 | 1.0 | 36 | 5 | 15.1 | 16.7 | 0.3 | 0.003 | 0.87 | |
| without treatment | EDAR | 16 | 2 | 13.6 | 17.6 | 0.1 | 0.002 | 1.0 | 22 | 3 | 13.3 | 15.4 | 0.2 | 0.0002 | 0.99 |
| with treatment, IL-12 | 1 | 5 | 12.5 | 16.5 | 0.1 | 1.0 | 5 | 3 | 13.6 | 17.5 | 0.1 | 0.0001 | 1.0 | ||
| with treatment, IL-12+G-CSF | 18 | 8 | 13.38 | 17.38 | 0.1 | <0.0001 | 1.0 | 9 | 3 | 13.1 | 16.2 | 0.1 | 0.001 | 1.0 | |
| all | 35 | 15 | 13.5 | 17.1 | 0.1 | <0.0001 | 0.99 | 36 | 9 | 13.3 | 16.4 | 0.1 | <0.0001 | 0.99 | |
| without treatment | FAM19A5 | 15 | 3 | 15.16 | 17.3 | 0.2 | 0.08 | 0.87 | 17 | 2 | 15.9 | 16.7 | 0.6 | 0.4 | 0.69 |
| with treatment, IL-12 | 1 | 5 | 12.5 | 16.1 | 0.1 | 1.0 | 3 | 3 | 17.2 | 17.1 | 1.0 | 0.95 | 0.5 | ||
| with treatment, IL-12+G-CSF | 15 | 9 | 15.9 | 17.1 | 0.4 | 0.012 | 0.83 | 8 | 3 | 15.2 | 17.1 | 0.3 | 0.11 | 0.88 | |
| all | 31 | 17 | 15.4 | 16.9 | 0.4 | 0.0021 | 0.78 | 28 | 8 | 15.8 | 17.0 | 0.4 | 0.004 | 0.75 | |
| without treatment | GAL3ST4 | 16 | 2 | 14.2 | 17.6 | 0.1 | 0.08 | 1.0 | 22 | 2 | 14.4 | 15.7 | 0.4 | 0.07 | 1.0 |
| with treatment, IL-12 | 1 | 4 | 13.95 | 17.4 | 0.1 | 1.0 | 5 | 1 | 14.9 | 17.1 | 0.2 | 1.0 | |||
| with treatment, IL-12+G-CSF | 18 | 5 | 14.8 | 17.5 | 0.2 | <0.0001 | 0.99 | 9 | 3 | 14.1 | 16.2 | 0.2 | 0.15 | ||
| all | 35 | 11 | 14.5 | 17.5 | 0.1 | <0.0001 | 0.99 | 36 | 6 | 14.4 | 16.2 | 0.3 | <0.0001 | 0.92 | |
| without treatment | GCNT4 | 17 | 6 | 11.2 | 15.4 | 0.1 | <0.0001 | 1.0 | 23 | 3 | 11.3 | 12.9 | 0.3 | 0.095 | 0.94 |
| with treatment, IL-12 | 1 | 6 | 9.9 | 14.3 | 0.0 | 1.0 | 5 | 3 | 11.0 | 15.2 | 0.1 | <0.0001 | 1.0 | ||
| with treatment, IL-12+G-CSF | 18 | 14 | 11.1 | 14.9 | 0.1 | <0.0001 | 1.0 | 9 | 6 | 10.6 | 14.9 | 0.1 | 0.004 | 1.0 | |
| all | 36 | 26 | 11.1 | 14.9 | 0.1 | <0.0001 | 37 | 12 | 11.1 | 14.5 | 0.1 | <0.0001 | 0.96 | ||
| without treatment | HBG2_HBG1 | 18 | 6 | 10.18 | 5.17 | 32.2 | 0.002 | 0.91 | 23 | 3 | 8.5 | 10.6 | 0.2 | 0.1 | 0.87 |
| with treatment, IL-12 | 1 | 6 | 7.5 | 5 | 5.7 | 5 | 3 | 9.8 | 4.8 | 32.0 | 0.035 | 1.0 | |||
| with treatment, IL-12+G-CSF | 18 | 14 | 8.75 | 6.6 | 4.4 | 0.0025 | 0.86 | 9 | 6 | 9.1 | 5.5 | 12.1 | 0.048 | 0.83 | |
| all | 37 | 26 | 9.4 | 5.9 | 11.3 | <0.0001 | 0.86 | 37 | 12 | 8.8 | 6.6 | 4.6 | 0.07 | 0.68 | |
| without treatment | LRRN1 | 21 | 5 | 12.24 | 15.52 | 0.1 | 0.0008 | 0.93 | 22 | 3 | 12.2 | 15.0 | 0.1 | 0.005 | 0.94 |
| with treatment, IL-12 | 1 | 6 | 12 | 14.95 | 0.1 | 0.83 | 5 | 3 | 12.6 | 14.6 | 0.3 | 0.029 | 1.0 | ||
| with treatment, IL-12+G-CSF | 18 | 11 | 12.8 | 16.3 | 0.1 | <0.0001 | 0.99 | 9 | 5 | 12.5 | 14.8 | 0.2 | 0.014 | 0.89 | |
| all | 40 | 22 | 12.5 | 15.77 | 0.1 | <0.0001 | 0.94 | 36 | 11 | 12.3 | 14.8 | 0.2 | <0.0001 | 0.92 | |
| without treatment | NOG | 20 | 6 | 11.64 | 15.96 | 0.1 | <0.0001 | 1.0 | 23 | 3 | 11.6 | 13.3 | 0.3 | 0.049 | 0.96 |
| with treatment, IL-12 | 1 | 6 | 10.8 | 15.5 | 0.0 | 1.0 | 5 | 3 | 11.5 | 16.1 | 0.0 | 0.0002 | 1.0 | ||
| with treatment, IL-12+G-CSF | 18 | 14 | 11.3 | 15.5 | 0.1 | <0.0001 | 1.0 | 9 | 6 | 10.7 | 15.2 | 0.0 | 0.004 | ||
| all | 39 | 26 | 11.4 | 15.4 | 0.1 | <0.0001 | 37 | 12 | 11.3 | 15.0 | 0.1 | <0.0001 | 0.96 | ||
| without treatment | SYT14 | 15 | 1 | 16.04 | 17.69 | 0.3 | 22 | 2 | 15.5 | 16.8 | 0.4 | 0.15 | 0.86 | ||
| with treatment, IL-12 | 1 | 2 | 15.5 | 16.4 | 0.5 | 1.0 | 5 | 1 | 15.8 | 18.0 | 0.2 | 1.0 | |||
| with treatment, IL-12+G-CSF | 18 | 4 | 15.9 | 17.2 | 0.4 | 0.008 | 0.9 | 9 | 3 | 15.4 | 16.2 | 0.6 | 0.2 | 0.72 | |
| all | 34 | 7 | 16 | 17.1 | 0.5 | 0.011 | 36 | 6 | 15.5 | 16.7 | 0.4 | 0.016 | 0.8 | ||
Overview of nine genes that were significantly different in post-exposure relative to pre-exposure samples and expressed in ≥ 50% of samples per group. Data are presented separately for male (left part) and female rhesus macaques (right part) and ordered starting with untreated, treated (IL-12 and IL-12 + G-CSF), and untreated and treated groups combined (all). The number of measurements and the mean gene expression (threshold cycle, Ct) values are provided by pre- and post-exposure samples as well as the fold-change difference (derived from Ct-values applying the ΔΔ-Ct-approach). For instance, fold-changes of 0.1 or 10 are referring to a 10-fold down-regulation or up-regulation, respectively. A t-test was applied for this group comparison. P-values (chisquare statistic) of the category “all” are presented in bold when surviving Bonferroni correction for multiple comparison. ROC areas were derived from logistic regression analysis.
Fig 3. The jitter plots represent fold-changes in gene expression calculated between post-exposure samples relative to the pre-exposure samples, with the latter arbitrarily set to the value one.
Single measurements (symbols with grey edging) as well as mean values (black edged symbols, error bars do show the standard error) of two representative genes are shown. Gene expression data from pre- and post-exposure samples used for screening (left side) or validation (right side) are shown in untreated, treated (IL12, IL12+G-CSF) and both groups combined “All”. The last plots to the right reveal gene expression measurements of all samples (screening as well as validation). Fold changes for the no treatment group were calculated based on untreated pre-exposure samples, for “all” they were calculated based on pre-exposure samples from the no treatment, IL12 and IL12+G-CSF group to ensure comparability.
P-values of the nine most promising, validated mRNAs in Table 2 further decreased after merging treated and untreated NHP and all except one mRNA (SYT14) in males and females and another mRNA (HBG2/1) in females survived Bonferroni correction for multiple comparison (p<0.0056; 0.05/9, Table 2).
Eight genes were significantly down-regulated (3-10-fold) and HBG2/1 (treatment group dependently) appeared about 10-32-fold up-regulated. Complete or almost complete separation of groups (ROC lying mostly between 0.8–1.0) was observed (Table 2).
Phase III: Inter-species validation
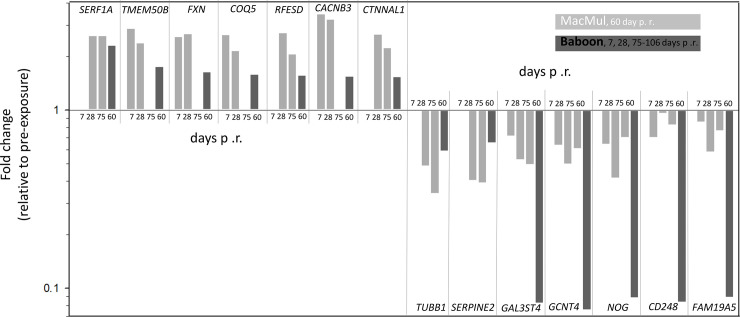
The nine successfully validated candidate genes (Table 2) from male rhesus macaques were compared to the whole genome microarray screening database from an earlier performed baboon study in males [19]. Vice versa, the 32 validated candidate genes from male baboons were compared to the NGS screening results of the current study on rhesus macaques. Overall, 34% of the genes (14 of the 41 genes (nine originating from rhesus macaques and 32 originating from a previous baboon study) showed a concordant DGE (FC) ≥ |1.5|; S2 Table), whereby nine baboon candidate genes and five rhesus macaques candidate genes could be validated in the other species (Fig 4). No inter-species validation could be performed in 23 candidate genes from both species together. Four genes were inversely regulated and 23 could not be validated between the two species (Fig 4).
Fig 4. Bar graphs represent the fold changes of 14 successfully validated candidate genes originating from Baboon (light gray, 1–3 time points after irradiation) respectively Macaca mulatta (dark grey, 60 days after irradiation) from S2 Table.
Fold changes are shown relative to pre-treatment samples on a logarithmic scale (fold change ≥ |1.5|). p. r., post irradiation.
Discussion
In developed countries about two-third of all cancer patients undergo radiotherapy and about half of them survive, so that a secondary tumor or chronic non-cancer late effects might develop [11, 23]. Individuals involved in the malevolent use of radiation sources or involved in an accident like a nuclear power plant disaster might be exposed to even higher, potentially lethal doses of radiation and survivor might develop delayed effects of acute radiation exposure (DEARE, [17–19]). Taking advantage of a non-human primate model with high genetic homology to humans, we examined for transcriptomic changes occurring in ARS survivor and compared our molecular results with published molecular changes known to be associated with later occurring health effects and DEARE.
In collaboration with Neumedicine Inc., we analyzed 137 RNA samples from 113 rhesus macaque originating from a clinical study to examine the effect of IL-12 and G-CSF on the survival of rhesus macaque after potentially lethal whole-body exposure. Male and female rhesus macaques were treated during this study, providing the opportunity to examine sex and treatment specific differences in gene-expression changes 60 days after exposure in comparison to the pre-exposure measurements.
Whole-genome bioinformatic analysis revealed a small overlap of sex-associated gene enrichment to certain gene ontology (GO) classifications. This overlap was mainly related to the WNT signaling pathway and immunological processes (Table 1). Previous studies already reported, that immunological alterations might play a role in post-radiation tumor development besides the direct carcinogenic effect of radiation [24]. Radiation exposure that led to long-term gene expression changes even weeks after exposure [25–27] or DEARE [17–19] were already described in previous studies.
Because our measurements were performed on peripheral whole blood, presumably our results either reflect altered hematopoietic stem cells or indicated a selection for less radiosensitive stem cells reconstituting the peripheral blood after radiation exposure. That would explain the rather high fold-changes observed in several of the mRNA species and their persistency in unirradiated cellular progeny. It can be speculated whether our WNT pathway results might relate to it, since the canonical WNT pathway is known to promote hematopoiesis via β-catenin [28] and reports indicate a radiation-induced high activity of the canonical WNT pathway associated with increased DNA damage in murine hematopoietic stem cells [29]. Moreover, for the gastrointestinal tract, two functional distinct pools of intestinal stem cells are proposed [30]. Actively cycling, radiation-sensitive, WNT pathway modulated stem cells are discriminated from slowly cycling, radiation-resistant, WNT pathway refractory and injury inducible stem cells. After radiation injury, a shift in the composition of these differently radiosensitive stem cells after irradiation is expected in the gastrointestinal tract [30]. A mechanism like that could be proposed for the hematopoetic system. This shift in the composition of hematopoetic stem cells would be reflected by strong fold-differences in gene expression as observed in our study 60 days after irradiation relative to the pre-exposure measurements.
From the 37 selected candidate genes, nine appeared significantly deregulated (Table 1). These changes at 60 days after exposure were found to be similar in males and females and independent of the treatment regimen. Some of the validated genes were described to be coding for proteins involved e.g. in chemokine/cytokine function (FAM19A5, NOG) and extracellular matrix function (CD248, GCNT4) (S3 Table). Others are known for their association with certain diseases (EDAR, GAL3ST4, LRRN1, SYT14). Corresponding to our findings, a de-regulation of HBG2/HBG1 was observed in peripheral blood of radiotherapy patients within 16 hours after 2 Gy radiation. Its role as potential predictor for radiation toxicity was discussed [31].
For further validation, we compared previously reported gene expression results from irradiated male baboons [19, 32] with male rhesus macaque data from the current study. Methodological differences in both experiments (microarrays in baboons versus NGS in this study) necessitated adjustments of the fold change to a less strict cut off value ((FC) ≥ |1.5|) to avoid false negative results. We earlier showed that even when organisms with a striking gene sequence homology are compared, inter-species differences might occur (inverse regulation of FDXR gene in baboons and humans after radiations [33]), which underlines this additional validation step. Altogether 34% of deregulated genes could be successfully validated in both species (Fig 4) [19], which further supports the significance of our findings. Examinations in baboons covered a period of about three months. As a limitation of our rhesus macaque study, we could perform gene expression measurements at 60 days after irradiation only. However, one third of the identified genes showed similar gene expression changes over the whole three months period of time in both species. This led us speculate, that the 60 days after irradiation identified rhesus macaque gene expression changes might reflect long lasting changes with impact on DEARE and late health effects as discussed above.
Because of sex and cytokine treatment differences (Fig 1) a separate analysis was performed. Similarities in gene expression independently of sex and treatment regimens observed in the majority (71–95%) of examined samples indicate that cytokine treatment (IL-12 and G-CSF) and sex differences for these genes do not affect gene expression changes at least 60 days after irradiation (Table 1). To our knowledge that has not been examined before in healthy irradiated NHP.
A higher radiosensitivity in females was discussed in the literature, especially in the context of second malignancies [34–36]. In our study, the number of deregulated genes was about 30–50% higher in females relative to males indicating females being more radio-responsive on the transcriptome level compared to males.
NGS results could be successfully validated with qRT-PCR in 13 but not all expressed 20 mRNAs. This might be improved in future studies when identifying with NGS not the radiation-induced gene, but the specific radiation-responsive exon-region of a gene contributing primarily to the radiation-induced gene expression changes and using a complementary TaqMan assay for q-RT-PCR validation, specifically covering this exon region.
Some limitations of our study should be kept in mind. Only one time point after irradiation (60 days) was available in this experiment and it is unclear whether the observed gene expression changes persist over time. However, the study design did not allow for further examinations on other time points [20], but corresponding changes of validated genes detected in baboons suggest a persistency over a larger time frame. The strength of our study certainly is a large number of animals as well as having males and females included. The validation was performed with high robustness by changing both, methodology and samples during the first validation step and performing an inter-species comparison as a second validation step.
In summary, 60 days after radiation exposure we identified (1) cytokine treatment independent transcriptional changes, (2) females with almost twice as much deregulated genes appeared more radio-responsive than males and (3) Panther analysis revealed a strong association with immunological processes as well as the WNT pathway for both sexes and these processes are known to be associated with secondary malignancies and other inflammatory associated diseases, e.g. lung fibrosis or cardiovascular diseases.
Supporting information
Mean fold-changes are presented for animals used for NGS (n = 20) and the remaining animals using qRT-PCR (n = 117). Genes where a successful validation failed are shown in the inserted graph.
(TIFF)
Gene numbers and fold-changes (first number represents male and second number female fold-changes) and sexes are presented.
(XLSX)
Data is sorted by successful inter-species validation (14/41), no inter-species validation (23/41) and inverse inter-species validation (4/41). Successfully validated time points are shown in bold.
(XLSX)
A summary of the currently known function is given. Results for a Pubmed search (http://www.pubmed.gov) for “gene name” + “radiation” are given.
(XLSX)
Acknowledgments
The carefully performed technical assistance of Eva Grumpelt, Thomas Müller, Oliver Wittmann and Sven Doucha-Senf are very much appreciated. The authors thank Jamie L. Tom and Alice Trinh who facilitated handling of blood samples.
Data Availability
NGS data is available under DOI: 10.17632/t2b295b4gc.1 https://data.mendeley.com/datasets/t2b295b4gc/2.
Funding Statement
This study was funded with a contract from the Biomedical Advanced Research and Development Authority (contract grant nos. HHSO100200800060C and HHSO100201100037C). The funding agency had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kanakoglou DS, Michalettou TD, Vasileiou C, Gioukakis E, Maneta D, Kyriakidis K V., et al. Effects of high-dose ionizing radiation in human gene expression: A meta-analysis. Int J Mol Sci. 2020;21. doi: 10.3390/ijms21061938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Scherthan H, Abend M, Müller K, Beinke C, Braselmann H, Zitzelsberger H, et al. Radiation-Induced Late Effects in Two Affected Individuals of the Lilo Radiation Accident. Radiat Res. 2007. doi: 10.1667/RR0774.1 [DOI] [PubMed] [Google Scholar]
- 3.Gregoire E, Sorokine-Durm I, Bertho J, Jacquet N, Delbos M, Demarquay C, et al. Follow-up of stable chromosomal aberrations in gamma-ray irradiated non-human primates. Int J Radiat Biol. 2006;82: 493–502. doi: 10.1080/09553000600840930 [DOI] [PubMed] [Google Scholar]
- 4.Bhatia S, Sklar C. Second cancers in survivors of childhood cancer. Nature Reviews Cancer. European Association for Cardio-Thoracic Surgery; 2002. pp. 124–132. doi: 10.1038/nrc722 [DOI] [PubMed] [Google Scholar]
- 5.Little MP. Cancer and non-cancer effects in Japanese atomic bomb survivors. J Radiol Prot. 2009;29: A43–59. doi: 10.1088/0952-4746/29/2A/S04 [DOI] [PubMed] [Google Scholar]
- 6.Shimizu Y, Kodama K, Nishi N, Kasagi F, Suyama A, Soda M, et al. Radiation exposure and circulatory disease risk: Hiroshima and Nagasaki atomic bomb survivor data, 1950–2003. BMJ. 2010;340: 193. doi: 10.1136/bmj.b5349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mulrooney DA, Yeazel MW, Kawashima T, Mertens AC, Mitby P, Stovall M, et al. Cardiac outcomes in a cohort of adult survivors of childhood and adolescent cancer: Retrospective analysis of the childhood cancer survivor study cohort. BMJ. 2009;339: 34. doi: 10.1136/bmj.b4606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krull KR, Minoshima S, Edelmann M, Morris B, Sabin ND, Brinkman TM, et al. Regional brain glucose metabolism and neurocognitive function in adult survivors of childhood cancer treated with cranial radiation. J Nucl Med. 2014;55: 1805–1810. doi: 10.2967/jnumed.114.142950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krull KR, Sabin ND, Reddick WE, Zhu L, Armstrong GT, Green DM, et al. Neurocognitive function and CNS integrity in adult survivors of childhood Hodgkin lymphoma. J Clin Oncol. 2012;30: 3618–3624. doi: 10.1200/JCO.2012.42.6841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quast U. Whole body radiotherapy: A TBI-guideline. Journal of Medical Physics. Medknow Publications and Media Pvt. Ltd; 2006. pp. 5–12. doi: 10.4103/0971-6203.25664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delaney G, Jacob S, Featherstone C, Barton M. The role of radiotherapy in cancer treatment: Estimating optimal utilization from a review of evidence-based clinical guidelines. Cancer. Cancer; 2005. pp. 1129–1137. doi: 10.1002/cncr.21324 [DOI] [PubMed] [Google Scholar]
- 12.Macvittie TJ, Farese AM. Defining the concomitant multiple organ injury within the ARS and DEARE in an animal model research platform. Health Physics. 2020. doi: 10.1097/HP.0000000000001327 [DOI] [PubMed] [Google Scholar]
- 13.Andrews RN, Bloomer EG, Olson JD, Hanbury DB, Dugan GO, Whitlow CT, et al. Non-Human Primates Receiving High-Dose Total-Body Irradiation are at Risk of Developing Cerebrovascular Injury Years Postirradiation. Radiat Res. 2020. doi: 10.1667/RADE-20-00051.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Macvittie TJ, Farese AM, Parker GA, Bennett AW, Jackson WE. Acute radiation-induced lung injury in the non-human primate: A review and comparison of mortality and co-morbidities using models of partial-body irradiation with marginal bone marrow sparing and whole thorax lung irradiation. Health Physics. 2020. doi: 10.1097/HP.0000000000001346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flynn DF, Goans RE. Nuclear Terrorism: Triage and Medical Management of Radiation and Combined-Injury Casualties. Surgical Clinics of North America. Surg Clin North Am; 2006. pp. 601–636. doi: 10.1016/j.suc.2006.03.005 [DOI] [PubMed] [Google Scholar]
- 16.Zablotska LB. 30 years After the Chernobyl Nuclear Accident: Time for Reflection and Re-evaluation of Current Disaster Preparedness Plans. J Urban Heal. 2016;93: 407–413. doi: 10.1007/s11524-016-0053-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaafar L, Podolsky RH, Dynan WS. Long-Term Effects of Ionizing Radiation on Gene Expression in a Zebrafish Model. PLoS One. 2013;8. doi: 10.1371/journal.pone.0069445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghandhi SA, Turner HC, Shuryak I, Dugan GO, Daniel Bourland J, Olson JD, et al. Whole thorax irradiation of non-human primates induces persistent nuclear damage and gene expression changes in peripheral blood cells. PLoS One. 2018;13. doi: 10.1371/journal.pone.0191402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Port M, Hérodin F, Valente M, Drouet M, Ostheim P, Majewski M, et al. Persistent mRNA and miRNA expression changes in irradiated baboons. Sci Rep. 2018. doi: 10.1038/s41598-018-33544-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gluzman-Poltorak Z, Vainstein V, Basile LA. Recombinant interleukin-12, but not granulocyte-colony stimulating factor, improves survival in lethally irradiated nonhuman primates in the absence of supportive care: Evidence for the development of a frontline radiation medical countermeasure. Am J Hematol. 2014. doi: 10.1002/ajh.23770 [DOI] [PubMed] [Google Scholar]
- 21.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2009. doi: 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mi H, Muruganujan A, Ebert D, Huang X, Thomas PD. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019;47: D419–D426. doi: 10.1093/nar/gky1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barton MB, Frommer M, Shafiq J. Role of radiotherapy in cancer control in low-income and middle-income countries. Lancet Oncology. Lancet Publishing Group; 2006. pp. 584–595. doi: 10.1016/S1470-2045(06)70759-8 [DOI] [PubMed] [Google Scholar]
- 24.Gasser S, Orsulic S, Brown EJ, Raulet DH. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 2005;436: 1186–1190. doi: 10.1038/nature03884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paul S, Ghandhi SA, Weber W, Doyle-Eisele M, Melo D, Guilmette R, et al. Gene Expression Response of Mice after a Single Dose of 137Cs as an Internal Emitter. Radiat Res. 2014;182: 380–389. doi: 10.1667/RR13466.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Datta K, Hyduke DR, Suman S, Moon BH, Johnson MD, Fornace AJ. Exposure to ionizing radiation induced persistent gene expression changes in mouse mammary gland. Radiat Oncol. 2012. doi: 10.1186/1748-717X-7-205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fält S, Holmberg K, Lambert B, Wennborg A. Long-term global gene expression patterns in irradiated human lymphocytes. Carcinogenesis. 2003. doi: 10.1093/carcin/bgg134 [DOI] [PubMed] [Google Scholar]
- 28.Mosimann C, Hausmann G, Basler K. β-Catenin hits chromatin: Regulation of Wnt target gene activation. Nature Reviews Molecular Cell Biology. Nat Rev Mol Cell Biol; 2009. pp. 276–286. doi: 10.1038/nrm2654 [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Cui H, Tao S, Zeng T, Wu J, Tao Z, et al. High Canonical Wnt/β-Catenin Activity Sensitizes Murine Hematopoietic Stem and Progenitor Cells to DNA Damage. Stem Cell Rev Reports. 2020;16: 212–221. doi: 10.1007/s12015-019-09930-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mah AT, Yan KS, Kuo CJ. Wnt pathway regulation of intestinal stem cells. Journal of Physiology. 2016. doi: 10.1113/JP271754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nie YH, Liu XD, Huang R, Xie DF, Yin WJ, Guan H, et al. Analysis of mRNA Expression Patterns in Peripheral Blood Cells of 3 Patients With Cancer After the First Fraction of 2 Gy Irradiation: An Integrated Case Report and Systematic Review. Dose-Response. SAGE Publications Inc.; 2019. doi: 10.1177/1559325819833474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Port M, Herodin F, Valente M, Drouet M, Lamkowski A, Majewski M, et al. First Generation Gene Expression Signature for Early Prediction of Late Occurring Hematological Acute Radiation Syndrome in Baboons. Radiat Res. 2016;186: 39–54. doi: 10.1667/RR14318.1 [DOI] [PubMed] [Google Scholar]
- 33.Port M, Majewski M, Herodin F, Valente M, Drouet M, Forcheron F, et al. Validating Baboon Ex Vivo and in Vivo Radiation-Related Gene Expression with Corresponding Human Data. Radiat Res. 2018;189: 389–398. doi: 10.1667/RR14958.1 [DOI] [PubMed] [Google Scholar]
- 34.Constine LS, Tarbell N, Hudson MM, Schwartz C, Fisher SG, Muhs AG, et al. Subsequent Malignancies in Children Treated for Hodgkin’s Disease: Associations With Gender and Radiation Dose. Int J Radiat Oncol Biol Phys. 2008;72: 24–33. doi: 10.1016/j.ijrobp.2008.04.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Narendran N, Luzhna L, Kovalchuk O. Sex difference of radiation response in occupational and accidental exposure. Frontiers in Genetics. 2019. doi: 10.3389/fgene.2019.00260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bluhm EC, Ronckers C, Hayashi RJ, Neglia JP, Mertens AC, Stovall M, et al. Cause-specific mortality and second cancer incidence after non-Hodgkin lymphoma: A report from the childhood cancer survivor study. Blood. 2008;111: 4014–4021. doi: 10.1182/blood-2007-08-106021 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Mean fold-changes are presented for animals used for NGS (n = 20) and the remaining animals using qRT-PCR (n = 117). Genes where a successful validation failed are shown in the inserted graph.
(TIFF)
Gene numbers and fold-changes (first number represents male and second number female fold-changes) and sexes are presented.
(XLSX)
Data is sorted by successful inter-species validation (14/41), no inter-species validation (23/41) and inverse inter-species validation (4/41). Successfully validated time points are shown in bold.
(XLSX)
A summary of the currently known function is given. Results for a Pubmed search (http://www.pubmed.gov) for “gene name” + “radiation” are given.
(XLSX)
Data Availability Statement
NGS data is available under DOI: 10.17632/t2b295b4gc.1 https://data.mendeley.com/datasets/t2b295b4gc/2.