Abstract
Background
Endometrial carcinoma (EC) has become a common gynecologic malignancy with a high mortality. The m6A regulators have been identified to be closely associated with multiple human cancers including EC. However, the CpG methylation signature related to m6A regulators in EC remains unclear.
Method
The methylation profiles of EC patients including cancer samples and adjacent normal samples were obtained from The Cancer Genome Atlas (TCGA) database. The CpG sites in 20 m6A regulators were identified. Univariate Cox regression and LASSO Cox regression analysis were used to screen key CpG sites which were located at m6A regulators and significantly related to the prognosis of EC. The predictive model for EC prognosis was constructed, and multivariate Cox regression analysis was applied to explore whether the risk score derived from the model could function as an independent signature for EC prognosis. Meanwhile, a nomogram model was constructed by combing the independent prognostic signatures for prediction of the long-term survival in EC patients.
Results
A total of 396 CpG sites located at 20 m6A regulators were identified. A specific predictive model for EC prognosis based on 7 optimal CpG sites was constructed, which presented good performance in prognosis prediction of EC patients. Moreover, risk score was determined to be an independent signature both in the training set and validation set. By bringing in three independent prognostic factors (age, risk score, and TNM stage), the nomogram was constructed and could effectively predict the 3- and 5-year survival rates of EC patients.
Conclusion
Our study suggested that the CpG sites located at m6A regulators might be considered as potential prognostic signatures for EC patients.
Keywords: CpG site, endometrial carcinoma, m6A regulators, methylation, nomogram, risk score
Highlights
A total of 15 CpG sites located at m6A regulators and significantly related to the prognosis of endometrial carcinoma were identified.
Risk score calculated by the predictive model based on 7 optimal CpGs sites was an independent prognostic signature.
Nomogram based on multiple independent prognostic factors could better predict the long-term survival of endometrial carcinoma patients.
1. Introduction
Endometrial carcinoma (EC) has become a common malignancy in the female reproductive system with high incidence and mortality.[1] It is estimated that over 140,000 women are diagnosed with EC globally, and more than 40,000 women die from it.[2] Most women who are diagnosed at an early stage have a long survival time; however, those diagnosed at advanced stage seriously exhibit poor prognosis.[3] Although the International Federation of Gynecology and Obstetrics (IFGO) staging system and histology contribute much to the control of EC, it remains insufficient to effectively predict the prognosis because of the molecular heterogeneity of this disease.[4] Therefore, it is more urgent to identify and develop sensitive and specific prognostic signatures to improve personalized treatment and clinical outcomes.
N6-methyladenosine (m6A) has been identified to be a more abundant internal modification of mRNA in the majority of eukaryotes.[5] The formation of m6A is a reversible process controlled by three classes of m6A regulators including methyl-transferase (writers), demethylase (erasers), and binding protein (readers).[6] It has been reported that m6A regulators were closely involved in the progression of several cancers including EC. METTL14, a methyl-transferase complex, promotes the proliferation and tumorigenicity of EC through regulating AKT activity.[7] In addition, m6A regulators were also proved to affect the prognosis of bladder cancer. Chen et al constructed a risk signature based on the methylation levels of three m6A regulators, FTO, YTHDC1, and WTAP and suggested that these three m6A regulators might be promising prognostic biomarkers of bladder cancer.[8]
Recently, increasing studies have identified a series of specific molecular changes in EC, such as mutations, DNA methylation, copy number alterations, and microsatellite instability.[9–13] DNA methylation in gene promoters, an epigenetic regulator of gene expression that often leads to gene silencing,[14] plays an important role in the progression and clinical significance of EC. Deng et al found that the rate of methylation in DACH1 gene promoter was higher in EC tissues than that in normal issues, and the methylation of DACH1 gene promoter was closely related to the pathological grade and histological type of EC.[15] The hypermethylation of CDH1 gene promoter, which resulted in a low level of E-cadherin in EC, was significantly related to both clinicopathological progress and 5-year clinical survival rate of EC.[16] These researches indicated that the DNA methylation of gene promoter exerts a certain prognostic value in EC. However, the prognostic value of m6A regulator methylation in EC has not been reported.
Here, our study aims to explore the prognostic value of CpG sites in m6A regulators for EC. Fifteen CpG sites, which were located at m6A regulators and significantly related to the prognosis of EC patients, were identified. A specific predictive model was constructed based on 7 optimal CpG sites out of the 15 CpGs and could efficiently predict the prognosis of EC patients. Taken together, our results suggested that the CpG sites in m6A regulators might be potential prognostic signatures in EC.
2. Materials and methods
2.1. Data collection
The methylation profiles of EC were obtained from TCGA (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga)database. We downloaded the methylation profiles of 483 EC samples (437 cancer samples and 46 adjacent normal samples) and the corresponding clinical information. All 483 samples had complete survival information (Table 1).
Table 1.
Clinicopathological characteristics of endometrial carcinoma samples from TCGA database.
| UCEC patients | ||||
| Parameters | Training cohort (N = 312) | Validation cohort (N = 171) | χ2 | P-value |
| Age (Mean ± SD) | 57.26 ± 8.22 | 56.57 ± 9.24 | 0.0041826 | .9484 |
| Gender | ||||
| Female | 312 (100%) | 171 (100%) | 0 | 1 |
| Pathologic stage | ||||
| i) | 191 (61.22%) | 91 (53.22%) | 1.5209 | .6775 |
| ii) | 34 (10.90%) | 23 (13.45%) | ||
| iii) | 71 (22.76%) | 49 (28.65%) | ||
| iv) | 16 (5.12%) | 8 (4.68%) | ||
| Grade | ||||
| 1 | 48 (15.38%) | 27 (15.79%) | 0.91265 | .8224 |
| 2 | 62 (19.87%) | 43 (25.15%) | ||
| 3 | 197 (63.14%) | 98 (57.31%) | ||
| High grade | 5 (1.61%) | 3 (1.75%) | ||
| OS status | ||||
| Dead | 54 (17.31%) | 40 (23.39%) | 0.7896 | .3742 |
| Alive | 258 (82.69%) | 131 (76.71%) | ||
OS = overall survival, SD = standard deviation, UCEC = uterine corpus endometrial carcinoma.
2.2. Selection of CpG sites
The m6A regulators were divided into three classes: m6A methyltransferase (writers), m6A demethylase (erasers), and m6A binding proteins (readers). In this study, we selected the CpG sites that were located at 20 major m6A regulators for subsequent analysis. Writers: METTL3, METTL14, WTAP, VIRMA, RBM15, RBM15B and ZC3H13; erasers: FTO and ALKBH5; readers: YTHDC1, YTHDC2, YTHDF1, YTHDF2, YTHDF3, IGF2BP1, IGF2BP2, IGF2BP3, RBMX, HNRNPC and HNRNPA2B1.
2.3. LASSO Cox regression analysis
We randomly grouped EC samples into training and validation sets with the sample size ratio of 2:1. Univariate Cox regression analysis of samples in the training set was performed according to the methylation level (beta value) of CpG site, and P < .05 was used as the threshold to screen the key CpGs which were significantly associated with the prognosis of EC. The glmnet package in R language[17] was used to perform LASSO Cox regression analysis to further screen optimal CpG sites. Risk score of each sample was computed using the optimal CpG sites based on the following formula:
Of which, Coefi represented the risk coefficient which was obtained by LASSO Cox model, Xi represented the expression value of each gene. Here, Xi was the β value of CpG site. After that, the optimal cutoff value of risk score was determined by survival and survminer packages and bilateral log-rank tests.
2.4. Survival analysis
The overall survival rate of EC patients was estimated by using the survival and survminer packages in R based on Kaplan–Meier method. Log-rank test was adopted to determine the difference in survival rate among different groups. The accuracy of the predictive model was verified by time-dependent receiver operating characteristic (ROC) curves plotted by “survivalROC” package in R. We conducted the Multivariate Cox regression analysis to explore whether risk score was an independent prognostic signature.
2.5. The construction of nomogram model
To predict the 1-, 3- and 5-year survival probabilities of EC patients, the nomogram model was constructed with the rms package in R by bringing in all independent prognostic factors obtained from multivariate Cox regression analysis. Meanwhile, the calibration curve of the nomogram was plotted to analyze the correlation between the predicted and actual survival probability.
2.6. Statistical analysis
The difference in survival probability between distinct groups was determined by log-rank test. All statistical analyses in our study were performed by R software v3.5.2.
3. Results
3.1. Selection of CpG sites for predictive model
According to the annotation information of methylation profiles, we analyzed all CpG sites in the 20 m6A regulators and a total of 396 CpG sites were identified. These CpG sites were located in different regions of the genes, of which, 39.2% were in the promoter regions (TSS200 and TSS1500) and 13.07% in the 1st exon region (Fig. 1A). Previous researches have shown that hyper-methylation in the promoter region was closely associated with gene inactivation[18] and methylation in the 1st exon was also related to gene silencing.[19] These reports provide a theoretical basis for our research on the CpG sites in the promoter and 1st exon.
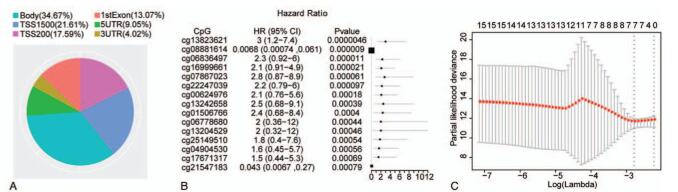
Figure 1.
The selection of CpG sites for predictive model. (A) The pie chart of CpG sites distribution. Different colors represent different regions, and the area size represents the number of CpG sites. (B) The forest plot of univariate Cox regression analysis with 15 CpG sites. HR: Hazard ratio; 95% CI: 95% confidence interval. (C) Determination of the tuning parameter lambda by LASSO Cox regression analysis. The horizontal axis denotes log (lambda), and the vertical axis denotes partial likelihood deviance.
Univariate Cox regression analysis was performed with the methylation level (beta value) of CpG site as the variable in the training set. The results suggested that there were 15 CpG sites that were significantly correlated to the prognosis of EC (Fig. 1B). Meanwhile, the hazard ratios (HRs) of cg0888161 and cg21547183 were less than 1, and the remaining variables were all greater than 1, indicating that the high methylation level of CpG sites cg08881614 and cg21547183 might lead to a low risk of death and these two CpG sites were protective sites. While high methylation level of the sites whose HRs were greater than 1 could lead to a high risk of death. Next, LASSO Cox regression analysis was performed based on the 15 CpG sites in the training set, and the optimal lambda value was determined by using the cross-validation method. Based on the optimal lambda value, we selected 7 CpG sites including cg13823621, cg08881614, cg07867023, cg22247039, cg00624976 cg06778680 and cg13204529 for the construction of the predictive model (Fig. 1C). These results suggested that these 7 CpG sites were potentially correlated with the prognosis of EC.
3.2. Construction of predictive model
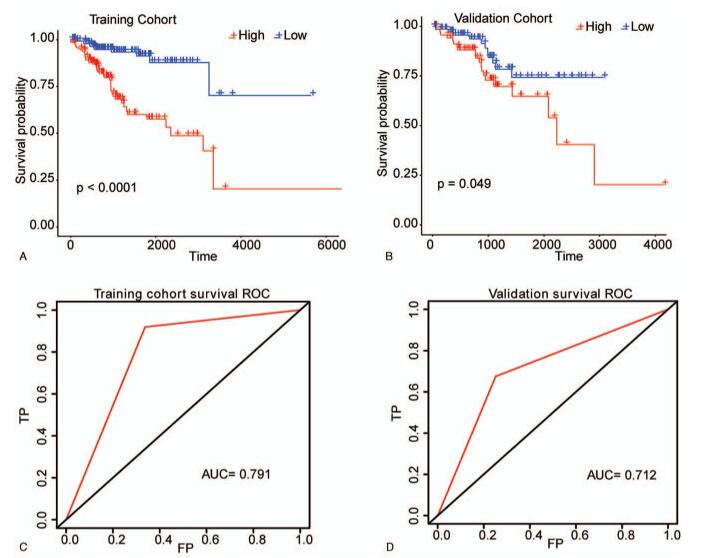
Based on the LASSO Cox regression model, the risk coefficients of these 7 CpG sites were 10.158488, -1.455201, 14.515869, 20.521320, 2.652900, 5.234852 and 7.498602, respectively. Then a predictive model was established according to the formula: Risk score = 10.158488 ∗ β value of cg13823621 - 1.455201 ∗ β value of cg08881614 + 14.515869 ∗ β value of cg07867023 + 20.521320 ∗ β value of cg22247039 + 2.652900 ∗ β value of cg00624976 + 5.234852 ∗ β value of cg06778680 + 7.498602 ∗ β value of cg13204529. The risk score of each patient was then computed based on above formula. Next, all samples in the training set were divided into high-risk group and low-risk group according to the median risk score, and the survival analysis indicated that the prognosis of high-risk group was more inferior than that in the low-risk group (P < .05 Fig. 2A). Meanwhile, the area under curve (AUC) value of this model was 0.791 (Fig. 2C). Furthermore, a similar method was applied to the validation set. The results also showed that the high-risk group had a worse prognosis compared with the low-risk group (P < .05, Fig. 2B) and the AUC value was 0.712 (Fig. 2D). These data indicated that the predictive model could efficiently predict the prognosis of EC patients.
Figure 2.
The construction of the predictive model for EC. (A) The Kaplan–Meier survival curve of samples in training set. (B) The survival curve of samples in validation set was performed by Kaplan–Meier method. The horizontal axis denotes time, the vertical axis denotes survival rate, and altered colors represent different groups. (C) The time-dependent ROC curve of samples in training set. (D) The time-dependent ROC curve of samples in validation set. The horizontal axis denotes false positive, and the vertical axis denotes True Positive.
3.3. Risk score was an independent biomarker for EC prognosis
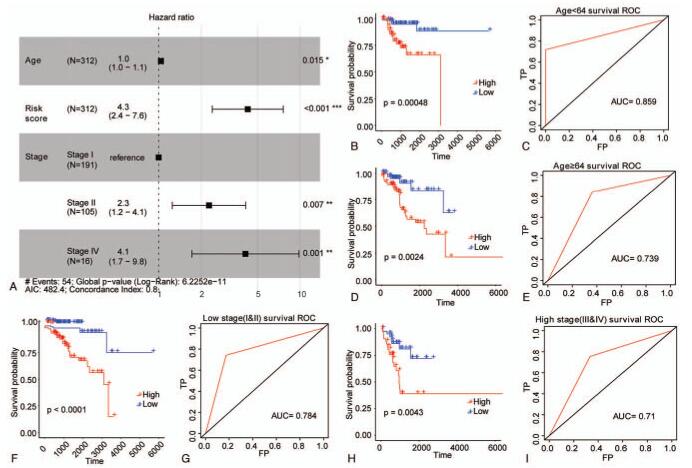
To explore whether risk score was an independent prognostic signature, multivariate Cox regression analysis was carried out by bringing in three independent factors in the training set. The results indicated that age, TNM stage, and risk score were still significantly related to the overall survival of EC patients. In addition, patients with high risk score exhibited a greater death risk (HR = 4.3, 95% CI: 2.4 – 7.6, P < .001) (Fig. 3A). To further investigate the prognostic value of risk score in EC, we regrouped the samples in the training set based on age and TNM stage, and performed the Kaplan–Meier survival analysis. For samples up to 64 years old (Fig. 3B and C), samples over 64 years old (Fig. 3D and E), samples at the early stage (Stage I+II) (Fig. 3F and G) and samples at the advanced stage (Stage III+IV) (Fig. 3H and I), the overall survival was obviously lower in the high-risk group than that in the low-risk group, and the AUC values of samples from different groups were all greater than 0.7, suggesting that the risk score in the training set could be considered as an independent prognostic signature for EC.
Figure 3.
Risk score was an independent prognostic signature for EC in training set. (A) The forest plot of multivariate Cox regression analysis. Samples with hazard ratio > 1 exhibited higher risk of death, and samples with hazard ratio < 1 exhibited lower risk of death. (B–E) The survival curves of EC patients with different ages were determined by Kaplan–Meier method. (F–I) The survival curves of EC patients with different TNM stages were determined by Kaplan–Meier method. The horizontal axis denotes time, the vertical axis denotes survival rate, and altered colors represent different groups. P value is computed by log-rank test.
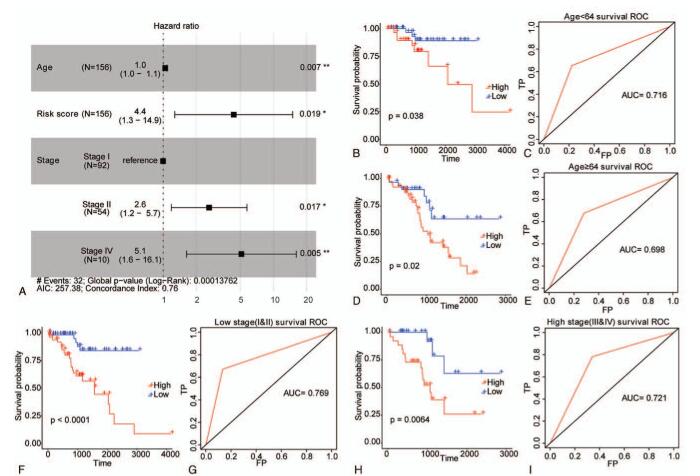
Similarly, in the validation set, we found that age, TNM stage, and risk score were also closely related to the overall survival of EC patients, and patients with a high risk score showed a greater risk of death (HR = 4.4, 95% CI: 1.3 – 14.9, P = .019) (Fig. 4A). We also regrouped the samples in the validation set and conducted the survival analysis. For samples up to 64 years old (Fig. 4B and C), samples over 64 years old (Fig. 4D and E), samples at stage I+II (Fig. 4F and G) and samples at stage III+IV (Fig. 4H and I), the overall survival of EC patients was lower in the high-risk group than that in the low-risk group, and the AUC values of samples from different groups were all greater than 0.7, suggesting that the risk score was also an independent prognostic signature in validation set of EC samples.
Figure 4.
Risk score was an independent prognostic signature for EC in validation set. (A) The forest plot of multivariate Cox regression analysis. Samples with hazard ratio > 1 exhibited higher risk of death, and samples with hazard ratio < 1 exhibited lower risk of death. (B–E) The survival curves of EC samples with different ages were determined by Kaplan–Meier method. (F–I) The survival curves of EC samples with different TNM Stages were determined by Kaplan–Meier method. The horizontal axis denotes time, the vertical axis denotes survival rate, and altered colors represent different groups. The P value is computed by log-rank test.
3.4. The nomogram model performed well in predicting the long-term survival of EC patients
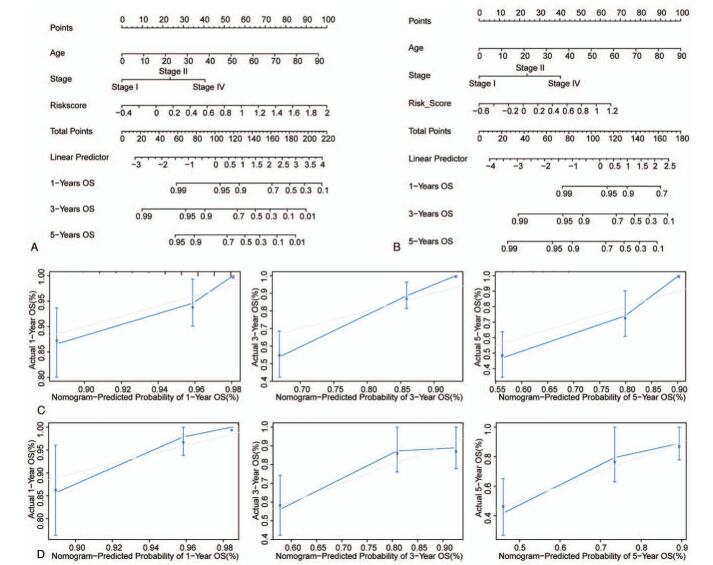
Finally, the nomogram model was established by bringing in three independent prognostic factors including age, TNM stage, and risk score in the training set (Fig. 5A) and validation set (Fig. 5B). We found that the calibration curve was close to the ideal curve both in the training set (Fig. 5C) and validation set (Fig. 5D), indicating that the prediction was consistent with the actual results. These data suggested that the constructed nomogram model performed well in predicting the long-term survival of EC patients.
Figure 5.
The nomogram could efficiently predict the long-term survival of EC patients. (A and B) The nomogram to predict the OS probability at 1, 3, and 5 years in training set (A) and validation set (B). (C and D) The calibration curves of OS probabilities at 1, 3, and 5 years predicted by nomogram in training set (C) and validation set (D).
4. Discussion
Despite the great advances in the pathogenesis research of EC, the clinical outcome of EC patients at advanced stage remains unsatisfactory, even with a lower 5-year overall survival rate of approximately 16% to 69%.[20] Hence, the identification of potential prognostic makers could not only contribute to the development of personalized therapy but also improve clinical outcomes. Although a series of previous studies have developed many prognostic biomarkers for EC, most work only focuses on mRNA or protein expression profiles.[21,22] In the last decades, the prognostic value of DNA methylation has attracted more attention on the prognosis of EC. TBX2, CHST11, and NID2 were found to serve as specific prognostic signatures, meanwhile a prognostic model containing 15 methylation markers was constructed, which could separate EC samples with a poor prognosis from normal subjects.[23,24] However, the biomarkers with higher accuracy in EC should be further explored.
DNA methylation is one of the commonly studied epigenetic modifications in eukaryotic cells and the covalent addition of a methyl group always occurs in cytosine in CpG dinucleotides called CpG island.[25] Changes in DNA methylation during cancer progression have been regarded as promising biomarkers for the development of effective diagnostic and prognostic biomarkers.[26] In EC, Powell et al suggested that ribosomal DNA methylation was an independent prognostic indicator for EC patients, which might be considered as effective biomarkers to identify women with EC at early stage.[27] Shan et al demonstrated that hyper-methylation of SOX11 was common in EC progression and could act as a biomarker in EC.[28]
The m6A regulators have been reported to participate in various biological processes and might have potential diagnostic or prognostic significance in cancer. METTL3-mediated m6A modification of HDGF gene promoter enhances gastric cancer development and has a certain prognostic value.[29] ALKBH5 suppresses pancreatic cancer motility through reducing the expression of lncRNA KCNK15-AS1 in a m6A dependent manner.[30] The m6A demethylase FTO has been found to enhance the proliferation of lung cancer cells by downregulating the mRNA level of USP7 in a m6A dependent manner.[31] Liu et al indicated that the levels of m6A RNA methylation regulators consisting of YTHDF1, HNRNPC, YTHDC2, YTHDC1, ZC3H13, and RBM15 were significantly different in colorectal adenocarcinoma samples compared with healthy controls, and among them, YTHDF1 and HNRNPC could be considered as prognostic biomarkers of colon cancer.[32] These reports all confirmed the important role of m6A regulators in the progression and clinical value of human cancers. However, little is known about the prognostic value of m6A regulator methylation in human cancers, especially in EC.
In this study, we selected 20 m6A regulators as the research objects and identified 396 CpG sites in the sequences of m6A regulators. Meanwhile, 7 optimal CpG sites located in multiple m6A regulators including METTL3, RBM15B, ALKBH5, and YTHDF1 were screened by using LASSO Cox regression analysis. Downregulation of METTL3 could promote the growth and tumorigenicity of EC cells, which might be achieved by activating the AKT pathway.[7] Although the roles of RBM15B, ALKBH5, and YTHDF1 in EC have not been studied, their roles in other cancers are well known. Zhang et al demonstrated that RBM15B was a potential pathogenic site using NGS-based genomic profiling analysis in ovarian cancer.[33] ALKBH5 can promote the progression of gastric cancer through inhibiting the methylation of lncRNA NEAT1.[34] YTHDF1 promotes the development of ovarian cancer via augmenting EIF3C translation,[35] and YTHDF1 overexpression is closely involved in the inferior prognosis of patients with hepatocellular carcinoma.[36] The above researches revealed that these four m6A regulators were closely associated with the development of human cancers. Here, a specific predictive model based on the 7 optimal CpG sites could efficiently predict the prognosis of EC, and the risk score derived from this model might act as an independent prognostic signature both in the training set and validation set.
Nomogram is a widely used method for prognosis prediction in human cancers, majorly due to their ability to reduce statistical predictive models to a single numerical estimate of the probability of an event including death or recurrence.[37] In the present study, the nomogram model was constructed by bringing in three independent prognostic factors including age, risk score, and TNM stage, and it was found that this nomogram model had better performance in predicting the 1-, 3- and 5-year overall survival of EC. However, there were a few limits in this study:
-
1.
Although our study screened 7 optimal CpG sites for the construction of a predictive model, the specific roles of their corresponding m6A regulators in EC are still needed to be explored in the future.
-
2.
Due to the limitation of clinical information in the dataset, our study only investigated the relationship between the predictive model and overall survival of EC patients.
More samples with complete clinical information should be collected to further explore the relationships of the predictive model with radiotherapy effects, chemotherapy drugs, progression-free survival, and other factors.
5. Conclusion
In summary, our study constructed a reliable protective model based on 7 significant CpG sites that were located at m6A regulators. This model could efficiently predict the prognosis of EC, suggesting that CpG sites might be potential prognostic signatures in EC.
Author contributions
Data curation: Xiang Zhang.
Formal analysis: Xuecheng Pang, Yue Huang.
Methodology: Xiang Zhang.
Project administration: Sumin Qian.
Writing – original draft: Xiang Zhang.
Writing – review & editing: Sumin Qian.
Footnotes
Abbreviations: EC = Endometrial carcinoma, IFGO = International Federation of Gynecology and Obstetrics, TCGA = The Cancer Genome Atlas.
How to cite this article: Zhang X, Pang X, Huang Y, Qian S. A seven-m6A regulator-related CpG site-based prognostic signature for endometrial carcinoma. Medicine. 2021;100:29(e26648).
The authors have no funding to disclose.
The authors have no conflicts of interest to disclose.
Ethics approval and consent to participate: Not applicable. This article does not contain any studies with human participants or animals performed by any of the authors.
Consent for publication: All co-authors agree to the publication of the article.
Availability of data and materials: Our data were downloaded from the TCGA (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga). We shared the data and could be accessed from the same website.
The datasets generated during and/or analyzed during the current study are publicly available.
References
- [1].Sorosky JI. Endometrial cancer. Obstet Gynecol 2012;120:383–97. [DOI] [PubMed] [Google Scholar]
- [2].Zhang C, Shao S, Zhang Y, et al. LncRNA PCAT1 promotes metastasis of endometrial carcinoma through epigenetical downregulation of E-cadherin associated with methyltransferase EZH2. Life Sci 2020;243:117295. [DOI] [PubMed] [Google Scholar]
- [3].Ueda SM, Kapp DS, Cheung MK, et al. Trends in demographic and clinical characteristics in women diagnosed with corpus cancer and their potential impact on the increasing number of deaths. Am J Obstet Gynecol 2008;198:218.e211-216. [DOI] [PubMed] [Google Scholar]
- [4].Piulats JM, Guerra E, Gil-Martin M, et al. Molecular approaches for classifying endometrial carcinoma. Gynecol Oncol 2017;145:200–7. [DOI] [PubMed] [Google Scholar]
- [5].Liu ZX, Li LM, Sun HL, Liu SM. Link between m6A modification and cancers. Front Bioeng Biotechnol 2018;6:89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Jia G, Fu Y, He C. Reversible RNA adenosine methylation in biological regulation. Trend Genet 2013;29:108–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Liu J, Eckert MA, Harada BT, et al. m(6)A mRNA methylation regulates AKT activity to promote the proliferation and tumorigenicity of endometrial cancer. Nature Cell Biol 2018;20:1074–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Chen M, Nie ZY, Wen XH, Gao YH, Cao H, Zhang SF. m6A RNA methylation regulators can contribute to malignant progression and impact the prognosis of bladder cancer. Biosci Rep 2019;39: [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jiang SW, Li J, Podratz K, Dowdy S. Application of DNA methylation biomarkers for endometrial cancer management. Expert Rev Mol Diagn 2008;8:607–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Cancer Genome Atlas Research N, Kandoth C, Schultz N, et al. Integrated genomic characterization of endometrial carcinoma. Nature 2013;497:67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zhou XC, Dowdy SC, Podratz KC, Jiang SW. Epigenetic considerations for endometrial cancer prevention, diagnosis and treatment. Gynecol Oncol 2007;107:143–53. [DOI] [PubMed] [Google Scholar]
- [12].Kinde I, Bettegowda C, Wang Y, et al. Evaluation of DNA from the Papanicolaou test to detect ovarian and endometrial cancers. Sci Transl Med 2013;5:ra164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wentzensen N, Bakkum-Gamez JN, Killian JK, et al. Discovery and validation of methylation markers for endometrial cancer. Int J cancer 2014;135:1860–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Vaissiere T, Sawan C, Herceg Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mut Res 2008;659:40–8. [DOI] [PubMed] [Google Scholar]
- [15].Deng XC, Li SR, Zhang Q, et al. [Analysis of the status of DACH1 gene promoter methylation in endometrial carcinoma and its clinical significance]. Zhonghua fu chan ke za zhi 2012;47:263–7. [PubMed] [Google Scholar]
- [16].Yi TZ, Guo J, Zhou L, et al. Prognostic value of E-cadherin expression and CDH1 promoter methylation in patients with endometrial carcinoma. Cancer Invest 2011;29:86–92. [DOI] [PubMed] [Google Scholar]
- [17].Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 2010;33:01–22. [PMC free article] [PubMed] [Google Scholar]
- [18].Zhang Z, Liu J, Kaur M, Krantz ID. Characterization of DNA methylation and its association with other biological systems in lymphoblastoid cell lines. Genomics 2012;99:209–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Brenet F, Moh M, Funk P, et al. DNA methylation of the first exon is tightly linked to transcriptional silencing. PLoS one 2011;6:e14524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:07–30. [DOI] [PubMed] [Google Scholar]
- [21].Yang JY, Werner HM, Li J, et al. Integrative protein-based prognostic model for early-stage endometrioid endometrial cancer. Clinical Can Res 2016;22:513–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Catasus L, D’Angelo E, Pons C, Espinosa I, Prat J. Expression profiling of 22 genes involved in the PI3K-AKT pathway identifies two subgroups of high-grade endometrial carcinomas with different molecular alterations. Mod Pathol 2010;23:694–702. [DOI] [PubMed] [Google Scholar]
- [23].Farkas SA, Sorbe BG, Nilsson TK. Epigenetic changes as prognostic predictors in endometrial carcinomas. Epigenetics 2017;12:19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ying J, Xu T, Wang Q, Ye J, Lyu J. Exploration of DNA methylation markers for diagnosis and prognosis of patients with endometrial cancer. Epigenetics 2018;13:490–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kulis M, Esteller M. DNA methylation and cancer. Adv Genet 2010;70:27–56. [DOI] [PubMed] [Google Scholar]
- [26].Koch A, Joosten SC, Feng Z, et al. Analysis of DNA methylation in cancer: location revisited. Nat Rev Clin Oncol 2018;15:467. [DOI] [PubMed] [Google Scholar]
- [27].Powell MA, Mutch DG, Rader JS, Herzog TJ, Huang TH, Goodfellow PJ. Ribosomal DNA methylation in patients with endometrial carcinoma: an independent prognostic marker. Cancer 2002;94:2941–52. [DOI] [PubMed] [Google Scholar]
- [28].Shan T, Uyar DS, Wang LS, et al. SOX11 hypermethylation as a tumor biomarker in endometrial cancer. Biochimie 2019;162:08–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wang Q, Chen C, Ding Q, et al. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut 2020;69:1193–205. [DOI] [PubMed] [Google Scholar]
- [30].He Y, Hu H, Wang Y, et al. ALKBH5 inhibits pancreatic cancer motility by decreasing long non-coding RNA KCNK15-AS1 methylation. Cell Physiol Biochem 2018;48:838–46. [DOI] [PubMed] [Google Scholar]
- [31].Li J, Han Y, Zhang H, et al. The m6A demethylase FTO promotes the growth of lung cancer cells by regulating the m6A level of USP7 mRNA. Biochem Biophys Res Commun 2019;512:479–85. [DOI] [PubMed] [Google Scholar]
- [32].Liu T, Li C, Jin L, Li C, Wang L. The prognostic value of m6A RNA methylation regulators in colon adenocarcinoma. Med Sci Monit 2019;25:9435–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Zhang L, Luo M, Yang H, Zhu S, Cheng X, Qing C. Next-generation sequencing-based genomic profiling analysis reveals novel mutations for clinical diagnosis in Chinese primary epithelial ovarian cancer patients. J Ovarian Res 2019;12:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Zhang J, Guo S, Piao HY, et al. ALKBH5 promotes invasion and metastasis of gastric cancer by decreasing methylation of the lncRNA NEAT1. J Physiol Biochem 2019;75:379–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Liu T, Wei Q, Jin J, et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res 2020;48:3816–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Zhao X, Chen Y, Mao Q, et al. Overexpression of YTHDF1 is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Biomark 2018;21:859–68. [DOI] [PubMed] [Google Scholar]
- [37].Iasonos A, Schrag D, Raj GV, Panageas KS. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol 2008;26:1364–70. [DOI] [PubMed] [Google Scholar]