Abstract
Introduction
Containing COVID-19 requires broad-scale testing. However, sample collection requires qualified personnel and protective equipment and may cause transmission. We assessed the sensitivity of SARS-CoV-2-rtPCR applying three self-sampling techniques as compared to professionally collected oro-nasopharyngeal samples (cOP/NP).
Methods
From 62 COVID-19 outpatients, we obtained: (i) multi-swab, MS; (ii) saliva sponge combined with nasal vestibula, SN; (iii) gargled water, GW; (iv) professionally collected cOP/NP (standard). We compared ct-values for E-gene and ORF1ab and analysed variables reducing sensitivity of self-collecting procedures.
Results
The median ct-values for E-gene and ORF1ab obtained in cOP/NP samples were 20.7 and 20.2, in MS samples 22.6 and 21.8, in SN samples 23.3 and 22.3, and in GW samples 30.3 and 29.8, respectively. MS and SN samples showed sensitivities of 95.2% (95%CI, 86.5-99.0) and GW samples of 88.7% (78.1-95.3). Sensitivity was inversely correlated with ct-values, and became <90% for samples obtained more than 8 days after symptom onset. For MS and SN samples, false negativity was associated with language problems, sampling errors, and symptom duration.
Conclusion
Conclusions from this study are limited to the sensitivity of self-sampling in mildly to moderately symptomatic patients. Still, self-collected oral/nasal/saliva samples can facilitate up-scaling of testing in early symptomatic COVID-19 patients if operational errors are minimized.
Keywords: Covid-19, Testing, Self-sampling, Screenings, rtRT-PCR sensitivity, Diagnostic accuracy
Introduction
Containment of the current COVID-19 pandemic(Lu et al., 2020, Zhu et al., 2020) requires broad-scale testing capacities(Zhu and Wong, 2020) for patients, potentially contagious persons and groups at risk of infection. Laboratory capacities for real-time reverse transcriptase polymerase chain reaction (rtRT-PCR) have been significantly increased in many countries, and are complemented by novel rapid test devices based on antigen detection(Rai et al., 2021). Still, professionally collected (oro-)nasopharyngeal samples are considered the gold standard(Marty et al., 2020, Pan et al., 2020). However, this approach remains challenging considering the needs of qualified medical personnel and protective equipment as well as the risk of potential virus transmission to health care workers or others at testing-sites(Zhu and Wong, 2020). Moreover, (oro-)nasopharyngeal sampling is being perceived as uncomfortable, and possibly deterring, by many patients. Simplified sampling techniques may help overcome these limitations. Reliable self-collecting procedures could be home-based and thus contribute to reduced virus transmission due to more rapid diagnosis and reduced mobility of potentially contagious persons. Self-collected samples from the oral cavity, e.g. saliva or from the nasal vestibule (anterior nasal cavity) are therefore being investigated as non-invasive, more comfortable and less resource-intensive alternatives and show variable reliability (Fernandez-Gonzalez et al., 2021, Tu Yuan-Po et al., 2020, Tu Y.P. et al., 2020).
We performed a prospective manufacturer-independent sensitivity study in symptomatic outpatients with confirmed SARS-CoV-2 infection to examine whether combinations of simple self-collection technics may be reliable alternatives to professionally collected oro-nasopharyngeal sampling. We obtained four simultaneous samples for rtRT-PCR testing from these patients: one professionally collected, oro-nasopharyngeal swab sample and three self-collected specimens using different simplified sampling procedures from more distal locations in the upper respiratory tract. Herein, we present the sensitivities using self-collected samples as compared to the gold standard.
Methods
Study Design and Participant Enrolment
We calculated that 60 patients with confirmed SARS-CoV-2 infection would be necessary to compare the sensitivity of rtRT-PCR assays in self-collected versus professionally collected samples assuming a true sensitivity of 98%.
Between 7th December 2020 and 11th January 2021, we prospectively enrolled 62 SARS-CoV-2 infected outpatients into the present sudy. On the day before enrolment, all patients had attended the central testing site of Charité – Universitaetsmedizin Berlin (Maechler et al., 2020), with symptoms compatible with COVID-19. Professionally collected, combined oro-nasophayryngeal swabs had been subjected to RT-PCR at the central Charité laboratory. Upon results communication and counselling via telephone, study participation was offered to patients. Symptomatic patients were eligible in case of a confirmed RT-PCR test result not older than 24 hours before phone contact, and location of residence enabling a home visit on the same day. The study was reviewed by the ethics committee at Charité-Universitätsmedizin Berlin, Germany (EA2/192/20), and written informed consent was obtained prior to study entry.
A medical study team visited the participants and handed written instructions and necessary materials for three self-collecting procedures. To assess independent patient-self-collection, the collections were observed without any additional verbal instructions or intervention, and performance and irregularities were documented by the study team. The three procedures of self-collection included (in order of application): (i) MS (multi-swab): a combination swab from the tongue, the inner cheek and both nasal vestibules (insertion 2-3 cm, twisting 4x), (ii) SN (saliva-nasal): insalivating of the swab for 10-15 sec. before swabbing both nasal vestibules (insertion 2-3 cm, twisting 4x), and (iii) GW (gargle water): collection of 10 ml of gargled tap water into a plastic container (SarstedR, L494-9). Alternating, before or after the gargling procedure (iii), a trained medical professional collected an oro-nasopharyngeal swab as reference sample. After completion of collections, participants were interviewed on perception of the procedures, their professional and linguistic background and competences, as well as on prior experience with swabs. Swabs used were nylon-flocked applicators with 1 ml of Amies preservation medium (ESwab® Copan, Italy).
Laboratory analyses
For this study, we performed a quadruplex RT-PCR assay to simultaneously detect the E-gene and ORF1ab of SARS-CoV-2, the c-myc gene representing human nucleic acid, and KoMa, an artificial sequence that has no significant homology to any sequence in GenBank. Total RNA was extracted with the QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany). PCR was performed using the AgPath-ID™ One-Step RT-PCR Reagents kit (Applied Biosystems, Foster City, CA USA) on a Bio-Rad CFX96 device. Cycling conditions were: 45°C for 15 min, 95°C for 10 min followed by 45 cycles of 95°C for 15 s and 60°C for 30 s. The E-Gene primer and probe sequences were taken from Corman et al.(Corman et al., 2020). The SARS-CoV-2 specific ORF1ab assay was designed based on the 72 sequences that were available at the time from Michel et al.(Michel et al., 2020). KoMa and human cell c-myc were detected to serve as quality and internal amplification (KoMa) control for potential inhibitors of RT-PCR and the respective sampling technique(Kirchner et al., 2010). The cycle threshold-value (ct-value), i.e. the PCR cycle at which the fluorescence signal crosses the detection threshold, was determined for each target sequence.
Probit analysis revealed the limit of detection for the quadruplex PCR under the above described conditions as 28.7 genome copies for the E-Gene assay and 32.0 genome copies for the ORF1ab assay. Tests with signals that crossed the detection threshold were considered positive. All samples were measured on the day of collection by using 140 µl aliquots for RNA extraction, respectively.
Statistical analyses
Descriptive statistics used proportions, means ± standard deviation (SD), or medians with interquartile ranges (IQR), as applicable. Categorical variables were compared by two-tailed Fisher´s exact test, numeric variables by a Mann Whitney U-test, and paired numeric data by a Wilcoxon Signed Rank test. Sensitivity and 95% confidence interval (CI) were calculated for each sampling method.
We assessed the linear dependence between ct-values of the professional collected swabs and ct-values of the self-collecting sampling methods by target gene, using Pearson correlation (r). A binominal regression model was used to determine the variables reducing sensitivity of the self-collecting procedures (negative RT-PCR) compared to the correspondent professional collected sample. All computations were performed using “R” version 3.6.3 for all analyses. P<0.05 was considered to reflect statistical significance.
Results
Comparison of sensitivities and ct-values by sampling technique
All 62 participants provided three self-collected samples (n = 186) in addition to the 62 newly obtained professionally collected oro-nasopharyngeal specimens. Half of the patients were female, and their median age was 31.5 (range, 17-66). The median time between onset of symptoms and enrolment was four days (range, 2-15). At the day of testing, the majority of patients reported mild to moderate symptomatic illness, no one was admitted to in-patient care. Symptoms and other clinical variables are presented in table 1 .
Table 1.
Patient characteristics and symptoms
| All patients (N=62) % (n) or median (range) | |
|---|---|
| Female | 50% (31) |
| Age (years) | 31.5 (17.0, 66.0) |
| Shortness of breath | 6.5% (4) |
| Chest pain/Chest tightness | 0% (0) |
| Fever in the last 48 hours | 30.6% (19) |
| Chills | 38.7% (24) |
| Fatigue | 80.6% (50) |
| Body aches | 66.1% (41) |
| Cough | 62.9% (39) |
| Rhinorrhea | 67.7% (42) |
| Diarrhea | 21% (13) |
| Sore throat | 41.9% (26) |
| Headache | 75.8% (47) |
| Impaired smell and taste | 50% (31) |
| Time between (study-) test and symptom onset (days) | 4.0 (2.0, 15.0) |
| Chronic lung disease | 9.7% (6) |
| Diabetes I/II | 1.6% (1) |
| Cardiovascular disease | 8.1% (5) |
| Obesity | 6.4% (4) |
| Contact to a confirmed SARS-CoV-2 case | 33.9% (21) |
| Time between (study-) test and last contact (days) | 6.0 (1.0, 10.0) |
| German NOT as first language | 30.6% (19) |
All 62 professionally collected samples tested positive for both SARS-CoV-2 target genes, the E-gene and the ORF1ab. In all samples, regardless of sampling technique, c-myc was detected, indicating that all contained human cells. This was done using the c-myc PCR assay in a singleplex reaction, as samples which are positive in the E-gene and ORF1ab PCR assays do not always allow the amplification of the KoMa and c-myc controls in the quadruplex reaction. No signs of PCR inhibition were detected.
Detection sensitivities for E-gene and ORF1ab differed depending on the self-collecting procedure (Table 2 ). For E-gene, MS samples were positive in 93.5% (95%CI: 84.3-98.2), SN samples in 95.2% (95%CI 86.5-99.0), and GW samples in 87.1% (95%CI76.1-94.3). For ORF1ab, both MS and SN samples were positive in 91.9% (82.2-97.3), and GW samples in 88.7% (78.1-95.3). Defining a sample as SARS-CoV-2 positive if either E-gene or ORF1ab was detected, the MS and SN samples showed a sensitivity of 95.2% (86.5-99.0) and GW samples of 88.7% (78.1-95.3).
Table 2.
Comparison of variables between negative and positive test result in patient collected samples separated by collection method
| Sample type | Variable | Negative | Positive | p-value |
|---|---|---|---|---|
| MS | N | 3 | 59 | |
| Ct value of professionally collected cOP/NP, median (IQR) | 27.2 (27.1, 30.4) | 20.4 (17.2, 22.9) | 0.008 | |
| Female | 1 (33.3%) | 30 (50.8%) | 1.000 | |
| Age in years, median (range) | 48.0 (35.0, 53.0) | 31.0 (17.0, 66.0) | 0.064 | |
| Number of symptoms, median (range) | 7.0 (7.0, 7.0) | 5.0 (2.0, 10.0) | 0.335 | |
| Symptom interval in days, median (range) | 11.0 (4.0, 15.0) | 4.0 (2.0, 12.0) | 0.083 | |
| German is not first language | 3 (100.0%) | 16 (27.1%) | 0.026 | |
| No. of mistakes during sampling procedure, median (range) | 2.0 (2.0, 3.0) | 0.0 (0.0, 3.0) | 0.005 | |
| SN | N | 3 | 59 | |
| Ct value of professionally collected cOP/NP, median (IQR) | 29.1 (28.1, 31.4) | 20.4 (17.2, 22.9) | 0.006 | |
| Female | 1 (33.3%) | 30 (50.8%) | 1.000 | |
| Age in years, median (range) | 35.0 (19.0, 48.0) | 31.0 (17.0, 66.0) | 0.948 | |
| Number of symptoms, median (range) | 3.0 (3.0, 3.0) | 5.0 (2.0, 10.0) | 0.127 | |
| Symptom interval in days, median (range) | 12.0 (4.0, 15.0) | 4.0 (2.0, 11.0) | 0.077 | |
| German is not first language | 3 (100.0%) | 16 (27.1%) | 0.026 | |
| No. of mistakes during sampling procedure, median (range) | 2.0 (1.0, 3.0) | 0.0 (0.0, 3.0) | 0.012 | |
| GW | N | 7 | 55 | |
| Ct value of professionally collected OP/NP, median (IQR) | 26.6 (23.4, 28.1) | 20.1(17.1, 22.3) | < 0.001 | |
| Female | 4 (57.1%) | 27 (49.1%) | 1.000 | |
| Age in years, median (range) | 34.0 (19.0, 48.0) | 31.0 (17.0, 66.0) | 0.798 | |
| Number of symptoms, median (range) | 6.0 (3.0, 9.0) | 5.0 (2.0, 10.0) | 0.794 | |
| Symptom interval in days, median (range) | 4.0 (3.0, 15.0) | 5.0 (2.0, 11.0) | 0.804 | |
| German is not first language | 4 (57.1%) | 15(27.3%) | 0.187 | |
| No. of mistakes during sampling procedure, median (range) | 0.0 (0.0, 2.0) | 0.0 (0.0, 2.0) | 0.426 |
MS – Multis-swab (cheek, tongue, nares), SN – Saliva-nasal (saliva, nasal vestibule), GW – gargle water. A patient sample was considered positive when either the e-gene or the ORF1ab-gene or both were detected.
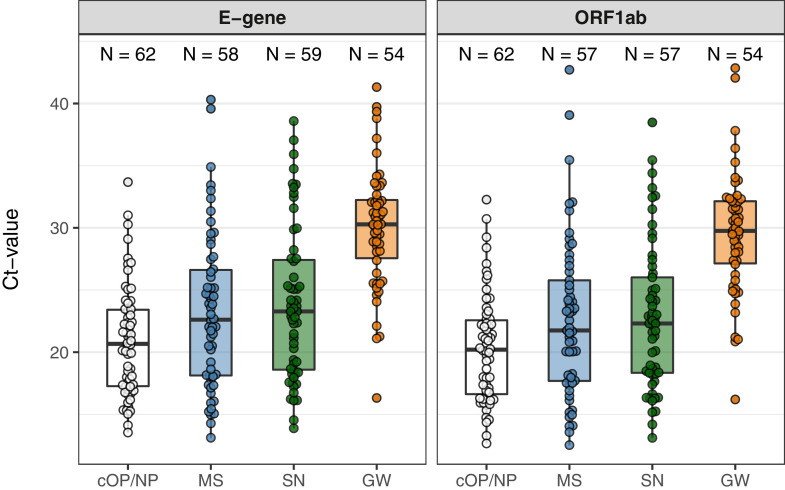
Figure 1 presents ct-values of all samples that tested positive for E-gene and for ORF1ab. Median ct-values of the professional collected samples were 20.7 for E-gene and 20.2 for ORF1ab. Median ct-values of the self-collected samples were slightly but significantly (p<0.05) higher for MS (22.6 and 21.8) and SN (23.3 and 22.3), and substantially so for GW (30.3 and 29.8).
Figure 1.
Ct values for E-gene and the ORF1ab of all positive samples by type of sample collection: professionally collected cOP/NP sample (white), patient-collected samples MS (blue), SN (green), and GW (orange)
The boxes in the plot depict the 25th, 50th and 75th percentiles.
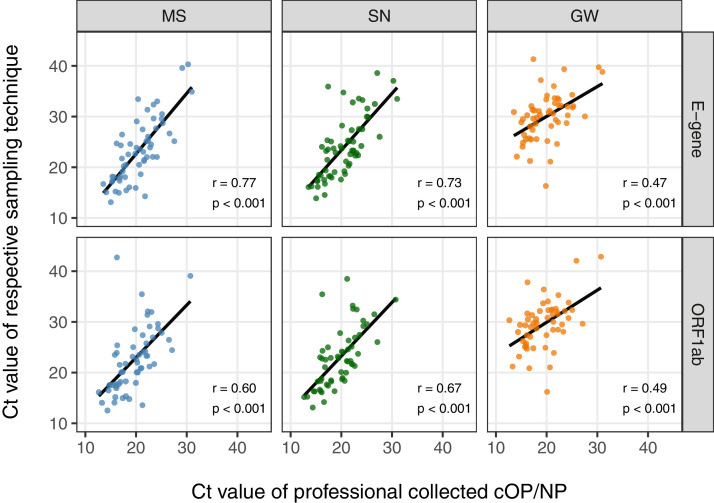
For both target genes, ct-values of all sampling techniques were significantly correlated with their respective ct-values for the cOP/NP (p<0.001 for all comparisons), with the strongest correlation for the E-gene in MS (r=0.77) and SN (r=0.73) (Figure 2 ).
Figure 2.
Ct-values of each patient-collected sample type (MS, SN, GW) compared with the ct-value of the diagnostic standard (medical professionally collected OP/NP sample, x-axis) shown for the used two target genes.
MS (Multi-swab – blue), SN (Saliva-nasal – green), GW (gargle water – orange)
Factors associated with false-negative results in patient-collected samples
In three (4.8%) MS and SN samples as well as in seven (11.3%) GW specimens, none of the two SARS-CoV-2 genes were detected. These false negative results were associated with high ct-values, i.e. low viral loads, in the corresponding professionally collected swab (Table 2). For MS and SN samples, but not for GW specimens, false negativity was also associated with a non-German mother-tongue, the number of sampling procedure mistakes, and as a trend, with symptom duration.
In a binomial logistic regression model fit on our data, for every day of symptom duration, the odds for a positive test decreased by 40% (OR, 0.6; 95%CI, 0.4-0.9; P=0.01, for both MS and SN samples). Following the model, sensitivity (as compared to oro-nasopharyngeal swabs) dropped below 90% for symptom duration longer than eight days. In addition, when only assessing patients with a symptom duration of less than 8 days in our study population, test sensitivity for SN samples was 98.2% (95%CI, 90.4, 100.0), for MS 96.4% (95%CI, 87.7, 99.6).
Discussion
Our findings indicate that during the early phase of clinical disease, self-collected samples provide only slightly reduced sensitivity in the detection of SARS-CoV-2 by RT-PCR as compared to professionally collected oro-nasopharyngeal samples. This is particularly true for sample collection using swabs (MS, SN) when operational errors are minimized using comprehensive instructions.
Acceptable sensitivity when using self-collected samples including gargling techniques has previously been reported(Fernandez-Gonzalez et al., 2021, Lee et al., 2021, Lindner et al., 2020, McCulloch et al., 2020, Migueres et al., 2020, Tu Yuan-Po et al., 2020, Tu Y.P. et al., 2020, Wehrhahn et al., 2020, Wyllie et al., 2020). Public health agencies like the US Centers for Disease Control and Prevention, the Infectious Diseases Society of America, and the German Robert Koch Institute consider self-collecting techniques for symptomatic patients as potential alternatives under certain circumstance. However, they emphasize the scarcity of available data and the potential of erroneous conduct and results (Centers for Disease Control and Prevention, 2021, Kojima et al., 2020, Robert Koch Institute, 2021).
The critical temporal roles of viral shedding and sample collection was recognized early during the pandemic(Woelfel et al., 2020, Zou et al., 2020). Our data confirm a decrease of sensitivity when upper-respiratory tract samples are collected during the second week of disease(Wyllie et al., 2020). The present data suggest that until day eight of symptom onset, self-collected samples in symptomatic patients may be similarly reliable as professional collected oro-nasopharyngeal samples with sensitivities of >98% (MS, SN), with some reservation for GW (sensitivity >90%).
The gargling procedure performed below expectations. It is likely that the 10 ml water used for gargling instead of 1 ml of transport medium for all other samples diluted viral material in the GW sample. However, it is recommended by others (Goldfarb et al., 2021, Kojima et al., 2020) and officially used in Austria.
Diagnostic tools in the hands of untrained people require education and comprehensive instructions. Indeed, the false negative results of the patient-collected procedures were associated with procedural errors and reduced German language competence (written instructions in German). This might indicate an even higher potential for sample self-collection when pictorial illustrations are offered, and in different languages.
Supervision and support of self-collection procedures may be provided directly by personnel through a window or via video consultation. Such would not require personal protective equipment and still reduce transmission risks at testing sites.
With respect to the slightly reduced sensitivity of self-collected samples (MS and SN) in this study, it needs to be taken into account that oro-nasopharyngeal swabbing was performed by very experienced medical professionals. In a scenario of massive up-scale of testing by public health systems this may not be the case, potentially shrinking the sensitivity differences between professional and self-collection of samples. This has particular significance for a central component of pandemic response globally, i.e., repetitive testing of groups, e.g., school attendees or employees as claimed also by the WHO(World Health Organization, 2021). For that, testing including sample-collection needs to be simplified, non-invasive to prevent refusal, efficient and safe. In this regard, we provide further evidence that (home) sample self-collection may help to reduce test restraints and transmission risks as well as human and material resources. This data refers only to self-collecting for RT-PCR and not to self-collecting for rapid tests. Still, the results might support the reliability of self-testing as a public-health tool.
As a major limitation, our study involved only mildly to moderately symptomatic patients. We cannot provide evidence regarding the sensitivity of patient-collected samples in populations of asymptomatic persons who are described to present with comparatively lower viral loads, i.e., with higher ct-values in some studies (Jones et al., 2021, Lee et al., 2020, Lescure et al., 2020, Pujadas et al., 2020, Zheng et al., 2020). Likewise, our symptomatic study population included only a small fraction of cases with comparably high ct-values (around 30), which interferes with a solid interpretation of the sensitivity of the self-sampling procedures in such patients while other authors also report about study populations with larger proportions of participants with comparably high ct-values. (Perchetti et al., 2021) It is possible that sensitivity would be smaller with a larger proportion of low-positives.
Still our findings may indicate reliability of self-collection of samples in the early symptomatic phase. Therefore we see self-collection first of all as a possibility to increase testing in newly symptomatic persons in the context of primary outpatient care without large investments in specialised test sites being necessary. Whether self-collection is sufficiently sensitive in asymptomatic persons is and was being investigated by others and results are heterogenous, ranging from moderately reduced to comparable sensitivity of the self-collection methods. (Lee RA et al. 2021). However, statistical modelling showed that smaller sensitivity in screenings of asymptomatic persons may be largely outweighed by the far broader scope of application.(Larremore et al., 2021)
Our study among SARS-CoV-2 positive outpatients could not produce specificity data. However, since specificity is predominantly determined by the diagnostic assay applied, we assume that this parameter is comparable for the sampling method. Moreover, potentially decreased specificity could still be addressed by confirming positivity via professional sample collections.
Conclusion
In outpatients presenting with an acute moderate clinical disease, self-collected samples based on combined oral and nasal swabs for the detection of SARS-CoV-2 by RT-PCR offer similar sensitivity compared to professionally collected oro-nasopharyngeal samples, if symptom duration does not exceed one week and operational errors are minimized using comprehensive instructions. Self-sampling could thus facilitate up-scaling of clinical diagnostic testing for symptomatic outpatients at the primary level of health care through more efficient use of human and material resources, and reduce transmission risks during sample collection and attending test sites.
Acknowledgments
Acknowledgements
Mia Wintel, Julia Macos
Author contributions to be completed
MG, AKL, UP, FPM, FH, designed the study and developed standard operating procedures. MG, UP, SB, NK, CH, FK, ON implemented the study design, enrolled patients. MG led the writing of the manuscript. EK, JM, AN were responsible for PCR testing and contributed to the interpretation of the data. FPM, MG, HR and JS coordinated and supervised the outpatient-testing center. CR, SB, FK, UP, NK and MG enrolled patients. WvL led the data analysis. All authors have reviewed the manuscript.
Data availability
All raw data and analysis code are available upon a request to the corresponding author.
Conflict of interest
None declared.
Support statement
The study was supported by Charité Universitaetsmedizin and the Senate of Berlin
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ijid.2021.07.047.
Appendix. Supplementary materials
References
- Centers for Disease Control and Prevention. Interim Guidelines for Collecting and Handling of Clinical Specimens for COVID-19 Testing; 2021. Available from: https://www.cdc.gov/coronavirus/2019-nCoV/lab/guidelines-clinical-specimens.html#handling-specimens-safely. [Accessed March 31, 2021].
- Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DK, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Gonzalez M, Agullo V, de la Rica A, Infante A, Carvajal M, Garcia JA, et al. Performance of saliva specimens for the molecular detection of SARS-CoV-2 in the community setting: does sample collection method matter? Journal of clinical microbiology. 2021 doi: 10.1128/JCM.03033-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldfarb DM, Tilley P, Al-Rawahi GN, Srigley JA, Ford G, Pedersen H, et al. Self-collected Saline Gargle Samples as an Alternative to Healthcare Worker Collected Nasopharyngeal Swabs for COVID-19 Diagnosis in Outpatients. Journal of clinical microbiology. 2021 doi: 10.1128/JCM.02427-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TC, Biele G, Mühlemann B, Veith T, Schneider J, Beheim-Schwarzbach J, et al. Estimating infectiousness throughout SARS-CoV-2 infection course. Science. 2021 doi: 10.1126/science.abi5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner S, Kraemer KM, Schulze M, Pauly D, Jacob D, Gessler F, et al. Pentaplexed Quantitative Real-Time PCR Assay for the Simultaneous Detection and Quantification of Botulinum Neurotoxin-Producing Clostridia in Food and Clinical Samples. Applied and Environmental Microbiology. 2010;76(13):4387–4395. doi: 10.1128/AEM.02490-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima N, Turner F, Slepnev V, Bacelar A, Deming L, Kodeboyina S, et al. Self-Collected Oral Fluid and Nasal Swab Specimens Demonstrate Comparable Sensitivity to Clinician-Collected Nasopharyngeal Swab Specimens for the Detection of SARS-CoV-2. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2020 doi: 10.1093/cid/ciaa1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larremore DB, Wilder B, Lester E, Shehata S, Burke JM, Hay JA, et al. Test sensitivity is secondary to frequency and turnaround time for COVID-19 screening. Sci Adv. 2021;7(1) doi: 10.1126/sciadv.abd5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RA, Herigon JC, Benedetti A, Pollock NR, Denkinger CM. Performance of Saliva, Oropharyngeal Swabs, and Nasal Swabs for SARS-CoV-2 Molecular Detection: A Systematic Review and Meta-analysis. Journal of clinical microbiology. 2021 doi: 10.1128/JCM.02881-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Kim T, Lee E, Lee C, Kim H, Rhee H, et al. Clinical Course and Molecular Viral Shedding Among Asymptomatic and Symptomatic Patients With SARS-CoV-2 Infection in a Community Treatment Center in the Republic of Korea. JAMA Internal Medicine. 2020;180(11):1447–1452. doi: 10.1001/jamainternmed.2020.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescure FX, Bouadma L, Nguyen D, Parisey M, Wicky PH, Behillil S, et al. Clinical and virological data of the first cases of COVID-19 in Europe: a case series. Lancet Infect Dis. 2020;20(6):697–706. doi: 10.1016/S1473-3099(20)30200-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindner AK, Nikolai O, Kausch F, Wintel M, Hommes F, Gertler M, et al. Head-to-head comparison of SARS-CoV-2 antigen-detecting rapid test with self-collected anterior nasal swab versus professionally collected nasopharyngeal swab. The European respiratory journal. 2020 doi: 10.1183/13993003.03961-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maechler F, Gertler M, Hermes J, van Loon W, Schwab F, Piening B, et al. Epidemiological and clinical characteristics of SARS-CoV-2 infections at a testing site in Berlin, Germany, March and April 2020-a cross-sectional study. Clin Microbiol Infect. 2020;26(12) doi: 10.1016/j.cmi.2020.08.017. 1685.e7-.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty FM, Chen K, Verrill KA. How to Obtain a Nasopharyngeal Swab Specimen. New England Journal of Medicine. 2020;382(22) doi: 10.1056/NEJMvcm2010260. [DOI] [PubMed] [Google Scholar]
- McCulloch DJ, Kim AE, Wilcox NC, Logue JK, Greninger AL, Englund JA, et al. Comparison of Unsupervised Home Self-collected Midnasal Swabs With Clinician-Collected Nasopharyngeal Swabs for Detection of SARS-CoV-2 Infection. Jama Network Open. 2020;3(7) doi: 10.1001/jamanetworkopen.2020.16382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel D, Danzer KM, Gross R, Conzelmann C, Muller JA, Freischmidt A, et al. Rapid, convenient and efficient kit-independent detection of SARS-CoV-2 RNA. J Virol Methods. 2020;286 doi: 10.1016/j.jviromet.2020.113965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migueres M, Mengelle C, Dimeglio C, Didier A, Alvarez M, Delotel P, et al. Saliva sampling for diagnosing SARS-CoV-2 infections in symptomatic patients and asymptomatic carriers. Journal of Clinical Virology. 2020;130 doi: 10.1016/j.jcv.2020.104580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Zhang D, Yang P, Poon LLM, Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020;20(4):411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perchetti GA, Pepper G, Shrestha L, LaTurner K, Yae Kim D, Huang ML, et al. Performance characteristics of the Abbott Alinity m SARS-CoV-2 assay. J Clin Virol. 2021;140 doi: 10.1016/j.jcv.2021.104869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pujadas E, Chaudhry F, McBride R, Richter F, Zhao S, Wajnberg A, et al. SARS-CoV-2 viral load predicts COVID-19 mortality. Lancet Respir Med. 2020;8(9):e70. doi: 10.1016/S2213-2600(20)30354-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai P, Kumar BK, Deekshit VK, Karunasagar I, Karunasagar I. Detection technologies and recent developments in the diagnosis of COVID-19 infection. Applied Microbiology and Biotechnology. 2021;105(2):441–455. doi: 10.1007/s00253-020-11061-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert Koch Institute. Hinweise zur Testung von Patienten auf Infektion mit dem neuartigen Coronavirus SARS-CoV-2; 2021. Available from: https://www.rki.de/DE/Content/InfAZ/N/Neuartiges_Coronavirus/Vorl_Testung_nCoV.html;jsessionid=CD421678C21E595557DF2DB07E1E590A.internet072?nn=13490888#doc13490982bodyText1. [Accessed March 31, 2021 2021].
- Tu Y-P, Jennings R, Berke EM. Swabs Collected by Patients or Health Care Workers for SARS-CoV-2 Testing. New England Journal of Medicine. 2020;383(5):494–496. doi: 10.1056/NEJMc2016321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu YP, Jennings R, Hart B, Cangelosi GA, Wood RC, Wehber K, et al. Patient-collected tongue, nasal, and mid-turbinate swabs for SARS-CoV-2 yield equivalent sensitivity to health care worker collected nasopharyngeal swabs. 2020.
- Wehrhahn MC, Robson J, Brown S, Bursle E, Byrne S, New D, et al. Self-collection: An appropriate alternative during the SARS-CoV-2 pandemic. Journal of clinical virology: the official publication of the Pan American Society for Clinical Virology. 2020;128 doi: 10.1016/j.jcv.2020.104417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woelfel R, Corman VM, Guggemos W, Seilmaier M, Zange S, Mueller MA, et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581(7809) doi: 10.1038/s41586-020-2196-x. 465-+ [DOI] [PubMed] [Google Scholar]
- World Health Organization . World Health Organization; Geneva: 2021. COVID-19 strategic preparedness and response plan: 1 February 2021 to 31 January 2022. [Google Scholar]
- Wyllie AL, Fournier J, Casanovas-Massana A, Campbell M, Tokuyama M, Vijayakumar P, et al. Saliva or Nasopharyngeal Swab Specimens for Detection of SARS-CoV-2. New England Journal of Medicine. 2020;383(13):1283–1286. doi: 10.1056/NEJMc2016359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Fan J, Yu F, Feng B, Lou B, Zou Q, et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January-March 2020: retrospective cohort study. BMJ. 2020;369:m1443. doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N, Wong PK. Advances in Viral Diagnostic Technologies for Combating COVID-19 and Future Pandemics. Slas Technology. 2020;25(6):513–521. doi: 10.1177/2472630320953798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. New England Journal of Medicine. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, et al. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. New England Journal of Medicine. 2020;382(12):1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data and analysis code are available upon a request to the corresponding author.