Abstract

Mass spectrometry imaging (MSI) of lipids within tissues has significant potential for both biomolecular discovery and histopathological applications. Conventional MSI technologies are, however, challenged by the prevalence of phospholipid regioisomers that differ only in the location(s) of carbon–carbon double bonds and/or the relative position of fatty acyl attachment to the glycerol backbone (i.e., sn position). The inability to resolve isomeric lipids within imaging experiments masks underlying complexity, resulting in a critical loss of metabolic information. Herein, ozone-induced dissociation (OzID) is implemented on a mobility-enabled quadrupole time-of-flight (Q-TOF) mass spectrometer capable of matrix-assisted laser desorption/ionization (MALDI). Exploiting the ion mobility region in the Q-TOF, high number densities of ozone were accessed, leading to ∼1000-fold enhancement in the abundance of OzID product ions compared to earlier MALDI-OzID implementations. Translation of this uplift into imaging resulted in a 50-fold improvement in acquisition rate, facilitating large-area mapping with resolution of phospholipid isomers. Mapping isomer distributions across rat brain sections revealed distinct distributions of lipid isomer populations with region-specific associations of isomers differing in double bond and sn positions. Moreover, product ions arising from sequential ozone- and collision-induced dissociation enabled double bond assignments in unsaturated fatty acyl chains esterified at the noncanonical sn-1 position.
Introduction
Lipids, the primary constituents of cell membranes,1 are key players in many biochemical functions including membrane structure,2 energy storage,3 signal transduction,4 and apoptosis,5 as well as cancer metabolism.6 The function and bioactivity of a lipid are dependent on its chemical structure. Subtle alterations in the molecular structure of glycerophospholipids, arising from acyl chain and/or double bond (db) positions can influence the physicochemical properties of membranes,7 and lipid–protein interactions can be specific to isomeric structures.8 Moreover, lipid synthesis is precisely regulated in different tissues9 and altered in cancer metabolism6,10 and diabetes.11
Elucidating the composition of tissue and cellular lipidomes is spearheaded by continually advancing mass spectrometry (MS) technologies.12 The workhorse of lipidomics is the analysis of lipid extracts via electrospray ionization (ESI) using either direct infusion (“shotgun”)12 or hyphenated chromatographic13 liquid chromatography–mass spectrometry (LC-MS) approaches. When coupled with high mass-resolving power and accuracy, such methods are capable of identifying many lipid species to the sum-composition level (i.e., identification of lipid class, total number of carbons, and degree of unsaturation).14 Tandem mass spectrometry, employing low-energy collision-induced dissociation (CID), provides additional structural information such as the length and degree of unsaturation of individual fatty acyl chains (i.e., the molecular lipid level). Despite the widespread and ever-growing popularity of these methods, they are still far from resolving all structural features. Fatty acyl stereospecific numbering (sn-) position in glycerophospholipids, carbon–carbon db location, and db geometry (i.e., cis or trans) are all unresolved in such methods. Considering the possibility of more than 180 000 unique lipid structures in nature,15 and the intimate link between lipid structure and function, the development of isomer resolution represents a frontier challenge for lipidomics.
The drive to improve structural specificity in lipid mass spectrometry has led to a rapid growth in the application of ion-activation and derivatization methods, with a particular focus toward assigning db and sn positions in glycerophospholipids. For direct infusion and LC-MS lipid analysis, options include ozone-induced dissociation (OzID),16 ultraviolet photodissociation (UVPD),17 ion–ion reactions,18 electron impact excitation of ions from organics (EIEIO),19−21 epoxidation,22 and the Paterno–Buchi (PB) reaction.23 These techniques identify db locations and can be used in combination with CID to support sn-positional assignments.17,24 In general, these methods involve either (i) the application of a derivatization agent prior to ionization within the mass spectrometer with subsequent CID (e.g., PB and epoxidation); or (ii) direct activation of mass-selected lipid ions within the mass spectrometer (e.g., OzID, UVPD, ion–ion reactions, and EIEIO).
Extraction-based techniques are the gold standard for lipid identification; however, they may mask localized changes in the lipidome of heterogeneous tissues. Recent work has shown the utility of combining mass spectrometry imaging (MSI) with PB,25−27 epoxidation,28 UVPD,29 OzID,30 ion–ion reactions,31 and EIEIO32 for isomer-resolved MSI. For PB and epoxidation, reaction times are typically on the order of several minutes, with conversion efficiencies up to 40%.33 One disadvantage, however, is that derivatization increases signal degeneracy—particularly for polyunsaturated lipids—dividing ion populations into derivatized, underivatized, and fragmented product ions.
UVPD-MSI has revealed db-positional isomers in multiple tissue types.29,34 Multiple diagnostic fragments of db-positional isomers are produced through UVPD, although overall sensitivity is low due to inefficient dissociation (less than 1% for db-specific fragments).34 When applied to MSI, only fatty acids34 and phosphatidylcholines (PC)29 have been analyzed through UVPD; however, the technique has also been demonstrated on other glycerophospholipids17 and sphingolipids35 from homogenized lipid extracts. Additionally, sequential CID and UVPD experiments can distinguish sn isomers; however, only db- or sn isomers have been distinguished in a single experiment.17,29,35 Comparatively, OzID can elucidate lipid db or sn structures.11,36 To establish fatty acyl db location(s), lipid precursor ions are first trapped in the presence of ozone. This causes fragmentation at the site(s) of unsaturation and thus, neutral loss masses that are diagnostic for specific db locations. Separately, fatty acyl sn position can be established through consecutive CID and OzID (CID/OzID) events. Lipid precursors first undergo CID, which leads to the loss of the phospho-headgroup and a cyclic ketal rearrangement including the formation of a new db anchoring the sn-2 fatty acyl chain to the glycerol backbone. Subsequent ozonolysis of this db selectively liberates the sn-2 chain enabling the explicit assignment of the composition of fatty acids at sn-1 and sn-2 positions.24
The main impediment to combining OzID with matrix-assisted laser desorption/ionization-MSI (MALDI-MSI) is the slow rate of reaction between ozone and the ionized lipid. In previous implementations, reaction times of up to 10 s per pixel for db position analysis have been required to produce sufficient diagnostic fragments, resulting in long acquisition times for even relatively small tissue areas.30 CID/OzID is several orders of magnitude faster due to the cyclic ketal activating the carbon–carbon double bond, allowing sn isomer identification in as little as 250 ms—a time frame that is more compatible with the sampling frequency of MSI experiments.30 Recent implementation of OzID in the high-pressure ion mobility region of SYNAPT mass spectrometers has provided up to 1000-fold enhancement in reaction rates.37,38 This configuration results in the production of abundant OzID product ions in the time needed for ions to transit the traveling-wave ion mobility region. This provides several benefits for MSI: (i) the ability to operate the MS using its native duty cycle, allowing for either imaging of larger areas or higher spatial resolution in practical time frames; (ii) detection of low-abundance isomers due to increased sensitivity; and (iii) the ability to generate OzID and CID/OzID fragments simultaneously, if precursor ion populations are preactivated by collisions prior to entering the ion-molecule reaction zone.
In this work, we report the implementation of isomer-resolved MALDI-MSI on a SYNAPT HDMS G2-Si Q-TOF mass spectrometer, leveraging fast OzID for the imaging of both db and sn isomers in biological tissues. This unique combination provided an ∼50-fold gain in acquisition speed while achieving higher product ion yields; the ability to perform simultaneous OzID and CID/OzID reactions; and the observation of third- and fourth-generation product ions (CID/OzID2). The latter afford explicit assignment of db positions associated with specific sn fatty acyls. This platform provides a key breakthrough toward the goal of studying the distributions of structurally defined lipids in tissues.
Methods
Materials
2,5-Dihydroxyacetophenone (DHA, 97% purity), sodium acetate (anhydrous, >99%), and chloroform (≥99%) were purchased from Sigma-Aldrich (Zwijndrecht, The Netherlands) and used without further purification. Methanol (ULC-MS grade), water (LC-MS grade), ethanol (LC-MS grade), and xylene (AR grade) were purchased from Biosolve (Valkenswaard, The Netherlands). Hematoxylin and Entellan were purchased from Merck (Darmstadt, Germany) and eosin Y from J.T. Baker (Center Valley). Indium tin oxide (ITO)-coated glass slides were purchased from Delta Technologies (Loveland). Lipid standards (PC 18:0/18:1n-9 and PC 16:0/18:1n-9) were purchased from Avanti Polar Lipids (Alabaster).
Biological Samples
Rats, housed and cared for at the Central Animal Facility of Maastricht University according to local standards, were provided ad libitum access to water and regular chow. Healthy rat brain was obtained in accordance with protocols approved by the Animal Care and Use Committee (DEC number 2014-120). Horizontal sections of thickness 10 μm were prepared using a cryo-microtome (Leica, Nussloch, Germany) at −20 °C, thaw-mounted on ITO-coated glass slides, and stored at −80 °C until matrix application and MSI analysis.
Sample Preparation
An automated TM-Sprayer (HTX Technologies, LLC, Chapel Hill) was used for sodium acetate deposition. First, 15 layers of sodium acetate solution (4 mg/mL in 2:1 CHCl3/MeOH (v/v)) were deposited at 0.12 mL/min, 30 s drying time between layers, at 30 °C, preferentially forming sodiated ions (Figure S1). Following this, matrix was applied by sublimation: 40 mg of DHA for 4 min at 160 °C with a pressure of <4 × 10–5 bar.
Lipid standards were prepared at 0.01 and 0.1 g/L concentrations in 2:1 CHCl3/MeOH and sprayed on clean ITO slides for 1–10 layers following the same protocol as for sodium acetate deposition, creating concentrations from 0.33 to 3.3 ng/mm2 (assuming equal dispersion). Samples were then treated equivalently to brain tissue sections.
Hematoxylin and eosin (H&E) staining was performed after MALDI imaging. Matrix was removed from tissue by immersion in 70% ethanol for 3 min. A standard H&E protocol was then used. High-resolution optical images of stained tissues were generated using an Aperio CS2 digital pathology slide scanner (Leica Biosystems, Wetzlar, Germany).
Mass Spectrometry Imaging Instrumentation
Tissue sections were analyzed using a Waters prototype μMALDI source mounted to a Waters SYNAPT HDMS G2-Si system (Waters Corporation, Manchester, U.K.), as previously described by Barré et al.39 Samples were analyzed in continuous raster mode using waters research enabled software (WRENS) to operate at 5 pixels/s, laser repetition rate of 1500 Hz, pixel size set to 50 μm, MS/MS isolation window set to ±1.5 m/z, and detection set to sensitivity mode (mass resolution: ∼15 000). The laser spot size was approximately 15 × 15 μm. Operation of the T-Wave was optimized to generate the greatest abundance of selected diagnostic fragment ions without totally depleting the precursor signal (wave velocity, 1200 m/s; wave height, 38 V). Optimization of the ion mobility traveling-wave parameters (see Supporting Information Explanation 1, Figure S2) increased the OzID efficiency, allowing the acquisition speed to be increased 50 times compared to earlier OzID implementations (cf., linear ion-trap performance Figure S3) with sensitivity as low as 2.2 fg/μm2 (Figure S4). The dominant factor in this increased performance was the greater number density of ozone (>10-fold) with gains from traveling-wave parameters facilitating small increases in reaction time giving a further 3- to 4-fold improvement. The trap and transfer collision energies were set at 4.0 and 2.0 V, respectively, for normal OzID, and trap energy was set to 40 V during CID/OzID experiments.
In-Line Ozone Generation
Ozone generation and delivery to the instrument are as described previously.40 Ozone was produced using a high-concentration ozone generator (TG-40; Ozone Solutions, Hull, IA) from UHP oxygen (5.0 grade, 20 psi @ 0.4 slm; Linde Gas Therapeutics Benelux bv, Eindhoven, The Netherlands). Ozone production was optimized to ∼275 gm−3 in O2 as measured by an in-line monitor (106-H; 2B Technologies, Boulder). The generated ozone/oxygen mixture was connected via a needle valve (SS-SS8; Swagelok) to the ion mobility cell gas inlet of the mass spectrometer, and the flow was adjusted to maintain a pressure of 2.9 mbar in the cell. Excess ozone was converted to oxygen using an unheated destruct catalyst (810-0008; IN USA, Inc., Norwood). Laboratory ambient ozone concentration was monitored (106-L; 2B Technologies, Boulder) and interlocked to shut off the generator if the background ozone level rose above 75 ppb. An instrument schematic is shown in Figure S5.
Data Analysis
WatersRawConverter (Waters Corporation, Manchester, U.K.) was used to convert WRENS data using a bin size of 1 Da. Data were visualized using in-house MATLAB scripts (version R2014a, MathWorks, Natick). Regions of interest (ROI) were manually selected to remove off-tissue regions when plotting images. Full-scan MS was visualized using total-ion-current normalization. Ratio images were created with the numerator being the sum of the aldehyde and Criegee OzID fragments of a single isomer and the denominator being the sum of the OzID fragments for all isomers. Spectra were averaged in MassLynx v4.1 and loaded into mMass software for offline recalibration and peak picking (S/N = 3). A total of 100 individual spectra were loaded into mMass to determine individual scan noise level to define the limit of detection and quantitation (3- and 10-fold of noise, respectively). Calibrant peaks were the most abundant OzID fragments (typically n-7 and n-9), along with phosphatidylcholine (m/z 184.07) and the ozonide of the precursor lipid.
Lipid Nomenclature
Lipid structure nomenclature is based on the recommendations of Liebisch et al.,41 whereby the site(s) of unsaturation are indicated by n-x, where x is the number of carbons relative to the methyl terminus of the acyl chain. The underscore “_” or slash “/” is used for unknown or known sn positions, respectively, of the fatty acids.
Results and Discussion
MALDI-OzID Imaging of Monounsaturated Phosphatidylcholines
Db-isomer distributions of the abundant monounsaturated phosphatidylcholines, PC 34:1 and PC 36:1, were investigated in rat brain tissue to assess the sensitivity and speed of the high-pressure MSI-OzID setup (Figure 1). PC 34:1 and PC 36:1 have previously been studied with isomer-resolved MSI using OzID,10,30 UVPD,17 and on-tissue PB reactions,27 which provide benchmarks for this approach. The MALDI-OzID spectrum of [PC 34:1 + Na]+ (Figure 1A) revealed product ions indicative of two db positions, namely, n-7 (m/z 700.45 and 716.45) and n-9 (m/z 672.42 and 688.42). Under optimized experiment conditions, OzID product ions were of comparable abundance to the mass-selected precursor ion, aiding isomer identification. Imaging of the intact precursor lipid distribution was performed in “TOF-only” mode on an adjacent tissue section from the same animal. The distribution of the [PC 34:1 + Na]+ ion at <2 ppm mass accuracy is shown in Figure 1B and represents the combined intensity distribution of signals arising from lipid isomers with this composition. The distributions of the individual PC 34:1n-7 and PC 34:1n-9 db isomers were extracted from the OzID data and were individually compared against the total abundance of product ions from all isomers to generate fractional distribution images (FDI), as shown in Figure 1C. Comparing the sum composition distribution with the isomer-resolved image reveals that although PC 34:1 shows higher abundance in the gray matter of the brain (Figure 1B), an underlying variation in the relative population of PC 34:1n-7 and PC 34:1n-9 is present (Figure 1C). A comparison between the isomer-resolved MSI and H&E-stained tissue revealed that the n-7 isomer is proportionately more abundant in gray matter and the cerebellum and less abundant in white matter and the cortex, consistent with previous isomer-resolved MSI studies.26−28,30 Compared to prior isomer-resolved MSI implementations, the acquisition time for the data reported here (∼4 h) is significantly shorter than would have been required to acquire equivalent data using earlier OzID-MSI30 (∼200 h) or UVPD-MSI29 (∼36 h) implementations, while simultaneously improving sensitivity for the diagnostic fragment ions. In addition, the faster reaction rate with our method enabled the ability to perform OzID-MSI experiments on non-alkali-adducted ions that typically exhibit lower reaction rates. The MALDI-OzID data of monounsaturated [PC 34:1 + H]+ is shown in Figure S7 and is similar to the results of [PC 34:1 + Na]+, showing that the n-7 isomer is proportionately more abundant in the gray matter of the cerebellum.
Figure 1.
MALDI-OzID of monounsaturated (A–C) [PC 34:1 + Na]+ and (D–F) [PC 36:1 + Na]+, showing different isomer distributions in rat brain. MALDI-OzID spectrum of (A) [PC 34:1 + Na]+ and (D) [PC 36:1 + Na]+, where labeled product ions indicate the presence of n-7 and n-9 db isomers. Distribution of precursor ions (B) [PC 34:1 + Na]+ at m/z 782.57 and (E) [PC 36:1 + Na]+ at m/z 810.61 extracted from TOF-only scans. Fractional distribution images of (C) PC 34:1n-7 and (F) PC 36:1n-7 obtained from MALDI-OzID, showing relative changes in n-7 isomers throughout the rat brain. In (B, C) and (E, F), H&E staining of the same tissue after the MSI experiments is shown on the left. Note that MALDI-OzID and MALDI-TOF images are acquired from different brain tissue sections from the same animal. Explanation of additional ions present in (A) is provided as Supporting Information Figure S6. A technical replicate of these experiments with MS1 and OzID measurements acquired on consecutive sections is provided in Figure S8.
A representative MALDI-OzID spectrum of mass-selected [PC 36:1 + Na]+ ions is shown in Figure 1D and reveals the presence of both n-7 (m/z 728.48 and 744.44) and n-9 (m/z 700.45 and 716.46) db isomers, in agreement with previous studies.30,40 The precursor ion distribution of m/z 810.61 showed that PC 36:1 was concentrated throughout the white matter (Figure 1E), in contrast to the correlation between PC 34:1 and gray matter. As was also observed in previous studies,30,42 the FDI (Figure 1F) revealed that the n-7 isomer was more prevalent in the gray matter and the cerebellum, while n-9 was more abundant in the white matter and the cortex.
MALDI-OzID Imaging of Polyunsaturated Phosphatidylcholines
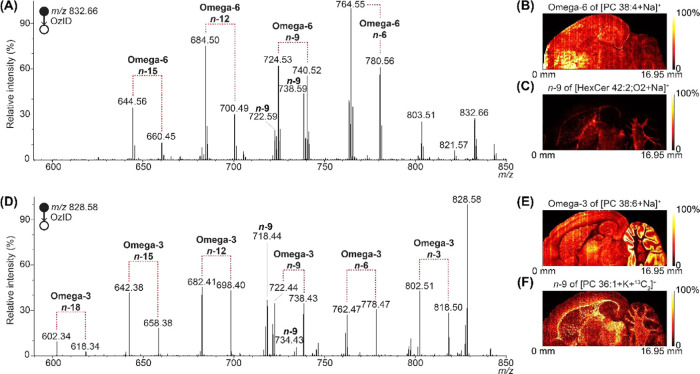
The increased sensitivity of the MALDI-MSI-OzID implementation is particularly important for polyunsaturated lipids because the diagnostic db-positional information is spread across multiple pairs of OzID fragments. OzID of mass-selected [PC 38:4 + Na]+ and [PC 38:6 + Na]+ produced spectra with sufficient S/N to allow spatial distributions to be extracted for each db-position (see Figures 2 and S9). The MALDI-OzID spectrum of [PC 38:4 + Na]+ revealed four double bonds at the n-6, 9, 12, and 15 positions (Figure 2A), which enables assignment of the lipid as PC 18:0_20:4n-6. Isobars around the n-9 neutral loss indicated the presence of an isobaric monounsaturated lipid, assigned as hexosylceramide HexCer 42:2;O2 by accurate mass. OzID imaging of [HexCer 42:2;O2 + Na]+ revealed that the distribution of this sphingolipid was distinct from PC 38:4 (Figure 2B,C). While PC 38:4 was widely distributed throughout the gray matter (Figure 2B), HexCer 42:2;O2 was highly localized to the white matter/myelin, which is known to be rich in glycosphingolipids (Figure 2C).
Figure 2.
MALDI-OzID of polyunsaturated PC lipids showing the distribution of the polyunsaturated phospholipid and an interfering lipid throughout the rat brain, showing different distributions in white/gray matter. (A) MALDI-OzID spectrum of [PC 38:4 + Na]+, revealing the presence of an omega-6 fatty acid. In addition, n-9 OzID fragments from an interference are visible in the spectrum, labeled as n-9. (B) Distribution images of the omega-6 and (C) the n-9 OzID fragments. (D) MALDI-OzID spectrum of [PC 38:6 + Na]+, revealing the presence of an omega-3 fatty acid. In addition, n-9 OzID fragments from an interference are visible in the spectrum, labeled as n-9. (E) Distribution images of the omega-3 and (F) the n-9 OzID fragments. For these images, 99th quantile hotspot removal was applied to the non-normalized data.
For [PC 38:6 + Na]+ (Figure 2D), the MALDI-OzID spectra showed six pairs of product ions corresponding to double bonds at the n-3, 6, 9, 12, 15, and 18 positions, assigning the lipid as PC 16:0_22:6n-3, which has been previously reported in rat brain30 and is shown here to be primarily present in the granular cell layer of the cerebellum (Figure 2E). As with PC 38:4, isobars around the n-9 neutral loss revealed the presence of an additional monounsaturated lipid, in this case, the [PC 36:1 + K + 13C2]+ isotopologue in the gray matter (Figure 2F). Interestingly, in these spectra, we observed an “inverted” isotopic distribution, created from the loss of either one or both 13C in the OzID neutral loss fragment. This could potentially be used as a diagnostic pattern for identifying isobaric and co-isolated 13C-containing lipids (Figure S10).
MALDI-COzID Imaging of Monounsaturated Phosphatidylcholines
CID/OzID has been used to identify sn-positional isomers of PC lipid species via an MS3 sequence. As only MS2-level activation could be performed with the SYNAPT, the CID products could not be mass-selected prior to ozonolysis. Instead, to generate CID/OzID product ions, a combined collision- and ozone-induced dissociation (COzID) strategy was employed.43 Mass-selected [M + Na]+ ions were activated in the trap region (i.e., prior to ozonolysis) with a 40 V collision energy, forming [M-183 + Na]+ head group loss fragment ions without significantly depleting the [M + Na]+ precursor. Subsequent reaction of these ions with ozone produced both CID/OzID and OzID product ions, providing db- and sn-positional isomer information in the same spectrum. This experiment sequence was performed for [PC 34:1 + Na]+ (Figure 3A), revealing product ions indicative of both n-7 (m/z 700.47 and 716.47) and n-9 (m/z 672.47 and 688.47) db positions in addition to CID/OzID fragments diagnostic for both 16:0 and 18:1 acyl chains at the sn-1 position (m/z 379.25/395.25 and 405.25/421.26, respectively; see Figure S11A). In accordance with previous investigations, the fractional distribution showed that the canonical PC 16:0/18:1 is elevated within the white matter, while the noncanonical PC 18:1/16:0 isomer is proportionately elevated in the gray matter (Figure 3B).30
Figure 3.
MALDI-COzID of (A–D) [PC 34:1 + Na]+ and (E–J) [PC 36:1 + Na]+ in rat brain obtained by preactivating the ions with CID (40 V) in the trap region prior to ozonolysis in the ion mobility cell. (A) COzID mass spectrum of [PC 34:1 + Na]+ showing diagnostic product ion pairs indicating the presence of db-positional (PC 34:1n-7 and PC 34:1n-9) and sn-positional (PC 16:0/18:1 and PC 18:1/16:0) isomers. Additional product ions observed in the spectrum are consistent with CID/OzID2 processes, which are regiospecific for the n-7 and n-9 db positions of the sn-1 18:1. (B–D) FDIs of the different isomers present, showing the intensity of signals specific for (B) PC 16:0/18:1, (C) PC 34:1n-7, and (D) [PC 18:1(n-7)/16:0 + Na]+ isomers. (E) COzID spectrum of [PC 36:1 + Na]+ showing diagnostic product ion pairs indicating the presence of db-positional (PC 36:1n-7 and PC 36:1n-9) and sn-positional (PC 18:0/18:1 and PC 18:1/18:0, PC 16:0/20:1 and PC 20:1/16:0) isomers. Additional product ions observed in the spectrum are consistent with CID/OzID2 processes, which are regiospecific for the n-7 and n-9 db positions of the sn-1 18:1 and sn-1 20:1. (F–J) FDIs of the different isomers present, showing the intensity of (F) PC 18:0/18:1, (G) PC 16:0/20:1, (H) PC 36:1(n-7) db position, (I) [PC 18:1(n-7)/18:0 + Na]+, and (J) [PC 20:1(n-7)/16:0 + Na]+ isomers. For the FDIs, an H&E staining of a consecutive section is shown on the left.
The observation of both db- and sn-specific product ions in a single spectrum (Figure 3A) supports the presence of at least three, and likely four, PC 34:1 isomers (i.e., PC 16:0/18:1n-7, PC 16:0/18:1n-9, PC 18:1n-7/16:0, and PC 18:1n-9/16:0). Additional information can be gleaned from low mass ions (i.e., m/z 250–350) in the same spectra, the presence of which indicates subsequent OzID reactions on CID/OzID fragment ions (i.e., CID/OzID2 product ions). These ions have been characterized previously by Pham et al. using explicit MS4 activation on an ion-trap mass spectrometer.24 Importantly, these product ions allow unambiguous assignment of db locations to sn-1 acyl chains based on the fragmentation sequence outlined in Figure S12. For example, in the case of [PC 34:1 + Na]+ (Figure 3A), the CID/OzID2 ions observed at m/z 295.12 and 323.15 reveal the presence of lipid isomers carrying both n-7 and n-9 db locations at the sn-1 position, namely, the noncanonical PC 18:1n-9/16:0 and PC 18:1n-7/16:0 lipids. Pham et al. also identified an alternative MS4 sequence, which uniquely assigns db isomers of the sn-2-substituted acyl chains. Unfortunately, these low m/z product ions were not detected under the COzID conditions used; however, sn-2 unsaturation can be inferred from the comparison between OzID and COzID spectra. In rat brain, CID/OzID2 product ions showed unique distributions of [PC 18:1n-7/16:0 + Na]+ and [PC 18:1n-9/16:0 + Na]+. Although the overall signal from PC 34:1n-7 was relatively more abundant in the gray matter of the cerebellum (Figure 3C), the noncanonical PC 18:1n-7/16:0 isomer appeared more abundant in the white matter of the brain (Figure 3D). Despite our inability to directly visualize sn-2-related db fragments in this experiment, a comparison between Figure 3C,D indicates that the location of double bonds in fatty acids is closely correlated with the sn position of the fatty acid.
The equivalent analysis for [PC 36:1 + Na]+ revealed the presence of both n-7 and n-9 isomers for each PC 18:0/18:1, PC 18:1/18:0, PC 16:0/20:1, and PC 20:1/16:0 (Figure 3E; magnification of m/z 250–500 is provided in Figure S11B). In line with previous investigations of [PC 36:1 + Na]+, these data reveal that PC 18:0/18:1 accounts for >90% of all PC 18:0_18:1 sn isomers (Figure 3F)30 based on the relative intensity of 18:0/18:1-specific peaks compared to the summed intensity of 18:0/18:1 and 18:1/18:0-specific signals, and the relative fractions of PC 18:0/18:1 and PC 18:1/18:0 do not change significantly throughout the brain. By comparison, PC 16:0/20:1 relative to the combined PC 16:0_20:1 signal is confined almost completely to the white matter (Figure 3G). The FDI of the db-locational isomers revealed PC 36:1n-7 to be relatively more abundant in the gray matter and cerebellum compared to the cortex (Figure 3H). Although the FDIs based on CID/OzID2 product ions had lower signal-to-noise ratios for PC 36:1, PC 18:1n-7/18:0 and PC 20:1n-7/16:0 isomers could still be observed; however, n-9 isomer species appeared entirely absent (Figure 3I–J). This indicates that the n-9 db is exclusively associated with the 18:1 or 20:1 chain substituted at the sn-2 position or was present below the detection limit of our analysis.
Conclusions
This work reports significant advancement for lipid isomer-resolved MSI by performing OzID in the high-pressure environment of the ion mobility region of a SYNAPT HDMS G2-Si mass spectrometer. Combining this approach with MALDI-MSI has provided: (i) 1000-fold shorter reaction times; (ii) OzID acquisition rates of up to 5 pixels s–1 (a 50-fold improvement from previous experiments); and (iii) simultaneous generation of OzID, CID/OzID, and CID/OzID2 product ions. Combined, these advances allow for OzID characterization of db-, sn-, and db+sn-positional isomers at suitable reaction times for MALDI-MSI.
The ability to probe both db- and sn-characteristic product ions creates the possibility to study regiospecific enzyme activity relating to both desaturation-elongation and remodeling processes.10,44 As lipids are both the substrates and the products of enzymatic processes, the spatial distribution of lipid isomers enables insight into the enzymatic processes occurring in distinct tissue types. Moreover, the enhanced sensitivity afforded by the OzID-MSI implementation reported within enables isomer-resolved imaging of less abundant glycerophospholipids, particularly from among the phospholipid classes. Direct observation of the similarities and differences between the spatial distribution of phospholipid classes and their isomer populations (including fatty acyl chain composition, regiochemistry, and db locations) will provide a signature for lipid modification and remodeling within different tissues. Isomer-resolved MSI is thus essential for providing holistic visualization of phospholipids populations across and between tissue samples. Ultimately, combining this technology with multiplexing techniques (such as SONAR45 or DDA30) will allow for a broader interrogation of tissue lipid metabolism by profiling multiple lipid precursor ions within a single imaging acquisition. Improvements in mass spectral sensitivity or source ionization could allow for the interrogation of even lesser abundant species or enable acquisitions with smaller pixel sizes. Taken together, this work represents a significant advance in capabilities for isomer-resolved MSI lipidomics, including the unambiguous imaging of lipid species at the full-structure level.41
Acknowledgments
This work was supported by the Dutch province of Limburg through the LINK program. B.S.R.C. and R.M.A.H. acknowledge funding from the National Cancer Institute of the NIH, Grant No. ROI1 CA213492. A.P.B, S.R.E., and R.M.A.H. acknowledge funding from Interreg V-A EMR and the Netherlands Ministry of Economic Affairs within the “Interreg Euro-Maas-Rijn” project (project number EMR23). B.L.J.P. and S.J.B. acknowledge funding from the Australian Research Council (LP180100238 and DP190101486). R.S.E.Y. acknowledges support through an RTP scholarship funded by the Australian Government and also extends gratitude toward the Central Analytical Research Facility at QUT for access to their facilities. S.R.E. acknowledges funding from the Australian Research Council Future Fellowship Scheme (grant number FT190100082) and the Netherlands Organization for Scientific Research VIDI scheme (grant number 198.011). The authors are grateful to Tiffany Porta-Siegel and Hang Nguyen for manuscript editing and to Martin R.L. Paine for sectioning the rat brains.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.analchem.1c01377.
Sodium acetate application effect; (Figure S1); instrument parameter optimization (Explanation S1); wave height and wave velocity optimization (Figure S2); comparison of OzID performed in the SYNAPT vs the ion trap of the LTQ Orbitrap Elite (Figure S3); sensitivity test (Figure S4); schematic of the modified μMALDI-Synapt-G2-Si system (Figure S5); additional ions observed in Figure 1A (Figure S6); MALDI-OzID of the monounsaturated [PC 34:1 + H]+ (Figure S7); technical replicate of MALDI-OzID of monounsaturated PCs (Figure S8); spatial distribution of the single db ion pairs of the polyunsaturated PC lipids (Figure S9); inverted isotopic distribution of [PC 36:1 + K + 13C2]+ (Figure S10); enlargement of m/z 250–500 of the COzID spectra (Figure S11); and proposed CID/OzID2 reaction scheme for [PC 16:0_18:1 + Na]+ (Figure S12) (PDF)
Author Contributions
# B.S.R.C. and A.P.B. contributed equally to this work.
The authors declare no competing financial interest.
Dedication
This work is dedicated to the memory of Dr. Martin R. L. Paine (1982–2021), a great friend, colleague, and true believer in OzID and MSI technologies. Marty’s insights and enthusiasm inspired this paper and continue to motivate the authors to do their best science with one eye always on the fun.
Supplementary Material
References
- van Meer G. Cellular lipidomics. EMBO J. 2005, 24, 3159–3165. 10.1038/sj.emboj.7600798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soteriou C.; Kalli A. C.; Connell S. D.; Tyler A. I. I.; Thorne J. L. Advances in understanding and in multi-disciplinary methodology used to assess lipid regulation of signalling cascades from the cancer cell plasma membrane. Prog. Lipid Res. 2020, 81, 101080 10.1016/j.plipres.2020.101080. [DOI] [PubMed] [Google Scholar]
- Dautel S. E.; Kyle J. E.; Clair G.; Sontag R. L.; Weitz K. K.; Shukla A. K.; Nguyen S. N.; Kim Y. M.; Zink E. M.; Luders T.; Frevert C. W.; Gharib S. A.; Laskin J.; Carson J. P.; Metz T. O.; Corley R. A.; Ansong C. Lipidomics reveals dramatic lipid compositional changes in the maturing postnatal lung. Sci. Rep. 2017, 7, 40555 10.1038/srep40555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jové M.; Pradas I.; Naudi A.; Rovira-Llopis S.; Banuls C.; Rocha M.; Portero-Otin M.; Hernandez-Mijares A.; Victor V. M.; Pamplona R. Lipidomics reveals altered biosynthetic pathways of glycerophospholipids and cell signaling as biomarkers of the polycystic ovary syndrome. Oncotarget 2018, 9, 4522–4536. 10.18632/oncotarget.23393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyurina Y. Y.; St Croix C. M.; Watkins S. C.; Watson A. M.; Epperly M. W.; Anthonymuthu T. S.; Kisin E. R.; Vlasova I. I.; Krysko O.; Krysko D. V.; Kapralov A. A.; Dar H. H.; Tyurin V. A.; Amoscato A. A.; Popova E. N.; Bolevich S. B.; Timashev P. S.; Kellum J. A.; Wenzel S. E.; Mallampalli R. K.; Greenberger J. S.; Bayir H.; Shvedova A. A.; Kagan V. E. Redox (phospho)lipidomics of signaling in inflammation and programmed cell death. J. Leukocyte Biol. 2019, 106, 57–81. 10.1002/JLB.3MIR0119-004RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos C. R.; Schulze A. Lipid metabolism in cancer. FEBS J. 2012, 279, 2610–2623. 10.1111/j.1742-4658.2012.08644.x. [DOI] [PubMed] [Google Scholar]
- Martinez-Seara H.; Rog T.; Pasenkiewicz-Gierula M.; Vattulainen I.; Karttunen M.; Reigada R. Interplay of unsaturated phospholipids and cholesterol in membranes: effect of the double-bond position. Biophys. J. 2008, 95, 3295–3305. 10.1529/biophysj.108.138123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzawa-Itoh K.; Aoyama H.; Muramoto K.; Terada H.; Kurauchi T.; Tadehara Y.; Yamasaki A.; Sugimura T.; Kurono S.; Tsujimoto K.; Mizushima T.; Yamashita E.; Tsukihara T.; Yoshikawa S. Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J. 2007, 26, 1713–1725. 10.1038/sj.emboj.7601618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradas I.; Huynh K.; Cabre R.; Ayala V.; Meikle P. J.; Jove M.; Pamplona R. Lipidomics Reveals a Tissue-Specific Fingerprint. Front. Physiol. 2018, 9, 1165 10.3389/fphys.2018.01165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. S. E.; Bowman A. P.; Williams E. D.; Tousignant K. D.; Bidgood C. L.; Narreddula V. R.; Gupta R.; Marshall D. L.; Poad B. L. J.; Nelson C. C.; Ellis S. R.; Heeren R. M. A.; Sadowski M. C.; Blanksby S. J. Apocryphal FADS2 activity promotes fatty acid diversification in cancer. Cell Rep. 2021, 34, 108738 10.1016/j.celrep.2021.108738. [DOI] [PubMed] [Google Scholar]
- Ståhlman M.; Pham H. T.; Adiels M.; Mitchell T. W.; Blanksby S. J.; Fagerberg B.; Ekroos K.; Boren J. Clinical dyslipidaemia is associated with changes in the lipid composition and inflammatory properties of apolipoprotein-B-containing lipoproteins from women with type 2 diabetes. Diabetologia 2012, 55, 1156–1166. 10.1007/s00125-011-2444-6. [DOI] [PubMed] [Google Scholar]
- Yang K.; Cheng H.; Gross R. W.; Han X. Automated lipid identification and quantification by multidimensional mass spectrometry-based shotgun lipidomics. Anal. Chem. 2009, 81, 4356–4368. 10.1021/ac900241u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holčapek M.; Liebisch G.; Ekroos K. Lipidomic Analysis. Anal. Chem. 2018, 90, 4249–4257. 10.1021/acs.analchem.7b05395. [DOI] [PubMed] [Google Scholar]
- Rustam Y. H.; Reid G. E. Analytical Challenges and Recent Advances in Mass Spectrometry Based Lipidomics. Anal. Chem. 2018, 90, 374–397. 10.1021/acs.analchem.7b04836. [DOI] [PubMed] [Google Scholar]
- Yetukuri L.; Ekroos K.; Vidal-Puig A.; Oresic M. Informatics and computational strategies for the study of lipids. Mol. BioSyst. 2008, 4, 121–127. 10.1039/B715468B. [DOI] [PubMed] [Google Scholar]
- Thomas M. C.; Mitchell T. W.; Harman D. G.; Deeley J. M.; Nealon J. R.; Blanksby S. J. Ozone-induced dissociation: elucidation of double bond position within mass-selected lipid ions. Anal. Chem. 2008, 80, 303–311. 10.1021/ac7017684. [DOI] [PubMed] [Google Scholar]
- Williams P. E.; Klein D. R.; Greer S. M.; Brodbelt J. S. Pinpointing Double Bond and sn-Positions in Glycerophospholipids via Hybrid 193 nm Ultraviolet Photodissociation (UVPD) Mass Spectrometry. J. Am. Chem. Soc. 2017, 139, 15681–15690. 10.1021/jacs.7b06416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randolph C. E.; Blanksby S. J.; McLuckey S. A. Toward Complete Structure Elucidation of Glycerophospholipids in the Gas Phase through Charge Inversion Ion/Ion Chemistry. Anal. Chem. 2020, 92, 1219–1227. 10.1021/acs.analchem.9b04376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J. L.; Baba T. Near-Complete Structural Characterization of Phosphatidylcholines Using Electron Impact Excitation of Ions from Organics. Anal. Chem. 2015, 87, 5837–5845. 10.1021/acs.analchem.5b01460. [DOI] [PubMed] [Google Scholar]
- Baba T.; Campbell J. L.; Le Blanc J. C. Y.; Baker P. R. S.; Ikeda K. Quantitative structural multiclass lipidomics using differential mobility: electron impact excitation of ions from organics (EIEIO) mass spectrometry. J. Lipid Res. 2018, 59, 910–919. 10.1194/jlr.D083261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T.; Campbell J. L.; Le Blanc J. C. Y.; Baker P. R. S. Distinguishing Cis and Trans Isomers in Intact Complex Lipids Using Electron Impact Excitation of Ions from Organics Mass Spectrometry. Anal. Chem. 2017, 89, 7307–7315. 10.1021/acs.analchem.6b04734. [DOI] [PubMed] [Google Scholar]
- Zhao Y.; Zhao H.; Zhao X.; Jia J.; Ma Q.; Zhang S.; Zhang X.; Chiba H.; Hui S. P.; Ma X. Identification and Quantitation of C=C Location Isomers of Unsaturated Fatty Acids by Epoxidation Reaction and Tandem Mass Spectrometry. Anal. Chem. 2017, 89, 10270–10278. 10.1021/acs.analchem.7b01870. [DOI] [PubMed] [Google Scholar]
- Cao W.; Cheng S.; Yang J.; Feng J.; Zhang W.; Li Z.; Chen Q.; Xia Y.; Ouyang Z.; Ma X. Large-scale lipid analysis with C=C location and sn-position isomer resolving power. Nat. Commun. 2020, 11, 375 10.1038/s41467-019-14180-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham H. T.; Maccarone A. T.; Thomas M. C.; Campbell J. L.; Mitchell T. W.; Blanksby S. J. Structural characterization of glycerophospholipids by combinations of ozone- and collision-induced dissociation mass spectrometry: the next step towards ″top-down″ lipidomics. Analyst 2014, 139, 204–214. 10.1039/C3AN01712E. [DOI] [PubMed] [Google Scholar]
- Su Y.; Ma X.; Page J.; Shi R.; Xia Y.; Ouyang Z. Mapping Lipid C=C Location Isomers in Organ Tissues by Coupling Photochemical Derivatization and Rapid Extractive Mass Spectrometry. Int. J. Mass Spectrom. 2019, 445, 116206 10.1016/j.ijms.2019.116206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wäldchen F.; Spengler B.; Heiles S. Reactive Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging Using an Intrinsically Photoreactive Paterno-Buchi Matrix for Double-Bond Localization in Isomeric Phospholipids. J. Am. Chem. Soc. 2019, 141, 11816–11820. 10.1021/jacs.9b05868. [DOI] [PubMed] [Google Scholar]
- Bednařík A.; Bolsker S.; Soltwisch J.; Dreisewerd K. An On-Tissue Paterno-Buchi Reaction for Localization of Carbon-Carbon Double Bonds in Phospholipids and Glycolipids by Matrix-Assisted Laser-Desorption-Ionization Mass-Spectrometry Imaging. Angew. Chem., Int. Ed. 2018, 57, 12092–12096. 10.1002/anie.201806635. [DOI] [PubMed] [Google Scholar]
- Kuo T.-H.; Chung H.-H.; Chang H.-Y.; Lin C.-W.; Wang M.-Y.; Shen T.-L.; Hsu C.-C. Deep Lipidomics and Molecular Imaging of Unsaturated Lipid Isomers: A Universal Strategy Initiated by mCPBA Epoxidation. Anal. Chem. 2019, 91, 11905–11915. 10.1021/acs.analchem.9b02667. [DOI] [PubMed] [Google Scholar]
- Klein D. R.; Feider C. L.; Garza K. Y.; Lin J. Q.; Eberlin L. S.; Brodbelt J. S. Desorption Electrospray Ionization Coupled with Ultraviolet Photodissociation for Characterization of Phospholipid Isomers in Tissue Sections. Anal. Chem. 2018, 90, 10100–10104. 10.1021/acs.analchem.8b03026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paine M. R. L.; Poad B. L. J.; Eijkel G. B.; Marshall D. L.; Blanksby S. J.; Heeren R. M. A.; Ellis S. R. Mass Spectrometry Imaging with Isomeric Resolution Enabled by Ozone-Induced Dissociation. Angew. Chem., Int. Ed. 2018, 57, 10530–10534. 10.1002/anie.201802937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Specker J. T.; Van Orden S. L.; Ridgeway M. E.; Prentice B. M. Identification of Phosphatidylcholine Isomers in Imaging Mass Spectrometry Using Gas-Phase Charge Inversion Ion/Ion Reactions. Anal. Chem. 2020, 92, 13192–13201. 10.1021/acs.analchem.0c02350. [DOI] [PubMed] [Google Scholar]
- Born M.-E. N.; Prentice B. M. Structural elucidation of phosphatidylcholines from tissue using electron induced dissociation. Int. J. Mass Spectrom. 2020, 452, 116338 10.1016/j.ijms.2020.116338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng J.; Yang Y.; Liu Y.; Fang L.; Lin L.; Luan T. Coupling Paterno-Buchi Reaction with Surface-Coated Probe Nanoelectrospray Ionization Mass Spectrometry for In Vivo and Microscale Profiling of Lipid C horizontal lineC Location Isomers in Complex Biological Tissues. Anal. Chem. 2019, 91, 4592–4599. 10.1021/acs.analchem.8b05803. [DOI] [PubMed] [Google Scholar]
- Feider C. L.; Macias L. A.; Brodbelt J. S.; Eberlin L. S. Double Bond Characterization of Free Fatty Acids Directly from Biological Tissues by Ultraviolet Photodissociation. Anal. Chem. 2020, 92, 8386–8395. 10.1021/acs.analchem.0c00970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan E.; Nguyen C. Q. N.; Shiea C.; Reid G. E. Detailed Structural Characterization of Sphingolipids via 193 nm Ultraviolet Photodissociation and Ultra High Resolution Tandem Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2017, 28, 1406–1419. 10.1007/s13361-017-1668-1. [DOI] [PubMed] [Google Scholar]
- Marshall D. L.; Pham H. T.; Bhujel M.; Chin J. S.; Yew J. Y.; Mori K.; Mitchell T. W.; Blanksby S. J. Sequential Collision- and Ozone-Induced Dissociation Enables Assignment of Relative Acyl Chain Position in Triacylglycerols. Anal. Chem. 2016, 88, 2685–2692. 10.1021/acs.analchem.5b04001. [DOI] [PubMed] [Google Scholar]
- Poad B. L. J.; Green M. R.; Kirk J. M.; Tomczyk N.; Mitchell T. W.; Blanksby S. J. High-Pressure Ozone-Induced Dissociation for Lipid Structure Elucidation on Fast Chromatographic Timescales. Anal. Chem. 2017, 89, 4223–4229. 10.1021/acs.analchem.7b00268. [DOI] [PubMed] [Google Scholar]
- Vu N.; Brown J.; Giles K.; Zhang Q. Ozone-induced dissociation on a traveling wave high-resolution mass spectrometer for determination of double-bond position in lipids. Rapid Commun. Mass Spectrom. 2017, 31, 1415–1423. 10.1002/rcm.7920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barré F.; Rocha B.; Dewez F.; Towers M.; Murray P.; Claude E.; Cillero-Pastor B.; Heeren R.; Porta Siegel T. Faster raster matrix-assisted laser desorption/ionization mass spectrometry imaging of lipids at high lateral resolution. Int. J. Mass Spectrom. 2019, 437, 38–48. 10.1016/j.ijms.2018.09.015. [DOI] [Google Scholar]
- Poad B. L. J.; Zheng X.; Mitchell T. W.; Smith R. D.; Baker E. S.; Blanksby S. J. Online Ozonolysis Combined with Ion Mobility-Mass Spectrometry Provides a New Platform for Lipid Isomer Analyses. Anal. Chem. 2018, 90, 1292–1300. 10.1021/acs.analchem.7b04091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebisch G.; Fahy E.; Aoki J.; Dennis E. A.; Durand T.; Ejsing C.; Fedorova M.; Feussner I.; Griffiths W. J.; Koefeler H.; Merrill A. H. Jr.; Murphy R. C.; O’Donnell V. B.; Oskolkova O. V.; Subramaniam S.; Wakelam M.; Spener F. Update on LIPID MAPS Classification, Nomenclature and Shorthand Notation for MS-derived Lipid Structures. J. Lipid Res. 2020, 61, 1539–1555. 10.1194/jlr.S120001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F.; Guo C.; Ma X.; Zhang J.; Su Y.; Tian R.; Shi R.; Xia Y.; Wang X.; Ouyang Z. Rapid In Situ Profiling of Lipid C=C Location Isomers in Tissue Using Ambient Mass Spectrometry with Photochemical Reactions. Anal. Chem. 2018, 90, 5612–5619. 10.1021/acs.analchem.7b04675. [DOI] [PubMed] [Google Scholar]
- Batarseh A. M.; Abbott S. K.; Duchoslav E.; Alqarni A.; Blanksby S. J.; Mitchell T. W. Discrimination of isobaric and isomeric lipids in complex mixtures by combining ultra-high pressure liquid chromatography with collision and ozone-induced dissociation. Int. J. Mass Spectrom. 2018, 431, 27–36. 10.1016/j.ijms.2018.05.016. [DOI] [Google Scholar]
- Hamilton B. R.; Marshall D. L.; Casewell N. R.; Harrison R. A.; Blanksby S. J.; Undheim E. A. B. Mapping Enzyme Activity on Tissue by Functional Mass Spectrometry Imaging. Angew. Chem., Int. Ed. 2020, 59, 3855–3858. 10.1002/anie.201911390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King A.; Baginski M.; Morikawa Y.; Rainville P. D.; Gethings L. A.; Wilson I. D.; Plumb R. S. Application of a Novel Mass Spectral Data Acquisition Approach to Lipidomic Analysis of Liver Extracts from Sitaxentan-Treated Liver-Humanized PXB Mice. J. Proteome Res. 2019, 18, 4055–4064. 10.1021/acs.jproteome.9b00334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.