Abstract
Gleditsia sinensis is an endemic species widely distributed in China with high economic and medicinal value. To explore the genomic evolution and phylogenetic relationships of G. sinensis, the complete mitochondrial (mt) genome of G. sinensis was sequenced and assembled, which was firstly reported in Gleditsia. The mt genome was circular and 594,121 bp in length, including 37 protein-coding genes (PCGs), 19 transfer RNA (tRNA) genes and 3 ribosomal RNA (rRNA) genes. The overall base composition of the G. sinensis mt genome was 27.4% for A, 27.4% for T, 22.6% for G, 22.7% for C. The comparative analysis of PCGs in Fabaceae species showed that most of the ribosomal protein genes and succinate dehydrogenase genes were lost. In addition, we found that the rps4 gene was only lost in G. sinensis, whereas it was retained in other Fabaceae species. The phylogenetic analysis based on shared PCGs of 24 species (22 Fabaceae and 2 Solanaceae) showed that G. sinensis is evolutionarily closer to Senna species. In general, this research will provide valuable information for the evolution of G. sinensis and provide insight into the phylogenetic relationships within the family Fabaceae.
Subject terms: Mitochondrial genome, Phylogenetics
Introduction
Mitochondria are semi-autonomous organelles in eukaryotic cells, and they have relatively independent transcription and translation systems1. Mitochondria can provide ATP and other energy required for life activities through oxidative phosphorylation2,3. At present, the serial endosymbiosis theory is the most popular theory explaining the origin of mitochondria, which suggests that mitochondria originated from an endosymbiotic α-proteobacteria4. Most of the published complete mt genomes are from animals, protists and fungi. In contrast, the available plants mt genomes are very scarce. In 1992, the first mt genome sequencing of the land plant Marchantia polymorpha was completed5. To date, NCBI (National Center for Biotechnology Information, https://www.ncbi.nlm.nih.gov/) has collected 333 complete plant mt genomes. There is no doubt that with the development of DNA sequencing technology, the number of available plant mitochondrial sequences will increase rapidly.
The higher plants mt genomes vary dramatically in size and structure organization6. The length ranges from 66 kbp of Viscum scurruloideum7 to 11.3 Mbp of Silene conica8. Paradoxically, in most plants, the mitochondrial sequences evolve very slowly9 and the mutation rate is quite low10. Compared with animal mt genomes, plant mt genomes are usually large and complex3,11. The complexity of the plant mt genome mainly due to the presence of a large number of non-coding regions and the introgression of foreign DNA from the chloroplast or nuclear genome12. Despite the plant mt genome is relatively large, it contains fewer genes than its plastid counterpart, and the number of known genes is usually between 50 and 601,13. The structure of the plant mt genome is usually circular, while linear form also exists in some species, such as the rice (Oryza sativa)14. The higher plants mt genome is characterized by repeat sequences15,16, which accounts for 2%-60% of the total genome size17. Some repeat sequences are species-specific and can be used as genome-specific genetic markers to study the evolutionary relationship between species18.
Fabaceae is the third-largest angiosperm family after Asteraceae and Orchidaceae19. Fabaceae plants are used in many aspects of human life, including food, wood, medicine, textiles, ornamental and horticultural plants20. G. sinensis, a kind of Fabaceae plant widely distributed throughout China21, provide a wide array of benefits. It plays an important role in the conservation and maintenance of soil and water resources due to its drought resistance and low requirements on the soil. In addition, G. Sinensis saponin is effective in decontamination, foaming, which is widely used in the production of cosmetics and detergents with high economic value22. The fruits and thorns of G. Sinensis with remarkable antioxidant, anti-tumor, antiviral, antibacterial, and anti-allergic activities 23, are used as medicinal herbs in China and have been used in the treatment of cancer, carbuncles, skin diseases as well as other diseases23,24. However, its mt genome has not been determined, which highly limits the process of molecular research on G. sinensis.
In this study, we assembled the complete mt genome of G. sinensis, which is the first mt genome for Gleditsia. We analyzed its gene content, repeat sequences, codon usage bias, synonymous and nonsynonymous substitution rate. Besides, gene loss and phylogenetic analyses were performed by comparisons with other Fabaceae plants mt genomes. Our data will provide valuable information for studying the evolutionary processes of the G. sinensis mt genome.
Results and discussion
Genome features
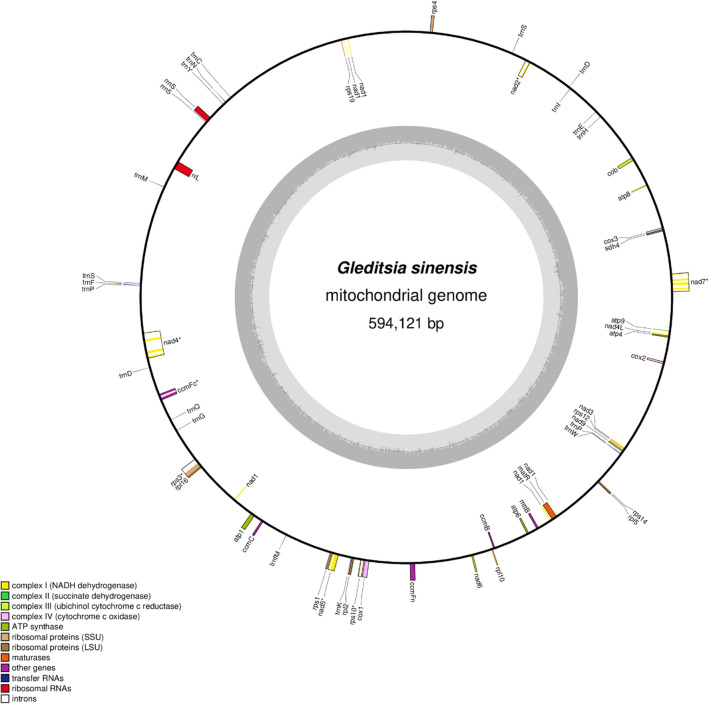
The complete mt genome of G. sinensis is 594,121 bp in length with a circular structure (Fig. 1), and its size is similar to the mt genomes of some Fabaceae plants, such as V. faba (588,000 bp)25, L. coriaria (601,574 bp)26 and T. indica (607,282 bp)26. The base composition is as follows: A (27.4%), T (27.4%), C (22.7%), G (22.6%), and GC content is 45.3%. A total of 57 genes were identified in the G. sinensis mt genome, including 37 PCGs, 19 transfer RNA genes and 3 ribosomal RNA genes (Table 1). As shown in Table 2, the PCGs in the G. sinensis mt genome account for 5.11% of the entire genome with a total length of 30,336, while non-coding regions account for 90.65% of the entire genome, with a total length of 538,550. The total length of tRNA genes and rRNA genes comprise 0.24% and 0.85% of the entire mt genome, respectively. There exist 12 introns in 37 PCGs, accounting for 3.16% of the genome. Among them, nad2, nad5, ccmFc, rps3 and rps10 contain one intron, and nad4 and nad7 contain three and four introns, respectively (Table 1). Additionally, a protein-coding gene (nad1) and three tRNA genes (trnP, trnD, trnS) were found to contain two copies (Table 1).
Figure 1.
Genome map of the G. sinensis mt genome. Genes belonging to the functional group are color-coded on the circle as transcribed clockwise (outside) and transcribed counter-clockwise (inside). The darker gray in the inner circle represents the GC content, while the lighter gray represents the AT content.
Table 1.
Gene annotation of the G. sinensis mt genome.
| Category | Group | Genes |
|---|---|---|
| Mitochondrial respiratory chain related genes | Complex I | nad1(× 2), nad2a, nad3, nad4b, nad4L, nad5a, nad6, nad7b, nad9 |
| Complex II | sdh4 | |
| Complex III | cob | |
| Complex IV | cox1, cox2, cox3 | |
| Complex V | atp1, atp4, atp6, atp8, atp9 | |
| Cytochrome c synthesis | ccmFn, ccmB, ccmC, ccmFca | |
| Transcription and translation related genes | Ribosomal proteins |
rpl2, rpl10, rpl5, rpl16, rps1, rps3a, rps4, rps10a, rps12, rps14, rps19 |
| RNA genes | Transfer RNA |
trnP(× 2), trnW, trnK, trnfM, trnG, trnQ, trnD(× 2), trnF, trnS(× 2), trnM, trnY, trnN, trnC, trnI, trnE, trnH |
| Ribosomal RNA | rrnL, rrn5, rrnS | |
| Other genes | Maturase | matR |
| Methyltransferase | mttB |
aGenes with one intron, b genes with at least two introns.
Table 2.
Genomic features of G. sinensis mt genome.
| Feature | Size (bp) | Proportion in Genome (%) |
|---|---|---|
| Whole genome | 594,121 | 100 |
| PCGsa | 30,336 | 5.11 |
| intronsa | 18,752 | 3.16 |
| tRNA genesa | 1,420 | 0.24 |
| rRNA genesa | 5,063 | 0.85 |
| Non-coding regions | 538,550 | 90.65 |
aPCGs, introns, tRNA genes, and rRNA genes belong to coding regions.
Codon usage analysis
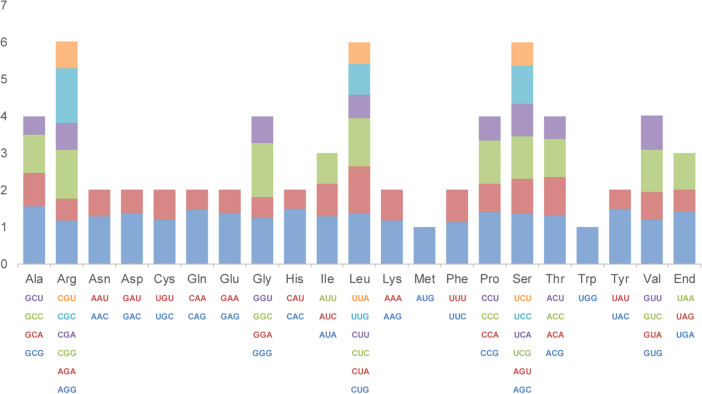
Relative synonymous codon usage (RSCU) refers to the relative probability of a specific codon between the synonymous codons encoding the corresponding amino acid27. RSCU = 1 indicates that there is no preference for codon usage, while RSCU > 1 indicates that the codon is a used relatively frequently codon28,29. The 37 PCGs of the G. sinensis mt genome contained 10,112 codons (Supplementary Table S1). Among them, 1057 (10.45%) encoded leucine (Leu) while only 147 (1.45%) encoded cysteine (Cys), which were the most and least used amino acids in the G. sinensis mt genome, respectively (Table S1). The AT content of the first, second, and third codon positions was 52.01%, 56.59% and 61.26%, respectively. The high AT content at the third codon position was similar to other reported higher plants mt genomes26,27. Apart from UGG, all preferred synonymous codons (RSCU > 1) end in either A or U (Fig. 2).
Figure 2.
RSCU based on PCGs of the mt genome of G. sinensis.
Repeat sequences
The angiosperms mt genomes are characterized by repeat sequences, which play an important role in biological evolution, genetic regulation and gene expression32,33. SSRs are tandem repeats with 1–6 nucleotides as the basic unit34, which are particularly abundant in plant genomes and have an important impact on the function and evolution of the genome21. SSRs are generally used as DNA markers for population genetic studies due to the advantages of high polymorphism35. In the present study, we identified 71 SSRs with a total length of 718 bp, including 11 dinucleotides (15.49%), two trinucleotides (2.82%), and 58 mononucleotides (84.51%), while tetranucleotides, pentanucleotides, and hexanucleotides were not identified in the mt genome (Fig. 3A). Among them, the most abundant repeat sequences were mononucleotides, which suggests that mononucleotide repeats may contribute more to genetic variation than other SSRs36. Further analysis of the repeat unit of SSRs showed that 80.28% of mononucleotides were A/T, while G/C only accounted for 4.23% (Table S2). The higher AT contents in mononucleotide repeats of G. sinensis mt genome was congruent with other reported Fabaceae plants37. The identification of featured SSRs in this study can provide valuable resources on developing markers for phylogenetic research and population studies of G. sinensis.
Figure 3.
Analyses of repeats in the G. sinensis mt genome. (A) The number of different types of SSRs. (B) The number of different types of long repeats.
The sequences with a repeat unit longer than 30 bp were regarded as long repeats, including forward repeats (F), palindromic repeats (P), reverse repeats (R) and complement repeats (C). We identified 50 long repeat sequences in G. sinensis mt genome, ranging from 86 to 270 bp, including 26 forward repeats and 24 palindromic repeats. Most long repeats were 80–119 bp in length, and only 7 repeats were longer than 150 bp (Fig. 3). Repeat sequences, especially long repeats, have important impacts on the structure of plant mt genomes, and they are positively correlated with the size of the genome12.
Synonymous and nonsynonymous substitution rate
The calculation of Ka/Ks ratio is important for understanding the dynamics of molecular evolution38,39. This ratio can infer whether the PCGs are under selective pressure. Ka/Ks = 1 indicates neutral mutation, Ka/Ks < 1 indicates negative (purifying) selection, and Ka/Ks > 1 indicates positive (diversifying) selection. In this study, all of the PCGs of the G. sinensis mt genome were used to calculate the Ka/Ks ratios. As shown in Fig. 4, the Ka/Ks ratios of most PCGs were less than 1, indicating that most of the PCGs were under purification selection. These mitochondrial genes that experienced purification selection may play a vital role in stabilizing the normal function of mitochondria37. In addition, the Ka/Ks ratios of atp4, atp6, atp8, cox1, matR, nad1, nad4, nad4L, nad5, rps1 were all greater than 1, and almost all of these genes belong to mitochondrial respiratory chain related genes category, indicating that they were under positive selection, which suggests that some advantages had emerged during evolution37.
Figure 4.
The Ka/Ks ratios for 37 PCGs of G. sinensis.
Gene loss
During the evolution of the angiosperm mt genome, the loss of PCGs occurred frequently40,41. In this study, we compared the distribution of PCGs in 22 Fabaceae plant mt genomes (Table S3). As shown in Fig. 5, most PCGs were conserved, especially for mitochondrial respiratory chain related genes, maturase and methyltransferase genes. In contrast, the ribosomal protein and succinate dehydrogenase genes were highly variable. The rpl2, rpl10, rpl14, rps7, rps11, rps19, sdh3, sdh4 genes were lost in most mt genomes, which is understandable because that ribosomal protein and succinate dehydrogenase genes are frequently lost or transferred to the nucleus during the evolution of angiosperm mt genomes (e.g. rps10, rpl2, sdh3, sdh4)26,41,42. A total of five genes were lost in the G. sinensis mt genome, including four ribosomal protein genes (rpl10, rps4, rps10, rps19) and one succinate dehydrogenase gene (sdh4). The rps10 gene was only lost in G. sinensis but it was retained in other Caesalpinioideae species. In addition, we found that the rps4 gene was only lost in G. sinensis but it was retained in other Fabaceae species. Interestingly, this gene has not been found lost in other plant mt genomes, yet. Therefore, it is an open question as to whether rps4 was lost for the reason that its function may no longer be needed for G. sinensis, or whether it was functionally transferred to the nucleus43,44.
Figure 5.
Distribution of PCGs in 22 Fabaceae plant mt genomes. White boxes indicate that the gene is not present in the mt genomes. Light yellow, golden, blue, purple, black, pink and red boxes indicate that one, two, three, four, five, six and twelve copies exist in the particular mt genomes, respectively. Light green, orange, brown and rose red boxes indicate that Papilionoideae, Caesalpinioideae, Detarioideae, Cercidoideae, respectively.
Phylogenetic analyses
The higher plant mt genomes evolve slowly, and its mutation rate is significantly low1,8,10, which makes it a useful tool for phylogenetic research45. In this study, phylogenetic analyses were performed based on the 24 plants mt genomes, including 22 Fabaceae (P. vulgaris, H. brasiletto, L. coriaria, T. indica, S. flavescens, A. ligulate, G. soja, L. trichandra, A. mongolicus, S. japonicum, S. occidentalis, S. tora, M. truncatula, G. max, G. sinensis, L. japonicus, P. pinnata, V. radiata, C. canadensis, C. austral, A. nanus, V. faba) and two Solanaceae (Capsicum annuum, Nicotiana tabacum ). Meanwhile, two Solanaceae species were used as outgroups. The ML tree and BI tree were constructed based on 17 shared PCGs (atp6, ccmB, ccmC, ccmFn, cox1, cox3, matR, nad2, nad4, nad5, nad6, nad7, nad9, rpl16, rps3, rps4, rps12). The ML and BI trees shared a consistent typology. As shown in Fig. 6, all Fabaceae plants were clustered within a lineage distinct from the outgroup. Most nodes in the ML and BI trees had high support values (bootstrap proportions ≥ 75, posterior probabilities ≥ 0.963), whereas the support value of the clade Detarioideae and Caesalpinioideae was only 53 in the ML tree. The phylogenetic relationship of the four subfamilies was described as (Cercidoideae + (Papilionoideae + (Detarioideae + Caesalpinioideae))). The tree strongly support the separation of Cercidoideae from the clade (Papilionoideae, Detarioideae, Caesalpinioideae), with bootstrap proportions = 100, posterior probabilities = 1, which was consistent with a previous study report26. It was worth noting that the G. sinensis and two Senna species were clustered into one clade with a bootstrap support value of 87 and a posterior probability of 1, which indicates that G. sinensis were evolutionarily closer to Senna species within the Fabaceae family. The phylogenetic tree constructed in this study could not reflect the true phylogenetic relationship of Fabaceae for the fact that few Fabaceae mt genomes have been sequenced. To illustrate more accurately the evolutionary relationship among Fabaceae species, it is necessary to use more species to analyze the phylogeny.
Figure 6.
Maximum likelihood phylogenies of G. sinensis within Fabaceae. Relationships were inferred using 17 conserved PCGs of 24 plant mt genomes. Numbers on each node are bootstrap support values and posterior probabilities. The scale indicates the number of nucleotide substitutions per site.
Methods
DNA extraction and sequencing
The fresh leaves of Gleditsia sinensis used in this study were collected from the Chinese Medicine Botanical Garden of Tianjin University of Traditional Chinese Medicine (117.06°E, 38.96°N), and it was identified by Prof. Tianxiang Li. The collection of Gleditsia Sinensis was approved by Tianjin University of Traditional Chinese Medicine and was conducted in accordance with the standards of "Medicine Mountain Collection of Tianjin University of Traditional Chinese Medicine". The voucher specimens were deposited in the State Key Laboratory of Component-Based Chinese Medicine, voucher No.G20191120. The collected leaves were quickly frozen in liquid nitrogen and then stored at − 80 °C until DNA extraction. Total genomic DNA was extracted by using the extract Plant DNA kit (QIAGEN, Germany). Truseq Nano DNA HT sample preparation kit (Illumina USA) was used to construct a 350 bp insert-sized DNA sequencing library, which was later sequenced with a paired-end read length of 2 × 150 bp on Illumina HiSeq X Ten platform following the standard Illumina protocols (Illumina, San Diego, CA).
Mitochondrial genome assembly and annotation
A total of 14,836,699 raw reads of G. sinensis were produced by Illumina pair-end sequencing, and 14,794,823 clean reads were retained after the quality checking by FastQC. The base quality value Q20 and Q30 were 94.16% and 86.35%, respectively. Subsequent analyses were based on the filtered high-quality sequences. For mt genome assembly, high-quality DNA sequencing reads were mapped to reference mt genome of Senna occidentalis (NCBI accession number NC_038221) using Geneious46 to get the sequence of cox1, the number of iterations was set to 5 times. Then, the G. sinensis mitochondrial genome was de novo assembled using NOVOPlasty3.7.247, with cox1 sequence set as seed and K-mer length of 39. The N50 and N90 of the obtained contigs were 64428 bp and 18438 bp, respectively. In order to obtain a high-quality mt genome, the base of the genome was corrected based on high-quality DNA sequencing data by using BWA software48–50, and a total of 96 bases were corrected. Finally, to determine whether the assembled contig is a circular structure, we designed primers based on the base sequence at the head and tail of the contig and performed PCR amplification (Table S4). The results confirmed that the G. sinensis mt genome was a typical circular molecule (Figure S1).
The mt genome was annotated using MITOFY12 (http://dogma.ccbb.utexas.edu/mitofy/) and GeSeq51 (https://chlorobox.mpimp-golm.mpg.de/geseq.html) and was manually checked and adjusted the annotation using Senna occidentalis as the reference sequence. The online tRNAscan-SE search server (http://lowelab.ucsc.edu/tRNAscan-SE) was used to annotate the tRNA gene to determine its position, and the parameter settings were default. The start and stop codons of protein-coding genes were manually adjusted to fit open reading frames. The mt genome of G. sinensis was visualized using OGDRAW52. The mt genome of G. sinensis was deposited in NCBI GenBank under accession number MT921986.
Codon usage and substitution rate calculation
The relative synonymous codon usage (RSCU) was calculated by MEGA X53. The Ka/Ks ratios were calculated individually on each protein-coding gene of G. sinensis by DnaSP v654, and Acacia ligulata (NCBI accession number NC_040998) was used as an outgroup.
Repeat sequence
The position and type of SSR (Simple Repeated Sequence) were detected using the microsatellite identification tool MISA-web55 (https://webblast.ipk-gatersleben.de/misa/) with parameters set to 10, 5, 4, 3, 3 and 3 for mono-, di-, tri- tetra-, penta-, and hexanucleotides, respectively. The size and position of long repeat sequences, including forward, palindromic, reverse and complement repeats, were detected by REPuter56 (http://bibiserv.tech--fak.uni-bielefeld.de/reputer/), with a minimal repeat size of 30, and a hamming distance of 3.
Phylogenetic analyses
To better infer the phylogenetic relationship within the Fabaceae family, 24species (22 Fabaceae species and 2 outgroups) were selected to construct a phylogenetic tree. We extracted the nucleotide sequences of shared PCGs from these mt genomes. The 17 shared PCGs were aligned individually using PhyloSuite v1.2.157, and the alignment was manually adjusted. All aligned PCGs were then concatenated. Maximum likelihood (ML) analysis was performed using IQ-TREE58 under the model automatically selected. The Bayesian inference (BI) was implemented with MrBayes 3.2.659 under JC + I + G model determined from the ModelFinder60.
Supplementary Information
Acknowledgements
This work is supported by the grants from the State Key Laboratory of Component-Based Chinese Medicine, Tianjin University of Traditional Chinese Medicine, Tianjin, 300193, China.
Author contributions
X.T, X.Y. designed the study; H.Y. and X.Z. assembled, annotated and analyzed the mt genome; H.Y. drafted the manuscript and W.L. revised and polished; W.L. and Z.Z. prepared Figs. 1–3, Y.L., W.W. prepared Figs. 4–6. All authors reviewed the manuscript.
Data availability
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no.MT921986. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA726335, SRR14368777 and SAMN18927823 respectively.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Hongxia Yang, Wenhui Li and Xiaolei Yu.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-93480-6.
References
- 1.Gualberto JM, et al. The plant mitochondrial genome: dynamics and maintenance. Biochimie. 2014;100:107–120. doi: 10.1016/j.biochi.2013.09.016. [DOI] [PubMed] [Google Scholar]
- 2.Burger, G. & Lang, B. F. Parallels in Genome Evolution in Mitochondria and Bacterial Symbionts. IUBMB Life (International Union of Biochemistry and Molecular Biology: Life)55, 205–212 (2003). [DOI] [PubMed]
- 3.Nielsen BL. Plant mitochondrial DNA. Front Biosci. 2017;22:1023–1032. doi: 10.2741/4531. [DOI] [PubMed] [Google Scholar]
- 4.Lang BF, Gray MW, Burger G. Mitochondrial genome evolution and the origin of eukaryotes. Annu. Rev. Genet. 1999;33:351–397. doi: 10.1146/annurev.genet.33.1.351. [DOI] [PubMed] [Google Scholar]
- 5.Oda K, et al. Gene organization deduced from the complete sequence of liverwort Marchantia polymorpha mitochondrial DNA. J. Mol. Biol. 1992;223:1–7. doi: 10.1016/0022-2836(92)90708-R. [DOI] [PubMed] [Google Scholar]
- 6.Backert S, Lynn Nielsen B, Börner T. The mystery of the rings: structure and replication of mitochondrial genomes from higher plants. Trends Plant Sci. 1997;2:477–483. doi: 10.1016/S1360-1385(97)01148-5. [DOI] [Google Scholar]
- 7.Skippington E, Barkman TJ, Rice DW, Palmer JD. Miniaturized mitogenome of the parasitic plant Viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes. Proc. Natl. Acad. Sci. USA. 2015;112:E3515–E3524. doi: 10.1073/pnas.1504491112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sloan DB, et al. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 2012;10:e1001241. doi: 10.1371/journal.pbio.1001241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiesel R, von Haeseler A, Brennicke A. Plant mitochondrial nucleic acid sequences as a tool for phylogenetic analysis. Proc. Natl. Acad. Sci. 1994;91:634–638. doi: 10.1073/pnas.91.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Christensen AC. Plant mitochondrial genome evolution can be explained by DNA repair mechanisms. Genome Biol. Evol. 2013;5:1079–1086. doi: 10.1093/gbe/evt069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kubo T, Newton KJ. Angiosperm mitochondrial genomes and mutations. Mitochondrion. 2008;8:5–14. doi: 10.1016/j.mito.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 12.Alverson, A. J. et al. RESEARCH ARTICLE: Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). 46. [DOI] [PMC free article] [PubMed]
- 13.Kitazaki K, Kubo T. Cost of having the largest mitochondrial genome: evolutionary mechanism of plant mitochondrial genome. J. Bot. 2010;2010:1–12. doi: 10.1155/2010/620137. [DOI] [Google Scholar]
- 14.Notsu Y, et al. The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol. Gen. Genomics. 2002;268:434–445. doi: 10.1007/s00438-002-0767-1. [DOI] [PubMed] [Google Scholar]
- 15.Liao X, et al. Complete sequence of kenaf (Hibiscus cannabinus) mitochondrial genome and comparative analysis with the mitochondrial genomes of other plants. Sci. Rep. 2018;8:12714. doi: 10.1038/s41598-018-30297-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alverson AJ, Zhuo S, Rice DW, Sloan DB, Palmer JD. The mitochondrial genome of the legume Vigna radiata and the analysis of recombination across short mitochondrial repeats. PLoS ONE. 2011;6:e16404. doi: 10.1371/journal.pone.0016404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arrieta-Montiel, M. P. & Mackenzie, S. A. Plant Mitochondrial Genomes and Recombination. in Plant Mitochondria (ed. Kempken, F.) 65–82 (Springer, 2011). 10.1007/978-0-387-89781-3_3.
- 18.Wu K, et al. Genetic analysis and molecular characterization of Chinese sesame (Sesamum indicum L.) cultivars using Insertion-Deletion (InDel) and Simple Sequence Repeat (SSR) markers. BMC Genet. 2014;15:35. doi: 10.1186/1471-2156-15-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.The Legume Phylogeny Working Group Legume phylogeny and classification in the 21st century: progress, prospects and lessons for other species-rich clades. Taxon. 2013;62:217–248. doi: 10.12705/622.8. [DOI] [Google Scholar]
- 20.Azani N, et al. A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny—The Legume Phylogeny Working Group (LPWG) Taxon. 2017;66:44–77. doi: 10.12705/661.3. [DOI] [Google Scholar]
- 21.Li J, Ye C. Genome-wide analysis of microsatellite and sex-linked marker identification in Gleditsia sinensis. BMC Plant Biol. 2020;20:338. doi: 10.1186/s12870-020-02551-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu L, Zhang Y, Guo W, Wang Q. Gleditsia sinensis : Transcriptome Sequencing, Construction, and Application of Its Protein-Protein Interaction Network. Biomed. Res. Int. 2014;2014:1–9. doi: 10.1155/2014/404578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y-B, et al. Chemical constituents from the thorns of Gleditsia sinensis and their cytotoxic activities. J. Asian Nat. Prod. Res. 2020;22:1121–1129. doi: 10.1080/10286020.2020.1731799. [DOI] [PubMed] [Google Scholar]
- 24.Jin S-K, Yang H-S, Choi J-S. Effect of Gleditsia sinensis Lam. extract on physico-chemical properties of emulsion-type pork sausages. Korean J. Food Sci. Anim.Resour. 2017;37:274–287. doi: 10.5851/kosfa.2017.37.2.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Negruk, V. Mitochondrial genome sequence of the legume Vicia faba. Front. Plant Sci.4, (2013). [DOI] [PMC free article] [PubMed]
- 26.Choi I-S, et al. Fluctuations in Fabaceae mitochondrial genome size and content are both ancient and recent. BMC Plant Biol. 2019;19:448. doi: 10.1186/s12870-019-2064-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang L, et al. Genome-wide analysis of codon usage bias in four sequenced cotton species. PLoS ONE. 2018;13:e0194372. doi: 10.1371/journal.pone.0194372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sau K, Gupta SK, Sau S, Mandal SC, Ghosh TC. Factors influencing synonymous codon and amino acid usage biases in Mimivirus. Biosystems. 2006;85:107–113. doi: 10.1016/j.biosystems.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 29.Wu M, Li Q, Hu Z, Li X, Chen S. The complete Amomum kravanh chloroplast genome sequence and phylogenetic analysis of the commelinids. Molecules. 2017;22:1875. doi: 10.3390/molecules22111875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou M, Li X. Analysis of synonymous codon usage patterns in different plant mitochondrial genomes. Mol. Biol. Rep. 2009;36:2039–2046. doi: 10.1007/s11033-008-9414-1. [DOI] [PubMed] [Google Scholar]
- 31.Liu, Q., Feng, Y. & Xue, Q. Analysis of factors shaping codon usage in the mitochondrion genome of Oryza sativa. 8 (2004). [DOI] [PubMed]
- 32.Wynn, E. L. & Christensen, A. C. Repeats of unusual size in plant mitochondrial genomes: identification, incidence and evolution. G3 g3.200948.2018 (2018) 10.1534/g3.118.200948. [DOI] [PMC free article] [PubMed]
- 33.Tanaka Y, Tsuda M, Yasumoto K, Terachi T, Yamagishi H. The complete mitochondrial genome sequence of Brassica oleracea and analysis of coexisting mitotypes. Curr Genet. 2014;60:277–284. doi: 10.1007/s00294-014-0433-2. [DOI] [PubMed] [Google Scholar]
- 34.Qin Z, et al. Evolution analysis of simple sequence repeats in plant genome. PLoS ONE. 2015;10:e0144108. doi: 10.1371/journal.pone.0144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kumar, M., Choi, J.-Y., Kumari, N., Pareek, A. & Kim, S.-R. Molecular breeding in Brassica for salt tolerance: importance of microsatellite (SSR) markers for molecular breeding in Brassica. Front. Plant Sci.6, (2015). [DOI] [PMC free article] [PubMed]
- 36.Xu, C. et al. Comparative analysis of six lagerstroemia complete chloroplast genomes. Front. Plant Sci.8, (2017). [DOI] [PMC free article] [PubMed]
- 37.Bi C, Lu N, Xu Y, He C, Lu Z. Characterization and analysis of the mitochondrial genome of common bean (Phaseolus vulgaris) by comparative genomic approaches. IJMS. 2020;21:3778. doi: 10.3390/ijms21113778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Z, Nielsen R. Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol. Biol. Evol. 2000;17:32–43. doi: 10.1093/oxfordjournals.molbev.a026236. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Z, et al. KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genom. Proteom. Bioinform. 2006;4:259–263. doi: 10.1016/S1672-0229(07)60007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adams, K. L., Daley, D. O., Qiu, Y.-L., Whelan, J. & Palmer, J. D. Repeated, recent and diverse transfers of a mitochondrial gene to the nucleus in owering plants. 408, 4 (2000). [DOI] [PubMed]
- 41.Adams KL, Qiu Y-L, Stoutemyer M, Palmer JD. Punctuated evolution of mitochondrial gene content: high and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc. Natl. Acad. Sci. 2002;99:9905–9912. doi: 10.1073/pnas.042694899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams KL, Ong HC, Palmer JD. Mitochondrial gene transfer in pieces: fission of the ribosomal protein gene rpl2 and partial or complete gene transfer to the nucleus. Mol. Biol. Evol. 2001;18:2289–2297. doi: 10.1093/oxfordjournals.molbev.a003775. [DOI] [PubMed] [Google Scholar]
- 43.Chang S, et al. The mitochondrial genome of soybean reveals complex genome structures and gene evolution at intercellular and phylogenetic levels. PLoS ONE. 2013;8:e56502. doi: 10.1371/journal.pone.0056502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adams K. Evolution of mitochondrial gene content: gene loss and transfer to the nucleus. Mol. Phylogenet. Evol. 2003;29:380–395. doi: 10.1016/S1055-7903(03)00194-5. [DOI] [PubMed] [Google Scholar]
- 45.Liu G, et al. The Complete mitochondrial genome of Gossypium hirsutum and evolutionary analysis of higher plant mitochondrial genomes. PLoS ONE. 2013;8:e69476. doi: 10.1371/journal.pone.0069476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kearse M, et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dierckxsens, N., Mardulyn, P. & Smits, G. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res gkw955 (2016) 10.1093/nar/gkw955. [DOI] [PMC free article] [PubMed]
- 48.Wang X, et al. Organellar genome assembly methods and comparative analysis of horticultural plants. Hortic. Res. 2018;5:3. doi: 10.1038/s41438-017-0002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Y, et al. Characterization of the complete chloroplast genome of Camellia brevistyla, an oil-rich and evergreen shrub. Mitochond. DNA B. 2020;5:386–387. doi: 10.1080/23802359.2019.1703607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 [q-bio] (2013).
- 51.Tillich M, et al. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017;45:W6–W11. doi: 10.1093/nar/gkx391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Greiner, S., Lehwark, P. & Bock, R. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res.47, W59–W64 (2019). [DOI] [PMC free article] [PubMed]
- 53.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rozas J, et al. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017;34:3299–3302. doi: 10.1093/molbev/msx248. [DOI] [PubMed] [Google Scholar]
- 55.Beier S, Thiel T, Münch T, Scholz U, Mascher M. MISA-web: a web server for microsatellite prediction. Bioinformatics. 2017;33:2583–2585. doi: 10.1093/bioinformatics/btx198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kurtz S. REPuter: the manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001;29:4633–4642. doi: 10.1093/nar/29.22.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang D, et al. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020;20:348–355. doi: 10.1111/1755-0998.13096. [DOI] [PubMed] [Google Scholar]
- 58.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ronquist F, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. System. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no.MT921986. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA726335, SRR14368777 and SAMN18927823 respectively.