Abstract
Cancer progression is dependent on heightened mechanical adaptation, both for the cells’ ability to change shape and to interact with varying mechanical environments. This type of adaptation is dependent on mechanoresponsive proteins that sense and respond to mechanical stress, as well as their regulators. Mechanoresponsive proteins are part of the mechanobiome, which is the larger network that constitutes the cell’s mechanical systems that are also highly integrated with many other cellular systems, such as gene expression, metabolism, and signaling. Despite the altered expression patterns of key mechanobiome proteins across many different cancer types, pharmaceutical targeting of these proteins has been overlooked. Here, we review the biochemistry of key mechanoresponsive proteins, specifically nonmuscle myosin II, α-actinins, and filamins, as well as the partnering proteins 14-3-3 and CLP36. We also examined a wide range of data sets to assess how gene and protein expression levels of these proteins are altered across many different cancer types. Finally, we determined the potential of targeting these proteins to mitigate invasion or metastasis and suggest that the mechanobiome is a goldmine of opportunity for anticancer drug discovery and development.
Keywords: α-actinin, filamin, mechanoresponse, metastasis, myosin
INTRODUCTION
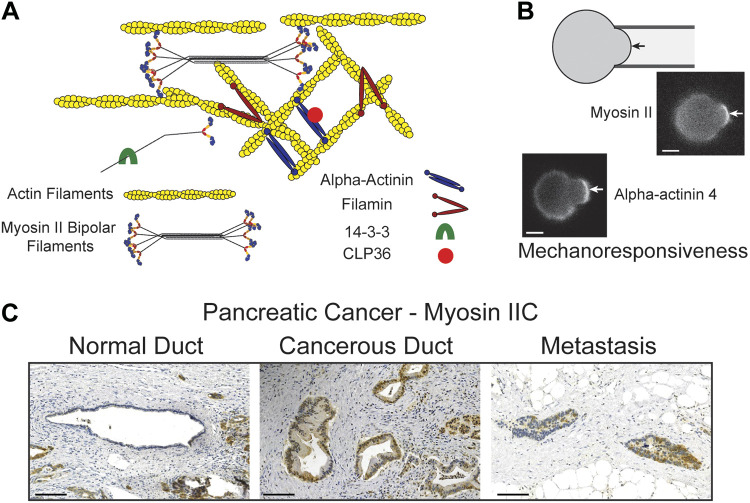
Cancer is fundamentally a disease of altered mechanics. Every step—from proliferation and growth of the original tumor to dissemination and intravasation for new metastatic niches—depends on evolving internal mechanical machinery. As with healthy cells, cancer cells must be able to integrate chemical and physical signals from their external environments. But, unlike their healthy counterparts, the survival of cancer cells is contingent upon their ability to adapt to mechanically distinct microenvironments. This adaptation requires the mechanobiome, which includes the collection of proteins that are uniquely poised to respond to mechanical stresses as well as other proteins, which help set the mechanical and force-producing activities of the cell. The full mechanobiome then constitutes a large integrated network that couples these mechanical systems with a host of other cellular systems, including gene expression, cell signaling, and metabolism, among others (1, 2). These mechanoresponsive proteins, defined as those that accumulate in response to applied mechanical stresses, include the force-generating motor protein nonmuscle myosin II and specific paralogs of the actin-crosslinking proteins α-actinin (ACTN) and filamin (1, 3) (Fig. 1, A and B). Many of these proteins have partnering proteins, such as α-actinin’s CLP36 and the 14-3-3 proteins. Both CLP36 and 14-3-3 s undergo significant expression changes during cancer development (Fig. 1A).
Figure 1.
Mechanoresponsive proteins provide cell structure and adaptability to mechanical stresses, and their expression levels are frequently elevated in cancer progression. A: mechanoresponsive proteins include paralogs of nonmuscle myosin II, α-actinin, and filamin. Myosin II assembles into bipolar filaments, which are then organized into the actin meshworks and stress fibers in the cells. α-Actinins are antiparallel dimers, which organize actin filaments into bundles. Filamins on the other hand are v-shaped dimers that also crosslink actin filaments. 14-3-3 proteins can bind to myosin II tails regions where they modulate myosin II bipolar filament assembly, in addition to other biochemical functions. CLP36 binds to α-actinin and associates with actin-rich structures including stress fibers and the cell cortex. B: mechanoresponsiveness (defined as the ability to accumulate locally in response to applied mechanical stresses) is well revealed by micropipette aspiration (3, 4). The protein accumulates in the region of the cortex deformed by the suction pressure (arrow), increasing in intensity relative to the opposing cortex. Scale bars, 7 µm. C: in pancreatic ductal adenocarcinoma, many mechanoresponsive proteins elevate in expression. As an example, myosin IIC is found at low expression levels in normal ductal epithelia. As the ductal adenocarcinoma forms, myosin IIC elevates in expression and persists in metastases (shown on the right is a metastasis found in a lymph node). Increases in the relative levels of low-expressing proteins, as compared with healthy tissue, can have dramatic effects on cancer cell behavior. Scale bars, 100 µm. [B and C adapted from Surcel et al. (4) with permission.]
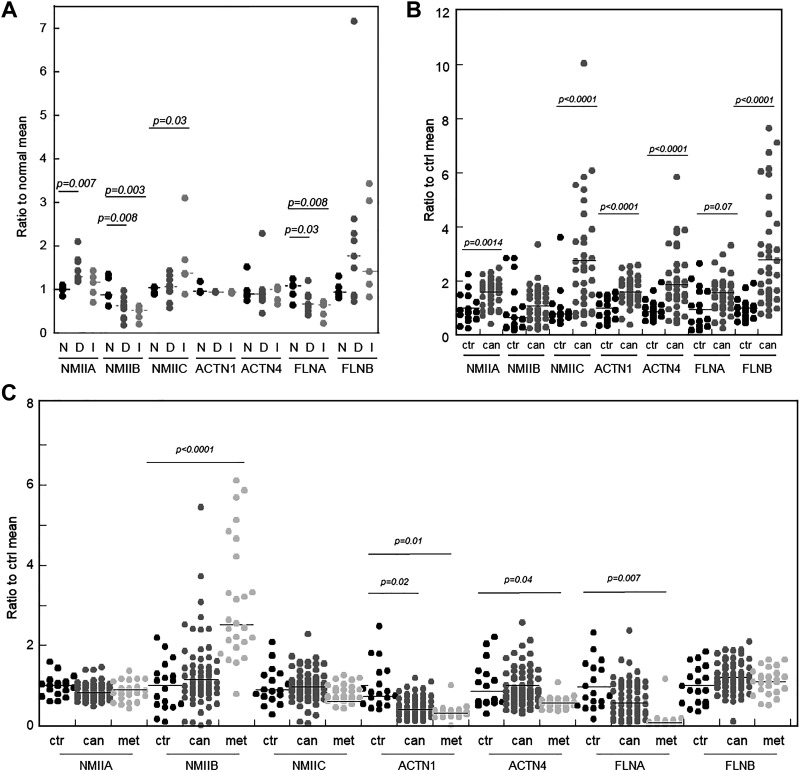
In numerous cancer types, the expression levels of mechanoresponsive protein families are often significantly altered in a highly paralog-specific manner—isoforms that are mechanoresponsive tend to have increased expression levels, whereas those that are not often show decreased or steady expression correlated with cancer progression. Changes in the expression level of even low-abundance mechanoresponsive isoforms reflects a reprogramming of cancer cells that favors increased adaptability required for efficacious growth and metastasis (4) (Fig. 1C).
Mechanoresponsive isoforms can be predicted based on their differential biochemistry, such as actin-binding affinities and myosin bipolar filament assembly dynamics (3, 5, 6). This differential biochemistry can be leveraged to develop anticancer chemical screens that are isoform specific. Although the development of most anticancer therapies relies on target inhibition, the mechanoresponsive machinery provides a target space in which activation of key components may reduce metastatic potential while protecting healthy tissue. This targeted push of the adaptive system out of its optimum (sweet spot) of activity—the position of the system that allows for maximal adaptability and ultimately, progrowth, invasion, and metastatic potential—requires a thorough understanding of the mechanobiome players. Here, we will review the mechanoresponsive protein families and what makes some paralogs mechanoresponsive and others not, their overall and relative protein concentrations in different cancer types, and some early indicators of pharmacological success in targeting of the mechanobiome for cancer therapies.
MECHANORESPONSIVE PLAYERS
Nonmuscle Myosin II
Nonmuscle myosin II (NMII) is a member of the myosin superfamily and the major active force generator on the actin cytoskeleton. Myosin II’s functional unit is the bipolar thick filament (Fig. 1A). For mammalian systems, nonmuscle myosin IIs are composed of 10–30 hexameric monomers, where each monomer consists of two heavy chains, two essential light chains, and two regulatory light chains. Myosin II head domains use ATP hydrolysis to induce conformational changes that are propagated through the lever arm, allowing the motor to pull on and generate force on the actin filament.
Mammals have three paralogs: NMIIA, NMIIB, and NMIIC, with unique mechanochemistry and cellular distributions. Of the three, NMIIA has the highest rate of ATP hydrolysis, pulling along actin filaments more rapidly than NMIIB or NMIIC (7), whereas NMIIB has a much higher duty ratio (the amount of time the motor domain is strongly bound to the actin filament in a force-generating state) than NMIIA (8, 9). In the majority of cells studied, NMIIA and NMIIB collectively form the largest pool of NMIIs, with NMIIC forming the smallest pool. All three isoforms can be mechanoresponsive (3, 4). Across multiple cell types, NMIIA and NMIIC always respond to internally and externally applied stresses, whereas NMIIB is situationally mechanoresponsive, dependent on its assembly and regulation by the protein kinase PKCζ (10).
In addition, each isoform has distinct roles in motility, adhesion, and other mechanically driven processes. In cancer, NMIIs play essential roles in tumor initiation, tumor formation and growth, and metastasis, all driven by their role in adhesion, mechanotransduction, motility, and contractility. During adhesion-dependent single-cell migration, for example, the localized activity of distinct NMIIs determines migratory speed and persistence, with NMIIA typically at the front promoting protrusions and adhesion maturation, and NMIIB in the back forming the contractile rear, promoting cellular detachment from the substrate, and impacting nuclear distortion [reviewed in (11)]. In collective cell migration, NMIIs are generally considered to generate restrictive forces that help limit protrusion formation at the sides and rear of the tissue mass, rectifying Arp2/3-mediated protrusion formation toward the front of the mass and promoting collective tissue movement [e.g. (12)]. However, it should be noted that in a colorectal cancer model study, the opposite was the case whereby inhibiting upstream regulators of myosin II and myosin II itself promoted collective migration (13). In glioma, NMII is required for contractility through the brain tissue’s submicrometer pores (14). Interestingly, inhibition of NMII in glioblastoma does block invasion but then leads to faster tumor growth, as myosin II is needed to inhibit progrowth pathways, such as the ERK pathway. These two cancer examples identify a tumor suppressor role for myosin II and may underscore the “optimum/sweet spot” concept where different systems can be uniquely poised as well as highlight how the various cellular systems are integrated with one another (myosin II, signaling, growth pathways, etc.) (15). Additionally, NMIIA and NMIIB are both found in stress fibers: in younger stress fibers, predominately transverse arcs and radial stress fibers, NMIIA is enriched, whereas in older stress fibers and longer-lived ventral ones, NMIIB is predominant (16–18). Less is known about the lower-in-abundance NMIIC, but all three isoforms are involved in the retrograde flow of actin (4, 19, 20).
The list of cancers with altered myosin II expression and/or regulation is extensive and includes breast, lung, prostate, bladder, pancreatic, melanoma, colorectal, ovarian, uterine, glioma, and squamous cell carcinomas (4, 14, 15, 21–27) (Fig. 2 and Table 1). Despite their ubiquitous presence, NMIIs and their regulators have been the target of some anticancer drug development. For example, Rhodblock6 inhibits Rho kinase (55), thiosemicarbazone iron chelators block rho-associated protein kinase/regulatory light chain (ROCK/RLC) (56), and BDP529 inhibits myotonic dystrophy kinase-related CDC42-binding kinase (MRCK), reducing breast cancer tumor cell motility and invasion (57). In addition, 17e inhibits the myosin light-chain phosphatase, impacting prostate cancer cell growth (58), and Berberine, a RhoA/ROCK inhibitor, decreases colorectal cancer growth but also impacts cardiac and smooth muscle (59, 60). Moreover, Fasudil, also a ROCK inhibitor, inhibited cellular contractility, decreasing pancreatic tumor stiffness and making the cancer cells more sensitive to cytotoxic drugs, gemcitabine and Abraxane, a standard of care for patients with pancreatic cancer (61). As a result, Fasudil increased cell death and reduced the metastatic potential of pancreatic cancer cells in mouse models. This example highlights the potential of synergizing myosin II regulator modulation with other treatment strategies.
Figure 2.
Pancreatic and prostate, but not breast, cancers have one or more mechanoresponsive protein-encoding genes upregulated as compared with healthy tissue. These data were collected from Gene Expression Omnibus (GEO) Repository, which assesses transcript levels. A: in human patient-derived ductal carcinoma in situ (DCIS; D), and invasive ductal carcinoma (IDC, I) breast cancer vs. normal tissue (N), some mechanoresponsive proteins have increased gene expression while nonmechanoresponsive paralog-encoding genes remain unchanged or are decreased (53) (BioProject: PRJNA126373). B: in pancreatic cancer, several mechanoresponsive protein transcript levels are increased in patient pancreatic cancer tissue relative to normal tissue (54) (BioProject: PRJNA116073). C: metastatic prostate cancer samples from patients with androgen-ablation-resistant metastatic tumors as well as primary tumor samples relative to normal prostate tissue revealed elevation of NMIIB gene expression (52) (BioProject: PRJNA104173, 104175, 104177, 104179). NMII, nonmuscle myosin II.
Table 1.
Expression trends of nonmuscle myosin II in various cancers
| Cancer Type | Myosin IIA |
Myosin IIB |
Myosin IIC |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Protein Cell Lines | Protein Human Tissue | mRNA Human Tissue or Cell Lines | Protein Cell Lines | Protein Human Tissue | mRNA Human Tissue | Protein Cell Lines | Protein Human Tissue | mRNA Human Tissue or Cell Lines | |
| Acute myeloid leukemia | Increased (28) | ||||||||
| Bladder | Increased (27) | Increased (27) | Increased (29) | Increased (30) | |||||
| Breast | Decreased (31)Increased (22) | Increased (31)Increased in cell lines (32) | Decreased (31) | No change (22) | Increased (31) | Increased in cell lines (32) | |||
| Cervical | |||||||||
| Colorectal | Increased (33) | Increased (34) | |||||||
| Endometrial | |||||||||
| Esophageal | Increased (35) | ||||||||
| Gastric | Increased (36–38) | Increased (36,37) | |||||||
| Glioma | Increased (14) | Variable (14) Increased (39) Decreased (40) | Variable (14) | Increased (39) | Variable (14) | Variable (14) | |||
| Head and neck,including tongue and larynx | Decreased (41) | ||||||||
| Liver | |||||||||
| Lung | Decreased (31) | Increased (42) | Decreased (31) | No change (31) | Increased (31) | ||||
| Melanoma | Increased, Spitz tumors (43) | Increased (44) | Increased (44) | ||||||
| Neuroblastomas | |||||||||
| Ovarian | Increased (45) | Increased (46) | Increased (45) | ||||||
| Pancreatic | Increased (4) | Increased (4, 47) | Decreased (4) | Decreased (4) | Increased (4) | Increased (4) | |||
| Prostate | Increased (48) | ||||||||
| Renal | Favorable prog. indicator (49) | MYH10-RET gene fusion (50) | Favorable prog. indicator (49) | ||||||
| Thyroid | Increased (51) | ||||||||
| Salivary gland | |||||||||
| Squamous cell carcinoma | Decreased (26) | ||||||||
| Stomach | Favorable prog. indicator (49) | ||||||||
| Urothelial | Unfavorableprog. indicator (49) | ||||||||
Prog., prognosis.
Most of the aforementioned small molecules show no isoform specificity. However, NMII distribution across a wide variety of cancers reflects differential expression patterns of NMIIA, IIB, and IIC, suggesting that isoform-specific targeting compounds may be a better approach. One such possibility is 4-hydroxyacetophenone (4-HAP). Identified in a screen for mechanical modulators, 4-HAP specifically activates NMIIB and NMIIC but not NMIIA. In pancreatic cancer, the mechanoresponsive isoforms NMIIA and NMIIC expression increases, whereas the conditionally mechanoresponsive NMIIB decreases [NMIIB is nonmechanoresponsive in pancreatic cancer cells (4)]. In both pancreatic and colorectal cancer models, treatment with 4-HAP leads to decreased dissemination in in vitro models and decreased metastatic tumor burden in hemisplenectomized mice (4, 62). In addition, 4-HAP impacts the mechanics of circulating tumor cells from breast cancer (63).
What this cluster of studies suggest is that targeting specific isoforms in the mechanobiome impacts cancer cell behavior and has potential therapeutic benefit. Understanding the characteristics that underlie the biochemical differences among mechano- and nonmechanoresponsive isoforms, can lead to better approaches for drug discovery platforms.
Actin Crosslinkers
α-Actinins.
α-Actinins are a family of cross-linking proteins that includes four paralogs. The muscle-associated actinins are α-actinin 2 and 3 and are encoded by ACTN2 and 3, whereas nonmuscle isoforms are encoded by ACTN1 and 4, respectively. The muscle isoforms are found in skeletal and smooth muscle and organize actin filaments into sarcomeric structures, which constitute the minimal contractile unit. The nonmuscle paralogs organize actin filaments into the networks that contribute to nonmuscle cell structure, migration, and adhesion (Fig. 1A). ACTN4 has been linked to metastasis in cancer through alterations in cellular features and cellular misbehavior (64–67).
Based upon stopped flow kinetic measurements and actin filament cosedimentation assays using single actin-binding domains, mammalian α-actinin 1 and α-actinin 4 have a ∼90-fold differential in binding affinities for actin, with Kds of 0.36 µM and 32 µM, respectively (68, 69). This major difference in binding affinity coupled with the force-dependent bond formation (catch-slip bond) observed for these actin crosslinking proteins was sufficient to predict which of the α-actinin paralogs would accumulate in response to mechanical stress (3). As predicted and confirmed experimentally, α-actinin 4 senses mechanical stress on the actin cytoskeleton, resulting in accumulation to sites of external stress whereas α-actinin 1 does not. Accumulation depends on catch-slip bond formation and an optimal actin-binding affinity, impacting cells that need to be highly adaptive like cancer cells.
Amazingly, these two ACTNs also differ in how their expression changes as a normal pancreatic ductal epithelia cell transforms into a ductal adenocarcinoma (4). ACTN1 is already abundant in normal pancreatic ductal epithelial cells and rises somewhat in the cancer cells and across the cancer stroma.
Meanwhile, ACTN4 is poorly expressed in healthy pancreatic ductal epithelial cells but then dramatically increases only in the ductal adenocarcinoma.
Expression level changes of ACTN genes and their proteins are observed in multiple types of cancers (Fig. 2 and Table 2). For example, ACTN4 has a decreased expression level in endometrial, neuroblastoma, and prostate cancer, relative to normal tissue. In contrast, the expression levels of ACTN4 are increased in breast and pancreatic cancer, among others, as compared with the relevant healthy tissue (Table 2). What might these expression changes mean? First, the reduced expression of ACTN4 impairs fibroblasts in cell migration, spreading adhesion and proliferation, suggesting that α-actinin 4 is necessary for normal cell morphology and motility (111). In contrast, the overexpression of ACTN4 promotes breast cancer tumorigenesis through enhanced cell motility (112). Gene expression profiles from patient primary lung cancer, adjacent benign tissue, and metastatic brain tumor show that ACTN4 is elevated in the metastatic brain tumor and leads to lung cancer metastasis to the brain (113). In colorectal cancer cells, α-actinin 4 provokes immature focal adhesions, which leads to cell motility and invasion, whereas α-actinin 1 does not (114). Additionally, patients with an increase in the copy number of ACTN4 have metastatic phenotypes leading to lower prognosis, like salivary gland carcinoma (109). Thus, ACTN4 appears to play a role in cancer progression and metastasis whereas ACTN1 appears not to.
Table 2.
Expression trends of α-actinins in various cancers
| Cancer Type | α-Actinin 1 |
α-Actinin 4 |
||||
|---|---|---|---|---|---|---|
| Protein Cell Lines | Protein Human Tissue | mRNA Human Tissue | Protein Cell Lines | Protein Human Tissue | mRNA Human Tissue | |
| Acute myeloid leukemia | Variable (70) | |||||
| Bladder | Increased (71) | Increased (71, 72) | Increased (71) | |||
| Breast | Increased (73) | Increased (32) | Distribution pattern correlates w/prognosis (65); Increased (74) | Increased (75, 76) | ||
| Cervical | Increased (77,78) | |||||
| Colorectal | Unfavorable prog. indicator (49) | Increased (79) | ||||
| Endometrial | Decreased (80, 81) | |||||
| Esophageal | Increased (82, 83) | |||||
| Gastric | Increased (84) | Increased (84) | ||||
| Glioma | Increased (85) | Increased (85, 86) | ||||
| Head and neck,including tongue and larynx | Unfavorable prog indicator (49) | Increased (87, 88) | ||||
| Liver | ||||||
| Lung | Unfavorable prog. indicator (49) | Increased (89) | Increased (90); Unfavorable prog. indicator (49) | Increased (91) | ||
| Melanoma | Increased (92) | Increased (93) | ||||
| Neuroblastomas | Decreased (94) | |||||
| Ovarian | Increased (95–98) | Increased (95, 97, 98) | ||||
| Pancreatic | No change (4) | Increased (4, 99–101); Unfavorable prog. indicator (49) | Increased (79) | |||
| Prostate | Decreased (102)Increased (103) | Decreased (104, 105) | ||||
| Renal | Unfavorable prog. indicator (49) | Favorable prog. indicator (49) | ||||
| Thyroid | Increased (106) | Increased (107) | Increased (108) | |||
| Salivary gland | Increased (109) | |||||
| Squamous cell carcinoma | Increased (110) | Increased (110) | ||||
| Stomach | ||||||
| Urothelial | Unfavorable prog. indicator (49) | |||||
Prog., prognosis.
Filamins.
The filamins are a family of actin-binding proteins that can associate with F-actin filaments in a solution to result in a dense gel meshwork (115). In humans, the filamin family includes three isoforms: filamin A (FLNA), filamin B (FLNB), and filamin C (FLNC) (116). FLNA and FLNB are widely expressed in various human tissues (117), whereas FLNC is primarily expressed in cardiac tissue and skeletal muscle (118, 119). Collectively, filamins are found in the cell cortex, the F-actin-rich region underlying the plasma membranes (116, 120–122) (Fig. 1A). In mammalian cells, nonmuscle filamin A and filamin B form dimers, crosslink actin filaments, and can link the actin filaments to integral membrane proteins, helping to couple the cortical actin meshwork to the plasma membrane (116).
Similar to α-actinins, filamin isoforms also have distinct actin-binding affinities though the range is not as broad [Filamin B, Kd = 7 µM (123); Filamin A, Kd = 17 µM (124)]. Filamin B is highly mechanoresponsive, whereas filamin A is much less so (3, 4). Filamin B and A have a flipped relationship between binding affinity and mechanoresponsiveness as compared with the α-actinins where the lower affinity paralog α-actinin 4 is the more mechanoresponsive isoform. This difference may reflect that filamins are more subtly tuned and also have a cooperativity component that contributes to their mechanism of mechanoresponsiveness (3, 5). Consistent with this, filamin A is necessary for the active stiffening of melanoma cells that are plated on collagen, but in response to large external forces, filamin A is not required for passive stiffening (125). In addition to the mechanoresponsiveness, other factors also help modulate filamin-actin-binding affinity. For example, filamins bind tropomyosin-bound F-actin with increased affinity (Kd = 0.13–3.2 μM) (126). The interaction between filamin and F-actin can also be regulated by phosphorylation (127), inositol phospholipids (128), and Ca2+-calmodulin (124).
In vitro, increasing the ratio of filamin to actin leads to tighter networks (129). In cells, increasing the filamin concentrations during cancer progression could shift the type of actin filament organization with differing effects on cell behavior, depending on the starting point. As a starting reference point, in melanoma cells, the ratio of filamin to actin has been measured to be around 1:80–140 (130). In these cells, increasing FLNA enhances invasive ability of human melanoma cells (131). In contrast, in many scenarios, FLNA inhibits cell growth and metastasis (132). For example, recent research found that the expression of the FLNA gene can inhibit the malignancy of prostate adenocarcinoma (133) and breast cancer cell migration and invasion (23, 134) and can regulate colorectal cancer cell growth and migration (135). Additionally, FLNB is upregulated in pancreatic cancerous ducts, whereas FLNA is upregulated across the pancreatic tissue and stroma (4). Among other human cancers (Table 3), B cell childhood acute lymphoblastic leukemia and Pro-B acute lymphoblastic leukemia have an increase of FLNB mRNA expression when compared with normal tissues (136).
Table 3.
Expression trends of filamins in various cancers
| Cancer Type | Filamin A |
Filamin B |
||||
|---|---|---|---|---|---|---|
| Protein Cell Lines | Protein Human Tissue | mRNA Human Tissue | Protein Cell Lines | Protein Human Tissue | mRNA Human Tissue | |
| Acute myeloid leukemia | ||||||
| Bladder | Increased (136) | |||||
| Breast | Increased (137, 138) | Increased (137) | Increased (136) | |||
| Cervical | Increased (139, 140) | |||||
| Colorectal | Increased (138, 141); Unfavorable prog. indicator (49) | Decreased (136) | ||||
| Endometrial | ||||||
| Esophageal | Increased (136) | |||||
| Gastric | Decreased (142) | Decreased (136) | ||||
| Glioma | Increased (143) | |||||
| Head and neck,including tongue and larynx | Increased (136) | |||||
| Liver | Increased (138) | |||||
| Lung | Increased (138, 144) | |||||
| Melanoma | Decreased (136) | |||||
| Neuroblastomas | ||||||
| Ovarian | Increased (138) | Increased (136) Decreased (136) | ||||
| Pancreatic | Increased (138) | Increased (4) Unfavorable Prog Indicator (49) | Increased (136) | |||
| Prostate | Increased (138, 145) | |||||
| Renal | Unfavorable prog. indicator (49) | Favorable prog. indicator (49) | ||||
| Thyroid | ||||||
| Salivary gland | ||||||
| Squamous cell carcinoma | Increased (136) | |||||
| Stomach | ||||||
| Urothelial | Unfavorable prog. indicator (49) | |||||
Prog., prognosis.
Overall, highly mechanoresponsive ACTN4 and FLNB paralogs are consistently associated with greater invasion and metastasis, whereas the non- or less-responsive paralogs are generally associated with less invasive and metastatic potential across many cancer types. Metastatic cells need to remodel their network to undergo shape change rapidly as they migrate through an ever-changing microenvironment. A shift in binding affinity in response to mechanical stress is a major driver for the mechanoresponsiveness and upregulation of ACTN4 and FLNB. Chemical screens could be developed that leverage isoform-specific biochemistry toward therapeutic treatments. Although inhibition is one possible strategy, compounds that alter cancer cells’ mechanoresponsiveness by shifting the force sensitivity of the crosslinker’s actin-binding activity, making α-actinin 4 or filamin B more α-actinin 1- or filamin A-like, respectively, may provide an alternate therapeutic strategy.
Other Interacting Players
Due to the isoform-specific activity of mechanoresponsive proteins, some of their cofactors, such as 14-3-3 and CLP36, may be of interest to help mediate invasive potential.
14-3-3
The human 14-3-3 family comprises seven isoforms (β, γ, ε, η, σ, θ, and ζ), each expressed by a different gene (146). 14-3-3s generate interest because of their roles in signal transduction pathways that control cell cycle checkpoints (147), MAP kinase activation (148), apoptosis (149), and DNA damage repair (150). 14-3-3s have been studied extensively as a drug target by either stabilizing or inhibiting protein-protein interactions. For example, 14-3-3-targeting drugs include fusicoccanes, which stabilize 14-3-3 binary structures (151), pyrrolidone1 and epibestatin, which were identified in a high-throughput screen for 14-3-3 stabilizers (152), and phosphonate-type inhibitors, PPI inhibitor 7 (153), among many more summarized in (154).
14-3-3 was originally identified as an interactor of myosin II through a combination of genetics, cellular biophysics, and proteomics assays in Dictyostelium discoideum (155). For the mammalian counterparts, based on a suite of in vitro assays, 14-3-3 paralogs, especially 14-3-3σ, inhibit myosin II filament assembly, whereas 14-3-3ζ had no effect on myosin IIB or IIC but promotes assembly of myosin IIA (156) (Fig. 1A). The mammalian 14-3-3-myosin II interaction has a relatively high affinity with an apparent Kd of 380 nM (156).
The 14-3-3 protein family plays a major role in cancer, and the majority of the isoforms are upregulated in a variety of disease states [reviewed in (157)] (Table 4). For example, 14-3-3σ is strongly upregulated in colorectal cancer cells (147) and functions as a tumor suppressor in breast (160) and gastric cancer (222). Additionally, 14-3-3σ is upregulated in lung cancer (223), head and neck squamous cell carcinomas (194), and chemoresistant pancreatic adenocarcinoma cells (224). In astrocytoma, 14-3-3β seems to have distinct tissue localization and increased protein expression (225). In several cancer types, the seven 14-3-3 isoforms exhibit different ratios of expression. For example, all the isoforms have increased cytoplasmic levels in vulvar carcinoma, with the exception of 14-3-3θ, which has decreased expression (226). Amazingly, expression levels of 14-3-3ε, ζ, and θ increased with the increase of pathological grade of meningioma, whereas the 14-3-3η, β, γ, and σ isoforms were reduced in expression (227). A group reported that tamoxifen treatment appears to induce 14-3-3ζ expression in breast cancer cells (228). A thorough understanding of isoform-specific function is needed, as proteins in the mechanobiome are interconnected and affect each other’s expression and/or activity. For example, overexpression of 14-3-3ε in a human gastric cancer cell line resulted in an increase in the total cellular level of filamin A and an increase in the subcellular localization of filamin A in the cytoplasm (229).
Table 4.
Expression trends of 14-3-3 in various cancers
| Cancer Type | 14-3-3 |
||
|---|---|---|---|
| Protein Cell Lines | Protein Human Tissue | mRNA Human Tissue | |
| Acute myeloid leukemia | |||
| Bladder | σ: Decreased (158); Increased (159) | σ: Decreased (158) | |
| Breast | β: No change (160)σ: Decreased (160, 161)ζ: No change (160) | ε: Increased (162, 163)θ: Increased (164, 165)σ: Decreased (160)ζ: Increased (166)β: Unfavorable Prog Indicator (49) | ε: Increased (167)σ: Decreased (161) |
| Cervical | β: Decreased (168)ε: Decreased (168) | σ: Increased (169)γ: Unfavorable Prog Indicator (49) | σ: Increased (169) |
| Colorectal | β: Increased (170)σ: Decreased (171) | β: Increased (172)ε: Decreased (173) | β: Increased (174)η: Decreased (175)σ: Decreased (175)ζ: Decreased (175) |
| Endometrial | σ: Decreased (176)β: Unfavorable prog. indicator (49)ε: Favorable prog. indicator (49)θ: Unfavorable prog. indicator (49) | ||
| ζ: Unfavorable prog. indicator (49) | |||
| Esophageal | σ: Increased (177)ζ: Increased (178) | σ: Decreased (177)ζ: Increased (178) | ζ: Increased (178) |
| Gastric | ζ: Increased (179) | β: Increased (180,181)ε: Increased (182)σ: Increased (183,184)ζ: Increased (179) | β: Increased (185)ε: Increased (185)η: Increased (185)γ: Increased (185)θ: Increased (185)ζ: Increased (185) |
| Glioma | β: Increased (186) | β: Increased (187–189)ε: No change (188)η: Increased (188)γ: No change (188); Increased (187)θ: No change (188)ζ: Decreased (188); Increased (187) | β: Increased (188)ε: Increased (188)η: Increased (188)γ: No change (188); Decreased (190)θ: No change (188)ζ: Decreased (188) |
| Head and neck,including tongue and larynx | β: Increased (191)ε: Decreased (192) | σ: Increased (193)ζ: Increased (193)β: Unfavorable prog. indicator (49) | σ: Increased (194)ζ: Increased (195) |
| Liver | ε: Increased (196)σ: Decreased, gene (171) | β: Increased (197)ε: Increased (196, 198)σ: Decreased (199)ζ: Increased (200)β: Unfavorable prog. indicator (49)η: Unfavorable prog. indicator (49)θ: Unfavorable prog. indicator (49)ζ: Unfavorable prog. indicator (49) | |
| Lung | β: Increased (201)ε: Increased (201)γ: Increased (202); No change (201)θ: Increased (201, 203)σ: Increased (201)ζ: Increased (201) | β:Increased (201)ε: Increased (201)γ: Increased (204, 205)θ: Increased (201)σ: No change NSCLC (206); Increased in NSCLC (207); Decreased in SCLC (206) ; Increased (201)ζ: Increased (201)β: Unfavorable prog. indicator (49)γ: Unfavorable prog. indicator (49)ζ: Unfavorable prog. indicator (49) | γ: Increased NSCLC (205)σ: Increased NSCLC (207) |
| Melanoma | |||
| Neuroblastomas | |||
| Ovarian | σ: Decreased (208); Increased (209)ζ: Increased (210) | σ: Decreased (208)σ: Increased (209)ζ: Increased (210) | |
| Pancreatic | σ: Variable depending on cell line (211) | σ: Increased (212)ζ: Increased (212,213)γ: Unfavorable prog. indicator (49)ζ: Unfavorable prog. indicator (49) | σ: Increased (211)ζ: Increased (212) |
| Prostate | β: Increased (214)σ: Decreased (215) | ε: Increased (216)θ: Increased (216)σ: Decreased (176, 217) | ε: Increased (216)σ: Decreased (215)θ: Increased (216) |
| Renal | β: No change (218)ε: Increased (218)η: No change (218)γ: No change (218)θ: No change (218)ζ: No change (218)β: Favorable prog. indicator (49)γ: Unfavorable prog. indicator (49)ζ: Unfavorable prog. Indicator (49) | β: Increased (218)ε: Increased (218)η: Increased (218)γ: No change (218)θ: Increased (218)ζ: No change (218) | |
| Thyroid | β: Increased (219) | ||
| Salivary gland | |||
| Squamous cell carcinoma | η: Increased (220) | ||
| Stomach | |||
| Urothelial | ζ: Increased (221) | ||
NSCLC, non-small cell lung cancer; Prog., prognosis.
Table 4.
Expression trends of 14-3-3 in various cancers, continued
| Lung | β: Increased (201)ε: Increased (201)γ: Increased (202); No change (201)θ: Increased (201, 203)σ: Increased (201)ζ: Increased (201) | β:Increased (201)ε: Increased (201)γ: Increased (204, 205) θ: Increased (201)σ: No change NSCLC (206); Increased in NSCLC (207); Decreased in SCLC (206) ; Increased (201)ζ: Increased (201) β: Unfavorable prog. indicator (49)γ: Unfavorable prog. indicator (49)ζ: Unfavorable prog. indicator (49) | γ: Increased NSCLC (205)σ: Increased NSCLC (207) |
|---|---|---|---|
| Melanoma | |||
| Neuroblastomas | |||
| Ovarian | σ: Decreased (208); Increased (209)ζ: Increased (210) | σ: Decreased (208)σ: Increased (209)ζ: Increased (210) | |
| Pancreatic | σ: Variable depending on cell line (211) | σ: Increased (212)ζ: Increased (212,213)γ: Unfavorable prog. indicator (49)ζ: Unfavorable prog. indicator (49) | σ: Increased (211)ζ: Increased (212) |
| Prostate | β: Increased (214)σ: Decreased (215) | ε: Increased (216)θ: Increased (216)σ: Decreased (176, 217) | ε: Increased (216) σ: Decreased (215) θ: Increased (216) |
| Renal | β: No change (218)ε: Increased (218)η: No change (218) γ: No change (218)θ: No change (218) ζ: No change (218)β: Favorable prog. indicator (49)γ: Unfavorable prog. indicator (49)ζ: Unfavorable prog. Indicator (49) | β: Increased (218)ε: Increased (218)η: Increased (218)γ: No change (218) θ: Increased (218)ζ: No change (218) | |
| Thyroid | β: Increased (219) | ||
| Salivary gland | |||
| Squamous cell carcinoma | η: Increased (220) | ||
| Stomach | |||
| Urothelial | ζ: Increased (221) |
Prog., progress.
CLP36/PDLIM1
CLP36 (also known as CLIM1 or Elfin) is a 38-kD protein that has an N-terminal PDZ domain and a C-terminal LIM domain. Due to the association between actin and α-actinins in cancer, CLP36 becomes of interest here because of its interactions with α-actinins (Fig. 1A). CLP36 associates with actin filaments during cell shape changes, migration, and during the contraction of endothelial cells. CLP36 is also expressed in most nonmuscle tissues though in some, such as the pancreas, expression can be very low (230). However, in situ hybridization analysis of mouse tissues revealed that CLP36 can be highly expressed, and when present, it localizes to actin stress fibers through the PDZ domain and interacts with α-actinin 1 and α-actinin 4 (231). During cell shape change events, including cell spreading, migration, and contraction, CLP36 associates with actin filaments and stress fibers (231, 232). ACTN4 is highly expressed in the colon, and again in this context, CLP36 interacts with α-actinin 4, forming stable α-actinin 4-CLP36 complexes, which extends throughout the actin stress fibers (231). CLP36 also showed significant changes in levels in breast cancer as compared with ovarian cancer patient plasma samples (233). Thus, the mechanoresponsive proteins and interacting players have potential for early detection of cancers. For example, patients with breast cancer can have increased tumor-associated autoantibodies to CLP36 (234). Understanding the protein interactions between CLP36 and α-actinin 4 may provide additional insight into how to modify their activities toward a therapeutic end.
Conclusions and Vision
The mechanoresponsive machinery plays a central role in many cell shape change events and is also heavily associated with cancer progression and metastasis. Further, many of these proteins have expression levels that can be altered in different ways in different cancer types that likely then lead to the altered cell mechanics and mechanoresponsiveness associated with cancer progression (4, 69, 113, 134, 235) (Fig. 2 and Tables 1–4). Despite their differential cancer expression levels, the mechanobiome proteins have been largely overlooked in drug development and trials. This oversight is caused by four primary assertions.
The first is that the classic view of these proteins often prescribes a singular major role in the cell, without taking into account the diversity of functions that can be attained by varying ratios between the paralogs. For example, nonmuscle myosin II’s textbook definition is that of contractility required for cytokinesis and motility. However, myosin II’s functions are much more extensive and include roles in mechanosensation, elasticity and viscoelasticity, cortical tension and fluidity, the modulation of cell adhesion to substrates as well as other cells, the integration between signaling and mechanical inputs, and the impact on overall cell mechanics on many other cell functions (3, 236–238). It is the interplay between the three myosin isoforms that skews cell behavior in disease states (4, 15); understanding how the isoforms participate fully in all NMII functions will shed light on how changes in their expression yield transformative cancer cells. Similarly, the preponderance of research on 14-3-3 in the cancer field has focused on its role in processes such as the DNA damage response [e.g., (161)], with less focus on its interactions with cytoskeletal components (156, 239). With seven isoforms that exhibit some overlapping function and differential expression in multiple cancer types, fully defining their function will allow for better targeting of individual 14-3-3 isoforms in cancer.
The second is that ubiquitous expression of mechanoresponsive proteins across multiple cell types has led to the assumption that their targeting would be toxic to human patients. This is despite the fact that proteins Kras, Rho, and Aurora kinase, which are also abundantly expressed, are the target of multiple drug trials for cancer [e.g., reviewed in (240, 241)]. In addition, the families of proteins that are mechanoresponsive are often treated as an aggregate of all of their isoforms or of the most abundantly expressed isoform. This is most starkly seen with the myosins, where the preponderance of research on nonmuscle myosin IIs is focused on IIA and IIB, with little consideration to IIC due to its low abundance compared with IIA and IIB. However, despite its lower amounts (18 nM vs. 565 nM of IIA in pancreatic ductal adenocarcinoma cancer cells), myosin IIC helps facilitate actin organization and retrograde flow, working in concert with myosin IIA to increase dissemination and metastasis (4, 62). These data are just one example of how low abundance proteins that are often disregarded in large data mining can actually be viable candidates for drug targeting.
Although the abundance of proteins needs to be reconsidered for defining the importance of a given protein for cancer progression, it is also worth noting that mechanoresponsiveness, as well as nearly all cellular processes, can also be regulated by posttranslational modifications. In one key example, NMIIB has exquisite cell-type-specific and even cell-cycle-stage-specific mechanoresponsiveness (10). Myosin II heavy-chain phosphorylation, carried out by PKCζ, mediates this differential effect in mechanoresponsiveness. No doubt, this example highlights the importance of considering the specific context of any disease, which will likely influence the best strategy for targeting the disease.
Third, and perhaps most important, proteins that are upregulated in cancers are often pursued for pharmacological inhibition. Knocking down or inhibiting mechanoresponsive proteins in cancerous systems can actually yield more disseminative behavior and animals with higher metastatic load (4, 15, 26, 242). Instead of relegating mechanoresponsive proteins as untargetable, data from several studies suggest that the pharmaceutical paradigm needs to be shifted away from inhibition alone. Mechanical adaptability exists on a continuum with cells occupying a sweet spot (optimum) between adaptability and stability for a given tissue environment. Cancer cells seek to favor all types of adaptability but have evolved to have multiple routes to ensure survival, such that inhibition alone is insufficient to stop cancerous cellular behavior, in some cases even increasing disseminative behaviors.
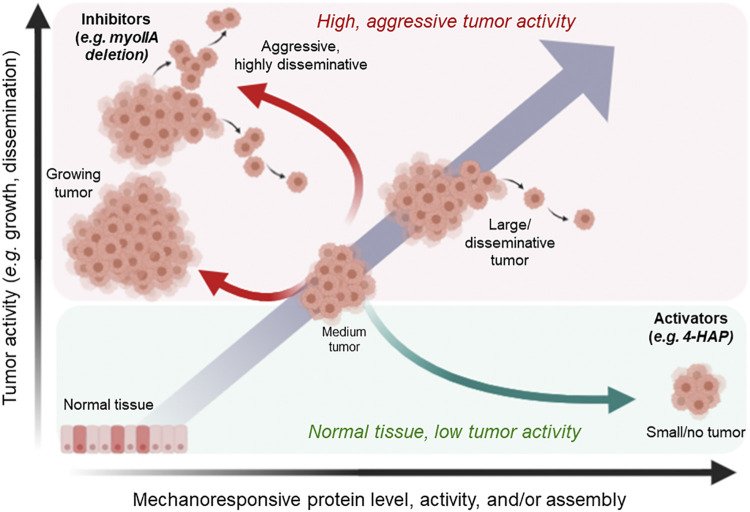
Instead, it is a viable strategy to develop small molecules that specifically activate mechanoresponsive proteins, pushing them into a regime where they act in a manner that is more stable than their nonmechanoresponsive sister paralogs. One way to generate this hyperactivation is to increase the binding affinity of the proteins for cytoskeletal binding partners (such as the actin filaments themselves), preventing their disassembly and reducing further diseased morphologies (Fig. 3). In fact, such a strategy has been successfully used with omecamtiv mecarbil, a selective activator of cardiac myosin currently in phase 3 clinical trials for hypertrophic cardiomyopathies (243). In two different studies, 4-hydroxyacetophenone (4-HAP), which activates specifically myosin IIC in pancreatic ductal adenocarcinoma and colorectal cancer models by locking it onto actin filaments, shows promise that skewing the activation/inhibition curve toward activation can curb cancer behaviors in mouse models (4, 62). Looking to mechanoresponsive proteins as targetable drug spaces (Fig. 3) can do much to change the cancer fighting landscape.
Figure 3.
Shifting the activation curve of mechanoresponsive proteins and their partners is a viable strategy for developing cancer therapeutics. Cancer progression and tumor formation is marked by altered expression in the mechanoresponsive proteins, α-actinin, filamin, and nonmuscle myosin II (NMII), as well as partnering proteins such as 14-3-3 and CLP36. Left unchecked, the altered in gene expression correlate with increased cell activity, specifically metastasis. Because mechanoresponsive proteins such as NMIIs also have tumor-suppressive roles as they inhibit pathways such as the ERK pathway, pharmacological inhibition can lead to enhanced tumor growth. In contrast, activators can lock in a cell, leading to anticancer behaviors. One such example is the activator 4-HAP, which increases nonmuscle myosin IIB and IIC assembly, leading to increased cortical tension and reduced tumor metastatic activity. Figure created with BioRender.com. 4-HAP, 4-hydroxyacetophenone.
One final hurdle is that development of antimetastatic cancer treatments typically depends on measurement of primary tumor size, looking for tumor size reduction. The logic is that imaging the primary tumor is easier because of the tumor’s larger mass and that tumor reduction can be a faster, suitable surrogate readout for long-term benefit of the drug for patients. However, this hurdle makes it more tempting to overlook the real potential of antimetastatic therapeutic strategies (244). Ironically, at least two traditional anticancer drugs, the DNA-damaging cisplatin and the microtubule stabilizer docetaxel, also modulate the cancer cell’s mechanical properties and help reduce invasiveness (245). Perhaps improved strategies that more precisely leverage underappreciated mechanoresponsive proteins can help promote the fortitude necessary to develop new drugs that target cancer metastasis. It is also possible that the upregulation of mechanoresponsive proteins could offer a molecular program for early detection of metastases. α-Actinin 4 may already provide such an opportunity for cervical cancer (246).
Collectively, the mechanobiome, particularly the mechanoresponsive proteins and their networks, offer enormous opportunity for cancer intervention. Strategically targeting this machinery may allow for the inhibition of a wider range of cancer types at their most lethal impact point—formation of metastases—while minimizing toxicity effects for patients.
GRANTS
Our research is supported by the NIH (National Institute of General Medical Sciences Grant R01 GM66817 and National Heart, Lung, and Blood Institute Grant R01 HL124099) and a Johns Hopkins Discovery grant.
DISCLOSURES
D. N. Robinson is currently exploring formation of a startup company named Amoibe Discovery. It is still in exploration phase. A patent has been granted on the use of 4-HAP to treat disease, and A. Surcel and D. N. Robinson are inventors on this patent. E. Parajon does not have any conflicts of interest, financial or otherwise, to disclose.
AUTHOR CONTRIBUTIONS
E.P., A.S., and D.N.R. prepared figures; E.P., A.S., and D.N.R. drafted manuscript; E.P., A.S., and D.N.R. edited and revised manuscript; E.P., A.S., and D.N.R. approved final version of manuscript.
ACKNOWLEDGMENTS
We thank the members of our lab for many helpful discussions.
REFERENCES
- 1.Kothari P, Johnson C, Sandone C, Iglesias PA, Robinson DN. How the mechanobiome drives cell behavior, viewed through the lens of control theory. J. Cell Sci 132: jcs234476, 2019. doi: 10.1242/jcs.234476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kothari P, Srivastava V, Aggarwal V, Tchernyshyov I, Van Eyk JE, Ha T, Robinson DN. Contractility kits promote assembly of the mechanoresponsive cytoskeletal network. J Cell Sci 132: jcs226704, 2019. doi: 10.1242/jcs.226704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schiffhauer ES, Luo T, Mohan K, Srivastava V, Qian X, Griffis ER, Iglesias PA, Robinson DN. Mechanoaccumulative elements of the mammalian actin cytoskeleton. Curr Biol 26: 1473–1479, 2016. doi: 10.1016/j.cub.2016.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Surcel A, Schiffhauer ES, Thomas DG, Zhu Q, DiNapoli KT, Herbig M, Otto O, West-Foyle H, Jacobi A, Krater M, Plak K, Guck J, Jaffee EM, Iglesias PA, Anders RA, Robinson DN. Targeting mechanoresponsive proteins in pancreatic cancer: 4-hydroxyacetophenone blocks dissemination and invasion by activating MYH14. Cancer Res 79: 4665–4678, 2019. doi: 10.1158/0008-5472.CAN-18-3131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luo T, Mohan K, Iglesias PA, Robinson DN. Molecular mechanisms of cellular mechanosensing. Nat Mater 12: 1064–1071, 2013. doi: 10.1038/nmat3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luo T, Mohan K, Srivastava V, Ren Y, Iglesias PA, Robinson DN. Understanding the cooperative interaction between myosin II and actin cross-linkers mediated by actin filaments during mechanosensation. Biophys J 102: 238–247, 2012. doi: 10.1016/j.bpj.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim K-Y, Kovács M, Kawamoto S, Sellers JR, Adelstein RS. Disease-associated mutations and alternative splicing alter the enzymatic and motile activity of nonmuscle myosins II-B and II-C. J Biol Chem 280: 22769–22775, 2005. doi: 10.1074/jbc.M503488200. [DOI] [PubMed] [Google Scholar]
- 8.Kovács M, Wang F, Hu A, Zhang Y, Sellers JR. Functional divergence of human cytoplasmic myosin II. Kinetic characterization of the non-muscle IIA isoform. J Biol Chem 278: 38132–38140, 2003. doi: 10.1074/jbc.M305453200. [DOI] [PubMed] [Google Scholar]
- 9.Wang F, Kovács M, Hu A, Limouze J, Harvey EV, Sellers JR. Kinetic mechanism of non-muscle myosin IIB: functional adaptations for tension generation and maintenance. J Biol Chem 278: 27439–27448, 2003. doi: 10.1074/jbc.M302510200. [DOI] [PubMed] [Google Scholar]
- 10.Schiffhauer ES, Ren Y, Iglesias VA, Kothari P, Iglesias PA, Robinson DN. Myosin IIB assembly state determines its mechanosensitive dynamics. J Cell Biol 218: 895–908, 2019. doi: 10.1083/jcb.201806058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Newell-Litwa KA, Horwitz R, Lamers ML. Non-muscle myosin II in disease: mechanisms and therapeutic opportunities. Dis Model Mech 8: 1495–1515, 2015. doi: 10.1242/dmm.022103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aranjuez G, Burtscher A, Sawant K, Majumder P, McDonald JA. Dynamic myosin activation promotes collective morphology and migration by locally balancing oppositional forces from surrounding tissue. Mol Biol Cell 27: 1898–1910, 2016. doi: 10.1091/mbc.E15-10-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Libanje F, Raingeaud J, Luan R, Thomas Z, Zajac O, Veiga J, Marisa L, Adam J, Boige V, Malka D, Goéré D, Hall A, Soazec J, Prall F, Gelli M, Dartigues P, Jaulin F. ROCK 2 inhibition triggers the collective invasion of colorectal adenocarcinomas. EMBO J 38: e99299, 2019. doi: 10.15252/embj.201899299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beadle C, Assanah MC, Monzo P, Vallee R, Rosenfeld SS, Canoll P. The role of myosin II in glioma invasion of the brain. Mol Biol Cell 19: 3357–3368, 2008. doi: 10.1091/mbc.e08-03-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Picariello HS, Kenchappa RS, Rai V, Crish JF, Dovas A, Pogoda K, McMahon M, Bell ES, Chandrasekharan U, Luu A, West R, Lammerding J, Canoll P, Odde DJ, Janmey PA, Egelhoff T, Rosenfeld SS. Myosin IIA suppresses glioblastoma development in a mechanically sensitive manner. Proc Natl Acad Sci USA 116: 15550–15559, 2019. doi: 10.1073/pnas.1902847116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hotulainen P, Lappalainen P. Stress fibers are generated by two distinct actin assembly mechanisms in motile cells. J Cell Biol 173: 383–394, 2006. doi: 10.1083/jcb.200511093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Svitkina TM, Verkhovsky AB, McQuade KM, Borisy GG. Analysis of the actin-myosin II system in fish epidermal keratocytes: mechanism of cell body translocation. J Cell Biol 139: 397–415, 1997. doi: 10.1083/jcb.139.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tee YH, Shemesh T, Thiagarajan V, Hariadi RF, Anderson KL, Page C, Volkmann N, Hanein D, Sivaramakrishnan S, Kozlov MM, Bershadsky AD. Cellular chirality arising from the self-organization of the actin cytoskeleton. Nat Cell Biol 17: 445–457, 2015. doi: 10.1038/ncb3137. [DOI] [PubMed] [Google Scholar]
- 19.Brown ME, Bridgman PC. Retrograde flow rate is increased in growth cones from myosin IIB knockout mice. J Cell Sci 116: 1087–1094, 2003. doi: 10.1242/jcs.00335. [DOI] [PubMed] [Google Scholar]
- 20.Shih W, Yamada S. Myosin IIA dependent retrograde flow drives 3D cell migration. Biophys J 98: L29–L31, 2010. doi: 10.1016/j.bpj.2010.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen R, Yi EC, Donohoe S, Pan S, Eng J, Cooke K, Crispin DA, Lane Z, Goodlett DR, Bronner MP, Aebersold R, Brentnall TA. Pancreatic cancer proteome: the proteins that underlie invasion, metastasis, and immunologic escape. Gastroenterology 129: 1187–1197, 2005. doi: 10.1053/j.gastro.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 22.Derycke L, Stove C, Vercoutter-Edouart A-S, De Wever O, Dollé L, Colpaert N, Depypere H, Michalski J-C, Bracke M. The role of non-muscle myosin IIA in aggregation and invasion of human MCF-7 breast cancer cells. Int J Dev Biol 55: 835–840, 2011. doi: 10.1387/ijdb.113336ld. [DOI] [PubMed] [Google Scholar]
- 23.Ji Z-M, Yang L-L, Ni J, Xu S-P, Yang C, Duan P, Lou L-P, Ruan Q-R. Silencing filamin A inhibits the invasion and migration of breast cancer cells by up-regulating 14-3-3σ. Curr Med Sci 38: 461–466, 2018. doi: 10.1007/s11596-018-1901-6. [DOI] [PubMed] [Google Scholar]
- 24.Makohon-Moore AP, Zhang M, Reiter JG, Bozic I, Allen B, Kundu D, Chatterjee K, Wong F, Jiao Y, Kohutek ZA, Hong J, Attiyeh M, Javier B, Wood LD, Hruban RH, Nowak MA, Papadopoulos N, Kinzler KW, Vogelstein B, Iacobuzio-Donahue CA. Limited heterogeneity of known driver gene mutations among the metastases of individual patients with pancreatic cancer. Nat Genet 49: 358–366, 2017. doi: 10.1038/ng.3764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mandruzzato S, Callegaro A, Turcatel G, Francescato S, Montesco MC, Chiarion-Sileni V, Mocellin S, Rossi CR, Bicciato S, Wang E, Marincola FM, Zanovello P. A gene expression signature associated with survival in metastatic melanoma. J Transl Med 4: 50, 2006. doi: 10.1186/1479-5876-4-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schramek D, Sendoel A, Segal JP, Beronja S, Heller E, Oristian D, Reva B, Fuchs E. Direct in vivo RNAi screen unveils myosin IIa as a tumor suppressor of squamous cell carcinomas. Science 343: 309–313, 2014. doi: 10.1126/science.1248627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiong D, Ye Y-L, Chen M-K, Qin Z-K, Li M-Z, Zhang H, Xu L-H, Xu Z-Z, Zeng M-S. Non-muscle myosin II is an independent predictor of overall survival for cystectomy candidates with early-stage bladder cancer. Oncol Rep 28: 1625–1632, 2012. doi: 10.3892/or.2012.1965. [DOI] [PubMed] [Google Scholar]
- 28.Yu M, Wang J, Zhu Z, Hu C, Ma Q, Li X, Yin X, Huang J, Zhang T, Ma Z, Zhou Y, Li C, Chen F, Chen J, Wang Y, Pan H, Wang D, Jin J. Prognostic impact of MYH9 expression on patients with acute myeloid leukemia. Oncotarget 8: 156–163, 2017. doi: 10.18632/oncotarget.10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu P-F, Wang Y-H, Cao Y-W, Jiang H-P, Yang X-C, Wang X-S, Niu H-T. Far from resolved: stromal cell-based iTRAQ research of muscle-invasive bladder cancer regarding heterogeneity. Oncol Rep 32: 1489–1496, 2014. doi: 10.3892/or.2014.3340. [DOI] [PubMed] [Google Scholar]
- 30.Lee H, Kim K, Woo J, Park J, Kim H, Lee KE, Kim H, Kim Y, Moon KC, Kim JY, Park IA, Shim BB, Moon JH, Han D, Ryu HS. Quantitative proteomic analysis identifies AHNAK (Neuroblast Differentiation-associated Protein AHNAK) as a novel candidate biomarker for bladder urothelial carcinoma diagnosis by liquid-based cytology. Mol Cell Proteomics 17: 1788–1802, 2018. doi: 10.1074/mcp.RA118.000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jana SS, Kawamoto S, Adelstein RS. A specific isoform of nonmuscle myosin II-C is required for cytokinesis in a tumor cell line. J Biol Chem 281: 24662–24670, 2006. doi: 10.1074/jbc.M604606200. [DOI] [PubMed] [Google Scholar]
- 32.Kallioniemi A, Kallioniemi OP, Piper J, Tanner M, Stokke T, Chen L, Smith HS, Pinkel D, Gray JW, Waldman FM. Detection and mapping of amplified DNA sequences in breast cancer by comparative genomic hybridization. Proc Natl Acad Sci USA 91: 2156–2160, 1994. doi: 10.1073/pnas.91.6.2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang B, Qi X, Liu J, Zhou R, Lin C, Shangguan J, Zhang Z, Zhao L, Li G. MYH9 promotes growth and metastasis via activation of MAPK/AKT signaling in colorectal cancer. J Cancer 10: 874–884, 2019. doi: 10.7150/jca.27635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma Y-S, Huang T, Zhong X-M, Zhang H-W, Cong X-L, Xu H, Lu G-X, Yu F, Xue S-B, Lv Z-W, Fu D. Proteogenomic characterization and comprehensive integrative genomic analysis of human colorectal cancer liver metastasis. Mol Cancer 17: 139, 2018. doi: 10.1186/s12943-018-0890-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xia Z-K, Yuan Y-C, Yin N, Yin B-L, Tan Z-P, Hu Y-R. Nonmuscle myosin IIA is associated with poor prognosis of esophageal squamous cancer. Dis Esophagus 25: 427–436, 2012. doi: 10.1111/j.1442-2050.2011.01261.x. [DOI] [PubMed] [Google Scholar]
- 36.Liang S, He L, Zhao X, Miao Y, Gu Y, Guo C, Xue Z, Dou W, Hu F, Wu K, Nie Y, Fan D. MicroRNA let-7f inhibits tumor invasion and metastasis by targeting MYH9 in human gastric cancer. PLoS One 6: e18409, 2011. doi: 10.1371/journal.pone.0018409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu D, Zhang L, Shen Z, Tan F, Hu Y, Yu J, Li G. Clinicopathological significance of NMIIA overexpression in human gastric cancer. Int J Mol Sci 13: 15291–15304, 2012. doi: 10.3390/ijms131115291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu T, Ye Y, Zhang X, Zhu A, Yang Z, Fu Y, Wei C, Liu Q, Zhao C, Wang G. Downregulation of non-muscle myosin IIA expression inhibits migration and invasion of gastric cancer cells via the c-Jun N-terminal kinase signaling pathway. Mol Med Rep 13: 1639–1644, 2016. doi: 10.3892/mmr.2015.4742. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Yang Q, Cheng Y, Gao M, Kuang L, Wang C. Myosin heavy chain 10 (MYH10) gene silencing reduces cell migration and invasion in the glioma cell lines U251, T98G, and SHG44 by inhibiting the wnt/β-catenin pathway. Med Sci Monit 24: 9110–9119, 2018. doi: 10.12659/MSM.911523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li H, Lei B, Xiang W, Wang H, Feng W, Liu Y, Qi S. Differences in protein expression between the U251 and U87 cell lines. Turk Neurosurg 27: 894–903, 2017. doi: 10.5137/1019-5149.JTN.17746-16.1. [DOI] [PubMed] [Google Scholar]
- 41.Coaxum SD, Tiedeken J, Garrett-Mayer E, Myers J, Rosenzweig SA, Neskey DM. The tumor suppressor capability of p53 is dependent on nonmuscle myosin IIA function in head and neck cancer. Oncotarget 8: 22991–23007, 2017. doi: 10.18632/oncotarget.14967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maeda J, Hirano T, Ogiwara A, Akimoto S, Kawakami T, Fukui Y, Oka T, Gong Y, Guo R, Inada H, Nawa K, Kojika M, Suga Y, Ohira T, Mukai K, Kato H. Proteomic analysis of stage I primary lung adenocarcinoma aimed at individualisation of postoperative therapy. Br J Cancer 98: 596–603, 2008. doi: 10.1038/sj.bjc.6604197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yeh I, Tee MK, Botton T, Shain AH, Sparatta AJ, Gagnon A, Vemula SS, Garrido MC, Nakamaru K, Isoyama T, McCalmont TH, LeBoit PE, Bastian BC. NTRK3 kinase fusions in Spitz tumours. J Pathol 240: 282–290, 2016. doi: 10.1002/path.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jacobs K, Van Gele M, Forsyth R, Brochez L, Vanhoecke B, De Wever O, Bracke M. P-cadherin counteracts myosin II-B function: implications in melanoma progression. Mol Cancer 9: 255, 2010. doi: 10.1186/1476-4598-9-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kapoor A, Barai A, Thakur B, Das A, Patwardhan SR, Monteiro M, Gaikwad S, Bukhari AB, Mogha P, Majumder A, De A, Ray P, Sen S. Soft drug-resistant ovarian cancer cells migrate via two distinct mechanisms utilizing myosin II-based contractility. Biochim Biophys Acta Mol Cell Res 1865: 392–405, 2018. doi: 10.1016/j.bbamcr.2017.11.012. [DOI] [PubMed] [Google Scholar]
- 46.Liu L, Yi J, Deng X, Yuan J, Zhou B, Lin Z, Zeng Z. MYH9 overexpression correlates with clinicopathological parameters and poor prognosis of epithelial ovarian cancer. Oncol Lett 18: 1049–1056, 2019. doi: 10.3892/ol.2019.10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou P, Li Y, Li B, Zhang M, Liu Y, Yao Y, Li D. NMIIA promotes tumor growth and metastasis by activating the Wnt/β-catenin signaling pathway and EMT in pancreatic cancer. Oncogene 38: 5500–5515, 2019. doi: 10.1038/s41388-019-0806-6. [DOI] [PubMed] [Google Scholar]
- 48.Chen C, Zhang L-G, Liu J, Han H, Chen N, Yao A-L, Kang S-S, Gao W-X, Shen H, Zhang L-J, Li Y-P, Cao F-H, Li Z-G. Bioinformatics analysis of differentially expressed proteins in prostate cancer based on proteomics data. Onco Targets Ther 9: 1545–1557, 2016. doi: 10.2147/OTT.S98807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, Benfeitas R, Arif M, Liu Z, Edfors F, Sanli K, Von Feilitzen K, Oksvold P, Lundberg E, Hober S, Nilsson P, Mattsson J, Schwenk JM, Brunnström H, Glimelius B, Sjöblom T, Edqvist P-H, Djureinovic D, Micke P, Lindskog C, Mardinoglu A, Ponten F. A pathology atlas of the human cancer transcriptome. Science 357: eaan2507, 2017. doi: 10.1126/science.aan2507. [DOI] [PubMed] [Google Scholar]
- 50.Davis JL, Vargas SO, Rudzinski ER, López Marti JM, Janeway K, Forrest S, Winsnes K, Pinto N, Yang SE, VanSandt M, Boyd TK, Corless CL, Liu YJ, Surrey LF, Harris MH, Church A, Al-Ibraheemi A. Recurrent RET gene fusions in paediatric spindle mesenchymal neoplasms. Histopathology 76: 1032–1041, 2020. doi: 10.1111/his.14082. [DOI] [PubMed] [Google Scholar]
- 51.Wang Y, He H, Li W, Phay J, Shen R, Yu L, Hancioglu B, de la Chapelle A. MYH9 binds to lncRNA gene PTCSC2 and regulates FOXE 1 in the 9q22 thyroid cancer risk locus. Proc Natl Acad Sci USA 114: 474–479, 2017. doi: 10.1073/pnas.1619917114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chandran UR, Ma C, Dhir R, Bisceglia M, Lyons-Weiler M, Liang W, Michalopoulos G, Becich M, Monzon FA. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer 7: 64, 2007. doi: 10.1186/1471-2407-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kretschmer C, Sterner-Kock A, Siedentopf F, Schoenegg W, Schlag PM, Kemmner W. Identification of early molecular markers for breast cancer. Mol Cancer 10: 15, 2011. doi: 10.1186/1476-4598-10-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pei H, Li L, Fridley BL, Jenkins GD, Kalari KR, Lingle W, Petersen G, Lou Z, Wang L. FKBP51 affects cancer cell response to chemotherapy by negatively regulating Akt. Cancer Cell 16: 259–266, 2009. doi: 10.1016/j.ccr.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Castoreno AB, Smurnyy Y, Torres AD, Vokes MS, Jones TR, Carpenter AE, Eggert US. Small molecules discovered in a pathway screen target the Rho pathway in cytokinesis. Nat Chem Biol 6: 457–463, 2010. doi: 10.1038/nchembio.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun J, Zhang D, Zheng Y, Zhao Q, Zheng M, Kovacevic Z, Richardson DR. Targeting the metastasis suppressor, NDRG1, using novel iron chelators: regulation of stress fiber-mediated tumor cell migration via modulation of the ROCK1/pMLC2 signaling pathway. Mol Pharmacol 83: 454–469, 2013. doi: 10.1124/mol.112.083097. [DOI] [PubMed] [Google Scholar]
- 57.Unbekandt M, Olson MF. The actin-myosin regulatory MRCK kinases: regulation, biological functions and associations with human cancer. J Mol Med (Berl) 92: 217–225, 2014. doi: 10.1007/s00109-014-1133-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grindrod S, Suy S, Fallen S, Eto M, Toretsky J, Brown ML. Effects of a fluorescent myosin light chain phosphatase inhibitor on prostate cancer cells. Front Oncol 1: 27, 2011. doi: 10.3389/fonc.2011.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu X, Ji Q, Ye N, Sui H, Zhou L, Zhu H, Fan Z, Cai J, Li Q. Berberine inhibits invasion and metastasis of colorectal cancer cells via COX-2/PGE2 mediated JAK2/STAT3 signaling pathway. PLoS One 10: e0123478, 2015. doi: 10.1371/journal.pone.0123478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu Z, Zhang M, Dou D, Tao X, Kang T. Berberine depresses contraction of smooth muscle via inhibiting myosin light-chain kinase. Pharmacogn Mag 13: 454–458, 2017. doi: 10.4103/pm.pm_205_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vennin C, Chin VT, Warren SC, Lucas MC, Herrmann D, Magenau A, , et al. Transient tissue priming via ROCK inhibition uncouples pancreatic cancer progression, sensitivity to chemotherapy, and metastasis. Sci Transl Med 9: eaai8504, 2017. doi: 10.1126/scitranslmed.aai8504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bryan DS, Stack M, Krysztofiak K, Cichoń U, Thomas DG, Surcel A, Schiffhauer ES, Beckett MA, Khodarev NN, Xue L, Poli EC, Pearson AT, Posner MC, Robinson DN, Rock RS, Weichselbaum RR. 4-hydroxyacetophenone modulates the actomyosin cytoskeleton to reduce metastasis. Proc Natl Acad Sci USA 117: 22423–22429, 2020. doi: 10.1073/pnas.2014639117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xin Y, Chen X, Tang X, Li K, Yang M, Tai WC-S, Liu Y, Tan Y. Mechanics and actomyosin-dependent survival/chemoresistance of suspended tumor cells in shear flow. Biophys J 116: 1803–1814, 2019. doi: 10.1016/j.bpj.2019.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Beggs AH, Byers TJ, Knoll JHM, Boyce FM, Bruns GAP, Kunkel LM. Cloning and characterization of two human skeletal muscle alpha-actinin genes located on chromosomes 1 and 11. J Biol Chem 267: 9281–9288, 1992. doi: 10.1016/S0021-9258(19)50420-3. [DOI] [PubMed] [Google Scholar]
- 65.Honda K, Yamada T, Endo R, Ino Y, Gotoh M, Tsuda H, Yamada Y, Chiba H, Hirohashi S. Actinin-4, a novel actin-bundling protein associated with cell motility and cancer invasion. J Cell Biol 140: 1383–1393, 1998[Erratum inJ Cell Biol5: 143, 1998]. doi: 10.1083/jcb.140.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Otey CA, Carpen O. Alpha-actinin revisited: a fresh look at an old player. Cell Motil Cytoskeleton 58: 104–111, 2004. doi: 10.1002/cm.20007. [DOI] [PubMed] [Google Scholar]
- 67.Youssoufian H, McAfee M, Kwiatkowski DJ. Cloning and chromosomal localization of the human cytoskeletal alpha-actinin gene reveals linkage to the beta-spectrin gene. Am J Hum Genet 47: 62–72, 1990. [PMC free article] [PubMed] [Google Scholar]
- 68.Ferrer JM, Lee H, Chen J, Pelz B, Nakamura F, Kamm RD, Lang MJ. Measuring molecular rupture forces between single actin filaments and actin-binding proteins. Proc Natl Acad Sci USA 105: 9221–9226, 2008. doi: 10.1073/pnas.0706124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weins A, Schlondorff JS, Nakamura F, Denker BM, Hartwig JH, Stossel TP, Pollak MR. Disease-associated mutant α-actinin-4 reveals a mechanism for regulating its F-actin-binding affinity. Proc Natl Acad Sci USA 104: 16080–16085, 2007. doi: 10.1073/pnas.0702451104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang X, Pang Y, Zhang J, Shi J, Zhang X, Zhang G, Yang S, Wang J, Hu K, Wang J, Jing H, Ke X, Fu L. High expression levels of ACTN1 and ACTN3 indicate unfavorable prognosis in acute myeloid leukemia. J Cancer 10: 4286–4292, 2019. doi: 10.7150/jca.31766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Koizumi T, Nakatsuji H, Fukawa T, Avirmed S, Fukumori T, Takahashi M, Kanayama H. The role of actinin-4 in bladder cancer invasion. Urology 75: 357–364, 2010. doi: 10.1016/j.urology.2009.09.037. [DOI] [PubMed] [Google Scholar]
- 72.Yoshii H, Ito K, Asano T, Horiguchi A, Hayakawa M, Asano T. Increased expression of α-actinin-4 is associated with unfavorable pathological features and invasiveness of bladder cancer. Oncol Rep 30: 1073–1080, 2013. doi: 10.3892/or.2013.2577. [DOI] [PubMed] [Google Scholar]
- 73.Kovac B, Mäkelä TP, Vallenius T. Increased α-actinin-1 destabilizes E-cadherin-based adhesions and associates with poor prognosis in basal-like breast cancer. PLoS One 13: e0196986, 2018. doi: 10.1371/journal.pone.0196986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Procházková I, Lenčo J, Fučíková A, Dresler J, Čápková L, Hrstka R, Nenutil R, Bouchal P. Targeted proteomics driven verification of biomarker candidates associated with breast cancer aggressiveness. Biochim Biophys Acta Proteins Proteom 1865: 488–498, 2017. doi: 10.1016/j.bbapap.2017.02.012. [DOI] [PubMed] [Google Scholar]
- 75.Sugano T, Yoshida M, Masuda M, Ono M, Tamura K, Kinoshita T, Tsuda H, Honda K, Gemma A, Yamada T. Prognostic impact of ACTN4 gene copy number alteration in hormone receptor-positive, HER2-negative, node-negative invasive breast carcinoma. Br J Cancer 122: 1811–1817, 2020. doi: 10.1038/s41416-020-0821-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang N, Wang Q, Tang H, Zhang F, Zheng Y, Wang S, Zhang J, Wang Z, Xie X. Direct inhibition of ACTN4 by ellagic acid limits breast cancer metastasis via regulation of β-catenin stabilization in cancer stem cells. J Exp Clin Cancer Res 36: 172, 2017. doi: 10.1186/s13046-017-0635-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.An H-T, Yoo S, Ko J. α-Actinin-4 induces the epithelial-to-mesenchymal transition and tumorigenesis via regulation of Snail expression and β-catenin stabilization in cervical cancer. Oncogene 35: 5893–5904, 2016. doi: 10.1038/onc.2016.117. [DOI] [PubMed] [Google Scholar]
- 78.Wang Q, Qin Q, Song R, Zhao C, Liu H, Yang Y, Gu S, Zhou D, He J. NHERF1 inhibits beta-catenin-mediated proliferation of cervical cancer cells through suppression of alpha-actinin-4 expression article. Cell Death Dis 9: 668, 2018. doi: 10.1038/s41419-018-0711-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Honda K, Yamada T, Hayashida Y, Idogawa M, Sato S, Hasegawa F, Ino Y, Ono M, Hirohashi S. Actinin-4 increases cell motility and promotes lymph node metastasis of colorectal cancer. Gastroenterology 128: 51–62, 2005. doi: 10.1053/j.gastro.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 80.Mittal P, Klingler-Hoffmann M, Arentz G, Winderbaum L, Kaur G, Anderson L, Scurry J, Leung Y, Stewart CJ, Carter J, Hoffmann P, Oehler MK. Annexin A2 and alpha actinin 4 expression correlates with metastatic potential of primary endometrial cancer. Biochim Biophys Acta Proteins Proteom 1865: 846–857, 2017. doi: 10.1016/j.bbapap.2016.10.010. [DOI] [PubMed] [Google Scholar]
- 81.Slater M, Cooper M, Murphy CR. The cytoskeletal proteins α-actinin, ezrin, and talin are de-expressed in endometriosis and endometrioid carcinoma compared with normal uterine epithelium. Appl Immunohistochem Mol Morphol 15: 170–174, 2007. doi: 10.1097/01.pai.0000194762.78889.26. [DOI] [PubMed] [Google Scholar]
- 82.Fu L, Yan RQ, Xie D, Hoi YC, Sai MN, Kwong DLW, Li Y, Xin YG. Identification of alpha-actinin 4 and 67 kDa laminin receptor as stage-specific markers in esophageal cancer via proteomic approaches. Cancer 110: 2672–2681, 2007. doi: 10.1002/cncr.23110. [DOI] [PubMed] [Google Scholar]
- 83.Hatakeyama H, Kondo T, Fujii K, Nakanishi Y, Kato H, Fukuda S, Hirohashi S. Protein clusters associated with carcinogenesis, histological differentiation and nodal metastasis in esophageal cancer. Proteomics 6: 6300–6316, 2006. doi: 10.1002/pmic.200600488. [DOI] [PubMed] [Google Scholar]
- 84.Liu X, Chu KM. α-Actinin-4 promotes metastasis in gastric cancer. Lab Invest 97: 1084–1094, 2017. doi: 10.1038/labinvest.2017.28. [DOI] [PubMed] [Google Scholar]
- 85.Quick Q, Skalli O. α-Actinin 1 and α-actinin 4: contrasting roles in the survival, motility, and RhoA signaling of astrocytoma cells. Exp Cell Res 316: 1137–1147, 2010. doi: 10.1016/j.yexcr.2010.02.011. [DOI] [PubMed] [Google Scholar]
- 86.Fukushima S, Yoshida A, Honda K, Maeshima AM, Narita Y, Yamada T, Shibui S, Tsuda H. Immunohistochemical actinin-4 expression in infiltrating gliomas: association with WHO grade and differentiation. Brain Tumor Pathol 31: 11–16, 2014. doi: 10.1007/s10014-013-0139-z. [DOI] [PubMed] [Google Scholar]
- 87.Berania I, Cardin GB, Clément I, Guertin L, Ayad T, Bissada E, Nguyen-Tan PF, Filion E, Guilmette J, Gologan O, Soulieres D, Rodier F, Wong P, Christopoulos A. Four PTEN-targeting co-expressed miRNAs and ACTN4- targeting miR-548b are independent prognostic biomarkers in human squamous cell carcinoma of the oral tongue. Int J Cancer 141: 2318–2328, 2017. doi: 10.1002/ijc.30915. [DOI] [PubMed] [Google Scholar]
- 88.Kakuya T, Mori T, Yoshimoto S, Watabe Y, Miura N, Shoji H, Onidani K, Shibahara T, Honda K. Prognostic significance of gene amplification of ACTN4 in stage I and II oral tongue cancer. Int J Oral Maxillofac Surg 46: 968–976, 2017. doi: 10.1016/j.ijom.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 89.Honda K. The biological role of actinin-4 (ACTN4) in malignant phenotypes of cancer. Cell Biosci . 5: 41, 2015. doi: 10.1186/s13578-015-0031-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Shiraishi H, Fujiwara Y, Kakuya T, Tsuta K, Motoi N, Miura N, Watabe Y, Watanabe S, Noro R, Nagashima K, Huang W, Yamada T, Asamura H, Ohe Y, Honda K. Actinin-4 protein overexpression as a predictive biomarker in adjuvant chemotherapy for resected lung adenocarcinoma. Biomark Med 11: 721–731, 2017. doi: 10.2217/bmm-2017-0150. [DOI] [PubMed] [Google Scholar]
- 91.Noro R, Honda K, Tsuta K, Ishii G, Maeshima AM, Miura N, Furuta K, Shibata T, Tsuda H, Ochiai A, Sakuma T, Nishijima N, Gemma A, Asamura H, Nagai K, Yamada T. Distinct outcome of stage I lung adenocarcinoma with ACTN4 cell motility gene amplification. Ann Oncol 24: 2594–2600, 2013. doi: 10.1093/annonc/mdt293. [DOI] [PubMed] [Google Scholar]
- 92.Hood BL, Grahovac J, Flint MS, Sun M, Charro N, Becker D, Wells A, Conrads TP. Proteomic analysis of laser microdissected melanoma cells from skin organ cultures. J Proteome Res 9: 3656–3663, 2010. doi: 10.1021/pr100164x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Duncan LM, Bouffard D, Howard C, Mihm MC, Byers HR. In situ distribution of integrin α2β1 and α-actinin in melanocytic proliferations [Online]. Mod Pathol 9: 938–943, 1996. [PubMed] [Google Scholar]
- 94.Nikolopoulos SN, Spengler BA, Kisselbach K, Evans AE, Biedler JL, Ross RA. The human non-muscle α-actinin protein encoded by the ACTN4 gene suppresses tumorigenicity of human neuroblastoma cells. Oncogene 19: 380–386, 2000. doi: 10.1038/sj.onc.1203310. [DOI] [PubMed] [Google Scholar]
- 95.Barbolina MV, Adley BP, Kelly DL, Fought AJ, Scholtens DM, Shea LD, Stack MS. Motility-related actinin alpha-4 is associated with advanced and metastatic ovarian carcinoma. Lab Invest 88: 602–614, 2008. doi: 10.1038/labinvest.2008.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yamamoto S, Tsuda H, Honda K, Kita T, Takano M, Tamai S, Inazawa J, Yamada T, Matsubara O. Actinin-4 expression in ovarian cancer: a novel prognostic indicator independent of clinical stage and histological type. Mod Pathol 20: 1278–1285, 2007. doi: 10.1038/modpathol.3800966. [DOI] [PubMed] [Google Scholar]
- 97.Yamamoto S, Tsuda H, Honda K, Onozato K, Takano M, Tamai S, Imoto I, Inazawa J, Yamada T, Matsubara O. Actinin-4 gene amplification in ovarian cancer: a candidate oncogene associated with poor patient prognosis and tumor chemoresistance. Mod Pathol 22: 499–507, 2009. doi: 10.1038/modpathol.2008.234. [DOI] [PubMed] [Google Scholar]
- 98.Yamamoto S, Tsuda H, Honda K, Takano M, Tamai S, Imoto I, Inazawa J, Yamada T, Matsubara O. ACTN4 gene amplification and actinin-4 protein overexpression drive tumour development and histological progression in a high-grade subset of ovarian clear-cell adenocarcinomas. Histopathology 60: 1073–1083, 2012. doi: 10.1111/j.1365-2559.2011.04163.x. [DOI] [PubMed] [Google Scholar]
- 99.Kikuchi S, Honda K, Tsuda H, Hiraoka N, Imoto I, Kosuge T, Umaki T, Onozato K, Shitashige M, Yamaguchi U, Ono M, Tsuchida A, Aoki T, Inazawa J, Hirohashi S, Yamada T. Expression and gene amplification of actinin-4 in invasive ductal carcinoma of the pancreas. Clin Cancer Res 14: 5348–5356, 2008. doi: 10.1158/1078-0432.CCR-08-0075. [DOI] [PubMed] [Google Scholar]
- 100.Pan S, Chen R, Reimel BA, Crispin DA, Mirzaei H, Cooke K, Coleman JF, Lane Z, Bronner MP, Goodlett DR, McIntosh MW, Traverso W, Aebersold R, Brentnall TA. Quantitative proteomics investigation of pancreatic intraepithelial neoplasia. Electrophoresis 30: 1132–1144, 2009. doi: 10.1002/elps.200800752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Welsch T, Keleg S, Bergmann F, Bauer S, Hinz U, Schmidt J. Actinin-4 expression in primary and metastasized pancreatic ductal adenocarcinoma. Pancreas 38: 968–976, 2009. doi: 10.1097/MPA.0b013e3181b28d6f. [DOI] [PubMed] [Google Scholar]
- 102.Shipitsin M, Small C, Choudhury S, Giladi E, Friedlander S, Nardone J, Hussain S, Hurley AD, Ernst C, Huang YE, Chang H, Nifong TP, Rimm DL, Dunyak J, Loda M, Berman DM, Blume-Jensen P. Identification of proteomic biomarkers predicting prostate cancer aggressiveness and lethality despite biopsy-sampling error. Br J Cancer 111: 1201–1212, 2014. doi: 10.1038/bjc.2014.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Myers JS, Vallega KA, White J, Yu K, Yates CC, Sang Q-XA. Proteomic characterization of paired non-malignant and malignant African-American prostate epithelial cell lines distinguishes them by structural proteins. BMC Cancer 17: 480, 2017. doi: 10.1186/s12885-017-3462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hara T, Honda K, Shitashige M, Ono M, Matsuyama H, Naito K, Hirohashi S, Yamada T. Mass spectrometry analysis of the native protein complex containing actinin-4 in prostate cancer cells. Mol Cell Proteomics 6: 479–491, 2007. doi: 10.1074/mcp.M600129-MCP200. [DOI] [PubMed] [Google Scholar]
- 105.Jasavala R, Martinez H, Thumar J, Andaya A, Gingras A-C, Eng JK, Aebersold R, Han DK, Wright ME. Identification of putative androgen receptor interaction protein modules: cytoskeleton and endosomes modulate androgen receptor signaling in prostate cancer cells. Mol Cell Proteomics 6: 252–271, 2007. doi: 10.1074/mcp.M600169-MCP200. [DOI] [PubMed] [Google Scholar]
- 106.Jin S, Bao W, Yang Y-T, Fu Q, Bai Y, Liu Y. Proteomic analysis of the papillary thyroid microcarcinoma. Ann Endocrinol (Paris) 80: 293–300, 2019. doi: 10.1016/j.ando.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 107.Tanaka N, Yamashita T, Yamamoto S, Matsunobu T, Tsuda H, Honda K, Yamada T, Tamai S, Shiotani A. Histological growth pattern of and alpha-actinin-4 expression in thyroid cancer. Anticancer Res 34: 3157–3164, 2014. [PubMed] [Google Scholar]
- 108.Wang Q, Shen Y, Ye B, Hu H, Fan C, Wang T, Zheng Y, Lv J, Ma Y, Xiang M. Gene expression differences between thyroid carcinoma, thyroid adenoma and normal thyroid tissue. Oncol Rep 40: 3359–3369, 2018. doi: 10.3892/or.2018.6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Watabe Y, Mori T, Yoshimoto S, Nomura T, Shibahara T, Yamada T, Honda K. Copy number increase of ACTN4 is a prognostic indicator in salivary gland carcinoma. Cancer Med 3: 613–622, 2014. doi: 10.1002/cam4.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yamada S, Yanamoto S, Yoshida H, Yoshitomi I, Kawasaki G, Mizuno A, Nemoto TK. RNAi-mediated down-regulation of α-actinin-4 decreases invasion potential in oral squamous cell carcinoma. Int J Oral Maxillofac Surg 39: 61–67, 2010. doi: 10.1016/j.ijom.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 111.Shao H, Wang JHC, Pollak MR, Wells A. α-actinin-4 is essential for maintaining the spreading, motility and contractility of fibroblasts. PLoS One 5: e13921, 2010. doi: 10.1371/journal.pone.0013921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Hsu KS, Kao HY. Alpha-actinin 4 and tumorigenesis of breast cancer. Vitam Horm 93: 323–351, 2013. doi: 10.1016/b978-0-12-416673-8.00005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gao Y, Li G, Sun L, He Y, Li X, Sun Z, Wang J, Jiang Y, Shi J. ACTN4 and the pathways associated with cell motility and adhesion contribute to the process of lung cancer metastasis to the brain. BMC Cancer 15: 277, 2015. doi: 10.1186/s12885-015-1295-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fukumoto M, Kurisu S, Yamada T, Takenawa T. α-Actinin-4 enhances colorectal cancer cell invasion by suppressing focal adhesion maturation. PLoS One 10: e0120616, 2015. doi: 10.1371/journal.pone.0120616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang K, Singer SJ. Interaction of filamin with f-actin in solution. Proc Natl Acad Sci USA 74: 2021–2025, 1977. doi: 10.1073/pnas.74.5.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Stossel TP, Condeelis J, Cooley L, Hartwig JH, Noegel A, Schleicher M, Shapiro SS. Filamins as integrators of cell mechanics and signalling. Nat Rev Mol Cell Biol 2: 138–145, 2001. doi: 10.1038/35052082. [DOI] [PubMed] [Google Scholar]
- 117.Xu Wf, Xie ZW, Chung DW, Davie EW. A novel human actin-binding protein homologue that binds to platelet glycoprotein Ib. Blood 92: 1268–1276, 1998. doi: 10.1182/blood.V92.4.1268. [DOI] [PubMed] [Google Scholar]
- 118.Fujita M, Mitsuhashi H, Isogai S, Nakata T, Kawakami A, Nonaka I, Noguchi S, Hayashi YK, Nishino I, Kudo A. Filamin C plays an essential role in the maintenance of the structural integrity of cardiac and skeletal muscles, revealed by the medaka mutant zacro. Dev Biol 361: 79–89, 2012. doi: 10.1016/j.ydbio.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 119.Maestrini E, Patrosso C, Mancini M, Rivella S, Rocchi M, Repetto M, Villa A, Frattini A, Zoppè M, Vezzoni P, Toniolo D. Mapping of two genes encoding isoforms of the actin binding protein ABP-280, a dystrophin like protein, to Xq28 and to chromosome 7. Hum Mol Genet 2: 761–766, 1993. doi: 10.1093/hmg/2.6.761. [DOI] [PubMed] [Google Scholar]
- 120.Feng Y, Walsh CA. The many faces of filamin: a versatile molecular scaffold for cell motility and signalling. Nat Cell Biol 6: 1034–1038, 2004. doi: 10.1038/ncb1104-1034. [DOI] [PubMed] [Google Scholar]
- 121.Glogauer M, Arora P, Chou D, Janmey PA, Downey GP, McCulloch CA. The role of actin-binding protein 280 in integrin-dependent mechanoprotection. J Biol Chem 273: 1689–1698, 1998. doi: 10.1074/jbc.273.3.1689. [DOI] [PubMed] [Google Scholar]
- 122.Zhou A-X, Hartwig JH, Akyürek LM. Filamins in cell signaling, transcription and organ development. Trends Cell Biol . 20: 113–123, 2010. doi: 10.1016/j.tcb.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 123.Sawyer GM, Clark AR, Robertson SP, Sutherland-Smith AJ. Disease-associated substitutions in the filamin B actin binding domain confer enhanced actin binding affinity in the absence of major structural disturbance: insights from the crystal structures of filamin B actin binding domains. J Mol Biol 390: 1030–1047, 2009. doi: 10.1016/j.jmb.2009.06.009. [DOI] [PubMed] [Google Scholar]
- 124.Nakamura F, Hartwig JH, Stossel TP, Szymanski PT. Ca2+ and calmodulin regulate the binding of filamin A to actin filaments. J Biol Chem 280: 32426–32433, 2005. doi: 10.1074/jbc.M502203200. [DOI] [PubMed] [Google Scholar]
- 125.Kasza KE, Nakamura F, Hu S, Kollmannsberger P, Bonakdar N, Fabry B, Stossel TP, Wang N, Weitz DA. Filamin A is essential for active cell stiffening but not passive stiffening under external force. Biophys J 96: 4326–4335, 2009. doi: 10.1016/j.bpj.2009.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Nomura M, Yoshikawa K, Tanaka T, Sobue K, Maruyama K. The role of tropomyosin in the interactions of F‐actin with caldesmon and actin‐binding protein (or filamin). Eur J Biochem 163: 467–471, 1987. doi: 10.1111/j.1432-1033.1987.tb10892.x. [DOI] [PubMed] [Google Scholar]
- 127.Ohta Y, Hartwig JH. Actin filament crosslinking by chicken gizzard filamin is regulated by phosphorylation in vitro. Biochemistry 34: 6745–6754, 1995. doi: 10.1021/bi00020a020. [DOI] [PubMed] [Google Scholar]
- 128.Furuhashi K, Inagaki M, Hatano S, Fukami K, Takenawa T. Inositol phospholipid - Induced suppression of F-actin-gelating activity of smooth muscle filamin. Biochem Biophys Res Commun 184: 1261–1265, 1992.doi: 10.1016/s0006-291x(05)80018-x. [DOI] [PubMed] [Google Scholar]
- 129.Niederman R, Amrein PC, Hartwig J. Three-dimensional structure of actin filaments and of an actin gel made with actin-binding protein. J Cell Biol 96: 1400–1413, 1983. doi: 10.1083/jcb.96.5.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Cunningham CC, Gorlin JB, Kwiatkowski DJ, Hartwig JH, Janmey PA, Byers HR, Stossel TP. Actin-binding protein requirement for cortical stability and efficient locomotion. Science 255: 325–327, 1992. doi: 10.1126/science.1549777. [DOI] [PubMed] [Google Scholar]
- 131.Zhang K, Zhu T, Gao D, Zhang Y, Zhao Q, Liu S, Su T, Bernier M, Zhao R. Filamin A expression correlates with proliferation and invasive properties of human metastatic melanoma tumors: implications for survival in patients. J Cancer Res Clin Oncol 140: 1913–1926, 2014. doi: 10.1007/s00432-014-1722-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Savoy RM, Ghosh PM. The dual role of filamin A in cancer: can’t live with (too much of) it, can’t live without it. Endocr Relat Cancer 20: R341–R356, 2013. doi: 10.1530/ERC-13-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Li X-C, Huang C-X, Wu S-K, Yu L, Zhou G-J, Chen L-J. Biological roles of filamin a in prostate cancer cells. Int Braz J Urol 45: 916–924, 2019. doi: 10.1590/S1677-5538.IBJU.2018.0535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Xu Y, Bismar TA, Su J, Xu B, Kristiansen G, Varga Z, Teng L, Ingber DE, Mammoto A, Kumar R, Alaoui-Jamali MA. Filamin A regulates focal adhesion disassembly and suppresses breast cancer cell migration and invasion. J Exp Med 207: 2421–2437, 2010. doi: 10.1084/jem.20100433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang K, Zhu T-N, Zhao R-J. Filamin A regulates EGFR/ERK/Akt signaling and affects colorectal cancer cell growth and migration. Mol Med Rep 20: 3671–3678, 2019. doi: 10.3892/mmr.2019.10622. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 136.Bandaru S, Zhou A-X, Rouhi P, Zhang Y, Bergo MO, Cao Y, Akyürek LM. Targeting filamin B induces tumor growth and metastasis via enhanced activity of matrix metalloproteinase-9 and secretion of VEGF-A. Oncogenesis 3: e119, 2014. doi: 10.1038/oncsis.2014.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Tian H-M, Liu X-H, Han W, Zhao L-L, Yuan B, Yuan C-J. Differential expression of filamin A and its clinical significance in breast cancer. Oncol Lett 6: 681–686, 2013. doi: 10.3892/ol.2013.1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Zhou A-X, Toylu A, Nallapalli RK, Nilsson G, Atabey N, Heldin C-H, Borén J, Bergo MO, Akyürek LM. Filamin a mediates HGF/c-MET signaling in tumor cell migration. Int J Cancer 128: 839–846, 2011. doi: 10.1002/ijc.25417. [DOI] [PubMed] [Google Scholar]
- 139.Jiang X, Yue J, Lu H, Campbell N, Yang Q, Lan S, Haffty BG, Yuan C, Shen Z. Inhibition of filamin-A reduces cancer metastatic potential. Int J Biol Sci 9: 67–77, 2013. doi: 10.7150/ijbs.5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Jin YZ, Pei CZ, Wen LY. FLNA is a predictor of chemoresistance and poor survival in cervical cancer. Biomark Med 10: 711–719, 2016. doi: 10.2217/bmm-2016-0056. [DOI] [PubMed] [Google Scholar]
- 141.Cheng M, Jiang Y, Yang H, Zhao D, Li L, Liu X. FLNA promotes chemoresistance of colorectal cancer through inducing epithelial-mesenchymal transition and smad2 signaling pathway. Am J Cancer Res 10: 403–423, 2020. [PMC free article] [PubMed] [Google Scholar]
- 142.Sun GG, Sheng SH, Jing SW, Hu WN. An antiproliferative gene FLNA regulates migration and invasion of gastric carcinoma cell in vitro and its clinical significance. Tumor Biol 35: 2641–2648, 2014. doi: 10.1007/s13277-013-1347-1. [DOI] [PubMed] [Google Scholar]
- 143.Chantaravisoot N, Wongkongkathep P, Loo JA, Mischel PS, Tamanoi F. Significance of filamin A in mTORC2 function in glioblastoma. Mol Cancer 14: 127, 2015. doi: 10.1186/s12943-015-0396-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Uramoto H, Akyurek LM, Hanagiri T. A positive relationship between filamin and VEGF in patients with lung cancer. Anticancer Res 30: 3939–3944, 2010. [PubMed] [Google Scholar]
- 145.Bedolla RG, Wang Y, Asuncion A, Chamie K, Siddiqui S, Mudryj MM, Prihoda TJ, Siddiqui J, Chinnaiyan AM, Mehra R, De Vere White RW, Ghosh PM. Nuclear versus cytoplasmic localization of filamin a in prostate cancer: immunohistochemical correlation with metastases. Clin Cancer Res 15: 788–796, 2009. doi: 10.1158/1078-0432.CCR-08-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Aitken A, Jones D, Soneji Y, Howell S. 14-3-3 proteins: biological function and domain structure. Biochem Soc Trans 23: 605–611, 1995. doi: 10.1042/bst0230605. [DOI] [PubMed] [Google Scholar]
- 147.Hermeking H, Lengauer C, Polyak K, He TC, Zhang L, Thiagalingam S, Kinzler KW, Vogelstein B. 14-3-3σ is a p53-regulated inhibitor of G2/M progression. Mol Cell 1: 3–11, 1997. doi: 10.1016/S1097-2765(00)80002-7. [DOI] [PubMed] [Google Scholar]
- 148.Xing H, Zhang S, Weinheimer C, Kovacs A, Muslin AJ. 14-3-3 proteins block apoptosis and differentially regulate MAPK cascades. EMBO J 19: 349–358, 2000. doi: 10.1093/emboj/19.3.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Zhang L, Chen J, Fu H. Suppression of apoptosis signal-regulating kinase 1-induced cell death by 14-3-3 proteins. Proc Natl Acad Sci USA 96: 8511–8515, 1999. doi: 10.1073/pnas.96.15.8511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ford JC, Al-Khodairy F, Fotou E, Sheldrick KS, Griffiths DJF, Carr AM. 14-3-3 protein homologs required for the DNA damage checkpoint in fission yeast. Science 265: 533–535, 1994. doi: 10.1126/science.8036497. [DOI] [PubMed] [Google Scholar]
- 151.Oecking C, Eckerskorn C, Weiler EW. The fusicoccin receptor of plants is a member of the 14-3-3 superfamily of eukaryotic regulatory proteins. FEBS Lett 352: 163–166, 1994. doi: 10.1016/0014-5793(94)00949-X. [DOI] [PubMed] [Google Scholar]
- 152.Rose R, Erdmann S, Bovens S, Wolf A, Rose M, Hennig S, Waldmann H, Ottmann C. Identification and structure of small-molecule stabilizers of 14-3-3 protein-protein interactions. Angew Chem Int Ed Engl 49: 4129–4132, 2010. doi: 10.1002/anie.200907203. [DOI] [PubMed] [Google Scholar]
- 153.Wu H, Ge J, Yao SQ. Microarray-assisted high-throughput identification of a cell-permeable small-molecule binder of 14-3-3 proteins. Angew Chem Int Ed Engl 49: 6528–6532, 2010. doi: 10.1002/anie.201003257. [DOI] [PubMed] [Google Scholar]
- 154.Stevers LM, Sijbesma E, Botta M, Mackintosh C, Obsil T, Landrieu I, Cau Y, Wilson AJ, Karawajczyk A, Eickhoff J, Davis J, Hann M, O’Mahony G, Doveston RG, Brunsveld L, Ottmann C. Modulators of 14-3-3 protein-protein interactions. J Med Chem 61: 3755–3778, 2018. doi: 10.1021/acs.jmedchem.7b00574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Zhou Q, Kee Y-S, Poirier CC, Jelinek C, Osborne J, Divi S, Surcel A, Will ME, Eggert US, Müller-Taubenberger A, Iglesias PA, Cotter RJ, Robinson DN. 14-3-3 coordinates microtubules, Rac, and myosin II to control cell mechanics and cytokinesis. Curr Biol 20: 1881–1889, 2010. doi: 10.1016/j.cub.2010.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.West-Foyle H, Kothari P, Osborne J, Robinson DN. 14-3-3 proteins tune non-muscle myosin II assembly. J Biol Chem 293: 6751–6761, 2018. doi: 10.1074/jbc.M117.819391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Fan X, Cui L, Zeng Y, Song W, Gaur U, Yang M. 14-3-3 proteins are on the crossroads of cancer, aging, and age-related neurodegenerative disease. Int J Mol Sci. 20: 3518, 2019. doi: 10.3390/ijms20143518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhang Z, Zhang G, Kong C. High expression of Cdc25B and low expression of 14-3-3σ is associated with the development and poor prognosis in urothelial carcinoma of bladder. Tumour Biol 35: 2503–2512, 2014. doi: 10.1007/s13277-013-1331-9. [DOI] [PubMed] [Google Scholar]
- 159.Mhawech-Fauceglia P, Fischer G, Alvarez V, Ahmed A, Herrmann FR. Predicting outcome in minimally invasive (T1a and T1b) urothelial bladder carcinoma using a panel of biomarkers: a high throughput tissue microarray analysis. BJU Int 100: 1182–1187, 2007. doi: 10.1111/j.1464-410X.2007.07090.x. [DOI] [PubMed] [Google Scholar]
- 160.Vercoutter-Edouart AS, Lemoine J, Le Bourhis X, Louis H, Boilly B, Nurcombe V, Révillion F, Peyrat JP, Hondermarck H. Proteomic analysis reveals that 14-3-3σ is down-regulated in human breast cancer cells. Cancer Res 61: 76–80, 2001. [PubMed] [Google Scholar]
- 161.Ferguson AT, Evron E, Umbricht CB, Pandita TK, Chan TA, Hermeking H, Marks JR, Lambers AR, Futreal PA, Stampfer MR, Sukumar S. High frequency of hypermethylation at the 14-3-3 σ locus leads to gene silencing in breast cancer. Proc Natl Acad Sci USA 97: 6049–6054, 2000. doi: 10.1073/pnas.100566997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Cimino D, Fuso L, Sfiligoi C, Biglia N, Ponzone R, Maggiorotto F, Russo G, Cicatiello L, Weisz A, Taverna D, Sismondi P, De Bortoli M. Identification of new genes associated with breast cancer progression by gene expression analysis of predefined sets of neoplastic tissues. Int J Cancer 123: 1327–1338, 2008. doi: 10.1002/ijc.23660. [DOI] [PubMed] [Google Scholar]
- 163.Yang Y-F, Lee Y-C, Wang Y-Y, Wang C-H, Hou M-F, Yuan S-SF. YWHAE promotes proliferation, metastasis, and chemoresistance in breast cancer cells. Kaohsiung J Med Sci 35: 408–416, 2019. doi: 10.1002/kjm2.12075. [DOI] [PubMed] [Google Scholar]
- 164.Hiraoka E, Mimae T, Ito M, Kadoya T, Miyata Y, Ito A, Okada M. Breast cancer cell motility is promoted by 14-3-3γ. Breast Cancer 26: 581–593, 2019[Erratum inBreast Cancer26: 594, 2019]. doi: 10.1007/s12282-019-00957-4. [DOI] [PubMed] [Google Scholar]
- 165.Wang B, Liu K, Lin H-Y, Bellam N, Ling S, Lin W-C. 14-3-3τ regulates ubiquitin-independent proteasomal degradation of p21, a novel mechanism of p21 downregulation in breast cancer. Mol Cell Biol 30: 1508–1527, 2010. doi: 10.1128/MCB.01335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zang L, Toy DP, Hancock WS, Sgroi DC, Karger BL. Proteomic analysis of ductal carcinoma of the breast using laser capture microdissection, LC-MS, and 16O/18O isotopic labeling. J Proteome Res 3: 604–612, 2004. doi: 10.1021/pr034131l. [DOI] [PubMed] [Google Scholar]
- 167.Santpere G, Alcaráz-Sanabria A, Corrales-Sánchez V, Pandiella A, Gyorffy B, Ocaña A. Transcriptome evolution from breast epithelial cells to basallike tumors. Oncotarget 9: 453–463, 2018. doi: 10.18632/oncotarget.23065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Xu T, Pang Q, Zhou D, Zhang A, Luo S, Wang Y, Yan X. Proteomic investigation into betulinic acid-induced apoptosis of human cervical cancer HeLa cells. PLoS One 9: e105768, 2014. doi: 10.1371/journal.pone.0105768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Sano T, Shimooka H, Weixa P, Segawa A, Jian Z, Motegi A, Nakayama H, Oyama T, Nakajima T. Immunohistochemical expression of 14-3-3 sigma protein in various histological subtypes of uterine cervical cancers. Pathol Int 54: 743–750, 2004. doi: 10.1111/j.1440-1827.2004.01747.x. [DOI] [PubMed] [Google Scholar]
- 170.O’Dwyer D, Ralton LD, O’Shea A, Murray GI. The proteomics of colorectal cancer: identification of a protein signature associated with prognosis. PLoS One 6: e27718, 2011. doi: 10.1371/journal.pone.0027718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Suzuki H, Itoh F, Toyota M, Kikuchi T, Kakiuchi H, Imai K. Inactivation of the 14-3-3 σ gene is associated with 5′ CpG island hypermethylation in human cancers. Cancer Res 60: 4353–4357, 2000. http://cancerres.aacrjournals.org/cgi/content/full/60/16/4353 [21 Aug. 2020]. [PubMed] [Google Scholar]
- 172.Kirana C, Peng L, Miller R, Keating JP, Glenn C, Shi H, Jordan TW, Maddern GJ, Stubbs RS. Combination of laser microdissection, 2D-DIGE and MALDI-TOF MS to identify protein biomarkers to predict colorectal cancer spread. Clin Proteomics 16: 3, 2019. doi: 10.1186/s12014-019-9223-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Wang H, Huang H, Li W, Jin X, Zeng J, Liu Y, Gu Y, Sun X, Wen G, Ding Y, Zhao L. Nuclear localization of 14-3-3epsilon inversely correlates with poor long-term survival of patients with colorectal cancer. J Surg Oncol 106: 224–231, 2012. doi: 10.1002/jso.22152. [DOI] [PubMed] [Google Scholar]
- 174.Ahluwalia P, Mondal AK, Bloomer C, Fulzele S, Jones K, Ananth S, Gahlay GK, Heneidi S, Rojiani AM, Kota V, Kolhe R. Identification and clinical validation of a novel 4 gene-signature with prognostic utility in colorectal cancer. Int J Mol Sci 20: 3818, 2019. doi: 10.3390/ijms20153818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Young GM, Radhakrishnan VM, Centuori SM, Gomes CJ, Martinez JD. Comparative analysis of 14-3-3 isoform expression and epigenetic alterations in colorectal cancer. BMC Cancer 15: 826, 2015. doi: 10.1186/s12885-015-1856-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Mhawech P, Benz A, Cerato C, Greloz V, Assaly M, Desmond JC, Koeffler HP, Lodygin D, Hermeking H, Herrmann F, Schwaller J. Downregulation of 14-3-3sigma in ovary, prostate and endometrial carcinomas is associated with CpG island methylation. Mod Pathol 18: 340–348, 2005. doi: 10.1038/modpathol.3800240. [DOI] [PubMed] [Google Scholar]
- 177.Qi YJ, Wang M, Liu R-M, Wei H, Chao W-X, Zhang T, Lou Q, Li X-M, Ma J, Zhu H, Yang Z-H, Liu H-Q, Ma Y-F. Downregulation of 14-3-3s correlates with multistage carcinogenesis and poor prognosis of esophageal squamous cell carcinoma. PLoS One 9: e95386, 2014. doi: 10.1371/journal.pone.0095386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Bajpai U, Sharma R, Kausar T, Dattagupta S, Chattopadhayay TK, Ralhan R. Clinical significance of 14-3-3 zeta in human esophageal cancer. Int J Biol Markers 23: 231–237, 2008. doi: 10.5301/jbm.2009.2112. [DOI] [PubMed] [Google Scholar]
- 179.Nishimura Y, Komatsu S, Ichikawa D, Nagata H, Hirajima S, Takeshita H, Kawaguchi T, Arita T, Konishi H, Kashimoto K, Shiozaki A, Fujiwara H, Okamoto K, Tsuda H, Otsuji E. Overexpression of YWHAZ relates to tumor cell proliferation and malignant outcome of gastric carcinoma. Br J Cancer 108: 1324–1331, 2013. doi: 10.1038/bjc.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Ma Y, Li Y-F, Wang T, Pang R, Xue Y-W, Zhao S-P. Identification of proteins associated with lymph node metastasis of gastric cancer. J Cancer Res Clin Oncol 140: 1739–1749, 2014. doi: 10.1007/s00432-014-1679-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Tseng C-W, Yang J-C, Chen C-N, Huang H-C, Chuang K-N, Lin C-C, Lai H-S, Lee P-H, Chang K-J, Juan H-F. Identification of 14-3-3β in human gastric cancer cells and its potency as a diagnostic and prognostic biomarker. Proteomics 11: 2423–2439, 2011. doi: 10.1002/pmic.201000449. [DOI] [PubMed] [Google Scholar]
- 182.Leal MF, Calcagno DQ, Demachki S, Assumpção PP, Chammas R, Burbano RR, Smith M de AC. Clinical implication of 14-3-3 epsilon expression in gastric cancer. World J Gastroenterol 18: 1531–1537, 2012. doi: 10.3748/wjg.v18.i13.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Mühlmann G, Öfner D, Zitt M, Müller HM, Maier H, Moser P, Schmid KW, Zitt M, Amberger A. 14-3-3 sigma and p53 expression in gastric cancer and its clinical applications. Dis Markers 29: 21–29, 2010. doi: 10.3233/DMA-2010-0722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Zhou WH, Tang F, Xu J, Wu X, Feng ZY, Li HG, Lin DJ, Shao CK, Liu Q. Aberrant upregulation of 14-3-3o′ expression serves as an inferior prognostic biomarker for gastric cancer. BMC Cancer 11: 397, 2011. doi: 10.1186/1471-2407-11-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Sun J, Long Y, Peng X, Xiao D, Zhou J, Tao Y, Liu S. The survival analysis and oncogenic effects of CFP1 and 14-3-3 expression on gastric cancer. Cancer Cell Int 19: 225, 2019. doi: 10.1186/s12935-019-0946-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Park S-G, Jung S, Ryu H-H, Jung T-Y, Moon K-S, Kim I-Y, Jeong Y Il,Pei J, Park S-J, Kang S-S. Role of 14-3-3-beta in the migration and invasion in human malignant glioma cell line U87MG. Neurol Res 34: 893–900, 2012. doi: 10.1179/1743132812Y.0000000087. [DOI] [PubMed] [Google Scholar]
- 187.Com E, Clavreul A, Lagarrigue M, Michalak S, Menei P, Pineau C. Quantitative proteomic isotope-coded protein label (ICPL) analysis reveals alteration of several functional processes in the glioblastoma. J Proteomics 75: 3898–3913, 2012. doi: 10.1016/j.jprot.2012.04.034. [DOI] [PubMed] [Google Scholar]
- 188.Liang S, Shen G, Liu Q, Xu Y, Zhou L, Xiao S, Xu Z, Gong F, You C, Wei Y. Isoform-specific expression and characterization of 14-3-3 proteins in human glioma tissues discovered by stable isotope labeling with amino acids in cell culture-based proteomic analysis. Proteomics Clin Appl 3: 743–753, 2009. doi: 10.1002/prca.200800198. [DOI] [PubMed] [Google Scholar]
- 189.Petri MK, Koch P, Stenzinger A, Kuchelmeister K, Nestler U, Paradowska A, Steger K, Brobeil A, Viard M, Wimmer M. PTPIP51, a positive modulator of the MAPK/Erk pathway, is upregulated in glioblastoma and interacts with 14-3-3ß and PTP1B in situ. Histol Histopathol 26: 1531–1543, 2011. doi: 10.14670/HH-26.1531. [DOI] [PubMed] [Google Scholar]
- 190.Wang H, Zhi H, Ma D, Li T. MiR-217 promoted the proliferation and invasion of glioblastoma by repressing YWHAG. Cytokine 92: 93–102, 2017. doi: 10.1016/j.cyto.2016.12.013. [DOI] [PubMed] [Google Scholar]
- 191.Chen W, Ye F, Cui M, Sikora AG, Wang X, Wang P, Cui X, Guo X, Zhu W, Zhang DY. Protein marker profiling in different T classification in laryngeal squamous cell carcinoma. Head Neck 37: 357–365, 2015. doi: 10.1002/hed.23607. [DOI] [PubMed] [Google Scholar]
- 192.Che X-H, Chen H, Xu Z-M, Shang C, Sun K-L, Fu W-N. 14-3-3epsilon contributes to tumour suppression in laryngeal carcinoma by affecting apoptosis and invasion. BMC Cancer 10: 306, 2010. doi: 10.1186/1471-2407-10-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Matta A, Desouza L V, Shukla NK, Gupta SD, Ralhan R, Michael Siu KW. Prognostic significance of head-and-neck cancer biomarkers previously discovered and identified using iTRAQ-labeling and multidimensional liquid chromatography-tandem mass spectrometry. J Proteome Res 7: 2078–2087, 2008. doi: 10.1021/pr7007797. [DOI] [PubMed] [Google Scholar]
- 194.Villaret DB, Wang T, Dillon D, Xu J, Sivam D, Cheever MA, Reed SG. Identification of genes overexpressed in head and neck squamous cell carcinoma using a combination of complementary DNA subtraction and microarray analysis. Laryngoscope 110: 374–381, 2000. doi: 10.1097/00005537-200003000-00008. [DOI] [PubMed] [Google Scholar]
- 195.Lin M, Morrison CD, Jones S, Mohamed N, Bacher J, Plass C. Copy number gain and oncogenic activity of YWHAZ/14-3-3ζ in head and neck squamous cell carcinoma. Int J Cancer 125: 603–611, 2009. doi: 10.1002/ijc.24346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wu C-Y, Jan Y-J, Ko B-S, Wu Y-J, Wu Y-J, Liou J-Y. Prognostic significance of 14-3-3ε, aldo-keto reductase family 1 B10 and metallothionein-1 in hepatocellular carcinoma. Anticancer Res 38: 6855–6863, 2018. doi: 10.21873/anticanres.13060. [DOI] [PubMed] [Google Scholar]
- 197.Liu T-A, Jan Y-J, Ko B-S, Chen S-C, Liang S-M, Hung Y-L, Hsu C, Shen T-L, Lee Y-M, Chen P-F, Wang J, Shyue S-K, Liou J-Y. Increased expression of 14-3-3β promotes tumor progression and predicts extrahepatic metastasis and worse survival in hepatocellular carcinoma. Am J Pathol 179: 2698–2708, 2011[Erratum inAm J Pathol180: 429, 2012]. doi: 10.1016/j.ajpath.2011.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Ko B-S, Chang T-C, Hsu C, Chen Y-C, Shen T-L, Chen S-C, Wang J, Wu K-K, Jan Y-J, Liou J-Y. Overexpression of 14-3-3ε predicts tumour metastasis and poor survival in hepatocellular carcinoma. Histopathology 58: 705–711, 2011. doi: 10.1111/j.1365-2559.2011.03789.x. [DOI] [PubMed] [Google Scholar]
- 199.Iwata N, Yamamoto H, Sasaki S, Itoh F, Suzuki H, Kikuchi T, Kaneto H, Iku S, Ozeki I, Karino Y, Satoh T, Toyota J, Satoh M, Endo T, Imai K. Frequent hypermethylation of CpG islands and loss of expression of the 14-3-3 σ gene in human hepatocellular carcinoma. Oncogene 19: 5298–5302, 2000. doi: 10.1038/sj.onc.1203898. [DOI] [PubMed] [Google Scholar]
- 200.Tang Y, Liu S, Li N, Guo W, Shi J, Yu H, Zhang L, Wang K, Liu S, Cheng S. 14-3-3ζ promotes hepatocellular carcinoma venous metastasis by modulating hypoxia-inducible factor-1α. Oncotarget 7: 15854–15867, 2016. doi: 10.18632/oncotarget.7493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Qi W, Liu X, Qiao D, Martinez JD. Isoform-specific expression of 14-3-3 proteins in human lung cancer tissues. Int J Cancer 113: 359–363, 2005. doi: 10.1002/ijc.20492. [DOI] [PubMed] [Google Scholar]
- 202.Qi W, Liu X, Chen W, Li Q, Martinez JD. Overexpression of 14-3-3γ causes polyploidization in H322 lung cancer cells. Mol Carcinog 46: 847–856, 2007. doi: 10.1002/mc.20314. [DOI] [PubMed] [Google Scholar]
- 203.Lee T-G, Jeong E-H, Kim S-Y, Kim H-R, Kim H, Kim C-H. Fhit, a tumor suppressor protein, induces autophagy via 14-3-3t in non-small cell lung cancer cells. Oncotarget 8: 31923–31937, 2017. doi: 10.18632/oncotarget.16652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Raungrut P, Wongkotsila A, Lirdprapamongkol K, Svasti J, Geater SL, Phukaoloun M, Suwiwat S, Thongsuksai P. Prognostic significance of 14-3-3γ overexpression in advanced non-small cell lung cancer. Asian Pac J Cancer Prev 15: 3513–3518, 2014. doi: 10.7314/apjcp.2014.15.8.3513. [DOI] [PubMed] [Google Scholar]
- 205.Wang P, Deng Y, Fu XN. MiR-509-5p suppresses the proliferation, migration, and invasion of non-small cell lung cancer by targeting YWHAG. Biochem Biophys Res Commun 482: 935–941, 2017. doi: 10.1016/j.bbrc.2016.11.136. [DOI] [PubMed] [Google Scholar]
- 206.Yatabe Y, Osada H, Tatematsu Y, Mitsudomi T, Takahashi T. Decreased expression of 14-3-3σ in neuroendocrine tumors is independent of origin and malignant potential. Oncogene 21: 8310–8319, 2002. doi: 10.1038/sj.onc.1206014. [DOI] [PubMed] [Google Scholar]
- 207.Radhakrishnan VM, Jensen TJ, Cui H, Futscher BW, Martinez JD. Hypomethylation of the 14-3-3σ promoter leads to increased expression in non-small cell lung cancer. Genes Chromosomes Cancer 50: 830–836, 2011. doi: 10.1002/gcc.20904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Akahira J-I, Sugihashi Y, Suzuki T, Ito K, Niikura H, Moriya T, Nitta M, Okamura H, Inoue S, Sasano H, Okamura K, Yaegashi N. Decreased expression of 14-3-3σ is associated with advanced disease in human epithelial ovarian cancer: its correlation with aberrant DNA methylation. Clin Cancer Res 10: 2687–2693, 2004. doi: 10.1158/1078-0432.CCR-03-0510. [DOI] [PubMed] [Google Scholar]
- 209.Ravi D, Chen Y, Karia B, Brown A, Gu TT, Li J, Carey MS, Hennessy BT, Bishop AJR. 14-3-3 σ expression effects G2/M response to oxygen and correlates with ovarian cancer metastasis. PLoS One 6: e15864, 2011. doi: 10.1371/journal.pone.0015864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Kim HJ, Sung SH, Kim CY, Bae MK, Cho MS, Kim YH, Kim SC, Ju W. 14-3-3ζ overexpression is associated with poor prognosis in ovarian cancer. Yonsei Med J 59: 51–56, 2018. doi: 10.3349/ymj.2018.59.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Neupane D, Korc M. 14-3-3σ modulates pancreatic cancer cell survival and invasiveness. Clin Cancer Res 14: 7614–7623, 2008. doi: 10.1158/1078-0432.CCR-08-1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Logsdon CD, Simeone DM, Binkley C, Arumugam T, Greenson JK, Giordano TJ, Misek DE, Hanash S. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res 63: 2649–2657, 2003. [PubMed] [Google Scholar]
- 213.Shen J, Person MD, Zhu J, Abbruzzese JL, Li D. Protein expression profiles in pancreatic adenocarcinoma compared with normal pancreatic tissue and tissue affected by pancreatitis as detected by two-dimensional gel electrophoresis and mass spectrometry. Cancer Res 64: 9018–9026, 2004. doi: 10.1158/0008-5472.CAN-04-3262. [DOI] [PubMed] [Google Scholar]
- 214.Singh AN, Sharma N. Quantitative SWATH-based proteomic profiling for identification of mechanism-driven diagnostic biomarkers conferring in the progression of metastatic prostate cancer. Front Oncol 10: 493, 2020. doi: 10.3389/fonc.2020.00493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Lodygin D, Diebold J, Hermeking H. Prostate cancer is characterized by epigenetic silencing of 14-3-3σ expression. Oncogene 23: 9034–9041, 2004. doi: 10.1038/sj.onc.1208004. [DOI] [PubMed] [Google Scholar]
- 216.Oh S, Shin S, Lightfoot SA, Janknecht R. 14-3-3 proteins modulate the ETS transcription factor ETV1 in prostate cancer. Cancer Res 73: 5110–5119, 2013. doi: 10.1158/0008-5472.CAN-13-0578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Cheng L, Pan C-X, Zhang J-T, Zhang S, Kinch MS, Li L, Baldridge LA, Wade C, Hu Z, Koch MO, Ulbright TM, Eble JN. Loss of 14-3-3 in prostate cancer and its precursors. Clin Cancer Res 10: 3064–3068, 2004. doi: 10.1158/1078-0432.ccr-03-0652. [DOI] [PubMed] [Google Scholar]
- 218.Liang S, Xu Y, Shen G, Liu Q, Zhao X, Xu Z, Xie X, Gong F, Li R, Wei Y. Quantitative protein expression profiling of 14-3-3 isoforms in human renal carcinoma shows 14-3-3 epsilon is involved in limitedly increasing renal cell proliferation. Electrophoresis, 30: 4152–4162, 2009. doi: 10.1002/elps.200900249. [DOI] [PubMed] [Google Scholar]
- 219.Musholt TJ, Brehm C, Hanack J, von Wasielewski R, Musholt PB. Identification of differentially expressed genes in papillary thyroid carcinomas with and without rearrangements of the tyrosine kinase receptors RET and/or NTRK1. J Surg Res 131: 15–25, 2006. doi: 10.1016/j.jss.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 220.Li X-H, Noguchi A, Nishida T, Takahashi H, Zheng Y, Yang X-H, Masuda S, Kikuchi K, Takano Y. Cytoplasmic expression of ING1b is correlated with tumorigenesis and progression of head and neck squamous cell carcinoma. Histol Histopathol 26: 597–607, 2011. doi: 10.14670/HH-26.597. [DOI] [PubMed] [Google Scholar]
- 221.Heidenblad M, Lindgren D, Jonson T, Liedberg F, Veerla S, Chebil G, Gudjonsson S, Borg Å, Månsson W, Höglund M. Tiling resolution array CGH and high density expression profiling of urothelial carcinomas delineate genomic amplicons and candidate target genes specific for advanced tumors. BMC Med Genomics 1: 3, 2008. doi: 10.1186/1755-8794-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Prasad GL, Valverius EM, McDuffie E, Cooper HL. Complementary DNA cloning of a novel epithelial cell marker protein, HME1, that may be down-regulated in neoplastic mammary cells. Cell Growth Differ 3: 507–513, 1992. [PubMed] [Google Scholar]
- 223.Nakanishi K, Hashizume S, Kato M, Honjoh T, Setoguchi Y, Yasumoto K. Elevated expression levels of the 14-3-3 family of proteins in lung cancer tissues. Hum Antibodies, 8: 189–194, 1997. doi: 10.3233/hab-1997-8404. doi:. [DOI] [PubMed] [Google Scholar]
- 224.Sinha P, Hütter G, Köttgen E, Dietel M, Schadendorf D, Lage H. Increased expression of epidermal fatty acid binding protein, cofilin, and 14-3-3-σ (stratifin) detected by two-dimensional gel electrophoresis, mass spectrometry and microsequencing of drug-resistant human adenocarcinoma of the pancreas. Electrophoresis 20: 2952–2960, 1999. doi:. [DOI] [PubMed] [Google Scholar]
- 225.Yang X, Cao W, Lin H, Zhang W, Lin W, Cao L, Zhen H, Huo J, Zhang X. Isoform-specific expression of 14-3-3 proteins in human astrocytoma. J Neurol Sci 276: 54–59, 2009. doi: 10.1016/j.jns.2008.08.040. [DOI] [PubMed] [Google Scholar]
- 226.Wang Z, Nesland JM, Suo Z, Trope CG, Holm R. The prognostic value of 14-3-3 isoforms in vulvar squamous cell carcinoma cases: 14-3-3β and ε are independent prognostic factors for these tumors. PLoS One 6: e24843, 2011. doi: 10.1371/journal.pone.0024843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Liu Y, Tian R-F, Li Y-M, Liu W-P, Cao L, Yang X-L, Cao W-D, Zhang X. The expression of seven 14-3-3 isoforms in human meningioma. Brain Res 1336: 98–102, 2010. doi: 10.1016/j.brainres.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 228.Bergamaschi A, Katzenellenbogen BS. Tamoxifen downregulation of miR-451 increases 14-3-3ζ and promotes breast cancer cell survival and endocrine resistance. Oncogene 31: 39–47, 2012. doi: 10.1038/onc.2011.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Zhao Y, Fang X, Fang H, Feng Y, Chen F, Xia Q. ATPR-induced G0/G1 phase arrest in gastric cancer cells by regulating the binding of 14-3-3ε and filamin A. Cancer Med 7: 3373–3384, 2018. doi: 10.1002/cam4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Kotaka M, Ngai SM, Garcia-Barcelo M, Tsui SKW, Fung KP, Lee CY, Waye MMY. Characterization of the human 36-kDa carboxyl terminal LIM domain protein (hCLIM1). J Cell Biochem 72: 279–285, 1999. doi:. [DOI] [PubMed] [Google Scholar]
- 231.Vallenius T, Luukko K, Mäkelä TP. CLP-36 PDZ-LIM protein associates with nonmuscle α-actinin-1 and α- actinin-4. J Biol Chem 275: 11100–11105, 2000. doi: 10.1074/jbc.275.15.11100. [DOI] [PubMed] [Google Scholar]
- 232.Bauer K, Kratzer M, Otte M, De Quintana KL, Hagmann J, Arnold GJ, Eckerskorn C, Lottspeich F, Siess W. Human CLP36, a PDZ-domain and LIM-domain protein, binds to α-actinin-1 and associates with actin filaments and stress fibers in activated platelets and endothelial cells. Blood 96: 4236–4245, 2000.doi: 10.1182/blood.V96.13.4236, . [DOI] [PubMed] [Google Scholar]
- 233.Dufresne J, Bowden P, Thavarajah T, Florentinus-Mefailoski A, Chen ZZ, Tucholska M, Norzin T, Ho MT, Phan M, Mohamed N, Ravandi A, Stanton E, Slutsky AS, Dos Santos CC, Romaschin A, Marshall JC, Addison C, Malone S, Heyland D, Scheltens P, Killestein J, Teunissen C, Diamandis EP, Siu KWM, Marshall JG. The plasma peptides of breast versus ovarian cancer. Clin Proteomics 16: 43, 2019. doi: 10.1186/s12014-019-9262-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Gupta P, Suman S, Mishra M, Mishra S, Srivastava N, Kumar V, Singh PK, Shukla Y. Autoantibodies against TYMS and PDLIM1 proteins detected as circulatory signatures in Indian breast cancer patients. Proteomics Clin Appl 10: 564–573, 2016. doi: 10.1002/prca.201500138. [DOI] [PubMed] [Google Scholar]
- 235.Cross SE, Jin Y-S, Rao J, Gimzewski JK. Nanomechanical analysis of cells from cancer patients. Nat Nanotechnol 2: 780–783, 2007. doi: 10.1038/nnano.2007.388. [DOI] [PubMed] [Google Scholar]
- 236.Bai H, Zhu Q, Surcel A, Luo T, Ren Y, Guan B, Liu Y, Wu N, Joseph NE, Wang TL, Zhang N, Pan D, Alpini G, Robinson DN, Anders RA. Yes-associated protein impacts adherens junction assembly through regulating actin cytoskeleton organization. Am J Physiol Gastrointest Liver Physiol 311: G396–G411, 2016. doi: 10.1152/ajpgi.00027.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.De Lozanne A, Spudich JA. Disruption of the dictyostelium myosin heavy chain gene by homologous recombination. Science 236: 1086–1091, 1987. doi: 10.1126/science.3576222. [DOI] [PubMed] [Google Scholar]
- 238.Halder D, Saha S, Singh RK, Ghosh I, Mallick D, Dey SK, Ghosh A, Das BB, Ghosh S, Jana SS. Nonmuscle myosin IIA and IIB differentially modulate migration and alter gene expression in primary mouse tumorigenic cells. Mol Biol Cell 30: 1463–1476, 2019. doi: 10.1091/mbc.E18-12-0790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Gohla A, Bokoch GM. 14-3-3 Regulates actin dynamics by stabilizing phosphorylated cofilin. Curr Biol 12: 1704–1710, 2002. doi: 10.1016/S0960-9822(02)01184-3. [DOI] [PubMed] [Google Scholar]
- 240.Cicenas J. The Aurora kinase inhibitors in cancer research and therapy. J Cancer Res Clin Oncol 142: 1995–2012, 2016. doi: 10.1007/s00432-016-2136-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Feng Y, Lograsso P V, Defert O, Li R. Rho kinase (ROCK) inhibitors and their therapeutic potential. J Med Chem 59: 2269–2300, 2016. doi: 10.1021/acs.jmedchem.5b00683. [DOI] [PubMed] [Google Scholar]
- 242.Conti MA, Saleh AD, Brinster LR, Cheng H, Chen Z, Cornelius S, Liu C, Ma X, Van Waes C, Adelstein RS. Conditional deletion of nonmuscle myosin II-A in mouse tongue epithelium results in squamous cell carcinoma. Sci Rep 5: 14068, 2015. doi: 10.1038/srep14068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Morgan BP, Muci A, Lu P-P, Qian X, Tochimoto T, Smith WW, Garard M, Kraynack E, Collibee S, Suehiro I, Tomasi A, Valdez SC, Wang W, Jiang H, Hartman J, Rodriguez HM, Kawas R, Sylvester S, Elias KA, Godinez G, Lee K, Anderson R, Sueoka S, Xu D, Wang Z, Djordjevic N, Malik FI, Morgans DJ Jr.. Discovery of omecamtiv mecarbil the first, selective, small molecule activator of cardiac myosin. ACS Med Chem Lett 1: 472–477, 2010. doi: 10.1021/ml100138q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Welch DR, Hurst DR. Defining the hallmarks of metastasis. Cancer Res 79: 3011–3027, 2019. doi: 10.1158/0008-5472.CAN-19-0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Raudenska M, Kratochvilova M, Vicar T, Gumulec J, Balvan J, Polanska H, Pribyl J, Masarik M. Cisplatin enhances cell stiffness and decreases invasiveness rate in prostate cancer cells by actin accumulation. Sci Rep 9: 1660, 2019. doi: 10.1038/s41598-018-38199-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Ma X, Xue H, Zhong J, Feng B, Zuo Y. Serum actinin-4 levels as a potential diagnostic and prognostic marker in cervical cancer. Dis Markers 2020: 1–6, 2020. doi: 10.1155/2020/5327378. [DOI] [PMC free article] [PubMed] [Google Scholar]