Abstract
Cytochrome P450 2D6 (CYP2D6) O-demethylates codeine to the active drug, morphine. However, the utility of testing for CYP2D6 metabolizer status in patients receiving codeine in real-world clinical practice is poorly defined. Using data from a DNA bank linked to de-identified electronic health records, we studied 157 patients with a baseline pain score higher than 4 (0–10 scale) who received codeine. Based on CYP2D6 genotyping, 69 were classified as poor/intermediate and 88 as normal/ultrarapid CYP2D6 metabolizers. Pain response was defined as a score of 4 or lower while receiving codeine. In a propensity-score adjusted model, poor/intermediate metabolizers had lower odds [OR=0.35, p=0.02] of achieving a pain response than normal/ultrarapid metabolizers. To discriminate between codeine responders and non-responders, a score including CYP2D6 phenotype and clinical variables was built. The response rate was 38.5% among patients in the high, 17.3% in the intermediate, and 9.4% in the low score groups, respectively (p=0.001).
Introduction:
Codeine, one of the most widely prescribed analgesics for pain, is an integral part of step 2 of the World Health Organization’s (WHO) analgesic ladder.(1) Codeine is an inactive pro-drug that is O-demethylated to morphine, the active drug, by cytochrome P450 2D6 (CYP2D6).(2) The amount of morphine produced from codeine is markedly affected by variation in the CYP2D6 gene,(3) which is highly polymorphic. There are over 130 variant alleles of CYP2D6(4) and this genetic variation has been translated into an activity a score (AS) that reflects CYP2D6 metabolizer phenotype. Approximately 6.5% of Caucasians have a genetic variation that results in no functional CYP2D6 enzyme and are termed poor metabolizers (PMs) and produce almost no morphine after the administration of codeine.(5–7) At the other extreme, a small number of Caucasians (approximately 3%) have multiple copies of CYP2D6 (ultrarapid metabolizers or UMs) and thus have an enhanced capacity to produce morphine from codeine. Additionally, approximately 40% of Caucasians have allelic variants that result in diminished enzyme activity – termed intermediate metabolizers or IMs and would to be expected to have attenuated benefit from codeine. The remaining individuals (approximately 50%) are considered normal metabolizers (NMs).
In keeping with the reduced production of morphine from codeine in CYP2D6 PMs and IMs, the Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines recommend alternative analgesics for all CYP2D6 PMs and those IMs who do not respond to codeine.(2), (7) Additionally the guidelines note: “despite these data, codeine continues to be widely used, and most patients receive codeine without CYP2D6 genotyping.” A lack of an analgesic effect of codeine in CYP2D6 PMs has been shown in small groups of healthy subjects subjected to experimental pain stimuli.(8) However, the clinical importance of the effect of CYP26 metabolizer status on pain control in patients receiving codeine is poorly defined, particularly in real-world settings where patients almost always receive codeine in combination with acetaminophen and are also often receiving other analgesics.
The availability of large DNA biobanks linked to electronic health records (EHRs) provides an opportunity to examine the clinical importance of CYP2D6 metabolizer status in relation to analgesia after codeine therapy in a real-world clinical setting. Thus, we examined the hypothesis that CYP2D6 PMs and IMs would have poorer pain control than patients with normal or ultrarapid CYP2D6 metabolism in real-world clinical practice; additionally, we hypothesized that a response score composed of select clinical variables derived from the EHR combined with genetics (namely, CYP2D6 metabolizer status) could predict the likelihood of response to codeine. We conducted a retrospective cohort study using data from BioVU, a DNA bank linked to de-identified EHRs, to test these hypotheses.
Methods:
The study was conducted using BioVU, a clinical practice-based biobank at Vanderbilt University Medical Center. This biobank stores DNA samples linked to a de-identified version of a patient’s EHR, with all Health Insurance Portability and Accountability Act (HIPAA) identifiers removed.(9, 10) Data, including diagnostic and procedure codes, demographic characteristics, clinical care notes, patient history, problem lists, and medications are available for research.(11) We reviewed all electronic health records of patients included in the study while blinded to CYP2D6 genotype and phenotype assignment. The Vanderbilt University Medical Center’s Institutional Review Board approved this study.
Study population:
We studied adults (age≥18 years), who had been prescribed codeine for the outpatient treatment of pain; they were required to have a moderate-severe or severe baseline pain score (defined as >4 on a scale of 0–10)(12, 13) and at least one subsequent pain score related to the codeine prescription. Baseline pain score was the closest score to the prescription in the 90 days before (and including the date of) the prescription. We considered a pain score to be related to the codeine prescription if it met either of the following criteria: 1) it was recorded within 14 days after the prescription, or 2) the pain score was recorded at a time when codeine had been prescribed at consecutive clinical visits occurring within 6 months. For patients who had multiple episodes of codeine use, we used the first episode. If a patient had more than one pain score related to the intake of codeine, we captured all the pain scores available and used the median score. We reviewed medical records and excluded patients who had been prescribed codeine for indications other than pain (e.g., cough, diarrhea).
Outcomes:
In local clinical practice, pain is routinely recorded using a numerical rating scale (an ordinal 0–10 scale), with 0 indicating “no pain” and 10 indicating the worst possible pain. The primary outcome was response to codeine, defined as a pain score of 4 or lower during the period of likely exposure to codeine, as defined above. The secondary outcome was the pain score, examined as continuous variable, while on codeine treatment.
Covariates:
We extracted demographic and covariate information with an integrated approach, using both bioinformatics and manual review of EHRs. Covariates included sex, race, age at codeine prescription, body mass index, concurrent use of strong CYP2D6 inhibitors (i.e., bupropion, fluoxetine, paroxetine, and terbinafine),(14) concurrent use of other pain medications (i.e., non-steroidal anti-inflammatories, non-codeine opioids, muscle relaxants, duloxetine, gabapentin, and pregabalin), and diagnosis related to the use of codeine (i.e., malignancy, back pain, joint pain, trauma/surgery, or other).
Genotyping and CYP2D6 phenotypes:
DNA was genotyped to determine CYP2D6 functional status. We selected 12 allelic variants that occur at a frequency >0.15% in patients of either European (EA) or African (AA) ancestry for testing: CYP2D6*2, *3, *4, *5, *6, *9, *10, *17, *29, *35, *41, and *42 (Supplemental Table 1).(6, 15) All genotyping was performed using TaqMan assays by the Vanderbilt Technologies for Advanced Genomics (VANTAGE). We subjected genotyping data to standard quality control (QC) procedures; these measures included removing SNPs with a call rate less than 95%. Copy number variation was assessed using TaqMan gene copy number assays targeting intron 6 and exon 9, following the manufacturer’s protocol.
We first examined response rates by activity score and phenotype. CYP2D6 genotype was translated into phenotype using the method recommended by the CPIC and Dutch Pharmacogenetics Working Group.(7) Briefly, each allele received a value (i.e., 0 for no function alleles; 0.25 for *10; 0.5 for all other decreased function alleles; 1 for normal function alleles; and for gene duplications, double the value of their single counterparts). The values of both alleles were summed to form the Activity Score (AS). AS were used to categorize individuals into one of four phenotype groups: UM (AS>2.25), NM (1.25≤AS≤2.25), IM (0<AS<1.25), and PM (AS=0). Due to the small number of patients in each individual category and in order to have sufficient power for analysis, IMs and PMs were pooled, as were NMs and UMs. This grouping is consistent with one of the primary discussion points raised in the 2019 guidelines,(7) which suggested that individuals with lower AS (e.g., 0.5) may, at least for certain drugs, behave more like PMs.
Statistical analyses:
Demographic and clinical characteristics data are presented as number and percentages for categorical variables and as median and interquartile range (IQR) for continuous variables. We used Fisher’s exact tests to compare categorical variables and Wilcoxon’s rank sum tests to compare continuous variables.
We performed analyses in two phases. First, we tested whether CYP2D6 metabolizer status was independently associated with pain response using three logistic regression models: 1) univariate, 2) adjusted for age, sex, and race; and 3) adjusted for propensity score. We calculated the propensity score for the third model using all clinical variables (age, sex, race, BMI, indication, concomitant use of pain medications, concomitant use of strong CYP2D6 inhibitors, baseline pain score, calendar year, and number of pain scores) and log-transformed the score for analysis. As a secondary analysis, we examined whether CYP2D6 metabolizer status was independently associated with the pain score as a continuous variable while on codeine. We used linear regression to test three models parallel to our primary analysis: 1) univariate, 2) adjusted for age, sex, and race; and 3) adjusted for propensity score.
Risk scores for response prediction:
In the second phase, we sought to determine if a penalized regression model could generate a predictive risk score for response. First, we built a multivariate model using ridge regression, a penalized regression that employs a standard cross-validated penalty to shrink the coefficients of the covariates. Second, using the beta coefficients generated from the ridge regression model, we built a weighted score for the discrimination of codeine response. More specifically, we estimated lambda.min (i.e., the regression penalty associated with the minimum mean cross-validated error) and lambda.1se (i.e., the largest penalty whose estimated error was within one standard error of this minimum); we used the regression coefficient estimates associated with lambda.1se to generate the risk scores. We then standardized the scores to a 0–100 scale and completed receiver-operating-characteristic (ROC) curve analysis to examine whether the risk score could discriminate between those patients who responded to codeine and those who did not. These results are reported as area under the curve (AUC). Using these standardized scores, we grouped patients’ risk scores into three mutually exclusive tertiles: high, intermediate, and low scores; we then compared the proportion of responders in each category, using a chi-squared test.
Sensitivity analysis:
We also conducted two pre-specified sensitivity analyses: 1) restricting analysis to patients who had a baseline pain score within a 7-day window before the codeine prescription (compared to the primary analysis window of 90 days); 2) repeating the analysis using the previous CPIC classification for CYP2D6 metabolizer status (i.e., only activity scores <1 were grouped as PM/IM, rather than activity scores <1.25).(2)
All analyses were conducted using STATA version 16.0 (College Station, TX) or R version 3.4.3 for Windows. Codes will be available to academic researchers upon request.
Sample size calculation/detectable difference:
We included everyone who qualified for the study. A post-hoc detectable difference analysis used PS, a power and Sample Size Calculations Computer Program.(16) Thus, with 69 PM/IM and 88 NM/UM patients and a probability of response to codeine of 27% among NM/UM, the study had 80% power to detect a 10% response rate among PM/IM.
Results:
Eighty-three patients were NM, five were UMs, nine PM and sixty were IMs (Supplemental Table 2); the number of patients in each category were smaller when grouped by activity score (Supplemental Table 3), As such, results were pooled into two groups, PM/IM and NM/UM, and their clinical characteristics are shown by CYP2D6 metabolizer status (Table 1). There were small, statistically non-significant differences with trends towards higher BMI, less use of concurrent pain medications and more pain scores in PM/IM than in NM/UM patients. Thirty-four patients were responders (with a median pain response score ≤4), while 123 patients did not respond to codeine (median pain response score >4).
Table 1:
Patient Characteristics at Time of First Codeine Prescription
| Normal/Ultrarapid CYP2D6 Metabolizer n=88 |
Poor/Intermediate CYP2D6 Metabolizer n=69 |
p-value | |
|---|---|---|---|
| Sex, female (%) | 60 (68) | 46 (67) | 0.87 |
| Age (years), median [IQR] | 57 [45–69] | 58 [47–70] | 0.59 |
| Race, European ancestry (%) | 69 (78) | 63 (91) | 0.03 |
| BMI (kg/m2) | 27.9 [23.5–32.6] | 29.5 [24.0–33.8] | 0.32 |
| Indication | |||
| Malignancy (%) | 7 (8) | 5 (7) | 1.00 |
| Back pain (%) | 17 (19) | 13 (19) | 1.00 |
| Joint pain (%) | 29 (33) | 23 (33) | 1.00 |
| Trauma/Surgery (%) | 16 (18) | 9 (13) | 0.51 |
| Other (%) | 19 (22) | 19 (28) | 0.45 |
| Concomitant pain medications* (%) | 73 (83) | 51 (74) | 0.17 |
| Concomitant strong CYP2D6 inhibitors** (%) | 7 (8) | 12 (17) | 0.09 |
| Baseline pain score, median [IQR] | 7 [6–8] | 7 [6–7] | 0.67 |
| Calendar year of codeine prescription, median [IQR] | 2010 [2008–2012] | 2011 [2008–2013] | 0.58 |
| Number of pain scores during exposure to codeine, median [IQR] | 1 [1–2] | 1 [1–1] | 0.05 |
concomitant pain medications include NSAIDs, muscle relaxants, amitriptyline, duloxetine, gabapentin, and pregabalin
concomitant strong inhibitors include bupropion, fluoxetine, paroxetine, and terbinafine
CYP2D6 phenotyping and pain response:
Among PM and IMs there were 2 (22%) and 8 (13%) responders, respectively, for an overall pooled response rate of 14% (10 out of 69), compared to 27% among NM/UM metabolizers (24 out of 88), p=0.08. After adjustment for a propensity score that included all clinical variables, the odds ratio of response was 0.35 (95% CI: 0.14–0.84, p=0.02) among PM/IMs (Table 2a). The secondary analysis of pain response to codeine as a continuous variable yielded results concordant with the primary analysis: the median (IQR) pain score during follow-up was 5.5 (4.0–7.0) among NM/UM metabolizers compared to 6.0 (4.0–7.0 cm) among PM/IMs (p=0.09). After adjustment for the propensity score, PM/IMs had an estimated 0.73 units (p=0.04) higher pain score than NM/UM patients (Table 2b).
Table 2a:
Association Between Pain Response (Score ≤4) and CYP2D6 Phenotype in Patients Taking Codeine
| Unadjusted | Adjusted – sex, race, age | Adjusted – PS* | |||
|---|---|---|---|---|---|
| Odds ratio | p-value | Odds ratio | p-value | Odds ratio | p-value |
| 0.45 (0.20–1.02) |
0.06 | 0.45 (0.20–1.04) |
0.06 | 0.35 (0.14–0.84) |
0.02 |
log transformed propensity score (PS) includes sex, race, age, BMI, indications, concomitant medications, baseline pain score, number of pain scores, and year of medication
Table 2b:
Association Between Pain Score and CYP2D6 Phenotype in Patients Taking Codeine
| Unadjusted | Adjusted – sex, race, age | Adjusted – PS* | |||
|---|---|---|---|---|---|
| Beta coefficient | p-value | Beta coefficient | p-value | Beta coefficient | p-value |
| 0.56 (−0.08–1.19) |
0.08 | 0.52 (−0.12–1.16) |
0.11 | 0.73 (0.03–1.43) |
0.04 |
log transformed propensity score (PS) includes sex, race, age, BMI, indications, concomitant medications, baseline pain score, number of pain scores, and year of medication
Scores for discrimination in codeine analgesic effects:
To discriminate pain response among patients prescribed codeine, we built a risk score for response that included the NM/UM and PM/IM status as well as all clinical variables. Figure 1 depicts the performance of the standardized risk scores derived from the penalized regression; the AUC was significant for the primary outcome of response (median pain response score ≤4) (AUC=0.70, 95% CI: 0.61–0.79). The median (IQR) standardized risk score for response among responders was 75.5 (64.0–80.9) compared to 60.0 (50.0–74.5) among non-responders. In the tertiles of risk score, the response rate was 38.5% (95% CI: 25.3–53.0%) among patients in the high-score tertile, 17.3% (95% CI: 8.2–30.3%) in the intermediate-score tertile, and 9.4% (95% CI: 3.1–20.7%) in the low-score tertile (Figure 2), p=0.001.
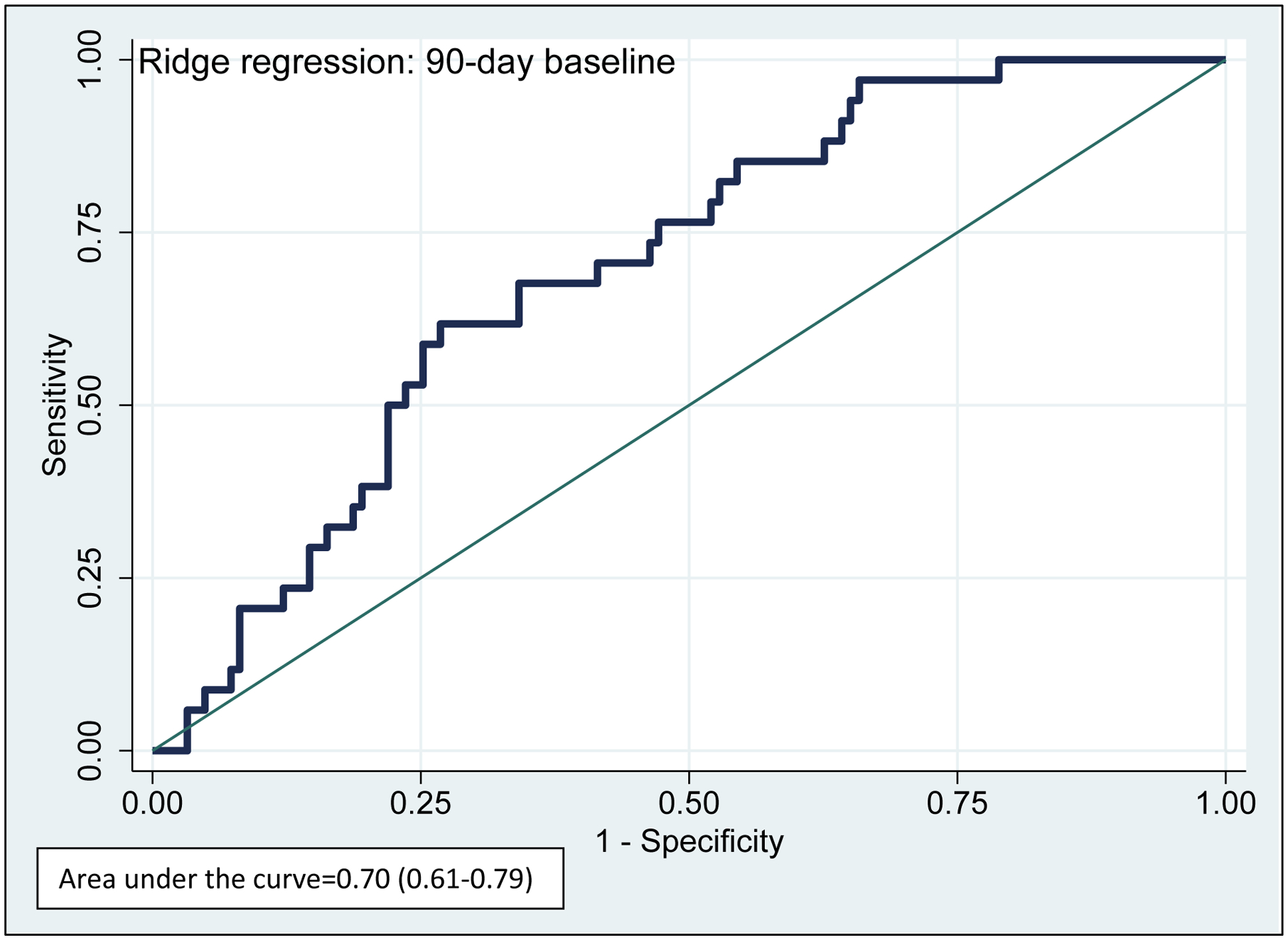
Figure 1: Performance of Discrimination Scores for Pain Response in Patients Taking Codeine
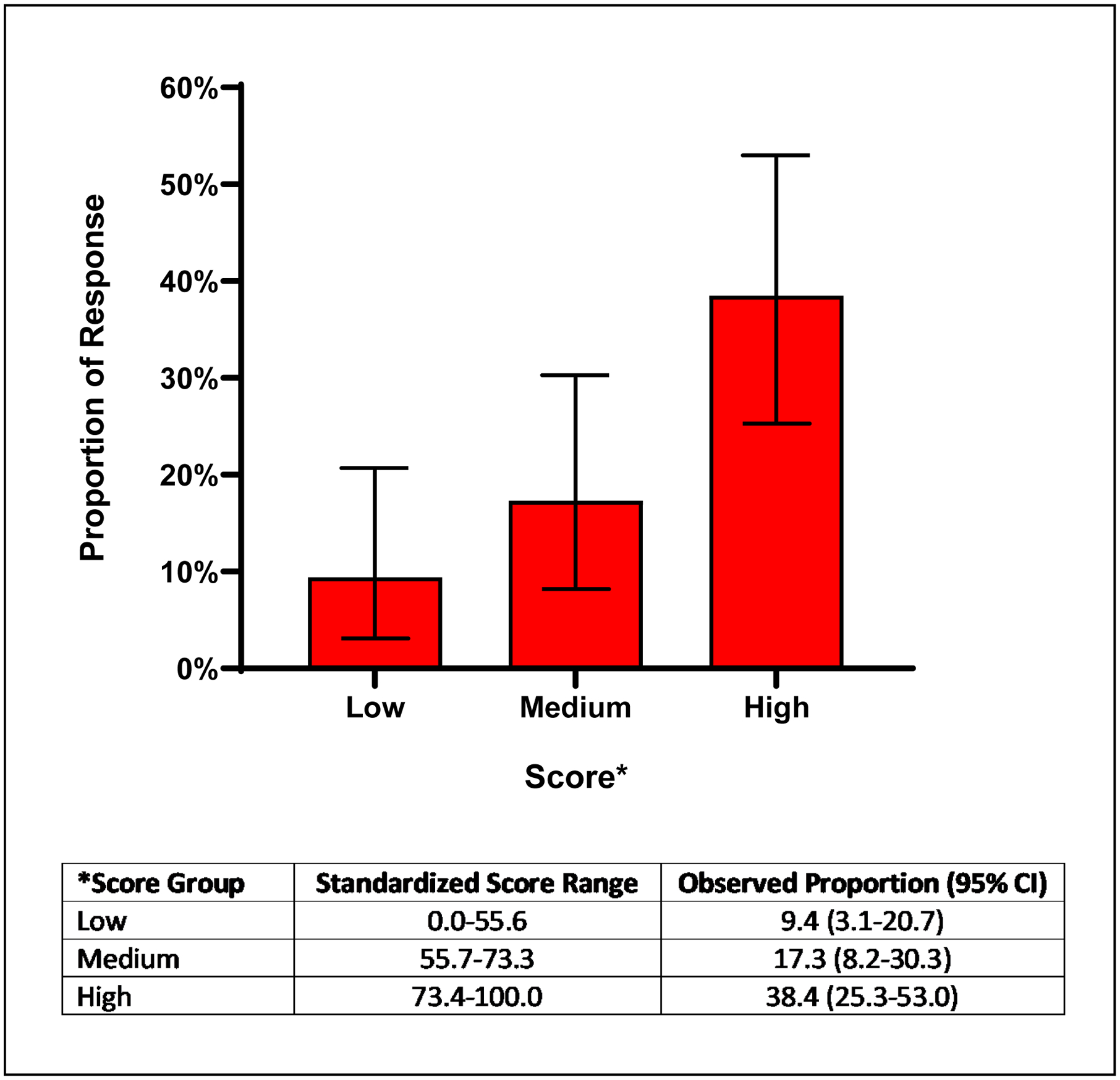
Figure 2: Pain Response Rates in Patients Prescribed Codeine: Results by Discriminant Scores
Sensitivity analyses:
The first sensitivity analysis, restricted to patients who had a baseline pain score within a 7-day window before the codeine prescription (Supplemental Table 4), yielded similar results regarding the association between metabolizer status and response rate (Supplemental Tables 5a and 5b). Likewise, when we calculated risk scores for this cohort, the AUC was significant and slightly higher than the primary outcome (AUC=0.77, 95% CI=0.66–0.88), suggesting increased discrimination capacity (Supplemental Fig 1). Additionally, the response rates were similar to those of the risk score tertiles in the primary analysis: 48.1% (95% CI: 28.7–68.1%) in the high-score group, 29.6% (95% CI: 13.8–50.2%) in the intermediate-score group, and 7.4% (95% CI: 0.9–24.3%) in the low-score group (Supplemental Fig 2), p=0.004. We also found similar trends, with somewhat lower discrimination capacity, in the sensitivity analysis which used the previous CPIC classification of metabolizer status based on genotype; the AUC was 0.67 (95% CI=0.56–0.77) for risk score prediction with a 90-day baseline pain score using the previous CPIC standard of classification for metabolizer status. Likewise, the response rates for this sensitivity analysis were consistent with our other findings: 32.7% (95% CI: 20.3–47.1%) in the high score group, 19.2% (95% CI: 9.6–32.5%) in the intermediate score group, and 13.2% (95% CI: 5.5–25.3%) in the low score group (p=0.046).
Discussion:
To the best of our knowledge, this is the first study to examine the effect of CYP2D6 metabolizer status on response to codeine using genetic testing combined with EHRs in real-world data. Our results, indicating that the patients with a CYP2D6 activity score of <1.25 (PM and IM groups) had lower rates of pain response and higher pain scores while receiving codeine compared to the NM/UM group. These results were consistent with those of a pragmatic clinical trial in which IMs and PMs receiving CYP2D6 genotype-guided therapy had better pain responses than those receiving usual care.(17) Moreover, the use of data from a real-world clinical setting allowed the integration of clinical variables in our analysis as well as the generation of a multivariable score to predict the proportion of response based on these characteristics.
These results remained robust beyond the pre-specified primary outcome. When we restricted our analysis to patients with a baseline pain score within 7 days of a codeine dose, we found similar associations between response and metabolizer status as well as the discrimination capacity of the risk scores generated by penalized regression. Additionally, a sensitivity analysis using the previous CPIC standard of classification for metabolizer status, yielded similar results.
As a polymorphic enzyme, CYP2D6 plays a critical role in not only the risk, but also the effectiveness of a wide-ranging set of drugs, accounting for the metabolism of approximately 20% of drugs used clinically.(18, 19) Thus, this analysis is a proof-of-concept of the utility of genotype and real-world data, in particular, when genetic data are combined with variables routinely obtained in the clinical setting such as concurrent use of CYP2D6 inhibitors, race, baseline pain, and specific indication.
Current recommendations by CPIC include avoidance of codeine in PMs and a trial of the recommended age- or weight-specific codeine dose in IMs. Our approach of calculating a score that integrates clinical variables could be of use, particularly in IMs or NMs who have additional factors such as high baseline pain and concurrent CYP2D6 inhibitors. While implementation of this approach for identification of patients likely to respond or not respond to codeine in routine clinical practice will require replication of our results in independent cohorts, our study underscores the potential role of pre-emptive over point of care genotyping in routine practice. In pre-emptive genotyping, tests are obtained and results stored in the electronic health records, so they are available at the time of prescription.(20) Our results suggest that with the availability of genetic and clinical data, providers could have the information needed to prescribe a different pain medication to patients with a score in the lowest tertile, for whom the anticipated pain response rate would be less than 10%. It is also important to consider that the overall response to codeine was low, and even with genotype and clinical information the highest tertile had a predicted response rate of 38.5%.
We acknowledge that this study has limitations. The study population was small; analysis of a larger patient group may provide additional nuances, particularly with a more racially diverse population. The multiplicity of indications given the sample size may also limit the application of our findings for any given indication, although this breadth indicates that our findings are relatively generalizable. Additionally, due to the limitations of EHRs, we were unable to adjust for codeine dose, as exact doses were not specified for most patients and the drug was prescribed pro re nata (i.e., as needed). Further, the pain score response difference between groups was small, and the point estimate of 0.73 units on a 0–10 scale may not be considered clinically important.(21, 22) Finally, a replication study would add weight to the findings.
Despite these limitations, these findings suggest that a combination of clinical and genetic characteristics could offer utility in a clinical setting.
Supplementary Material
ACKNOWLEDGEMENTS
Supported by grant R01GM109145 and R35GM131770. CPC and ALD were also supported by grants R01AR073764 and the Veterans Health Administration Merit Award 1I01CX001741. The dataset(s) used for the analyses described were obtained from Vanderbilt University Medical Center’s BioVU which is supported by numerous sources: institutional funding, private agencies, and federal grants. These include the NIH funded Shared Instrumentation Grant S10RR025141; and CTSA grants UL1TR002243, UL1TR000445, and UL1RR024975. Genomic data are also supported by investigator-led projects that include U01HG004798, R01NS032830, RC2GM092618, P50GM115305, U01HG006378, U19HL065962, R01HD074711; and additional funding sources listed at https://victr.vumc.org/biovu/index.html?sid=229. The funding sources had no role in the collection, analysis, or interpretation of data, writing of the manuscript, or decision to submit for publication.
Footnotes
CONFLICT OF INTEREST
The authors do not have any conflicts of interest to report.
References
- 1.Forbes K Pain in patients with cancer: the World Health Organization analgesic ladder and beyond. Clin Oncol (R Coll Radiol). 2011;23(6):379–80. [DOI] [PubMed] [Google Scholar]
- 2.Crews KR, Gaedigk A, Dunnenberger HM, Klein TE, Shen DD, Callaghan JT, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for codeine therapy in the context of cytochrome P450 2D6 (CYP2D6) genotype. Clin Pharmacol Ther. 2012;91(2):321–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kirchheiner J, Schmidt H, Tzvetkov M, Keulen JT, Lotsch J, Roots I, et al. Pharmacokinetics of codeine and its metabolite morphine in ultra-rapid metabolizers due to CYP2D6 duplication. Pharmacogenomics J. 2007;7(4):257–65. [DOI] [PubMed] [Google Scholar]
- 4.Nofziger C, Turner AJ, Sangkuhl K, Whirl-Carrillo M, Agundez JAG, Black JL, et al. PharmVar GeneFocus: CYP2D6. Clin Pharmacol Ther. 2020;107(1):154–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poulsen L, Brosen K, Arendt-Nielsen L, Gram LF, Elbaek K, Sindrup SH. Codeine and morphine in extensive and poor metabolizers of sparteine: pharmacokinetics, analgesic effect and side effects. Eur J Clin Pharmacol. 1996;51(3–4):289–95. [DOI] [PubMed] [Google Scholar]
- 6.Consortium CPI. CPIC Guideline for Codeine and CYP2D6 2020 [Available from: https://cpicpgx.org/guidelines/guideline-for-codeine-and-cyp2d6/.
- 7.Caudle KE, Sangkuhl K, Whirl-Carrillo M, Swen JJ, Haidar CE, Klein TE, et al. Standardizing CYP2D6 Genotype to Phenotype Translation: Consensus Recommendations from the Clinical Pharmacogenetics Implementation Consortium and Dutch Pharmacogenetics Working Group. Clinical and Translational Science. 2020;13(1):116–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desmeules J, Gascon MP, Dayer P, Magistris M. Impact of environmental and genetic factors on codeine analgesia. Eur J Clin Pharmacol. 1991;41(1):23–6. [DOI] [PubMed] [Google Scholar]
- 9.Pulley J, Clayton E, Bernard GR, Roden DM, Masys DR. Principles of human subjects protections applied in an opt-out, de-identified biobank. Clin Transl Sci. 2010;3(1):42–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roden DM, Pulley JM, Basford MA, Bernard GR, Clayton EW, Balser JR, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008;84(3):362–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wei WQ, Denny JC. Extracting research-quality phenotypes from electronic health records to support precision medicine. Genome Med. 2015;7(1):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kannampallil T, Galanter WL, Falck S, Gaunt MJ, Gibbons RD, McNutt R, et al. Characterizing the pain score trajectories of hospitalized adult medical and surgical patients: a retrospective cohort study. Pain. 2016;157(12):2739–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woo A, Lechner B, Fu T, Wong CS, Chiu N, Lam H, et al. Cut points for mild, moderate, and severe pain among cancer and non-cancer patients: a literature review. Ann Palliat Med. 2015;4(4):176–83. [DOI] [PubMed] [Google Scholar]
- 14.FDA. Drug Development and Drug Interactions: Table of Substrates, Inhibitors and Inducers http://www.fda.gov/Drugs/DevelopmentApprovalProcess/DevelopmentResources/DrugInteractionsLabeling/ucm093664.htm#classInhibit2011 [updated June 17, 2020. Available from: http://www.fda.gov/Drugs/DevelopmentApprovalProcess/DevelopmentResources/DrugInteractionsLabeling/ucm093664.htm#classInhibit.
- 15.PharmGKB. Gene-specific Information Tables for CYP2D6 2020 [Available from: https://www.pharmgkb.org/page/cyp2d6RefMaterials.
- 16.Dupont WD, Plummer WD Jr. PS power and sample size program available for free on the internet. Controlled Clinical Trials. 1997;18(3):274. [Google Scholar]
- 17.Smith DM, Weitzel KW, Elsey AR, Langaee T, Gong Y, Wake DT, et al. CYP2D6-guided opioid therapy improves pain control in CYP2D6 intermediate and poor metabolizers: a pragmatic clinical trial. Genet Med. 2019;21(8):1842–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saravanakumar A, Sadighi A, Ryu R, Akhlaghi F. Physicochemical Properties, Biotransformation, and Transport Pathways of Established and Newly Approved Medications: A Systematic Review of the Top 200 Most Prescribed Drugs vs. the FDA-Approved Drugs Between 2005 and 2016. Clin Pharmacokinet. 2019;58(10):1281–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang B, Yang LP, Zhang XZ, Huang SQ, Bartlam M, Zhou SF. New insights into the structural characteristics and functional relevance of the human cytochrome P450 2D6 enzyme. Drug Metab Rev. 2009;41(4):573–643. [DOI] [PubMed] [Google Scholar]
- 20.Roden DM, Van Driest SL, Mosley JD, Wells QS, Robinson JR, Denny JC, et al. Benefit of Preemptive Pharmacogenetic Information on Clinical Outcome. Clin Pharmacol Ther. 2018;103(5):787–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olsen MF, Bjerre E, Hansen MD, Hilden J, Landler NE, Tendal B, et al. Pain relief that matters to patients: systematic review of empirical studies assessing the minimum clinically important difference in acute pain. BMC Med. 2017;15(1):35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelly AM. The minimum clinically significant difference in visual analogue scale pain score does not differ with severity of pain. Emerg Med J. 2001;18(3):205–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


