Abstract
Emerging evidence suggests that viral infection modifies host plant traits that in turn alter behaviour and performance of vectors colonizing the plants in a way conducive for transmission of both nonpersistent and persistent viruses. Similar evidence for semipersistent viruses like cauliflower mosaic virus (CaMV) is scarce. Here we compared the effects of Arabidopsis infection with mild (CM) and severe (JI) CaMV isolates on the feeding behaviour (recorded by the electrical penetration graph technique) and fecundity of the aphid vector Myzus persicae. Compared to mock‐inoculated plants, feeding behaviour was altered similarly on CM‐ and JI‐infected plants, but only aphids on JI‐infected plants had reduced fecundity. To evaluate the role of the multifunctional CaMV protein P6‐TAV, aphid feeding behaviour and fecundity were tested on transgenic Arabidopsis plants expressing wild‐type (wt) and mutant versions of P6‐TAV. In contrast to viral infection, aphid fecundity was unchanged on all transgenic lines, suggesting that other viral factors compromise fecundity. Aphid feeding behaviour was modified on wt P6‐CM‐, but not on wt P6‐JI‐expressing plants. Analysis of plants expressing P6 mutants identified N‐terminal P6 domains contributing to modification of feeding behaviour. Taken together, we show that CaMV infection can modify both aphid fecundity and feeding behaviour and that P6 is only involved in the latter.
Keywords: aphid vector, plant virus, vector behaviour, vector modification, vector transmission, viral factors
The viral protein P6, a major determinant of symptoms in infected plants, affects aphid behaviour in a way that could be favourable for transmission of cauliflower mosaic virus.

1. INTRODUCTION
Most plant viruses rely on arthropod vectors for their transmission to a new host. Among these, insects with a piercing‐sucking feeding behaviour such as aphids are the most efficient vectors, because they can, with great precision and without inflicting major damage to plant cells, acquire and inoculate viruses in distinct plant tissues. Evidence accumulates that viruses interact with both hosts and vectors and alter some of their characteristics to optimize transmission. The interactions may be direct, that is, changes in vector behaviour or fitness following virus acquisition by and retention in the vector, or indirect, that is, plant traits like odour, colour, and nutritive value modified by viral infection impact vector behaviour and fitness (for review see Dáder et al., 2017; Fereres & Moreno, 2009; Mauck et al., 2018). The ways viruses modify or even manipulate plant hosts and vectors depend on the transmission mode and tissue tropism of the virus (Mauck et al., 2012). Transmission modes are classified by two criteria: retention time in the vector (persistence) and vector interaction, with circulative viruses cycling through the vector body before being inoculated as a saliva component into a new host plant and noncirculative viruses interacting only with the vector mouthparts. Circulative and persistent viruses are most often phloem‐restricted and characterized by long acquisition and retention times in the vector, resulting in prolonged, often lifelong transmissibility. General predictions assume that these viruses would benefit from fast and prolonged access of vectors to the phloem, which would facilitate virus acquisition. The circulative and persistent viruses also tend to increase food quality of the host, resulting in an improvement in vector fitness and an increase in vector population (Dáder et al., 2017; Fereres & Moreno, 2009; Mauck et al., 2018). In contrast, noncirculative and nonpersistent viruses are often tissue generalists, with fast acquisition and short retention times. Plants infected with noncirculative and nonpersistent viruses may attract vectors for virus acquisition (Fereres & Moreno, 2009) and subsequently encourage them to leave the plants rapidly for fast dispersal of the virus, as shown for cucumber mosaic virus (Mauck et al., 2010). Vector departure is often related to the poor taste and low nutritive value of infected plants (Mauck et al., 2014). While many reports support these models, there are also a number of examples that do not follow the expectations. Outcome of virus–host–vector interactions may vary depending on the specific virus–host–vector combination and probably other factors (Mauck & Chesnais, 2020; Mauck et al., 2018).
Cauliflower mosaic virus (CaMV) (genus Caulimovirus, family Caulimoviridae) has features of nonpersistent and persistent viruses and is often classified as a semipersistent virus. Being a tissue generalist, CaMV can infect all cell types and is acquired from epidermis and mesophyll cells like nonpersistent viruses, but also from the phloem sap like persistent viruses (Palacios et al., 2002). With regard to vector interaction, CaMV is a noncirculative virus that binds to stylin receptors in the stylet tips of its aphid vectors (Uzest et al., 2010; Webster et al., 2018). Virus particles require the virus‐encoded transmission helper protein P2 for vector interaction that forms a complex with the virions (Leh et al., 1999; Moreno et al., 2005). Formation of this P2–virus complex, mandatory for transmission, can occur in infected plant cells or in the stylets, allowing simultaneous or sequential binding of P2 and virus to the vector (Drucker et al., 2002). Simultaneous binding is associated with fast CaMV acquisition from epidermis and mesophyll, whereas sequential binding favours uptake of CaMV from the phloem sieve tubes that contain virions but are devoid of P2 (Palacios et al., 2002).
Whereas the molecular and cellular mechanisms of CaMV acquisition are well studied, information on the effect of CaMV infection on transmission‐relevant changes in aphid behaviour and performance is scarce. Chesnais et al. (2019) reported that the green peach aphid Myzus persicae and the cabbage aphid Brevicoryne brassicae did not show any preference for CaMV‐infected Camelina sativa plants. Both aphid species created more intracellular punctures, ingested less phloem sap, and displayed reduced fecundity on infected plants compared to healthy plants, as expected for a nonpersistent virus.
In this study, we tested how different isolates of CaMV affect M. persicae feeding behaviour and fitness on the model plant Arabidopsis thaliana. We then addressed the specific role of the multifunctional viral protein P6‐TAV in modification of aphid traits. P6 is a key player in CaMV infection (for review see Pooggin & Ryabova, 2018; Schoelz et al., 2016). It is required for translation of the polycistronic CaMV 35S RNA and interacts, among others, with TOR kinase. TOR is involved in translation initiation, but plays also a pivotal role in controlling cell homeostasis of catabolic and anabolic processes. P6 binding triggers TOR phosphorylation, which then promotes protein translation and simultaneously suppresses potential antiviral autophagy and innate immunity (Hafrén et al., 2017; Zvereva et al., 2016). P6 is also a suppressor of RNA silencing and sufficient to induce typical symptoms of infection by itself (Yu et al., 2003). Finally, P6 is the matrix protein of the viral factories, where replication occurs and virus particles are stored (Schoelz & Leisner, 2017). The P6 functions in dampening plant defences and in inducing symptom expression could impact aphid–plant interactions. P6 is, therefore, a perfect candidate to modify aphid behaviour and performance.
2. RESULTS
2.1. Effect of viral infection on M . persicae
We first compared the impact of CaMV infection on M. persicae feeding behaviour and performance. For this, we chose the well‐characterized CaMV isolates Cabb B‐JI (referred to as JI) and Cm1841 that accumulate in infected Brassicaceae at high and low levels (Lung & Pirone, 1973) and cause severe and mild symptoms in Arabidopsis, respectively (Cecchini et al., 1998). Unlike JI, Cm1841 is a nontransmissible CaMV isolate, due to a mutation of amino acid 94 in the transmission helper protein P2 (Woolston et al., 1987). For better comparison, we reverted this mutation to wild type (wt) and carried out all experiments with the revertant virus, Cm1841‐Rev, hereafter referred to as CM. Figure 1a–c shows that, at 21 days postinoculation, JI‐infected Arabidopsis Col‐0 plants displayed stronger symptoms compared to CM‐infected plants with a reduced growth, intense yellowing, and leaf curling. In addition, we observed lower accumulation of P2, P4 capsid protein, and P6‐TAV protein in CM‐infected plants (Figure 1d).
FIGURE 1.
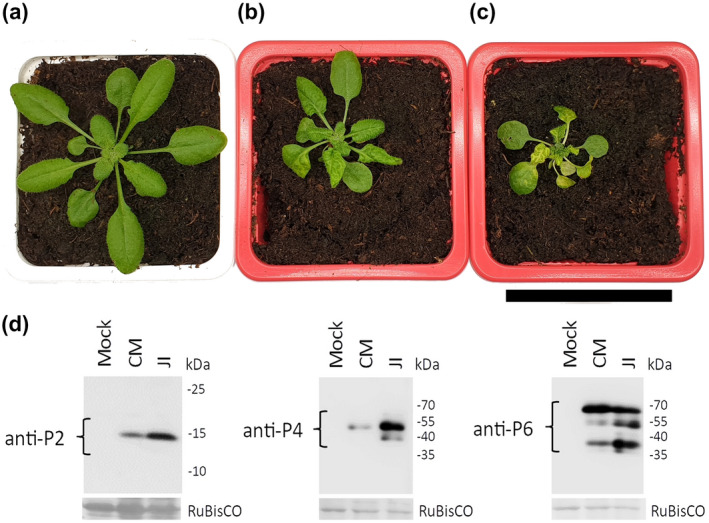
Symptoms of CaMV infection and viral protein accumulation in Arabidopsis Col‐0 plants. (a) Mock‐inoculated, (b) CM‐infected, and (c) JI‐infected plant 21 days postinoculation with virus‐free or viruliferous aphids. Scale bar = 5 cm. (d) Western blot analysis of accumulation of CaMV proteins in CM‐ and JI‐infected Arabidopsis 21 days postinoculation. The membranes were stained for P2 (left panel), P4 (middle panel), and P6 (right panel). Ponceau red staining of the large RuBisCO subunit is shown as a loading control. Molecular weights are indicated on the right of the blots
We next evaluated aphid fecundity on infected and mock‐inoculated plants by counting the number of offspring of synchronized adult wingless aphids after 5 days of infestation. Fecundity was significantly lower on JI‐infected Arabidopsis compared to mock‐inoculated control plants and CM‐infected plants (generalized linear model [GLM], df = 2, χ2 = 15.542, p < .001; Figure 2a).
FIGURE 2.
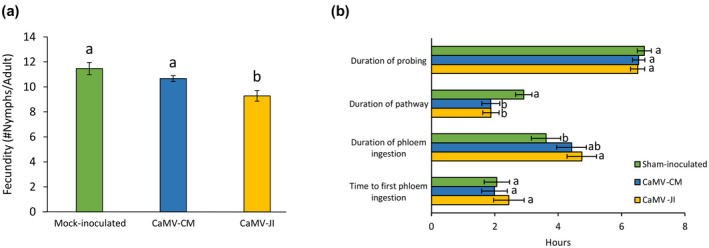
Fecundity and feeding behaviour of Myzus persicae on mock‐inoculated or CaMV‐infected Arabidopsis. Two‐week‐old plants were inoculated with the indicated isolate and used 3 weeks later for the experiments. (a) Aphid fecundity after 5 days of infestation (n = 28–33). (b) Aphid feeding behaviour parameters recorded with the electrical penetration graph (EPG) technique (n = 22–27). Letters show significant differences between plant infection status as tested by GLM followed by pairwise comparisons using “emmeans” (p < .05; method: Tukey)
Then we assayed aphid feeding behaviour by the electrical penetration graph (EPG) technique. Behaviour of individual aphids was recorded for 8 hr on mock‐inoculated or infected Arabidopsis. The total duration of probing in plant tissue and the time to the first phloem phase were not affected by either CaMV isolate (GLM, df = 2, χ2 = 0.952, p = .621; Cox model, χ2 = 0.07, p = .96; Figure 2b). The duration of the pathway phase was significantly reduced on infected Arabidopsis for both isolates (GLM, df = 2, χ2 = 11.14, p = .004). The duration of phloem ingestion was significantly longer on JI‐infected Arabidopsis compared to CM‐infected and mock‐inoculated plants (GLM, df = 2, χ2 = 6.27, p = .04; Figure 2b).
2.2. Effect of P6 on aphids
We used transgenic Arabidopsis expressing wt and mutant P6 proteins from the constitutive 35S promoter to determine whether P6 is involved in plant–aphid interactions. Figure 3a–h shows the phenotype of Col‐0 plants expressing the P6 proteins 5 weeks after germination: Plants expressing wt P6 from JI or CM or P6 from CM with an N‐terminal HA‐tag (P6‐CM‐HA) were smaller and leaves showed yellowing but no leaf curling as CaMV‐infected plants (Figure 3b,d,g). Plants expressing P6 from CaMV isolate D4 resembled untransformed plants (Figure 3c). Arabidopsis plants expressing mutated versions of P6 from isolate JI, JI‐Eki (bearing three amino acid substitutions at P6’s N‐terminus) and JI‐ΔdsR (containing a deletion of the TOR‐ and RNA‐binding region), displayed no or only a weak phenotype compared to untransformed plants (Figure 3e,f). Also, the P6 CM‐Δd23‐HA mutant (containing an N‐terminal HA‐tag and a deletion of the virulence/avirulence region Vir/Avr) induced no visible phenotype (Figure 3h). Western blotting indicated strong accumulation of P6‐D4 and P6‐JI‐ΔdsR and low accumulation of P6‐CM and P6‐JI in transgenic plants (Figure 3i). P6‐CM‐HA was visible after prolonged exposure of the blot, and P6‐JI‐Eki and P6‐CM‐Δd23‐HA only after redoing the blots (Figure 3j).
FIGURE 3.
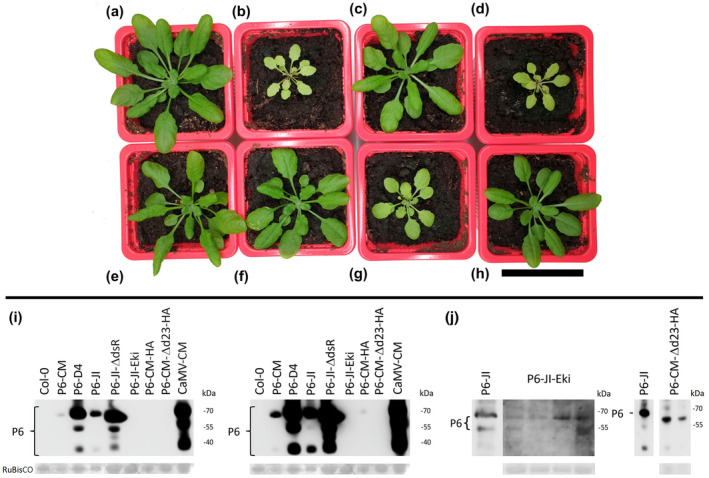
Phenotype of 5‐week‐old transgenic Arabidopsis plants expressing various P6 proteins. The images show (a) an untransformed Col‐0 plant and (b–h) transgenic Col‐0 plants expressing (b) P6 from the CaMV isolate CM; (c) P6 from isolate D4; (d) P6 from isolate JI; (e) the P6 dsR domain deletion mutant from isolate JI, referred to as JI‐ΔdsR; (f) the P6 Eki mutant from isolate JI, referred to as JI‐Eki; (g) HA‐tagged wild‐type P6 from CM (CM‐HA), and (h) the HA‐tagged P6 Vir/Avr domain deletion mutant from isolate CM, referred to as CM‐Δd23‐HA. Scale bar = 5 cm. (i) Western blot analysis of P6 protein accumulation in 5‐week‐old transgenic plants expressing the indicated P6 mutants. The figure shows the same blot revealed with a short (left) or a longer (right) exposure time to visualize weak bands. Extracts prepared from untransformed Col‐0 and from CM‐infected leaves were loaded as negative and positive controls, respectively. (j) Detection of P6‐JI‐Eki and P6‐CM‐Δd23‐HA by western blot. The two proteins that were not detected in the blot shown in (i) could be revealed in a different experiment using more concentrated extracts. Each lane presents extract from a different transgenic plant. Signals from wild‐type P6 loaded on the same blots are shown to the left of the panels and either the blots were exposed much shorter (P6‐JI‐Eki) or extracts were diluted (P6‐CM‐Δd23‐HA). Note that the mutant P6 concentration varied considerably. Ponceau red staining of the large RuBisCO subunit is shown as a loading control
M. persicae fecundity was assayed on the P6‐expressing transgenic plants. In contrast to CaMV‐infected plants, aphid fecundity was not affected on any transgenic plant when compared to untransformed plants (GLM, df = 7, χ2 = 6.61, p = .478; Figure 4).
FIGURE 4.
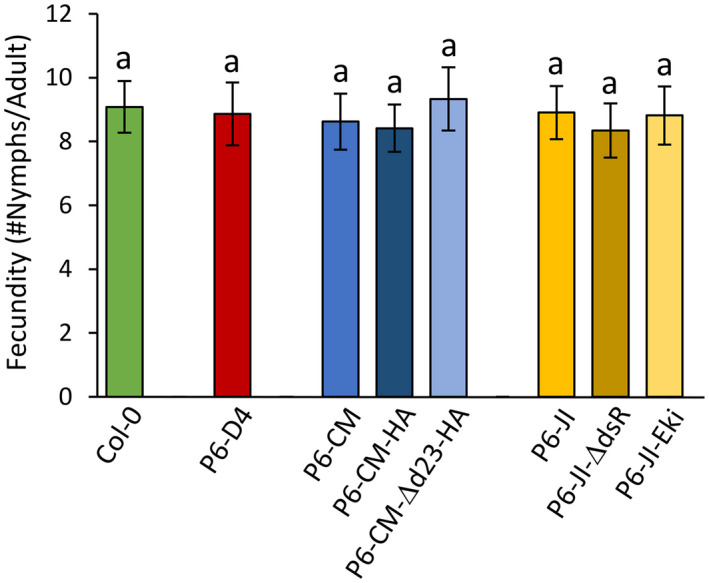
Myzus persicae fecundity 5 days after deposit on untransformed or transgenic 5‐week‐old Arabidopsis plants expressing P6 proteins. Different letters show significant differences between plants as tested by GLM followed by pairwise comparisons using “emmeans” (p < .05; method: Tukey, n = 21–24). No statistically significant differences were recorded
Then, the feeding behaviour of M. persicae on the P6‐transgenic plants was analysed by EPG (Figure 5). All aphid behaviour parameters on P6‐D4 plants were similar to those on untransformed Col‐0 plants. We detected significant differences in aphid feeding behaviour on the other transgenic plants. Compared to plants expressing P6‐CM and P6‐CM‐HA, aphids probed for significantly shorter times on P6‐CM‐Δd23‐HA and P6‐JI plants (GLM, df = 7, χ2 = 23.07, p = .002; Figure 5). The pathway phase was significantly reduced on P6‐CM‐HA plants, compared to P6‐CM‐Δd23‐HA, JI, and untransformed Col‐0 plants (GLM, df = 7, χ2 = 24.33, p < .001). The duration of phloem ingestion was significantly longer on P6‐CM‐HA plants compared to P6‐JI and untransformed plants (GLM, df = 7, χ2 = 25.12, p < .001). Aphids reached the first phloem phase significantly faster on plants expressing P6‐CM compared to untransformed Col‐0 or plants expressing P6‐JI and P6‐CM‐Δd23‐HA (GLM, df = 7, χ2 = 20.76, p = .04).
FIGURE 5.
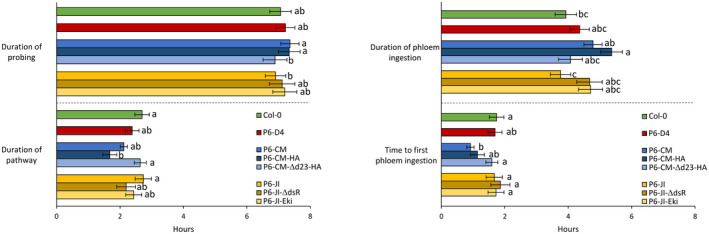
Aphid feeding behaviour parameters recorded by EPG on untransformed Col‐0 and transgenic P6‐expressing 5‐week‐old Arabidopsis. Different letters indicate significant differences between plants as tested by GLM followed by pairwise comparisons using “emmeans” (p < .05; method: Tukey, n = 25–31)
3. DISCUSSION
Many studies report changes in vector behaviour and performance after infection of plants with viruses, but only a few identify viral determinants involved in vector manipulation. Also, most work centres on persistent and nonpersistent viruses; studies on semipersistent viruses are scarce. To fill this gap, we studied the feeding behaviour and fecundity of aphids on Arabidopsis infected with a severe and a mild isolate of CaMV, a virus with transmission properties of both nonpersistent and persistent viruses. Then, we studied the potential role of the multifunctional viral protein P6 in virus‐induced alterations of host‐plant traits and vector responses.
3.1. Effect of CaMV infection on aphid fecundity
The severe and high‐accumulating CaMV isolate JI reduced aphid fecundity on infected Arabidopsis significantly (c.2.5 fewer nymphs produced per adult after 5 days compared to mock‐inoculated plants), while infection with the mild and low‐accumulating isolate CM had only a moderate impact on aphid fecundity, below the significance threshold (c.1 nymph less produced per adult after 5 days compared to mock‐inoculated plants; Figure 2a). Thus, both increased symptom severity and higher viral accumulation correlated with reduced aphid fecundity. Whether symptom severity is related to CaMV accumulation is still unclear and seems to depend on the host–virus association (Doumayrou et al., 2013). Lower fecundity and other negative effects of plant infection on vector performance such as reduced vector survival or delayed development are predicted for nonpersistent viruses. These modifications may promote rapid dispersion of vectors and subsequently virus transmission (Mauck et al., 2018). Indeed, mathematical models show that increasing departure rates from hosts lead to increased vector movements between host plants, which is beneficial for virus dissemination (Carr et al., 2020; Shaw et al., 2017). Whether this applies for CaMV on Arabidopsis remains to be determined because although reduced fecundity is a proxy for aphid fitness, it does not indicate that aphids will really leave CaMV‐infected plants faster than healthy plants, as for example documented for M. persicae on cucumber mosaic virus‐infected squash (Mauck et al., 2010). A negative effect of CaMV JI on M. persicae fecundity was already observed by Chesnais et al. (2019), albeit on another host plant, the Brassicaceae member Camelina sativa. This suggests that the effect on fecundity is not host plant‐specific, but rather virus isolate‐specific.
3.2. Effect of CaMV infection on aphid feeding behaviour
M. persicae on CaMV‐JI‐infected plants spent significantly less time in the pathway phase in the leaf tissue and more time ingesting phloem sap than aphids on healthy plants (Figure 2b). This is expected to be counterproductive for transmission of nonpersistent viruses (Carr et al., 2020; Eigenbrode et al., 2018; Mauck et al., 2012). In fact, typical nonpersistent viruses like potyviruses or cucumber mosaic virus (single‐stranded RNA viruses of families Potyviridae and Bromoviridae, respectively) are acquired during short intracellular stylet punctures in plant epidermis and mesophyll (Martin et al., 1997). They are lost from the stylets when aphids stay on plants for longer times and when the stylets reach the sieve tubes that do not contain transmission‐competent virus forms (Kloth & Kormelink, 2020; Wang & Ghabrial, 2002). The case is different for CaMV that is acquired efficiently from both cells and phloem sap and after both short and long acquisition periods (Bouchery et al., 1990; Markham et al., 1987; Palacios et al., 2002). Therefore, both improved palatability (characterized by few probes, rapid access to the phloem, and long phloem ingestion) and reduced palatability (characterized by many probes, impairment in phloem access, and reduced phloem ingestion) of infected plants can be conducive for CaMV transmission. In fact, Chesnais et al. (2019) observed, in contrast to our results here, a significantly lower number of probes and an increased sap ingestion of M. persicae feeding on CaMV‐JI‐infected Camelina plants. Yet transmission efficiency is high using either plant species as a virus source (Dáder et al., 2019; Verdier, 2020).
In summary, CaMV is, with regard to transmission, a special case combining characteristics of nonpersistent and persistent transmission modes. We propose that the particular acquisition mode of CaMV optimizes transmission by lowering its dependency on specific aphid feeding behaviours. Most noncirculative viruses (i.e., nonpersistent and semipersistent viruses) are transmitted by a large number of vectors (e.g., 27 for CaMV, 89 for TuMV; Edwardson & Christie, 2018; Kennedy et al., 1962) while circulative viruses are generally transmitted by less than a dozen. This means that these noncirculative viruses need to interact with multiple vectors, and it is unlikely that they can engage in manipulations that are specific for each vector species. Rather, they must target a common and accessible vector feature, for example a conserved receptor in the aphid stylets (Webster et al., 2018), to which they can bind easily and already after short vector–plant contact. This enables most aphids, even noncolonizers, to engage in behaviours conducive with nonpersistent virus acquisition and inoculation (e.g., epidermis and mesophyll probing). As a consequence, there may be little (or no) advantage and no strong selection pressure for CaMV and other noncirculative viruses to manipulate vector feeding behaviours (Mauck & Chesnais, 2020).
3.3. A role for P6 in aphid manipulation
Aphid fecundity was unchanged on Arabidopsis plants expressing wt P6 from JI or CM (Figure 4), although these plants displayed dwarfing and bleaching, indicative of physiological modifications (Figure 3b,d). Three nonexclusive explanations are possible. The first one is that P6 alone is not involved in modifying aphid fecundity. This would mean that other viral determinants (proteins or RNAs), either independently or in concert with P6, induce changes in plants that reduce aphid fecundity. Indeed, it has been suggested that the CaMV proteins P1–5 are mainly responsible for leaf malformation, while P6 causes chlorosis and dwarfism (Yu et al., 2003). The second explanation is that P6 levels in the transgenic plants were too low to reduce aphid fecundity. Indeed, we observed that P6 levels were considerably lower and variable in transgenic plants compared to infected plants (Figure 3i,j). However, in CaMV‐infected plants a major fraction of P6 protein is located in inclusion bodies/replication factories (Schoelz & Leisner, 2017) and only a small fraction of P6 may be available in a soluble form to fulfil its other functions (reviewed by Pooggin & Ryabova, 2018). Finally, it is also possible that phloem sap composition (e.g., amino acid and/or sugar concentration and composition) was changed in infected but not in transgenic plants. Further experiments are required to solve this issue.
Some aphid feeding behaviour parameters were significantly altered on transgenic plants expressing P6 (Figure 5). Compared to control plants, the time to first phloem ingestion was reduced on transgenic P6‐CM plants. Interestingly, no such effect was observed on plants infected with CaMV‐CM (Figure 2b). This might indicate that other viral determinants counteracted the effect of P6. A second significantly changed parameter was the duration of phloem ingestion, which was longer on P6‐CM plants than on P6‐JI plants. Opposite results were obtained with infected plants, where the duration of phloem ingestion was longer on JI‐infected than on mock‐inoculated plants, with the duration of CM‐infected Arabidopsis being intermediate. Again, this observation indicates that P6 contributes to the modifications in phloem sap ingestion, but that other viral determinants are involved as well.
3.4. P6 domains involved in aphid manipulation
The comparison of the aphid feeding behaviour on transgenic Arabidopsis expressing various P6 versions yielded evidence for the involvement of its N‐terminus in modification of plant–aphid interactions (Figure 5). P6 from the D4 isolate, although accumulating to much higher levels in transgenic plants than P6‐JI or P6‐CM (Figure 3i), had no effect on aphid behaviour, and plants displayed a wt phenotype (Figure 3c), as previously reported by Yu et al. (2003). Because P6‐D4’s RNA silencing suppression domains are functional in Arabidopsis (Shivaprasad et al., 2008; Zvereva et al., 2016), we conclude that the silencing suppression domain (probably located in the C‐terminal portion of P6; see Figure 6 for an overview of P6’s functional domains) has no impact on aphid behaviour or on symptom expression. P6‐D4’s Vir/Avr domain, responsible for symptom expression, and the mini‐TAV domain, required for translation transactivation and TOR‐mediated immunity, are nonfunctional in Arabidopsis (Yu et al., 2003; Zvereva et al., 2016), precluding any definitive conclusions but leaving a possibility that corresponding domains of P6‐CM and P6‐JI can contribute to modification of aphid feeding behaviour. Indeed, aphids spent more time in the pathway phase on P6‐CM‐Δd23‐HA plants (containing an N‐terminal HA‐tag and a deletion of the Vir/Avr domain), compared to P6‐CM‐HA plants, despite similar low accumulation in transgenic plants. Compared to untransformed plants, aphids had a shorter pathway phase and longer phloem sap ingestion on P6‐CM‐HA but not on P6‐CM‐Δd23‐HA plants. This is in favour of a role of the Vir/Avr region in altering aphid feeding behaviour. Neither P6‐JI nor P6‐JI‐ΔdsR plants (functional Vir/Avr domain, no TOR interaction) had a significant effect on aphid behaviour, despite higher protein accumulation in transgenic plants compared to P6‐CM‐Δd23‐HA. This might indicate that the effect of the Vir/Avr domain on aphid behaviour is CaMV isolate‐specific. In fact, the Vir/Avr domain is one of the most variable domains of P6 among CaMV isolates. Notably, despite previous controversial evidence (Laird et al., 2013), the P6‐CM Vir/Avr domain was found to be crucial for virus infectivity and virulence but not suppression of RNA silencing in Arabidopsis (Zvereva et al., 2016). Moreover, this domain contributes to suppression of salicylic acid‐dependent autophagy and effector‐triggered innate immunity in Arabidopsis, although it was not absolutely essential for this TOR‐dependent function of P6‐CM (Zvereva et al., 2016). We speculate P6‐mediated dampening of pattern‐ and/or effector‐triggered innate immunity may explain the observed changes in aphid feeding behaviour. Indeed, aphids are known to induce innate immunity responses in Arabidopsis and deliver effector proteins to suppress these responses (Mugford et al., 2016; Prince et al., 2014). A role for the P6 mini‐TAV region with TOR‐binding domain in modification of aphid behaviour remains disputed. Despite its high accumulation levels in transgenic plants, comparable to those in virus‐infected plants, P6‐JI‐ΔdsR (no TOR interaction) only slightly affected aphid behaviour. This indicates that the TOR‐binding domain is not required but other elements of the mini‐TAV region preserved in the mutant might still be important for modification of aphid behaviour. Finally, P6‐JI‐Eki did not change aphid behaviour significantly, suggesting that the extreme N‐terminus of P6 may not be involved in aphid interactions. However, in this case the mutant protein accumulated in transgenic plants at extremely low levels, precluding any definitive conclusion.
FIGURE 6.
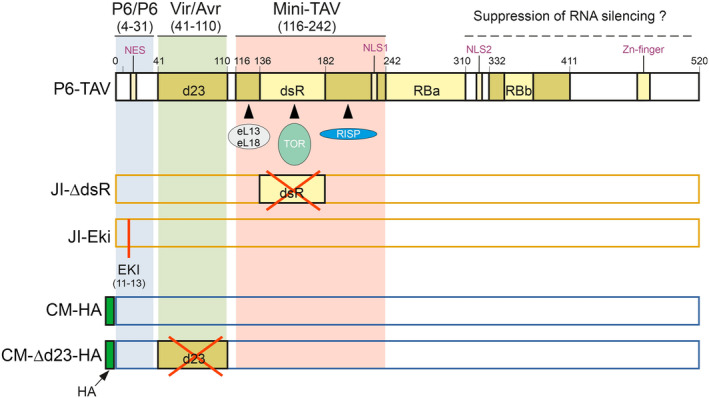
Functional domains of P6 and P6 mutants used in this study. Only relevant domains and interacting proteins are shown. The N‐terminal half of P6 contains three major regions: The N‐terminus (amino acids 4–31) contains one of several P6/P6 self‐interaction domains required for P6’s function as the matrix protein of virus inclusions. This region is followed by the Vir/Avr domain delimited by the d23 region, which is involved in chlorosis induction and dwarfism. The mini‐TAV domain comprises amongst others an N‐terminal region binding to ribosomal proteins eL13 and eL18, the central double‐stranded RNA‐binding (dsR) region that interacts with TOR kinase, and a C‐terminal region that interacts with reinitiation supporting protein (RISP) and contains also a nuclear localization signal (NLS1). The mini‐TAV region is important for antiviral autophagy, innate immunity, and polycistronic translation. One of the functions of the C‐terminal half of P6 is probably suppression of RNA silencing, in which also the N‐terminal nuclear export signal (NES) and the two nuclear localization signals (NLS1 in the mini‐TAV domain and NLS2 in the C‐terminal region) play a role. The dsR region is deleted in P6‐JI‐ΔdsR. In P6‐JI‐Eki, the conserved Eki motif just preceding the NES is substituted by three alanines. CM‐HA contains an N‐terminal HA‐tag as modification and CM‐Δd23‐HA in addition a deletion of the d23 region. RBa, RBb, RNA‐binding regions. Figure adapted from Pooggin and Ryabova (2018)
Taken together, this report shows that aphid responses to CaMV infection depend on the virus isolate, and that part of the effects, but not the entire response, relies on P6.
4. EXPERIMENTAL PROCEDURES
4.1. Cloning
Cm1841‐Rev, where the mutation of amino acid 94 in P2 in the Cm1841 genome is reverted to wt, was obtained by site‐directed mutagenesis using the QuikChange Lightning Kit (Agilent) following the manufacturer’s instructions and using the oligonucleotides QuikReve‐F (5′‐GTCAGTTTTTAATACTGCAAAAAACATTTTTAAAAGTGGGGGGGTTGATTACTCG‐′) and QuikReve‐R (5′‐CGAGTAATCAACCCCCCCACTTTTAAAAATGTTTTTTGCAGTATTAAAAACTGAC‐3′). The two oligonucleotides contain a silent G → A mutation at nucleotide 277 of the P2 sequence to create a DraI restriction site for easy identification of recombinants and an A → G mutation at nucleotide 280 of P2 to revert amino acid R94 to wt G94. The pCa122 plasmid containing 1.2 copies of the Cm1841 genome (Tsuge et al., 1994) was used as a matrix. The cycling programme was 2 min at 95 °C; followed by 18 cycles of 20 s at 95 °C, 10 s at 60 °C, and 7 min at 69 °C; and a final extension step of 5 min at 68 °C. The plasmid pCa122rev was verified by sequencing the mutated region.
4.2. Virus inoculation
Initial inoculation was performed with the pCa122rev plasmid or the pGreen‐BJI plasmid (Khelifa et al., 2010) coding for the Cabb B‐JI genome. The plasmids were rub‐inoculated with carborundum into the first true leaf of turnip (Brassica napus ‘Just Right’) seedlings (20 μg plasmid per seedling at the 2‐leaf stage). When disease symptoms were well developed, leaves were harvested and stored at −20 °C. The virus was passaged once by mechanical inoculation of Arabidopsis ecotype Col‐0 plants at the 4‐ to 8‐leaf stage. For this, frozen turnip leaves were thawed and ground in a mortar with carborundum in 10 mM HEPES buffer pH 8.0 at a ratio of 1 g leaf per 0.5 ml buffer, and used for rub inoculation. All further inoculations were by aphid transmission using infected Arabidopsis plants as virus source. Aphids were deposited on CaMV‐infected plants overnight, and then five or six viruliferous aphids were placed on 2‐week‐old Arabidopsis for a few hours before being removed manually.
Plants infected with Cm1841‐Rev (CM) or with Cabb B‐JI (JI) were used 3 weeks after inoculation for experiments. Growth conditions were 8 hr light/16 hr dark at 22/20 °C.
4.3. Arabidopsis mutant lines
The transgenic lines P6‐CM‐HA and P6‐CM‐Δd23‐HA are described by Zvereva et al. (2016), P6‐CM1841 (as lines CM‐6 and CM‐8) and P6‐D4 (as D4‐2) by Yu et al. (2003), and P6‐JI (as AT7) and P6‐JI‐ΔdsR (as AT7ΔdsR) by Schepetilnikov et al. (2011). The P6‐JI‐Eki line is described as TAVm3 by Haas et al. (2008). Wt and mutant lines were all in the Col‐0 background.
4.4. Aphid rearing
The M. persicae green peach aphid clone was originally isolated in the Netherlands. Aphids were reared on Chinese cabbage (Brassica rapa pekinensis) in a growth chamber at 20 ± 1 °C, under a 16 hr photoperiod. Only wingless forms were used in assays. For synchronization, we placed adults on detached Chinese cabbage leaves that were spread on 1% agarose in a Petri dish. The adults were removed 24 hr later and the newborn larvae used in experiments after another 7 days when they had reached the adult stage.
4.5. Aphid feeding behaviour
We used the electrical penetration graph DC‐system as described by Tjallingii (1988) to investigate the effects of plant CaMV infection on the feeding behaviour of M. persicae. Eight aphids were connected to the Giga‐8 DC‐EPG amplifier and each one was placed on the leaf of an individual A. thaliana plant. The recordings were performed continuously for 8 hr during the photophase. Each aphid–plant system was placed inside a Faraday cage at 21 ± 1 °C. Acquisition and analysis of the EPG waveforms were carried out with PROBE 3.5 software (EPG Systems). Relevant aphid behaviour EPG parameters were calculated with EPG‐Calc 6.1 software (Giordanengo, 2014) and were based on different EPG waveforms described by Tjallingii and Hogen Esch (1993). The following parameters were selected because they are relevant and important for the acquisition of CaMV by aphids: the total duration of “probing time”, “pathway phase”, and “phloem sap ingestion phase” and the time needed by the aphid to reach the phloem. For each condition (healthy or infected and wt or mutant, respectively), EPGs of 25–30 individual aphids were analysed. Aphids that produced signals (i.e., total duration of probing time) for fewer than 5 hr were excluded from the analysis.
4.6. Aphid fecundity
Synchronized wingless adults (8 ± 1 days old) were randomly selected from the aphid pools and transferred onto Arabidopsis plants (one aphid per plant) to study adult fecundity. The number of nymphs produced were recorded after 5 days. Adult aphids that died before day 5 were excluded from the analysis. Data on CaMV‐infected or mock‐inoculated Arabidopsis were collected in three repetitions, and data on Arabidopsis mutants were collected in four repetitions, comprising altogether 21–33 aphids per condition (infected or mock‐inoculated and wt or mutant, respectively).
4.7. Statistical analysis
We used GLMs with a likelihood ratio and the chi‐square (χ2) test to assess whether there was an effect of plant infection or Arabidopsis mutants on M. persicae feeding behaviour. When a significant effect was detected, a pairwise comparison using estimated marginal means (R package “emmeans”; p value adjustment with Tukey method) at the .05 significance level was used to test for differences between treatments. Data on aphid feeding behaviour (probing, pathway, and phloem sap ingestion phases) were not normally distributed; accordingly we carried out a GLM using a gamma (link = “inverse”) distribution. Data on aphid feeding behaviour (t1 < E2) were modelled using the Cox proportional hazards model and we treated cases where the given event did not occur as censored. The assumption of validity of proportional hazards was checked using the functions “coxph” and “cox.zph”, respectively (R package “survival”). When a significant effect of one of the main factors was detected or when an interaction between factors was significant, a pairwise comparison using estimated marginal means (R package “emmeans”) (p value adjustment with Tukey method) at the .05 significance level was used to test for differences between treatments. The fit of all GLMs was controlled by inspecting residuals and QQ plots. All statistical analyses were performed using R software v. 3.3.2 (www.r‐project.org/).
4.8. Western blot
Total leaf extracts were prepared as follows: Leaves were frozen in a mortar with liquid N2 and ground with a pistil to a fine powder. The powder was transferred to a 1.5 ml reaction tube and 2× Laemmli buffer (Laemmli, 1970) was added at a ratio of 1:1 (wt/vol). The samples were heated for 5 min at 80 °C and centrifuged for 10 min at 16,000 × g, and aliquots of the supernatants were charged on 6/12.5% discontinuous sodium dodecyl sulphate (SDS)‐polyacrylamide gels. Proteins were transferred after SDS polacrylamide gel electrophoresis onto nitrocellulose membranes using the wet blotting technique (Towbin et al., 1979). Efficiency of transfer was controlled by Ponceau red staining. Membranes were blocked for 30 min with 5% low‐fat milk powder in Tris‐buffered saline (TBS) and incubated overnight at 4 °C with primary antibodies. After three washes with TBS, membranes were incubated for 3–4 hr at room temperature with secondary antibodies. After another three washes with TBS, protein bands were visualized by enhanced chemiluminescence using a G‐Box. The following 1:2,000 dilutions of primary antibodies were used: anti‐P2 (Blanc et al., 1993), anti‐P4 (Champagne et al., 2004), and anti‐P6 (Khelifa et al., 2010). Secondary antibodies were horseradish peroxidase conjugates, which were used at a 1:25,000 dilution.
ACKNOWLEDGEMENTS
The authors are grateful to Claire Villeroy for aphid care. This work was funded by Human Frontier Science Program grant RGP0013/2015 (to M.D.) and Agence Nationale de la Recherche grant ANR‐18‐CE20‐0017‐01 (to M.D. and M.P.). The funding sources had no role in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; and in the decision to submit the article for publication. The authors declare no conflict of interest.
Chesnais Q, Verdier M, Burckbuchler M, Brault V, Pooggin M, Drucker M. Cauliflower mosaic virus protein P6‐TAV plays a major role in alteration of aphid vector feeding behaviour but not performance on infected Arabidopsis . Mol Plant Pathol. 2021;22:911–920. 10.1111/mpp.13069
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Blanc, S. , Cerutti, M. , Usmany, M. , Vlak, J.M. & Hull, R. (1993) Biological activity of cauliflower mosaic virus aphid transmission factor expressed in a heterologous system. Virology, 192, 643–650. [DOI] [PubMed] [Google Scholar]
- Bouchery, Y. , Givord, L. & Monestiez, P. (1990) Comparison of short‐ and long‐feed transmission of the cauliflower mosaic virus Cabb‐S strain and S delta II hybrid by two species of aphid: Myzus persicae (Sulzer) and Brevicoryne brassicae (L.). Research in Virology, 141, 677–683. [DOI] [PubMed] [Google Scholar]
- Carr, J.P. , Tungadi, T. , Donnelly, R. , Bravo‐Cazar, A. , Rhee, S.‐J. , Watt, L.G. et al. (2020) Modelling and manipulation of aphid‐mediated spread of non‐persistently transmitted viruses. Virus Research, 277, 197845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchini, E. , Al‐Kaff, N.S. , Bannister, A. , Giannakou, M.E. , McCallum, D.G. , Maule, A.J. et al. (1998) Pathogenic interactions between variants of cauliflower mosaic virus and Arabidopsis thaliana . Journal of Experimental Botany, 49, 731–737. [Google Scholar]
- Champagne, J. , Benhamou, N. & Leclerc, D. (2004) Localization of the N‐terminal domain of cauliflower mosaic virus coat protein precursor. Virology, 324, 257–262. [DOI] [PubMed] [Google Scholar]
- Chesnais, Q. , Couty, A. , Uzest, M. , Brault, V. & Ameline, A. (2019) Plant infection by two different viruses induce contrasting changes of vectors fitness and behavior. Insect Science, 26, 86–96. [DOI] [PubMed] [Google Scholar]
- Dáder, B. , Burckbuchler, M. , Macia, J.‐L. , Alcon, C. , Curie, C. , Gargani, D. & et al. (2019) Split green fluorescent protein as a tool to study infection with a plant pathogen, cauliflower mosaic virus. PLoS One, 14, e0213087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dáder, B. , Then, C. , Berthelot, E. , Ducousso, M. , Ng, J.C.K. & Drucker, M. (2017) Insect transmission of plant viruses: Multilayered interactions optimize viral propagation. Insect Science, 24, 929–946. [DOI] [PubMed] [Google Scholar]
- Doumayrou, J. , Avellan, A. , Froissart, R. & Michalakis, Y. (2013) An experimental test of the transmission‐virulence trade‐off hypothesis in a plant virus. Evolution: International Journal of Organic Evolution, 67, 477–486. [DOI] [PubMed] [Google Scholar]
- Drucker, M. , Froissart, R. , Hébrard, E. , Uzest, M. , Ravallec, M. , Espérandieu, P. et al. (2002) Intracellular distribution of viral gene products regulates a complex mechanism of cauliflower mosaic virus acquisition by its aphid vector. Proceedings of the National Academy of Sciences of the United States of America, 99, 2422–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwardson, J.R. & Christie, R.G. (2018) CRC handbook of viruses infecting legumes. Boca Raton: CRC Press. [Google Scholar]
- Eigenbrode, S.D. , Bosque‐Pérez, N.A. & Davis, T.S. (2018) Insect‐borne plant pathogens and their vectors: ecology, evolution, and complex interactions. Annual Review of Entomology, 63, 169–191. [DOI] [PubMed] [Google Scholar]
- Fereres, A. & Moreno, A. (2009) Behavioural aspects influencing plant virus transmission by homopteran insects. Virus Research, 141, 158–168. [DOI] [PubMed] [Google Scholar]
- Giordanengo, P. (2014) EPG‐Calc: a PHP‐based script to calculate electrical penetration graph (EPG) parameters. Arthropod‐Plant Interactions, 8, 163–169. [Google Scholar]
- Haas, G. , Azevedo, J. , Moissiard, G. , Geldreich, A. , Himber, C. , Bureau, M. et al. (2008) Nuclear import of CaMV P6 is required for infection and suppression of the RNA silencing factor DRB4. EMBO Journal, 27, 2102–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafrén, A. , Macia, J.‐L. , Love, A.J. , Milner, J.J. , Drucker, M. & Hofius, D. (2017) Selective autophagy limits cauliflower mosaic virus infection by NBR1‐mediated targeting of viral capsid protein and particles. Proceedings of the National Academy of Sciences of the United States of America, 114, E2026–E2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy, J.S. , Day, M.F. & Eastop, V.F. (1962) A conspectus of aphids as vectors of plant viruses. London: Eastern Press Ltd. [Google Scholar]
- Khelifa, M. , Massé, D. , Blanc, S. & Drucker, M. (2010) Evaluation of the minimal replication time of cauliflower mosaic virus in different hosts. Virology, 396, 238–245. [DOI] [PubMed] [Google Scholar]
- Kloth, K.J. & Kormelink, R. (2020) Defenses against virus and vector: a phloem‐biological perspective on RTM‐ and SLI1‐mediated resistance to potyviruses and aphids. Viruses, 12, 129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Laird, J. , McInally, C. , Carr, C. , Doddiah, S. , Yates, G. , Chrysanthou, E. et al. (2013) Identification of the domains of cauliflower mosaic virus protein P6 responsible for suppression of RNA silencing and salicylic acid signalling. Journal of General Virology, 94, 2777–2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leh, V. , Jacquot, E. , Geldreich, A. , Hermann, T. , Leclerc, D. , Cerutti, M. et al. (1999) Aphid transmission of cauliflower mosaic virus requires the viral PIII protein. EMBO Journal, 18, 7077–7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lung, M.C.Y. & Pirone, T.P. (1973) Studies on the reason for differential transmissibility of cauliflower mosaic virus isolates by aphids. Phytopathology, 63, 910–914. [Google Scholar]
- Markham, P.G. , Pinner, M.S. , Raccah, B. & Hull, R. (1987) The acquisition of a caulimovirus by different aphid species: comparison with a potyvirus. Annals of Applied Biology, 111, 571–587. [Google Scholar]
- Martin, B. , Collar, J.L. , Tjallingii, W.F. & Fereres, A. (1997) Intracellular ingestion and salivation by aphids may cause the acquisition and inoculation of non‐persistently transmitted plant viruses. Journal of General Virology, 78, 2701–2705. [DOI] [PubMed] [Google Scholar]
- Mauck, K. , Bosque‐Pérez, N.A. , Eigenbrode, S.D. , De Moraes, C.M. & Mescher, M.C. (2012) Transmission mechanisms shape pathogen effects on host–vector interactions: evidence from plant viruses. Functional Ecology, 26, 1162–1175. [Google Scholar]
- Mauck, K.E. & Chesnais, Q. (2020) A synthesis of virus–vector associations reveals important deficiencies in studies on host and vector manipulation by plant viruses. Virus Research, 285, 197957. [DOI] [PubMed] [Google Scholar]
- Mauck, K.E. , Chesnais, Q. & Shapiro, L.R. (2018) Evolutionary determinants of host and vector manipulation by plant viruses. Advances in Virus Research, 101, 189–250. [DOI] [PubMed] [Google Scholar]
- Mauck, K.E. , De Moraes, C.M. & Mescher, M.C. (2010) Deceptive chemical signals induced by a plant virus attract insect vectors to inferior hosts. Proceedings of the National Academy of Sciences of the United States of America, 107, 3600–3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauck, K.E. , De Moraes, C.M. & Mescher, M.C. (2014) Biochemical and physiological mechanisms underlying effects of cucumber mosaic virus on host‐plant traits that mediate transmission by aphid vectors. Plant, Cell and Environment, 37, 1427–1439. [DOI] [PubMed] [Google Scholar]
- Moreno, A. , Hébrard, E. , Uzest, M. , Blanc, S. & Fereres, A. (2005) A single amino acid position in the helper component of cauliflower mosaic virus can change the spectrum of transmitting vector species. Journal of Virology, 79, 13587–13593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugford, S.T. , Barclay, E. , Drurey, C. , Findlay, K.C. & Hogenhout, S.A. (2016) An immuno‐suppressive aphid saliva protein is delivered into the cytosol of plant mesophyll cells during feeding. Molecular Plant‐Microbe Interactions, 29, 854–861. [DOI] [PubMed] [Google Scholar]
- Palacios, I. , Drucker, M. , Blanc, S. , Leite, S. , Moreno, A. & Fereres, A. (2002) Cauliflower mosaic virus is preferentially acquired from the phloem by its aphid vectors. Journal of General Virology, 83, 3163–3171. [DOI] [PubMed] [Google Scholar]
- Pooggin, M.M. & Ryabova, L.A. (2018) Ribosome shunting, polycistronic translation, and evasion of antiviral defenses in plant pararetroviruses and beyond. Frontiers in Microbiology, 9, 644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prince, D.C. , Drurey, C. , Zipfel, C. & Hogenhout, S.A. (2014) The leucine‐rich repeat receptor‐like kinase BRASSINOSTEROID INSENSITIVE1‐ASSOCIATED KINASE1 and the cytochrome P450 PHYTOALEXIN DEFICIENT3 contribute to innate immunity to aphids in Arabidopsis. Plant Physiology, 164, 2207–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov, M. , Kobayashi, K. , Geldreich, A. , Caranta, C. , Robaglia, C. , Keller, M. et al. (2011) Viral factor TAV recruits TOR/S6K1 signalling to activate reinitiation after long ORF translation. EMBO Journal, 30, 1343–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoelz, J.E. , Angel, C.A. , Nelson, R.S. & Leisner, S.M. (2016) A model for intracellular movement of cauliflower mosaic virus: the concept of the mobile virion factory. Journal of Experimental Botany, 67, 2039–2048. [DOI] [PubMed] [Google Scholar]
- Schoelz, J.E. & Leisner, S. (2017) Setting up shop: the formation and function of the viral factories of cauliflower mosaic virus. Frontiers in Plant Science, 8, 1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, A.K. , Peace, A. , Power, A.G. & Bosque‐Pérez, N.A. (2017) Vector population growth and condition‐dependent movement drive the spread of plant pathogens. Ecology, 98, 2145–2157. [DOI] [PubMed] [Google Scholar]
- Shivaprasad, P.V. , Rajeswaran, R. , Blevins, T. , Schoelz, J. , Meins, F. , Hohn, T. et al. (2008) The CaMV transactivator/viroplasmin interferes with RDR6‐dependent trans‐acting and secondary siRNA pathways in Arabidopsis . Nucleic Acids Research, 36, 5896–5909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjallingii, W.F. (1988) Electrical recording of stylet penetration activities. In: Minks, A.K. & Harrewijn, P. (eds). Aphids, their biology, natural enemies and control. Amsterdam: Elsevier Science Publishers, pp. 95–108. [Google Scholar]
- Tjallingii, W.F. & Hogen Esch, T. (1993) Fine structure of aphid stylet routes in plant tissues in correlation with EPG signals. Physiological Entomology, 18, 317–328. [Google Scholar]
- Towbin, H. , Staehelin, T. & Gordon, J. (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Biotechnology, 1992, 145–149. [PubMed] [Google Scholar]
- Tsuge, S. , Kobayashi, K. , Nakayashiki, H. , Okuno, T. & Furusawa, I. (1994) Replication of cauliflower mosaic virus ORF I mutants in turnip protoplasts. Japanese Journal of Phytopathology, 60, 27–35. [Google Scholar]
- Uzest, M. , Gargani, D. , Dombrovsky, A. , Cazevieille, C. , Cot, D. & Blanc, S. (2010) The “acrostyle”: a newly described anatomical structure in aphid stylets. Arthropod Structure & Development, 39, 221–229. [DOI] [PubMed] [Google Scholar]
- Verdier, M. (2020) Etude de la transmission des virus de plantes par puceron. Comment le virus manipule sa plante hôte (et son puceron vecteur) pour optimiser sa transmission? Master’s thesis. Paris, France: Muséum National D’Histoire Naturelle. [Google Scholar]
- Wang, R.Y. , & Ghabrial, S.A. (2002) Effect of aphid behavior on efficiency of transmission of soybean mosaic virus by the soybean‐colonizing aphid, Aphis glycines . Plant Disease, 86, 1260–1264. [DOI] [PubMed] [Google Scholar]
- Webster, C.G. , Pichon, E. , van Munster, M. , Monsion, B. , Deshoux, M. , Gargani, D. et al. (2018) Identification of plant virus receptor candidates in the stylets of their aphid vectors. Journal of Virology, 92, e00432–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolston, C. , Czaplewski, L. , Markham, P. , Goad, A. , Hull, R. & Davies, J. (1987) Location and sequence of a region of cauliflower mosaic‐virus gene‐2 responsible for aphid transmissibility. Virology, 160, 246–251. [Google Scholar]
- Yu, W. , Murfett, J. & Schoelz, J.E. (2003) Differential induction of symptoms in Arabidopsis by P6 of cauliflower mosaic virus. Molecular Plant‐Microbe Interactions, 16, 35–42. [DOI] [PubMed] [Google Scholar]
- Zvereva, A.S. , Golyaev, V. , Turco, S. , Gubaeva, E.G. , Rajeswaran, R. , Schepetilnikov, M.V. et al. (2016) Viral protein suppresses oxidative burst and salicylic acid‐dependent autophagy and facilitates bacterial growth on virus‐infected plants. New Phytologist, 211, 1020–1034. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


