Abstract
Purpose
Orbital fat hyperplasia commonly occurs in thyroid-associated orbitopathy (TAO). To understand molecular mechanisms underlying orbital adipogenesis, we used transcriptomics to compare gene expression in controls and patients with TAO, as well as in orbital fibroblasts (OFs) undergoing adipogenic differentiation.
Methods
We performed bulk RNA sequencing (RNA-Seq) on intraconal orbital fat from controls and patients with TAO. We treated cultured OFs derived from patients with TAO with adipogenic media to induce adipogenesis. We used single nucleus RNA-Seq (snRNA-Seq) to profile treated and control OFs, identifying genes that are dynamically expressed during orbital adipogenesis in vitro, and compared these results to data from control and TAO orbital fat.
Results
Gene expression profiles in control and TAO orbital fat are distinct. Signaling pathways including PI3K-Akt signaling, cAMP signaling, AGE-RAGE signaling, regulation of lipolysis, and thyroid hormone signaling are enriched in orbital fat isolated from patients with TAO. SnRNA-Seq of orbital fibroblasts undergoing adipogenesis reveals differential expression of the adipocyte-specific genes FABP4/5, APOE, PPARG, and ADIPOQ during adipogenic differentiation. The insulin-like growth factor-1 receptor and Wnt signaling pathways appear to be enriched early in adipogenesis. Gene modules that are enriched in TAO orbital fat are upregulated in orbital adipocytes during differentiation in vitro, whereas genes that are enriched in control orbital fat are enriched in undifferentiated OFs.
Conclusions
We identified pathways enriched in TAO orbital fat, and dynamic changes in gene expression that occur during adipogenic differentiation of orbital fibroblasts. These findings may help guide functional studies of genes and pathways critical for orbital adipogenesis.
Keywords: thyroid associated orbitopathy, Graves’ disease, orbital fat, adipogenesis, orbital fibroblasts
Thyroid-associated orbitopathy (TAO) is a form of autoimmune thyroid disease in which shared auto-antigens in the thyroid gland and orbit are targets of the immune response.1–3 About 25% of patients with autoimmune thyroid disease will develop ocular disease, which can be vision-threatening. It is thought that inflammatory signaling causes expansion of the orbital soft tissues due to fibrosis and orbital fat hyperplasia which causes ocular morbidity due to corneal exposure and/or compressive optic neuropathy.4–6
The orbital soft tissue expansion that causes vision loss and other ocular morbidity associated with TAO is thought to be the final step of an inflammatory cascade that results in fibrosis and adipogenesis. Details of the mechanisms underlying orbital adipogenesis are not well understood. Generally, adipogenesis is initiated by transcription factor cascades involving transient early expression of CCAAT/enhancer-binding protein (C/EBP) -β and -δ followed by induction of CEBP-α and peroxisome proliferator-activated receptor γ, transcription factors, which in turn induce expression of genes involved in terminal adipocyte differentiation.7,8 These pathways contribute to a generic adipocyte differentiation program. However, adipocyte metabolic function, morphology, preadipocyte proliferation, and capacity for adipogenesis differ among adipose tissue depots.9 These molecular and physiological differences, which have been explored in subcutaneous, visceral, and brown adipose tissue depots, are thought to be intrinsic to adipocytes and may be related to their embryonic origins.9 The differences in orbital adipose tissue, and the way in which they may contribute to the TAO phenotype, however, have not yet been explored.
To identify molecular mechanisms controlling adipogenesis in TAO, we have conducted bulk RNA sequencing (RNA-Seq) analysis of primary orbital fat from both controls and patients with TAO, and also used single nucleus RNA-Seq (snRNA-Seq) to profile orbital fibroblasts undergoing adipogenesis in vitro.
Methods
Bulk RNA-Seq
Human intraconal orbital fat was obtained from patients with TAO undergoing orbital decompression or controls undergoing routine resection of prolapsed orbital fat (Table), which has been shown to be intraconal fat,10 or primary enucleation for uveal melanoma using a protocol approved by the Johns Hopkins University Institutional Review Board and following the tenets of the Declaration of Helsinki. Patients with TAO were inactive at the time of surgery, defined by a clinical activity score (CAS) less than 4.11 Samples were placed in RNAlaterTM (#AM7020; ThermoFisher Scientific, Waltham, MA, USA), ice, or frozen fresh on dry ice, depending on the intended subsequent use. RNA was extracted from each sample using Trizol (#15596018; ThermoFisher) and RNeasy mini kits (#74104; Qiagen, Germany). Bioanalyzer (Agilent) analysis was used to perform quality control. The cDNA libraries were prepared for samples with RIN >6. RNA-Seq libraries were made using TruSeq Stranded Total RNA kit (Illumina, San Diego, CA, USA) and libraries were sequenced using Illumina NextSeq 500 (Illumina), paired-end read of 75 bp, 50 million reads per library. Illumina adapters of libraries after sequencing were removed using Cutadapt (version 1.18)12 with default parameters. Sequenced libraries were then aligned to GRCh38 using STAR (version 2.42a)13 with –twopassMode Basic. RSEM (version 1.3.1)14 was used for transcript quantification, with rsem-calculate-expression (–forwad-prob 0.5).
Table.
Characteristics of TAO and Control Patients Undergoing Orbital Surgery
| Control S1 | Control S2 | Control S3 | TAO S1 | TAO S2 | TAO S3 | |
|---|---|---|---|---|---|---|
| Age (years) | >70 | 58 | >70 | 37 | 51 | 51 |
| Gender | M | M | F | F | F | M |
| Race | Asian | C | C | C | AA | AA |
| Duration of thyroid disease prior to surgery (approx. mo) | N/A | N/A | N/A | 48 | 13 | 13 |
| Duration of TED prior to surgery (approx. mo) | N/A | N/A | N/A | 18 | 13 | 10 |
| Previous treatment for Graves’ disease | N/A | N/A | N/A | Armor thyroid | Methimazole | Carbimazole |
| Previous treatment for TED | N/A | N/A | N/A | N/A | N/A | Prednisone, Methylprednisolone, Azathioprine |
| Smoking history | >10 years ago | No | No | Former | No | No |
| Exophthalmometry, Hertel (mm) | N/A | N/A | N/A | 21 | 27.5 | 30 |
| Presence of compressive optic neuropathy | N/A | N/A | N/A | No | No | Yes |
| Surgery | Excision of orbital fat prolapse | Excision of orbital fat prolapse | Primary Enucleation (UM) | Orbital decompression | Orbital decompression | Orbital decompression |
| Clinical Activity Score (0-7) | N/A | N/A | N/A | 0 | 1 | 3 |
AA, African American; C, Caucasian; N/A, not applicable; UM, uveal melanoma.
DESeq2 (version 1.24.0)15 was used to analyze the bulk RNA-Seq dataset using the standard pipeline, filtering low counts (<10) and choosing genes with adjusted P value < 0.05. ClusterProfiler (version 3.12.0)16 enrichKEGG function was used to identify pathways that are enriched in the TAO group.
In Vitro Differentiation
Orbital fibroblast cell lines were derived from TAO intraconal fat as previously described.17 Briefly, tissue explants were obtained from patients undergoing surgical decompression for TAO. Patients were inactive, with a CAS less than 4, at the time of surgery.11 Explants were placed on the bottom of culture plates and covered with Eagle's medium containing 10% FBS, antibiotics, and glutamine. They were incubated at 37°C, 5% CO2, in a humidified environment. Media was changed weekly until the cells reached confluence. The resulting fibroblast monolayers were passaged serially by gentle treatment with TrypLE (#12604039; ThermoFisher Scientific). Strains were stored in liquid N2 until needed and were used between the fourth and eighth passages.
Orbital fibroblasts were induced to undergo adipogenesis as previously described.17 Briefly, fibroblasts between passages four and eight were seeded on plastic tissue culture plates and allowed to proliferate to near-confluence in DMEM containing 10% FBS and antibiotics. The cells were then treated with adipogenic medium consisting of DMEM:F-10 (1:1, #11320033; ThermoFisher Scientific) supplemented with 3% FBS, 100 nmol/liter insulin (#12585014; ThermoFisher Scientific), 1 µmol/liter dexamethasone (#D4902; Sigma, St. Louis, MO, USA), and for the first week only, 1 µmol/liter rosiglitazone (#R2408; Sigma) and 0.2 mmol/liter IBMX (#13630S;, Cell Signalling Technology, Danvers, MA, USA). Media was changed every other day for the first week, and then twice weekly for the remaining weeks. Cultures were maintained in this medium for 21 days. Control cultures were treated with DMEM:F-10 (1:1), supplemented with 3% FBS and vehicle. Differentiation was observed using a Nikon microscope (Japan). Each time point was analyzed in triplicate.
Quantification of Adipocytes
On days 0, 5, 9, and 21, cells were washed with PBS and fixed using 4% paraformaldehyde in PBS for 10 minutes at room temperature. Cells were washed with PBS or stored in PBS at 4°C. To stain adipocytes, cells were washed with 60% isopropanol and stained with freshly prepared 0.3% (w/v) Oil red O (Sigma) at room temperature for 15 minutes. Cells were rinsed with 60% isopropanol, stained lightly with hematoxylin (Sigma) for 5 minutes, and then rinsed with PBS. Cells were imaged by a masked study team member using a Keyence BZ-X710 microscope (Keyence, Japan). Four high-power field images were obtained per replicate. Cell counts and lipid vacuole measurements were performed using ImageJ.18 Oil red O-positive cells were counted as adipocytes and nuclei in cells that did not stain with Oil red O, which represent fibroblasts or preadipocytes, were not included in the adipocyte cell count. To quantify lipid vacuole area, images were converted to eight8-bit, and made binary using an automated algorithm, scaled to pixels and analyzed using the Analyze Particles in ImageJ.18
Single-Nucleus RNA-Seq
Nuclei from the treated groups were isolated at days 0, 5, 9, and 21 using the methods described in the 10x Genomics Sample Preparation Demonstrated Protocol (10x Genomics, Pleasanton, CA, USA). Nuclei from control groups were isolated at day 0, 5, and 31 using the same methods. Briefly, cells were washed with chilled PBS and lysed in chilled lysis buffer consisting of 10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2, and 0.1% Nonidet P40 Substitute (MP Biomedicals, Solon, OH, USA) in nuclease-free water at 4°C. Cells were scraped from the plate bottom and centrifuged at 500 RCF for 5 minutes at 4°C. Cells were washed twice in nuclei wash and resuspension buffer consisting of PBS with 0.1% BSA and 0.2 U/ul RNase inhibitor (#N2615; Promega, Madison, WI, USA). Cells were passed through a 50 um CellTrics filter (#NC9491906; Sysmex, Germany) and centrifuged at 500 RCF for 5 minutes at 4°C prior to resuspension in wash and resuspension buffer. Isolated nuclei were counted manually via hemocytometer with Trypan Blue staining, and nucleus concentration was adjusted following the 10x Genomics guideline. The 10x Genomics Chromium Single Cell system (10x Genomics) using V2 chemistry (days 0, 5, 9, and 31) or V3 chemistry (day 21) per the manufacturer's instructions, generating a total of 10 libraries. Day 5 and day 21 were run with technical replicates. Libraries were sequenced on Illumina NextSeq 500 mid-output or NovaSeq 6000 (150 million reads). Sequencing data were first preprocessed through the Cell Ranger pipeline (10x Genomics, Cellranger version 5.0.0) with default parameters, using GRCh38-2020-A genome with include-introns.
Matrix files generated from the Cellranger run were used for subsequent analysis. Seurat version 319 was used to perform downstream analysis, only including cells with more than 500 genes, 1000 UMI, to process control and treated samples separately. Seurat SCTransform function was used to normalize the dataset20 and UMAP was used to reduce dimensions derived from the Harmony21 output.
In order to identify cells that show adipocyte signatures, key adipocyte-enriched genes were first extracted from GTEx dataset from ascot.cs.jhu.edu,22 relying on both robustness (NAUC > 20) and specificity (expressed only in both adipocytes dataset or in tissues that are enriched with adipocytes, such as mammary tissues). These genes were then used to train the dataset using Garnett23 to identify differentiated adipocytes.
RNA velocity24 was used to verify in vitro adipocyte differentiation trajectories. Kallisto and Bustools25 were used to obtain splicing values, and Scanpy26 and scVelo27 were used to obtain velocity trajectory. Pseudotime analysis was performed using Monocle version 328 and genes that are significant (q value < 0.001) along pseudotime trajectory were used to run Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis, as described above. Genes that were involved across KEGG signaling pathways were then used to identify signaling pathways that are specific to different stages of in vitro differentiation. SCENIC29 was used to calculate regulons controlling gene expressions across adipocytes differentiation stages, as previously described.30,31 Enriched genes from control and TAO bulk RNA-Seq datasets were superimposed on in vitro datasets using the Seurat AddModuleScore function.
Data Availability
All sequencing data are available on GEO as GSE158464 (bulk RNA-Seq dataset) and GSE174139 (bulk RNA-Seq dataset and snRNA-Seq).
Results
Bulk Transcriptome Analysis of Intraconal Fat from TAO and Control Patients
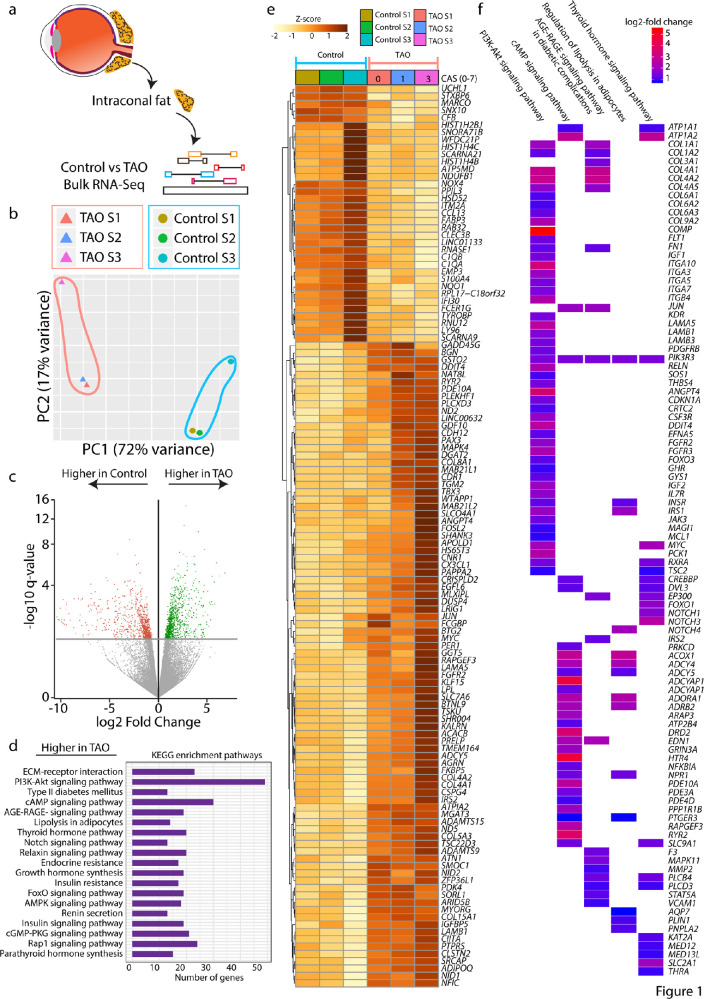
Intraconal fat was collected from patients undergoing orbital surgery for TAO or other indications (Table). To compare gene expression in orbital fat in patients with TAO and controls, bulk RNA-Seq was performed (Fig. 1a). Principal component analysis (PCA) revealed that patterns of gene expression in TAO and control orbital fat segregated, with control and TAO replicates clustering together (Fig. 1b). We observed 902 genes that are enriched in control orbital fat, and 964 genes enriched in TAO orbital fat (Fig. 1c). The KEGG pathway analysis of genes enriched in TAO orbital fat revealed numerous signaling pathways that are highly enriched, including PI3K-Akt signaling, cAMP signaling, AGE-RAGE signaling, regulation of lipolysis in adipocytes, and thyroid hormone signaling pathway (Fig. 1d). Heat map analysis of genes expressed demonstrated significant differences (adjusted P value < 0.05; Table S1) between TAO and control orbital fat. Correlation analysis also demonstrated that differences in gene expression levels correlate with the patient's CAS at the time of surgery (Supplementary Fig. S1, see the Table). Heat map analysis of individual genes within the above-mentioned pathways demonstrated the degree to which genes, such as THRA and IGF1, are enriched in TAO orbital fat (Fig. 1f).
Figure 1.
Bulk RNA-Seq analysis comparing intraconal fat of control and TAO patients. (a) Schematic diagram showing the overall experiment pipeline. (b) PCA plot showing the distribution of control and TAO replicates along PC1, where the axis represents a total of 72% variance, and PC2, where the axis represents a total of 17% variance. (c) Volcano plot of genes that are higher in control or TAO orbital fat. (d) KEGG pathway analysis showing pathways that are enriched in the TAO group. (e) Heatmap demonstrating gene expression differences between the control and TAO. (f) Top KEGG pathways in 1d and genes that are expressed at a higher level in TAO than in the control group log2 (Fold change). CAS = Clinical activity score.
snRNA-Seq of Orbital Fibroblasts Undergoing Adipogenesis Reveals Induction of Adipocyte-Specific Markers
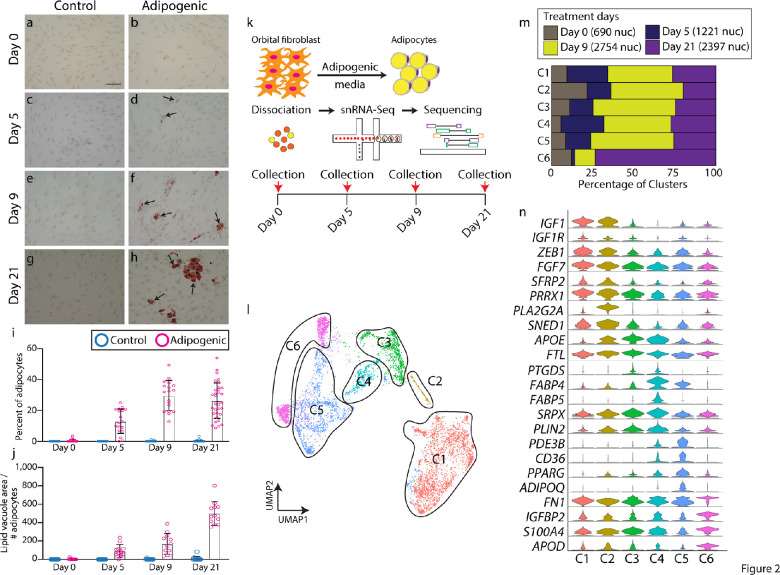
In order to identify genes that are involved in adipogenesis in orbital fibroblasts, which may mediate the orbital fat hyperplasia seen in TAO, we used snRNA-Seq to profile differentiating orbital fibroblasts in vitro. Orbital fibroblasts derived from case #2 were treated to induce adipogenesis over a 21-day time course. Cells treated with control media did not undergo adipogenesis (Figs. 2a,c,e,g). Based on histologic changes in cells treated with adipogenic media, we chose to analyze adipogenesis and gene expression at days 0, 5, 9, and 21 (Figs. 2a–h). Adipocyte induction first occurred between days 0 and 5, with the maximum density of adipocytes, noted at day 9 (Fig. 2i). Adipocyte maturation, as quantified by the lipid vacuole area, increased most between days 9 and 21 (Fig. 2j). We performed snRNA-Seq on orbital fibroblasts treated with control media, and did not observe any adipogenic-like cells at 0, 5 and 31 days of culture (Supplementary Fig. S2a–c, Supplementary Table S2). Furthermore, day 0 snRNA-Seq only detected fibroblast-like cells, indicating the purity of our culture condition (Supplementary Fig. S2d, Supplementary Table S3).
Figure 2.
snRNA-Seq shows adipogenesis trajectory in vitro. (a–h) Oil Red O staining (arrows) of orbital fibroblasts treated with control (a, c, e, g) and adipogenic (b, d, f, h) medium orbital at day 0 (a, b), day 5 (c, d); day 9 (e, f), and day 21 (g, h). (i) Bar plot showing the percentage of adipocytes seen after treatment with control or adipogenic medium on days 0, 5, 9, and 21. (j) Bar plot showing lipid vacuole area / the number of adipocytes between orbital fibroblasts treated with control or adipogenic medium on days 0, 5, 9, and 21. (k) Schematic diagram showing snRNA-Seq pipeline. (l) UMAP plot showing key six clusters of cells undergoing adipogenesis from snRNA-Seq. (m) Bar plot showing the percentage of clusters that are occupied across treatment days. (n) Violin plot showing key cluster markers. Scale bar = 100 µM.
Based on the observed histologic changes, we collected nuclei at days 0, 5, 9, and 21 treatments for analysis by snRNA-Seq (Fig. 2k). We identified unique clusters that were enriched from day 5 onward and expressed unique sets of markers that were absent from the other clusters, such as IGF1 (Supplementary Fig. S2, Supplementary Table S4). These appear to give rise to a cluster that expressed high levels of adipocyte signatures, including ADIPOQ (Supplementary Fig. S3, Supplementary Table S2). We identified these clusters as fibroblasts undergoing adipogenesis.
UMAP analysis of differentiating orbital fibroblasts revealed 6 distinct clusters defined by a unique set of genes (Fig. 2l). Overall, differentiating cells appeared to progress from cluster 1 (C1) to C6 over time (Fig. 2m). Classic adipocyte stem cell markers, such as FABP4, APOE, and FABP5, were enriched in C3 to C4, whereas mature adipocyte markers, such as PPARG and ADIPOQ, were enriched in C5, demonstrating that the clusters corresponded to a developmental progression from orbital fibroblast, through adipocyte stem cells, to mature adipocytes (Fig. 2n, Supplementary Table S5). At day 21, a new cell type was present that demonstrated high expression of APOD, but had lost expression of other adipogenic markers, such as FABP4/5 but shared expression of many of the other markers expressed in other clusters were represented in cluster C6 (Fig. 2n). To identify genes that were induced early in the process of adipogenesis, we focused on genes that were specifically expressed in C1 and C2, which differed from both quiescent, control-treated orbital fibroblasts, and adipocyte stem cells. This population showed enrichment of IGF1, IGF1R, ZEB1, FGF7, and SFRP2 expression (Fig. 2n). Enrichment of IGF1 expression within differentiating orbital fibroblasts suggests that paracrine signaling may be involved in the process of adipogenesis.
Analysis of Key Pathways Involved in Orbital Adipogenesis in Vitro
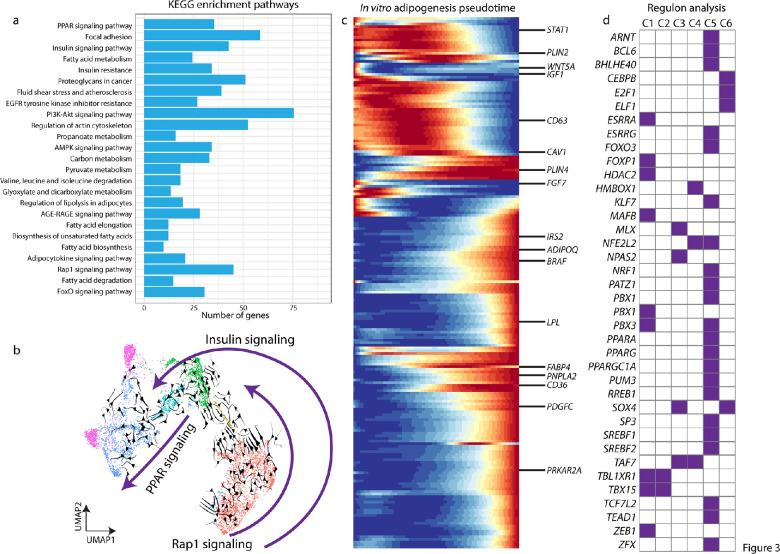
We next focused on identifying key signaling pathways and potential regulatory factors that are differentially expressed during orbital adipogenesis in vitro. Using KEGG analysis of the snRNA-Seq data, we observed that differentiating adipocytes express genes involved in multiple signaling pathways, including the PI3-AKT, AGE-RAGE, and insulin resistance. These pathways were also enriched in TAO orbital fat compared to control orbital fat, as measured by bulk RNA-Seq analysis (Figs. 1c, 3a). Other pathways that are specifically involved in adipogenesis, such as PPAR signaling, fatty acid biosynthesis, and fatty acid elongation (Fig. 3a) were also enriched in differentiating orbital fibroblasts.
Figure 3.
snRNA-Seq reveals adipogenesis signaling pathway. (a) KEGG plot based on pseudotime analysis from snRNA-Seq data in Figure 2. (b) UMAP plot with RNA velocity showing adipocyte differentiation trajectory, and three pathways that are specific across differentiation stages. (c) Pseudotime trajectory of adipogenesis and key genes. (d) Regulon analysis demonstrates key transcription factors that might be involved in each step of adipogenesis differentiation.
RNA velocity analysis demonstrated that components of the Rap1 signaling pathway are active early in orbital fibroblasts in C1 (Fig. 3b). Components of the insulin signaling pathway were expressed throughout the course of adipocyte differentiation in C1 to C5 (Fig. 3b), whereas peroxisome proliferator-activated receptor (PPAR) signaling, which is known to be active late in adipogenesis, was active in C4 to C5 (Fig. 3b). Pseudotime analysis showed the progression of gene expression through C1 to C5 for all differentially expressed genes (Fig. 3c). Genes with particularly strong differential expression are highlighted, such as IGF1, SFRP2, and WNT5A (Fig. 3c, Supplementary Table S6). Regulon analysis identified transcription factors that are differentially expressed in each cluster (Fig. 3d).
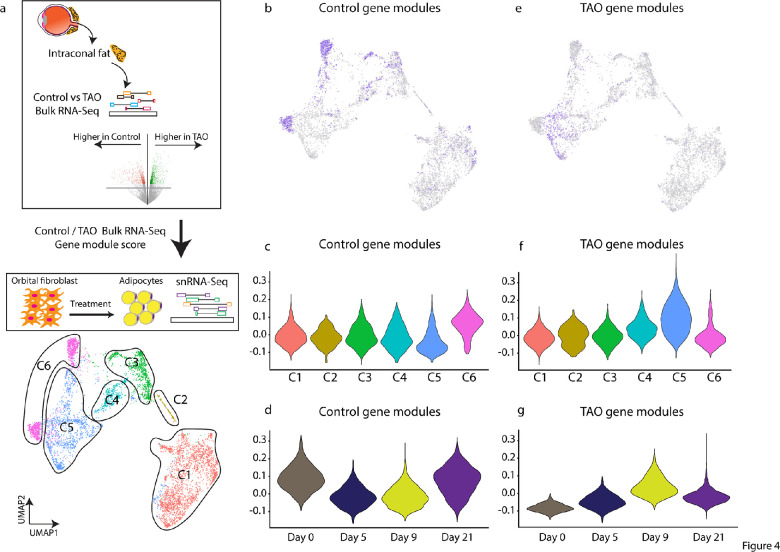
The TAO Orbital Fat Transcriptome Resembles that of in Vitro Differentiated Adipocytes
In order to determine the similarity between in vitro derived orbital adipocytes and orbital fat in vivo, we compared gene expression from control and TAO orbital fat, analyzed by bulk RNA-Seq, to gene expression during orbital adipogenesis as assessed by snRNA-Seq (Fig. 4a). Control gene modules were found to be expressed most highly in C1 and C6. These modules were most strongly expressed in day 0 cells (C1) and a subset of day 21 cells that do not express mature adipocyte markers (C6; Figs. 4b–d). TAO gene modules were expressed most highly in C5, and at days 9 and 21 (Figs. 4e,f). Interestingly, although the orbital fibroblast cell line was derived from a patient with TAO, the gene expression pattern resembled control gene modules prior to treatment with adipogenic media (Figs. 4c,d), and changes to resemble TAO genes modules following treatment with adipogenic media (Figs. 4f,g).
Figure 4.
In vitro differentiated adipocytes express high levels of TAO tissue-enriched markers. (a) Schematic showing that genes that are higher in control or TAO orbital fat were projected into snRNA-Seq dataset to obtain gene module score. (b–d) Control intraconal fat-enriched genes demarcate cluster six in snRNA-Seq dataset (b, d), expressed higher at day 0 (c). (e–g) TAO intraconal fat-enriched genes demarcate cluster five adipocytes in snRNA-Seq dataset (e, f), expressed higher at day 0 (g).
Discussion
In this study, we demonstrate that orbital fibroblasts derived from patients with TAO can be used as a model to study adipogenesis in vitro. Histologic studies and transcriptome profiling clearly show that, although they are derived from patients with TAO, the gene expression profile of undifferentiated orbital fibroblasts closely resembles that of control orbital fat cells, while those treated to undergo adipogenesis display a gene expression profile more similar to that of TAO orbital fat.
We have identified signaling pathways and transcription factors that are upregulated early in orbital adipogenesis, including the insulin-like growth factor-1 (IGF-1) signaling pathway, which has previously shown to contribute to TAO pathophysiology by increasing auto-antigen display, cytokine synthesis, and hyaluronan production by orbital fibroblasts.32,33 The novel TAO therapy teprotumumab targets the IGF-1 receptor (IGF1-R) to improve proptosis, diplopia, CAS, and quality of life in patients with TAO.34,35 Although it is thought to exert its exerts via modulation of the immune response, our data raise the possibility that inhibition of the IGF-1R pathway may also impact TAO by reducing adipogenesis in the orbit.
As with all transcriptomic analyses, there are limitations to this study. In particular, with the use of human tissue, there is heterogeneity in the patients’ genetic background and other characteristics, which likely affect disease such as age, gender, smoking status, and CAS. Furthermore, these studies have identified very large numbers of differentially expressed genes between both control and TAO samples, and that are dynamically expressed over the course of orbital adipogenesis. Although we have focused our discussion to pathways and genes of interest based on our knowledge of TAO, there may be other pathways and genes with significant function in orbital adipogenesis and TAO.
Prior transcriptome profiling of cultured orbital fibroblasts derived from controls and patients with TAO has demonstrated significantly increased expression of homeobox transcription factors and decreased expression of Wnt signaling pathways components in TAO orbital fibroblasts.36 In profiling TAO orbital fibroblasts undergoing adipogenesis, we find that WNT5A expression is high early in the course of adipogenesis, but decreases as differentiation occur, whereas homeobox gene expression increases later (Figs. 3c,d). Genes and pathways that demonstrate strong differential expression, particularly early in the process of adipogenesis, likely serve critical functions in adipogenesis, and merit further study.
The model that we have used to study gene expression changes during orbital adipogenesis depends on the treatment of orbital fibroblasts with drugs including insulin and rosiglitazone. For example, whereas cells are treated with insulin throughout the experiment, and the insulin pathway is active throughout the differentiation process, cells are also treated with the ppar-gamma agonist rosiglitazone early (during the first week) in the experiment, but PPAR signaling is active only later in the differentiation process (Fig. 3b). Thus, the dynamic gene expression that we describe is likely reflective of functional changes occurring in intracellular signaling during adipogenesis and not simply artifacts of treatment with the adipogenic media.
Microarray comparisons of orbital fat from TAO and control patients have demonstrated increased expression in Wnt signaling pathway genes, such as SFRPs, IGF-1 pathway genes, and adipogenic genes, consistent with our bulk RNA-Seq results.37,38 In our heat map analysis of TAO-enriched genes, we note differences in the level of gene expression that correlate with CAS. Case 1, with a CAS of 0, has lower levels of TAO-enriched genes than case 2, from a patient with a CAS of 1, which in turn has lower levels of TAO-enriched genes than case 3, with CAS of 3 (Fig. 1e). However, even in the least severely affected patient, TAO-enriched genes are more highly expressed than in controls (Fig. 1e).
Other studies have compared RNA-Seq data of orbital fat from patients with active TAO compared to blepharoplasty fat and observed stronger upregulation of inflammatory signaling pathways, which is consistent with the higher CAS in their patients compared to those profiled in our study.39 Conversely, in our patients, pathways including insulin and thyroid signaling, as well as adipogenic pathways, were more prominent, likely reflective of the patients being in earlier stages of the disease. In addition, we used intraconal fat as our control orbital adipose sample, rather than blepharoplasty fat, which may account for some of the other observed differences in gene expression. A potential confounding factor in comparing these data sets may be that, in our analysis, we have excluded all genes with low counts due to low expression levels. Nonetheless, we also found large changes in Wnt signaling such as (DKK2 and NKD1) and other pathways (HOX3D) in TAO orbital fat compared to control orbital fat in post hoc analysis (see Table S1), although these genes were expressed at low levels. One limitation of the transcriptomic approach is that we cannot determine the functional significance of these low levels of gene expression.
We further have identified factors that have not been previously implicated in the biology of orbital fibroblasts, including cell-autonomous pathways, such as Rap1 signaling, which have been shown to drive adipogenesis in human bone mesenchymal stem cells.40,41 In addition, transcription factors that drive adipogenesis are of particular interest in understanding the regulation of orbital adipogenesis. ZEB1 has been identified as a critical mediator of adipogenesis in mouse pre-adipocytes and is induced by IGF-1R activation in prostate cancer cell lines.42,43 Finally, the use of in vitro analysis of differentiating cultured orbital fibroblasts to identify pathways that regulate orbital adipogenesis, and that are enriched in TAO orbital fat, opens the possibility of using high-throughput screening to identify drug and gene-based approaches to modulating pathological adipogenesis that may ultimately be useful in treating TAO-related disorders.
Supplementary Material
Acknowledgments
The authors thank the Transcriptomics and Deep Sequencing Core (Johns Hopkins) for sequencing of snRNA-Seq libraries. We also thank Lizhi Jiang for technical assistance.
Supported by an unrestricted departmental grant to the Wilmer Eye Institute from Research to Prevent Blindness. D.W.K. is supported by the Maryland Stem Cell Research Fund (2019-MSCRFF-5124). S.B. is supported by NIH Grant R01EY020560. F.R. is supported by the National Eye Institute of the National Institutes of Health under award number K08EY027093.
Disclosure: D.W. Kim, None; K. Taneja, None; T. Hoang, None; C.P. Santiago, None; T.J. McCulley, None; S.L. Merbs, None; N.R. Mahoney, None; S. Blackshaw, CDI Labs (I, P, R), Genentech (F), Third Rock Ventures (C); F. Rajaii, Horizon Therapeutics (C, I)
References
- 1. Bahn RS, Dutton CM, Natt N, Joba W, Spitzweg C, Heufelder AE.. Thyrotropin receptor expression in Graves’ orbital adipose/connective tissues: potential autoantigen in Graves' ophthalmopathy. J Clin Endocrinol Metab. 1998; 83(3): 998–1002. [DOI] [PubMed] [Google Scholar]
- 2. Crisp M, Lane C, Halliwell M, Wynford-Thomas D, Ludgate M.. Thyrotropin Receptor Transcripts in Human Adipose Tissue [Internet]. The Journal of Clinical Endocrinology & Metabolism. 1997:82: 2003–2005. Available from: 10.1210/jcem.82.6.2003. [DOI] [PubMed] [Google Scholar]
- 3. Porcellini A, Ruggiano G, Pannain S, et al.. Mutations of thyrotropin receptor isolated from thyroid autonomous functioning adenomas confer TSH-independent growth to thyroid cells. Oncogene. 1997; 15(7): 781–789. [DOI] [PubMed] [Google Scholar]
- 4. Kumar S, Coenen MJ, Scherer PE, Bahn RS.. Evidence for enhanced adipogenesis in the orbits of patients with Graves’ ophthalmopathy. J Clin Endocrinol Metab. 2004; 89(2): 930–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kumar S, Iyer S, Bauer H, Coenen M, Bahn RS.. A stimulatory thyrotropin receptor antibody enhances hyaluronic acid synthesis in Graves’ orbital fibroblasts: inhibition by an IGF-I receptor blocking antibody [Internet]. J Clin Endocrinol Metab. 2012; 97: 1681–1687. Available from: 10.1210/jc.2011-2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang L, Grennan-Jones F, Lane C, Rees DA, Dayan CM, Ludgate M.. Adipose tissue depot-specific differences in the regulation of hyaluronan production of relevance to Graves’ orbitopathy. J Clin Endocrinol Metab. 2012; 97(2): 653–662. [DOI] [PubMed] [Google Scholar]
- 7. Rosen ED, Walkey CJ, Puigserver P, Spiegelman BM.. Transcriptional regulation of adipogenesis. Genes Dev. 2000; 14(11): 1293–1307. [PubMed] [Google Scholar]
- 8. Rosen ED, MacDougald OA.. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol. 2006; 7(12): 885–896. [DOI] [PubMed] [Google Scholar]
- 9. Hilton C, Karpe F, Pinnick KE.. Role of developmental transcription factors in white, brown and beige adipose tissues. Biochim Biophys Acta. 2015; 1851(5): 686–696. [DOI] [PubMed] [Google Scholar]
- 10. Tejo CR, da Costa PA, Batista RM, Rocha YRR, Borba MA.. Subconjunctival fat prolapse: a disease little known to radiologists. Radiol Bras. 2017; 50(4): 272–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mourits MP, Prummel MF, Wiersinga WM, Koornneef L.. Clinical activity score as a guide in the management of patients with Graves’ ophthalmopathy. Clin Endocrinol . 1997; 47(1): 9–14. [DOI] [PubMed] [Google Scholar]
- 12. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011; 17(1): 10–12. [Google Scholar]
- 13. Dobin A, Davis CA, Schlesinger F, et al.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29(1): 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li B, Dewey CN.. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011; 12: 323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Love MI, Huber W, Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15(12): 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yu G, Wang L-G, Han Y, He Q-Y.. clusterProfiler: an R Package for comparing biological themes among gene clusters [Internet]. OMICS: A Journal of Integrative Biology. 2012; 16: 284–287. Available from: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Smith TJ, Koumas L, Gagnon A, et al.. Orbital fibroblast heterogeneity may determine the clinical presentation of thyroid-associated ophthalmopathy. J Clin Endocrinol Metab. 2002; 87(1): 385–392. [DOI] [PubMed] [Google Scholar]
- 18. Schneider CA, Rasband WS, Eliceiri KW.. NIH Image to ImageJ: 25 years of image analysis. Nat Methods. 2012; 9(7): 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stuart T, Butler A, Hoffman P, et al.. Comprehensive Integration of Single-Cell Data. Cell. 2019; 177(7): 1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hafemeister C, Satija R.. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019; 20(1): 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Korsunsky I, Millard N, Fan J, et al.. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019; 16(12): 1289–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ling JP, Wilks C, Charles R, et al.. ASCOT identifies key regulators of neuronal subtype-specific splicing. Nat Commun. 2020; 11(1): 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pliner HA, Shendure J, Trapnell C.. Supervised classification enables rapid annotation of cell atlases. Nat Methods. 2019; 16(10): 983–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. La Manno G, Soldatov R, Zeisel A, et al.. RNA velocity of single cells. Nature. 2018; 560(7719): 494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Melsted P, Booeshaghi AS, Liu L, et al.. Modular, efficient and constant-memory single-cell RNA-seq preprocessing. Nat Biotechnol [Internet]. 2021; Available from: 10.1038/s41587-021-00870-2. [DOI] [PubMed] [Google Scholar]
- 26. Wolf FA, Angerer P, Theis FJ.. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018; 19(1): 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bergen V, Lange M, Peidli S, Wolf FA, Theis FJ.. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol. 2020; 38(12): 1408–1414. [DOI] [PubMed] [Google Scholar]
- 28. Trapnell C, Cacchiarelli D, Grimsby J, et al.. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat Biotechnol. 2014; 32(4): 381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aibar S, González-Blas CB, Moerman T, et al.. SCENIC: single-cell regulatory network inference and clustering. Nat Methods. 2017; 14(11): 1083–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim DW, Washington PW, Wang ZQ, et al.. The cellular and molecular landscape of hypothalamic patterning and differentiation from embryonic to late postnatal development. Nat Commun. 2020; 11(1): 4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim DW, Liu K, Wang ZQ, et al.. Gene regulatory networks controlling differentiation, survival, and diversification of hypothalamic Lhx6-expressing GABAergic neurons. Commun Biol. 2021; 4(1): 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen H, Mester T, Raychaudhuri N, et al.. Teprotumumab, an IGF-1R blocking monoclonal antibody inhibits TSH and IGF-1 action in fibrocytes. J Clin Endocrinol Metab. 2014; 99(9): E1635–E1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen H, Shan SJC, Mester T, Wei Y-H, Douglas RS.. TSH-mediated TNFα production in human fibrocytes is inhibited by teprotumumab, an IGF-1R antagonist. PLoS One. 2015; 10(6): e0130322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Smith TJ, Kahaly GJ, Ezra DG, et al.. Teprotumumab for thyroid-associated ophthalmopathy. N Engl J Med. 2017; 376(18): 1748–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Douglas RS, Kahaly GJ, Patel A, et al.. Teprotumumab for the treatment of active thyroid eye disease. N Engl J Med. 2020; 382(4): 341–352. [DOI] [PubMed] [Google Scholar]
- 36. Tao W, Ayala-Haedo JA, Field MG, Pelaez D, Wester ST.. RNA-sequencing gene expression profiling of orbital adipose-derived stem cell population implicate HOX genes and WNT signaling dysregulation in the pathogenesis of thyroid-associated orbitopathy. Invest Ophthalmol Vis Sci. 2017; 58(14): 6146–6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kumar S, Leontovich A, Coenen MJ, Bahn RS.. Gene expression profiling of orbital adipose tissue from patients with Graves’ ophthalmopathy: a potential role for secreted frizzled-related protein-1 in orbital adipogenesis. J Clin Endocrinol Metab. 2005; 90(8): 4730–4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ezra DG, Krell J, Rose GE, Bailly M, Stebbing J, Castellano L.. Transcriptome-level microarray expression profiling implicates IGF-1 and Wnt signalling dysregulation in the pathogenesis of thyroid-associated orbitopathy. J Clin Pathol. 2012; 65(7): 608–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lee BW, Kumar VB, Biswas P, et al.. Transcriptome analysis of orbital adipose tissue in active thyroid eye disease using next generation RNA sequencing technology. Open Ophthalmol J. 2018; 12: 41–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wu J, Cai P, Lu Z, et al.. Identification of potential specific biomarkers and key signaling pathways between osteogenic and adipogenic differentiation of hBMSCs for osteoporosis therapy. J Orthop Surg Res. 2020; 15(1): 437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yeung F, Ramírez CM, Mateos-Gomez PA, et al.. Nontelomeric role for Rap1 in regulating metabolism and protecting against obesity. Cell Rep. 2013; 3(6): 1847–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gubelmann C, Schwalie PC, Raghav SK, et al.. Identification of the transcription factor ZEB1 as a central component of the adipogenic gene regulatory network. Elife. 2014; 3: e03346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Graham TR, Zhau HE, Odero-Marah VA, et al.. Insulin-like growth factor-I–dependent up-regulation of ZEB1 drives epithelial-to-mesenchymal transition in human prostate cancer cells [Internet]. Cancer Research. 2008; 68: 2479–2488. Available from: 10.1158/0008-5472.can-07-2559. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data are available on GEO as GSE158464 (bulk RNA-Seq dataset) and GSE174139 (bulk RNA-Seq dataset and snRNA-Seq).