Abstract
Pseudomonas aeruginosa, an opportunistic human pathogen, causes fatal effects in patients with cystic fibrosis and immunocompromised individuals and leads to around 1000 deaths annually. The quorum sensing mechanism of P. aeruginosa plays a major role in promoting biofilm formation and expression of virulent genes. Hence, quorum sensing inhibition is a promising novel approach to treat these bacterial infections as these organisms show a wide range of antibiotic resistance. Among the interconnected quorum sensing network of P. aeruginosa, targeting the las system is of increased interest as its principal receptor protein LasR is the earliest activated gene. It is also shown to be involved in the regulation of other virulence-associated genes. In this study, we have applied high-throughput virtual screening, an in silico computational method to identify a new class of LasR inhibitors that could serve as potent antagonists to treat P. aeruginosa-associated infections. Three-tire structure-based virtual screening was performed on the Schrödinger small molecule database, which resulted in 12 top hit compounds with docking scores lesser than −11.0 kcal/mol. Three of these best-scored compounds CACPD2011a-0001928786 (C1), CACPD2011a-0001927437 (C2), and CACPD2011a-0000896051 (C3) were further analyzed. The binding free energies of these compounds in complex with the target protein LasR (3IX4) were evaluated, and the pharmacokinetic properties were determined. The stability of the docked complexes was assessed by running a molecular dynamics simulation for 100 ns. Molecular dynamics simulation analysis revealed that all three compounds were found to be in stable contact with the protein over the entire simulation period. The antagonistic effect of these compounds was validated using the LasR reporter gene assay in the presence of acyl homoserine lactone. Significant reduction in the β-galactosidase enzyme activity was achieved at 100 nM concentration for all three compounds pursued. Hence, the present study provides strong evidence that these three compounds could serve as quorum sensing inhibitors of P. aeruginosa LasR protein and can be a probable candidate to treat Pseudomonas-associated infections.
1. Introduction
Pseudomonas aeruginosa, a pathogenic Gram-negative rod-shaped bacterium, is a major causative microorganism responsible for biofilm formation and causes secondary infections in conditions like cystic fibrosis, nosocomial infections, and in immunocompromised individuals.1,2 According to the National Institute of Health (NIH), these bacterial biofilms constitute about 65% of the microbial diseases and are responsible for worsening by contributing to more than 80% of chronic infections in humans.3 For more than a decade, P. aeruginosa has been among the “top 10” common hospital “superbugs” majorly attributed to several antimicrobial-resistant strains that cause life-threatening complications.4 The antibiotic-resistant strains of P. aeruginosa contribute to around 11% of the hospital-acquired infections and leads to 61% of the mortality rate.5−7 It is the need of the hour to look at novel approaches to treat P. aeruginosa infections.
In its aggregated form, quorum sensing (QS), an intercellular cell-to-cell communication system, coordinates bacterial behavior by utilizing signaling molecules such as autoinducers produced in response to environmental changes and population density alterations.8 The QS system of P. aeruginosa is an interconnected network consisting of las, rhl, Pseudomonas quinolone signal (pqs), and integrated QS (iqs) systems regulated by various QS signaling molecules.9 Of these, the signaling molecules produced by las and rhl QS systems are sensed by their cognate LuxR-type receptors, namely, LasR and RhlR, respectively.10 The PQS system binds to the receptor protein PqsR, a transcriptional factor that is not related to LuxR-type receptors.11 Among these, the las system was found to be the most crucial one in regulating other QS systems. The las system includes LasI and LasR, where its autoinducer molecule N-(3-oxo-dodecanoyl)-l-homoserine lactone (OdDHL) formed by LasI binds to LasR and activates the transcriptional regulation of multiple genes.12,13
LasR is the principal protein involved in the regulation of virulence phenotypes such as protease production, biofilm formation, and pyocyanin and rhamnolipid production.14−16 LasR consists of two domains that are folded independently, namely, an amino-terminal ligand binding domain (LBD) and a C-terminal DNA binding domain (DBD).17,18 When the native acyl homoserine lactone (AHL), 3-oxo-C12 HSL, binds to LasR, the monomeric form stabilizes and dimerizes to two LasR subunits. The resultant ligand-bound homodimer then acquires the potential to bind DNA and activates transcriptional changes.19,20 Hence, targeting LasR may be of increased interest as it is significantly involved in QS-associated virulence in P. aeruginosa.21
Different approaches have been attempted over the past years to identify a potent lead molecule with anti-QS and antibiofilm activities for treating P. aeruginosa infections.22 Yet, the literature related to experimental and modeling studies have reported that the spread of resistance in bacteria for the identified quorum sensing inhibitors (QSIs) is still at its rise.23,24 In silico computational methods offer various tools for identifying novel drug candidates from which the chemical and biological information about the ligands and target can be derived.25 The efficacy of the compounds can be observed by approaches such as molecular docking, dynamics, and simulation.26 Molecular docking analysis paves the way for “virtual” or “structure-based” screening of ligands that fit into the target protein structure.27
Structure-based virtual screening (SBVS) can be applied to discover new QSIs using the crystal structure of the QS receptor. In silico discovery of QSIs against the LasR receptor has already been studied by a few groups.28−31 However, validation of these compounds by a combined molecular dynamics (MD) simulation and in vitro studies have not been attempted.
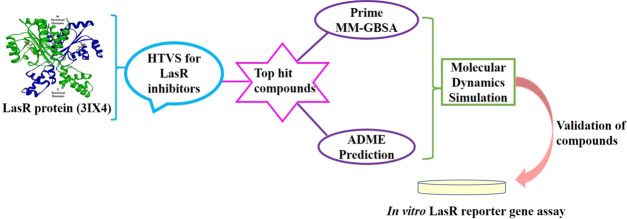
In the present study, novel inhibitors of LasR were identified by executing the following steps: high-throughput virtual screening (HTVS) was performed to screen for potent antagonist compounds. The compounds with relatively higher docking scores were selected, and their pharmacokinetic profiles were analyzed. The stability of their interactions with the target protein was determined by an MD simulation study. Finally, the selected compounds were validated using the in vitro LasR reporter gene assay.
2. Results
2.1. Structural Analysis of the Target Protein LasR
The three-dimensional (3D) crystal structure of the P. aeruginosa LasR (Protein Data Bank (PDB) ID: 3IX4) protein at 1.8 Å resolution was retrieved from PDB. The cocrystal structure of the LasR protein was found to have five strands of the antiparallel β-sheet flanked on both sides by six α-helices.17 Analysis of the ligand binding site in LasR revealed that the cocrystallized agonist molecule TP-1 (triphenyl mimic of the natural inducer) forms five direct and one water-mediated intermolecular hydrogen bond interactions involving Tyr56, Trp60, Arg61, Asp73, Thr75, and Ser129. The residues Tyr56, Trp60, Asp73, and Ser129 are highly conserved among LuxR homologues.18 TP-1 is a signal mimic of the natural LasR activator and is more specific to LasR.32 The stable and precise binding of ligands to the active site of the receptor protein depends on the hydrogen bond interaction with these residues as they provide directionality and specificity.33−35
2.2. Identification of Antagonists Targeting LasR by High-Throughput Virtual Screening
Virtual screening was carried out with 3 034 496 compounds from the Schrödinger small molecule database to identify compounds that showed interactions with LasR. Among them, 1 942 018 compounds are docked in the HTVS mode; the top 10% of the compounds from HTVS screening was subjected to standard precision (SP) docking, which ranked molecules based on docking scores. A total of 19 420 compounds were docked, and the top 10% of the SP docked compounds were further taken for flexible extra precision (XP) docking. Finally, 1942 compounds were filtered from the XP docking mode. The top 12 hit compounds with a score less than −11.0 kcal/mol were selected and are shown in Table 1. To set a cutoff score for the docking study, we docked various reported antagonists.36−41 The results showed a Gscore that ranged between −11.0 and −6.0 kcal/mol (Table S1). Hence, for screening potent inhibitors of LasR, a Gscore cutoff value of −11.0 kcal/mol was set as the filtering criterion in virtual screening.
Table 1. Docking Scores and Hydrogen Bond Interactions of Hit Compounds.
| compounds | XP docking score (kcal/mol) | no. of H-bonds | H-bond interaction residues |
|---|---|---|---|
| native ligand (autoinducer -OdDHL) | –7.5 | 4 | Tyr56, Asp73, Ser129, Trp60 |
| TP-1 (triphenyl mimic of autoinducer) | –12.8 | 4 | Tyr56, Asp73, Ser129, Trp60 |
| CACPD2011a-0001928786 | –13.0 | 3 | Tyr56, Ser129, Asp73 |
| CACPD2011a-0001927437 | –12.2 | 3 | Tyr56, Ser129, Asp73 |
| CACPD2011a-0000896051 | –11.7 | 0 | |
| CACPD2011a-0001779781 | –11.7 | 2 | Tyr56, Ser129 |
| CACPD2011a-0001734913 | –11.6 | 0 | |
| CACPD2011a-0002367758 | –11.4 | 3 | Tyr56, Ser129, Trp60 |
| CACPD2011a-0002145356 | –11.3 | 1 | Tyr56 |
| CACPD2011a-0001893654 | –11.2 | 0 | |
| CACPD2011a-0002017222 | –11.1 | 2 | Tyr56, Ser129 |
| CACPD2011a-0002215130 | –11.1 | 3 | Tyr56, Asp73, Thr115 |
| CACPD2011a-0002142621 | –11.0 | 1 | Ser129 |
| CACPD2011a-0000661063 | –11.0 | 3 | Tyr56, Asp73, Leu110 |
The native autoinducer ligand (OdDHL) and the cocrystallized triphenyl mimic of the autoinducer form four H-bond interactions with LasR active site residues Tyr56, Trp60, Asp73, and Ser129 (Table 1). The docking study also showed that the top-ranked ligands did not show the same pattern of H-bond interactions as that of the inducer molecules OdDHL and TP-1. The molecular docking study of the reported antagonist also showed that out of the 36 antagonist molecules, only six molecules formed H-bond interaction with all four active site residues Tyr56, Trp60, Asp73, and Ser129 (Table S1).
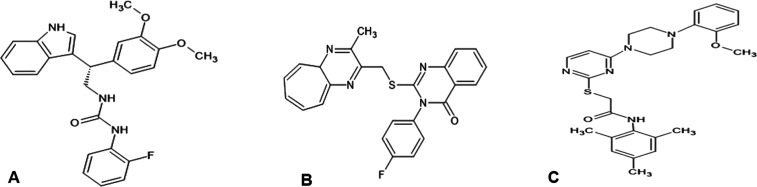
Further analysis was carried out with the three top hit compounds (Figure 1), namely, CACPD2011a-0001928786 (3-[2-(3,4-dimethoxyphenyl)-2-(1H-indol-3-yl) ethyl]-1-(2-fluorophenyl)urea), CACPD2011a-0001927437 (N-(2-fluorophenyl)-2-{[2-(1-naphthyl) pyrazolo[1,5-a] pyrazin-4-yl] thio}acetamide), and CACPD2011a-0000896051 (2-({4-[4-(2-methoxyphenyl) piperazin-1-yl] pyrimidin-2-yl} sulfanyl)-N-(2,4,6-trimethyl phenyl)acetamide), which were selected based on their docking scores and designated as C1, C2, and C3, respectively. Among them, CACPD2011a-0001928786 (C1) was found to have a chiral center, but the other form of the isomer present in the initial screening deck was not present in the final list of compounds filtered from the XP docking mode, which indicates the strong stereospecificity of this compound toward LasR.
Figure 1.
Two-dimensional (2D) structures of the top three selected compounds: (A) CACPD2011a-0001928786, (B) CACPD2011a-0001927437, and (C) CACPD2011a-0000896051.
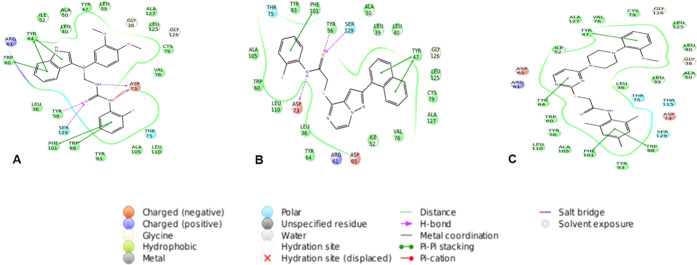
The protein–ligand interactions provide an insight into the interaction pattern of selected compounds (Figure 2), the native ligand (OdDHL), and the cocrystallized ligand TP-1 with the LasR protein (Figure 3). An in-depth analysis of all of these docked complexes suggests nonbonding interactions like hydrogen bonding, hydrophobic interactions, π–π stacking, polar interactions, and charged positive interactions have a crucial role in the binding of compounds to the active site. Docking studies revealed that CACPD2011a-0001928786 (C1) (Figure 2A) was involved in hydrogen bond interaction with amino acid residues Tyr56, Ser129, and Asp73. Hydrophobic contacts were established with amino acid residues, namely Tyr47, Leu39, Ala127, Leu125, Cys79, Val76, Leu110, Ala105, Tyr93, Trp88, Phe101, Tyr56, Leu36, Trp60, Tyr64, Leu40, Ile52, and Ala50. It also showed π–π stacking with residues Tyr64, Trp60, Phe101, and Trp88.
Figure 2.
Ligand interaction diagram for the top three compounds docked with LasR (PDB:3IX4): (A) CACPD2011a-0001928786 (C1), (B) CACPD2011a-0001927437 (C2), and (C) CACPD2011a-0000896051 (C3).
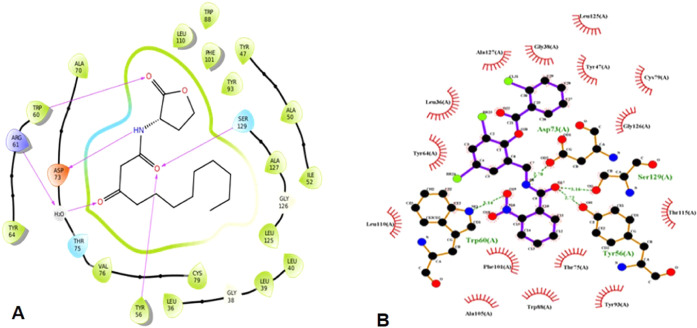
Figure 3.
Ligand interactions of the (A) native ligand with LasR and (B) TP-1 (triphenyl mimic of the autoinducer) with LasR.
In the case of the CACPD2011a-0001927437 (C2) complex (Figure 2B), the residues engaged in hydrogen bond networking were Tyr56, Ser129, and Asp73. Two amino acids, namely, Phe101 and Tyr47, were involved in π–π interaction. Tyr93, Phe101, Tyr56, Ala50, Leu39, Leu40, Tyr47, Leu125, Cys79, Ala127, Val76, Ile52, Tyr64, Leu36, Leu110, Trp60, and Ala105 amino acid residues contributed to hydrophobic contacts.
The third compound, CACPD2011a-0000896051 (C3) (Figure 2C), exceptionally did not make any hydrogen bond interaction but showed hydrophobic interactions with residues Ala127, Tyr47, Val76, Cys79, Leu125, Leu40, Ala50, Leu39, Leu36, Trp88, Tyr93, Phe101, Ala105, Leu110, Tyr56, Trp60, Tyr64, and Ile52. In addition, π–π stacking interactions were seen involving amino acids Tyr47, Trp88, Phe101, and Tyr64. Arg61 was primarily intricated in charged positive interactions with all three docked ligands. On the other hand, amino acids Thr75 and Ser129 exhibited polar interactions with C1 and C2, whereas C3 formed polar interactions through Thr75, Thr119, and Ser129.
Analysis of the ligand interaction profile of the native ligand (OdDHL) and TP-1 with LasR reported that both showed a similar pattern of hydrogen bonding involving four residues, namely, Tyr56, Asp73, Ser129, and Trp60 (Figure 3A,B). Since hydrogen bond interactions are important for stable binding of ligands to the protein, we took into account the hydrogen bond interaction profile of the three selected compounds and compared it with OdDHL and TP-1. On observation, we found that the selected compounds C1 and C2 shared only three out of four residues to make hydrogen bonds with LasR, whereas C3 did not show any hydrogen bond contacts. The pattern of interaction exhibited by the three selected compounds and their stabilities were verified by molecular dynamics simulation prior to further investigation.
2.3. Prime MM-GBSA Binding Free Energy Calculation
The binding free energy (ΔGBind) of the selected compounds could be dissociated into various energy terms and ranged from −92.2 to −112.2 kcal/mol. MM-GBSA studies revealed that the native ligand and TP-1 have binding free energy scores of −93.7 and −147.4 kcal/mol, respectively. Among the three compounds, the free binding energy (ΔGBind) of C2 was the highest and C3 had the lowest free binding energy (Table 2).
Table 2. Prime MM-GBSA Binding Energy Calculation of the Docked Complexesa.
| compounds | ΔGBind | ΔGCoul | ΔGLipo | ΔGvdW | ΔGSolvGB | SELig |
|---|---|---|---|---|---|---|
| native ligand | –93.7 | –16.7 | –44.2 | –49.3 | 16.0 | 13.5 |
| TP-1 | –147.4 | –20.8 | –63.2 | –78.0 | 21.7 | 3.3 |
| C1 | –103.6 | –59.2 | –49.9 | –61.0 | 65.4 | 14.9 |
| C2 | –112.2 | –19.9 | –54.3 | –61.7 | 28.2 | 16.3 |
| C3 | –92.2 | –3.0 | –68.6 | –65.7 | 35.6 | 37.3 |
ΔGBind, MM-GBSA free binding energy; ΔGCoul, Coulomb energy of the complex; ΔGLipo, lipophilic energy of the complex; ΔGvdW, van der Waals energy of the complex; ΔGSolvGB, solvation energy of the complex; SELig, strain energy of the ligands.
The postdocking analysis of the docked complexes concerning various energies, namely, Coulomb energy, lipophilic energy, van der Waals energy, solvation energy, and strain energy of the ligands, revealed that the three compounds pursued have a good binding affinity toward the protein active site by exhibiting the most favorable free energy binding scores.
ΔGCoul denotes the Coulomb energy of the complexes, with C1 showing the highest Coulomb energy of −59.2 kcal/mol compared to the others. The solvation energy of the complexes is distributed into either nonpolar or polar energy terms and calculated using the default solvation model. Here, the polar solvation energy (ΔGSolvGB) was computed and it was inferred that C1 exhibited the highest solvation energy with a value of 65.4 kcal/mol among the three compounds pursued.
From the parameters calculated for each compound, the most favored binding energy exhibited by C1 was with Coulomb energy and solvation energy. C2 possessed the highest free binding energy, whereas C3 showed a better binding affinity in terms of lipophilic energy (ΔGLipo), van der Waals energy (ΔGvdW), and strain energy of ligands (SELig) with values of −68.6, −65.7, and 37.3 kcal/mol, respectively. Therefore, in terms of various binding energies computed, it can be reported that all three compounds selected for the study possess significant binding affinity toward the target protein LasR.
2.4. ADMET and Drug-Likeness Prediction
Pharmacokinetics and drug-likeness properties of the selected compounds were determined by the QikProp v5.8 module of Schrödinger.60 The ADMET and drug-likeness profiles of the compounds are presented in Table 3.
Table 3. ADMET and Drug-Likeness Profiles of Selected Compounds.
| ADMET properties | C1 | C2 | C3 | optimal range (in 95% drugs) |
|---|---|---|---|---|
| molecular weight | 433.481 | 442.51 | 477.623 | 130.0–725.0 |
| no. of hydrogen bond donors | 3 | 0 | 1 | 0–6.0 |
| no. of hydrogen bond acceptors | 3.5 | 6.0 | 6.75 | 2.0–20.0 |
| predicted aqueous solubility (QP log S) | –6.3 | –7.2 | –7.7 | –6.5–0.5 |
| predicted polarizability in cubic angstroms (QPPolrz) | 46.4 | 49.8 | 53.4 | 13.0–70.0 |
| predicted hexane/gas partition coefficient (QP log PC16) | 14.4 | 13.9 | 15.1 | 4.0–18.0 |
| predicted octanol/gas partition coefficient (QP log Poct) | 21.9 | 20.1 | 23.2 | 8.0–35.0 |
| predicted water/gas partition coefficient (QP log PW) | 12.0 | 9.9 | 10.9 | 4.0–45.0 |
| predicted octanol/water partition coefficient (QP log PO/W) | 5.2 | 5.4 | 6.0 | –2.0–6.5 |
| conformation-independent predicted aqueous solubility (CIQP logS) | –6.9 | –6.9 | –7.4 | –6.5–0.5 |
| predicted IC50 value for blockage of HERG K+ channels (QP log HERG) | –5.2 | –7.0 | –6.3 | concern = <−5 |
| predicted apparent Caco-2 cell permeability in nm/s (QPPCaco) | 1551.8 | 2414.1 | 3749.9 | <25 = poor; >500 = great |
| no. of primary metabolites (#metab) | 4 | 3 | 6 | 1.0–8.0 |
| predicted brain/blood partition coefficient (QP log BB) | –0.5 | –0.2 | –0.1 | –3.0–1.2 |
| predicted apparent MDCK cell permeability in nm/s (QPPMDCK) | 1566.7 | 2610.7 | 3581.6 | <25 = poor; >500 = great |
| predicted skin permeability (QP log Kp) | –0.8 | –1.0 | –0.9 | –8.0 to −1.0 |
| prediction of binding to human serum albumin (QP log Khsa) | 0.7 | 0.8 | 1.1 | –1.5–1.5 |
| predicted human oral absorption (pHOA) | 100 | 100 | 100 | <25% is poor |
| predicted central nervous system activity (CNS) | –1 | 0 | 0 | –2 to +2 |
| number of violations of the 95% range (#stars) | 0 | 1 | 1 | |
| number of violations of Lipinski’s rule of five (VR05) | 1 | 1 | 1 | maximum is 4 |
| number of violations of Jorgensen’s rule of three (VR03) | 1 | 1 | 1 | maximum is 3 |
For orally active, a druggable candidate should not have more than one violation in Lipinski’s “Rule of Five”.42 The selected compounds in the present study were found to have one violation in Lipinski’s Rule of 5 and could be considered druglike. The bioavailability of the drug depends on the absorption of the drug and the first-pass metabolism of the liver.43 QikProp uses a set of parameters for predicting the bioavailability of the drug, which includes Jorgensen’s Rule of Three (RO3), conformation-independent aqueous solubility (ClQP log S), human oral absorption percentage (pHOA), and qualitative human oral absorption. RO3 includes QP log S, #metab, and QPPCaco, which are used to detect oral and intestinal absorption of the drug.
One of the factors to predict intestinal absorption or permeation is the Caco-2 cell permeability (QPPCaco), which is used as a model for the gut–blood barrier. QP log S and # metab are used to predict the aqueous solubility and number of likely metabolic reactions.44 The results indicated that all three compounds were in the recommended ranges of QPPCaco and # metab. In the case of CIQP log S, all three compounds were not found to be in the acceptable range. For QP log S, excluding C1, the other two compounds C2 and C3 were not within the normal limits. However, all of the selected compounds were found to possess the highest human oral absorption percentage (pHOA).
The Qikprop parameter QP log Khsa is used to predict the ability of the compounds to bind to human serum albumin (HSA). All of the selected compounds were found to be within the permissible range, indicating that these compounds would have a lower affinity to HSA; therefore, the compound will be available for binding to the target molecule. One of the important parameters to detect cardiac toxicity and neurotoxicity is the QP log HERG value. The drug molecule that binds to the HERG K+ channel may lead to heart failure. The results showed that all three selected compounds have a log IC50 value for blockage of the HERG K+ channel as less than −5, which was in the concerned range. The blood–brain barrier (BBB) prediction coefficient and the central nervous system (CNS) activity are the parameters for computing the prediction of the BBB permeability. All three compounds were found to penetrate the BBB and possess CNS activity.
The #stars parameter reveals the number of properties of each compound that fall outside the recommended range of values of 95% of the known drugs. C1 was found to have zero violation, whereas C2 and C3 showed one violation (Table 3). The predicted ADME properties revealed that the selected three compounds possess good pharmacokinetic profiles.
2.5. Molecular Dynamics Simulation
MD simulations were carried out using the Desmond v5.6 package implemented in the Schrödinger module.61 MD simulations were performed to explore the stability of the protein–ligand complexes. MD simulations were performed for 100 ns on compounds C1, C2, and C3 to understand their binding mode. The overall stability was analyzed by root-mean-square deviation (RMSD), root-mean-square fluctuations (RMSFs), radius of gyration (ROG), and intramolecular hydrogen bonds.
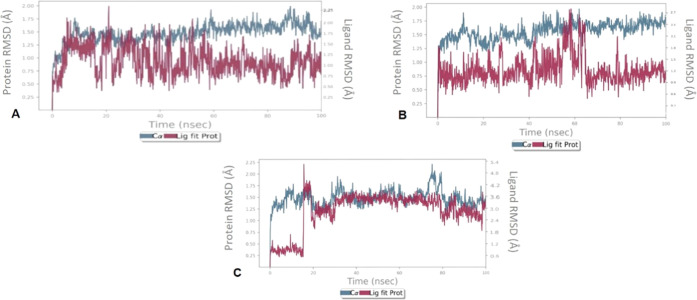
To assess the structural stability of the protein–ligand complex and to compare the conformational changes of the protein in its apo state and in the presence of the ligand, RMSD was calculated (Figure 4).
Figure 4.
Protein–ligand RMSD (Å) observed during the course of a 100 ns simulation study for (A) C1, (B) C2, and (C) C3 in complex with LasR.
The RMSD of the backbone atoms with respect to the initial docking structure determines the stability of the protein. The RMSD of protein in the presence of C1 was stable around 1.25 Å throughout the simulation time of 100 ns. The RMSD of the protein–C2 complex fluctuated up to 60 ns, and then, it was quite stable around 1.2 Å. In the case of C3, the RMSD of the holoprotein was found to be around 1.0 Å up to 18 ns, after which it leaped to 4.2 Å. However, after 20 ns, it remained constant around 3.0 Å until the end of the simulation process. Although some deviations in RMSD were observed in this study, those were not very significant ones (on the order of 1–3 Å).
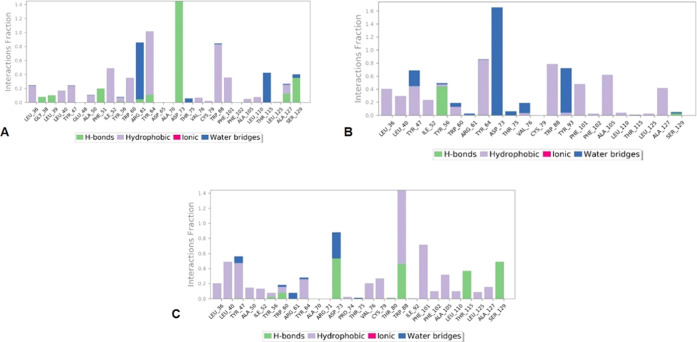
From Figure 5, it is evident that the hydrophobic and hydrogen bond interactions played significant roles in the binding of these compounds into the active site of the LasR protein. C1 showed hydrogen bond interaction with the residues Gly38, Leu39, Phe51, Arg61, Tyr64, Asp73, Ala127, and Ser129. In particular, Asp73 maintained a stable interaction throughout the simulation. On the other hand, hydrophobic interactions were seen with residues Leu36, Leu40, Tyr47, Ala50, Ile52, Try56, Trp60, Tyr64, Val76, Cys79, Trp88, Phe101, Ala105, Leu110, and Ala127. C2 formed hydrogen bonds with only two residues Tyr56 and Ser129. Among them, Tyr56 was engaged in hydrogen bond interaction with the active site over 45% of the simulation process and residues Leu36, Leu40, Tyr47, Ile52, Tyr56, Trp60, Tyr64, Val76, Trp88, Tyr93, Phe101, Phe102, Ala105, Leu110, Leu125, and Ala127 were engaged in hydrophobic interactions.
Figure 5.
Protein–ligand interaction histogram plots for LasR and (A) C1, (B) C2, and (C) C3.
The compound C3 exhibited hydrogen bond interaction with many residues, out of which Asp73, Trp88, and Ser129 showed interactions over 50% of the simulation period. C3 made hydrophobic interactions with the amino acids Leu36, Leu40, Tyr47, Ala50, Ile52, Tyr56, Trp60, Tyr64, Pro74, Val76, Cys79, Phe101, Phe102, Ala105, Leu110, Leu125, and Ala127. In the event of water bridge formation, C1 formed water bridges with residues Trp60, Thr75, and Thr115. Significant water bridge formation was exhibited by C2 with Asp73 and Tyr93 residues, followed by C3, showing water bridges involving residues Tyr47, Trp60, Arg61, Tyr64, Asp73, and Thr75 during the course of the simulation.
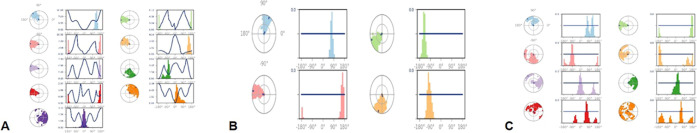
The ligand torsion profiles of C1, C2, and C3 during the course of the simulation period of 100 ns are depicted in Figure 6.
Figure 6.
Ligand torsion profiles of (A) C1, (B) C2, and (C) C3 during the simulation process.
The plot explains the conformational evolution of the rotatable bonds in the ligand until the end of the simulation period. Each of the rotatable bond torsions is described by a dial plot and a bar plot. The dial plot shows the conformation of torsion seen through the period of simulation, and the bar plot explains the probability density of each torsional rotation. There are nine, four, and eight rotatable bonds present in C1, C2, and C3, respectively. These relationships are essential toward gaining knowledge about the conformational strain all of the ligands undergo to maintain protein-bound conformation.
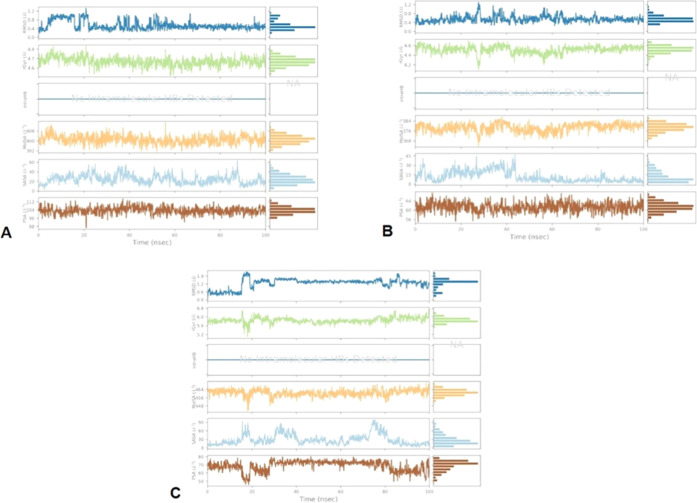
The stability of the compounds in the active site of the LasR receptor during the simulation study of 100 ns was observed by examining the properties depicted in Figure 7.
Figure 7.
Variations in the ligand properties observed during the course of 100 ns simulation: (A) C1, (B) C2, and (C) C3.
Here (Figure 7), RMSD, radius of gyration, which measures the extendedness of the compounds, molecular surface area calculation with 1.4 Å probe radius, number of intramolecular hydrogen bonds detected, surface area of a molecule accessible by a water molecule, and solvent surface area in a molecule contributed only by O and N atoms are identified. From the plot depicted in Figure 6, C1 was found to be slightly fluctuating in terms of all parameters for up to 25 ns; however, it was consistent after 25 ns. In the context of C2, there was an initial fluctuation in terms of radius of gyration and molecular surface area until 25 ns but eventually became constant. Similarly, C3 revealed fluctuations up to 20 ns with respect to the radius of gyration and the molecular surface area, but after this, it was stable until the end of the simulation trajectory. No intramolecular hydrogen bonds were detected for any of the compounds. Parameters like solvent-accessible surface area (SASA) and polar surface area (PSA) also showed an initial fluctuation in the graph, but later on, they remained consistent, which proves the stability of these compounds in the receptor active site over the 100 ns simulation process.
2.6. In Vitro LasR Reporter Gene Assay
Suneby et al.45 reported that the function of antagonists is to bind LasR and stabilize its conformation in such a way that it prevents the DNA binding. When LasR couples to its native ligand and binds to the DNA, it leads to its interaction with RNA polymerase and hence regulates gene expression.46 Thus, numerous antagonists are specifically designed to cause disturbance in this LasR–ligand interaction.47,48 The efficacy of antagonists can be measured by cell-based reporter gene assays.49
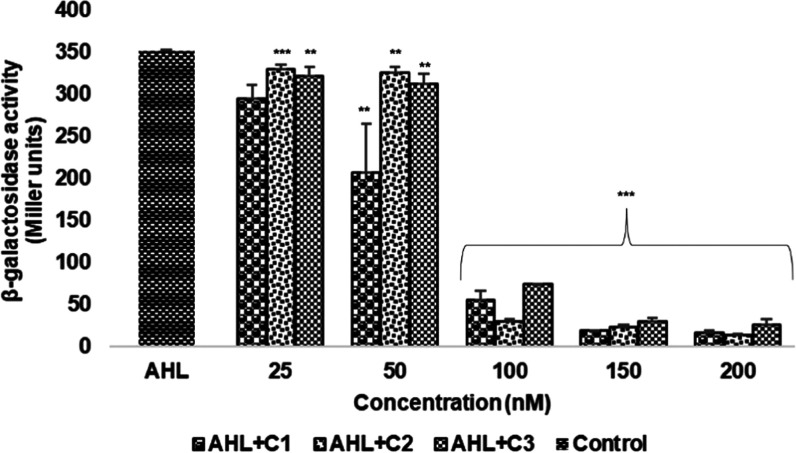
The antagonistic effects of the selected compounds (C1–C3) were evaluated in the presence of AHL, a native autoinducer molecule of LasR, which activates the QS-related transcriptional changes. A total of 50 nM AHL was taken as the positive control for the study. The activity of the β-galactosidase enzyme was calculated in Miller units and is presented in Figure 8.
Figure 8.
Inhibitory effect of compounds on β-galactosidase enzyme production assayed using the in vitro LasR reporter gene assay. The assay was performed in triplicates, and values are given as the mean ± standard deviation (SD). *p < 0.05 vs control, **p < 0.01 vs control, ***p < 0.001 vs control.
Our findings have shown that (Figure 8) in the presence of AHL, the compounds decreased the production of the β-galactosidase enzyme with an increase in concentration. A significant reduction in the β-galactosidase enzyme activity was observed at a concentration of 100 nM. The β-galactosidase activities (expressed in Miller units) at 100 nM concentration of C1, C2, and C3 were found to be 54.46 ± 11.07, 29.18 ± 2.25, and 74.21 ± 0.23, respectively. The basal level activity of the β-galactosidase enzyme was found to be 7.8 ± 0.21 Miller units. Thus, the results suggest that these compounds might inhibit LasR by preventing it from binding to its target lasB promoter, which is necessary for LasR expression.
In concordance with the findings of Kafle et al.50 our compounds have also shown a decrease in the enzyme activity in the presence of AHL. LasR antagonists get involved in interaction with the native ligand directly to produce insoluble or unstable LasR by disrupting protein folding.18 This provides evidence that these compounds might affect the DNA binding affinity of LasR by interacting with its native ligand.
3. Discussion
LasR, a transcriptional regulator, plays a promising role in the pathogenesis of P. aeruginosa by mediating the expression of QS genes. The pathogenicity of P. aeruginosa can be controlled by inhibiting the LasR protein, which holds the topmost position in the hierarchical QS network.51 The native LasR signaling molecule N-(3-oxo-dodecanoyl)-l-homoserine lactone (OdDHL) when present at a quorate concentration binds to LasR and activates the QS pathway. The polar head group of the autoinducer OdDHL forms hydrogen bonds with the residues Tyr56, Trp60, Asp73, and Ser129 in the ligand binding pocket of LasR (Figure 3A). The nonpolar tail group of OdDHL binds with hydrophobic residues including Leu36, Leu40, Ile52, Val76, and Leu125 present at the other side of the ligand binding pocket of LasR.
These interactions make the autoinducer shield the ligand binding pocket from bulk solvents and sequester the hydrophobic residues near the active site pocket into a favorable hydrophobic environment. This leads to the stabilization of the LasR in its active homodimer state.17 The LasR homodimer–ligand complex binds to certain promoters and activates the QS gene expression. Hence, LasR has gained significant interest as an antivirulence target. Different research groups have identified LasR inhibitors from natural sources, combinatorial chemistry, and computer-based drug designing. This provides an opportunity to design inhibitors for LasR that can decrease its resistance, pathogenicity, and virulence without directly retarding the bacterial growth.
The main objective of this study was to find a potent LasR inhibitor through the high-throughput virtual screening approach. Twelve compounds were shortlisted from around 3 034 496 compounds from the Schrödinger small molecule library using the HTVS mode of screening. The top three compounds (C1–C3) were taken for further analysis. The docking score of C1 was found to be even greater than the score of the cocrystallized ligand TP-1 bound to LasR. The compounds were then checked for their binding affinity with the ligand, and the stability of their binding interactions was analyzed by molecular dynamics simulation studies. Simulation was carried out for 100 ns, which revealed that the compounds formed a stable complex with LasR mediated by favorable interactions with key amino acid residues.
Analysis of the active site residues in the native ligand and TP-1 showed the involvement of crucial hydrogen bond interactions with amino acids, namely, Tyr56, Trp60, Asp73, and Ser129 (Figure 3A,B). In the molecular docking study, C1 and C2 formed hydrogen bond interactions (Figure 2A,B), whereas C3 did not show any hydrogen bond interactions (Figure 2C). However, molecular simulation revealed that only C1 forms stable hydrogen bond interaction with Asp73. In silico analysis suggested that the three compounds did not form stable hydrogen bond interactions with Trp60. These results were consistent with the previous findings, in which Tan et al.31 identified quorum sensing inhibitors (QSIs), namely, 2-amino-3-(3-fluorophenyl) propanoic acid, 2-amino-3-hydroxy-3-phenyl propanoic acid, and indole-3-carboxylic acid, and reported that they did not show hydrogen bond interaction with Trp60. A study by Kim et al.52 also observed that amino acids, namely, Trp60 and Arg61 as the key residues, were involved in hydrogen bond interactions with the X-ray crystal ligand OdDHL.
Another study by O’Reilly et al.39 has suggested that the compounds that did not form hydrogen bond interaction with Trp60 served as potent antagonists. In addition, a study by Müh et al.32 stated that although both the inhibitors and the activators of LasR bind to the same active site, only those compounds that make significant hydrogen bond contacts with both Trp60 and Asp73 can serve as activators of LasR. A study by Bottomley et al.18 reported a model of the LasR–patulin complex in which patulin that mimics lactone of AHLs and can serve as the activator of LasR forms a canonical hydrogen bond with Trp60. In concordance with the above literature reports, our docking and molecular dynamics simulation results also report that all three compounds selected for analyses were not capable of forming any significant hydrogen bond interaction with Trp60 and, hence, these might act as inhibitors of LasR. Our in vitro LasR reporter gene assay further confirms that the selected compounds could serve as potential lead compounds against LasR. The predicted ADME profile revealed that the selected compounds could serve as effective lead compounds toward drug development. Further studies with these compounds warrant a novel and promising antibiofilm agent against P. aeruginosa.
4. Materials and Methods
All of the computational analyses were performed using Maestro v11.8 (Schrödinger, LLC, New York, NY)53 installed on the CentOS 6.10 Linux platform.
4.1. Protein Preparation
The crystal structure of the LasR protein (PDB ID: 3IX4) was retrieved from Protein Data Bank (PDB) (https://www.rcsb.org/).17 The PDB structure of the protein was subjected to preparation by the Protein Preparation Wizard module of Schrödinger software.54 During preparation, the missing hydrogen atoms were incorporated, actual bond orders were assigned, and proper ionization states were generated using the OPLS-2005 force field. Finally, restrained minimization was done until the heavy atoms converged to an RMSD of 0.30 Å for performing the optimal docking study.
4.2. Receptor Grid Generation
The Glide v8.1 module of Schrödinger55 comprises a receptor grid generation panel for generating a grid box around the cocrystallized ligand in the receptor protein that allows docking into the active site. The cubic boxes with coordinates of 3.336, 15.597, and 4.868 in x, y, and z directions, respectively, centered on the centroid of the cocrystallized ligand were created. The van der Waals radius scaling factor was set to 1.0 Å, and the partial charge cutoff was maintained at 0.25 with no constraints.
4.3. High-Throughput Virtual Screening
The structure-based HTVS can identify potent lead molecules in the case of a drug or inhibitor design.56 It screens for a large ligand database containing millions of compounds to identify molecules that interact and fit perfectly into the target protein’s active site.57 HTVS was carried out on the Schrödinger small molecule database in three stages, namely, structure-based virtual screening, standard precision (SP) docking, and extra precision (XP) docking using the Schrödinger module.
The Glide v8.1 module of Schrödinger55 contains a workflow for virtual screening and all docking calculations. It employs a grid-based ligand docking method that generates a series of ligand poses. The ligand interactions with the receptor are then evaluated by passing through a number of hierarchical filters. The spatial fit of the ligand into the active site is tested by the initial filters, and they also examine the complementarity of the ligand–receptor interaction based on the empirical ChemScore function. Only those ligand poses that pass the screen enter the final stage of the algorithm.
The SP docking of the Glide module was initially employed to identify probable binders from a large set of compounds. Thereafter, the high scoring 10–30% of compounds was more intensively investigated using Glide XP docking. XP docking opts out false positives that SP lets in, and ligands that do not fit well to the receptor conformation were penalized. The final scoring was done on energy minimized poses, and the compounds with the best docked pose and the highest negative score values were taken for further study. The ligand interaction pattern of the TP-1 docked complex was analyzed using Ligplot+ software.58
4.4. Prime MM-GBSA Binding Free Energy Calculation
To validate the binding free energy of the docked complexes, the postdocking binding affinity calculation was performed using the Prime Molecular Mechanics/Generalized Born and Surface Area (MM-GBSA) method in Maestro v11.8 of the Schrödinger module.59 The binding affinities of the complexes were obtained using the OPLS_2005 force field, a VSGB 2.0 polar solvation model, a nonpolar solvation term that consists of solvent accessible surface area (SASA), and van der Waals interactions. Finally, the structures with good binding free energies were taken for simulation studies. Based on the docked complex, the binding affinity was calculated based on the following equation
4.5. ADMET and Drug-Likeness Properties of Screened Compounds
The ADMET and drug-likeness properties of the top-scoring QSIs were identified with the QikProp v5.8 module of Schrödinger.60 The most significant pharmacokinetic properties were analyzed to evaluate the druggable effects and acceptability of the screened compounds.
4.6. Molecular Dynamics Simulation
The stability of the compounds docked into the active site of LasR was studied using the Desmond v5.6 package61 for MD simulations by applying the OPLS-AA force field with a single point charge (SPC) water model in an explicit solvent. It also provides information about the movements of the atoms in the complexes in a predefined environment. The SPC water molecules were added and complex systems were pre-equilibrated and minimized using the default relaxation routine found in Desmond. The distance between the complex and the box wall was kept at 10 Å. The equilibrated systems were used for simulations with a time step of 100 ns for each simulation at a temperature of 300 K and a constant pressure of 1 atm with 4.8 ps intervals for achieving a stabilized system. The model system was relaxed before simulation. The stability of the complexes was expressed in terms of the root-mean-square deviation (RMSD) and root-mean-square fluctuation (RMSF) for the backbone and side chain of the protein. The compactness of the protein was determined by calculating the radius of gyration throughout the simulation. Hydrogen bonds play a major role in precise and stable binding; hence, the number of hydrogen bonds in the complex was also observed.
4.7. In Vitro LasR Reporter Gene Assay
To evaluate whether the compounds can stimulate or antagonize LasR-dependent transcription, Escherichia coli DH5α cells transformed with the plasmid pKDT17 (gift from Peter Greenberg (Addgene plasmid #27503; http://n2t.net/addgene:27503; RRID:Addgene_27503)) were used in the reporter assay. This plasmid serves as a reporter by linking the LasR gene to a lasB::lacZ translational fusion.62 The β-galactosidase reporter gene assay was performed as per the procedure reported by Kim et al.63 The LasR activity was identified as a function of the β-galactosidase enzyme activity and is expressed in Miller units.64
Acknowledgments
The authors wholeheartedly thank Mr. Raghu Rangaswamy and Ms. Shelvia Malik, Schrödinger team, for support with the Schrödinger software licensing and Mr. Vinod Devaraji, Senior Scientist, Schrödinger team, for helping with molecular dynamics simulation studies.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c02191.
Docking results of reported antagonists (PDF)
(PDF)
The authors declare no competing financial interest.
Notes
All in silico analyses were carried out using commercial software, Schrödinger, obtained from https://www.schrodinger.com/downloads/releases. The authors have used Schrödinger Release 2018-4: Maestro v11.8, Schrodinger, LLC, New York, NY, 2018, for performing all computational analyses. The software can be accessed only by obtaining a request-based license from the Schrödinger team. The requisition for trial license details is given in the link https://www.schrodinger.com/license-information.
Supplementary Material
References
- Mulcahy L. R.; Isabella V. M.; Lewis K. Pseudomonas aeruginosa biofilms in disease. Microb. Ecol. 2014, 68, 1–12. 10.1007/s00248-013-0297-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch E. B.; Tam V. H. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev. Pharmacoecon Outcomes Res. 2010, 10, 441–451. 10.1586/erp.10.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamal M.; Ahmad W.; Andleeb S.; Jalil F.; Imran M.; Nawaz M. A.; Hussain T.; Ali M.; Rafiq M.; Kamil M. A. Bacterial biofilm and associated infections. J. Chin. Med. Assoc. 2018, 81, 7–11. 10.1016/j.jcma.2017.07.012. [DOI] [PubMed] [Google Scholar]
- Streeter K.; Katouli M. Pseudomonas aeruginosa: A review of their pathogenesis and prevalence in clinical settings and the environment. Infect., Epidemiol. Microbiol. 2016, 2, 25–32. 10.18869/modares.iem.2.1.25. [DOI] [Google Scholar]
- Lila G.; Mulliqi G.; Raka L.; Kurti A.; Bajrami R.; Azizi E. Molecular epidemiology of Pseudomonas aeruginosa in University clinical center of Kosovo. Infect. Drug Resist. 2018, 11, 2039–2046. 10.2147/IDR.S174940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazeli H.; Akbari R.; Moghim S.; Narimani T.; Arabestani M. R.; Ghoddousi A. R. Pseudomonas aeruginosa infections in patients, hospital means and personnel’s specimens. J. Res. Med. Sci. 2012, 17, 332–337. [PMC free article] [PubMed] [Google Scholar]
- Andhale J. D.; Misra R. N.; Gandham N. R.; Angadi K. M.; Jadhav S. V.; Vyawahare C. R.; Pawar M.; Hatolkar S. Incidence of Pseudomonas aeruginosa with special reference to drug resistance and biofilm formation from clinical samples in tertiary care hospitals. J. Pharm. Biomed. Sci. 2016, 6, 387–391. [Google Scholar]
- Li Z.; Nair S. K. Quorum sensing: how bacteria can coordinate activity and synchronize their response to external signals?. Protein Sci. 2012, 21, 1403–1417. 10.1002/pro.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K.; Yoon S. S. Pseudomonas aeruginosa biofilm, a programmed bacterial life for fitness. J. Microbiol. Biotechnol. 2017, 27, 1053–1064. 10.4014/jmb.1611.11056. [DOI] [PubMed] [Google Scholar]
- Schuster M.; Greenberg E. P. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int. J. Med. Microbiol. 2006, 296, 73–81. 10.1016/j.ijmm.2006.01.036. [DOI] [PubMed] [Google Scholar]
- Welsh M. A.; Blackwell H. E. Chemical genetics reveals environment-specific roles for quorum sensing circuits in Pseudomonas aeruginosa. Cell Chem. Biol. 2016, 23, 361–369. 10.1016/j.chembiol.2016.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattmann M. E.; Shipway P. M.; Heth N. J.; Blackwell H. E. Potent and selective synthetic modulators of a quorum sensing repressor in Pseudomonas aeruginosa identified from second-generation libraries of N-acylated L-homoserine lactones. ChemBioChem 2011, 12, 942–949. 10.1002/cbic.201000708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattmann M. E.; Blackwell H. E. Small molecules that modulate quorum sensing and control virulence in Pseudomonas aeruginosa. J. Org. Chem. 2010, 75, 6737–6746. 10.1021/jo101237e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asfahl K. L.; Schuster M. Additive effects of quorum sensing anti-activators on Pseudomonas aeruginosa virulence traits and transcriptome. Front. Microbiol. 2018, 8, 2654 10.3389/fmicb.2017.02654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh M. A.; Blackwell H. E. Chemical probes of quorum sensing: from compound development to biological discovery. FEMS Microbiol. Rev. 2016, 40, 774–794. 10.1093/femsre/fuw009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekimpe V.; Déziel E. Revisiting the quorum-sensing hierarchy in Pseudomonas aeruginosa: the transcriptional regulator RhlR regulates LasR-specific factors. Microbiology 2009, 155, 712–723. 10.1099/mic.0.022764-0. [DOI] [PubMed] [Google Scholar]
- Zou Y.; Nair S. K. Molecular basis for the recognition of structurally distinct autoinducer mimics by the Pseudomonas aeruginosa LasR quorum sensing signaling receptor. Chem. Biol. 2009, 16, 961–970. 10.1016/j.chembiol.2009.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottomley M. J.; Muraglia E.; Bazzo R.; Carfi A. Molecular insights into quorum sensing in the human pathogen Pseudomonas aeruginosa from the structure of the virulence regulator LasR bound to its autoinducer. J. Biol. Chem. 2007, 282, 13592–13600. 10.1074/jbc.M700556200. [DOI] [PubMed] [Google Scholar]
- Papenfort K.; Bassler B. Quorum sensing signal-response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill M. E. A.; Chen L. Structural basis of acyl-homoserine lactone-dependent signaling. Chem. Rev. 2011, 111, 68–85. 10.1021/cr1000817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway W. R. J. D.; Hodgkinson J. T.; Bowden S.; Welch M.; Spring D. R. Applications of small molecule activators and inhibitors of quorum sensing in Gram-negative bacteria. Trends Microbiol. 2012, 20, 449–458. 10.1016/j.tim.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Rasamiravaka T.; Labtani Q.; Duez P.; El Jaziri M. The formation of biofilms by Pseudomonas aeruginosa: a review of the natural and synthetic compounds interfering with control mechanisms. BioMed Res. Int. 2015, 759348 10.1155/2015/759348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Contreras R.; Maeda T.; Wood T. K. Resistance to quorum-quenching compounds. Appl. Environ. Microbiol. 2013, 79, 6840–6846. 10.1128/AEM.02378-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Defoirdt T.; Boon N.; Bossier P. Can bacteria evolve resistance to quorum sensing disruption?. PLoS Pathog. 2010, 6, e1000989 10.1371/journal.ppat.1000989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapetanovic I. M. Computer-aided drug discovery and development (CADDD): in silico-chemico-biological approach. Chem.-Biol. Interact. 2008, 171, 165–176. 10.1016/j.cbi.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokhande K. B.; Nagar S.; Swamy K. V. Molecular interaction studies of deguelin and its derivatives with cyclin D1 and cyclin E in cancer cell signalling pathway: The computational approach. Sci. Rep. 2019, 9, 1778 10.1038/s41598-018-38332-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb P.; Ferreira R. S.; Irwin J. J.; Shoichet B. K. Docking and chemoinformatic screens for new ligands and targets. Curr. Opin. Biotechnol. 2009, 20, 429–436. 10.1016/j.copbio.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadiq S.; Rana N. F.; Zahid M. A.; Zargaham M. K.; Tanweer T.; Batool A.; Naeem A.; Nawaz A.; Rizwan-ur-Rehman; Muneer Z.; Siddiqi A. R. Virtual screening of FDA-approved drugs against LasR of Pseudomonas aeruginosa for antibiofilm potential. Molecules 2020, 25, 3723 10.3390/molecules25163723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nain Z.; Sayed S. B.; Karim M. M.; Islam M. A.; Adhikari U. K. Energy-optimized pharmacophore coupled virtual screening in the discovery of quorum sensing inhibitors of LasR protein of Pseudomonas aeruginosa. J. Biomol. Struct. Dyn. 2020, 38, 5374–5388. 10.1080/07391102.2019.1700168. [DOI] [PubMed] [Google Scholar]
- Kalia M.; Singh P. K.; Yadav V. K.; Yadav B. S.; Sharma D.; Narvi S. S.; Mani A.; Agarwal V. Structure based virtual screening for identification of potential quorum sensing inhibitors against LasR master regulator in Pseudomonas aeruginosa. Microb. Pathog. 2017, 107, 136–143. 10.1016/j.micpath.2017.03.026. [DOI] [PubMed] [Google Scholar]
- Tan S. Y.; Chua S.; Chen Y.; Rice S. A.; Kjelleberg S.; Nielsen T. E.; Yang L.; Givskov M. Identification of five structurally unrelated quorum-sensing inhibitors of Pseudomonas aeruginosa from a natural-derivative database. Antimicrob. Agents Chemother. 2013, 57, 5629–5641. 10.1128/AAC.00955-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müh U.; Hare B. J.; Duerkop B. A.; Schuster M.; Hanzelka B. L.; Heim R.; Olson E. R.; Greenberg E. P. A structurally unrelated mimic of a Pseudomonas aeruginosa acyl-homoserine lactone quorum-sensing signal. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 16948–16952. 10.1073/pnas.0608348103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu S.; Azofra L. M.; MacFarlane D. R.; Sun C. Hydrogen bonding effect between active site and protein environment on catalysis performance in H2-producing [NiFe] hydrogenases. Phys. Chem. Chem. Phys. 2018, 20, 6735–6743. 10.1039/C7CP07685A. [DOI] [PubMed] [Google Scholar]
- Hubbard R. E.; Haider M. K.. Hydrogen bonds in proteins: Role and strength. In Encyclopedia of Life Sciences.; John Wiley & Sons, Ltd: Chichester, 2010; pp 1–7. [Google Scholar]
- Sigala P. A.; Tsuchida M. A.; Herschlag D. Hydrogen bond dynamics in the active site of photoactive yellow protein. Proc. Natl. Acad. Sci. U.S.A. 2009, 106, 9232–9237. 10.1073/pnas.0900168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geske G. D.; O’Neill J. C.; Miller D. M.; Wezeman R. J.; Mattmann M. E.; Lin Q.; Blackwell H. E. Comparative analyses of N-acylated homoserine lactones reveal unique structural features that dictate their ability to activate or inhibit quorum sensing. Chembiochem 2008, 9, 389–400. 10.1002/cbic.200700551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien K. T.; Noto J. G.; Nicholos-O’Neill L.; Perez L. J. Potent irreversible inhibitors of LasR quorum sensing in Pseudomonas aeruginosa. ACS Med. Chem. Lett. 2014, 6, 162–167. 10.1021/ml500459f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi H.; Ham S. Y.; Cha E.; Shin Y.; Kim H. S.; Bang J. K.; Son S. H.; Park H. D.; Byun Y. Structure-activity relationships of 6- and 8- gingerol analogs as anti-biofilm agents. J. Med. Chem. 2017, 60, 9821–9837. 10.1021/acs.jmedchem.7b01426. [DOI] [PubMed] [Google Scholar]
- O’Reilly M. C.; Dong S. H.; Rossi F. M.; Karlen K. M.; Kumar R. S.; Nair S. K.; Blackwell H. E. Structural and biochemical studies of non-native agonists of the LasR quorum-sensing receptor reveal an L3 loop “out” conformation for LasR. Cell Chem. Biol. 2018, 25, 1128–1139. 10.1016/j.chembiol.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K. M.; Bu Y.; Suga H. Induction and inhibition of Pseudomonas aeruginosa quorum sensing by synthetic autoinducer analogs. Chem. Biol. 2003, 10, 81–89. 10.1016/S1074-5521(03)00002-4. [DOI] [PubMed] [Google Scholar]
- Ni N.; Chou H. T.; Wang J.; Li M.; Lu C. D.; Tai P. H.; Wang B. Identification of boric acids as antagonists of bacterial quorum sensing in Vibrio harveyi. Biochem. Biophys. Res. Commun. 2008, 369, 590–594. 10.1016/j.bbrc.2008.02.061. [DOI] [PubMed] [Google Scholar]
- Lipinski C. A.; Lombardo F.; Dominy B. W.; Feeney P. J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Delivery Rev. 1997, 23, 3–25. 10.1016/S0169-409X(96)00423-1. [DOI] [PubMed] [Google Scholar]
- Pond S. M.; Tozer T. N. First-pass elimination basic concepts and clinical consequences. Clin. Pharmacokinet. 1984, 9, 1–25. 10.2165/00003088-198409010-00001. [DOI] [PubMed] [Google Scholar]
- Shahbazi S.; Kuanar A.; Gade D. R.; Kar D.; Shrivastava A.; Kunala P.; Mahto M. K. Semiemperical investigation of the postmenopausal breast cancer treatment potential of xanthone derivatives. Nat. Prod. Chem. Res. 2016, 4, 206 10.4172/2329-6836.1000206. [DOI] [Google Scholar]
- Suneby E. G.; Herndon L. R.; Schneider T. L. Pseudomonas aeruginosa LasR-DNA Binding is Directly Inhibited by Quorum Sensing Antagonists. ACS Infect. Dis. 2017, 3, 183–189. 10.1021/acsinfecdis.6b00163. [DOI] [PubMed] [Google Scholar]
- Gilbert K. B.; Kim T. H.; Gupta R.; Greenberg E. P.; Schuster M. Global position analysis of the Pseudomonas aeruginosa quorum-sensing transcription factor LasR. Mol. Microbiol. 2009, 73, 1072–1085. 10.1111/j.1365-2958.2009.06832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amara N.; Gregor R.; Rayo J.; Dandela R.; Daniel E.; Liubin N.; Willems H. M. E.; Ben-Zvi A.; Krom B. P.; Meijler M. M. Fine-tuning covalent inhibition of bacterial quorum sensing. ChemBioChem. 2016, 17, 825–835. 10.1002/cbic.201500676. [DOI] [PubMed] [Google Scholar]
- O’Brien K. T.; Noto J. G.; Nichols-O’Neill L.; Perez L. J. Potent irreversible inhibitors of LasR quorum sensing in Pseudomonas aeruginosa. ACS Med. Chem. Lett. 2015, 6, 162–167. 10.1021/ml500459f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J. D.; Rossi F. M.; Welsh M. A.; Nyffeler K. E.; Blackwell H. E. A comparative analysis of synthetic quorum sensing modulators in Pseudomonas aeruginosa: new insights into mechanism, active efflux susceptibility, phenotypic response, and next-generation ligand design. J. Am. Chem. Soc. 2015, 137, 14626–14639. 10.1021/jacs.5b06728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafle P.; Amoh A. N.; Reaves J. M.; Suneby E. G.; Tutunjian K. A.; Tyson R. L.; Schneider T. L. Molecular insights into the impact of oxidative stress on the quorum-sensing regulator protein LasR. J. Biol. Chem. 2016, 291, 11776–11786. 10.1074/jbc.M116.719351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.; Zhang L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 2015, 6, 26–41. 10.1007/s13238-014-0100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H.; Lee S.; Byun Y.; Park H. 6-Gingerol reduces Pseudomonas aeruginosa biofilm formation and virulence via quorum sensing inhibition. Sci. Rep. 2015, 5, 8656 10.1038/srep08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrödinger Release 2018-4. Maestro; Schrödinger, LLC: New York, NY, 2018.
- Schrödinger Release 2018-4. Protein Preparation Wizard; Epik; Schrödinger, LLC: New York, NY, 2018.
- Schrödinger Release 2018-4. Glide; Schrödinger, LLC: New York, NY, 2018.
- Dhasmana A.; Raza S.; Jahan R.; Lohani M.; Arif J. M.. High-throughput virtual screening (HTVS) of natural compounds and exploration of their biomolecular mechanisms. In New Look to Phytomedicine; Academic Press: Cambridge, MA, 2019; pp 523–548. [Google Scholar]
- Subramaniam S.; Mehrotra M.; Gupta D. Virtual high throughput screening (vHTS)-a perspective. Bioinformation. 2008, 3, 14–17. 10.6026/97320630003014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R. A.; Swindells M. B. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- Schrödinger Release 2018-4. Prime; Schrödinger, LLC: New York, NY, 2018.
- Schrödinger Release 2018-4. QikProp; Schrödinger, LLC: New York, NY, 2018.
- Schrödinger Release 2018-4. Desmond Molecular Dynamics System, D. E. Shaw Research, New York, NY, 2018. Maestro-Desmond Interoperability Tools; Schrödinger: New York, NY, 2018.
- Pearson J. P.; Gray K. M.; Passador L.; Tucker K. D.; Eberhard A.; Iglewski B. H.; Greenberg E. P. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. U.S.A. 1994, 91, 197–201. 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim E.; Wang W.; Deckwer W.; Zeng A. Expression of the quorum-sensing regulatory protein LasR is strongly affected by iron and oxygen concentrations in cultures of Pseudomonas aeruginosa irrespective of cell density. Microbiology 2005, 151, 1127–1138. 10.1099/mic.0.27566-0. [DOI] [PubMed] [Google Scholar]
- Miller J. H.Experiments in Molecular Genetics; Cold Spring Harbor Laboratory: NY, 1972. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.