Abstract
Antibiotics are highly successful against microbial infections. However, current challenges include rising antibiotic resistance rates and limited efficacy against intracellular pathogens. A novel form of a nanomaterial-based antimicrobial agent is investigated for efficient treatment of an intracellular Salmonella enterica sv Typhimurium infection. A known antimicrobial polysaccharide, chitosan, is engineered to be readily soluble under neutral aqueous conditions for systemic administration. The modified biologic, named acid-transforming chitosan (ATC), transforms into an insoluble, antimicrobial compound in the mildly acidic intracellular compartment. In cell culture experiments, ATC is confirmed to have antimicrobial activity against intracellular S. Typhimurium in a concentration- and pH-dependent manner, without affecting the host cells, RAW264.7 macrophages. For improved cellular uptake and pharmacokinetic/pharmacodynamic properties, ATC is further complexed with fragment DNA (fDNA), to form nano-size spherical polyplexes. The resulting ATC/fDNA polyplexes efficiently eradicated S. Typhimurium from RAW264.7 macrophages. ATC/fDNA polyplexes may bind with microbial wall and membrane components. Consistent with this expectation, transposon insertion sequencing of a complex random mutant S. Typhimurium library incubated with ATC did not reveal specific genomic target regions of the antimicrobial. This study demonstrates the utility of a molecularly engineered nanomaterial as an efficient and safe antimicrobial agent, particularly against an intracellular pathogen.
Keywords: antimicrobial polymers, nanoparticles, stimuli-responsive transformations, antibacterial treatments, Salmonella Typhimurium infections
Graphical Abstract
In this manuscript, we detail the use of acid-transforming chitosan, a polysaccharide that has been previously reported to have excellent antimicrobial properties, to eradicate intracellular S. Typhimurium. This presents an application of nanoparticle-based antibiotics that take advantage of their innate antimicrobial properties and focuses them against difficult to treat intracellular infections.
1. Introduction
Despite the need for new antimicrobial agents to combat the rise of drug resistant bacterial infections, new discoveries have been scarce in the past few years, and the global pipelines for new antibiotics are woefully limited. [1,2] The microbes’ capability to quickly evolve and develop resistance in response to environmental pressures limits the utility of conventional approaches to identify and develop additional antimicrobial compounds from other microbial organisms. [3] A new, alternative strategy to address the challenge is to utilize the unique biophysical properties of emerging materials against which microbes are unlikely to develop resistance. [4,5] Nanoantibiotics are nano-scale materials with capabilities to kill microbes via broad and versatile modes of action on vital structures and biological processes. [6] Recent advances in nanotechnology enable further engineering of nanoantibiotics to be molecularly tuned for activation in response to an environmental signal of microbial infection.
Intracellular pathogenic microbes replicate inside host cells, often in mononuclear phagocytes (MPs), [7] thereby evading the host’s humoral defense and establishing persistent infection. [8] Salmonella enterica sv Typhimurium, a pathogenic Gram-negative intracellular bacterium, infects humans and other mammalian species, including domestic animals. [9] S. Typhimurium injects effector proteins (e.g., SipA, SopA, SopB, SopD, SopE2) into host cells for cytoskeletal rearrangement before cellular uptake into the endosome. [10] Escape from the mildly acidic (~pH 5.0) endosomal compartment to a neutral vacuole (~pH 7.4) makes S. Typhimurium infection persistent and difficult to treat by antibiotics or vaccines. [11,12]
An effective and safe antibiotic agent that is activated in response to the intracellular environment, ideally triggered by the presence of S. Typhimurium, would be highly desirable to combat infection. In this study, a novel polymeric nanomaterial, acid-transforming chitosan (ATC), was engineered to transform in the endosome/lysosome, and its antimicrobial activities against S. Typhimurium were investigated. Chitosan is a natural polysaccharide, derived from the exoskeleton of arthropods and the fungal cell wall, with well-known broad-spectrum antimicrobial efficacy against both Gram-positive and Gram-negative bacteria. [13] Broad application of chitosan as an antimicrobial is limited by its solubility only at a pH of 6.0 or lower. ATC was designed to improve the molecular limitations of chitosan, including its limited solubility in aqueous solution (which makes it incompatible with systemic administration) and its inability to respond to an intracellular stimulus (i.e., non-specific activation of antimicrobial activities). [14] ATC was confirmed to be amenable to systemic administration and its acid-transformation in the target intracellular compartment restored its antimicrobial activities, making it suitable to treat both extracellular and intracellular infections of S. Typhimurium. For optimized cellular uptake and pharmacokinetic/pharmacodynamic properties, ATC was further complexed with short, non-coding fragment DNA (fDNA) [15] (Figure 1). Here, we show that the resulting ATC/fDNA polyplexes efficiently eradicate S. Typhimurium in macrophages and investigate by transposon insertion sequencing (Tn-Seq) of a mutant S. Typhimurium library whether ATC’s antimicrobial activity was associated with specific genes in Salmonella.
Figure 1.
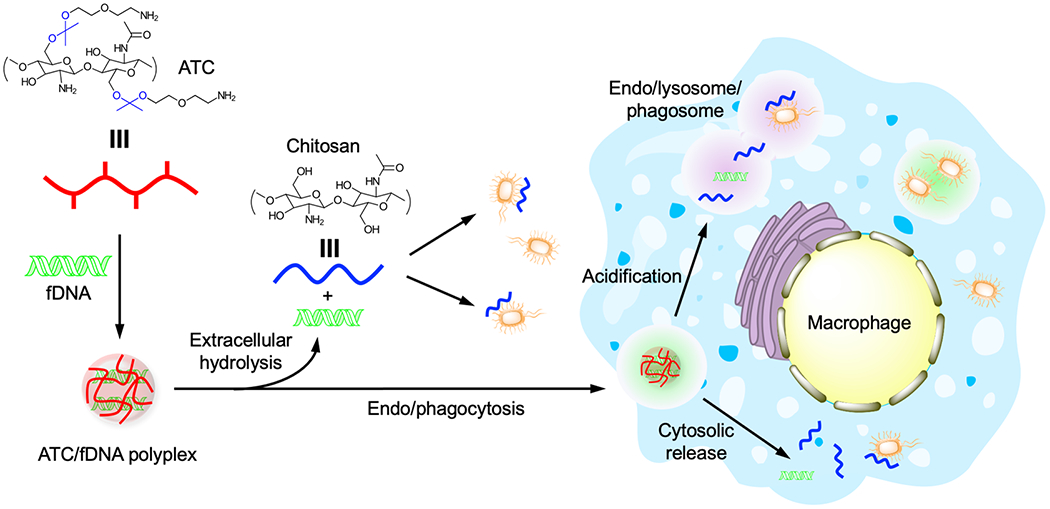
Preparation, acid-responsive transformation, and eradication of intracellular infection by ATC/fDNA polyplexes. ATC (red line) is complexed with fDNA to form a polyplex. Upon extracellular and intracellular hydrolysis at acidic pH, ATC transforms into chitosan (blue line), leading to the eradication of infections in both compartments.
2. Results and Discussion
2.1. Acid-responsive antimicrobial activity of ATC with low toxicity
When internalized by a cell, several microbial pathogens (e.g., S. Typhimurium, C. trachomatis, M. tuberculosis) reside in intracellular compartments such as early and late endosomes or vacuoles, [7,16,17] thereby avoiding the bactericidal mechanisms in the host cells exerted by lysozymes, proteases, and reactive oxygen species (ROS). For example, S. Typhimurium has been shown to transition from the mildly acidic late endosome/lysosome to a specialized vacuole with neutral pH within the host cell. [18,19] Therefore, an effective antibiotic agent against S. Typhimurium should be active at a wide range of pHs, including those present in extracellular environments and intracellular compartments where the bacteria are vulnerable.
The antimicrobial efficacy of ATC against S. Typhimurium at varying pH was assessed (Figure 2A). When incubated with 500 μg/mL ATC at pH 5.5 or 7.4 for 24 h, less than 50% of S. Typhimurium microbes survived (Figure 2A) at both pHs, although survival was higher at pH 7.4. At pH 5.5, the survival of S. Typhimurium decreases more profoundly, even at ATC concentrations of ≤ 100 μg/mL. The key advantage of ATC compared to chitosan in this environment is its observed antimicrobial efficacy at pH 7.4, which is likely due to its improved solubility. Chitosan only displays antimicrobial activity at a low pH due to its limited solubility under neutral conditions. [20] In an acidic environment, the ketal functional group hydrolyzes and the amino group of the D-glucosamine unit with a pKa value of ~ 6.5 becomes protonated, making ATC, which has now transformed into chitosan, more favorably interact with microbial wall and membrane components than at a neutral pH. [21] It is desirable for an antimicrobial agent to effectively eradiate infections but minimally affecting the host cells. Both chitosan and ATC showed no toxicity against RAW 264.7 macrophage cells at all tested concentrations (Figure 2B). Unmodified chitosan is known for its minimal cytotoxicity in vitro, [22] but it was toxic enough to kill mice when intravenously administered at a dose of 50 mg/kg or higher, while ATC was well tolerated by mice at doses of up to 100 mg/kg (Figure S1). Moreover, chitosan was reported to rapidly aggregate in blood and likely cause embolisms in animals. [23] Originally derived from chitosan, ATC was engineered for substantially improved aqueous solubility, [24] minimizing aggregation in blood and hence toxicity in vivo. The observed lack of acute toxicity in vivo at doses of up to 100 mg/kg is promising for the potential utility of ATC for treating antimicrobial infections that require systemic administrations. However, ATC’s dose-dependent, long-term toxicity during the treatment of chronic infections should also be verified as it eventually transforms back to the original structure of chitosan. [25,26] Early studies indicated in vivo chronic toxicity in zebrafish at a dose of 250 mg/L, close to a lethal dose of 280 mg/L,[25] which is much higher than doses of potential clinical use.
Figure 2.
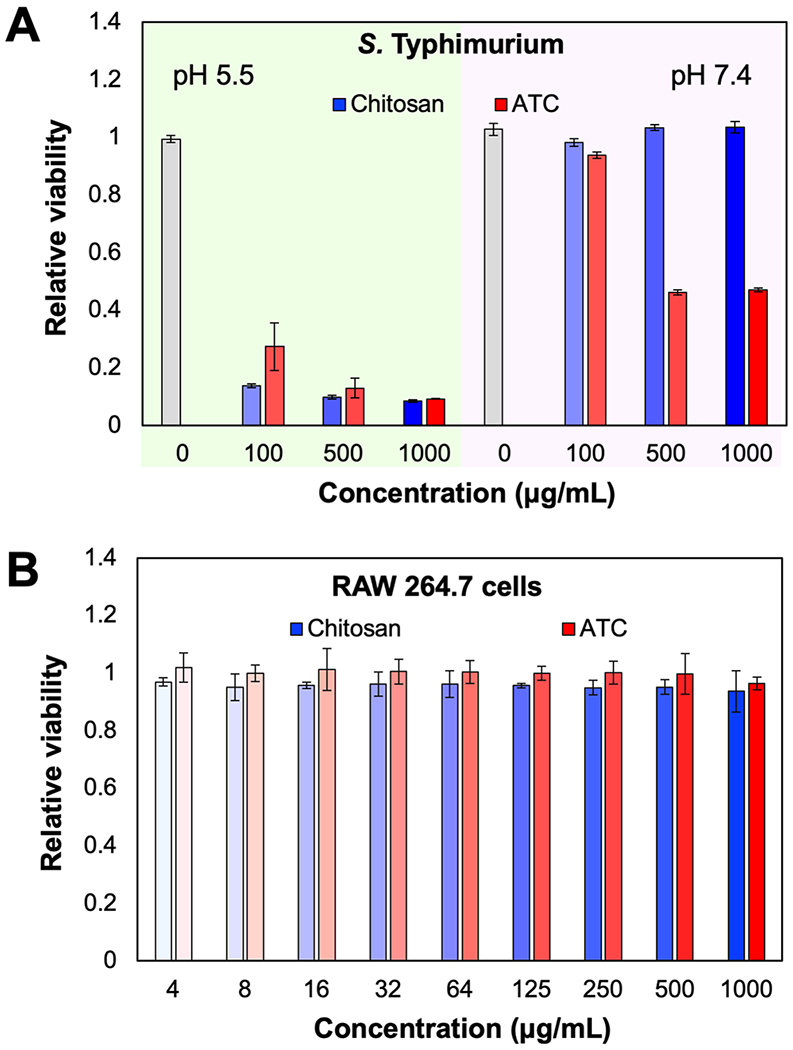
(A) Eradication of S. Typhimurium treated by ATC at varying concentrations at pH 5.5. and 7.4. (B) Viability of RAW 264.7 cells incubated with chitosan or ATC. The relative viability was quantified by OD600 (bacteria) and MTT assay (RAW 264.7 cells).
2.2. Design, preparation, and characterization of ATC/fDNA polyplexes
ATC displayed excellent antimicrobial activity, especially at lower pH, to potentially eradicate S. Typhimurium in intracellular compartments (Figure 2). However, macromolecules and nanoparticles have advantages regarding biodistribution and cellular penetration. [27,28] To address these anticipated limits, ATC was complexed with fragment DNA (fDNA). Pure fDNA is currently used in tissue repair, anti-ischemic therapy, and anti-inflammatory therapy. In these cases, fDNA is extracted from salmon or trout via multi-step purification and sterilization processes, including repeated de-hybridization and re-hybridization. [29] This DNA is largely devoid of intact gene-coding sequences and serves as a biodegradable material that is eventually processed and salvaged in the body. [29,30] Cationic ATC and anionic fDNA bind together via electrostatic interactions and form nano-size spherical polyplexes. These polyplexes are expected to be stable in a condensed form, circulate longer in vivo, disperse well in blood without aggregation, avoid premature or off-target acid-transformation of DNA, be efficiently endocytosed, and sustainably disassemble in cells. The obtained polyplexes are homogenously spherical nanoparticles with a size of ~ 200 nm in diameter with a polydispersity index (PDI) of 0.04 and a zeta potential of +13 mV (Figure 3). Complexation with ATC also substantially reduced the hydrodynamic size of fDNA, resulting in polyplexes that are stable at a physiological pH but rapidly disassemble in the mildly acidic phagolysomal environment (Figure S2). When chitosan was complexed with fDNA, its limited aqueous solubility resulted in heterogenous particles (PDI ~ 0.27). The efficient complexation of ATC with fDNA did not require additional purification of ATC/fDNA polyplexes. However, a large-scale preparation for clinical applications may result in incomplete complexation and a measurable amount of uncomplexed materials that could be removed by conventional purification methods such as ion-exchange column chromatography.
Figure 3.
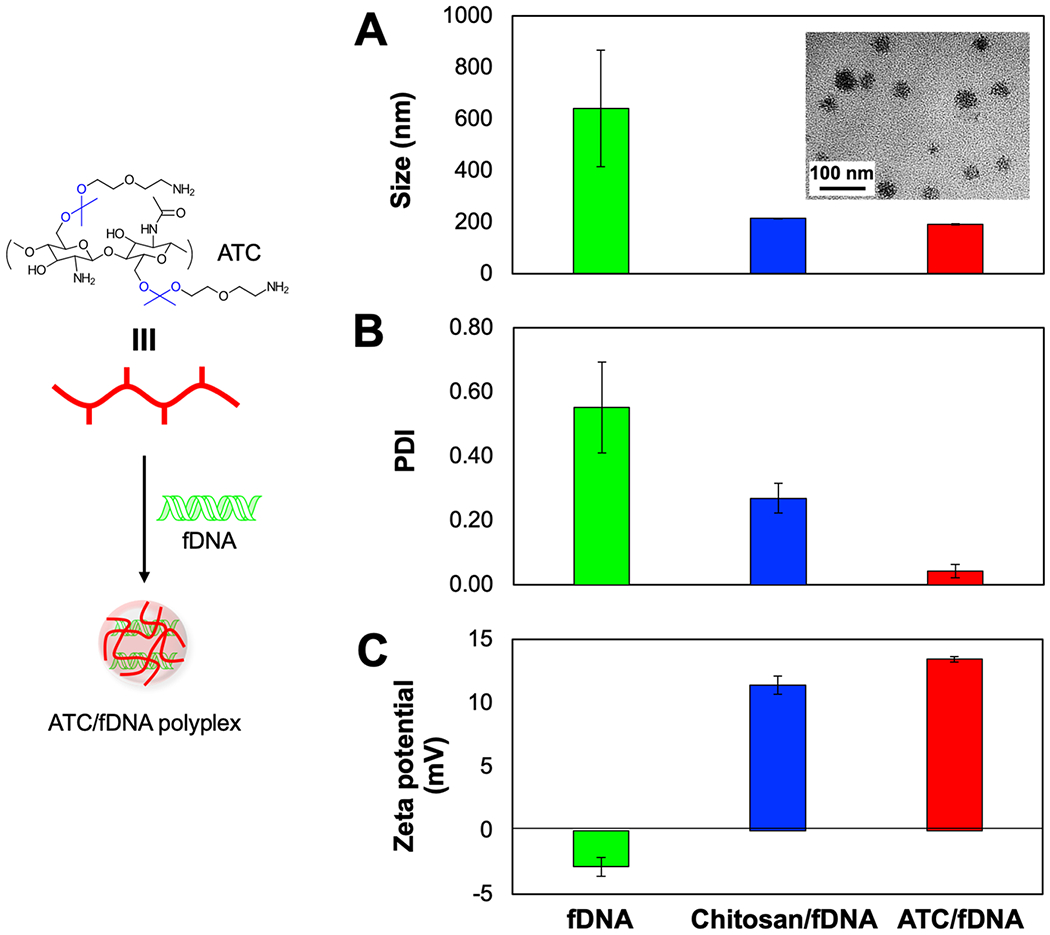
Characterizations of ATC/fDNA polyplexes. (A) Size (inset: TEM image of the polyplex morphology), (B) polydispersity, and (C) zeta potential. The polyplexes of ATC/fDNA and chitosan/fDNA were prepared at an N/P ratio (molecular ratio of deacetylated D-glucosamine groups of ATC and chitosan to the phosphate groups of fDNA) of 100.
2.3. Eradication of intracellular S. Typhimurium infection by ATC/fDNA polyplexes
Intracellular infections can occur in many cell types. Macrophages can be infected when bacteria survive phagocytosis and subsequently reside in intracellular compartments. [31] Murine macrophage RAW 264.7 cells were infected by GFP-expressing S. Typhimurium SL1344 for 1 h, followed by an overnight incubation with ATC/fDNA polyplexes that were efficiently internalized compared with ATC (Figure S3). Quantified by flow cytometry, substantial eradication of intracellular S. Typhimurium by ATC/fDNA was demonstrated in a concentration-dependent manner. As shown in Figure 4, 81%, 23%, and 7% RAW 264.7 cells were found to be infected by S. Typhimurium when treated with 100, 500, and 1,000 μg/mL of ATC/fDNA polyplexes, respectively, demonstrating efficient suppression at the highest concentration. Incubation with chitosan/fDNA polyplexes kept 80-90% RAW 264.7 cells infected with S. Typhimurium, regardless of polyplex concentration (Figure 5). Under a physiological condition such as extracellular environment, the antimicrobial activity of ATC/fDNA polyplexes was negligible (Figure S4). ATC is molecularly designed to rapidly transform into its antimicrobial form in the mildly acidic phagolysosomal environment, [24] followed by its relocation into the cytoplasm or vacuole where S. Typhimurium often resides for proliferation. [18,19] Therefore, ATC/fDNA polyplexes are capable of eradicating S. Typhimurium not only in the concomitantly located phagolyosomes but also in the cytoplasm.
Figure 4.
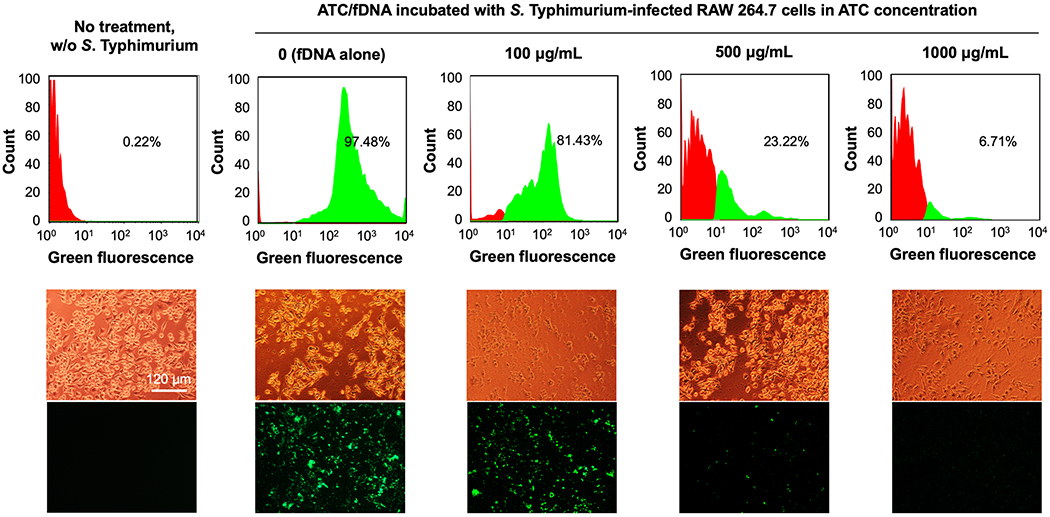
Intracellular S. Typhimurium infection in RAW 264.7 cells treated with fDNA and ATC/fDNA polyplexes at varying concentrations. RAW 264.7 cells were incubated with S. Typhimurium SL1344 for 1 h prior to overnight treatment with fDNA or polyplexes. Intracellular loading of S. typhimurium in RAW 264.7 cells was quantified by flow cytometry and visualized in corresponding fluorescence micrographs. Representative bright field and fluorescence micrographs are shown.
Figure 5.
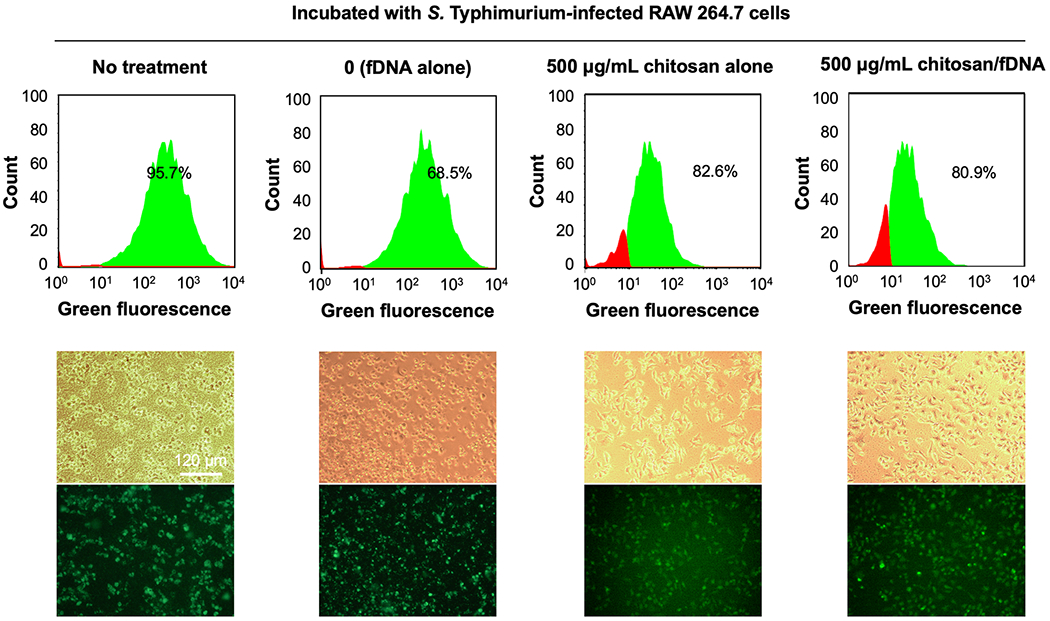
Intracellular S. Typhimurium infection in RAW 264.7 cells treated with fDNA, chitosan or chitosan/fDNA polyplexes. RAW 264.7 cells were incubated with GFP-expressing S. Typhimurium for 1 h, before overnight treatment. The relative number of RAW 264.7 cells infected with S. Typhimurium was quantified by GFP-expressing cells counted by flow cytometry. Representative bright field and fluorescence micrographs are shown.
The intracellular loading of S. Typhimurium in RAW 264.7 cells was also observed under a fluorescence microscope (Figure S5) but remaining fluorescence may also stem from inactivated bacteria. In order to quantify actively proliferating intracellular Salmonella, the lysates of infected and treated RAW 264.7 cells were incubated on LB agar plates. Evidenced by bacterial colony counts obtained from these lysed infected RAW264.7 cells after treatment, ATC/fDNA at a concentration of 1,000 μg/mL eradicated 99% of proliferating S. Typhimurium in RAW 264.7 cells (Figure S6). About 90% of culturable intracellular S. Typhimurium cells were killed when 500 μg/mL of the antimicrobial polyplexes was used. Even at a low concentration of 100 μg/mL, ATC/fDNA polyplexes reduced the intracellular loading of S. Typhimurium in RAW 264.7 cells to 32%, compared to the cells treated with fDNA alone. To investigate the efficacy of the polyplexes against prolonged infection, RAW 264.7 cells were infected with S. Typhimurium for 16 h before being treated with ATC/fDNA polyplexes (Figure S7). When RAW 264.7 cells were fully saturated with S. Typhimurium, ATC/fDNA polyplexes were not able to fully eradicate all bacterial cells but substantially reduced intracellular bacterial loads in a concentration-dependent manner (Figure S7). For example, the intracellular S. Typhimurium load in RAW 264.7 cells was reduced by about 62% when treated with ATC/fDNA polyplexes at a concentration of 1000 μg/mL, compared to treatment with fDNA only. However, at a concentration of 100 μg/mL, there was only a 25% reduction in intracellular S. Typhimurium loading in RAW 264.7 cells (Figure S7).
2.4. Antimicrobial mechanisms of chitosan and ATC
Two of the suggested mechanisms to explain the antimicrobial activity of chitosan, and hence ATC, involve charge-mediated transmembrane pore formation on bacteria [32] and disrupted microbial intracellular processes. [13,33,34] In attempts to identify targets of chitosan and ATC, a random transposon mutant library of S. Typhimurium growing either logarithmically or in stationary phase was exposed to the polymers at various concentrations (100 μg/ml, 500 μg/ml, 1000 μg/ml) at pH 5.5 or pH 7. Surviving bacteria were processed by Tn-Seq, as outlined in Experimental section. As shown in Figure 6, unlike a traditional small-molecule antibiotic (e.g., gentamicin), which interacts with identifiable genetic regions, both chitosan and ATC polyplexes did not appear to target mutants in specific genes and did not yield the same dramatic changes in representation of specific mutants as gentamicin. These observations may indicate a mode of action largely independent from bacterial genetic determinants. However, at low statistical relevance, several underrepresented transposon mutants after ATC or chitosan treatment were disrupted in genes involved in lipopolysaccharide (LPS) formation. This observation may suggest LPS to represent a certain barrier to entry of ATC or chitosan into the bacterial cell. The relative lack of genetic targeting by ATC and chitosan may suggest that their mode of action could be difficult to counteract by the bacterium via genetic modification. Such ubiquitous antibiotics could be effective against most pathogenic bacteria, which needs to be further investigated in subsequent studies.
Figure 6.
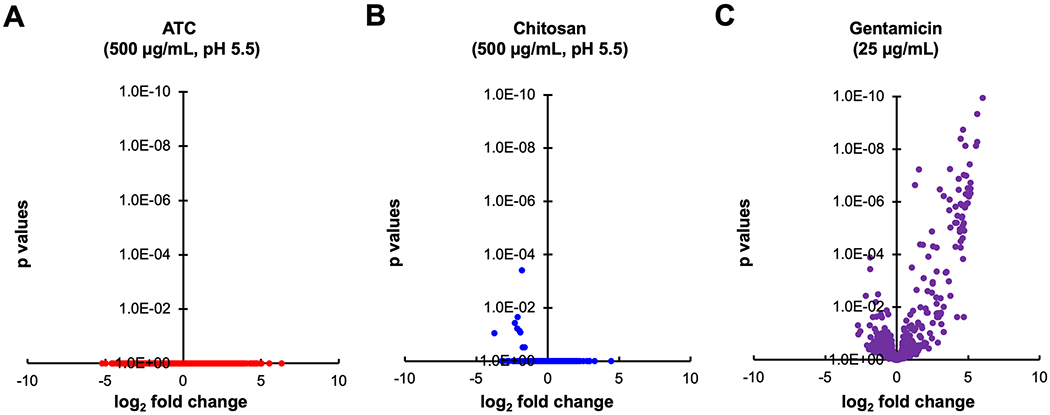
The effects of antimicrobial treatments on the representation of transposon mutants in complex S. Typhimurium Tn5 libraries. (A) Treatment with 500 μg/ml ATC for 1h at pH 5.5. (B) Treatment with 500 μg/ml chitosan for 1h at pH 5.5. (C) Treatment with 25 μg/ml gentamicin for 4h at physiological pH. See Experimental Section for experimental details. Data for (C) have been obtained with a lower complexity (125,000 mutants) library in S. Typhimurium 14028 Δhmp. Treatment conditions in (A) and (B) mimicked those where bacterial growth was found to be most suppressed (Figure 2A), although those tests had not been performed in growth medium. Volcano plots show the summed differences of Tn mutants per strand-specific genomic feature before and after treatment, and the statistical probability of regulation. A minimum of five transposon insertions per feature was required for inclusion in the plot. Data are from at least three biological replicates. All tested conditions for chitosan and ATC (dosages of 100, 500, and 1000 μg/ml, pH 5.5 or 7, logarithmically growing or stationary cultures) resulted in similar plots, with no statistically relevant dose-dependent effects observed on mutant representation in specific genes. Gentamicin affects representation of mutants in many genes at a much higher probability, compared with ATC or chitosan.
3. Conclusions
In this study, RAW 264.7 cells infected with an intracellular pathogen, S. Typhimurium, were treated with novel nanomaterials. Acid-transforming chitosan (ATC) was designed to transform its aqueous soluble structure to a native structure of chitosan, a known natural polymer, in mildly acidic intracellular compartments where S. Typhimurium often resides and proliferates. ATC was found to be antimicrobial in a concentration- and pH-dependent manner, while macrophages, common host cells of S. Typhimurium, remained unaffected. For improved stability and cellular uptake, ATC was further complexed with fragmented DNA (fDNA) lacking gene-encoding sequence in a condensed form, and the resulting ATC/fDNA polyplexes efficiently eradicated intracellular S. Typhimurium, leaving no detectable proliferating bacteria when treated at a high dose. Subsequent studies may reveal more detailed disassembly kinetics of ATC/fDNA polyplexes, ATC’s transformation to chitosan in infected cells, and the molecular interactions of ATC with bacterial proteins, leading to improved designs of ATC and to formulation optimization. Assessing antimicrobial activity of ATC and its polyplexes against S. Typhimurium infection using an animal model will demonstrate the feasibility of clinical translation. Overall, ATC and its polyplex with fDNA are promising antimicrobial agents to treat infections of S. Typhimurium and possibly other intracellular pathogens.
4. Experimental Section
Materials:
All chemicals were purchased from commercially available sources and used as received. Chitosan (Mw 18 - 44 kDa, 95% deacetylated) was purchased from Heppe Medical Chitosan (Halle, Germany) and 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium bromide (MTT), RIPA buffer and Gentamicin were purchased from Sigma-Aldrich (Milwaukee, WI). Fragment DNA (fDNA) as a mixture of segments 50-2,000 bps in size was received as a gift from Pharma Research Products, Co., Ltd. (Seongnam-si, South Korea). Acid-transforming chitosan (ATC) designed for improved aqueous solubility and a molecular change to native chitosan upon degradation at a mildly acidic pH was synthesized as previously reported. [24] Murine macrophage RAW 264.7 cells (ATCC, Rockville, MD) were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (MediaTech, Herndon, VA) with 10% fetal bovine serum (FBS) (Hyclone, Logan, UT) and 1% antibiotics (100 units/mL penicillin; 100 μg/mL streptomycin) (MediaTech, Herdon, VA). Complex transposon libraries of S. Typhimurium ATCC14028 [9,35] were grown in LB broth at 37°C, supplemented with 60 μg/mL kanamycin.
A complex Tn5 transposon insertion library in S. Typhimurium strain ATCC14028 (STM200K TN5) was prepared with the Epicentre EZ-Tn5 <T7/Kan2> promoter insertion kit. Details of library preparation and mapping of the insertion sites were previously described. [35] The library had a complexity of approximately 200,000 independent transposon insertions.
pH-dependent antimicrobial efficacy of ATC:
To study the antimicrobial effects of ATC, a Tn5 transposon insertion library aliquot of 100 μL of 1 ×109 CFU/mL was added to 10 mL of LB broth with 10 μL of 60 mg/mL kanamycin and grown overnight at 37°C. In a 24-well plate, 5 μL of 1 M acetic acid buffer at pH 5.5 or 1 M phosphate buffer at pH 7.4 was mixed with 100 μL of ATC prepared at concentrations of 50, 100, 500, and 1000 μg/mL in DI water, followed by incubation for 5 min on an orbital shaker. Then, the overnight STM200K TN5 was diluted to 1 ×107 CFU/mL, and 100ul of this dilution was added to each well, followed by incubation for 24 h at 37°C. The density of STM200K TN5 in each well was determined by OD600 measurements.
Cytotoxicity of ATC and chitosan on RAW 264.7 cells:
RAW 264.7 cells in DMEM with 10% FBS were seeded at a density of 9,000 cells/well in a 96-well plate and incubated overnight. The culture medium was replaced with 200 μL of ATC or chitosan at concentrations of 4-1,000 μg/mL in DI water by serial dilution in DMEM and incubated for 12 h at 37 °C. The medium was replaced with 200 μL of 1 mg/mL MTT solution in FBS-free DMEM. After 2 h of incubation at 37 °C, the MTT solution was discarded from each well, the cells were rinsed with PBS once, 200 μL of DMSO was added to each well to dissolve the MTT formazan crystals formed by live cells, and the plate was incubated at 37 °C for 5 min. The absorbance of formazan products was then measured at 561 nm wavelength using a Synergy HT plate reader (BioTEK, Winooski, VT, USA) and the relative viability was determined by comparing the absorbance of the cells incubated without the polymers to those incubated with the polymers.
Preparation of ATC/fDNA polyplexes:
We observed that cationic ATC and anionic fragmented DNA (fDNA) bind together via electrostatic interactions and form nano-size spherical polyplexes. Polyplexes of ATC and fDNA at an N/P ratio (a molecular ratio of nitrogen in ATC to phosphates in fDNA) of 100 were created as follows. 500 μL ATC at concentrations of 100, 500, and 1000 μg/mL were added dropwise to 500 μL of 34.07 μg of fDNA in DI water, followed by a vortex and incubation at room temperature for 30 min to form stable complexes. Mean particle diameters (Z-average) with polydispersity index (PDI) and zeta-potentials were measured by dynamic light scattering (DLS) particle analysis using a Zetasizer Nano ZS (Malvern, UK). The measurements were obtained at 25 °C and an angle of 90°, and a viscosity and a refractive index of water at 25 °C (0.887 mPA/s and 1.33) were used for analyses.
Antimicrobial treatment of S. Typhimurium-infected RAW 264.7 cells:
An aliquot (10 μL) of GFP-expressing S. Typhimurium strain SL1344 grown overnight (1 × 109 CFU/mL in 10 mL of LB broth with 60 μg/mL of kanamycin) was diluted 1000-fold with LB broth, followed by adding 20 μL of the diluted aliquot to 20 mL of DMEM with 10% FBS, making a final density of 1 × 105 CFU/mL. In each well of a 24-well plate inoculated with 40,000 RAW 264.7 cells per well, 400 μL of inoculant was added and incubated for 1 h or 16 h at 37 °C, followed by media removal and rinsing the well plate twice with DMEM to remove excess extracellular bacteria. Subsequently, 400 μL of DMEM with 100 μg/mL gentamicin was added to each well and the plate was incubated for 30 min at 37 °C. Then, the media was removed, and the plate was rinsed once with DMEM. ATC/fDNA polyplexes in 400 μL of DMEM at concentrations of 100, 500, 1000 μg/mL were added to each well and incubated for 1 h or overnight at 37 °C. After subsequent media removal, the well plate was rinsed twice with DMEM, 400 μL of DMEM with 10% FBS was added per well and incubated overnight at 37 °C. For comparison, RAW 264.7 cells were also treated with chitosan or chitosan/fDNA polyplexes at an equivalent chitosan concentration of 500 μg/mL and an equivalent fDNA concentration of 34.07 μg/mL. Intracellular density of GFP-expressing S. Typhimurium in RAW 264.7 cells was observed and assessed by fluorescence microscopy (FV10-ASW, Olympus America, Melville, NY) and flow cytometry using a Guava EasyCyte Plus (Millipore-Sigma, Burlington, MA).
To assess the number of culturable intracellular bacteria, infected RAW 264.7 cells were treated with ATC/fDNA polyplexes at concentrations of 100, 500, 1000 μg/mL overnight, as described earlier. Then, the cells were rinsed twice with 1 mL of PBS, and 400 μL of RIPA buffer was added to each well, followed by an incubation on ice for 5 min. The cell lysates collected from the 24-well plates were transferred into individual 1 mL centrifuge tubes and pelleted at 1200 rpm for 10 minutes. The cell pellet was rinsed twice with 500 μL of PBS and resuspended in 400 μL of FBS. A 50 μL aliquot from each suspension was serially diluted, and 10 μL of the 3rd to 5th serial dilutions were plated for colony counts.
Transposon screening of S. Typhimurium incubated with antimicrobial agents[ISP CHK ALL]:
The effects of ATC and chitosan on S. Typhimurium were observed at both log and stationary phases of growth. For log phase experimentation, 100 μL of STM200k TN5 grown overnight at 37°C was added to 10 mL of LB with 60 μg/mL kanamycin and grown to an OD600 of 0.3. A 100 μL aliquot was then mixed with 40 μL of 50% glycerol (20% f.c.) and frozen in −80 °C for future titration. In addition, a 500 μL bacterial aliquot was added to wells of a 24-well plate, 500 μL of ATC or chitosan (500μg/mL in DI water) was further added to each “seeded” well, followed by a 1h incubation at 37 °C at 200 rpm. A 100 μL aliquot of each well in 20% glycerol was then stored for future titration. The remaining bacterial culture in each well was centrifuged at 1,500 rpm for 10 min and the resulting pellet was rinsed with 5 mL LB broth by resuspension and centrifugation. Finally, the bacterial pellet was resuspended in 3 mL of LB broth with 60 μg/mL kanamycin and incubated overnight at 37°C. A 1 mL bacterial aliquot in 20% glycerol was then stored for sequencing. For stationary phase experiments, a 500 μL aliquot of the STM 200k TN5 library grown overnight was added to wells of a 24-well plate and exposed to chitosan or ATC exactly as described above.
Transposon insertion sequencing (Tn-Seq) was then completed by applying a nested PCR regimen to amplify the transposon flanks including the barcode regions, the products of which were then sequenced on an Illumina sequencer, as previously described, [35] Sequences were analyzed by custom Perl scripts that enumerated the abundance of each unique 18-mer. These N18 barcodes were then mapped strand-specifically to annotated genome features.
Statistical analysis:
All data were presented as mean +/− standard deviation. One-way analysis of variance (ANOVA) was used to evaluate data for significant differences between means, with p < 0.05 as threshold. For sequencing data, N18 barcode abundances were statistically analyzed using DESeq2 within the Bioconductor package, [36] and summed per feature, to identify differences in Tn abundance between input and output samples on single-gene scale. Log2 fold changes and adjusted p-values were then reported.
Supplementary Material
Acknowledgements
This work was supported by the California Institute of Regenerative Medicine Training Grant (TG2-01152) and the National Science Foundation Graduate Research Fellowship (DGE-1321846). We gratefully acknowledge Dr James Till and Andres Vazquez-Torres (University of Colorado, Denver) for allowing us to utilize unpublished data obtained in gentamicin assays performed in their laboratory. MM, SP, and WC were supported, in part by grants R03 AI139557, USDA 2015-67017-23360, 2017-67015-26085, an NIFA Hatch grant (CA-D-PLS-2327-H), and an NIFA-BARD award (2017-67017-26180).
Footnotes
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author. The information includes systemic administration of chitosan and ATC at varying doses to mice, stability of polyplexes under physiological and phagolysosomal conditions, cellular uptake of ATC/fDNA polyplexes compared with ATC, antimicrobial activity of ATC/fDNA polyplexes at physiological pH, magnified bright field and fluorescence micrographs of S. Typhimurium-infected RAW 264.7 cells after the treatment with or without ATC/fDNA polyplexes, colony count of surviving intracellular S. Typhimurium in RAW 264.7 cells treated with ATC/fDNA polyplexes at varying concentrations, and flow cytometry data and fluorescence images of RAW 264.7 cells infected with S. Typhimurium for 16 h, before the treatment with ATC/fDNA polyplexes.
Contributor Information
Julius A. Edson, Department of Chemical Engineering and Materials Science, University of California, Irvine, CA 92697, United States
Weiping Chu, Department of Microbiology and Molecular Genetics, University of California, Irvine, CA 92697, United States.
Steffen Porwollik, Department of Microbiology and Molecular Genetics, University of California, Irvine, CA 92697, United States.
Kaycee Tran, Department of Pharmaceutical Sciences, University of California, Irvine, CA 92697, United States.
Nathalie Iribe, Department of Pharmaceutical Sciences, University of California, Irvine, CA 92697, United States.
Michael McClelland, Department of Microbiology and Molecular Genetics, University of California, Irvine, CA 92697, United States.
Young Jik Kwon, 132 Sprague Hall, Irvine, CA 92697, United States.
References
- [1].Levy SB, Bonnie M, Nat. Med 2004, 10, S122. [DOI] [PubMed] [Google Scholar]
- [2].Piddock LJV, Lancet Infect. Dis 2012, 12, 249. [DOI] [PubMed] [Google Scholar]
- [3].Rello J, Bunsow E, Perez A, Expert Rev. Clin. Pharmacol 2016, 9, 1547. [DOI] [PubMed] [Google Scholar]
- [4].Gabriel GJ, Som A, Madkour AE, Eren T, Tew GN, Mater. Sci. Eng. R Reports 2007, 57, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zhou Z, Zheng A, Zhong J, Acta Biochim. Biophys. Sin. (Shanghai) 2011, 43, 729. [DOI] [PubMed] [Google Scholar]
- [6].Huh AJ, Kwon YJ, Control J. Release 2011, 156, 128. [DOI] [PubMed] [Google Scholar]
- [7].Ribet D, Cossart P, Microbes Infect 2015, 17, 173. [DOI] [PubMed] [Google Scholar]
- [8].Kaufmann SHE, Annu. Rev. Immunol 1993, 11, 129. [DOI] [PubMed] [Google Scholar]
- [9].McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, et al. , Nature 2001, 413, 852. [DOI] [PubMed] [Google Scholar]
- [10].V Gart E, Suchodolski JS, Welsh TH, Alaniz RC, Randel RD, Lawhon SD, Lawhon SD, Front. Microbiol 2016, 7, 1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Fàbrega A, Vila J, Clin. Microbiol. Rev 2013, 26, 308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Hopkins S, Kraehenbuhl JP, Schödel F, Potts A, Peterson D, de Grandi P, Nardelli-Haefliger D, Infect. Immun 1995, 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Verlee A, Mincke S, Stevens CV, Carbohydr. Polym 2017, 164, 268. [DOI] [PubMed] [Google Scholar]
- [14].Du H, Yang X, Zhai G, Nanomedicine 2014, 9, 723. [DOI] [PubMed] [Google Scholar]
- [15].Rajendra Y, Kiseljak D, Manoli S, Baldi L, Hacker DL, Wurm FM, Biotechnol. Bioeng 2012, 109, 2271. [DOI] [PubMed] [Google Scholar]
- [16].Cossart P, Sansonetti PJ, Science 2004, 304, 242. [DOI] [PubMed] [Google Scholar]
- [17].García-Del Portillo F, Microbes Infect 2001, 3, 1305. [DOI] [PubMed] [Google Scholar]
- [18].Buchmeier NA, Heffron F, Infect. Immun 1991, 59, 2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Rathman M, Barker LP, Falkow S, Infect. Immun 1997, 65, 1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].No HK, Young Park N, Ho Lee S, Meyers SP, Int. J. Food Microbiol 2002, 74, 65. [DOI] [PubMed] [Google Scholar]
- [21].Friedman AJ, Phan J, Schairer DO, Champer J, Qin M, Pirouz A, Blecher-Paz K, Oren A, Liu PT, Modlin RL, et al. , J. Invest. Dermatol 2013, 133, 1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kean T, Thanou M, Adv. Drug Deliv. Rev 2010, 62, 3. [DOI] [PubMed] [Google Scholar]
- [23].Yang J, Tian F, Wang Z, Wang Q, Zeng Y-J, Chen S-Q, J. Biomed. Mater. Res. Part B Appl. Biomater 2008, 84B, 131. [DOI] [PubMed] [Google Scholar]
- [24].Edson JA, Ingato D, Wu S, Lee B, Kwon YJ, Biomacromolecules 2018, 19, 1508. [DOI] [PubMed] [Google Scholar]
- [25].Wang Y, Zhou J, Liu L, Huang C, Zhou D, Fu L, Carbohydr. Polym 2016, 141, 204. [DOI] [PubMed] [Google Scholar]
- [26].Yuan Z, Li Y, Hu Y, You J, Higashisaka K, Nagano K, Tsutsumi Y, Gao J, Int. J. Pharm 2016, 515, 644. [DOI] [PubMed] [Google Scholar]
- [27].Jiang Z, He H, Liu H, Thayumanavan S, Biomacromolecules 2019, 20, 4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Hiruta Y, Funatsu T, Matsuura M, Wang J, Ayano E, Kanazawa H, Sensors Actuators B Chem 2015, 207, 724. [Google Scholar]
- [29].Squadrito F, Bitto A, Irrera N, Pizzino G, Pallio G, Minutoli L, Altavilla D, Front. Pharmacol 2017, 8, 224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Bitto A, Polito F, Altavilla D, Minutoli L, Migliorato A, Squadrito F, J. Vasc. Surg 2008, 48, 1292. [DOI] [PubMed] [Google Scholar]
- [31].Pizarro-Cerdá J, Cossart P, Cell 2006, 124, 715. [DOI] [PubMed] [Google Scholar]
- [32].Liu H, Du Y, Wang X, Sun L, Int. J. Food Microbiol 2004, 95, 147. [DOI] [PubMed] [Google Scholar]
- [33].Chung Y, Su Y, Chen C, Jia G, Wang H, Wu JCG, Lin J, Acta Pharmacol. Sin 2004, 25, 932. [PubMed] [Google Scholar]
- [34].Zheng LY, Zhu JF, Carbohydr. Polym 2003, 54, 527. [Google Scholar]
- [35].de Moraes MH, Desai P, Porwollik S, Canals R, Perez DR, Chu W, McClelland M, Teplitski M, Appl. Environ. Microbiol 2017, 83, DOI 10.1128/AEM.03028-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Love MI, Huber W, Anders S, Genome Biol 2014, 15, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


