Abstract
Background
In response to the current COVID-19 pandemic, multiple companies marketed serological tests. Rigorous, independent and comparative performances of these assays on defined clinical specimens are needed.
Methods
In a first preliminary phase, we investigated 16 IgG, IgM, IgA and pan Ig serological ELISA using a panel of 180 sera, comprising 97 sera from patients with a positive RT-PCR, and 83 negative sera sampled before November 1, 2019. In a second phase and to complete the evaluation on the full panel (100 positive and 300 negative), tests that passed pre-defined exclusion criteria of 90% sensitivity and 97% specificity were further evaluated on 220 additional sera chosen to assess possible cross-reactivity with other human viral infections.
Results
Among the 16 tests evaluated in the preliminary phase, two were excluded due to insufficient sensitivity at 15 days post-symptom onset and one was excluded due to poor specificity. Of the 13 tests evaluated using the full panel comprised of a diverse pool of sera including those reactive against known respiratory viruses, no systematic cross-reactivity was observed. However, heterogeneities across tests were found. Consistent with kinetics of antibody expression, maximal sensitivity was found two weeks post-symptom onset.
Conclusion
In this independent evaluation, we compared the performance of 16 SARS-CoV-2 serological tests using well-characterized sera and found 13 tests with more than 90% sensitivity at 15 days post-symptom onset and 97% specificity across a diverse range of negative samples.
Keywords: SARS-CoV-2, COVID-19, Serological test, Evaluation, Specificity, Sensitivity
1. Introduction
Early January 2020, a viral respiratory disease caused by a new coronavirus was reported in Wuhan City, Hubei Province (China). This new disease and its associated virus named respectively COVID-19 (Coronavirus Disease 2019) and SARS-CoV-2 (Severe Acute Respiratory Syndrome Coronavirus 2) rapidly spread within China and then to all continents, causing a pandemic with more than 67 million cases and 3.1 million deaths worldwide end of April, 2020 [1]. The RT-PCR (Reverse Transcription Polymerase Chain Reaction) detecting SARS-CoV-2 RNA in clinical specimens is a key tool to diagnose and manage the current pandemic [2,3]. More recently, numerous lateral flow antigen assays have been developed as rapid point-of-care tests to detect active COVID-19 infection. However, the gain in time to result of these “while you wait” tests was often accompanied with a lower sensitivity compared to RT-PCRs [4]. While both tests can be efficiently used in the early phase of the disease, their diagnostic performances decrease progressively in the course of infection, especially after resolution of the symptoms associated with viral clearance [[5], [6], [7], [8]]. In contrast, the immune response mounted during the course of COVID-19 is expected to be a durable marker of infection especially when the viral surface Spike glycoprotein (S) is used as antigen [9]. Most patients infected with COVID-19 develop specific IgM, IgA, and IgG responses within 5–15 days, which persist for several months in the case of IgG and for at least 3 to 6 weeks for IgM and IgA [[10], [11], [12], [13]]. Therefore, these circulating antibodies could be used to investigate SARS-CoV-2 infections [6,[14], [15], [16], [17], [18]]. This can be especially useful in atypical clinical presentations of the disease where antigen tests or RT-PCR results are not conclusive [19]. Furthermore, serology is the method of choice to conduct prevalence studies among the population and may help verify effectiveness of vaccines. Indeed, antibody titers could be used as markers of protective immunity even though there is currently absence of robust immunological correlates of protection. Additionally, serological investigations could help the selection of recovered patients for convalescent plasma donation [10,20].
The viral surface Spike glycoprotein (S) and the nucleocapsid protein (N) appear to be the main targets of the humoral immune response in coronavirus infections, including SARS-CoV-2 [10]. The N that encapsulates the viral genome is the most abundant viral protein whereas the S envelope is composed of two subunits, S1 and S2 cleaved by the host protease furin [21,22]. The S1 subunit contains the receptor-binding domain (RBD) needed for binding to the host angiotensin-converting enzyme 2 (ACE2) receptor, whereas the S2 subunit contains the element for cell membrane fusion [23,24]. Many serological tests based on the detection of antibodies specific for SARS-CoV-2 antigens were rapidly developed and commercialized. These tests are mainly composed of either full or specific domains of the S or N proteins, or alternatively composed of whole viral lysate. Accuracy of these tests may be highly dependent on the subdomain chosen, the resulting successful protein folding and for some antigens the conserved protein glycosylation. In this study, we evaluated several SARS-CoV-2 serological tests on well-defined positive and negative sera. This independent evaluation identified high quality tests and reported sensitivity measures relative to days post-symptom onset.
2. Materials and methods
2.1. Samples
The pool of sera used in this evaluation was selected from the sera collection used in our previous study [25]. All sera were collected at the Lausanne University Hospital (CHUV), Switzerland. A pool of 180 sera (97 positive and 83 negative) were used during the preliminary phase of the evaluation (Tables 1 and S1). An extended evaluation phase was completed on additional samples to analyze the different tests on a complete total panel for the two phases of 400 sera (100 positive and 300 negative). SARS-CoV-2 negative sera were collected before November 1, 2019, assumed to be prior to the SARS-CoV-2 pandemic. Possible cross-reactivity was assessed through testing of sera known to be positive for other microorganisms or auto-immune disease (lupus). The 100 expected-positive sera were sampled during the first 2 months post-symptoms from hospitalized patients with a documented positive SARS-CoV-2 RT-PCR and presenting moderate to severe symptoms. Date of first symptoms was extracted from their medical electronic records.
Table 1.
Characteristics of the sera used in the preliminary and the extended evaluations.
| Sera | Positive | Negative |
| Preliminary Evaluation | ||
| COVID-19 d0-7 | 19 | |
| COVID-19 d8-14 | 38 | |
| COVID-19 d>15 | 40 | |
| Hcov | 16 | |
| Lupus | 11 | |
| undefined | 56 | |
| Total = 180 | 97 | 83 |
| Extended Evaluation | ||
| COVID-19 d0-7 | 3 | |
| B19 | 13 | |
| CMV | 33 | |
| EBV | 22 | |
| FSME | 13 | |
| HSV1 | 14 | |
| HSV1-2 | 3 | |
| HSV2 | 4 | |
| Influ A | 27 | |
| Influ A/B | 1 | |
| Influ A/RSV | 2 | |
| Influ B | 9 | |
| Influ B/RSV | 1 | |
| Measles | 12 | |
| Measles-Mumps | 3 | |
| Mumps | 11 | |
| RSV | 15 | |
| Rubella | 17 | |
| undefined | 6 | |
| Varicella | 11 | |
| Total = 400 | 100 | 300 |
Positive sera: The categories d0-7, d8-14, >15d represent SARS-CoV-2 RT-PCR positive patients with expected-positive serum samples at different days post-symptoms onset. All Negative sera were sampled before November 1, 2019, anterior to the SARS-CoV-2 pandemic. Possible cross-reactions were investigated with sera positive for other known viruses. Undefined represent negative sera without association to other microorganism. Human coronaviruses (Hcov, 4x E229, 2x NL63, 6x OC43, 4x HKU1), Parvovirus (B19), Cytomegalovirus (CMV), Epstein-Barr virus (EBV), Tick-borne encephalitis virus (FSME), Herpes simplex virus 1 and 2 (HSV-1 and 2), Influenza A and B (Influ A and B), Respiratory Syncytial Virus (RSV), Measles virus, Mumps virus, Rubella, Varicella-zoster virus. Sera of patients with lupus were also included.
2.2. Serological ELISA tests
All 16 IgG, IgM, IgA and pan-Ig tests with their characteristics and references are listed in Table S2. Each assay was performed according to the manufacturers’ instructions. Samples were tested in duplicate and all steps were performed manually to diminish dead volume. The washing steps were performed with a microplate washer (PW40, Bio-Rad, France). Optical densities (OD) were measured with a microplate reader (800 TSI, BioTek, USA).
2.3. Statistical analysis
Sensitivity was evaluated on the expected positive sera and according to days post-symptom onset. Specificity was determined on the expected negative sera sampled before November 1, 2019. Equivocal results were excluded from the analysis. The R version 3.5.3 was used for data processing using the package Meta-Analysis of Diagnostic Accuracy (mada_0.5.10) for calculations of specificities, sensitivities and their corresponding 95% CI. The package irr was used for positive and negative agreement calculations along with heatmap.2 for representation. Graphpad prism 8.3.0 was used for graphical representation.
3. Results
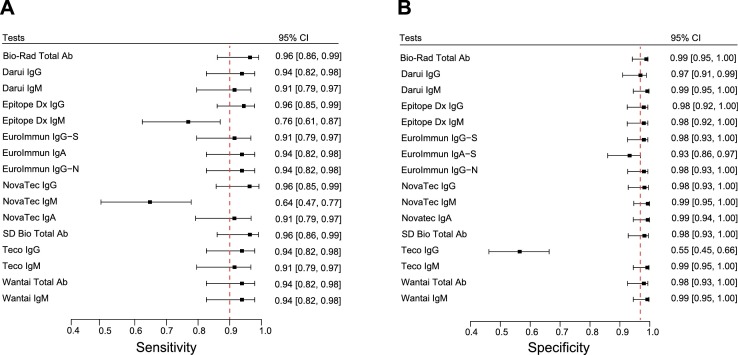
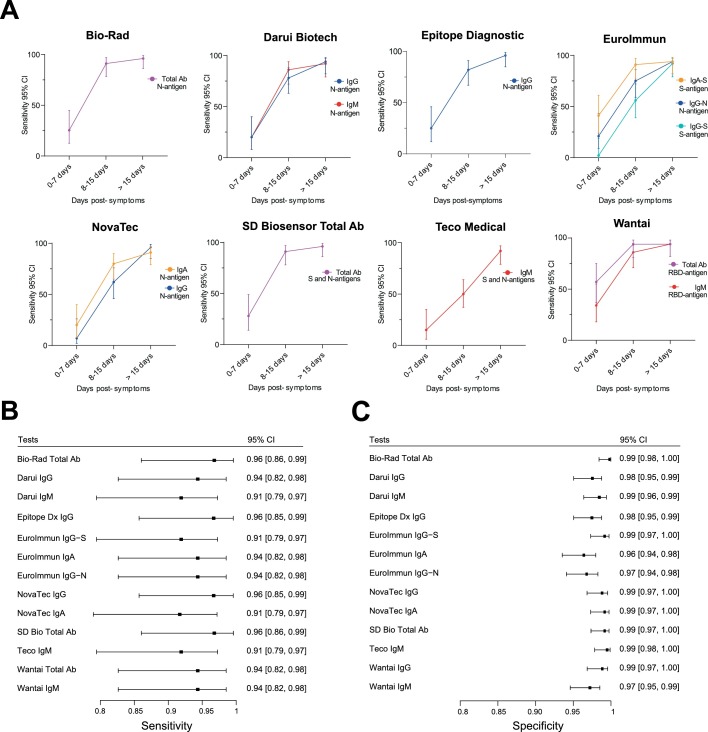
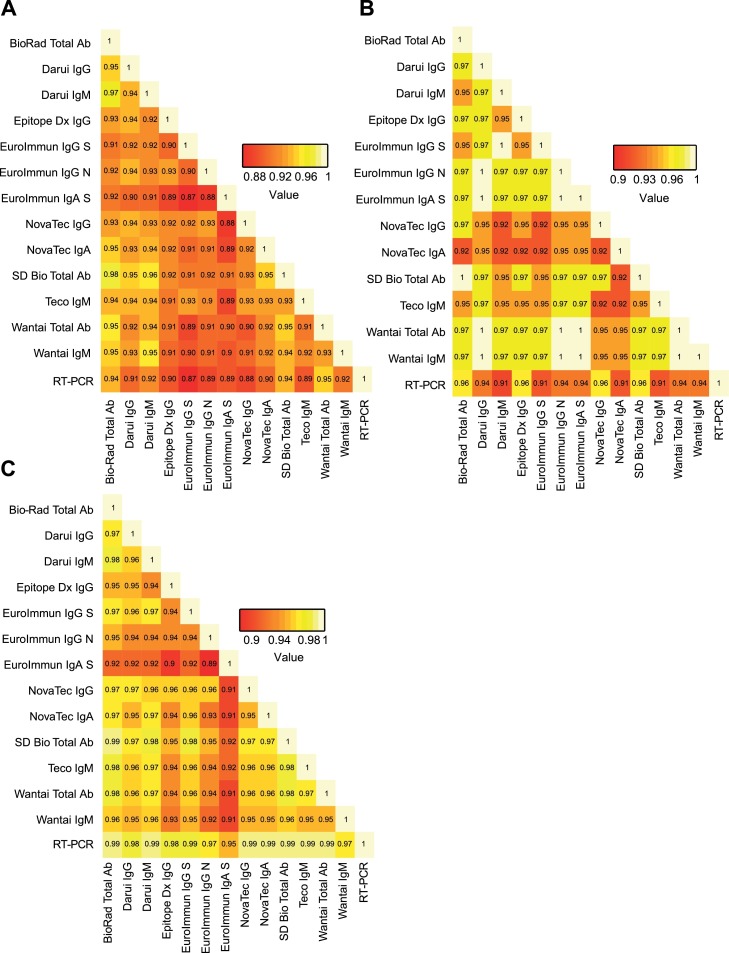
A preliminary evaluation on 180 sera (97 positive and 83 negative) of all 16 SARS-CoV-2 serologic tests was performed to discard any test presenting insufficient performances of sensitivity < 90% and/or specificity < 97% taking into account the upper values of the 95% confidence interval (CI) of sensitivity and specificity. Results were stratified according to the time between symptoms onset and sera sampling and are presented as three categories: 0–7 days (n = 19), 8–14 days (n = 38), and > 15 days (n = 40) post-symptom onset. Sensitivities and corresponding 95% CI intervals were calculated for each test at each time point (Figs. S1 and S2 and Table S3). Sensitivities and corresponding 95% CI intervals at >15 days post-symptom onset is presented in Fig. 1 A. Specificities of the different tests and corresponding 95% CI intervals were calculated and reported in Fig. 1B and Table S4. As expected, sensitivity increased over time to reach > 90% after 15 days. Among the 16 tests, the Epitope Dx IgM and Novatec IgM showed sensitivities at > 15 days below 90% whereas Teco IgG presented a poor specificity of 55.4% (Fig. 1A). These three assays were therefore discarded for the extended evaluation based on pre-defined exclusion criteria. The EuroImmun IgA−S showed an upper specificity of the 95% CI at the pre-defined exclusion criteria of 97% and was kept for the extended evaluation also due to its good sensitivity at early time points (Fig. 1B). The extended evaluation was performed on additional 220 sera (3 additional positive and 217 additional negative sera) for a total of 400 sera (100 positive and 300 negative) for the two phases. All tests showed excellent sensitivity at > 15 days (Figs. 2 A,B, S3, Table S5) with four assays showing more than 96% sensitivity: Biorad Platelia, Epitope Dx IgG, SD Bio Total Ab and Novatec IgG. Only three tests, EuroImmun IgA−S, the Teco IgG and Wantai IgG showed sensitivity over 90% already at 8-15 days. All tests displayed high specificities (Fig. 2C and Table S6) with only three tests below 98% (Wantai IgM, EuroImmun IgG−N and EuroImmun IgA−S). An extended negative sera collection was chosen to assess possible cross-reactivity with other human viral infections and contained: four other circulating human coronaviruses (E229, NL63, OC43,HKU1), Parvovirus B19, Cytomegalovirus, Epstein-Barr virus, Tick-borne encephalitis virus, Herpes simplex virus 1 and 2, Influenza A and B, Respiratory Syncytial Virus, Measles virus, Mumps virus, Rubella virus, Varicella-zoster virus. Sera of patients with lupus were also included as control. No particular cross-reactions were observed (Table S7). Overall agreement analyses was performed between all tests (Fig. 3 A). Among expected positive sera (with a positive SARS-CoV-2 RT-PCR), the Novatec IgA showed the lowest overall positive agreement between all tests at >15 days post-symptom onset (Fig. 3B). In the expected negative sera, the EuroImmun IgA showed the lowest overall negative agreement in accordance with the highest number of cross-reactions observed among control sera (Fig. 3C and Table S7).
Fig. 1.
A: Forest plots of the sensitivity at > 15 days post-symptom onset with the corresponding 95% CI for assays investigated during the preliminary phase of the evaluation. Dashed line represents the pre-defined exclusion criteria of 90% sensitivity. B: Forest plots of the specificity with the corresponding 95% CI. Dashed line represents the pre-defined exclusion criteria of 97% specificity.
Fig. 2.
A: Percentage sensitivity with 95% CI of the different tests according to days from symptoms onset investigated during the extended phase of the evaluation. B: Corresponding Forest plots of the sensitivity at > 15 days post-symptom onset with the corresponding 95% CI. C: Forest plots of the specificity with the corresponding 95% CI.
Fig. 3.
A: Overall percentage agreement between all tests and against the gold standard RT-PCR. B: Overall positive percentage agreement at > 15 days post-symptom onset between all tests and against the gold standard RT-PCR C: Overall negative percentage agreement between all tests and against the gold standard RT-PCR.
4. Conclusions
In this study, we evaluated 16 SARS-CoV-2 serological tests using sera from three groups of patients: confirmed SARS-CoV-2 infected patients, healthy negative controls sampled before the pandemic, and an extended set of patients with other viral infections and diseases to address possible cross-reactions. Sensitivity calculations were stratified by days post-symptom onset to monitor rising antibody levels. Overall, an important heterogeneity was observed across all tests, likely explained by the SARS-CoV-2 antigen targeted and the ELISA assay used.
IgM and IgG response showed similar kinetics with no benefit for IgM-specific assays whereas the IgG assays performed slightly better than IgM and IgA both in term of sensitivity and specificity. The observed kinetics of IgG and IgM is in accordance with most studies of laboratory-confirmed COVID-19 cases, where both IgM and IgG antibodies appears simultaneously and start to be detectable around 5–7 days after symptoms onset, with a median seroconversion at day 13 for both antibodies [14]. Detection of IgA showed mixed results, with either good sensitivity but reduced specificity or vice versa. Interestingly, Pan Ig based tests were among the best performing tests consistent with previous observations [[27], [28], [29], [30]].
The sera investigated here were collected at a maximum of 38 days post-symptom onset from hospitalized patients presenting moderate to severe symptoms. The measure of the production and persistence of anti-SARS-CoV-2 antibodies is depending on many factors including (1) the targeted antigens used in the different assays (protein N, protein S or domains of protein S), (2) the disease severity and (3) time of serum collection post-symptom onset [30,31]. In particular, the performances observed in this study, with sera collected during the first 38 days post-symptoms from patients presenting moderate to severe symptoms, showed no specific differences between N-based and S-based assays. This was in contrast to what was observed on convalescent sera taken more than 2 months post-symptoms where the N-assays exhibited an overall lower sensitivity due to anti-N antibodies decay; already 6 to 8 weeks post infection [30]. Thus, the present results cannot be extrapolated to sera collected from low severity or asymptomatic patients nor to sera collected several months post-symptoms. As shown by Fenwick et al. [30], additional studies should be performed to evaluate the performance of these tests from different sera collection obtained from different populations and different time collection. In addition, these tests provide no information on the characteristic of the detected antibodies classes including their affinity, avidity and neutralization potential that have a differential production kinetics post-infection.
In conclusion, this study identified several serological test showing high performances for sera obtained from hospitalized patients. This independent evaluation on clinical specimens showed that IgG or pan-Ig might be preferred over IgM that are not contributive during the acute to subacute phase of the disease.
Ethical statement
The data were obtained during a quality enhancement project at our institution. According to national law the performance and publishing the results of such a quality project can be done without asking the permission of the competent research ethics committee.
Funding information
Foundation for Innovative New Diagnostics (FIND) supported this study by funding the salary of one laboratory technician and by providing the serological tests consumables and reagents.
CRediT authorship contribution statement
Damien Jacot: Writing – original draft, Formal analysis, Writing – review & editing. Milo Moraz: Writing – original draft, Formal analysis, Writing – review & editing. Alix T. Coste: Writing – original draft, Formal analysis, Conceptualization, Writing – review & editing. Christele Aubry: Writing – original draft. Jilian A. Sacks: Writing – review & editing. Gilbert Greub: Writing – review & editing. Antony Croxatto: Writing – original draft, Formal analysis, Conceptualization, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
CHUV, Audrey Moulin, Sarah Chappuis, Fabienne Di Paola, Carine Pintér Pont, Claudine Gostely, Emilie Rüegger, Sylvie Caillon-Bouchez, Alexandre Mamin, and Benjamin Gayton, for their wonderful implication in this project, and the early implementation of SARS-CoV-2 serologic testing in our hospital already since 14th April 2020. We would like to thank the laboratory of immunology to provide us some sera, especially those positive for Lupus. We thank Pr Marchetti Oscar and Pr Pache Antoine from the Ensemble Hospitalier de la Cote, Morges, Switzerland, Dr Caroline Chapuis-Taillard and Dr De Vallière Serge from Clinique de la Source, Lausanne, Switzerland and, Dre Aurélie Jayol-Virely from Clinical laboratory of the Etablissement Hospitalier Nord-Vaudois, Yverdon, Switzerland, to have shared information about the symptoms date of some patients.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.jcv.2021.104931.
Appendix. Supplementary materials
References
- 1.WHO . WHO; 2021. WHO Coronavirus Disease (COVID-19) Dashboard.https://covid19.who.int/ [Google Scholar]
- 2.Corman V.M., et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25(3) doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Opota O., et al. Comparison of SARS-CoV-2 RT-PCR on a high-throughput molecular diagnostic platform and the cobas SARS-CoV-2 test for the diagnostic of COVID-19 on various clinical samples. Pathog. Dis. 2020;78(8) doi: 10.1093/femspd/ftaa061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vandenberg O., et al. Considerations for diagnostic COVID-19 tests. Nat. Rev. Microbiol. 2020;19(3) doi: 10.1038/s41579-020-00461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Long Q.X., et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26(6):845–848. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 6.Peeling R.W., et al. Serology testing in the COVID-19 pandemic response. Lancet Infect. Dis. 2020;20(9):e245–e249. doi: 10.1016/S1473-3099(20)30517-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He X., et al. Temporal dynamics in viral shedding and transmissibility of COVID-19. Nat. Med. 2020;26(5) doi: 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- 8.Liu Y., et al. Viral dynamics in mild and severe cases of COVID-19. Lancet Infect. Dis. 2020;20(6) doi: 10.1016/S1473-3099(20)30232-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caruana G., et al. Diagnostic strategies for SARS-CoV-2 infection and interpretation of microbiological results. Clin. Microbiol. Infect. 2020;26(9) doi: 10.1016/j.cmi.2020.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Poland G.A., Ovsyannikova I.G., Kennedy R.B. SARS-CoV-2 immunity: review and applications to phase 3 vaccine candidates. Lancet. 2020;396(10262):1595–1606. doi: 10.1016/S0140-6736(20)32137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sethuraman N., Jeremiah S.S., Ryo A. Interpreting Diagnostic Tests for SARS-CoV-2. JAMA. 2020;323(22):2249–2251. doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- 12.Ghaffari A., Meurant R., Ardakani A. COVID-19 serological tests: how well do they actually perform? Diagnostics. 2020;10(7) doi: 10.3390/diagnostics10070453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee C.Y., et al. Serological approaches for COVID-19: epidemiologic perspective on surveillance and control. Front. Immunol. 2020;11:879. doi: 10.3389/fimmu.2020.00879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long Q.X., et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020;26(6):845. doi: 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- 15.Zhao J., et al. Antibody responses to SARS-CoV-2 in patients of novel coronavirus disease 2019. Clin. Infect. Dis. 2020;71(16) doi: 10.1093/cid/ciaa344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krammer F., Simon V. Serology assays to manage COVID-19. Science, 2020;368(6495):1060–1061. doi: 10.1126/science.abc1227. [DOI] [PubMed] [Google Scholar]
- 17.Longchamp A., et al. Serum antibody response in critically ill patients with COVID-19. Intensive Care Med. 2020;46(10) doi: 10.1007/s00134-020-06171-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sethuraman N., Jeremiah S.S., Ryo A. Interpreting diagnostic tests for SARS-CoV-2. JAMA. 2020;323(22) doi: 10.1001/jama.2020.8259. [DOI] [PubMed] [Google Scholar]
- 19.Coste, A.T., et al., Indication for SARS-CoV-2 serology: first month follow-up. 2020: p. 2020.06.30.20140715.
- 20.Okba N.M.A., et al. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg. Infect. Dis. 2020;26(7):1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cantuti-Castelvetri L., et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370(6518):856. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Daly J.L., et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science. 2020;370(6518):861. doi: 10.1126/science.abd3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bangaru, S., et al., Structural analysis of full-length SARS-CoV-2 spike protein from an advanced vaccine candidate. bioRxiv, 2020. [DOI] [PMC free article] [PubMed]
- 24.Chen C.Y., et al. Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA. J. Mol. Biol. 2007;368(4):1075–1086. doi: 10.1016/j.jmb.2007.02.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coste A.T., et al. Comparison of SARS-CoV-2 serological tests with different antigen targets. J. Clin. Virol. 2020;134 doi: 10.1016/j.jcv.2020.104690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Espejo A.P., et al. Review of current advances in serologic testing for COVID-19. Am. J. Clin. Pathol. 2020;154(3):293–304. doi: 10.1093/ajcp/aqaa112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lisboa Bastos M., et al. Diagnostic accuracy of serological tests for covid-19: systematic review and meta-analysis. BMJ. 2020;370:m2516. doi: 10.1136/bmj.m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitman J.D., et al. Evaluation of SARS-CoV-2 serology assays reveals a range of test performance. Nat. Biotechnol. 2020;38(10):1174–1183. doi: 10.1038/s41587-020-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fenwick C., et al. Changes in SARS-CoV-2 spike versus nucleoprotein antibody responses impact the estimates of infections in population-based seroprevalence studies. J. Virol. 2020;95(3) doi: 10.1128/JVI.01828-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seow J., et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat. Microbiol. 2020;5(12):1598–1607. doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.