Abstract
The small, regulatory RNA RepG (Regulator of polymeric G-repeats) regulates the expression of the chemotaxis receptor TlpB in Helicobacter pylori by targeting a variable G-repeat in the tlpB mRNA leader. Here, we show that RepG additionally controls lipopolysaccharide (LPS) phase variation by also modulating the expression of a gene (hp0102) that is co-transcribed with tlpB. The hp0102 gene encodes a glycosyltransferase required for LPS O-chain biosynthesis and in vivo colonization of the mouse stomach. The G-repeat length defines a gradual (rather than ON/OFF) control of LPS biosynthesis by RepG, and leads to gradual resistance to a membrane-targeting antibiotic. Thus, RepG-mediated modulation of LPS structure might impact host immune recognition and antibiotic sensitivity, thereby helping H. pylori to adapt and persist in the host.
Subject terms: Bacterial immune evasion, Bacterial genetics, Pathogens, Small RNAs
The small RNA RepG modulates expression of chemotaxis receptor TlpB in Helicobacter pylori by targeting a length-variable G-repeat in the tlpB mRNA. Here, Pernitzsch et al. show that RepG also gradually controls lipopolysaccharide biosynthesis, antibiotic susceptibility, and in-vivo colonization of the stomach, by regulating a gene that is co-transcribed with tlpB.
Introduction
Lipopolysaccharide (LPS) is essential for the physiological integrity and functionality of the outer membrane of most Gram-negative bacteria. As a main surface antigen, it also plays an important role in the interaction between bacterial pathogens and their host. LPS is one of the most potent stimulators of the host immune system and its recognition is essential for the host organism to clear bacterial infections1. During the course of infection, many pathogens modify their LPS synthesis and structure to adapt to diverse microenvironments and evade recognition by the host’s immune system2.
Helicobacter pylori is a Gram-negative pathogen that colonizes the stomach of 50% of the world’s population, which can lead to peptic ulcer disease and gastric cancer3. H. pylori produces a highly modified LPS, unique in both its structure and function, that is essential for establishing colonization and persistence within the human stomach4–6. Like in most Gram-negative bacteria, H. pylori LPS is composed of lipid A, a core oligosaccharide, and the hypervariable O-specific polysaccharide repeats (O-chain)7. H. pylori LPS lacks a canonical inner and outer core organization, and was recently reported to have a short core and a longer O-antigen domain, including structures that were previously assigned to the outer core8. This long O-antigen comprises a trisaccharide (Trio), a glucan, a DD-heptan, and terminal Lewis antigens. Despite its important roles in immune evasion and persistence, a complete picture of the LPS biosynthesis pathway in H. pylori is only being revealed stepwise4,9,10. This might be due to the dispersed genomic locations of LPS genes throughout the H. pylori genome.
Compared to enterobacterial LPS, H. pylori LPS has a reduced immunostimulatory effect (~1,000-fold reduced) due to an under-acetylated and dephosphorylated lipid A2. Moreover, the H. pylori O-chain contains fucosylated oligosaccharides that mimic the structure of mammalian histo-blood group antigens, including Lewis x/y antigens2,5. Both the low endotoxicity of the lipid A and the molecular mimicry of human Lewis antigens contribute to H. pylori host immune surveillance and establishment of long-lasting infection7,11,12. H. pylori LPS is highly variable13. Phase-variable ON/OFF expression of LPS biosynthetic genes, such as Lewis antigen-producing fucosyltransferases, has been shown to increase the diversity of LPS phenotypes, thereby enabling H. pylori to adapt to its individual host and/or changing environments in the gastric mucosa during infection14,15. Different H. pylori clinical isolates were found to extensively alter their Lewis antigen expression in vivo, probably reflecting bacterial adaptation to intra-individual host environments16.
Hypervariable simple sequence repeats (SSRs) are a major source of so-called phase variation, which facilitates adaptation to changing environments and immune escape of pathogens17,18. Length variation of SSRs occurring during replication can either affect translation through the introduction of frameshift mutations (intragenic) or transcription (intergenic) by changing the spacing of promoter elements or transcription factor binding sites17. We previously uncovered that SSRs can also serve as targeting sites for small regulatory RNAs (sRNAs), an important class of post-transcriptional regulators controlling gene expression in response to various stress conditions or during infection19,20. We previously demonstrated that the conserved and abundant H. pylori sRNA RepG (Regulator of polymeric G-repeats) directly base-pairs with its C/U-rich terminator loop to a phase-variable homopolymeric G-repeat in the 5′ untranslated region (UTR) of the tlpB mRNA, encoding the chemotaxis receptor TlpB21. The length of this SSR determines the outcome of post-transcriptional control (activation/repression) of TlpB by RepG and thereby gradually modulates tlpB expression. Although TlpB is assumed to play a role in quorum sensing, biofilm dispersal, acid-/urea-sensing, and pH-taxis22–25, its role in host colonization by H. pylori is controversial23,26,27. Thus, the biological rationale of controlling TlpB via a variable SSR remained unclear.
Using sequence conservation analysis, we noticed that H. pylori tlpB is typically encoded upstream of hp0102, a gene with previously unknown function. Here, we show that the encoded HP0102 protein is required for LPS O-chain production as well as Lewis x antigen display and is essential for murine stomach colonization. Furthermore, we show that Δhp0102 mutants display an increased sensitivity to high-salt stress and antibiotics treatment. Our study demonstrates that RepG co-regulates expression of tlpB and hp0102, which are transcribed in a bicistronic mRNA, and that the length of the homopolymeric G-repeat in the tlpB-hp0102 leader determines the outcome of RepG-mediated control of tlpB-hp0102 mRNA. The post-transcriptional regulation of hp0102 expression by antisense base-pairing of a sRNA to a variable SSR allows for a gradual modulation of LPS biosynthesis as well as Lewis x antigen display, a more scalable mechanism than the previously reported ON/OFF control. In turn, the fine-tuning of LPS O-chain expression also mediates gradual resistance of H. pylori to a membrane-targeting antibiotic.
Results
Expression of the tlpB-hp0102 operon is regulated by RepG
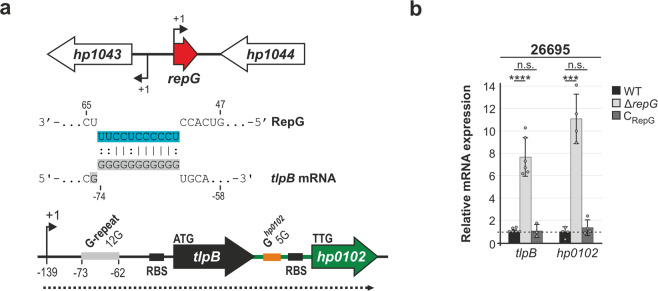
The H. pylori RepG sRNA directly interacts with a 12G-repeat in the 5′ UTR of tlpB mRNA and thereby post-transcriptionally represses tlpB in strain 26695 (Fig. 1a, ref. 21). RepG is highly conserved among different H. pylori strains, and homologs are also present in the related gastric Helicobacter species, H. acinonychis, H. cetorum, and H. mustelae21. Our previous dRNA-seq study28 suggested tlpB is transcribed as a bicistronic mRNA together with its downstream gene hp0102, which we could validate by RT-PCR analysis (Supplementary Fig. 1a, b). We found a highly conserved genetic organization with adjacent tlpB and hp0102 genes in 86 H. pylori genomes (Supplementary Fig. 1c). Notably, in the closely related H. acinonychis and H. cetorum species, the tlpB and hp0102 homologous genes are also adjacent and preceded by a G-repeat sequence. The conserved genetic organization of tlpB and hp0102 was suggestive of their co-expression and co-regulation by RepG.
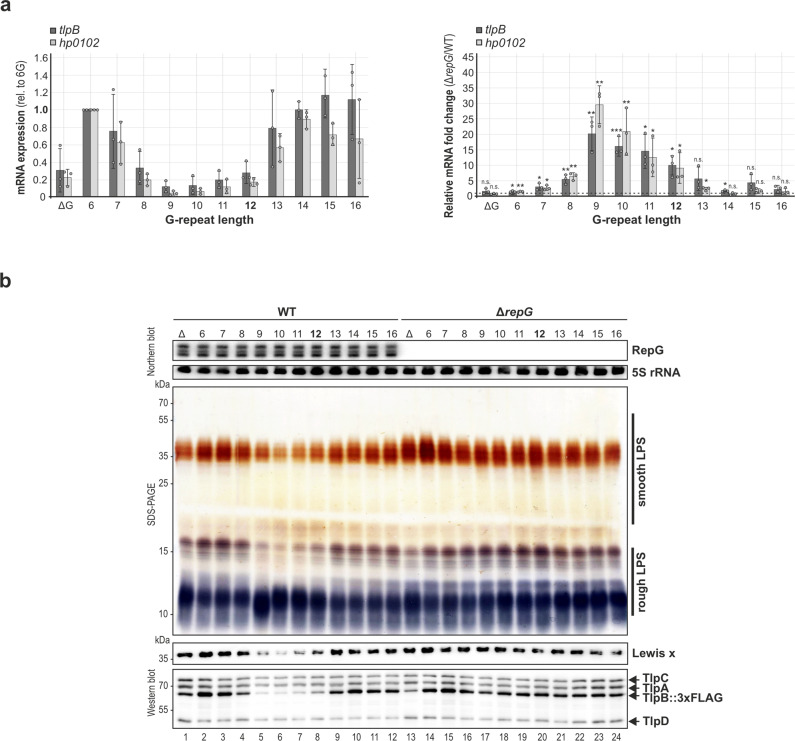
Fig. 1. RepG sRNA represses expression of the tlpB-hp0102 mRNA.
a In H. pylori strain 26695, RepG sRNA is transcribed from the intergenic region between hp1043 and hp1044, encoding an orphan response regulator and a protein of unknown function. The C/U-rich terminator loop of RepG (blue) binds to a homopolymeric G-repeat (12G, light gray) in the 5′ UTR of the bicistronic tlpB-hp0102 mRNA (dotted line), encoding a chemotaxis receptor and a protein of unknown function21. Transcriptional start sites (TSS, +1; ref. 28) and ribosome binding sites (RBS) are indicated by arrows and black bars, respectively. Numbers designate the distance to the tlpB start codon. A G-rich sequence (Ghp0102, orange) was identified in the tlpB-hp0102 IGR. b RT-qPCR analysis of RepG-dependent regulation of tlpB and hp0102 in H. pylori strain 26695 wildtype (WT), repG deletion (∆repG), and complementation (CRepG) mutants. Wild-type mRNA levels of each gene were set to 1, and relative mRNA levels in mutants are shown as bars. Values are shown as mean ± standard deviations (s.d.) for n = 6 (tlpB; exception CRepG n = 3) and n = 4 (hp0102) biologically independent experiments. ****—highly significant, p-value < 0.0001; ***—highly significant, p-value < 0.001; n.s.—not significant; Student’s t-test, two-tailed. Source data underlying (b) is provided as a Source data file.
RT-qPCR analysis showed about eight- and ten-fold increase in mRNA levels of the tlpB and hp0102 genes, respectively, in the ΔrepG mutant when compared to the wildtype (WT) of H. pylori strain 26695 (Fig. 1b). This co-regulation is in line with tlpB and hp0102 being encoded in an operon. Complementation of the ΔrepG mutant with ectopically expressed RepG under control of its native promoter from the neutral rdxA locus (CRepG) restored wild-type mRNA levels of both genes. This indicates that not only tlpB, but also hp0102 is repressed by RepG in H. pylori strain 26695.
RepG represses HP0102 by targeting an upstream G-repeat
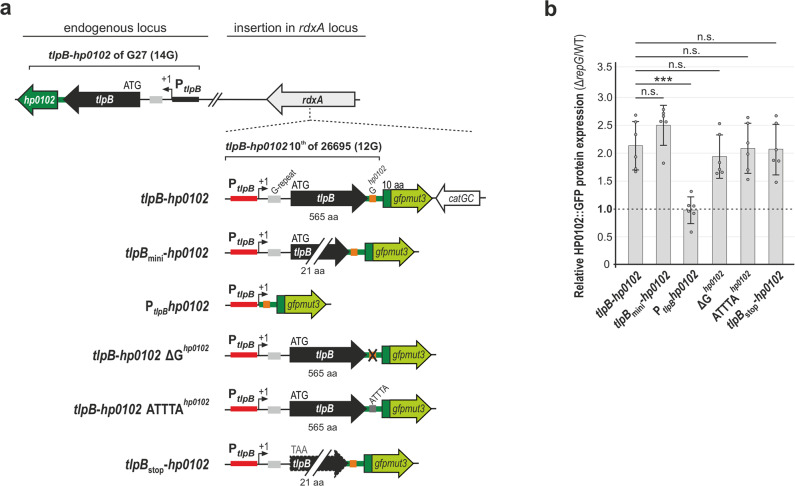
To investigate whether RepG also controls HP0102 protein synthesis, we used a translational gfpmut3 reporter system established in strain G2721. Here, the first ten amino acids of the 26695 hp0102 coding region were fused to gfpmut3 and introduced together with tlpB, including native promoter (PtlpB) and 5′ UTR (12G), into the rdxA locus of G27 (tlpB-hp0102, Fig. 2a). HP0102::GFP protein levels were increased about two-fold upon repG deletion, demonstrating that RepG represses expression of HP0102 (Fig. 2b and Supplementary Fig. 3). Full-length tlpB expression seemed to be dispensable for RepG-mediated HP0102 regulation because HP0102::GFP expression was also increased more than two-fold in H. pylori G27 carrying a tlpBmini-hp0102 reporter fusion upon repG deletion. The latter comprises a non-functional 21 codon-long tlpB mini-gene, in which the middle region (544 codons) was deleted (Fig. 2a, b).
Fig. 2. RepG represses HP0102 protein expression through targeting the G-repeat upstream of tlpB-hp0102.
a Schematic representation of translational hp0102::gfpmut3 reporter fusion constructs originating from H. pylori strain 26695 (12G) that were integrated into the rdxA locus in H. pylori G27 wildtype and ΔrepG. The endogenous tlpB-hp0102 operon in G27 carries a 14G repeat. Red bar: tlpB promoter region; gray: G-repeat upstream of tlpB; black: tlpB coding region; orange: G-repeat upstream of hp0102; dark green: HP0102 coding region; light green: gfpmut3 coding sequence. Arrows indicate the transcriptional start site. b H. pylori G27 WT and ΔrepG strains that carry the indicated GFP reporter fusions were grown to exponential phase and HP0102::GFP protein levels were determined by western blot analysis. The relative HP0102::GFP protein levels in the ΔrepG mutant (vs. the respective wild-type background) are indicated by bars with corresponding s.d. (n = 6 biologically independent experiments; corresponding western blot representative is shown in Supplementary Fig. 3). ***—highly significant, p-value < 0.001; n.s.—not significant; Student’s t-test, two-tailed. Source data underlying (b) is provided as a Source data file.
The hp0102 TTG start codon in the 30-nt-long intergenic region (IGR) between tlpB and hp0102 is preceded by a conserved potential AAGGGT ribosome binding site (RBS) (Fig. 1a and Supplementary Fig. 4b), suggesting co-transcription but not necessarily co-translation of both genes. A short, variable G-rich sequence (Ghp0102, 3–6G) is found upstream of the hp0102 RBS, which might also be targeted by the C/U-rich loop of RepG sRNA (Supplementary Fig. 4c). To investigate whether the Ghp0102-repeat is involved in RepG-mediated HP0102 regulation, the 30-nt-long tlpB-hp0102 IGR followed by the hp0102 10th::gfpmut3 coding region were fused to the tlpB promoter (PtlpBhp0102, Fig. 2a). Equally expressed HP0102::GFP fusion protein levels in the wildtype and the ΔrepG mutant carrying the PtlpBhp0102 reporter fusion indicated that the Ghp0102-repeat is not sufficient for RepG-mediated HP0102 expression control (Fig. 2b and Supplementary Fig. 3). While slight differences in basal expression levels of HP0102::GFP were observed in the wildtype, neither deletion of the Ghp0102-repeat (ΔGhp0102) nor its exchange to an ATTTA-stretch (ATTTAhp0102) did significantly affect RepG-mediated repression of HP0102::GFP (Fig. 2b and Supplementary Fig. 3).
We had previously shown that RepG regulates tlpB expression mainly at the level of translation21. Therefore, although harboring a separate RBS, HP0102 expression control through RepG might still be coupled to tlpB mRNA translation. Replacement of the tlpB start codon by a stop codon in the tlpBmini-hp0102 reporter fusion (tlpBstop-hp0102, Fig. 2a) neither affected HP0102::GFP protein expression nor RepG-mediated repression thereof (Fig. 2b and Supplementary Fig. 3). This shows that hp0102 translation and regulation is not directly influenced by tlpB translation. Taken together, these data demonstrate that targeting of the homopolymeric G-repeat in the tlpB mRNA leader by the RepG sRNA mediates a coordinated regulation of the tlpB-hp0102 operon, at the transcript and protein level.
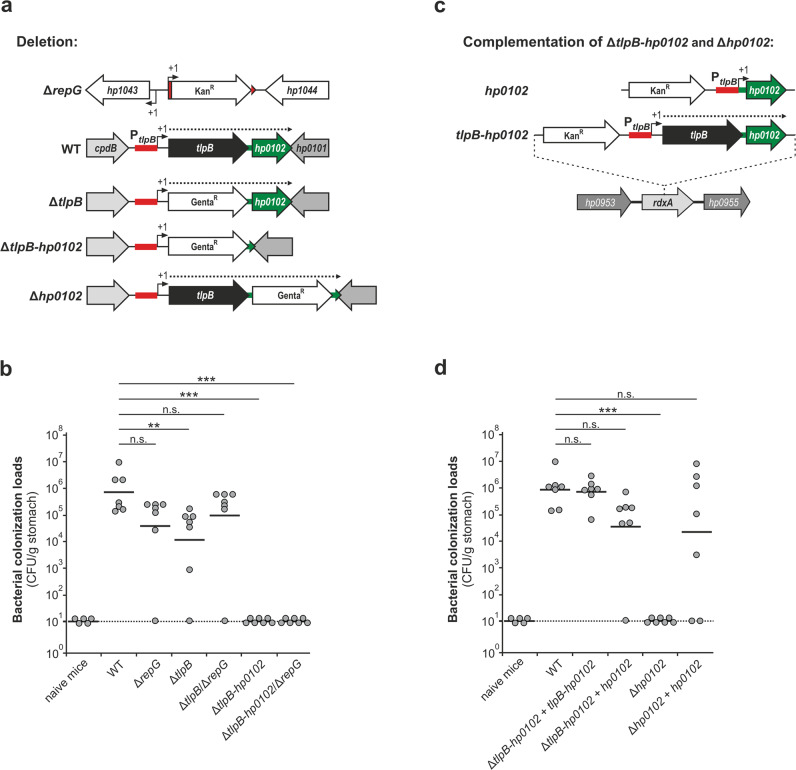
HP0102 is required for mouse stomach colonization
Variable SSRs are often associated with genes important for host-pathogen interactions18,29. To investigate whether RepG sRNA and/or its targets, tlpB and hp0102, contribute to virulence and/or survival, we infected mice with either wildtype or non-polar deletion mutant strains of the mouse-adapted strain H. pylori X47-2AL (as in ref. 30). While the syntenic organization of the tlpB-hp0102 operon in strain X47-2AL is the same as in strain 26695 (Supplementary Fig. 1c), the G-repeat in the tlpB mRNA leader is only 7Gs long in X47-2AL compared to 12Gs in 26695 (Supplementary Fig. 4a). An approximately two-fold repression of tlpB through RepG was observed in strain X47-2AL compared to an about five-fold repression in strain 26695 (Supplementary Fig. 2). This is in line with the previously described strain-specific, RepG-mediated tlpB regulation depending on G-repeat length21.
While no significant change was observed for the ΔrepG or ΔtlpB/ΔrepG double deletion mutants, in vivo infection studies revealed that the H. pylori X47-2AL ΔtlpB single mutant is slightly attenuated (~1.5-log lower CFU/g of stomach ) in its ability to colonize the murine stomach compared to the wildtype (Fig. 3b). This minor effect suggests that both RepG and TlpB are largely dispensable for H. pylori mouse stomach colonization. In contrast, no bacteria could be recovered from the stomachs of mice infected with the ∆tlpB-hp0102, ΔtlpB-hp0102/∆repG, and ∆hp0102 deletion mutants (Fig. 3b, d), indicating that hp0102 is essential for H. pylori to colonize mice. Accordingly, stomach colonization was restored in independent infection experiments, in which either the single Δhp0102 or double ΔtlpB-hp0102 mutants were complemented with either the whole tlpB-hp0102 operon or only hp0102 expressed from the tlpB promoter (PtlpB) in the rdxA locus (Fig. 3c, d). A slightly reduced colonization level compared to wildtype (WT) was still observed in the Δhp0102 + hp0102 strain, which might be due to altered hp0102 expression, e.g., caused by the fusion of the tlpB-hp0102 intergenic region (IGR) to PtlpB. Overall, our in vivo infection studies identified hp0102 as an essential factor for colonization of the murine stomach by H. pylori. Therefore, we aimed to explore a potential link between the variable G-repeat and host colonization that would rely on the function of HP0102.
Fig. 3. HP0102 is essential for mice stomach colonization with H. pylori strain X47-2AL.
a, c Schematic representation of the construction of H. pylori X47-2AL ∆repG and ∆tlpB/hp0102 deletion mutant and complementation strains. TSS are denoted as +1 (black arrows). The repG gene and the tlpB promoter (PtlpB) are shown in red. The hp0102 gene is shown in green. b, d About 108 bacteria of H. pylori X47-2AL WT or indicated mutant strains were orogastrically administered to NMRI Swiss mice. As a control, mice were infected with peptone broth only. Four weeks post infection, mice were sacrificed and colony-forming units (CFUs) per gram of stomach weight were calculated by serial dilutions and plating assays. Each circle indicates the colonization titer in the stomach of a single mouse. The horizontal bars represent the geometric mean for each group of data (n = 5/4 non-infected control animals for panel b/d, respectively; n = 7 for mice infected with indicated H. pylori strains). ***—highly significant, p-value < 0.001; **—very significant, p-value < 0.01; n.s.—not significant; Mann–Whitney test (Prism), two-tailed. Source data underlying (b, d) are provided as a Source data file.
HP0102 is involved in O-chain biosynthesis
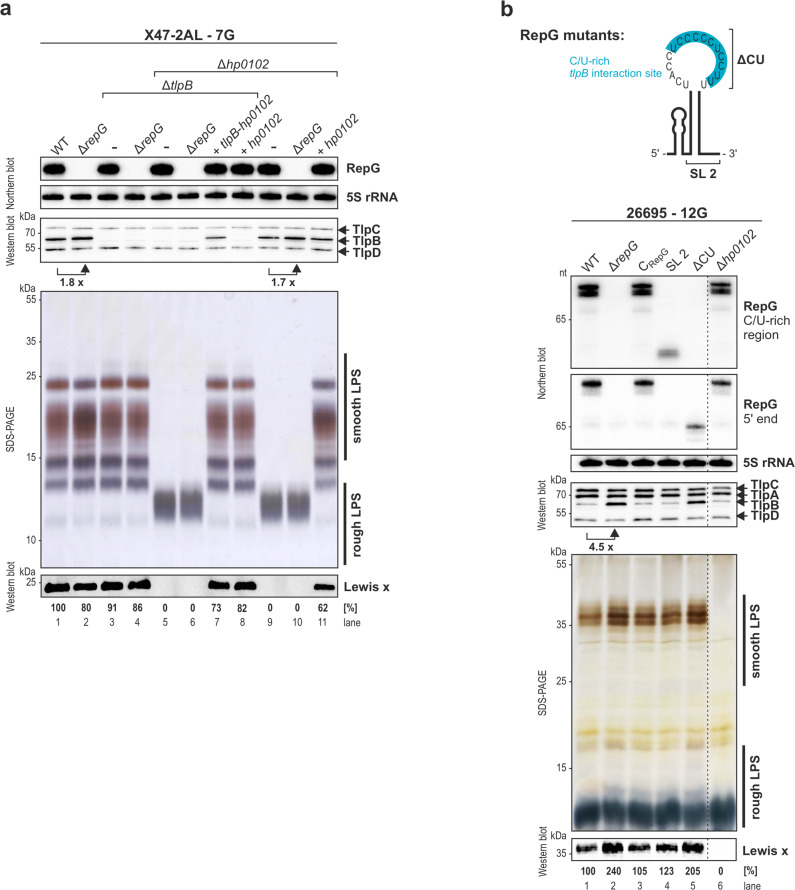
The HP0102 protein sequence contains several motifs of the glycosyltransferase family 2 (GT-2; www.kegg.jp, http://pfam.sanger.ac.uk/, http://www.cazy.org/), suggesting HP0102 is involved in LPS biosynthesis. While at the time of this study, the precise function of HP0102 was unknown, it was recently described to act as a fucosyltransferase involved in O-chain biosynthesis in H. pylori strain G2710. In line with the study by Li and colleagues10, we also observed that H. pylori X47-2AL ∆hp0102 deletion mutants have only rough-form LPS, i.e. lack the O-chains, and no Lewis x antigens could be detected on western blots (Fig. 4a, lanes 5–6 and 9–10). In contrast, the ∆repG, ∆tlpB, and ∆tlpB/∆repG mutant strains showed the same smooth LPS profiles (composed of lipid A-core and O-antigens) as wildtype, with comparable Lewis x antigens levels (Fig. 4a, lanes 1–4). Complementation of ∆tlpB-hp0102 and ∆hp0102 with either the tlpB-hp0102 operon or with hp0102 alone restored smooth LPS patterns and wild-type Lewis x expression (Fig. 4a, lanes 7–8 and 11). Next, we further validated this function of HP0102 in LPS biosynthesis in the most commonly utilized H. pylori laboratory strains, namely 26695 (Fig. 4b, lanes 1 and 6), J99, and G27 (Supplementary Fig. 5). Variations in overall LPS patterns were observed among different WT strains supporting previous reports on strain-specific LPS variations8. However, irrespective of the parental strain background, mutants lacking hp0102 express only rough LPS. This confirms a conserved function of HP0102 essential for LPS O-chain biosynthesis and Lewis x antigen display, which is also in agreement with HP0102 homologs universally present in different H. pylori strains10. The LPS O-chains contribute to antigenicity and serospecificity of native LPS and play an important role in H. pylori virulence11,31,32. Accordingly, the complete abrogation of mouse colonization by the H. pylori X47-2AL hp0102 mutant strain most probably results from its loss of the LPS O-chains.
Fig. 4. The hp0102 gene is required for LPS O-antigen biosynthesis and is regulated by RepG sRNA.
The WT and indicated mutant strains of H. pylori strains X47-2AL (a) and 26695 (b) were grown to exponential growth phase, and RNA and protein samples were analyzed by northern and western blot, respectively. b In 26695, the ΔrepG mutant was complemented with wild-type RepG (CRepG) or mutant sRNAs. SL 2 consists of only the second stem-loop and in ΔCU the tlpB binding site (marked in blue) was replaced by an extra-stable tetraloop21. RepG sRNA was detected with CSO-0003 (C/U-rich terminator loop) and JVO-2134 (5’ end). 5S rRNA was used as loading control (JVO-0485). Expression of the chemotaxis receptors TlpA, TlpB, TlpC, and TlpD was analyzed using a polyclonal anti-TlpA22 antiserum. Please note that the Δhp0102 mutant was loaded/analyzed on the same northern and western blots like the WT and indicated sRNA mutant strains; however, samples were not loaded directly next to each other (indicated by dotted line). a, b LPS samples of the indicated strains were separated on 15% SDS-PAGE gels and either directly visualized by silver staining, or electro-blotted to PVDF membrane and probed with a Lewis x antigen-specific antibody. The results shown are representative of at least three independent experiments. Source data underlying (a, b) are provided as a Source data file.
In order to narrow down how HP0102 affects LPS O-chain biosynthesis, we constructed several additional LPS mutants (Supplementary Figs. 6 and 7a) in H. pylori strains 26695 and X47-2AL and compared their LPS profiles with the ∆hp0102 mutants. These deletions affected LPS core oligosaccharide (HP12848), various parts of the O-antigen (HP103933; HP158133; HP015934 HP082635), lipid A modulation (HP0579-058036), or LPS O-chain translocation (HP120633). Comparing their LPS profiles and Lewis x/y antigen expression to the patterns of the corresponding ∆hp0102 mutants showed that the deep rough LPS phenotype of the HP0102 deletion mutant most closely resembled the pattern observed for the HP1039 and HP1581 mutant strains (Supplementary Fig. 7b, c). In addition, deletions of HP0102, HP1039, HP1581, HP0159, HP0826, and HP1206 resulted in the loss of both Lewis x and y antigens. These observations were consistent in both H. pylori strain backgrounds. These mutational analyses together with a previously missing enzyme for the conserved trisaccharide of the O-antigen (previously referred to as the outer core)8, indicated HP0102 is involved in the biosynthesis of the trio (Supplementary Fig. 7). Indeed, and in line with our observations, Li and colleagues revealed HP0102 as the fucosyltransferase of the LPS trisaccharide using mass spectrometry-based structural LPS analyses10. Because both, X47-2AL and 26695, showed stronger signals for Lewis x than Lewis y, all following experiments regarding LPS profiling and Lewis antigen expression were focused on western blot analyses of Lewis x.
RepG regulates tlpB-hp0102 and consequently LPS biosynthesis
To investigate whether RepG sRNA regulates expression of hp0102 (in addition to tlpB) and in turn LPS biosynthesis, we examined LPS patterns and Lewis x antigen display in H. pylori strain 26695, in which tlpB is more strongly regulated by RepG than in strain X47-2AL (Supplementary Fig. 2). Deletion of repG resulted in increased band intensities in smooth LPS and Lewis x antigen levels in strain 26695 (Fig. 4b, lanes 1–2), indicating regulation of hp0102 by RepG. Complementation of the ΔrepG mutant with wild-type (CRepG) or a mutant RepG sRNA expressing only the second stem-loop with the C/U-rich tlpB binding site (SL2) restored wild-type LPS and Lewis x (Fig. 4b, lanes 3–4). In contrast, deletion of the previously identified C/U-rich tlpB interaction site in RepG (∆CU)21, led to increased O-antigen and Lewis x similar as observed for the repG deletion mutant (Fig. 4b, lane 5). These RepG mutant analyses demonstrate that the C/U-rich terminator loop of RepG is sufficient to repress both tlpB and hp0102.
The G-repeat in the tlpB-hp0102 leader determines LPS production
While RepG is highly conserved, the length of its G-repeat target in the tlpB leader varies among H. pylori isolates, resulting in strain-specific tlpB regulation21. RepG represses TlpB protein expression in H. pylori strains X47-2AL (7G, two-fold; Fig. 4a) and 26695 (12G, five-fold; Fig. 4b). In contrast, TlpB protein levels were unaffected or increased upon repG deletion in strains J99 (13G) and G27 (14G, two-fold), respectively (Supplementary Fig. 5). Deletion of repG did not significantly affect LPS profiles in strains X47-2AL, J99, and G27 (Fig. 4a, lane 2 and Supplementary Fig. 5), but resulted in increased band intensities in smooth LPS and Lewis x antigen levels in strain 26695 (Fig. 4b, lanes 1–2), confirming a strain-specific G-repeat-dependent tlpB-hp0102 regulation.
To more systematically investigate whether the G-repeat length impacts RepG-mediated control of hp0102, the G-repeat was either deleted (ΔG) or mutated from 6 to 16 guanines (6–16G) in the tlpB mRNA leader of H. pylori 26695 expressing TlpB::3xFLAG (as in ref. 21). RT-qPCR analysis of tlpB and hp0102 mRNA levels in the tlpB leader variants (∆G, 6–16G) of H. pylori 26695 revealed that expression of both genes is indeed dependent on the G-repeat length (Fig. 5a, left panel). While deletion of the G-repeat (ΔG) had only a minor influence on tlpB and hp0102 mRNA levels when compared to wildtype (12G), expression of both genes was increased in the 6G-variant. A gradual decrease in tlpB and hp0102 mRNA levels was observed with an increasing number of guanines in the tlpB mRNA leader, reaching a minimum level for 8–12Gs. Further extension of the G-stretch from 13–16Gs resulted again in elevated transcript abundances. Whereas deletion of repG affected neither tlpB nor hp0102 expression in ∆G, 6G, and 13–16G, significantly increased mRNA levels were observed in the 7–12G leader mutants upon repG deletion (Fig. 5a, right panel).
Fig. 5. The G-repeat length impacts RepG-mediated tlpB-hp0102 co-regulation and in turn smooth LPS production.
a (Left panel) RT-qPCR of relative tlpB and hp0102 mRNA levels of different tlpB leader mutants (ΔG, 6–16G) in the H. pylori 26695 wild-type background. The mRNA level of each gene in the tlpB 6G-leader mutant was used as reference and set to 1. (Right panel) Relative fold changes of tlpB and hp0102 mRNA levels upon repG deletion in H. pylori 26695 tlpB leader mutants when compared to the respective wild-type backgrounds. The fold changes are shown as mean of three biological replicates with corresponding error bars (s.d.). ***—highly significant, p-value < 0.001; **—very significant, p-value < 0.01; *—significant, p-value < 0.05; n.s.—not significant; Student’s t-test, two-tailed. b The LPS patterns and Lewis x antigen levels of tlpB leader mutants in H. pylori 26695 wild-type and ∆repG mutant backgrounds were analyzed by silver staining and western blot analysis with anti-Lewis x antigen antibody, respectively. Expression of the chemotaxis receptors was analyzed by an anti-TlpA22 antiserum. Note that tlpB leader mutants express FLAG-tagged TlpB21. A representative silver-stained PAA-gel and western blot are shown (out of three independent experiments). Source data underlying (a, b) are provided as a Source data file.
Next, we investigated whether SSR-dependent regulation of the tlpB-hp0102 operon also affects LPS biosynthesis. Every tlpB mRNA leader variant (∆G, 6–16G) of the wild-type background expresses smooth LPS. However, similar to gradual TlpB expression levels depending on the G-repeat length, varying O-chain and Lewis x profiles were detected (Fig. 5b, lanes 1–12). Whereas tlpB leader length variants of 6–8G and 13–16G displayed increased expression levels, decreased amounts of O-chains and Lewis x levels were detected for 9–11G variants in comparison to WT (12G; Fig. 5b and Supplementary Fig. 8). The gradual TlpB and LPS/Lewis x patterns were lost upon repG deletion, confirming that the G-repeat length indeed affects sRNA-mediated regulation. The strongest effects were observed in tlpB mRNA leader mutants that comprise a G-repeat of 9–12Gs, in which deletion of repG resulted in significantly increased O-chain and Lewis x levels. In contrast, LPS patterns remained almost unaltered in the ∆G, 6–8G, and 13–16G variants.
In conclusion, the amounts of Lewis x antigen and TlpB protein levels closely correlate in the varying G-repeat strains. RepG significantly represses TlpB protein and smooth LPS biosynthesis/O-antigen levels in tlpB leader variants that comprise a 9–12G-long repeat, in line with the previously defined optimal window for RepG-mediated repression21. Overall, these data show that the length of the G-repeat in the tlpB mRNA leader impacts RepG-mediated co-regulation of tlpB and hp0102 and consequently, smooth LPS production.
Deletion of hp0102 leads to reduced growth upon salt stress
LPS is essential for the integrity and functionality of the bacterial outer membrane and protects the bacteria against surface stress37. For example, divalent cation bridging of phosphates on lipid A and/or the core oligosaccharide of LPS contributes to the stabilization of the outer membrane and ensures its function as an effective permeability barrier. Bacteria with incomplete LPS, e.g., rough LPS, are more sensitive to environmental stresses, including osmotic stress38. To examine whether, or to which extent, the hp0102-dependent rough LPS phenotype affects H. pylori survival under osmotic stress, the growth of the wildtype and mutant strains of H. pylori X47-2AL, 26695, J99, and G27 was examined under elevated sodium chloride (NaCl) concentrations, an often-used method to simulate osmotic stress conditions. A drastic decrease in viable bacterial counts was observed on high-salt plates for all tested H. pylori mutants lacking hp0102 (Fig. 6a and Supplementary Fig. 9a). Hence, HP0102-dependent smooth LPS production is required to maintain the integrity of the H. pylori cell envelope. Complementation of the H. pylori X47-2AL Δhp0102 mutant only partially restored wild-type growth (Fig. 6a), which might be due to a slightly different expression of HP0102 in the complementation strain. In line with hp0102 being repressed by RepG in H. pylori strain 26695, deletion of repG rendered H. pylori more resistant to high-salt conditions (Fig. 6a). In contrast, an increased sensitivity toward osmotic stress was observed for the H. pylori G27 ΔrepG mutant (Supplementary Fig. 9a). Because smooth LPS biosynthesis seems to be unaffected by RepG in G27 (Supplementary Fig. 5), this phenotype might be linked to the regulation of other, so far undefined, targets of RepG.
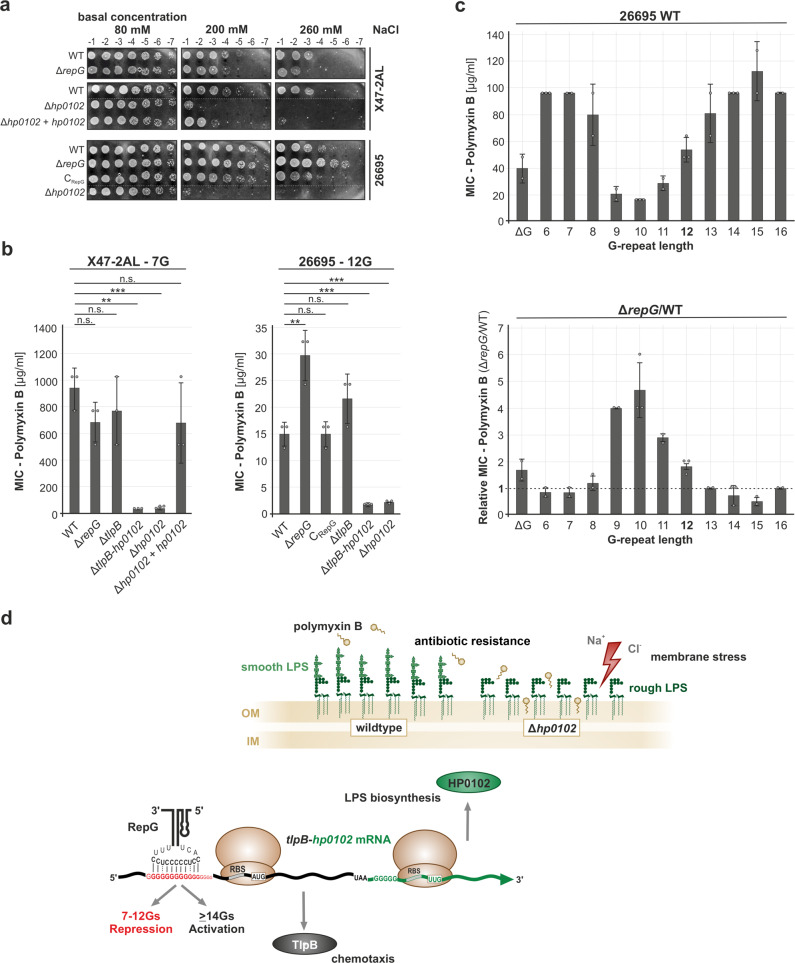
Fig. 6. RepG affects sensitivity to salt stress and polymyxin B via regulation of hp0102.
a H. pylori X47-2AL and 26695 WT and indicated mutant strains were grown in liquid culture to exponential growth phase (OD600 of 1). Indicated ten-fold dilutions of the bacterial suspensions were spotted on GC-agar plates containing 80 mM (basal concentration), 200 mM (mild stress), or 260 mM (harsh stress) sodium chloride. Plates were incubated for 3–5 days at 37 °C under microaerobic conditions. The results shown are representative of at least two independent experiments. b Polymyxin B sensitivity testing for H. pylori X47 and 26695 WT and indicated mutant strains using E-tests. Error bars indicate s.d. for n = 3 independent biological experiments. ****— highly significant, p-value < 0.0001; **—very significant, p-value < 0.01; n.s.—not significant; Student’s t-test, two-tailed. c (Upper panel) Polymyxin B MICs of H. pylori 26695 tlpB mRNA leader variants in the wild-type background. (Lower panel) Relative fold changes in polymyxin B sensitivity (MICs) upon repG deletion in different tlpB leader variants when compared to the respective wild-type backgrounds (n = 2 for WT/∆repG_∆G, ∆repG_6G, WT/∆repG_7-9G, WT/∆repG_11G, WT/∆repG_13G, WT/∆repG_15-16G; n = 3 for WT_6G, WT/∆repG_10G, WT/∆repG_12G, WT/∆repG_14G). d Model of RepG-mediated regulation of TlpB and HP0102. The genes encoding the chemotaxis receptor TlpB (dark gray) and the fucosyltransferase HP0102 (green) are transcribed as a bicistronic mRNA and play a role in chemotaxis25 and smooth LPS biosynthesis (light green) as well as protection against membrane stress (indicated by red arrow), respectively. Deletion of hp0102 leads to loss of O-antigens and a rough LPS phenotype (dark green). Antisense base-pairing of the C/U-rich terminator loop of RepG sRNA to the homopolymeric G-repeat in the 5′ UTR of the tlpB-hp0102 mRNA results in post-transcriptional co-regulation of tlpB and hp0102 at the transcript and protein level. Depending on the G-repeat length, RepG mediates both repression (7–12Gs) and activation (≥14Gs) of tlpB and hp0102. OM/IM—outer/inner membrane, RBS—ribosome binding site, AUG/UUG—start codons, UAA—stop codon. Source data underlying (b, c) are provided as a Source data file.
RepG affects polymyxin B sensitivity by repression of hp0102
Modifications in the LPS structure have been shown to affect H. pylori resistance to cationic antimicrobial peptides (CAMPs9,12), which represent critical components of the human innate immune system. CAMPs interact with negatively charged phospholipid groups or the LPS lipid A anchor, inducing bacterial lysis and cell death. Using E-test strips, we measured the resistance of H. pylori X47-2AL, 26695, J99, and G27 wildtype and chosen mutant strains to polymyxin B (PxB), a membrane-targeting antibiotic and surrogate for host CAMPs. Irrespective of the parental strain background, Δhp0102 mutants displayed reduced minimal inhibitory concentrations (MICs) to PxB compared to WT (Fig. 6b and Supplementary Fig. 9b–d, Supplementary Table 1). Complementation of Δhp0102 in X47-2AL restored wild-type PxB susceptibility, suggesting that the alteration of the LPS structure through hp0102 is associated with antibiotic sensitivity in H. pylori. While no significant effect was observed upon repG deletion in X47-2AL, J99, and G27, the 26695 ΔrepG mutant was twice as resistant to PxB as the wildtype. Deletion of tlpB in X47-2AL leads to a negligible reduction in MIC and a non-significant increase in strain 26695. Thus, the observed effects on PxB sensitivity seem to be mediated primarily by modulation of hp0102 expression (Fig. 6b, Supplementary Table 1). Moreover, deletion of hp0102 affects PxB sensitivity more strongly compared to mutants defective in O-chain length (HP0826 and HP0159) but less pronounced when compared to deletion mutants completely devoid of O-chain (HP1039 and HP1581), the core oligosaccharide (HP1284), and lipid A (HP0579/80) (Supplementary Fig. 9c, d and Supplementary Table 1). This additionally supports the recently reported function of HP0102 as the LPS trisaccharide fucosyltransferase10.
Next, we characterized PxB sensitivity of tlpB-hp0102 G-repeat leader variants of 26695 WT and ΔrepG. While sensitivity to PxB was not affected in ΔG, the 6–8G and 13–16G variants were more resistant to PxB than the wildtype (12G; Fig. 6c, upper panel). Consistent with reduced smooth LPS expression (Fig. 5b), 9–11G mutants were more susceptible to PxB. In the ∆repG background, all tlpB mRNA leader variants displayed similar MICs (data not shown). Accordingly, deletion of repG resulted in two- to four-fold increased resistance to PxB for tlpB leader variants 9–12G (Fig. 6c, lower panel), whereas MICs remained unaltered in almost all other leader mutants. Taken together, these data demonstrate that RepG-mediated expression control of hp0102 leads to gradual control of LPS O-chain synthesis and thereby contributes to the integrity and permeability of the bacterial membrane and H. pylori susceptibility to PxB.
Discussion
Here, we showed that the H. pylori HP0102 protein is involved in LPS O-chain production, essential for murine stomach colonization and important for resistance to the membrane-targeting antibiotic PxB. Moreover, we demonstrated that hp0102 is post-transcriptionally regulated by RepG sRNA through base-pairing to a variable, homopolymeric G-repeat in the 5′ UTR of the bicistronic tlpB-hp0102 mRNA. In contrast to SSR-based phase-variable ON/OFF gene expression switches, this unique mode of regulation allows for a fine-tuned control of hp0102 and in turn smooth LPS biosynthesis, which might be required for H. pylori adaptation to different host niches.
Binding of RepG to the G-repeat upstream of the tlpB open reading frame is sufficient to regulate tlpB and hp0102 both at the transcript and protein level (Fig. 2). Our GFP reporter assay showed that hp0102 translation is likely independent of tlpB, suggesting that RepG-mediated repression of hp0102 is rather based on ribosomal recruitment and/or destabilization of the entire mRNA than on direct translational coupling. RepG regulates tlpB expression at the translational level and induces structural rearrangements within the tlpB mRNA when binding to the G-repeat21. Sequestration of the G-repeat, which is located relatively far upstream of the RBS and might function as a translational enhancer and/or ribosome standby site, could affect both tlpB and hp0102 translation similarly as previously shown for other sRNAs39,40. Furthermore, structural rearrangements caused by RepG binding might also lead to the inhibition of translation elongation and/or whole transcript destabilization by RNase recruitment41.
Bacterial adaptive changes, including modulation of LPS synthesis and structure, play an important role during infection. In our study, we observed H. pylori Δhp0102 mutants display rough LPS phenotypes, indicative of O-antigen loss, supporting its recent identification as the trisaccharide fucosyltransferase10. H. pylori LPS can mimic host structures and is essential for resistance to host-derived CAMPs6, making it a key surface determinant required for colonization and persistence as shown in rodent models12,31,32. In line with a previous transposon mutagenesis screen in strain G27, which indicated hp0102 is a candidate gene required for colonization in mice42, we observed that mice stomach colonization of an H. pylori X47-2AL ∆hp0102 mutant is completely abolished. Similarly, other H. pylori glycosyltransferases involved in LPS biosynthesis are also essential for colonization of mice8,43. As LPS core and variable Lewis antigens facilitate immune escape11,44 and promote adherence to the gastric epithelium45–47, the Δhp0102 colonization defect might be associated with an enhanced susceptibility of this mutant to the inflammatory host response and membrane stress within the host and/or its impaired ability to bind to the gastric epithelium. Other functions might also be affected in the H. pylori ∆hp0102 mutant, such as a recently reported modulation of the expression of the major virulence factor CagA as well as H. pylori chemotaxis48.
While chemotaxis in H. pylori is important for proper bacterial orientation and localization in the gastric glands25,49, the X47-2AL ∆tlpB mutant presented only a mildly reduced murine stomach colonization (Fig. 3) and no defect during competitive co-infection experiments26,27. Together with our data, this indicates that the TlpB chemotaxis receptor is not necessarily required for bacterial growth in the rodent stomach. Further studies are required to evaluate the importance of the tlpB-hp0102 operon co-regulation and its functional implications for H. pylori persistence.
Diverse pathogens benefit from SSR-mediated phase variation of surface structures as these allow them to rapidly adapt to changes in their host environment50,51. The H. pylori genome contains variable SSRs in a number of outer membrane proteins, adhesins, or LPS-biosynthetic or -modifying enzymes (Supplementary Table 2)52,53. When sequentially isolated from human patients54–58 or re-isolated from animal colonization experiments59,60, even H. pylori strains within the same host exhibited varying G-repeat length in the tlpB 5′ UTR (Supplementary Table 3). This suggests that tlpB-hp0102 is associated with a phase-variable SSR. So far, Lewis antigen-producing fucosyltransferases have been described to be controlled by SSRs in an ON/OFF manner14–16,60–62. In contrast, we uncovered a gradual regulation of an operon encoding two proteins important for H. pylori colonization, namely the chemotaxis receptor TlpB and HP0102 required for O-chain display, by a sRNA that targets a SSR in the mRNA leader sequence, resulting in gradual changes of smooth LPS. This “LPS fine-tuning” might help H. pylori to balance the fine line between host immune evasion and lack of sufficient adherence potential63–65. The here described gradual display of LPS O-chain might increase heterogeneity in the population, enabling H. pylori to adapt and survive in the gastric mucosa of individual host niches and thus contributing to persistence. HP0102-mediated structural LPS changes can support this adaptation as demonstrated by an increased sensitivity of a hp0102 mutant to high salt concentrations and to membrane-targeting antibiotics such as PxB, which is a surrogate of host-derived CAMPs. The regulation of hp0102 by the RepG sRNA thereby adds to a number of bacterial sRNAs that impact antibiotic resistance, e.g., by regulating expression of drug transporters or efflux proteins, biofilm formation, or metabolic enzymes involved in cell envelope synthesis or LPS biosynthesis66.
The RepG-dependent regulatory mechanism presents several unique features. We demonstrated that the G-repeat length mediates repression (7–12Gs) or activation (≥14Gs) of the tlpB-hp0102 operon by RepG. This is a unique regulatory mechanism linking post-transcriptional gene expression control to phase variation. To the best of our knowledge, intergenic SSRs that affect full-length LPS O-chain display in a reversible and rheostat-like fashion at the post-transcriptional level, as described in this study, have not been identified in other bacterial pathogens so far. Besides hp0102, three other LPS-modifying enzymes are associated with intergenic SSRs in H. pylori strain 26695 (Supplementary Table 2). Whether these intergenic SSRs affect promoter strength and/or are targeting sites for sRNAs still needs to be examined. At the transcriptional level, for example, length variation of a promoter-associated, intergenic SSR has been described to result in low, mediate, and high expression levels of the major H. pylori adhesin SabA67.
Future experiments are needed to evaluate if the presence or expression of RepG sRNA itself might provide a selective force toward a certain length of the G-repeat, thereby influencing the rate of SSR polymorphism. Phase variation is usually stochastic and random; however, gene regulation has been shown to be integrated with phase variation events. For example, phase variation of methyltransferases can modulate bacterial gene expression via epigenetic mechanisms68 and Dam methylation can impact phase-variable ON/OFF switching69. Moreover, sRNA-mediated regulation of the P-fimbriae phase regulator papI affects the ON/OFF switch and, thus, expression of P-fimbriae on the surface of uropathogenic E. coli70. And transcription of a cis-encoded sRNA antisense to the pilE promoter impacts pilin antigenic variation in Neisseria meningitidis71. Recently, we and others also uncovered that sRNAs themselves can be associated with SSRs and identified a phase-variable sRNA, NikS, that acts as a global post-transcriptional regulator of the major virulence genes of H. pylori72,73.
In H. pylori, alterations in the LPS structure of H. pylori have been described at low pH74. The post-transcriptional regulation of the tlpB-hp0102 operon through RepG might provide an additional layer of LPS gene expression control under varying environmental conditions. As antisense transcription has been identified to various genes involved in H. pylori LPS biosynthesis28, regulation of surface structures by cis- or trans-encoded antisense RNAs might be a more general theme in H. pylori gene expression control. In conclusion, our study has unraveled a rheostat-like type of phase variation-dependent, post-transcriptional regulation of surface structures that are important for the colonization capacity of a bacterial pathogen.
Methods
Bacterial strains, oligonucleotides, and plasmids
Helicobacter pylori and Escherichia coli strains are listed in Supplementary Table 4. DNA oligonucleotides and plasmids are summarized in Supplementary Tables 5 and 6, respectively. Sequence information on 5′ UTRs or coding sequences of tlpB and hp0102 used in the GFP-reporter assays and tlpB leader variants are shown in Supplementary Tables 7 and 8, respectively.
Bacterial growth
E. coli strains were grown in Luria Bertani (LB) medium supplemented with 100 µg/ml ampicillin, 20 µg/ml chloramphenicol, 20 µg/ml kanamycin, and/or 10 µg/ml gentamicin if applicable. H. pylori strains were grown on GC-agar (Oxoid) plates supplemented with 10% horse serum (DHS, Biochrom AG), 1% vitamin mix, 10 µg/ml vancomycin, 5 µg/ml trimethoprim, and 1 µg/ml nystatin. For transformant selection and growth of mutant strains, 20 µg/ml kanamycin, 20 µg/ml chloramphenicol, 10 µg/ml gentamicin or 10 µg/ml erythromycin were added. For liquid cultures, 15 or 50 ml Brain Heart Infusion medium (BHI, Roth) supplemented with 10% FBS (Biochrom AG) and 10 µg/ml vancomycin, 5 µg/ml trimethoprim, and 1 µg/ml nystatin were inoculated with H. pylori strains from plates to a final OD600 of 0.02–0.04 and grown under agitation at 140 rpm in 25 cm3 or 75 cm3 cell culture flasks. Bacteria were grown at 37 °C in a HERAcell 150i incubator (Thermo Scientific) in a microaerobic environment (10% CO2, 5% O2, and 85% N2).
Construction of Helicobacter pylori mutant strains
All mutant strains are listed in Supplementary Table 4. Mutants were constructed by homologous recombination and natural transformation of PCR-amplified constructs carrying either the aphA-3 kanamycin75, the catGC chloramphenicol76, the aac(3)-IV apramycin/gentamicin77 or rpsL-erm erythromycin78 resistance cassette flanked by ~500 bp homology regions up- and downstream of the respective genomic locus. Briefly, H. pylori was grown from frozen stocks, passaged twice, then streaked on a fresh GC-agar plate and grown for 6–8 h at 37 °C under microaerobic conditions. For transformation, 500 ng up to 1 µg purified PCR product was added to the cells. After incubation for 14–16 h at 37 °C, cells were re-streaked on selective plates with the corresponding antibiotic. Genomic DNA (gDNA) of mutants was isolated using NucleoSpin Plasmid Kit according to the manufacturer’s instructions and mutants were checked by PCR and sequencing.
The H. pylori 26695 sRNA mutant (ΔrepG, CRepG, SL 2, ΔCU) as well as ΔtlpB deletion strains, and tlpB mRNA leader variants (ΔG, 6–16G in wild-type and ΔrepG mutant strain backgrounds), and the repG deletion mutants in diverse H. pylori strains (G27 and J99) were constructed in our previous study21.
Construction of X47-2AL mutant strains for mouse infections
To delete repG in the mouse-adapted H. pylori strain X47-2AL, a ΔrepG deletion construct (aphA-3 flanked by 500 nt up- and downstream of repG) was amplified by PCR using JVO-5070/-5072 and gDNA from JVS-7014 (H. pylori 26695 ΔrepG, ref. 28). The purified PCR product was transformed into CSS-0996 (H. pylori X47-2AL wildtype). Deletion of repG was verified by PCR using JVO-5069/-5257 on gDNA, resulting in strain CSS-0997 (X47-2AL ΔrepG).
The tlpB gene was deleted from strain CSS-0996 (H. pylori X47-2AL wildtype) by insertion of the aac(3)-IV cassette. To avoid potential polar effects of ΔtlpB on the expression of the downstream gene hp0102, the tlpB coding region was replaced by a non-polar gentamicin resistance cassette (aac(3)-IV), leaving the tlpB promoter and 5′ UTR intact. Therefore, a plasmid containing the aac(3)-IV cassette flanked by 500 nt up- and downstream of the H. pylori strain X47-2AL tlpB locus was cloned into E. coli. 500 nt up- and downstream of the tlpB open reading frame were amplified from gDNA of strain CSS-0996 (H. pylori X47-2AL wildtype) using CSO-0039/-0040. The resulting PCR product was XbaI/XhoI digested and introduced into likewise digested pJV752-1. Next, the resulting plasmid (pBA5-4) was used as a template for a PCR with CSO-0942/-1745 to replace the aac(3)-IV cassette with the tlpB open reading frame using an EcoRI restriction site. In parallel, a PCR with phosphorylated CSO-0263 and CSO-0293 on pUC1813apra77 was performed and the resulting PCR product was digested with EcoRI. Both EcoRI-digested PCR products (plasmid with tlpB up- and downstream region as well as aac(3)-IV) were ligated and transformed into E. coli. Insertion of the aac(3)-IV cassette was verified by colony PCR (pZE-A/CSO-0293), resulting in pBA13-5. A PCR product amplified with CSO-0039/-0040 from pSP13-5 was transformed into H. pylori X47-2AL (CSS-0996). Positive gentamicin-resistant mutants were confirmed by PCR on gDNA using CSO-0051/-0293, resulting in strain CSS-1123 (X47-2AL ΔtlpB).
An analogous cloning strategy was used for the construction of the tlpB-hp0102 double deletion mutant (ΔtlpB-hp0102) and deletion of hp0102 alone (Δhp0102). Please note that in these mutants, the aac(3)-IV cassette was inserted 77 nt upstream of the hp0102 stop codon to avoid interference with hp0101 expression. For deletion of the entire tlpB-hp0102 operon, a PCR-fragment containing 500 nt downstream of the H. pylori X47-2AL hp0102 open reading frame (CSO-0871/-1359 on gDNA of CSS-0996) was inserted into pBA13-5 (CSO-0874/0293 on pBA13-5) using oligo-introduced EcoRI/XhoI restriction sites. The ligated PCR-products were transformed into E. coli, resulting in pSP127-3. Insertion of the 500 nt downstream of hp0102 was verified by colony PCR using pZE-A/CSO-1359. The tlpB(500up)-aac(3)-IV-hp0102(500down) deletion construct was amplified from pSP127-3 with CSO-0040/-1359 and transformed into H. pylori X47-2AL (CSS-0996). Positive gentamicin-resistant mutants were confirmed by PCR on gDNA using CSO-0051/-0293, resulting in strain CSS-1743 (X47-2AL ΔtlpB-hp0102).
In order to delete hp0102 alone, a PCR-product containing 500 nt upstream of the hp0102 open reading frame was amplified using CSO-1737/-1739 on gDNA of CSS-0996. This PCR product was XbaI/BamHI digested and ligated together with a likewise digested PCR-product amplified from pSP127-3 using CSO-0873/3195. The resulting plasmid pSP186-2 was checked by PCR using pZE-A and CSO-1739. A PCR product (hp0102(500up)-aac(3)-IV-hp0102(500down)) amplified from pSP186-2 with CSO-1737/-1359 was transformed into H. pylori X47-2AL wildtype (CSS-0996). Positive gentamicin-resistant mutants were confirmed by PCR on gDNA (CSO-1738/0293), resulting in CSS-2019 (X47-2AL Δhp0102).
For construction of double deletion mutants ΔtlpB/ΔrepG, ΔtlpB-hp0102/ΔrepG, and Δhp0102/ΔrepG, the sRNA deletion construct (aphA-3 flanked by 500 nt up- and downstream of repG) amplified by PCR using JVO-5070/-5072 on gDNA from JVS-7014 (H. pylori 26695 ΔrepG) was transformed into CSS-1123 (X47-2AL ΔtlpB), CSS-1743 (X47-2AL ΔtlpB-hp0102), and CSS-2019 (X47-2AL Δhp0102). The double deletion strains CSS-1769 (X47-2AL ΔtlpB/ΔrepG), CSS-1773 (X47-2AL ΔtlpB-hp0102/ΔrepG) and CSS-2022 (X47-2AL Δhp0102/ΔrepG) were verified by PCR using JVO-5069/-5257.
The ΔtlpB-hp0102 and Δhp0102 deletion mutants were complemented with either the entire tlpB-hp0102 operon or hp0102 alone in the unrelated rdxA locus. Therefore, plasmids containing rdxA(500up)-aphA-3-tlpB-hp0102-rdxA(500down) or rdxA(500up)-aphA-3-PtlpBhp0102-rdxA(500down) were constructed in E. coli. First, ClaI/NdeI-digested PCR products amplified with CSO-1740/-1741 on gDNA of CSS-0996 (H. pylori X47-2AL) and CSO-0146/-0147 on pBA4-2 (pSP39-3 carrying aac(3)-IV gentamicin resistance cassette) were ligated and transformed into E. coli, resulting in pSP189-4. The aac(3)-IV gentamicin resistance cassette from pSP189-4 was exchanged with the aphA-3 kanamycin resistance cassette by ligation of BamHI/NheI-digested PCR products amplified with CSO-0940/-0941 on pSP189-4 and CSO-1813/-1814 on gDNA of JVS-7014 (H. pylori 26695 ΔrepG). The resulting plasmid pSP190-1 (rdxA(500up)-aphA-3-tlpB-hp0102-rdxA(500down)) was used as template for cycle-PCR using CSO-1743/-1742. This PCR product was DpnI-digested and directly transformed into E. coli, resulting in pSP192-1 (rdxA(500up)-aphA-3-PtlpBhp0102-rdxA(500down)). PCR products amplified by CSO-0017/-0018 on pSP190-1 and pSP192-1 were used for transformation in CSS-1743 (X47-2AL ΔtlpB-hp0102) and CSS-2019 (X47-2AL Δhp0102). The obtained, kanamycin-resistant strains CSS-2046 (X47-2AL ΔtlpB-hp0102 + tlpB-hp0102), CSS-2080 (X47-2AL ΔtlpB-hp0102 + hp0102), and CSS-2087 (X47-2AL Δhp0102 + hp0102) were verified by PCR using oligos CSO-0207/-1813 and sequencing by CSO-0206/-0086.
Construction of Δhp0102 in H. pylori 26695, G27, and J99
The hp0102 gene was deleted from strain CSS-0004 (H. pylori 26695 wildtype), CSS-0010 (H. pylori G27 wildtype) and CSS-0001 (H. pylori J99 wildtype) by insertion of the rpsL-erm cassette, which confers dominant streptomycin susceptibility and erythromycin resistance78. A plasmid containing the rpsL-erm cassette flanked by 500 nt up- and downstream of the hp0102 open reading frame was generated in E. coli. A PCR product corresponding to 500 nt upstream of hp0102 (CSO-0869/-0870 on gDNA of CSS-0004) was BamHI/XhoI-digested and ligated into likewise digested pSP60-221, resulting on pBA1-1. About 500 nt downstream of hp0102 were introduced into pBA1-1 by ligation of EcoRI/XbaI-digested PCR products amplified by CSO-0871/-0872 on gDNA of CSS-0004 and CSO-0309/-0873 on pBA1-1. The resulting plasmid pBA7-4 was used for PCR with oligonucleotides CSO-0870/-0872 and the obtained PCR product was transformed into CSS-0004, CSS-0010, and CSS-0001. Positive erythromycin-resistant clones were checked by PCR on gDNA using CSO-0051 and CSONIH-0033, resulting in strain CSS-1000 (26695 Δhp0102), CSS-1007 (G27 Δhp0102), and CSS-1019 (J99 Δhp0102).
Construction of H. pylori LPS mutants in 26695 and X47-2AL
Deletion of genes involved in various steps of LPS biosynthesis in H. pylori strains 26695 and X47-2AL were constructed by overlap PCR and subsequent double-crossover homologous recombination. In addition, for strain 26695, new deletion mutants for tlpB, tlpB-hp0102, and hp0102 carrying the aphA-3 instead of the rpsL-erm cassette were constructed. Overlap PCR products carried resistance cassettes flanked by ~500 nt of homologous sequence up- and downstream of the gene to be deleted. Resistance cassettes used for cloning were either aphA-3 (kanamycin) for strain 26695 or aac(3)-IV (apramycin/gentamicin) for X47-2AL. Non-polar resistance cassettes were amplified with HPK1/HPK2 from gDNA of H. pylori strain 26695 CRepG (CSS-0046)21, or with CSO-SP008 and CSO-SP009 from pUC1813apra77.
As an example, deletion of hp1284 in H. pylori strain 26695 will be described in detail. About 500 nt up- and downstream of the hp1284 coding region were amplified using CSO-SP010 × CSO-SP066 and CSO-SP067 × CSO-SP013, respectively. The antisense oligonucleotide of the hp1284 upstream region (CSO-SP066) and the sense oligonucleotide of the hp1284 downstream region (CSO-SP013) contained 21 and 25 nt overlap, respectively, with the sense and antisense oligonucleotide used to amplify the aphA-3 resistance cassette (HPK1 × HPK2). The final overlap PCR product was amplified with CSO-SP010 and CSO-SP013 using a 1:1:2 ratio of up-/downstream region of hp1284 and aphA-3 cassette. The program for the overlap PCR was as follows: 1 cycle of [98 °C, 1 min; 61 °C, 1 min; 72 °C, 10 min; 98 °C, 1 min], 40 cycles of [98 °C, 15 s; 57 °C, 30 s; 72 °C, 1 min], followed by 72 °C for 10 min. After size confirmation via agarose gel electrophoresis, the purified PCR product (Macherey-Nagel NucleoSpin PCR cleanup kit) was naturally transformed into the recipient H. pylori 26695 wild-type background (CSS-0004). Kanamycin-resistant clones were verified via colony PCR with CSO-SP014 and HPK2 resulting in the final hp1284 deletion mutant (CSS-5928; ∆hp1284). Deletion mutants for tlpB::aphA-3 (CSS-5924; ∆tlpB (Kan)), tlpB-hp0102::aphA-3 (CSS-5926; ∆tlpB-hp0102), hp0102::aphA-3 (CSS-5942; ∆hp0102 (Kan)), hp1039::aphA-3 (CSS-5930; ∆hp1039), hp1581::aphA-3 (CSS-5932; ∆hp1581), hp0159::aphA-3 (CSS-5934; ∆hp0159), hp0826::aphA-3 (CSS-5936; ∆hp0826), hp0579-0580::aphA-3 (CSS-5938; ∆hp0579-580), hp1206::aphA-3 (CSS-5940; ∆hp1206) in H. pylori strain 26695, and hp1284::aac(3)-IV (CSS-5910; X47-2AL ∆hp1284), hp1039::aac(3)-IV (CSS-5912; X47-2AL ∆hp1039), hp1581::aac(3)-IV (CSS-5914; X47-2AL ∆hp1581), hp0159::aac(3)-IV (CSS-5916; X47-2AL ∆hp0159), hp0826::aac(3)-IV (CSS-5918; X47-2AL ∆hp0826), hp0579-0580::aac(3)-IV (CSS-5920; X47-2AL ∆hp0579-580), hp1206::aac(3)-IV (CSS-5922; X47-2AL ∆hp1206) in H. pylori strain X47-2AL were constructed analogously.
Cloning of translational reporter fusions to gfpmut3
For the generation of hp0102 translational reporter fusions, the N-terminal coding region of hp0102 was fused to gfpmut3 and introduced together with the catGC resistance cassette76 into the rdxA locus of H. pylori strain G27. The first ten amino acids of the hp0102 coding region together with the upstream-encoded gene tlpB, including the tlpB promoter and 5′ UTR (26695, 12G), were amplified from gDNA of H. pylori 26695 (CSS-0004) using oligos CSO-0581/-1803. The purified PCR product was digested with ClaI/NheI and ligated with a likewise digested PCR product, which was amplified from pMA5-2 (cagA 28th::gfpmut3) using CSO-0146/-0683, resulting in pSP195-6 (tlpB-hp0102 10th::gfpmut3). Additional hp0102 10th::gfpmut3 reporter fusions were generated by cycle-PCR on pSP195-6 with CSO-1984/-1985 (tlpBmini-hp0102), CSO-2055/-2056 (PtlpBhp0102), CSO-2052/-2053 (tlpB-hp0102 ΔGhp0102), CSO-2053/-2061 (tlpB-hp0102 ATTTAhp0102) and CSO-2274/-2275 (tlpBstop-hp0102), respectively. The obtained PCR products were DpnI-digested, self-ligated, and transformed into E. coli. Positive clones were selected on plates with 100 μg/ml ampicillin and 20 µg/ml chloramphenicol, and confirmed by colony PCR using oligos pZE-XbaI/CSO-0581. The resulting plasmids pSP197-3 (tlpBmini-hp0102), pSP198-4 (PtlpBhp0102), pSP200-2 (tlpB-hp0102 ΔGhp0102), pSP201-1 (tlpB-hp0102 ATTTAhp0102), and pSP205-17 (tlpBstop-hp0102) were validated by sequencing with CSO-0206/JVO-0155. Afterwards, these plasmids were used as templates for PCR with CSO-0017/-0018. The PCR products with different hp0102::gfpmut3 reporter fusions were transformed into H. pylori G27 wildtype (CSS-0010) and/or ΔrepG (CSS-0169). Positive transformants were checked by PCR with CSO-0205/-0207 and the in-frame fusion of hp0102 to gfpmut3, as well as tlpB, was verified by sequencing with CSO-0206/JVO-0155. The corresponding H. pylori G27 strains are CSS-2104 (tlpB-hp0102 10th::gfpmut3), CSS-2107 (tlpB-hp0102 10th::gfpmut3/ΔrepG), CSS-2116 (tlpBmini-hp0102 10th::gfpmut3), CSS-2119 (tlpBmini-hp0102 10th::gfpmut3/ΔrepG), CSS-2138 (PtlpBhp0102 10th::gfpmut3), CSS-2141 (PtlpBhp0102 10th::gfpmut3/ΔrepG), CSS-2150 (tlpB-hp0102 ΔGhp0102 10th::gfpmut3), CSS-2153 (tlpB-hp0102 ΔGhp0102 10th::gfpmut3/ΔrepG), CSS-2156 (tlpB-hp0102 ATTTAhp0102 10th::gfpmut3), CSS-2159 (tlpB-hp0102 ATTTAhp0102 10th::gfpmut3/ΔrepG), CSS-3241 (tlpBstop-hp0102 10th::gfpmut3) and CSS-3244 (tlpBstop-hp0102 10th::gfpmut3/ΔrepG).
RNA preparation, northern blot, and RT-qPCR
If not mentioned otherwise, total RNA from H. pylori grown in liquid culture to mid-exponential growth phase (OD600 nm of ~0.7–1) was extracted using the hot phenol method as described previously21. For northern blot analysis, 5–10 µg total RNA was separated on 6% polyacrylamide (PAA) gels containing 7 M urea and blotted to Hybond-XL membranes (GE-Healthcare). After blotting, total RNA was UV cross-linked to the membrane and hybridized with 5′ end labeled (γ32P) DNA oligonucleotides as described previously21.
For RT-qPCR analysis, RNA samples were digested with DNaseI (Fermentas) to remove genomic DNA. All RT-qPCR experiments were carried out in at least technical and biological triplicates on a CFX96 system (Biorad) using Power SYBR Green RNA-to-CTTM 1-Step kit (Applied Biosystems by Thermo Fisher Scientific) according to the manufacturer’s instructions. Fold-changes were determined by the 2(−ΔΔCT)-method79. The specific primer sets are JVO-5267/-5268 for tlpB mRNA, CSO-0867/-0868 for hp0102 mRNA, and CSO-1173/-1174 for 6S rRNA, which served as internal standard.
RT-PCR for verification of the tlpB-hp0102 operon structure
For RT-PCR analysis, a DNaseI-digested RNA sample of H. pylori wild-type strain 26695 was reverse transcribed using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems by Thermo Fisher Scientific, #4368814) according to the manufacturer’s instruction. Briefly, reverse transcriptions were run with 1 µg of RNA ± MultiScribe™ Reverse Transcriptase using the following thermal cycling conditions: [25 °C, 10 min; 37 °C, 120 min; 85 °C, 5 min]. PCR reactions were performed using gene-specific primer sets for tlpB mRNA (JVO-5267/-5268), hp0102 mRNA (CSO-0867/-0868), and for the tlpB-hp0102 operon spanning junction (CSO-SP0061/-0062). PCR amplifications on gDNA of H. pylori wild-type strain 26695 served as a positive control. Subsequently, PCR products were run on a 2% agarose gel.
Mouse infection studies
H. pylori strain X47-2AL wild-type strain and mutants were used to infect 5-weeks old NMRI-specific pathogen-free female mice (Charles River Laboratories) as published previously in ref. 80 and as described below. The housing conditions of the mice were optimal to reduce their stress. This included half/half light/dark cycle, a constant temperature of 20–23 °C, about 50% humidity, and ad libitum access to water and food. Before infection, the animals underwent an acclimation period of one week. For infections, about 108 colony-forming units (CFUs) of H. pylori strains prepared in 100 μl of peptone broth were orogastrically administered to groups of seven mice per strain. In each experiment, four to five mice were inoculated with peptone broth alone as a naive mice negative control. One month after inoculation, mice were sacrificed and stomachs were crushed in peptone broth. CFUs of viable H. pylori bacteria per gram of stomach weight were calculated by serial dilutions and plating on blood-agar plates supplemented with the usual antibiotic-fungicide mixture with, in addition, 200 μg/ml bacitracin and 10 μg/ml nalidixic acid.
SDS-PAGE and immunoblotting
Cells corresponding to an OD600 of 1 from H. pylori cells grown to mid-exponential growth phase were collected by centrifugation (16,000 × g at 4 °C for 2 min), resuspended in 100 µl 1× protein loading buffer (62.5 mM Tris-HCl pH 6.8, 100 mM DTT, 10% glycerol, 2% SDS, 0.01% bromophenol blue) and boiled at 95 °C for 8 min.
For SDS-PAGE, protein samples corresponding to a total OD600 of 0.1 were separated by 12% SDS-PAA gels and stained by Coomassie (Fermentas, #R0571). For western blot analysis, protein samples corresponding to an OD600 of 0.1 or 0.01 were separated by 10–12% SDS-PAGE and transferred to a PVDF membrane by semi-dry blotting. Membranes were blocked for 1 h with 10% milk powder/TBS-T and incubated overnight with primary antibody at 4 °C. After washing with TBS-T, membranes were incubated for 1 h with secondary antibody linked to horseradish peroxidase. Signals were detected using ECL-reagent and Image Quant LAS 4000.
To detect the four chemotaxis receptors TlpA, B, C, and D, a polyclonal rabbit TlpA22-antiserum (1:2,000 in 3%BSA/TBS-T) that recognizes a conserved cytosolic domain (kindly provided by Karen Ottemann, University of California, Santa Cruz, CA) and secondary goat anti-rabbit IgG (1:10,000 in 3% BSA/TBS-T; GE-Healthcare, #RPN4301, RRID: AB_2650489) were used. TlpB::3×FLAG was detected by a monoclonal anti-FLAG antibody (1:1,000 in 3% BSA/TBS-T; Sigma-Aldrich, #F1804, RRID: AB_262044) and secondary sheep anti-mouse IgG (1:10,000 in 3% BSA/TBS-T; GE-Healthcare, #RPN4201). GroEL, was visualized by monoclonal anti-GroEL antibody (1:10,000 in 3% BSA/TBS-T; Sigma-Aldrich, #G6532, RRID: AB_259939) and goat anti-rabbit IgG (1:10,000 in 3%BSA/TBS-T). GFP reporter fusion proteins were detected by an anti-GFP antibody (1:1,000 in 3% BSA/TBS-T; Roche, #11814460001) and secondary sheep anti-mouse IgG (1:10,000 in 3% BSA/TBS-T).
LPS silver-staining and Lewis x antigen analysis
For LPS silver-staining, protein samples were digested with Proteinase K solution (20 mg/ml) for 90–180 min at 60 °C, followed by boiling of the samples for 10 min at 98 °C. Lipopolysaccharide structures from H. pylori were visualized by silver-staining according to ref. 74. Briefly, LPS samples corresponding to an OD600 of 0.1 were separated on 15% one dimensional SDS-PAA gel and fixed overnight in 25% isopropanol and 7% acetic acid (1× fixing solution). After incubation in sodium periodate solution (0.7% (w/v) Na5IO6 in 1× fixing solution) for 15 min, gels were washed with water (3 × 30 min). Silver staining solution (0.35% ammonia, 0.02 N NaOH, 0.4% (w/v) silver nitrate) was applied with vigorous agitation for 10 min. Following additional washing steps, gels were developed using a 2.5% (w/v) sodium carbonate solution containing 0.01% (v/v) formaldehyde (37%). Upon completion, 50 mM EDTA was used to stop the development.
For western blot analysis of Lewis x/y antigens, LPS samples corresponding to an OD600 of 0.02 were separated on 15% SDS-PAGE and transferred to a PVDF membrane by semi-dry blotting. Western blot analysis was performed using monoclonal anti-Lewis x antibody (1:500 in 3% BSA/TBS-T, Calbiochem, #434631) and monoclonal anti-Lewis y antibody (1:500 in 3% BSA/TBS-T, Merck, #434636). Chemiluminescence detection was performed with a secondary sheep anti-mouse IgG antibody conjugated to horseradish peroxidase (1:1,000 in 3% BSA/TBS-T, GE Healthcare, #RPN4201).
Growth/survival under high salt conditions
H. pylori strains were grown in liquid medium (BHI) to exponential growth phase (OD600 of ~0.7–1.0). Cells were adjusted to an OD600 of 1.0 in 0.5 ml BHI and serial dilutions of the indicated strains were spotted on GC-agar plates containing sodium chloride at 85 mM (no stress, normal salt content), 200 mM (mild stress), or 260 mM (harsh stress). Plates were incubated for 3–5 days at 37 °C under microaerobic conditions.
Antibiotics sensitivity tests
The MICs of H. pylori wild-type and mutant strains to polymyxin B were determined in triplicates by Epsilometer (E)-tests (bioMérieux, Inc., #533400) according to the manufacturer’s instructions. Briefly, H. pylori cells corresponding to an OD600 of 0.1 (exponential growth phase) were spread on GC-agar plates and E-stripes (polymyxin B concentration from 0.064 to 1024 µg/ml) were applied onto the dried surface of the agar plates. MIC values were read from the scale at the intersection of the elliptical zone of inhibition with the graded E-strip after 3 days of incubation under microaerobic conditions at 37 °C. E-test MICs that were between the standardized two-fold dilution steps were rounded to the next higher concentration.
Biocomputational search for repG, tlpB, and hp0102 homologs
Using BLAST (Basic Local Alignment Search Tool, NCBI) and SyntTax webserver (https://archaea.i2bc.paris-saclay.fr/SyntTax/), biocomputational searches for repG, tlpB, and hp0102 homologs were performed in diverse Helicobacter species (spp.) and other related Epsilonproteobacteria, including Campylobacter spp., Wolinella spp., Sulfuricurvum spp., Arcobacter spp., Sulfurospirillum spp., and Nautilia profundicola (~120 genomes). Genes were considered to be homologous when they share more than 30% percent sequence similarity with repG, tlpB, and hp0102 of H. pylori strain 26695. No homologs for repG, tlpB, and hp0102 could be identified outside of the Helicobacter spp. and thus, are not further mentioned in the main text.
Ethics Statement
Experiments in mice were carried out in strict accordance with the recommendations in the Specific Guide for the Care and the Use of Laboratory Animals of the Institut Pasteur, according to the European Directive (2010/63/UE) and the corresponding French law on animal experimentation (Arrêtés de 1988). The protocol has been approved by the Committee of Central Animal Facility Board of the Institut Pasteur. To follow the new European directives, the project was approved by the CETEA, Comité d’éthique en Expérimentation Animale of the Institut Pasteur (#2013-0051) and by the Ministère de l’Enseignement Supérieur et de la recherche (#751501).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
We thank members of the Sharma lab for fruitful discussions on this study. Moreover, we are grateful to D.S. Merrell for gfpmut3 plasmids and K. Ottemann for antisera against H. pylori chemotaxis receptors.
Source data
Author contributions
S.R.P., C.M.S., M.A., and H.d.R. wrote the manuscript. S.R.P., M.A., and C.M.S. designed the experiments, S.R.P., M.A., and B.U.B. performed experiments. Mouse infections studies were planned and done by H.d.R. and M.S.-R., respectively.
Funding
Open Access funding enabled and organized by Projekt DEAL. This publication was supported by the Open Access Publication Fund of the University of Würzburg. The Sharma lab received financial support from the Deutsche Forschungsgemeinschaft (DFG) project Sh580/1-1, the Young Academy program of the Bavarian Academy of Sciences and Humanities (BAdW), and the Bavarian Research Network bayresq.net. The De Reuse’s lab was funded by the Agence National de la Recherche [ANR-12-BSV5-0025-Bactox1] and the Pasteur-Weizmann consortium “The roles of noncoding RNAs in regulation of microbial life styles and virulence.”
Data availability
All data supporting the findings of this study are available within the article and its Supplementary information files. All data is available from the corresponding author upon reasonable request. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Peer review information Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Sandy R. Pernitzsch, Mona Alzheimer.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-021-24689-2.
References
- 1.Miller SI, Ernst RK, Bader MW. LPS, TLR4 and infectious disease diversity. Nat. Rev. Microbiol. 2005;3:36–46. doi: 10.1038/nrmicro1068. [DOI] [PubMed] [Google Scholar]
- 2.Maldonado RF, Sá-Correia I, Valvano MA. Lipopolysaccharide modification in Gram-negative bacteria during chronic infection. FEMS Microbiol. Rev. 2016;40:480–493. doi: 10.1093/femsre/fuw007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cover TL, Blaser MJ. Helicobacter pylori in health and disease. Gastroenterology. 2009;136:1863–1873. doi: 10.1053/j.gastro.2009.01.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H, et al. Insights from the redefinition of Helicobacter pylori lipopolysaccharide O-antigen and core-oligosaccharide domains. Microb. Cell. 2017;4:175–178. doi: 10.15698/mic2017.05.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chmiela M, Miszczyk E, Rudnicka K. Structural modifications of Helicobacter pylori lipopolysaccharide: an idea for how to live in peace. World J. Gastroenterol. 2014;20:9882–9897. doi: 10.3748/wjg.v20.i29.9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salama NR, Hartung ML, Müller A. Life in the human stomach: persistence strategies of the bacterial pathogen Helicobacter pylori. Nat. Rev. Microbiol. 2013;11:385–399. doi: 10.1038/nrmicro3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moran AP. Lipopolysaccharide in bacterial chronic infection: insights from Helicobacter pylori lipopolysaccharide and lipid A. Int. J. Med. Microbiol. 2007;297:307–319. doi: 10.1016/j.ijmm.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 8.Li H, et al. The redefinition of Helicobacter pylori lipopolysaccharide O-antigen and core-oligosaccharide domains. PLoS Pathog. 2017;13:e1006280. doi: 10.1371/journal.ppat.1006280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li H, et al. Lipopolysaccharide structure and Biosynthesis in Helicobacter pylori. Helicobacter. 2016;21:445–461. doi: 10.1111/hel.12301. [DOI] [PubMed] [Google Scholar]
- 10.Li H, et al. East-Asian Helicobacter pylori strains synthesize heptan-deficient lipopolysaccharide. PLoS Genet. 2019;15:e1008497. doi: 10.1371/journal.pgen.1008497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moran AP. Relevance of fucosylation and Lewis antigen expression in the bacterial gastroduodenal pathogen Helicobacter pylori. Carbohydr. Res. 2008;343:1952–1965. doi: 10.1016/j.carres.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 12.Cullen TW, et al. Helicobacter pylori versus the host: remodeling of the bacterial outer membrane is required for survival in the gastric mucosa. PLoS Pathog. 2011;7:e1002454. doi: 10.1371/journal.ppat.1002454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Appelmelk, B. J. & Vandenbroucke-Grauls, C. M. J. E. in Helicobacter pylori: Physiology and Genetics (eds. Mobley, H. L., Mendz, G. L. & Hazell, S. L.) (ASM Press, 2001). [PubMed]
- 14.Nilsson C, et al. Lipopolysaccharide diversity evolving in Helicobacter pylori communities through genetic modifications in fucosyltransferases. PLoS ONE. 2008;3:e3811. doi: 10.1371/journal.pone.0003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nilsson C, et al. An enzymatic ruler modulates Lewis antigen glycosylation of Helicobacter pylori LPS during persistent infection. Proc. Natl Acad. Sci. USA. 2006;103:2863–2868. doi: 10.1073/pnas.0511119103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skoglund A, et al. A changing gastric environment leads to adaptation of lipopolysaccharide variants in Helicobacter pylori populations during colonization. PLoS ONE. 2009;4:e5885. doi: 10.1371/journal.pone.0005885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou K, Aertsen A, Michiels CW. The role of variable DNA tandem repeats in bacterial adaptation. FEMS Microbiol. Rev. 2014;38:119–141. doi: 10.1111/1574-6976.12036. [DOI] [PubMed] [Google Scholar]
- 18.Moxon R, Bayliss C, Hood D. Bacterial contingency loci: the role of simple sequence DNA repeats in bacterial adaptation. Annu. Rev. Genet. 2006;40:307–333. doi: 10.1146/annurev.genet.40.110405.090442. [DOI] [PubMed] [Google Scholar]
- 19.Caldelari I, Chao Y, Romby P, Vogel J. RNA-mediated regulation in pathogenic bacteria. Cold Spring Harb. Perspect. Med. 2013;3:a010298. doi: 10.1101/cshperspect.a010298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol. Cell. 2011;43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pernitzsch SR, Tirier SM, Beier D, Sharma CM. A variable homopolymeric G-repeat defines small RNA-mediated posttranscriptional regulation of a chemotaxis receptor in Helicobacter pylori. Proc. Natl Acad. Sci. USA. 2014;111:E501–E510. doi: 10.1073/pnas.1315152111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rader BA, et al. Helicobacter pylori perceives the quorum-sensing molecule AI-2 as a chemorepellent via the chemoreceptor TlpB. Microbiol. (Read., Engl.) 2011;157:2445–2455. doi: 10.1099/mic.0.049353-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Croxen MA, Sisson G, Melano R, Hoffman PS. The Helicobacter pylori chemotaxis receptor TlpB (HP0103) is required for pH taxis and for colonization of the gastric mucosa. J. Bacteriol. 2006;188:2656–2665. doi: 10.1128/JB.188.7.2656-2665.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson JK, et al. Chemorepulsion from the quorum signal autoinducer-2 promotes Helicobacter pylori biofilm dispersal. MBio. 2015;6:e00379. doi: 10.1128/mBio.00379-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang JY, et al. Chemodetection and destruction of host urea allows Helicobacter pylori to locate the epithelium. Cell Host Microbe. 2015;18:147–156. doi: 10.1016/j.chom.2015.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGee DJ, et al. Colonization and inflammation deficiencies in Mongolian Gerbils infected by Helicobacter pylori chemotaxis mutants. Infect. Immun. 2005;73:1820–1827. doi: 10.1128/IAI.73.3.1820-1827.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Williams SM, et al. Helicobacter pylori chemotaxis modulates inflammation and bacterium-gastric epithelium interactions in infected mice. Infect. Immun. 2007;75:3747–3757. doi: 10.1128/IAI.00082-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sharma CM, et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010;464:250–255. doi: 10.1038/nature08756. [DOI] [PubMed] [Google Scholar]
- 29.Wisniewski-Dyé F, Vial L. Phase and antigenic variation mediated by genome modifications. Antonie Van. Leeuwenhoek. 2008;94:493–515. doi: 10.1007/s10482-008-9267-6. [DOI] [PubMed] [Google Scholar]
- 30.Vinella D, et al. Evolution of Helicobacter: acquisition by gastric species of two histidine-rich proteins essential for colonization. PLoS Pathog. 2015;11:e1005312. doi: 10.1371/journal.ppat.1005312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moran AP, Sturegård E, Sjunnesson H, Wadström T, Hynes SO. The relationship between O-chain expression and colonisation ability of Helicobacter pylori in a mouse model. FEMS Immunol. Med. Microbiol. 2000;29:263–270. doi: 10.1111/j.1574-695X.2000.tb01532.x. [DOI] [PubMed] [Google Scholar]
- 32.Logan SM, Conlan JW, Monteiro MA, Wakarchuk WW, Altman E. Functional genomics of Helicobacter pylori: identification of a beta-1,4 galactosyltransferase and generation of mutants with altered lipopolysaccharide. Mol. Microbiol. 2000;35:1156–1167. doi: 10.1046/j.1365-2958.2000.01784.x. [DOI] [PubMed] [Google Scholar]
- 33.Hug I, et al. Helicobacter pylori lipopolysaccharide is synthesized via a novel pathway with an evolutionary connection to protein N-glycosylation. PLoS Pathog. 2010;6:e1000819. doi: 10.1371/journal.ppat.1000819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Altman E, et al. Effect of the HP0159 ORF mutation on the lipopolysaccharide structure and colonizing ability of Helicobacter pylori. FEMS Immunol. Med. Microbiol. 2008;53:204–213. doi: 10.1111/j.1574-695X.2008.00416.x. [DOI] [PubMed] [Google Scholar]
- 35.Altman E, Chandan V, Li J, Vinogradov E. Lipopolysaccharide structures of Helicobacter pylori wild-type strain 26695 and 26695 HP0826::Kan mutant devoid of the O-chain polysaccharide component. Carbohydr. Res. 2011;346:2437–2444. doi: 10.1016/j.carres.2011.06.036. [DOI] [PubMed] [Google Scholar]
- 36.Stead CM, Zhao J, Raetz CRH, Trent MS. Removal of the outer Kdo from Helicobacter pylori lipopolysaccharide and its impact on the bacterial surface. Mol. Microbiol. 2010;78:837–852. doi: 10.1111/j.1365-2958.2010.07304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nikaido H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003;67:593–656. doi: 10.1128/MMBR.67.4.593-656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yethon JA, et al. Salmonella enterica serovar Typhimurium waaP mutants show increased susceptibility to polymyxin and loss of virulence in vivo. Infect. Immun. 2000;68:4485–4491. doi: 10.1128/IAI.68.8.4485-4491.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma CM, Darfeuille F, Plantinga TH, Vogel J. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev. 2007;21:2804–2817. doi: 10.1101/gad.447207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Darfeuille F, Unoson C, Vogel J, Wagner EGH. An antisense RNA inhibits translation by competing with standby ribosomes. Mol. Cell. 2007;26:381–392. doi: 10.1016/j.molcel.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 41.Dutta T, Srivastava S. Small RNA-mediated regulation in bacteria: a growing palette of diverse mechanisms. Gene. 2018;656:60–72. doi: 10.1016/j.gene.2018.02.068. [DOI] [PubMed] [Google Scholar]
- 42.Baldwin DN, et al. Identification of Helicobacter pylori genes that contribute to stomach colonization. Infect. Immun. 2007;75:1005–1016. doi: 10.1128/IAI.01176-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hiratsuka K, et al. Identification of a D-glycero-D-manno-heptosyltransferase gene from Helicobacter pylori. J. Bacteriol. 2005;187:5156–5165. doi: 10.1128/JB.187.15.5156-5165.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Monteiro MA. Helicobacter pylori: a wolf in sheep’s clothing: the glycotype families of Helicobacter pylori lipopolysaccharides expressing histo-blood groups: structure, biosynthesis, and role in pathogenesis. Adv. Carbohydr. Chem. Biochem. 2001;57:99–158. doi: 10.1016/S0065-2318(01)57016-X. [DOI] [PubMed] [Google Scholar]
- 45.Fowler M, Thomas RJ, Atherton J, Roberts IS, High NJ. Galectin-3 binds to Helicobacter pylori O-antigen: it is upregulated and rapidly secreted by gastric epithelial cells in response to H. pylori adhesion. Cell. Microbiol. 2006;8:44–54. doi: 10.1111/j.1462-5822.2005.00599.x. [DOI] [PubMed] [Google Scholar]
- 46.Valkonen KH, Wadström T, Moran AP. Interaction of lipopolysaccharides of Helicobacter pylori with basement membrane protein laminin. Infect. Immun. 1994;62:3640–3648. doi: 10.1128/iai.62.9.3640-3648.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edwards NJ, et al. Lewis X structures in the O antigen side-chain promote adhesion of Helicobacter pylori to the gastric epithelium. Mol. Microbiol. 2000;35:1530–1539. doi: 10.1046/j.1365-2958.2000.01823.x. [DOI] [PubMed] [Google Scholar]
- 48.Bhattacharya S, et al. A conserved Helicobacter pylori Gene, HP0102, is induced upon contact with gastric cells and has multiple roles in pathogenicity. J. Infect. Dis. 2016;214:196–204. doi: 10.1093/infdis/jiw139. [DOI] [PubMed] [Google Scholar]
- 49.Huang JY, Goers Sweeney E, Guillemin K, Amieva MR. Multiple acid sensors control Helicobacter pylori colonization of the stomach. PLoS Pathog. 2017;13:e1006118. doi: 10.1371/journal.ppat.1006118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lerouge I, Vanderleyden J. O-antigen structural variation: mechanisms and possible roles in animal/plant-microbe interactions. FEMS Microbiol. Rev. 2002;26:17–47. doi: 10.1111/j.1574-6976.2002.tb00597.x. [DOI] [PubMed] [Google Scholar]
- 51.Lukácová M, Barák I, Kazár J. Role of structural variations of polysaccharide antigens in the pathogenicity of Gram-negative bacteria. Clin. Microbiol. Infect. 2008;14:200–206. doi: 10.1111/j.1469-0691.2007.01876.x. [DOI] [PubMed] [Google Scholar]
- 52.Salaün L, Linz B, Suerbaum S, Saunders NJ. The diversity within an expanded and redefined repertoire of phase-variable genes in Helicobacter pylori. Microbiol. (Reading, Engl.) 2004;150:817–830. doi: 10.1099/mic.0.26993-0. [DOI] [PubMed] [Google Scholar]
- 53.Saunders NJ, Peden JF, Hood DW, Moxon ER. Simple sequence repeats in the Helicobacter pylori genome. Mol. Microbiol. 1998;27:1091–1098. doi: 10.1046/j.1365-2958.1998.00768.x. [DOI] [PubMed] [Google Scholar]
- 54.Devi SH, et al. Genome of Helicobacter pylori strain 908. J. Bacteriol. 2010;192:6488–6489. doi: 10.1128/JB.01110-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Avasthi TS, et al. Genomes of two chronological isolates (Helicobacter pylori 2017 and 2018) of the West African Helicobacter pylori strain 908 obtained from a single patient. J. Bacteriol. 2011;193:3385–3386. doi: 10.1128/JB.05006-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kennemann L, et al. Helicobacter pylori genome evolution during human infection. Proc. Natl Acad. Sci. USA. 2011;108:5033–5038. doi: 10.1073/pnas.1018444108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Linz B, et al. A mutation burst during the acute phase of Helicobacter pylori infection in humans and rhesus macaques. Nat. Commun. 2014;5:4165. doi: 10.1038/ncomms5165. [DOI] [PubMed] [Google Scholar]
- 58.Nell S, et al. Genome and methylome variation in Helicobacter pylori with a cag pathogenicity island during early stages of human infection. Gastroenterology. 2018;154:612–623 e7. doi: 10.1053/j.gastro.2017.10.014. [DOI] [PubMed] [Google Scholar]
- 59.Behrens W, et al. Role of energy sensor TlpD of Helicobacter pylori in gerbil colonization and genome analyses after adaptation in the gerbil. Infect. Immun. 2013;81:3534–3551. doi: 10.1128/IAI.00750-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Salaün L, Ayraud S, Saunders NJ. Phase variation mediated niche adaptation during prolonged experimental murine infection with Helicobacter pylori. Microbiol. (Reading, Engl.) 2005;151:917–923. doi: 10.1099/mic.0.27379-0. [DOI] [PubMed] [Google Scholar]
- 61.Bergman MP, et al. Helicobacter pylori modulates the T helper cell 1/T helper cell 2 balance through phase-variable interaction between lipopolysaccharide and DC-SIGN. J. Exp. Med. 2004;200:979–990. doi: 10.1084/jem.20041061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Khamri W, et al. Variations in Helicobacter pylori lipopolysaccharide to evade the innate immune component surfactant protein D. Infect. Immun. 2005;73:7677–7686. doi: 10.1128/IAI.73.11.7677-7686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.D’Elios MM, Bergman MP, Amedei A, Appelmelk BJ, Del Prete G. Helicobacter pylori and gastric autoimmunity. Microbes Infect. 2004;6:1395–1401. doi: 10.1016/j.micinf.2004.10.001. [DOI] [PubMed] [Google Scholar]
- 64.Monteiro MA, et al. Helicobacter pylori from asymptomatic hosts expressing heptoglycan but lacking Lewis O-chains: Lewis blood-group O-chains may play a role in Helicobacter pylori induced pathology. Biochem. Cell Biol. 2001;79:449–459. doi: 10.1139/o01-035. [DOI] [PubMed] [Google Scholar]
- 65.Eaton KA, et al. Helicobacter pylori with a truncated lipopolysaccharide O chain fails to induce gastritis in SCID mice injected with splenocytes from wild-type C57BL/6J mice. Infect. Immun. 2004;72:3925–3931. doi: 10.1128/IAI.72.7.3925-3931.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dersch P, Khan MA, Mühlen S, Görke B. Roles of regulatory RNAs for antibiotic resistance in bacteria and their potential value as novel drug targets. Front. Microbiol. 2017;8:803. doi: 10.3389/fmicb.2017.00803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Åberg A, et al. A repetitive DNA element regulates expression of the Helicobacter pylori sialic acid binding adhesin by a rheostat-like mechanism. PLoS Pathog. 2014;10:e1004234. doi: 10.1371/journal.ppat.1004234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Atack JM, Tan A, Bakaletz LO, Jennings MP, Seib KL. Phasevarions of bacterial pathogens: methylomics sheds new light on old enemies. Trends Microbiol. 2018;26:715–726. doi: 10.1016/j.tim.2018.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zaleski P, Wojciechowski M, Piekarowicz A. The role of Dam methylation in phase variation of Haemophilus influenzae genes involved in defence against phage infection. Microbiol. (Read., Engl.) 2005;151:3361–3369. doi: 10.1099/mic.0.28184-0. [DOI] [PubMed] [Google Scholar]
- 70.Khandige S, Kronborg T, Uhlin BE, Møller-Jensen J. sRNA-mediated regulation of P-fimbriae phase variation in uropathogenic Escherichia coli. PLoS Pathog. 2015;11:e1005109. doi: 10.1371/journal.ppat.1005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cahoon LA, Seifert HS. Transcription of a cis-acting, noncoding, small RNA is required for pilin antigenic variation in Neisseria gonorrhoeae. PLoS Pathog. 2013;9:e1003074. doi: 10.1371/journal.ppat.1003074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Eisenbart SK, et al. A repeat-associated small RNA controls the major virulence factors of Helicobacter pylori. Mol. Cell. 2020;80:210–226. doi: 10.1016/j.molcel.2020.09.009. [DOI] [PubMed] [Google Scholar]
- 73.Kinoshita-Daitoku, R. et al. A bacterial small RNA regulates the adaptation of Helicobacter pylori to the host environment. Nat Commun.12, 2085 (2021). [DOI] [PMC free article] [PubMed]
- 74.Moran AP, et al. Phenotypic variation in molecular mimicry between Helicobacter pylori lipopolysaccharides and human gastric epithelial cell surface glycoforms. Acid-induced phase variation in Lewis(x) and Lewis(y) expression by H. Pylori lipopolysaccharides. J. Biol. Chem. 2002;277:5785–5795. doi: 10.1074/jbc.M108574200. [DOI] [PubMed] [Google Scholar]
- 75.Skouloubris S, Thiberge JM, Labigne A, De Reuse H. The Helicobacter pylori UreI protein is not involved in urease activity but is essential for bacterial survival in vivo. Infect. Immun. 1998;66:4517–4521. doi: 10.1128/IAI.66.9.4517-4521.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Boneca IG, et al. Development of inducible systems to engineer conditional mutants of essential genes of Helicobacter pylori. Appl. Environ. Microbiol. 2008;74:2095–2102. doi: 10.1128/AEM.01348-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bury-Moné S, et al. Presence of active aliphatic amidases in Helicobacter species able to colonize the stomach. Infect. Immun. 2003;71:5613–5622. doi: 10.1128/IAI.71.10.5613-5622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dailidiene D, Dailide G, Kersulyte D, Berg DE. Contraselectable streptomycin susceptibility determinant for genetic manipulation and analysis of Helicobacter pylori. Appl. Environ. Microbiol. 2006;72:5908–5914. doi: 10.1128/AEM.01135-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 80.Bury-Moné S, et al. Roles of alpha and beta carbonic anhydrases of Helicobacter pylori in the urease-dependent response to acidity and in colonization of the murine gastric mucosa. Infect. Immun. 2008;76:497–509. doi: 10.1128/IAI.00993-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the article and its Supplementary information files. All data is available from the corresponding author upon reasonable request. Source data are provided with this paper.