Abstract
Over the past decade, scientific discovery games (SDGs) have emerged as a viable approach for biomedical research, engaging hundreds of thousands of volunteer players and resulting in numerous scientific publications. After describing the origins of this novel research approach, we review the scientific output of SDGs across molecular modeling, sequence alignment, neuroscience, pathology, cellular biology, genomics, and human cognition. We find compelling results and technical innovations arising in problem-oriented games such as Foldit and Eterna and in data-oriented games such as EyeWire and Project Discovery. We discuss emergent properties of player communities shared across different projects, including the diversity of communities and the extraordinary contributions of some volunteers, such as paper writing. Finally, we highlight connections to artificial intelligence, biological cloud laboratories, new game genres, science education, and open science that may drive the next generation of SDGs.
Keywords: scientific discovery games, crowdsourcing, video games, citizen science, interactive media, cloud laboratory
1. INTRODUCTION
Games are an ancient medium for human creativity and exploration that have only recently been harnessed for biomedical research. Scientific discovery games (SDGs) bring together researchers looking for novel ways to approach unsolved problems with a global population of volunteers interested in contributing to scientific research but without the traditional means of doing so. The field is quite new, with reviews so far primarily focusing on the promise of SDGs (1-4) rather than critically evaluating their performance relative to prior Internet-scale volunteer initiatives, paid crowdsourcing initiatives, or computational approaches.
This review emerged from discussions that began in 2010 between two of the authors—R. Das, who has developed early SDGs (5), and I.H. Riedel-Kruse, who is designing platforms for playful interactions with living microbiology (6). In 2010, the most basic questions on the scientific effectiveness, player communities, and costs of development of SDGs were unknown. In recent years, the launch and release of publications from Foldit, Eterna, EyeWire, Phylo, and numerous other SDGs (Figure 1) have provided answers to many of these questions. The successes of these and other projects on nontrivial research problems demonstrate that SDGs can deliver genuine scientific value. Here, we discuss the relationship between games and previous modes of scientific discovery, classify and critically evaluate the scientific potential of current SDGs, identify similarities across different game player communities, and outline best practices for creating SDGs. After this overview, we then identify new areas of research that might be addressed in the next generation of SDGs.
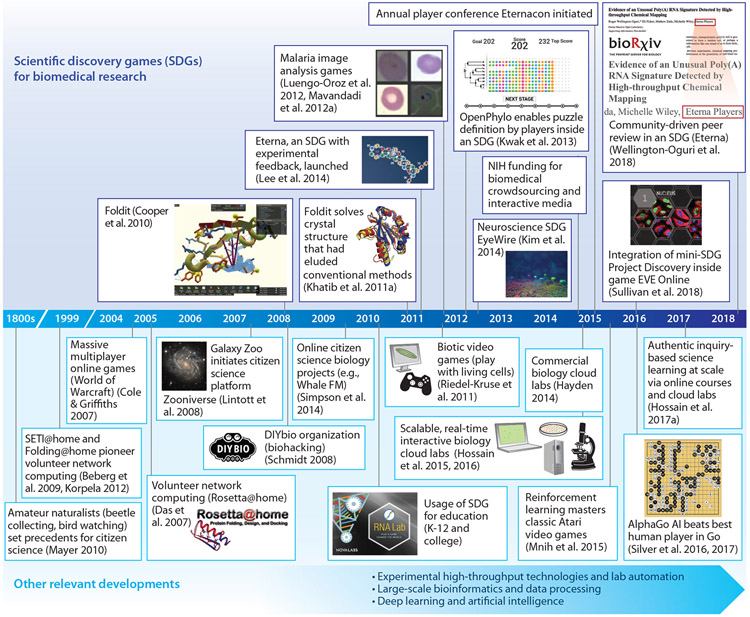
Figure 1.
A history of SDGs in biomedical research. (Top) Selected key events concerning the emergence and maturation of SDGs in biomedical research. (Bottom) Selected key events that have influenced the development of SDGs or may impact SDGs in the future. Key developments not explicitly listed include Wikipedia, the Internet, online games, virtual and augmented reality, mobile computing, and social media. Screenshots and logos are taken from the SDGs themselves; other graphics are used with permission from the NASA Image Library (Galaxy Zoo), Reference 16 (Foldit crystal), Reference 18 (malaria image), the Seung Lab Flickr photostream (EyeWire), and Wikipedia (AlphaGo). Abbreviations: AI, artificial intelligence; NIH, National Institutes of Health; SDG, scientific discovery game.
2. ROOTS OF SCIENTIFIC DISCOVERY GAMES
SDGs emerged from several prior approaches to engage large communities. In this section, we briefly review how citizen science, crowdsourcing, and video games set critical precedents for SDGs (Figure 1).
2.1. Citizen Science
Projects that reach out to nonexpert volunteers to carry out biological research extend back to at least the nineteenth century. Milestones of citizen science include the North American Bird Phenology Program (1890–1970) (7), the Audubon Christmas Bird Count (1900–present) (30), and the Continuous Plankton Recorder (1931–present) (31), which have all leveraged volunteer efforts to track natural phenomena over large geographical areas and long time frames—across continents and decades. The development of new technologies, particularly the Internet, allowed these projects to achieve even larger scales (32-34). A new kind of citizen science, involving the Internet-scale classification of images, was pioneered in the 2000s in the astronomy community (12, 15, 35). NASA’s ClickWorkers (2000) was an effort to classify large image data sets of moon craters and attracted over 100,000 online volunteers (35). The Zooniverse citizen science web portal has engaged nearly a million volunteers in citizen science, beginning with morphology classification of galaxies from the Sloan Digital Sky Survey [Galaxy Zoo, 2007–present; (12)] and extending to numerous other areas. For example, the Zooniverse citizen science project Whale FM enabled the categorization of sounds made by killer whales and followed the travels of individual animals around the oceans (15).
Within biomedical research, large-scale volunteer projects largely trailed behind the citizen science efforts in the ecology and astronomy communities, except in one field, volunteer-distributed computing networks. In 1999–2000, two screensaver projects, Folding@home (8) and SETI@home (9), demonstrated that hundreds of thousands of volunteers were willing to donate unused computer cycles to help process or generate data for large-scale projects for the protein folding and astronomy communities. In 2002, the Berkeley Open Infrastructure for Network Computing (36) accelerated the development of numerous other distributed computing projects, including Rosetta@home (11), which inspired the first SDG, Foldit (13), described below. Furthermore, various DIY biology approaches emerged that aimed to increase lay citizens’ access to biological experimentation (14).
2.2. Crowdsourcing
“Crowdsourcing,” a term first coined in 2006 (37), refers to projects that divide work among a very large number of people. Although citizen science projects inspired the term “crowdsourcing,” it now encompasses other kinds of projects where the participants are not volunteers but are instead paid for their efforts, such as Amazon Mechanical Turk (38). As exemplified by Wikipedia (2001) and later biomedically focused variants like Gene Wiki (39) and Rfam Wiki (40), crowdsourcing promises cost-efficient ways of classifying and organizing large biomedical data sets, which are arriving at a spectacular pace due to the rise of next-generation sequencing (41), medical imaging (42), medical robotics (43), and automated microscopy (44).
Beyond its applications to large-scale data curation, crowdsourcing offers the prospect of solving difficult biomedical problems that require unusual human insight (1). In this context, the term “crowdsourcing” often includes global community-wide challenges for predicting DNA–protein affinities [DREAM (Dialogue on Reverse Engineering Assessment and Methods)] (45), protein structure [CASP (Critical Assessment of Structure Prediction)] (46), and genotype-to-phenotype relationships [CAGI (Critical Assessment of Genome Interpretation)] (47), as well as commercial platforms like Kaggle and InnoCentive (48). Most of these crowdsourcing approaches involve competition, although these projects typically are not further gamified.
2.3. Video Games
The cultural and commercial impact of video games is hard to overstate. By sales, revenues, and hours spent by audiences, the video game industry has already surpassed the literature and film media industries (49). The distribution of web services and mobile applications provides significant game markets that reach billions of people around the developed and developing world (49). Academic research on games has also exploded in recent years, with many universities founding departments and student programs focusing on the topic (50). The terms “serious games” and “games with a purpose” have been coined to emphasize that there can be other objectives in games beyond player enjoyment (51). Online video games have also evolved in structure and form, with emerging properties such as social interactions in massive multiplayer online games like World of Warcraft, released in 2004 (10). The massive amount of interest in video games and the successes of crowdsourced scientific projects in the early 2000s set the stage for video games to become a medium for biomedical research.
2.4. Emergence of Scientific Discovery Games
Foldit is generally considered the first SDG (13). Launched in 2008 from the University of Washington, Foldit integrated developments in all three fields described above. First, Foldit was intellectually and technologically grounded in the volunteer computing paradigm. Rosetta@home (11) participants who dedicated their idle computing time to explore protein folding configurations could watch these simulations displayed via screensavers and would occasionally suggest to Rosetta developers that they could intuit better folding paths than what they saw on their screens. The Zooniverse projects provided a strong precedent for the value of crowdsourcing scientific research, and community-wide challenges like CASP further suggested that bringing disparate teams into rigorous competition might spur progress on the protein structure prediction problem (46). Lastly, the success of video games had established a set of standard game elements that could draw players in. Foldit merged these core game elements into a large-scale, volunteer-driven scientific project (Figure 2), defining the SDG paradigm for biomedical research and beyond.
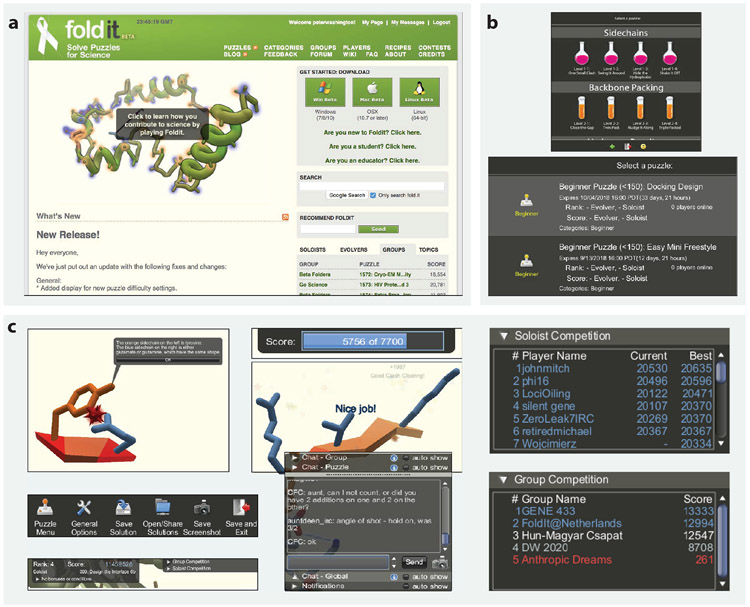
Figure 2.
Anatomy of a scientific discovery game, illustrated by the paradigm-setting Foldit. (a) First-time players of Foldit first navigate to the project home page, where a link can take the user to a FAQ page. The FAQ contains an explanation of protein folding, a description of the scientific problems Foldit is tackling, a description of how game playing contributes to curing diseases, and a list of scientific publications. On either page, the user can download Foldit applications for Windows, Mac, or Linux. (b) Players can participate in published science puzzles and contests created by other players and can see top players and top player groups. Player groups can complete protein folding challenges together. (c) Foldit gamifies certain biological concepts while introducing the relevant science. For example, in the first tutorial, the player is told that “the blue sidechain on the right is either glutamate or glutamine, which have the same shape.” The player goes through 38 folding challenges in the tutorial sequence. During game play, the score updates on the top of the screen based on how well the protein folded. The goal of each puzzle is to create a protein conformation with the lowest energy. After completing a puzzle, a congratulatory animation is played. The player can save a solution or share their solutions with other players. The player’s global rank and score are displayed at the top of the graphical user interface. The interface also contains a public chat display, which allows players to communicate with and receive help from other players. The puzzles can either be solved by individual players in the soloist competition or by teams that work collaboratively on a solution in the group competition.
3. CURRENT SCIENTIFIC DISCOVERY GAMES FOR BIOMEDICAL RESEARCH
This section surveys scientific accomplishments of SDGs, starting with protein modeling results in Foldit and then extending to other SDGs and areas of biomedical research. We propose grouping SDGs broadly into two categories, problem-oriented games and data-oriented games, which we discuss below.
3.1. Problem-Oriented Games
Problem-oriented games address the most basic research gap: the inability to find solutions to a general scientific problem, even after repeated attempts using state-of-the-art techniques. Problem-oriented games both inside and outside of biomedical research often explicitly cite Foldit as their inspiration. Topics vary from molecular modeling and molecular design problems to the sequence alignment problem. These problems have intrinsic game-like qualities: They have a set of rules that constrain play, immense amounts of complexity, and no clearly defined or automatable path toward success. They are also all open problems that have been the subject of intense scientific interest and research.
3.1.1. Molecular structure modeling problems.
The initial focus of Foldit was to model the three-dimensional (3D) structures of proteins at atomic resolution, guided by the computational macromolecule energy function provided by the Rosetta software (52). Finding the lowest-energy conformations of a protein in silico is challenging, and even simple subproblems, such as protein side-chain packing, have been proven to be NP-hard (53).
The Foldit graphical user interface contains several gamified elements, including the score of the fold, a player leaderboard that shows the scores of other players and groups, and a chatbox for interacting with other players (Figure 2). To manipulate proteins, Foldit players can launch tools from Rosetta to help them solve the problem. Scientific and technical terms are replaced with more colloquial terms throughout the game to make the game more accessible to citizen scientists. The puzzles can either be solved by individual players in the soloist competition or by teams working collaboratively on a solution in the group competition.
Early results from Foldit demonstrated that human players could find unusual routes from initially incorrect protein conformations to refined configurations (13, 54). Human players and computational approaches solved these problems in different ways. Players outperformed Rosetta’s computational procedures when they could see that unraveling the structure of the protein, which might appear detrimental to solving the problem, would ultimately lead to more favorable foldings, whereas the computational approaches embedded in Rosetta could not do this. In other cases, however, computational algorithms bested human players. For example, human players had trouble reaching a native conformation when given only an extended protein chain. Because humans and computers excel in different subtasks of the protein folding problem, an ideal approach would combine computational and human techniques, a theme repeated for SDGs beyond Foldit.
The first biomedical research discovery from Foldit—and of any SDG in the life sciences— was an accurate homology model for the Mason–Pfizer monkey virus retroviral protease (Figure 3a), which enabled the successful crystallographic phasing of a diffraction data set that had been previously refractory to state-of-the-art approaches, including automated Rosetta structure modeling and phasing protocols (16). Applications to crystallographic model building (55) and the CASP worldwide protein structure prediction challenges (56) have further demonstrated applications of Foldit to protein structure modeling. Foldit has also inspired another SDG to tackle crystallographic phasing, CrowdPhase (57), in which player moves are integrated with a genetic selection algorithm, as well as HiRE-RNA, an interactive 3D molecular dynamics game for RNA (58). These projects have not yet been tested in realistic scenarios.
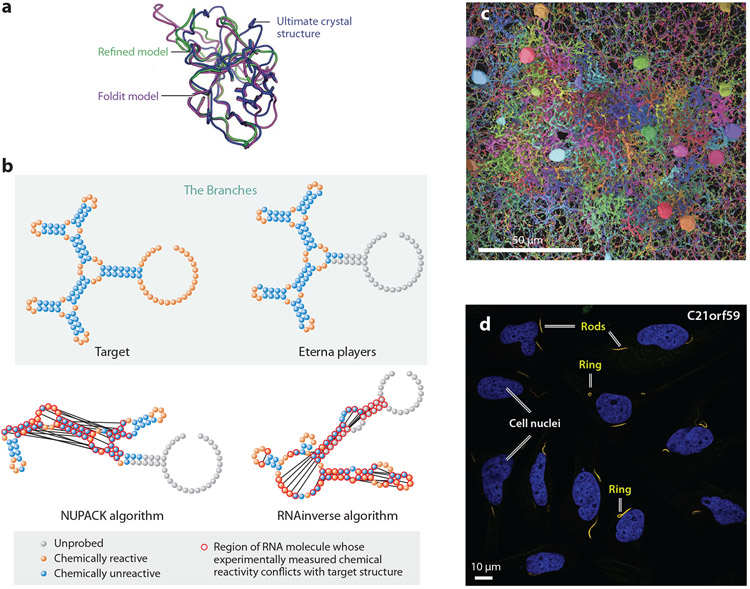
Figure 3.
Successes of scientific discovery games. (a) Foldit-refined models of a Mason–Pfizer monkey virus retroviral protease (purple and green) that led to successful crystallographic phasing and an experimental solution of the enzyme’s structure (blue). (b) RNA sequences designed by Eterna players achieve a secondary structure termed The Branches (top panels), while algorithmic solutions do not (bottom panels). (c) An EyeWire reconstruction of circuitry of OFF-type starburst amacrine cells and bipolar cells in the mouse retina. (d) Project Discovery players identify novel localizations to cytoophidia (red; also known as “rings and rods”) of the protein C21orf59 (green), which appear together as yellow in the panel due to colocalization; cell nuclei are stained in blue (25). Panels adapted with permission from (a) Reference 16, (b) Reference 5, (c) Reference 20, and (d) the Human Protein Atlas.
3.1.2. Molecular design problems.
A major goal of modern biomedical research is to go beyond determining the structures of natural macromolecules and to instead design novel sequences that might fold up into a target structure (the inverse folding problem) or even carry out new functions. Foldit again pioneered the use of SDGs for molecular design, introducing new tools that allowed insertions, deletions and sequence substitutions into its protein manipulation interface. The resulting sequence changes led to an 18-fold improvement of a Diels–Alderase enzyme’s activity and included a player-designed 24-residue insertion whose conformation was tested by crystallography (59).
Macromolecule design was extended from proteins to RNA with the web video game Eterna, released in 2011 (5). The problem of designing RNA sequences that fold into desired Watson–Crick base pairing patterns (secondary structures) is NP-hard even using simplified energy functions to simulate RNA folding, due to the large number of alternative structures that any sequence can form (60). The problem becomes particularly difficult due to a reality gap: RNA secondary structure modeling methods have assumptions and missing parameters that lead to inaccuracies in predicting how RNAs actually fold in vitro. To address this reality gap, Eterna pioneered the integration of experimental design and feedback into the gameplay of an SDG, a paradigm dubbed a massive open laboratory.
After playing starter puzzles and gaining entry to Eterna’s lab, players virtually design RNA sequences on-screen. Their designs are synthesized and experimentally analyzed. The data are returned directly to players, providing feedback on whether their submissions fold into the predicted structures. Eterna players outperformed existing computational prediction algorithms in designing sequences that folded into novel target secondary structures (Figure 3b), and the heuristic rules discovered by players were turned into a predictive algorithm called EteRNAbot, which also outperformed all existing algorithms (5). The increasing experimental throughput in testing designs and the increasing participation of players in defining game content (see Section 4.4 below) suggest new models of experimental science through massive open laboratories with unusual rigor and reproducibility (2). Another SDG working on the design of nucleic acid molecules is Nanocrafter, which seeks collections of interacting DNA strands for complex nanotechnology applications (61).
3.1.3. Sequence alignment.
Vast amounts of DNA sequencing data have inundated many fields of biomedical research. Leveraging these data for biological insight typically requires the alignment of homologous sequences, but even in simplified form, this problem is NP-hard (62). Phylo is a web and mobile game centered around improving the accuracy of multiple DNA sequence alignments drawn from species spanning the tree of life, and Fraxinus is a Facebook-embedded game created to analyze a fungal pathogen that has devastated populations of ash trees across Europe (63). Both projects translate the DNA sequence alignment problem into puzzle games. In Phylo, player-refined alignments of sequence subsets improved independently computed scores in a notable fraction of cases (64). In most Fraxinus puzzles, the players gave alignments distinct from the tested baseline computational method, and encouragingly, alignments by different high-scoring players agreed with each other for 98% of these contributions, confirming high precision. For both Phylo and Fraxinus, whether players outperformed computers depended on which computational scoring method was used to assess accuracy (see Reference 19). A challenge for sequence alignment SDGs is to define gold standards through, for example, expert-curated alignments that have been validated through prospective experiments. The field of RNA comparative modeling has such alignments for ribosomal, intron, and other RNAs (65); encouragingly, an RNA extension to Phylo called Ribo has been prototyped (66).
3.1.4. Future problem-oriented games: prediction and design of patient treatments.
Problem-oriented SDGs so far largely focus on molecular-level problems, but their areas of application could extend to larger biological scales. Perhaps the most important problem in biomedical research involves the interpretation of diagnostic data from patients and the design of effective treatments for the diagnosed disease. Proof-of-concept studies suggest that (nongame) crowdsourcing can predict the course of multiple lateral sclerosis in patients (67). The recently released Cancer Crusade game explores treatment of solid tumors through detailed mathematical modeling, although it does not yet include actual patients (which would probably require a new ethical framework; see the sidebar titled Ethical Questions in Scientific Discovery Games). At scales between molecular and whole human patients, several SDGs have also been launched, but these largely follow a distinct data-oriented paradigm, as described next.
ETHICAL QUESTIONS IN SCIENTIFIC DISCOVERY GAMES.
There are currently few guidelines for the ethical treatment of players in SDGs, but there is extensive literature in the related fields of citizen science and online video games. Designers of SDGs should consider several ethical questions (68):
Are players provided with a meaningful and joyful experience, i.e., is the SDG an enjoyable game?
How are players compensated? Intrinsic fun, learning about science, and contributing to the greater benefit of society might seem sufficient, but should other, more extrinsic rewards with real world value be incorporated as well, such as receiving college credit (69) or scientific coauthorship by name?
How can SDGs ensure that players are adequately compensated rather than becoming underpaid or even exploited crowdworkers (70), especially in the context of the growing number of temporary workers and micropayments in the gig economy (71)?
When discoveries are made in an SDG, should they be patentable, and how would the player receive credit or royalties (72, 73)?
Are the games fair, and do they have to be fair? For example, players without sufficient computing resources might be disadvantaged, likely affecting both the intrinsic and extrinsic rewards discussed above.
How is the well-being of players ensured (e.g., preventing game addiction, harassment, or feelings of incompetence) in an online community (74–76)?
How can SDGs provide an all-inclusive, nondiscriminating community? For example, do players feel discouraged due to language barriers, gender, or disabilities (76, 77)?
How is the privacy of the player ensured (as well as any potential protected patient information in the game) (78)?
How can players who wish to contribute at the highest levels to an SDG gain legal access to scientific journal articles that require payment to access?
Does a game generate biosafety, biosecurity, or animal rights issues, in particular when players’ actions affect experimental procedures, e.g., through cloud labs?
Do we need special IRB guidelines for SDGs, and what should they include without becoming overly restrictive?
To help address these questions, SDGs offer the prospect of motivating large communities—and perhaps even specialized citizen ethicists within such communities—to continuously provide diverse perspectives on what is acceptable or not for each project. Such crowdsourcing of ethical decisions has not yet been instantiated in an SDG.
3.2. Data-Oriented Games
Data-oriented games address a different kind of research gap from problem-oriented games. In numerous areas of biomedical research, large data sets are needed to solve fundamental problems but are difficult to create or curate. Data-oriented SDGs ask players to gather, classify, or otherwise annotate data; these projects include game elements that are often inspired by problem-oriented games like Foldit but also draw from the longer lineage of citizen science and crowdsourcing projects that predate SDGs. Here, we describe the contributions of data-oriented games in several disciplines.
3.2.1. Pathology.
Throughout biomedical research, increased automation of optical microscopy is resulting in large collections of images at the cellular and subcellular level. These data sets pose challenges for classification and annotation, and several SDGs have piloted the gamification of these tasks for human players. Two SDGs have tested whether players might replace expert pathologists in malaria detection based on microscope images of blood smears. In proof-of-concept studies, MalariaSpot players have been able to accurately carry out parasite count (17) and even discriminate between Plasmodium species and intraerythrocytic forms (79), with excellent accuracies achieved upon taking the consensus across players. Mavandadi et al. (18, 80) have used the BioGames platform to demonstrate that gamers can reach sensitivities and accuracies comparable to experts. These platforms have not yet been compared to newer deep learning methods for automated computational malaria detection (81). Many nongamified crowdsourcing platforms for annotating optical microscopy images have also been described (82-84); it remains unclear whether gamifying the image classification experience offers research advantages beyond these nongamified crowdsourcing platforms.
3.2.2. Neuroscience.
Beyond applications to pathology, visual classification and annotation problems arise in numerous large-scale cellular biology surveys. Here, the data are not from different patients but have been collected through systematic, deep surveys on one particular animal or one particular cell line. EyeWire, the first SDG in neuroscience, provides an impressive example of a data annotation project (20). The EyeWire interface presents 3D data based on serial electron microscopy of animal brains and trains players to identify subvolumes of the data that correspond to separate neurons (Figure 3c). When compared to gold standard segmentations determined through community-wide consensus and expert refinement, experienced individual players achieved higher precision and recall than a deep convolutional neural network algorithm developed for the problem (20). EyeWire-traced reconstructions of OFF-type starburst amacrine cells and bipolar cells informed a model of how directions of moving stimuli are integrated from retinal inputs (20). Other projects that crowdsource the annotation of neurological microscopy data sets have been launched, including the SDG Mozak and the citizen science project Stall Catchers. As with the pathology examples discussed above, it will be important to compare results to more recent algorithmic methods for the same visual tasks, which have been improving at an impressive pace (85).
3.2.3. Cellular biology.
Recent studies are beginning to pilot rigorous comparisons and integration of deep learning computational methods and data-oriented SDGs. Project Discovery rewarded players of the massively multiplayer role-playing game (MMRPG) EVE Online for classifying subcellular localization patterns of many proteins, based on public images in the Cell Atlas of the Human Protein Atlas (25). When compared to expert consensus classifications, player classifications were similar in accuracy to a deep neural network, Localization Cellular Automation Tool (Loc-CAT), developed for the same task. However, both players and Loc-CAT—even when integrated together through a novel transfer learning framework—were less accurate than individual experts from the Cell Atlas project in randomized blind annotation tests. Nevertheless, Project Discovery players identified several new classes of organelle localization beyond the 20 classes predefined by experts for the task, including a cytoophidium cellular structure (Figure 3d). This organelle class and four other Project Discovery classes are being included in future releases of the Cell Atlas (25).
3.2.4. Biomedical knowledge organization.
The biomedical literature is vast. The online game Dizeez has shown that scientifically literate participants in an SDG might establish gene-to-disease links that are not yet curated into resources like Gene Wiki (86). Mark2Cure seeks further volunteer help in combing the literature (87). These projects have not yet been compared head-to-head to algorithmic literature-mining approaches.
3.2.5. Related work: data gathering.
Decades of citizen science projects in ecology and geography have demonstrated how large research data sets can be collected by large networks of volunteers. Newer projects make use of not just human observations but state-of-the-art biotechnologies, including molecular genotyping of flora and fauna (the International Barcode of Life) (88) and genome sequencing and electron microscopy in the SEA-PHAGES (Science Education Alliance Phage Hunters Advancing Genomics and Evolutionary Science) project (89). Sea Hero Quest is the first project that explicitly gamifies large-scale scientific data collection, asking 2.5 million players to play a mobile game testing their spatial ability (90). The project has provided researchers with initial global benchmarks on human navigational skills, demonstrating, for example, that such skills are clustered according to economic wealth and gender inequalities globally (90). Asking players of future SDGs to generate or donate data about themselves is an intriguing frontier, but one that also challenges the ethical framework for biomedical research, especially in the context of for-profit ventures (see the sidebar titled Ethical Questions in Scientific Discovery Games).
3.2.6. Future data-oriented games: genomics.
Large-scale genomics efforts generate petabytes of DNA and RNA sequencing data (91), and SDGs have emerged to investigate these data. Colony B asks players to cluster bacterial 16S ribosomal RNA collections sequenced from different people in the American Gut open science project (92). There are crowdsourcing projects aiming to accelerate discovery of gene signatures from publicly available data sets, including CREEDS (Crowd Extracted Expression of Differential Signatures) (93) and STAR-GEO (Search Tag Analyze Resource for the Gene Expression Omnibus) (94), but there are not yet efforts to turn these into discovery games.
4. EMERGENT BEHAVIOR OF PLAYER COMMUNITIES
For SDGs that have successfully solved scientific problems, how do the players actually do it? Thanks to detailed reports of player demographics and specific contributions, common themes are emerging across SDG player communities that we discuss in this section.
4.1. Sizes of Player Communities
Many thousands of active players can be engaged in an SDG, well beyond the size of typical biomedical research efforts. For example, Foldit, EyeWire, and Eterna each attained more than 100,000 registered players within a few years of launch. More recently, Project Discovery has leveraged the MMRPG community of EVE Online; embedding classification tasks for high-resolution immunofluorescence cell images as an in-game mission brought in 322,000 players within one year (25). As has been reported by projects ranging from Fraxinus to EyeWire, players tend to arrive in bursts timed with press releases or other media events such as TV programs, podcasts, and features in YouTube science channels (Figure 4a) (20, 63); readers are also referred to Reference 95. The audiences for interactive discovery games are similar in size to the networks of hundreds of thousands of volunteers for distributed computing projects.
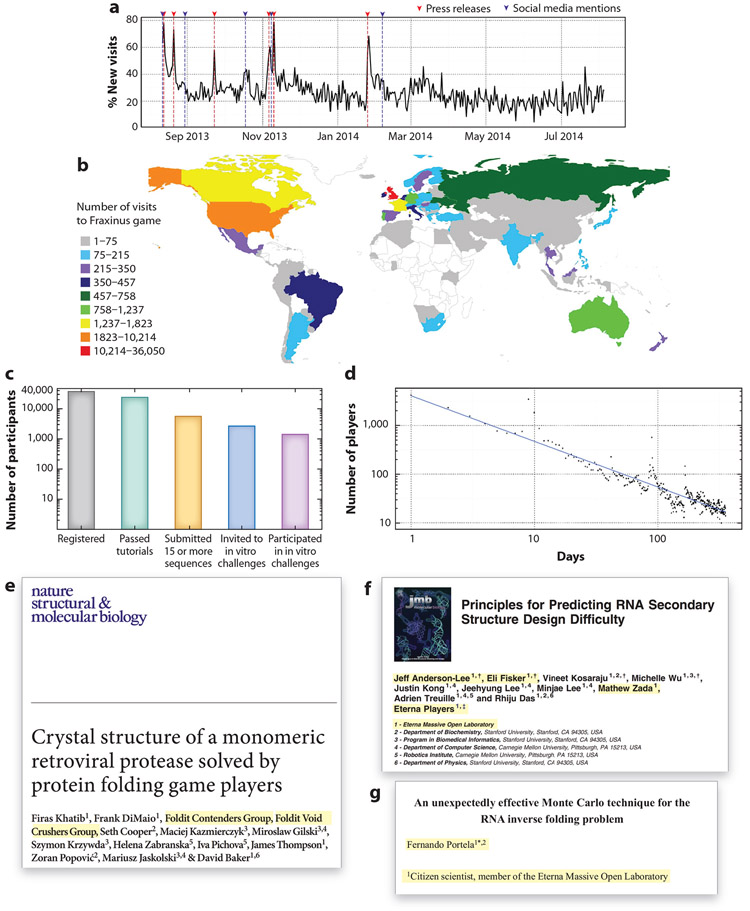
Figure 4.
Emergent properties of scientific discovery game (SDG) player communities. (a) Spikes in new visits to the SDG Fraxinus correspond to press releases (red) and social media mentions (blue). (b) The global distribution of Fraxinus game players. (c) Drops in player counts for progressively more involved tasks in the SDG Eterna (5). (d) Drops in activity in days since Fraxinus game release. (e–g) Examples of player contributions highlighted (yellow) in author lists and affiliations of scientific manuscripts (16, 96, 97). Panels a, b, and d adapted with permission from Reference 63.
4.2. Diversity of Scientific Discovery Game Players
The demographic diversity within SDG player communities can be compared to analogous numbers in nongame citizen science projects and video games. Surveys of Eterna and EyeWire players show that their communities skew male, but with significant female participation (25% and 38% female, respectively) (98). Notably, female players form the majority of the top 100 players of EyeWire (20). These female participation numbers are somewhat higher than the percentage of players in the (nongame) Zooniverse citizen science projects (17%) (99) and closer to industry estimates of the percentage of US video gamers who are female (41%) (100).
The ages of SDG players are widely distributed. In Eterna, players under the age of 20 and over the age of 50 constitute significant fractions of the community, at both entry-level stages and at highly experienced levels (those who have gained lab privileges in Eterna). There is a similarly large spread of ages among participants of Zooniverse citizen science projects, which has an almost flat distribution of ages between 20 and 55 (99), and also among US players of video games in general, which show roughly equal representation in the age categories of under 18 years, 18–35 years, 36–49 years, and over 50 years (100). Anecdotally, a significant fraction—perhaps the majority—of high-level SDG players are not otherwise connected to scientific research (e.g., see Reference 101), but a systematic study of the backgrounds or career tracks of SDG participants has not yet been carried out. Counterbalancing these signs of diversity, SDGs have so far reported player communities dominated by users logging in from the United States and Europe (Figure 4b). This global geographic imbalance in SDGs is particularly noteworthy given the rise of popular (nonscientific) mobile games developed in China for Chinese-speaking users and the recent purchase of Western video game companies by Chinese technology giants like Tencent.
Why do players participate in SDG platforms? Surveys suggest various motivations. In a 1,505-player EyeWire survey, the dominant motivation was “contribution to the project” with “improvement of personal science knowledge,” “fun,” and “learning” cited nearly as often (98). A survey of 38 Foldit players suggested that “contributing to scientific research” and “interest in science” were their most important motivations; motivations relating to puzzle solving or gaming were significantly less important (102). These top motivations are similar to those seen among 11,000 volunteers of the (nongame) Galaxy Zoo project (99), where “contribute” was the most important motivation, followed by “astronomy” and “discovery,” again with “fun” being less important.
4.3. Widely Varying Extents of Player Participation
In every SDG where player participation has been evaluated, a few players typically dominate the scientific output of every project. For example, Figure 4c shows how the size of the Eterna player community dropped by an order of magnitude when going from the number of players who register to the number of players who play enough to gain the privilege of submitting solutions for synthesis. The number of scientifically valuable solutions on Eterna is dominated by less than 1% of the players who register (Figure 4c). Similarly, Fraxinus reported that 49 of 25,614 players (0.7%) contributed to half of the 74,356 sequence alignments (Figure 4d) (63). Each of these examples bears out the so-called 1% rule concerning user participation seen in crowdsourcing platforms like Wikipedia and across the Web (103). Similarly, the top 100 EyeWire players (out of over 100,000; i.e., the top 0.1% of contributors) contributed about half of all challenges completed for its first paper (20). Despite this seeming disparity between the very top contributors and more casual players, it is worth pointing out that casual players can provide a powerful sense of community and long-term memory that can sustain scientific innovation for an SDG, and anecdotally, long-term players transition between modes of casual or intense engagement. While a longitudinal study of player contributions in SDGs has not yet been carried out, Project Discovery has shown that their churn rate (32% loss of players over 3 months) appears similar to or lower than those of other challenges in the MMRPG EVE Online (25, 104).
4.4. Evolution of New Roles for Scientific Discovery Game Players
Beyond their puzzle-solving contributions, the most strongly engaged players in SDGs have often taken on special roles that were not expected at each SDG’s launch. In projects with chat interfaces such as Eterna, Foldit, and EyeWire, chat moderators now include volunteer players. For its first enzyme design project (59), Foldit established an advanced player as an intermediary between the player community and the experimental researchers, who presented players with puzzles at each stage of the design process. EyeWire awards its most dedicated players with the statuses of Scout and then Mystic; players who achieve the latter status are able to curate other players’ solutions (20). Similarly, the scripting languages for Foldit (Lua) and Eterna (EternaScript) are widely accessible to all players, but the majority of scripts are created by a subset of players comfortable with coding who progressively modify each other’s scripts and then share these tools with the larger community (54, 105).
The contributions of devoted SDG players can go beyond these specialized roles. In terms of contributions to publications, most SDGs have included players as consortium authors on publications (typically with usernames listed in supplemental materials), but in several cases, player authors or teams have been given special credit by name (Figure 4e-g). Recently, SDG players have been not just coauthors but rather lead authors of a discovery manuscript that used puzzle creation on Eterna to uncover principles of RNA secondary structure design difficulty (Figure 4f) (96). At a further extreme, two papers from Eterna have been submitted for peer review with players as the only authors (29, 97). One of these papers is a single-author manuscript in which a player coded an algorithm for automated RNA design that dramatically outperforms deep learning methods that were being developed simultaneously by professional scientists (Figure 4g; compare Reference 97 to References 96 and 106). Since 2014, Eterna has largely replaced its original academic developer team with player developers, and since 2015, Eterna players have also organized an annual academic conference (Eternacon), with approximately 30–40 attendees at each event. Overall, SDGs appear to be diverse communities that engage scientifically inclined nonexperts to take part in, and even lead, scientific research efforts.
5. CREATING AND SUSTAINING SCIENTIFIC DISCOVERY GAMES
With the launch of numerous SDGs in the last decade, some guidelines for the next generation of SDGs are emerging.
5.1. Elements of a Scientific Discovery Game
Since SDGs combine elements of large volunteer networks (citizen science), crowdsourcing, and video games, SDG development requires building and integrating each of these component platforms. In terms of citizen science elements, a comparison of relative successes across the different (nongame) Zooniverse projects documents the importance of communication between players and developers, especially through social media (107). The US government has also created a toolkit for citizen science and crowdsourcing (https://www.citizenscience.gov). For a biomedical researcher, probably the most unconventional aspect of SDG development involves transforming a research problem into a game. This transformation involves deriving game mechanics (e.g., possible moves) customized to a research problem. Beyond those specialized aspects, badges, points, and leaderboards are game elements shared across SDGs since the launch of Foldit (Figure 2). Several studies have evaluated the effectiveness of specific gaming elements within SDGs, such as awarding stars for solution quality (108) and real-time chat (109). Baaden et al. (110) have proposed concise rules for serious games in life sciences, with examples drawn from successful structural biology games.
5.2. Personnel Required to Develop a Scientific Discovery Game
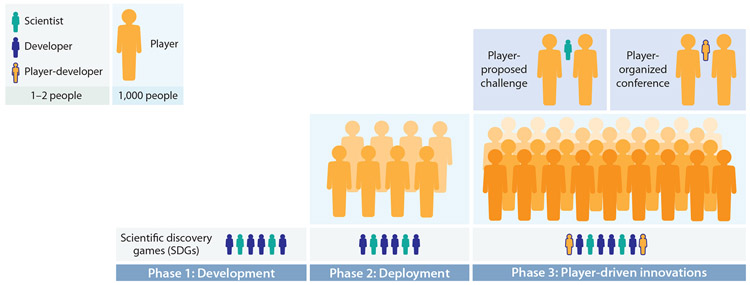
Despite the possibility of reusing prior infrastructure, the costs of developing a new SDG are still large. Figure 5 illustrates the time line for sustaining and creating Eterna, which has involved over 40 individuals in developer roles to recruit 150,000 players. Approximately 30 people were involved in the development of Eterna before and during launch, from 2009 to 2011. This team involved software developers, scientists, and two principal investigators to coordinate these teams, press releases, and funding. Some contributed through volunteering or game design coursework at Carnegie Mellon University, and others contributed through prototyping wet-lab experimental methods at Stanford University (Figure 5, phase 1). For the eight years from launch to present, a team of approximately ten developers has been active in puzzle deployment, analysis of project results, and writing of scientific papers, again with most volunteering their time (Figure 5, phase 2). Since Eterna is an academic project, nearly all of its original contributors completed their time on the project as their university training finished. Therefore, for major updates to Eterna, it has been critical in later years to recruit new developers. As noted above, these contributors have included dedicated Eterna players (Figure 5, phase 3) who design and evaluate potential changes to the game’s online interface in addition to implementing those changes as code. While Eterna’s personnel requirements are larger than most SDGs due to its experimental components, it is hard to envision launching and sustaining a successful SDG without contributions from at least ten developers.
Figure 5.
The life cycle of a scientific discovery game (SDG). Each symbol represents approximately 1,000 people (for players) or 1–2 people (for game developers, scientists, experimentalists, and player-developers) during development (phase 1), deployment (phase 2), and ongoing development (phase 3), with numbers representative of the Eterna SDG.
5.3. Financial Costs
Along with the time and personnel commitments, the financial costs of creating SDGs appear as high as or higher than typical scientific research projects. For example, funding to create and launch Eterna came mainly from laboratory startup funds to two principal investigators (R. Das and A. Treuille); the project has been sustained through grants from the US National Institutes of Health (NIH), private foundations, and various seed grants available at Stanford University. The total amount of money spent over nine years has been approximately $5 million, including indirect costs (overhead). This amount is as high as or higher than typical grants in biomedical research. For example, several teams have each been awarded $500,000 through an NIH Big Data to Knowledge initiative, but the scope of most of those projects has been to prototype proofs-of-concept rather than to deploy entire SDGs. The economic case for Internet-scale projects has been discussed for (nongamified) crowdsourcing projects (1, 95,111) but has not yet been analyzed for SDGs vis-à-vis more conventional approaches.
5.4. Practical Considerations when Starting a Scientific Discovery Game
Future SDG developers should note the possibility of reusing prior, openly accessible SDG infrastructure. Open-Phylo pioneered the reuse of an SDG architecture for researchers seeking to solve problems not envisioned by the original Phylo developers (19). In terms of repurposing existing social media networks, Fraxinus was deployed within a preexisting platform (Facebook), which provides a sophisticated infrastructure for social media and communication between developers and players. Another example of reusing infrastructure is the Project Discovery image classification project, which was embedded in EVE Online and brought in hundreds of thousands of players within one year (25). This platform was designed to enable simple reuse for other scientific projects to minimize future development costs. The initial cell biology project has now been replaced with a project for the discovery of exoplanets. New web technologies like WebAssembly (112) also enable rapid deployment of C/C++ scientific code onto the web. Finally, initiatives like Science Game Lab (113) are prototyping platforms that could share badges, points, and other gaming elements across multiple projects. Such platforms with prebuilt gaming elements could substantially lower costs of development of new SDGs in the near future, similar to how the Berkeley Open Infrastructure for Network Computing (36) and the Zooniverse platform (15) have accelerated development of distributed computing and online citizen science projects, respectively. Finally, developers are advised to consider ahead of time how they will evaluate success in their SDG. A success matrix has been proposed to help define and measure public engagement in science, resource savings, and other features of citizen science projects (107). In addition, each SDG should be able to state what constitutes the win condition for the project in terms of science, and this will be different for each project.
6. SYNERGISTIC DEVELOPMENTS FROM OTHER FIELDS
Given their highly interdisciplinary nature, SDGs are intertwined with numerous scientific and cultural trends. In this section, we highlight six newer trends that provide unique opportunities for future SDGs.
6.1. Biology Cloud Experimentation Labs
The design and interpretation of prospective wet-lab experiments are key components of biomedical science, but currently the only SDG with player-controlled experiments and integrated feedback is the massive open laboratory Eterna (2, 5). The lack of experiments in SDGs could change through emerging biology cloud labs (21, 22, 114). Commercial cloud labs enable molecular and cellular synthesis within days to weeks (21). Other biology cloud labs developed for educational applications provide feedback within seconds (23). Remote experimenters, for example, can interact with slime molds via the robot-controlled dispensing of food droplets (22), or they can remotely control directional light stimuli to evoke and study cellular phototaxis behaviors (23). These technologies can scale to millions of annual experiments at costs of cents per experiment (23). Biology cloud labs have been successfully deployed for K–12, college, and online education (22, 23, 28, 115, 116). As part of a MOOC (massive open online course), a biology cloud lab enabled the execution of key scientific practices, including experimentation, hypothesis formulation, and genuine discovery (28). This proof-of-concept suggests that real-time interactive cloud labs could be used inside an SDG, e.g., to study microswimmer biophysics (117). We propose that cloud labs that utilize highly domain-specific, high-throughput assays (e.g., References 5 and 23) are more effective for SDGs than general-purpose ones (114). Hence, while early SDGs are built on big data science and virtual interactions, we predict that direct design, execution, and evaluation of true experiments by the players will become prevalent, especially given the ever ongoing technological advances and price reduction in automated, high-throughput life science instrumentation (21, 114)—similar to the development of cloud computing (118-121).
6.2. Biotic (Video) Games
While biology cloud labs have not yet been incorporated into Internet-scale SDGs, biotic (video) games (6) show that nonexperts can engage in playful interactions with actual microscopic living matter and processes (6, 122). Biotic games are facilitated by advances in life science instrumentation, in the same way that electronic video games advance along with computer technology. For example, akin to Pac-Man, a human player can use light stimuli to manipulate the swimming direction of phototactic Euglena cells in real time while collecting virtual objects overlaid on a screen (6, 122). These games provide novel player experiences and emotions that are distinct from what is achievable with purely digital simulation games, particularly due to the semideterministic, noisy behaviors of living cells and materials (123). The design space for biotic games has been explored (123) using different biological materials (molecules, cells, and colonies) and stimuli (light, electric fields, and chemicals) (6, 122, 124-129). For the continued development of these games and potentially SDGs, researchers have conceptualized and engineered a variety of biological and digital infrastructures, such as biotic processing units (6, 23, 123). Domain-specific programming languages for interacting with living matter have also been developed (128), including cloud-based development environments for interactive biotic programs (129). It has been argued that technological milestones in biological experimentation trail equivalent milestones in electronic computation by about five decades (123). We therefore expect a variety of biotic games to emerge over coming decades. Allowing hundreds of thousands of SDG players to access these tools could radically change the SDG experience and the experimental problems that could be tackled.
6.3. Underutilized Video Game Genres
Underutilized video game genres offer another opportunity for future SDGs. Existing SDGs rely on a handful of simple genres (puzzle games; simple reward systems like points, badges, and leaderboards; and embedding the task in a story), but other game principles are possible. Project Discovery (25) has explored embedding an SDG in the MMRPG EVE Online. Such partnerships between scientific researchers and game companies can be mutually beneficial, since SDGs benefit from gaining large audiences quickly, while MMRPGs and other games are typically in continuous need of new content to retain players (104). Further extensions of SDG-MMRPG connections might take advantage of the observation that MMRPG players are notoriously good at exploring and exploiting what is possible in the game, including complex economies involving game currencies (130). In real-time strategy (RTS) games like StarCraft, players make a series of strategic decisions as they compete against other players. RTS games already present researchers with an artificial intelligence (AI) challenge (131); in principle, this rich genre could also be used to help scientists find optimal strategies for solving intractable problems. Finally, new game genres are arising with the commercialization of technologies like augmented reality, virtual reality, and passive wearables. The rise of these new user interfaces and simulation technologies could provide interesting new directions for future SDGs.
6.4. Machine Learning and Artificial Intelligence
AI offers both a challenge to and an opportunity for SDGs. Over the past decade, AI and machine learning approaches have excelled at solving diverse tasks, especially those that have been formalized into games. AI agents outperform human players in several instances, e.g., classic Atari video games (24) and the board game Go (26, 27). Importantly, some algorithms now perfect themselves through trial and error without relying on human-generated game data at all (27). For scientific discovery, we expect that as AI advances, certain types of problems will be more effectively solved directly through computer algorithms rather than through players in an SDG. Some data-oriented games may be obviated entirely. In problem-oriented games, AI agents could take over tasks from the player, such as dimensionality reduction of high-throughput data sets (132), allowing human players to focus on more conceptual tasks like defining the global structures and the desired functions of macromolecules or updating generalizable biophysical models, both of which remain difficult for AI methods. Ultimately, generating genuine scientific insight might be one of the hardest tasks for AI (133), and SDGs could provide a stimulating test bed for AI development.
6.5. Global Life Science and Biomedical Education
Population-wide scientific literacy is a key goal of modern society, and SDGs could uniquely contribute to and benefit from efforts to reach this goal. Many countries have special initiatives to promote STEM learning (134-137). For example, the United States has introduced the Next Generation Science Standards to promote inquiry-based learning, asking students to practice realistic science activities (138). Delivering such scientific practices to schools in a realistic and scaleable way is an unsolved challenge, but SDGs may provide unique and large-scale opportunities. Some SDGs, for example, have been used in K–12 and college classrooms; the PBS science program NOVA created specialized versions of SDGs tailored to middle school classrooms (http://www.pbs.org/wgbh/nova/labs/). SDGs typically have several introductory challenges that teach players about the scientific questions and available tools to make them effective in the game. Therefore, SDGs are intrinsically educational with respect to biomedical research. SDG research has involved college classroom use, e.g., for Foldit protein structure refinement (55) and HiRE-RNA fold prediction studies (58). However, there are not yet systematic studies that evaluate the impact of SDGs on formal or informal education in the way that education-focused video games are being rigorously assessed (139, 140).
6.6. Open Science
The open science movement offers numerous developments that are highly complementary to SDGs. As SDG players become experts, it becomes imperative that they have convenient access to the primary literature. The trend toward open access publishing aims to do just that: to make the scientific literature more accessible to both professionals and lay people (141). The do-it-yourself biology and biohacker movements (e.g., 142, 143) promote the decentralization of biological experimentation, which could give SDG players more nuanced control over their experiments. SDG players may also be empowered through new approaches to research funding, such as crowdfunding (144). Such mechanisms would enable groups of players to sponsor research that they could carry out themselves.
7. PERSPECTIVE
We conclude with a summary and speculations about future developments for SDGs in biomedical research.
7.1. What We Have Learned
SDGs constitute a new medium for biomedical research. Building on the previous decade’s advances in distributed computing, citizen science projects, crowdsourcing, and online games, SDGs launched since 2008 engage hundreds of thousands of volunteer players in biomedical research. Genuine research contributions now span numerous fields of biology and bioengineering: protein crystallographic applications (16, 55), RNA design refined through wet-lab experiments (5), and the segmentation of high-resolution 3D neuronal images (20). In each of these cases, publications have reported that an SDG platform found novel solutions to difficult problems that previously eluded expert scientists, computational algorithms, or both. For areas including protein sequence alignment (63, 64) and interpretation of pathology slides (17, 18), there are proofs-of-concept that SDGs may be useful, but there is not yet strong evidence of improvements relative to computational methods or nongame citizen science approaches.
SDG player communities share certain features. Players appear to be diverse in gender, age, and motivation, although the concentration of players in English-speaking countries indicates that SDGs remain restricted geographically. Many players are casual users, while a much smaller subset of enthusiasts drive the majority of scientific progress. These particularly enthusiastic players have made surprising contributions, sometimes meriting authorship and even lead authorship in scientific peer-reviewed papers (96). In some SDGs, game content and even core development tasks have been largely taken over by players, further demonstrating the involvement and ownership of SDGs by volunteer players.
Biomedical researchers interested in gamifying new scientific problems can build on the growing knowledge of what it takes to create and sustain an SDG. The number of personnel, the time, and the financial cost required to develop SDGs appear similar to or greater than typical biomedical research projects. At the same time, there is a canon of gaming elements common across most SDGs, including tutorials, leaderboards, badges, points, chats, and forums. Shared programming tools, database infrastructures, and game reward infrastructures may accelerate the development of the next generation of SDGs.
7.2. What is Next?
The most successful video games reach tens of millions of players. Can a well-designed SDG reach the same number, perhaps by integrating new technologies, new game genres, or existing platforms? How can a community form around SDGs given that it involves very interdisciplinary expertise, e.g., from biomedical researchers, game designers, gaming academics, human–computer interaction design, and citizen scientists and volunteers? Given advances in biological–digital integration, might the next neuroscience SDGs include actual optogenetic control of neuronal circuits? Could the next cancer-modeling SDGs make recommendations for patient therapies in real time? What are the ethical aspects for SDGs (see the sidebar titled Ethical Questions in Scientific Discovery Games)? These are just a few of the many open questions in this nascent field.
SDGs offer unique opportunities for extraordinarily ambitious biomedical research. Entire game genres, modes of interaction between large human communities and AI systems, and remote-controlled cloud laboratories remain to be explored. While some of these prospects sound like science fiction now, we expect that the next generation of SDGs will hold many surprises.
SUMMARY POINTS.
Multiple scientific discovery games (SDGs) have been successfully developed and sustained in the ten years following the 2008 launch of Foldit.
SDGs have published discoveries for numerous biomedical research problems; some of these advances in macromolecule structure modeling, macromolecule design, and sequence alignment could not be obtained with prior computational or expert approaches.
Massive open laboratories like Eterna show that SDGs can include prospective wet-lab experiments as part of their gameplay.
SDGs are tackling the curation and annotation of biomedical research data sets, including large corpora of cellular images, genomic data, and areas of the scientific literature.
Communities of SDG players appear to be diverse in gender and age but are still largely limited to English-speaking areas of the world.
A small percentage of SDG players contribute at unusually high levels, providing most of the solutions in problem-oriented games, organizing conferences, and even leading the writing of manuscripts for formal peer review.
A standard canon of game elements including tutorials, scores, leaderboards, badges, chats, and forums is shared across numerous successful SDGs and provides a parts list for researchers wanting to launch new SDGs.
The costs of developing SDGs are similar to or higher than conventional biomedical research projects, but scientific funding agencies are recognizing their promise with specialized requests for applications.
FUTURE ISSUES.
SDGs impact individual participants’ scientific knowledge and training for future science careers as well as the public understanding of science. How can these broader educational impacts be evaluated?
What are the relevant ethical frameworks for SDGs regarding player attribution, the protection of human subjects’ rights, player compensation, biosecurity, and open access to science?
Could participation in games provide a completely nonacademic route to engaging in biomedical research and supporting open science?
What new toolboxes, guides, academic journals, and conferences might make the development of new SDGs easier?
Can SDGs have the same social and economic impact as predecessor technologies such as Internet-scale video games (which reach billions of people) and crowdsourcing (Wikipedia)?
Can SDGs implement more complex gaming genres such as real-time strategy games?
What is the optimal synergy between constantly advancing artificial intelligence techniques and large numbers of human volunteers?
Can biology cloud labs make real-time experiments available in SDGs?
ACKNOWLEDGMENTS
We thank S. Cooper and M. Zada for early discussions and J. Anderson-Lee, E. Fisker, M. Gotrik, E. Lundberg, R. Moretti, K. Shih, and R. Starr for insightful comments on the manuscript. We thank Claire Baert for putting together www.citizensciencegames.com, which we used as a resource. We acknowledge support from the Stanford Discovery Innovation Fund (to I.H.R.-K. and R.D.), a Stanford School of Medicine Teaching Innovation Grant (to R.D.), and the US National Institutes of Health grant R35 GM122579 (to R.D.).
Glossary
- Game
generally, a challenging human activity that has rules, goals, and uncertain outcomes
- Scientific discovery game (SDG)
an interactive platform (curated by scientific domain experts) where lay people contribute to research through game-like activities, thereby leveraging human creativity at a massive scale
- Crowdsourcing
outsourcing a task from within an institution to an undefined, large group of external people, often in the form of an open call
- Citizen science
scientific activities carried out by volunteers who are not formally employed in or trained by professional research institutions
- Screensaver
a program that updates display pixels with animations when computers are idle to prevent burnout of cathode-ray tubes and other types of display screens
- Gamification
the application of elements of game playing (e.g., point scoring, competition with others, rules of play) to other areas of activity
- NP-hard
a class of computational problems at least as hard as the problems that can be solved with computational complexity scaling as a polynomial function of the problem length
- Crystallographic phasing
diffraction data on crystals enable protein structure determination but miss half the information (the scattering phases); a sufficiently accurate model can enable recovery of these missing phases through molecular replacement
- Reality gap
the gap between computational models of the real world and experimental reality
- Massive open laboratory
an Internet platform that enables anyone to participate in wet-lab experiments, steered through play of a scientific discovery game
- IRB
the Institutional Review Board is an administrative body that protects the rights and welfare of participants in scientific studies
- Biology cloud lab
a laboratory enabling users to perform biology experiments without direct physical contact; cloud labs can be fully or partially automated, and turnaround time depends on the biological assay
- Biotic (video) games
games using modern biotechnology to enable human players to interact with real biological materials or processes at micro- or nanoscales
- Biotic processing unit (BPU)
biotechnological analog of digital microprocessors that houses, actuates, and measures microbiological systems robustly over the long term and where digital stimulus instructions convert analog biological behavior to digital output
- Machine learning
computational and statistical techniques that allow computer systems to progressively improve performance on a specific task (learn) from data without being explicitly programmed
- STEM
science, technology, engineering, and mathematics; the acronym is typically used in educational contexts, sometimes even extended to “STEAM” to include arts
- Formal and informal education
education can be categorized as formal (at the K-12, college, and PhD level) or informal (after-school programs, museums, adult learners, and do-it-yourself biology and hobbyist communities)
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Good BM, Su AI. 2013. Crowdsourcing for bioinformatics. Bioinformatics 29:1925–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Treuille A, Das R. 2014. Scientific rigor through videogames. Trends Biochem. Sci 39:507–9 [DOI] [PubMed] [Google Scholar]
- 3.Curtis V 2014. Online citizen science games: opportunities for the biological sciences. Appl. Transl. Genom 3:90–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Roskams J, Popović Z. 2016. Power to the people: addressing big data challenges in neuroscience by creating a new cadre of citizen neuroscientists. Neuron 92:658–64 [DOI] [PubMed] [Google Scholar]
- 5.Lee J, Kladwang W, Lee M, Cantu D, Azizyan M, et al. 2014. RNA design rules from a massive open laboratory. PNAS 111:2122–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riedel-Kruse IH, Chung AM, Dura B, Hamilton AL, Lee BC. 2011. Design, engineering and utility of biotic games. Lab Chip 11:14–22 [DOI] [PubMed] [Google Scholar]
- 7.Mayer A 2010. Phenology and citizen science: Volunteers have documented seasonal events for more than a century, and scientific studies are benefiting from the data. BioScience 60:172–75 [Google Scholar]
- 8.Beberg AL, Ensign DL, Jayachandran G, Khaliq S, Pande VS. 2009. Folding@ home: lessons from eight years of volunteer distributed computing. In 2009 IEEE International Symposium on Parallel & Distributed Processing. Los Alamitos, CA: IEEE Comput. Soc. [Google Scholar]
- 9.Korpela EJ. 2012. Seti@ home, BOINC, and volunteer distributed computing. Annu. Rev. Earth Planet. Sci 40:69–87 [Google Scholar]
- 10.Cole H, Griffiths MD. 2007. Social interactions in massively multiplayer online role-playing gamers. Cyberpsychol. Behav 10:575–83 [DOI] [PubMed] [Google Scholar]
- 11.Das R, Qian B, Raman S, Vernon R, Thompson J, et al. 2007. Structure prediction for CASP7 targets using extensive all-atom refinement with Rosetta@ home. Proteins 69:118–28 [DOI] [PubMed] [Google Scholar]
- 12.Lintott CJ, Schawinski K, Slosar A, Land K, Bamford S, et al. 2008. Galaxy Zoo: morphologies derived from visual inspection of galaxies from the Sloan Digital Sky Survey. Mon. Not. R. Astron. Soc 389:1179–89 [Google Scholar]
- 13.Cooper S, Khatib F, Treuille A, Barbero J, Lee J, et al. 2010. Predicting protein structures with a multiplayer online game. Nature 466:756–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt M 2008. Diffusion of synthetic biology: a challenge to biosafety. Syst. Synth. Biol 2:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simpson R, Page KR, De Roure D. 2014. Zooniverse: observing the world’s largest citizen science platform. In Proceedings of the 23rd International Conference on World Wide Web, pp. 1049–54. New York: Assoc. Comput. Mach. [Google Scholar]
- 16.Khatib F, DiMaio F, Cooper S, Kazmierczyk M, Gilski M, et al. 2011a. Crystal structure of a monomeric retroviral protease solved by protein folding game players. Nat. Struct. Mol. Biol 18:1175–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luengo-Oroz MA, Arranz A, Frean J. 2012. Crowdsourcing malaria parasite quantification: an online game for analyzing images of infected thick blood smears. J. Med. Internet Res 14:e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mavandadi S, Dimitrov S, Feng S, Yu F, Sikora U, et al. 2012a. Distributed medical image analysis and diagnosis through crowd-sourced games: a malaria case study. PLOS ONE 7:e37245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwak D, Kam A, Becerra D, Zhou Q, Hops A,et al. 2013. Open-Phylo: a customizable crowd-computing platform for multiple sequence alignment. Genome Biol. 14:R116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim JS, Greene MJ, Zlateski A, Lee K, Richardson M, et al. 2014. Space–time wiring specificity supports direction selectivity in the retina. Nature 509:331–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hayden EC. 2014. The automated lab. Nature 516:131–32 [DOI] [PubMed] [Google Scholar]
- 22.Hossain Z, Jin X, Bumbacher EW, Chung AM, Koo S, et al. 2015. Interactive cloud experimentation for biology: an online education case study. In Proceedings of the 33rd Annual ACM Conference on Human Factors in Computing Systems, pp. 3681–90. New York: Assoc. Comput. Mach. [Google Scholar]
- 23.Hossain Z, Bumbacher EW, Chung AM, Kim H, Litton C, et al. 2016. Interactive and scalable biology cloud experimentation for scientific inquiry and education. Nat. Biotechnol 34:1293–98 [DOI] [PubMed] [Google Scholar]
- 24.Mnih V, Kavukcuoglu K, Silver D, Rusu AA, Veness J, et al. 2015. Human-level control through deep reinforcement learning. Nature 518:529–33 [DOI] [PubMed] [Google Scholar]
- 25.Sullivan DP, Winsnes CF, Åkesson L, Hjelmare M, Wiking M, et al. 2018. Deep learning is combined with massive-scale citizen science to improve large-scale image classification. Nat. Biotechnol 36:820–28 [DOI] [PubMed] [Google Scholar]
- 26.Silver D, Huang A, Maddison CJ, Guez A, Sifre L, et al. 2016. Mastering the game of Go with deep neural networks and tree search. Nature 529:484–89 [DOI] [PubMed] [Google Scholar]
- 27.Silver D, Schrittwieser J, Simonyan K, Antonoglou I, Huang A, et al. 2017. Mastering the game of Go without human knowledge. Nature 550:354–59 [DOI] [PubMed] [Google Scholar]
- 28.Hossain Z, Bumbacher E, Brauneis A, Diaz M, Saltarelli A, et al. 2017a. Design guidelines and empirical case study for scaling authentic inquiry-based science learning via open online courses and interactive biology cloud labs. Int. J. Artif. Intell. Educ 28:478–507 [Google Scholar]
- 29.Wellington-Oguri R, Fisker E, Wiley M, Zada M. 2018. Evidence of an unusual poly(A) RNA signature detected by high-throughput chemical mapping. bioRxiv 281147. 10.1101/281147 [DOI] [PubMed] [Google Scholar]
- 30.Nat. Audubon Soc 2019. Annual summaries of the Christmas Bird Count, 1901–present. https://www.audubon.org/content/american-birds-annual-summary-christmas-bird-count
- 31.Reid P, Colebrook J, Matthews J, Aiken J, Contin. Plankton Rec. Team. 2003. The Continuous Plankton Recorder: concepts and history, from plankton indicator to undulating recorders. Prog. Oceanogr 58:117–73 [Google Scholar]
- 32.Vezzulli L, Reid P. 2003. The CPR survey (1948–1997): a gridded database browser of plankton abundance in the North Sea. Prog. Oceanogr 58:327–36 [Google Scholar]
- 33.Sullivan BL, Wood CL, Ilif FMJ, Bonney RE, Fink D, Kelling S. 2009. eBird: a citizen-based bird observation network in the biological sciences. Biol. Conserv 142:2282–92 [Google Scholar]
- 34.Robinson KL, Luo JY, Sponaugle S, Guigand C, Cowen RK. 2017. A tale of two crowds: public engagement in plankton classification. Front. Mar. Sci 4:82 [Google Scholar]
- 35.Kanefsky B, Barlow NG, Gulick VC. 2001. Can distributed volunteers accomplish massive data analysis tasks? Paper presented at the 32nd Annual Lunar and Planetary Science Conference, Houston, Texas, March 12–16, Abstr. 1272 [Google Scholar]
- 36.Anderson DP. 2004. BOINC: a system for public-resource computing and storage. In Fifth IEEE/ACM International Workshop on Grid Computing, ed. Buyya R, pp. 4–10. Los Alamitos, CA: IEEE Comput. Soc. [Google Scholar]
- 37.Howe J 2006. The rise of crowdsourcing. Wired Magazine, Vol. 14. https://www.wired.com/2006/06/crowds/ [Google Scholar]
- 38.Ipeirotis PG. 2010. Analyzing the Amazon Mechanical Turk marketplace. XRDS: Crossroads, The ACM Magazine for Students,Vol. 17, pp. 16–21 [Google Scholar]
- 39.Good BM, Clarke EL, de Alfaro L, Su AI. 2011. The Gene Wiki in 2011: community intelligence applied to human gene annotation. Nucleic Acids Res. 40:D1255–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gardner PP, Daub J, Tate J, Moore BL, Osuch IH, et al. 2010. Rfam: Wikipedia, clans and the “decimal” release. Nucleic Acids Res. 39:D141–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schuster SC. 2007. Next-generation sequencing transforms today’s biology. Nat. Methods 5:16–18 [DOI] [PubMed] [Google Scholar]
- 42.Pham DL, Xu C, Prince JL. 2000. Current methods in medical image segmentation. Annu. Rev. Biomed. Eng 2:315–37 [DOI] [PubMed] [Google Scholar]
- 43.Dogangil G, Davies B, Rodriguez y Baena F. 2010. A review of medical robotics for minimally invasive soft tissue surgery. Proc. Inst. Mech. Eng. H 224:653–79 [DOI] [PubMed] [Google Scholar]
- 44.Thul PJ, Åkesson L, Wiking M, Mahdessian D, Geladaki A, et al. 2017. A subcellular map of the human proteome. Science 356:eaal3321. [DOI] [PubMed] [Google Scholar]
- 45.Prill RJ, Saez-Rodriguez J, Alexopoulos LG, Sorger PK, Stolovitzky G. 2011. Crowdsourcing network inference: the DREAM predictive signaling network challenge. Sci. Signal 4:mr7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moult J 2005. A decade of CASP: progress, bottlenecks and prognosis in protein structure prediction. Curr. Opin. Struct. Biol 15:285–89 [DOI] [PubMed] [Google Scholar]
- 47.Daneshjou R, Wang Y, Bromberg Y, Bovo S, Martelli PL, et al. 2017. Working toward precision medicine: predicting phenotypes from exomes in the critical assessment of genome interpretation (CAGI) challenges. Hum. Mutat 38:1182–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saez-Rodriguez J, Costello JC, Friend SH, Kellen MR, Mangravite L, et al. 2016. Crowdsourcing biomedical research: leveraging communities as innovation engines. Nat. Rev. Genet 17:470–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marchand A, Hennig-Thurau T. 2013. Value creation in the video game industry: industry economics, consumer benefits, and research opportunities. J. Interact. Mark 27:141–57 [Google Scholar]
- 50.Aarseth E 2001. Computer game studies, year one. Game Stud. 1. http://www.gamestudies.org/0101/editorial.html [Google Scholar]
- 51.Von Ahn L 2006. Games with a purpose. Computer 39:92–94 [Google Scholar]
- 52.Das R, Baker D. 2008. Macromolecular modeling with Rosetta. Annu. Rev. Biochem 77:363–82 [DOI] [PubMed] [Google Scholar]
- 53.Pierce NA, Winfree E. 2002. Protein design is NP-hard. Protein Eng. 15:779–82 [DOI] [PubMed] [Google Scholar]
- 54.Khatib F, Cooper S, Tyka MD, Xu K, Makedon I, et al. 2011b. Algorithm discovery by protein folding game players. PNAS 108:18949–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Horowitz S, Koepnick B, Martin R, Tymieniecki A, Winburn AA, et al. 2016. Determining crystal structures through crowdsourcing and coursework. Nat. Commun 7:12549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khoury GA, Liwo A, Khatib F, Zhou H, Chopra G, et al. 2014. Wefold: a coopetition for protein structure prediction. Proteins 82:1850–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jorda J, Sawaya MR, Yeates TO. 2014. Crowdphase: crowdsourcing the phase problem. Acta Crystallogr. D 70:1538–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mazzanti L, Doutreligne S, Gageat C, Derreumaux P, Taly A, et al. 2017. What can human-guided simulations bring to RNA folding? Biophys. J. 113:302–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eiben CB, Siegel JB, Bale JB, Cooper S, Khatib F, et al. 2012. Increased Diels-Alderase activity through backbone remodeling guided by Foldit players. Nat. Biotechnol 30:190–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bonnet É, Rzążewski P, Sikora F. 2018. Designing RNA secondary structures is hard. In Proceedings of the 22nd Annual International Conference on Research in Computational Molecular Biology (RECOMB), ed. BJ Raphael, pp. 248–50. Cham, Switz.: Springer Int. [Google Scholar]
- 61.Barone J, Bayer C, Copley R, Barlow N, Burns M, et al. 2015. Nanocrafter: design and evaluation of a DNA nanotechnology game. In Proceedings of the 10th International Conference on the Foundations of Digital Games (FDG 2015). http://www.fdg2015.org/papers/fdg2015_paper_74.pdf [Google Scholar]
- 62.Wang L, Jiang T. 1994. On the complexity of multiple sequence alignment. J. Comput. Biol 1:337–48 [DOI] [PubMed] [Google Scholar]
- 63.Rallapalli G, Saunders DG, Yoshida K, Edwards A, Lugo CA, et al. 2015. Cutting edge: lessons from Fraxinus, a crowd-sourced citizen science game in genomics. eLife 4:e07460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kawrykow A, Roumanis G, Kam A, Kwak D, Leung C, et al. 2012. Phylo: a citizen science approach for improving multiple sequence alignment. PLOS ONE 7:e31362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D’Souza LM, et al. 2002. The Comparative RNA Web (CRW) Site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinform. 3:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Waldispühl J, Kam A, Gardner PP. 2014. Crowdsourcing RNA structural alignments with an online computer game. Pac. Symp. Biocomput 20:330–41 [PubMed] [Google Scholar]
- 67.Tacchella A, Romano S, Ferraldeschi M, Salvetti M, Zaccaria A, et al. 2017. Collaboration between a human group and artificial intelligence can improve prediction of multiple sclerosis course: a proof-of-principle study. F1000Research 6:2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Harvey H, Havard M, Magnus D, Cho MK, Riedel-Kruse IH. 2014. Innocent fun or “microslavery”? An ethical analysis of biotic games. Hastings Center Rep. 44:38–46 [DOI] [PubMed] [Google Scholar]
- 69.Tsueng G, Kumar A, Nanis M, Su AI. 2018. Aligning interests: integrating citizen science efforts into schools through service requirements. bioRxiv 304766. 10.1101/304766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hughes AJ, Mornin JD, Biswas SK, Beck LE, Bauer DP, et al. 2018. Quanti.us: a tool for rapid, flexible, crowd-based annotation of images. Nat. Methods 15:587–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Alkhatib A, Bernstein MS, Levi M. 2017. Examining crowd work and gig work through the historical lens of piecework. In Proceedings of the 2017 CHI Conference on Human Factors in Computing Systems, pp. 4599–616. New York: Assoc. Comput. Mach. [Google Scholar]
- 72.Guerrini CJ, Majumder MA, Lewellyn MJ, McGuire AL. 2018. Citizen science, public policy. Science 361:134–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Scassa T, Chung H. 2015. Typology of citizen science projects from an intellectual property perspective. Policy Memo, Commons Lab, Wilson Cent., Washington, DC. https://www.wilsoncenter.org/publication/typology-citizen-science-projects-intellectual-property-perspective [Google Scholar]
- 74.Kuss DJ, Griffiths MD, Karila L, Billieux J. 2014. Internet addiction: a systematic review of epidemiological research for the last decade. Curr. Pharm. Des 20:4026–52 [DOI] [PubMed] [Google Scholar]
- 75.Anderson EL, Steen E, Stavropoulos V. 2017. Internet use and problematic Internet use: a systematic review of longitudinal research trends in adolescence and emergent adulthood. Int. J. Adolesc. Youth 22:430–54 [Google Scholar]
- 76.Mortensen TE. 2016. Anger, fear, and games: the long event of # GamerGate. Games Cult. 13(8):787–806 [Google Scholar]
- 77.Newman L, Browne-Yung K, Raghavendra P, Wood D, Grace E. 2017. Applying a critical approach to investigate barriers to digital inclusion and online social networking among young people with disabilities. Inf. Syst. J 27:559–88 [Google Scholar]
- 78.Martínez-Pérez B, De La Torre-Díez I, López-Coronado M. 2015. Privacy and security in mobile health apps: a review and recommendations. J. Med. Syst 39:181. [DOI] [PubMed] [Google Scholar]
- 79.Ortiz-Ruiz A, Postigo M, Gil-Casanova S, Cuadrado D, Bautista JM, et al. 2018. Plasmodium species differentiation by non-expert on-line volunteers for remote malaria field diagnosis. Malaria J. 17:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mavandadi S, Dimitrov S, Feng S, Yu F, Yu R, et al. 2012b. Crowd-sourced biogames: managing the big data problem for next-generation lab-on-a-chip platforms. Lab Chip 12:4102–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Poostchi M, Silamut K, Maude R, Jaeger S, Thoma G. 2018. Image analysis and machine learning for detecting malaria. Transl. Res 194:36–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.dos Reis FJC, Lynn S, Ali HR, Eccles D, Hanby A, et al. 2015. Crowdsourcing the general public for large scale molecular pathology studies in cancer. EBioMedicine 2:681–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Irshad H, Oh EY, Schmolze D, Quintana LM, Collins L, et al. 2017. Crowdsourcing scoring of immunohistochemistry images: evaluating performance of the crowd and an automated computational method. Sci. Rep 7:43286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lawson J, Robinson-Vyas RJ, McQuillan JP, Paterson A, Christie S, et al. 2017. Crowdsourcing for translational research: analysis of biomarker expression using cancer microarrays. Br. J. Cancer 116:237–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.LeCun Y, Bengio Y, Hinton G. 2015. Deep learning. Nature 521:436–44 [DOI] [PubMed] [Google Scholar]
- 86.Loguercio S, Good BM, Su AI. 2013. Dizeez: an online game for human gene-disease annotation. PLOS ONE 8:e71171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tsueng G, Nanis M, Fouquier J, Good B, Su A. 2016. Citizen science for mining the biomedical literature. Citiz. Sci 1:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ratnasingham S, Hebert PD. 2007. BOLD: the Barcode of Life Data System (www.barcodinglife.org). Mol. Ecol. Notes 7:355–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hatfull GF. 2015. Innovations in undergraduate science education: going viral. J. Virol 89:8111–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Coutrot A, Silva R, Manley E, de Cothi W, Sami S,et al. 2018. Global determinants of navigation ability. Curr. Biol 28:2861–66 [DOI] [PubMed] [Google Scholar]
- 91.Cochrane G, Akhtar R, Bonfield J, Bower L, Demiralp F, et al. 2008. Petabyte-scale innovations at the European Nucleotide Archive. Nucleic Acids Res. 37:D19–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McDonald D, Hyde E, Debelius JW, Morton JT, Gonzalez A, et al. 2018. American Gut: an open platform for citizen science microbiome research. mSystems 3:e00031–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang Z, Monteiro CD, Jagodnik KM, Fernandez NF, Gundersen GW, et al. 2016. Extraction and analysis of signatures from the Gene Expression Omnibus by the crowd. Nat. Commun 7: 12846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hadley D, Pan J, El-Sayed O, Aljabban J, Aljabban I, et al. 2017. Precision annotation of digital samples in NCBI’s Gene Expression Omnibus. Sci. Data 4:170125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Franzoni C, Sauermann H. 2014. Crowd science: the organization of scientific research in open collaborative projects. Res. Policy 43:1–20 [Google Scholar]
- 96.Anderson-Lee J, Fisker E, Kosaraju V, Wu M, Kong J, et al. 2016. Principles for predicting RNA secondary structure design difficulty. J. Mol. Biol 428:748–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Portela F 2018. An unexpectedly effective Monte Carlo technique for the RNA inverse folding problem. bioRxiv 345587. 10.1101/345587 [DOI] [Google Scholar]
- 98.Tinati R, Luczak-Roesch M, Simperl E, Hall W. 2016. Because science is awesome: studying participation in a citizen science game. In Proceedings of the 8th ACM Conference on Web Science (WebSci ’16), pp. 45–54. New York: Assoc. Comput. Mach. [Google Scholar]
- 99.Raddick JM, Bracey G, Gay PL, Lintott CJ, Murray P, et al. 2010. Galaxy Zoo: exploring the motivations of citizen scientist volunteers. Astron. Educ. Rev 9(1):010103–1 [Google Scholar]
- 100.Entertain. Softw. Assoc. 2017. Essential facts about the computer and video game industry. Surv. Rep, Entertain. Softw. Assoc., Washington, DC [Google Scholar]
- 101.Rowles TA. 2013. Power to the people: Does Eterna signal the arrival of a new wave of crowd-sourced projects? BMC Biochemistry 14:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Curtis V 2015. Motivation to participate in an online citizen science game: a study of Foldit. Sci. Commun 37:723–46 [Google Scholar]
- 103.Cooper C 2013. Zen in the art of citizen science: apps for collective discovery and the 1 percent rule of the web. Scientific American Blog, September. 11. https://blogs.scientificamerican.com/guest-blog/zen-in-the-art-of-citizen-science-apps-for-collective-discovery-and-the-1-percent-rule-of-the-web/
- 104.Feng WC, Brandt D, Saha D. 2007. A long-term study of a popular MMORPG. In Proceedings of the 6th ACM SIGCOMM Workshop on Network and System Support for Games, pp. 19–24. New York: Assoc. Comput. Mach. [Google Scholar]
- 105.Cooper S, Khatib F, Makedon I, Lu H, Barbero J, et al. 2011. Analysis of social gameplay macros in the Foldit cookbook. In Proceedings of the 6th International Conference on Foundations of Digital Games. New York: Assoc. Comput. Mach. [Google Scholar]
- 106.Eastman P, Shi J, Ramsundar B, Pande VS. 2018. Solving the RNA design problem with reinforcement learning. PLOS Comput. Biol 14:e1006176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cox J, Oh EY, Simmons B, Lintott C, Masters K, et al. 2015. Defining and measuring success in online citizen science: a case study of Zooniverse projects. Comput. Sci. Eng 17:28–41 [Google Scholar]
- 108.Gaston J, Cooper S. 2017. To three or not to three: improving human computation game onboarding with a three-star system. In Proceedings of the 2017 CHI Conference on Human Factors in Computing Systems, pp. 5034–39. New York: Assoc. Comput. Mach. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tinati R, Luczak-Roesch M, Simperl E, Shadbolt N, Hall W. 2015. ‘/Command’ and conquer: analysing discussion in a citizen science game. In Proceedings of the ACM Web Science Conference, Pap. 26. New York: Assoc. Comput. Mach. [Google Scholar]
- 110.Baaden M, Delalande O, Ferey N, Pasquali S, Waldispühl J, Taly A. 2018. Ten simple rules to create a serious game, illustrated with examples from structural biology. PLOS Comput. Biol 14:e1005955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Bruggemann J, Lander GC, Su AI. 2018. Exploring applications of crowdsourcing to cryo-EM. J. Struct. Biol 203:37–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Haas A, Rossberg A, Schuff DL, Titzer BL, Holman M, et al. 2017. Bringing the web up to speed with WebAssembly. In Proceedings of the 38th ACM SIGPLAN Conference on Programming Language Design and Implementation, pp. 185–200. New York: Assoc. Comput. Mach. [Google Scholar]
- 113.Good B, Santini S, Wallace M, Fortugno N, Szeder J, et al. 2017. Science Game Lab: tool for the unification of biomedical games with a purpose. bioRxiv 156141. 10.1101/156141 [DOI] [Google Scholar]
- 114.Hossain Z, Riedel-Kruse IH. 2018. Life-science experiments online: technological frameworks and educational use cases. In Cyber-Physical Laboratories in Engineering and Science Education, ed. ME Auer, Azad A, Edwards A, de Jong T, pp. 271–304. Cham, Switz.: Springer Int. [Google Scholar]
- 115.Hossain Z, Bumbacher E, Blikstein P, Riedel-Kruse I. 2017b. Authentic science inquiry learning at scale enabled by an interactive biology cloud experimentation lab. In Proceedings of the Fourth (2017) ACM Conference on Learning @ Scale, pp. 237–40. New York: Assoc. Comput. Mach. [Google Scholar]
- 116.Riedel-Kruse IH. 2018. Incorporating a commercial biology cloud lab into online education. In Online Engineering & Internet of Things, pp. 331–43. Cham, Switz.: Springer [Google Scholar]
- 117.Tsang AC, Lam AT, Riedel-Kruse IH. 2018. Polygonal motion and adaptable phototaxis via flagellar beat switching in the microswimmer Euglena gracilis. Nat. Phys 14:1216–22 [Google Scholar]
- 118.Corbató FJ, Merwin-Daggett M, Daley RC. 1962. An experimental time-sharing system. In Proceedings of the May 1–3, 1962, Spring Joint Computer Conference, pp. 335–44. New York: Assoc. Comput. Mach. [Google Scholar]
- 119.Murty J 2008. Programming Amazon web services: S3, EC2, SQS, FPS, and SimpleDB. Sebastopol, CA: O’Reilly Media [Google Scholar]
- 120.Zahariev A 2009. Google app engine. Paper presented at TKKT-110.5190 Seminar on Internetworking, Helsinki Univ. Technol. http://www.cse.hut.fi/en/publications/B/5/papers/1Zahariev_final.pdf [Google Scholar]
- 121.Wilder B 2012. Cloud Architecture Patterns: Using Microsoft Azure. Sebastopol, CA: O’Reilly Media [Google Scholar]
- 122.Kim H, Gerber LC, Chiu D, Lee SA, Cira NJ, et al. 2016. LudusScope: accessible interactive smartphone microscopy for life-science education. PLOS ONE 11:e0162602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gerber LC, Kim H, Riedel-Kruse IH. 2016. Interactive biotechnology: design rules for integrating biological matter into digital games. In Proceedings of the First International Joint Conference of DiGRA and FDG. http://www.digra.org/digital-library/publications/interactive-biotechnology-design-rules-for-integrating-biological-matter-into-digital-games/ [Google Scholar]
- 124.Lee SA, Chung AM, Cira N, Riedel-Kruse IH. 2015. Tangible interactive microbiology for informal science education. In Proceedings of the Ninth International Conference on Tangible, Embedded, and Embodied Interaction, pp. 273–80. New York: Assoc. Comput. Mach. [Google Scholar]
- 125.Lee SA, Bumbacher E, Chung AM, Cira N, Walker B, et al. 2015. Trap it!: a playful human-biology interaction for a museum installation. In Proceedings of the 33rd Annual ACM Conference on Human Factors in Computing Systems, pp. 2593–602. New York: Assoc. Comput. Mach. [Google Scholar]
- 126.Cira NJ, Chung AM, Denisin AK, Rensi S, Sanchez GN, et al. 2015. A biotic game design project for integrated life science and engineering education. PLOS Biol. 13:e1002110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gerber LC, Doshi MC, Kim H, Riedel-Kruse IH. 2016. Biographr: science games on a biotic computer. In Proceedings of the First International Joint Conference of DiGRA and FDG. http://www.digra.org/digital-library/publications/biographr-science-games-on-a-biotic-computer/ [Google Scholar]
- 128.Lam AT, Samuel-Gama KG, Griffin J, Loeun M, Gerber LC, et al. 2017. Device and programming abstractions for spatiotemporal control of active micro-particle swarms. Lab Chip 17:1442–51 [DOI] [PubMed] [Google Scholar]
- 129.Washington P, Samuel-Gamaa KG, Goyala S, Ramaswamia A, Riedel-Kruse IH. 2019. Interactive programming paradigm for real-time experimentation with remote living matter. PNAS 116(12):5411–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Chambers C 2011. Howvirtual are virtual economies? An exploration into the legal, social and economic nature of virtual world economies. Comput. Law Secur. Rev 27:377–84 [Google Scholar]
- 131.Buro M 2003. Real-time strategy games: a new AI research challenge. In Proceedings of the 18th International Joint Conference on Artificial Intelligence, 1534–35. San Francisco: Morgan Kaufmann [Google Scholar]
- 132.van der Maaten L, Hinton G. 2008. Visualizing data using t-SNE. J. Mach. Learn. Res 9:2579–605 [Google Scholar]
- 133.Marcus G 2018. Deep learning: a critical appraisal. arXiv:1801.00631 [cs.AI] [Google Scholar]
- 134.Chi MT. 2009. Active-constructive-interactive: a conceptual framework for differentiating learning activities. Top. Cogn. Sci 1:73–105 [DOI] [PubMed] [Google Scholar]
- 135.Comm. Concept. Framew. New K-12 Sci. Educ. Stand. 2012. A Framework for K-12 Science Education: Practices, Crosscutting Concepts, and Core Ideas. Washington, DC: Natl. Acad. Press [Google Scholar]
- 136.Williams JJ. 2013. Improving learning in MOOCs with cognitive science. In Proceedings of the Workshops at the 16th International Conference on Artificial Intelligence in Education (AIED 2013), ed. Walker E, Looi C-K, pp. 49–54. http://ceur-ws.org/Vol-1009/ [Google Scholar]
- 137.Pedaste M, Maeots M, Siiman LA, De Jong T, Van Riesen SA, et al. 2015. Phases of inquiry-based learning: definitions and the inquiry cycle. Educ. Res. Rev 14:47–61 [Google Scholar]
- 138.Stock A 2015. Guide to Implementing the Next Generation Science Standards by National Research Council. Sci. Scope 39:84–85 [PubMed] [Google Scholar]
- 139.Qian M, Clark KR. 2016. Game-based learning and 21st century skills: a review of recent research. Comput. Hum. Behav 63:50–58 [Google Scholar]
- 140.Boyle EA, Hainey T, Connolly TM, Gray G, Earp J, et al. 2016. An update to the systematic literature review of empirical evidence of the impacts and outcomes of computer games and serious games. Comput. Educ 94:178–92 [Google Scholar]
- 141.McKiernan EC. 2017. Imagining the “open” university: sharing scholarship to improve research and education. PLOS Biol. 15:e1002614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Baden T, Chagas AM, Gage G, Marzullo T, Prieto-Godino LL, Euler T. 2015. Open labware: 3-D printing your own lab equipment. PLOS Biol. 13:e1002086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kuiken T 2016. Learn from DIY biologists: the citizen-science community has a responsible, proactive attitude that is well suited to gene-editing. Nature 531:167–69 [DOI] [PubMed] [Google Scholar]
- 144.Scheifele LZ, Burkett T. 2016. The first three years of a community lab: lessons learned and ways forward. J. Microbiol. Biol. Educ 17:81–85 [DOI] [PMC free article] [PubMed] [Google Scholar]