Abstract
Grapevine leafroll‐associated virus (GLRaV) infections are accompanied by symptoms influenced by host genotype, rootstock, environment, and which individual or combination of GLRaVs is present. Using a dedicated experimental vineyard, we studied the responses to GLRaVs in ripening berries from Cabernet Franc grapevines grafted to different rootstocks and with zero, one, or pairs of leafroll infection(s). RNA sequencing data were mapped to a high‐quality Cabernet Franc genome reference assembled to carry out this study and integrated with hormone and metabolite abundance data. This study characterized conserved and condition‐dependent responses to GLRaV infection(s). Common responses to GLRaVs were reproduced in two consecutive years and occurred in plants grafted to different rootstocks in more than one infection condition. Though different infections were inconsistently distinguishable from one another, the effects of infections in plants grafted to different rootstocks were distinct at each developmental stage. Conserved responses included the modulation of genes related to pathogen detection, abscisic acid (ABA) signalling, phenylpropanoid biosynthesis, and cytoskeleton remodelling. ABA, ABA glucose ester, ABA and hormone signalling‐related gene expression, and the expression of genes in several transcription factor families differentiated the effects of GLRaVs in berries from Cabernet Franc grapevines grafted to different rootstocks. These results support that ABA participates in the shared responses to GLRaV infection and differentiates the responses observed in grapevines grafted to different rootstocks.
Keywords: Closteroviridae, leafroll disease, plant–virus interaction, rootstock–scion interaction, Vitis vinifera
GLRaVs elicit changes in gene expression, ripening‐related metabolites, and hormone metabolism in berries; one set of responses is observed across infections and host genotypes while other responses differ based on rootstock.

1. INTRODUCTION
Grapevine leafroll‐associated viruses (GLRaVs) are among the most consequential pathogens affecting grapevine and have considerable economic impact (Atallah et al., 2012; Fuchs et al., 2021; Ricketts et al., 2015). GLRaVs are diverse and belong to the family Closteroviridae, with six species and numerous strains in three genera (Fuchs, 2020; Naidu et al., 2015). Grapevines are often infected with several of these viruses simultaneously (Prosser et al., 2007; Rwahnih et al., 2009). Given their impact and global distribution, efforts to manage the spread of GLRaVs, characterize their effects, and understand the interaction between the vine and GLRaVs have been undertaken.
Generally, plant responses to viruses include numerous changes in gene expression, gene regulation, and metabolism (Alazem & Lin, 2015; Bester et al., 2017; Blanco‐Ulate et al., 2017; Moon & Park, 2016). Pathogens and stresses elicit conserved responses from their hosts (Amrine et al., 2015; Jiang et al., 2015; Postnikova & Nemchinov, 2012; Rodrigo et al., 2012; Shaik & Ramakrishna, 2013). Infections with GLRaVs have been associated with poorer fruit quality, lower yield, and leaves that curl, redden, and become brittle. Gene expression studies that implicate regulatory systems in the leafroll disease phenotype are few in number and have focused on the impact of GLRaV‐3, highlighting changes in the expression of senescence‐associated and flavonoid biosynthetic pathway genes (Espinoza, Medina, et al., 2007, Espinoza, Vega, et al., 2007; Gutha et al., 2010; Vega et al., 2011). Additional transcriptomic study could help generate novel hypotheses concerning the controls that are fundamental to GLRaV responses (Gaiteri et al., 2014; Mandadi & Scholthof, 2013; Moon & Park, 2016).
Though common responses might be expected in infected plants given the relatedness of GLRaVs, there is considerable variability in the severity of GLRaV infections. Some GLRaV infections appear without symptoms or are mild (Kovacs et al., 2001; Montero et al., 2016; Poojari et al., 2013), but others cause significant changes in photosynthesis, metabolism, and gas exchange in leaves (Bertamini et al., 2004; Endeshaw et al., 2014; Guidoni et al., 1997, 2000; Pereira et al., 2012). Changes in fruit yield, organic and amino acids, titratable acidity, potassium, sugars, and flavonoids are also observed (Alabi et al., 2016; Cabaleiro et al., 1999; Kliewer & Lider, 1976; Lee et al., 2009; Lee & Martin, 2009). These are influenced by host genotype (Kovacs et al., 2001; Montero et al., 2016), which virus or combination of viruses is present (Credi, 1997; Guidoni et al., 2000; Komar et al., 2010), and environmental conditions (Cui et al., 2016). Leaf reddening, for example, is only observed in red‐fruited grapevines (Naidu et al., 2015).
Evidence relating GLRaV responses to rootstock is mixed (Golino, 1993; Komar et al., 2010). In a study of Cabernet Franc vines grafted to different rootstocks, the effect of GLRaV infection on pruning weight depended on rootstock and the largest effects were observed in Kober 5BB‐grafted vines (Rowhani et al., 2015). Similarly, fruit yield was influenced by both infection type and rootstock, with Kober 5BB‐grafted vines most severely affected by a mixed infection with GLRaV‐2, GLRaV‐3, and grapevine fleck virus (Golino, Wolpert, et al., 2008). In another report, Red Globe scion buds infected with a strain of GLRaV‐2 were used to inoculate Cabernet Sauvignon plants grafted to 18 different rootstocks; the infection was lethal in plants grafted to several rootstock genotypes, including Kober 5BB (Alkowni et al., 2011; Uyemoto et al., 2001).
This study used Cabernet Franc grapevines infected with zero, one, or two GLRaVs and grafted to two different rootstocks to (a) identify leafroll effects in ripening berries that were conserved across experimental conditions, and (b) determine whether or not GLRaV responses could be distinguished in berries from plants grafted to different rootstocks. Grapevines were grown in a single experimental vineyard and evaluated in four consecutive years. Vine growth and several measures of fruit composition were taken in the first two years. Total soluble solids (TSS) were measured in all four years. RNA sequencing (RNA‐Seq), hormone, and metabolite data were collected from Cabernet Franc berries at four stages during ripening in the third and fourth years. RNA‐Seq reads were mapped to the Cabernet Franc genome, which was sequenced in long PacBio reads, assembled using the FALCON‐Unzip pipeline, and scaffolded using Hi‐C data. The same samples were used to measure the levels of stress and ripening‐associated hormones and metabolites. Among these were abscisic acid (ABA), jasmonic acid (JA), and salicylic acid (SA). Though many of the GLRaV effects occurred in individual years, a subset of reproducible conserved responses and rootstock‐differentiating responses were discovered.
2. RESULTS
2.1. GLRaV species and rootstock influence canopy density, cluster weight, and fruit composition
Cabernet Franc grapevines infected with individual and pairs of GLRaVs (GLRaV‐1, GLRaV‐1 [+]; GLRaV‐3, GLRaV‐3 [+]; GLRaV‐4 strain 5, GLRaV‐4 [+]; GLRaV‐1 and GLRaV‐2, GLRaV‐1,2 [+]; GLRaV‐1 and GLRaV‐3, GLRaV‐1,3 [+]) and grafted to different rootstocks (Millardet et de Grasset [MGT] 101‐14 and Kober 5BB) were studied during grape berry ripening in a dedicated experimental vineyard at the University of California, Davis.
Typical grapevine leafroll disease symptoms (i.e., leaf reddening and curling) were observed by mid‐ripening (Figure 1a). In addition, there was a visible, stark reduction in canopy density and cluster size in GLRaV‐1,2 (+) versus GLRaV (−) in vines grafted to Kober 5BB that was not readily apparent in vines grafted to MGT 101‐14 with the same infection status (Figure 1b).
FIGURE 1.
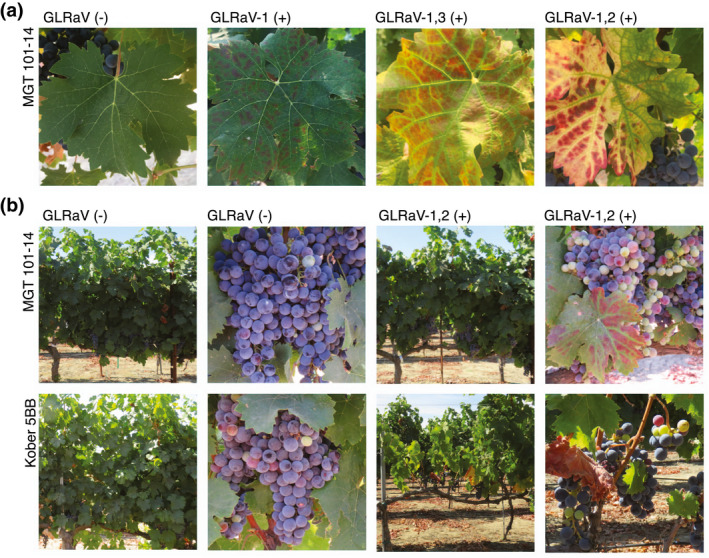
Examples of the effects of GLRaVs on Cabernet Franc leaves, canopy density, and cluster size. (a) Photographs depicting leaves from Cabernet Franc grapevines grafted to MGT 101‐14. The photographed leaves were from (left to right) GLRaV (−), GLRaV‐1 (+), GLRaV‐1,3 (+), and GLRaV‐1,2 (+). The photograph of the GLRaV (−) leaf was taken on 2018‐08‐13. The photographs of the leaves from GLRaV (+) were taken on 2018‐08‐08. The purpose of these photographs is to depict the range of leafroll disease symptoms in leaves observed in the study. The symptoms should not be construed as specific to certain infections. (b) Canopy and berry clusters from GLRaV (−) and GLRaV‐1,2 (+) in different rootstock conditions on 2017‐08‐14
Vine growth, cluster weight, and other measures were collected in 2015 and 2016 (Figure 2, Figure S1). The effect of GLRaV infection on dormant pruning weight, berry weight, pH, and tartaric acid content in 2015 and on moisture content, total anthocyanin content, and titratable acidity in 2016 differed significantly based on the rootstock present (analysis of variance [ANOVA], p < .05). This interaction was significant for malic acid in 2015 and 2016. Significant differences in dormant pruning weight, total cluster weight, and tartaric acid were observed in plants with different GLRaV infection status and rootstock (Tukey honestly significant difference [HSD] test, p < .05). In contrast, few or no significant differences between GLRaV (+) given the same rootstock were observed for total anthocyanins, moisture content, malic acid content, pH, titratable acidity, weight per berry, or yeast assimilable nitrogen and overwhelmingly in a single year if at all (Figure S1).
FIGURE 2.
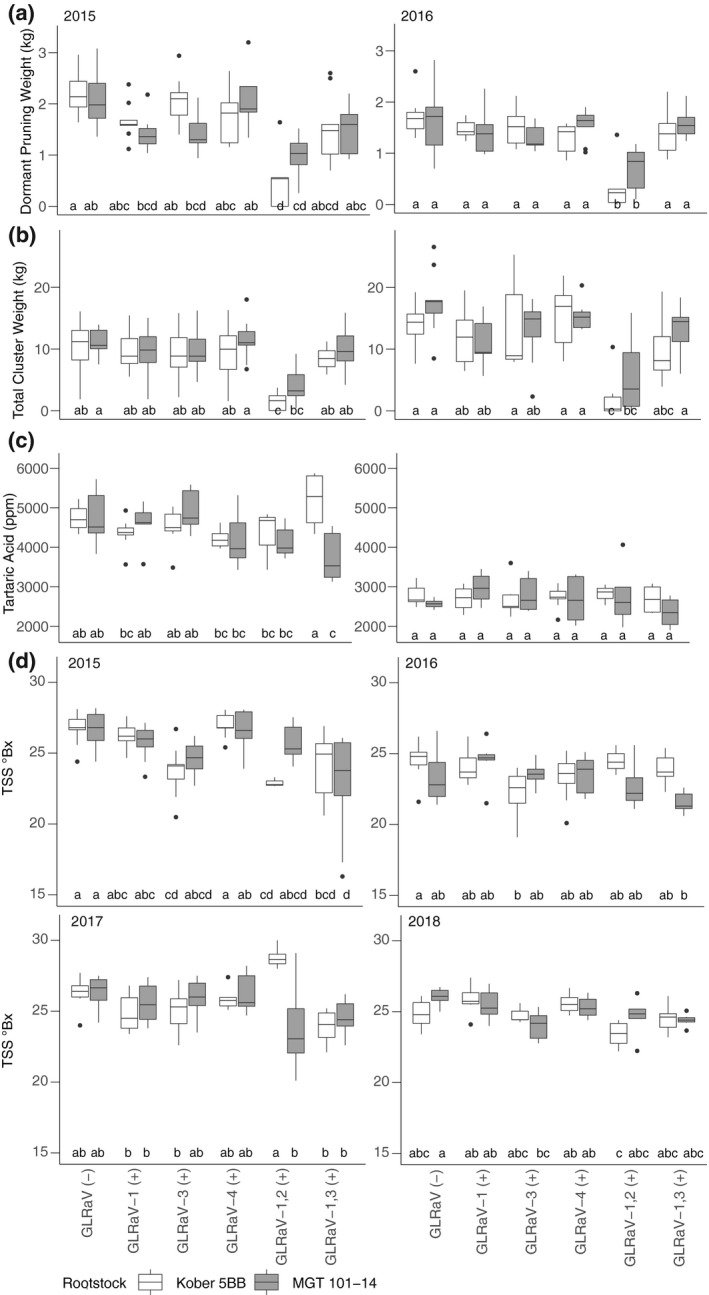
Effects of GLRaV infection on (a) dormant pruning weight, 5 ≤ n ≤ 9, (b) total cluster weight, 5 ≤ n ≤ 9, and (c) tartaric acid content, 3 ≤ n ≤ 9, in 2015 and 2016. (d) Total soluble solids (TSS) at harvest in four consecutive years. 2015–2016, 5 ≤ n ≤ 9; 2017–2018, n = 6. Group differences are indicated with nonoverlapping letters (Tukey HSD, p < .05)
Overall, GLRaV infection tended to reduce dormant pruning weight and cluster weight. The dormant pruning weights and cluster weights of GLRaV‐1,2 (+) was significantly lower than those of GLRaV (−) and other GLRaV (+) (Figure 2a,b); this was observed for both rootstock genotypes. Significant differences in fruit tartaric acid levels were observed only in 2015 and were between GLRaV‐1,3 (+) grafted to different rootstocks and between plants with different GLRaV infection status (Figure 2c). In each year except 2015, there was a significant interaction between rootstock and GLRaV infection status in terms of TSS at harvest (ANOVA, p < .05, Figure 2d). This interaction was significant at each other developmental stage in 2017 and at prevéraison in 2018 (Figure S1). Significant differences in TSS at harvest were found between GLRaV (−) and GLRaV (+) in each year except 2017 (Figure 2d). Overall, significant reductions in TSS relative to GLRaV (−) were limited to the dual infections and GLRaV‐3 (+). Significant differences were observed between rootstocks in GLRaV 1,2 (+) at every developmental stage, albeit only in 2017 (Figure 2d, Figure S1).
These data provide limited evidence that (a) different GLRaV infections may or may not affect various aspects of vine growth and fruit composition, (b) some of these differences are rootstock‐specific, and (c) although some of these effects are observed across years, year‐to‐year differences may impact whether or not effects occur.
2.2. GLRaVs elicit reproducible changes in gene expression across infection types and rootstocks
We used RNA‐Seq to sequence the transcriptome of 384 Cabernet Franc berry samples collected from plants grafted to different rootstocks (Kober 5BB or MGT 101‐14), with different GLRaV infection status, at four developmental stages (prevéraison, véraison, mid‐ripening, and harvest), and in two consecutive years (2017 and 2018).
Because of the remarkable structural and gene content variability among grape cultivars (Da Silva et al., 2013; Minio et al., 2019; Venturini et al., 2013), we built a genome reference specifically for the analysis of these RNA‐Seq data. The Cabernet Franc genome was assembled into 504 primary contigs (N50 = 5.74 Mb) for a total assembly size of 570 Mb. This is comparable to the size of the Zinfandel (591 Mb; Vondras et al., 2019), Cabernet Sauvignon (590 Mb; Chin et al., 2016), Chardonnay (490 Mb; Roach et al., 2018), and Pinot Noir PN40024 (487 Mb; Jaillon et al., 2007) genomes. In total, 3,085 additional haplotigs were assembled with an N50 of 184 kb (Table S3). The primary assembly and haplotigs were annotated with 33,563 and 19,146 protein‐coding genes, respectively (Table S3).
Ripening was associated with transcriptomically distinct developmental stages. Samples clustered primarily by developmental stage and secondarily by year, though samples at harvest clustered separately (Figure S2). Genes with comparable, significant responses (consistently upregulated or downregulated, p < .05) in both years of the study were selected to identify reproducible responses to GLRaVs during ripening. Gene expression in GLRaV (+) was compared to gene expression in GLRaV (−) grafted to the same rootstock at the same developmental stage (Figure 3). In addition, the effects of each GLRaV infection on gene expression at each developmental stage were compared in plants grafted to different rootstocks (Figure S3).
FIGURE 3.
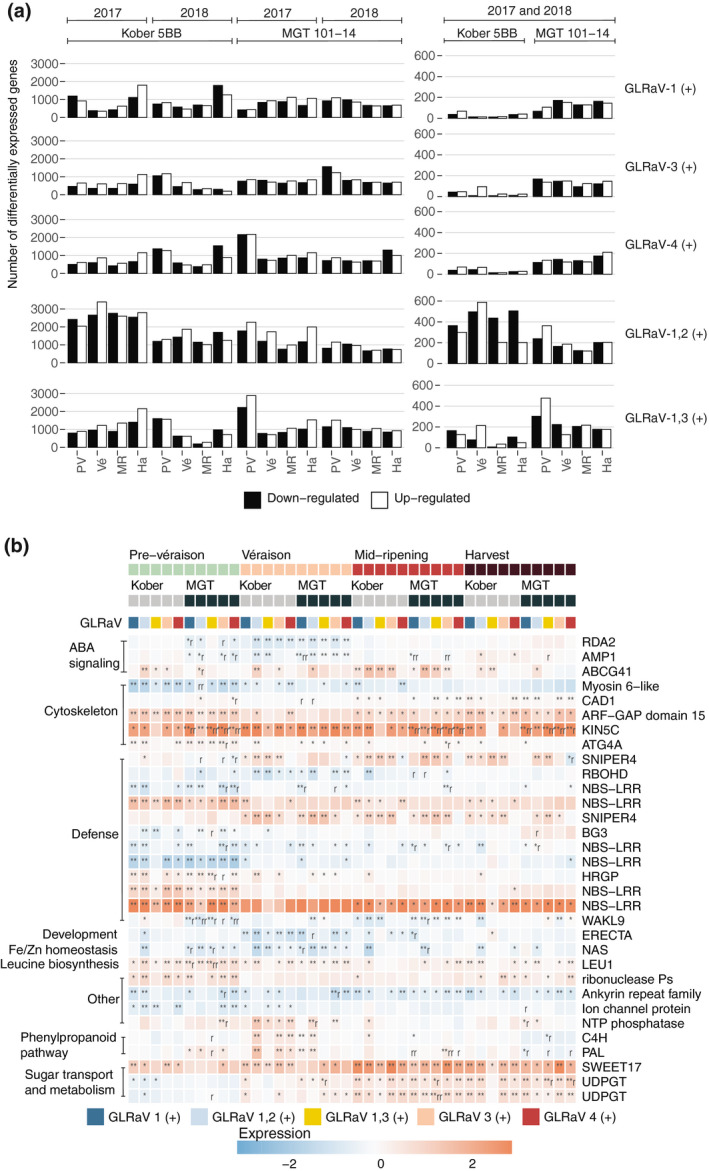
The conserved responses of ripening Cabernet Franc berries to GLRaV infection(s). (a) Barplots showing the number of differentially expressed (p < .05) genes up‐ and downregulated in GLRaV (+) versus GLRaV (−) in berries from Cabernet Franc grapevines grafted to different rootstocks (Kober 5BB [Kober] and MGT 101‐14 [MGT]) at each developmental stage (prevéraison [PV], véraison [Vé], mid‐ripening [MR], and harvest [Ha]) in 2017, in 2018, and in both years. (b) Heatmap showing the responses to GLRaVs (p < .05) in both rootstock conditions and in more than one GLRaV infection condition. *Differentially expressed in 1 year; **differentially expressed in both years. One or two letters “r” indicate that the effect of a particular GLRaV infection differs between rootstocks at the same developmental stage in 1 or 2 years. Any notation requires the direction (up/downregulation) of the effect to be consistent in both years, even if a significant change occurred in only 1 year
On average, 7.1% of the genes differentially expressed between GLRaV (−) and GLRaV (+) were reproduced in both years (Figure 3a). This percentage was slightly above average for plants with dual, relatively more severe, infections (8.6%, GLRaV‐1,3 [+]; 8.9%, GLRaV‐1,2 [+]) and below average for individual infections (5.8%, GLRaV‐1 [+]; 6% GLRaV‐4 [+]; 6.2% GLRaV‐3 [+]). A subset of 32 genes significantly changed their expression level in two or more GLRaV (+) infection conditions, in both rootstock conditions, and at least one developmental stage (Figure 3b). These genes constitute the “conserved” responses to GLRaVs in Cabernet Franc berries during ripening.
The majority of these differentially expressed genes are associated with defence, ABA signalling, and cytoskeleton organization and biogenesis (Figure 3b; Table S4). Six of these were genes encoding nucleotide‐binding site and leucine‐rich repeat‐containing (NBS‐LRR) proteins; half of these were upregulated. Two F‐box genes encoding SNIPER4 were upregulated (Huang et al., 2018), as was a gene encoding a hydroxyproline‐rich glycoprotein (HRGP). HRGP and NBS‐LRR proteins are associated with pathogen detection (DeYoung & Innes, 2006). Genes encoding a respiratory burst oxidase protein D (RBOHD), a wall‐associated kinase‐like protein (WAKL), and a β‐glucosidase 3 (BG3) were downregulated. RBOHD participates in the production of reactive oxygen species (ROS) and hypersensitive responses (HRs) to pathogens (Otulak‐Kozieł et al., 2019). RBOH family proteins are targeted by Snf1‐related kinase 2 (SnRK2) phosphorylation, a key component of the ABA signalling pathway. Likewise, a WAKL gene in citrus participates in JA and ROS signalling (Li et al., 2020). Among the functions of β‐glucosidases are the activation of ABA and SA by freeing them from the conjugates that render them inactive (Jia et al., 2016; Morant et al., 2008; Seo et al., 1995; Sun et al., 2015; Zhang et al., 2013). Several ABA‐related genes were among the conserved GLRaV responses, including an upregulated ABC transporter and two downregulated genes, AMP1 and RDA2. AMP1 negatively regulates ABA sensitivity (Shi et al., 2013). RDA2 participates in the inhibition of ABA signalling and the promotion of MAPK signalling (Park et al., 2019).
Five genes related to cytoskeleton organization were sensitive to GLRaV infection. Only one of these, a myosin VI motor protein‐coding gene, was downregulated. The four others were an autophagy gene (ATG4A) and constitutively activated cell death 1 (CAD1), which function in autophagy, lytic pore formation, and HRs (Haxim et al., 2017; Morita‐Yamamuro et al., 2005; Yoshimoto et al., 2004), Kinesin‐like 5C (KIN5C), which encodes a microtubule motor protein (Reddy & Day, 2001), and an ARF‐GAP encoding ADP‐ribosylation factor GTPase‐activating protein domain 15, which helps efficiently load vesicles and remodel the actin cytoskeleton (Inoue & Randazzo, 2007).
Several additional general functional categories were present among the 32 genes that exhibited conserved responses to GLRaVs (Figure 3b). Genes encoding phenylalanine ammonia‐lyase (PAL) and cinnamate 4‐hydroxylase (C4H), which catalyse the first two steps of the phenylpropanoid pathway, two genes encoding UDP glucosyltransferases (UDPGTs), which conjugate sugars, and SWEET17, encoding a sugar transporter, were upregulated, as was a gene encoding 3‐isopropylmalate dehydratase, an enzyme in the leucine biosynthetic pathway. Two genes, encoding an LRR receptor‐like kinase (LRR‐RLK) called ERECTA and nicotianamine synthase (NAS), were downregulated. ERECTA participates in organ development and resistance to bacterial and fungal pathogens (Goff & Ramonell, 2007). NAS expression increases Fe and Zn abundance in rice (Moreno‐Moyano et al., 2016; Nozoye, 2018).
Generally, these genes and their changes in expression suggest that a common response to GLRaVs in Cabernet Franc berries during ripening includes the modulation of pathogen‐detecting genes, an increase in ABA transport and signalling, a decrease in ROS‐related signalling, and an enhancement of cytoskeleton remodelling, vesicle trafficking, phenylpropanoid metabolism, sugar transport and conjugation, and leucine biosynthesis.
2.3. Rootstock influences the impact of GLRaV on ABA metabolism
The same berry samples used for RNA‐Seq were used to measure the levels of three hormones associated with ripening and/or stress, including SA, JA, and ABA, and additional metabolites, including xanthoxin, a precursor to ABA, and ABA glucose ester (ABA‐GE), a conjugate of ABA implicated in its long‐distance transport (Jiang & Hartung, 2008). The mean levels of SA and JA were significantly influenced by year and/or by interactions between year, rootstock, and GLRaV at prevéraison (ANOVA, p < .05), but no significant differences were observed between individual groups (Tukey HSD, p > .05) (Figure S4). In contrast, year alone had a significant impact on the levels of ABA and related metabolites measured at each developmental stage, but largely did not interact with rootstock or GLRaV infection type to affect the abundance of ABA and related metabolites (Figure S4). In addition, the effect of GLRaV infection on ABA and ABA‐GE content significantly differed based on rootstock (Figure S4). Significant differences in ABA and ABA‐GE content were observed between rootstocks in plants with identical infection status and between plants with different GLRaV status grafted to the same rootstock (Tukey HSD, p < .05). Such differences were scarcely observed for xanthoxin, a precursor to ABA (Figure 4a, Figure S4).
FIGURE 4.
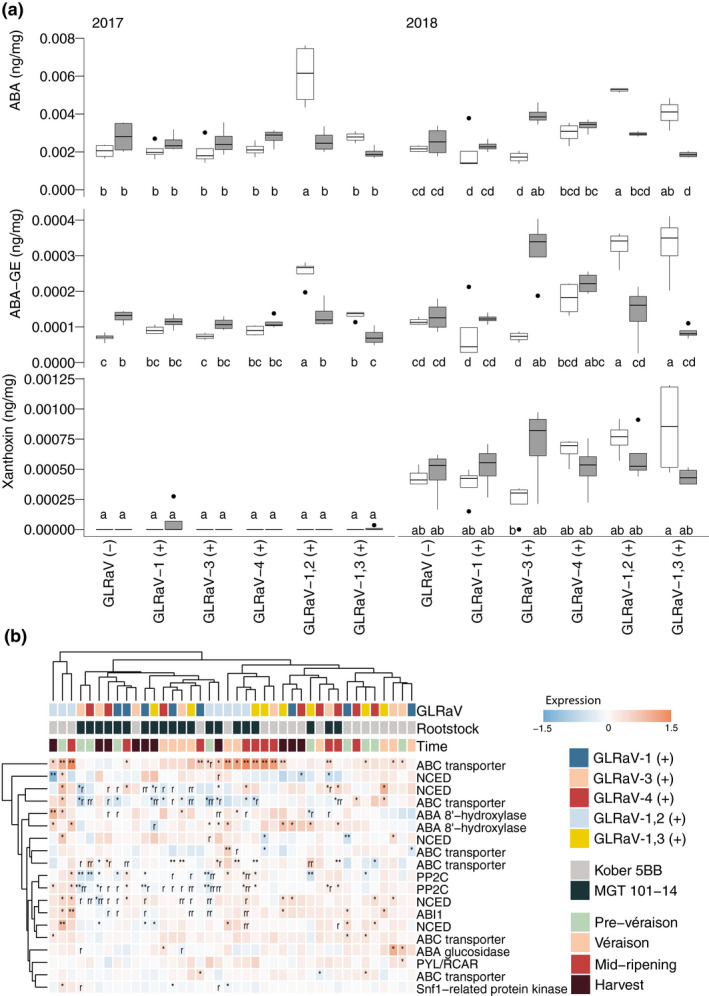
ABA metabolism and signalling pathways are sensitive to GLRaV infection. (a) Boxplots showing the abundance of abscisic acid (ABA), ABA glucose ester (ABA‐GE), and xanthoxin in 2017 and 2018 at prevéraison. Groups with nonoverlapping letters are significantly different (Tukey HSD, p < .05). (b) The effect of GLRaV infection(s) and rootstock on ABA biosynthesis and signalling genes. *Differentially expressed in 1 year; **differentially expressed in both years. One or two letters “r” indicate that the effect of a particular GLRaV infection differs between rootstocks at the same developmental stage in 1 or 2 years. Any notation requires the direction (up/downregulation) of the effect to be consistent in both years, even if a significant change occurred in only 1 year
Significant differences between rootstock genotypes in the abundance of these metabolites were observed most at prevéraison and in GLRaV (−), GLRaV‐3 (+), and dual infections (Figure 4a, Figure S4). In GLRaV (−) and most single infection conditions, the levels of all three metabolites tended to be higher in berries from plants grafted to MGT 101‐14 than in berries from plants grafted to Kober 5BB. The opposite tended to be true when two GLRaVs were present. With one exception, significant changes in the abundance of ABA and ABA‐GE in GLRaV (+) versus GLRaV (−) were typically increases and were most often observed in berries from Kober 5BB‐grafted plants (Tukey HSD, p < .05). Though slight reductions in these metabolites were observed versus GLRaV (−), these were almost always nonsignificant.
We further investigated ABA biosynthesis and signalling using a previously published, curated set of genes (Pilati et al., 2017). Their expression largely clustered according to rootstock, with some exceptions; Kober 5BB GLRaV‐1,2 (+), for example, tended to cluster separately from other Kober 5BB‐grafted plants. Nonetheless, the effects of GLRaVs differed between rootstocks for many of these genes (Figure 4b). One of these, encoding an ABC transporter (VITVvi_vCabFran04_v1_P438.ver1.0.g381930), is also included in Figure 3; significant increases in its expression were observed in both years, in both rootstock conditions, and for several GLRaV infections. All other significant changes in GLRaV (+) versus GLRaV (−) that were reproduced in both years occurred in only one rootstock or the other. These changes were sparse. However, significant differences between rootstocks in identical GLRaV (+) were reproduced in both years for 9 out of these 19 genes. Significant differences between rootstocks in at least one year were observed for 16 out of these 19 genes. On average, three genes encoding 9‐cis‐epoxycarotenoid dioxygenases (NCEDs), both ABA 8′‐hydroxylase genes, three ABC transporter genes, one gene encoding PP2C, and PYL/RCAR were upregulated in berries from plants infected with GLRaV in both rootstock conditions. One ABC transporter gene was downregulated in both rootstock conditions. Of the remaining eight genes, most were downregulated across development only in berries from MGT 101‐14‐grafted plants.
2.4. Gene expression, hormone, and other metabolite data distinguish the effects of GLRaVs in different rootstocks
Differential expression analysis identified 1,809 genes (a) that were differentially expressed (p < .05) in at least 1 year, (b) that were differentially expressed in only one rootstock condition and more than one GLRaV infection type versus GLRaV (−), and/or (c) for which the effects of more than one GLRaV infection significantly differed between rootstocks (Figure S5). RNA‐Seq, hormone, and metabolite data from ripening Cabernet Franc berries were integrated in a multiple factor analysis (MFA) to relate these variables and distinguish the effects of GLRaVs given different rootstocks (Figure 5, Figure S6, Table S5).
FIGURE 5.
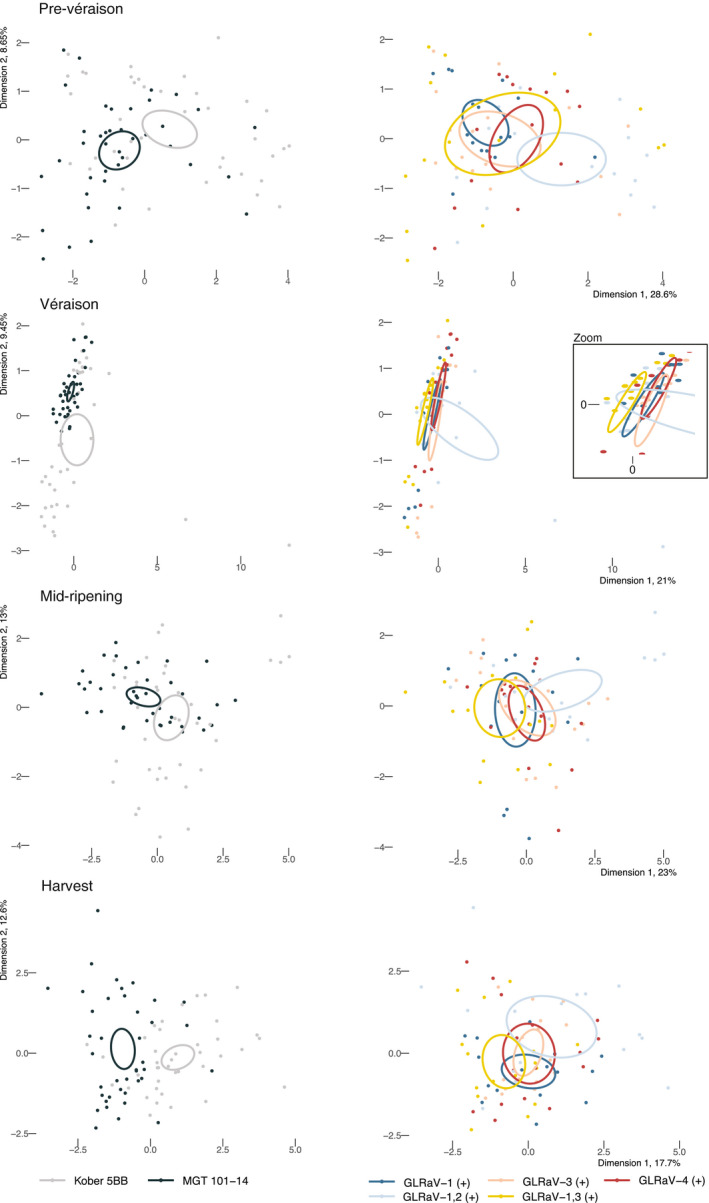
Multiple factor analysis (MFA) of the effects of GLRaV infection. This scatterplot shows the distribution of samples along the first two MFA dimensions at each developmental stage. For each rootstock (left) and each GLRaV infection condition (right), 95% confidence ellipses are drawn
As input for the MFA, all genes differentially expressed between GLRaV (−) and GLRaV (+) or between rootstocks were used, plus all hormones and metabolites measured. Overall, the rootstocks were distinct at each developmental stage (Figure 5). Some of the GLRaV (+) conditions could be distinguished from others at prevéraison, véraison, and harvest. At prevéraison, GLRaV‐1,2 (+) differed overall from GLRaV‐1 (+). At véraison, GLRaV‐1,3 (+) differed from every other GLRaV (+) condition except GLRaV‐1,2 (+). At harvest, the two dual infections were different than one another and GLRaV‐1,2 (+) differed from GLRaV‐1 (+) (Figure 5).
Next, we identified which variables were best correlated with each rootstock‐differentiating MFA dimension. At each developmental stage, ABA and/or ABA‐related metabolites were correlated with at least one of the first two MFA dimensions (Figure 6a). The rootstock‐dependent disparity in ABA levels and ABA‐related gene expression (Figure 4) is consistent with the observation that ABA and related metabolites tended to be highly correlated with rootstock‐differentiating MFA dimensions over time and that ripening initiates earliest in Kober 5BB plants with dual infections in terms of TSS (Figure S1).
FIGURE 6.
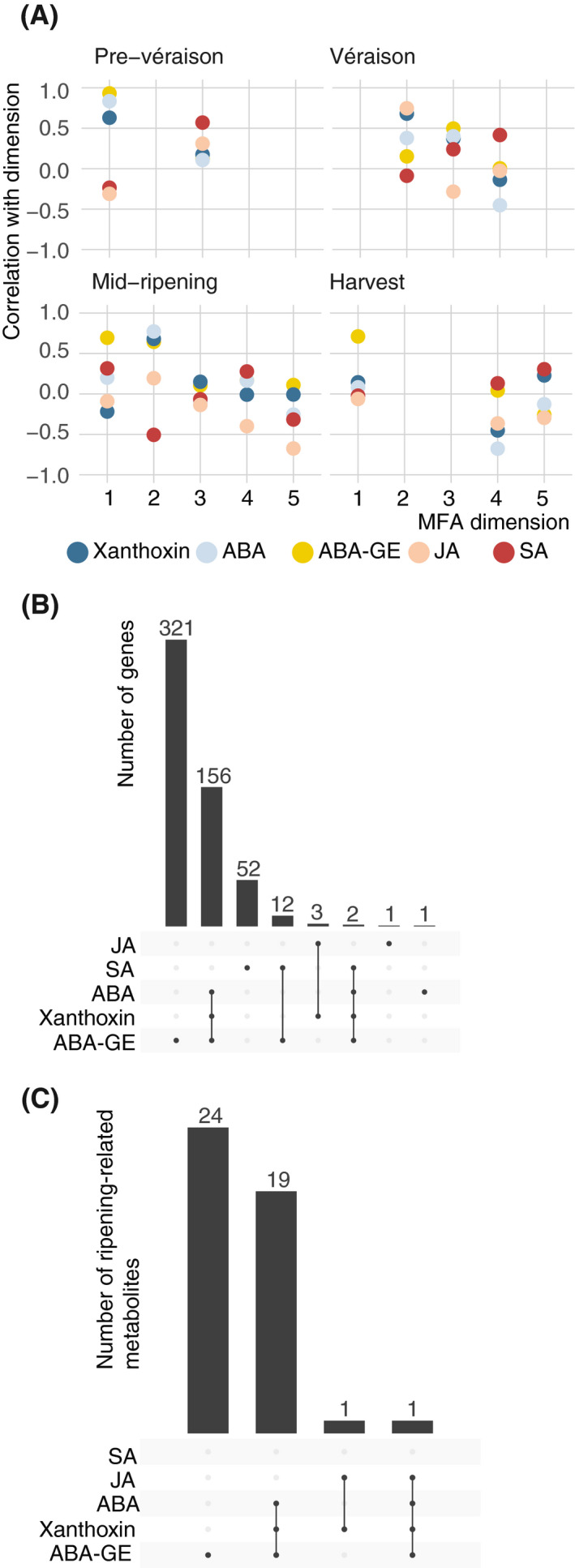
The roles of genes, hormones, and metabolites in a multiple factor analysis (MFA) of GLRaV effects. (a) Correlation between hormones and hormone‐related metabolites to rootstock‐differentiating MFA dimensions. If two variables both have either a strong positive or a strong negative correlation (|corr| > 0.5) to the same rootstock‐differentiating MFA dimension, their relationship is counted in the UpSet plots in(b) and (c), which are analogous to Venn diagrams. (b and c) The numbers of (b) genes and (c) ripening‐related metabolites with similar relationships to rootstock‐differentiating MFA dimensions as the hormone(s) and/or hormone‐related metabolites indicated below each bar. ABA, abscisic acid; ABA‐GE, ABA glucose ester; JA, jasmonic acid; SA, salicylic acid
There were 548 genes that shared high correlation (|corr| > .5) to rootstock‐differentiating MFA dimensions with hormones or hormone‐related metabolites. Most of these genes had shared positive or negative correlations to the same dimensions as ABA, xanthoxin, and/or ABA‐GE (Figure 6b). Categories of genes with functionally relevant relationships to each hormone or hormone‐related metabolite were over‐represented (hypergeometric test, p < .05 and n > 1 gene) among the genes that shared high correlation to rootstock‐differentiating MFA dimensions with each hormone. ABA signalling, starch biosynthesis and catabolism, and C2C2‐DOF transcription factor‐encoding genes were over‐represented among the genes correlated to the same dimensions as ABA and xanthoxin. The latter two categories were significantly over‐represented among the genes correlated with the same dimensions as ABA‐GE. Genes related to heat shock protein (HSP)‐mediated protein folding, chaperone‐mediated protein folding, the cation channel‐forming HSP‐70, channels and pores, the reductive carboxylate cycle, and carbon fixation were over‐represented among those correlated with the same MFA dimensions as SA. Similarly, most ripening‐related metabolites measured were correlated with the same MFA dimensions as ABA, xanthoxin, and/or ABA‐GE (Figure 6c).
Overall, the effects of GLRaVs differ between rootstocks primarily in terms of ABA and related metabolites. This finding is especially salient because of the role that ABA plays as a ripening promoter near véraison, in root–scion communication, and in plant stress. ABA, metabolites, and genes that were well correlated to rootstock‐differentiating MFA dimensions and were differentially expressed were scrutinized more closely.
2.5. Rootstock influences the impact of GLRaV on hormone signalling genes and transcriptional controls
There were 548 genes in 85 functional categories that were well correlated with rootstock‐differentiating MFA dimensions and differentially expressed between GLRaV (+) grafted to different rootstocks or in GLRaV (+) versus GLRaV (−) in only one rootstock condition (Figure 7). These functional categories were generally related to hormone and other types of signalling, amino acid and other metabolic pathways, transcription factors, transport, and cellular organization and biogenesis (Figure 7). Most of these genes coincided with ABA and related metabolites along rootstock‐differentiating MFA dimensions (Figure 7).
FIGURE 7.

Functional categories of genes correlated (|corr| > 0.5) to the same rootstock‐differentiating multiple factor analysis dimensions as hormones and/or hormone‐related metabolites. The counts of genes, per category, related to each hormone and metabolite are shown. ABA, abscisic acid; ABA‐GE, ABA glucose ester; JA, jasmonic acid; SA, salicylic acid
The distribution of expression for four transcription factor families differed significantly between rootstocks at all four developmental stages (Figure S7, Kolmogorov–Smirnov test, p < .05). This included bHLH, C2C2‐DOF, FHA, and homeobox domain transcription factors. The distribution of expression of 39 hormone signalling‐related genes differed significantly between rootstocks (Figure 7, Figure S7, File S1, Kolmogorov–Smirnov test, p < .05). This was true at each developmental stage for ABA, gibberellin (GA), and auxin signalling genes and at three developmental stages for JA/SA, cytokinin (CK), and ethylene signalling genes (Figure S7). Genes related to all of these hormone families had similar roles in MFA and were associated with ABA, including the JA/SA signalling genes (Figure 7). This may reflect interactions between hormone signalling pathways. In addition, the effects of GLRaV infections (dual infections and GLRaV‐4 [+]) on histone H1 expression were not equal in plants grafted to both rootstocks (Figure S8). Linker histone H1 contributes to higher‐order chromatin structure (Hill, 2001).
There were seven ABA signalling pathway genes (excluding VITVvi_vCabFran04_v1_P495.ver1.0.g468110, which was also associated with GA signalling) that differentiated GLRaV effects in plants grafted to different rootstocks (Figure 8). The effect of GLRaV on expression for all of these differed between rootstocks (Figure S7). This included SOS2, KEG, three PP2C genes (HAB1, AHG3, DBP), and two genes encoding ABA‐responsive element (ABRE)‐binding proteins (AREB2, ABI5). SOS2 is a kinase appreciated for its role in the salt stress response, seed germination, GA signalling (Trupkin et al., 2017), and ABA signal transduction via its interaction with ABI2 and ABI5 (Ji et al., 2013; Zhou et al., 2015). SOS2 was upregulated in both rootstock conditions before and at véraison and downregulated after véraison. KEG is a negative regulator of ABA signalling; it maintains low levels of ABI5 in the absence of stress by ubiquitination and degradation (Lyzenga et al., 2013; Stone et al., 2006) and helps regulate endocytic trafficking and the formation of signalling complexes on vesicles during stress (Gu & Innes, 2011). KEG was downregulated in Kober 5BB and downregulated in MGT 101‐14 at véraison and mid‐ripening.
FIGURE 8.
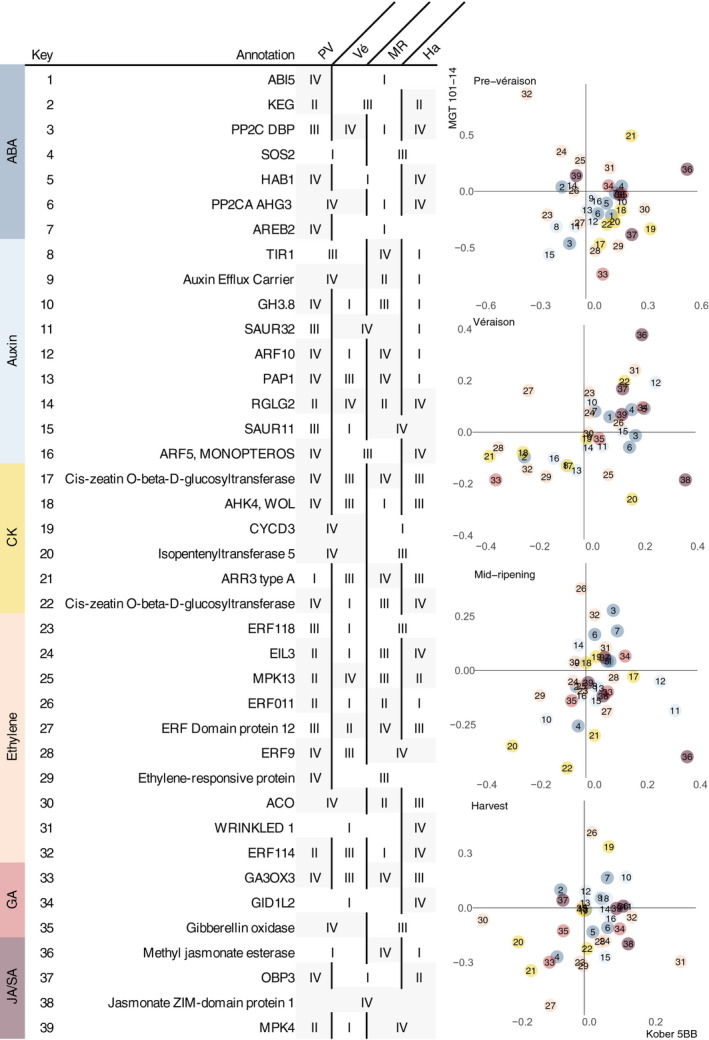
The effect of GLRaVs on hormone signalling gene expression in grape berries from plants grafted to different rootstocks. Quadrants are numbered counterclockwise from top right (I) to bottom right (IV). Individual genes are numbered 1–39. The key (left) indicates in which quadrant each gene can be found at each developmental stage. Developmental stages are abbreviated. PV, prevéraison; Vé, véraison; MR, mid‐ripening; Ha, harvest. ABA, abscisic acid; CK, cytokinin; GA, gibberellin; JA, jasmonic acid; SA, salicylic acid
In the presence of ABA, ABA receptors (PYR/PYL/RCAR family proteins) bind PP2Cs like HAB1 and AHG3 to inhibit their phosphatase activity. As a result, ABA signal transduction is permitted via SnRK2 phosphorylation of ABRE‐binding proteins (Hirayama & Shinozaki, 2010). ABI5 and AREB2 are bZIP transcription factors that bind to ABREs to drive ABA signalling and ABI5 can integrate signals across hormone signalling pathways (Skubacz et al., 2016). The effects of GLRaVs on these genes in Kober 5BB were consistent with an enhancement of ABA signalling during ripening. In Kober 5BB, HAB1, AHG3, AREB2, and ABI5 were upregulated. In MGT 101‐14, the PP2Cs were downregulated at two or more developmental stages; AREB2 and ABI5 were upregulated at and after véraison.
2.6. Rootstock influences the impact of GLRaV on the flavonoid biosynthetic pathway
In addition to analysing hormones and hormone‐related metabolites, we analysed metabolites associated with the shikimate, phenylpropanoid, and flavonoid pathways (Figure 9a,b) and their biosynthetic and regulatory genes (Figure 9c) in Cabernet Franc berries during ripening (Figure S9). Significant differences in expression versus GLRaV (−) were detected, as well as significant differences in the effects of GLRaV infection between different rootstock conditions (Figure 9c). The effects of GLRaV infection on the genes associated with this pathway were generally consistent with the change in abundance of corresponding metabolites (Figure 9b). Overall, these genes tended to be upregulated in GLRaV (+) at véraison (Figure 9c). After véraison, the amount of upregulation tended to decrease, or genes were downregulated (Figure 9c). The three amino acids examined (phenylalanine, tryptophan, and tyrosine) tended to be less abundant in GLRaV (+) across the developmental stages and the largest decreases were observed at harvest (Figure 9b). Mixed effects of GLRaVs were observed on the abundance of hydroxycinnamic acids (caftaric and coutaric acid), t‐resveratrol, and anthocyanins. Significant changes versus GLRaV (−) tended to occur in only 1 year. During ripening, these were significantly more abundant in Kober 5BB GLRaV‐1,2 (+), Kober 5BB GLRaV‐4 (+), and/or MGT 101‐13 GLRaV‐3 (+) (Figure 9b). Significant decreases were observed for GLRaV‐1 (+) and GLRaV‐1,3 (+). Though nonsignificant, the size of the downward effect of some GLRaV infections on these metabolites tended to increase towards harvest. Finally, flavanols (epigallocatechin and catechin) and flavonol (quercetin) glycosides tended to be elevated in GLRaV (+) (Figure 9b). The size of this effect tended to be greatest before and at véraison and decreased towards harvest. Significant differences between rootstocks were observed for GLRaV‐1,2 (+) in both years and for GLRaV‐1 (+), GLRaV‐1,3 (+), and GLRaV‐3 (+) in individual years; the increase in flavonols and flavanols tended to be greater in berries from Kober 5BB GLRaV (+) than in those from MGT 101‐14 GLRaV (+).
FIGURE 9.
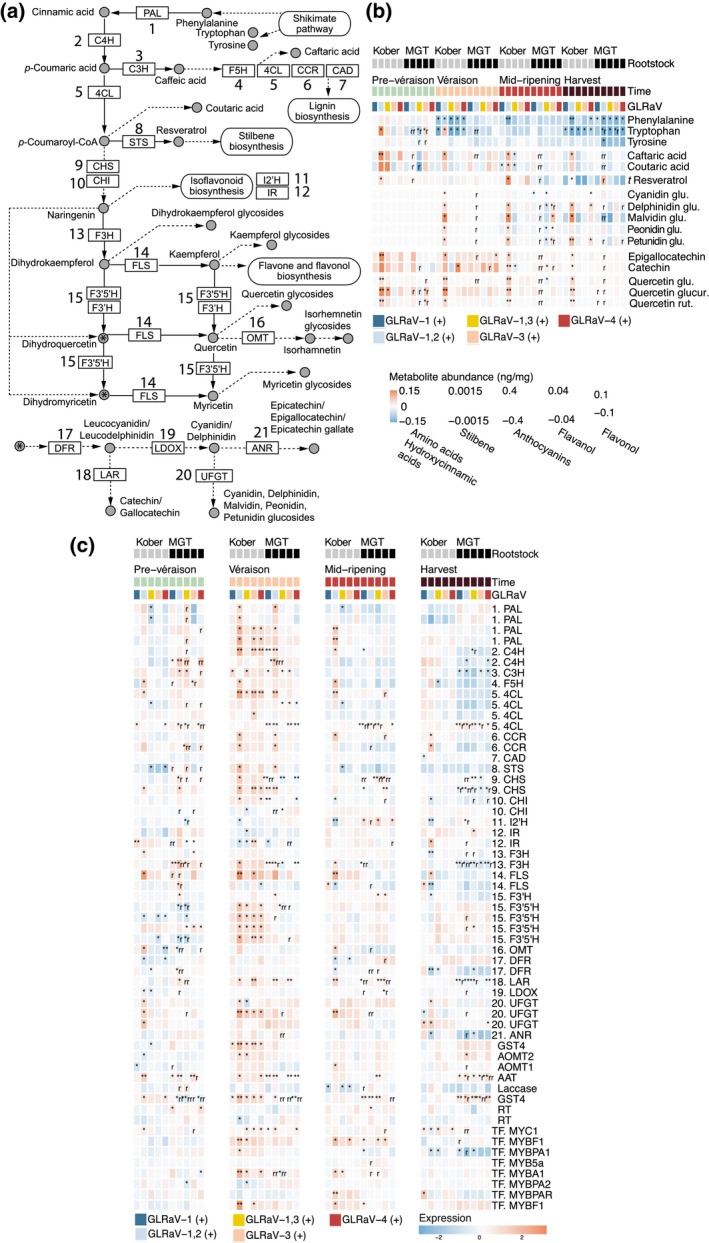
Differentially expressed genes and selected metabolites produced in the shikimate, phenylpropanoid, and flavonoid pathways. (a) Pathway diagram. (b) Metabolite abundances and (c) related biosynthetic and regulatory gene expression relative to GLRaV (−) in identical rootstock and at the same developmental stage. Notation requires the direction (up/downregulation) of the effect to be consistent in both years, even if a significant change occurred in only a single year. Glycosides are abbreviated: 3‐O‐glucoside [glu], 3‐O‐glucuronide [glucur], and 3‐O‐rutinoside [rut]. *Differentially expressed/abundant in 1 year; **differentially expressed/abundant in both years. One or two letters “r” indicate that the effect of a particular GLRaV infection differs between rootstocks at the same developmental stage. Kober, Kober 5BB rootstock; MGT, MGT 101‐14 rootstock. PAL, phenylalanine ammonia‐lyase; C4H, trans‐cinnamate 4‐monooxygenase; C3H, p‐coumarate 3‐hydroxylase; F5H, ferulate‐5‐hydroxylase; 4CL, 4‐coumaroyl‐CoA ligase; CCR, cinnamoyl‐CoA reductase; CAD, cinnamyl alcohol dehydrogenase; STS, stilbene synthase; CHS, chalcone synthase; CHI, chalcone isomerase; I2′H, isoflavone 2′‐hydroxylase; IR, isoflavone reductase; F3H, flavonone 3‐hydroxylase; FLS, flavonol synthase; F3′H, flavonoid 3′‐monooxygenase; F3′5′H, flavonoid 3′,5′‐hydroxylase; OMT, O‐methyltransferase; DFR, dihydroflavanol 4‐reductase; LAR, leucoanthocyanidin reductase; LDOX, leucoanthocyanidin dioxgenase; UFGT, UDP‐glucose:anthocyanidin/flavonoid 3‐O‐glucosyltransferase; ANR, anthocyanidin reductase; GST4, glutathione S‐transferase 4; AOMT, anthocyanin O‐methyltransferase; AAT, anthocyanin acyl‐transferase; RT, UDP‐rhamnose:rhamnosyltransferase. The pathway annotation is based on KEGG pathways (www.genome.jp/kegg/pathway.html, accessed 13 February 2021) and Blanco‐Ulate et al. (2017)
3. DISCUSSION
GLRaVs affect viticulture on nearly every continent (Akbaş et al., 2007; Charles et al., 2009; Fiore et al., 2008; Golino, Weber, et al., 2008; Habili & Nutter, 1997; Jooste et al., 2015; Mahfoudhi et al., 2008) and can a have considerable economic impact on a major crop. The presence and severity of symptoms in GLRaV‐infected grapevines is influenced by host genotype, rootstock, which GLRaV is present, and environmental conditions. In addition to the assembly and annotation of the Cabernet Franc genome, a valuable resource that might be applied for the larger purpose of understanding grapevine genomic diversity and evolution, the dedicated experimental vineyard used in this study is a tremendous asset for the study of GLRaV infections over time in a common environment. This work identified responses to GLRaVs in grape berries during ripening, including those that are conserved across experimental conditions and responses that differ based on the rootstock present. We propose which hormones and signalling pathways at least partially govern the responses observed and likely influence leafroll disease symptoms.
The effects of dual infections, particularly GLRaV‐1,2 (+), were most distinctive. All of the leafroll viruses selected for this study belong to the Closteroviridae family and all but one belong to the Ampelovirus genus; GLRaV‐2 belongs to the genus Closterovirus. All of the GLRaVs used in this study contain a conserved replication gene block (RGB) but are diverse outside of the RGB (Naidu et al., 2015). In addition to host genotype and environment, the sequences downstream of the RGB may account for the disparities in responses observed between infection types. These sequences encode a quintuple gene block and/or viral suppressors of host RNA silencing (VSRs; Naidu et al., 2015). Some VSRs are characterized (Gouveia & Nolasco, 2012), but most are not. Further research might determine their specific effects and relationship to host cellular machinery (Chapman et al., 2004; Wu et al., 2010).
Changes in the expression of NBS‐LRR genes were among the conserved responses to GLRaVs and were the single largest category of genes among them. NBS‐LRR genes confer resistance to powdery and downy mildew in grapevine (Riaz et al., 2011; Zini et al., 2019). The abundance of these genes varies among Vitis species and are particularly dense at resistance loci (Cochetel et al., 2021). The HR to viruses is mediated by resistance (R) genes. SA level and pathogenesis‐related gene expression increase for systemic acquired resistance. HR is a means of prohibiting pathogen spread and can confer resistance when a corresponding dominant avirulence protein is produced by the pathogen (Balint‐Kurti, 2019; Moffett et al., 2002). However, GLRaV infections are systemic and persist over time, and SA does not seem to play a preeminent role in the response to GLRaV infections. In a previous study of GLRaV‐3 infections in Cabernet Sauvignon and Carmenère, the authors also remarked on the induction of expression of defence genes but their inability to impede systemic infection (Espinoza, Vega, et al., 2007). Both SA and ABA can participate in the response to viruses, though considerably less is understood about the role of ABA (Alazem & Lin, 2015; Koornneef & Pieterse, 2008; Kunkel & Brooks, 2002) and its relationship to NBS‐LRRs. Notably, however, ABA deficiency is associated with an increase in R gene efficacy in incompatible interactions with Pseudomonas syringae and in a manner independent of SA (Mang et al., 2012).
Hormones have been implicated in mediating defence‐ and development‐related networks and are over‐represented at network hubs (Amrine et al., 2015; Jiang et al., 2015; Müller & Munné‐Bosch, 2015; Vandereyken et al., 2018). Hormones like ABA, SA, and JA act as important signalling molecules during ripening and defence. The pathways engaged under stress are often tailored to particular pathogens. This entails coordination between hormone pathways (Gao et al., 2011; Vos et al., 2015). Interestingly, the effects of GLRaVs on gene expression in several hormone signalling pathways differed between rootstocks. A subsequent effort could be made to measure the abundances of hormones not quantified here, like cytokinins, GAs, and ethylene. Of the hormones considered in this study, however, the abundance of ABA and ABA‐GE tended to increase in GLRaV (+) and this was influenced by rootstock. ABA can antagonize SA and JA signalling pathways and suppress ROS signalling (Alazem & Lin, 2015). WRKY transcription factors regulate and/or are regulated by ABA, SA, and JA (Gao et al., 2011; Jiang & Deyholos, 2009; Li, 2004; Liu et al., 2015; Xin et al., 2016). SA and ABA both can interact with RNAi, which is a fundamental component of antiviral defence. AGO1 expression is positively correlated with ABA levels and the expression of miR168a, which regulates AGO1, and contains ABREs in its promoter (Alazem & Lin, 2015). Levels of ABA and ABA‐GE increase in tobacco mosaic virus‐infected leaves. One way in which ABA might aid plant defence is by increasing callose deposition to impair virus movement (Alazem & Lin, 2015); a gene encoding callose synthase is upregulated in the leaves of grapevine virus B‐infected plants (Chitarra et al., 2018).
Relatively more is known about ABA’s function as a ripening promoter (Koyama et al., 2010; Wheeler et al., 2009), in response to drought stress (Cochetel et al., ; Deluc et al., 2009), and in transmitting long‐distance signals from roots to aerial organs and vice versa (Ferrandino & Lovisolo, 2014; Manzi et al., 2015). In a study of the impact of GLRaV‐3 infection, drought stress, and a combination of both on grapevine plantlets in vitro, individual stresses both induced increases in ABA levels (Cui et al., 2016). Drought stress increases ABA levels and induces the flavonoid pathway in both tea plants (Gai et al., 2020) and grapes (Deluc et al., 2009). Our findings, in which ABA abundance tends to increase in GLRaV (+), are different than that observed for red blotch virus‐infected berries, in which ABA abundance and NCED expression decrease in infected fruits (Blanco‐Ulate et al., 2017).
The results of our analysis of metabolites associated with the phenylpropanoid and flavonoid pathways are mixed in their consistency with previous work. Though nonsignificant decreases in anthocyanin levels were observed, anthocyanin levels significantly increased in several GLRaV (+) conditions, albeit usually in individual years. These findings differed from others; some observed significant decreases in anthocyanin levels in fruits from GLRaV‐infected plants (Lee & Martin, 2009; Vega et al., 2011) and others observed no significant changes in anthocyanin at harvest (Alabi et al., 2016; Endeshaw et al., 2014). In agreement with the results by Vega et al. (2011), flavonol levels were elevated in GLRaV (+) and the largest differences versus GLRaV (−) occurred at the first two stages, FLS expression was downregulated in GLRaV (+) at harvest, and CHS and MYBPA1 were upregulated at véraison and generally downregulated at harvest.
In the present study, changes in the abundance of ABA and related metabolites distinguished the effects of GLRaVs between rootstocks. The parentage of the two rootstocks used in this study, Kober 5BB and MGT 101‐14, includes Vitis riparia. The other parents of Kober 5BB and MGT 101‐14 are Vitis berlandieri and Vitis rupestris, respectively. These rootstocks were developed at different times. MGT 101‐14 originated in France in 1882 and Kober 5BB originated in Austria in 1930 (https://fps.ucdavis.edu/). Rootstocks are chosen for the advantages they confer to the scion given a particular set of circumstances, often having to do with resistance to Phylloxera, nematodes, scion vigour, soil type, and abiotic stress tolerance (Corso & Bonghi, 2014; Tramontini et al., 2013; Warschefsky et al., 2016). V. riparia, V. rupestris, and V. berlandieri are asymptomatic hosts of GLRaVs (Naidu et al., 2014). Yet, the particularly severe response to GLRaV‐1,2 (+) was not entirely unexpected in Kober 5BB‐grafted vines (Alkowni et al., 2011; Golino, Wolpert, et al., 2008; Uyemoto et al., 2001). It would be interesting to determine (a) whether differences in viral titre exist between Kober 5BB, MGT 101‐14, and Cabernet Franc and between different infection conditions and (b) whether such differences, if they exist, influence the severity of leafroll disease. Differences in wood abnormalities given different rootstocks (including Kober 5BB) have been observed for particular isolates causing grapevine rugose wood disease (Credi, 1997).
Notably, nutritional deficiencies in phosphorus, magnesium, and potassium produce symptoms that resemble those typically observed in GLRaV‐infected plants (Gohil et al., 2016). Magnesium deficiency tolerance (Livigni et al., 2019), the impact of phosphorus deficiency on canopy growth (Grant & Matthews, 1996), and potassium uptake and channels are influenced by rootstock (Wolpert et al., 2005). Potassium uptake, channel activity, and related gene expression are also regulated by ABA (Blatt, 2000; Köhler et al., 2003; Rogiers et al., 2017; Song et al., 2016), and the application of ABA to tomato roots by drip irrigation affects fruit mineral composition (Barickman et al., 2019). Furthermore, elevated levels of potassium are observed in leafroll virus‐infected Burger and Sultana fruits (Hale & Woodham, 1979; Kliewer & Lider, 1976) and in leaf petioles but potassium levels are lower in leaf blades (Cook & Goheen, 1961). Perhaps potassium deficient and GLRaV (+) phenotypes are similarly governed by ABA and fine‐tuned by rootstocks. If some portion of scion ABA originates in roots and/or if rootstock can influence scion ABA levels and signalling genes, as observed here and by others (Chitarra et al., 2017), then perhaps this partially accounts for the variation in response observed between rootstocks. This experiment did not include a comprehensive survey of phytohormones, which would be beneficial, but ABA’s function in root–shoot communication, its role in ripening, and the results here make it a good candidate around which to study the basis of leafroll disease symptom variability going forward. In addition, the transport of RNAs across the graft junction may perform some function that affects scion disease severity, but this remains to be seen in the particular case of GLRaV (Chitarra et al., 2017).
Together, these data support several conclusions. (a) The majority of genes differentially expressed as a consequence of infection or between GLRaV (+) plants with different rootstocks were year‐specific. A small subset of effects was consistently observed across experimental conditions and in both years. These shared changes in expression involved genes associated with pathogen detection, ABA signalling and transport, ROS‐related signalling, cytoskeleton remodelling, vesicle trafficking, phenylpropanoid metabolism, sugar transport and conjugation, and leucine biosynthesis. (b) The impacts of GLRaV‐1,2 dual infection on Kober 5BB‐grafted vines were the most distinctive and severe. Though there was variation between GLRaV infections observed, only the effects of GLRaV‐1,2 were distinguishable overall from those of other infections. (c) The particular effects of GLRaVs in plants grafted to different rootstocks were distinguishable overall at every developmental stage. ABA‐related variables were among those that best distinguished the responses to GLRaVs in different rootstock conditions. This included the abundance of ABA, the abundance of ABA‐GE, and the expression of genes associated with ABA and other hormone signalling pathways. Finally, this work alone is insufficient to recommend the use of one rootstock or another, but the disparity in sensitivity and symptom severity observed in berries from Cabernet Franc vines grafted to different rootstocks suggests that rootstock selection should be further explored as a strategy to mitigate some of the negative consequences of leafroll virus infections, should vectors of the virus encroach upon a vineyard.
4. EXPERIMENTAL PROCEDURES
4.1. Vineyard establishment
The experimental vineyard used in this study was established in 2010 and consists of Cabernet Franc clone 04 (UC Davis, Foundation Plant Services, https://fps.ucdavis.edu/fgrdetails.cfm?varietyid=355; accessed 2 March 2021) grapevines grafted on different rootstocks and infected with zero, individual, or pairs of GLRaVs. The rootstock portion of these plants was inoculated with chip buds carrying each virus in 2009 (Rowhani et al., 2015). All rootstocks and Cabernet Franc scions were tested for grapevine pathogens. Total nucleic acid (TNA) extracts were prepared from all rootstocks and Cabernet Franc scions as described by Al Rwahnih et al. (2017). Extracted TNA samples were analysed by reverse transcription quantitative PCR (RT‐qPCR) using TaqMan probes on the QuantStudio 6 Flex Real‐Time PCR System (Thermo Fisher Scientific) as described previously (Klaassen et al., 2011; Osman et al., 2008; Rwahnih et al., 2017). The samples were screened for the following pathogens: GLRaV‐1, GLRaV‐3, GLRaV‐4 (plus strains 5, 6, 9, Pr, and Car; genus Ampelovirus); GLRaV‐2 (plus strain 2RG; genus Closterovirus); GLRaV‐7 (genus Velarivirus); grapevine fleck virus (genus Maculavirus); grapevine rupestris vein feathering virus (genus Marafivirus); grapevine fanleaf virus, tobacco ringspot virus, and tomato ringspot virus (genus Nepovirus); grapevine virus A, grapevine virus B, grapevine virus D, grapevine virus E, and grapevine virus F (genus Vitivirus); grapevine red blotch virus (genus Grablovirus); and grapevine rupestris stem pitting‐associated virus (genus Foveavirus), phytoplasmas, and Xylella fastidiosa.
In autumn 2008, Cabernet Franc grapevines were bench‐grafted onto rootstocks, including MGT 101‐14 and Kober 5BB. These plants were subsequently grown in a greenhouse. Between 2009 and 2011, the rootstock portions of these plants were inoculated with two chip buds from single leafroll‐infected plants. Infected plants used for chip buds were reconfirmed by RT‐qPCR. Plants infected with a single species of GLRaV received two identical chip buds. Plants infected with two species of GLRaVs also received two chip buds, each carrying a single virus. Plants infected with GLRaV‐1, GLRaV‐2, and/or GLRaV‐3 were inoculated with two or more isolates of each species of GLRaV. The inoculated plants were kept in a greenhouse for approximately 1 month, acclimatized, and then planted in the field. Healthy controls included nonchip budded plants and plants chip budded from a healthy source.
The vines were planted in a randomized complete block design, with 7 feet (2.1 m) between vines and 9 feet (2.7 m) between rows. One group of five vines was planted per rootstock × infection condition in each of three blocks. Healthy vines were distributed throughout each block to monitor the spread of viruses, and experimental vines were sampled yearly to test and reaffirm the vines’ infection status. A buffer zone of healthy vines was planted as a barrier between the leafroll vineyard and other vineyards in the area. Vines were trained with a bilateral cordon and spur pruned.
4.2. Cabernet Franc genome sequencing and assembly
High‐quality genomic DNA was isolated from grape leaves using the method described in Chin et al. (2016). SMRTbell libraries were prepared for Cabernet Franc clone 04 as described by Massonnet et al. (2020). Final libraries were evaluated for quantity and quality using a Bioanalyzer 2100 (Agilent Technologies) and sequenced on a PacBio RS II (DNA Technology Core Facility, University of California, Davis).
De novo assembly of Cabernet Franc clone 04 (UC Davis, Foundation Plant Services, https://fps.ucdavis.edu/fgrdetails.cfm?varietyid=355; accessed 2 March 2021) was performed using FALCON‐Unzip (v. 1.7.7; Chin et al., 2016) as described in Minio et al. (2019a). Repetitive sequences were masked before and after read error correction using the TANmask and REPmask modules in Damasker (Myers, 2014). Contigs were polished with Quiver (Pacific Biosciences, bundled with FALCON‐Unzip v. 1.7.7). The primary assembly was scaffolded to reduce sequence fragmentation. First, primary contigs were scaffolded with SSPACE‐LongRead v. 1.1; (Boetzer & Pirovano, 2014). Junctions supported by at least 20 reads (“‐l 20”) were allowed. Hi‐C data and the proprietary HiRise software (v. 1.3.0‐1233267a1cde) were used for hybrid scaffolding. A Dovetail Hi‐C library was prepared by Dovetail Genomics (Scotts Valley, CA, USA) as described in Lieberman‐Aiden et al. (2009) and sequenced on an Illumina platform, generating 2 × 150‐bp paired‐end reads. The repeat and gene annotation were performed as reported in Vondras et al. (2019). Briefly, RepeatMasker (v. open‐4.0.6; Smit et al., 2015) and a custom V. vinifera repeat library (Minio et al., 2019b) were applied to identify repetitive elements in the genome. Publicly available data sets were used as evidence for gene prediction. Transcriptional evidence included Vitis expressed sequence tags, Cabernet Sauvignon corrected Iso‐Seq reads (Minio et al., 2019b), Tannat (Da Silva et al., 2013), Corvina (Venturini et al., 2013), and Cabernet Sauvignon transcriptomes (Massonnet et al., 2020), and previously published RNA‐Seq data (PRJNA260535). Swissprot Viridiplantae data and Vitis data were used as experimental evidence. Each RNA‐Seq sample was trimmed with Trimmomatic v. 0.36 (Bolger et al., 2014), assembled with Stringtie v. 1.3.3 (Pertea et al., 2015), and mapped onto the genome using Exonerate v. 2.2.0 (transcripts and proteins; Slater & Birney, 2005) and PASA v. 2.1.0 (transcripts; Haas et al., 2003). Alignments and ab initio predictions generated with SNAP v. 2006‐07‐28 (Korf, 2004), Augustus v. 3.0.3 (Stanke et al., 2006), and GeneMark‐ES v. 4.32 (Lomsadze et al., 2005) were used as input for EVidenceModeler v. 1.1.1 (Haas et al., 2008). EVidenceModeler was used to identify consensus gene structures. A functional annotation was obtained using the RefSeq plant protein database (ftp://ftp.ncbi.nlm.nih.gov/refseq, retrieved 17 January 2017; Jones et al., 2014) as in Minio et al. (2019a).
4.3. Sampling and sample preparation
Berries from Cabernet Franc grapevines grafted to Kober 5BB or MGT 101‐14 and infected with GLRaV‐1 (GLRaV‐1 [+]), GLRaV‐3 (GLRaV‐3 [+]), GLRaV‐4 (strain 5; GLRaV‐4 [+]), GLRaV‐1 and GLRaV‐2 (GLRaV‐1,2 [+]), or GLRaV‐1 and GLRaV‐3 (GLRaV‐1,3 [+]) were sampled during ripening in 2017 and 2018. In both years, fruits were sampled at prevéraison, véraison, mid‐ripening, and at commercial harvest. These stages correspond to modified Eichhorn–Lorenz (Coombe, 1995) stages 34 (green berries begin to soften and sugar content [unit, °Brix] starts increasing), 35 (berries begin to change colour and enlarge, c.50% of berries within clusters show colour transition), 36/37 (berries with intermediate sugar content/berries are not quite ripe), and 38 (berries are harvest‐ripe). Fruits in 2018 were sampled at developmental stages comparable to 2017 as determined by TSS. In 2017, berries were sampled on 7 July, 31 July, 14 August, and 31 August. In 2018, berries were sampled on 28 June, 30 July, 13 August, and 30 August.
On each sampling date, six biological replicates were taken, with berries from one plant constituting one biological replicate. Two biological replicates per condition were sampled from each of three blocks. There were two exceptions. For Kober 5BB‐grafted GLRaV‐1,2 (+), three biological replicates were drawn from each of two plants in one block. For Kober 5BB‐grafted GLRaV‐3 (+), two of the six biological replicates were drawn from one plant. Approximately 20 berries were sampled per plant, with equal numbers of berries sampled from each side of the vine. Samples were then temporarily cooled on ice. Next, berries were rinsed with deionized water, deseeded, and snap‐frozen in liquid nitrogen. Berries were stored at −80 °C until being crushed into a fine powder while frozen using a mechanical mill. TSS were measured in technical triplicate using a digital refractometer. Four of six biological replicates, a total of 192 samples per year, were used for RNA‐Seq and liquid chromatography coupled with mass spectrometry (LC‐MS).
4.4. RNA extraction and sequencing library preparation
Total RNA was extracted from 2 g of finely ground berry pericarp tissue as previously described (Blanco‐Ulate et al., 2013). RNA purity was evaluated with a NanoDrop 2000 spectrophotometer (Thermo Scientific), quantity with a Qubit 2.0 fluorometer (Life Technologies), and integrity by electrophoresis. RNA‐Seq libraries were prepared using the Illumina TruSeq RNA sample preparation kit v. 2 (Illumina) and barcoded individually following the manufacturer's protocol. Final libraries were evaluated for quantity and quality with the High Sensitivity chip in a Bioanalyzer 2100 (Agilent Technologies). Libraries were sequenced as 100‐bp, single‐end reads, using an Illumina HiSeq 4000 sequencer (DNA Technology Core Facility, University of California, Davis), producing an average of 18.07 ± 4.56 million reads per sample in 2017 and 14.69 ± 2.11 million reads per sample in 2018.
4.5. RNA‐Seq data analysis
RNA‐Seq reads were trimmed using Trimmomatic v. 0.36 (Bolger et al., 2014) and the following settings: leading: 2, trailing: 2, sliding window: 4:20, min length: 70. Reads were mapped onto the primary assembly of the Cabernet Franc genome using HISAT2 (‐k 1; v. 2.0.5; Kim et al., 2015) and counts were generated using htseq‐count with default parameters (v. 0.9.0; Anders et al., 2015).
All subsequent analyses were done using R (R Core Team, 2020) in the R Studio environment (R Studio Team, 2020). Data normalization and differential expression analyses were performed using DESeq2 v. 1.24.0 (Love et al., 2014). A variance‐stabilizing transformation (VST) was applied to expression data using DESeq2. VST data were centred in each GLRaV (+) condition relative to the mean expression per gene in GLRaV (−) given the same time, rootstock, and year.
Centred data were used for MFA with the FactoMineR R package (Le et al., 2008). Genes were included in the MFA if (a) they were differentially expressed versus GLRaV (−), and/or the effects of an infection differed between rootstocks (b) in at least 1 year (p < .05), and (c) the direction of the effect relative to GLRaV (−) was consistent in both years, even if a significant effect was only observed in 1 year. An MFA was repeated for each developmental stage separately. All hormone and metabolite data were included.
Statistical over‐representation tests were done using the clusterProfiler R package (Yu et al., 2012) and VitisNet functional categories (Grimplet et al., 2009). To make use of the VitisNet functional annotations, Cabernet Franc genes were used to query Pinot Noir PN40024 sequences with BLASTp (Altschul et al., 1990). The best hits, with no less than 80% reciprocal identity and coverage, were retained.
A curated list of ABA biosynthesis and signalling genes annotated in PN40024 was retrieved from Pilati et al., 2017 (Table S1). As described above, Cabernet Franc genes were used to query Pinot Noir PN40024 sequences. All target–query pairs had no less than reciprocal 97% identity and 86% reciprocal coverage (median coverage, 99%).
4.6. Hormone extraction and LC‐MS/MS
Approximately 50 mg (mean 50.86 ± 3.33 mg) of berry powder was weighed for the extraction and quantitation of ABA, SA, and JA. Exact weights were recorded to later calculate the exact amount of each of these analytes per milligram of fresh tissue. The same samples used for RNA‐Seq were used for this analysis. Four biological replicates were used. Extractions and analyses were randomized and performed in technical duplicate.
Hormones were extracted following a method described by Pan et al. (2010) with a few modifications. Samples were subjected to 500 ml 2‐propanol:H2O:HCl (2:1:0.002) and spiked with 50 ng of d6 ABA, d5 JA, and d4 SA (CDN Isotopes). Samples were vortexed, placed in an ultrasonic ice bath for 30 min, washed with dichloromethane, and centrifuged at 16,100 × g for 5 min. Nine‐hundred microlitres of the lower phase was taken and dried. Samples were reconstituted in 100 µl 15% methanol and stored at −20 °C until analysis. A detailed description of the chromatographic separation, mass spectrometry data acquisition methodology, and multiple reaction monitoring (MRM) transitions is included in Table S2.
4.7. Water‐soluble metabolite extraction and LC‐MS
The same samples used for RNA‐Seq and hormone analyses were used to measure water‐soluble metabolites by LC‐MS. Approximately 200 mg (mean 202.12 ± 4.87 mg) of frozen berry powder was weighed. Extractions and analyses were randomized and performed in technical duplicate. Hormones were extracted in 1 ml 1% HCl in HPLC‐grade water (prepared in‐house, final resistance 18 MΩ, 0.2 µm filtered) and spiked with salicin (Sigma Aldrich) as an internal standard (50 µg/L final concentration). The samples were vortexed, placed in an ultrasonic ice bath for 30 min, and centrifuged. The supernatants were collected and frozen at −80 °C until analysis. A detailed description of the chromatographic separation, mass spectrometry data acquisition methodology, and MRM transitions is included in Table S2.
4.8. Data visualization
Figures depicting quantitative data were built using the following R packages: ggplot2 (Wickham, 2016), hrbrthemes (Rudis, 2020), wesanderson (Ram & Wickham, 2018), pheatmap (Kolde, 2019), and UpSetR (Gehlenborg, 2019).
Supporting information
FIGURE S1 Malic acid, moisture content, pH, titratable acidity, weight per berry, yeast assimilable nitrogen (YAN), and total anthocyanins in 2015 and 2016. For most measures in both years, 3 ≤ n ≤ 9. For total anthocyanin and moisture content in 2015, 2 ≤ n ≤ 9. For total soluble solids before harvest in 2017 and 2018, n = 6. Differences between groups are indicated with nonoverlapping letters (Tukey HSD, p < .05)
FIGURE S2 Pearson correlation between RNA‐Seq samples
FIGURE S3 Barplots showing the number of differentially expressed (p < .05) genes up‐ and downregulated in MGT 101‐14‐grafted plants versus Kober 5BB‐grafted plants for each GLRaV condition in each year and in both years
FIGURE S4 Abundance of abscisic acid (ABA), ABA glucose ester (ABA‐GE), and xanthoxin in 2017 and 2018 at véraison, mid‐ripening, and harvest. Abundance of SA and JA in 2017 and 2018 at all four developmental stages. Analysis of variance tables for all five hormones and hormone‐related metabolites. Groups with nonoverlapping letters are significantly different (Tukey HSD, p < .05)
FIGURE S5 Rootstock‐differentiating genes
FIGURE S6 Percentage of variation explained by each of the first five multiple factor analysis components at each developmental stage
FIGURE S7 Distribution of centred expression for genes, per functional category, correlated to the same rootstock‐differentiating multiple factor analysis dimensions as hormones and hormone‐related metabolites. Results of Kolmogorov–Smirnov tests are shown
FIGURE S8 Centred expression of VITVvi_vCabFran04_v1_P478.ver1.0.g430590, histone H1, in berries from GLRaV (+) plants grafted to two different rootstocks
FIGURE S9 All metabolite products measured that are associated with the shikimate, phenylpropanoid, and flavonoid pathways. *Differentially expressed in 1 year; **differentially expressed in both years. One or two letters “r” indicate that the effect of a particular GLRaV infection differs between rootstocks at the same developmental stage in 1 or 2 years
TABLE S1 Curated list of abscisic acid‐related genes Pilati et al. (2017)
TABLE S2 Hormone and metabolite retention time, mass information, and additional method‐related information
TABLE S3 Cabernet Franc assembly and annotation statistics
TABLE S4 Conserved responses to GLRaVs depicted in Figure 3b
TABLE S5 Number of multiple factor analysis variables positively and negatively correlated with each rootstock‐differentiating multiple factor analysis dimension
FILE S1 Additional description of hormone signalling genes in Figure 8
ACKNOWLEDGEMENTS
This work was funded by the Pierce's Disease and Glassy‐winged Sharpshooter Research Board of the California Department of Food and Agriculture (project 17‐0417‐000‐SA). D.C. was partially supported by the Louis P. Martini Endowment in Viticulture. The authors thank all past and present Cantu laboratory members who assisted with the laborious process of sampling, deseeding, crushing, and weighing the tissue used in this study, including Abraham Morales‐Cruz, Eric Tran, Jerry Lin, Barbara Blanco‐Ulate, and Lucero K. Espinoza. The authors also thank Vicki A. Klaassen and Lori Leong from Foundation Plant Services for their help with virus testing.
Vondras AM, Lerno L, Massonnet M, et al. Rootstock influences the effect of grapevine leafroll‐associated viruses on berry development and metabolism via abscisic acid signalling. Mol Plant Pathol. 2021;22:984–1005. 10.1111/mpp.13077
DATA AVAILABILITY STATEMENT
The DNA (BioProject PRJNA701939) and RNA (BioProject PRJNA701940) sequencing data that support these findings are available at the National Center for Biotechnology Information (NCBI). A Cabernet Franc genome browser and on‐line BLAST tool are available at http://www.grapegenomics.com. The assembly, annotations, and raw RNA‐Seq counts per library and per gene are available at Zenodo (http://doi.org/10.5281/zenodo.4555977).
REFERENCES
- Akbaş, B. , Kunter, B. & Ilhan, D. (2007) Occurrence and distribution of grapevine leafroll‐associated viruses 1, 2, 3 and 7 in Turkey. Journal of Phytopathology, 155, 122–124. [Google Scholar]
- Alabi, O.J. , Casassa, L.F. , Gutha, L.R. , Larsen, R.C. , Henick‐Kling, T. , Harbertson, J.F. et al. (2016) Impacts of grapevine leafroll disease on fruit yield and grape and wine chemistry in a wine grape (Vitis vinifera L.) cultivar. Journal of Plant Pathology, 11, e0149666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alazem, M. & Lin, N.‐S. (2015) Roles of plant hormones in the regulation of host–virus interactions. Molecular Plant Pathology, 16, 529–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkowni, R. , Zhang, Y.‐P. , Rowhani, A. , Uyemoto, J.K. & Minafra, A. (2011) Biological, molecular, and serological studies of a novel strain of grapevine leafroll‐associated virus 2. Virus Genes, 43, 102–110. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F. , Gish, W. , Miller, W. , Myers, E.W. & Lipman, D.J. (1990) Basic local alignment search tool. Journal of Molecular Biology, 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Amrine, K.C.H. , Blanco‐Ulate, B. & Cantu, D. (2015) Discovery of core biotic stress responsive genes in Arabidopsis by weighted gene co‐expression network analysis. PLoS One, 10, e0118731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders, S. , Pyl, P.T. & Huber, W. (2015) HTSeq‐A Python framework to work with high‐throughput sequencing data. Bioinformatics, 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atallah, S.S. , Gomez, M.I. , Fuchs, M.F. & Martinson, T.E. (2012) Economic impact of grapevine leafroll disease on Vitis vinifera cv. Cabernet Franc in Finger Lakes vineyards of New York. American Journal of Enology and Viticulture, 63, 73–79. [Google Scholar]
- Balint‐Kurti, P. (2019) The plant hypersensitive response: concepts, control and consequences. Molecular Plant Pathology, 20, 1163–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barickman, T.C. , Kopsell, D.A. & Sams, C.E. (2019) Applications of abscisic acid and increasing concentrations of calcium affect the partitioning of mineral nutrients between tomato leaf and fruit tissue. Horticulturae, 5, 49. [Google Scholar]
- Bertamini, M. , Muthuchelian, K. & Nedunchezhian, N. (2004) Effect of grapevine leafroll on the photosynthesis of field grown grapevine plants (Vitis vinifera L. cv. Lagrein). Journal of Phytopathology, 152, 145–152. [Google Scholar]
- Bester, R. , Burger, J.T. & Maree, H.J. (2017) Differential expression of miRNAs and associated gene targets in grapevine leafroll‐associated virus 3‐infected plants. Archives of Virology, 162, 987–996. [DOI] [PubMed] [Google Scholar]
- Blanco‐Ulate, B. , Hopfer, H. , Figueroa‐Balderas, R. , Ye, Z. , Rivero, R.M. , Albacete, A. et al. (2017) Red blotch disease alters grape berry development and metabolism by interfering with the transcriptional and hormonal regulation of ripening. Journal of Experimental Botany, 68, 1225–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco‐Ulate, B. , Vincenti, E. , Powell, A.L.T. & Cantu, D. (2013) Tomato transcriptome and mutant analyses suggest a role for plant stress hormones in the interaction between fruit and Botrytis cinerea . Frontiers in Plant Science, 4, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blatt, M.R. (2000) Ca2+ signalling and control of guard‐cell volume in stomatal movements. Current Opinion in Plant Biology, 3, 196–204. [PubMed] [Google Scholar]
- Boetzer, M. & Pirovano, W. (2014) SSPACE‐LongRead: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinformatics, 15, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger, A.M. , Lohse, M. & Usadel, B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabaleiro, C. , Segura, A. & García‐Berrios, J.J. (1999) Effects of grapevine leafroll‐associated virus 3 on the physiology and must of Vitis vinifera L. cv. Albariño following contamination in the field. American Journal of Enology and Viticulture, 50, 40–44. [Google Scholar]
- Chapman, E.J. , Prokhnevsky, A.I. , Gopinath, K. , Dolja, V.V. & Carrington, J.C. (2004) Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes and Development, 18, 1179–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charles, J.G. , Froud, K.J. , van den Brink, R. & Allan, D.J. (2009) Mealybugs and the spread of grapevine leafroll‐associated virus 3 (GLRaV‐3) in a New Zealand vineyard. Australasian Plant Pathology, 38, 576–583. [Google Scholar]
- Chin, C.‐S. , Peluso, P. , Sedlazeck, F.J. , Nattestad, M. , Concepcion, G.T. , Clum, A. et al. (2016) Phased diploid genome assembly with single‐molecule real‐time sequencing. Nature Methods, 13, 1050–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitarra, W. , Cuozzo, D. , Ferrandino, A. , Secchi, F. , Palmano, S. , Perrone, I. et al. (2018) Dissecting interplays between Vitis vinifera L. and grapevine virus B (GVB) under field conditions. Molecular Plant Pathology, 19, 2651–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitarra, W. , Perrone, I. , Avanzato, C.G. , Minio, A. , Boccacci, P. , Santini, D. et al. (2017) Grapevine grafting: scion transcript profiling and defense‐related metabolites induced by rootstocks. Frontiers in Plant Science, 8, 654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochetel, N. , Ghan, R. , Toups, H.S. , Degu, A. , Tillett, R.L. , Schlauch, K.A. , et al. (2020) Drought tolerance of the grapevine, Vitis champinii cv. Ramsey, is associated with higher photosynthesis and greater transcriptomic responsiveness of abscisic acid biosynthesis and signaling. BMC Plant Biology, 20, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochetel, N. , Vondras, A.M. , Minio, A. , Massonnet, M. , Figueroa‐Balderas, R. & Cantu, D. (2021) Diploid chromosome‐scale assembly of the Muscadinia rotundifolia genome supports chromosome fusion and disease resistance gene expansion during Vitis and Muscadinia divergence. G3 Genes Genomes Genetics, 11, jkab033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook, J.A. & Goheen, A.C. (1961) The effect of a virus disease, leafroll, on the mineral composition of grape tissue and a comparison of leafroll and potassium deficiency symptoms. American Institute of Biological Sciences, 8, 338–354. [Google Scholar]
- Coombe, B.G. (1995) Growth stages of the grapevine: adoption of a system for identifying grapevine growth stages. Australian Journal of Grape and Wine Research, 1, 100–110. [Google Scholar]
- Corso, M. & Bonghi, C. (2014) Grapevine rootstock effects on abiotic stress tolerance. Plant Science Today, 1, 108–113. [Google Scholar]
- Credi, R. (1997) Characterization of grapevine rugose wood disease sources from Italy. Plant Disease, 81, 1288–1292. [DOI] [PubMed] [Google Scholar]
- Cui, Z.‐H. , Bi, W.‐L. , Hao, X.‐Y. , Xu, Y. , Li, P.‐M. , Walker, M.A. et al. (2016) Responses of in vitro‐grown plantlets (Vitis vinifera) to grapevine leafroll‐associated virus‐3 and PEG‐induced drought stress. Frontiers in Physiology, 7, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Silva, C. , Zamperin, G. , Ferrarini, A. , Minio, A. , Dal Molin, A. , Venturini, L. et al. (2013) The high polyphenol content of grapevine cultivar Tannat berries is conferred primarily by genes that are not shared with the reference genome. The Plant Cell, 25, 4777–4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluc, L.G. , Quilici, D.R. , Decendit, A. , Grimplet, J. , Wheatley, M.D. , Schlauch, K.A. et al. (2009) Water deficit alters differentially metabolic pathways affecting important flavor and quality traits in grape berries of Cabernet Sauvignon and Chardonnay. BMC Genomics, 10, 212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeYoung, B.J. & Innes, R.W. (2006) Plant NBS‐LRR proteins in pathogen sensing and host defense. Nature Immunology, 7, 1243–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endeshaw, S.T. , Sabbatini, P. , Romanazzi, G. , Schilder, A.C. & Neri, D. (2014) Effects of grapevine leafroll associated virus 3 infection on growth, leaf gas exchange, yield and basic fruit chemistry of Vitis vinifera L. cv. Cabernet Franc. Scientia Horticulturae, 170, 228–236. [Google Scholar]
- Espinoza, C. , Medina, C. , Somerville, S. & Arce‐Johnson, P. (2007) Senescence‐associated genes induced during compatible viral interactions with grapevine and Arabidopsis . Journal of Experimental Botany, 58, 3197–3212. [DOI] [PubMed] [Google Scholar]
- Espinoza, C. , Vega, A. , Medina, C. , Schlauch, K. , Cramer, G. & Arce‐Johnson, P. (2007) Gene expression associated with compatible viral diseases in grapevine cultivars. Functional and Integrative Genomics, 7, 95–110. [DOI] [PubMed] [Google Scholar]
- Ferrandino, A. & Lovisolo, C. (2014) Abiotic stress effects on grapevine (Vitis vinifera L.): focus on abscisic acid‐mediated consequences on secondary metabolism and berry quality. Environmental and Experimental Botany, 103, 138–147. [Google Scholar]
- Fiore, N. , Prodan, S. , Montealegre, J. , Aballay, E. , Pino, A.M. & Zamorano, A. (2008) Survey of grapevine viruses in Chile. Journal of Plant Pathology, 90, 125–130. [Google Scholar]
- Fuchs, M. (2020) Grapevine viruses: a multitude of diverse species with simple but overall poorly adopted management solutions in the vineyard. Journal of Plant Pathology, 102, 643–653. [Google Scholar]
- Fuchs, M. , Almeyda, C.V. , Al Rwahnih, M. , Atallah, S.S. , Cieniewicz, E.J. , Farrar, K. et al. (2021) Economic studies reinforce efforts to safeguard specialty crops in the United States. Plant Disease, 105, 14–26. [DOI] [PubMed] [Google Scholar]
- Gai, Z. , Wang, Y.U. , Ding, Y. , Qian, W. , Qiu, C. , Xie, H. et al. (2020) Exogenous abscisic acid induces the lipid and flavonoid metabolism of tea plants under drought stress. Scientific Reports, 10, 12275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaiteri, C. , Ding, Y. , French, B. , Tseng, G.C. & Sibille, E. (2014) Beyond modules and hubs: the potential of gene coexpression networks for investigating molecular mechanisms of complex brain disorders. Genes, Brain and Behavior, 13, 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, Q.M. , Venugopal, S. , Navarre, D. & Kachroo, A. (2011) Low oleic acid‐derived repression of jasmonic acid‐inducible defense responses requires the WRKY50 and WRKY51 proteins. Plant Physiology, 155, 464–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehlenborg, N. (2019) UpSetR: A more scalable alternative to Venn and Euler diagrams for visualizing intersecting sets. R package version 1.4.0. https://CRAN.R‐project.org/package=UpSetR [Accessed 19 April 2021]. [Google Scholar]
- Goff, K.E. & Ramonell, K.M. (2007) The role and regulation of receptor‐like kinases in plant defense. Gene Regulation and Systems Biology, 1, 167–175. [PMC free article] [PubMed] [Google Scholar]
- Gohil, H. , Pavlis, G.C. , Ward, D. & Nita, A.M. (2016) Red leaves in the vineyard: biotic and abiotic causes. Fact Sheet FS1260. New Brunswick: Rutgers New Jersey Agricultural Experimental Station Cooperative Extension. [Google Scholar]
- Golino, D.A. (1993) Potential interactions between rootstocks and grapevine latent viruses. American Journal of Enology and Viticulture, 44, 148–152. [Google Scholar]
- Golino, D.A. , Weber, E. , Sim, S. & Rowhani, A. (2008) Leafroll disease is spreading rapidly in a Napa Valley vineyard. California Agriculture, 62, 156–160. [Google Scholar]
- Golino, D.A. , Wolpert, J. , Sim, S.T. , Benz, J. , Anderson, M. & Rowhani, A. (2008) Virus effects on vine growth and fruit components of Cabernet Sauvignon on six rootstocks. In: Proceedings of the 2nd Annual National Viticulture Research Conference, July 9‐11. Davis: University of California, Davis, pp. 28–29. [Google Scholar]
- Gouveia, P. & Nolasco, G. (2012) The p19.7 RNA silencing suppressor from grapevine leafroll‐associated virus 3 shows different levels of activity across phylogenetic groups. Virus Genes, 45, 333–339. [DOI] [PubMed] [Google Scholar]
- Grant, R.S. & Matthews, M.A. (1996) The influence of phosphorus availability and rootstock on root system characteristics, phosphorus uptake, phosphorus partitioning, and growth efficiency. American Journal of Enology and Viticulture, 47, 403–409. [Google Scholar]
- Grimplet, J. , Cramer, G.R. , Dickerson, J.A. , Mathiason, K. , Van Hemert, J. & Fennell, A.Y. (2009) VitisNet: ‘Omics’ integration through grapevine molecular networks. PLoS One, 4, e8365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Y. & Innes, R.W. (2011) The keep on going protein of Arabidopsis recruits the enhanced disease resistance1 protein to Trans‐Golgi network/early endosome vesicles. Plant Physiology, 155, 1827–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidoni, S. , Mannini, F. , Ferrandino, A. , Argamante, N. & Di Stefano, R. (1997) The effect of grapevine leafroll and rugose wood sanitation on agronomic performance and berry and leaf phenolic content of a Nebbiolo clone (Vitis vinifera L.). American Journal of Enology and Viticulture, 48, 438–442. [Google Scholar]
- Guidoni, S. , Mannini, F. , Ferrandino, A. , Argamante, N. & Di Stefano, R. (2000) Effect of virus status on leaf and berry phenolic compounds in two wine grapevine Vitis vinifera cultivars. Acta Horticulturae, 526, 445–452. [Google Scholar]
- Gutha, L.R. , Casassa, L.F. , Harbertson, J.F. & Naidu, R.A. (2010) Modulation of flavonoid biosynthetic pathway genes and anthocyanins due to virus infection in grapevine (Vitis vinifera L.) leaves. BMC Plant Biology, 10, 187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, B.J. , Delcher, A.L. , Mount, S.M. , Wortman, J.R. , Smith, R.K. Jr. , Hannick, L.I. et al. (2003) Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Research, 31, 5654–5666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas, B.J. , Salzberg, S.L. , Zhu, W. , Pertea, M. , Allen, J.E. , Orvis, J. et al. (2008) Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biology, 9, R7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habili, N. & Nutter, F.W. Jr. (1997) Temporal and spatial analysis of grapevine leafroll‐associated virus 3 in Pinot Noir grapevines in Australia. Plant Disease, 81, 625–628. [DOI] [PubMed] [Google Scholar]
- Hale, C.R. & Woodham, R.C. (1979) Effect of grapevine leafroll disease on the acid and potassium composition of sultana grapes. American Journal of Enology and Viticulture, 30, 91–92. [Google Scholar]
- Haxim, Y. , Ismayil, A. , Jia, Q. , Wang, Y. , Zheng, X. & Elife, T.C. (2017) Autophagy functions as an antiviral mechanism against geminiviruses in plants. eLife, 6, e23897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, D.A. (2001) Influence of linker histone H1 on chromatin remodeling. Biochemistry and Cell Biology, 79, 317–324. [PubMed] [Google Scholar]
- Hirayama, T. & Shinozaki, K. (2010) Research on plant abiotic stress responses in the post‐genome era: past, present and future. The Plant Journal, 61, 1041–1052. [DOI] [PubMed] [Google Scholar]
- Huang, J. , Zhu, C. & Li, X. (2018) SCFSNIPER4 controls the turnover of two redundant TRAF proteins in plant immunity. The Plant Journal, 95, 504–515. [DOI] [PubMed] [Google Scholar]
- Inoue, H. & Randazzo, P.A. (2007) Arf GAPs and their interacting proteins. Traffic, 8, 1465–1475. [DOI] [PubMed] [Google Scholar]
- Jaillon, O. , Aury, J.‐M. , Noel, B. , Policriti, A. , Clepet, C. , Casagrande, A. et al. (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature, 449, 463–467. [DOI] [PubMed] [Google Scholar]
- Ji, H. , Pardo, J.M. , Batelli, G. , Van Oosten, M.J. , Bressan, R.A. & Li, X. (2013) The salt overly sensitive (SOS) pathway: established and emerging roles. Molecular Plant, 6, 275–286. [DOI] [PubMed] [Google Scholar]
- Jia, H. , Wang, C. , Zhang, C. , Haider, M.S. , Zhao, P. , Liu, Z. et al. (2016) Functional analysis of VvBG1 during fruit development and ripening of grape. Journal of Plant Growth Regulation, 35, 987–999. [Google Scholar]
- Jiang, F. & Hartung, W. (2008) Long‐distance signalling of abscisic acid (ABA): the factors regulating the intensity of the ABA signal. Journal of Experimental Botany, 59, 37–43. [DOI] [PubMed] [Google Scholar]
- Jiang, Y. & Deyholos, M.K. (2009) Functional characterization of Arabidopsis NaCl‐inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Molecular Biology, 69, 91–105. [DOI] [PubMed] [Google Scholar]
- Jiang, Z. , Dong, X. & Zhang, Z. (2015) Network‐based comparative analysis of Arabidopsis immune responses to Golovinomyces orontii and Botrytis cinerea infections. Scientific Reports, 6, 19149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, P. , Binns, D. , Chang, H.‐y. , Fraser, M. , Li, W. , McAnulla, C. et al. (2014) InterProScan 5: genome‐scale protein function classification. Bioinformatics, 30, 1236–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jooste, A.E.C. , Molenaar, N. , Maree, H.J. , Bester, R. , Morey, L. , de Koker, W.C. et al. (2015) Identification and distribution of multiple virus infections in grapevine leafroll diseased vineyards. European Journal of Plant Pathology, 142, 363–375. [Google Scholar]
- Kim, D. , Langmead, B. & Salzberg, S.L. (2015) HISAT: a fast spliced aligner with low memory requirements. Nature Methods, 12, 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaassen, V.A. , Sim, S.T. , Dangl, G.S. , Osman, F. , Rwahnih Al, M. , Rowhani, A. et al. (2011) Vitis californica and Vitis californica × Vitis vinifera hybrids are hosts for grapevine leafroll‐associated virus‐2 and ‐3 and grapevine virus A and B. Plant Disease, 95, 657–665. [DOI] [PubMed] [Google Scholar]
- Kliewer, W.M. & Lider, L.A. (1976) Influence of leafroll virus on composition of Burger fruits. American Journal of Enology and Viticulture, 27, 118–124. [Google Scholar]
- Köhler, B. , Hills, A. & Blatt, M.R. (2003) Control of guard cell ion channels by hydrogen peroxide and abscisic acid indicates their action through alternate signaling pathways. Plant Physiology, 131, 385–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde, R. (2019) pheatmap: pretty heatmaps. R package version 1.0.12. Available at: https://CRAN.R‐project.org/package=pheatmap [Accessed 19 April 2021]. [Google Scholar]
- Komar, V. , Vigne, E. , Demangeat, G. , Lemaire, O. & Fuchs, M. (2010) Comparative performance of virus‐infected Vitis vinifera cv. Savagnin Rose grafted onto three rootstocks. American Journal of Enology and Viticulture, 61, 68–73. [Google Scholar]
- Koornneef, A. & Pieterse, C.M.J. (2008) Cross talk in defense signaling. Plant Physiology, 146, 839–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf, I. (2004) Gene finding in novel genomes. BMC Bioinformatics, 5, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs, L.G. , Hanami, H. , Fortenberry, M. & Kaps, M.L. (2001) Latent infection by leafroll agent GLRaV‐3 is linked to lower fruit quality in French‐American hybrid grapevines Vidal Blanc and St. Vincent. American Journal of Enology and Viticulture, 52, 254–259. [Google Scholar]
- Koyama, K. , Sadamatsu, K. & Goto‐Yamamoto, N. (2010) Abscisic acid stimulated ripening and gene expression in berry skins of the Cabernet Sauvignon grape. Functional and Integrative Genomics, 10, 367–381. [DOI] [PubMed] [Google Scholar]
- Kunkel, B.N. & Brooks, D.M. (2002) Cross talk between signaling pathways in pathogen defense. Current Opinion in Plant Biology, 5, 325–331. [DOI] [PubMed] [Google Scholar]
- Lê, S. , Josse, J. & Husson, F. (2008) FactoMineR: an R package for multivariate analysis. Journal of Statistical Software, 25, 1–18. [Google Scholar]
- Lee, J. , Keller, K.E. , Rennaker, C. & Martin, R.R. (2009) Influence of grapevine leafroll associated viruses (GLRaV‐2 and ‐3) on the fruit composition of Oregon Vitis vinifera L. cv. Pinot Noir: free amino acids, sugars, and organic acids. Food Chemistry, 117, 99–105. [Google Scholar]
- Lee, J. & Martin, R.R. (2009) Influence of grapevine leafroll associated viruses (GLRaV‐2 and ‐3) on the fruit composition of Oregon Vitis vinifera L. cv. Pinot noir: phenolics. Food Chemistry, 112, 889–896. [Google Scholar]
- Li, J. (2004) The WRKY70 transcription factor: a node of convergence for jasmonate‐mediated and salicylate‐mediated signals in plant defense. The Plant Cell, 16, 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q. , Hu, A. , Qi, J. , Dou, W. , Qin, X. , Zou, X. et al. (2020) CsWAKL08, a pathogen‐induced wall‐associated receptor‐like kinase in sweet orange, confers resistance to citrus bacterial canker via ROS control and JA signaling. Horticulture Research, 7, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman‐Aiden, E. , van Berkum, N.l. , Williams, L. , Imakaev, M. , Ragoczy, T. , Telling, A. et al. (2009) Comprehensive mapping of long‐range interactions reveals folding principles of the human genome. Science, 326, 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S. , Kracher, B. , Ziegler, J. , Birkenbihl, R.P. & Somssich, I.E. (2015) Negative regulation of ABA signaling by WRKY33 is critical for Arabidopsis immunity towards Botrytis cinerea 2100. eLife, 4, e07295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livigni, S. , Lucini, L. , Sega, D. , Navacchi, O. , Pandolfini, T. , Zamboni, A. et al. (2019) The different tolerance to magnesium deficiency of two grapevine rootstocks relies on the ability to cope with oxidative stress. BMC Plant Biology, 19, 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomsadze, A. , Ter‐Hovhannisyan, V. , Chernoff, Y.O. & Borodovsky, M. (2005) Gene identification in novel eukaryotic genomes by self‐training algorithm. Nucleic Acids Research, 33, 6494–6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I. , Huber, W. & Anders, S. (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biology, 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyzenga, W.J. , Liu, H. , Schofield, A. , Muise‐Hennessey, A. & Stone, S.L. (2013) Arabidopsis CIPK26 interacts with KEG, components of the ABA signalling network and is degraded by the ubiquitin–proteasome system. Journal of Experimental Botany, 64, 2779–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahfoudhi, N. , Digiaro, M. & Dhouibi, M.H. (2008) Incidence and distribution of grapevine leafroll‐associated viruses in Tunisian vineyards. Journal of Phytopathology, 156, 556–558. [Google Scholar]
- Mandadi, K.K. & Scholthof, K.‐B.‐G. (2013) Plant immune responses against viruses: how does a virus cause disease? The Plant Cell, 25, 1489–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mang, H.‐G. , Qian, W. , Zhu, Y. , Qian, J. , Kang, H.‐G. , Klessig, D.F. et al. (2012) Abscisic acid deficiency antagonizes high‐temperature inhibition of disease resistance through enhancing nuclear accumulation of resistance proteins SNC1 and RPS4 in Arabidopsis . The Plant Cell, 24, 1271–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manzi, M. , Lado, J. , Rodrigo, M.J. , Zacarías, L. , Arbona, V. & Gómez‐Cadenas, A. (2015) Root ABA accumulation in long‐term water‐stressed plants is sustained by hormone transport from aerial organs. Plant and Cell Physiology, 56, 2457–2466. [DOI] [PubMed] [Google Scholar]
- Massonnet, M. , Cochetel, N. , Minio, A. , Vondras, A.M. , Lin, J. , Muyle, A. et al. (2020) The genetic basis of sex determination in grapes. Nature Communications, 11, 2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minio, A. , Massonnet, M. , Figueroa‐Balderas, R. , Castro, A. & Cantu, D. (2019a) Diploid genome assembly of the wine grape Carménère. G3 Genes Genomes Genetics, 9, 1331–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minio, A. , Massonnet, M. , Figueroa‐Balderas, R. , Vondras, A.M. , Blanco‐Ulate, B. & Cantu, D. (2019b) Iso‐Seq allows genome‐independent transcriptome profiling of grape berry development. G3 Genes Genomes Genetics, 9, 755–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett, P. , Farnham, G. , Peart, J. & Baulcombe, D.C. (2002) Interaction between domains of a plant NBS‐LRR protein in disease resistance‐related cell death. The EMBO Journal, 21, 4511–4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montero, R. , Mundy, D. , Albright, A. , Grose, C. , Trought, M. , Cohen, D. et al. (2016) Effects of grapevine leafroll associated virus 3 (GLRaV‐3) and duration of infection on fruit composition and wine chemical profile of Vitis vinifera L. cv. Sauvignon Blanc. Food Chemistry, 197, 1177–1183. [DOI] [PubMed] [Google Scholar]
- Moon, J.Y. & Park, J.M. (2016) Cross‐talk in viral defense signaling in plants. Frontiers in Microbiology, 7, 2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morant, A.V. , Jørgensen, K. , Jørgensen, C. , Paquette, S.M. , Sánchez‐Pérez, R. , Møller, B.L. et al. (2008) β‐glucosidases as detonators of plant chemical defense. Phytochemistry, 69, 1795–1813. [DOI] [PubMed] [Google Scholar]
- Moreno‐Moyano, L.T. , Bonneau, J.P. , Sánchez‐Palacios, J.T. , Tohme, J. & Johnson, A.A.T. (2016) Association of increased grain iron and zinc concentrations with agro‐morphological traits of biofortified rice. Frontiers in Plant Science, 7, 1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita‐Yamamuro, C. , Tsutsui, T. , Sato, M. , Yoshioka, H. , Tamaoki, M. , Ogawa, D. et al. (2005) The Arabidopsis gene CAD1 controls programmed cell death in the plant immune system and encodes a protein containing a MACPF domain. Plant and Cell Physiology, 46, 902–912. [DOI] [PubMed] [Google Scholar]
- Moutinho‐Pereira, J. , Correia, C.M. , Gonçalves, B. , Bacelar, E.A. , Coutinho, J.F. , Ferreira, H.F. et al. (2012) Impacts of leafroll‐associated viruses (GLRaV‐1 and ‐3) on the physiology of the Portuguese grapevine cultivar ‘Touriga Nacional’ growing under field conditions. Annals of Applied Biology, 160, 237–249. [Google Scholar]
- Müller, M. & Munné‐Bosch, S. (2015) Ethylene response factors: a key regulatory hub in hormone and stress signaling. Plant Physiology, 169, 32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers, G. (2014) Efficient local alignment discovery amongst noisy long reads. In: Brown, D. & Morgenstern, B. (Eds.) Algorithms in bioinformatics. WABI 2014. Lecture notes in computer science, vol. 8701. Wroclaw, Poland: Springer, pp. 52–67. [Google Scholar]
- Naidu, R.A. , Maree, H.J. & Burger, J.T. (2015) Grapevine leafroll disease and associated viruses: a unique pathosystem. Annual Review of Phytopathology, 53, 613–634. [DOI] [PubMed] [Google Scholar]
- Naidu, R.A. , Rowhani, A. , Fuchs, M. , Golino, D. & Martelli, G.P. (2014) Grapevine leafroll: a complex viral disease affecting a high‐value fruit crop. Plant Disease, 98, 1172–1185. [DOI] [PubMed] [Google Scholar]
- Nozoye, T. (2018) The nicotianamine synthase gene is a useful candidate for improving the nutritional qualities and Fe‐deficiency tolerance of various crops. Frontiers in Plant Science, 9, 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman, F. , Leutenegger, C. , Golino, D. & Rowhani, A. (2008) Comparison of low‐density arrays, RT‐PCR and real‐time TaqMan RT‐PCR in detection of grapevine viruses. Journal of Virological Methods, 149, 292–299. [DOI] [PubMed] [Google Scholar]
- Otulak‐Kozieł, K. , Kozieł, E. & Valverde, R.A. (2019) The respiratory burst oxidase homolog D (RbohD) cell and tissue distribution in potato–potato virus Y (PVYNTN) hypersensitive and susceptible reactions. International Journal of Molecular Sciences, 20, 2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, X. , Welti, R. & Wang, X. (2010) Quantitative analysis of major plant hormones in crude plant extracts by high‐performance liquid chromatography–mass spectrometry. Nature Protocols, 5, 986–992. [DOI] [PubMed] [Google Scholar]
- Park, J. , Kim, T.‐H. , Takahashi, Y. , Schwab, R. , Dressano, K. , Stephan, A.B. et al. (2019) Chemical genetic identification of a lectin receptor kinase that transduces immune responses and interferes with abscisic acid signaling. The Plant Journal, 98, 492–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea, M. , Pertea, G.M. , Antonescu, C.M. , Chang, T.‐C. , Mendell, J.T. & Salzberg, S.L. (2015) StringTie enables improved reconstruction of a transcriptome from RNA‐seq reads. Nature Biotechnology, 33, 290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilati, S. , Bagagli, G. , Sonego, P. , Moretto, M. , Brazzale, D. , Castorina, G. et al. (2017) Abscisic acid is a major regulator of grape berry ripening onset: new insights into ABA signaling network. Frontiers in Plant Science, 8, 1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poojari, S. , Alabi, O.J. & Naidu, R.A. (2013) Molecular characterization and impacts of a strain of Grapevine leafroll‐associated virus 2 causing asymptomatic infection in a wine grape cultivar. Virology Journal, 10, 324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postnikova, O.A. & Nemchinov, L.G. (2012) Comparative analysis of microarray data in Arabidopsis transcriptome during compatible interactions with plant viruses. Virology Journal, 9, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prosser, S.W. , Goszczynski, D.E. & Meng, B. (2007) Molecular analysis of double‐stranded RNAs reveals complex infection of grapevines with multiple viruses. Virus Research, 124, 151–159. [DOI] [PubMed] [Google Scholar]
- R Core Team (2020) R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. Available at: https://www.R‐project.org/ [Accessed 19 April 2021]. [Google Scholar]
- Ram, K. & Wickham, H. (2018) wesanderson: a Wes Anderson palette generator. R package version 0.3.6. Available at: https://CRAN.R‐project.org/package=wesanderson [Accessed 19 April 2021]. [Google Scholar]
- Reddy, A.S. & Day, I.S. (2001) Kinesins in the Arabidopsis genome: a comparative analysis among eukaryotes. BMC Genomics, 2, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riaz, S. , Tenscher, A.C. , Ramming, D.W. & Walker, M.A. (2011) Using a limited mapping strategy to identify major QTLs for resistance to grapevine powdery mildew (Erysiphe necator) and their use in marker‐assisted breeding. Theoretical and Applied Genetics, 122, 1059–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricketts, K.D. , Gomez, M.I. , Atallah, S.S. , Fuchs, M.F. , Martinson, T.E. , Battany, M.C. et al. (2015) Reducing the economic impact of grapevine leafroll disease in California: identifying optimal disease management strategies. American Journal of Enology and Viticulture, 66, 138–147. [Google Scholar]
- Roach, M.J. , Johnson, D.L. , Bohlmann, J. , van Vuuren, H.J.J. , Jones, S.J.M. , Pretorius, I.S. et al. (2018) Population sequencing reveals clonal diversity and ancestral inbreeding in the grapevine cultivar Chardonnay. PLoS Genetics, 14, e1007807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigo, G. , Carrera, J. , Ruiz‐Ferrer, V. , del Toro, F.J. , Llave, C. , Voinnet, O. et al. (2012) A meta‐analysis reveals the commonalities and differences in Arabidopsis thaliana response to different viral pathogens. PLoS One, 7, e40526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogiers, S.Y. , Coetzee, Z.A. , Walker, R.R. , Deloire, A. & Tyerman, S.D. (2017) Potassium in the grape (Vitis vinifera L.) berry: transport and function. Frontiers in Plant Science, 8, 1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowhani, A. , Golino, D.A. , Klaassen, V. , Sim, S.T. , Gouran, M. & Al Rwahnih, M. (2015) Grapevine leafroll associated virus‐3: effects on rootstocks, vine performance, yield and berries. In: Ertunç, F. (Ed.) 18th Congress of the international council for the study of virus and virus‐like diseases of the grapevine, 7‐11 September. Ankara: Ankara University, pp. 161–162. [Google Scholar]
- RStudio Team (2020) RStudio: integrated development for R. Available at: http://www.rstudio.com/ [Accessed 19 April 2021]. [Google Scholar]
- Rudis, B. (2020) hrbrthemes: additional themes, theme components and utilities for 'ggplot2'. R package version 0.8.0. Available at: https://CRAN.R‐project.org/package=hrbrthemes [Accessed 19 April 2021]. [Google Scholar]
- Rwahnih Al, M. , Alabi, O.J. , Westrick, N.M. , Golino, D. & Rowhani, A. (2017) Description of a novel monopartite geminivirus and its defective subviral genome in grapevine. Phytopathology, 107, 240–251. [DOI] [PubMed] [Google Scholar]
- Rwahnih Al, M. , Daubert, S. , Golino, D. & Rowhani, A. (2009) Deep sequencing analysis of RNAs from a grapevine showing Syrah decline symptoms reveals a multiple virus infection that includes a novel virus. Virology, 387, 395–401. [DOI] [PubMed] [Google Scholar]
- Seo, S. , Ishizuka, K. & Ohashi, Y. (1995) Induction of salicylic acid β‐glucosidase in tobacco leaves by exogenous salicylic acid. Plant Cell Physiology, 36, 447–453. [Google Scholar]
- Shaik, R. & Ramakrishna, W. (2013) Genes and co‐expression modules common to drought and bacterial stress responses in Arabidopsis and rice. PLoS One, 8, e77261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi, Y. , Wang, Z. , Meng, P. , Tian, S. , Zhang, X. & Yang, S. (2013) The glutamate carboxypeptidase AMP1 mediates abscisic acid and abiotic stress responses in Arabidopsis . New Phytologist, 199, 135–150. [DOI] [PubMed] [Google Scholar]
- Skubacz, A. , Daszkowska‐Golec, A. & Szarejko, I. (2016) The role and regulation of ABI5 (ABA‐Insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Frontiers in Plant Science, 7, 1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater, G.S.C. & Birney, E. (2005) Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics, 6, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit, A. , Hubley, R. , & Green, P. (2015) RepeatMasker Open‐4.0. Available at: http://www.repeatmasker.org [Accessed 19 April 2021].
- Song, M. , Wang, S. , Chai, L. , Zhang, S. & Shen, Y. (2016) Characterization of an ABA‐induced and K+ channel gene FaKAT1 that regulates strawberry fruit ripening. Journal of Plant Growth Regulation, 36, 312–322. [Google Scholar]
- Stanke, M. , Keller, O. , Gunduz, I. , Hayes, A. , Waack, S. & Morgenstern, B. (2006) AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Research, 34, W435–W439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, S.L. , Williams, L.A. , Farmer, L.M. , Vierstra, R.D. & Callis, J. (2006) KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. The Plant Cell, 18, 3415–3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, J.‐H. , Dong, Y.‐H. , Li, C.‐L. & Shen, Y.‐Y. (2015) Transcription and enzymatic analysis of β‐glucosidase VvBG1 in grape berry ripening. Plant Growth Regulation, 75, 67–73. [Google Scholar]
- Tramontini, S. , Vitali, M. , Centioni, L. , Schubert, A. & Lovisolo, C. (2013) Rootstock control of scion response to water stress in grapevine. Environmental and Experimental Botany, 93, 20–26. [Google Scholar]
- Trupkin, S.A. , Auge, G.A. , Zhu, J.K. , Sanchez, R.A. & Botto, J.F. (2017) SALT OVERLY SENSITIVE 2 (SOS2) and interacting partners SOS3 and ABSCISIC ACID–INSENSITIVE 2 (ABI2) promote red‐light‐dependent germination and seedling deetiolation in Arabidopsis . International Journal of Plant Science, 178, 485–493. [Google Scholar]
- Uyemoto, J. , Rowhani, A. , Luvisi, D. & Krag, C. (2001) New closterovirus in ‘Redglobe’ grape causes decline of grafted plants. California Agriculture, 55, 28–31. [Google Scholar]
- Vandereyken, K. , Van Leene, J. , De Coninck, B. & Cammue, B.P.A. (2018) Hub protein controversy: taking a closer look at plant stress response hubs. Frontiers in Plant Science, 9, 694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega, A. , Gutiérrez, R.A. , Peña‐Neira, A. , Cramer, G.R. & Arce‐Johnson, P. (2011) Compatible GLRaV‐3 viral infections affect berry ripening decreasing sugar accumulation and anthocyanin biosynthesis in Vitis vinifera . Plant Molecular Biology, 77, 261–274. [DOI] [PubMed] [Google Scholar]
- Venturini, L. , Ferrarini, A. , Zenoni, S. , Tornielli, G.B. , Fasoli, M. , Santo, S.D. et al. (2013) De novo transcriptome characterization of Vitis vinifera cv. Corvina unveils varietal diversity. BMC Genomics, 14, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vondras, A.M. , Minio, A. , Blanco‐Ulate, B. , Figueroa‐Balderas, R. , Penn, M.A. , Zhou, Y. et al. (2019) The genomic diversification of grapevine clones. BMC Genomics, 20, 972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos, I.A. , Moritz, L. , Pieterse, C.M.J. & Van Wees, S.C.M. (2015) Impact of hormonal crosstalk on plant resistance and fitness under multi‐attacker conditions. Frontiers in Plant Science, 6, 639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warschefsky, E.J. , Klein, L.L. , Frank, M.H. , Chitwood, D.H. , Londo, J.P. , von Wettberg, E.J.B. et al. (2016) Rootstocks: diversity, domestication, and impacts on shoot phenotypes. Trends in Plant Science, 21, 418–437. [DOI] [PubMed] [Google Scholar]
- Wheeler, S. , Loveys, B. , Ford, C. & Davies, C. (2009) The relationship between the expression of abscisic acid biosynthesis genes, accumulation of abscisic acid and the promotion of Vitis vinifera L. berry ripening by abscisic acid. Australian Journal of Grape and Wine Research, 15, 195–204. [Google Scholar]
- Wickham, H. (2016) ggplot2: elegant graphics for data analysis. New York: Springer‐Verlag. [Google Scholar]
- Wolpert, J.A. , Smart, D.R. & Anderson, M. (2005) Lower petiole potassium concentration at bloom in rootstocks with Vitis berlandieri genetic backgrounds. American Journal of Enology and Viticulture, 56, 163–169. [Google Scholar]
- Wu, Q. , Wang, X. & Ding, S.‐W. (2010) Viral suppressors of RNA‐based viral immunity: host targets. Cell Host and Microbe, 8, 12–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin, P.‐F. , Gao, C.‐S. , Cheng, C.‐H. , Tang, Q. , Dong, Z.‐X. , Zhao, L.‐N. et al. (2016) Identification and characterization of hemp WRKY transcription factors in response to abiotic stresses. Biologia Plantarum, 60, 489–495. [Google Scholar]
- Yoshimoto, K. , Hanaoka, H. , Sato, S. , Kato, T. , Tabata, S. , Noda, T. et al. (2004) Processing of ATG8s, ubiquitin‐like proteins, and their deconjugation by ATG4s are essential for plant autophagy. The Plant Cell, 16, 2967–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G. , Wang, L.‐G. , Han, Y. & He, Q.‐Y. (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS: A Journal of Integrative Biology, 16, 284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, G. , Duan, C. , Wang, Y.A. , Wang, Y. , Ji, K. , Xu, H. et al. (2013) The expression pattern of β‐glucosidase genes (VvBGs) during grape berry maturation and dehydration stress. Plant Growth Regulation, 70, 105–114. [Google Scholar]
- Zhou, X. , Hao, H. , Zhang, Y. , Bai, Y. , Zhu, W. , Qin, Y. et al. (2015) SOS2‐LIKE PROTEIN KINASE5, an SNF1‐RELATED PROTEIN KINASE3‐type protein kinase, is important for abscisic acid responses in Arabidopsis through phosphorylation of ABSCISIC ACID‐INSENSITIVE5. Plant Physiology, 168, 659–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zini, E. , Dolzani, C. , Stefanini, M. , Gratl, V. , Bettinelli, P. , Nicolini, D. et al. (2019) R‐loci arrangement versus downy and powdery mildew resistance level: a Vitis hybrid survey. International Journal of Molecular Sciences, 20, 3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
FIGURE S1 Malic acid, moisture content, pH, titratable acidity, weight per berry, yeast assimilable nitrogen (YAN), and total anthocyanins in 2015 and 2016. For most measures in both years, 3 ≤ n ≤ 9. For total anthocyanin and moisture content in 2015, 2 ≤ n ≤ 9. For total soluble solids before harvest in 2017 and 2018, n = 6. Differences between groups are indicated with nonoverlapping letters (Tukey HSD, p < .05)
FIGURE S2 Pearson correlation between RNA‐Seq samples
FIGURE S3 Barplots showing the number of differentially expressed (p < .05) genes up‐ and downregulated in MGT 101‐14‐grafted plants versus Kober 5BB‐grafted plants for each GLRaV condition in each year and in both years
FIGURE S4 Abundance of abscisic acid (ABA), ABA glucose ester (ABA‐GE), and xanthoxin in 2017 and 2018 at véraison, mid‐ripening, and harvest. Abundance of SA and JA in 2017 and 2018 at all four developmental stages. Analysis of variance tables for all five hormones and hormone‐related metabolites. Groups with nonoverlapping letters are significantly different (Tukey HSD, p < .05)
FIGURE S5 Rootstock‐differentiating genes
FIGURE S6 Percentage of variation explained by each of the first five multiple factor analysis components at each developmental stage
FIGURE S7 Distribution of centred expression for genes, per functional category, correlated to the same rootstock‐differentiating multiple factor analysis dimensions as hormones and hormone‐related metabolites. Results of Kolmogorov–Smirnov tests are shown
FIGURE S8 Centred expression of VITVvi_vCabFran04_v1_P478.ver1.0.g430590, histone H1, in berries from GLRaV (+) plants grafted to two different rootstocks
FIGURE S9 All metabolite products measured that are associated with the shikimate, phenylpropanoid, and flavonoid pathways. *Differentially expressed in 1 year; **differentially expressed in both years. One or two letters “r” indicate that the effect of a particular GLRaV infection differs between rootstocks at the same developmental stage in 1 or 2 years
TABLE S1 Curated list of abscisic acid‐related genes Pilati et al. (2017)
TABLE S2 Hormone and metabolite retention time, mass information, and additional method‐related information
TABLE S3 Cabernet Franc assembly and annotation statistics
TABLE S4 Conserved responses to GLRaVs depicted in Figure 3b
TABLE S5 Number of multiple factor analysis variables positively and negatively correlated with each rootstock‐differentiating multiple factor analysis dimension
FILE S1 Additional description of hormone signalling genes in Figure 8
Data Availability Statement
The DNA (BioProject PRJNA701939) and RNA (BioProject PRJNA701940) sequencing data that support these findings are available at the National Center for Biotechnology Information (NCBI). A Cabernet Franc genome browser and on‐line BLAST tool are available at http://www.grapegenomics.com. The assembly, annotations, and raw RNA‐Seq counts per library and per gene are available at Zenodo (http://doi.org/10.5281/zenodo.4555977).


