Abstract
We report an aerobic, growth-based selection platform founded on NADP(H) redox balance restoration in Escherichia coli, and we demonstrate its application in the high-throughput evolution of an oxygenase. A single round of selection followed by a facile growth assay enabled Pseudomonas aeruginosa 4-hydroxybenzoate hydroxylase (PobA) to efficiently hydroxylate both 4-hydroxybenzoic acid (4-HBA) and 3,4-dihydroxybenzoic acid (3,4-DHBA), two consecutive steps in gallic acid biosynthesis. Structural modeling suggests precise reorganization of active site hydrogen bond network, which is difficult to obtain without deep navigation of combinatorial sequence space. We envision universal application of this selection platform in engineering NADPH-dependent oxidoreductases.
Keywords: 4-hydroxybenzoate hydroxylase; NADPH-dependent monooxygenase; 3,4-dihydroxybenzoic acid; redox balance; directed evolution
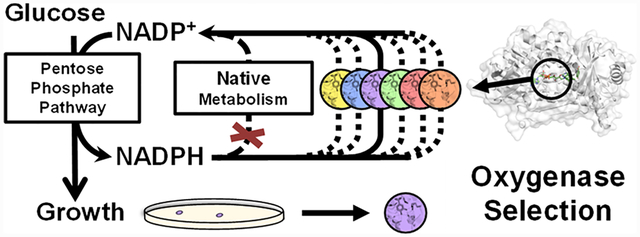
Graphical Abstract
INTRODUCTION
Oxygenases direct the strong oxidative power in molecular oxygen to activate inert C–H bonds with high regiospecificity and stereospecificity, which is difficult in traditional chemical synthesis.1 This makes them valuable catalysts for the production of industrially important and medicinal molecules such as ε-caprolactone,2 omeprazole,3 and artemisinin.4 However, engineering oxygenases is challenging, because of their highly complex catalytic mechanisms, which can obscure structure–function relationships.5,6
Recently, a “click” chemistry-based high-throughput mass spectrometric assay was established to facilitate P450 enzyme discovery.7 However, throughput remained insufficient (~1000 variants screened for a protein library with a theoretical library size of 400) for libraries routinely generated in directed evolution (104–106, a theoretical library size). To improve throughput, techniques using single cell or single enzyme analysis such as Fluorescence Activated Cell Sorting (FACS)8 and microfluidic assays9 have been developed. However, these methods are often limited by a need for colorimetric reactions,10 substrate labeling,7 or indirect activity readouts such as GFP fluorescence.11 In addition, these platforms often require extensive expertise and costly equipment.
We sought to address these limitations by developing a facile and universal selection scheme that employs E. coli growth as a simple readout. Specifically, we created an E. coli strain with overly high levels of NADPH/NADP+, which only grows on glucose in aerobic conditions when the desired oxygenase activity is present to recycle NADPH. This selection scheme is inspired by Nature: previously, natural evolution was observed under high NADPH stress, which enabled E. coli’s respiratory chain to oxidize NADPH using oxygen.12,13 Similar growth-based selection platforms based on redox balance principles have been developed previously.14–16 However, the metabolic pathway designs in these selection strains limited their functionality to anaerobic conditions. Therefore, they are not compatible with engineering oxygenases, which require oxygen for catalysis.
We applied this selection platform for directed evolution of an NADPH-dependent monooxygenase Pseudomonas aeruginosa 4-hydroxybenzoate hydroxylase (PobA) to engineer high activity for a non-native substrate, 3,4-dihydroxybenzoic acid (3,4-DHBA),6,17 which is the precursor of a natural product antioxidant, gallic acid (GA). Through a single round of selection of ~3 × 105 variants from a four-residue, site-saturated mutagenesis library (theoretical library size of 204 = 1.6 × 105), we obtained variants with roughly 8-fold improved apparent catalytic efficiency (kcat/KM) for 3,4-DHBA, compared to the wild type.6 Importantly, a subsequent screen using the same selection platform allowed rapid identification of variants which also retained ~50% catalytic efficiency with the native substrate 4-hydroxybenzoic acid (4-HBA), compared to the wild type. While the two consecutive hydroxylations of 4-HBA and 3,4-DHBA are required for GA biosynthesis, enzyme catalysts with dual activity have been difficult to obtain through rational design.6,17 Notably, Rosetta modeling suggests that the selected PobA variants form intricate hydrogen bonding networks at the active site to recognize 3,4-DHBA and coordinate catalysis, which is difficult to recapitulate in engineered enzymes without extensive sampling of sequence space.
RESULTS AND DISCUSSION
Development and Validation of the Selection Platform.
The selection process relies on an engineered E. coli strain (MX203) with a growth deficiency linked to NADPH/NADP+ imbalance. The perturbed redox state results from deletion of central metabolism genes, glucose-6-phosphate isomerase pgi and phosphogluconate dehydratase edd; a critical rebalancing tool, soluble pyridine nucleotide transhydrogenase udhA; and a significant sink for reduced nicotinamide cofactors, NAD(P)H:quinone oxidoreductase qor (Figure 1A).12 These disruptions cause MX203 to exhibit poor growth in glucose minimal media (Figure S1 in the Supporting Information), and we sought to restore growth through heterologous expression of NADPH-consuming enzymes. We employed water-producing oxidases, Lb NOX and TP NOX18 (on plasmids pLS101 and pLS102, respectively) (Figure 1B), derived from Lactobacillus brevis. When introduced into selection strain MX203, the NADPH-specific TP NOX restored growth, while the NADH-specific Lb NOX did not (Figure 1C). Desirable growth behavior demonstrated in both liquid and solid media provides flexibility in selection.
Figure 1.
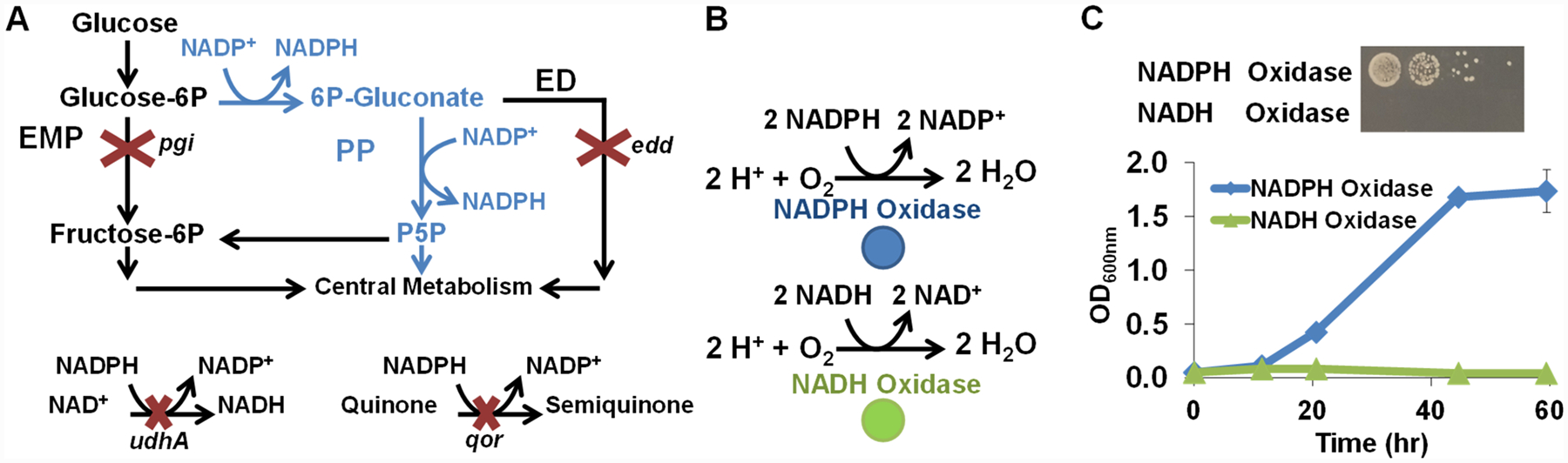
Development and validation of the selection strain with high NADPH accumulation. (A) Central metabolism redirection through pentose phosphate (PP, blue) pathway and disruption of Embden-Meyerhof-Parnas (EMP) and Entner Doudoroff (ED) pathways increased NADP+ reduction. Deletion of NADPH reoxidation transhydrogenase udhA and quinone oxidoreductase qor reduce NADPH sinks. (B) Complementary oxidases use reduced cofactors, NADPH (blue) or NADH (green), and oxygen to generate water. (C) In M9 minimal glucose media, growth restoration was achieved by heterologous expression of the NADPH-specific oxidase but not the NADH-specific oxidase in both solid and liquid media.
Construction and Selection of PobA Library.
Gallic acid has broad application in food and pharmaceutical industries19 and as the biosynthetic precursor of a platform chemical, pyrogallol.20 Biosynthesis of gallic acid from simple carbon sources is hindered by the absence of a natural enzyme that can catalyze two consecutive hydroxylation steps, namely, from 4-hydroxybenzoic acid (4-HBA) to 3,4-DHBA, and subsequently to gallic acid (Figure 2A). The NADPH-dependent 4-HBA hydroxylase from P. aeruginosa (Pa PobA) natively catalyzes the first step, and previous work engineered additional activity for the second step.6,17 While promising variants obtained from rational design, namely, Y385F, Y385F/T294A, and Y385F/L199 V, enabled 3,4-DHBA hydroxylation,6,17,20 they also had severely decreased native activity for the first hydroxylation step.6,17 We hypothesized that achieving high activity with both 4-HBA and 3,4-DHBA would require the active site to flexibly accommodate the 3-hydroxyl of 3,4-DHBA away from the C4a of flavin adenine dinucleotide (FAD) (Figure 2B). We inferred this mode of substrate binding would be more catalytically competent because oxygen transfer must occur between the flavin-C4a-hydroperoxide intermediate and a position on the aromatic ring with excess electron density, C5.21 Guided by this hypothesis, we targeted three residues within 4 Å of the 3-hydroxyl for site-saturated mutagenesis (V47, L199, Y385 in 2B). Position T294 was selected for its proximity to the 4-hydroxyl of 4-HBA and potential role in the hydrogen bonding network of the active site.17
Figure 2.
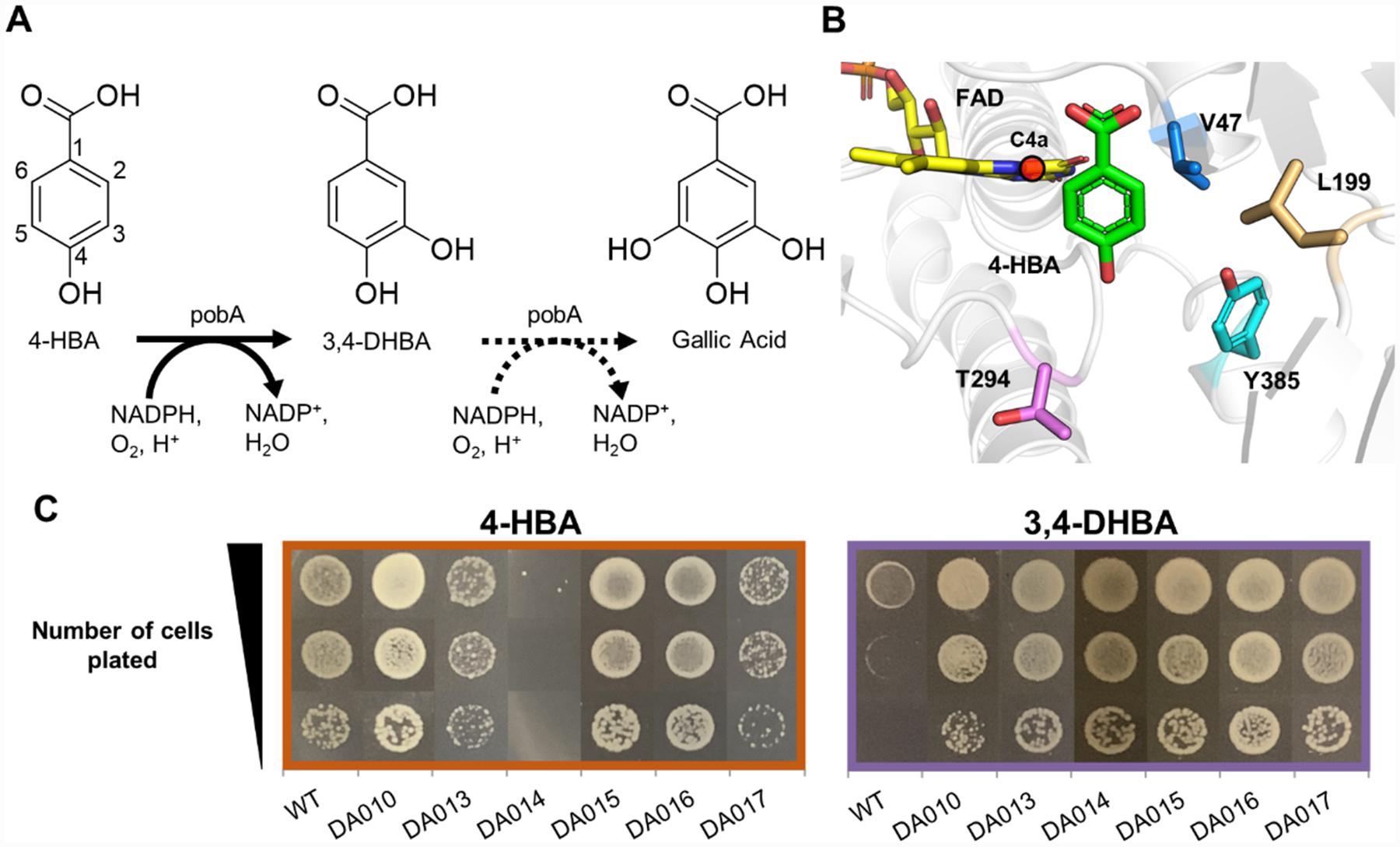
Construction and selection of the PobA library for 3,4-DHBA utilization. (A) Biosynthetic pathway of gallic acid involves two sequential hydroxylation steps. (B) Wild-type Pa PobA crystal structure with 4-HBA bound (PDB: 1IUW). Residues highlighted were targeted for site-saturated mutagenesis. The reactive FAD-C4a is highlighted by red sphere. (C) Growth restoration phenotype for wild-type PobA and variants identified in selection, with 4-HBA or 3,4-DHBA supplementation.
We simultaneously randomized residues 47, 199, 294, and 385 using NNK degenerate codons (Figure 2B). The resulting DNA library (pDA008) was transformed to selection strain MX203, yielding ~3 × 105 independent transformants (see the Methods section in the Supporting Information), which was sufficient to cover 1.8-fold of the theoretical library size of 204 = 1.6 × 105. Selection was performed on agar plates with 2 g/L d-glucose in M9 minimal medium and 1 g/L 3,4-DHBA, at 30 °C for 60 h. MX203 cells harboring wild-type Pa PobA served as a negative control. After incubation, ~600 colonies had formed on selection plates.
Characterization of PobA Variants Obtained from Selection.
We picked eight colonies with robust growth and extracted the mutant Pa PobA plasmids. Sequencing the eight candidates revealed six unique residue combinations (Table S2). Repetitive hits found within eight samples suggests that only a small fraction of the variants in the library were able to alleviate redox imbalance. Discussion on observed consensus is included in the Supporting Information.
We next examined growth restoration by retransforming the plasmids into selection strain MX203 and characterizing growth with 3,4-DHBA or 4-HBA as a substrate (Figure 2C). While the negative control, wild-type PobA, showed little to no growth with 3,4-DHBA, all six unique variants exhibited robust growth on this non-native substrate.
Four of the six unique variants were further characterized using purified proteins to inspect the mechanisms underlying their distinct growth restoration patterns on the two substrates. All four variants demonstrated improved 3,4-DHBA activity as measured in vitro by enzyme assay (Figure 3), indicating that growth was coupled to 3,4-DHBA activity. Interestingly, DA014, which struggled to grow on plates containing 4-HBA, exhibited high specific activities for 4-HBA (Figure 3). As noted in previous work, this discrepancy might arise from differences between measured in vitro specific activity and in vivo activity.14 For example, while the specific activity assays were performed with 1 mM substrates, the in vivo concentrations may be lower.
Figure 3.
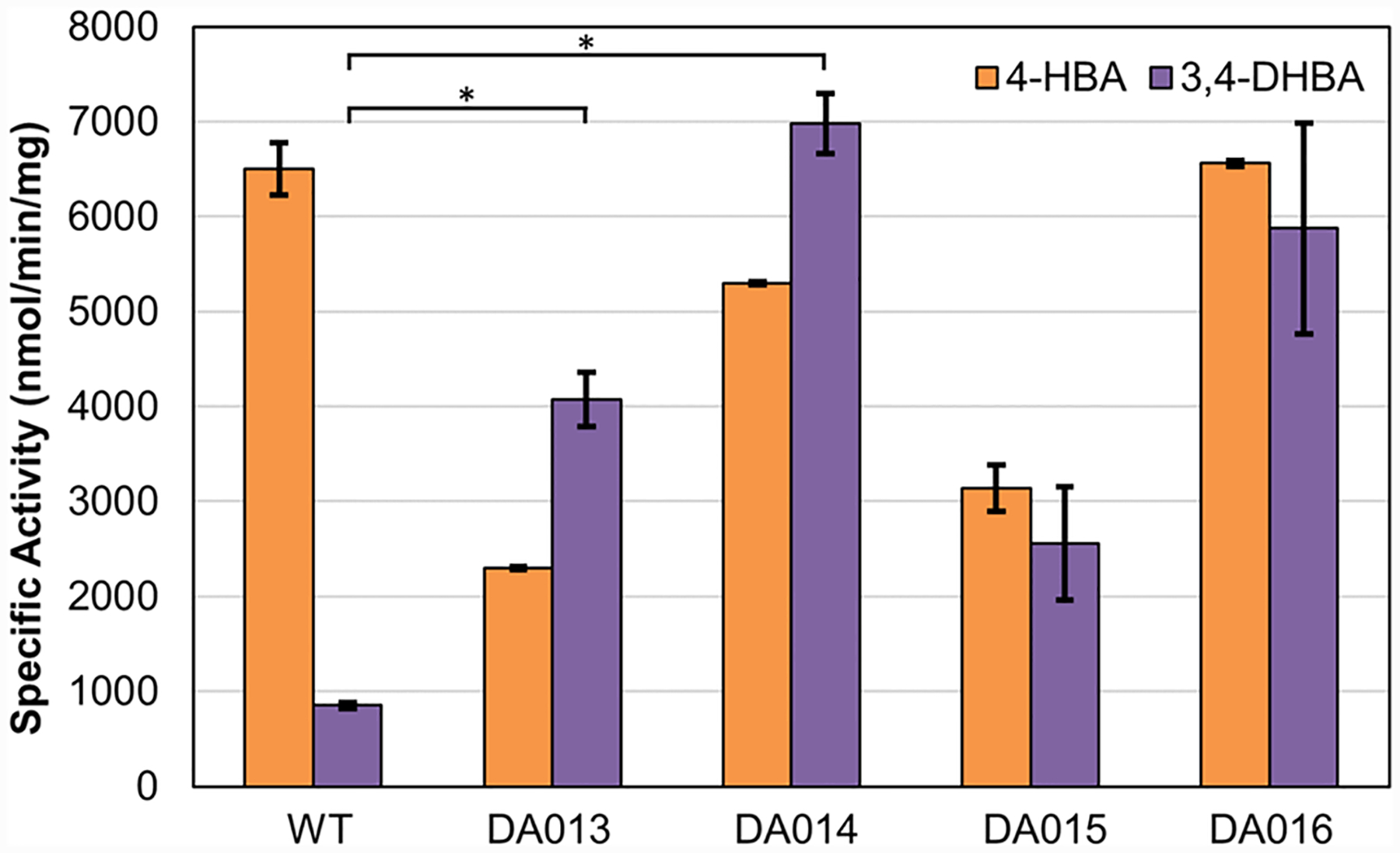
In vitro specific activity of four PobA variants identified in growth-based selection. All variants exhibited improved activity toward 3,4-DHBA. Detailed assay conditions are described in the Methods section in the Supporting Information. Statistical significance indicated by an asterisk was determined by two-tailed t-tests (P < 0.05).
While DA013 and DA014 exhibited the most significant improvement, compared to wild type for 3,4-DHBA specific activity (Figure 3), we chose to further characterize DA015 (L199R-T294C–Y385M) and DA016 (V47I-L199N-T294A-Y385I) for their robust growth with both 4-HBA and 3,4-DHBA (Figure 2C, Tables 1 and 2). As shown in Table 1, the best variant DA015 showed 7.9-fold improved apparent catalytic efficiency (kcat/KM), reaching ~0.062 μM–1 s–1 for 3,4-DHBA, compared to the wild type (~0.0078 μM–1 s–1, as reported by Moriwaki et al.6). Notably, the 4-HBA hydroxylating activity of DA015 reached ~50% of that of the wild-type enzyme, which is ~4-fold higher, compared to previously engineered variants, Y385F/T294A17 and Y385F/L199 V.6 DA016 has a high apparent kcat for 3,4-DHBA and 4-HBA (~4 s–1 for both), which is over 50% of wild-type PobA’s apparent kcat value (~7.6 s–1) toward the native substrate 4-HBA. This high turnover rate may be instrumental for in vitro catalysis.
Table 1.
Apparent Kinetic Parameters for Substrates
| 4-HBA | 3,4-DHBA | ||||||
|---|---|---|---|---|---|---|---|
| variant | KMa (μM) | kcata (s−1) | kcat/KMa(μM−1 s−1) | KMa (μM) | kcata (s−1) | kcat/KMa(μM−1 s−1) | |
| this study | wild-type | 48 ± 7 | 7.6 ± 0.1 | 0.16 ± 0.03 | − | − | − |
| DA015b | 24 ± 10 | 1.9 ± 0.2 | 0.079 ± 0.04 | 48 ± 10 | 3.0 ± 0.2 | 0.062 ± 0.01 | |
| DA016c | 120 ± 10 | 4.0 ± 0.1 | 0.034 ± 0.001 | 120 ± 10 | 4.1 ± 0.2 | 0.033 ± 0.004 | |
| Chen et al.17 | Y385F/T294A | 48 ± 3 | 0.90 ± 0.03 | 0.019 ± 0.001 | 160 ± 30 | 1.7 ± 0.2 | 0.012 ± 0.003 |
| Moriwaki et al.6 | Y385F/L199 V | 22 ± 5 | 0.45 ± 0.02 | 0.021 ± 0.004 | 52 ± 16 | 4.4 ± 0.4 | 0.085 ± 0.02 |
| wild-type | 39 ± 6 | 8.9 ± 0.5 | 0.23 ± 0.03 | 42 ± 17 | 0.33 ± 0.04 | 0.0078 ± 0.002 | |
Table 2.
Apparent Kinetic Parameters for NADPH
| variant | substrate | KMa (μM) | kcata (s−1) | kcat/KMa(μM−1 s−1) |
|---|---|---|---|---|
| wild-type | 4-HBA | 250 ± 40 | 7.8 ± 0.8 | 0.031 ± 0.006 |
| DA015b | 4-HBA | 47 ± 7 | 2.1 ± 0.2 | 0.044 ± 0.008 |
| DA016c | 4-HBA | 140 ± 40 | 4.4 ± 0.8 | 0.032 ± 0.01 |
| wild-type | 3,4-DHBA | − | − | − |
| DA015b | 3,4-DHBA | 76 ± 10 | 2.5 ± 0.2 | 0.033 ± 0.006 |
| DA016c | 3,4-DHBA | 120 ± 4 | 4.1 ± 0.1 | 0.033 ± 0.001 |
Apparent kinetic parameters shown for constant substrate concentration of 0.5 mM.
DA015, L199R-T294C-Y385M.
DA016, V47I-L199N-T294A-Y385I.
Because the variants are selected under the physiological concentration of NADPH, we hypothesize that the apparent catalytic efficiency (kcat/KM) of NADPH for DA015 and DA016 would be comparable to that of the wild-type enzyme, which has been shown to function in E. coli for gallic acid production in vivo. This is indeed the case, as shown in Table 2. Notably, both variants have lower apparent KM (ranging from 47 μM to 120 μM) for NADPH when using either 4-HBA or 3,4-DHBA as the substrate, compared to that of the wild-type enzyme using the native substrate 4-HBA (~250 μM). These results highlight the potential of the growth-based selection to directly discover enzyme variants that are compatible with in vivo biotransformation.
Mechanistic Study of the PobA Variants by Rosetta Modeling.
Molecular modeling has been established as an important tool in understanding the effect of mutations on catalytic function.22,23 We modeled the 3,4-DHBA binding pose with wild-type, DA015, and DA016 to identify structural features contributing to enhanced activity over the wild-type and previously reported variants. Wild-type PobA crystal structure with 4-HBA bound (PDB: 1IUW) indicates 4-HBA binding mediated by a densely packed hydrogen bond network (Figure 4A). Docking of 3,4-DHBA into wild-type suggests that the ligand is unable to bind in a catalytically active pose with the reactive carbon facing the FAD, because of the lack of favorable polar contacts in the interior of the pocket for the 3-hydroxyl to interact with (see Figure S3 in the Supporting Information). Previously reported PobA variants contained rationally designed mutations to disrupt hydrogen-bond networks, aiming to increase backbone flexibility and substrate promiscuity;17,24 additional mechanistic studies determined that, for 3,4-DHBA activity, individual L199 and Y385 replacement with hydrophilic residues reduces the relative activity and suggested that the formation of additional hydrogen bonds with Y201, Y385, or the substrate would be detrimental for the reaction.6 In contrast, variants obtained in this study are suggested to feature a remodeled hydrogen-bond network tailored for 3,4-DHBA (see Figures 4B and 4C).
Figure 4.
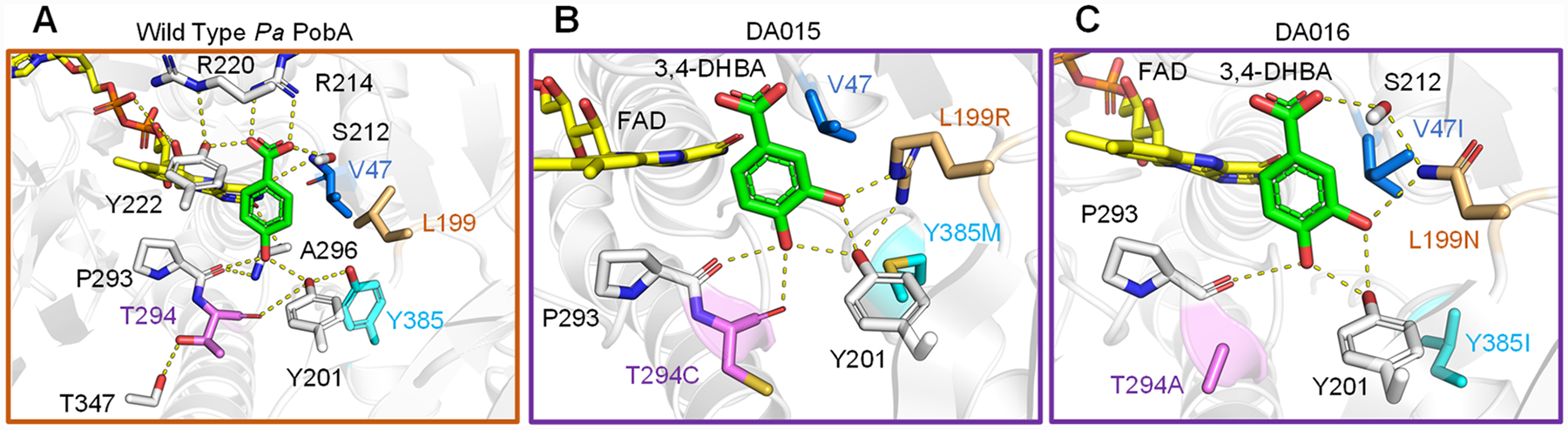
4-HBA and 3,4-DHBA binding poses. (A) In wild-type PobA (PDB: 1IUW), the productive binding mode is described by the hydrogen-bonding networks stabilizing 4-HBA and positioning the 3-carbon toward FAD for hydroxylation. (B) In the DA015 model, L199R supports Y201 and forms a new contact to the ligand 3-hydroxyl. Y385M makes space, no substitution occurs at V47, which maintains close hydrophobic packing against L199R, and T294C loosens the helix for increased flexibility and improved backbone hydrogen bonding to the 4-hydroxyl. (C) In the DA016 model, L199N forms interactions stabilizing S212 and to the ligand 3-hydroxyl. Y385I creates space, V47I braces L199N to minimize side-chain mobility, and T294A allows P293 to move closer to 3,4-DHBA. Importantly, the mutations orient 3,4-DHBA such that the 5-carbon is optimally exposed to FAD for hydroxylation in both variants. Detailed methods of Rosetta modeling are included in the Supporting Information.
Specifically, in the DA015 (L199R-T294C-Y385M) model, the L199R guanidinium extends to form two novel contacts: one to the Y201 hydroxyl and another to the 3,4-DHBA 3-hydroxyl. T294C eliminates a hydrogen bond to T347, allowing another new hydrogen bond to form between the backbone carbonyl to the 3,4-DHBA 4-hydroxyl (see Figure 4B). In the DA016 (V47I-L199N-T294A-Y385I) model, the L199N amide acts as a bridge stabilizing the ligand 3-hydroxyl and the S212 hydroxyl associated with the ligand carboxylic acid; V47I packs against and reinforces L199N; T294A, similarly to T294C, breaks a hydrogen bond to T347 and allows expanded loop mobility for improved backbone hydrogen bonding to the ligand 4-hydroxyl (Figure 4C). Comparison to wild-type structure shows a binding pocket volume increase from 55 Å3 to 69 Å3 in DA015 and 72 Å3 in DA016 (see Figure S2). The additional space accommodates the extra hydroxyl in 3,4-DHBA binding.
The discovered mutations are predicted to restructure the central hydrogen bonding array for a more-productive 3,4-DHBA binding orientation. Rational design to achieve the same effect is challenging, because of the multiple, simultaneous substitutions required to reach this functionality. Designing hydrogen-bonding networks is especially difficult, because of their high cooperativity and sensitivity to small perturbations in length and angle between hydrogen-bonding partners. For instance, singular L199 mutation to a polar residue contacting the ligand 3-hydroxyl would likely be ineffective without compensatory substitutions at V47 and Y385 relieving steric clash and reinforcing optimal hydrophobic packing. This highlights the effectiveness of our high-throughput selection approach in comprehensively exploring the sequence space and discovering highly synergistic mutations.
CONCLUSION
In summary, we established a high throughput screening platform with potential applications for directed evolution of various NADPH-dependent oxygenases. This aerobic in vivo system demonstrated strict cofactor dependence and simple growth-based selection. We successfully engineered PobA for enhanced activity toward the non-native substrate 3,4-DHBA. In one round of selection, we identified a variant, DA015, with greatly improved activity for both 4-HBA and 3,4-DHBA, compared to variants previously designed with restricted exploration of the available sequence space. Rosetta modeling suggests that enhanced activity is attributed to concerted changes in binding pocket volume, shape complementarity, and, more importantly, formation of a fully connected hydrogen-bond network in the active site. Future work is needed to overcome some of the limitations of this selection platform. For example, the target substrates may have cellular toxicity or low solubility. Therefore, a two-phase culturing system, which combines an organic solvent phase with the culture medium, may be needed.25 Because NADPH provides reducing power for a variety of oxygen-dependent industrially important enzyme classes, such as Baeyer–Villiger monooxygenases and cytochrome P450s, we envision applications of this platform for engineering a broad range of catalysts.
Supplementary Material
ACKNOWLEDGMENTS
H.L. acknowledges support from University of California, Irvine, the National Science Foundation (NSF) (Award No. 1847705), and the National Institutes of Health (NIH) (Award No. DP2 GM137427). S.M. acknowledges support from the NSF Graduate Research Fellowship Program (Grant No. DGE-1839285). D.A. acknowledges support from the Federal Work Study Program funded by the U.S. Department of Education.
Footnotes
Supporting Information
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acscatal.0c01892.
Experimental methods, plasmids and strains used in this study (Table S1); detailed analysis of the sequences of PobA variants obtained from selection (Table S2); growth phenotypes of E. coli strains with modified NADPH metabolism (Figure S1); active site volumes of wild type, DA015, and DA016 PobA (Figure S2); and model of PobA wild type with 3,4-DHBA bound (Figure S3) (PDF)
The authors declare no competing financial interest.
Complete contact information is available at: https://pubs.acs.org/10.1021/acscatal.0c01892
Contributor Information
Sarah Maxel, Department of Chemical and Biomolecular Engineering, University of California, Irvine, Irvine, California 92697, United States.
Derek Aspacio, Department of Chemical and Biomolecular Engineering, University of California, Irvine, Irvine, California 92697, United States.
Edward King, Department of Molecular Biology and Biochemistry, University of California, Irvine, Irvine, California 92697, United States.
Linyue Zhang, Department of Chemical and Biomolecular Engineering, University of California, Irvine, Irvine, California 92697, United States.
Ana Paula Acosta, Department of Chemical and Biomolecular Engineering, University of California, Irvine, Irvine, California 92697, United States.
Han Li, Department of Chemical and Biomolecular Engineering, University of California, Irvine, Irvine, California 92697, United States;.
REFERENCES
- (1).Hollmann F; Arends IWCE; Holtmann D Enzymatic Reductions for the Chemist. Green Chem. 2011, 13, 2285–2314. [Google Scholar]
- (2).Schmidt S; Scherkus C; Muschiol J; Menyes U; Winkler T; Hummel W; Gröger H; Liese A; Herz HG; Bornscheuer UT An Enzyme Cascade Synthesis of ε-Caprolactone and Its Oligomers. Angew. Chem., Int. Ed 2015, 54 (9), 2784–2787. [DOI] [PubMed] [Google Scholar]
- (3).Zhang Y; Wu YQ; Xu N; Zhao Q; Yu HL; Xu JH Engineering of Cyclohexanone Monooxygenase for the Enantioselective Synthesis of (S)-Omeprazole. ACS Sustainable Chem. Eng 2019, 7 (7), 7218–7226. [Google Scholar]
- (4).Dietrich JA; Yoshikuni Y; Fisher KJ; Woolard FX; Ockey D; McPhee DJ; Renninger NS; Chang MCY; Baker D; Keasling JD A Novel Semi-Biosynthetic Route for Artemisinin Production Using Engineered Substrate-Promiscuous P450BM3. ACS Chem. Biol 2009, 4 (4), 261–267. [DOI] [PubMed] [Google Scholar]
- (5).Whitehouse CJC; Bell SG; Wong LL P450 BM3 (CYP102A1): Connecting the Dots. Chem. Soc. Rev 2012, 41 (3), 1218–1260. [DOI] [PubMed] [Google Scholar]
- (6).Moriwaki Y; Yato M; Terada T; Saito S; Nukui N; Iwasaki T; Nishi T; Kawaguchi Y; Okamoto K; Arakawa T; Yamada C; Fushinobu S; Shimizu K Understanding the Molecular Mechanism Underlying the High Catalytic Activity of P-Hydroxybenzoate Hydroxylase Mutants for Producing Gallic Acid. Biochemistry 2019, 58 (45), 4543–4558. [DOI] [PubMed] [Google Scholar]
- (7).de Rond T; Gao J; Zargar A; de Raad M; Cunha J; Northen TR; Keasling JD A High-Throughput Mass Spectrometric Enzyme Activity Assay Enabling the Discovery of Cytochrome P450 Biocatalysts. Angew. Chem., Int. Ed 2019, 58 (30), 10114–10119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Siedler S; Schendzielorz G; Binder S; Eggeling L; Bringer S; Bott M SoxR as a Single-Cell Biosensor for NADPH-Consuming Enzymes in Escherichia Coli. ACS Synth. Biol 2014, 3 (1), 41–47. [DOI] [PubMed] [Google Scholar]
- (9).Debon A; Pott M; Obexer R; Green AP; Friedrich L; Griffiths AD; Hilvert D Ultrahigh-Throughput Screening Enables Efficient Single-Round Oxidase Remodelling. Nat. Catal 2019, 2 (9), 740–747. [Google Scholar]
- (10).Li QS; Ogawa J; Schmid RD; Shimizu S Engineering Cytochrome P450 BM-3 for Oxidation of Polycyclic Aromatic Hydrocarbons. Appl. Environ. Microbiol 2001, 67 (12), 5735–5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Lim JB; Sikes HD Use of a Genetically Encoded Hydrogen Peroxide Sensor for Whole Cell Screening of Enzyme Activity. Protein Eng., Des. Sel 2015, 28 (3), 79–83. [DOI] [PubMed] [Google Scholar]
- (12).Auriol C; Bestel-Corre G; Claude J; Soucaille P; Meynial-Salles I Stress-Induced Evolution of Escherichia Coli Points to Original Concepts in Respiratory Cofactor Selectivity. Proc. Natl. Acad. Sci. U. S. A 2011, 108 (4), 1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Frick K; Schulte M; Friedrich T Reactive Oxygen Species Production by Escherichia Coli Respiratory Complex I. Biochemistry 2015, 54, 2799–2801. [DOI] [PubMed] [Google Scholar]
- (14).Zhang L; King E; Luo R; Li H Development of a High-Throughput, In Vivo Selection Platform for NADPH-Dependent Reactions Based on Redox Balance Principles. ACS Synth. Biol 2018, 7, 1715–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Shen CR; Lan EI; Dekishima Y; Baez A; Cho KM; Liao JC Driving Forces Enable High-Titer Anaerobic 1-Butanol Synthesis in Escherichia Coli. Appl. Environ. Microbiol 2011, 77 (9), 2905–2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Atsumi S; Cann AF; Connor MR; Shen CR; Smith KM; Brynildsen MP; Chou KJY; Hanai T; Liao JC Metabolic Engineering of Escherichia Coli for 1-Butanol Production. Metab. Eng 2008, 10 (6), 305–311. [DOI] [PubMed] [Google Scholar]
- (17).Chen Z; Shen X; Wang J; Wang J; Yuan Q; Yan Y Rational Engineering of P-Hydroxybenzoate Hydroxylase to Enable Efficient Gallic Acid Synthesis via a Novel Artificial Biosynthetic Pathway. Biotechnol. Bioeng 2017, 114 (11), 2571–2580. [DOI] [PubMed] [Google Scholar]
- (18).Cracan V; Titov D V; Shen, H.; Grabarek, Z.; Mootha, V. K A Genetically Encoded Tool for Manipulation of NADP+/NADPH in Living Cells. Nat. Chem. Biol 2017, 13 (10), 1088–1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Kahkeshani N; Farzaei F; Fotouhi M; Alavi SS; Bahramsoltani R; Naseri R; Momtaz S; Abbasabadi Z; Rahimi R; Farzaei MH; Bishayee A Pharmacological Effects of Gallic Acid in Health and Disease: A Mechanistic Review. Iran. J. Basic Med. Sci 2019, 22 (3), 225–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Huo YX; Ren H; Yu H; Zhao L; Yu S; Yan Y; Chen Z CipA-Mediating Enzyme Self-Assembly to Enhance the Biosynthesis of Pyrogallol in Escherichia Coli. Appl. Microbiol. Biotechnol 2018, 102 (23), 10005–10015. [DOI] [PubMed] [Google Scholar]
- (21).Entsch B; Van Berkel WJH Structure and Mechanism of Para-hydroxybenzoate Hydroxylase. FASEB J. 1995, 9 (7), 476–483. [DOI] [PubMed] [Google Scholar]
- (22).Mak WS; Tran S; Marcheschi R; Bertolani S; Thompson J; Baker D; Liao JC; Siegel JB Integrative Genomic Mining for Enzyme Function to Enable Engineering of a Non-Natural Biosynthetic Pathway. Nat. Commun 2015, 6 (1), 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Siegel JB; Smith AL; Poust S; Wargacki AJ; Bar-Even A; Louw C; Shen BW; Eiben CB; Tran HM; Noor E; Gallaher JL; Bale J; Yoshikuni Y; Gelb MH; Keasling JD; Stoddard BL; Lidstrom ME; Baker D Computational Protein Design Enables a Novel One-Carbon Assimilation Pathway. Proc. Natl. Acad. Sci. U. S. A 2015, 112 (12), 3704–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Entsch B; Palfey BA; Ballou DP; Massey V Catalytic Function of Tyrosine Residues in Para-Hydroxybenzoate Hydroxylase as Determined by the Study of Site-Directed Mutants. J. Biol. Chem 1991, 266 (26), 17341–17349. [PubMed] [Google Scholar]
- (25).Alonso-Gutierrez J; Chan R; Batth TS; Adams PD; Keasling JD; Petzold CJ; Lee TS Metabolic Engineering of Escherichia Coli for Limonene and Perillyl Alcohol Production. Metab. Eng 2013, 19, 33–41. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


