Abstract
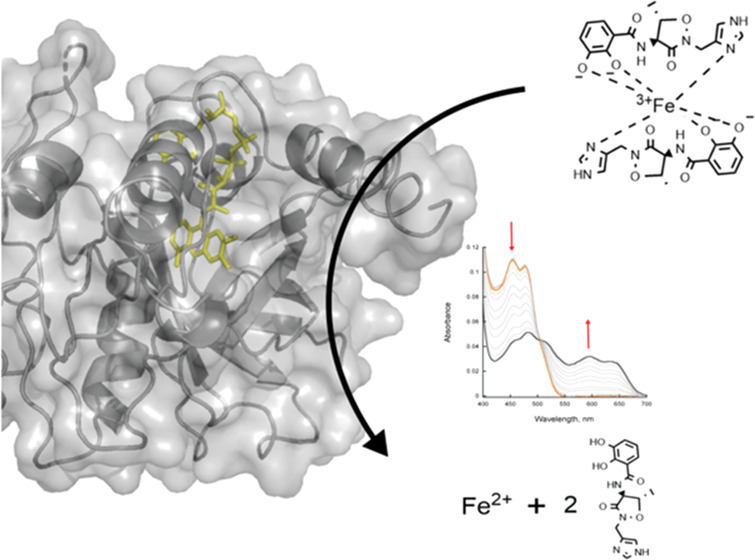
Acinetobacter baumannii is an opportunistic pathogen with a high mortality rate due to multi-drug-resistant strains. The synthesis and uptake of the iron-chelating siderophores acinetobactin (Acb) and preacinetobactin (pre-Acb) have been shown to be essential for virulence. Here, we report the kinetic and structural characterization of BauF, a flavin-dependent siderophore-interacting protein (SIP) required for the reduction of Fe(III) bound to Acb/pre-Acb and release of Fe(II). Stopped-flow spectrophotometric studies of the reductive half-reaction show that BauF forms a stable neutral flavin semiquinone intermediate. Reduction with NAD(P)H is very slow (kobs, 0.001 s–1) and commensurate with the rate of reduction by photobleaching, suggesting that NAD(P)H are not the physiological partners of BauF. The reduced BauF was oxidized by Acb-Fe (kobs, 0.02 s–1) and oxazole pre-Acb-Fe (ox-pre-Acb-Fe) (kobs, 0.08 s–1), a rigid analogue of pre-Acb, at a rate 3–11 times faster than that with molecular oxygen alone. The structure of FAD-bound BauF was solved at 2.85 Å and was found to share a similarity to Shewanella SIPs. The biochemical and structural data presented here validate the role of BauF in A. baumannii iron assimilation and provide information important for drug design.
Introduction
Acinetobacter baumannii is an opportunistic Gram-negative bacterial pathogen, which has become a serious public health concern due to the emergence of multi-drug-resistant (MDR) strains.1−4 Rapid spread of MDR genes in A. baumannii and other microbial populations occurs by horizontal gene transfer.5,6 Furthermore, drug resistance in A. baumanii is enhanced by production of biofilms and tolerance to extreme environments.2−4 Infections occur primarily in the respiratory tract, bloodstream, urinary tract, skin, and soft tissues, leading to severe illness and death.1 Its high survivability in hospital environments makes A. baumannii a recurrent cause of nosocomial illnesses, being responsible for 7% of ICU infections in the United States and up to 19% in other nations.2,7 Developing therapeutic strategies outside of the conventional antibiotics is paramount to effectively treat these MDR infections.
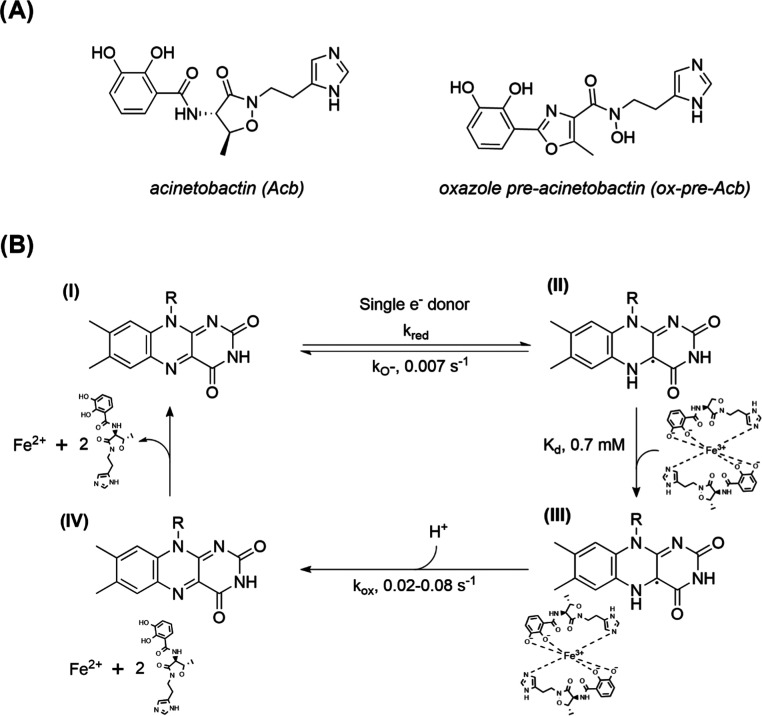
One major virulence factor utilized by some pathogenic microorganisms is the production of siderophores—iron-chelating compounds essential for iron uptake under nutrient-limiting conditions.8−10 This process is especially important during host invasion due to the tight regulation of iron homeostasis.11 When siderophore production or uptake is inhibited, it has been shown that virulence is significantly reduced or prevented in several microorganisms, including A. baumannii.12−14A. baumannii produces several siderophores; however, only acinetobactin (Acb) (Scheme 1A) and its isomer preacinetobactin (pre-Acb) are essential for virulence.15,16 Pre-Acb is produced biosynthetically and undergoes irreversible isomerization to Acb nonenzymatically in a pH-dependent equilibrium.17 Genetic disruption of the integral membrane receptor BauA involved in Acb/pre-Acb uptake resulted in A. baumannii mutants that failed to grow under iron-limiting conditions emulating the human host.18 In addition, it has been shown that an oxidized pre-Acb analogue, oxazole preacinetobactin (ox-pre-Acb) (Scheme 1A), does not undergo isomerization to Acb and inhibits the growth of A. baumannii under iron restrictive conditions.19 These results suggest that targeting the enzymes responsible for iron assimilation in A. baumannii can lead to the development of new antibiotics.
Scheme 1. Siderophore Structures and General Mechanism.
(A) Siderophores Acb and ox-pre-Acb without iron complexed. (B) Proposed mechanism of BauF. The catalytic cycle begins with an oxidized enzyme (I). A single electron donor then binds and reduces BauF to a neutral flavin semiquinone (II). The enzyme returns to the oxidized state, likely through formation of superoxide, or proceeds to the next step of the reaction by binding to the siderophore complexed with Fe(III) represented by a 2:1 Acb/Fe(III) complex34 (III). The electron from the neutral flavin semiquinone coenzyme reacts with ferric iron, resulting in its reduction to Fe(II) and the regeneration of oxidized flavin (IV). The reduced iron and the apo-siderophores are then released.
The pathway for siderophore uptake and iron assimilation in A. baumannii is well understood.16,20 Upon binding of ferric iron, the loaded siderophore is recognized by BauA and transported into the periplasm.16,18,21 BauB then shuttles the siderophore to a membrane-embedded multiprotein complex consisting of two permease proteins BauC and BauD and the ATP-binding protein BauE, which transfers it to the cytosol.16 BauF, a siderophore-interacting protein (SIP), is then predicted to reduce the iron–siderophore complex to facilitate iron release. While the extensive characterization of BauA and BauB has been done, the final step of iron release remains uncharacterized.22,23 In addition, BauF operates as the only known mechanism for iron release from Acb in A. baumannii, so it is likely that inhibiting this enzyme would have a significant effect on virulence.20 This has been shown to be true in other systems, where gene deletion of the SIP from Riemerella anatipestifer resulted in a significant reduction of virulence.14
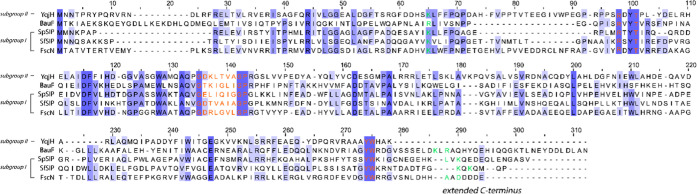
SIPs are flavin-dependent enzymes that catalyze the release of iron from the siderophore complex by facilitating the reduction of Fe(III) to Fe(II).24 Structural and kinetic studies of these enzymes have identified two different families: the ferric siderophore reductases (FSRs) and the SIPs. FSRs contain an iron–sulfur cluster, which is absent in SIPs.25−31 SIPs can be further classified into two subgroups, which differ in cofactor preference. Subgroup I is characterized by having a longer α-helical C-terminus and preferring NADH, while subgroup II SIPs have an extended N-terminus and utilize NADPH.24−26 Amino acid sequence analysis of BauF to other FSRs and SIPs shows that this protein belongs to SIP subgroup I (Figure 1). This work presents the structural and kinetic characterization of BauF.
Figure 1.
Sequence analysis of characterized SIPs. Multiple sequence alignment of BauF compared to the subgroup I SIPs FscN from Thermobifida fusca (NCBI: WP_011292284), SpSIP from Shewanella putrefaciens (NCBI: ABP73812), and SfSIP from Shewanella frigidimarina (NCBI: WP_011637846) and the subgroup II SIP YqjH from Escherichia coli (NCBI: WP_001066494). Residues shown in orange are involved in FAD binding. Residues depicted in green are the proposed ferric-siderophore binding pockets. The server T-Coffee was used for alignment and visualized with Jalview.52
Results
Protein Purification
BauF was heterologously expressed in E. coli ArcticExpress (DE3) RIL as an N-terminal hexahistidine fusion. Recombinant BauF was purified using immobilized metal affinity chromatography (IMAC) to >90% homogeneity as determined by SDS-PAGE (Figure S1). The protein yield was 3 ± 0.5 mg protein per 1 g of cell pellet and had FAD incorporation of 64 ± 5%. The extinction coefficient of FAD bound to BauF at pH 7.5 is 10.8 mM–1 cm–1 at 450 nm (Figure S2).
Curiously, after cell lysis, the supernatant did not show the yellow color characteristic of an overexpressed flavoenzyme in its oxidized state. Instead, the supernatant was a blue-gray color (Figure S3). Further spectral analysis showed that this was due to the fact that BauF had reduced to a neutral flavin semiquinone (Figure S4). This species was air-stable, but when the supernatant was vortexed the enzyme quickly oxidized (Figure S5). The solution returned to a blue-gray color a few minutes after vortexing. The additives 1% Triton X-100, 1% TWEEN 20, and 10 mM TCEP had no effect (Figure S5). BauF was yellow upon elution from the metal ion affinity column, indicating oxidation (Figure S3B). When the protein was purified anaerobically, reduced BauF was isolated at similar yields to the aerobic purification.
BauF Steady-State Activity
Under anaerobic conditions, the steady-state activity of BauF was determined by measuring the concentration of free Fe(II) with FerroZine (Figure S6). Under apparent saturating conditions, BauF exhibited slow initial velocities with NAD(P)H and Acb-Fe (NADH: 1 × 10–3 ± 3 × 10–4 s–1; NADPH: 1 × 10–3 ± 1 × 10–4 s–1). These apparent kcat values were ∼20-fold slower than those reported for other SIPs.24,26,27
Reductive Half-Reaction
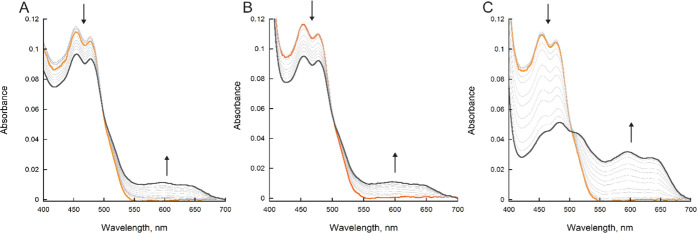
The reaction of BauF when mixed with NAD(P)H or sodium dithionite (DT) was measured using stopped-flow spectrophotometry inside of an anaerobic chamber. In the presence of excess NAD(P)H (1 mM) a slow reduction (∼0.001 s–1) was observed resulting in the formation of a neutral flavin semiquinone (Figures 2 and 3). A lower concentration of NAD(P)H could not be analyzed because photobleaching was faster than the reaction with NAD(P)H (Figure 2A,B). The reaction with 1 mM DT exhibited a fourfold increase in the rate resulting in the complete formation of neutral flavin semiquinone (Figures 2C and 3).
Figure 2.
BauF reduction. (A) Spectral changes of BauF over 900 s, showing photoreduction. (B) Spectral changes of BauF reduction with 1 mM NADH over 900 s. (C) Spectral changes of BauF reduction with 1 mM DT over 900 s. Oxidized BauF is represented as orange and semiquinone as gray. Spectral changes of BauF reduction with 1 mM NADPH were similar to NADH (not shown).
Figure 3.
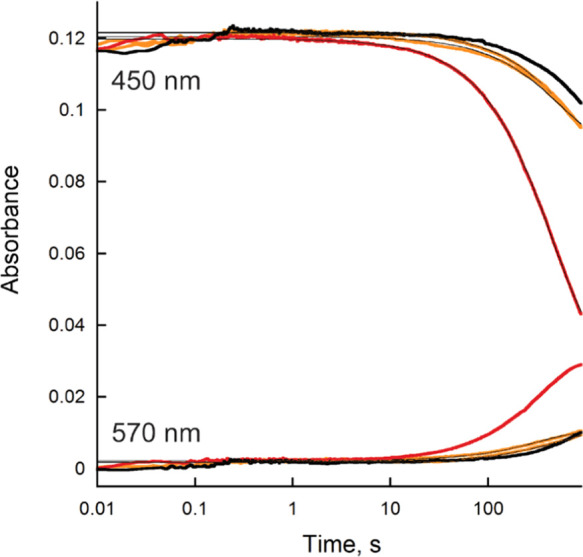
BauF reduction. Changes in the absorbance at 450 and 570 nm of BauF during photobleaching (black) and when reacted with DT (red) or NAD(P)H (orange). Traces at 450 nm were fit to eq 1 and 570 nm to eq 2, respectively. The rate of reduction at 450 nm was 1 × 10–3 ± 2 × 10–4 s–1 with photobleaching, 1 × 10–3 ± 2 × 10–4 s–1 for NAD(P)H, and 4 × 10–3 ± 1 × 10–4 s–1 for DT.
Oxidative Half-Reaction
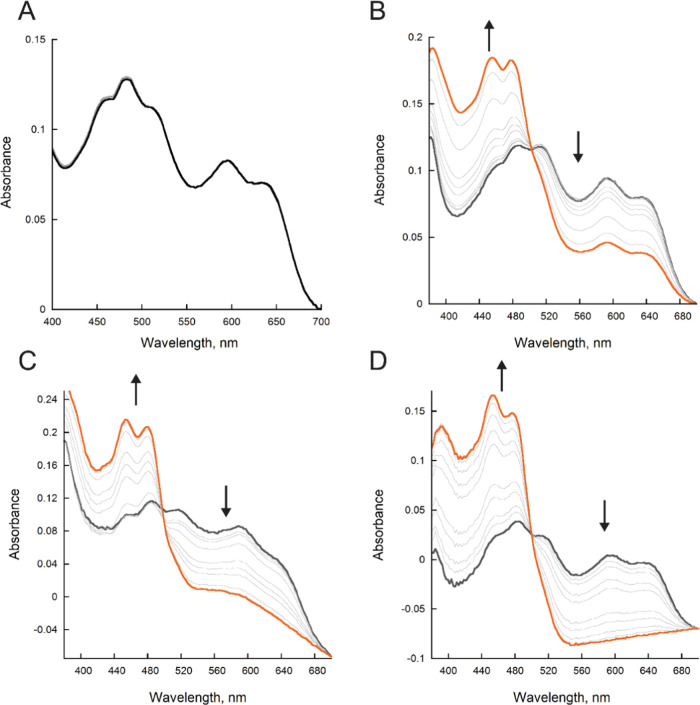
The reaction of reduced BauF with Acb-Fe, Fe(III)-bound ox-pre-Acb (ox-pre-Acb-Fe), and oxygen was determined. We used ox-pre-Acb-Fe as a stable surrogate for pre-Acb to avoid the spontaneous isomerization of pre-Acb to Acb during enzymatic assays.19 When no substrate was present, the flavin spectra were unchanged over the period of 450 s (Figure 4A). When reduced BauF was reacted with molecular oxygen (600 μM), there was a slow decay of the neutral flavin semiquinone (kobs = 0.007 ± 0.0007 s–1) (Figures 4B and 5). Oxidation occurred up to 10-fold faster in the presence of 100 μM Acb-Fe and 100 μM ox-pre-Acb-Fe with observed rates of 0.02 ± 0.001 and 0.080 ± 0.003 s–1 respectively (Figures 4C,D and 5).
Figure 4.
BauF oxidation. (A) Spectral changes of BauF under anaerobic conditions. (B) Spectral changes of BauF in the presence of 600 μM oxygen. (C) Spectral changes of BauF in the presence of 100 μM Acb-Fe. (D) Spectral changes of BauF in the presence of 100 μM ox-pre-Acb-Fe. The reduced enzyme is represented as gray and the final oxidize spectra are orange. All assays were recorded for 450 s.
Figure 5.
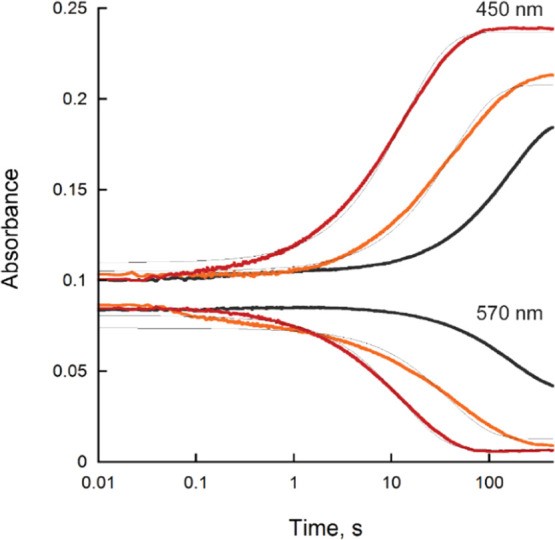
Changes in absorbance of BauF oxidation at 450 and 570 nm of oxygen (black), Acb-Fe (orange), and ox-pre-Acb-Fe (red) over 450 s. The traces at 450 nm were fit to eq 2 and 570 nm to eq 1.
Thermal Shift Assays
BauF exhibits a melting temperature of 59.6 ± 0.3 °C when no ligand is present. A thermal stabilization is observed with oxidized BauF at increasing concentrations of Acb-Fe and ox-pre-Acb-Fe (Figure 6). From this, the apparent KD values were calculated to be 0.70 ± 0.02 and 0.70 ± 0.04 mM for Acb-Fe and ox-pre-Acb-Fe, respectively. When the enzyme was incubated with 1 mM NAD(P)+ in the presence or absence of 10 mM DT, the Tm was unchanged (data not shown).
Figure 6.
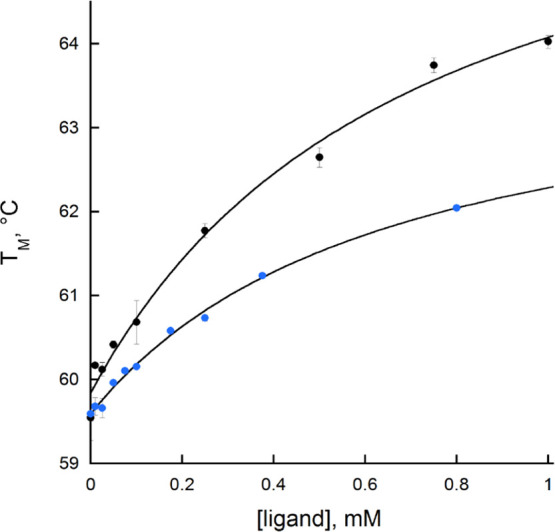
Change in the thermal stability of BauF at an increasing concentration of Acb (black) and ox-pre-Acb (blue). The data were fit to eq 4. The apparent KD value for Acb-Fe was 0.7 ± 0.02 mM and for ox-pre-Acb-Fe it was 0.7 ± 0.03 mM.
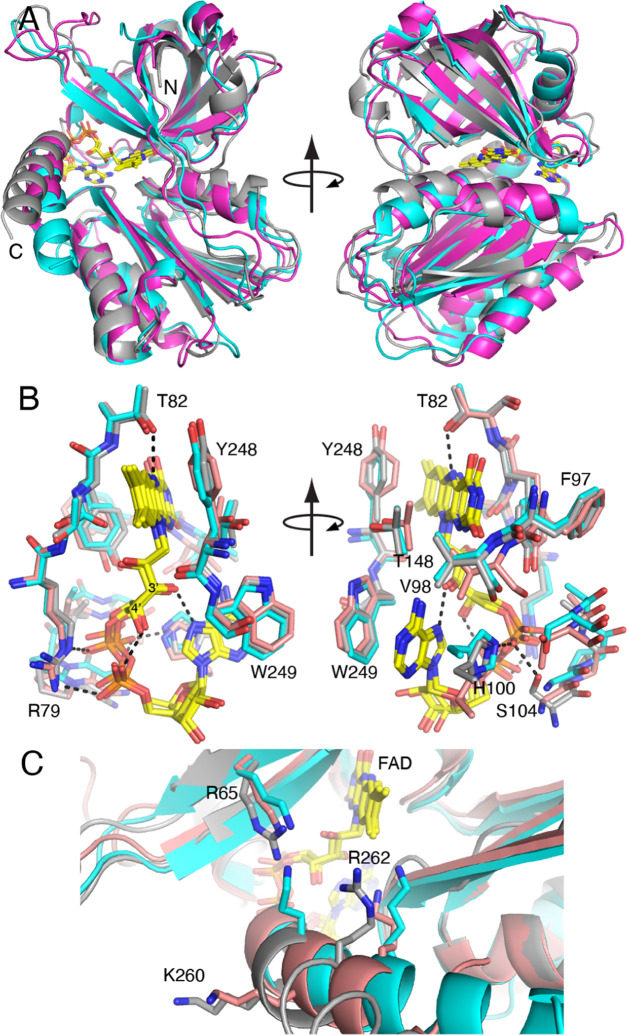
Crystal Structure of BauF
The structure of the BauF complexed with FAD was determined at 2.85 Å resolution (Figure 7A). Analysis of the structure with the CATH server32 identified two domains. The N-terminal domain (residues 23–127) has a β-barrel architecture, and the C-terminal domain (residues 128–273) has a three-layer αβα architecture with a Rossmann fold topology.
Figure 7.
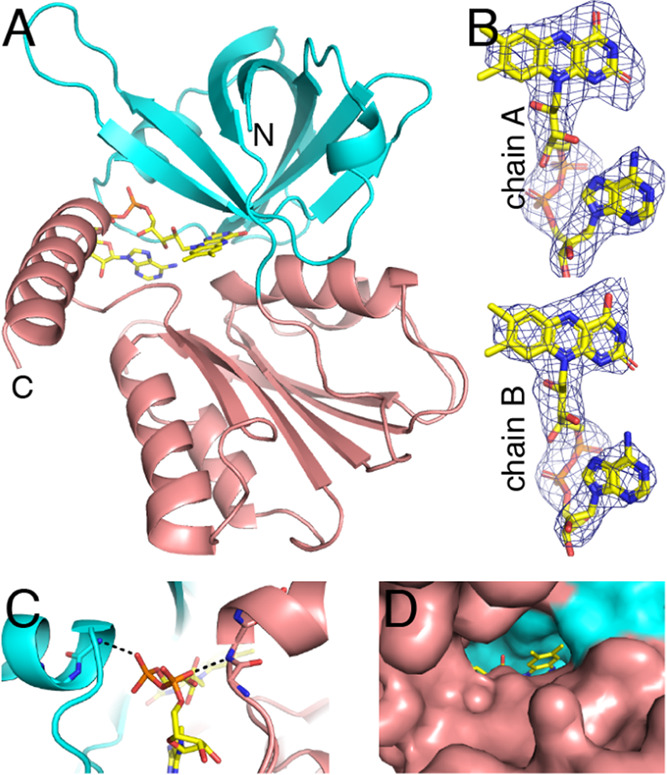
Structure of BauF. (A) Fold of BauF. The N- and C-terminal domains are colored cyan and salmon, respectively. The FAD is in yellow. (B) Polder omit electron density for the FAD (3.5σ). (C) Stabilization of the pyrophosphate of the FAD by the N-termini of two α-helices. (D) Surface representation showing the accessibility of dimethylbenzene edge of FAD.
Electron density for the FAD was strong in both chains (Figure 7B). The FAD binds in a crevice at the intersection of the two domains of the protomer (Figure 7A). The pyrophosphate of the FAD is flanked by the N-termini of two α-helices, one from each domain. This arrangement allows the pyrophosphate to form hydrogen bonds to the backbone of the N-terminal residues of the helices, while the helix macrodipoles stabilize the negative charge of the pyrophosphate (Figure 7C). The dimethylbenzene edge of the FAD is solvent-exposed (Figure 7D), suggesting that electron transfer to ferric siderophore substrates occurs at this location in the structure, in agreement with an analysis of the structure of Aeromonas hydrophila SIP.29
The PDB was searched using PDBeFold33 to identify the closest structural neighbors of BauF. This revealed several SIPs.24,26,29 The best matches were two subgroup I SIPs from Shewanella (PDB IDs: 2GPJ, 6GEH). The BauF protein sequence has 22–27% global identity and 26–29% local identity to the Shewanella SIPs. The BauF structure aligns with the Shewanella SIP structures with RMSDs of 1.8–1.9 Å. Superposition of the structures demonstrates the high similarity of the folds (Figure 8A). Only a few variations in the protein conformation are observed, such as the length and orientation of the C-terminal helix and a long loop in the N-terminal domain. Analysis of the interfaces in the crystal structure of BauF suggested that the enzyme is monomeric in solution, like the related Shewanella SIPs.
Figure 8.
Comparison of BauF (gray) with two Shewanella SIPs (PDB IDs: 2GPJ (cyan), 6GEH (salmon)). (A) Comparison of the folds. (B) Comparison of the FAD conformations and environments (BauF residue numbers are listed). (C) Proposed triad of basic residues of the ferric-siderophore binding pocket (BauF residue numbers are listed).
Both the conformations and the environments of the FAD are very similar in the three structures (Figure 8B). The similarity in conformation may be seen in the preservation of intra-FAD hydrogen bonds, where the ribityl O3′ and O4′ hydroxyl groups hydrogen bond with the adenine and pyrophosphate, respectively. Most of the side chains near the FAD (within 3.9 Å) are identically conserved among these proteins despite the relatively low overall sequence identity. Of particular note are conserved side chains that form hydrogen bonds and ion pairs with the FAD: Arg79, Thr82, and His100 (BauF numbering). One difference is that BauF has a serine that hydrogen bonds to the FAD pyrophosphate (Ser104); this residue is a glycine in the Shewanella SIPs (Figure 8B).
The structural basis of substrate recognition remains an open question due to the lack of structures of SIPs complexed with ferric siderophores. Nevertheless, a triad of basic residues has been proposed to form a ferric-siderophore binding pocket in SIPs.24,27 BauF residues Arg65, Lys260, and Arg262 align well with the triads observed in other SIP structures (Figure 8C).
Discussion
Understanding the steps of biological redox reactions associated with microbial siderophore-mediated iron acquisition is a significant knowledge gap. SIPs play an important role in this process by shuttling electrons from a bound flavin cofactor to the substrate ferric iron–siderophore complexes. Single electron transfer from a flavin semiquinone is proposed to reduce ferric iron to ferrous iron, resulting in destabilization of the siderophore chelation complex and entrance of ferrous iron into the intracellular pool of bioavailable metals. The kinetic and structural characterization of SIPs from bacterial and eukaryotic organisms has provided insight into their function.24,25,27,28 SIPs are divided into subgroups I or II. Subgroup I SIPs are identified by their extended C-terminus and activity with NADH, while subgroup II SIPs have a longer N-terminal region and a preference with NADPH.24−26 BauF was originally misannotated as a putative siderophore hydrolase.18 Comparative protein sequence analysis predicts BauF to be a flavin-containing member of the SIP family.16 Here, we confirmed through structural and biochemical characterization that BauF is a flavin-dependent SIP capable of catalyzing single electron transfer to the ferric complexes of native siderophore Acb and the pre-Acb surrogate ox-pre-Acb.
BauF shares the closest structural similarity to SIP members of subgroup I (Figure 8).24,26 To determine its cofactor preference, we measured the rate of reduction of BauF at 1 mM concentration of NAD(P)H using stopped-flow spectrophotometry. This experiment shows that in the absence of other components, NAD(P)H has little to no effect on enzyme reduction that is not related to photobleaching (Figure 3A,B). The enzyme also had no shift in thermal stability when incubated with 1 mM NAD(P)H, consistent with no specific/tight binding. The lack of reactivity and binding with NAD(P)H suggest that these cofactors are not the physiological reductants for BauF. The apparent kcat value (1 × 10–3 s–1) is very similar to that of the kobs of reduction with NAD(P)H (1 × 10–3 s–1), suggesting that this process remains unchanged in the presence of Acb-Fe.
A neutral semiquinone-reduced BauF was obtained by anaerobic purification, and oxidation occurs faster when reacted with the ferric siderophores Acb and ox-pre-Acb compared to molecular oxygen with a distinct preference for ox-pre-Acb (Figures 4 and 5). Oxidation with Acb-Fe and ox-pre-Acb-Fe was 5–20-fold faster than the rate of reduction with DT (Figure 5). BauF activity with ox-pre-Acb-Fe is not too surprising as it was shown that its ferric iron complex promotes A. baumannii growth.19 Thermal shift experiments show that these ligands bind to an oxidized enzyme with a similar apparent KD value of ∼0.7 mM (Figure 6). BauF exhibits some preference for ox-pre-Acb-Fe compared to Acb-Fe as it is reoxidized at a rate 4-fold faster with this substrate.
We propose a mechanism for BauF that is similar to other SIPs (Scheme 1B).27,28 Oxidized enzyme interacts with a reductive partner that is likely a single electron donor (i.e., reduced ferredoxin), forming a neutral flavin semiquinone. The enzyme either slowly reoxidizes as part of an uncoupled reaction or binds to a 2:1 siderophore/iron complex,23,34 leading to Fe(III) reduction to Fe(II). The ferrous iron and metal-free siderophores are then released from the active site priming the enzyme for a new catalytic cycle.
As previously mentioned, BauF is structurally similar to subgroup I SIPs and has the extended C-terminus typical of this subgroup (Figure 8).24,26 However, it shows no preference for NADH, which was a commonly associated feature to this subgroup. Similar observations were made with SfSIP, which showed limited activity with NAD(P)H even though it was classified as a subgroup I SIP.24 Instead, SfSIP was most active when reduced ferredoxin was used as the electron donor. These results in addition to what we report with BauF suggest that a third SIP subgroup exists, which uses a different electron donor other than NAD(P)H. The residues involved in FAD binding are well conserved, maintaining a GXGXXG motif as well as a tyrosine stacking with the isoalloxazine ring (Figures 1 and 8B).
BauF was isolated as a stable neutral flavin semiquinone under aerobic conditions (Figures S3 and S4). To our knowledge, this is the first report of this behavior from a SIP. Gentle shaking or brief vortexing of the cell lysate resulted in enzyme reoxidation, which returned to its reduced form over a period of a few minutes (Figure S5). The reduced protein was oxidized while bound to the column during the wash step of the purification procedure. It is possible that the reoxidation that occurs after vortexing and column washing is due to oxygen being introduced; however, the enzyme remains reduced after being stirred for several hours under aerobic conditions making this unlikely. Another explanation is that BauF is weakly interacting with a molecule that is stabilizing the neutral flavin semiquinone until the complex is broken through mechanical stress.
In summary, we show that BauF is capable of reducing the ferric-siderophores Acb and ox-pre-Acb, a crucial step in iron uptake. Its preference for ox-pre-Acb further supports the hypothesis that the initial biosynthetic product pre-Acb plays a major role in A. baumannii virulence. BauF exhibits limited or no activity with NAD(P)H and is likely to depend on a single electron donor for flavin neutral semiquinone formation. This observation is consistent with previous work that has shown reduced ferredoxin as an effective electron donor for SfSIP.25 Furthermore, BauF is structurally similar to other SIPs with a conserved basic triad forming a pocket near the isoalloxazine ring that could serve as the ferric-siderophore binding site.
Materials and Methods
Materials
The gene coding for full-length BauF (NCBI: WP_000160885) was synthesized in a codon-optimized form by GenScript and subsequently cloned into the pET28a vector in frame at the NdeI-HindIII sites for expression with an N-terminal 6×His tag. ArticExpress (DE3) RIL E. coli cells purchased from Agilent (Santa Clara, CA) were used for protein expression, and Top10 E. coli cells purchased from Thermo Fisher Scientific (Waltham, MA) were used for DNA amplification. Acb, ox-pre-Acb, and the respective ferric complexes were prepared by Dr. Wencewicz’s lab following previously published procedures.17,19,35 Reagents for BauF crystallization were from Hampton Research (Aliso Viejo, CA). Gases of nitrogen, 4% hydrogen/nitrogen mix, argon, and oxygen were purchased from Airgas (Radnor, PA).
Expression and Purification
Protein expression was performed using an autoinduction media as previously described.36 Protein expression was performed by growing 3 × 1 L cultures at 37 °C to an optical density of ∼4.0. The temperature was then decreased to 10 °C, and the cultures were incubated overnight. The cells were harvested by centrifugation at 5000g and stored at −70 °C.
For protein purification, cell paste (∼15 g) was suspended in 150 mL of Buffer A (25 mM HEPES pH 7.5, 300 mM NaCl, and 5 mM imidazole) containing with 1 mg/mL lysozyme, 1 mg/mL DNase, 1 mg/mL RNase, 1 mM phenyl methyl sulfonyl fluoride, and 150 μM FAD. After 30 min of constant stirring at 4 °C, cell lysis was performed by sonication at 70% amplitude with cycles of 5 s on and 10 s off for 15 min. Unbroken cells and insoluble proteins were separated by centrifugation at 30 000g for 1 h at 4 °C. The supernatant was loaded onto two in-tandem 5 mL nickel IMAC columns equilibrated with Buffer A using an AKTA prime system. The columns were then washed with a mixture of 90% Buffer A and 10% Buffer B (25 mM HEPES pH 7.5, 300 mM NaCl, and 300 mM imidazole). BauF was eluted with 100% Buffer B and the fractions containing protein were pooled and dialyzed in the storage buffer (25 mM HEPES pH 7.5 and 100 mM NaCl) overnight. The protein was flash-frozen with liquid nitrogen and stored at −70 °C.
For anaerobic protein purification, the same buffer compositions were used and made oxygen-free with 10 cycles of vacuum/argon degassing (2 min vacuum/2 min argon) before transferring to an anaerobic chamber 18 h before purification. The procedure for cell lysis was performed aerobically as described above. After centrifugation, the supernatant was transferred into a COY anaerobic chamber (Grass Lake, MI) and supplemented with 1 μM glucose oxidase and 30 mM dextrose. The solution was stirred under anaerobic conditions for at least an hour, to remove all oxygen. After incubation, the supernatant was loaded onto two in-tandem nickel-affinity columns on an AKTA prime system stored inside the chamber. The columns were washed and the protein was eluted following the described procedure above. BauF was then buffer exchanged into anaerobic storage buffer, concentrated, and frozen in 0.5 or 1 mL aliquots.
Steady-State Activity Assays
The activity of BauF was determined by measuring the formation of Fe2+ using the colorimetric indicator 3-(2-pyridyl)-5,6-diphenyl-1,2,4-triazine-p,p′-disulfonic acid (FerroZine) (Sigma-Aldrich, St. Louis, MO). All assays were performed anaerobically inside of a COY anaerobic chamber. Solutions were degassed through 15 cycles of 5 s vacuum and 60 s ultrapure argon before being transferred into the chamber. Assays were performed in triplicate using 96-well conical plates. The reactions were performed at a total volume of 90 μL with 15 μM BauF and 1 mM FerroZine in 100 mM potassium phosphate pH 7.5 at 1 mM NADH and 0.4 mM Acb-Fe. At each time point, the reaction was quenched with 88 μL of 0.5 M perchloric acid before being centrifuged. A 150 μL aliquot of the reaction mix was transferred to a clear 96-well plate. The absorbance was recorded at 562 nm using a SpectraMax M5 microplate reader (Molecular Devices, San Jose, CA). A standard curve constructed with ferrous sulfate and FerroZine was used to calculate the concentration of Fe2+ in the assays. The initial velocity of the reaction was determined from the slope of concentration of Fe2+ (μM) as a function of time (s) divided by the concentration of enzyme (μM).
Stopped-Flow Spectrophotometry
The reductive and oxidation half-reactions of BauF were studied using a stopped-flow spectrophotometer (Applied Photophysics, Surrey, U.K.) equipped with a photodiode array detector located inside the anaerobic chamber. The equipment and assay buffers were prepared following previously described procedures.37
For the reductive half-reaction, 20 μM BauF was mixed with 2 mM NAD(P)H or 2 mM sodium dithionite (DT) in 50 mM HEPES, pH 7.5 supplemented with 0.75 μM glucose oxidase and 20 mM dextrose. The change in absorbance from 190 to 850 nm was recorded for 900 s. The stock solution of DT was prepared anaerobically and its concentration was determined using potassium ferricyanide (ε420 = 1.04 mM–1 cm–1). The change in absorbance at 450 nm was fit with a single exponential decay model (eq 1), where A1 is the amplitude of change, kobs is the observed rate of change, t is the time, and C is the final absorbance at the specified wavelength.
| 1 |
The absorbance changes at 570 nm were fit to an exponential rise model (eq 2), where all variables are the same as in eq 1 with the exception of D, which represents the initial absorbance.
| 2 |
For the oxidative half-reaction, reduced BauF obtained by anaerobic purification was used. Under the same buffer conditions, 30 μM of reduced BauF was mixed with 200 μM Acb-Fe or ox-pre-Acb-Fe and the reaction was measured for 180 s. The spectra of each substrate were subtracted from the data to determine spectral changes of BauF. The change in absorbance at 450 nm was fit with eq 2 and changes at 570 nm used eq 1. The rate constants were very similar at both wavelengths. The reported rate constants are the averages of three different experiments.
Thermal Shift Assays
Thermal shift experiments were based on a previously established protocol.38 Assays were performed in a Hard-Shell 96-well PCR plate (Bio-Rad, Hercules, CA). Solutions were prepared by mixing 2 μL of ×10 SYPRO Orange protein gel stain (Thermo Fisher Scientific, Waltham, MA) and 1 mg/mL BauF in 100 mM potassium phosphate, pH 7.5, to a total volume of 20 μL. For ligand-binding studies, the protein was incubated for 10 min with 0.01–1 mM Acb-Fe, 0.01–0.8 mM ox-pre-Acb-Fe, 1 mM NAD(P)+, or 10 mM DT before analysis. The plate was sealed with the MicroAmp optical adhesive film (Thermo Fisher Scientific) and analyzed using a CFX qPCR (Bio-Rad) programmed to heat from 20 to 90 °C at a rate of 2 °C/min. Changes in the fluorescence were measured every 30 s with a λex 450–490 nm and λem 610–650 nm. The fluorescence emissions were analyzed to determine the enzyme melting temperature (Tm) using the Boltzmann sigmoidal curve with Graphpad Prism (Huynh, 2015) (eq 3).
| 3 |
where y is the fluorescent emission, x is the temperature, A1 is the initial fluorescence, A2 is the maximal fluorescence after enzyme melting, A3 is the steepness of the curve, and Tm is the temperature of protein melting. The Tm values were plotted against the concentration of ligand and the KD was determined using eq 4.39
| 4 |
where y is the Tm, L is the Tm when no ligand is present, H is the Tm at saturating concentration of ligand, P is the protein concentration in the same units as ligand concentration, KD is the dissociation constant, and x is the ligand concentration.
Crystallization
Conditions for BauF crystal formation were identified using crystal kits 1 and 2 from Hampton Research. Crystals were first observed in 2 M ammonium sulfate, 0.05 M NaCl, 5% PEG 400, and 0.1 M HEPES, Na pH 7.5 at 30 mg/mL of BauF. These conditions were optimized using microseeding. Large rectangular crystals were formed within 4 weeks when the droplets were prepared in a ratio of 1:3.5:0.5 (BauF/mother liquor/microseed) with the conditions 1.6 M ammonium sulfate, 0.1 M NaCl, 5% PEG 400, and 0.1 M HEPES–Na pH 7.5 at 17.5 mg/mL of BauF incubated at 25 ° C. In preparation for low-temperature data collection, the crystals were soaked in a cryogenic solution containing 25% glycerol, 1.65 M ammonium sulfate, 0.1 M NaCl, 5.5% PEG 400, and 0.1 M HEPES–Na pH 7.5 for 2 min. They were then flash-cooled in liquid nitrogen.
X-ray Diffraction Data Collection and Refinement
X-ray diffraction data were recorded in the shutterless mode on a Pilatus 6M detector at beamline 24-ID-C of the Advanced Photon Source. The data were integrated and scaled using XDS.40 Intensities were converted to amplitudes using Aimless.41 The space group is C2221 with the unit cell dimensions a = 86.79 Å, b = 127.25 Å, and c = 150.30 Å. The asymmetric unit contains two chains of BauF. The estimated solvent content is 59%, based on a VM of 3.0 Å3/Da.42 Data processing statistics are listed in Table 1.
Table 1. X-ray Diffraction and Data Collection Statistics.
| beamline | APS (24-ID-C) |
|---|---|
| space group | C2221 |
| unit cell parameters (Å) | a = 86.79, b = 127.25, c = 150.30 |
| mols. in asu. | 2 |
| wavelength (Å) | 0.97910 |
| resolution (Å) | 150.0–2.85 (3.00–2.85) |
| observationsa | 145 243 (21 131) |
| unique reflectionsa | 19 818 (2832) |
| Rmerge(I)a | 0.177 (2.4767) |
| Rmeas(I)a | 0.191 (2.652) |
| Rpim(I)a | 0.070 (0.963) |
| mean I/σa | 7.6 (0.8) |
| CC1/2 | 0.997 (0.321) |
| completeness (%)a | 100.0 (100.0) |
| multiplicitya | 7.3 (7.5) |
| no. of protein residues | 496 |
| no. of protein atoms | 3824 |
| no. of FAD atoms | 106 |
| Rcrysta | 0.218 (0.324) |
| Rfreeab | 0.264 (0.350) |
| rmsd bonds (Å) | 0.009 |
| rmsd angles (deg) | 1.11 |
| Ramachandran plotc | |
| favored (%) | 94.08 |
| outliers (%) | 0.00 |
| Clashscore (PR)c | 8.8 (97) |
| MolProbity score (PR)c | 2.39 (95) |
| average B—protein (Å2) | 85.4 |
| average B—FAD (Å2) | 67.8 |
| coord. error (Å)d | 0.42 |
| PDB code | 7LRN |
Values for the outer resolution shell of data are given in parenthesis.
5% test set.
From MolProbity. The percentile ranks (PR) for Clashscore and MolProbity score are given in parentheses.
Maximum likelihood-based coordinate error estimate from PHENIX.
The low sequence identity of BauF to structures in the PDB caused molecular replacement phasing to be challenging. Initial phases were generated using the BALBES automated molecular replacement pipeline server.43 The amino acid sequence of BauF and the structure factor amplitudes from Aimless were input. The best solution generated by BALBES was obtained with a search model derived from a structure of S. putrefaciens SIP SPUTCN32_0076 (PDB ID: 2GPJ). The BALBES search model was then used for molecular replacement in PHASER,44 and the initial phases from molecular replacement were improved by density modification and automated ab initio model building using PHENIX.45 The model and map from the PHASER, the experimental structure factor amplitudes, and the BauF sequence were input to phenix.autobuild44 for automated ab initio model building with density modification. The model from phenix.autobuild was used as the starting point for several iterative rounds of model building in COOT46−48 and refinement in PHENIX. Structure validation was performed using MolProbity and the wwPDB validation service.49,50 The model contains residues 23–273 (out of the 286 residues) and two noncovalently bound FAD molecules. Due to the modest resolution, noncrystallographic symmetry restraints were used in refinement, and water molecules were not included in the model. PDBePISA was used to analyze the protein–protein interfaces in the crystal structure.51 Refinement statistics can be found in Table 1.
Glossary
Abbreviations
- BauF
acinetobactin utilization protein from Acinetobacter baumannii
- SfSIP
siderophore utilization protein from Shewanella putrefaciens
- Acb
acinetobactin
- pre-Acb
preacinetobactin
- ox-pre-Acb
oxazole preacinetobactin
- Acb-Fe
Fe(III)-bound Acb
- ox-pre-Acb
Fe(III)-bound ox-pre-Acb
- MDR
multiple drug resistance
- FAD
flavin adenine dinucleotide
- NADH
nicotinamide adenine dinucleotide hydride
- NADPH
nicotinamide adenine dinucleotide phosphate hydride
- NAD(P)H
nicotinamide adenine dinucleotide (phosphate) hydride
- NAD(P)+
nicotinamide adenine dinucleotide (phosphate)
- DT
sodium dithionite
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c03047.
SDS-PAGE of purification samples (Figure S1); UV–vis spectrum of oxidized BauF (Figure S2); supernatant and wash samples with reduced BauF (Figure S3); UV–vis spectrum of blue-gray protein elution (Figure S4); supernatant exposed to different conditions (Figure S5); and BauF steady-state activity with NADH (Figure S6) (PDF)
Accession Codes
BauF protein, WP_000160885 (NCBI); BauF crystal structure, 7LRN (PDB).
Author Contributions
Project conceptualization was done by P.S. and T.A.W. H.V. expressed and purified protein. H.V. performed kinetic characterization and grew crystals for X-ray diffraction. D.A.K. and J.J.T. performed structural analysis. T.J.B. and J.A.S. cloned BauF, synthesized pure ox-pre-Acb, and purified Acb from A. baumannii ATCC 17978 cultures.
Research reported in this publication was supported by the National Science Foundation Grants CHE-2003658 (to P.S.), CHE-2003986 (to J.J.T.), and CAREER-1654611 (to T.A.W.).
The authors declare no competing financial interest.
Supplementary Material
References
- Howard A.; O’Donoghue M.; Feeney A.; Sleator R. D. Acinetobacter baumannii: an emerging opportunistic pathogen. Virulence 2012, 3, 243–250. 10.4161/viru.19700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchanda V.; Sanchaita S.; Singh N. Multidrug resistant Acinetobacter. J. Global Infect. Dis. 2010, 2, 291–304. 10.4103/0974-777X.68538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinal P.; Pantel A.; Rolo D.; López-Rojas R.; Smani Y.; Pachón J.; Vila J.; Lavigne J. P.; et al. Relationship Between Different Resistance Mechanisms and Virulence in Acinetobacter baumannii. Microb. Drug Resist. 2019, 25, 752–760. 10.1089/mdr.2018.0182. [DOI] [PubMed] [Google Scholar]
- Vrancianu C.; Gheorghe I.; Czobor I.; Chifiriuc Antibiotic Resistance Profiles, Molecular Mechanisms and Innovative Treatment Strategies of Acinetobacter baumannii. Microorganisms 2020, 8, 935 10.3390/microorganisms8060935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachino J.; Jin W.; Kimura K.; Arakawa Y. Intercellular Transfer of Chromosomal Antimicrobial Resistance Genes between Acinetobacter baumannii Strains Mediated by Prophages. Antimicrob. Agents Chemother. 2019, 63, e00334-19 10.1128/AAC.00334-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright G. Molecular mechanisms of antibiotic resistance. Chem. Commun. 2011, 47, 4055–4061. 10.1039/c0cc05111j. [DOI] [PubMed] [Google Scholar]
- Lynch J.; Zhanel G.; Clark N. Infections Due to Acinetobacter baumannii in the ICU: Treatment Options. Semin. Respir. Crit. Care Med. 2017, 38, 311–325. 10.1055/s-0037-1599225. [DOI] [PubMed] [Google Scholar]
- Wilson B.; Bogdan A.; Miyazawa M.; Hashimoto K.; Tsuji Y. Siderophores in Iron Metabolism: From Mechanism to Therapy Potential. Trends Mol. Med. 2016, 22, 1077–1090. 10.1016/j.molmed.2016.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leventhal G.; Ackermann M.; Schiessl K. Why microbes secrete molecules to modify their environment: the case of iron-chelating siderophores. J. R. Soc., Interface 2019, 16, 20180674 10.1098/rsif.2018.0674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A.; Singh P.; Srivastava A. Synthesis, Nature, and Utility of Universal Iron Chelator-Siderophore: A Review. Microbiol. Res. 2018, 212–213, 103–111. 10.1016/j.micres.2017.10.012. [DOI] [PubMed] [Google Scholar]
- Bogdan A.; Miyazawa M.; Hashimoto K.; Tsuji Y. Regulators of Iron Homeostasis: New Players in Metabolism, Cell Death, and Disease. Trends Biochem. Sci. 2016, 41, 274–286. 10.1016/j.tibs.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel K.; Butala S.; Khan T.; Suvarna V.; Sherje A.; Dravyakar B. Mycobacterial siderophore: A review on chemistry and biology of siderophore and its potential as a target for tuberculosis. Eur. J. Med. Chem. 2018, 157, 783–790. 10.1016/j.ejmech.2018.08.030. [DOI] [PubMed] [Google Scholar]
- Tonziello G.; Caraffa E.; Pinchera B.; Granata G.; Petrosillo N. Present and future of siderophore-based therapeutic and diagnostic approaches in infectious diseases. Infect. Dis. Rep. 2019, 11, 8208 10.4081/idr.2019.8208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu J.; Lu F.; Miao S.; Ni X.; Jiang P.; Yu H.; Xing L.; Yu S.; Ding C.; Hu Q. The siderophore-interacting protein is involved in iron acquisition and virulence of Riemerella anatipestifer strain CH3. Vet. Microbiol. 2014, 168, 395–402. 10.1016/j.vetmic.2013.11.027. [DOI] [PubMed] [Google Scholar]
- Sheldon J.; Skaar E. Acinetobacter baumannii can use multiple siderophores for iron acquisition, but only acinetobactin is required for virulence. PLoS Pathog. 2020, 16, e1008995 10.1371/journal.ppat.1008995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W.; Kim H. Current biochemical understanding regarding the metabolism of acinetobactin, the major siderophore of the human pathogen Acinetobacter baumannii, and outlook for discovery of novel ant infectious agents based thereon. Nat. Prod. Rep. 2020, 37, 477–487. 10.1039/C9NP00046A. [DOI] [PubMed] [Google Scholar]
- Shapiro J.; Wencewicz T. Acinetobactin Isomerization Enables Adaptive Iron Acquisition in Acinetobacter baumannii through pH-Triggered Siderophore Swapping. ACS Infect. Dis. 2016, 2, 157–168. 10.1021/acsinfecdis.5b00145. [DOI] [PubMed] [Google Scholar]
- Mihara K.; Tanabe T.; Yamakawa Y.; Funahashi T.; Nakao H.; Narimatsu S.; Yamamoto S. Identification and transcriptional organization of a gene cluster involved in biosynthesis and transport of acinetobactin, a siderophore produced by Acinetobacter baumannii ATCC 19606T. Microbiology 2004, 150, 2587–2597. 10.1099/mic.0.27141-0. [DOI] [PubMed] [Google Scholar]
- Bohac T.; Shapiro J.; Wencewicz T. Rigid Oxazole Acinetobactin Analog Blocks Siderophore Cycling in Acinetobacter baumannii. ACS Infect. Dis. 2017, 3, 802–806. 10.1021/acsinfecdis.7b00146. [DOI] [PubMed] [Google Scholar]
- Hasan T.; Choi C.; Oh M. H. Genes Involved in the Biosynthesis and Transportof Acinetobactin in Acinetobacter baumannii. Genomics Inf. 2015, 13, 2–6. 10.5808/GI.2015.13.1.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen B.; Skaar E. The contribution of nutrient metal acquisition and metabolism to Acinetobacter baumannii survival within the host. Front. Cell. Infect. Microbiol. 2013, 3, 95 10.3389/fcimb.2013.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohac T.; Fang L.; Giblin D.; Wencewiz T. Fimsbactin and Acinetobactin Compete for the Periplasmic Siderophore Binding Protein BauB in Pathogenic Acinetobacter baumannii. ACS Chem. Biol. 2019, 14, 674–687. 10.1021/acschembio.8b01051. [DOI] [PubMed] [Google Scholar]
- Moynié L.; Serra I.; Scorciapino M. A.; Oueis E.; Page M. G.; Ceccarelli M.; Naismith J. H. Preacinetobactin not acinetobactin is essential for iron uptake by the BauA transporter of the pathogen Acinetobacter baumannii. eLife 2018, 7, e42270 10.7554/eLife.42270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trindade I.; Silva J.; Fonseca B.; Catarino T.; Fujita M.; Matias P.; Moe E.; Luoro R. Structure and reactivity of a siderophore-interacting protein from the marine bacterium Shewanella reveals unanticipated functional versatility. J. Biol. Chem. 2019, 294, 157–167. 10.1074/jbc.RA118.005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzanke B.; Anemüller S.; Schünemann V.; Trautwein A.; Hantke K. (2004) FhuF, Part of a Siderophore-Reductase System. Biochemistry 2004, 43, 1386–1392. 10.1021/bi0357661. [DOI] [PubMed] [Google Scholar]
- Li K.; Chen W.; Bruner S. Structure and Mechanism of the Siderophore-Interacting Protein from the Fuscachelin Gene Cluster of Thermobifida fusca. Biochemistry 2015, 54, 3989–4000. 10.1021/acs.biochem.5b00354. [DOI] [PubMed] [Google Scholar]
- Miethke M.; Hou J.; Marahiel M. The siderophore-interacting protein YqjH acts as a ferric reductase in different iron assimilation pathways of Escherichia coli. Biochemistry 2011, 50, 10951–10964. 10.1021/bi201517h. [DOI] [PubMed] [Google Scholar]
- Kobylarz M.; Heies G.; Loutet S.; Murphy M. Iron Uptake Oxidoreductase (IruO) Uses a Flavin Adenine Dinucleotide Semiquinone Intermediate for Iron-Siderophore Reduction. ACS Chem. Biol. 2017, 12, 1778–1786. 10.1021/acschembio.7b00203. [DOI] [PubMed] [Google Scholar]
- Shang F.; Lan J.; Wang L.; Liu W.; Chen Y.; Chen J.; Ha N.; Quan C.; Nam K. H.; Xu Y. Crystal structure of the Siderophore-interacting protein SIP from Aeromonas hydrophila. Biochem. Biophys. Res. Commun. 2019, 519, 23–28. 10.1016/j.bbrc.2019.08.085. [DOI] [PubMed] [Google Scholar]
- Butterton J.; Calderwood S. Identification, Cloning, and Sequencing of a Gene Required for Ferric Vibriobactin Utilization by Vibrio cholerae. J. Bacteriol. 1994, 176, 5631–5638. 10.1128/jb.176.18.5631-5638.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y.; Zang K.; Liu C.; Li Y.; Ma Q. The putative siderophore-interacting protein from Vibrio anguillarum: protein production, analysis, crystallization and X-ray crystallographic studies. Acta Crystallogr., Sect. F: Struct. Biol. Commun. 2018, 74, 283–287. 10.1107/S2053230X18005125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson N. L.; Lewis T. E.; Das S.; Lees J. G.; Lee D.; Ashford P.; Orengo C. A.; Sillitoe I. CATH: an expanded resource to predict protein function through structure and sequence. Nucleic Acids Res. 2017, 45, D289–D295. 10.1093/nar/gkw1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E.; Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004, 60, 2256–2268. 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- Bailey D.; Bohac T.; Shapiro J.; Giblin D.; Wencewicz T.; Gulick A. Crystal structure of the siderophore binding protein BauB bound to an unusual 2: 1 complex between acinetobactin and ferric iron. Biochemistry 2018, 57, 6653–6661. 10.1021/acs.biochem.8b00986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J.; Wencewicz T. Structure–function studies of acinetobactin analogs. Metallomics 2017, 9, 463–470. 10.1039/C7MT00064B. [DOI] [PubMed] [Google Scholar]
- Chocklett S. W.; Sobrado P. Aspergillus fumigatus SidA is a highly specific ornithine hydroxylase with bound flavin cofactor. Biochemistry 2010, 49, 6777–6783. 10.1021/bi100291n. [DOI] [PubMed] [Google Scholar]
- Valentino H.; Sobrado P. Performing anaerobic stopped-flowspectrophotometry inside of an anaerobic chamber. Methods Enzymol. 2019, 620, 51–88. [DOI] [PubMed] [Google Scholar]
- Huynh K.; Partch C. L. Analysis of protein stability and ligand interactions by thermal shift assay. Curr. Protoc. Protein Sci. 2015, 79, 28.9.1–28.9.14. 10.1002/0471140864.ps2809s79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivoli M.; Novak H.; Littlechild J.; Harmer N. Determination of protein-ligand interactions using differential scanning fluorimetry. J. Visualized Exp. 2014, 51809 10.3791/51809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 125–132. 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans P. R.; Murshudov G. N. How good are my data and what is the resolution?. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2013, 69, 1204–1214. 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews B. W. Solvent content of protein crystals. J. Mol. Biol. 1968, 33, 491–497. 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- Long F.; Vagin A. A.; Young P.; Murshudov G. N. BALBES: a molecular-replacement pipeline. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2008, 64, 125–132. 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart P. H.; Afonine P. V.; Grosse-Kunstleve R. W.; Hung L. W.; Ioerger T. R.; McCoy A. J.; McKee E.; Moriarty N. W.; Read R. J.; Sacchettini J. C.; Sauter N. K.; Storoni L. C.; Terwilliger T. C.; Adams P. D. Automated structure solution with the PHENIX suite. Methods Mol. Biol. 2008, 426, 419–435. [DOI] [PubMed] [Google Scholar]
- Adams P. D.; Afonine P. V.; Bunkoczi G.; Chen V. B.; Davis I. W.; Echols N.; Headd J. J.; Hung L. W.; Kapral G. J.; Grosse-Kunstleve R. W.; McCoy A. J.; Moriarty N. W.; Oeffner R.; Read R. J.; Richardson D. C.; Richardson J. S.; Terwilliger T. C.; Zwart P. H. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- a Emsley P.; Lohkamp B.; Scott W. G.; Cowtan K. Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]; b Emsley P.; Lohkamp B.; Scott W. G.; Cowtan K. Features and development of Coot. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2004, 60, 2126–2132. 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Afonine P. V.; Grosse-Kunstleve R. W.; Echols N.; Headd J. J.; Moriarty N. W.; Mustyakimov M.; Terwilliger T. C.; Urzhumtsev A.; Zwart P. H.; Adams P. D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2012, 68, 352. 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V. B.; Arendall W. B. 3rd; Headd J. J.; Keedy D. A.; Immormino R. M.; Kapral G. J.; Murray L. W.; Richardson J. S.; Richardson D. C. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 12–21. 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gore S.; Sanz Garcia E.; Hendrickx P. M. S.; Gutmanas A.; Westbrook J. D.; Yang H.; Feng Z.; Baskaran K.; Berrisford J. M.; Hudson B. P.; Ikegawa Y.; Kobayashi N.; Lawson C. L.; Mading S.; Mak L.; Mukhopadhyay A.; Oldfield T. J.; Patwardhan A.; Peisach E.; Sahni G.; Sekharan M. R.; Sen S.; Shao C.; Smart O. S.; Ulrich E. L.; Yamashita R.; Quesada M.; Young J. Y.; Nakamura H.; Markley J. L.; Berman H. M.; Burley S. K.; Velankar S.; Kleywegt G. J. Validation of Structures in the Protein Data Bank. Structure 2017, 25, 1916–1927. 10.1016/j.str.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E.; Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Waterhouse A.; Procter J.; Martin D. M. A.; Clamp M.; Barton G. Jalview Version 2 - a multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.