Abstract
Background
The B3 superfamily (B3s) represents a class of large plant-specific transcription factors, which play diverse roles in plant growth and development process including flowering induction. However, identification and functional surveys of B3 superfamily have not been reported in ethylene-induced pineapple flowering (Ananas comosus).
Results
57 B3 genes containing B3 domain were identified and phylogenetically classified into five subfamilies. Chromosomal localization analysis revealed that 54 of 57 AcB3s were located on 21 Linkage Groups (LG). Collinearity analysis demonstrated that the segmental duplication was the main event in the evolution of B3 gene superfamily, and most of them were under purifying selection. The analysis of cis-element composition suggested that most of these genes may have function in response to abscisic acid, ethylene, MeJA, light, and abiotic stress. qRT-PCR analysis of 40 AcB3s containing ethylene responsive elements exhibited that the expression levels of 35 genes were up-regulated within 1 d after ethephon treatment and some were highly expressed in flower bud differentiation period in stem apex, such as Aco012003, Aco019552 and Aco014401.
Conclusion
This study provides a basic information of AcB3s and clues for involvement of some AcB3s in ethylene-induced flowering in pineapple.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-021-07854-1.
Keywords: Pineapple, Flowering, Ethylene, B3
Background
Flowering has a direct impact on the reproduction and yield of plants [1]. Many transcription factors (TFs) are involved in flowering process, including APETALA2 (AP2), basic helix-loop-helix (bHLH), basic region leucine zipper (bZIP), and minichromosome maintenance1, agamous, deficiens and serum response factor (MADS) et al. [2, 3]. Pineapple [Ananas comosus (Linn.) Merr] is one of the four tropical fruits, which is widely planted in more than 80 countries or regions around world [4]. In practice, there are mainly two ways to induce the pineapple flowering. One is natural flowering induction that needs short day-length and cold night temperatures to produce endogenous ethylene, and the other is artificial flowering induction that adopts chemicals such as ethephon or calcium carbide to release ethylene [5–7]. Due to the non-uniformity of flowering in natural environment, artificial flowering induction is often used in production to effectively improve flowering rate of pineapple [8]. At present, some genes related to ethylene or flowering in pineapple have been studied, but the underlying mechanisms of ethylene-induced pineapple flowering remain largely unclear.
Exclusively presented in plants, the B3-type DNA binding domain (DBD) is firstly identified in VIVIPAROUS1 (VP1) in Zea mays and found in all members of B3 superfamily (B3s) [9, 10]. The number of B3s varies among plants, and there are 118, 91, 81, 108 members in Arabidopsis thaliana, Oryza sativa, Zea mays, Glycine max, respectively [11, 12]. Based on the structure and function of the proteins, they can be divided into five subgroups: auxin response factor (ARF), Leafy Cotyledon 2 (LEC2)-Abscisic Acid Insensitive 3(ABI3)-VAL (LAV), high-level expression of sugar inducible (HSI), related to ABI3/VP1 (RAV) and reproductive meristem (REM) [13, 14]. In A. thaliana, ABI3 (one B3 domain), HSI (one B3 domain and zf-CW domain), ARF (one B3 domain, auxin response factor, and AUX/IAA domain), RAV (one B3 domain and AP2 domain), and REM (two B3 domains) are characterized by having different typical domains [15]. Each subgroup of B3s plays diverse roles in plant growth, development, and stress responses [16, 17]. For example, ARFs have the functions in the development of flowers and leaves, vascular tissue differentiation and root initiation [18, 19]. In A. thaliana, either arf1 or arf2 mutant exhibits delayed development, including flowering initiation and rosette leaf senescence [20], while the double mutant arf7 arf19 influences root growth and leaf expansion [21]. The study of LAV is few, mainly focusing on the regulation of seed germination, embryo development, and stress response [22–25]. AtABI3 induces lateral root primordia by auxin, and the root carrying loss-of-function abi3 alleles is insensitive to auxin [26]. The LEC2 induces the somatic embryo formation and promotes the accumulation of seed storage protein and oil bodies [27]. As a transcriptional repressor, HSI has functions in repressing the expression of seed maturation genes [28]. Recently, a study demonstrated that HSI2 represses AGL15 (relate to seed development) by the PRC2 component MSI1, contributing to seed maturation regulation [29]. Many studies indicated that the RAV is related to stress response and overexpression of cotton RAV1 gene in Arabidopsis increases the sensitivity of salinity and drought stresses [30]. From another perspective, TEM (TEMPRANILLO) gene (a member of the RAV family) downregulates the expression of FT (Flowering Locus T), so as to repress flowering [31]. While AtREM34 was the first identified REM member, VRN1 (VERNALIZATION1/REM5), was the first being functionally characterized to be related to vernalization and promotion of flowering [3, 32]. Silencing both REM34 and REM35 in Arabidopsis influenced its female and male gametophyte development, indicating that REMs play roles in flowering [33]. Despite the fact that B3s plays critical roles in regulating flowering, the genome-wide analysis of this superfamily in pineapple has not been reported. What’s more, it is unclear whether B3s involve in ethylene response and flowering in pineapple. In this study, we identified 57 B3s in pineapple, and explored features of their structure and expression. The results provides a reference for further understanding of the physiological function and mechanism of B3s in the process of ethephon-induced pineapple flowering.
Results
Genome-wide identification and phylogenetic analysis of AcB3s
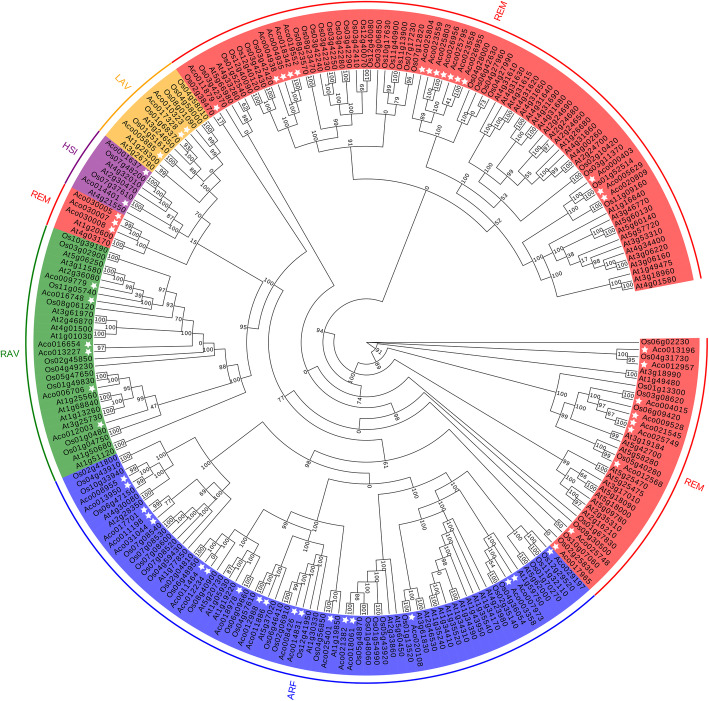
A total of putative 57 B3 proteins were identified in pineapple by HMMER 3.0, CD-search and SMART analysis. As shown in Table S1, the number of amino acid residues of 57 B3 proteins ranged from 137 to 997 aa, and their molecular weights (MWs) varied from 15.71 to 111.32 KDa. All predicted B3 proteins were hydrophobic proteins with isoelectronic point (PI) values ranging from 4.61 to 9.9. The prediction of subcellular localization demonstrated that they were mainly located in nuclear (42), chloroplast (8), cytoplasmic (5), extracellular (1), and vacuole (1). A phylogenetic tree was constructed using PhyML 3.0 to investigate the evolutionary relationship of B3s among pineapple. The results indicated that 57 B3 proteins could be divided into five distinct subfamilies (REM, ARF, RAV, HSI and LAV) (Fig. 1). REM which clustered into three branches was the largest with 27 AcB3s, while ARF followed by REM with 19 AcB3s. Remainder subfamilies of RVA, HSI, and LAV had 6, 2, and 3 AcB3s, respectively.
Fig. 1.
The phylogenetic tree of B3s from pineapple (Ac), Arabidopsis (At), and rice (Os). 87 in Arabidopsis [15], 86 in rice [15], and 57 in pineapple were utilized for the phylogenetic analysis. The different colour areas indicated different subfamlies. The white asterisk represented B3s from pineapple
Genomic location and gene duplication analysis of AcB3s
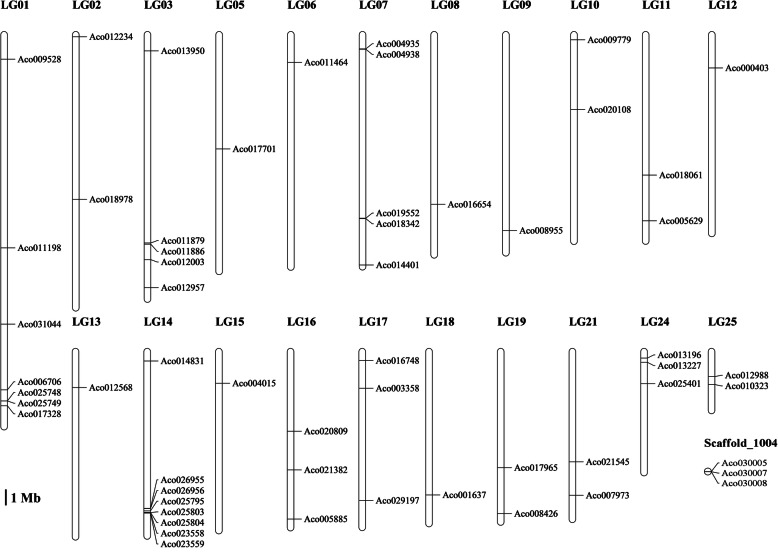
To examine the chromosomal distribution of the AcB3s, the gnomic sequence of each AcB3 was utilized to search against the pineapple genome database with MapChart software (Fig. 2). 54 AcB3s were distributed on 21 out of the 25 Linkage Groups (LG), and the rest (Aco030005, Aco030007 and Aco030008) were located on scaffold_1004. However, the distribution of AcB3s in each chromosomal was uneven. LG14 contained 8 AcB3s (7 of them highly concentrated on 9.94 Mb–10.24 Mb of LG14), followed by 7 on LG01. Only one gene was observed on LG5, 6, 8, 9, 12, 13, 15 and 18.
Fig. 2.
Distribution of AcB3 genes on linkage groups. All 57 B3 genes are mapped onto 21 linkage groups and one scaffold. “LG number” indicates the chromosome number. The scale is in megabases (Mb)
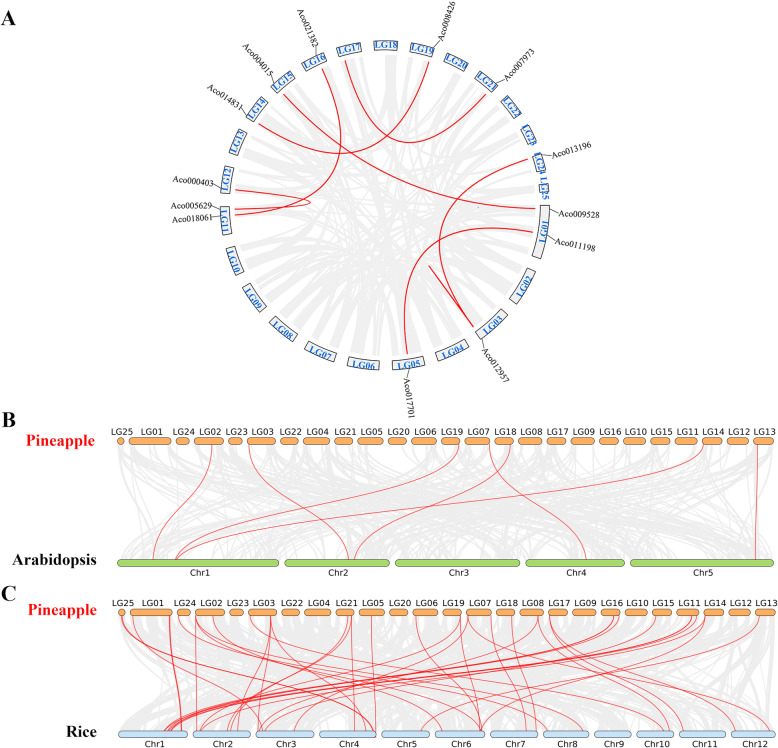
Both BLASTP and MCScanX methods were used to identify the collinearity and potential duplication events of the AcB3s in pineapple. A total of 8 segmental duplication events were identified, and the synteny blocks of AcB3 were on 12 LGs, including LG1, 3, 5, 11, 12, 14, 15, 16, 17, 19, 21 and 24 (Fig. 3A). Additionally, 5 tandem duplication events were identified in LG14 and scaffold_1044 (Table S2). Calculating the nonsynonymous (Ka) and synonymous (Ks) substitution rates is useful for the study of evolutionary. Among 13 duplication AcB3 pairs, 2 of them were “Ka/Ks > 1”, suggesting that those had evolved under the effect of positive selection; 10 of them were “Ka/Ks < 1”, demonstrating that those had evolved under the effect of purifying selection, and the only one pair had Ks value equal to 0, which imply that they may be the redundant gene (Table S2). To further explore the potential evolutionary mechanisms of the AcB3s, we compared the collinear relationships between the pineapple and the other two species Arabidopsis (dicotyledon) and rice (monocotyledon) (Fig. 3B and C). 7 collinear gene pairs between pineapple and Arabidopsis and 39 collinear gene pairs between pineapple and rice were identified. The number of orthologous events of AcB3s-OsB3s was far more than that of AcB3s-AtB3s, indicating that the genetic relationship between pineapple and rice is closer compared to Arabidopsis. The details were illustrated in Table S3.
Fig. 3.
Collinearity analysis of AcB3s. A Collinearity analysis of AcB3s in pineapple. B Collinearity analysis between pineapple and Arabidopsis. C Collinearity analysis between pineapple and rice. Gray lines suggest all segmental duplications and the red lines suggest duplicated AcB3 pairs. The chromosome number and gene ID are illustrated
Gene structural and protiein motif analysis of AcB3s
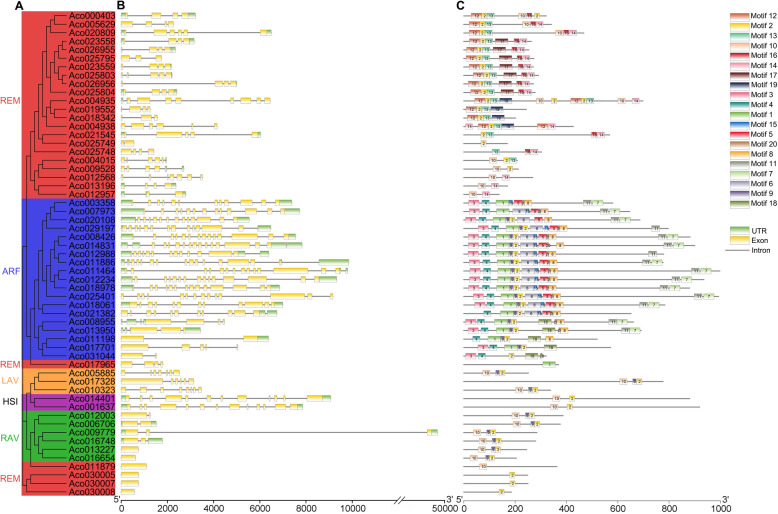
Gene structure analysis is helpful to understand the functional evolution of genes in adapting to environmental changes [34]. Among 57 AcB3s divided into 5 subfamilies (Fig. 4A), most ARF subfamily genes had more than 11 exons, and more than half REM subfamily genes only had 3–5 exons. The LAV subfamily genes had 6–8 exons, the HSI subfamily genes had 12 or 13 exons, and the RAV subfamily genes had 1 or 2 exons (Fig. 4B). Otherwise the same subfamily also has the similar domains (Fig. S1). Additionally, motif analysis identified 20 divergent motifs in the protein sequences of AcB3s (Fig. 4C; Table S4). As demonstrated in the result, all AcB3 genes had motifs annotated as B3 domain. The same group had similar motifs while motifs were diverse among different groups. For example, both HSI genes had motif 2 and 10 while all ARF genes had motif 3 and 4. Meanwhile, the motif 12, 13, 14 16, 17 and motif 19 only existed in the REM (Fig. 4C). The special motifs in groups may imply diverse functions of AcB3 superfamily.
Fig. 4.
The exon-intron structure and motif organization of AcB3s. A Phylogenetic relationships of AcB3s. B The intron and exon structure of B3 genes. The green and yellow boxes represent exons and UTRs, respectively, and the lines going through the boxes represent introns. C The motif identification of AcB3 proteins using MEME
Cis-acting elements and gene ontology analysis of AcB3s
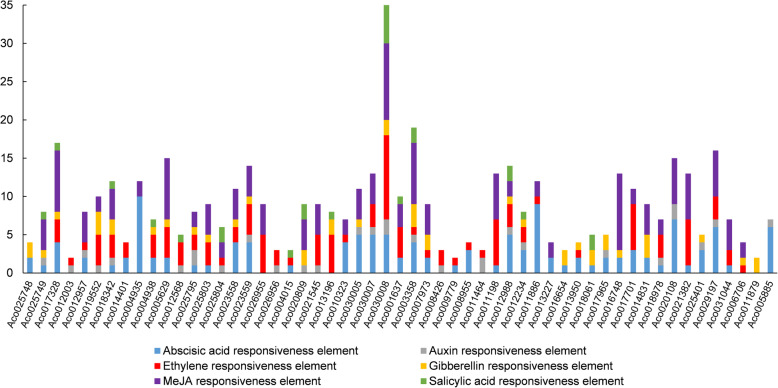
In order to understand the possible regulatory roles of plant hormones in AcB3s, the 2000 bp-length promoter sequences before ATG were analyzed. As shown in Fig. 5, there were 42, 26, 40, 28, 35 and 15 AcB3 promoter sequences with cis-elements related to abscisic acid, auxin, ethylene, gibberellin, methyl jasmonate and salicylic acid, respectively. Both Aco009528 and Aco000403 did not contain plant hormone-related cis-elements. Other cis-elements such as light responsiveness element, defense, and stress responsiveness element were also found (Table S5). Moreover, all AcB3 promoter sequences had ERF binding sites (Table S6). We also executed a gene ontology (GO) enrichment analysis of the AcB3s (Fig. S2). The prediction of cellular component suggested that AcB3s participated in cell, organelle and protein-containing complex. The prediction of the biological process indicated that most AcB3s participated in DNA binding, and then transcription regulator or catalytic activity. Moreover, the prediction of molecular function demonstrated that AcB3s were mainly involved in metabolic process, cellular process, regulation of the biological process, and biological regulation. Additionally, two genes (Aco13950 and Aco006706) were predicted to function in the process of flowering in the terms on level 3 (Fig. S3; Table S7).
Fig. 5.
The prediction of hormone-related cis-elements in AcB3 promoters. The promoter is 2.0 kb in length
Expression analysis of AcB3 genes after ethephon treatment
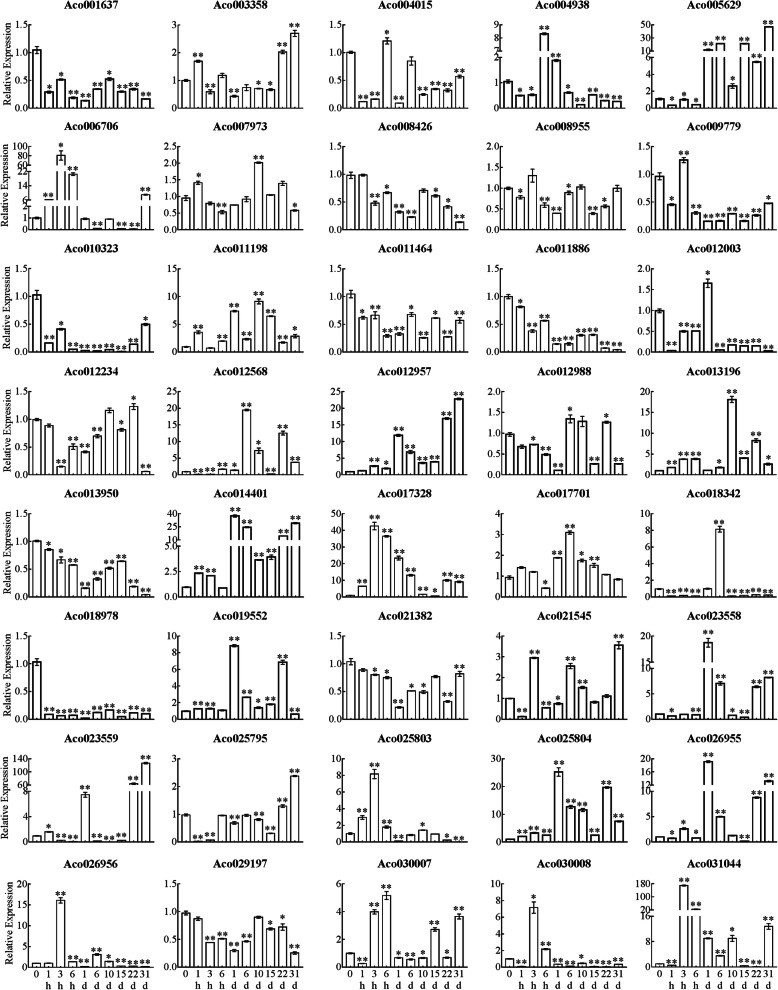
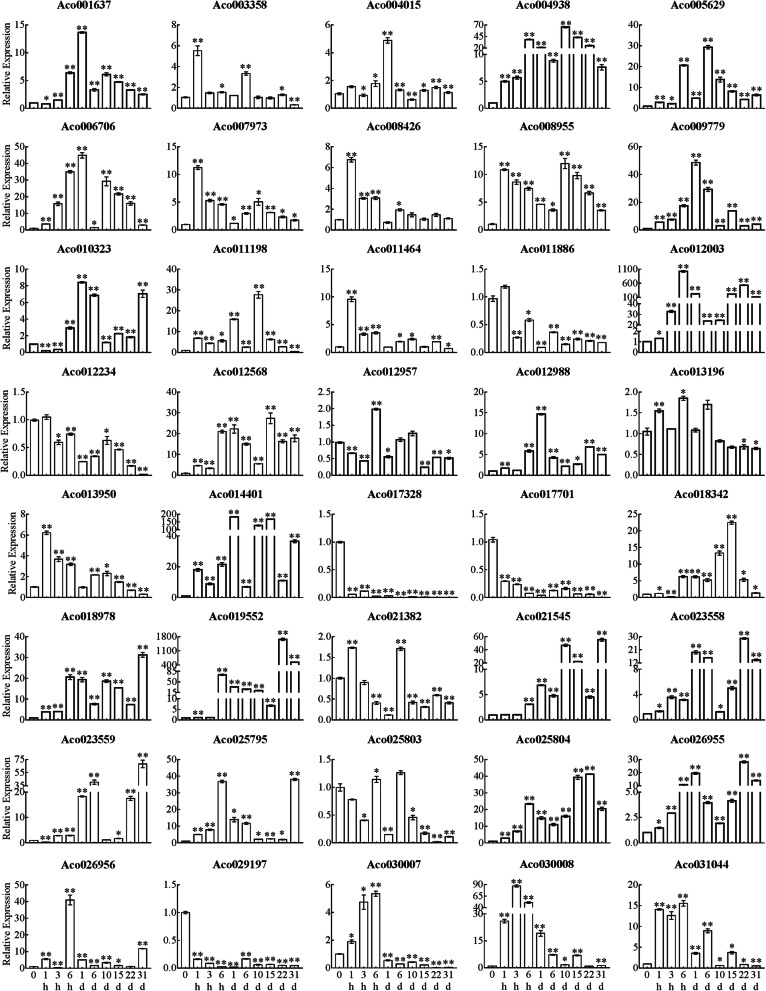
A previous study showed that flower buds of pineapple began to differentiate after 31 d of ethephon treatment [35]. In order to explore the dynamic expression pattern of AcB3s in response to ethylene within 31 d after ethylene treatment, 40 AcB3s covering all subfamilies and containing ethylene responsive elements were screened to analyze their expression changes by qRT-PCR. Generally, the expression of all the selected genes were significantly changed in response to ethephon. In leaves, the expression levels of 32 genes were significantly up-regulated while the expression levels of 8 genes were significantly down-regulated during the whole processing time. Especially, the expression levels of 16 genes (such as Aco012957, Aco014401, Aco005629, Aco025804, Aco023558, Aco026955 and so on) were significantly up-regulated within 1 d and were highly expressed in 31d after ethephon treatment (Fig. 6). Compared with leaves, ethephon treatment induced up-regulated expression of more genes in stem apex. The expression of 35 of 40 genes were up-regulated from 0 to 1 d after ethephon treatment, of which 7, 6 and 7 genes expression reached a peak at 1 h, 6 h and 1d, respectively (Fig. 7). 25 genes in stem apex were significantly up-regulated in 31d after ethephon treatment. Particularly, the transcripts of 14 genes (Aco01637, Aco004938, Aco005629, Aco007973, Aco008955, Aco009779, Aco012003, Aco012568, Aco014401, Aco018978, Aco023558, Aco025795, Aco025804 and Aco026955) in stem apex were at a high level all the time, while the expression level of Aco019552 was the highest in stem apex.
Fig. 6.
Expression profile of AcB3s in leaves after ethephon treatment. The AcEF1 gene was used as an internal control. Error bars indicated standard error (SE) based on three replicates. *P < 0.05; **P < 0.01, student’s t test
Fig. 7.
Expression profile of AcB3s in stem apex after ethephon treatment. The AcEF1 gene was used as an internal control. Error bars indicated standard error (SE) based on three replicates. *P < 0.05; **P < 0.01, student’s t test
Discussion
B3s have various functions in regulating the development of root, seed and flower, abiotic stresses responses and involving in hormone signaling pathways [9, 14]. Genome-wide characterization and expression analysis of B3s were investigated in some plants, and discovered a lot of meaningful information [10, 17, 36, 37]. In this study, a total of 57 AcB3s were identified by a comprehensive genome-wide analysis. Although having the same five subfamilies, the number of B3s in pineapple were less than Arabidopsis and rice, demonstrating that the parallel evolutionary events of B3s also existed in the plants of Crassulaceae family. All 57 identified AcB3s had the typical B3 domains characteristic of B3 genes. Consistent with the phylogenetic analysis, most genes had similar classification and number of motifs in the same subfamily, supporting the reliability of our subfamily classification.
57 AcB3 genes were classified into five subfamilies by phylogenetic analysis. Like other species [11], the REM subfamily in pineapple also had the most genes followed by ARF subfamily (Fig. 1). It was reported that there are 20 ARFs in pineapple [38]. However, only 19 ARF genes were identified in our study, and Aco015073 did not have the typical B3 domain although it had sequence similarity to one of the ARFs. Of the 19 AcARFs, 14 genes contained the B3, Aux_resp, and Aux_IAA domains, while the remaining genes had no Aux_IAA domain, indicating that conserved evolution in both B3 and Aux_resp domains. In addition, Aco009779, Aco016748, Aco013227, and Aco016654 without AP2 domain were also classified as RAV subfamily. The RAV subfamily usually have one AP2 and one B3 domains, but not all RAV proteins contain AP2 domains [15, 39]. The number of B3 domains of REM subfamily in pineapple varies from 1 to 4, which may be caused by domain duplication events.
Tandem duplication, segmental duplication, and transposition are considered as the main gene duplication events promoting the expansion of gene families, among which segmental duplication contributes more than tandem duplication in plants [40, 41]. In this study, 8 segmental duplication events and 5 tandem duplication events were identified in AcB3s. Among them, the Ka/Ks value of 10 genes was less than 1 while the value of two genes was more than 1 (Table S2), demonstrating that positive selection happened in AcB3s.
As illustrated by previous studies, AcFT was highly expressed within 1 d and in flower bud differentiation period in stem apex after ethephon treatment, and overexpression of AcFT gene resulted in early flowering [37, 42]. In this study, 35 of 40 AcB3s were observed to be responsive to ethylene in stem apex within 1d after ethephon treatment, and 25 exhibited the significant up-regulation in 31d after ethephon treatment (Fig. 7), implying that these AcB3s may play roles in the process of ethephon-induced pineapple flowering. Most of REM subfamily genes are targets of key flowing transcription factors, which are preferentially expressed in reproductive meristems, such as AP1, AP3, PI, LEAFY and SVP [3, 43, 44]. In Arabidopsis, AtREM22 is related to the stamen development while AtREM5 (VRN1) is involved in flowering time control [45, 46]. In this study, it was revealed that the expression levels of most AcREMs in stem apex were higher than in leaves after ethephon treatment. Among them, the AcREM genes (Aco005629 and Aco020809) had the close genetic distance and similar structure with AtREM17 (At4g34400), a target of LEAFY expressed preferentially in flowers [3, 10]. Thus, it is speculated that they may have the similar functions in the flower development of pineapple. AtVAL1 (At2g30470) also named as AtHSI2 is a target of FLC (Flowering Locus C) in the flowering process [47]. Aco001637 having closely clusters with At2g30470 was largely up-regulated in stem apex after 6 h ethephon treatment and reached to the higher level when treated for 31d (Fig. 1; Fig. 7), indicating that Aco001637 may function in the process of ethephon-induced flowering in pineapple. Auxin response mediated by ARF genes participates in flowering process [13]. In tomato, most ARFs had the higher expression levels in flower buds and flowers [48]. Arabidopsis ARF6 and ARF8 (At5g37020) coordinate the transition from immature to mature fertile flowers [49]. Interestingly, we found that ethylene up-regulated Aco012988 was classified together with At5g37020. In addition, phosphorus is necessary for plant growth and development, and the concentration of phosphorus has an influence on flowering [50]. OsARF16 (Os06g09660) in rice involves in phosphate transport [51]. Intriguingly, Aco018978 was classified together with OsARF16 in rice, and up-regulated significantly in 31d after ethephon treatment. It is reasonably to speculate that Aco018978 in pineapple may participate in the regulation of flowering by increasing the phosphate transport. The variety used in the published genome was F-153 which belonged to Cayenne, and this study we used ‘Tainong 16’ which comes from Smooth Cayenne (Cayenne) × Rough (Queen) [52, 53]. Based on the genome sequences, we preliminary studied the expression of AcB3 genes in ‘Tainong 16’, in order to demonstrate the expression divergence with other varieties, more experiments need to be carried out next.
Conclusions
In this study, 57 AcB3 genes were identified in the pineapple genome, and classified into five major classes (REM, ARF, RAV, HSI and LAV). Uneven distribution and segmental duplications on chromosomes contributed to the expansion of AcB3s. Several candidate genes that contain ethylene response elements and are significantly up-regulated by ethylene in stem apex were identified. These results will provide a reference for future research on the molecular mechanisms of ethylene-induced pineapple flowering.
Methods
Identification and analysis of B3s in pineapple
To identify AcB3s, the genome of Ananas comosus were downloaded from the PGD (Pineapple Genomics database) (http://pineapple.angiosperms.org/pineapple/html/index.html) [54]. The HMM (hidden Markov model) matrix of B3 domain (PF02362) was downloaded from Pfam, and then B3s were retrieved by Hmmer 3.0 software. These predicted B3s proteins were further confirmed and analyzed using the CD-search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) and SMART (http://smart.embl-heidelberg.de) web server. All pineapple B3s contained B3 domain.
Phylogenetic analysis
Multiple sequence alignments and phylogenetic tree of full-length B3s proteins from pineapple, Arabidopsis and rice were performed using PhyML 3.0 with maximum likelihood (ML) method, ‘AIC’ criterion and ‘aLRT Chi2-based’ as fast likelihood-based method [55]. The classifications of AcB3s were given according to the lists of AtB3s and OsB3s came from previous study [15]. The sequences of AtB3s and OsB3s were downloaded from Phytozome 12.1.6 (https://phytozome.jgi.doe.gov/pz/portal.html).
Chromosomal locations and synteny analysis
The physical locations of AcB3s were obtained from the PGD. The location images of AcB3s were drawn by MapChart software [56]. Multiple Collinearity Scan toolkit (MCscanX) was employed to analyze the tandem and segmental duplications of B3s [57]. The diagrams of synteny analysis were drawn by TBtools [58]. The value of Ks and Ka were calculated by DnaSP 5 software [59].
Structural and motif analysis of AcB3s
The exon/intron structures of the AcB3s were drawn by TBtools. The conserved motifs were analyzed by MEME (http://meme-suite.org/tools/meme) [60], searching up to 20 conserved motifs and each motif was set from 6 to 50 residues. Protein domains were identified using the NCBI CD-search and SMART.
Analysis of cis-acting elements and gene ontology
The 2 kb promoter region upstream of the start codon of each gene was downloaded from PGD to analyze the possible cis-acting elements of B3s by Plant CARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) online server. The ERF binding sites were predicted by JASPAR (http://jaspar.genereg.net/), and the relative profile score threshold was ‘80’. WEGO (http://wego.genomics.org.cn/) was used to analyze Gene ontology of B3 proteins.
Plant materials and treatments
The pineapple hybrid variety was “Tainong 16”. The temperature, humidity and other environmental conditions in the greenhouse which controlled in the appropriate range for pineapple flowering. The pineapple which grew 25 mature leaves (more than 30 cm in length) were treated by 80 mL of 1600 mg/L ethephon solution, and three biological replicates were sampled at 0, 1 h, 3 h, 6 h, 1 d, 6 d, 10 d, 15d, 22 d and 31 days, including leaves and stem apex. The leaves (medial green part) and stem apex were collected from three individual plants as a replication. The sample treated with water of the same volume as the contrast (0), all of them were frozen in liquid nitrogen and stored in refrigerator at − 80 °C until used.
RNA isolation and AcB3s expression analysis
Total RNA was extracted from samples using Total RNA was extracted from samples using a universal plants RNA extraction kit for HuaYueyang, according to the manufacturer’s guidelines. 1 μg RNA were used to synthesize the first-strand cDNA by MonScript™ RTIII All-in-One Mix with dsDNase. Primers were designed for real-time quantitative PCR (qRT-PCR) using Primer 5.0 software, and the primer sequences were shown in detail in Table S8. The cDNA was used as the template, the results [ΔΔCT = (Cttarget gene-CtAcEF1) - (Ct0-CtAcEF1)] were normalized against the Ct value of AcEF1 based on three biological replicates. The reaction was carried out on Roche Lightcyler® 480 instrument using SYBR Green Master Mix (Roche). The reaction system of real-time quantitative PCR was 10 μL, including 1 μL of cDNA, 5 μL of SYBR Green I master, 0.5 μL of 10 μmol/L of forward and reverse primers, and 3 μL of ddH2O. The relative expression data were analyzed by 2-ΔΔ CT method [61], and by Excel and Graphpad prism 7.0 software. The t test was analysed by SPSS 21 software.
Supplementary Information
Additional file 1: Fig. S1. Structural domain diagram of B3 genes. Fig. S2 Gene ontology analysis of AcB3 proteins. Three categories (cellular component, biological process and molecular function) and terms on level 2 were exhibited by different colors. Fig. S3. GO terms on level 3.
Additional file 2: Table S1. Physical and chemical properties of B3 genes predictions. Table S2. Ka/Ks calculation of the duplicated pineapple B3 gene pairs. Table S3. One-to-one orthologous relationships between pineapple and Arabidopsis, pineapple and rice. Table S3. One-to-one orthologous relationships between pineapple and Arabidopsis, pineapple and rice. Table S4. Seauence and characteristics of conserved motifs identified in pineapple. Table S5. The cis-element sites of 57 B3 promoters. Table S6. The prediction of ERF binding site. Table S7. GO ID in transcriptome data. Table S8. The primers of qRT-PCR.
Acknowledgements
The authors thank to all people who provided help for this article.
Authors’ contributions
Conceptualization: CCR, ZC and ZLZ; Methodology: CCR, ZC; Resources: XHW, LJG; Software Formal analysis: HYF, FCH; Visualization: ZWL; Writing - Original Draft: CCR; Writing - Review & Editing: ZLZ and WF. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (31960589 and 31260460), Science and Technology Innovation Program of Hainan Association for Science and Technology for Youths (QCXM201803).
Availability of data and materials
All data generated or analysed during this study are included in this published article and its supplementary information files. Phylogeny data are submitted into the Treebase repository (http://purl.org/phylo/treebase/phylows/study/TB2:S28162?x-access-code=e939af3376849ed9741d964ab1b9b1bc&format=html).
Declarations
Ethics approval and consent to participate
The pineapples used in this study were cultivated in the pineapple planting base of Cheng Mai County, Hainan Province, China. Collection of plant materials complied with the institutional, national and international guidelines. No specific permits were required.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflicts of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Imaizumi T, Kay SA. Photoperiodic control of flowering: not only by coincidence. Trends Plant Sci. 2006;11(11):550–8. 10.1016/j.tplants.2006.09.004. [DOI] [PubMed]
- 2.Riechmann JL, Ratcliffe OJ. A genomic perspective on plant transcription factors. Curr Opin Plant Biol. 2000;3(5):423–34. 10.1016/S1369-5266(00)00107-2. [DOI] [PubMed]
- 3.Mantegazza O, Gregis V, Mendes MA, Morandini P, Alves-Ferreira M, Patreze CM, et al. Analysis of the arabidopsis REM gene family predicts functions during flower development. Ann Bot. 2014;114(7):1507–15. 10.1093/aob/mcu124. [DOI] [PMC free article] [PubMed]
- 4.Fassinou Hotegni VN, Lommen WJ, Agbossou EK, Struik PC. Trade-offs of flowering and maturity synchronisation for pineapple quality. PLoS One. 2015;10(11):e0143290. doi: 10.1371/journal.pone.0143290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friend D. Effect of night temperature on flowering and fruit size in pineapple (Ananas comosus [L.] Merrill). Bot Gaz. 1981;142(2):188–90. 10.1086/337211.
- 6.Friend DJ, Lydon J. Effects of daylength on flowering, growth, and CAM of pineapple (Ananas comosus [L.] Merrill). Bot Gaz. 1979;140(3):280–3. 10.1086/337086.
- 7.GAPd C. Applied aspects of pineapple flowering. Bragantia. 2005;64(4):499–516. doi: 10.1590/S0006-87052005000400001. [DOI] [Google Scholar]
- 8.Zhang ZL, Fan HY, Hua M, He F. Induction of pineapple flowering and underlying physiological and molecular bases. Chin J Trop Crop. 2012;33(5):950–5.
- 9.Swaminathan K, Peterson K, Jack T. The plant B3 superfamily. Trends Plant Sci. 2008;13(12):647–55. 10.1016/j.tplants.2008.09.006. [DOI] [PubMed]
- 10.Peng FY, Weselake RJ. Genome-wide identification and analysis of the B3 superfamily of transcription factors in Brassicaceae and major crop plants. Theor Appl Genet. 2013;126(5):1305–19. 10.1007/s00122-013-2054-4. [DOI] [PubMed]
- 11.Wang YJ, Deng DX, Zhang R, Wang SX, Bian YL, Yin ZT. Systematic analysis of plant-specific B3 domain-containing proteins based on the genome resources of 11 sequenced species. Mol Biol Rep. 2012;39(5):6267–82. 10.1007/s11033-012-1448-8. [DOI] [PubMed]
- 12.Sun TT, Wang DW, Gong DP, Chen L, Chen YQ, Sun YH. Genome-wide identification and bioinformatic analysis of B3 superfamily in tomato. J Plant Genet Res. 2015;16(4):806–14.
- 13.Liu YH, Dong ZP. The function and structure of plant B3 domain transcription factor. Mol Plant Breed. 2017;15(05):1868–73.
- 14.Suzuki M, McCarty DR. Functional symmetry of the B3 network controlling seed development. Curr Opin Plant Biol. 2008;11(5):548–53. 10.1016/j.pbi.2008.06.015. [DOI] [PubMed]
- 15.Romanel EA, Schrago CG, Couñago RM, Russo CA, Alves-Ferreira M. Evolution of the B3 DNA binding superfamily: new insights into REM family gene diversification. PLoS One. 2009;4(6):e5791. doi: 10.1371/journal.pone.0005791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu JJ, Yu SC, Liu ZG, FU Y, Zhou MB. Genome identification and expression pattern analysis of Phyllostachys edulis B3 family. Chin J Agric Biotechol. 2019;27(1):43–54. [Google Scholar]
- 17.Verma S, Bhatia S. A comprehensive analysis of the B3 superfamily identifies tissue-specific and stress-responsive genes in chickpea (Cicer arietinum L.) 3 Biotech. 2019;9(9):346. doi: 10.1007/s13205-019-1875-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chandler JW. Auxin response factors. Plant Cell Environ. 2016;39(5):1014–28. 10.1111/pce.12662. [DOI] [PubMed]
- 19.Li SB, Xie ZZ, Hu CG, Zhang JZ. A review of auxin response factors (ARFs) in plants. Front Plant Sci. 2016;7:47. doi: 10.3389/fpls.2016.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellis CM, Nagpal P, Young JC, Hagen G, Guilfoyle TJ, Reed JW. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development. 2005;132(20):4563–74. 10.1242/dev.02012. [DOI] [PubMed]
- 21.Wilmoth JC, Wang SC, Tiwari SB, Joshi AD, Hagen G, Guilfoyle TJ, et al. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 2005;43(1):118–30. 10.1111/j.1365-313X.2005.02432.x. [DOI] [PubMed]
- 22.Stone SL, Kwong LW, Yee KM, Pelletier J, Lepiniec L, Fischer RL, et al. LEAFY COTYLEDON2 encodes a B3 domain transcription factor that induces embryo development. P Natl Acad Sci USA. 2001;98(20):11806–11. 10.1073/pnas.201413498. [DOI] [PMC free article] [PubMed]
- 23.Freitas NC, Barreto HG, Torres LF, Freire LL, Rodrigues LAZ, Diniz LEC, et al. In silico and in vivo analysis of ABI3 and VAL2 genes during somatic embryogenesis of Coffea arabica: competence acquisition and developmental marker genes. Plant Cell Tiss Org. 2019;137(3):599–611. 10.1007/s11240-019-01594-7.
- 24.Sugliani M, Rajjou L, Clerkx EJ, Koornneef M, Soppe WJ. Natural modifiers of seed longevity in the Arabidopsis mutants abscisic acid insensitive3-5 (abi3-5) and leafy cotyledon1-3 (lec1-3) New Phytol. 2009;184(4):898–908. doi: 10.1111/j.1469-8137.2009.03023.x. [DOI] [PubMed] [Google Scholar]
- 25.Khandelwal A, Cho SH, Marella H, Sakata Y, Perroud P-F, Pan A, et al. Role of ABA and ABI3 in desiccation tolerance. Science. 2010;327(5965):546. 10.1126/science.1183672. [DOI] [PubMed]
- 26.Brady SM, Sarkar SF, Bonetta D, McCourt P. The ABSCISIC ACID INSENSITIVE 3 (ABI3) gene is modulated by farnesylation and is involved in auxin signaling and lateral root development in Arabidopsis. Plant J. 2003;34(1):67–75. doi: 10.1046/j.1365-313X.2003.01707.x. [DOI] [PubMed] [Google Scholar]
- 27.Stone SL, Braybrook SA, Paula SL, Kwong LW, Meuser J, Pelletier J, et al. Arabidopsis LEAFY COTYLEDON2 induces maturation traits and auxin activity: implications for somatic embryogenesis. P Natl Acad Sci USA. 2008;105(8):3151–6. 10.1073/pnas.0712364105. [DOI] [PMC free article] [PubMed]
- 28.Veerappan V, Wang J, Kang M, Lee J, Tang YH, Jha AK, et al. A novel HSI2 mutation in Arabidopsis affects the PHD-like domain and leads to derepression of seed-specific gene expression. Planta. 2012;236(1):1–17. 10.1007/s00425-012-1630-1. [DOI] [PubMed]
- 29.Chen NC, Veerappan V, Abdelmageed H, Kang M, Allen RD. HSI2/VAL1 silences AGL15 to regulate the developmental transition from seed maturation to vegetative growth in Arabidopsis. Plant Cell. 2018;30(3):600–19. 10.1105/tpc.17.00655. [DOI] [PMC free article] [PubMed]
- 30.Li XJ, Li M, Zhou Y, Hu S, Hu R, Chen Y, et al. Overexpression of cotton RAV1 gene in Arabidopsis confers transgenic plants high salinity and drought sensitivity. PLoS One. 2015;10(2):e0118056. 10.1371/journal.pone.0118056. [DOI] [PMC free article] [PubMed]
- 31.Castillejo C, Pelaz S. The balance between CONSTANS and TEMPRANILLO activities determines FT expression to trigger flowering. Curr Biol. 2008;18(17):1338–43. 10.1016/j.cub.2008.07.075. [DOI] [PubMed]
- 32.Mylne JS, Barrett L, Tessadori F, Mesnage S, Johnson L, Bernatavichute YV, et al. LHP1, the Arabidopsis homologue of HETEROCHROMATIN PROTEIN1, is required for epigenetic silencing of FLC. P Natl Acad Sci USA. 2006;103(13):5012–17. 10.1073/pnas.0507427103. [DOI] [PMC free article] [PubMed]
- 33.Caselli F, Beretta VM, Mantegazza O, Petrella R, Leo G, Guazzotti A, et al. REM34 and REM35 control female and male gametophyte development in Arabidopsis thaliana. Front Plant Sci. 2019;10:1351. 10.3389/fpls.2019.01351. [DOI] [PMC free article] [PubMed]
- 34.Shang HH, Li W, Zou CS, Yuan YL. Analyses of the NAC transcription factor gene family in Gossypium raimondii Ulbr.: chromosomal location, structure, phylogeny, and expression patterns. J Integr Plant Biol. 2013;55(7):663–76. 10.1111/jipb.12085. [DOI] [PubMed]
- 35.Ruan CC, Nian YW, Hu FC, Wang XH, Fan HY, Wu FZ, et al. Expression and analysis of FT genes in pineapple flowering induced by ethephon. J Fruit Sci. 2019;36(12):1648–57.
- 36.Ahmad B, Zhang SL, Yao J, Rahman MU, Hanif M, Zhu YX, et al. Genomic organization of the B3-domain transcription factor family in grapevine (Vitis vinifera L.) and expression during seed development in seedless and seeded cultivars. Int J Mol Sci. 2019;20(18):4553. [DOI] [PMC free article] [PubMed]
- 37.Xia F, Sun TT, Yang SJ, Wang X, Chao JT, Li XX, et al. Insight into the B3 transcription factor superfamily and expression profiling of B3 genes in axillary buds after topping in tobacco (Nicotiana tabacum L.). Genes. 2019;10(2):164. [DOI] [PMC free article] [PubMed]
- 38.Su ZX, Wang LL, Li WM, Zhao LH, Huang XY, Azam SM, et al. Genome-wide identification of auxin response factor (ARF) genes family and its tissue-specific prominent expression in pineapple (Ananas comosus). Trop Plant Biol. 2017;10(2-3):86–96. 10.1007/s12042-017-9187-6.
- 39.Alvarez JP, Goldshmidt A, Efroni I, Bowman JL, Eshed Y. The NGATHA distal organ development genes are essential for style specification in Arabidopsis. Plant Cell. 2009;21(5):1373–1393. 10.1105/tpc.109.065482. [DOI] [PMC free article] [PubMed]
- 40.Gu ZL, Steinmetz LM, Gu X, Scharfe C, Davis RW, Li W-H. Role of duplicate genes in genetic robustness against null mutations. Nature. 2003;421(6918):63–66. 10.1038/nature01198. [DOI] [PubMed]
- 41.Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4(1):10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lv LL, Duan J, Xie JH, Wei CB, Liu YG, Liu SH, et al. Isolation and characterization of a FLOWERING LOCUS T homolog from pineapple (Ananas comosus (L.) Merr). Gene. 2012;505(2):368–73. 10.1016/j.gene.2012.06.011. [DOI] [PubMed]
- 43.Villarino GH, Hu QW, Manrique S, Flores-Vergara MA, Sehra B, Robles L, et al. Temporal and spatial domain-specific transcriptomic analysis of a vital reproductive meristem in Arabidopsis thaliana. Plant Physiol. 2016;171(1):42–61. 10.1104/pp.15.01845. [DOI] [PMC free article] [PubMed]
- 44.Franco-Zorrilla JM, Cubas P, Jarillo JA, Fernández-Calvın B, Salinas J, Martınez-Zapater JM. AtREM1, a member of a new family of B3 domain-containing genes, is preferentially expressed in reproductive meristems. Plant Physiol. 2002;128(2):418–27. 10.1104/pp.010323. [DOI] [PMC free article] [PubMed]
- 45.Romanel E, Das P, Amasino RM, Traas J, Meyerowitz E, Alves-Ferreira M. Reproductive Meristem22 is a unique marker for the early stages of stamen development. Int J Dev Biol. 2011;55(6):657–64. 10.1387/ijdb.113340er. [DOI] [PubMed]
- 46.Levy YY, Mesnage S, Mylne JS, Gendall AR, Dean C. Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science. 2002;297(5579):243–6. 10.1126/science.1072147. [DOI] [PubMed]
- 47.Wu BX, Zhang MM, Su SC, Liu HH, Gan JH, Ma JB. Structural insight into the role of VAL1 B3 domain for targeting to FLC locus in Arabidopsis thaliana. Biochem Bioph Res Co. 2018;501(2):415–422. doi: 10.1016/j.bbrc.2018.05.002. [DOI] [PubMed] [Google Scholar]
- 48.Kumar R, Tyagi AK, Sharma AK. Genome-wide analysis of auxin response factor (ARF) gene family from tomato and analysis of their role in flower and fruit development. Mol Gen Genomics. 2011;285(3):245–60. 10.1007/s00438-011-0602-7. [DOI] [PubMed]
- 49.Nagpal P, Ellis CM, Weber H, Ploense SE, Barkawi LS, Guilfoyle TJ, et al. Auxin response factors ARF6 and ARF8 promote jasmonic acid production and flower maturation. Development. 2005;132(18):4107–18. 10.1242/dev.01955. [DOI] [PubMed]
- 50.Scagel CF, Schreiner RP. Phosphorus supply alters tuber composition, flower production, and mycorrhizal responsiveness of container-grown hybrid Zantedeschia. Plant Soil. 2006;283(1-2):323–37. 10.1007/s11104-006-0022-3.
- 51.Shen CJ, Wang SK, Zhang SN, Xu YX, Qian Q, Qi YH, et al. OsARF16, a transcription factor, is required for auxin and phosphate starvation response in rice (Oryza sativa L.). Plant Cell Environ. 2013;36(3):607–20. 10.1111/pce.12001. [DOI] [PubMed]
- 52.Kato CY, Nagai C, Moore PH, Zee F, Kim MS, Steiger DL, et al. Intra-specific DNA Polymorphism in pineapple (Ananas comosus (L.) Merr.) assessed by AFLP markers. Genet Resour Crop Evol. 2005;51(8):815–25. 10.1007/s10722-005-0005-x.
- 53.Lin Y, Kuan C, Weng I, Tsai C. Cultivar identification and genetic relationship of pineapple (Ananas comosus) cultivars using SSR markers. Genet Mol Res. 2015;14(4):15035–43. 10.4238/2015.November.24.11. [DOI] [PubMed]
- 54.Xu HM, Yu QY, Shi Y, Hua XT, Tang HB, Yang L, et al. PGD: pineapple genomics database. Hortic Res. 2018;5(1):1–9. [DOI] [PMC free article] [PubMed]
- 55.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–21. 10.1093/sysbio/syq010. [DOI] [PubMed]
- 56.Voorrips RE. MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered. 2002;93(1):77–78. 10.1093/jhered/93.1.77. [DOI] [PubMed]
- 57.Wang YP, Tang HB, DeBarry JD, Tan X, Li JP, Wang XY, et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40(7):e49. 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed]
- 58.Chen CJ, Chen H, Zhang Y, Thomas HR, Frank MH, He YH, et al. TBtools-an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13(8):1194–202. 10.1016/j.molp.2020.06.009. [DOI] [PubMed]
- 59.Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25(11):1451–2. 10.1093/bioinformatics/btp187. [DOI] [PubMed]
- 60.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37(2):W202–W208. 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed]
- 61.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25(4):402–8. 10.1006/meth.2001.1262. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Fig. S1. Structural domain diagram of B3 genes. Fig. S2 Gene ontology analysis of AcB3 proteins. Three categories (cellular component, biological process and molecular function) and terms on level 2 were exhibited by different colors. Fig. S3. GO terms on level 3.
Additional file 2: Table S1. Physical and chemical properties of B3 genes predictions. Table S2. Ka/Ks calculation of the duplicated pineapple B3 gene pairs. Table S3. One-to-one orthologous relationships between pineapple and Arabidopsis, pineapple and rice. Table S3. One-to-one orthologous relationships between pineapple and Arabidopsis, pineapple and rice. Table S4. Seauence and characteristics of conserved motifs identified in pineapple. Table S5. The cis-element sites of 57 B3 promoters. Table S6. The prediction of ERF binding site. Table S7. GO ID in transcriptome data. Table S8. The primers of qRT-PCR.
Data Availability Statement
All data generated or analysed during this study are included in this published article and its supplementary information files. Phylogeny data are submitted into the Treebase repository (http://purl.org/phylo/treebase/phylows/study/TB2:S28162?x-access-code=e939af3376849ed9741d964ab1b9b1bc&format=html).