Abstract
In the development of novel and more effective anti-cancer approaches, combined treatments appear to be of great interest, based on the possibility of obtaining relevant biological or therapeutic effects using lower concentrations of single drugs. Combination therapy may prove to be of utmost significance in the management of glioblastoma (GBM), a lethal malignancy that accounts for 42% of cancer cases of the central nervous system, with a median survival rate of 15 months. As regards novel therapeutic approaches, the authors have recently demonstrated that peptide nucleic acids (PNAs) that target microRNA (miRNA/miR)-221 are very active in inducing the apoptosis of glioma cells. Furthermore, in a recent study, the authors described two novel series of tubulin polymerization inhibitors based on the 4,5,6,7-tetrahydrothieno[2,3-c]pyridine and 4,5,6,7-tetrahydrobenzo[b]thiophene scaffold, which exerted a potent anti-proliferative effect on a variety of tumor cell lines. The present study aimed to verify the activity on glioblastoma cancer cell lines of one of the most active compounds tested, corresponding to 2-(3′, 4′, 5′-trimethoxyanilino)-3-cyano/alkoxycarbonyl-6-substituted-4 5,6,7-tetrahydrothiene[2,3-c] pyridine (compound 3b), used in combination with an anti-miR-221-3p PNA, already demonstrated to be able to induce high levels of apoptosis. To the best of our knowledge, the results obtained herein demonstrate for the first time a 'combination therapy' performed by the combined use of a PNA targeting miR-221 and the tetrahydrothiene[2,3-c]pyridine derivative 3b, supporting the concept that the combined treatment of GBM cells with a PNA against a specific upregulated oncomiRNA (in the present study a PNA targeting miR-221-3p was used) and anti-tubulin agents (in the present study derivative 3b was used) is an encouraging strategy which may be used to enhance the efficacy of anticancer therapies and at the same time, to reduce side-effects.
Keywords: microRNA; anti-miR; peptide nucleic acid; miR-221; glioma; cell cycle; apoptosis; tubulin; tetrahydrothieno[2,3-c] pyridine; combination therapy
Introduction
In the development of anticancer approaches, combined treatments appear to be of great interest (1-5). The idea of combined treatments is based on the possibility to obtain the same biological or therapeutic effect with two or more drugs, using lower concentrations of single drugs. In this case, the side-effects of the single drug are expected to be limited (1). In addition, the combined therapy of cancer may have an impact on acquired resistance (1,2,5). Furthermore, patient-to-patient variability and independent drug action are sufficient to explain the superior efficacy of drug combinations in the absence of drug synergy or additivity (6). The hypothesis is that in a combination of drugs exhibiting specific mechanisms of action, each patient benefits solely from the drug to which the tumor is most sensitive, with no added benefit from other drugs. In any case, the impact and the overall interest in anticancer combined therapy are very high (2).
Combined therapy may be of utmost significance in the management of glioblastoma (GBM), a lethal malignant tumor accounting for 42% of the tumors of the central nervous system, with the median survival being 15 months (7-9). It should be emphasized that there is currently no effective pharmacological treatment available, and the first-line drug used, temozolomide (TMZ), is on average able to prolong the life expectancy of treated patients by only a few months (10). Additionally, a number of forms of GBM are or become resistant to TMZ over time (10-12).
Therefore, there is an urgent need to identify and develop new drugs and novel therapeutic approaches (such as combined therapy) in order to develop more effective anti-glioma therapies than those currently available, particularly for TMZ-resistant cells.
As regards the combined therapy of glioma cells, the authors have recently reported that antitumor drugs can be combined with molecules targeting microRNAs (miRNAs or miRs) (5). miRNAs are short non-coding RNA sequences which, owing to their mechanisms of action, function as gene regulators by repressing the translation or causing the cleavage of the RNA transcripts they target (13-15). There is currently ample evidence to indicate that the altered expression of miRNAs may be involved in the pathogenesis of cancer (16,17). In particular those miRNAs that are upregulated in cancer and that cause the downregulation of target tumor suppressor RNAs are defined as 'oncomiRNAs' and 'metastamiRNAs' (18). Conversely, tumor suppressor miRNAs may be downregulated in cancer, leading to the overexpression of target oncogenes (18). In summary, the rationale for this approach is the following: i) Some miRNAs are deeply involved in the regulation of cell apoptotic mechanisms and more generally in carcinogenesis, functioning as 'oncomiRNAs' and 'metastamRNAs' (13-18); and ii) synergistic effects between anticancer molecules and antagomiRNA molecules targeting specific oncomiRNAs have been highlighted by different research groups (4,19-21). There is strong evidence sustaining the concept that oncomiRNAs are potent anti-apoptotic agents (22-25). As regards possible candidates for anti-miRNA treatment, peptide nucleic acids (PNAs) (26,27) have been proposed as very useful bioactive molecules (28-34). PNAs are DNA analogues in which the sugar-phosphate backbone has been replaced by N-(2-aminoethyl)-glycine units (26,27). These intriguing molecules were first described by Nielsen et al (26) and, despite the general structure of the nucleic acid molecule becomes more peptide-like, they can hybridize with complementary DNA or RNA sequences with exceptional efficiency and specificity (27).
The present study employed a PNA targeting miR-221-3p, functionalized with an octaarginine peptide (R8) for maximizing cellular uptake, as previously reported (32). The reason for selecting miR-221-3p as PNA-based targeting was based on the following observations: i) miR-221-3p is overexpressed in glioma patients (35-38); and ii) miR-221-3p targeting decreases cell migration and metastasis when used in in vivo glioblastoma model systems (39). The oncogenic role of miR-221-3p has also been confirmed in other tumor types, including colon, liver, pancreatic, non-small cell lung cancer (40-43).
In a recent study, the authors tested and reported two novel series of active anti-tubulin agents based on the 4,5,6,7-tetrahydrothieno[2,3-c]pyridine and 4,5,6,7-tetrahydrobenzo[b]thiophene molecular skeleton (44). These compounds were found to interfere with the microtubule-tubulin equilibrium in cancer cells and were demonstrated to retain anti-proliferative activity on a panel of cancer cell lines. Compounds targeting tubulin are of great interest for the treatment of cancer cells (45-48).
The aim of the present study was to verify the activity on the glioma U251 and T98G tumor cell lines (49,50) of the 2-(3′,4′,5′-trimethoxyanilino)-3-cyano/alkoxycarbonyl-6-substit uted-4,5,6,7-tetrahydrothieno[2,3-c]pyridine 3b used in combination with an anti-miR-221-3p PNA, already demonstrated to be able to induce high levels of apoptosis (32).
Materials and methods
Chemicals and reagents
The anti-tubulin compound 3b and R8-PNA-a221 were synthesized by the research groups of the author RR (University of Ferrara) and Professor Roberto Corradini (University of Parma), respectively; the procedure for the synthesis of both molecules has been previously reported (32,44). For all cell cultures, RPMI-1640 medium supplemented with 10% FBS and 100 mg/ml streptomycin and 100 IU/ml penicillin was employed. RPMI-1640 medium (cat. no. BE12-702F) and PBS (cat. no. BE17-516F) were purchased from Lonza Biosciences, trypsin-EDTA solution (cat. no. 59428C) and 50,000 IU/ml streptomycin and 50 mg/ml penicillin (cat. no. 11074440001) were from Sigma-Aldrich; Merck KGaA and FBS (cat. no. S1400) was obtained from Biowest. For flow cytometric assays, the Muse® Annexin V & Dead Cell kit (cat. no. MCH100105), Muse® Caspase-3/7 kit (cat. no. MCH100108) and the Muse® Cell Cycle kit (cat. no. MCH100106) were purchased from Luminex Corporation. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) powder (cat. no. M5655) for MTT assay, crystal violet staining solution (cat. no. V5265) for clonogenic assay, DMSO (cat. no. D2650) used to resuspend compound 3b and the TRI Reagent (cat. no T9424) used for RNA extraction were purchased from Sigma-Aldrich; Merck KGaA, while methanol (cat. no. 309001) was supplied by CARLO ERBA Reagents.
Cell lines, cell growth conditions and anti-proliferative assays
The human glioma U251 (51) and T98G (49) cells were employed in the present study; both cell lines were purchased from Sigma-Aldrich; Merck KGaA. For anti-proliferative assay using the U251 cell line, 8×104 cells were seeded in a 12-well plate, and after 4 h the cells were treated with a serial dilution of the test compounds (compound 3b was used at the 1, 2, 4, 6 and 8 µM concentrations and R8-PNA-a221 at the 1, 2, 4 and 8 µM concentrations). The cells were incubated for 72 h at 37°C in a humidified 5% CO2 atmosphere. Following 72 h of incubation, cells were detached from the plate by trypsinization and counted using a BECKMAN COULTER® Z2 cell counter (Beckman Coulter, Inc.). The IC50 value (50% inhibitory concentration) is defined as the concentration of compound that inhibits cell proliferation by 50% (44). The IC50 values presented (± standard deviation) are the average values derived from three independent experiments.
Morphological analysis
Following each treatment, the cells were observed and representative images were acquired using a Nikon Eclipse 80i microscope (Nikon Corporation), in order to observe whether any morphological changes occurred in the cells following treatment.
Cytotoxicity assay
For determining the cytotoxic effects of the test compounds, MTT assay was performed using the U251 cell line (50). Cells were seeded in a 96-multi-well plate at a density of 8×103 cells/well and after 4 h, treated with the lower concentrations of compound 3b (4 µM) and R8-PNA-a221 (2 µM) individually and in combination. Cells were incubated at 37°C for a further 72 h and at the end of the incubation period, MTT was added to each well at a final concentration of 0.5 mg/ml. Following 3 h of incubation at 37°C, the medium was discarded, and dimethyl sulfoxide (DMSO) was added; the plate was stirred for 30 min to fully dissolve the formazan crystals formed at the bottom of the wells. The absorbance was measured at 570 nm using the SUNRISE microplate reader (Tecan Group, Ltd.).
Clonogenic assay
Cells were seeded into 6-well plates (400 cells per well) in RPMI-1640 medium with 10% FBS and incubated at 37°C for 24 h prior to treatment. The plates were incubated at 37°C for a further 10 days undisturbed in the incubator. Each well was then washed twice with PBS and covered with a methanol containing fixation/staining solution that allows the simultaneous coloration and fixation of the cells (crystal violet aqueous solution 0.5%/methanol, 1:1 ratio) for 15 min at room temperature and washed four times with tap water; the plate was air-dried for 1 day prior to obtaining images. Images were acquired using a Nikon SMZ1000 stereo zoom microscope (Nikon Corporation). Stained colonies consisting of >50 cells were manually counted, and the number was recorded (52).
RNA extraction
Cells were detached by trypsinization (cat. no. 59428C; Sigma-Aldrich; Merck KGaA), collected by centrifugation at 1,200 rpm (8 min at room temperature), and lysed with Tri-Reagent (Sigma-Aldrich; Merck KGaA) according to manufacturer's instructions. The isolated RNA was washed once with cold 75% ethanol and stored at −80°C until use. The obtained RNA was dried and dissolved in nuclease-free water prior to use (32).
Quantitative analyses of miRNAs
The miRNA levels were assayed using the TaqMan MicroRNA Reverse Transcription kit (cat. no. 43-665-96, Applied Biosystems; Thermo Fisher Scientific, Inc.) with RT-qPCR and miRNA-specific primers and probes (listed in Table I) obtained from Thermo Fisher Scientific, Inc.. All samples were run in duplicate using TaqMan Universal PCR Master Mix, no AmpErase UNG 2X (cat. no. 4324018; Thermo Fisher Scientific, Inc.) and the CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories, Inc.).
Table I.
List of assays employed for miRNA detection.
| miRNA name | Assay ID (Applied Biosystems; Thermo Fisher Scientific, Inc.) |
|---|---|
| hsa-miR-221-3p | 000524 |
| hsa-snRNA U6 | 001973 |
| hsa-let-7c-5p | 000379 |
For PCR reactions, the following protocol was employed: 95°C for 10 min, 95°C for 15 sec, followed by a step at 60°C for 1 min (last two steps repeated for 50 cycles). Data were collected and analyzed using Bio-Rad CFX Manager Software (Bio-Rad Laboratories, Inc.). Relative gene expression was calculated using the 2−ΔΔCq method and data normalization was performed using snRNA U6 and hsa-let-7c as reference (53).
Effects on the cell cycle
The cells were treated with the lower and higher concentrations of compound 3b (4 and 6 µM) and R8-PNA-a221 (2 and 4 µM) individually and in combination at the lower concentration of both. Following 72 h of incubation at 37°C, the cells were detached by trypsinization, washed once in PBS and fixed with 70% EtOH for 24 h. For analysis 5×105 cells were washed in PBS and resuspended in 200 µl of Muse® Cell Cycle Reagent and incubated for 30 min at room temperature protected from light. Finally, the cell suspension was transferred into a new 1.5 ml tube without cap and the samples were analyzed by flow cytometry using Guava® Muse® Cell Analyzer (Luminex Corp.) (44).
Cell apoptosis assay
Apoptosis assays were performed with the Guava® Muse® Cell Analyzer instrument, and its relative kits according to the instructions supplied by the manufacturer. Muse® Annexin V & Dead Cell Kit utilizes Annexin V to detect Phosphatidyl Serine (PS), a common apoptotic marker that is exposed out of the external membrane of apoptotic cells, and 7-ADD (7-aminoactinomycin D) a DNA intercalating molecule used as an indicator of cell membrane integrity (it can bind DNA only in cells undergoing late apoptosis/death stage, when the membrane integrity is lost). Following 72 h of treatment with compound 3b (4 and 6 µM) and R8-PNA-a221 (2 and 4 µM) administered individually and the combination of the lower concentration of both, the cells were washed with sterile PBS, trypsinized and resuspended in RPMI medium supplemented with 10% FBS. Finally, 50 µl of cell suspension were incubated with 100 µl Muse® Annexin V & Dead Cell reagent at room temperature and protected from light for 20 min. Samples were then analyzed using the Guava® Muse® Cell Analyzer (Luminex Corp.) and data acquired utilizing the Annexin V and Dead Cell Software Module (Luminex Corp.) (54).
Caspase-3/7 activity assay
For the analysis of caspase-3/7 activity following treatments, the Muse® Caspase-3/7 kit was employed, which is based on a DNA binding dye linked to a DEVD peptide substrate. When pro-apoptotic caspase-3/7 are activated, they cleave the dye and the binding of the dye to DNA results in high fluorescence signal. This kit also contains a fluorescent DNA intercalator (7-ADD) as indicator of cell membrane integrity. Following the same treatments indicated in the Cell apoptosis assay, trypsinization was performed and 50 µl of cell suspension were incubated with 5 µl of caspase-3/7 reagent for 30 min (under strict protection from light). Following 25 min of incubation at 37°C, 150 µl of 7-AAD working solution were added to each tube and incubated for 5 min at room temperature before reading the samples. The samples were then analyzed using Guava® Muse® Cell Analyzer (Luminex Corp.) and data acquired utilizing the Caspase-3/7 Software Module (Luminex Corp.) (54).
Statistical analysis
Results are expressed as the mean ± standard error of the mean (SEM). Comparisons between groups were made by using ordinary one-way ANOVA followed by a post hoc multiple comparison tests. Dunnett's test was used to compare groups against a single control and Tukey's test was used to make comparisons against more than one group. P<0.05 was considered to indicate a statistically significant difference.
Results
Structure of molecules employed in the present study
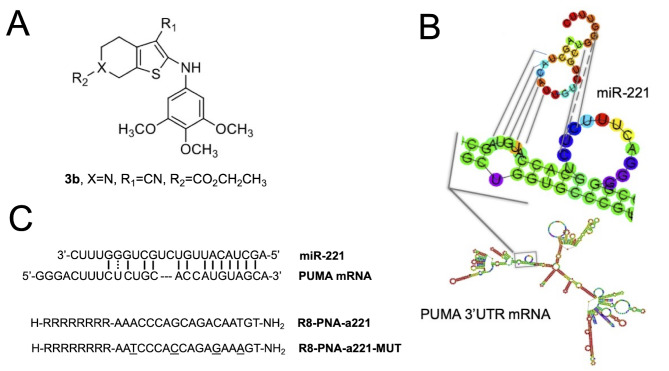
The chemical structures of the compounds used in the present study are presented in Fig. 1 [tetrahydrothieno[2,3-c]pyridine 3b (Fig. 1A) and PNA-a221 targeting miR-221 (Fig. 1C); these have been shown to exert a potent anti-apoptotic effect (34). Among the possible apoptotic-associated mRNAs, ATG10, CDKN1B/p27, BMF, APAF-1, PTEN, p27(kip1), p57(kip2) and PUMA have been validated as miR-221 targets (55-62). The example of PUMA mRNA is presented in Fig. 1B, exhibiting a functional miR-221 binding site in its 3′UTR sequence (61,62).
Figure 1.
Chemical structure of (A) compound 3b and (C) PNA anti-miR-221, and (B) graphic display of the miR-221 binding site on 3′ UTR of PUMA mRNA. The octaarginine (R8) was employed to maximize the uptake of the PNA, and the mutant PNA-a221 (R8-PNA-a221-MUT) was synthesized in order to perform experiments focusing on the specificity of the PNA-a221. PNA, peptide nucleic acid.
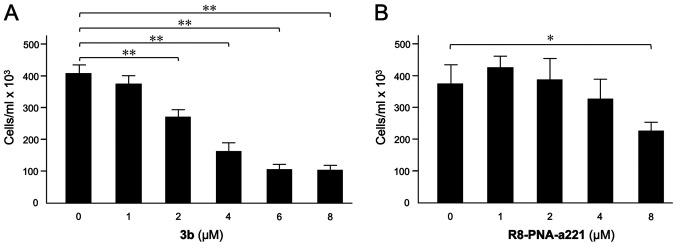
Effects of compound 3b and R8-PNA-a221 on U251 cell growth
The effects of compound 3b (Fig. 2A) and R8-PNA-a221 (Fig. 2B) on the proliferation of U251 cells were examined. Following 72 h of cell culture in the indicated experimental conditions, the cell number/ml was determined. The data indicated that the inhibitory effect of compound 3b on U251 cell growth reached maximum values when 4-8 µM concentrations of compound 3b were employed. The anti-proliferative effects of R8-PNA-a221 were lower and detectable in the same range of concentrations, reaching 50% inhibition of cell growth only at the 8 µM concentration. In order to obtain preliminary information on the combinatory effect of the two drugs, two different concentrations for 3b (4 and 6 µM) and R8-PNA-a221 (2 and 4 µM) were selected.
Figure 2.
(A) Effects of (A) tetrahydrothieno[2,3-c]pyridine 3b and (B) R8-PNA-a221 on the growth of human glioma U251 cells. A summary of three independent experiments is presented. The data represent the mean ± SD. *P<0.05, **P<0.01. PNA, peptide nucleic acid.
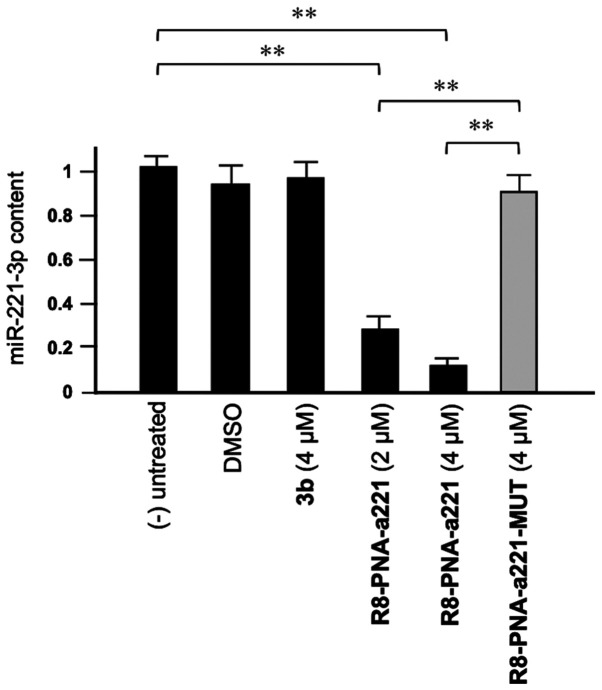
Compound 3b does not affect miR-221-3p expression
As shown in Fig. 3, tetrahydrothieno[2,3-c]pyridines 3b did not affect the miR-221-3p content. This experiment was conducted by exposing, for 72 h, human glioma U251 cells to the indicated concentrations of compound 3b and R8-PNA-a221 and R8-PNA-a221-MUT; the mutated PNA contains 4 mismatches in the sequence (Fig. 1) which prevents it from hybridizing with the target mir-221-3p sequence, demonstrating the enhanced sensitivity of R8-PNA-a221 (32). Following this period of cell culture, RNA was isolated and miR-221-3p sequences were quantified by RT-qPCR. As was expected, the R8-PNA-a221 (but not the mutated R8-PNA-a221-MUT) inhibited the production of miR-221-3p. On the contrary, no inhibitory effects were observed with compound 3b treatment at 4 µM.
Figure 3.
Effects of the tetrahydrothieno[2,3-c]pyridine 3b, R8-PNA-a221 and mutated R8-PNA-a221-MUT on the miR-221 content in human glioma U251 cells. Cells were cultured as indicated for 72 h, RNA was extracted and miR-221 quantified. A summary of three independent experiments is presented. The data represent the mean ± SD. **P<0.01. PNA, peptide nucleic acid.
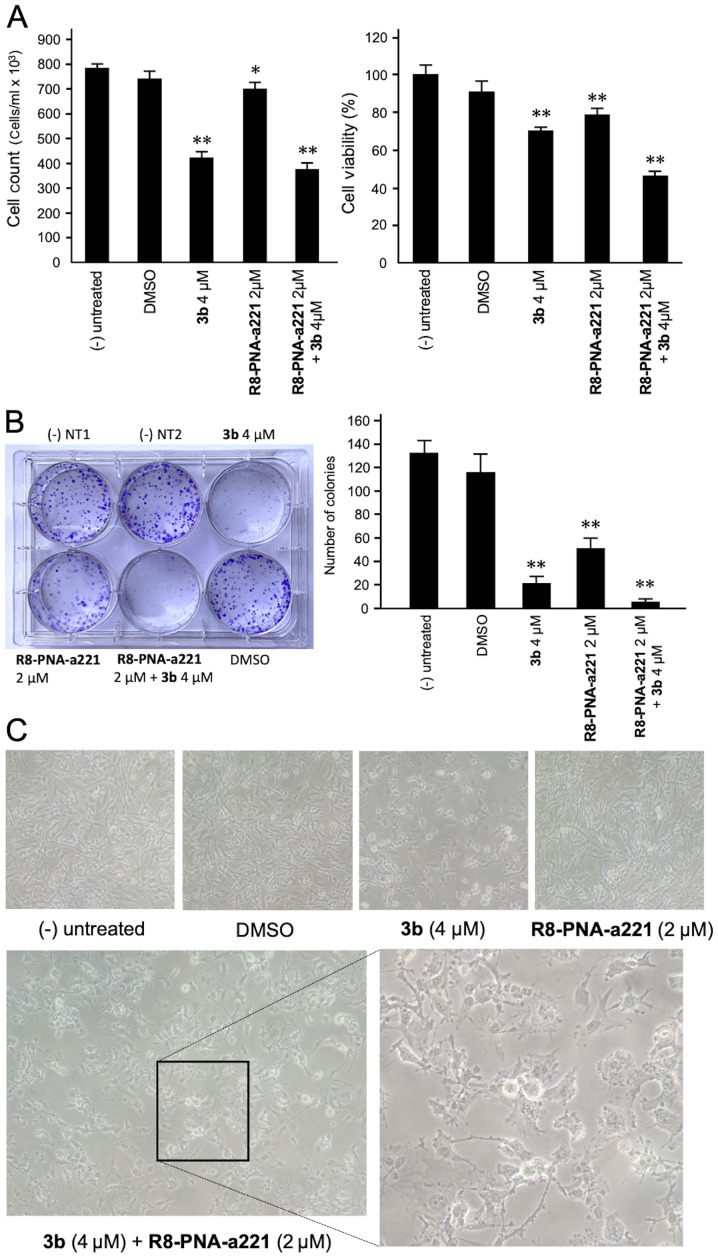
Co-treatment of U251 cells with 3b and R8-PNA-a221: Effects on cell growth, viability, and colony formation
The anti-proliferative and cytotoxic effects of single and combined treatments are shown in Fig. 4. The anti-proliferative effects were assayed by employing a cell counter (Fig. 4A, left panel), while cytotoxicity was evaluated by MTT assay (Fig. 4A, right panel). A representative image of the clonogenic assay is presented in Fig. 4B. The results obtained clearly indicated that co-treatment with compound 3b and R8-PNA-a221 was associated with the lowest number of colonies, and the highest anti-proliferative and cytotoxic effects.
Figure 4.
Effects of tetrahydrothieno[2,3-c]pyridine 3b and PNA-a221 on cell growth and viability. U251 cells were treated with the indicated concentrations of compound 3b and R8-PNA-a221 and (A) cell growth and cytotoxicity were determined. The anti-proliferative effects were assayed by employing a cell counter; cytotoxicity was evaluated by MTT assay. (B) Representative image of the clonogenic assay performed on U251 cells, and a summary of three independent experiments. Cells were fixed and stained with crystal violet after 10 days of treatment. (C) Morphological alteration of the treated U251 cells. Cells were treated as indicated and the morphological phenotype of the cells was analyzed by microphotographic analysis [magnification, ×100; combined treatment magnification, ×200 (left panel) and ×400 (right panel)]. PNA, peptide nucleic acid. *P<0.05, **P<0.01, vs. untreated cells.
A morphological analysis was also performed (Fig. 4C) demonstrating that, in agreement with the enhanced effects on cell growth and vitality, the alterations of the morphology of the U251 glioma cells occurred following co-treatment with compound 3b and R8-PNA-a221. In particular the combined use of the two compounds led to major morphology alterations, including the loss of the original 'astrocytic' shape, and typical hallmarks of apoptosis, such as membrane blebbing.
Cell cycle effects
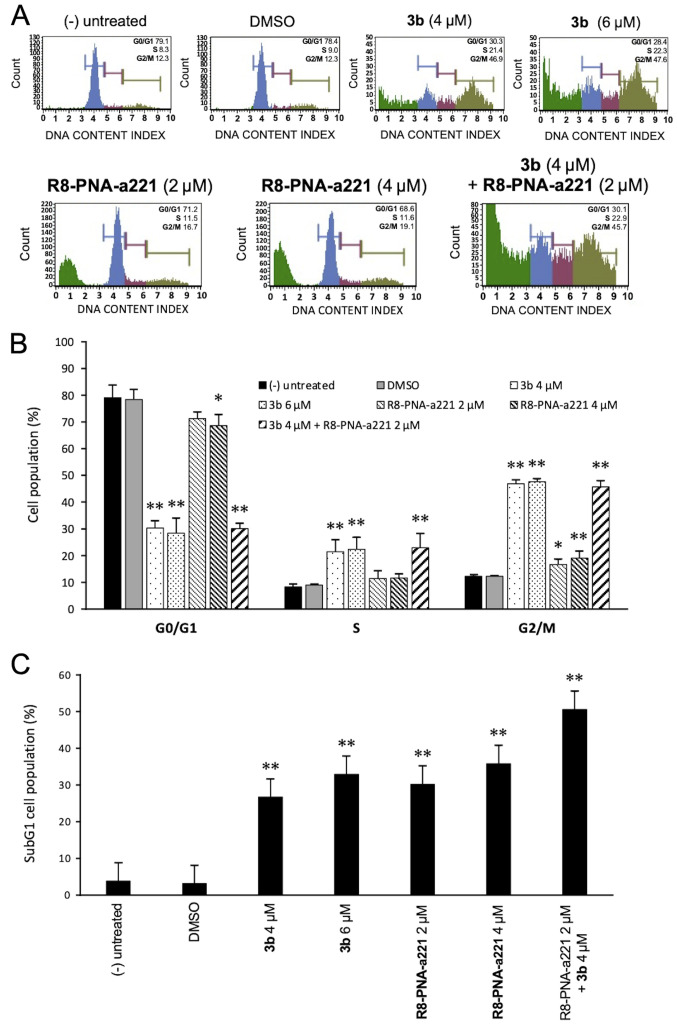
Since the mechanisms of action of anti-tubulin compounds, such as compound 3b involved causing cell cycle alterations, the effects of compound 3b and PNA-a221 on the cell cycle were investigated by flow cytometric analysis in the present study. The cells were treated and incubated for 72 h as indicated in the experiment illustrated in Fig. 5A. Compound 3b induced a marked increase in the number of cells in the G2/M phase of the cell cycle, associated with a decrease in the number of cells in the G0/G1 phase. As shown in Fig. 5A, treatment of the U251 cells with compound 3b at 4 and 6 µM increased the percentage of cells in the G2/M phase from 12.3% (control group) to 46.9 and 47.6%, respectively. This finding is in agreement with data obtained using human K562 leukemia cells previously reported (44) and confirms that compound 3b affects cell growth through cell cycle blockade. On the contrary, the R8-PNA-a221 exerted only minor effects on cell growth, inducing a very low increase in the percentage of cells in the G2/M phase from 12.3% (control group) to 16.7-19.1%. The minimal effect of 8-PNA-a221 on the cell cycle was confirmed by the fact that combined treatment with 4 µM compound 3b and 2 µM R8-PNA-a221 induced a cell cycle distribution very similar to that induced by treatment with compound 3b alone. The quantitative data of the effects of the treatments on the cell cycle are presented in Fig. 5B. When the results of FACS analyses shown in Fig. 5A were considered, it was evident that an increase in the sub-G1 cell population occurred when the cells were treated with compound 3b, R8-PNA-a221 or the combination of compound 3b and R8-PNA-a221. As shown in Fig. 5C, a marked increase in the number of cells in the sub-G1 phase was observed when the U251 cells were treated with compound 3b. As in the case of alteration of cell growth and cytotoxicity, the highest proportion of sub-G1 cells was observed when the cells were treated with compound 3b plus R8-PNA-a221. Since the appearance of a sub-G1 population is a hallmark of the activation of apoptosis, an analysis was performed using two apoptosis-associated assays, Annexin V and caspase-3/7 assays.
Figure 5.
Effects of the tetrahydrothieno[2,3-c]pyridine 3b and PNA-a221 on the cell cycle and on the proportion of the sub-G1 population. U251 cells were treated with the indicated concentrations of compound 3b and R8-PNA-a221 and the cell cycle distribution was determined. (A) Representative cell cycle analyses. (B) Proportion of U251 cells in the G1, S and G2/M phase of the cell cycle (sub-G1 cells excluded from the calculations). (C) Increase in the sub-G1 cell population following treatment of the U251 cells with compound 3b, PNA-a221 or both 3b and PNA-a221 used in combination. PNA, peptide nucleic acid. *P<0.05, **P<0.01, vs. untreated cells.
Apoptosis study of U251 glioma cells
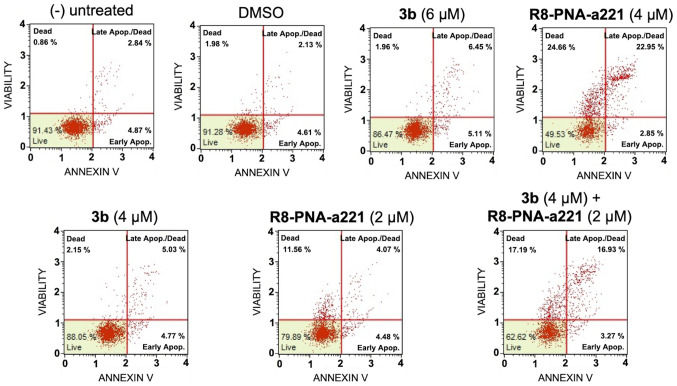
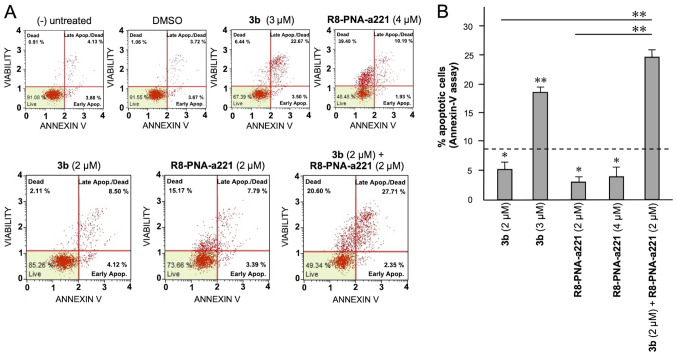
In order to examine the induction potential of compound 3b and R8-PNA-a221 to induce cell apoptosis and death when used individually and to verify a possible synergistic effect when used in combination, two different apoptosis detection kits were used. Representative plots demonstrating the effects of various concentrations of compound 3b and R8-PNA-a221 administered alone or in combination are shown in Fig. 6. Considering the increase in total apoptotic cells (early and late apoptotic cells of the treated minus the early and late apoptotic cells of the untreated sample or DMSO-treated cells), both agents induced an increase in the number of Annexin V-positive cells in comparison with the controls after 3 days, and this increase occurred in a concentration-dependent manner, reaching a maximum of a 4.82% increase with respect to the control DMSO-treated cells with 6 µM compound 3b and 18.09% with 4 µM R8-PNA-a221 (compared to the untreated cells). Combined treatments with sub-optimal concentrations of compound 3b and R8-PNA-a221 (4 and 2 µM, respectively) led to a sharp induction of apoptosis (13.46%), a proportion which was much higher than the sum of the effects of the singularly added agents (3.06+0.84=4.70%). It should be noted that the increase in the proportion of apoptotic cells was particularly evident in the 'late apoptotic cell' fraction. The increase of the proportion of the 'dead cell' fraction was fully in agreement with the cytotoxicity data depicted in Fig. 4.
Figure 6.
Effects of the tetrahydrothieno[2,3-c]pyridine 3b and PNA-a221 on the apoptosis of U251 cells, part I. U251 cells were treated with the indicated concentrations of compound 3b and R8-PNA-a221 and apoptosis was analyzed by Annexin V assay. Representative apoptosis analyses are reported. A summary of three independent experiments is presented in Fig. 8A. PNA, peptide nucleic acid.
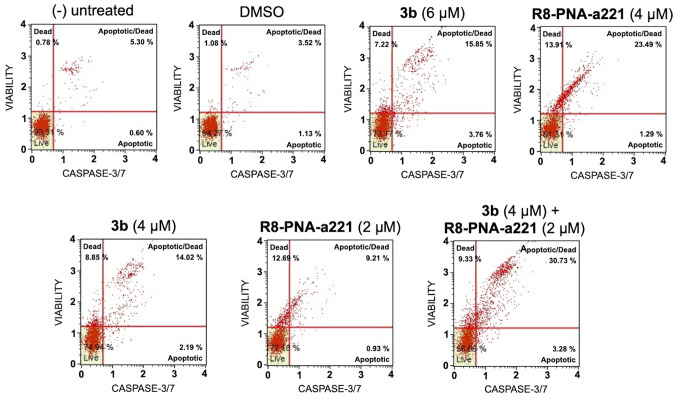
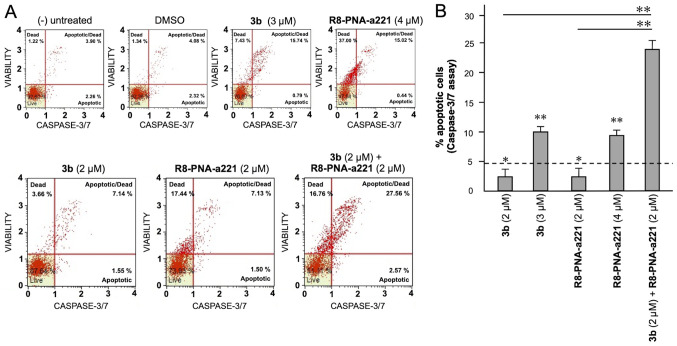
The same conclusion derived from the experiments performed using the Annexin V assay can be gathered on the basis of the caspase-3/7 assay shown in Fig. 7. In this case, always considering the increase in total apoptotic cells (early and late apoptotic cells of the treated minus the early and late apoptotic cells of the untreated sample or DMSO-treated cells) obtained following combined treatments with sub-optimal concentrations of compound 3b and R8-PNA-a221 (4 and 2 µM, respectively) was 29.36%, a proportion which was much higher than the sum of the effects of singularly added agents (11.56+4.24%=15.80).
Figure 7.
Effects of the tetrahydrothieno[2,3-c]pyridine 3b and PNA-a221 on the apoptosis of U251 cells, part II. U251 cells were treated with the indicated concentrations of compound 3b and R8-PNA-a221 and apoptosis was analyzed by caspase-3/7 assay. Representative apoptosis analyses are reported. A summary of three independent experiments is shown in Fig. 8B. PNA, peptide nucleic acid.
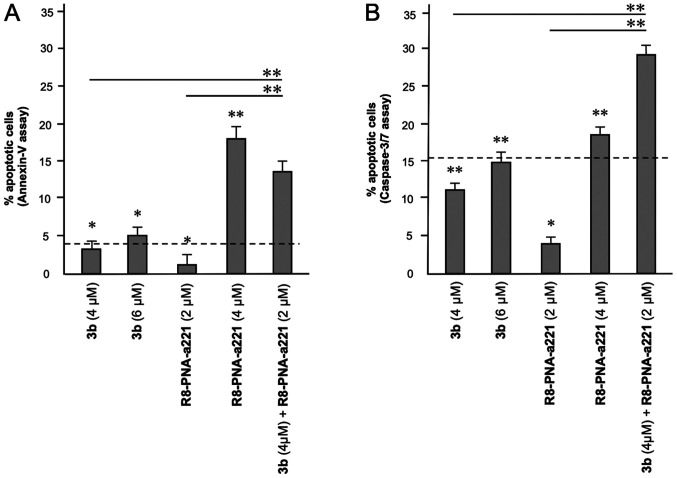
A summary of three independent experiments analyzing the effects of the tetrahydrothieno[2,3-c]pyridine compound 3b and PNA-a221 on cell apoptosis is presented in Fig. 8. In the histogram, only apoptotic cells (early and late) were reported, without including necrotic/dead cells in the calculations; moreover, the percentage of apoptotic cells already present in the untreated samples or in the samples treated with DMSO only was subtracted, in order to better demonstrate the exact increase in apoptotic cells following the treatments. It is evident that the combined treatment led to an increase in apoptosis, which was found to be higher than the sum of the effects obtained using single administration of compound 3b and R8-PNA-a221 (outlined by the dashed lines in the graphs in Fig. 8).
Figure 8.
Summary of three independent experiments analyzing the effects of the tetrahydrothieno[2,3-c]pyridine 3b and PNA-a221 on apoptosis, determined by (A) Annexin V and (B) caspase-3/7 assays. U251 cells were treated with the indicated concentrations of compound 3b and R8-PNA-a221 and apoptosis was analyzed. Representative data are shown in Figs. 6 and 7. The dotted line represents the sum of the effects obtained using a single administration of compound 3b and R8-PNA-a221 at the lower concentration. PNA, peptide nucleic acid. *P<0.05, **P<0.01.
Apoptosis of TMZ-resistant T98G glioma cells
To verify whether the synergistic effect between compound 3b (IC50, 2 µM) and R8-PNA-a221 can also occur in TMZ-resistant T98G cells, the protocol employed on U251 cells and presented in Figs. 6-8 was used. The results shown in Figs. 9 and 10 demonstrated that a synergistic effect between compound 3b and R8-PNA-a221 was also observed on T98G cells. In fact, in both Annexin V (Fig. 9) and caspase-3/7 (Fig. 10) assays, combined treatment led to an increase in apoptosis which was found to be higher than the sum of the effects obtained using single treatment with compound 3b or R8-PNA-a221 (outlined by the dashed lines in Figs. 9B and 10B).
Figure 9.
Effects of the tetrahydrothieno[2,3-c]pyridine 3b and PNA-a221 on the apoptosis of T98G cells, part I. T98G cells were treated with the indicated concentrations of compound 3b and R8-PNA-a221 and apoptosis was analyzed using by Annexin V assay. (A) Representative apoptosis analyses are shown. (B) Summary of three independent experiments is shown. The dotted line represents the sum of the effects obtained using a single administration of compound 3b and R8-PNA-a221 at the lower concentration. PNA, peptide nucleic acid. *P<0.05, **P<0.01.
Figure 10.
Effects of the tetrahydrothieno[2,3-c]pyridine 3b and PNA-a221 on the apoptosis of T98G cells, part II. T98G cells were treated with the indicated concentrations of compound 3b and R8-PNA-a221 and the apoptosis was analyzed by caspase-3/7 assay. (A) Representative apoptosis analyses are reported. (B) A summary of three independent experiments is shown. The dotted line represents the sum of the effects obtained using a single administration of compound 3b and R8-PNA-a221 at the lower concentration. PNA, peptide nucleic acid. *P<0.05, **P<0.01.
Discussion
Patients with GBM express high levels of miR-221-3p, which exerts anti-apoptotic effects and promotes malignant progression (35-39). The involvement of miR-221-3p in GBM is supported by the study by Xu et al (39), which demonstrated that the inhibition of both miR-221 and miR-222 diminished the proliferation, invasion, migration and angiogenesis of GBM cells in vitro and in vivo. The inhibition of the miR-221/222 cluster reduced the activation of the p-JAK2/JAK2 and p-STAT3/STAT3 pathway (39), the levels of different matrix metalloproteinases (MMPs; MMP-2 and MMP-9) and the levels of vascular endothelial growth factor (VEGF).
Considering these findings, it appears evident that the inhibition of the miR-221/222 cluster is an interesting therapeutic approach aimed at inhibiting the tumorigenesis and invasiveness of GBM cells. On the other hand, compounds interfering with the microtubule-tubulin equilibrium in glioma cells have been demonstrated to retain a potent anti-proliferative activity, suggesting that compounds targeting tubulin are of great interest for the treatment of GBM (63,64). In this context, in a recent study, the authors described two novel series of compounds based on the 4,5,6,7-tetrahydrothieno[2,3-c] pyridine and 4,5,6,7-tetrahydrobenzo[b]thiophene molecular skeleton exerting potent anti-proliferative effects and capable of inducing a preferential block of the cell cycle in the G2/M phase (44).
It should be underlined that combined treatments appear of great interest in the development of anticancer approaches (1-5), since they are expected to obtain the same biological or therapeutic effect using lower concentrations of two or more drugs, thereby limiting side effects (1). Notably, combined therapy may be of significance in the management of GBM, a lethal malignant tumor needing of novel therapeutic options. This is mainly due to the fact that, at present, there is no effective pharmacological approach in the treatment of glioblastoma. The first-line drug currently used is TMZ; however, this is able to extend the life expectancy of patients with GBM by only an average of a few months; moreover, very often, this type of tumor is or becomes resistant to this chemotherapeutic agent (7-10).
The present study described for the first time, to the best of our knowledge, a 'combo-therapy' performed by the combined use of a PNA targeting miR-221 and tetrahydrothieno[2,3-c] pyridine 3b, one of the most active compounds described by Romagnoli et al (44). The combined treatments were associated with more pronounced anti-proliferative and cytotoxic effects, and a major increase in the sub-G1 cell population. In agreement, the combined treatment induced the highest level of 'late apoptotic' and 'dead' cells following Annexin V analysis. The low level of 'early apoptotic' cells can be tentatively explained by a rapid transition to the late stage of apoptosis. The data from Annexin V assay were fully confirmed by the functional caspase-3/7 assay, which employs a specific substrate (N-Ac-DEVD-AFC), cleaved by active caspase-3 to contribute in generating a highly fluorescent activity. It has been well-established that this assay generates data consistent with data generated by western blot analyses focusing on the quantification of cleaved PARP (65,66). With respect to western blot analysis, caspase-3/7 activity assay allows researchers to obtain a distribution of cells in different classes, facilitating a comparison with the data obtained by the Annexin V assay.
In conclusion, the results of the present study support the concept that the combined treatment of GBM cells with a PNA targeting a specific upregulated 'oncomiRNA' (in the present study, miR-221-3p) and an anticancer agent (in the present study, the anti-tubulin agent, tetrahydrothiene [2,3-c] pyridine 3b) is a promising strategy in the field of developing effective anti-GBM therapeutic approaches.
Acknowledgments
Not applicable.
Funding Statement
The authors wish to thank the 'Associazione Tutti per Chiara' (Montagnana, Italy) and AIRC/FIRC for supporting MZ and JG, respectively with research fellowships. The present study was supported by the Associazione Italiana per la Ricerca sul Cancro (AIRC) (IG #13575 to RG), FAR (University Fund for Scientific Research) and FIR (University Fund for the Incentivation of Research). The present study was also supported by the European Union (EU) Horizon 2020 Research and Innovation Programme [GA #633937, project ULTRAsensitive PLAsmonic devices for early CAncer Diagnosis (ULTRAPLACAD)] and by the Interuniversity Consortium for the Biotechnology, Italy.
Availability of data and materials
The datasets generated and/or analyzed during the present study are available from the corresponding author upon reasonable request.
Authors' contributions
RG, AF and RR were involved in the conceptualization, supervision and experimental design of the study. MZ, JG, RR and PO performed the experiments. MZ and JG were involved in the conceptualization and production of the figures. RG, AF and RR were involved in the writing, reviewing and editing of the manuscript. RG and AF were involved in project administration and funding acquisition. RG and MZ confirm the authenticity of all the raw data. All authors have read and approved the final version of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bayat Mokhtari R, Homayouni TS, Baluch N, Morgatskaya E, Kumar S, Das B, Yeger H. Combination therapy in combating cancer. Oncotarget. 2017;23:38022–38043. doi: 10.18632/oncotarget.16723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tolcher AW, Mayer LD. Improving combination cancer therapy: The CombiPlex® development platform. Future Oncol. 2018;14:1317–1332. doi: 10.2217/fon-2017-0607. [DOI] [PubMed] [Google Scholar]
- 3.Bozic I, Reiter JG, Allen B, Antal T, Chatterjee K, Shah P, Moon YS, Yaqubie A, Kelly N, Le DT, et al. Evolutionary dynamics of cancer in response to targeted combination therapy. Elife. 2013;2:e00747. doi: 10.7554/eLife.00747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun X, Xu H, Huang T, Zhang C, Wu J, Luo S. Simultaneous delivery of anti-miRNA and docetaxel with supramolecular self-assembled 'chitosome' for improving chemosensitivity of triple negative breast cancer cells. Drug Deliv Transl Res. 2021;11:192–204. doi: 10.1007/s13346-020-00779-4. [DOI] [PubMed] [Google Scholar]
- 5.Gasparello J, Gambari L, Papi C, Rozzi A, Manicardi A, Corradini R, Gambari R, Finotti A. High Levels of apoptosis are induced in the human colon cancer HT-29 cell line by co-administration of sulforaphane and a peptide nucleic acid targeting miR-15b-5p. Nucleic Acid Ther. 2020;30:164–174. doi: 10.1089/nat.2019.0825. [DOI] [PubMed] [Google Scholar]
- 6.Palmer AC, Sorger PK. Combination cancer therapy can confer benefit via patient-to-patient variability without drug additivity or synergy. Cell. 2017;171:1678–1691.e13. doi: 10.1016/j.cell.2017.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.von Neubeck C, Seidlitz A, Kitzler HH, Beuthien-Baumann B, Krause M. Glioblastoma multiforme: Emerging treatments and stratification markers beyond new drugs. Br J Radiol. 2015;88:20150354. doi: 10.1259/bjr.20150354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buczkowicz P, Hawkins C. Pathology, molecular genetics, and epigenetics of diffuse intrinsic pontine glioma. Front Oncol. 2015;5:147. doi: 10.3389/fonc.2015.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pace A, Dirven L, Koekkoek JAF, Golla H, Fleming J, Rudà R, Marosi C, Le Rhun E, Grant R, Oliver K, et al. European association for neuro-oncology (EANO) guidelines for palliative care in adults with glioma. Lancet Oncol. 2017;18:e330–e340. doi: 10.1016/S1470-2045(17)30345-5. [DOI] [PubMed] [Google Scholar]
- 10.Anjum K, Shagufta BI, Abbas SQ, Patel S, Khan I, Shah SAA, Akhter N, Hassan SSU. Current status and future therapeutic perspectives of glioblastoma multiforme (GBM) therapy: A review. Biomed Pharmacother. 2017;92:681–689. doi: 10.1016/j.biopha.2017.05.125. [DOI] [PubMed] [Google Scholar]
- 11.Santangelo A, Rossato M, Lombardi G, Benfatto S, Lavezzari D, De Salvo GL, Indraccolo S, Dechecchi MC, Prandini P, Gambari R, et al. A molecular signature associated with prolonged survival in glioblastoma patients treated with regorafenib. Neuro Oncol. 2021;23:264–276. doi: 10.1093/neuonc/noaa156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Touat M, Idbaih A, Sanson M, Ligon KL. Glioblastoma targeted therapy: Updated approaches from recent biological insights. Ann Oncol. 2017;28:1457–1472. doi: 10.1093/annonc/mdx106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sontheimer EJ, Carthew RW. Silence from within: Endogenous siRNAs and miRNAs. Cell. 2005;122:9–12. doi: 10.1016/j.cell.2005.06.030. [DOI] [PubMed] [Google Scholar]
- 14.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 15.He L, Hannon GJ. MicroRNAs: Small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- 16.Fabbri M, Ivan M, Cimmino A, Negrini M, Calin GA. Regulatory mechanisms of microRNAs involvement in cancer. Expert Opin Biol Ther. 2007;7:1009–1019. doi: 10.1517/14712598.7.7.1009. [DOI] [PubMed] [Google Scholar]
- 17.Taylor MA, Schiemann WP. Therapeutic opportunities for targeting microRNAs in cancer. Mol Cell Ther. 2014;2:1–13. doi: 10.1186/2052-8426-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gambari R, Brognara E, Spandidos DA, Fabbri E. Targeting oncomiRNAs and mimicking tumor suppressor miRNAs: New trends in the development of miRNA therapeutic strategies in oncology (review) Int J Oncol. 2016;49:5–32. doi: 10.3892/ijo.2016.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miroshnichenko S, Patutina O. Enhanced inhibition of tumorigenesis using combinations of miRNA-targeted therapeutics. Front Pharmacol. 2019;10:488. doi: 10.3389/fphar.2019.00488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gajda E, Godlewska M, Mariak Z, Nazaruk E, Gawel D. Combinatory treatment with miR-7-5p and drug-loaded cubosomes effectively impairs cancer cells. Int J Mol Sci. 2020;21:5039. doi: 10.3390/ijms21145039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghasabi M, Majidi J, Mansoori B, Mohammadi A, Shomali N, Shirafkan N, Baghbani E, Kazemi T, Baradaran B. The effect of combined miR-200c replacement and cisplatin on apoptosis induction and inhibition of gastric cancer cell line migration. J Cell Physiol. 2019;234:22581–22592. doi: 10.1002/jcp.28823. [DOI] [PubMed] [Google Scholar]
- 22.He JQ, Zheng MX, Ying HZ, Zhong YS, Zhang HH, Xu M, Yu CH. PRP1, a heteropolysaccharide from platycodonis radix, induced apoptosis of HepG2 cells via regulating miR-21-mediated PI3K/AKT pathway. Int J Biol Macromol. 2020;158:542–551. doi: 10.1016/j.ijbiomac.2020.04.193. [DOI] [PubMed] [Google Scholar]
- 23.Tao Y, Zhan S, Wang Y, Zhou G, Liang H, Chen X, Shen H. Baicalin, the major component of traditional Chinese medicine Scutellaria baicalensis induces colon cancer cell apoptosis through inhibition of oncomiRNAs. Sci Rep. 2018;8:14477. doi: 10.1038/s41598-018-32734-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, Duan J, Qu Y, Deng T, Liu R, Zhang L, Bai M, Li J, Ning T, Ge S, et al. Onco-miR-24 regulates cell growth and apoptosis by targeting BCL2L11 in gastric cancer. Protein Cell. 2016;7:141–151. doi: 10.1007/s13238-015-0234-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu Z, Liu K, Wang Y, Xu Z, Meng J, Gu S. Upregulation of microRNA-96 and its oncogenic functions by targeting CDKN1A in bladder cancer. Cancer Cell Int. 2015;15:107. doi: 10.1186/s12935-015-0235-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 27.Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, Berg RH, Kim SK, Norden B, Nielsen PE. PNA hybridizes to complementary oligonucleotides obeying the watson-crick hydrogen-bonding rules. Nature. 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 28.Fabani MM, Gait MJ. miR-122 targeting with LNA/2′-O-methyl oligonucleotide mixmers, peptide nucleic acids (PNA), and PNA-peptide conjugates. RNA. 2008;14:336–346. doi: 10.1261/rna.844108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brown PN, Yin H. PNA-based microRNA inhibitors elicit anti-inflammatory effects in microglia cells. Chem Commun (Camb) 2013;49:4415–4417. doi: 10.1039/C2CC36540E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fabani MM, Abreu-Goodger C, Williams D, Lyons PA, Torres AG, Smith KG, Enright AJ, Gait MJ, Vigorito E. Efficient inhibition of miR-155 function in vivo by peptide nucleic acids. Nucleic Acids Res. 2010;38:4466–4475. doi: 10.1093/nar/gkq160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fabbri E, Manicardi A, Tedeschi T, Sforza S, Bianchi N, Brognara E, Finotti A, Breveglieri G, Borgatti M, Corradini R, et al. Modulation of the biological activity of microRNA-210 with peptide nucleic acids (PNAs) ChemMedChem. 2011;6:2192–2202. doi: 10.1002/cmdc.201100270. [DOI] [PubMed] [Google Scholar]
- 32.Brognara E, Fabbri E, Bazzoli E, Montagner G, Ghimenton C, Eccher A, Cantù C, Manicardi A, Bianchi N, Finotti A, et al. Uptake by human glioma cell lines and biological effects of a peptide-nucleic acids targeting miR-221. J Neurooncol. 2014;118:19–28. doi: 10.1007/s11060-014-1405-6. [DOI] [PubMed] [Google Scholar]
- 33.Gambari R, Fabbri E, Borgatti M, Lampronti I, Finotti A, Brognara E, Bianchi N, Manicardi A, Marchelli R, Corradini R. Targeting microRNAs involved in human diseases: A novel approach for modification of gene expression and drug development. Biochem Pharmacol. 2011;82:1416–1429. doi: 10.1016/j.bcp.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 34.Fabbri E, Tamanini A, Jakova T, Gasparello J, Manicardi A, Corradini R, Sabbioni G, Finotti A, Borgatti M, Lampronti I, et al. A peptide nucleic acid against MicroRNA miR-145-5p enhances the expression of the cystic fibrosis transmembrane conductance regulator (CFTR) in Calu-3 cells. Molecules. 2017;23:71. doi: 10.3390/molecules23010071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Swellam M, Ezz El Arab L, Al-Posttany AS, B Said S. Clinical impact of circulating oncogenic MiRNA-221 and MiRNA-222 in glioblastoma multiform. J Neurooncol. 2019;144:545–551. doi: 10.1007/s11060-019-03256-2. [DOI] [PubMed] [Google Scholar]
- 36.Chen YY, Ho HL, Lin SC, Ho TD, Hsu CY. Upregulation of miR-125b, miR-181d, and miR-221 predicts poor prognosis in MGMT promoter-unmethylated glioblastoma patients. Am J Clin Pathol. 2018;149:412–417. doi: 10.1093/ajcp/aqy008. [DOI] [PubMed] [Google Scholar]
- 37.Yang JK, Yang JP, Tong J, Jing SY, Fan B, Wang F, Sun GZ, Jiao BH. Exosomal miR-221 targets DNM3 to induce tumor progression and temozolomide resistance in glioma. J Neurooncol. 2017;131:255–265. doi: 10.1007/s11060-016-2308-5. [DOI] [PubMed] [Google Scholar]
- 38.Xie Q, Yan Y, Huang Z, Zhong X, Huang L. MicroRNA-221 targeting PI3-K/Akt signaling axis induces cell proliferation and BCNU resistance in human glioblastoma. Neuropathology. 2014;34:455–464. doi: 10.1111/neup.12129. [DOI] [PubMed] [Google Scholar]
- 39.Xu CH, Liu Y, Xiao LM, Chen LK, Zheng SY, Zeng EM, Li DH, Li YP. Silencing microRNA-221/222 cluster suppresses glioblastoma angiogenesis by suppressor of cytokine signaling-3-dependent JAK/STAT pathway. J Cell Physiol. 2019;234:22272–22284. doi: 10.1002/jcp.28794. [DOI] [PubMed] [Google Scholar]
- 40.Tao K, Yang J, Guo Z, Hu Y, Sheng H, Gao H, Yu H. Prognostic value of miR-221-3p miR-342-3p and miR-491-5p expression in colon cancer. Am J Transl Res. 2014;6:391–401. [PMC free article] [PubMed] [Google Scholar]
- 41.Dong Y, Zhang N, Zhao S, Chen X, Li F, Tao X. miR-221-3p and miR-15b-5p promote cell proliferation and invasion by targeting Axin2 in liver cancer. Oncol Lett. 2019;18:6491–6500. doi: 10.3892/ol.2019.11056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yin G, Zhang B, Li J. miR-221-3p promotes the cell growth of non-small cell lung cancer by targeting p27. Mol Med Rep. 2019;20:604–612. doi: 10.3892/mmr.2019.10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li F, Xu JW, Wang L, Liu H, Yan Y, Hu SY. MicroRNA-221-3p is up-regulated and serves as a potential biomarker in pancreatic cancer. Artif Cells Nanomed Biotechnol. 2018;46:482–487. doi: 10.1080/21691401.2017.1315429. [DOI] [PubMed] [Google Scholar]
- 44.Romagnoli R, Prencipe F, Oliva P, Cacciari B, Balzarini J, Liekens S, Hamel E, Brancale A, Ferla S, Manfredini S, et al. Synthesis and biological evaluation of new antitubulin agents containing 2-(3′,4′,5′-trimethoxyanilino)-3,6-disubstituted-4,5,6,7-tetrahydrothieno[2,3-c]pyridine scaffold. Molecules. 2020;25:1690. doi: 10.3390/molecules25071690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khodyuk RGD, Bai R, Hamel E, Lourenço EMG, Barbosa EG, Beatriz A, Dos Santos EDA, de Lima DP. Diaryl disulfides and thiosulfonates as combretastatin A-4 analogues: Synthesis, cytotoxicity and antitubulin activity. Bioorg Chem. 2020;101:104017. doi: 10.1016/j.bioorg.2020.104017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu H, Fu Q, Lu Y, Zhang W, Yu P, Liu Z, Sun X. Anti-tubulin agent vinorelbine inhibits metastasis of cancer cells by regulating epithelial-mesenchymal transition. Eur J Med Chem. 2020;200:112332. doi: 10.1016/j.ejmech.2020.112332. [DOI] [PubMed] [Google Scholar]
- 47.Wang G, Liu W, Gong Z, Huang Y, Li Y, Peng Z. Design, synthesis, biological evaluation and molecular docking studies of new chalcone derivatives containing diaryl ether moiety as potential anticancer agents and tubulin polymerization inhibitors. Bioorg Chem. 2020;95:103565. doi: 10.1016/j.bioorg.2019.103565. [DOI] [PubMed] [Google Scholar]
- 48.Yang F, Yu LZ, Diao PC, Jian XE, Zhou MF, Jiang CS, You WW, Ma WF, Zhao PL. Novel [1,2,4]triazolo[1,5-a]pyrimidine derivatives as potent antitubulin agents: Design, multicomponent synthesis and antiproliferative activities. Bioorg Chem. 2019;92:103260. doi: 10.1016/j.bioorg.2019.103260. [DOI] [PubMed] [Google Scholar]
- 49.Pen A, Durocher Y, Slinn J, Rukhlova M, Charlebois C, Stanimirovic DB, Moreno MJ. Insulin-like growth factor binding protein 7 exhibits tumor suppressive and vessel stabilization properties in U87MG and T98G glioblastoma cell lines. Cancer Biol Ther. 2011;12:634–646. doi: 10.4161/cbt.12.7.17171. [DOI] [PubMed] [Google Scholar]
- 50.Twentyman PR, Luscombe M. A study of some variables in a tetrazolium dye (MTT) based assay for cell growth and chemosensitivity. Br J Cancer. 1987;56:279–285. doi: 10.1038/bjc.1987.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cao X, Gu Y, Jiang L, Wang Y, Liu F, Xu Y, Deng J, Nan Y, Zhang L, Ye J, Li Q. A new approach to screening cancer stem cells from the U251 human glioma cell line based on cell growth state. Oncol Rep. 2013;29:1013–1018. doi: 10.3892/or.2012.2206. [DOI] [PubMed] [Google Scholar]
- 52.Munshi A, Hobbs M, Meyn RE. Clonogenic cell survival assay. Methods Mol Med. 2005;110:21–28. doi: 10.1385/1-59259-869-2:021. [DOI] [PubMed] [Google Scholar]
- 53.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 54.Milani R, Brognara E, Fabbri E, Manicardi A, Corradini R, Finotti A, Gasparello J, Borgatti M, Cosenza LC, Lampronti I, et al. Targeting miR-155-5p and miR-221-3p by peptide nucleic acids induces caspase-3 activation and apoptosis in temozolomide-resistant T98G glioma cells. Int J Oncol. 2019;55:59–68. doi: 10.3892/ijo.2019.4810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shen H, Lin Z, Shi H, Wu L, Ma B, Li H, Yin B, Tang J, Yu H, Yin X. MiR-221/222 promote migration and invasion, and inhibit autophagy and apoptosis by modulating ATG10 in aggressive papillary thyroid carcinoma. 3 Biotech. 2020;10:339. doi: 10.1007/s13205-020-02326-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hu XH, Zhao ZX, Dai J, Geng DC, Xu YZ. MicroRNA-221 regulates osteosarcoma cell proliferation, apoptosis, migration, and invasion by targeting CDKN1B/p27. J Cell Biochem. 2019;120:4665–4674. doi: 10.1002/jcb.27755. [DOI] [PubMed] [Google Scholar]
- 57.Xie X, Huang Y, Chen L, Wang J. miR-221 regulates proliferation and apoptosis of ovarian cancer cells by targeting BMF. Oncol Lett. 2018;16:6697–6704. doi: 10.3892/ol.2018.9446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li J, Li Q, Huang H, Li Y, Li L, Hou W, You Z. Overexpression of miRNA-221 promotes cell proliferation by targeting the apoptotic protease activating factor-1 and indicates a poor prognosis in ovarian cancer. Int J Oncol. 2017;50:1087–1096. doi: 10.3892/ijo.2017.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhou L, Jiang F, Chen X, Liu Z, Ouyang Y, Zhao W, Yu D. Downregulation of miR-221/222 by a microRNA sponge promotes apoptosis in oral squamous cell carcinoma cells through upregulation of PTEN. Oncol Lett. 2016;12:4419–4426. doi: 10.3892/ol.2016.5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sarkar S, Dubaybo H, Ali S, Goncalves P, Kollepara SL, Sethi S, Philip PA, Li Y. Down-regulation of miR-221 inhibits proliferation of pancreatic cancer cells through up-regulation of PTEN, p27(kip1), p57(kip2), and PUMA. Am J Cancer Res. 2013;3:465–477. [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang CZ, Zhang JX, Zhang AL, Shi ZD, Han L, Jia ZF, Yang WD, Wang GX, Jiang T, You YP, et al. MiR-221 and miR-222 target PUMA to induce cell survival in glioblastoma. Mol Cancer. 2010;9:229. doi: 10.1186/1476-4598-9-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang C, Zhang J, Zhang A, Wang Y, Han L, You Y, Pu P, Kang C. PUMA is a novel target of miR-221/222 in human epithelial cancers. Int J Oncol. 2010;37:1621–1626. doi: 10.3892/ijo_00000816. [DOI] [PubMed] [Google Scholar]
- 63.Döbber A, Phoa AF, Abbassi RH, Stringer BW, Day BW, Johns TG, Abadleh M, Peifer C, Munoz L. Development and biological evaluation of a photoactivatable small molecule microtubule-targeting agent. ACS Med Chem Lett. 2017;8:395–400. doi: 10.1021/acsmedchemlett.6b00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cherry AE, Haas BR, Naydenov AV, Fung S, Xu C, Swinney K, Wagenbach M, Freeling J, Canton DA, Coy J, et al. ST-11: A new brain-penetrant microtubule-destabilizing agent with therapeutic potential for glioblastoma multiforme. Mol Cancer Ther. 2016;15:2018–2029. doi: 10.1158/1535-7163.MCT-15-0800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nam GH, Jo KJ, Park YS, Kawk HW, Kim SY, Kim YM. In vitro and in vivo induction of p53-dependent apoptosis by extract of euryale ferox salisb in A549 human caucasian lung carcinoma cancer cells is mediated through Akt signaling pathway. Front Oncol. 2019;9:406. doi: 10.3389/fonc.2019.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kim EJ, Kim GT, Kim BM, Lim EG, Kim SY, Kim YM. Apoptosis-induced effects of extract from artemisia annua linné by modulating PTEN/p53/PDK1/Akt/signal pathways through PTEN/p53-independent manner in HCT116 colon cancer cells. BMC Complement Altern Med. 2017;17:236. doi: 10.1186/s12906-017-1702-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and/or analyzed during the present study are available from the corresponding author upon reasonable request.