Abstract
The impact of repeated sample collection on COVID-19 test performance is unknown. The FDA and CDC currently recommend the primary collection of diagnostic samples to minimize the perceived risk of false-negative findings. We therefore evaluated the association between repeated sample collection and test performance among 325 symptomatic patients undergoing COVID-19 testing in Atlanta, GA. High concordance was found between consecutively collected mid-turbinate samples with both molecular (n = 74, 100% concordance) and antigen-based (n = 147, 97% concordance, kappa = 0.95, CI = 0.88–1.00) diagnostic assays. Repeated sample collection does not decrease COVID-19 test performance, demonstrating that multiple samples can be collected for assay validation and clinical diagnosis.
Subject terms: Viral infection, Laboratory techniques and procedures
Introduction
Diagnostic testing for SARS-CoV-2 is essential for guiding acute-phase clinical management, initiation or continuation of patient isolation, and the need for epidemiologic surveillance and community response. Ensuring accurate test results is of utmost importance to combat the COVID-19 pandemic. Standard of care (SOC) COVID-19 testing in the acute-phase relies on real-time reverse transcription polymerase chain reaction (rRT-PCR) or antigen detection. Per Centers for Disease Control and Prevention (CDC) guidance, such tests can be performed on a variety of samples, including nasopharyngeal (NP), nasal [mid-turbinate (MT) and anterior nares], and oropharyngeal swabs. SARS-CoV-2 detection is highly dependent on the type and quality of specimen obtained1. In all cases, specimen collection involves removing potentially limited diagnostic material from the patient2. If multiple samples are required for testing, there is concern that viral detection will be significantly reduced in the second sample, leading to false-negative results for certain methods or different pathogens.
The issue of repeat testing comes to the fore in two important scenarios: device evaluation and clinical testing. New SARS-CoV-2 diagnostics under evaluation are frequently designed for near-care, point-of-care, or in-home use. As a result, these come as test kits with engineered swabs and reaction vessels that may not be compatible with routine swabs collected for SOC methods. Because of the possibility of “taking away the virus/sample”, the Federal Drug Administration (FDA) and CDC recommended that SOC NP swabs be collected first during a clinical comparison study between a new device and the SOC. Clinically, the etiology of respiratory tract infections are often indistinguishable, and testing may require multiple tests from separate specimens of the same anatomical site. The FDA/CDC recommendation for the order of sample collection raises the dilemma for clinicians regarding which sample, for which pathogen, to collect first and if diagnostic yield in the second sample will be affected. This will be a consideration whenever multiple swabs are necessary for SOC testing.
The current study sought to address concerns regarding the reproducibility of both SARS-CoV-2 rRT-PCR and antigen testing on paired MT swabs obtained during a single encounter for both clinical care and as a center in the NIH Rapid Acceleration of Diagnostics program tasked with evaluating new rapid tests. We hypothesized that repeated collection of consecutive MT samples does not impact test sensitivity for either amplification or antigen-based assays.
Results
Patient population
There were 325 participants. Demographics including participant age, sex, race and number of days since symptom onset are in Table 1.
Table 1.
Overall, MT rRT-PCR group and MT antigen group participant demographics.
| Categories | Overall (N = 325) | MT rRT-PCR (N = 178) | MT antigen (N = 147) |
|---|---|---|---|
| Total, n (%) | |||
| Adult | 196 (60.3%) | 100 (56.2%) | 96 (65.3%) |
| Pediatric | 129 (39.7%) | 78 (43.8%) | 51 (34.7%) |
| Age, years | |||
| Mean (min–max) | 33.1 (0.1–82.6) | 32.1 (0.1–82.6) | 34.3 (0.4–78.3) |
| Median (25th–75th) | 33.6 (9.3–54.8) | 28.8 (9.5–55.2) | 36.8 (9.0–54.2) |
| Gender, n (%) | |||
| Male | 164 (50.5%) | 91 (51.1%) | 73 (49.7%) |
| Female | 161 (49.5%) | 87 (48.9%) | 74 (50.3%) |
| Race, n (%) | |||
| White | 110 (33.8%) | 69 (38.8%) | 41 (27.9%) |
| Black/African American | 164 (50.5%) | 81 (45.5%) | 83 (56.5%) |
| Asian | 17 (5.2%) | 12 (6.7%) | 5 (3.4%) |
| Other | 34 (10.5%) | 16 (9.0%) | 18 (12.2%) |
| Ethnicity, n (%) | |||
| Hispanic | 44 (13.5%) | 24 (13.5%) | 20 (13.6%) |
| Non-hispanic | 281 (86.5%) | 154 (86.5%) | 127 (86.4%) |
| Days post-symptom onset | |||
| Mean (min–max) | 3.5 (0.0–7.0) | 3.4 (0.0–7.0) | 3.6 (0.0–7.0) |
| Median (25th–75th) | 3.0 (2.0–5.0) | 3.0 (2.0–5.0) | 3.0 (2.0–5.0) |
| NP result, n (%) | |||
| Positive | 128 (39.4%) | 74 (41.6%) | 54 (36.7%) |
| Negative | 197 (60.6%) | 104 (58.4%) | 93 (63.3%) |
Missing: n = 1 for race/ethnicity (refused to answer).
Molecular
There was 100% concordance of rRT-PCR results among 74 NP positive patients between MT1 and MT2 tests (72 positive and 2 negative pairs). Additionally, the average Ct value did not differ significantly between MT1 and MT2 samples [median (IQR): MT1 27.09 (20.87–31.30), MT2 26.03 (20.89–31.57), p = 0.21; Fig. 1A]. Ct values among pediatric participants were, on average, lower than Cts for adults even after adjusting for days post-symptom onset [median (IQR): pediatric 22.96 (19.53–29.64), adult 28.69 (23.84–34.84), p < 0.01; Fig. 1B]. This did not impact concordance between MT1 and MT2 rRT-PCR results. 97.3% (72/74) concordance was demonstrated between the results of molecular tests at MT and NP sites.
Figure 1.
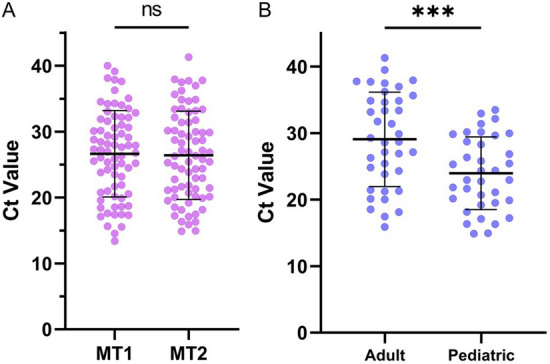
Repeated mid-turbinate sampling does not impact test performance. (A) MT1 vs. MT2 mean Ct values did not differ significantly (p = 0.21). (B) MT1 Ct values for adult and pediatric participants; pediatric Ct values were significantly lower than adult Ct values (p < 0.01, adjusted for days post-symptom onset).
Antigen
There was 97% concordance among 147 patients with MT1 and MT2 antigen tests, including 39 positive and 104 negative pairs (95% positive concordance, 98% negative concordance; Table 2). The sequence of results among the four discordant pairs were as follows: two MT1 positive/MT2 negative, one MT1 negative/MT2 positive and one MT1 indeterminate/MT2 positive. Among discordant MT antigen pairs, the mean associated RT-PCR Ct value was 25.61 (range 19.04–31.4). After excluding samples with discordant MT1 and MT2 antigen results (n = 4), 97.3% correlation was found between test results from from MT and NP sources.
Table 2.
Impact of repeated sample collection on test performance.
| MT2 positive | MT2 negative | MT2 indeterminate | Total | |
|---|---|---|---|---|
| a. Concordance between paired MT rRT-PCR samples (N = 74) | ||||
| MT1 positive | 72 | 0 | 0 | 72 |
| MT1 negative | 0 | 2 | 0 | 2 |
| MT1 indeterminate | 0 | 0 | 0 | 0 |
| Total | 72 | 2 | 0 | 74 |
| b. Concordance between paired MT antigen samples (N = 147) | ||||
| MT1 positive | 39 | 2 | 0 | 41 |
| MT1 negative | 1 | 104 | 0 | 105 |
| MT1 indeterminate | 1 | 0 | 0 | 1 |
| Total | 41 | 106 | 0 | 147 |
| NP rRT-PCR positive | NP rRT-PCR negative | Total | ||
|---|---|---|---|---|
| c. Concordance between paired MT antigen samples and NP rRT-PCR (N = 143a) | ||||
| MT1 + MT2 antigen positive | 37 | 2 | 39 | |
| MT1 + MT2 antigen negative | 14 | 90 | 104 | |
| Total | 51 | 92 | 143 | |
a: McNemar’s test for agreement of positive/negative results: N/A No discordant pairs.
a: Kappa coefficient (exact 95% CI): 1.00 (N/A).
a: Overall agreement (exact 95% CI): 100% (N/A).
b: McNemar’s test for agreement of positive/negative results: p = 0.56.
b: Kappa coefficient (exact 95% CI): 0.95 (0.88–1.00).
b: Overall agreement (exact 95% CI): 97% (0.93–0.99).
c: Sensitivity (exact 95% CI): 0.73 (0.58–0.84); Specificity (exact 95% CI): 0.98 (0.92–1.00).
c: McNemar’s test for agreement of positive/negative results: p = 0.003.
c: Kappa coefficient (exact 95% CI): 0.74 (0.63–0.86).
c: Overall agreement (exact 95% CI): 88.8% (0.82–0.93).
aDiscordant MT1 and MT2 antigen results excluded (n = 4).
Discussion
Both the FDA and CDC have expressed concern that the sensitivity of SARS-CoV-2 detection may decline in sequentially collected samples. Previous device evaluations employed a number of study designs to address this issue, including the use of different nares3, distinct anatomical sites4,5, or alternating the order of sample collection6. However, in clinical practice, such measures are not possible and may introduce different biases, such as diagnostic yield between different nares or specimen types7. The current study demonstrates high concordance between sequential MT samples collected during a single clinical encounter, including complete concordance of results among 74 paired samples tested by rRT-PCR and 97% concordance among paired samples tested for SARS-CoV-2 antigen. These data demonstrate there is ample SARS-CoV-2 virus across a range of viral loads (based on Ct values) to allow for repeated MT sampling if required for multiple tests. In addition, these data support the use of MT sampling as a less invasive collection method for point-of-care and potentially at home testing, with 88.8% concordance (exact CI = 0.82–0.93) between MT antigen test and NP rRT-PCR results.
The current study focused on MT samples, which are being increasingly used for SARS-CoV-2 diagnostic testing3. These results cannot necessarily be generalized to the collection of other nasal or oral specimens. Additionally, we did not vary the order of the nasal sampling with SOC NP sampling. In pediatric patients, NP samples were always last and in adults they were always first. Despite these limitations, this study provides proof that SARS-CoV-2 diagnostic test performance is maintained among multiple MT samples when taken at the same encounter. These data have implications for clinical testing algorithms and the design of future device evaluation studies.
Methods
Clinical samples
Paired, sequential MT swabs were obtained from 325 participants from COVID-19 testing centers utilized by the Atlanta Center for Microsystems Engineered Point-of-Care Technologies (ACME-POCT) network and affiliated with Emory University, Grady Memorial Hospital and Children’s Healthcare of Atlanta. Inclusion criteria were symptomatic respiratory illness for ≤ 7 days and a documented SARS-CoV-2 molecular test obtained within 24 h of study enrollment. SOC molecular tests differed by site and included the Roche Cobas 6800, Abbott Alinity, and Panther Fusion. NP swabs obtained for SOC testing were collected prior to MT swabs in adults and after in children. Exclusion criteria included inability to tolerate MT swabs or provide informed consent. Clinical and demographic variables were collected in web-based databases (REDCap, Nashville, TN). The study protocol was approved by the Emory University Institutional Review Board, Children’s Healthcare of Atlanta and the Grady Research Oversight Committee. All experiments were performed in accordance with relevant guidelines and regulations. Informed consent was obtained from all participating subjects, including from a parent and/or legal guardian if the patient was under the age of 18 years.
SARS-CoV-2 testing
Each participant gave two MT samples plus a NP. Of the first 178 participants, 74 tested positive by NP rRT-PCR and had MT pairs tested by rRT-PCR for the current study. MT pairs from the remaining 147 participants were tested for antigen (Supplementary Figure 1). MT swabs for rRT-PCR testing were collected using flocked tapered swabs (Copan FLOQSwab, Ref # 520CS01)8. Swabs were placed in 1 mL of saline in a sterile tube. Samples were stored at 4 °C for up to 72 h or − 80 °C if longer delays were expected. Samples were extracted from 500 µL of saline on an eMAG instrument (bioMeriéux) and eluted in 50 µL of buffer. Eluates were tested in an internally controlled, duplex rRT-PCR for the SARS-CoV-2 N2 target and RNase P. Oligonucleotide concentrations, master mix preparation, and cycling conditions were maintained from a previously published, triplex assay that included these two targets9. Paired MT swabs were tested side-by-side on a single run.
Sample collection was the same for antigen tests and were completed using the Abbott BinaxNOW™ COVID-19 Ag Card Home Test (Abbott Laboratories, Abbott Park, IL). Results were recorded for both completed tests.
Statistical analysis
Statistical analyses were performed using SAS 9.4 (Cary, NC). Agreement between the first (MT1) and second MT (MT2) antigen results was assessed using Cohen’s Kappa coefficient with exact 95% confidence intervals. McNemar’s test evaluated overall differences in MT1 vs. MT2 antigen results. Paired t-tests were used for MT1 vs. MT2 Ct comparisons and Wilcoxon signed-rank tests were used for overall adult vs. pediatric Ct comparisons.
Supplementary Information
Author contributions
J.M.L.—Trial oversight, coordination, analysis, drafting of manuscript. J.F.K.—Trial oversight, coordination, analysis, drafting of manuscript. E.A.T.—Coordination, drafting of manuscript. A.W.—Data analysis and interpretation, drafting of manuscript. J.F.—Data analysis and interpretation, drafting of manuscript. R.R.K.—Patient enrollment, oversight and manuscript review. P.A.R.—Patient enrollment, oversight and manuscript review. M.D.G.—Data analysis, interpretation and manuscript review. J.S.—Patient enrollment, oversight and manuscript review. M.B.V.—Data analysis, interpretation and manuscript review. J.O.—Patient enrollment, oversight and manuscript review. G.S.M.—Trial oversight, coordination, data analysis, manuscript review. W.A.L.—Trial oversight, coordination, data analysis, manuscript review. J.J.W.—Trial oversight, coordination, data analysis, manuscript review.
Funding
RADx, Grant/Award Number: U54 EB027690 02S1, UL1 TR002378.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Joshua M. Levy and Jennifer K. Frediani
Contributor Information
Wilbur A. Lam, Email: Wilbur.lam@emory.edu
Jesse J. Waggoner, Email: jjwaggo@emory.edu
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-94547-0.
References
- 1.Binnicker MJ. Challenges and controversies to testing for Covid-19. J. Clin. Microbiol. 2020 doi: 10.1128/Jcm.01695-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zou L, et al. Sars-Cov-2 viral load in upper respiratory specimens of infected patients. N. Engl. J. Med. 2020;382:1177–1179. doi: 10.1056/Nejmc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lindner AK, et al. Head-to-head comparison of Sars-Cov-2 antigen-detecting rapid test with self-collected anterior nasal swab versus professional-collected nasopharyngeal swab. Eur. Respir. J. 2020 doi: 10.1183/13993003.03961-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prince-Guerra JL, et al. Evaluation of Abbott Binaxnow rapid antigen test for Sars-Cov-2 infection at two community-based testing sites—Pima County, Arizona, November 3–17, 2020. MMWR Morb. Mortal. Wkly. Rep. 2021;70:100–105. doi: 10.15585/Mmwr.Mm7003e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callahan CJ, et al. Open development and clinical validation of multiple 3d-printed nasopharyngeal collection swabs: Rapid resolution of a critical Covid-19 testing bottleneck. J. Clin. Microbiol. 2020 doi: 10.1128/Jcm.00876-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.James AE, et al. Performance of the Binaxnow Coronavirus Disease 2019 (Covid-19) antigen card test relative to the severe acute respiratory coronavirus virus 2 (Sars-Cov-2) real-time reverse transcriptase polymerase chain reaction (Rrt-Pcr) assay among symptomatic and asymptomatic healthcare employees. Infect. Control Hosp. Epidemiol. 2021 doi: 10.1017/Ice.2021.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang W, et al. Detection of Sars-Cov-2 in different types of clinical specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/Jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention, Interim Guidelines for Collecting and Handling of Clinical Specimens for COVID-19 Testing. https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html (2021)
- 9.Waggoner JJ, et al. Triplex real-time Rt-Pcr for severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020;26:1633–1635. doi: 10.3201/Eid2607.201285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


