Highlights
-
•
Reducing the context around the tumor results in better segmentations with CNNs.
-
•
Combining multiple MRI sequences as input can be beneficial to segmentation networks.
-
•
A semi-automatic approach is fast and can potentially improve the accuracy of segmentations.
Keywords: Oropharyngeal cancer, Convolutional neural network, Segmentation, MRI, Radiotherapy, Semi-automatic approach
Abstract
Background and purpose
Segmentation of oropharyngeal squamous cell carcinoma (OPSCC) is needed for radiotherapy planning. We aimed to segment the primary tumor for OPSCC on MRI using convolutional neural networks (CNNs). We investigated the effect of multiple MRI sequences as input and we proposed a semi-automatic approach for tumor segmentation that is expected to save time in the clinic.
Materials and methods
We included 171 OPSCC patients retrospectively from 2010 until 2015. For all patients the following MRI sequences were available: T1-weighted, T2-weighted and 3D T1-weighted after gadolinium injection. We trained a 3D UNet using the entire images and images with reduced context, considering only information within clipboxes around the tumor. We compared the performance using different combinations of MRI sequences as input. Finally, a semi-automatic approach by two human observers defining clipboxes around the tumor was tested. Segmentation performance was measured with Sørensen–Dice coefficient (Dice), 95th Hausdorff distance (HD) and Mean Surface Distance (MSD).
Results
The 3D UNet trained with full context and all sequences as input yielded a median Dice of 0.55, HD of 8.7 mm and MSD of 2.7 mm. Combining all MRI sequences was better than using single sequences. The semi-automatic approach with all sequences as input yielded significantly better performance (p < 0.001): a median Dice of 0.74, HD of 4.6 mm and MSD of 1.2 mm.
Conclusion
Reducing the amount of context around the tumor and combining multiple MRI sequences improved the segmentation performance. A semi-automatic approach was accurate and clinically feasible.
1. Introduction
Worldwide, there are more than 679,000 new cases of head and neck cancer (HNC) per year and 380,000 of those cases result in death [1]. Radiotherapy (RT) is indicated for 74% of head and neck cancer patients, and up to 100% in some subsites [2]. Tumor delineation is needed for RT planning. In clinical practice, tumor contouring is done manually, which is time consuming and suffers from interobserver variability. Thus, accurate automatic segmentation is desirable.
Convolutional neural networks (CNNs) are considered the current state of the art for computer vision techniques, such as automatic segmentation. Specifically for tumor segmentation, promising results have been obtained for various tumor sites such as brain [3], lung [4], liver [5] and rectum [6].
For HNC, previous literature [7], [8] focused on the segmentation of other RT-related target volumes rather than the primary tumor and without special focus on any particular HNC subsite, such as nasopharyngeal or oropharyngeal cancer. However, anatomy and imaging characteristics of tumors and their surrounding tissue vary greatly across subsites. Nasopharyngeal tumors are bounded by the surrounding anatomy and thus they present with lower spatial variability. Men et al. [9] proposed an automatic segmentation method for nasopharyngeal primary tumors. To the best of our knowledge, no studies have been published on automatic segmentation of primary tumors in oropharyngeal squamous cell cancer (OPSCC). Tumors in this category are quite variable in shape, size and location compared to other subsites in head and neck cancer and their delineation suffers from high interobserver variability [10].
The modalities of choice in other works for HNC automatic segmentation are PET and/or CT [7], [8]. PET presents low spatial resolution and only shows the metabolically active part of the tumor while CT has low soft tissue contrast. MRI is now becoming a modality of interest in RT and provides improved soft tissue contrast compared to other modalities, being better suitable for oropharyngeal tumor segmentation. In line with this, previous works have suggested that the use of MRI for head and neck cancer delineation provides unique information compared to PET/CT or CT [11].
We investigated the effect on segmentation performance of different MRI sequences and its combination as inputs to the model. We hypothesized that by decreasing the amount of context around the tumor, thereby simplifying the task, the performance of the segmentation model would improve. Hence, we proposed a semi-automatic approach in which a clipbox around the tumor is used to crop the input image. We demonstrated its clinical applicability by having two observers (including one radiation oncologist) manually selecting the clipbox. The aim of this study was to develop a CNN model for segmenting OPSCC on MRI images.
2. Materials and methods
2.1. Data
A cohort of 171 patients treated at our institute between January 2010 and December 2015 was used for this project. Mean patient age was 60 (Standard deviation ± 7 years) and 62% of the patients were male. Further details on tumor stage and HPV status can be found in the Supplemental Material (table S.1). All patients had histologically proven primary OPSCC and pre-treatment MRI, acquired for primary staging. The institutional review board approved the study (IRBd18047). Informed consent was waived considering the retrospective design. Any identifiable information was removed.
All MRI scans were acquired on 1.5 T (n = 79) or 3.0 T (n = 92) MRI scanners (Achieva, Philips Medical System, Best, The Netherlands). The imaging protocol included: 2D T1-weighted fast spin-echo (T1w), 2D T2-weighted fast spin-echo with fat suppression (T2w) and 3D T1-weighted high-resolution isotropic volume excitation after gadolinium injection with fat suppression (T1gd). Further details on the MRI protocols are given in the Supplemental Material (table S.2). The primary tumors were manually contoured in 3D Slicer (version 4.8.0, https://www.slicer.org/) by one observer with 1 year of experience (P.B.). Afterwards, they were reviewed and adjusted, if needed, by a radiologist with 7 years of experience (B.J.). All tumor volumes were delineated on the T1gd but observers were allowed to consult the other sequences.
For the experimental set-up, we split the data set in three subsets: training set (n = 131), validation set (n = 20) and test set (n = 20). The test set was not used for training or hyper-parameter tuning. We stratified the three subsets for tumor volume, subsite, and aspect ratio since these features are likely relevant for segmentation. Subsites were defined as tonsillar tissue, soft palate, base of tongue and posterior wall. Aspect ratio was defined as the ratio between the shortest and the longest axis of the tumor. All images were resampled to a voxel size of 0.8 mm × 0.8 mm × 0.8 mm.
2.2. Model architecture
The UNet architecture was chosen as the basis for our experiments because of the promising results on segmentation of medical structures [5], [12], [13], [14], [15]. Given the 3D nature of the images, we chose a 3D UNet as the architecture in this work [12], [16]. We used Dice as loss function [17], the Adam optimizer [18] and early stopping. Dropout and data augmentation were used for regularization. Further details on the training procedure can be found in the Supplemental Material (Tables S.3. and S.4.).
2.3. Fully automatic approach
We trained the 3D UNet using the full 3D scans. We studied the effect of incorporating multiple MRI sequences into the training by introducing the available MRI sequences as input channels. Five networks were trained for the following MRI sequences and combinations thereof: T1w, where the tumor is hypo-intense but homogeneous; T2w, where the tumor is hyper-intense; T1gd, since the tumor presents with clearer boundaries; combining T1gd and T2w, and combining all sequences together (T1gd, T2w and T1w), to explore all the available information.
2.4. Semi-automatic approach
We proposed a semi-automatic approach in which we trained the networks with only the information within a clipbox around the tumor instead of with the full image as input.
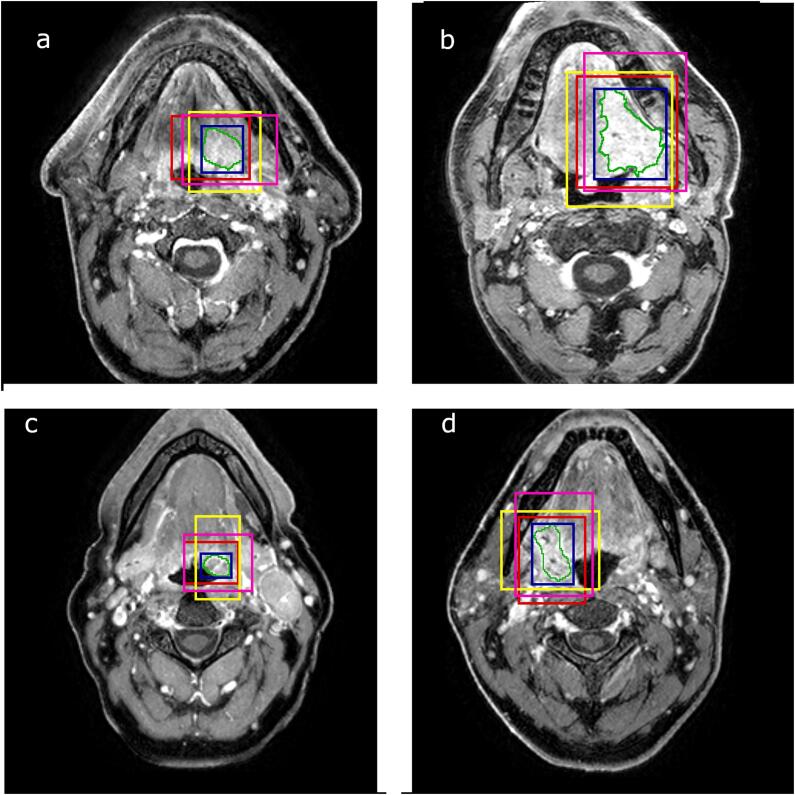
During training, the clipbox was computed from the tumor delineations. First, the bounding box was calculated (i.e. the minimal box around the tumor). Then, random shifts of up to 25 mm were applied to all of the six directions to make clipboxes of different sizes and allow off-centered positioning of the tumors. We considered that shifts of more than 25 mm would represent unrealistic errors during clipbox selection. Examples of inputs possibly seen by the network are shown in Fig. 1.
Fig. 1.
Original MRI image with the manual segmentation (green) of the oropharyngeal tumor. The blue boxes are the bounding boxes of the tumor. The rest of the boxes are used as inputs to the network during training. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To study the clinical feasibility of this semi-automatic approach, two human observers were asked to manually select a clipbox around the tumor for each test set patient. The clipboxes were selected using 3D Slicer on the T1gd with access to the other sequences. The first observer (P.B.) had delineated the tumors two years earlier. The second observer was a radiation oncologist with 16 years of experience (A.A.) and had no information about the tumor delineations. To mitigate the risk of the observers defining too small clipboxes, cropping the tumor, the clipboxes were dilated 5 mm so as to ensure that they encompass the tumors. We consider it unlikely that a human observer would crop the tumor by more than 5 mm.
2.5. Experiments
For the fully automatic approach, the performance of the networks trained with different sequences (T1w, T2w, T1gd, T1gd/T2w, and all sequences combined) was compared for the patients on the separate test set.
Because of memory constraints, scans were resized to a lower resolution by a factor of ~2.5 to 1.9 mm × 1.9 mm × 1.9 mm. Thus, even the smallest tumors were seen by the network. As a control experiment, to assess the impact of the resulting loss of resolution, we additionally trained a 2D UNet with full resolution axial slices. We checked for significant differences in performance of both approaches.
For the semi-automatic approach, one network was trained with all the sequences as input. The results with the clipboxes of the two observers were compared to the fully automatic approach experiment when combining all sequences as input (baseline).
To evaluate the robustness of the semi-automatic approach to off-centered tumors inside the clipboxes, we presented the trained model with increasingly shifted versions of the clipboxes, starting from the bounding box. The artificially induced shifts were applied in the 6 possible directions of the clipbox and expressed as two metrics: the centroid displacement and the relative difference in clipbox diagonal length before and after the shifts.
2.6. Statistics
To confirm that the three subsets were balanced in subsite, volume and aspect ratio, we used a Kruskal-Wallis test for continuous variables (volume and aspect ratio) and a chi-square test for independence for the categorical data (subsite).
Automatic contours were compared against the delineations from the human experts using common segmentation metrics: Sørensen–Dice coefficient (Dice), 95th Hausdorff Distance (HD) and Mean surface distance (MSD), implemented using the Python package from DeepMind (https://github.com/deepmind/surface-distance). Differences among experiments were assessed by the Wilcoxon signed-ranked test. P-values below 0.05 were considered statistically significant. Statistical analyses were performed with the SciPy package (version 1.1.0) and Python 3.6. Other relevant libraries can be found in the Supplemental Material (Table S.5.). The code is publicly available and can be found in: https://github.com/RoqueRouteiral/oroph_segmentation.git
3. Results
3.1. Summary of tumor characteristics
Tumor characteristics (location, volume and aspect ratio) of our cohort are described in Table S.6. No significant differences were found in the distributions of subsite, volume and aspect ratio between the training, validation and test sets.
3.2. Fully automatic approach
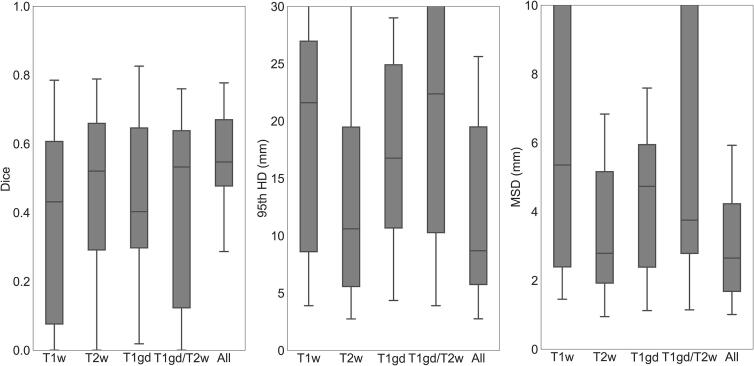
As shown in Fig. 2, combining all MR sequences resulted in the best performance, with a median Dice of 0.55 (range 0–0.78), median 95th HD of 8.7 mm (range 2.8–84.8 mm) and median MSD of 2.7 mm (range 1.0–26.8 mm), and the least variability among patients. The control experiment showed that by training a 2D UNet with full resolution scans the results were not significantly better than when using its 3D counterpart (Table S.7).
Fig. 2.
Segmentation performance in terms of Dice, 95th HD and MSD for the 3D. The different boxes show different MRI sequences as input: T1w (T1 weighted), T2w (T2 weighted), T1gd (T1 3D after gadolinium injection), T1gd and T2w (T1 3D after gadolinium injection and T2 weighted) and combining all sequences (All). The box includes points within the interquartile range (IQR) while the whiskers show points within 1.5 times the IQR.
3.3. Semi-automatic approach
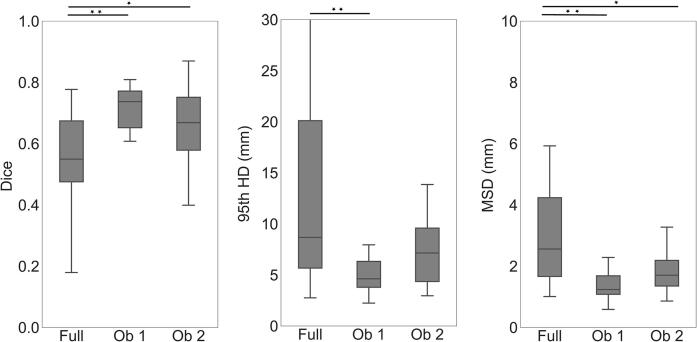
In Fig. 3, it is observed that the semi-automatic approach using the boxes of the first observer achieved a median Dice score of 0.74 (range 0.32–0.80), HD of 4.6 mm (range 2.2 mm–10.5 mm) and MSD of 1.2 mm (range 0.6 mm- 2.9 mm). For the second observer, the network achieved a median Dice score of 0.67 (range 0.28–0.87), HD of 7.2 mm (range of 3.0 mm–19.9 mm) and MSD of 1.7 mm (range of 0.9 mm–4.9 mm).
Fig. 3.
Segmentation performance of the semi-automatic approach with boxes drawn by two human observers. We compare the semi-automatic results (Ob 1 and Ob 2) to the fully automatic approach (Full). The box includes points within the interquartile range (IQR) while the whiskers show points within 1.5 times the IQR. Significance is represented as one star (*) for p < 0.01 and two stars (**) for p < 0.001.
The semi-automatic approach significantly outperformed the fully automatic approach in all of the metrics for the first observer (p < 0.001) and in Dice and MSD for the second observer (p < 0.01). These results were expressed for 19 out of the 20 patients in the test set (also for the fully automatic approach - equivalent to “All’ in Fig. 2), as one of the observers did not detect one of the tumors when asked to draw the clipbox.
The average time to draw the boxes was of 7.5 min per patient for the first observer and 2.8 min for the second observer.
3.4. Robustness to shifts
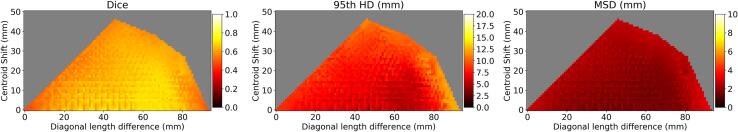
Fig. 4 shows the segmentation performance of the network trained for the semi-automatic approach as a function of the artificially induced shifts applied to the tumor within the clipbox. For centroid displacements below 20 mm and diagonal length differences of between 25 mm and 60 mm the Dice was consistently greater than 0.70, the HD was lower than 6.5 mm and the MSD was lower than 1.7 mm.
Fig. 4.
Robustness analysis. Segmentation performance in terms of median Dice, 95th HD and MSD for the semi-automatic approach as a function of the tumor centroid displacement and the clipbox diagonal length difference. The grey areas correspond to undetermined values due to the geometric constraints (i.e. no combination of shifts can achieve those values of centroid displacement and diagonal length difference).
3.5. Qualitative results
Fig. 5a and 5b show examples in which the shape of the semi-automatic approach output and ground truth segmentation agreed while the fully automatic approach oversegmented (a) or undersegmented (b) the tumor. Fig. 5c shows a case where the segmentation by the network trained with the fully automatic approach showed a similar shape to the ground truth segmentation but there were additional false positive volumes on the image.
Fig. 5.
Comparison of the oropharyngeal segmentations in three different patients (a, b, c) trained with the fully automatic approach (red), with the semi-automatic approach (blue) and the manual delineation (green). The yellow boxes are the boxes drawn by the observer. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
It was shown that using multiple MRI sequences yielded better results compared to using a single sequence as input. Also, decreasing the amount of context given to the CNN improved the segmentation performance. Finally, a functional semi-automatic approach that outperformed the fully automatic baseline was proposed and it was shown to be robust to clipbox selection errors, suggesting its potential clinical applicability.
Our network resulted in worse performance in terms of Dice compared to other tumor sites as reported by Sahiner et al. [19], where the authors provide a comparison of CNN segmentations for different tumor/lesions (Dices: 0.51–0.92). However, lower performance for oropharyngeal tumor segmentation is consistent with what is known about the inter-observer variability for this subsite: Blinde et al. [10] have shown differences in volume of up to 10 times among observers when segmenting OPSCC on MR, indicating the complexity of this task even for human observers. In this study, the mean Dice between our observers was 0.8. However, this number is an overestimation of the interobserver variability, considering that one of the observers corrected the other’s delineation.
No significant differences were found between training the network with full context in 3D compared to its 2D counterpart. This shows that reducing the resolution due to memory constrains in the 3D case is not critical for the segmentation performance when the full image is used as input.
When restricting the context, the network outperformed significantly the full context approach for all metrics. This means that local textural differences between tumor and immediate surrounding tissues are sufficient for delineation.
Using clipboxes drawn by human observers demonstrates the feasibility of a semi-automatic approach for OPSCC primary tumor segmentation. Additionally, these boxes were drawn by two independent observers with different backgrounds and levels of expertise, suggesting that the method is not highly sensitive to the observer. This is supported by the results of our robustness analysis, which showed that when training with shifted versions of the clipbox, the networks were fairly robust to these shifts. More concretely, the network was robust centroid displacements below 20 mm and diagonal length differences of between 25 mm and 60 mm, which we consider a fair estimate of the maximum error an observer can make when selecting the clipbox.
A fully manual segmentation can take from 30 min to almost 2 h (depending on the shape and size of the tumor), the average time between our two observers for the semi-automatic approach can take an average of 5 min (average of our two observers). Although after the proposed semi-automatic approach, some manual adaptations may be needed by a radiation oncologists to make the contours clinically acceptable, the overall process is expected to be less labor-intensive. Additionally, in the clinic it would be possible to use software designed to draw the clipboxes faster. Consequently, a functional semi-automatic system is not only feasible in terms of segmentation performance but also relevant for speeding up the radiotherapy workflow.
There are limitations in this study. First, given the high interobserver variability of OPSCC delineation, we are likely training the network with imperfect ground truths. However, we palliated the possible errors on the delineations by having the second observer correcting the first observer’s delineation. Secondly, we used a standard 3D UNet in our studies. Despite the extensive literature on deep learning architecture modifications, investigating the best architecture for this task is outside of our scope. Thirdly, our results would need validation with an independent cohort in a multi-center study. Furthermore, the scan protocols were not standardized in our dataset. Arguably, that makes the network robust to such differences (e.g. TR/TE), given that the network has learnt from a diverse dataset. Finally, our work can still be improved by adding other MRI sequences into the training (such as DWI) or by fully automatizing our semi-automatic approach, but we leave that as future work.
There is an increasing interest in the literature about differences on the tumors depending on their HPV status. According to Bos et al. [20], HPV positive tumors present on MRI post contrast with rounder shapes, lower maximum intensity values, and texture homogeneity. One strength of our work is that we include both HPV positive and HPV negative tumors in the training set, making the networks able to segment both subtypes of OPSCC. To check that the network is not biased to the HPV status, we compared the performance of the network stratified per HPV status and found non-significant results. We also did not find any relationship between performance and size.
In conclusion, this is the first study of primary tumor segmentation in the OPSCC site on MRI images with CNNs to the best of our knowledge. We trained a standard 3D UNet architecture using full MRI images as input. We showed that combining MRI sequences is beneficial for OPSCC segmentation with CNNs. Additionally, the CNN trained with reduced context around the tumor outperformed the fully automatic baseline and approaches that of other tumor sites reported in the literature. Hence, our proposed semi-automatic approach can save time in the clinic while achieving competitive performance and being robust to the choice of observer and manual clipbox selection errors.
Disclosure statement
The authors report no conflicts of interest.
Data statement
The data that has been used in this study is confidential. The institutional review board approved the study (IRBd18047). Informed consent was waived considering the retrospective design.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.phro.2021.06.005.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Fitzmaurice C., Allen C., Barber R.M., Barregard L., Bhutta Z.A., Brenner H. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015. JAMA Oncol [Internet] 2017;3:524. doi: 10.1001/jamaoncol.2016.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Delaney G., Jacob S., Barton M. Estimation of an optimal external beam radiotherapy utilization rate for head and neck carcinoma. Cancer. 2005;103:2216–2227. doi: 10.1002/cncr.21084. [DOI] [PubMed] [Google Scholar]
- 3.D Souza AM, Chen L, Wu Y, Abidin AZ, Xu C, Wismüller A. MRI tumor segmentation with densely connected 3D CNN. 2018. SPIE Medical Imaging Proceedings, 10574. doi: 10.1117/12.2293394.
- 4.Li J., Chen H., Li Y., Peng Y. ACM international conference proceeding series. 2019. A novel network based on densely connected fully convolutional networks for segmentation of lung tumors on multi-modal MR images; pp. 1–5. doi: 10.1145/3358331.3358400. [Google Scholar]
- 5.Li X., Chen H., Qi X., Dou Q., Fu C.W., Heng P.A. H-DenseUNet: Hybrid Densely Connected UNet for Liver and Tumor Segmentation from CT Volumes. IEEE Trans Med Imaging. 2018;37:2663–2674. doi: 10.1109/tmi.2018.2845918. [DOI] [PubMed] [Google Scholar]
- 6.Trebeschi S., van Griethuysen J.J.M., Lambregts D.M.J., Lahaye M.J., Parmar C., Bakers F.C.H. Deep Learning for Fully-Automated Localization and Segmentation of Rectal Cancer on Multiparametric MR. Sci Rep. 2017;7 doi: 10.1038/s41598-017-05728-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo Z., Guo N., Gong K., Zhong S., Li Q. Gross tumor volume segmentation for head and neck cancer radiotherapy using deep dense multi-modality network. Phys Med Biol. 2019;64:205015. doi: 10.1088/1361-6560/ab440d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cardenas C.E., McCarroll R.E., Court L.E., Elgohari B.A., Elhalawani H., Fuller C.D. Deep learning algorithm for auto-delineation of high-risk oropharyngeal clinical target volumes with built-in dice similarity coefficient parameter optimization function. Int J Radiat Oncol Biol Phys. 2018;101:468–478. doi: 10.1016/j.ijrobp.2018.01.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Men K., Chen X., Zhang Y., Zhang T., Dai J., Yi J. Deep deconvolutional neural network for target segmentation of nasopharyngeal cancer in planning computed tomography images. Front Oncol. 2017;7(315) doi: 10.3389/fonc.2017.00315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blinde S., Mohamed A.S.R., Al-Mamgani A., Newbold K., Karam I., Robbins J.R. Large interobserver variation in the international MR-LINAC oropharyngeal carcinoma delineation study. Int J Radiat Oncol. 2017;99:E639–E640. [Google Scholar]
- 11.Anderson C.M., Sun W., Buatti J.M., Maley J.E., Policeni B., Mott S.L. Interobserver and intermodality variability in GTV delineation on simulation CT, FDG-PET, and MR Images of Head and Neck Cancer. Jacobs J Radiat Oncol. 2014;1:006. [PMC free article] [PubMed] [Google Scholar]
- 12.Çiçek Ö, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation BT - Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016. 2016. 424–32.
- 13.Zeng G, Yang X, Li J, Yu L, Heng PA, Zheng G. 3D U-net with multi-level deep supervision: Fully automatic segmentation of proximal femur in 3D MR images. Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). 2017. 274–82. doi: 10.1007/978-3-319-67389-9_32.
- 14.Gordienko Y., Gang P., Hui J., Zeng W., Kochura Y., Alienin O. Deep learning with lung segmentation and bone shadow exclusion techniques for chest X-ray analysis of lung cancer. Adv Intell Syst Comput. 2019;754:638–647. doi: 10.1007/978-3-319-91008-6_63. [DOI] [Google Scholar]
- 15.Norman B., Pedoia V., Majumdar S. Use of 2D U-net convolutional neural networks for automated cartilage and meniscus segmentation of knee MR imaging data to determine relaxometry and morphometry. Radiology. 2018;288:177–185. doi: 10.1148/radiol.2018172322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation. In: Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). 2015. 234–41. doi: 10.1007/978-3-319-24574-4_28.
- 17.Sudre CH, Li W, Vercauteren T, Ourselin S, Jorge Cardoso M. Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. In: Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). 2017. 10553. doi: 10.1007/978-3-319-67558-9_28. [DOI] [PMC free article] [PubMed]
- 18.Kingma DP, Ba JL. Adam: A method for stochastic optimization. 3rd International Conference on Learning Representations, ICLR 2015 - Conference Track Proceedings. 2015.
- 19.Sahiner B., Pezeshk A., Hadjiiski L.M., Wang X., Drukker K., Cha K.H. Deep learning in medical imaging and radiation therapy. Med Phys. 2019;46:e1–e36. doi: 10.1002/mp.13264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bos P., Brekel M.W.M., Gouw Z.A.R., Al‐Mamgani A., Waktola S., Aerts H.J.W.L. Clinical variables and magnetic resonance imaging-based radiomics predict human papillomavirus status of oropharyngeal cancer. Head Neck. 2021;43:485–495. doi: 10.1002/hed.26505. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.