Abstract
Nearly all bacteria are encased in peptidoglycan, an extracytoplasmic matrix of polysaccharide strands crosslinked through short peptide stems. In the Gram-n egative model organism Escherichia coli, more than 40 synthases and autolysins coordinate the growth and division of the peptidoglycan sacculus in the periplasm. The precise contribution of many of these enzymes to peptidoglycan metabolism remains unclear due to significant apparent redundancy, particularly among the autolysins. E. coli produces three major LytC-type-N-acetylmuramoyl-L-alanine amidases, which share a role in separating the newly formed daughter cells during cytokinesis. Here, we reveal two of the three amidases that exhibit growth medium-dependent changes in activity. Specifically, we report acidic growth conditions stimulate AmiB—and to a lesser extent, AmiC—amidase activity. Combining genetic, biochemical, and computational analyses, we demonstrate that low pH-dependent stimulation of AmiB is mediated through the periplasmic amidase activators NlpD, EnvC, and ActS (formerly known as YgeR). Although NlpD and EnvC promote amidase activity across pH environments, ActS preferentially stimulates AmiB activity in acidic conditions. Altogether, our findings support partially overlapping roles for E. coli amidases and their regulators in cell separation and illuminate the physiochemical environment as an important mediator of cell wall enzyme activity.
Escherichia coli produces three semi-redundant peptidoglycan amidases involved in cell separation during cytokinesis. Here, we report that two of these amidases, AmiB and AmiC, have enhanced activity in acidic medium. Low pH-dependent AmiB stimulation requires two known amidase activators, EnvC and NlpD, as well as the previously unknown amidase activator ActS. These findings reveal a previously unappreciated role for the growth environment in regulating the active repertoire of extracellular cell wall enzymes and modulatory factors.
Keywords: amidases, cytokinesis, morphogenesis, peptidoglycan, pH
Graphical Abstract
1 |. INTRODUCTION
A cell wall made of peptidoglycan (PG) surrounds nearly all bacterial cells. PG consists of glycan strands of repeating disaccharides of N-acetylglucosamine and N-acetylmuramic acid sugars. Adjacent glycan strands are crosslinked to one another through short peptide stems to form a continuous matrix, referred to as the sacculus, which protects cells from osmotic rupture and confers cell shape. The growth and division of the sacculus must be coordinated with cell replication to prevent lethal breaches in the PG. Accordingly, small molecules that disrupt this process, including β-lactam and glycopeptide antibiotics, are among the oldest and most effective weapons against bacterial pathogens.
In Gram-negative bacteria, the reactions involved in PG synthesis occur in both the cytoplasm and periplasm. Cytoplasmic reactions generate lipid-linked disaccharide precursors, which are translocated to the outer leaflet of the plasma membrane by the flippase MurJ (Sham et al., 2014). In the periplasm, enzymes involved in cell wall synthesis (PG synthases) and degradation (PG autolysins) assemble and modify the new PG material. PG synthases polymerize and crosslink the glycan strands. PG autolysins, on the other hand, exhibit diverse biochemical activities, and their cumulative activity can break nearly every bond in the PG sacculus (Do, Page, et al., 2020). The activities of PG synthases and autolysins must be tightly coupled to one another and to the cell cycle to preserve PG integrity (Egan et al., 2020). Spatiotemporal regulation of these enzymes is particularly important during cytokinesis, which requires concomitant synthesis and hydrolysis of cell wall material to form and separate the new daughter cells.
The precise cellular function of many periplasmic enzymes remains unclear due to significant enzymatic redundancy, particularly among PG autolysins (Heidrich et al., 2001, 2002; Nelson & Young, 2000; Singh et al., 2012; Suzuki et al., 1978). As many as eight enzymes are capable of catalyzing each autolytic reaction in PG metabolism in E. coli (Pazos et al., 2017). In some cases, apparently redundant PG enzymes can be distinguished by differences in substrate specificity (Do, Schaefer, et al., 2020; Yunck et al., 2016), subcellular localization (Banzhaf et al., 2012; Bernhardt & de Boer, 2003; Bertsche et al., 2006), and interaction partners (Banzhaf et al., 2020; Bertsche et al., 2006; Müller et al., 2007; Paradis-Bleau et al., 2010; Typas et al., 2010). Additionally, recent work from our group and others revealed the activity of certain “redundant” PG synthases and autolysins changes based on the physiochemical properties of the growth medium (Castanheira et al., 2017, 2020; Lonergan et al., 2019; Mueller et al., 2019; Murphy et al., 2019; Park et al., 2020; Peters et al., 2016). For example, we discovered two semi-redundant E. coli PG synthases, PBP1a and PBP1b, that are preferentially required for PG integrity in opposing pH environments, in part due to pH-dependent differences in enzymatic activity (Mueller et al., 2019). Collectively, this work suggests the active repertoire of periplasmic PG enzymes is modular with respect to the growth environment (Mueller & Levin, 2020; Pazos et al., 2017), with some enzymes only active in a subset of growth conditions (Castanheira et al., 2017, 2020; Mueller et al., 2019; Murphy et al., 2019; Peters et al., 2016). We hypothesize these enzymes, henceforth referred to as condition-dependent “specialists,” ensure robustness in the periplasmic steps of PG metabolism, which are exposed to the physiochemical properties of the extracellular medium (Chakraborty et al., 2017; Rosenbusch, 1990; Wilks & Slonczewski, 2007). In support of this model, pH specialist enzymes have been identified in every major class of E. coli PG autolysins except one—the amidases (Mueller et al., 2019; Peters et al., 2016; Ursinus & Höltje, 1994).
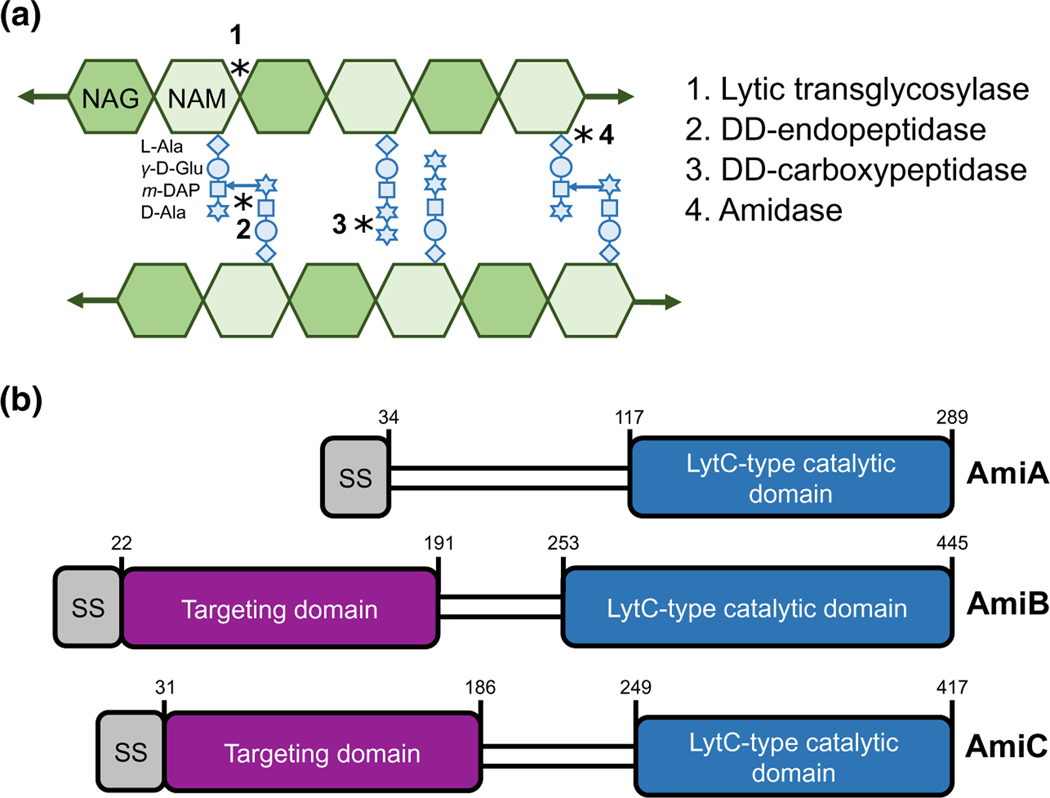
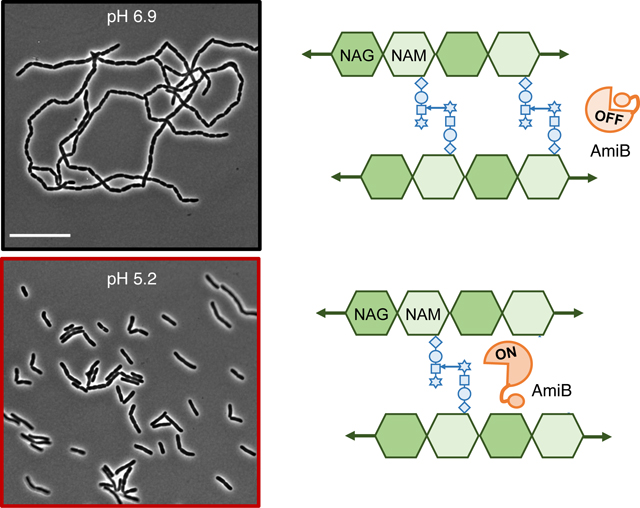
Amidases cleave the peptide stem from the glycan backbone of PG, producing stem-less or “denuded” glycans (Vollmer et al., 2008) (Figure 1a). This reaction is required to separate newly formed daughter cells following cross-wall synthesis during cytokinesis. E. coli produces three major LytC-type-N-acetylmuramoyl-L-alanine amidases with overlapping roles in cell separation: AmiA, AmiB, and AmiC (Figure 1b). Several pieces of evidence support functional redundancy between these three enzymes. Cells separate normally in the absence of any individual amidase, but loss of multiple amidases leads to the formation of chains of unseparated cells that retain distinct cytoplasmic compartments but share an outer membrane (Heidrich et al., 2001; Priyadarshini et al., 2007). At the same time, cytological and interaction studies indicate these enzymes have overlapping, but not necessarily redundant, activities. Notably, the amidases differ in subcellular localization profiles. AmiB and AmiC are enriched at the invaginating septum during cross-wall synthesis, whereas AmiA is uniformly distributed around the cell periphery throughout the cell cycle (Bernhardt & de Boer, 2003; Peters et al., 2011). Moreover, unique interacting partners stimulate amidase activity. Although all three amidases possess an autoinhibitory helix that prevents aberrant autoactivation (Yang et al., 2012), different LytM protein activators are required to relieve autoinhibition and license their cognate amidases’ cleavage activity at midcell during cytokinesis (Peters et al., 2013). The periplasmic protein EnvC activates AmiA and AmiB, whereas the outer membrane-anchored NlpD promotes AmiC activity (Tsang et al., 2017; Uehara et al., 2009, 2010; Yang et al., 2011). Whether the amidases or their regulators also exhibit condition-dependent differences in activity remains unknown.
FIGURE 1.
E. coli produces three LytC-type-N-acetylmuramoyl-L-alanine amidases. (a) A model of the peptidoglycan sacculus. N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) sugars conjoined by a β-(1,4) linkage form glycan strands. Peptide stems emanate from the MurNAc and may be crosslinked to adjacent glycan strands. Peptidoglycan hydrolases cleave the bonds indicated by asterisks. (b) E. coli produces three LytC-type-N-acetylmuramoyl-L-alanine amidases involved in cell separation. Schematic depicts signal sequence (grey), septum targeting domain (purple), and LytC-type catalytic domain (blue). Note that AmiA and AmiC are targeted through the periplasm through the twin arginine translocation (Tat) protein export pathway, whereas AmiB is targeted through the Sec protein translocation pathway
Here, we evaluated the impact of environmental pH on E. coli amidase activity. By comparing the morphology of variants producing a single amidase at pH 6.9 and 5.2, we reveal that AmiB and AmiC have heightened activity in acidic medium, whereas AmiA activity is unaffected across this pH range. AmiB, in particular, is unable to support cell separation in neutral medium but becomes sufficient for cell separation in acidic conditions. Our data indicate that AmiB is not directly activated by acidic pH but instead by the concerted action of EnvC, NlpD, and YgeR, a third LytM protein. Although EnvC and NlpD contribute to cell separation and amidase activity across pH environments, YgeR promotes cell separation in vivo and AmiB activation in vitro specifically in acidic conditions. Based on these finding and data in the accompanying report in this issue of Molecular Microbiology by Gurnani and colleagues indicating YgeR activity is sensitive to outer membrane perturbation (Gurnani et al., 2021), we have renamed it ActS, for amidase activator during stress. Altogether, our work supports a central role for the extracellular environment in dictating the active repertoire of cell wall enzymes and modulatory proteins.
2 |. RESULTS
2.1 |. Acidic pH alleviates cell separation defect of amidase mutants
To assess the impact of pH on the activity of individual amidases, we compared the growth and morphological phenotypes of cells deficient for one or a combination of the three major E. coli LytC-type-N-acetylmuramoyl-L-alanine amidases—AmiA, AmiB, and AmiC. Deletion alleles were sourced from the Keio deletion collection (Baba et al., 2006) with the exception of ΔamiB (a gift from Daniel Kahne, (Chung et al., 2009)). We were unable to verify the Keio version of ΔamiB by Sanger sequencing and hence elected not to use it. All alleles were transduced into the wild-type parental strain MG1655 (Table S1A). We cultured the resulting bacterial strains to steady state in LB medium buffered to pH 5.2 or 6.9 and then imaged the cells by phase-contrast microscopy (see Experimental Procedures). We recorded the growth rate and chain length for each mutant as a function of environmental pH (Figure 2; Table S2).
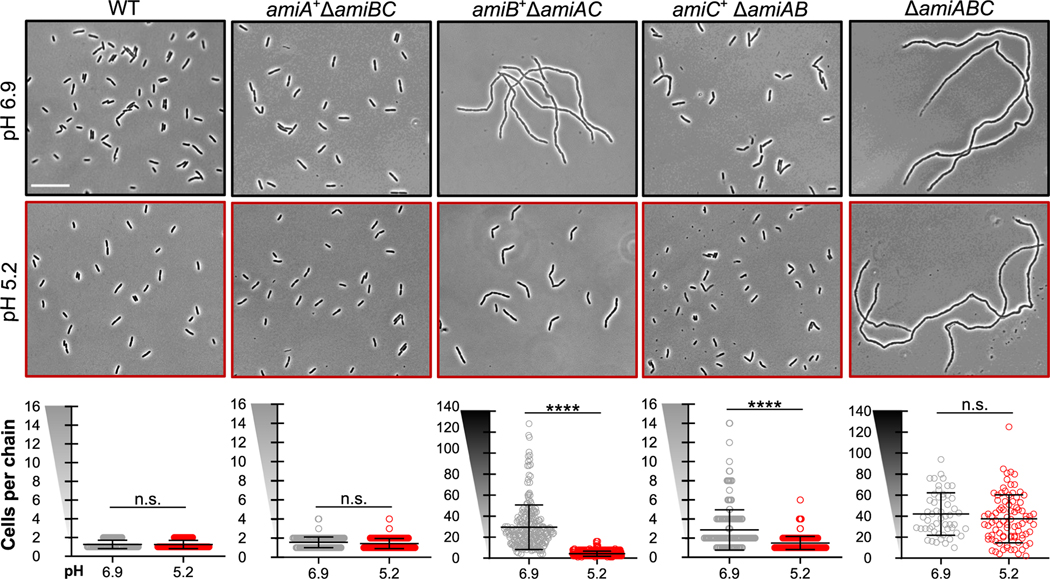
FIGURE 2.
Acidic pH alleviates cell separation defect of amidase mutants. Representative micrographs of double [ΔamiAB (EAM1379), ΔamiAC (EAM927), and ΔamiBC (EAM1381)] and triple amidase (ΔamiABC, EAM1385) mutants during steady-state growth in LB medium buffered to pH 6.9 or 5.2 compared with the parental strain (MG1655). Cells were grown to mid-exponential phase (OD600 ~ 0.2–0.6) at the indicated pH, sub-cultured into the same medium at an OD600 ~ 0.005, then sampled and fixed at OD600 ~ 0.1–0.2 for microscopy. Scale bar denotes 20 μm. Quantification for each mutant is shown below the corresponding set of micrographs. Points indicate individual chain length measurements from at least two independent experiments, and error bars denote standard deviation. Y-axis gradient emphasizes the difference in scale used between mutants. Significance was determined by Kolmogorov–Smirnov test with a threshold of statistical significance set to p < .01
Although none of the single amidase mutants exhibited pH-dependent changes in growth rate or morphology (Figure S1, and Table S2), analysis of the double mutants indicates that two of E. coli’s three amidases, AmiB and AmiC, are sensitive to pH. Double mutants encoding only amiB or amiC (amiB+ ΔamiAC or amiC+ ΔamiAB, respectively) displayed pH-dependent chaining phenotypes (Figure 2; Table S2). Consistent with previous studies (Chung et al., 2009; Heidrich et al., 2001), each of these strains formed chains at pH 6.9, albeit to differing extents. amiB+ ΔamiAC cells formed long chains of 25 ± 1 cells in length, whereas amiC+ ΔamiAB formed short chains of 2–4 cells in length. As expected, normal cell separation could be restored by ectopic expression of the missing amidase alleles from a plasmid (Figure S3). More surprisingly, culturing the double mutants in acidic medium was also sufficient to reduce chaining by up to 86% (amiB+ ΔamiAC, mean of 4 ± 1 cells per chain; mean of amiC+ ΔamiAB, 1 ± 1 cell per chain). The dramatic reduction in chaining was accompanied by only a minimal reduction in growth rate at low pH (Table S2).
To validate the findings for the amiB+ ΔamiAC mutant, we took advantage of mutants defective in the twin-arginine translocation (Tat) protein export pathway (ΔtatB and ΔtatC), which is required for the secretion of AmiA and AmiC into the periplasm (Bernhardt & de Boer, 2003; Ize et al., 2003). In agreement with previous reports (Bernhardt & de Boer, 2003; Ize et al., 2003), ΔtatB and ΔtatC mutants were phenotypically identical to the amiB+ ΔamiAC mutant at pH 6.9. Likewise, growth in acidic medium dramatically reduced the chain length of ΔtatB and ΔtatC mutants (Figure S2), highlighting acid-dependent stimulation of AmiB activity. These findings reinforce a model in which AmiB is largely sufficient for cell separation during growth in acidic, but not neutral, pH conditions.
In contrast to AmiB and AmiC, AmiA appears sufficient for normal cell separation during growth in both neutral and acidic conditions. An amiA+ ΔamiBC-double mutant separated normally when cultured at pH 6.9 or 5.2. Conversely, the ΔamiABC-triple mutant formed long chains of unseparated cells at both acidic and neutral pH (pH 6.9, mean of 44 ± 2 cells per chain; pH 5.2, mean of 37 ± 5 cells per chain) (Table S2).
2.2 |. Acidic pH restores cell envelope integrity of amidase mutants
Defects in cell separation are typically associated with compromised cell envelope permeability and render cells sensitive to hydrophobic compounds, detergents, and certain antibiotics (Heidrich et al., 2002; Ize et al., 2003; Tsang et al., 2017; Weaver et al., 2019; Yakhnina et al., 2015). We reasoned that because pH influences cell separation, the amidase mutants may also exhibit differential envelope permeability across pH conditions. To assess cell envelope permeability, we compared the amidase-double mutants’ sensitivity to the commonly used detergent Triton-X during growth in neutral (pH 6.9) or acidic (pH 5.2) medium. Relative sensitivity was assessed by determining the minimum inhibitory concentration of Triton-X for each strain.
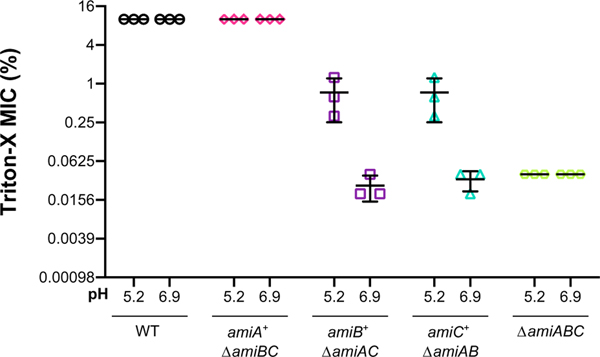
Supporting the pH sensitivity of AmiB and AmiC, amiB+ ΔamiAC and amiC+ ΔamiAB strains were up to 64-fold more resistant to Triton-X when cultured in acidic medium compared with neutral medium (Figure 3). By contrast, wild-type cells and amiA+ ΔamiBC mutants, both of which exist primarily as individual cells (Figure 2), were resistant to Triton-X at both pH conditions (MIC of 10% Triton-X). The ΔamiABC-triple mutant remained sensitive to the detergent at both pH conditions (MIC of 0.039% Triton-X). Altogether, our data indicate that acidic pH partially alleviates the cell separation defect and cell envelope permeability of ΔamiAC and ΔamiAB-double mutants, presumably through activation of AmiB and AmiC.
FIGURE 3.
Acidic pH restores cell envelope integrity of amidase mutants. Comparison of the minimum inhibitory concentration (MIC) of Triton-X for indicated mutants when cultured in buffered LB medium at pH 6.9 or 5.2 (wild type [MG1655], ΔamiAB [EAM1379], ΔamiAC [EAM927], ΔamiBC [EAM1381], and ΔamiABC [EAM1385]). Cells were grown to mid-exponential phase (OD600 ~ 0.2–0.6) in LB medium at pH 6.9, then sub-cultured into 96-well plates to 1 × 105 CFU/ml in LB medium buffered to either pH 6.9 or 5.2 with varying percentages of Triton-X detergent. Cells were grown with aeration at 37 °C for 20 hr before MIC was determined by visual inspection. Error bars denote standard deviation
2.3 |. Acidic pH stimulates AmiB and AmiC activity in vivo
Our genetic data are consistent with two models: 1) AmiB and AmiC activity is sufficiently enhanced at low pH to support cell separation in the absence of other amidases; 2) cell separation in acidic conditions may be mediated in part by an unknown amidase or by another class of cell wall autolysins. Endopeptidases and lytic transglycosylases also play roles in cell separation under standard culture conditions (Heidrich et al., 2002; Priyadarshini et al., 2006; Weaver et al., 2019). Consistent with the first model, cells deficient for all three amidases (ΔamiABC) produced long chains of similar cell lengths and displayed equivalent sensitivity to Triton-X when cultured under either pH condition (Figures 2 and 3; Table S2). Moreover, overexpression of the wild-type gene, but not a catalytically dead variant of amiB (H269A), from an arabinose-inducible promoter restored cell separation in amiB+ ΔamiAC and ΔamiABC mutants at pH 6.9 (Figures S3 and S4). Together these data support the first model, indicating that increases in basal AmiB activity are sufficient to alleviate chaining.
To further assess the impact of pH on amidase activity in vivo and exclude indirect effects by another class of PG autolysins, we leveraged the finding that the products of amidase activity—“denuded” glycans lacking stem peptides—are specifically recognized by “sporulation-related repeat” (SPOR) domain-containing proteins. These proteins include the E. coli cell division proteins DamX, DedD, RlpA, and FtsN, which rely on the SPOR domain to localize to midcell during cytokinesis (Yahashiri et al., 2015, 2017). As amidase activity is concentrated at sites of septation, there should be an increase in denuded glycans at medial and polar positions in growing cells. Thus, the midcell binding of a fluorescent protein-SPOR-domain fusion should reflect levels of septal amidase activity in vivo. To this end, we incubated purified His6-GFP-DamX SPOR—which binds denuded glycans in isolated sacculi (Yahashiri et al., 2015)—and incubated the fusion protein with fixed wild-type, amiA+ ΔamiBC, amiB+ ΔamiAC, or amiC+ ΔamiAB cells (see Experimental Procedures; Figure S5a). Notably, while cells were cultured at either neutral or acidic pH prior to fixation, the SPOR domain-binding assay was performed on fixed cells equilibrated to pH 7.4 to ensure uniform reactions conditions. Note that fixation typically permeabilizes the cell envelope (Levin & Losick, 1996), thus both wild type and mutant cells should be equivalent in this regard.
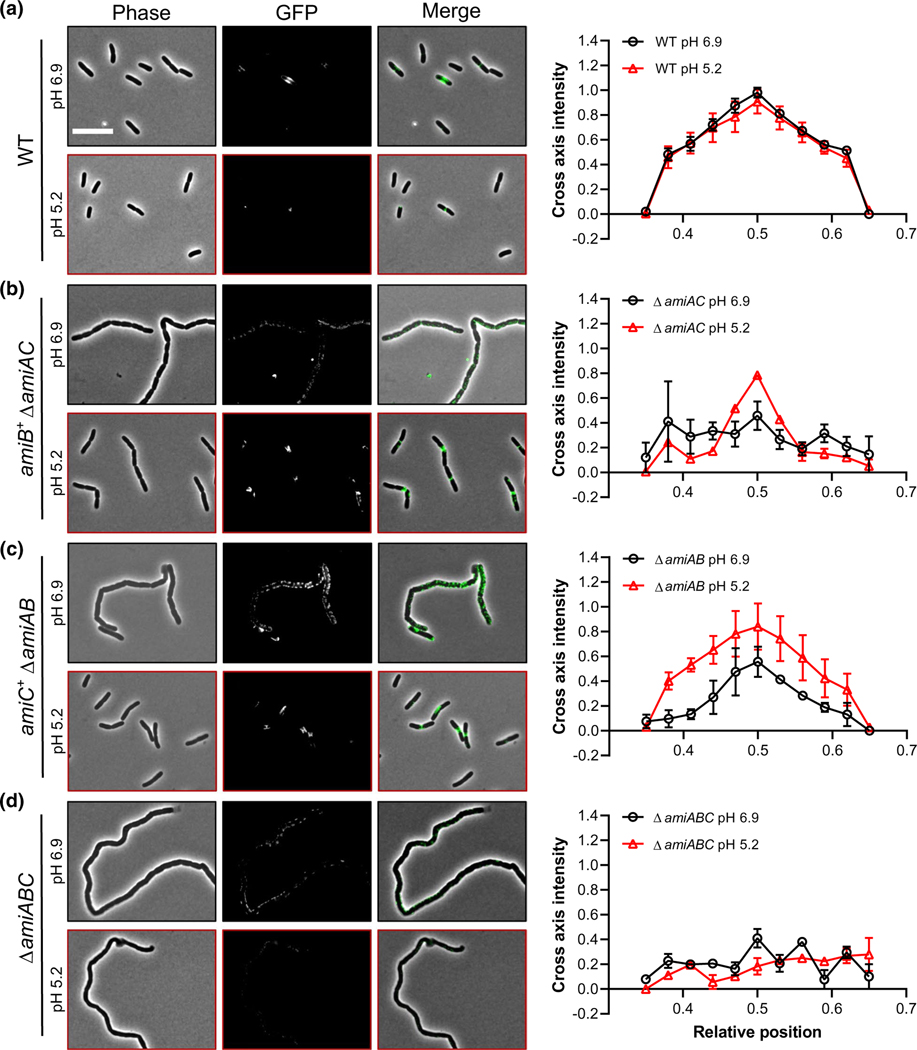
Consistent with SPOR domain-binding serving as a bone fide reporter of denuded glycans in fixed cells, His6-GFP-DamXSPOR fluorescence was concentrated at midcell in wild-type E. coli cultured at neutral and acidic pH (Figure 4a; Figure S5b,c). As expected, we did not observe midcell enrichment of His6-GFP-DamXSPOR localization in the ΔamiABC mutant (Yahashiri et al., 2015; Yakhnina & Bernhardt, 2020) (Figure S5b,c). We did, however, observe foci of His6-GFP-DamXSPOR binding along the cell body of the ΔamiABC mutant. Conversely, medial fluorescence from His6-GFP-DamXSPOR was enhanced four-fold over wild type in a strain that lacks six lytic transglycosylases (Δ6LT) (Figure S5b,c); this strain was previously shown to have elevated levels of denuded glycans (Yahashiri et al., 2015), possibly due to reduced glycan processing (Yakhnina & Bernhardt, 2020). Importantly, chain length is not necessarily correlated with His6-GFP-DamXSPOR intensity at the septum, as the Δ6LT mutant forms chains 2–8 cells in length (Figure S5d).
FIGURE 4.
SPOR domain-binding assay indicates elevated AmiB and AmiC activity at midcell in acidic medium. Representative micrographs and normalized intensity profiles for wild type (MG1655, a), ΔamiAC (EAM927, b), ΔamiBC (EAM1381, c), and ΔamiABC (EAM1385, d) after labeling with His6-GFP- DamXSPOR protein. Cells were cultured to steady-state in LB medium (pH 6.9 or 5.2) and then fixed at OD600 ~ 0.1–0.2. Fixed cells were immediately labeled with 100 ng/ml of purified His6-GFP-DamXSPOR protein for 30 min on ice, washed, and then visualized by fluorescence microscopy. Fluorescence intensity profiles were generated from at least 50 cells across two biological replicates and normalized to the maximum intensity of the wild type at pH 6.9. Error bars represent standard deviation. Scale bar denotes 10 μm
In support of elevated AmiB and AmiC activity at low pH, amiB+ ΔamiAC or amiC+ ΔamiAB strains cultured at pH 5.2 prior to fixation displayed a ~50% increase in His6-GFP-DamXSPOR at midcell relative to their counterparts cultured at 6.9 (Figure 4b,c). This phenotype was particularly stark for the amiB+ ΔamiAC cells, which only exhibited nonspecific probe binding in neutral medium (Figure 4b). Conversely, wild-type cells and cells producing only AmiA (ΔamiBC) bound GFP-DamX SPOR with similar affinities at midcell regardless of whether they were cultured at pH 6.9 or 5.2 (Figure 4a; Figure S5e). Overall, our overexpression and cytological studies support a model in which low pH stimulates AmiB and AmiC amidase activity in vivo to promote cell separation.
2.4 |. Acidic pH does not directly stimulate AmiB and AmiC activity in vitro
To address whether low pH stimulates AmiB or AmiC hydrolytic activity directly, we purified AmiA, AmiB, and AmiC and incubated the enzymes individually with purified E. coli PG sacculi for 1 hr at pH 5.2 or pH 7.5. We digested the reaction products with the muramidase cellosyl and resolved the muropeptides by reverse-phase high-performance liquid chromatography as previously described (Glauner et al., 1988). A reduction in the quantity of monomeric and dimeric muropeptides, referred to as Tetra and TetraTetra muropeptides, indicates enhanced amidase activity in a particular reaction condition. Consistent with previous reports, all of the amidases had low activity in vitro in the absence of their cognate LytM protein activator (Uehara et al., 2010), and acidic pH did not significantly enhance hydrolytic activity for any enzyme (Figure 5a). This result suggests that pH does not directly modulate AmiB or AmiC enzymatic activity and instead acts through other modulatory factors.
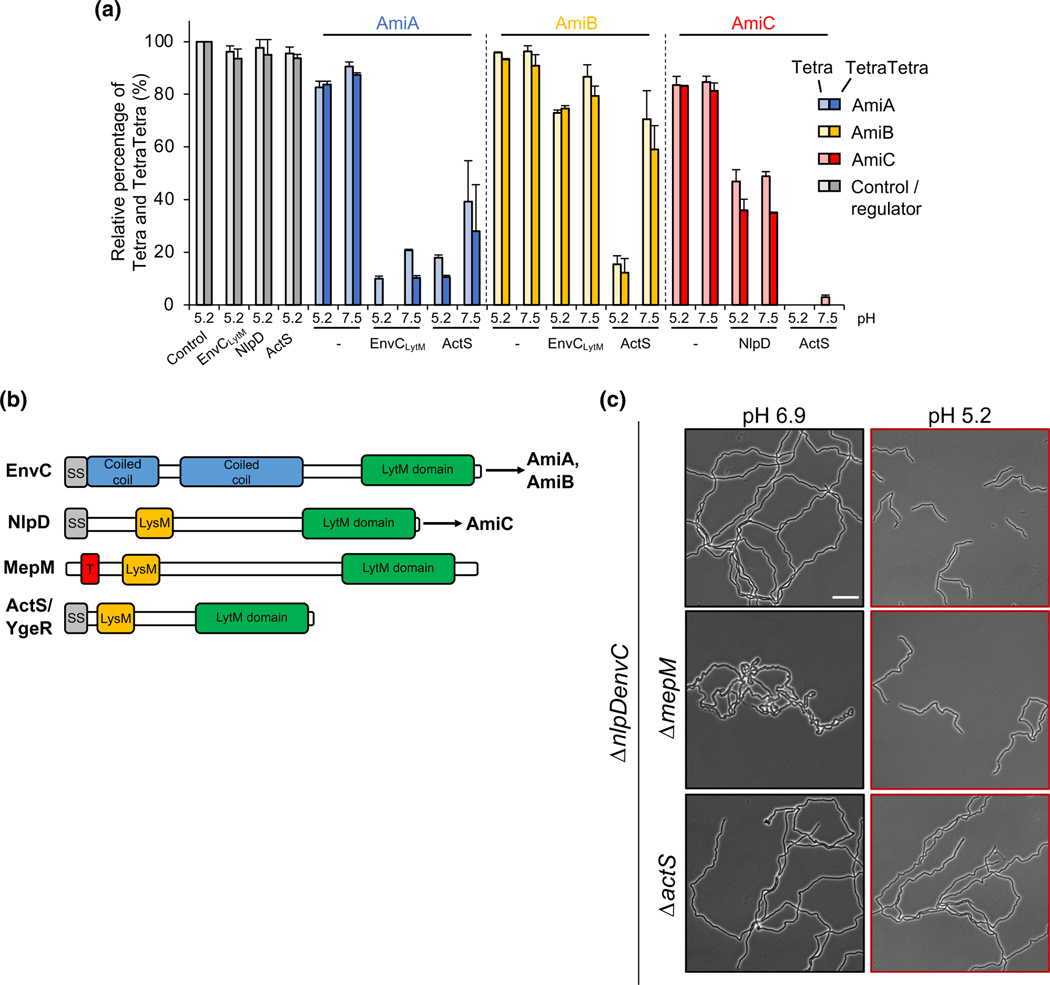
FIGURE 5.
LytM-domain protein ActS contributes to cell separation at acidic pH. (a) PG was incubated with AmiA, AmiB or AmiC, in the presence or absence of their cognate activator or ActS, digested with the muramidase cellosyl and the resulting PG fragments were reduced with sodium borohydride and separated by HPLC. The peaks of the major muropeptides, Tetra (monomer) and TetraTetra (dimer), were quantified from the chromatograms and their amount in control samples were set as 100%. Amidase activity causes a reduction in Tetra and TetraTetra. At pH 5.2, ActS activated AmiB and AmiC better than EnvCLytM and NlpD, respectively. Representative HPLC chromatograms are shown in Figure S6. Values are mean ± variation of two independent samples. EnvCLytM, LytM-domain of EnvC. B) E. coli produces four LytM-domain proteins: EnvC, NlpD, MepM, and ActS. Schematic depicts conserved and unique domains among the proteins, including the signal sequence (grey), coiled coil regions (blue), LysM domain (orange), and the conserved LytM domain (green). The current model suggests EnvC specifically activates AmiA and AmiB, whereas NlpD specifically activates AmiC. C) Representative micrographs for ΔnlpDenvC (EAM1372), ΔnlpDenvCmepM (EAM1477), and ΔnlpDenvCactS (EAM1521) during steady-state growth in LB medium buffered to pH 6.9 or 5.2. Cells were grown to mid-exponential phase (OD600 ~ 0.2–0.6) at the indicated pH, sub-cultured into the same medium at an OD600 ~ 0.005, then sampled at OD600 ~ 0.1–0.2 for live cell microscopy. Scale bar denotes 20 μm
2.5 |. pH-dependent AmiB and AmiC stimulation is partially independent of amidase activators EnvC and NlpD
Amidase activity is spatiotemporally controlled in vivo and enhanced in vitro through direct interaction with the LytM proteins EnvC and NlpD. The periplasmic protein EnvC (Bernhardt & de Boer, 2004; Hara et al., 2002) and the outer membrane lipoprotein NlpD (Tsang et al., 2017) are required for efficient amidase activity in vitro and in vivo and restrict amidase activation to the midcell after the onset of cytokinesis (Uehara et al., 2009, 2010). At neutral pH, EnvC specifically stimulates AmiA and AmiB, whereas NlpD is responsible for AmiC activation (Uehara et al., 2010) (Figure 5b), although cross-activation has been observed in some genetic backgrounds in vivo (Tsang et al., 2017; Yang et al., 2012). In accordance with their requirement for amidase activation, cells defective for EnvC and NlpD production phenotypically mimic an ΔamiABC mutant and form long chains of unseparated cells under standard culture conditions (Uehara et al., 2009). To assess whether either one of these regulators mediate AmiB or AmiC activation in acidic medium, we compared cell chaining of an ΔnlpDenvC mutant across pH conditions.
Our data indicate acidic pH stimulates the amidases through a mechanism partially independent of EnvC and NlpD. At neutral pH, the ΔnlpDenvC mutant formed chains with an average of 45 ± 3 cells (Figures 5c and 7a; Table S2), similar to the chain length observed for the ΔamiABC-triple mutant (Figure 2). However, in contrast to the pH-insensitive ΔamiABC-triple mutant, the average chain length for the ΔnlpDenvC mutant was reduced by more than four-fold to 11 ± 1 cells per chain during growth in acidic medium (Figures 5c and 6a; Table S2). The reduction in chaining in acidic medium was dependent on amiB and amiC, but not amiA (Figure S7). Altogether, the retention of pH sensitivity by the ΔnlpDenvC mutant suggests that AmiB and AmiC activation during growth at low pH is partially independent of EnvC or NlpD.
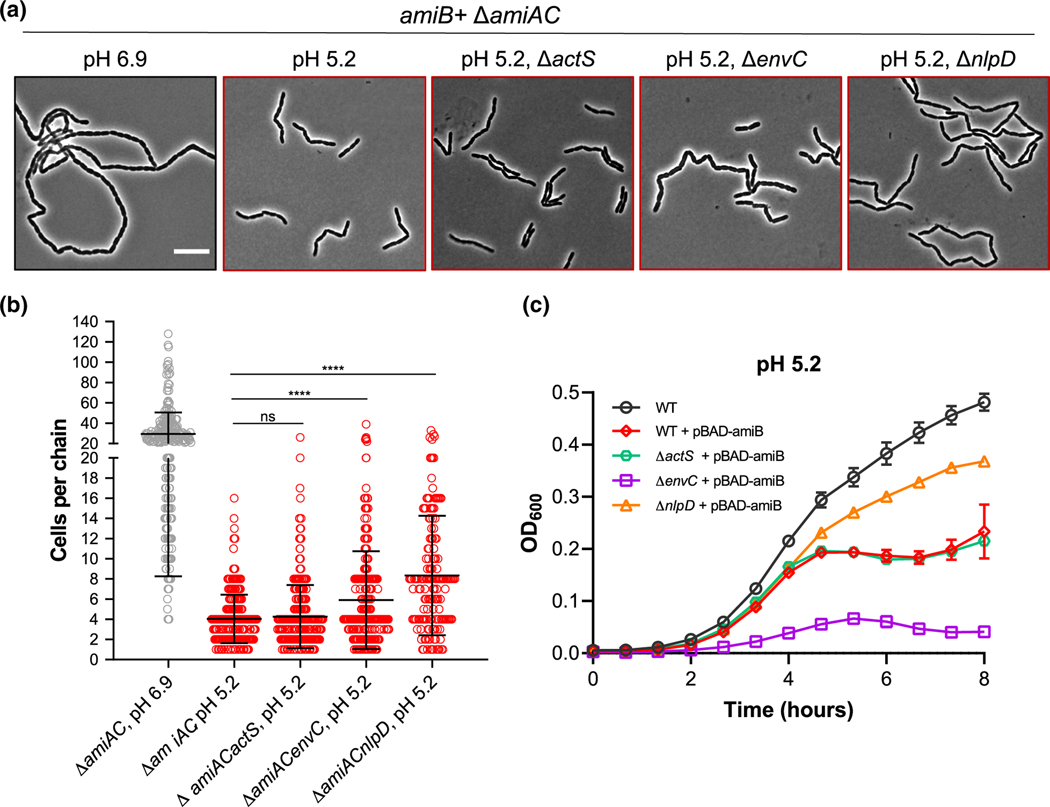
FIGURE 7.
Promiscuous AmiB activation by LytM-domain proteins at acidic pH. (a) Representative micrographs for ΔamiAC (EAM1000), ΔamiACactS (EAM1549), ΔamiACenvC (EAM1551), and ΔamiACnlpD (EAM1553) during steady-state growth in LB medium buffered to pH 6.9 or 5.2. Cells were grown to mid-exponential phase (OD600 ~ 0.2–0.6) at the indicated pH, sub-cultured into the same medium at an OD600 ~ 0.005, then sampled at OD600 ~ 0.1–0.2 for live cell microscopy. Scale bar denotes 10 μm. (b) Quantification of cell chaining for mutants described in panel A. Points indicate individual chain length measurements from at least two independent experiments, and error bars denote standard deviation. Significance was determined by Kolmogorov–Smirnov test with Dunn’s correction for multiple comparison with a threshold of statistical significance set to p < .01. (c) Growth curves of wild type (MG1655), ΔactS (EAM1514), ΔenvC (EAM1370), and ΔnlpD (EAM1364) overexpressing amiB from a plasmid in the presence or absence of inducer (0.04% arabinose) when cultured at pH 5.2. Cells were grown to mid-exponential phase (OD600 ~ 0.2–0.6) in LB medium at pH 6.9, then sub-cultured into 96-well plates at an OD600 ~ 0.005 in LB medium buffered to 5.2. Cells were grown with aeration at 37 °C, and the optical density was measured every 10 min. Curves represent the average ± standard deviation of two independent experiments
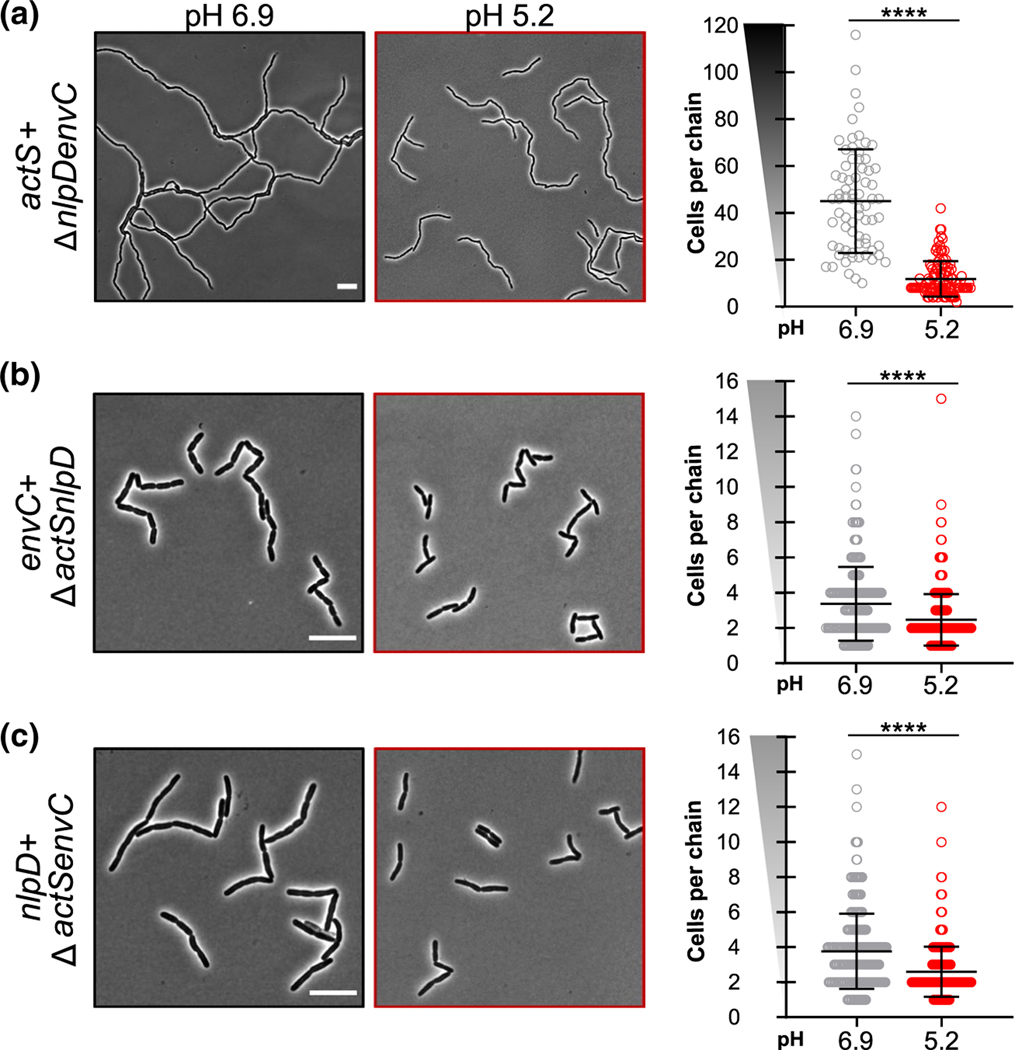
FIGURE 6.
LytM-domain protein activation at acidic pH. Representative micrographs (left) and quantification of cell chaining (right) for ΔnlpDenvC (EAM1372, a), ΔactSnlpD (EAM1516, b), ΔactSenvC (EAM1540, c) during steady-state growth in LB medium buffered to pH 6.9 or 5.2. Cells were grown to mid-exponential phase (OD600 ~ 0.2–0.6) at the indicated pH, sub-cultured into the same medium at an OD600 ~ 0.005, then sampled at OD600 ~ 0.1–0.2 for live cell microscopy. Points indicate individual chain length measurements from at least two independent experiments, and error bars denote standard deviation. Y-axis gradient emphasizes the difference in scale used between mutants. Significance was determined by Kolmogorov–Smirnov test with a threshold of statistical significance set to p < .01. Scale bar denotes 10 μm
2.6 |. The LytM protein ActS contributes to amidase-dependent cell separation in acidic medium
Several models could explain the ΔnlpDenvC mutant’s enhanced cell separation in acidic medium. First, lysis of cells internal to the chain may be sufficient to separate cells and reduce overall chain length at low pH. However, unlike their counterparts at pH 6.9, few ΔnlpDenvC chains terminated with lysed cells at pH 5.2 (Figure 5c), as would be expected if chain breakage was the predominant mechanism for cell separation (Priyadarshini et al., 2006). Alternatively, acidic pH may stimulate AmiB or AmiC through an interaction with an unknown amidase activator.
Apart from EnvC and NlpD, E. coli produces two additional proteins with LytM domains: 1) ActS, a lipoprotein formerly known as YgeR with unknown function, and 2) MepM, an enzyme with DD-endopeptidase activity also known as YebA (Figure 5b). Inactivation of either or both genes modestly enhances the chaining of ΔnlpDenvC mutants during growth in standard culture conditions (Uehara et al., 2009), suggesting a minor role in cell separation in these conditions. To investigate whether either ActS or MepM play a more predominant role in cell separation in acidic medium, we inactivated each gene in the ΔnlpDenvC background and compared chaining across pH conditions.
Our results suggest ActS functions as an amidase activator at low pH. Loss of ActS, but not MepM, abolished cell separation of ΔnlpDenvC cells in acidic medium (Figure 5c). ΔnlpDenvCactS cells formed long chains of >40 cells in length at both neutral and acidic pH, indicating this protein is responsible for the residual amidase activity in ΔnlpDenvC cells. Results from Gurnani et al. in this issue of Molecular Microbiology further support a role for ActS in amidase activation (Gurnani et al., 2021). ΔnlpDenvCmepM cells, on the other hand, displayed a similar cell separation pattern as the ΔnlpDenvC parental strain with enhanced separation in acidic medium (Figure 5c). Although its absence did not affect pH-dependent chaining, loss of MepM dramatically altered cell morphology at neutral pH. ΔnlpDenvCmepM chains exhibit atypical bulges and twists in neutral medium, but these morphological aberrations are largely absent when cells were cultured at pH 5.2. These data implicate MepM in the preservation of normal rod shape during growth in neutral pH medium.
2.7 |. Promiscuous amidase activation by LytM proteins in acidic medium
Although ActS contributes to cell separation at low pH, our data demonstrate this protein does not function as the sole amidase activator in this condition. ΔnlpDenvC cells still form chains greater in length than the amiB+ ΔamiAC mutant in acidic medium (Figures 2, 5c and 6a; Table S2). Moreover, single ΔactS mutants separate normally when cultured at low pH (Figure S8a). Together, this evidence suggests EnvC and/or NlpD may also have heightened activation capacity in acidic medium. To isolate the pH sensitivity of each activator, we generated additional mutants that only produce a single LytM activator and compared cell chaining across pH conditions.
Our data indicate acidic pH augments both EnvC and NlpD’s ability to promote cell separation in vivo. Similar to the actS+ ΔnlpDenvC mutant, cells only expressing envC (ΔnlpDactS) or nlpD (ΔactSenvC) also exhibited reductions in chaining in acidic medium. Both mutants formed short chains of 3–4 cells in length when cultured in neutral medium but existed predominately as single cells or pairs of cells at low pH (Figure 6; Table S2). These results imply that heightened amidase activity in acidic medium could be a result of promiscuous activation by LytM-domain proteins, as opposed to cognate activation of each amidase by its specific activator as previously demonstrated in vitro at neutral pH (Uehara et al., 2010). To differentiate between these possibilities, we inactivated each LytM activator in the amiB+ ΔamiAC background, which exhibits the most dramatic pH-dependent cell chaining phenotype (Figure 2). If multiple LytM activators stimulate AmiB in acidic medium, as our model predicts, then we should still observe low pH-dependent reductions in chaining in these strains.
In accordance with promiscuous activation, inactivation of ActS, EnvC, or NlpD alone did not abolish the sensitivity of amiB+ ΔamiAC cells to low pH (Figure 7a,b; Table S2). Strikingly, loss of EnvC—the cognate activator for AmiB at neutral pH (Uehara et al., 2010)—had only a modest effect on chain length (ΔamiAC, mean of 4 ± 1 cells per chain; ΔamiACenvC, mean of 6 ± 1 cells per chain). Loss of NlpD, in contrast, had the strongest effect (ΔamiACnlpD, mean of 8 ± 1 cells per chain). This result was surprising considering NlpD does not stimulate AmiB activity in vitro at neutral pH (Uehara et al., 2010). To independently verify a dominant role for NlpD in AmiB activation at low pH, we compared cell lysis during amiB overexpression for mutants defective for a single LytM protein. At intermediate inducer concentrations (0.04% arabinose), amiB overexpression in the wild type background induces cell lysis only in acidic growth conditions (Figure S10). Cells defective for EnvC or ActS exhibited similar lysis kinetics to the parental strain upon amiB overexpression in acidic medium. Although we note that ΔenvC mutant has a significant growth rate defect, lysis kinetics during amiB overexpression remains unchanged. Loss of NlpD, in contrast, alleviated toxicity (Figure 7c). Overall, our findings argue acidic conditions stimulate AmiB in vivo through multiple LytM activators with NlpD having the greatest individual effect.
2.8 |. Computational analysis suggests pH activates AmiB by promoting its interaction with activators
We next sought to understand the mechanistic basis for promiscuous AmiB stimulation by the LytM activators in acidic medium. Structural modeling and genetic studies of EnvC and NlpD suggest that they stimulate their cognate amidase(s) through a direct interaction between their degenerate (noncatalytic) LytM domain and the target amidase’s autoinhibitory helix, resulting in the displacement of the helix from the enzyme’s active site (Peters et al., 2013). The proposed interaction interface is highly charged, implying that electrostatic forces drive the interaction between these domains. At neutral pH, the degenerate LytM domains are positively charged (isoelectric point between 8.80 and 8.90 for EnvC, NlpD, and ActS), whereas the autoinhibitory helices are either neutral (isoelectric points: 6.27 for AmiA and 6.08 for AmiC) or negatively charged (isoelectric point 4.32 for AmiB). Consistent with an interaction based in part on electrostatics, mutations leading to charge inversion in the autoinhibitory helix of AmiB or the LytM domain of EnvC impact amidase activity in vivo and in vitro (Peters et al., 2013; Yang et al., 2012).
With these data in mind, we hypothesized that acidic pH may modulate the amidase–activator interaction through one or both of the following mechanisms: 1) increase in the electrostatic complementation between amidase and activator at the interaction interface and 2) decrease in the affinity of the amidase autoinhibitory helix for the enzyme’s active site, which would facilitate interaction with the activator. To examine the effect of environmental pH on electrostatic complementation, we modeled the LytM domains of EnvC, NlpD, and ActS and the autoinhibitory helices of AmiA, AmiB, and AmiC (see Experimental Procedures). We generated two models: one leveraged a docked structure of AmiB with EnvC that is supported by mutational analyses of both proteins (Peters et al., 2013; Yang et al., 2012) and structural studies of the catalytic cleft of the S. aureus LytM protein (Grabowska et al., 2015) (Figure 8a); the second is a structure-based holistic analysis, which is agnostic to the specific LytM domain-binding cleft prediction (Figure S9). We then calculated the electrostatic potential of each modeled structure using the APBS-PDB2PQR software suite (Baker et al., 2001; Dolinsky et al., 2007). Interestingly, the autoinhibitory helices of AmiA and AmiC remain neutral also at pH 5.2, whereas that of AmiB becomes more negatively charged (Figure 8a and Figure S9). At the same time, all three LytM domains become more positively charged at pH 5.2 compared with pH 6.9 (Figure 8a and Figure S9), albeit for EnvC the change is minor.
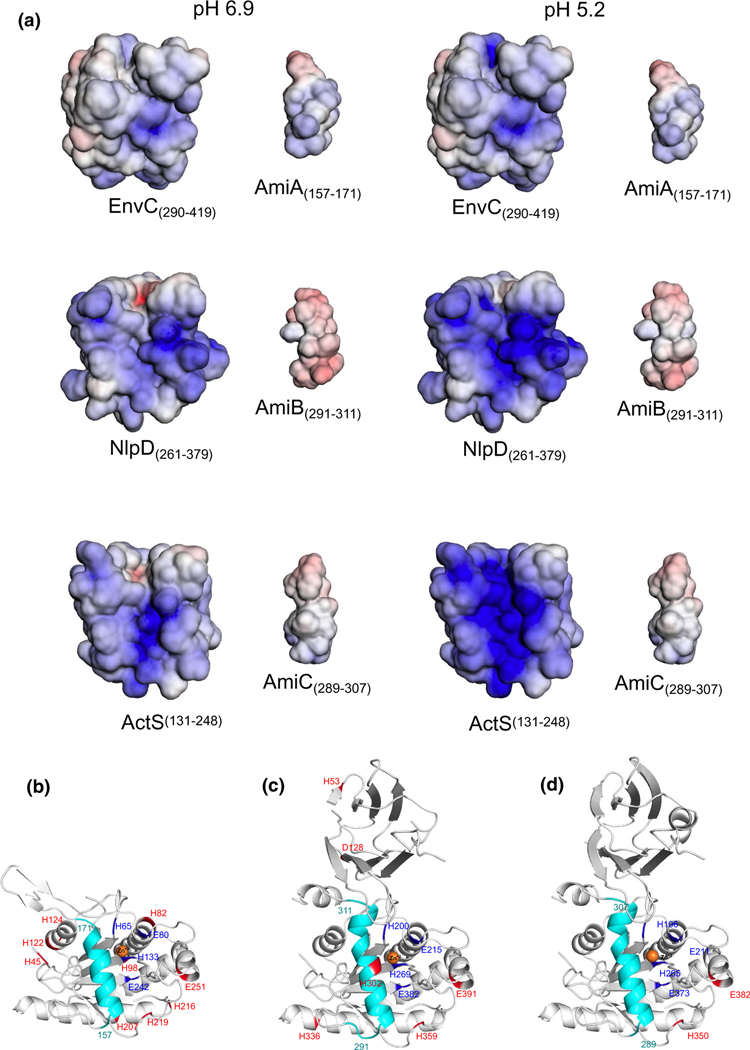
FIGURE 8.
Computational analysis of electrostatic potentials and protonation state shifting residues of E. coli LytM and Ami proteins from pH 6.9–5.2. (a) Minimum (−5 kT/e; red) and maximum (5 kT/e; blue) electrostatic potentials of the LytM domains of EnvC (residues 290–419), NlpD (residues 261–379), ActS (residues 131–248), AmiA (157–171), AmiB (291–311) and AmiC (289–307), calculated using the APBS-PDB2PQR software (Baker et al., 2001; Dolinsky et al., 2007) at pH 6.9 and 5.2. (b–d) Residues that change protonation state from pH 6.9 to 5.2 are shown and highlighted in red for AmiA (residues 41–276) (b), AmiB (residues 19–416) (c), and AmiC (residues 30–408) (d) structures. Zn+2 is shown as orange sphere, active site in dark blue and LytM domain-interacting helices in cyan (AmiA: residues 157–171; AmiB: residues 291–311; AmiC: residues 289–307). The figure was generated with PyMOL (The PyMOL Molecular Graphics System, Version 1.8.4.0, Schrödinger, LLC [software], n.d.)
Next, to analyze the impact of pH on the autoinhibitory helix of the three amidases, we applied patcHwork, a webserver allowing for the automatic identification of residues that undergo charge shifts in two user-defined pH values (Schmitz et al., manuscript in preparation; https://patchwork.biologie.uni-freiburg.de:2000), patcHwork revealed a putative pH-sensitive histidine (H302) in AmiB’s autoinhibitory helix (Figure 8c). This helix, which abuts the enzyme’s catalytic core, has previously been implicated in AmiB autoinhibition (Yang et al., 2012). AmiA and AmiC, in contrast, were not predicted to possess pH-sensitive residues within their autoinhibitory helices (Figure 8b,d). Taken together, our computational analyses predict AmiB to be the most pH sensitive among the three amidases and NlpD and ActS to be the strongest activators of AmiB at low pH.
2.9 |. Acidic pH augments amidase stimulation by ActS
Our genetic data suggest 1) ActS is an amidase activator with enhanced stimulatory capacity at low pH and 2) enhanced AmiB activity at low pH is a result of promiscuous stimulation by the three LytM amidase activators. Our computational modeling indicates both of these may be driven in part by pH-dependent changes at the amidase–activator interaction interface. To test these predictions biochemically, we purified ActS, EnvCLytM, and NlpD and compared their ability to enhance amidase activity in vitro pH 5.2 or 7.5 as described previously.
Co-incubation with ActS enhanced amidase activity at both pH conditions, but consistent with our genetic data, the stimulation was greatest at acidic pH for all three amidases. The pH-dependence of ActS’ activation capacity was pronounced for AmiB. At pH 7.5, ActS was capable of stimulating all three amidases’ activity at levels comparable to or greater than the amidase’s cognate regulator (Figure 5a), a result reinforcing its classification as an amidase activator. Similar to AmiB’s cognate activator EnvC, ActS only modestly enhanced AmiB activity at neutral pH. At pH 5.2, however, ActS stimulated AmiB at least five-fold more than EnvC in the same reaction conditions (Figure 5a). These findings complement our genetic and computational analyses (Figures 2 and 5c) and reinforce a model in which amidase activation in acidic medium is driven at least in part by ActS. We also observed enhancement of EnvC stimulatory capacity at low pH in vitro. Acidic pH improved EnvCLytM-mediated stimulation of cognate enzymes AmiA and AmiB (Figure 5a) but not of the noncognate enzyme AmiC (Figure S11). Surprisingly, in contradiction to our genetic analysis, acidic pH did not promote NlpD-mediated stimulation of its cognate amidase AmiC (Figure 5a, Figure S11) or either of the noncognate enzymes AmiA or AmiB, suggesting additional factors are required for NlpD activation at low pH (see Discussion).
3 |. DISCUSSION
Here, we report two of E. coli’s three LytC-type-N-acetylmuramoyl-L-alanine amidases that exhibit pH-dependent changes in activity. Although AmiB or AmiC alone cannot support normal cell separation or cell envelope integrity in neutral medium, low pH increases their activity by at least two-fold in vivo (Figure 4), permitting formation of short chains of two to four cells and single bacteria (Figure 2). The pH dependence of amidase activity is most stark for AmiB. Our genetic, cytological, and biochemical data indicate this enzyme has only minimal activity in vivo in neutral medium when it is specifically stimulated by its cognate LytM activator EnvC (Uehara et al., 2010). In acidic medium, promiscuous activation by three LytM proteins—EnvC, NlpD, and ActS—augments AmiB’s ability to support cell separation (Figure 7). This finding builds upon our previous work (Mueller et al., 2019) and challenges the perception of redundancy (i.e., completely overlapping activity profiles) within periplasmic PG enzymes. Instead of being redundant, PG enzymes exhibit complementary activities with some specialized for distinct environmental conditions. This mechanism may ensure robust cell wall construction across environmental niches in which cells naturally grow outside the laboratory (Mueller & Levin, 2020).
3.1 |. LytM proteins enhance amidase activity in acidic environments
Our in vitro and in vivo data support a model in which acidic conditions stimulate AmiB through the LytM activators EnvC, NlpD, and ActS. Importantly, in combination with the accompanying report by Gurnani et al. in this issue of Molecular Microbiology (Gurnani et al., 2021), our work firmly establishes ActS as a condition-dependent amidase activator. Although EnvC and NlpD have long been implicated in amidase activation in E. coli (Bernhardt & de Boer, 2004; Hara et al., 2002; Lenz et al., 2016; Peters et al., 2011, 2013; Rocaboy et al., 2013; Tsang et al., 2017; Uehara et al., 2009, 2010; Yang et al., 2011) and other Gammaproteobacteria (Möll et al., 2014; Nakamura et al., 2020), it remained unclear whether ActS or MepM stimulates amidase activity like their counterparts. Loss of either factor only modestly exacerbates the cell separation phenotypes of mutants defective for EnvC and/or NlpD at neutral pH (Uehara et al., 2009). Here, we demonstrate ActS—but not MepM—preferentially participates in cell separation in acidic conditions (Figure 5), possibly due to a more favorable electrostatic complementation with the amidases in this environment (Figure 8a). Similar to the bone fide amidase regulators EnvC and NlpD (Peters et al., 2013), ActS does not likely possess any intrinsic hydrolytic activity. The LytM domain of ActS contains substitutions at key amino acid residues implicated in zinc coordination and hydrolytic activity among LytM family metallopeptidases (e.g., LytM of Staphylococcus aureus) (Figure S8b) (Browder et al., 1965; Cohen et al., 2009; Firczuk & Bochtler, 2007; Firczuk et al., 2005; Lu et al., 2006). MepM, in contrast, shares conserved catalytic residues with hydrolytic members of the LytM family and accordingly possesses metalloendopeptidase activity (Singh et al., 2012). Curiously, although MepM does not participate in cell separation, our data indicate that its activity may still be pH-sensitive, at least in the absence of EnvC and NlpD. Inactivation of MepM confers morphological aberrations in neutral, but not acidic, medium (Figure 5). Future work will be necessary to uncover the basis for this condition-dependent phenotype.
Our findings also challenge the prevailing model of amidase activation in which the activity of an individual amidase is specifically stimulated by a cognate LytM regulator in vivo (Peters et al., 2013; Tsang et al., 2017; Uehara et al., 2009, 2010; Yang et al., 2012). Although collectively required for low pH-mediated cell separation (Figure 5), neither EnvC, NlpD, or ActS alone account for full AmiB activation in vivo during growth in acidic medium (Figure 7a,b). Unexpectedly, NlpD has the greatest individual impact on AmiB hyperactivation at low pH: loss of NlpD increases the average chain length of amiB+ ΔamiAC and relieves lysis during AmiB overproduction in acidic medium (Figure 7). In vitro, however, NlpD alone is not sufficient to stimulate AmiB activity at acidic pH (Figure S11). Although our computational analysis suggests that the pH sensitivity of AmiB’s activation by NlpD might be modulated via complementing electrostatic interactions, it does not preclude the existence of further modulators. The biochemical data suggest that acid-mediated stimulation of NlpD-AmiB interaction requires additional cellular components. Loss of EnvC, in contrast, only minimally affects these phenotypes despite its critical role in AmiB activation at neutral pH (Uehara et al., 2010). We note that there is precedent for nonspecific amidase activation by LytM regulators in the Gammaproteobacteria. In Vibrio cholerae, both EnvC and NlpD modulate the activity of AmiB, the pathogen’s sole amidase, and NlpD is preferentially required for intestinal colonization in an infant mouse model and during exposure to bile salts (Möll et al., 2014).
It remains unclear whether the enhanced stimulatory capacity of EnvC and ActS in acidic medium in vivo is solely a product of increased affinity for their substrates (Figure 8) or requires additional upstream signals, as in the case for NlpD. Work from other groups suggests the ability of EnvC and NlpD to stimulate the amidases is tied to the activity of the cell division apparatus, albeit through distinct mechanisms. EnvC activation is dependent on the FtsEX complex (Du et al., 2020; Yang et al., 2011), an ABC transporter implicated in constriction initiation, whereas NlpD activation requires DolP (YraP), a recently discovered division protein of unknown function (Bryant et al., 2020; Tsang et al., 2017). Together, these extra activation criteria may explain why we did not obtain pH-dependent changes in NlpD activity in vitro as DolP was not included in our reactions. Although the ActS activation cascade remains unknown, this protein is also modestly enriched at midcell (Uehara et al., 2009) in proximity to the division machinery, suggesting it may be sensitive to cytokinesis cues. Consistent with a role for the cell division apparatus in the stimulation of amidase activity, we recently demonstrated that acidic pH promotes activation of cytokinesis in E. coli. Specifically, the cell division protein “triggers” FtsN hyperaccumulates at midcell during growth at low pH and stimulates cytokinesis at a reduced cell volume in this condition (Mueller et al., 2020). It is tempting to speculate hyperactivation of division at low pH increases the stimulatory capacity of the LytM activators. AmiB and AmiC midcell localization is likewise responsive to cell division activation and in particular to FtsN accumulation at the cytokinetic ring. A small amount of FtsN and both EnvC and NlpD accumulate at midcell prior to constriction initiation (Du et al., 2020; Peters et al., 2011); however, AmiB and AmiC require active septal cell wall synthesis and midcell localization of additional FtsN for their recruitment to the cytokinetic machinery (Bernhardt & de Boer, 2003; Peters et al., 2011). It is conceivable that low pH-mediated septal enrichment of FtsN leads to hyperaccumulation of AmiB and AmiC at the cytokinetic ring, increasing the proportion of enzymes available for activation by the LytM regulators. FtsN-mediated amidase recruitment may be self-reinforcing: medial FtsN localization itself is enhanced by a C-terminal periplasmic SPOR domain, which binds denuded glycans—the products of amidase activity (Yahashiri et al., 2015, 2017). Compellingly, this model may also explain why AmiA appears insensitive to pH. Unlike AmiB and AmiC, AmiA does not exhibit midcell localization and is instead peripherally distributed (Bernhardt & de Boer, 2003).
3.2 |. Widespread condition-dependent activity of extracytoplasmic PG enzymes
Our present work adds E. coli AmiB, AmiC, and the three LytM-domain amidase activators to a growing list of pH-sensitive PG enzymes and regulators (Castanheira et al., 2017, 2020; Mueller et al., 2019, 2020; Peters et al., 2016; Rico-Pérez et al., 2015; Ursinus & Höltje, 1994; Vandal et al., 2008; Wang et al., 2019). Remarkably, Although E. coli PG composition remains nearly unchanged during growth in different pH environments (Peters et al., 2016), pH “specialist” enzymes are present in every major category of its cell wall autolysins (Mueller et al., 2019; Peters et al., 2016; Straaten et al., 2005). From a technical standpoint, widespread conditional activity of PG enzymes underscores the need to report and standardize culture conditions when assessing the growth and morphological phenotypes of cell wall mutants. The chaining phenotype of amidase mutants is enhanced during exponential growth in nutrient-rich medium, providing a potential explanation for discrepancies between chaining phenotypes in previous studies (Chung et al., 2009; Heidrich et al., 2001). More broadly, conditional activation of PG enzymes supports the hypothesis that the active repertoire of extracytoplasmic PG enzymes in a given cell changes based on the characteristics of its growth environment (Mueller & Levin, 2020; Pazos et al., 2017). Attractively, this model explains how the extracytoplasmic reactions in PG synthesis remain robust in spite of the unstable chemical and physical properties of the periplasm (Wilks & Slonczewski, 2007). It is tempting to speculate that specialization of PG enzymes evolved to allow environmental generalists like E. coli to grow across a wide range of conditions.
4 |. EXPERIMENTAL PROCEDURES
4.1 |. Bacterial strains, plasmids, and growth conditions
Unless otherwise indicated, all chemicals, media components, and antibiotics are purchased from Sigma–Aldrich (St. Louis, MO). All deletion alleles, with the exception of the ΔamiB::kan (a gift from Daniel Kahne (Chung et al., 2009)) and ΔactS::cm (generated by lambda red recombineering Baba et al., 2006; Datsenko & Wanner, 2000)), were originally supplied by the Coli Genetic Stock from the Keio collection (Baba et al., 2006) and transduced via P1 transduction into the MG1655 background, referred to as “wild type” (WT) in the text. Transductants have been confirmed with diagnostic PCR and sequencing. Plasmids were constructed using the pBAD TOPO TA Expression Kit. Unless otherwise indicated, strains are grown in lysogeny broth (LB) medium (1% tryptone, 1% NaCl, 0.5% yeast extract) supplemented with 100 mM filter-sterilized MMT buffer (1:2:2 molar ratio of D,L-malic acid, MES, and Tris base) to vary media pH values. When selection was necessary, cultures were supplemented with 50 μg/ml kanamycin (Kan), 25–100 μg/ml ampicillin (Amp), or 15–30 μg/ml chloramphenicol. Cells were grown at 37°C in glass culture tubes with agitation at 200 rpm for aeration unless otherwise stated in the figure legends.
4.2 |. Growth rate measurements
Strains were grown from single colonies in glass culture tubes in LB + MMT buffer (pH 6.9) to mid-log phase (OD600 ~ 0.2–0.6), pelleted, and re-suspended to an OD600 1.0 (~1 × 109 CFU/ml). With the exception of ΔamiABC, cells were diluted and inoculated into fresh LB + MMT buffer at various pH values in 12-well (1 ml final volume) or 96-well plates (150 μl final volume) at 1 × 103 CFU/ml. Plates were grown at 37°C shaking for 20 hr in a BioTek Eon microtiter plate reader, measuring the OD600 of each well every 10 min. For the ΔamiABC mutant (which clumps extensively and cannot be reliably measured in a plate reader), cells were sub-cultured into glass test tubes (7 ml final volume), sampled at various time points, and vortexed for 30 s prior to measuring OD600. Doublings per hour was calculated by least squares fitting of early exponential growth (OD600 between 0.005 and 0.1). At least two biological replicates were performed.
4.3 |. Microscopy
Phase-contrast and fluorescence images of live or fixed cells were acquired from samples on 1% agarose/phosphate-buffered saline or H2O pads with a Nikon TiE inverted microscope equipped with a 100X Plan N (N.A. = 1.25) objective (Nikon), SOLA SE Light Engine (Lumencor), heated control chamber (OKO Labs), and ORCA-Flash4.0 sCMOS camera (Hammamatsu Photonics). Filter sets for fluorescence were purchased from Chroma Technology Corporation. Nikon Elements software (Nikon Instruments) was used for image capture.
4.4 |. Cell chaining measurements
Cells were initially cultured from a single colony in LB medium + MMT buffer (pH 5.2 or 6.9) to the exponential growth phase (OD600 ~ 0.2–0.6). Cultures were then back-diluted into fresh media to an OD600 = 0.005 and grown to early exponential phase (OD600 ~ 0.1–0.2). When necessary, 0.5 μM Mitotracker (Thermo Fisher, Waltham, MA) was added to the cultures upon back dilution to visualize septa in chains without clear constriction sites. Live cells were washed two times in 1x phosphate-buffered saline then mounted onto 1% agarose (in either H2O or phosphate buffered saline) pads and allowed to dry. Cells (500 μl) were fixed by adding 20 μl of 1 M NaPO4 (pH 7.4) and 100 μl of fixative (16% paraformaldehyde + 8% glutaraldehyde). Samples were incubated at room temperature for 15 min, then on ice for 30 min. Fixed cells were pelleted, washed three times in 1 ml 1X phosphate-buffered saline (PBS, pH 7.4) then resuspended in GTE buffer (glucose-tris-EDTA) and stored at 4°C. Images were acquired for analysis within 48 hr of fixation. Chain length (i.e., number of cells per chain) was determined by eye or by detection of septa in MicrobeJ (Ducret et al., 2016). We only counted chains that were within the micrograph field of view, so the presented data underestimate chain length. At least 50 chains across at least two biological replicates were counted.
4.5 |. Triton-X sensitivity
For determination of minimum inhibitory concentrations, cells were grown from a single colony in LB medium + MMT buffer (pH 5.2, 6.9) at the indicated pH to mid-exponential phase (OD600 ~ 0.2–0.6) at 37°C with aeration and then inoculated at 1 × 105 CFU/ml into the same medium in sterile 96-well plates with a range of two-fold dilutions of Triton-X (final volume, 150 μl). Plates were incubated at 37°C shaking for 20 hr before determination of the well with the lowest concentration of the antibiotic that had prevented growth by visual inspection. Three biological replicates were performed.
4.6 |. Lysis curves
For overexpression experiments, wild-type cells harboring the indicated plasmids were grown from single colonies in glass culture tubes in LB + MMT buffer (pH 6.9) to mid-log phase (OD600 ~ 0.2–0.6) in the absence of inducer, pelleted, and re-suspended to an OD600 1.0. Cells were diluted and inoculated into fresh LB + MMT buffer (pH 6.9 or pH 5.2) with various concentrations of inducer in 96-well plates (150 μl final volume). For strains harboring plasmids, the culture medium was supplemented with 100 μg/ml ampicillin. Plates were grown at 37°C shaking for 20 hr in a BioTek Eon microtiter plate reader.
4.7 |. SPOR domain-binding assay
His6-GFP-DamXSPOR purification was performed as previously described (Yahashiri et al., 2015) with minor modifications. Briefly, 1L of culture (BL21/pDSW1171) was grown in LB medium at 37°C to OD600 ~ 0.5. Cells were transferred to 18°C and induced by the addition of isopropyl-β-D-thiogalactopyranoside to 1 mM overnight. Cells were harvested and broken by sonication, and the His6-tagged protein was purified by cobalt-affinity chromatography. Purity was assessed by SDS–PAGE, and protein concentration was determined by Bradford assay using bovine serum albumin (BSA) standards.
To label the cells, 100 ng/ml of His6-GFP-DamXSPOR and 100 ng/ml of BSA were added to ~2.5 × 106 fixed cells (see “Cell chaining measurements”) in phosphate-buffered saline (pH 7.4). Reactions were incubated on ice for 30 min, washed two times in phosphate-buffered saline, and resuspended in GTE buffer. Cells were spotted on to 1% agarose pads (H2O) and imaged by fluorescence microscopy. Two independent biological replicates were performed. Cell segmentation and fluorescence profiles were generated in MicrobeJ (Ducret et al., 2016). For each replicate, relative fluorescence intensity was normalized to the maximum value for wild-type cells at pH 6.9.
4.8 |. Protein purification
GSHM-ActS(27–251) was purified from LOBSTR-BL21(DE3) pET28a-His-ActS cells as described previously (Gurnani et al., 2021). The amidases AmiA, AmiB, AmiC and their regulators NlpD and EnvCLytM were purified as described previously (Uehara et al., 2010).
4.9 |. ActS in vitro activity assays
PG was isolated from E. coli MC1061 (Glauner et al., 1988) and used as a substrate in assays. PG (∼100 μg) was incubated with an amidase (AmiA, AmiB or AmiC) alone, or in the presence of its cognate activator (NlpD or EnvCLytM) or ActS (concentration of proteins: 2 μM) in 20 mM HEPES/NaOH pH 7.5, 100 mM NaCl, 1 mM ZnCl2, 0.05% Triton X-100 or 20 mM sodium acetate pH 5.2, 100 mM NaCl, 1 mM ZnCl2, 0.05% Triton X-100, in a total volume of 50 μl at 37°C for 1 hr. Reactions were terminated by boiling for 15 min at 100°C, and the remaining PG was digested overnight at 37°C with the muramidase cellosyl (1 μM). PG fragments were reduced with sodium borohydride and analyzed by reversed-phase HPLC ((Glauner et al., 1988).
4.10 |. Molecular modeling and determination of residues changing protonation state upon pH shift
The Phyre web-server (Kelley et al., 2015) was used to model AmiA (Uniprot ID: P36548, residues 41–276, template PDB id: 4BIN [Rocaboy et al., 2013]), AmiB (Uniprot ID: P26365, residues 19–416, template PDB id: 4BIN [Rocaboy et al., 2013]) and AmiC (Uniprot ID: P63883; X-ray crystal structure was downloaded from the RCSB PDB database [Berman et al., 2000] residues 30–408, PDB id: 4BIN [Rocaboy et al., 2013]). The PDB2PQR software (Baker et al., 2001; Dolinsky et al., 2007) was used for atomic charge assignments at pH 6.9 and 5.2 with the PARSE force field (Sitkoff et al., 1994) and the PROPKA tool (Olsson et al., 2011). Then atomic charges were converted to residue charges by summing the atomic charges of each residue. Residue charge changes upon pH shift were calculated by subtraction of residue charges at pH 6.9 and 5.2.
4.11 |. Molecular modeling of the LytM domains of EnvC, NlpD and ActS
The EnvC LytM domain (residues 290–419) was extracted using PyMOL (The PyMOL Molecular Graphics System, Version 1.8.4.0, Schrödinger, LLC) from the X-ray structure downloaded from the RCSB PDB database (Berman et al., 2000) (Uniprot ID: P37690, residues 278–419 PDB id: 4BH5 [Peters et al., 2013]). The 3D structures of NlpD (Uniprot ID: P0ADA3) and ActS (Uniprot ID: Q46798) were modeled using the Phyre web-server (Kelley et al., 2015) using PDB id: 6U2A (Shin et al., 2020) as template for both structures. The LytM domains of NlpD (residues 261–379) and ActS (residues 131–248) were then extracted using PyMOL (The PyMOL Molecular Graphics System, Version 1.8.4.0, Schrödinger, LLC).
4.12 |. Electrostatic potential calculations for LytM-domains and amidase inhibitory helices at pH 6.9 and 5.2
The structures of the LytM domains of EnvC, NlpD, and ActS, and of the autoinhibitory helices of the amidases (AmiA: residues 157–171; AmiB: residues 291–311, and AmiC: residues 289–307) were analyzed using the APBS-PDB2PQR software suite (Baker et al., 2001; Dolinsky et al., 2007) using the PARSE force field (Sitkoff et al., 1994) and the PROPKA tool (Olsson et al., 2011) for pH 5.2 and 6.9 with N- and C-terminal neutralization options. Individual electrostatic potential snapshots were saved using the web-embedded molecular visualization tool for minimum and maximum potential limits of −5 kT/e and 5 kT/e.
Supplementary Material
ACKNOWLEDGMENTS
We thank David Weiss for the gift of pDSW1171 and EC3708, Daniel Kahne for the gift of HSC078, Corey Westfall for helpful discussions, and Sarah Beagle and Navaneethan Palanisamy for technical assistance. This work was supported by NIH GM127331 to PAL, a Washington University Biology Summer Undergraduate Research Fellowship (BioSURF) to AGI, NSF graduate research fellowship DGE-1745038 to EAM, and a Center for Science and Engineering of Living Systems (CSELS) graduate scholar fellowship to EAM. This project also received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No 721484 awarded to WV.
Funding information
National Science Foundation Division of Graduate Research, Grant/Award Number: DGE-1745038; National Institute of General Medical Sciences, Grant/Award Number: GM127331; H2020 Marie Skłodowska-Curie Actions, Grant/Award Number: 721484
Footnotes
DATA AVAILABILITY STATEMENT
The data that supports the findings of this study are available in the supplementary material of this article and from the corresponding author upon request.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section.
REFERENCES
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M. et al. (2006) Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Molecular Systems Biology, 2, 2006–0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ & McCammon JA (2001) Electrostatics of nanosystems: Application to microtubules and the ribosome. Proceedings of the National Academy of Sciences, 98, 10037–10041. 10.1073/pnas.181342398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banzhaf M, van den Berg van Saparoea B, Terrak M, Fraipont C, Egan A, Philippe J. et al. (2012) Cooperativity of peptidoglycan synthases active in bacterial cell elongation. Molecular Microbiology, 85, 179–194. 10.1111/j.1365-2958.2012.08103.x [DOI] [PubMed] [Google Scholar]
- Banzhaf M, Yau HC, Verheul J, Lodge A, Kritikos G, Mateus A. et al. (2020) Outer membrane lipoprotein NlpI scaffolds peptidoglycan hydrolases within multi-enzyme complexes in Escherichia coli. EMBO Journal, 39, e102246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. (2000) The protein data bank. Nucleic Acids Research, 28, 235–242. 10.1093/nar/28.1.235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt TG & de Boer PAJ (2003) The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway: Amidase transport and localization. Molecular Microbiology, 48, 1171–1182. 10.1046/j.1365-2958.2003.03511.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt TG & de Boer PAJ (2004) Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity. Molecular Microbiology, 52, 1255–1269. 10.1111/j.1365-2958.2004.04063.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertsche U, Kast T, Wolf B, Fraipont C, Aarsman MEG Kannenberg K. et al. (2006) Interaction between two murein (peptidoglycan) synthases, PBP3 and PBP1B, in Escherichia coli. Molecular Microbiology, 61, 675–690. 10.1111/j.1365-2958.2006.05280.x [DOI] [PubMed] [Google Scholar]
- Browder HP, Zygmunt WA, Young JR & Tavormina PA (1965) Lysostaphin: Enzymatic mode of action. Biochemical and Biophysical Research Communications, 19, 383–389. 10.1016/0006-291X(65)90473-0 [DOI] [PubMed] [Google Scholar]
- Bryant JA, Morris FC, Knowles TJ, Maderbocus R, Heinz E, Boelter G. et al. (2020) Structure of dual-BON domain protein DolP identifies phospholipid binding as a new mechanism for protein localization. Elife, 9, e62614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanheira S, Cestero JJ, Rico-Pérez G, García P, Cava F, Ayala JA et al. (2017) A specialized peptidoglycan synthase promotes salmonella cell division inside host cells. MBio, 8, e01685–17. 10.1128/mBio.01685-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castanheira S, López-Escarpa D, Pucciarelli MG, Cestero JJ, Baquero F. & Portillo FG (2020) An alternative penicillin-binding protein involved in Salmonella relapses following ceftriaxone therapy. Ebiomedicine, 55, 102771. 10.1016/j.ebiom.2020.102771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Winardhi RS, Morgan LK, Yan J. & Kenney LJ (2017) Non-canonical activation of OmpR drives acid and osmotic stress responses in single bacterial cells. Nature Communications, 8, 1587. 10.1038/s41467-017-02030-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HS, Yao Z, Goehring NW, Kishony R, Beckwith J, Kahne D. (2009) Rapid Beta-lactam-induced lysis requires successful assembly of the cell division machinery. Proceedings of the National Academy of Sciences, 106 (51), 21872–21877. 10.1073/pnas.0911674106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen DN, Sham YY, Haugstad GD, Xiang Y, Rossmann MG, Anderson DL & et al. (2009) Shared catalysis in virus entry and bacterial cell wall depolymerization. Journal of Molecular Biology, 387, 607–618. 10.1016/j.jmb.2009.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA & Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proceedings of the National Academy of Sciences, 97, 6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do T, Page JE & Walker S. (2020) Uncovering the activities, biological roles, and regulation of bacterial cell wall hydrolases and tailoring enzymes. Journal of Biological Chemistry, 295, 3347–3361. 10.1074/jbc.REV119.010155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do T, Schaefer K, Santiago AG, Coe KA, Fernandes PB, Kahne D. et al. (2020) Staphylococcus aureus cell growth and division are regulated by an amidase that trims peptides from uncrosslinked peptidoglycan. Nature Microbiology, 5, 291–303. 10.1038/s41564-019-0632-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G. & et al. (2007) PDB2PQR: Expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Research, 35, W522–W525. 10.1093/nar/gkm276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du S, Pichoff S. & Lutkenhaus J. (2020) Roles of ATP Hydrolysis by FtsEX and interaction with FtsA in regulation of septal peptidoglycan synthesis and hydrolysis. MBio, 11. 10.1128/mBio.01247-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducret A, Quardokus EM & Brun YV (2016) MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nature Microbiology, 1, 16077. 10.1038/nmicrobiol.2016.77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan AJF, Errington J. & Vollmer W. (2020) Regulation of peptidoglycan synthesis and remodelling. Nature Reviews Microbiology, 18, 446–460. 10.1038/s41579-020-0366-3 [DOI] [PubMed] [Google Scholar]
- Firczuk M. & Bochtler M. (2007) Folds and activities of peptidoglycan amidases. FEMS Microbiology Reviews, 31, 676–691. 10.1111/j.1574-6976.2007.00084.x [DOI] [PubMed] [Google Scholar]
- Firczuk M, Mucha A. & Bochtler M. (2005) Crystal structures of active LytM. Journal of Molecular Biology, 354, 578–590. 10.1016/j.jmb.2005.09.082 [DOI] [PubMed] [Google Scholar]
- Glauner B, Höltje JV & Schwarz U. (1988) The composition of the murein of Escherichia coli. Journal of Biological Chemistry, 263, 10088–10095. 10.1016/S0021-9258(19)81481-3 [DOI] [PubMed] [Google Scholar]
- Grabowska M, Jagielska E, Czapinska H, Bochtler M. & Sabala I. (2015) High resolution structure of an M23 peptidase with a substrate analogue. Scientific Reports, 5, 14833. 10.1038/srep14833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurnani Serrano CK, Winkle M, Martorana AM, Biboy J, Morè N, Moynihan P, Banzhaf M, Vollmer W. and Polissi A. (2021) ActS activates peptidoglycan amidases during outer membrane stress in Escherichia coli. Molecular Microbiology, 10.1111/mmi.14712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara H, Narita S, Karibian D, Park JT, Yamamoto Y. & Nishimura Y. (2002) Identification and characterization of the Escherichia coli envC gene encoding a periplasmic coiled-coil protein with putative peptidase activity. FEMS Microbiology Letters, 212, 229–236. [DOI] [PubMed] [Google Scholar]
- Heidrich C, Templin MF, Ursinus A, Merdanovic M, Berger J, Schwarz H. et al. (2001) Involvement of N-acetylmuramyl-l-alanine amidases in cell separation and antibiotic-induced autolysis of Escherichia coli: Cell separation and antibiotic-induced autolysis of E. coli. Molecular Microbiology, 41, 167–178. 10.1046/j.1365-2958.2001.02499.x [DOI] [PubMed] [Google Scholar]
- Heidrich C, Ursinus A, Berger J, Schwarz H. & Holtje J-V (2002) Effects of multiple deletions of murein hydrolases on viability, septum cleavage, and sensitivity to large toxic molecules in Escherichia coli. Journal of Bacteriology, 184, 6093–6099. 10.1128/JB.184.22.6093-6099.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ize B, Stanley NR, Buchanan G. & Palmer T. (2003) Role of the Escherichia coli Tat pathway in outer membrane integrity. Molecular Microbiology, 48, 1183–1193. 10.1046/j.1365-2958.2003.03504.x [DOI] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN & Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nature Protocols, 10, 845–858. 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz JD, Stohl EA, Robertson RM, Hackett KT, Fisher K, Xiong K. et al. (2016) Amidase activity of AmiC controls cell separation and stem peptide release and is enhanced by NlpD in Neisseria gonorrhoeae. Journal of Biological Chemistry, 291, 10916–10933. 10.1074/jbc.M116.715573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin PA & Losick R. (1996) Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes & Development, 10, 478–488. 10.1101/gad.10.4.478 [DOI] [PubMed] [Google Scholar]
- Lonergan ZR, Nairn BL, Wang J, Hsu Y-P, Hesse LE, Beavers WN et al. (2019) An Acinetobacter baumannii, zinc-regulated peptidase maintains cell wall integrity during immune-mediated nutrient sequestration. Cell Reports, 26, 2009–2018.e6. 10.1016/j.celrep.2019.01.089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JZ, Fujiwara T, Komatsuzawa H, Sugai M. & Sakon J. (2006) Cell wall-targeting domain of glycylglycine endopeptidase distinguishes among peptidoglycan cross-bridges. Journal of Biological Chemistry, 281, 549–558. 10.1074/jbc.M509691200 [DOI] [PubMed] [Google Scholar]
- Möll A, Dörr T, Alvarez L, Chao MC, Davis BM, Cava F. & et al. (2014) Cell separation in Vibrio cholerae is mediated by a single amidase whose action is modulated by two nonredundant activators. Journal of Bacteriology, 196, 3937–3948. 10.1128/JB.02094-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller EA, Egan AJ, Breukink E, Vollmer W. & Levin PA (2019) Plasticity of Escherichia coli cell wall metabolism promotes fitness and antibiotic resistance across environmental conditions. Elife, 8, e40754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller EA & Levin PA (2020) Bacterial cell wall quality control during Environmental Stress. MBio, 11. 10.1128/mBio.02456-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller EA, Westfall CS & Levin PA (2020) pH-dependent activation of cytokinesis modulates Escherichia coli cell size. PLoS Genetics, 16, e1008685. 10.1371/journal.pgen.1008685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller P, Ewers C, Bertsche U, Anstett M, Kallis T, Breukink E. et al. (2007) The essential cell division protein FtsN interacts with the murein (peptidoglycan) synthase PBP1B in Escherichia coli. Journal of Biological Chemistry, 282, 36394–36402. 10.1074/jbc.M706390200 [DOI] [PubMed] [Google Scholar]
- Murphy SG, Alvarez L, Adams MC, Liu S, Chappie JS, Cava F. & et al. (2019) Endopeptidase regulation as a novel function of the Zur-dependent zinc starvation response. MBio, 10. 10.1128/mBio.02620-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N, Hoshino Y, Shiga T, Haneda T, Okada N. & Miki T. (2020) A peptidoglycan amidase activator impacts Salmonella enterica serovar Typhimurium gut infection. Infection and Immunity, 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DE & Young KD (2000) Penicillin binding protein 5 affects cell diameter, contour, and morphology of Escherichia coli. Journal of Bacteriology, 182, 1714–1721. 10.1128/JB.182.6.1714-1721.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsson MHM, Søndergaard CR, Rostkowski M. & Jensen JH (2011) PROPKA3: Consistent treatment of internal and surface residues in empirical pKa predictions. Journal of Chemical Theory and Computation, 7, 525–537. [DOI] [PubMed] [Google Scholar]
- Paradis-Bleau C, Markovski M, Uehara T, Lupoli TJ, Walker S, Kahne DE & et al. (2010) Lipoprotein cofactors located in the outer membrane activate bacterial cell wall polymerases. Cell, 143, 1110–1120. 10.1016/j.cell.2010.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Kim YJ, Lee HB, Seok Y-J & Lee C-R (2020) Genetic evidence for distinct functions of peptidoglycan endopeptidases in Escherichia coli. Frontiers in Microbiology, 11, 565767, 10.3389/fmicb.2020.565767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazos M, Peters K. & Vollmer W. (2017) Robust peptidoglycan growth by dynamic and variable multi-protein complexes. Current Opinion in Microbiology, 36, 55–61. 10.1016/j.mib.2017.01.006 [DOI] [PubMed] [Google Scholar]
- Peters K, Kannan S, Rao VA, Biboy J, Vollmer D, Erickson SW et al. (2016) The redundancy of peptidoglycan carboxypeptidases ensures robust cell shape maintenance in Escherichia coli. MBio, 7, e00819–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters NT, Dinh T. & Bernhardt TG (2011) A fail-safe mechanism in the septal ring assembly pathway generated by the sequential recruitment of cell separation amidases and their activators. Journal of Bacteriology, 193, 4973–4983. 10.1128/JB.00316-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters NT, Morlot C, Yang DC, Uehara T, Vernet T. & Bernhardt TG (2013) Structure-function analysis of the LytM domain of EnvC, an activator of cell wall remodelling at the Escherichia coli division site. Molecular Microbiology, 89, 690–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyadarshini R, de Pedro MA & Young KD (2007) Role of peptidoglycan amidases in the development and morphology of the division septum in Escherichia coli. Journal of Bacteriology, 189, 5334–5347. 10.1128/JB.00415-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priyadarshini R, Popham DL & Young KD (2006) Daughter cell separation by penicillin-binding proteins and peptidoglycan amidases in Escherichia coli. Journal of Bacteriology, 188, 5345–5355. 10.1128/JB.00476-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico-Pérez G, Pezza A, Pucciarelli MG, de Pedro MA, Soncini FC & Portillo FG (2015) A novel peptidoglycan D, L-endopeptidase induced by Salmonella inside eukaryotic cells contributes to virulence. Molecular Microbiology, 99, 546–556. [DOI] [PubMed] [Google Scholar]
- Rocaboy M, Herman R, Sauvage E, Remaut H, Moonens K, Terrak M. et al. (2013) The crystal structure of the cell division amidase AmiC reveals the fold of the AMIN domain, a new peptidoglycan binding domain. Molecular Microbiology, 90, 267–277. 10.1111/mmi.12361 [DOI] [PubMed] [Google Scholar]
- Rosenbusch JP (1990) Structural and functional properties of porin channels in E. coli outer membranes. Experientia, 46, 167–173. [PubMed] [Google Scholar]
- Sham L-T, Butler EK, Lebar MD, Kahne D, Bernhardt TG & Ruiz N. (2014) Bacterial cell wall. MurJ is the flippase of lipid-l inked precursors for peptidoglycan biogenesis. Science, 345, 220–222. 10.1126/science.1254522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J-H, Sulpizio AG, Kelley A, Alvarez L, Murphy SG, Fan L. et al. (2020) Structural basis of peptidoglycan endopeptidase regulation. Proceedings of the National Academy of Sciences, 117, 11692–11702. 10.1073/pnas.2001661117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, SaiSree L, Amrutha RN & Reddy M. (2012) Three redundant murein endopeptidases catalyse an essential cleavage step in peptidoglycan synthesis of Escherichia coli K12. Molecular Microbiology, 86, 1036–1051. [DOI] [PubMed] [Google Scholar]
- Sitkoff D, Sharp KA & Honig B. (1994) Accurate calculation of hydration free energies using macroscopic solvent models. Journal of Physical Chemistry, 98, 1978–1988. 10.1021/j100058a043 [DOI] [Google Scholar]
- Suzuki H, Nishimura Y. & Hirota Y. (1978) On the process of cellular division in Escherichia coli: A series of mutants of E. coli altered in the penicillin-binding proteins. Proceedings of the National Academy of Sciences, 75, 664–668. 10.1073/pnas.75.2.664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The PyMOL Molecular Graphics System, Version 1.8.4.0, Schrödinger, LLC (software) undefined. [Google Scholar]
- Tsang M-J, Yakhnina AA & Bernhardt TG (2017) NlpD links cell wall remodeling and outer membrane invagination during cytokinesis in Escherichia coli. PLoS Genetics, 13, e1006888. 10.1371/journal.pgen.1006888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A. Banzhaf M, van den Berg van Saparoea B, Verheul J, Biboy J, Nichols RJ et al. (2010) Regulation of peptidoglycan synthesis by outer-membrane proteins. Cell, 143, 1097–1109. 10.1016/j.cell.2010.11.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Dinh T. & Bernhardt TG (2009) LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. Journal of Bacteriology, 191, 5094–5107. 10.1128/JB.00505-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Parzych KR, Dinh T. & Bernhardt TG (2010) Daughter cell separation is controlled by cytokinetic ring-activated cell wall hydrolysis. EMBO Journal, 29, 1412–1422. 10.1038/emboj.2010.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursinus A. & Höltje JV (1994) Purification and properties of a membrane-bound lytic transglycosylase from Escherichia coli. Journal of Bacteriology, 176, 338–343. 10.1128/JB.176.2.338-343.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Straaten KE, Dijkstra BW, Vollmer W. & Thunnissen A-M-W-H (2005) Crystal structure of MltA from Escherichia coli reveals a unique lytic transglycosylase fold. Journal of Molecular Biology, 352, 1068–1080. 10.1016/j.jmb.2005.07.067 [DOI] [PubMed] [Google Scholar]
- Vandal OH, Roberts JA, Odaira T, Schnappinger D, Nathan CF & Ehrt S. (2008) Acid-susceptible mutants of Mycobacterium tuberculosis share hypersusceptibility to cell wall and oxidative stress and to the host environment. Journal of Bacteriology, 191, 625–631. 10.1128/JB.00932-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vollmer W, Joris B, Charlier P. & Foster S. (2008) Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiology Reviews, 32, 259–286. 10.1111/j.1574-6976.2007.00099.x [DOI] [PubMed] [Google Scholar]
- Wang R, Kreutzfeldt K, Botella H, Vaubourgeix J, Schnappinger D. & Ehrt S. (2019) Persistent Mycobacterium tuberculosis infection in mice requires PerM for successful cell division. Elife, 8, e49570. 10.7554/eLife.49570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver AI, Jiménez-Ruiz V, Tallavajhala SR, Ransegnola BP, Wong KQ & Dörr T. (2019) Lytic transglycosylases RlpA and MltC assist in Vibrio cholerae daughter cell separation. Molecular Microbiology, 112, 1100–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilks JC & Slonczewski JL (2007) pH of the cytoplasm and periplasm of Escherichia coli: Rapid measurement by green fluorescent protein fluorimetry. Journal of Bacteriology, 189, 5601–5607. 10.1128/JB.00615-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahashiri A, Jorgenson MA & Weiss DS (2015) Bacterial SPOR domains are recruited to septal peptidoglycan by binding to glycan strands that lack stem peptides. Proceedings of the National Academy of Sciences, 112, 11347–11352. 10.1073/pnas.1508536112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahashiri A, Jorgenson MA & Weiss DS (2017) The SPOR domain, a widely conserved peptidoglycan binding domain that targets proteins to the site of cell Division. Journal of Bacteriology, 199, e00118–e217., 10.1128/JB.00118-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakhnina A. & Bernhardt T. (2020) The Tol-Pal system is required for peptidoglycan-cleaving enzymes to complete bacterial cell division. Proceedings of the National Academy of Sciences, 117(12), 6777–6783. 10.1073/pnas.1919267117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakhnina AA, McManus HR & Bernhardt TG (2015) The cell wall amidase AmiB is essential for Pseudomonas aeruginosa cell division, drug resistance and viability. Molecular Microbiology, 97, 957–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DC, Peters NT, Parzych KR, Uehara T, Markovski M. & Bernhardt TG (2011) An ATP-binding cassette transporter-like complex governs cell-wall hydrolysis at the bacterial cytokinetic ring. Proceedings of the National Academy of Sciences, 108, E1052–E1060. 10.1073/pnas.1107780108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DC, Tan K, Joachimiak A. & Bernhardt TG (2012) A conformational switch controls cell wall-remodelling enzymes required for bacterial cell division. Molecular Microbiology, 85, 768–781. 10.1111/j.1365-2958.2012.08138.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunck R, Cho H. & Bernhardt TG (2016) Identification of MltG as a potential terminase for peptidoglycan polymerization in bacteria. Molecular Microbiology, 99, 700–718. 10.1111/mmi.13258 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.