Abstract
The spectrum of germline predisposition in pediatric cancer continues to be realized. Here we report 751 solid tumor patients who underwent prospective matched tumor-normal DNA sequencing and downstream clinical use (clinicaltrials.gov NCT01775072). Germline pathogenic and likely pathogenic (P/LP) variants were reported. One or more P/LP variants were found in 18% (138/751) of individuals when including variants in low, moderate, and high penetrance dominant or recessive genes, or 13% (99/751) in moderate and high penetrance dominant genes. 34% of high or moderate penetrance variants were unexpected based on the patient’s diagnosis and previous history. 76% of patients with positive results completed a clinical genetics visit, and 21% had at least one relative undergo cascade testing as a result of this testing. Clinical actionability additionally included screening, risk reduction in relatives, reproductive use, and use of targeted therapies. Germline testing should be considered for all children with cancer.
Introduction
Pediatric cancer is rare, with less than 10,000 solid tumors diagnosed in children annually in the US.[1] Previous studies interrogating germline predisposition broadly across pediatric cancer types have found heritable germline predisposition in 8–12% of patients. The yield of germline predisposition detected is dependent on the genes included for analysis and variant interpretation, as well as the ascertainment biases found in each cohort. Iterative data is required to expand upon the understanding of susceptibility to pediatric cancer and determine the extent to which germline data translates to clinical practice.[2–7]
Certain pediatric cancer diagnoses have well established associations with germline mutations in specific genes and should automatically prompt clinical suspicion for a cancer predisposition, for example retinoblastoma (RB1), pleuropulmonary blastoma (DICER1), optic pathway gliomas (NF1), atypical teratoid/rhabdoid tumors (SMARCB1), small cell hypercalcemic ovarian tumors (SMARCA4), adrenal cortical tumors (TP53), and hypodiploid acute lymphoblastic leukemia (TP53).[8–10] Germline testing can also be critical to distinguishing between conditions like neurofibromatosis type 1 (NF1) and constitutional mismatch repair deficiency (CMMRD), which can be phenocopies of each other. For example, a child presenting with numerous café au lait spots and leukemia may have either of these conditions, but treatment and screening recommendations for the proband and family members will be significantly different depending on the germline diagnosis.[11]
Besides the known associations of causal germline mutations, broad tumor-normal sequencing has revealed many novel associations.[9, 12] While some of these findings likely represent population detection and do not play a role in the pathogenesis of the cancer in question,[13] other novel associations are likely causal. Population detection is expected, especially in large studies, given the frequency of common germline predisposition; BRCA1/2 mutations are found in 2–3% (1 in 40) of individuals of Ashkenazi Jewish ancestry, and Lynch syndrome is detected in approximately 0.3% (1 in 300) of the general population.[14] Even in the case of detecting apparently unrelated germline mutations, this information can be useful for adult family members through cascade testing and initiation of recommended screening and prevention measures for relatives who may not otherwise have come to medical attention.[15, 16] In cases where functional assessment and/or enrichment analysis has supported causality, these findings have resulted in an expansion of the cancer spectrum of known syndromes (i.e. SDHx mutations with neuroblastoma and BRCA2 and PALB2 mutations with medulloblastoma).[9, 12] However, for many observed associations, functional assessment and/or enrichment analysis have not yet been done, and it remains unknown what role, if any, the germline mutation played in tumorigenesis.
Therapeutic options have expanded for certain adult cancer patients with BRCA1, BRCA2, MLH1, MSH2, MSH6, and PMS2 germline mutation driven tumors.[17, 18] In pediatric oncology, the main successes of germline targeted therapies so far have been CMMRD-related tumors responding to immunotherapy, NF1-associated inoperable plexiform neurofibromas responding to selumetinib, and NF1-associated optic pathway gliomas responding to MEK inhibitors.[19–21] The efficacy of other targeted therapies, for example against BRCA1/2-associated pediatric tumors (i.e. subsets of medulloblastoma) remains pre-clinical and theoretical at this time.[22]
We have previously reported that the MSK-IMPACT custom matched tumor-normal sequencing platform can identify a wide spectrum of germline mutations in adult cancer patients and reveal novel genetic associations.[23] Here, we report findings from our pediatric population who underwent testing via the MSK-IMPACT platform, including the prevalence and spectrum of pathogenic/likely pathogenic (P/LP) germline variants in cancer predisposition genes, associations between germline status and somatic profile, and subsequent clinical use of this information.
Results
Patient Characteristics
During the study period, 751 patients had tumor-normal testing with consent for return of germline results. This represents an addition of 44% to the total number of cases of germline data reported from large sequencing studies of pediatric cancer (Table 1). Clinical characteristics of patients are listed in Table 2. Patients were relatively evenly distributed between males and females (54% vs 46%) and had a mean age at diagnosis of 8.2 years. Age at diagnosis and sex were not significantly associated with likelihood of detecting a P/LP variant.
Table 1:
Broad germline sequencing studies of pediatric cancer with >100 patients
| Study | Hematologic | Central nervous system | Solid tumor (non-CNS) | Percent reported with germline P/LP variants in high/moderate penetrance cancer predisposition genes |
|---|---|---|---|---|
| Michigan – Mody et al. JAMA 2015 | 30 | 8 | 64 | 10% |
| St. Jude – Zhang et al. NEJM 2015 | 588 | 245 | 287 | 8% |
| Baylor – Parsons et al. JAMA Oncology 2016 | 56 | 94 | 10% | |
| Columbia – Oberg et al. Genomic Medicine 2016 | 36 | 16 | 49 | 10% |
| Australia – Wong et al. 2020 | 43 | 92 | 112 | 11% |
| MSKCC – present study | 139 | 612 | 13% |
Table 2:
Patient characteristics
| Characteristic | Mutation Carriers, N = 1381 | Non-Mutation Carriers, N = 6131 | Overall, N = 751 | p-value2 |
|---|---|---|---|---|
| Sex | 0.65 | |||
| Female | 67 (49%) | 282 (46%) | 349 (46%) | |
| Male | 71 (51%) | 331 (54%) | 402 (54%) | |
| Age at diagnosis (years) | 7.4 ± 6.8 (0.0–19.0) | 8.4 ± 6.3 (0.0–19.0) | 8.2 ± 6.4 (0.0–19.0) | 0.11 |
| Multiple diagnoses | 0.0001519 | |||
| No | 124 (90%) | 595 (97%) | 719 (96%) | |
| Yes | 14 (10%) | 18 (3%) | 32 (4%) | |
| Deceased | ||||
| No | 117 (85%) | 519 (85%) | 636 (85%) | |
| Yes | 21 (15%) | 94 (15%) | 115 (15%) | |
Statistics presented: n (%); mean ± SD (minimum-maximum)
Statistical tests performed: chi-square test of independence and t-test, all two-sided
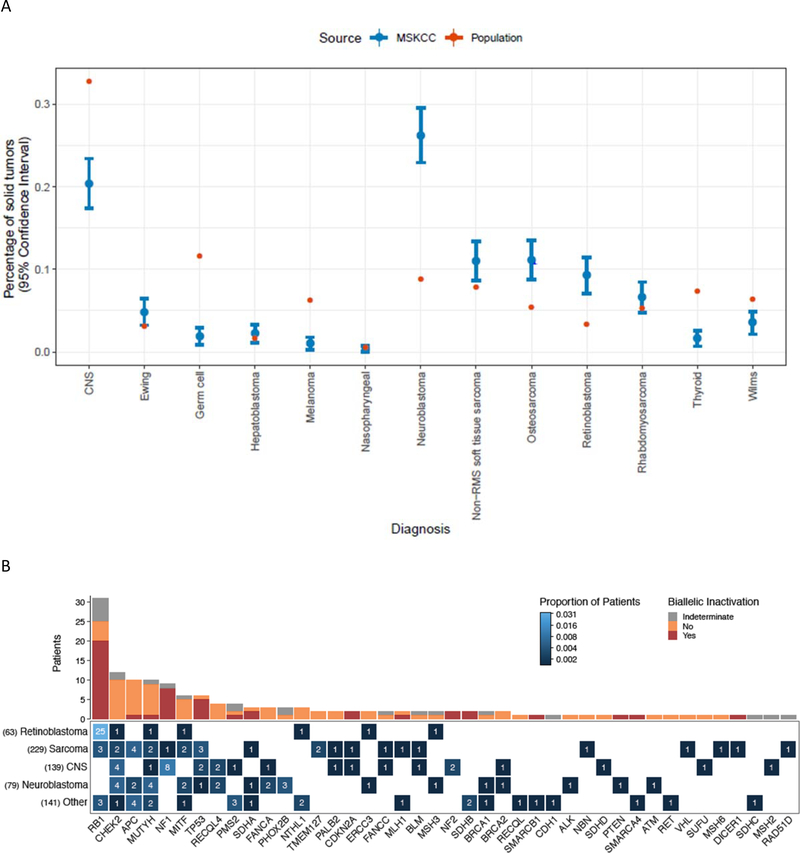
Cancer distribution in our cohort included sarcoma 30% (n=229), neuroblastoma 24% (n=182), central nervous system (CNS) tumors 19% (n=143), retinoblastoma 9% (n=70), and other rare solid tumors 19% (n=143). Thirty-two individuals (4%) had multiple tumors, hence the percentages total >100%. Seven patients in our cohort had leukemia in addition to a solid tumor, either prior to their solid tumor that they were in remission from or that developed after IMPACT was sent for their solid tumor. In comparison with the International Childhood Cancer Consortium distribution of solid tumors, our cohort is widely representative of all major groups, with enrichment of neuroblastoma, retinoblastoma, and osteosarcoma, and relatively few germ cell tumors, CNS tumors, thyroid cancers, and melanomas, which reflects the patient population seen at our hospital (Figure 1A).
Figure 1.
(A) Cancer types represented in the current study (blue) compared to the Annual International Childhood Cancer Classification (AICCC) distribution (red). Current study data is based on n=751 patients and shown with 95% confidence intervals. US census data (https://www.census.gov/quickfacts/fact/table/US/PST0452190) was used with AICCC data to calculate population percentages. (B) Germline P/LP variants and biallelic inactivation across 138 patients in the cohort. Bottom, the heat map shows the occurrence of germline P/LP variants across genes and cancer types. The color indicates the proportion of patients with a particular cancer type and germline alteration, and the number on the tiles indicate the absolute count. The total number of patients in the cohort for each of the cancer types is displayed on the left. Top, the absolute count of patients with biallelic inactivation in the genes shown on the x-axis. Biallelic inactivation includes loss of heterozygosity and second somatic hits. Patients for whom the allele status in tumor could not be assessed due to insufficient tumor purity, unavailability of sufficient tumor tissue, or low sequencing coverage in the tumor are indicated as indeterminate for biallelic inactivation.
Results Reporting
Our process for analyzing and returning germline results involved expertise from Diagnostic Molecular Pathology and the Clinical Genetics Service, with input from a multidisciplinary molecular tumor board when disease or treatment context was important. An example illustrating the value of this approach was in the assessment of the TP53 variant p.Arg196Leu. Based on the existing annotation in ClinVar, this variant was previously classified as a VUS. However, taking into account hotspot data from the cBioPortal database and tumor histology (choroid plexus carcinoma), we used the ClinGen framework for integrating somatic tumor data for germline variant interpretation to consider this variant as LP and return to the family.[28]
Frequency and Spectrum of Germline Variants
A total of 138 patients were positive for one or more P/LP variants (18%) when including low, moderate and high penetrance variants in recessive and dominant genes, or 13% (99/751) when including moderate and high penetrance mutations in dominant genes. P/LP variants were reported in 49% (34/70) of patients with retinoblastoma, 21% (30/143) of patients with central nervous system tumors, 15% (28/182) of patients with neuroblastoma, 12% (28/229) of patients with sarcoma, and 19% (27/143) of patients with other tumor types. Spectrum of variants found in patients with specific disease types is shown in Table 3. All variants were heterozygous with the exception of one individual with a homozygous PMS2 likely pathogenic variant, and two RB1 pathogenic variants that were mosaic. A full description of all patients and variants is in Supplementary Table 1.
Table 3.
Percentage of positive cases by selected tumor types. Genes in which P/LP variants were detected are listed by disease type. Note that some patients had multiple P/LP variants, hence some tumor types have more genes with P/LP variants listed than positive cases.
| Tumor type | Total cases | Positive cases | Genes with P/LP variants |
|---|---|---|---|
| Astrocytoma | 27 | 5 (19%) | BRCA2, SDHD, FANCC, CHEK2, RECQL4 |
| Desmoplastic Small Round Cell Tumor | 16 | 1 (6%) | PALB2 |
| Diffuse Intrinsic Pontine Glioma | 7 | 0 | |
| Ependymoma | 14 | 3 (21%) | NF1, NF2, FANCA |
| Ewing sarcoma | 31 | 5 (16%) | MLH1, CHEK2, TMEM127, MITF, APC I1307K (2) |
| Ganglioglioma | 7 | 1 (14%) | SUFU |
| Gastrointestinal stromal tumor | 4 | 3 (75%) | SDHA, SDHB, SDHC |
| Germ cell tumor | 17 | 2 (12%) | PMS2, APC I1307K |
| Glioblastoma | 13 | 3 (23%) | PMS2 (homozygous), NF1, CHEK2, MUTYH |
| Hepatoblastoma | 4 | 1 (25%) | PMS2 |
| Hepatocellular carcinoma | 11 | 1 (9%) | MUTYH |
| Malignant Peripheral Nerve Sheath Tumor | 7 | 7 (100%) | NF1 (6), MSH2 |
| Medulloblastoma | 14 | 3 (21%) | TP53, CHEK2, RECQL4 |
| Meningioma | 4 | 2 (50%) | NF2, RB1 |
| Neuroblastoma | 182 | 28 (15%) | PHOX2B (3), ALK, TP53, SDHA, |
| BRCA1, BRCA2, ATM, PTEN, MITF (2), CHEK2 (4), FANCA (2), MUTYH (4), RECQL4 (2), MSH3, ERCC3, APC I1307K (2) | |||
| Osteosarcoma | 74 | 7 (9%) | RB1 (3), TP53, RAD51D, CDKN2A, MUTYH |
| Retinoblastoma | 70 | 34 (49%) | RB1 (31), ERCC3, MITF, CHEK2, MUTYH (2), MSH3, NTHL1 |
| Rhabdomyosarcoma | 45 | 9 (20%) | DICER1, TP53, MSH6, VHL, NBN, APC I1307K (2), FANCC |
| Wilms tumor | 25 | 2 (8%) | APC I1307K, MUTYH |
The most common P/LP variants in high or moderate penetrance genes were in RB1 (n=31, 4%), NF1 (n=9, 1%), and TP53 (n=6, 1%) in patients with retinoblastoma, previous clinical diagnoses of NF1, and Li Fraumeni syndrome (LFS) associated tumors, respectively (Table 3). Moderate or high penetrance P/LP variants were also detected in DNA-damage repair genes n=24, 3.2% of patients (ATM n=1, BRCA1 n=2, BRCA2 n=2, CDKN2A n=2, CHEK2 n=6, ERCC3 n=2, MLH1 n=2, MSH2 n=1, MSH6 n=1, PMS2 n=4, RAD51D n=1), the RAS/MEK or mTOR/PTEN pathway n=12, 1.6% of patients (NF1 n=9, NF2 n=2, PTEN n=1), and metabolic pathways related to cancer n=8, 1.1% of patients (SDHA n=3, SDHB n=2, SDHC n=1, SDHD n=1, VHL n=1). Seven individuals (0.9%) harbored a second P/LP variant in a different cancer predisposition gene. One individual was found to have two variants in FANCC. Her phenotype was not consistent with Fanconi anemia (tumor type was a pilocytic astrocytoma, no extreme reaction to chemotherapy), but she did not complete a Clinical Genetics visit, so it was not determined if these variants are in cis or trans.
34% (34/101) of high/moderate penetrance variants were unexpected based on the patient’s diagnosis and previous history, or 50% (35/70) when excluding RB1 P/LP variants (Supplementary Table 1). Individuals who tested positive for a P/LP variant were more likely than those who tested negative to have had multiple primary cancer diagnoses (10% vs 3%, p<0.001, Table 2). Of 14 mutation carriers with multiple diagnoses, 7 individuals (50%) had multiple cancers associated with their germline mutation (hereditary retinoblastoma, NF1, and NF2), and 2 were known to be therapy-related, including one child with LFS who developed a therapy-related leukemia. Family history of childhood cancer was reported in 13% (14/105) of individuals in our cohort with P/LP variants who completed a Clinical Genetics visit.
Correlation Between Germline Genotype and Tumor Phenotype
We were able to evaluate LOH status for 120 P/LP variants, including 80 high or moderate penetrance variants. Variants were considered associated with biallelic loss in the tumor if LOH or a second somatic hit was present, or in the case of the germline homozygous LP PMS2 variant. Germline P/LP variants in moderate or high penetrance genes were associated with biallelic loss in 55% (44/80) (Figures 1B and 2A, Supplementary Table 1). Of 53 germline P/LP variants with somatic data available that were considered expected based on the patient’s tumor type or previous history, 43 (81%) had either LOH, a second somatic variant, or germline biallelic loss. In comparison, 27 high/moderate penetrance germline P/LP variants with somatic data available were considered unexpected, and just 2 (7%) of these were associated with LOH (both CDKN2A variants, one in a case of osteosarcoma and one in a low-grade neuroepithelial tumor).
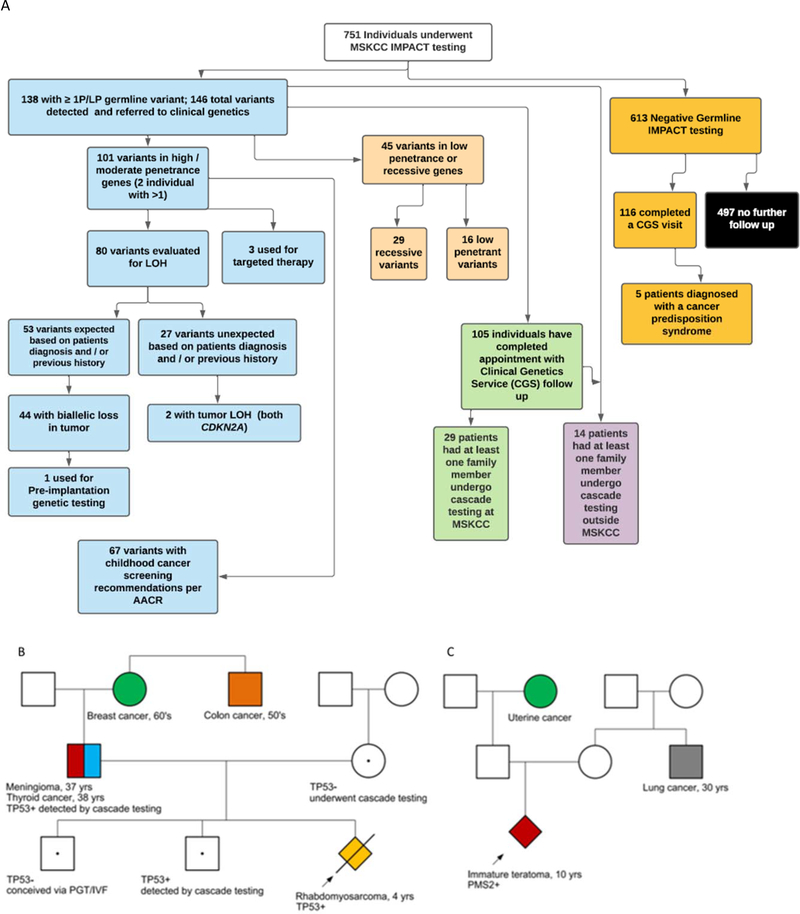
Figure 2:
(A) Universal germline sequencing in 751 patients undergoing tumor-normal sequencing using MSK-IMPACT with tumor status of alternate allele if available, screening recommendations when indicated, cascade testing in relatives and examples of clinical translation. One family reported back undergoing IVF/PGT which resulted in a pregnancy and birth of a child unaffected with LFS. While using germline results in this manner was counseled on by the Clinical Genetics team, the number of families using this information for reproductive purposes was not systematically followed.
Examples of clinical use of germline results. (B) A proband with embryonal rhabdomyosarcoma was detected to have a germline TP53 mutation which was subsequently found in her father through cascade testing. He was found through screening to have thyroid cancer and a meningioma. (C) A proband with a teratoma was found to have a germline PMS2 mutation and was started on immunotherapy. Cascade testing has not been performed in this family.
Clinical Genetics Follow-Up
All 138 patients with positive results were referred to the Clinical Genetics Service, and 105 (76%) have completed an appointment to date (Figure 2A). Reasons for declining an appointment, when given, included previous knowledge and counseling on the mutation identified, declining patient status/death, or being overwhelmed by disease/treatment and wishing to defer discussion until completion of therapy. Appointments were conducted in person (or via telemedicine during COVID-19) for all autosomal dominant mutations and by patient/clinician preference, or by phone for carrier status of autosomal recessive mutations. Additionally, 116/613 (19%) patients with negative results completed a visit with the Clinical Genetics Service. Five patients (including two sisters) with negative IMPACT testing were diagnosed with a cancer predisposition from additional testing (Figure 2A).
Utility of Germline Results
Cascade testing was completed as a result of IMPACT testing in at least one family member in 29 of 138 individuals (21%) with a P/LP variant detected (Figure 2A). This resulted in the detection of 27 mutation carriers who were not previously aware of their status. In 14 additional cases (10%, all of which were high/moderate penetrance genes), there was documentation in the chart that family member testing had previously taken place, either because the variant detected on IMPACT was previously detected with initiation of cascade testing, or because the family members had previously been referred for genetic testing due to their own personal and/or family history of cancer. It should be noted that this number only reflects cascade testing that was completed through our clinic, or done at an outside institution with records provided. Many families were provided with contact information for local genetic counseling services, and may have pursued testing elsewhere.
Likelihood to undergo cascade testing was associated with penetrance of mutation detected and varied by gene; 37/99 (37%) patients with a high/moderate penetrance variant had or had previously had at least one family member undergo cascade testing compared to 6/39 (15%) of patients who only had a low penetrance or autosomal recessive variant (p = 0.01). All six patients with germline TP53 P/LP variants were found to be inherited from a parent, half of which were detected following IMPACT testing and half of which were previously known to the family. Cascade testing was completed less often in the families of probands identified to have P/LP variants in RB1 (7/31 cases; 23%) and NF1 (2/9 cases; 22%). Six patients were found to have expected de novo mutations (confirmation of paternity was not performed), in RB1 (two cases), PHOX2B, SMARCA4, NF1, and, surprisingly, MLH1.
Patients identified to have mutations predisposing to cancer in their age range were referred to our surveillance clinic, or additional screening was discussed with the primary attending if the patient was actively being treated. 67 patients had mutations associated with risk of cancer in childhood and thus screening recommendations in childhood (although 36 of these were variants in RB1 in individuals with bilateral retinoblastoma or variants in NF1 in individuals meeting clinical criteria, hence not changing recommendations)[33, 34]. Family members identified to carry P/LP variants through cascade testing were similarly referred. This resulted in the detection of multiple tumors. A patient identified to have a germline SDHD mutation was found to have a carotid body paraganglioma on screening MRI. In a family identified to have a TP53 mutation following the proband’s diagnosis of rhabdomyosarcoma, the father was subsequently diagnosed with thyroid carcinoma and meningioma on whole-body MRI (Figure 2B). Additionally, in one family without a prior cancer predisposition diagnosis, identifying the germline TP53 mutation led to the birth of an unaffected child who was conceived via in vitro fertilization (IVF) and pre-implantation testing (PGT).
Seven patients (0.9%) were identified in our cohort with a heterozygous P/LP variant in a mismatch repair (MMR) gene (MLH1, MSH2, MSH6, and PMS2), which is significantly higher than the population frequency previously reported in the Healthy Nevada Project of 0.03% (p=0.0095) (Supplementary Table 4).[14] In addition, one patient with a glioblastoma had a homozygous LP variant in PMS2 diagnostic of CMMRD. Her tumor was MSI-H and had a very high tumor mutational burden of 243 mutations per megabase, consistent with CMMRD-related disease. She was treated with pembrolizumab, but unfortunately progressed while on this and died. Microsatellite analysis was available for 6 of the 7 patients with heterozygous mutations. One patient with colon adenocarcinoma had an MSI-H tumor, while the other 5 were microsatellite-stable (MSS) (none of these five tumors are associated with Lynch syndrome). Four patients with heterozygous mutations in the MMR genes had advanced disease. Two of these patients with advanced disease were prescribed immunotherapy based on their germline finding. One patient with an immature teratoma that had developed into a mature teratoma with glioma and spindle cell sarcoma and a PMS2 mutation exhibited a partial response to immunotherapy (Figure 2C). Another patient with a de novo MLH1 mutation and Ewing sarcoma died of disease.
Discussion
Cancer predisposition is found in high or moderate penetrance genes in 8–13% of patients with pediatric cancer according to this and multiple previous pan-pediatric cancer studies.[2, 3, 5, 6] As with previous studies, our study has an ascertainment bias based on the diagnoses of patients presenting to our center (Figure 1A), and these numbers do not represent a truly unselected pediatric cancer cohort. The clinical utility of detecting a germline predisposition will depend on the individual patient and variant found, but has not previously been studied in a systematic way across pediatric cancer. Our cohort underwent prospective CLIA-validated in-house testing enabling clinical actionability through our multidisciplinary molecular tumor board, Clinical Genetics Service, and Pediatric Surveillance Clinic.
Even in the context of large pan-cancer NGS panels, it is important to recognize limitations of the assay, and to recognize which patients warrant additional testing. Many patients with negative results on MSK-IMPACT were seen by Clinical Genetics for this reason; in fact, more patients with negative results (116) were seen than patients with positive results (105). NGS-based assays are not designed to detect imprinting disorders or disorders of telomere biology in patients without a detectable mutation in a gene included on the panel.[8] Four patients in our cohort with negative IMPACT had epimutations associated with Beckwith-Wiedemann syndrome, three of whom did not have prior diagnoses of this condition (as previously reported).[35] Certain genetic changes will also be missed by panel-based testing, such as balanced translocations, some copy number alterations, and repeat expansions (such as in PHOX2B). Genes with known pathogenic mutations in non-exonic regions (promoter or deep intronic mutations) need to have these regions specifically targeted in order to detect them. For example, one patient in our cohort with a clinical diagnosis of NF1 but whose IMPACT testing only showed a germline CHEK2 mutation underwent additional clinical testing of NF1 which revealed the deep intronic splicing pathogenic variant c.5749+332A>G. In some cases, lower-level mosaicism may be detected by a clinical test developed to have particularly high sequencing depth of one particular gene. Two patients (twin sisters) with juvenile xanthogranulomas in our cohort with negative IMPACT testing were found to be mosaic for an NF1 pathogenic variant that was below the level of detection of IMPACT. Knowledge of clinically relevant limitations for an individual patient is critical to guide appropriate follow-up testing, when needed, to maximize detection of predisposition.
Some limitations of NGS panels may be addressed by incorporating additional technologies into the clinical workflow. RNA-based testing has recently been shown to both detect pathogenic variants that would be missed altogether by DNA sequencing, and to help clarify the pathogenicity of VUS’s in a substantial portion of cases.[36, 37]
Recommendations for germline testing of pediatric cancer patients are often tumor-specific, or may take additional factors such as family history into account (i.e., Chompret criteria for LFS).[38] However, consistent with previous studies[23], we found a substantial number of patients with high/moderate penetrance germline P/LP variants in genes not expected given their cancer type or previous history. The role of these mutations in tumorigenesis is often not known, but some evidence for or against causality may be inferred from the patient’s accompanying tumor data (i.e., MSI status, second somatic hit, LOH, tumor signature, etc.), or from further study (i.e., functional studies or enrichment studies). Our finding of two unexpected CDKN2A variants associated with tumor LOH is interesting and warrants further study to determine if these rare tumor types represent an expansion of the phenotype, especially in the case of osteosarcoma as an enrichment of CDKN2A germline mutations in this disease type has previously been reported.[39] These results underscore the importance of assessing tumor LOH, which may help point towards novel causal associations in the case of unexpected germline findings.
On a broader scale, enrichment data may suggest a causal relationship, such as our finding of more Lynch syndrome (heterozygous MMR P/LP variants) than would be expected as a population frequency. However, much further study of this potential association in additional cohorts is warranted, especially given that the tumors here were MSS, with the exception of the colorectal cancer. More robust data on the response of such patients to immunotherapy may also guide our understanding of the significance of these unexpected findings in pediatric cancers.
Targeted therapy informed by germline data has resulted in rare clinical responses in pediatric cancer (e.g. CMMRD). However, the clinical utility of germline data informing therapy requires much further exploration. Data from adult studies of targeted therapies for germline mutations has shown somewhat improved clinical outcomes in a larger number of cases.[18] It is very likely that the list of germline mutations associated with clinical trial eligibility and approved therapies will continue to grow. In addition to targeted therapies, germline data may also be useful in predicting which patients will experience increased therapeutic toxicity and may benefit from reduced dosing. In the past this has been mainly considered in patients with rare bone marrow failure syndromes (i.e. Fanconi anemia and dyskeratosis congenita), but recently certain heterozygous germline mutations in DNA repair genes (including in genes only associated with autosomal recessive disease) have been shown to be associated with increased risk of secondary cancers following radiation therapy or alkylating chemotherapy.[40] If this association continues to be supported in further studies, there could be increasing clinical relevance to the patient themselves when detecting carrier status for certain autosomal recessive genes in terms of dosing considerations.
Additionally, cascade testing in families can multiply the benefits of detecting a germline mutation. Surveillance, risk reduction, and reproductive planning can become available to family members in addition to the proband.[15, 16] Despite the high pre-test probability of detecting a P/LP variant, the relatively low cost, and the straightforward interpretation (without variants of uncertain significance), uptake of cascade testing in family members at risk of a cancer predisposition has typically been suboptimal. Previous studies in adult cancer predisposition have generally found that approximately half of at-risk first-degree relatives undergo cascade testing[41], with more recent initiatives investigating practices to identify and reduce burdens on family members in order to increase uptake.[16, 42–44] Uptake in our cohort was similarly low, although variable based on penetrance of gene as well as the gene itself (i.e., a low cascade testing rate for NF1 is unsurprising given that the diagnosis, or absence of a diagnosis, can typically be made clinically in relatives).
An important consideration of germline testing in pediatric cancer is cost-effectiveness, which has yet to be rigorously studied in this area. While the hope is that appropriate germline testing can be clinically utilized in a way that is overall cost-effective to the larger system, evaluating this will take much more long-term follow up of patients than we have performed in this study. Potential cost savings exist through cancer screening and early detection, prevention, pre-implantation genetic testing, and potentially more effective therapeutics; however, there are also significant costs associated with each of these, in addition to the costs of sequencing and clinical genetics visits. Studies of cost-effectiveness in this area to date have focused on individual pieces of this process, for example cancer screening in patients with Li Fraumeni syndrome.[45] The full analysis of the effects of pan-pediatric cancer germline sequencing will require long-term follow-up, as costs may be largely up front (i.e., sequencing), while cost savings may not be realized for many years (i.e., early tumor detection associated with need for less treatment).
While disease and family history-based testing guidelines have utility in detecting a portion of children with underlying predisposition, it is critical to recognize that a significant proportion of germline mutations will not be predicted based on these guidelines and will be missed if testing is restricted to only those meeting criteria. The potential for cancer predisposition should be considered for every child with cancer. Outside of guidelines-based testing, the potential additional utility of broader germline testing needs to be considered in the context of the patient’s clinical condition (i.e., potential to enroll on a clinical trial if certain mutations were detected) as well as the patient and family’s desire for information that could primarily be used for other family members in the immediate future (i.e., the finding of a BRCA1/2 mutation in a young child). Broad pan-cancer predisposition testing is a key part of research to appreciate the full spectrum associated with many germline mutations, but it also has increasing clinical utility for many patients.
Methods
Patient Cohort
From July 2015 to July 2020, pediatric patients with solid tumors seen at Memorial Sloan Kettering Cancer Center (MSKCC) were offered matched tumor-normal DNA sequencing under an institutional protocol (ClinicalTrials.gov identifier: NCT01775072). Patients with sufficient tumor tissue available were offered enrollment at physician discretion and were shown a pre-test video describing the testing, or were seen by Clinical Genetics prior to having testing sent. Appropriate consents were obtained.
Patients were ascertained for this cohort based being ≤19 years old at diagnosis with the tumor that was used for sequencing. Clinical characteristics were collected from institutional electronic medical records. This study was approved by the MSKCC Institutional Review Board/Privacy Board.
Sequencing, Variant Interpretation, and Referral to Clinical Genetics
Tumor and blood samples were obtained and sequenced using the MSK-IMPACT platform, a capture-based next-generation sequencing (NGS) assay capable of identifying sequence mutations, copy number alterations, and select gene fusions in 468 genes (341 and 410 genes in earlier versions), as described previously.[24, 25] Germline data was analyzed in 88 genes (76 in the first version) (Supplemental Tables 2–3).
Variants were identified as described previously.[24, 26] Germline variants were interpreted based on American College of Medical Genetics and Genomics (ACMG) guidelines by a clinical molecular geneticist or molecular pathologist.[27] When applicable, somatic data was utilized in accordance with ClinGen’s framework for integrating somatic data for germline variant curation in cancer predisposition genes.[28] P/LP variants were reported, while variants of unknown significance (VUS) were not.
Variants classified as uncertain significance but with a high suspicion for pathogenicity were reviewed as part of a multidisciplinary molecular tumor board including molecular geneticists, molecular pathologists, clinical geneticists, oncologists, and genetic counselors. In certain cases, after careful review of all molecular and clinical data, special interpretation was given to highly suspicious variants of uncertain significance, which were treated as likely pathogenic and thus returned to these individuals with clinical recommendations.
Results were disclosed to the patient/parent by the ordering clinician. Patients with germline P/LP findings were referred for genetic counseling through the Clinical Genetics Service. Cascade testing was offered to family members of the proband as appropriate. Patients with negative results were referred to the Clinical Genetics Service at the discretion of the primary treating physician. Consultation was requested when there was suspicion of a predisposition condition that would not be detected via MSK-IMPACT (e.g., Beckwith-Wiedemann syndrome), desire for clinical-grade testing of a particular gene or genes (to obtain VUS’s or known P/LP variants that would not be detected on MSK-IMPACT), concerning family history of cancer, or based on the family’s desire for genetic counseling. Additional clinical testing was sent if deemed appropriate by the Clinical Genetics Service and desired by the patient/family.
Statistics & Reproducibility
No statistical method was used to predetermine sample size. No data were excluded from the analyses, and the experiment was not randomized.
According to known disease risks and prior modeling, P/LP variants were classified at the gene level as high penetrance (relative risk [RR] of disease > 4), moderate penetrance (RR 2–4), low penetrance (RR < 2), or they were associated with autosomal recessive inheritance.[29] For CHEK2 and APC, penetrance classification was performed on the variant level: APC p.Ile1307Lys and CHEK2 p.Ile157Thr were considered low penetrance; if detected, other APC mutations were considered high penetrance and other CHEK2 mutations were considered moderate penetrance.[23, 29]
Somatic mutation data was also reported and considered to assess causality of identified germline variants. Estimates of tumor purity and somatic zygosity of all germline variants were evaluated using the FACETS algorithm (version 0.5.12) as described previously.[30, 31] Tumor purity was estimated using mutant allele fractions or pathologist curation for cases where FACETS was unable to estimate a purity. Biallelic inactivation was evaluated by inferring loss of heterozygosity (LOH), defined as a loss of the wild-type allele in the tumor at the locus of a germline mutation. In the absence of LOH in tumor, germline pathogenic variants were also considered biallelically inactivated if a second clonal or subclonal truncating mutation was observed in the same gene in the tumor, or in the case of germline biallelic loss. Biallelic inactivation was considered indeterminate if 1) tumor tissue was not available for zygosity assessment; 2) tumor zygosity could not be evaluated because of germline loss of heterozygosity; 3) tumor purity could neither be estimated by FACETS nor be estimated based on variant allele fraction or pathology curation; 4) read depth in the specimen at the locus of interest was less than 50; or 5) allele-specific copy number could not be evaluated by FACETS. Microsatellite instability-high (MSI-H) was defined as evidence of microsatellite instability at more than 10% of analyzed loci.[32]
P/LP variants were considered expected or unexpected based on the patient’s current diagnosis as well as any previous history (i.e., history of previous tumors or of previous diagnosis of the germline predisposition detected). Family history was collected for patients who were seen by the Clinical Genetics Service.
The distribution of the age at diagnosis, sex, and presence of multiple diagnoses were compared between mutation carriers and non-mutation carriers using chi-square test of independence and t-test. Cascade testing rates were compared between genes with moderate/high penetrance and genes with low penetrance or autosomal recessive inheritance using Fisher’s exact test. The frequency of Lynch syndrome detected in our cohort was compared to the population frequency previously reported in the Healthy Nevada Project[14] using Fisher’s exact test.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data Availability
Deidentified clinical and molecular data for all patients reported in this study are available in the Supplementary Table. P/LP variants reported here have been submitted to ClinVar with submission number SUB8689141.
Supplementary Material
Acknowledgments and Funding:
This work was funded in part by the Robert and Kate Niehaus Center for Inherited Cancer Genomics, the Pediatric Translational Medicine Program, the Marie-Josée and Henry R. Kravis Center for Molecular Oncology, and the National Cancer Institute Cancer Center Core Grant No. P30-CA008748.
Competing Interests Statement:
PAM- Takeda: uncompensated speaking, Amgen: stock ownership, Ipsen: mock ODAC panel, Salarius: mock ODAC panel; PAM’s spouse-Boehringer Ingelheim: consulting, Genentech: speakers bureau, Eastern Pulmonary Society. NKC holds ownership interest/equity in Y-mAbs Therapeutics Inc, reports receiving commercial research grants from Y-mAbs Therapeutics, Inc. and Abpro-Labs, Inc., holding ownership interest/equity in Abpro-Labs, and owning stock options in Eureka Therapeutics, Inc. NKC is a scientific advisory board member of Abpro Labs and Eureka Therapeutics. NKC is the inventor and owner of issued patents licensed by MSK to Y-mAbs Therapeutics, Biotec Pharmacon, and Abpro Labs. IJD has done consulting and advisory board activities with: Apexigen, Astra-Zeneca, Bayer, Bristol-Myers Squibb/Celgene, Fennec, Roche. IJD has served as institutional PI for pharma sponsored clinical trials at MSKCC from Bristol-Myers Squibb, Genentech, and Novartis. MK has consultant agreements with AstraZeneca, Bayer, CereXis, QED Therapeutics and Recursion Pharma (personal fees received). SM is a consultant to YmAbs Therapeutics Inc., Illumina Inc., and Progenics Pharmaceuticals. ZKS’s immediate family member serves as a consultant for Allergan, Adverum Biotechnologies, Alimera Sciences, Fortress Biotech, Genentech/Roche, Novartis, Optos, Regeneron, Regenxbio, and Spark Therapeutics.
References
- 1.Howlader N, et al. , Cronin KA (eds). SEER Cancer Statistics Review, 1975–2014, National Cancer Institute. Bethesda, MD. 2017. [Google Scholar]
- 2.Zhang J, et al. , Germline Mutations in Predisposition Genes in Pediatric Cancer. N Engl J Med, 2015. 373(24): p. 2336–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mody RJ, et al. , Integrative Clinical Sequencing in the Management of Refractory or Relapsed Cancer in Youth. JAMA, 2015. 314(9): p. 913–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harris MH, et al. , Multicenter Feasibility Study of Tumor Molecular Profiling to Inform Therapeutic Decisions in Advanced Pediatric Solid Tumors: The Individualized Cancer Therapy (iCat) Study. JAMA Oncol, 2016. 2(5): p. 608–615. [DOI] [PubMed] [Google Scholar]
- 5.Oberg JA, et al. , Implementation of next generation sequencing into pediatric hematology-oncology practice: moving beyond actionable alterations. Genome Med, 2016. 8(1): p. 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parsons DW, et al. , Diagnostic Yield of Clinical Tumor and Germline Whole-Exome Sequencing for Children With Solid Tumors. JAMA Oncol, 2016. 2(5): p. 616–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wong M, et al. , Whole genome, transcriptome and methylome profiling enhances actionable target discovery in high-risk pediatric cancer. Nat Med, 2020. [DOI] [PubMed] [Google Scholar]
- 8.Walsh M, Cadoo K, Salo-Mullins EE, Dubard-Gault Marianne, Stadler ZS, Offit K, Abeloff’s Clinical Oncology. 2020(Sixth edtiion): p. 180–208. [Google Scholar]
- 9.Holmfeldt L, et al. , The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet, 2013. 45(3): p. 242–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scollon S, et al. , A Comprehensive Review of Pediatric Tumors and Associated Cancer Predisposition Syndromes. J Genet Couns, 2017. 26(3): p. 387–434. [DOI] [PubMed] [Google Scholar]
- 11.Suerink M, et al. , Constitutional mismatch repair deficiency as a differential diagnosis of neurofibromatosis type 1: consensus guidelines for testing a child without malignancy. J Med Genet, 2019. 56(2): p. 53–62. [DOI] [PubMed] [Google Scholar]
- 12.Gault MD, et al. , Germline SDHA mutations in children and adults with cancer. Cold Spring Harbor Molecular Case Studies, 2018. 4(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oak N, et al. , Ancestry-specific predisposing germline variants in cancer. Genome Med, 2020. 12(1): p. 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grzymski JJ, et al. , Population genetic screening efficiently identifies carriers of autosomal dominant diseases. Nat Med, 2020. 26(8): p. 1235–1239. [DOI] [PubMed] [Google Scholar]
- 15.Walsh MF, et al. , Germline BRCA2 mutations detected in pediatric sequencing studies impact parents’ evaluation and care. Cold Spring Harb Mol Case Stud, 2017. 3(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Offit K, et al. , Cascading After Peridiagnostic Cancer Genetic Testing: An Alternative to Population-Based Screening. J Clin Oncol, 2020: p. JCO1902010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le DT, et al. , PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N Engl J Med, 2015. 372(26): p. 2509–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore K, et al. , Maintenance Olaparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N Engl J Med, 2018. 379(26): p. 2495–2505. [DOI] [PubMed] [Google Scholar]
- 19.Bouffet E, et al. , Immune Checkpoint Inhibition for Hypermutant Glioblastoma Multiforme Resulting From Germline Biallelic Mismatch Repair Deficiency. J Clin Oncol, 2016. 34(19): p. 2206–11. [DOI] [PubMed] [Google Scholar]
- 20.Gross AM, et al. , Selumetinib in Children with Inoperable Plexiform Neurofibromas. N Engl J Med, 2020. 382(15): p. 1430–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fangusaro J, et al. , Selumetinib in paediatric patients with BRAF-aberrant or neurofibromatosis type 1-associated recurrent, refractory, or progressive low-grade glioma: a multicentre, phase 2 trial. Lancet Oncol, 2019. 20(7): p. 1011–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Vuurden DG, et al. , PARP inhibition sensitizes childhood high grade glioma, medulloblastoma and ependymoma to radiation. Oncotarget, 2011. 2(12): p. 984–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandelker D, et al. , Mutation Detection in Patients With Advanced Cancer by Universal Sequencing of Cancer-Related Genes in Tumor and Normal DNA vs Guideline-Based Germline Testing. JAMA, 2017. 318(9): p. 825–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng DT, et al. , Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J Mol Diagn, 2015. 17(3): p. 251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zehir A, et al. , Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med, 2017. 23(6): p. 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng DT, et al. , Comprehensive detection of germline variants by MSK-IMPACT, a clinical diagnostic platform for solid tumor molecular oncology and concurrent cancer predisposition testing. BMC Med Genomics, 2017. 10(1): p. 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Richards S, et al. , Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med, 2015. 17(5): p. 405–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walsh MF, et al. , Integrating somatic variant data and biomarkers for germline variant classification in cancer predisposition genes. Hum Mutat, 2018. 39(11): p. 1542–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tung N, et al. , Counselling framework for moderate-penetrance cancer-susceptibility mutations. Nat Rev Clin Oncol, 2016. 13(9): p. 581–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen R and Seshan VE, FACETS: allele-specific copy number and clonal heterogeneity analysis tool for high-throughput DNA sequencing. Nucleic Acids Res, 2016. 44(16): p. e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jonsson P, et al. , Tumour lineage shapes BRCA-mediated phenotypes. Nature, 2019. 571(7766): p. 576–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niu B, et al. , MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics, 2014. 30(7): p. 1015–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kratz CP, et al. , Cancer Screening Recommendations for Individuals with Li-Fraumeni Syndrome. Clin Cancer Res, 2017. 23(11): p. e38–e45. [DOI] [PubMed] [Google Scholar]
- 34.Rednam SP, et al. , Von Hippel-Lindau and Hereditary Pheochromocytoma/Paraganglioma Syndromes: Clinical Features, Genetics, and Surveillance Recommendations in Childhood. Clin Cancer Res, 2017. 23(12): p. e68–e75. [DOI] [PubMed] [Google Scholar]
- 35.Fiala EM, et al. , 11p15.5 epimutations in children with Wilms tumor and hepatoblastoma detected in peripheral blood. Cancer, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Benayed R, et al. , High Yield of RNA Sequencing for Targetable Kinase Fusions in Lung Adenocarcinomas with No Mitogenic Driver Alteration Detected by DNA Sequencing and Low Tumor Mutation Burden. Clin Cancer Res, 2019. 25(15): p. 4712–4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karam R, et al. , Assessment of Diagnostic Outcomes of RNA Genetic Testing for Hereditary Cancer. JAMA Netw Open, 2019. 2(10): p. e1913900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Knapke S, et al. , Hereditary cancer risk assessment in a pediatric oncology follow-up clinic. Pediatr Blood Cancer, 2012. 58(1): p. 85–9. [DOI] [PubMed] [Google Scholar]
- 39.Mirabello L, et al. , Frequency of Pathogenic Germline Variants in Cancer-Susceptibility Genes in Patients With Osteosarcoma. JAMA Oncol, 2020. 6(5): p. 724–734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qin N, et al. , Pathogenic Germline Mutations in DNA Repair Genes in Combination With Cancer Treatment Exposures and Risk of Subsequent Neoplasms Among Long-Term Survivors of Childhood Cancer. Journal of Clinical Oncology, 2020. 38(24): p. 2728–2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sharaf RN, et al. , Uptake of genetic testing by relatives of lynch syndrome probands: a systematic review. Clin Gastroenterol Hepatol, 2013. 11(9): p. 1093–100. [DOI] [PubMed] [Google Scholar]
- 42.Courtney E, et al. , Impact of free cancer predisposition cascade genetic testing on uptake in Singapore. NPJ Genom Med, 2019. 4: p. 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frey MK, et al. , Prospective Feasibility Trial of a Novel Strategy of Facilitated Cascade Genetic Testing Using Telephone Counseling. Journal of Clinical Oncology, 2020. 38(13): p. 1389–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Srinivasan S, et al. , Stakeholder Perspectives on Overcoming Barriers to Cascade Testing in Lynch Syndrome: A Qualitative Study. Cancer Prev Res (Phila), 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tak CR, et al. , Cost-effectiveness of early cancer surveillance for patients with Li-Fraumeni syndrome. Pediatr Blood Cancer, 2019. 66(5): p. e27629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Deidentified clinical and molecular data for all patients reported in this study are available in the Supplementary Table. P/LP variants reported here have been submitted to ClinVar with submission number SUB8689141.