Abstract
Purpose
To systematically review and evaluate the methodological quality of studies using radiomics for diagnostic and predictive purposes in patients with intracranial meningioma. To perform a meta-analysis of machine learning studies for the prediction of intracranial meningioma grading from pre-operative brain MRI.
Methods
Articles published from the year 2000 on radiomics and machine learning applications in brain imaging of meningioma patients were included. Their methodological quality was assessed by three readers with the radiomics quality score, using the intra-class correlation coefficient (ICC) to evaluate inter-reader reproducibility. A meta-analysis of machine learning studies for the preoperative evaluation of meningioma grading was performed and their risk of bias was assessed with the Quality Assessment of Diagnostic Accuracy Studies tool.
Results
In all, 23 studies were included in the systematic review, 8 of which were suitable for the meta-analysis. Total (possible range, −8 to 36) and percentage radiomics quality scores were respectively 6.96 ± 4.86 and 19 ± 13% with a moderate to good inter-reader reproducibility (ICC = 0.75, 95% confidence intervals, 95%CI = 0.54–0.88). The meta-analysis showed an overall AUC of 0.88 (95%CI = 0.84–0.93) with a standard error of 0.02.
Conclusions
Machine learning and radiomics have been proposed for multiple applications in the imaging of meningiomas, with promising results for preoperative lesion grading. However, future studies with adequate standardization and higher methodological quality are required prior to their introduction in clinical practice.
Supplementary Information
The online version contains supplementary material available at 10.1007/s00234-021-02668-0.
Keywords: Systematic review, Meta-analysis, Machine learning, Meningioma, Magnetic resonance imaging
Introduction
Meningiomas are the most common primary intracranial tumor in adults, being more frequent in middle-aged women [1]. The average age-adjusted yearly incidence rate is 7.86 cases per 100.000 individuals, which has increased during the past 30 years due to the improvement of diagnostic imaging [2]. Magnetic resonance imaging (MRI) is the modality of choice for their radiological diagnosis and follow-up, whereas computed tomography (CT) is used when patients cannot undergo MRI. The World Health Organization (WHO) classification of central nervous system tumors of 2016 grades meningiomas into three groups: grade I (slowly growing tumors), grade II (atypical meningioma), and grade III (anaplastic or malignant meningioma) [3]. Among these, grade II and III meningiomas are associated with high rates of recurrence and premature mortality [4]. Although conventional imaging is usually reliable for meningioma evaluation, it still presents some limitations, in particular in determining pathological grading from preoperative scans [5].
The term radiomics includes different quantitative radiological image analysis techniques, ranging from first order statistics to texture analysis [6]. These produce large amounts of data that can be challenging to process with classical statistical methods but may contribute novel imaging biomarkers. Machine learning (ML), a subfield of artificial intelligence, has seen growing interest in medicine and especially in radiology for numerous applications [7–10]. In particular, supervised learning, based on labeling of data by an expert, is mainly employed for classification and regression tasks. Among the promises of ML for clinical practice, there are automatic detection and characterization of lesions and the possibility to predict response to therapy and risk of recurrence [11–13]. Regarding neuroradiology, it has shown good results in different applications, especially in the field of neuro-oncology [14–16]. In recent years, the number of investigations based on these techniques published allows for data pooling potentially achieving higher levels of evidence through systematic reviews and/or meta-analyses.
Aim of this systematic review is to analyze the methodological quality of prospective and retrospective studies published on radiomics analyses of intracranial meningiomas. Furthermore, a meta-analysis of those employing ML algorithms for the MRI preoperative assessment of meningioma grading has been performed.
Materials and methods
Literature search
The PRISMA-DTA (Preferred Reporting Items for Systematic Reviews and Meta-analysis for Diagnostic Test Accuracy) statement was used for this systematic review [17]. Primary publications in English using radiomics and/or ML in MRI exams of meningioma patients, published between 01/01/2000 and 30/06/2020, were searched for on multiple electronic databases (PubMed, Scopus, and Web of Science). The search terms consisted of machine learning OR artificial intelligence OR radiomics OR texture AND meningioma; the detailed search string is presented in the supplementary materials.
Two researchers determined the eligibility of the articles though title and abstract evaluation. Case reports, non-original investigations (e.g., editorials, letters, reviews), and studies not focused on the topic of interest were excluded. The full text of articles in which radiomics was employed on CT or MRI images of intracranial meningiomas were obtained for further evaluation. The reference lists of included studies were also screened for potentially eligible articles and those evaluating the grading of meningioma through ML were selected to perform a meta-analysis.
Data collection and study evaluation
The radiomics quality score (RQS) was used to evaluate the methodological quality of the studies included in the systematic review whereas the Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) was used to assess the risk of bias of the studies included in the meta-analysis [18, 19]. For studies included in the meta-analysis, the predictive accuracy was quantified using the AUC for the receiver operator characteristic (ROC) analysis [20]. The number of low (grade I) and high (grade II–III) lesions used to test the model, the source of the dataset, MRI sequences employed to extract the features, ML algorithm, and type of validation were also recorded.
The RQS is a tool developed to assess the methodological quality of studies using radiomics. It evaluates image acquisition, radiomics features extraction, data modeling, model validation, and data sharing. Each of the 16 items it comprises is rated, and the summed total score ranges from −8 to 36, converted to a percentage score where −8 to 0 is defined as 0% and 36 as 100% [18] (Table 1). Three readers with previous experience in radiomics independently assigned an RQS score to each article included in the systematic review.
Table 1.
Overview of radiomics quality score items and mode of the respective scores in the reviewed studies
| RQS checkpoint | RQS item number and name | Description and (points) | Mode |
|---|---|---|---|
| First | Item 1: image protocol quality | Well-documented protocol (+1) AND/OR publicly available protocol (+1) | 1 |
| Second | Item 2: multiple segmentation | Testing feature robustness to segmentation variability, e.g., different physicians/algorithms/software (+1) | 0 |
| Item 3: phantom study | Testing feature robustness to scanner variability, e.g., different vendors/scanners (+1) | 0 | |
| Item 4: multiple time points | Testing feature robustness to temporal variability, e.g., organ movement/expansion/shrinkage (+1) | 0 | |
| Third | Item 5: feature reduction | Either feature reduction OR adjustment for multiple testing is implemented (+ 3); otherwise, (−3) | 3 |
| Item 6: multivariable analysis | Non-radiomic feature are included in/considered for model building (+1) | 0 | |
| Item 7: Biological correlates | Detecting and discussing correlation of biology and radiomic features (+1) | 0 | |
| Item 8: cut-off analysis | Determining risk groups by either median, pre-defined cut-off, or continuous risk variable (+1) | 0 | |
| Item 9: discrimination statistics | Discrimination statistic and its statistical significance are reported (+ 1); a resampling technique is also applied (+1) | 2 | |
| Item 10: calibration statistics | Calibration statistic and its statistical significance are reported (+ 1); a resampling technique is also applied (+1) | 0 | |
| Item 11: prospective design | Prospective validation of a radiomics signature in an appropriate trial (+7) | 0 | |
| Item 12: validation | Validation is missing (−5) OR internal validation (+2) OR external validation on single dataset from one institute (+3) OR external validation on two datasets from two distinct institutes (+4) OR validation of a previously published signature (+4) validation is based on three or more datasets from distinct institutes (+5) | 2 | |
| Item 13: comparison to “gold standard” | Evaluating model’s agreement with/superiority to the current “gold standard” (+2) | 0 | |
| Item 14: potential clinical application | Discussing model applicability in a clinical setting (+2). | 2 | |
| Item 15: cost-effectiveness analysis | Performing a cost-effectiveness of the clinical application (+1) | 0 | |
| Item 16: open science and data | Open source scans (+1) AND/OR open source segmentations (+1) AND/OR open source code (+1) AND/OR open source representative features and segmentations (+1) | 0 |
RQS radiomics quality score
The QUADAS-2 evaluates the risk of bias in different domains (“patient selection,” “index test,” “reference standard,” and “flow and timing”) and can be personalized according to the specific research question [21]. It was assessed in consensus by two readers for each of the studies selected for the meta-analysis.
Statistical analysis
Continuous variables are presented as mean ± standard deviation. Following previous experiences both with RQS and other scoring systems [22, 23], inter-reader reproducibility was evaluated by calculating the intraclass correlation coefficient (ICC) for the total RQS score obtained by each study. In accordance with recent guidelines, a two-way, random-effects, single-rater, absolute agreement ICC model was used [24]. For the remaining descriptive statistics, RQS score assigned by the most expert reader is reported.
Regarding the meta-analysis, the AUC standard error was calculated from the total number of positive and negative meningiomas patients. The I2 value was used to assess statistical heterogeneity, providing an estimate of the percentage of variability among included studies. I2 values of 0–25%, 25–50%, 50–75%, and >75% represent very low, low, medium, and high heterogeneity, respectively. The I2 statistic describes the percentage of variation across studies that is due to heterogeneity rather than chance [25]. I2 was calculated as follows: I2 = 100% × (Q − df)/Q. The weight of each study was calculated with the inverse variance method [26]. The results from all included studies were pooled, and an overall estimate of effect size was evaluated using a random effect model. This approach helped in reducing heterogeneity among studies. Publication bias was examined using the effective sample size funnel plot described by Egger et al. [27]. Two-sided p values ≤ 0.05 were considered statistically significant.
The described statistical analyses were performed using R (v3.6.2, “irr” and “auctestr” packages) and MedCalc Statistical Software (version 16.4.3, Ostend, Belgium; https://www.medcalc.org) [28].
Results
Literature search
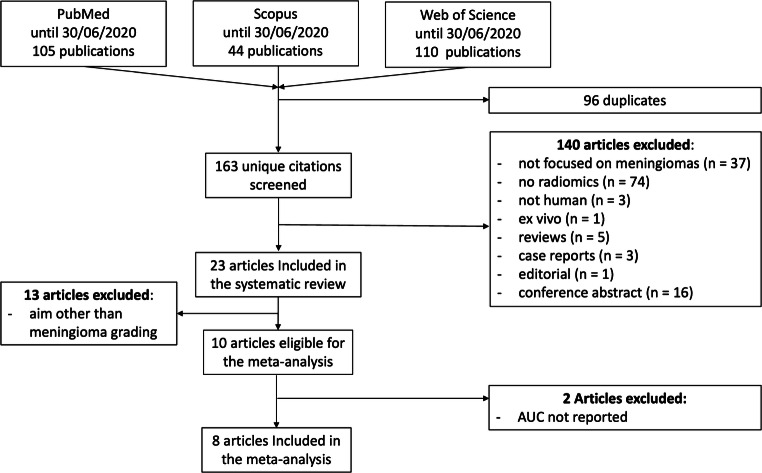
In total, 256 articles were obtained from the initial search, of which 96 were duplicates. Of the remaining 163, 140 were rejected based on the selection criteria. Finally, 23 articles were included in the systematic review, 8 of which were eligible for the meta-analysis. The described flowchart is represented in Fig. 1, whereas Table 2 contains details on study aim, ML method, and performance.
Fig. 1.
Study selection process flowchart
Table 2.
Overview of study aim, ML method, and performance for the included studies
| Authors | Study aim | ML methodology | Performance |
|---|---|---|---|
| AlKubeyyer et al. 2020 [29] | Development of a computer-aided detection of the meningioma tumor firmness |
• Support vector machine • k-nearest neighbor |
• F-score=0.95 • Balanced accuracy= 0.87 • AUC=0.87 |
| Arokia Jesu Prabhu et al. 2018 [30] | Automatic classification of parasagittal meningioma | Support vector machine | Accuracy= 0.92 |
| Chen et al. 2019 [31] | Automatic classification of meningiomas |
• Linear discriminant analysis • Support vector machine |
Accuracy=0.76 |
| Chu et al. 2020 [11] | Prediction of meningiomas grade | Logistic regression |
• Accuracy= 0.95 (training group) and 0.93 (test group) • Sensitivity= 0.94 training group) and 0.92 (test group) |
| Florez et al. 2018 [32] | Differentiation of vasogenic from tumor cell infiltration edema for radiotherapy | Linear regression | AUC>0.71 |
| Hamerla et al. 2019 [33] | Differentiation of low grade from high grade meningioma |
• Random forest • Extreme gradient boosting • Support vector machine • Multilayer perceptron |
AUC= 0.97 (Extreme gradient boosting) |
| Kanazawa et al. 2018 [34] | Distinction of solitary fibrous tumor/hemangiopericytoma from angiomatous meningioma | Texture analysis |
• Positive predictive value=0.63 • Specificity=0.63 |
| Ke et al. 2019 [35] | Differentiation between benign and non-benign meningiomas | • Support vector machine |
• AUC= 0.91 • Accuracy= 0.89 • Sensibility=0.93 • Specificity=0.87 |
| Laukamp et al. 2018 [36] | Automatic detection and segmentation of meningioma | Deep learning |
• Detection accuracy=0.98 • Mean Dice coefficient for total tumor volume =0.81 ± 0.10 |
| Laukamp et al. 2019 [37] | Prediction of meningioma grade | Multivariate logistic regression model | AUC=0.91 |
| Li et al. 2019 [38] | Automatic differentiation of malignant hemangiopericytoma from angiomatous meningioma | Texture analysis | AUC=0.90 |
| Lu et al. 2018 [39] | Prediction of meningioma grade using ADC maps |
• Classic decision tree • Conditional inference • Decision forest |
Accuracy= 0.62 |
| Morin et al. 2019 [40] | Prediction of meningioma grade, local failure and overall survival | Random forest |
• Grade= Accuracy 0.65; AUC 0.71 • Local Failure= Accuracy 0.61, AUC=0.68 • Overall Survival= accuracy 0.67, AUC= 0.75 |
| Niu et al. 2019 [41] | Differentiation of meningioma subtypes | Fisher discriminant analysis | Accuracy= 0.99-0.1 |
| Park et al. 2018 [42] | Prediction of grade and histological subtype |
• Support vector machine • Random forest |
AUC= 0.86 |
| Speckter et al. 2018 [13] | Prediction of response after radiosurgery | Texture analysis | Correlation coefficient=−0.64 |
| Tian et al. 2020 [43] | Contrastive analysis between craniopharyngioma and meningioma | Binary logistic regression | AUC>0.70 |
| Wei et al. 2020 [44] | Differentiation of hemangiopericytoma from meningioma | Logistic regression model | AUC= 0.92–0.99 |
| Yan et al. 2017 [45] | Prediction of meningioma grade |
• Logistic regression • Naïve Bayes • Support vector machine |
• AUC= 0.73–0.88 • Sensitivity= 0.48–0.91 • Specificity= 0.70–0.96 |
| Zhang et al. 2019 [12] | Prediction of recurrence in skull base meningiomas | Random forest | Accuracy= 0.90 |
| Zhang et al. 2020 [46] | Discrimination of lesions located in the anterior skull base |
• Linear discriminant analysis • Support vector machine • Random forest • Adaboos • K-nearest neighbor • GaussianNB • Logistic regression • gradient • boosting decision tree • Decision tree |
AUC>0.80 |
| Zhu et al. 2019 [47] | Automatic prediction of meningioma grade | Convolutional neural network | AUC= 0.83 |
| Zhu et al. 2019 [48] | Automatic prediction of meningioma grade | Deep learning |
• AUC= 0.81 • Sensitivity= 0.8 • Specificity=0.9 |
AUC area under the receiver operating characteristic curve
Study evaluation
The RQS total and percentage scores were respectively 6.96 ± 4.86 and 19 ± 13% (Figs. 2, 3). A detailed report of the RQS item score by the most expert reader is shown in Table 3. Inter-reader reproducibility resulted moderate to good, with an ICC = 0.75 (95% confidence intervals, 95% CI = 0.54–0.88). RQS scores assigned by the other readers are presented in the supplementary materials.
Fig. 2.
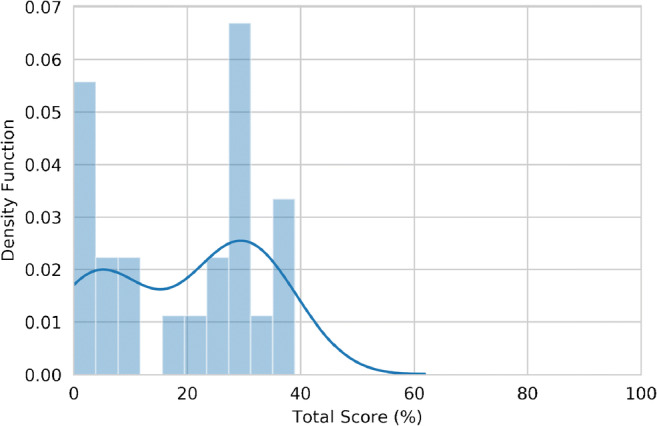
Histogram (bars, bin number = 10) and kernel density estimation (line) plot of RQS percentage score distribution
Fig. 3.
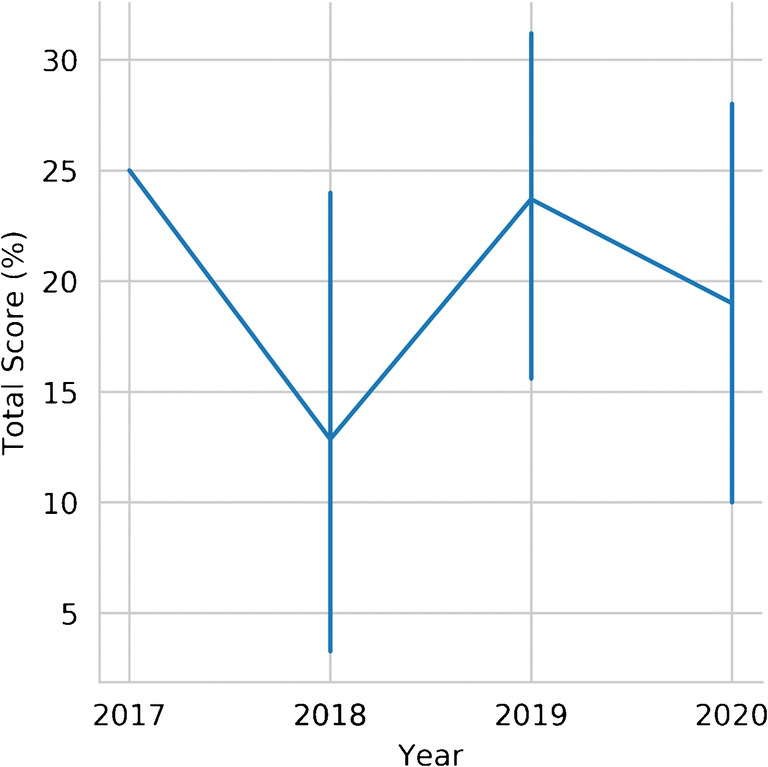
RQS percentage score line plot in relation to publication year. Bars represent 95% confidence intervals, calculated with bootstrapping (1000 iterations)
Table 3.
Radiomics quality scores for all included articles
| First author | Year | Item 1 | Item 2 | Item 3 | Item 4 | Item 5 | Item 6 | Item 7 | Item 8 | Item 9 | Item 10 | Item 11 | Item 12 | Item 13 | Item 14 | Item 15 | Item 16 | RQS (total) | RQS (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alkubeyyer | 2020 | 0 | 0 | 0 | 0 | −3 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 3 | 8 |
| Arokia Jesu Prabhu | 2018 | 0 | 0 | 0 | 0 | −3 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 |
| Chen | 2019 | 1 | 0 | 0 | 0 | 3 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 10 | 28 |
| Chu | 2020 | 1 | 0 | 0 | 0 | 3 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 10 | 28 |
| Florez | 2018 | 1 | 1 | 0 | 0 | 3 | 0 | 0 | 0 | 1 | 0 | 0 | −5 | 0 | 0 | 0 | 0 | 1 | 3 |
| Hamerla | 2019 | 1 | 0 | 0 | 0 | 3 | 0 | 1 | 0 | 2 | 0 | 0 | 5 | 0 | 2 | 0 | 0 | 14 | 39 |
| Kanazawa | 2018 | 1 | 0 | 0 | 0 | −3 | 0 | 1 | 1 | 1 | 0 | 0 | −5 | 0 | 2 | 0 | 0 | 0 | 0 |
| Ke | 2019 | 1 | 0 | 0 | 0 | 3 | 0 | 1 | 0 | 1 | 0 | 0 | 3 | 0 | 2 | 0 | 0 | 11 | 31 |
| Laukamp | 2018 | 1 | 0 | 0 | 0 | −3 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 2 | 2 | 0 | 0 | 6 | 17 |
| Laukamp | 2019 | 1 | 0 | 0 | 0 | 3 | 0 | 1 | 0 | 1 | 0 | 0 | −5 | 0 | 2 | 0 | 0 | 3 | 8 |
| Li | 2019 | 1 | 0 | 0 | 0 | 3 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 2 | 2 | 0 | 0 | 12 | 33 |
| Lu | 2018 | 1 | 0 | 0 | 0 | 3 | 1 | 1 | 0 | 1 | 0 | 0 | 2 | 2 | 2 | 0 | 0 | 13 | 36 |
| Morin | 2019 | 0 | 0 | 0 | 0 | 3 | 1 | 1 | 0 | 1 | 0 | 0 | 3 | 2 | 2 | 0 | 0 | 13 | 36 |
| Niu | 2019 | 1 | 0 | 0 | 0 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 9 | 25 |
| Park | 2018 | 1 | 0 | 0 | 0 | 3 | 0 | 1 | 0 | 2 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 11 | 31 |
| Speckter | 2018 | 0 | 0 | 0 | 0 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | −5 | 0 | 2 | 0 | 0 | 1 | 3 |
| Tian | 2020 | 0 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 2 | 0 | 0 | −5 | 0 | 2 | 0 | 0 | 2 | 6 |
| Wei | 2020 | 1 | 1 | 0 | 1 | 3 | 0 | 0 | 0 | 2 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 11 | 31 |
| Yan | 2017 | 1 | 0 | 0 | 0 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 9 | 25 |
| Zhang | 2019 | 1 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | −5 | 0 | 2 | 0 | 0 | 1 | 3 |
| Zhang | 2020 | 1 | 0 | 0 | 0 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 8 | 22 |
| Zhu H | 2019 | 0 | 0 | 0 | 0 | −3 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 2 | 6 |
| Zhu Y | 2019 | 1 | 0 | 0 | 0 | 3 | 0 | 1 | 0 | 1 | 0 | 0 | 2 | 0 | 2 | 0 | 0 | 10 | 28 |
RQS radiomics quality score
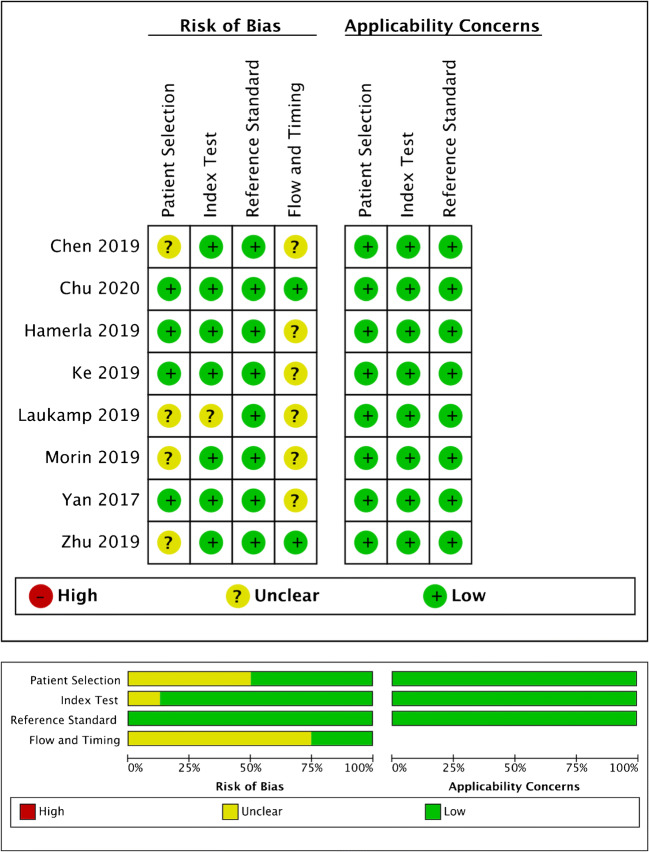
Regarding the evaluation of the risk of bias through the QUADAS-2, the number of studies with high, unclear, and low risk of bias was respectively 0, 7, and 2, for the four domains (patient selection, index test, reference standard, and flow and timing) (Fig. 4). In particular, 4 studies scored an unclear risk of bias in the patient selection domain as the authors did not clearly report the steps of patient selection process [31, 37, 40, 47]. One study scored an unclear risk of bias in index test domain as the radiomics feature extraction was performed from both diffusion-weighted images (DWI) and apparent diffusion coefficient (ADC) maps [37]. Finally, the time elapsed between MRI and neurosurgery was not reported in 6 studies, thus scoring an unclear risk of bias in the flow and timing domain [31–45, 49]. All the studies included in the meta-analysis had low concerns regarding applicability for the three domains (patient selection, index test, and reference standard).
Fig. 4.
Methodological quality of the studies included in the meta-analysis according to the QUADAS 2 tool for risk of bias and applicability concerns. Green, yellow, and red circles represent low, unclear, and high risk of bias, respectively
Meta-analysis
The articles included in the meta-analysis are reported in Table 4. The ML models for meningioma characterization showed an overall pooled AUC = 0.88 (95% CI = 0.84–0.93) with a standard error of 0.02 (Figs. 5 and 6). Study heterogeneity was 82.5% (p < 0.001).
Table 4.
Characteristics of the studies included in the meta-analysis
| Paper | AUC | Low grade | High grade | Data source | Sequences | Model | Validation |
|---|---|---|---|---|---|---|---|
| Chen et al. | 0.93 | 12 | 18 | Single institution | CE T1 | LDA | CV |
| Chu et al. | 0.95 | 24 | 4 | Single institution | CE T1 | Logistic regression | Test set |
| Hamerla et al. | 0.97 | 102 | 45 | Multicenter | CE T1+others | XGBoost | CV |
| Ke et al. | 0.83 | 60 | 19 | Multicenter | CE T1+others | SVM | CV + test set |
| Laukamp et al. | 0.91 | 46 | 25 | Multicenter | CE T1+others | Logistic regression | CV |
| Morin et al. | 0.78 | 67 | 18 | Multicenter | CE T1 | RF | Test set |
| Yan et al. | 0.87 | 110 | 21 | Single institution | CE T1 | SVM | CV |
| Zhu et al. | 0.82 | 69 | 13 | Single institution | CE T1 | LDA | CV + test set |
AUC area under the receiver operating characteristic curve, CE T1 contrast-enhanced T1-weighted sequence, LDA linear discriminant analysis, SVM support vector machine, RF random forest, CV cross validation
Fig. 5.
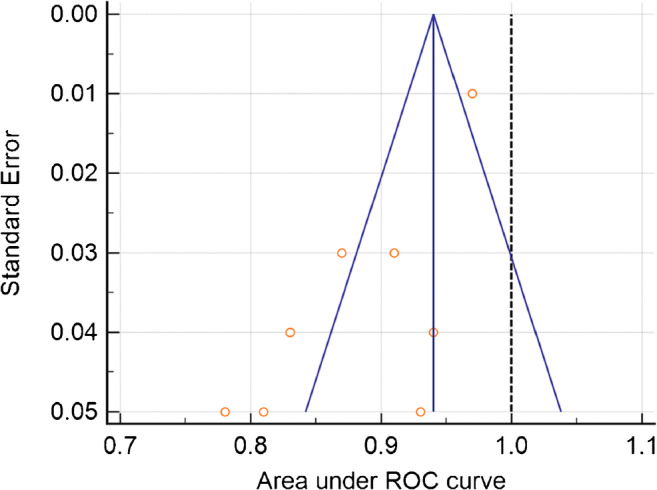
Funnel plot asymmetry test for publication bias in the literature evaluation for high-grade meningioma characterization
Fig. 6.
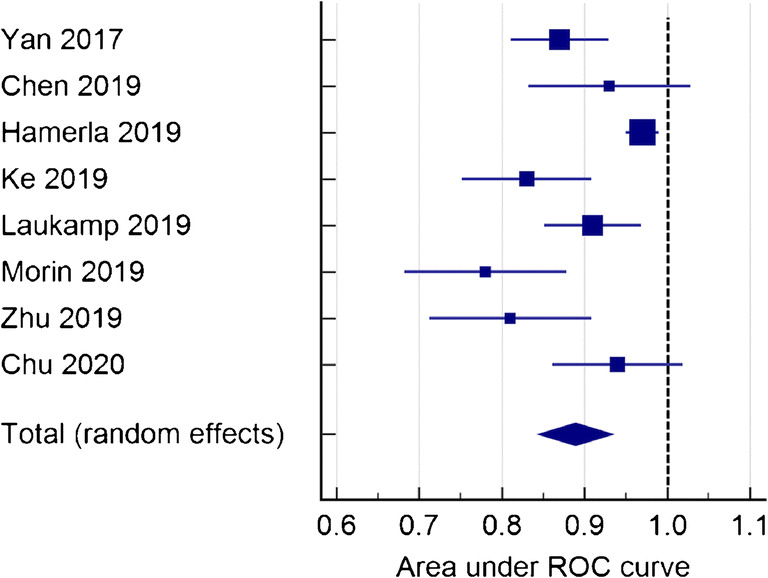
Forest plot of single studies for the pooled area under the curve (AUC) and 95% CI of high-grade meningioma characterization
Subgroup analysis was performed to compare studies evaluating the performance of ML for meningioma characterization using patients from a single institution (n = 4) and from multiple centers (n = 4). The pooled AUC was 0.88 (95% CI = 0.84–0.92), standard error 0.02, and heterogeneity 42.17% (p < 0.001) in the single institution group and the pooled AUC was 0.88 (95% CI = 0.81–0.95), standard error 0.03, and heterogeneity 88.60% (p < 0.001) in the multi-center group.
Of the included studies, 5 used only post-contrast T1-weighted. Their pooled AUC was 0.87 (95% CI = 0.82–0.92), standard error 0.02, and heterogeneity 56.34% (p = 0.05). On the other hand, 3 studies also used conventional MR sequences, including T1-weighted and T2-weighted imaging, in addition to contrast-enhanced T1-weighted imaging. Their pooled AUC was 0.91 (95% CI = 0.85–0.97), standard error 0.03, and heterogeneity 85.94% (p < 0.001).
In a subgroup analysis based on pre-processing image type, the pooled AUC of 6 studies included in the analysis was 0.89 (95% CI = 0.85–0.94), standard error 0.02, and heterogeneity 83.01% (p < 0.001). The remaining studies reported an AUC value respectively of 0.93 and 0.78.
Four studies applied exclusively k-fold cross-validation for training and testing of the model. Their pooled AUC was 0.92 (95% CI = 0.88–0.97), standard error 0.02, and heterogeneity 76.52% (p = 0.005). The remaining studies (n = 4) employed a test set, in 2 cases paired with k-fold cross-validation. Their pooled AUC was 0.84 (95% CI = 0.78–0.90), standard error 0.03, and heterogeneity 62.09% (p < 0.005). The corresponding plots of subgroup analyses are presented in the supplementary materials.
Discussion
Radiomics has numerous potential applications in neuroradiology and could help in obtaining additional quantitative information from routine medical images. Even though there are ongoing efforts to standardize radiomic feature extraction, their use is not yet justified outside of the research field [50]. The RQS is a recently introduced score whose aim is to evaluate the methodological quality of radiomics-based investigations. It could help identifying high-quality results among the large number of publications in this field as well as issues limiting their value and applicability. The average RQS of the articles included in our systematic review was low (6.96, 19%), reflecting a lacking overall methodological quality. This finding is in line with previous systematic reviews performing a quality assessment with the RQS tool in other fields of radiology. In detail, Ursprung et al reported a total RQS score of 3.41 ± 4.43 (9.4% average) in a review of renal cell carcinoma radiomics CT and MRI studies, Stanzione et al 7.93 ± 5.13 (23 ± 13%) for prostate MRI, and Granzier et al 20.9% for breast MRI [22, 51, 52]. Therefore, the problems affecting radiomics studies and limiting the RQS score seem to be general and not restricted to a specific application. The current situation can be at least in part explained by an exponential growth in interest and number of papers submitted using radiomics, a dynamic also experienced in the wider field of ML [7]. On the other hand, the RQS scoring system is relatively new and has been used in a limited number of occasions [18, 22, 51–53]. Therefore, further revisions and improvements after initial feedback may produce a different weighting of each item and/or modifications in the items themselves. In our review, we wish to highlight that all studies collected 0 points on items 3, 4, 10, 11, and 15. In detail, feature robustness to scanner or temporal variability was never tested, also due to the retrospective nature of all the investigations. Similarly, a prospective validation of the radiomics signature in appropriate trials was missing as well as a cost-effectiveness analysis.
Regarding the studies included in the meta-analysis, the QUADAS-2 assessment revealed an overall low risk of bias but also highlighted some critical issues. In particular, in one paper, DWI was used for feature extraction together with ADC maps [37]. As ADC maps are derived from DWI, it would be more appropriate to only use one of the two for feature extraction and probably ADC maps are preferable due to their qualitative nature. Furthermore, only two studies reported time elapsed between the MRI exam and surgery, a possible source of bias that should always be specified [11, 48] None of the articles selected scored a high risk of bias in relation to the reference standard as histopathological grading was always employed. Overall, radiomics features analyzed with a ML approach turned out to be promising for meningioma grading, with an AUC of 0.88. All the included studies used handcrafted radiomics except for Zhu et al. who employed deep learning [48]. This is understandable given that deep learning requires a large amount of data to be advantageous over other ML algorithms, often not available in this setting. Almost all studies (n = 7) performed a 3D segmentation of the lesion, though it is still not clear whether this approach is clearly better than 2D segmentation [48]. Only Morin et al. trained a model using radiomics features together with demographic data [40]. Despite this, its AUC value is among the lowest (0.78) suggesting that these may not be essential in the preoperative definition of meningioma grading. It is also interesting to note that most (n = 5) of the studies used linear ML models [11, 31, 37, 48, 49] while only one included a data augmentation technique [33].
In the subgroup analyses, AUC was higher (0.91 vs 0.87) for studies (n = 3) that paired T1 contrast-enhanced sequences with other sequences [11, 31, 40]. This finding supports the use of multiple imaging sequences rather than relying exclusively on T1 contrast-enhanced sequences for future investigations. Similarly, the good accuracy (AUC = 0.89) obtained by studies (n = 6) who included image pre-processing in their pipeline also suggests the usefulness of this step [11, 33, 37, 45, 48, 49]. While the AUC for single institution (n = 4) and multicenter studies was equally high (AUC = 0.88), external testing of ML models is always preferable to demonstrate their ability to generalize. Similarly, while k-fold cross-validation helps in extracting more information and reliable results from small datasets, its exclusive use may present some issues as there is no final model whose performance can be tested on unseen data. In all, 4 studies only employed cross-validation, with better results than the remaining (AUC = 0.92 vs 0.84) [11, 40, 48, 49]. Ideally, it would be preferable to use cross-validation for model tuning and initial testing followed by further assessment on new data, as done in 2 cases (AUC = 0.82 and 0.83). This approach combines the advantages of both testing strategies [48, 49].
As previously reported, the presentation of accuracy metrics in radiomics and ML studies is often inconsistent and incomplete [21]. Due to this situation, our meta-analysis could only employ AUC values as these were the most commonly reported. However, sensitivity and specificity analysis could have provided additional insights if feasible.
Indeed, ROC AUC treats sensitivity and specificity as equally important overall when averaged across all thresholds. For example, poor sensitivity could mean missed diagnosis and delayed treatment or even death, whereas poor specificity means unnecessary test. ROC AUC ignores clinical differentials in “misclassification cost” and, therefore, risks finding a new test worthless when patients and physicians would consider otherwise. ROC AUC weighs changes in sensitivity and specificity equally only where the curve slope equals one. Other points assign different weights, determined by curve shape and without considering clinically meaningful information, e.g., a 5 % improvement in sensitivity contributes less to AUC at high specificity than at low specificity. Thus, AUC can consider a test that increases sensitivity at low specificity superior to one that increases sensitivity at high specificity [54].
Greater care should be given in future research to avoid this issue, ideally confusion matrices should always be reported if possible.
The ability to distinguish low-grade from high-grade meningiomas based on preoperative MR images could influence personalized treatment decisions. In particular, in patients with meningiomas at certain locations where biopsy is difficult to obtain due to a high risk of mortality and morbidity (e.g., petroclival meningiomas), a tailored radiation treatment in the high-grade forms may be recommended [55]. Furthermore, in asymptomatic patients with small meningiomas, radiotherapy may be avoided for benign lesions, while high-grade meningiomas could undergo radiation treatment before resection [56]. Therefore, noninvasive MRI prediction of meningioma grading could address in the future small meningioma treatment strategy, also without histological confirmation. However, radiomics are not currently ready for clinical implementation due to the issues found in RQS.
Our study has some limitations that should be acknowledged. The RQS is relatively recent and a purely methodological scoring system and does not consider differences in study aim. Regarding the meta-analysis, a relatively low number of papers met the selection criteria. While the QUADAS-2 analysis presented some unclear elements, no high-risk sources of bias were identified. Study heterogeneity was high, but this is in line with other machine learning meta-analyses and diagnostic meta-analyses in general [21, 57, 58]. Finally, not all articles were specified if the WHO 2016 classification of central nervous system tumors was used. However, meningioma grading did not change substantially compared to the previous version, except for the introduction of brain invasion as a criterion for the diagnosis of grade II lesions [3].
In conclusion, radiomics studies show promising results for improving management of intracranial meningiomas, though they require more methodological rigor. The prediction of meningioma grading from preoperative brain MRI also demonstrated good results in our meta-analysis. Well-designed, prospective trials are necessary to demonstrate their validity and reporting of methods and results has to be standardized prior to their use in daily clinical practice.
Supplementary Information
(PNG 550 kb)
(PNG 560 kb)
(PNG 300 kb)
(PNG 545 kb)
(DOCX 22 kb)
Acknowledgements
Open access funding provided by Università degli Studi di Napoli Federico II within the CRUI-CARE Agreement.
Abbreviations
- AUC
Area under the receiver operating characteristic curve
- CI
Confidence intervals
- ICC
Inter-rater correlation coefficient
- ML
Machine learning
- QUADAS-2
Quality Assessment of Diagnostic Accuracy Studies 2 tool
- RQS
Radiomics quality score
Author contribution
Each author has contributed to all of the following areas:
- Conception and design, or acquisition of data, or analysis and interpretation of data
- Drafting the article or revising it critically for important intellectual content
- Final approval of the version to be published
- Agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved
Declarations
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Ethical approval
Ethical approval was not required for this study because the article type is a systematic review and meta-analysis.
Informed consent
Written informed consent was not required for this study because the article type is a systematic review and meta-analysis.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Buerki RA, Horbinski CM, Kruser T, Horowitz PM, James CD, Lukas RV. An overview of meningiomas. Future Oncol. 2018;14:2161–2177. doi: 10.2217/fon-2018-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thust S, Kumar A. Clinical Neuroradiology. Cham: Springer International Publishing; 2019. Extra-axial tumors; pp. 1115–1150. [Google Scholar]
- 3.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 4.Lee JH, Kim OL, Seo YB, Choi JH. Prognostic factors of atypical meningioma: overall survival rate and progression free survival rate. J Korean Neurosurg Soc. 2017;60:661–666. doi: 10.3340/jkns.2017.0303.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang RY, Bi WL, Griffith B, Kaufmann TJ, la Fougère C, Schmidt NO, Tonn JC, Vogelbaum MA, Wen PY, Aldape K, Nassiri F, Zadeh G, Dunn IF, International Consortium on Meningiomas. Aldape K, Au K, Barnhartz-Sloan J, Bi WL, Brastianos PK, Butowski N, Carlotti C, Cusimano MD, DiMeco F, Drummond K, Dunn IF, Galanis E, Giannini C, Goldbrunner R, Griffith B, Hashizume R, Hanemann CO, Herold-Mende C, Horbinski C, Huang RY, James D, Jenkinson MD, Jungk C, Kaufman TJ, Krischek B, Lachance D, Lafougère C, Lee I, Liu JC, Mamatjan Y, Mansouri A, Mawrin C, McDermott M, Munoz D, Nassiri F, Noushmehr H, Ng HK, Perry A, Pirouzmand F, Poisson LM, Pollo B, Raleigh D, Sahm F, Saladino A, Santarius T, Schichor C, Schultz D, Schmidt NO, Selman W, Sloan A, Spears J, Snyder J, Suppiah S, Tabatabai G, Tatagiba M, Tirapelli D, Tonn JC, Tsang D, Vogelbaum MA, Deimling A, Wen PY, Walbert T, Westphal M, Workewych AM, Zadeh G. Imaging and diagnostic advances for intracranial meningiomas. Neuro-Oncology. 2019;21:i44–i61. doi: 10.1093/neuonc/noy143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cuocolo R, Caruso M, Perillo T, Ugga L, Petretta M. Machine learning in oncology: a clinical appraisal. Cancer Lett. 2020;481:55–62. doi: 10.1016/j.canlet.2020.03.032. [DOI] [PubMed] [Google Scholar]
- 8.Zaharchuk G, Gong E, Wintermark M, Rubin D, Langlotz CP. Deep Learning in Neuroradiology. Am J Neuroradiol. 2018;39:1776–1784. doi: 10.3174/ajnr.A5543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cuocolo R, Perillo T, De Rosa E, et al. Current applications of big data and machine learning in cardiology. J Geriatr Cardiol. 2019;16:601–607. doi: 10.11909/j.issn.1671-5411.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niel O, Bastard P. Artificial intelligence in nephrology: core concepts, clinical applications, and perspectives. Am J Kidney Dis. 2019;74:803–810. doi: 10.1053/j.ajkd.2019.05.020. [DOI] [PubMed] [Google Scholar]
- 11.Chu H, Lin X, He J et al (2020) Value of MRI radiomics based on enhanced T1WI images in prediction of meningiomas grade. Acad Radiol 1–7. 10.1016/j.acra.2020.03.034 [DOI] [PubMed]
- 12.Zhang Y, Chen J-H, Chen T-Y, Lim SW, Wu TC, Kuo YT, Ko CC, Su MY. Radiomics approach for prediction of recurrence in skull base meningiomas. Neuroradiology. 2019;61:1355–1364. doi: 10.1007/s00234-019-02259-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Speckter H, Bido J, Hernandez G, Rivera D, Suazo L, Valenzuela S, Miches I, Oviedo J, Gonzalez C, Stoeter P. Pretreatment texture analysis of routine MR images and shape analysis of the diffusion tensor for prediction of volumetric response after radiosurgery for meningioma. J Neurosurg. 2018;129:31–37. doi: 10.3171/2018.7.GKS181327. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X, Yan L-F, Hu Y-C et al (2017) Optimizing a machine learning based glioma grading system using multi-parametric MRI histogram and texture features. Oncotarget 8. 10.18632/oncotarget.18001 [DOI] [PMC free article] [PubMed]
- 15.Lotan E, Jain R, Razavian N, Fatterpekar GM, Lui YW. State of the art: machine learning applications in glioma imaging. Am J Roentgenol. 2019;212:26–37. doi: 10.2214/AJR.18.20218. [DOI] [PubMed] [Google Scholar]
- 16.Cuocolo R, Ugga L, Solari D, Corvino S, D’Amico A, Russo D, Cappabianca P, Cavallo LM, Elefante A. Prediction of pituitary adenoma surgical consistency: radiomic data mining and machine learning on T2-weighted MRI. Neuroradiology. 2020;62:1649–1656. doi: 10.1007/s00234-020-02502-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McInnes MDF, Moher D, Thombs BD, et al. Preferred Reporting Items for a Systematic Review and Meta-analysis of Diagnostic Test Accuracy Studies. JAMA. 2018;319:388. doi: 10.1001/jama.2017.19163. [DOI] [PubMed] [Google Scholar]
- 18.Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, Sanduleanu S, Larue RTHM, Even AJG, Jochems A, van Wijk Y, Woodruff H, van Soest J, Lustberg T, Roelofs E, van Elmpt W, Dekker A, Mottaghy FM, Wildberger JE, Walsh S. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 19.Whiting PF. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 20.Steyerberg EW, Vickers AJ, Cook NR, Gerds T, Gonen M, Obuchowski N, Pencina MJ, Kattan MW. Assessing the performance of prediction models. Epidemiology. 2010;21:128–138. doi: 10.1097/EDE.0b013e3181c30fb2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cuocolo R, Cipullo MB, Stanzione A, Romeo V, Green R, Cantoni V, Ponsiglione A, Ugga L, Imbriaco M. Machine learning for the identification of clinically significant prostate cancer on MRI: a meta-analysis. Eur Radiol. 2020;30:6877–6887. doi: 10.1007/s00330-020-07027-w. [DOI] [PubMed] [Google Scholar]
- 22.Stanzione A, Gambardella M, Cuocolo R, Ponsiglione A, Romeo V, Imbriaco M. Prostate MRI radiomics: a systematic review and radiomic quality score assessment. Eur J Radiol. 2020;129:109095. doi: 10.1016/j.ejrad.2020.109095. [DOI] [PubMed] [Google Scholar]
- 23.Romeo V, Stanzione A, Cocozza S, Ugga L, Cuocolo R, Brunetti A, Bisdas S. A critical appraisal of the quality of head and neck cancer imaging guidelines using the AGREE II tool: a EuroAIM initiative. Cancer Med. 2019;8:209–215. doi: 10.1002/cam4.1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koo TK, Li MY. A guideline of selecting and reporting intraclass correlation coefficients for reliability research. J Chiropr Med. 2016;15:155–163. doi: 10.1016/j.jcm.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Higgins JPT. Measuring inconsistency in meta-analyses. BMJ. 2003;327:557–560. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cleophas TJ, Zwinderman AH. Meta-analysis. Circulation. 2007;115:2870–2875. doi: 10.1161/CIRCULATIONAHA.105.594960. [DOI] [PubMed] [Google Scholar]
- 27.Egger M, Smith GD, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.R Core Team (2020) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL http://www.R-project.org/
- 29.AlKubeyyer A, Ben Ismail MM, Bchir O, Alkubeyyer M. Automatic detection of the meningioma tumor firmness in MRI images. J Xray Sci Technol. 2020;28:659–682. doi: 10.3233/XST-200644. [DOI] [PubMed] [Google Scholar]
- 30.Arokia Jesu Prabhu L, Jayachandran A. Mixture model segmentation system for parasagittal meningioma brain tumor classification based on hybrid feature vector. J Med Syst. 2018;42:251. doi: 10.1007/s10916-018-1094-3. [DOI] [PubMed] [Google Scholar]
- 31.Chen C, Guo X, Wang J, Guo W, Ma X, Xu J. The diagnostic value of radiomics-based machine learning in predicting the grade of meningiomas using conventional magnetic resonance imaging: a preliminary study. Front Oncol. 2019;9:1–9. doi: 10.3389/fonc.2019.01338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Florez E, Nichols T, Parker E et al (2018) Multiparametric magnetic resonance imaging in the assessment of primary brain tumors through Radiomic features: a metric for guided radiation treatment planning. Cureus. 10.7759/cureus.3426 [DOI] [PMC free article] [PubMed]
- 33.Hamerla G, Meyer HJ, Schob S, Ginat DT, Altman A, Lim T, Gihr GA, Horvath-Rizea D, Hoffmann KT, Surov A. Comparison of machine learning classifiers for differentiation of grade 1 from higher gradings in meningioma: a multicenter radiomics study. Magn Reson Imaging. 2019;63:244–249. doi: 10.1016/j.mri.2019.08.011. [DOI] [PubMed] [Google Scholar]
- 34.Kanazawa T, Minami Y, Jinzaki M, et al. Preoperative prediction of solitary fibrous tumor/Hemangiopericytoma and Angiomatous meningioma using magnetic resonance imaging texture analysis. World Neurosurg. 2018;120:e1208–e1216. doi: 10.1016/j.wneu.2018.09.044. [DOI] [PubMed] [Google Scholar]
- 35.Ke C, Chen H, Lv X, et al. Differentiation between benign and nonbenign Meningiomas by using texture analysis from multiparametric MRI. J Magn Reson Imaging. 2020;51:1810–1820. doi: 10.1002/jmri.26976. [DOI] [PubMed] [Google Scholar]
- 36.Laukamp KR, Thiele F, Shakirin G, et al. Fully automated detection and segmentation of meningiomas using deep learning on routine multiparametric MRI. Eur Radiol. 2019;29:124–132. doi: 10.1007/s00330-018-5595-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laukamp KR, Shakirin G, Baeßler B, Thiele F, Zopfs D, Große Hokamp N, Timmer M, Kabbasch C, Perkuhn M, Borggrefe J. Accuracy of radiomics-based feature analysis on multiparametric magnetic resonance images for noninvasive meningioma grading. World Neurosurg. 2019;132:e366–e390. doi: 10.1016/j.wneu.2019.08.148. [DOI] [PubMed] [Google Scholar]
- 38.Li X, Lu Y, Xiong J, et al. Presurgical differentiation between malignant haemangiopericytoma and angiomatous meningioma by a radiomics approach based on texture analysis. J Neuroradiol. 2019;46:281–287. doi: 10.1016/j.neurad.2019.05.013. [DOI] [PubMed] [Google Scholar]
- 39.Lu Y, Liu L, Luan S, et al. The diagnostic value of texture analysis in predicting WHO grades of meningiomas based on ADC maps: an attempt using decision tree and decision forest. Eur Radiol. 2019;29:1318–1328. doi: 10.1007/s00330-018-5632-7. [DOI] [PubMed] [Google Scholar]
- 40.Morin O, Chen WC, Nassiri F, Susko M, Magill ST, Vasudevan HN, Wu A, Vallières M, Gennatas ED, Valdes G, Pekmezci M, Alcaide-Leon P, Choudhury A, Interian Y, Mortezavi S, Turgutlu K, Bush NAO, Solberg TD, Braunstein SE, Sneed PK, Perry A, Zadeh G, McDermott MW, Villanueva-Meyer JE, Raleigh DR. Integrated models incorporating radiologic and radiomic features predict meningioma grade, local failure, and overall survival. Neuro-Oncology Adv. 2019;1:1–15. doi: 10.1093/noajnl/vdz011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Niu L, Zhou X, Duan C, et al. Differentiation researches on the meningioma subtypes by Radiomics from contrast-enhanced magnetic resonance imaging: a preliminary study. World Neurosurg. 2019;126:e646–e652. doi: 10.1016/j.wneu.2019.02.109. [DOI] [PubMed] [Google Scholar]
- 42.Park YW, Oh J, You SC, et al. Radiomics and machine learning may accurately predict the grade and histological subtype in meningiomas using conventional and diffusion tensor imaging. Eur Radiol. 2019;29:4068–4076. doi: 10.1007/s00330-018-5830-3. [DOI] [PubMed] [Google Scholar]
- 43.Tian Z, Chen C, Zhang Y, et al. Radiomic analysis of Craniopharyngioma and meningioma in the Sellar/Parasellar area with MR images features and texture features: a feasible study. Contrast Media Mol Imaging. 2020;2020:1–9. doi: 10.1155/2020/4837156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wei J, Li L, Han Y et al (2020) Accurate preoperative distinction of intracranial Hemangiopericytoma from meningioma using a multihabitat and multisequence-based Radiomics diagnostic technique. Front Oncol 10. 10.3389/fonc.2020.00534 [DOI] [PMC free article] [PubMed]
- 45.Yan PF, Yan L, Hu TT, Xiao DD, Zhang Z, Zhao HY, Feng J. The potential value of preoperative MRI texture and shape analysis in grading meningiomas: a preliminary investigation. Transl Oncol. 2017;10:570–577. doi: 10.1016/j.tranon.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Y, Shang L, Chen C et al (2020) Machine-learning classifiers in discrimination of lesions located in the anterior Skull Base. Front Oncol 10. 10.3389/fonc.2020.00752 [DOI] [PMC free article] [PubMed]
- 47.Zhu H, Fang Q, He H, Hu J, Jiang D, Xu K. Automatic prediction of meningioma grade image based on data amplification and improved convolutional neural network. Comput Math Methods Med. 2019;2019:1–9. doi: 10.1155/2019/7289273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu Y, Man C, Gong L, Dong D, Yu X, Wang S, Fang M, Wang S, Fang X, Chen X, Tian J. A deep learning radiomics model for preoperative grading in meningioma. Eur J Radiol. 2019;116:128–134. doi: 10.1016/j.ejrad.2019.04.022. [DOI] [PubMed] [Google Scholar]
- 49.Ke C, Chen H, Lv X, Li H, Zhang Y, Chen M, Hu D, Ruan G, Zhang Y, Zhang Y, Liu L, Feng Y. Differentiation between benign and nonbenign meningiomas by using texture analysis from multiparametric MRI. J Magn Reson Imaging. 2020;51:1810–1820. doi: 10.1002/jmri.26976. [DOI] [PubMed] [Google Scholar]
- 50.Zwanenburg A, Vallières M, Abdalah MA et al (2020) The image biomarker standardization initiative: Standardized quantitative radiomics for high-throughput image-based phenotyping. Radiology 191145. 10.1148/radiol.2020191145 [DOI] [PMC free article] [PubMed]
- 51.Granzier RWY, van Nijnatten TJA, Woodruff HC, Smidt ML, Lobbes MBI. Exploring breast cancer response prediction to neoadjuvant systemic therapy using MRI-based radiomics: a systematic review. Eur J Radiol. 2019;121:108736. doi: 10.1016/j.ejrad.2019.108736. [DOI] [PubMed] [Google Scholar]
- 52.Ursprung S, Beer L, Bruining A, Woitek R, Stewart GD, Gallagher FA, Sala E. Radiomics of computed tomography and magnetic resonance imaging in renal cell carcinoma—a systematic review and meta-analysis. Eur Radiol. 2020;30:3558–3566. doi: 10.1007/s00330-020-06666-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanduleanu S, Woodruff HC, de Jong EEC, van Timmeren JE, Jochems A, Dubois L, Lambin P. Tracking tumor biology with radiomics: a systematic review utilizing a radiomics quality score. Radiother Oncol. 2018;127:349–360. doi: 10.1016/j.radonc.2018.03.033. [DOI] [PubMed] [Google Scholar]
- 54.Halligan S, Altman DG, Mallett S. Disadvantages of using the area under the receiver operating characteristic curve to assess imaging tests: a discussion and proposal for an alternative approach. Eur Radiol. 2015;25:932–939. doi: 10.1007/s00330-014-3487-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maurer A, Safavi-Abbasi S, Cheema A, Glenn C, Sughrue M. Management of petroclival meningiomas: a review of the development of current therapy. J Neurol Surg Part B Skull Base. 2014;75:358–367. doi: 10.1055/s-0034-1373657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mohammad MH, Chavredakis E, Zakaria R, Brodbelt A, Jenkinson MD. A national survey of the management of patients with incidental meningioma in the United Kingdom. Br J Neurosurg. 2017;31:459–463. doi: 10.1080/02688697.2017.1297765. [DOI] [PubMed] [Google Scholar]
- 57.Cronin P, Kelly AM, Altaee D, Foerster B, Petrou M, Dwamena BA. How to perform a systematic review and meta-analysis of diagnostic imaging studies. Acad Radiol. 2018;25:573–593. doi: 10.1016/j.acra.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 58.Lee J, Kim KW, Choi SH, Huh J, Park SH. Systematic review and meta-analysis of studies evaluating diagnostic test accuracy: a practical review for clinical researchers-part II. Statistical methods of meta-analysis. Korean J Radiol. 2015;16:1188–1196. doi: 10.3348/kjr.2015.16.6.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PNG 550 kb)
(PNG 560 kb)
(PNG 300 kb)
(PNG 545 kb)
(DOCX 22 kb)