Abstract
Background
Triple-negative breast cancers (TNBC), comprising about 20% of breast cancers, have a poor prognosis. Currently, there is no effective target therapy for TNBC. LncRNA TUSC7 has been identified as a tumor suppressor in osteosarcoma and colorectal cancer. In this study, we investigated the clinical significance and the biological function of TUSC7 in breast cancer.
Methods
We retrospectively evaluated the expression level and clinical significance of TUSC7 in 90 paired breast cancer tissues and normal tissues. The proliferation, migration, and invasion assays were performed to investigate the biological function of TUSC7 in breast cancer. Finally, microarray, a luciferase reporter assay, and quantitative real-time polymerase chain reaction (qPCR) were used to explore the potential underlying mechanism of tumor suppressor role of TUSC7.
Results
Low TUSC7 expression was found to be an independent prognostic factor of poor overall survival (OS) in TNBC patients. Ectopic expression of TUSC7 inhibited tumor cell growth both in vitro and in vivo. TUSC7 overexpression significantly promoted the sensitivity of MDA-MB-468 cells to paclitaxel and carboplatin. In terms of the mechanism, TUSC7 might perform its biological function through binding with miR-1224-3P and regulating its expression level. Besides, genes in cell cycle pathways, such as BUB3 (budding uninhibited by benzimidazoles 3) and TGF-ß (targeting transforming growth factor β) pathways were downregulated, and genes involved in the MAPK (mitogen-activated protein kinase) (TGFBR2, transforming growth factor-beta receptor 2), PI3K-AKT (phosphoinositide 3-kinase- AKT serine/threonine kinase 1) and NF-κB (nuclear factor-kappa B subunit) pathways were upregulated in TUSC7 knockdown MDA-MB-231 cells.
Conclusion
The low TUSC7 expression is an independent prognostic factor of poor OS of TNBC patients. TUSC7 might inhibit breast cancer cell growth and metastasis both in vitro and vivo through binding with miR-1224-3P and regulating MAPK, PI3K/AKT, and NF-κB signaling pathways.
Keywords: TUSC7, long non-coding RNA, MiR-1224-3P, triple-negative breast cancer, prognosis
Introduction
Dysregulation of long non-coding RNAs (lncRNAs) of lncRNAs was involved in various human diseases, including cancer.1–5 A previous report demonstrated the potential role of lncRNA in the progression of breast cancer.6
Triple-negative breast cancer (TNBC) makes up about 20% of whole breast cancers.7,8 Compared with other types of breast cancer, TNBC is associated with early recurrence and significantly shorter survival.9–11 The genes of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2) were not expressed in TNBC. While trastuzumab targets HER2 and endocrine therapy targets the ER and PR, no therapeutic target has been identified for TBNC and thus the prognosis of TNBC patients is worse than that of patients with other types of breast cancer.12 Thus, the identification of targets specific to TNBC is critical for the development of future therapies and improving patient prognosis.
The TUSC7 gene, located in chr3q13.31, is 2105nt in length and consists of four exons.13 The overexpression of TUSC7 inhibits the proliferation of normal osteoblasts via the regulation of cell cycle, apoptotic, and the transcripts of VEGF/VEGFR1.14 Besides, TUSC7 was related to poor survival of osteosarcoma,15 colorectal cancer,16 and pancreatic ductal adenocarcinoma patients.17 These data indicated the potential tumor suppressor role of TUSC7 in cancer. Our previous microarray results revealed significantly decreased TUSC7 expression at cancer sites in TNBC than at adjacent cancer sites (data not shown). But the definitive relation between TUSC7 and TNBC remained unspecific.
In this study, the expression patterns in TNBC tissues and cell lines were evaluated. Besides, the clinical significance of TUSC7 in TNBC was investigated in 90 patients. Finally, we also investigated the biological role of TUSC7 in BC and the underlying mechanism of TUSC7 function as a tumor suppressor.
Patients and Methods
Clinical Samples
90 paired breast cancer samples (bulk samples) were obtained from the Department of General Surgery, Zhongshan Hospital, Fudan University in 2013. The specimens were frozen in liquid nitrogen in time and then kept at a temperature of −80°C. This study was permitted by the Institutional Review Board of Fudan University Zhongshan Hospital, and all participants involved were willing to be committed to this research. Written informed consent was obtained from all patients. All samples were collected according to the Declaration of Helsinki. No patient received chemotherapy or radiotherapy before surgery. All TNBC patients meet the following criteria: 1) no neoadjuvant chemotherapy; 2) all patients received modified radical mastectomy; 3) all TNBC patients received postoperative chemotherapy.
Cell Culture and Real-Time Polymerase Chain Reaction
Details of cell culture and real-time polymerase chain reaction were listed in the Supplementary Materials.
Vector Construction and Lentivirus Packaging
Details were listed in the Supplementary Materials.
CCK8 (Cell Counting Kit-8) Assays, Migration, Invasion, and Cell Cycle Analysis
Details were listed in the Supplementary Materials.
In vivo Tumorigenesis Experiments
All animal studies were approved by the Fudan University Animal Care and Use Committee and carried out according to the policy of the committee. 24 female nude mice, four weeks old (BALB/cA-nu (nu/nu)), were selected from the Shanghai Experimental Animal Center (Chinese Academy of Sciences, China). 24 mice were divided into four groups at random. TUSC7-scrambled shRNA control MDA-MB-231 cells (scrambled shRNA), TUSC7-shRNA MDA-MB-231 cells (TUSC7 shRNA), TUSC7-empty vector (scramble) or TUSC7-DOX-overexpressing (TUSC Over) MDA-MB-468 cells were suspended with the density of 107 cells/mL, and 100 μL were bilaterally injected subcutaneously into the flank of nude mice (n = 6, each group, 107 cells/100μL per flank). One week after the injection, tumor formation was observed. For each group, the tumorigenesis rate reached 100%. Measurements of bidimensional tumor took calipers for use every 4 days. The tumor volume was worked out through the formula (width2 × length)/2. After 32 days, mice were sacrificed and the tumors were removed and weighed.
Cytotoxic Assay
Cells were cultivated at the density of 2×103 cells/well in 96-well plates and treated with 10−2 to 105 umol/L of paclitaxel and carboplatin. After 72 h, viable cells were counted to determine the IC50 (half maximal inhibitory concentration). Each condition was tested in duplicate and repeated three times.
miRNA Microarray and mRNA Microarray
Details were listed in Supplementary Materials.
TargetScan
TargetScan (http://www.targetscan.org/vert_72/) was used to determine the putative miRNA binding site of TUSC7.
Luciferase Reporter Assays
Details were listed in Supplementary Materials.
Statistical Analyses
Continuous data were presented in the mean ± standard deviation (SD) form. The Student’s t-test (two-tailed) was performed to evaluate the statistical importance of the differences among the groups. The correlation between TUSC7 expression level and clinicopathological characteristics was determined by using the person’s χ2 test. The prognostic significance was evaluated via the Kaplan–Meier method. The Cox proportional hazards model was carried out for univariate and multivariate analysis. A two-tail P < 0.05 was regarded as statistically significant. All statistical analyses were carried out using SPSS 20.0 software (SPSS Inc., Chicago, Illinois, USA) for Windows.
Results
The Baseline Clinical Characteristics of the Patients
90 BC patients were enrolled in this study. The clinicopathological characteristics of patients were listed in Table 1. Among 90 enrolled patients, 21 patients were luminal A type, 34 patients were luminal B type, 18 patients were HER2 type and 17 patients were TNBC type.
Table 1.
Relationship Between TUSC7 Expression and Clinicopathological Features in Breast Cancer Patients (N=90)
| Characteristics | Total (Cases) | TUSC7 Expression [Cases (%)] | P valuea | |
|---|---|---|---|---|
| High | Low | |||
| Total | 90 | 45 | 45 | |
| Age (years) | 0.876 | |||
| <50 | 49 | 24 | 25 | |
| ≥50 | 41 | 21 | 20 | |
| Tumor size(cm) | ||||
| T1 | 30 | 16 | 14 | 0.537 |
| T2 | 56 | 28 | 28 | |
| T3 | 4 | 1 | 3 | |
| Node status | ||||
| N0 | 46 | 23 | 23 | 0.988 |
| N1 | 26 | 14 | 12 | |
| N2 | 9 | 4 | 5 | |
| N3 | 9 | 4 | 5 | |
| Metastasis | ||||
| No | 88 | 43 | 45 | 0.135 |
| Yes | 2 | 2 | 0 | |
| Histological grade | ||||
| I | 17 | 8 | 9 | 0.752 |
| II | 49 | 27 | 22 | |
| III | 24 | 10 | 14 | |
| ER status | ||||
| Negative | 35 | 22 | 13 | 0.064 |
| Positive | 55 | 23 | 32 | |
| PR status | ||||
| Negative | 49 | 34 | 15 | 0.000* |
| Positive | 41 | 11 | 30 | |
| HER-2 status | ||||
| Negative | 58 | 28 | 30 | 0.451 |
| Positive | 32 | 17 | 15 | |
| Ki67 | ||||
| <14% | 26 | 9 | 17 | 0.046* |
| >14% | 64 | 36 | 28 | |
| Molecular subtype | ||||
| Luminal A | 21 | 12 | 9 | 0.003* |
| Luminal B | 34 | 18 | 16 | |
| HER2 | 18 | 13 | 5 | |
| TNBC | 17 | 2 | 15 | |
Notes: aStatistical analyses were performed by the Pearson χ2 test. *P < 0.05 is considered significant.
The Expression Level of TUSC7 in Breast Cancer Tissues and Cell Lines
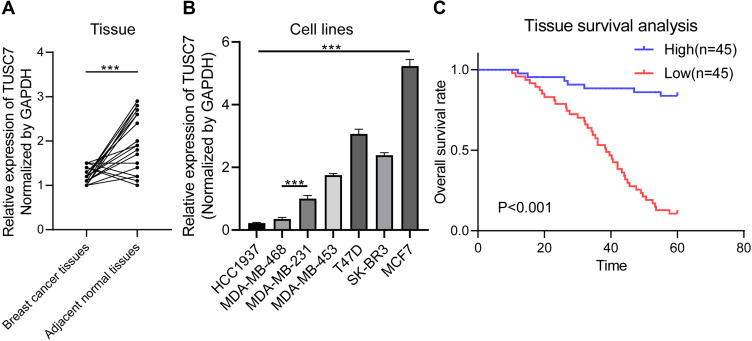
Compared with adjacent normal tissues, qPCR results indicated that the expression level of TUSC7 was significantly lower in breast cancer tissues (P = 0.002; Figure 1A). Besides, qPCR results indicated a significantly higher expression of TUSC7 in ER/PR positive breast cancer cells (T47D and MCF-7) compared with TNBC cell lines (MDA-MB-231, MDA-MB-468, and HCC1937) (P < 0.05, Figure 1B). Meanwhile, the expression level of TUSC7 was higher in HER2 positive breast cancer cells (SK-BR-3 and MDA-MB-453) when compared with TNBC cells, but lower when compared with ER/PR positive breast cancer cells (T47D and MCF-7).
Figure 1.
The expression pattern of TUSC7 in breast cancer. (A) RT-qPCR of TUSC7 expression in 43 breast cancer tissues compared with adjacent normal tissues. GADPH was used as normalization control. (B) RT-qPCR of TUSC7 expression in a panel of breast cancer cell lines: HCC1937, MDA-MB-468, MDA-MB-231, MDA-MB-453, T47D, SK-BR-3, MCF-7 cells. GADPH was used as normalization control. (C) The low expression level of TUSC7 is correlated with poor overall survival in TNBC patients. Kaplan–Meier curve for overall survival in patients with low (n=45) versus high (n=45) TUSC7 expression. Corresponding P values were analyzed by Log rank test. ***P<0.0001.
The Low TUSC7 Expression Level Was Associated with the Luminal B and TNBC Subtype
The median of TUSC7 expression level was identified as the cut-off value, then patients have divided into high and low groups accordingly. The low TUSC7 level (n = 45) was associated with higher Ki-67 scores (Table 1, n = 45; P = 0.022). The low TUSC7 expression was positively associated with TNBC (P = 0.019) and luminal B subtype.
The Low Expression of TUSC7 Was Associated with Poor Overall Survival in TNBC Patients
Further investigation was carried out to evaluate the prognostic role of this lncRNA. Kaplan-Meier analysis indicated that the low expression level of TUSC7 was associated with poorer postoperative overall survival (P<0.001, Figure 1C). Next, multivariate analysis indicated that TUSC7 expression level was an independent prognostic factor of TNBC patients (Table 2). All these data indicated that low TUSC7 expression was an independent predictor of poor overall survival for TNBC patients.
Table 2.
Results of the Multivariate Analyses of TUSC7 in 90 Patients with Breast Cancer
| Multivariate Analyses | ||||
|---|---|---|---|---|
| Variable | HR | 95% CI | P value | |
| Age (<50 vs ≥50 (Ref)) | 0.972 | 0.927 | 0.968 | 0.003 |
| Tumor size | 1.168 | 0.642 | 2.167 | 0.669 |
| Lymph node status (Metastasis vs No Metastasis (Ref)) | 4.743 | 2.736 | 8.336 | 0 |
| Metastasis (Present vs Absent (Ref)) | 4.858 | 2.858 | 8.359 | 0 |
| Histological Grade | 1.632 | 0.95 | 2.746 | 0.083 |
| ER | 0.662 | 0.478 | 1.136 | 0.164 |
| PR | 0.749 | 0.446 | 1.341 | 0.425 |
| HER2 | 1.343 | 0.034 | 1.568 | 0.543 |
| Ki-67 (High vs Low (Ref)) | 1.572 | 1.887 | 2.787 | 0.021 |
| TUSC7 (High vs Low (Ref)) | 0.574 | 0.673 | 0.848 | 0.034 |
Note: Bold values are significant (P<0.05).
Abbreviations: ER, estrogen receptor; PR, progesterone receptor; HR, hazard ratio; HER2, human epidermal growth factor receptor-2.
The Overexpression of TUSC7 Suppresses the Growth, Migration, and Invasion of Breast Cancer Cell Lines
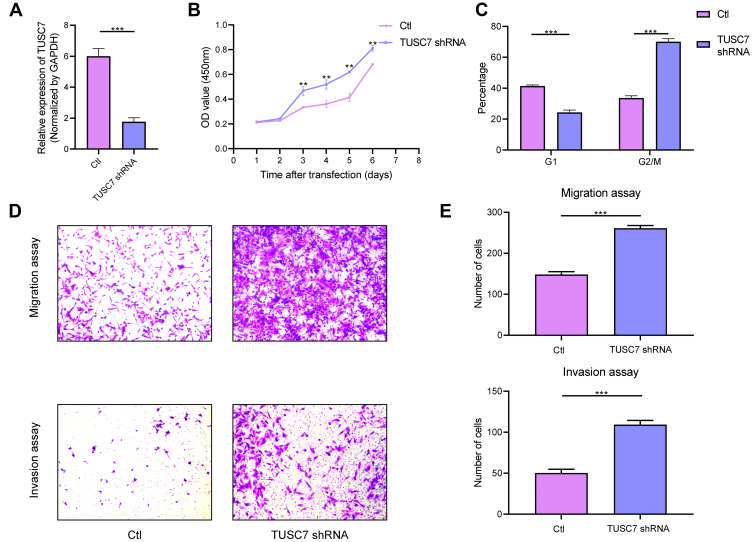
To further explore the biological role of TUSC7 in breast cancer cells, shRNA was constructed to suppress TUSC7 expression. The knock-down efficiency in MDA-MB-231 cell lines has been confirmed by using qPCR (P = 0.023, Figure 2A). TUSC7 knockdown resulted in increased cell growth in MDA-MB-231 cells (Figure 2B). Meanwhile, the downregulation of TUSC7 increased the percentage of cells in the G2/M phase (Figure 2C). A 3.73-fold increase in cell migration and a 6.30-fold increase in cell invasion were also found after the knockdown of TUSC7 (Figure 2D and E).
Figure 2.
The effects of TUSC7 knockdown on the growth, migration and invasion of triple negative breast cancer (TNBC) cells. (A) The knockdown of TUSC7 by shRNA was performed using shRNA sequences. TUSC7 shRNAs and scrambled sequences (scramble) were transfected into MDA-MB-231 cells and knockdown was confirmed by qRT-PCR. (B) CCK8 assays. Cell growth of MDA-MB-231 cells transfected with TUSC7 shRNAs or control shRNAs for 5 days. (C) Cell cycle analysis of MDA-MB-231 cells transfected with TUSC7 shRNAs or scrambled shRNAs for five days. (D and E) Transwell migration and invasion assays of MDA-MB-231 cells transfected with TUSC7 shRNAs or scrambled shRNAs for five days. Magnification, 100X. **P<0.001; ***P<0.0001.
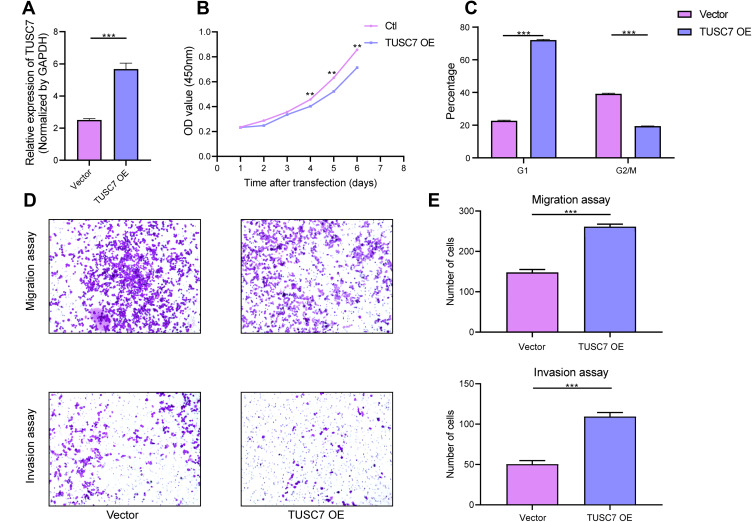
The overexpression efficiency in MDA-MB-468 cell lines has been confirmed by using qPCR (MDA-MB-468 cells transfected with DOX-inducible vector) (Figure 3A). The overexpression of TUSC7 in MDA-MB-468 cells resulted in decreased cell growth (Figure 3B), decreased percentage of cells in G2/M, and increased numbers of cells in G1 (Figure 3C). Meanwhile, a 2.73-fold decrease in cell migration and a 2.30-fold decrease in cell invasiveness were detected after overexpression of TUSC7 (Figure 3D and E). Taken together, these data indicated that TUSC7 might be involved in inhibiting growth, migration, and invasion in MDA-MB-468 cells in vitro.
Figure 3.
The effects of TUSC7 overexpression on the growth, migration and invasion of TNBC cells. (A) The Overexpression of TUSC7 was performed by transfecting MDA-MB-468 cells with the TUSC7-DOX vector (TUSC7 Over). Expression was confirmed by qRT-PCR. (B) CCK8 assays. MDA-MB-231 cells were transfected with TUSC7-DOX (TUSC7 Over) or scramble vector for five days. (C) Cell cycle analysis of MDA-MB-231 cells transfected with TUSC7-DOX (TUSC7 Over) or scramble vector for 5 days. (D and E) Transwell migration and invasion assays of MDA-MB-231 cells transfected with TUSC7-DOX (TUSC7 Over) or scramble vector for 5 days. Magnification, 100X. **P<0.001; ***P<0.0001.
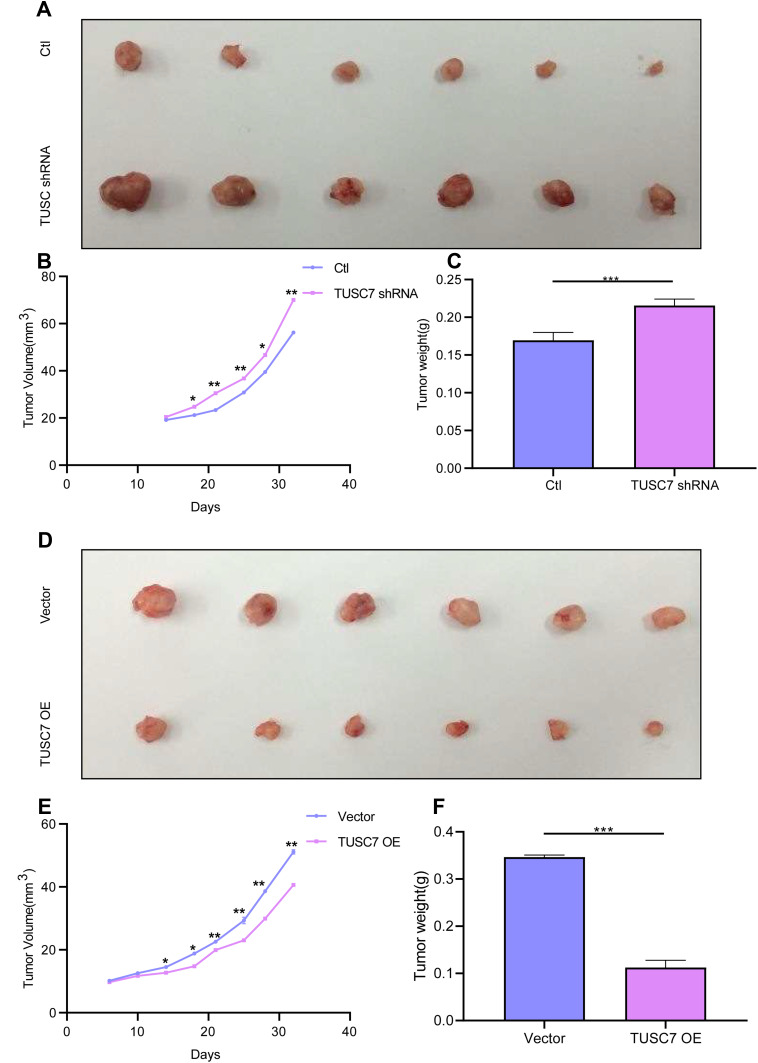
To clarify whether TUSC7 exhibits a tumor-suppressive role in breast cancer, MDA-MB-231 cells transfected with vector as control or TUSC7 shRNAs were injected into nude mice. Mice harboring cells with TUSC7 shRNA showed a significantly increased tumor volume (P = 0.021, Figure 4A and B) and final tumor weight compared with control mice (P = 0.023, Figure 4C). We also performed similar experiments in mice injected with MDA-MB-468 cells transfected with vector control or the vector drives DOX-inducible TUSC7 expression. We found that TUSC7 overexpression significantly decreased tumor based on tumor volume (P = 0.025, Figure 4D and E) and final tumor weight compared with controls (P = 0.028, Figure 4F). However, overexpression/knockdown of TUSC7 did not affect the body weight of mice significantly. All these data suggested that TUSC7 suppressed tumor cell growth both in vitro and in vivo, further demonstrated that TUSC7 could suppress the tumor growth in breast cancer.
Figure 4.
TUSC7 inhibits tumor growth in a xenograft mouse model. Scrambled shRNA control, TUSC7 shRNA, TUSC7 scramble vector or TUSC7-DOX-expressing MDA-MB-468 cells were injected into the flank of nude mice (n = 6, each group). After 32 days, the tumors were removed. The tumorigenesis rate was 100% for each group. (A) Tumor volume. (B) Tumor growth curves. Data are presented as the average ± s.d. of 6 tumors for each group. *P < 0.05; **P<0.001; P<0.0001. (C) Tumor weights were measured when tumors were harvested. Error bars represent SEM, n = 6. ***P<0.0001. (D) Tumor volume. (E) Tumor growth curves. Data are presented as the average ± s.d. of six tumors for each group. *P < 0.05; **P<0.001; P<0.0001. (F) Tumor weights were measured when tumors were harvested. Error bars represent SEM, n = 10. ***P<0.0001.
The Overexpression of TUSC7 in TNBC Decreases Chemoresistance in Breast Cancer Cells
We found that MDA-MB-468 cells overexpressing TUSC7 were significantly less resistant to paclitaxel and carboplatin; the IC50 values for paclitaxel and carboplatin in MDA-MB-468 cells overexpressing TUSC7 were 0.002 and 0.013 mmol (P < 0.0001) (Figure S1).
The Expression Level of TUSC7 is Associated with the p53
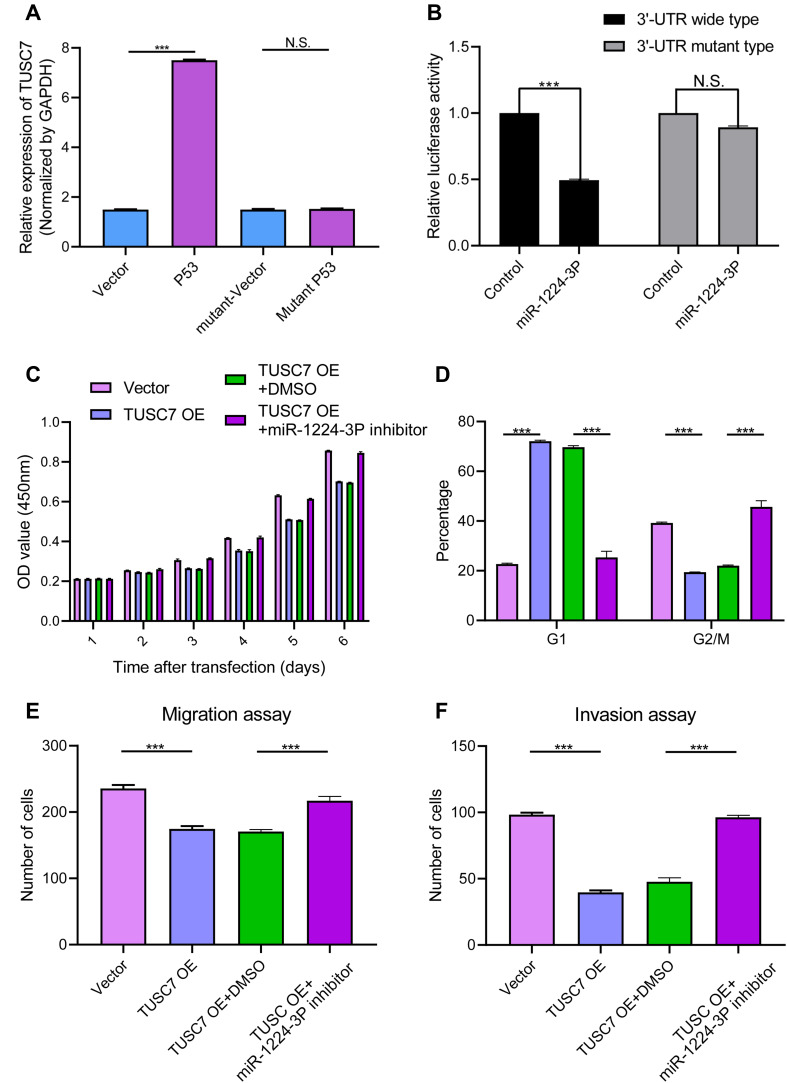
MDA-MB-231 cell line was transfected with a plasmid vector overexpressing wild-type p53 and a 7.3-fold increase in TUSC7 levels was detected compared with controls (Figure 5A). Notably, a transcriptionally inactive point mutation (R175H) of p53, a frequent mutant in the DNA-binding domain in cancer,18 did not affect TUSC7 levels. These results suggest that TUSC7 is a p53-regulated tumor suppressor in breast cancer cell lines.
Figure 5.
(A) Induction of TUSC7 by ectopically expressed p53. MDA-MB-231 cells were transfected with empty vector or plasmids expressing wild-type p53 or mutant p53 (R175H) overnight. After changing medium, the cells were cultured for another 24 h before qRT-PCR for TUSC7. Error bars represent SEM, n = 3. ***P<0.0001, NS, non significant. (B) TUSC7 is a direct target of miR-1224-3P. MDA-MB-231 cells were transfected with luciferase reporters with the wild-type TUSC7 3′-UTR or TUSC7 mutant 3′-UTR together with the miR-1224-3P mimics or control oligonucleotides. Renilla luciferase activity was normalized to firefly luciferase activity. Error bars represent SEM, n = 3. NS, non significant, ***P<0.0001. (C) CCK8 assays. The miR-1224-3P inhibitor could reverse the effect of TUSC7 on the growth of MDA-MB-231 cell lines. Error bars represent SEM, n = 3. (D) Cell cycle assays. The miR-1224-3P inhibitor could reverse the effect of TUSC7 on the cell cycle of MDA-MB-231 cell lines. Error bars represent SEM, n = 3. ***P<0.0001. (E) Migration assays. The miR-1224-3P inhibitor could reverse the effect of TUSC7 on the migration ability of MDA-MB-231 cell lines. Error bars represent SEM, n = 3. ***P<0.0001. (F) Invasion assays. The miR-1224-3P inhibitor could reverse the effect of TUSC7 on the invasion ability of MDA-MB-231 cell lines. Error bars represent SEM, n = 3. ***P<0.0001.
TUSC7 Might Perform Its Biological Function Through Binding with miR-1224-3P and Regulating Its Expression Level
To investigate the potential underlying mechanism of the biological role of TUSC7, the miRNA and mRNA expression profile of TUSC7 knockdown MDA-MB-231 cells were compared with the control cells. Accordingly, 29 TUSC7-associated miRNAs were identified: 13 miRNAs were downregulated while 16 miRNAs were upregulated after the knockdown of TUSC7 (Table S1). Then, TargetScan results indicated possible binding sites of the 3ʹ-UTR sequence of TUSC7 and miR-1224-3P, miR-920, miR-7113-5p, miR-608, miR-637, hsa-miR-6770-5p, hsa-miR-1207-5p, hsa-miR-4451 (Table S2), which turned out that TUSC7 has two binding sites of miR-1224-3P (Figure S3). To verify whether TUSC7 was acting as a molecular sponge for these miRNAs through close attachment to its 3ʹ-UTR, Luciferase reporter assay was conducted with the panel of eight identified TUSC7 associated miRNAs. Of note, we, therefore, observed that TUSC7 might act as a candidate target of miR-1224-3P. A fragment of TUSC7 3′-UTR encompassing the binding site of miR-1224-3P (Supplementary Materials) was inserted downstream of the luciferase open reading frame in the reporter plasmid. The reporter vector was co-transfected in MDA-MB-231 cells with Mir-1224-3P mimic or negative control. In the presence of miR-1224-3P mimic, luciferase activity decreased by approximately 45.78% (Figure 5B). However, miR-1224-3P did not result in the decreased luciferase activity of the TUSC7 3′-UTR mutant type. Meanwhile, the correlation between the TUSC7 and miR-1224-3P has been evaluated through qPCR analysis in tumor tissues, which turned out that the expression level of TUSC7 was positively correlated with the expression level of miR-1244-3P (Figure S2A, R=0.86, P<0.001). Besides, compared with adjacent normal tissue, the miR-1224-3P expression level was lower in tumor tissue (Figure S2B, P<0.001). To conclude, TUSC7 might perform its biological function through binding with miR-1224-3P and regulating its expression level in breast cancer cell lines.
MiR-1224-3P Inhibitor Could Reverse the Biological Function of TUSC7
Further experiments were conducted to explore whether TUSC7 performed its biological function through binding with miR-1224-3P. The miR-1224-3P inhibitor was added into the TUSC7 overexpression cell lines, which turned out that the inhibitor could reverse the biological function caused by the overexpression of TUSC7, including the growth (Figure 5C), cell cycle (Figure 5D), migration (Figure 5E), and invasion (Figure 5F). All these results indicated that TUSC7 might carry out its function through binding with miR-1224-3P.
The Potentially Involved Signaling Pathway of TUSC7
In terms of the mRNA microarray results analysis, 32 pathways were down-regulated (involving 127 genes) and seven pathways were significantly up-regulated (involving 23 genes) in TUSC7 downregulated MDA-MB-231 cells (Figure S4A). All the 23 significantly upregulated genes were examined in TUSC7 knockdown MDA-MB-231 breast cancer cells by using qPCR and our gene validation results show that genes in the cell cycle pathways (BUB3) and TGF-β pathways (Rho-associated coiled-coil containing protein kinase 1/2, ROCK1/2) were downregulated (Figure S4B) and genes in the MAPK (TGFBR2), PI3K-AKT and NF-KB pathways were upregulated in TUSC7 knockdown MDA-MB-231 breast cancer cells (Figure S4C). These results indicated that MAPK, PI3K-AKT, and NF-κB pathways might be involved in the biological function of TUSC7 in breast cancer, and further investigation should be carried out.
Discussion
As a new discovery of non-coding genes, lncRNAs have been proved to be widely transcribed in the genome. Modification in the expression levels, or structure, together with their cognate RNA-binding proteins are often related to the progressions of cancer.19 In this study, we observed that the lncRNA TUSC7 was downregulated in breast cancer tissues compared with adjacent normal tissues. Importantly, the low TUSC7 expression was positively correlated with poor overall survival of TNBC patients. Consistent with these results, the overexpression of TUSC7 suppressed breast tumor cell growth in vitro and in vivo.
Despite the importance of chemotherapy in the treatment of TNBC,20 no optimal regimen has been identified and established. The combination of paclitaxel and carboplatin was reported to have high response rates in metastatic TNBC.21 Here, we found out that TUSC7 overexpression could significantly enhance the sensitivity to paclitaxel and carboplatin in the MDA-MB-468 cell line.
As for lncRNA’s strong influences on cellular functions through diversified mechanisms, including reactions with chromatin remodeling proteins, they can still regulate other microRNAs.22,23 MiR-1224-3P is a novel miRNA that has been identified by extensive cloning.24 The high expression of this microRNA was observed in mouse spleen, kidney, and lung tissues. This microRNA was a negative regulator of TNF-α in an Sp1-dependent manner, which can decrease the basal tumor necrosis factor-α (TNF-α) promoter-reporter gene activity, and might be involved in regulating the LPS-mediated inflammatory responses.25,26 In our study, we discovered that TUSC7 might perform its biological function via binding with the miR-1224-3P in breast cancer cell lines.
TNBC presents a large number of DNA aberrations and were relatively sensible to platins.18,19 As a well-known tumor suppressor, p53 is activated by multiple upstream signals, including DNA damage, and influences a variety of cellular functions and disease processes, in part through transcriptional regulation of specific genes.27,28 As the main regulator for gene expression, p53 can directly or indirectly regulate a huge number of protein-coding genes and non-coding genes, like microRNAs. For instance, in a p53-dependent manner, the miR-34 family (miR-34a, miR-34b, and miR-34c) are stimulated by DNA damage and oncogenic stress.29 Besides, it is reported that TUS7 is a p53-regulated gene in gastric cancer.30 Consistent with the previous study, the present study indicates that TUSC7 is a p53-regulated tumor suppressor. Therefore, further characterization of potential lncRNAs will reinforce the idea that, like protein-coding genes, lncRNAs are also an integral part of the p53 regulatory network. Obviously, more than two significant lncRNAs, mouse lincRNA-p21 and human PANDA (promoter of CDKN1A antisense DNA damage activated RNA), have been identified as transcription target of p53,31,32 and involved in the p53-related cellular processes. Thus, the identification of TUSC7 as a p53 inducible lncRNA expands the repertoire of p53-regulated genes.33
The PI3K/AKT and MAPK pathways are involved in a lot of biological processes, including cell apoptosis, proliferation, and cell cycle.34,35 Our results suggest that TUSC7 mediates the proliferation of TNBC cells by influencing the PI3K/AKT and MAPK and signaling pathways, which requires further investigation.
This finding is limited to a retrospective study in a single center, and thus, further studies performed in a multicenter or prospective manner are necessary to validate the clinical usage of TUSC7 as a prognostic marker for breast cancer. Besides, our result indicates that TUSC7 is a p53 related lncRNA, further experiments are needed to investigate how the p53 regulates the expression of TUSC7. Meanwhile, we found that TUSC7 might perform its function through binding with miR-1224-3P and regulating its expression. Further investigation should be performed to investigate how the miR-1224-3P influence the biological function of breast cancer cell lines. Moreover, although downstream signaling pathways related to TUSC7 have been found, no clear mechanism of action has been clarified. Herein, further investigations focusing on the downstream signaling pathways of TUSC7 should be carried out.
Conclusions
In the present study, we discovered that the low expression of TUSC7 was an independent biomarker of poor OS of TNBC patients. Also, our in vitro and in vivo experiments revealed that TUSC7 inhibited tumor cell growth through binding with miR-1224-3P. Besides, PI3K/AKT and MAPK signaling pathways might be involved in the biological function of TUSC7. Our results indicated that TUSC7 might serve as a potential biomarker and therapeutic target for TNBC patients.
Funding Statement
The study was supported by grants from the National Natural Science Foundation of China (81702586); the National Natural Science Foundation of China (81902687); and the 2016 Zhongshan Hospital, Fudan University, Qingnian Funds (2016ZSQN59); and Shanghai key clinical medical center and construction of key disciplines.
Abbreviations
ER, estrogen receptor; HER2, human epidermal growth receptor-2; TNBC, triple-negative breast cancer; LncRNAs, long non-coding RNAs; OS, overall survival; HR, hazards ratios; PR, progesterone receptor; CI, confidence interval; TUSC7, tumor suppressor candidate 7.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author Xiao-Jian Ni on reasonable request.
Ethics Approval and Consent to Participate
This study was approved by the institutional review board of Zhongshan Hospital, Fudan University. Every patient provided written informed consent before enrollment.
Disclosure
The authors reported no conflicts of interest for this work.
References
- 1.Parikshak NN, Swarup V, Belgard TG, et al. Genome-wide changes in lncRNA, splicing, and regional gene expression patterns in autism. Nature. 2016;540(7633):423–427. doi: 10.1038/nature20612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316(5830):1484–1488. doi: 10.1126/science.1138341 [DOI] [PubMed] [Google Scholar]
- 3.Calore F, Lovat F, Garofalo M. Non-coding RNAs and cancer. Int J Mol Sci. 2013;14(8):17085–17110. doi: 10.3390/ijms140817085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Engreitz JM, Haines JE, Perez EM, et al. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature. 2016;539(7629):452–455. doi: 10.1038/nature20149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JT. Epigenetic regulation by long noncoding RNAs. Science. 2012;338(6113):1435–1439. doi: 10.1126/science.1231776 [DOI] [PubMed] [Google Scholar]
- 6.Jiang YZ, Liu YR, Xu XE, et al. Transcriptome analysis of triple-negative breast cancer reveals an integrated mRNA-lncRNA signature with predictive and prognostic value. Cancer Res. 2016;76(8):2105–2114. doi: 10.1158/0008-5472.CAN-15-3284 [DOI] [PubMed] [Google Scholar]
- 7.Carey LA, Dees EC, Sawyer L, et al. The triple negative paradox: primary tumor chemosensitivity of breast cancer subtypes. Clin Cancer Res. 2007;13(8):2329–2334. doi: 10.1158/1078-0432.CCR-06-1109 [DOI] [PubMed] [Google Scholar]
- 8.Dent R, Trudeau M, Pritchard KI, et al. Triple-negative breast cancer: clinical features and patterns of recurrence. Clin Cancer Res. 2007;13(15 Pt 1):4429–4434. doi: 10.1158/1078-0432.CCR-06-3045 [DOI] [PubMed] [Google Scholar]
- 9.Tischkowitz M, Brunet J-S, Bégin LR, et al. Use of immunohistochemical markers can refine prognosis in triple-negative breast cancer. BMC Cancer. 2007;7:134. doi: 10.1186/1471-2407-7-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyle P. Triple-negative breast cancer: epidemiological considerations and recommendations. Ann Oncol. 2012;23(Suppl 6):vi7–12. doi: 10.1093/annonc/mds187 [DOI] [PubMed] [Google Scholar]
- 11.Harris LN, Broadwater G, Lin NU, et al. Molecular subtypes of breast cancer in relation to paclitaxel response and outcomes in women with metastatic disease: results from CALGB 9342. Breast Cancer Res. 2006;8(6):R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Omarini C, Guaitoli G, Pipitone S, et al. Neoadjuvant treatments in triple-negative breast cancer patients: where we are now and where we are going. Cancer Manag Res. 2018;10:91–103. doi: 10.2147/CMAR.S146658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Q, Huang J, Zhou N, et al. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res. 2013;41(9):4976–4987. doi: 10.1093/nar/gkt182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pasic I, Shlien A, Durbin AD, et al. Recurrent focal copy-number changes and loss of heterozygosity implicate two noncoding RNAs and one tumor suppressor gene at chromosome 3q13.31 in osteosarcoma. Cancer Res. 2010;70(1):160–171. doi: 10.1158/0008-5472.CAN-09-1902 [DOI] [PubMed] [Google Scholar]
- 15.Cong M, Li J, Jing R, et al. Long non-coding RNA tumor suppressor candidate 7 functions as a tumor suppressor and inhibits proliferation in osteosarcoma. Tumour Biol. 2016;37(7):9441–9450. doi: 10.1007/s13277-015-4414-y [DOI] [PubMed] [Google Scholar]
- 16.Qi P, Xu MD, Ni SJ, et al. Low expression of LOC285194 is associated with poor prognosis in colorectal cancer. J Transl Med. 2013;11:122. doi: 10.1186/1479-5876-11-122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ding YC, Yu W, Ma C, et al. Expression of long non-coding RNA LOC285194 and its prognostic significance in human pancreatic ductal adenocarcinoma. Int J Clin Exp Pathol. 2014;7(11):8065–8070. [PMC free article] [PubMed] [Google Scholar]
- 18.Soussi T, Ishioka C, Claustres M, et al. Locus-specific mutation databases: pitfalls and good practice based on the p53 experience. Nat Rev Cancer. 2006;6(1):83–90. doi: 10.1038/nrc1783 [DOI] [PubMed] [Google Scholar]
- 19.Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21(6):354–361. doi: 10.1016/j.tcb.2011.04.001 [DOI] [PubMed] [Google Scholar]
- 20.Hsu DS, Balakumaran BS, Acharya CR. Retraction. Pharmacogenomic strategies provide a rational approach to the treatment of cisplatin-resistant patients with advanced cancer. J Clin Oncol. 2007;25(28):4350–4357. doi: 10.1200/JCO.2007.11.0593 [DOI] [PubMed] [Google Scholar]
- 21.Yardley DA, Coleman R, Conte P, et al. nab-Paclitaxel plus carboplatin or gemcitabine versus gemcitabine plus carboplatin as first-line treatment of patients with triple-negative metastatic breast cancer: results from the tnAcity trial. Ann Oncol. 2018;29(8):1763–1770. doi: 10.1093/annonc/mdy201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rinn JL, Kertesz M, Wang JK, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129(7):1311–1323. doi: 10.1016/j.cell.2007.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chu C, Qu K, Zhong FL, et al. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol Cell. 2011;44(4):667–678. doi: 10.1016/j.molcel.2011.08.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berezikov E, van Tetering G, Verheul M, et al. Many novel mammalian microRNA candidates identified by extensive cloning and RAKE analysis. Genome Res. 2006;16(10):1289–1298. doi: 10.1101/gr.5159906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dolganiuc A, Petrasek J, Kodys K, et al. MicroRNA expression profile in Lieber-DeCarli diet-induced alcoholic and methionine choline deficient diet-induced nonalcoholic steatohepatitis models in mice. Alcohol Clin Exp Res. 2009;33(10):1704–1710. doi: 10.1111/j.1530-0277.2009.01007.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Niu Y, Mo D, Qin L, et al. Lipopolysaccharide-induced miR-1224 negatively regulates tumour necrosis factor-alpha gene expression by modulating Sp1. Immunology. 2011;133(1):8–20. doi: 10.1111/j.1365-2567.2010.03374.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137(4):609–622. doi: 10.1016/j.cell.2009.04.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kastenhuber ER, Lowe SW. Putting p53 in context. Cell. 2017;170(6):1062–1078. doi: 10.1016/j.cell.2017.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447(7148):1130–1134. doi: 10.1038/nature05939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qi P, Xu M-D, Shen X-H, et al. Reciprocal repression between TUSC7 and miR-23b in gastric cancer. Int J Cancer. 2015;137(6):1269–1278. doi: 10.1002/ijc.29516 [DOI] [PubMed] [Google Scholar]
- 31.Raver-Shapira N, Marciano E, Meiri E, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26(5):731–743. doi: 10.1016/j.molcel.2007.05.017 [DOI] [PubMed] [Google Scholar]
- 32.Hung T, Wang Y, Lin MF, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43(7):621–629. doi: 10.1038/ng.848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baugh EH, Ke H, Levine AJ, et al. Why are there hotspot mutations in the TP53 gene in human cancers? Cell Death Differ. 2018;25(1):154–160. doi: 10.1038/cdd.2017.180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peluso I, Yarla NS, Ambra R, Pastore G, Perry G. MAPK signalling pathway in cancers: olive products as cancer preventive and therapeutic agents. Semin Cancer Biol. 2017;56:185–195. doi: 10.1016/j.semcancer.2017.09.002 [DOI] [PubMed] [Google Scholar]
- 35.Matsuda S, Ichimura M, Ogino M, et al. Effective PI3K modulators for improved therapy against malignant tumors and for neuroprotection of brain damage after tumor therapy (review). Int J Oncol. 2016;49(5):1785–1790. doi: 10.3892/ijo.2016.3710 [DOI] [PubMed] [Google Scholar]