Abstract
Introduction
Klebsiella pneumoniae (K. pneumoniae) is one of the leading causes of hospital-acquired and community-acquired infections in the world. This study was conducted to investigate the molecular epidemiology of drug resistance in clinical isolates of K. pneumoniae in Azerbaijan, Iran.
Materials and Methods
A total of 100 nonduplicated isolates were obtained from the different wards of Azerbaijan state hospitals, Iran, from 2019 to 2020. Antibiotic susceptibility testing was done. The DNA was extracted, and the PCR for evaluation of the resistance genes was carried out.
Results
The highest antibiotic resistance was shown to ampicillin (96%), and the highest susceptibility was shown to tigecycline (9%), and 85% of isolates were multidrug resistant. The most frequent ESBL gene in the tested isolates was blaSHV-1 in 58%, followed by blaCTXM-15 (55%) and blaSHV-11(42%). The qepA, oqxB, and oqxA genes were found to be 95%, 87.5%, and 70%, respectively. We detected tetB in 42%, tetA in 32%, tetD in 21%, and tetC in 16%. Seventy isolates were resistant to co-trimoxazole, and the rate of resistance genes was sul1 in 71%, followed by sul2 (43%), dfr (29%), and sul3 (7%). The most common aminoglycoside resistance genes were ant3Ia, aac6Ib, aph3Ib, and APHs in 44%, 32%, 32%, and 31.4%, respectively. The most frequent resistance gene to fosfomycin was fosA (40%) and fosX (40%) followed by fosC (20%).
Conclusion
The results of this study indicate the high frequency of drug resistance among K. pneumoniae isolated from hospitals of Azerbaijan state. The present study shows the presence of high levels of drug-resistant genes in various antibiotics, which are usually used in the treatment of infections due to K. pneumoniae.
1. Introduction
Klebsiella pneumoniae (K. pneumoniae) is one of the prominent causes of hospital-acquired and community-acquired infections worldwide [1–4]. This bacterium is the second most common cause of urinary tract infections [1]. In comparison to classic nonvirulent isolates, hypervirulent K. pneumoniae strains affect healthy individuals with dangerous and invasive infections, while classic strains have usually ESBL producers that affect hospitalized patients and are leading causes of hospital-acquired infections [5]. K. pneumoniae is one of the three major drug-resistant bacteria on the WHO priority list that requires more research and production of novel antibiotics for the treatment [1, 2]. Extensive drug resistance (XDR) against different classes of antibiotics had converted this organism to a big bug in health-care settings, which in turn limits treatment options of infections due to K. pneumoniae [1, 4]. This bacterium uses various resistance mechanisms including destructive enzyme production, efflux pumps, porin loss, and target alteration to counteract the effects of antibiotics [1, 3, 6]. So, nosocomial infections with multidrug-resistant (MDR) strains of K. pneumoniae are occurring with high mortality and morbidity [3, 4]. K. pneumoniae similar to other opportunistic pathogens affect patients who have predisposing debilitating backgrounds and are mainly reported from ICUs, urinary tract infections, ventilator-associated pneumonia, and sepsis [1, 3, 5, 7, 8].
This bacterium has to carry a collection of antibiotic genes with chromosomal and plasmid origins. Acquiring antibiotic resistance genes with plasmid and other transposable agents and mutations after antibiotic pressure has produced superresistant strains of high-risk MDR and XDR K. pneumoniae isolates in clinical settings especially in the two past decades which have high distribution potential worldwide [4]. The correct and prompt treatment of infections due to K. pneumoniae is vital. The resistance mechanisms epidemiology should be evaluated comprehensively in hospital settings worldwide. Based on the critical literature reviews in the different data sources, the antibiotic resistance profile of K. pneumoniae isolates as a worldwide threat in the northwest of Iran (Azerbaijan province) was not clear. Therefore, we conducted this study to consider the molecular epidemiology of drug resistance in clinical isolates of K. pneumoniae.
2. Materials and Methods
2.1. Bacterial Isolates
A total of 100 nonduplicated K. pneumoniae isolates were obtained from the different wards of Azerbaijan state hospitals, Iran, from 2019 to 2020. All clinical samples were isolated aseptically from the different organs. Presumptive colonies were further studied by the standard conventional methods [9]. The current study was approved by the research ethics committee (IR.TBZMED.REC.1399.745) at Tabriz University of Medical Sciences, Tabriz, Iran.
2.2. Antibiotic Susceptibility Testing
Antibiotic susceptibility testing was prepared by the disk diffusion agar method (DDA) based on the CLSI guidelines. The disks from two groups of beta-lactam and non-beta-lactam drugs were used including cefoxitin (30 µg), ceftazidime (30 µg), cefotaxime (30 µg), ertapenem (10 µg), imipenem (10 µg), meropenem (10 µg), gentamicin (10 µg), amikacin (30 µg), ciprofloxacin (5 µg), ofloxacin (5 µg), gatifloxacin (5 µg), kanamycin (30 µg), tobramycin (10 µg), ampicillin (10 µg), amoxicillin-clavulanate (20/10 µg), tetracycline (30 µg), doxycycline (30 µg), tigecycline (15 µg), trimethoprim-sulfamethoxazole (1.25/23.75 µg), fosfomycin (200 µg), aztreonam (30 µg), levofloxacin (5 µg), moxifloxacin (30 µg), ceftriaxone (30 µg), and piperacillin-tazobactam (100/10 µg) (Mast, UK). The results of the DDA method on Muller–Hinton agar plates were analyzed according to the Clinical and Laboratory Standards Institute (CLSI) guidelines [10].
To determine the MICs, the agar dilution was used for ciprofloxacin, levofloxacin, gentamicin, kanamycin, tobramycin, amikacin, cefotaxime, ampicillin, tetracycline, co-trimoxazole, fosfomycin, and imipenem recommended by CLSI [11]. In the current study, the results of susceptibility testing were confirmed by the Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC 27853 as positive controls of susceptibility testing.
2.3. ESBL by the Phenotypic Tests
CLSI recommended combination disk with ceftazidime (30 µg) and cefotaxime (30 µg) disks alone and in mixture with clavulanate (10 µg). A ≥ 5 mm increase in the zone diameter for cefotaxime or ceftazidime in combination with clavulanate versus the zone diameter of the corresponding antimicrobial agent alone defined an ESBL producer [9].
2.4. Determination of Carbapenemases by the Phenotypic Tests
To detect carbapenemases, a phenotypic technique was carried out according to the previous described study [12]. All the carbapenem-resistant isolates were selected for more assessment by two methods, the modified Hodge Test (MHT) and the Carba NP test [13, 14].
2.5. DNA Extraction and the PCR for Evaluation of the Resistance Genes
The DNA was taken out by the CTAB method as defined previously [15]. PCR assays were carried out by primers specific for the blaNDM,blaVIM, blaIMP, blaKPC, and blaOXA-48-like carbapenemases-encoding genes and ompK35 and ompK36 as porin-coding genes. Resistance genes of different classes including ESBLs (blaTEM, blaSHV, and blaCTX-M), carbapenemases (KPC, MBL, or both MBL and KPC, AmpC); fluoroquinolones, tetracycline, sulfonamides/trimethoprim, aminoglycosides, and fosfomycin were detected by the PCR. The PCR products were analyzed by electrophoresis in 1.5% agarose gel, and after staining with 0.5 µg/mL safe stain, they were imagined underneath ultraviolet (UV) light. Pseudomonas aeruginosa, E. coli, and K. pneumoniae were used as controls in the current study. Primers used in the current study selected from previous studies [16–31].
2.6. Statistical Analysis
The results were studied by the descriptive statistics in the SPSS software for Windows (version 23 SPSS Inc., Chicago, IL, USA).
3. Results
3.1. Bacterial Isolates
From 2019 to 2020, a total of 100 nonduplicated K. pneumoniae isolates were collected from the different wards of Azerbaijan state hospitals, Iran. The number of bacteria isolated according to each clinical specimen included the wound (23%), blood (14%), urinary tract (49%), and pulmonary (14%). The mean age of patients was 38 ± 21. These patients (47 males and 53 females) were hospitalized in the pediatrics wards (5%), burn (12%), surgery (14%), ICU (25%), and internal (44%).
3.2. Antibiotic Susceptibility Patterns
According to the disk diffusion agar, the frequency of resistance to ciprofloxacin, levofloxacin, ofloxacin, moxifloxacin, gatifloxacin, gentamicin, kanamycin, tobramycin, amikacin, cefotaxime, ceftriaxone, ceftazidime, cefoxitin, ertapenem, meropenem, imipenem, ampicillin, amoxicillin- clavulanate, tetracycline, doxycycline, tigecycline, co-trimoxazole, fosfomycin, aztreonam and, piperacillin-tazobactam was 67%, 39%, 42%, 54%, 47%, 51%, 49%, 51%, 14%, 68%, 67%, 68%, 28%, 33%, 25%, 25%, 96%, 77%, 41%, 54%, 9%, 63%, 11%, 65, and 72%. In the present study, 85% of isolates were resistant to at least three classes of antimicrobial agents considered as an MDR. Of the MDR isolates, sixty-five (65%) were ESBLs-producer. The results of MIC method were consistent with the DDA method except slightly different (Figure 1). Wholly, the carbapenem-resistant isolates were confirmed by two methods, the modified Hodge test and Carba NP test.
Figure 1.
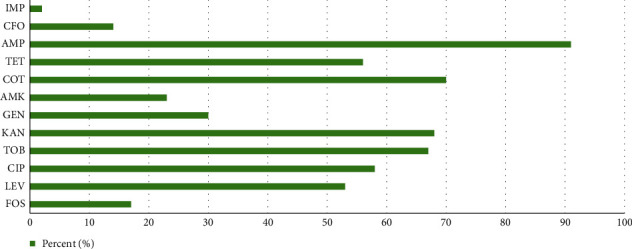
Antibiotic susceptibility testing by the agar dilution. CIP: ciprofloxacin; LEV: levofloxacin; GEN: gentamicin; KAN: kanamycin; TOB: tobramycin; AMK: amikacin; CFO: cefoxitin; IMP: imipenem; AMP: ampicillin; TET: tetracycline; COT: co-trimoxazole; and FOS: fosfomycin.
3.3. Molecular Epidemiology
The fluoroquinolone resistance gene was found in 49 (49%) isolates, and the most common qnr gene was qepA in 95% (46 of 49), followed by oqxB in 87.5% (43 of 49) and oqxA in 70% (34 of 49). Table 1 shows the distribution of qnr genes among K. pneumoniae isolates. Among the 100 K. pneumoniae isolates, 52 (52%) isolates were resistant to tetracycline, as determined via the DDA technique. The PCR detected tetB in 42% (22 of 52) isolates, tetA in 32% (17 of 52), tetD in 21% (11 of 52), and tetC in 16% (8 of 52). Seventy isolates (70%) were resistant to sulfonamides/trimethoprim. We discovered sul1 in 71% (50 of 70) isolates, sul2 in 43% (30 of 70), dfr in 29% (20 of 70), and sul3 in 7% (5 of 70). In the current study, 54 (54%) isolates were resistant to aminoglycosides. The most common resistance gene was ant3Ia in 44% (24 of 54), followed by aac6Ib (32%, 17 of 54), aph3Ib (32%, 17 of 54), and APHs (31.4%, 16 of 54). Sixty-five isolates (65%) were phenotypically detected as ESBL. The most frequent ESBL gene was blaSHV-1 58% (38 of 65), followed by blaCTX-15 55% (36 of 65) and blaSHV-11 42% (27 of 65). Sixteen (16%) isolates were resistant to fosfomycin. The most frequent resistance genes in the tested isolated were fosA (40%, 6 of 16) and fosX (40%, 6 of 16), followed by fosC (20%, 4 of 16) (Table 1).
Table 1.
Distribution of antibiotic resistance genes in the current study.
| Antibiotics | Genes conferring resistance (%) |
|---|---|
|
Fluoroquinolone
Ciprofloxacin, ofloxacin, gatifloxacin, levofloxacin, and moxifloxacin |
qepA (95%), oqxB (87.5%), oqxA (70%), qnrA (5%), qnrB (27.5%), qnrD (25%), and qnrS (17.5%) |
|
| |
|
Tetracycline
Tetracycline, doxycycline, and tigecycline |
tetA (32%), tetB (42%), tetC (16%), and tetD (21%) |
|
| |
| Co-trimoxazole | dfr (29%), Sul3 (7%), Sul2 (43%), and Sul1 (71%) |
|
| |
|
Aminoglycosides
Gentamicin, amikacin, kanamycin, and tobramycin |
APHs (31.4%), ANTs (30.7%), rmtB (20%), aac3Ib (10%), aac3IIa (24%), aac6Ib (32%), ant3Ia (44%), ant4IIa (5%), aph3Ia (10%), and aph3Ib (32%) |
|
| |
|
Beta-lactamases
Cefoxitin, ceftazidime, cefotaxime, ertapenem, imipenem, meropenem, ampicillin, ceftriaxone, amoxicillin-clavulanate, piperacillin-tazobactam, and aztreonam |
bla TEM-1 (40%), blaTEM-16 (33%), blaTEM-24 (3%), blaSHV-1 (58%), blaSHV-11 (42%), blaCTXM-15 (55%), blaCTXM-3 (27%), blaCTXM-1 (5%), blaCTXM-79 (9%), blaCTXM-27 (5%), blaMBL (16%), blaKPC (7%), blaOXA-48 (4%), porin loss (35%), omp35 (80%), omp36 (77%), and ampC (12%) |
|
| |
| Fosfomycin | fosA (40%), fosX (40%), and fosC (20%) |
4. Discussion
Nosocomial infections have been a major challenge for clinicians worldwide, especially in developing countries. K. pneumoniae is one of the most common nosocomial infection agents in hospitals. Increasing resistance to antimicrobial agents has not only increased the duration of the hospitalization but also increases the cost of treatment. Despite a huge number of studies on antimicrobial profiles of resistance and MDR from clinical settings in different provinces of Iran, the comprehensive epidemiology of K. pneumoniae resistance genes is not clear yet. Literature review of resistance patterns of K. pneumoniae isolates indicates that different rates of resistance and related genes existing in health-care settings in the world. Certainly, this study could help to prepare a consensus on the extensive analysis of resistance gene maps and implement control and treatment programs against such threatening infections in the future.
In the current study, positive ESBLs phenotype was observed in 65%, and the MDR producing K. pneumoniae isolates was 85%. The most frequent ESBL gene in the tested isolates was blaSHV-1 in 58% followed by blaCTXM-15 in 55% and blaSHV-11 in 42%. In a study from South Africa, 202 Klebsiella spp. were collected; 83.7% were identified as K. pneumoniae and 57.9% as other Klebsiella spp. The isolates were studied for their resistance patterns and presence of ESBLs and carbapenemases genes. They reported that blaSHV (77.1%) was the most highly prevalent ESBL gene, followed by blaTEM (66.9%) and blaCTXM (56.7%) [32]. Ferreira et al. in a study on the clinical K. pneumoniae isolates in the ICU explained that 84% of isolates were MDR with a high rate of resistance against β-lactams, quinolones, aminoglycosides, tigecycline, and colistin. They found that all of the isolates were as ESBLs and carbapenemases-producers which harboring blaKPC (100%), blaTEM (100%), blaSHV variants (96%), blaOXA-1 group (84%), and blaCTXM-1 group (72%) genes [3]. In the present study, 85% of isolates were resistant to as a minimum of three antibiotics classes considered as an MDR, and ESBLs phenotype was observed in 65 isolates, similar to previous studies [3, 32]. The high frequency of ESBL-producing K. pneumoniae may be due to arbitrary consumption of third-generation cephalosporin in the society [33]. According to the results of the antibiotic susceptibility patterns, the highest frequency of resistance was found to ampicillin. This finding is similar to another study from Iran [34] and Russia [5]. Tigecycline and fosfomycin were the most effective antibiotics in this study. Carbapenem has been reflected as an important option for the treatment of resistant Enterobacteriaceae particularly MDR and ESBLs producing isolates. The emergence of carbapenem-resistant Enterobacteriaceae was reported following the increased use of carbapenem [35]. In our study, the resistance to meropenem and imipenem was observed in 25% by the DDA. Other studies also reported carbapenem-resistant Enterobacteriaceae isolates from China and Iran [7, 34, 36]. Xu et al. in a study on 89 K. pneumoniae isolated from ventilator-associated pneumonia reported that 33.7% were ESBL producers which were simultaneously MDR. These isolates had genes of blaSHV, blaCTXM, blaOXA, and blaTEM with 70%, 70%, 3.3%, and 66.6%, respectively. Interestingly, blaCTXM-15 was reported as the highest frequent ESBL gene in China [8]. The resistance profile of K. pneumoniae isolates from Iranian provinces is relatively different. According to the Kiaei et al. from south of Iran (Kerman) from 175 clinical K. pneumoniae isolates, 41.1%, 6.8%, and 21.1% were positive as ESBLs, AmpC, and carbapenemases, respectively; also, 4.5% was both ESBL and AmpC positive. Very interestingly, the resistance rate against carbapenem including imipenem (25.7%) and meropenem (18.9%) was higher than ciprofloxacin at 17.7%. The frequency of ESBL genes was reported as follows blaCTXM (46.28%), blaSHV (41.1%), blaTEM (38.9%), and blaOXA-1 (21.7%). blaNDM was the only carbapenemases gene reported in 21.14% of isolates [6]. The occurrence of blaSHV and blaCTXM genotypes greatly varies geographically among ESBL-producing isolates. A previous study revealed that blaSHV and blaCTXM variants were the most common among the ESBLs gene [6, 8, 32]. The results of the present study are also close to the previously mentioned studies.
Tetracycline resistance can be mediated by ribosomal protection, efflux pumps, or chemical modification. In the present study, the resistance rate to tetracycline and doxycycline by the DDA assay was 41% and 54%, respectively. The frequency of tetracycline resistance was reported differently. Tetracycline resistance rate was reported to be 27.5 to 50% among Enterobacteriaceae from Saudi Arabia [37]. In a previous study, the tetracycline resistance among K. pneumoniae was reported at 41.3% from Tabriz, Iran [34]. In another study from Tehran hospital, 64% of K. pneumoniae were isolated from UTIs carrying tet gene (highest antibiotic resistance), and the lowermost resistance (2%) was reported to imipenem and gentamicin [37]. The difference in the frequency of resistance to tetracycline may be due to the geographic regions, amount of antibiotic usage, continued prescribing of tetracycline in veterinary, and a difference in a program of infection control. In the present study, tetA, tetB, tetC, and tetD were detected in 32%, 42%, 16%, and 21% of resistance isolates, respectively. Another study from Tabriz, Iran, reported tetA, tetB, tetC, and tetD in 14.4%, 18.4%, 2%, and 4.4%, respectively [34]. Both tetA and tetB genes generally were reported as the most common tet genes among tetracycline-resistant Enterobacteriaceae [38–40].
The previous studies indicated that fosfomycin is a broad-spectrum antibiotic against Gram-negative and Gram-positive bacteria [41]. Recently, much attention has been paid to fosfomycin for high clinical value in the treatment of MDR Enterobacteriaceae [42]. However, with the increased application of fosfomycin, resistant isolates have been reported. Multiple mechanisms of fosfomycin resistance can be due to antimicrobial-modifying enzymes, target site modification, or decreased permeability [43]. In the current research, the resistance rate to fosfomycin was 16%. In a previous study from Iran, the resistance rate to fosfomycin in K. pneumoniae was 10.9% [44], and these findings are similar to our study. Also, the obtained results of resistance to fosfomycin in both the DDA and agar dilution methods in the current study are in accordance, only the slight and neglectable difference was observed. In the present study, the dominant founded resistance genes to fosfomycin were fosA, fosC, and fosX in 40%, 20%, and 40%, respectively. These results contradict some previous studies in Iran [44], and the percentage differs slightly in the world. The trivial difference in the obtained statistics can be explained according to the number of specimens, the health status of countries, and the pattern of prescribed antibiotics in the various reagents. On the other hand, the identification of fosX gene is of the utmost importance in our study because of the scarcity of this gene in the previous investigations. Although fosfomycin has dramatically antibacterial activity, the trivial identified resistance cases are an alarm because of the determinant locations on mobile genetic elements like the plasmids.
Co-trimoxazole is a combination of the antifolate compounds trimethoprim and sulfamethoxazole, which have a synergistic effect, and has been used as a broad-spectrum antibiotic [45]. Co-trimoxazole has been used for several decades as an efficient antibiotic for the treatment of enteric bacterial and urinary tract infections [46]. The injudicious use of co-trimoxazole in developing countries has been a major factor in the emergence of decreased susceptibility to this antibiotic [47, 48]. Three plasmid-mediated dihydropteroate synthase genes encoding sulfonamide resistance, sul1, sul2, and sul3, have been identified in Gram-negative bacteria and more than 30 dfr gene encoding resistant variants to trimethoprim have been described [46, 49]. In the current research, co-trimoxazole is the second most common drug-resistant (70%) which was consistent with previous studies in Iran and another country [5, 34]. Based on this study, the sul1 gene has the highest frequency in K. pneumoniae resistance to co-trimoxazole. The rate of sul1 (71%) was higher than sul2 (43%) and sul3 (7%), which is in accordance with a study from Tanzania [50]. The frequency of sul genes in a previous study in Iran on Escherichia coli isolates was close to our study [51]. The existence of sul genes in different environmental and clinical isolates shows that these genes have a general role in carrying and distributing sulfonamide resistance in bacteria [52–54]. The dfr gene in our research was detected in 29% of K. pneumoniae isolates. The high rate of sul genes and high occurrence of co-trimaxazole resistance in K. pneumoniae isolates points toward that uninterrupted investigation programs should be applied in clinical settings to better control and manage associated diseases and monitor the trends of co-trimaxazole resistance in Gram-negative bacteria.
Fluoroquinolone is the most significant antibiotic used for the management of bacterial infections [55]. Newly, fluoroquinolone resistance has increased in clinical isolates. The most common fluoroquinolone resistance mechanism is due to the chromosomal mutations in quinolone resistance determining regions (QRDR) and plasmid-mediated quinolone resistance (PMQR) [56]. In the present study, 49 isolates contain fluoroquinolone resistance genes. As formerly revealed, PMQR genes play a significant role in resistance to quinolone due to parallel transferability [57]. In our study, the most prevalent PMQR gene was qepA (95%) followed by oqxB (87.5%), oqxA (70%), and qnrB (27.5%). Contrary to our study, another study in Iran on K. pneumoniae reported that the most predominant PMQR gene was oqxA followed by oqxB and qepA [58]. In this study, qnrB gene (27.5%) was more prevalent among qnr genes and the lowest rate was qnrA (5%). Interestingly, in some studies, qnrA was not found. Our results are constant with the results of previous studies [59–61]. In our study like some research, all three resistant genes (qnrB, qnrA, and qnrS) have also been originated in clinical isolates [60, 62]. The types of qnr genes may vary in different geographical locations [63]. One of the important features of this research is the observation of qnrD (25%) in clinical isolates. The present study established a high prevalence of qepA (95%), oqxB (87.5%), and oqxA (70%) genes among PMQR factors. The transferability amount of these factors is high. This is a reason for worry since the horizontal transfer of PMQR genes can raise the spread of resistance to fluoroquinolone.
Aminoglycosides are one the most essential treatments used for K. pneumoniae infections. The mechanisms of resistance to aminoglycosides include modification of the ribosomal target, enzymatic modification, and diminished intracellular antibiotic accumulation by changes of the outer membrane permeability, reduced inner membrane transport, or active efflux pumps. Adenylation and acetylation are two main mechanisms of aminoglycoside resistance, catalyzed by actyl-CoA in need of N-acetyltransferase and ATP-dependent O-nucleotidyltransferase, respectively [64, 65]. In the current study, a total of 54 aminoglycoside-resistant K. pneumoniae strains were found among 100 clinical isolates. Our results showed that the resistance of K. pneumoniae isolates to gentamicin, kanamycin, tobramycin, and amikacin was 51%, 49%, 51%, and 14%, respectively. Our results showed that the rate of resistance to gentamicin, tobramycin, and amikacin was consistent with another research from Iran [66]. Interestingly, the results of Hesam et al. study about the rate of resistance to gentamicin and tobramycin were very close to our study [66]. In our research, according to the DDA method, the highest aminoglycosides resistance among K. pneumoniae isolates was related to gentamicin and the lowest resistance was related to amikacin. These results were consistent with El-Badawy et al., but the rate of resistance to amikacin was approximately doubled in their study [61]. The findings of this study showed that the most frequent aminoglycosides resistant genes were ant (3) Ia (44%) followed by aac (6) Ib (32%), aph (3) Ib (32%), and APHs (31.4%). In another study on Klebsiella spp. and E. coli isolated from Egypt, the most dominant aminoglycoside-modifying enzymes (AMEs) were aac (3) IIa and aac (6) Ib. On the other hand, their research specified that none of aac (3) Ia and rmt55 genes were distinguished in any of Klebsiella spp. and E. coli isolated from Egyptian patients [67]. Our results were consistent with a study in Egypt, but contrary to their research, the highest presence of AME genes in the current study was related to ant (3) Ia. In the study of Iraj et al., the aac (3) IIa, aac (6) Ib, and rmtB genes were identified in 55.8%, 80.8%, and 1.9% of the isolates, respectively. The aac (3) Ia and armA were not found in any of the isolates [68]. In the previous studies from Spain by Elisenda et al., the most frequent AME genes in Enterobacteriaceae were aph (3) Ib (65.4%) and ant (3) Ia (37.5%) [69], whereas in the recent study, the most frequent AME genes in K. pneumoniae were ant (3) Ia (44%) followed by aac (6) Ib (32%) and aph (3) Ib (32%). This could be due to the diversity of bacterial species used in the Spanish study. The rate of AAC (3)-II has diminished. In 1993, Shaw et al. defined an occurrence of AAC (3)-II in 60.3% of 2445 Gram-negative bacteria [70]. However, we found a prevalence of AAC (3)-II in (24%). In a previous study, the most predominant enzyme was AAC (6)-Ib [70]. However, these enzymes are essential for the reason that they are the few genes that confer resistance to amikacin and quinolones. The results of our study also confirmed the issue. Our results showed high frequencies of aminoglycosides resistance genes in K. pneumoniae isolates which could be the results of unnecessary use of antibiotics, and these outcomes discovered the status of the transfer of antibiotic resistance genes.
In this study, the patterns of antibiotic resistance and the frequency of resistance coding genes in K. pneumoniae isolated from Azerbaijan state hospitals by phenotypic and molecular methods were studied. So, the determination of the patterns of antibiotic resistance in common pathogenic bacteria including K. pneumoniae is important to guide empirical therapy against a particular pathogen; also, it helps reduce treatment costs and length of hospital stay; moreover, it may be used as a warning for officials and policymakers in the national and provincial health basin to take more seriously the issue of the emergence incidence of drug resistance among bacteria especially K. pneumoniae. The routine usage of antibiotics in the food business, especially in the agricultural and livestock industry, is recommended monitor by health officials. Furthermore, choosing an antibiotic by antibiotic susceptibility testing and continuous monitoring of drug resistance could have a key role in the management of infectious diseases and reduce antibiotic resistance. Due to the high occurrence of drug resistance in Azerbaijan hospitals, it is recommended to conduct a national study in Iran, to evaluate the rate of antimicrobial resistance in K. pneumoniae.
5. Conclusion
Antimicrobial resistance in K. pneumoniae has become one of the most challenging issues faced in daily clinical practice. Our results indicated a high occurrence of drug resistance among K. pneumoniae isolates in Azerbaijan hospitals. The resistance to co-trimoxazole is the most common, followed by aminoglycosides, tetracycline, fluoroquinolone, and fosfomycin. This is due to the presence of high levels of drug-resistant genes in various antibiotics, which are frequently used in the treatment of infections caused by K. pneumoniae.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Disclosure
This article was written based on a dataset of a Ph.D. thesis (no. 65885) registered with Tabriz University of Medical Sciences, Tabriz, Iran.
Conflicts of Interest
The authors declare that they have no conflicts of interest regarding the publication of this paper.
Authors' Contributions
Design was carried out by R. Ghotaslou, writing by M. Kashefieh and R. Ghotaslou, statistical analysis by H. Hosseinzadegan, and review and editing by S. Baghbani.
References
- 1.Caneiras C., Lito L., Melo-Cristino J., Duarte A. Community- and hospital-acquired Klebsiella pneumoniae urinary tract infections in Portugal: virulence and antibiotic resistance. Microorganisms. 2019;7(5):p. 138. doi: 10.3390/microorganisms7050138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tacconelli E., Magrini N. Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. Geneva, Switzerland: WHO; 2017. https://www.who.int/medicines/publications/global-priority-list-antibiotic-resistant-bacteria/en/ [Google Scholar]
- 3.Ferreira R. L., Da Silva B. C. M., Rezende G. S., et al. High prevalence of multidrug-resistant Klebsiella pneumoniae harboring several virulence and β-lactamase encoding genes in a Brazilian intensive care unit. Frontiers in Microbiology. 2019;9:3198. doi: 10.3389/fmicb.2018.03198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Navon-Venezia S., Kondratyeva K., Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiology Reviews. 2017;41(3):252–275. doi: 10.1093/femsre/fux013. [DOI] [PubMed] [Google Scholar]
- 5.Khaertynov K. S., Anokhin V. A., Rizvanov A. A., et al. Virulence factors and antibiotic resistance of Klebsiella pneumoniae strains isolated from neonates with sepsis. Frontiers in Medicine. 2018;5(225):p. 225. doi: 10.3389/fmed.2018.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiaei S., Moradi M., Hosseini-Nave H., Ziasistani M., Kalantar-Neyestanaki D. Endemic dissemination of different sequence types of carbapenem-resistant Klebsiella pneumoniae strains harboring bla (NDM) and 16S rRNA methylase genes in Kerman hospitals, Iran, from 2015 to 2017. Infection and Drug Resistance. 2018;12:45–54. doi: 10.2147/IDR.S186994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu L., Zhong Q., Shang Y., et al. The prevalence of carbapenemase genes and plasmid-mediated quinolone resistance determinants in carbapenem-resistant enterobacteriaceae from five teaching hospitals in Central China. Epidemiology and Infection. 2014;142(9):1972–1977. doi: 10.1017/s0950268813002975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu H., Huo C., Sun Y., et al. Emergence and molecular characterization of multidrug-resistant Klebsiella pneumoniae isolates harboring blaCTX-M-15 extended-spectrum β-lactamases causing ventilator-associated pneumonia in China. Infection and Drug Resistance. 2018;12:33–43. doi: 10.2147/idr.s189494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahon C. R., Lehman D. C., Manuselis G. Text Book of Diagnostic Microbiology. Philadelphia, PA, USA: Saunders; 2007. [Google Scholar]
- 10.CLSI. Performance Standards for Antimicrobial Susceptibility Testing. CLSI Supplement M100. 27th. Wayne, PA, USA: Clinical and Laboratory Standards Institute; 2017. [Google Scholar]
- 11.CLSI. Performance Standards for Antimicrobial Susceptibility Testing. CLSI Supplement M100. 27th. Wayne, PA, USA: Clinical and Laboratory Standards Institute; 2019. [Google Scholar]
- 12.Tsakris A., Poulou A., Pournaras S., et al. A simple phenotypic method for the differentiation of metallo-beta-lactamases and class A KPC carbapenemases in enterobacteriaceae clinical isolates. Journal of Antimicrobial Chemotherapy. 2010;65(8):1664–1671. doi: 10.1093/jac/dkq210. [DOI] [PubMed] [Google Scholar]
- 13.CLSI. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fourth Informational Supplement. M100-S24 January. 2014. Wayne, PA, USA: Clinical and Laboratory Standard Institute; 2014. [Google Scholar]
- 14.Akhi M. T., Khalili Y., Ghotaslou R., et al. Carbapenem inactivation: a very affordable and highly specific method for phenotypic detection of carbapenemase-producing Pseudomonas aeruginosa isolates compared with other methods. Journal of Chemotherapy. 2017;29(3):144–149. doi: 10.1080/1120009x.2016.1199506. [DOI] [PubMed] [Google Scholar]
- 15.Akhi M. T., Ghotaslou R., Alizadeh N., pirzadeh T., Beheshtirouy S., Memar M. Y. High frequency of MRSA in surgical site infections and elevated vancomycin MIC. Wound Medicine. 2017;17:7–10. doi: 10.1016/j.wndm.2017.01.002. [DOI] [Google Scholar]
- 16.Nüesch-Inderbinen M. T., Kayser F. H., Hächler H. Survey and molecular genetics of SHV beta-lactamases in enterobacteriaceae in Switzerland: two novel enzymes, SHV-11 and SHV-12. Antimicrobial Agents and Chemotherapy. 1997;41(5):943–949. doi: 10.1128/aac.41.5.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Noppe-Leclercq I., Wallet F., Haentjens S., Courcol R., Simonet M. PCR detection of aminoglycoside resistance genes: a rapid molecular typing method for acinetobacter baumannii. Research in Microbiology. 1999;150(5):317–322. doi: 10.1016/s0923-2508(99)80057-6. [DOI] [PubMed] [Google Scholar]
- 18.Aminov R. I., Chee-Sanford J. C., Garrigues N., et al. Development, validation, and application of PCR primers for detection of tetracycline efflux genes of gram-negative bacteria. Applied and Environmental Microbiology. 2002;68(4):1786–1793. doi: 10.1128/aem.68.4.1786-1793.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kerrn M. B., Klemmensen T., Frimodt-Møller N., Espersen F. Susceptibility of Danish Escherichia coli strains isolated from urinary tract infections and bacteraemia, and distribution of sul genes conferring sulphonamide resistance. Journal of Antimicrobial Chemotherapy. 2002;50(4):513–516. doi: 10.1093/jac/dkf164. [DOI] [PubMed] [Google Scholar]
- 20.Hammerum A. M., Sandvang D., Andersen S. R., et al. Detection of sul1, sul2 and sul3 in sulphonamide resistant Escherichia coli isolates obtained from healthy humans, pork and pigs in Denmark. International Journal of Food Microbiology. 2006;106(2):235–237. doi: 10.1016/j.ijfoodmicro.2005.06.023. [DOI] [PubMed] [Google Scholar]
- 21.Cattoir V., Poirel L., Rotimi V., Soussy C.-J., Nordmann P. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. Journal of Antimicrobial Chemotherapy. 2007;60(2):394–397. doi: 10.1093/jac/dkm204. [DOI] [PubMed] [Google Scholar]
- 22.Grape M., Motakefi A., Pavuluri S., Kahlmeter G. Standard and real-time multiplex PCR methods for detection of trimethoprim resistance dfr genes in large collections of bacteria. Clinical Microbiology and Infection. 2007;13(11):1112–1118. doi: 10.1111/j.1469-0691.2007.01807.x. [DOI] [PubMed] [Google Scholar]
- 23.Lachmayr K. L., Kerkhof L. J., DiRienzo G., Cavanaugh C. M., Ford T. E. Quantifying nonspecific TEM beta-lactamase (blaTEM) genes in a wastewater stream. Applied and Environmental Microbiology. 2008;75 doi: 10.1128/AEM.01254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dallenne C., Da Costa A., Decré D., Favier C., Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important β-lactamases in Enterobacteriaceae. Journal of Antimicrobial Chemotherapy. 2010;65(3):490–495. doi: 10.1093/jac/dkp498. [DOI] [PubMed] [Google Scholar]
- 25.Chen X., Zhang W., Pan W., et al. Prevalence of qnr, aac (6′)-Ib-cr, qepA, and oqxABin Escherichia coli isolates from humans, animals, and the environment. Antimicrobial Agents and Chemotherapy. 2012;56(6):3423–3427. doi: 10.1128/aac.06191-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Candan E. D., Aksöz N. Klebsiella pneumoniae: characteristics of carbapenem resistance and virulence factors. Acta Biochimica Polonica. 2015;62(4):867–874. doi: 10.18388/abp.2015_1148. [DOI] [PubMed] [Google Scholar]
- 27.Wasfi R., Elkhatib W. F., Ashour H. M. Molecular typing and virulence analysis of multidrug resistant Klebsiella pneumoniae clinical isolates recovered from Egyptian hospitals. Scientific Reports. 2016;6(1):p. 38929. doi: 10.1038/srep38929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bi W., Li B., Song J., et al. Antimicrobial susceptibility and mechanisms of fosfomycin resistance in extended-spectrum β-lactamase-producing Escherichia coli strains from urinary tract infections in Wenzhou, China. International Journal of Antimicrobial Agents. 2017;50(1):29–34. doi: 10.1016/j.ijantimicag.2017.02.010. [DOI] [PubMed] [Google Scholar]
- 29.Ghotaslou R., Yeganeh Sefidan F., Akhi M. T., Asgharzadeh M., Mohammadzadeh Asl Y. Dissemination of genes encoding aminoglycoside-modifying enzymes andarm among enterobacteriaceae isolates in Northwest Iran. Microbial Drug Resistance. 2017;23(7):826–832. doi: 10.1089/mdr.2016.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yeganeh Sefidan F., Mohammadzadeh-Asl Y., Ghotaslou R. High-level resistance to aminoglycosides due to 16S rRNA methylation in enterobacteriaceae isolates. Microbial Drug Resistance. 2019;25(9):1261–1265. doi: 10.1089/mdr.2018.0171. [DOI] [PubMed] [Google Scholar]
- 31.Alizadeh N., Ahangarzadeh Rezaee M., Samadi Kafil H., et al. Evaluation of resistance mechanisms in carbapenem-resistant enterobacteriaceae. Infection and Drug Resistance. 2020;13:1377–1385. doi: 10.2147/idr.s244357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vasaikar S., Obi L., Morobe I., Bisi-Johnson M. Molecular characteristics and antibiotic resistance profiles of Klebsiella isolates in mthatha, Eastern Cape Province, South Africa. International Journal of Microbiology. 2017;2017:7. doi: 10.1155/2017/8486742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Paterson D. L., Bonomo R. A. Extended-spectrum β-lactamases: a clinical update. Clinical Microbiology Reviews. 2005;18(4):657–686. doi: 10.1128/cmr.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sheykhsaran E., Bannazadeh Baghi H., Soroush Barhaghi M. H., et al. The rate of resistance to tetracyclines and distribution of tetA, tetB, tetC, tetD, tetE, tetG, tetJ and tetY genes in Enterobacteriaceae isolated from Azerbaijan, Iran during 2017. Physiology and Pharmacology. 2018;22(3):205–212. [Google Scholar]
- 35.Gupta N., Limbago B. M., Patel J. B., Kallen A. J. Carbapenem-resistant enterobacteriaceae: epidemiology and prevention. Clinical Infectious Diseases. 2011;53(1):60–67. doi: 10.1093/cid/cir202. [DOI] [PubMed] [Google Scholar]
- 36.Sadeghi M. R., Ghotaslou R., Akhi M. T., Asgharzadeh M., Hasani A. Molecular characterization of extended-spectrum β-lactamase, plasmid-mediated AmpC cephalosporinase and carbapenemase genes among enterobacteriaceae isolates in five medical centres of East and West Azerbaijan, Iran. Journal of Medical Microbiology. 2016;65(11):1322–1331. doi: 10.1099/jmm.0.000356. [DOI] [PubMed] [Google Scholar]
- 37.Tavakol M., Momtaz H. Determination of antibiotic resistance profile in Klebsiella pneumonia strains isolated from urinary tract infections of patients hospitalized in Peyambaran hospital (Tehran-Iran) Feyz Journal of Kashan University of Medical Sciences. 2017;21(1):74–82. [Google Scholar]
- 38.Tao R., Ying G.-G., Su H.-C., Zhou H.-W., Sidhu J. P. S. Detection of antibiotic resistance and tetracycline resistance genes in enterobacteriaceae isolated from the Pearl rivers in South China. Environmental Pollution. 2010;158(6):2101–2109. doi: 10.1016/j.envpol.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 39.Momtaz H., Rahimi E., Moshkelani S. Molecular detection of antimicrobial resistance genes in E. coli isolated from slaughtered commercial chickens in Iran. Veterinární Medicína. 2012;57(4):193–197. doi: 10.17221/5916-vetmed. [DOI] [Google Scholar]
- 40.Miranda C. D., Kehrenberg C., Ulep C., Schwarz S., Roberts M. C. Diversity of tetracycline resistance genes in bacteria from Chilean salmon farms. Antimicrobial Agents and Chemotherapy. 2003;47(3):883–888. doi: 10.1128/aac.47.3.883-888.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suárez J. E., Mendoza M. C. Plasmid-encoded fosfomycin resistance. Antimicrobial Agents and Chemotherapy. 1991;35(5):791–795. doi: 10.1128/aac.35.5.791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Falagas M. E., Kastoris A. C., Kapaskelis A. M., Karageorgopoulos D. E. Fosfomycin for the treatment of multidrug-resistant, including extended-spectrum β-lactamase producing, enterobacteriaceae infections: a systematic review. The Lancet Infectious Diseases. 2010;10(1):43–50. doi: 10.1016/s1473-3099(09)70325-1. [DOI] [PubMed] [Google Scholar]
- 43.Castañeda-García A., Blázquez J., Rodríguez-Rojas A. Molecular mechanisms and clinical impact of acquired and intrinsic fosfomycin resistance. Antibiotics. 2013;2(2):217–236. doi: 10.3390/antibiotics2020217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lalehzadeh A., Soroush M. H., Sadeghi J., et al. Determination of fosfomycin resistant enterobacteriaceae in isolates and frequency of fos genes in Tabriz Hospitals during 2018. Journal of Biochemical Technology. 2019;10(2):143–148. [Google Scholar]
- 45.Rubin R. H., Swartz M. N. Trimethoprim-sulfamethoxazole. The New England Journal of Medicine. 1980;303(8):426–432. doi: 10.1056/NEJM198008213030804. [DOI] [PubMed] [Google Scholar]
- 46.Huovinen P., Sundström L., Swedberg G., Sköld O. Trimethoprim and sulfonamide resistance. Antimicrobial Agents and Chemotherapy. 1995;39(2):279–289. doi: 10.1128/aac.39.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morgan D. J., Okeke I. N., Laxminarayan R., Perencevich E. N., Weisenberg S. Non-prescription antimicrobial use worldwide: a systematic review. The Lancet Infectious Diseases. 2011;11(9):692–701. doi: 10.1016/s1473-3099(11)70054-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Okeke I. N., Lamikanra A., Edelman R. Socioeconomic and behavioral factors leading to acquired bacterial resistance to antibiotics in developing countries. Emerging Infectious Diseases. 1999;5(1):18–27. doi: 10.3201/eid0501.990103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gündoğdu A., Long Y. B., Vollmerhausen T. L., Katouli M. Antimicrobial resistance and distribution of sul genes and integron-associated intI genes among uropathogenic Escherichia coli in Queensland, Australia. Journal of Medical Microbiology. 2011;60(Pt 11):1633–1642. doi: 10.1099/jmm.0.034140-0. [DOI] [PubMed] [Google Scholar]
- 50.Manyahi J., Tellevik M. G., Ndugulile F., Moyo S. J., Langeland N., Blomberg B. Molecular characterization of cotrimoxazole resistance genes and their associated integrons in clinical isolates of gram-negative bacteria from Tanzania. Microbial Drug Resistance. 2017;23(1):37–43. doi: 10.1089/mdr.2016.0074. [DOI] [PubMed] [Google Scholar]
- 51.Arabi H., Pakzad I., Nasrollahi A., et al. Sulfonamide resistance genes (sul) M in extended spectrum beta lactamase (ESBL) and non-ESBL producing Escherichia coli isolated from Iranian hospitals. Jundishapur Journal of Microbiology. 2015;8(7):p. e19961. doi: 10.5812/jjm.19961v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Infante B., Grape M., Larsson M., et al. Acquired sulphonamide resistance genes in faecal Escherichia coli from healthy children in Bolivia and Peru. International Journal of Antimicrobial Agents. 2005;25(4):308–312. doi: 10.1016/j.ijantimicag.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 53.Toleman M. A., Bennett P. M., Bennett D. M. C., Jones R. N., Walsh T. R. Global emergence of trimethoprim/sulfamethoxazole resistance in stenotrophomonas maltophilia mediated by acquisition of sul genes. Emerging Infectious Diseases. 2007;13(4):559–565. doi: 10.3201/eid1304.061378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Trobos M., Christensen H., Sunde M., et al. Characterization of sulphonamide-resistant Escherichia coli using comparison of sul2 gene sequences and multilocus sequence typing. Microbiology. 2009;155(3):831–836. doi: 10.1099/mic.0.024190-0. [DOI] [PubMed] [Google Scholar]
- 55.Cattoir V., Nordmann P. Plasmid-mediated quinolone resistance in gram-negative bacterial species: an update. Current Medicinal Chemistry. 2009;16(8):1028–1046. doi: 10.2174/092986709787581879. [DOI] [PubMed] [Google Scholar]
- 56.Correia S., Poeta P., Hébraud M., Capelo J. L., Igrejas G. Mechanisms of quinolone action and resistance: where do we stand? Journal of Medical Microbiology. 2017;66(5):551–559. doi: 10.1099/jmm.0.000475. [DOI] [PubMed] [Google Scholar]
- 57.Nordmann P., Poirel L. Emergence of plasmid-mediated resistance to quinolones in Enterobacteriaceae. Journal of Antimicrobial Chemotherapy. 2005;56(3):463–469. doi: 10.1093/jac/dki245. [DOI] [PubMed] [Google Scholar]
- 58.Dehnamaki M., Ghane M., Babaeekhou L. Detection of OqxAB and QepA efflux pumps and their association with antibiotic resistance in Klebsiella pneumoniae isolated from urinary tract infection. International Journal of Infection. 2020;7(4):p. e107397. doi: 10.5812/iji.107397. [DOI] [Google Scholar]
- 59.Yang H. Y., Nam Y. S., Lee H. J. Prevalence of plasmid-mediated quinolone resistance genes among ciprofloxacin-nonsusceptible escherichia coliand Klebsiella pneumoniae isolated from blood cultures in Korea. Canadian Journal of Infectious Diseases and Medical Microbiology. 2014;25(3):163–169. doi: 10.1155/2014/329541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Araújo B. F., Campos P. A. d., Royer S., et al. High frequency of the combined presence of QRDR mutations and PMQR determinants in multidrug-resistant Klebsiella pneumoniae and Escherichia coli isolates from nosocomial and community-acquired infections. Journal of Medical Microbiology. 2017;66(8):1144–1150. doi: 10.1099/jmm.0.000551. [DOI] [PubMed] [Google Scholar]
- 61.El-Badawy M. F., Tawakol W. M., El-Far S. W., et al. Molecular identification of aminoglycoside-modifying enzymes and plasmid-mediated quinolone resistance genes among Klebsiella pneumoniae clinical isolates recovered from Egyptian patients. International Journal of Microbiology. 2017;2017 doi: 10.1155/2017/8050432.8050432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vali L., Dashti A. A., Jadaon M. M., El-Shazly S. The emergence of plasmid mediated quinolone resistance qnrA2 in extended spectrum β-lactamase producing Klebsiella pneumoniae in the Middle East. DARU Journal of Pharmaceutical Sciences. 2015;23(1):p. 34. doi: 10.1186/s40199-015-0116-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meteab Alshammari M. M., Alkhattat H. Detection of plasmid-mediated quinolone resistance genes in clinical and environmental hospital isolates of Klebsiella pneumoniae in Al-Najaf city pneumoniae Klebsiella. Kufa Journal for Nursing Sciences. 2015;5 [Google Scholar]
- 64.Magnet S., Blanchard J. S. Molecular insights into aminoglycoside action and resistance. Chemical Reviews. 2005;105(2):477–498. doi: 10.1021/cr0301088. [DOI] [PubMed] [Google Scholar]
- 65.Ramirez M. S., Tolmasky M. E. Aminoglycoside modifying enzymes. Drug Resistance Updates. 2010;13(6):151–171. doi: 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mokhtari H., Eslami G., Zandi H., Dehghan-Banadkouki A., Vakili M. Evaluating the frequency of aac (6’)-IIa, ant (2″)-I, intl1, and intl2 genes in aminoglycosides resistant Klebsiella pneumoniae isolates obtained from hospitalized patients in Yazd, Iran. Avicenna Journal of Medical Biotechnology. 2018;10(2):115–119. [PMC free article] [PubMed] [Google Scholar]
- 67.Abo-State M. A. M., Saleh Y. E.-S., Ghareeb H. M. Prevalence and sequence of aminoglycosides modifying enzymes genes among E.coli and Klebsiella species isolated from Egyptian hospitals. Journal of Radiation Research and Applied Sciences. 2018;11(4):408–415. doi: 10.1016/j.jrras.2018.08.005. [DOI] [Google Scholar]
- 68.Pakzad I., Samadi N., Imanieini M., et al. Assessment of 16srRNA methylase genes among non-ESBL and ESBL-producing Klebsiella pneumoniae isolates. Archives of Clinical Infectious Diseases. 2019;14(6)e84372 [Google Scholar]
- 69.Miró E., Grünbaum F., Gómez L., et al. Characterization of aminoglycoside-modifying enzymes in enterobacteriaceae clinical strains and characterization of the plasmids implicated in their diffusion. Microbial Drug Resistance. 2013;19(2):94–99. doi: 10.1089/mdr.2012.0125. [DOI] [PubMed] [Google Scholar]
- 70.Shaw K. J., Rather P. N., Hare R. S., Miller G. H. Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiological Reviews. 1993;57(1):138–163. doi: 10.1128/mr.57.1.138-163.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.


