Abstract
Well-defined tunable nanostructures formed through the hierarchical self-assembly of peptide building blocks have drawn significant attention due to their potential applications in biomedical science. Artificial protein polymers derived from elastin-like polypeptides (ELPs), which are based on the repeating sequence of tropoelastin (the water-soluble precursor to elastin), provide a promising platform for creating nanostructures due to their biocompatibility, ease of synthesis, and customizable architecture. By designing the sequence and composition of ELPs at the gene level, their physicochemical properties can be controlled to a degree that is unmatched by synthetic polymers. A variety of ELP-based nanostructures are designed, inspired by the self-assembly of elastin and other proteins in biological systems. The choice of building blocks determines not only the physical properties of the nanostructures, but also their self-assembly into architectures ranging from spherical micelles to elongated nanofibers. This review focuses on the molecular determinants of ELP and ELP-hybrid self-assembly and formation of spherical, rod-like, worm-like, fibrillar, and vesicle architectures. A brief discussion of the potential biomedical applications of these supramolecular assemblies is also included.
Keywords: bioinspired materials, elastin-like polypeptides, hierarchical self-assembly, partially ordered polypeptides, recombinant proteins
1. Introduction
Advances in biotechnology and material science have stimulated the development of nature-imitating materials in the past few decades. The ability to fabricate biological macromolecules such as collagen[1] and exosomes[2] in their native hierarchical form has emerged from an improved understanding of the requirements for recreating cellular and extracellular environments in the laboratory. Mimicry of biological macromolecules has produced self-assembling nanostructures and scaffolds useful in biomedical applications such as tissue engineering,[3–5] drug delivery,[6–8] and theranostics.[2,9]
Molecular self-assembly is a thermodynamic process in which molecules assemble into an ordered structure due to attractive and repulsive intramolecular and/or intermolecular interactions.[10,11] These interactions are noncovalent and include hydrogen bonding, electrostatic interactions, hydrophobic and hydrophilic interactions, and van der Waals forces.[10–13] Molecular self-assembly can be categorized as either static or dynamic.[14] The more common and extensively studied is static self-assembly, in which the systems involved approach local or global equilibrium and do not dissipate energy. Formation of an ordered structure through static self-assembly may require energy, but the product is stable. Examples of static self-assembly include lipid bilayer, and self-assembled monolayers. In dynamic self-assembly, an ordered equilibrium state occurs only when the system dissipates energy.[12] Prototypical examples of dynamic self-assembly include dynamic macro- and mesoscopic structures, organization of cellular organelle.
Nature has evolved an array of biological nanostructures through self-assembly, including: i) higher-order polypeptide structures (secondary, tertiary, quaternary); ii) the DNA double helix; iii) lipid bilayers; and iv) RNA-ribosome complexes. Of these, polypeptide self-assembly has gained the most momentum for creating nature-imitating materials due to the stability and diversity of polypeptide structures, as well as their intrinsic biocompatibility, high yield, and applicability in the biomedical field.[15–17] In particular, elastin-like polypeptides (ELPs)—a class of self-assembling peptides derived from the repeated amino acid sequence in the hydrophobic domain of human tropoelastin—have received significant attention.[6,8,18,19]
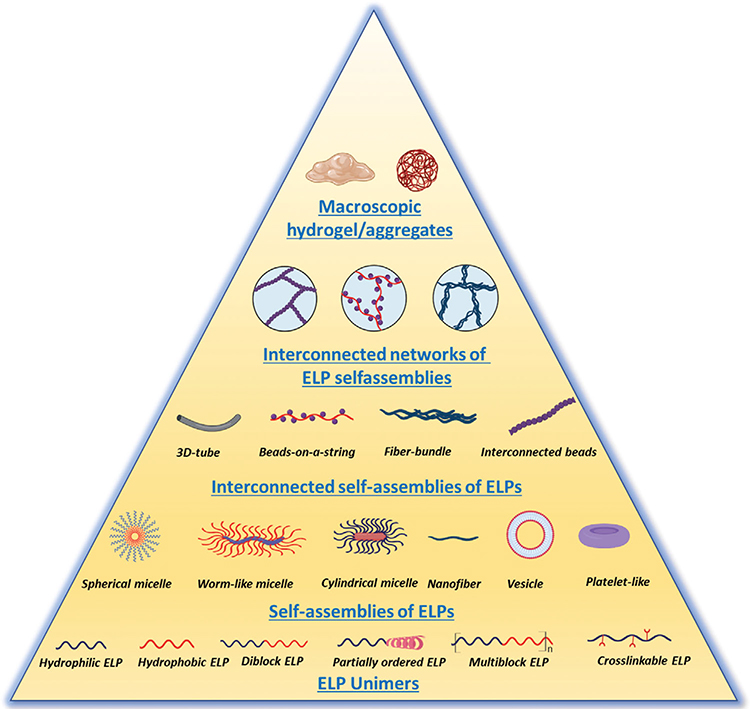
The most common ELP sequence motif is a multimeric repeat of the pentapeptide unit VPGXG, where X is any canonical amino acid except proline. This class of biopolymers undergoes a lower critical solution temperature (LCST) phase separation in aqueous solution above a transition temperature (Tt), to form an inhomogeneous coacervate—a viscous liquid phase that is immiscible in water. The LCST transition is reversible: the ELP coacervate is resolubilized when the solution temperature is lowered below Tt. In addition to temperature, a variety of other stimuli can be used to trigger this phase transition, including kosmotropic salts,[20–22] light,[23] crosslinking,[24] pH,[25] and redox potential.[26] Throughout this review we will explore how intrinsic and extrinsic parameters influence the phase transition behavior of ELP and ELP-hybrid materials and their organization into hierarchical supramolecular structures (Figure 1). We will focus on the synthesis of these materials and how the choice of molecular building blocks determines their self-organization into higher-order structures. Potential biomedical applications will also be discussed briefly.
Figure 1.
Various hierarchical self-assemblies of elastin-like-polypeptide and their hybrids. Created with BioRender.com.
2. A Brief Overview of ELPs
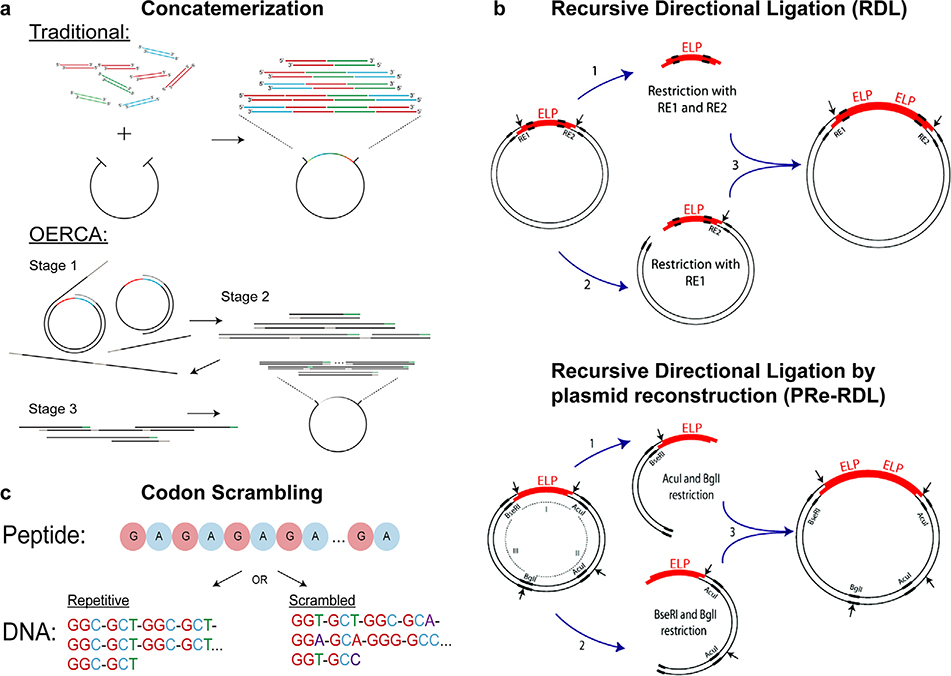
ELPs can be synthesized using several approaches. ELPs have been created through chemical synthesis (solid-state peptide synthesis) methods,[27,28] but this approach poses several challenges.[8,29] For example, during Boc-protected solid-state synthesis, the peptide can be degraded by hydrogen fluoride.[29] In addition, it is costly to synthesize long, complex peptides using solid-state synthesis.[30] ELPs are more affordably synthesized by recombinant DNA methods (Figure 2), which also allow more precise control of molecular weight and architecture.[31] There are two general approaches to using recombinant DNA methods to synthesize ELPs: concatemerization and directional ligation. Concatemerization involves ligating DNA oligomers with sticky ends successively using a ligase or thermal cycling (Figure 2a).[32] Concatemerization is straightforward and quick, but has two disadvantages: low yield and no control of ELP molecular weight.[21] As a result, directional ligation is more popular (Figure 2d). One type of directional ligation, called recursive directional ligation (RDL),[21] involves inserting an ELP oligomer into a linearized vector with a repetitive gene of interest to dimerize the original gene.
Figure 2.
Different methods to assemble genes encoding ELPs: a) concatemerization; b) OERCA; c) codon scrambling; d) recursive directional ligation (RDL); e) recursive directional ligation by plasmid reconstruction (PRe-RDL). (a–c) Reproduced with permission.[36] Copyright 2017, Wiley. (d,e) Reproduced with permission.[46] Copyright 2010, American Chemical Society.
Using RDL to synthesize ELP genes poses two challenges: i) circularization of the insert can prevent insertion into the vector and cause low-efficiency ligation; and ii) the symmetric Type II endonucleases required for RDL limit the peptide sequence that can be oligomerized due to overlap required between the recognition sequence of the enzyme and coding region.[33] Consequently, a variation of this approach called “RDL by plasmid reconstruction” (PRe-RDL) was developed (Figure 2e). In PReRDL, two halves of a parent plasmid, each with an oligomer, are ligated to generate the desired ELP product. This method places no restriction on the peptide sequence because it uses Type II’s restriction enzymes, which cleave at a specific position located away from the recognition sequence. In addition, these enzymes create overlaps that are complementary, and the two halves of the plasmid ligate to join genes seamlessly (with no extraneous nucleotides at the ligation junction). Furthermore, the plasmid is reconstructed only after successful ligation of the two gene products, ensuring that the vector contains both of the desired genes fused together.[33]
Another variation of RDL, overlap extension rolling circle amplification (OERCA), uses PCR to amplify repetitive sequences from a circular gene template (Figure 2b).[34] In OERCA, antisense primers bind to and extend the template—a circular, single-stranded DNA encoding repeats of the motif of interest— generating linear oligomers by rolling circle amplification. Then, sense primers bind to the extended linear sequences to produce double-stranded products of different lengths. Through a single cloning step, these products are blunt ligated into an expression vector and transformed into a host cell for protein production.[35] OERCA offers an advantage over other concatemerization methods because the use of a circular template provides significantly longer products than is possible with concamerization and overlap extension PCR for polymers with both short and long repeat units.[34,36] While OERCA generates double-stranded DNA oligomers with an expansive and adjustable range of DNA repeats simply by changing primer concentration and the number of PCR cycles, the technique’s stochastic nature remains limiting. In particular, if the polymer is made up of more than one type of monomer, the random distribution of monomers precludes precise control over the sequence.[36]
Until recently, PCR-based methods were not useful for engineering ELPs.[37] The characteristics of ELPs, notably their high GC content and repetitive nature, result in errors during PCR. The high GC content of ELPs increases their stability, and thereby, facilitates formation of secondary structures (i.e., hairpin loops), which impedes their denaturation during PCR. Furthermore, the repetitive nature of ELPs makes its gene fragments highly complementary to one another, leading to annealing of ELP fragments at multiple sites and generation of polydisperse products.[38] Tang and Chilkoti addressed these issues by creating a codon-scrambling algorithm that enables amplification of DNA sequences encoding repetitive polypeptides.[38] Based on the degeneracy of the genetic code (codon redundancy), this algorithm mathematically generates the least repetitive DNA sequence that encodes a repetitive polypeptide. The DNA sequence can then be chemically synthesized as oligonucleotides that are used to assemble long, repetitive gene sequences (Figure 2c).
Following the synthesis of genes that encode ELPs, the protein polymers can be expressed in Escherichia coli and purified using a simple non-chromatographic separation process called inverse transition cycling (ITC).[22] ITC exploits the ELP’s ability to undergo LCST phase transition at its inverse transition temperature (Tt).[20] Below its Tt, an ELP adopts a random coil conformation and is well solvated; as temperature increases above its Tt, the solution phase separates into an insoluble, ELP-rich coacervate phase and an ELP-poor aqueous phase.[20,21] This phase separation is reversible and can serve as an effective strategy to purify ELP (and ELP fusions) from contaminants by using temperature and salt to trigger the LCST phase transition.[22] When fused to a protein of interest, an ELP can act as a stimulus-responsive tag to enable purification of the fused protein by ITC.[20] An alternative approach to ELP purification is indirect ITC, in which ITC is combined with affinity capture methods. For example, Kim et al. tagged staphylococcal protein A (SpA), an antibody-binding protein, with an ELP, allowing ITC for the ELP-SpA fusion protein to be used to purify antibodies and antibody–antigen complexes.[39]
Urry et al. carried out the first biophysical studies on ELPs and determined that the (VPGVG)n pentapeptide at n > 200, like crosslinked elastin, exhibits unique temperature-dependent behavior that is unlike that of classical rubber.[40] In the absence of a load and under a constant applied force, both elastin and ELPs decrease in length as temperature is increased. Urry et al. determined that the phase transition temperature (Tt) of ELPs could be controlled by altering the hydrophobicity of the guest residue, X, in the VPGXG motif.[41] Building on this work, McMillan et al. determined that hydrophobic groups lower Tt, while hydrophilic groups increase Tt.[42] Meyer and Chilkoti later devised an equation to account for the effect of such sequence changes as well as two other important variables—molecular weight and concentration—on transition temperature.[43] This equation suggests that at a fixed pH, the chain length of the ELP is inversely related to Tt. Therefore, the ELP has a lower Tt at a higher molecular weight given the same composition and concentration. The transition temperature can be further tuned by incorporating noncanonical or unnatural amino acids as the guest residue.[44] In an extension of this line of enquiry, MacKay et al. developed a quantitative model using the Henderson–Hasselbalch relationship to explain the effect of pH—along with molecular weight and solution concentration—on the Tt of an ELP that contains ionizable guest residues. They designed two libraries of ELPs with pH-responsive phase behavior—one library with basic (histidine) guest residues, and the other library with acidic (glutamic acid) guest residues—to validate the model by evaluating phase separation of these ELPs. Their model predicts that the Tt of ELPs containing basic amino acids, such as histidine, decreases above the pKa of the basic residue, while the Tt of ELPs containing acidic amino acids, such as glutamic acid, increases above the pKa of the acidic residue. This model is useful to design ELPs that exhibit phase transition behavior due to a specific change in pH.[25] Finally, McDaniel proposed a quantitative model that predicts the Tt of a family of ELPs based on their composition, chain length, and concentration. Unlike the previous models described that do not incorporate composition in the model, this model accounts for the effect of composition on the Tt, and thus provides—as the output—ELPs with a specific amino acid sequence and chain length based on two inputs—the desired Tt at a specified concentration.[45]
3. Self-Assembly of ELP Block Copolymers
ELP block copolymers have been used extensively to create self-assembling micelles due to their ease of synthesis, biocompatibility, solubility, stimuli responsiveness, and potential applications in drug delivery.[21,47,48] Most previous work in self-assembly of protein polymers has focused on ELP diblocks. ELP diblocks are amphiphilic, with a hydrophobic block (containing a hydrophobic guest residue) and a hydrophilic block (containing a hydrophilic guest residue). The Tt values of each block are designed to be sufficiently different to allow independent desolvation of each block. At temperatures below the critical micelle temperature (CMT), the diblock ELP is soluble. Upon raising the temperature above the CMT, the hydrophobic block selectively desolvates, which turns the diblock ELP into an amphiphile and drives its self-assembly into a micelle, in which the more hydrophobic, desolvated block forms the core of a micelle, and the solvated hydrophilic block forms the corona.[37] These ELP micelles are stable over a range of temperature above the CMT. As the temperature is increased above a critical temperature, the hydrophilic block desolvates and undergoes hydrophobic collapse, resulting in a micelle-to-coacervate transition.[49] This critical—bulk transition—temperature is referred to the Tt of the diblock ELP. Changing the length or guest residue of either block allows control of the CMT and the Tt.
Conticello and colleagues provided the first example of an ELP diblock copolymer that exhibited temperature-triggered micelle assembly.[47] The ELP diblock was composed of a hydrophilic [VPGEG-(IPGAG)4]14 block and a hydrophobic [VPGFG-(IPGVG)4]16 block. The guest residues (in bold) were glutamic acid (E) and alanine (A) in the hydrophilic block, and phenylalanine (F) and valine (V) in the hydrophobic block. The large difference in polarity between the guest residues in the two blocks allowed temperature-triggered self-assembly into mostly spherical—and in some instances—cylindrical, micelles above the CMT. The presence of the ionizable glutamic acid also allowed the use of pH to control the size and shape of the micelles, making them sensitive to both pH and temperature.
Chilkoti and colleagues expanded the field of ELP diblock copolymers by systematically studying the behavior of ELP diblocks by varying the molecular weight and the molar ratio of hydrophilic and hydrophobic blocks.[50] Using a series of ten ELP diblocks, Dreher et al. showed (Figure 3a,b) that the unimer-to-micelle transition temperature is controlled by the length of the hydrophobic block, while the micelle size is controlled by both the length of the ELP diblock copolymer and the ratio of hydrophilic and hydrophobic blocks. They also showed that the ELP diblock copolymers exhibited monodisperse spherical micelles only within a certain range of hydrophilic-to-hydrophobic block ratios, while outside this range, a unimer-to-bulk transition occurred.
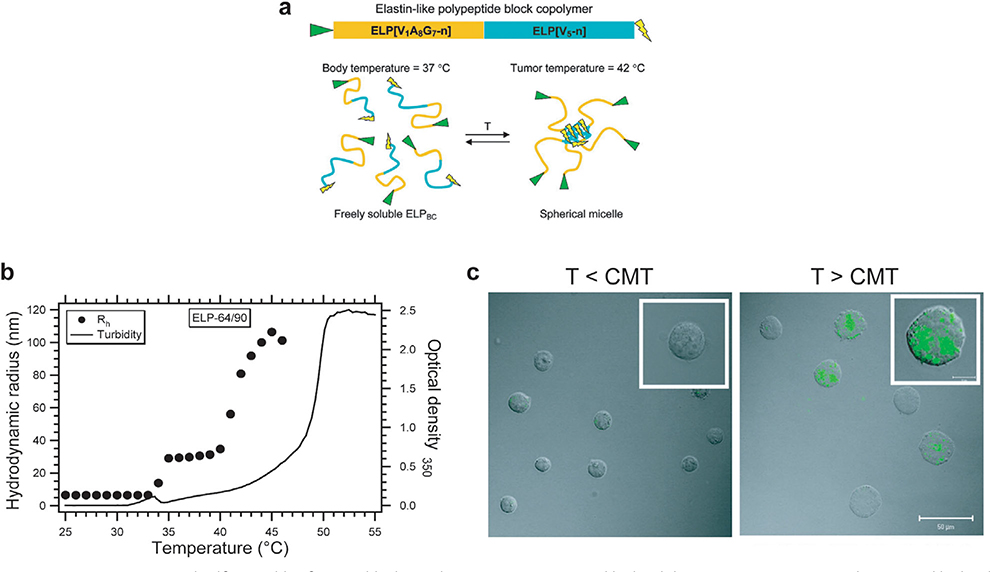
Figure 3.
Temperature-triggered self-assembly of an ELP block copolymers. a) An N-terminal hydrophilic ELP[V1A8G7-n] gene and C-terminal hydrophobic ELP[V5-n] gene are seamlessly fused together to create a gene that encodes an ELP block copolymer. When the size and ratio of the blocks are correctly selected, the ELP block copolymer self-assembles into a micelle close to body temperature. b) Hydrodynamic radius (Rh) and optical density profile at 350 nm of ELP[V1A8G7-64]/[[V5-90] block copolymer at 25 μm in PBS shows that ELPs exist as unimers (Rh of 5–10 nm) below CMT at 35 °C and form micelles (Rh 30–80 nm) above their CMT. c) Confocal fluorescence images of K562/αvβ3 cells treated with 10 μm ELP block copolymer with N terminal RGD ligand (green). Fluorescence images showed that there was no binding/uptake of RGD-ELP below the CMT but above the CMT there was significant binding/uptake of RGD-ELP due to the temperature-triggered multivalent display of RGD ligands. (a,b) Reproduced with permission.[50] Copyright 2007, American Chemical Society. (c) Reproduced with permission.[56] Copyright 2010, American Chemical Society.
Janib and colleagues furthered our understanding of design parameters that control temperature-triggered ELP diblock assembly by developing a mathematical model that can predict the CMT and the bulk transition temperature of ELP diblocks from the Tt values of their individual blocks.[51] Hassouneh et al. studied the assembly of six ELP diblocks and developed a theoretical—polymer physics—model that explains the mechanism of ELP diblock self-assembly into micelles.[52] They found that ELP diblocks assemble into “weak” spherical micelles with dense cores and unstretched coronas, making them distinct from micelles formed by analogous synthetic polymers that also undergo self-assembly.
Self-assembly of ELP block copolymers containing domains that target peptides, proteins, and receptor ligands allows multivalent display of the targeting moiety.[50,53–63] These biofunctional ELP block copolymers can be engineered to self-assemble in response to clinically relevant stimuli such as temperature or pH,[50,64] allowing modulation of the multivalency, affinity, and avidity of the targeting moiety to optimize cellular uptake.[65] Dynamic affinity modulation (DAM), developed by Chilkoti and co-workers, enables the use of a clinical stimulus to create drug carriers with a high-affinity “on” state and a low-affinity “off” state, for selective accumulation at a diseased site with low nonspecific activity. Simnick and co-workers demonstrated the proof-of-concept behind DAM by using a small tripeptide motif, RGD, that binds the αvb3 integrin receptor expressed in the tumor vasculature.[56] They showed that incorporating RGD in the hydrophilic sequence of the ELP block copolymer does not disrupt its self-assembly, and that multivalent display of RGD ligands on the corona of ELP micelles resulted in an increase in αvb3-mediated cell uptake of micelles compared to unimers of the diblock ELP that present a monovalent RGD peptide below the CMT (Figure 3c). The increased uptake was due to higher-affinity interactions between cells and multivalent micelles than between cells and monovalent micelles. Simnick et al. then compared cellular uptake of ELP diblock micelles displaying multivalent ligands with ELP monoblock aggregates after bulk transition and showed that the uptake of micelles was greater than the uptake of the ELP monoblock aggregates (Figure 3c). These results demonstrate the benefit of multivalent targeting in cellular uptake using nanostructures such as micelles.
Applying the DAM phenomenon, MacEwan et al. developed a “nanopeptifier” system that provides controlled intracellular delivery of anti-cancer peptide drugs by using the CMT of ELP diblocks to modulate the density of cancer cell-penetrating peptides displayed on the micelle corona.[57,61] Temperature-triggered self-assembly of micelles can be used to improve the accumulation of ELP-based therapeutics at diseased sites, especially for tumors that can be heated externally by mild hyperthermia. By using a tumor-targeting ELP diblock that exists as a unimer at body temperature (37 °C) and self-assembles at a clinically relevant hyperthermic temperature (42 °C), self-assembly can enhance accumulation of drugs delivered with ELP diblocks due to specific, high-affinity interactions between the tumor and multivalent micelles.
Adding proteins to ELP diblocks is well tolerated in nanoparticle formation.[58,60] Hassouneh et al. showed that adding proteins of 95–110 residues (≈10 kDa) at the hydrophilic end of ELP diblocks does not significantly affect their self-assembly.[60] They genetically appended two proteins, thioredoxin and the fibronectin Type III (Fn3) domain, to the hydrophilic block of two different ELP diblocks, and found that upon increasing the temperature to 39 °C, the diblocks self-assembled into micelles with a hydrodynamic radius (Rh) of 24–36 nm, and displayed multiple copies of the protein in the corona. Multivalent display of Fn3 resulted in increased avidity for αvb3 integrin. Similarly, Costa et al. developed an ELP micelle in which the hydrophilic block contained a nanobody—a small antibody fragment derived from camelid single-chain antibodies—to target human epidermal growth factor receptor.[58] In this system, the hydrophobic block incorporated the unnatural amino acid p-acetylphenylalanine for doxorubicin attachment. They found that the ELP diblocks retained their temperature-triggered self-assembly behavior and displayed the nanobody on the micelle corona, with doxorubicin sequestered in the core. These studies showed that addition of proteins of around 100 amino acids to the corona of ELP diblock micelles is well tolerated, demonstrating significant structural flexibility in the self-assembly of ELP diblocks into micelles.
To create ELP-based tumor drug carriers, Callahan et al. designed ELP diblocks that assemble at 37 °C and disassemble at the low pH found in the extracellular environment of solid tumors (≈pH 6).[64] To achieve this, they used a histidine-rich hydrophobic block. The pKa of histidine is near physiological pH (7.4). These diblocks self-assemble into micelles at pH 7.4, but dissemble when the pH drops to pH 6, as found in tumor tissue. At pH 6, the histidines are ionized, causing an increase in the Tt of the histidine-rich block, bringing its Tt value closer to that of the hydrophilic block. When the Tt values of the two blocks are no longer sufficiently different, micelle formation is energetically unfavorable, leading to micelle disassembly. Callahan et al. used autoradiographic imaging to show that these ionizable ELP nanoparticles disassemble at the low pH in tumor tissue, resulting in enhanced accumulation and penetration of ELP polymers in the tumor. They studied the intratumoral distribution of ELP polymers following intravenous administration of radiolabeled, pH-responsive micelles, and found enhanced and homogeneous tumor distribution in mice versus pH-insensitive micelles, possibly due to the increased diffusivity of the single ELP chains from the dissembled histidine-rich micelle. The pH-insensitive micelles accumulated at the periphery of the tumor, possibly due to a diffusion barrier. This is a promising work as it shows that control of micelle disassembly can address the tumor penetration limitation faced by many nanocarriers. The use of self-assembled ELPs as drug carriers is discussed in the next section.
An alternative to block copolymers to drive ELP self-assembly is fusing a hydrophobic peptide to the ELP C-terminus to create an amphiphile.[66] McDaniel et al. fused peptides of the repeat sequence (XGy)z (Figure 4), where X is hydrophobic, to an ELP C-terminus, and showed that self-assembly could be controlled by varying X and y (the number of glycines).[67] This is interesting and unusual because the length of the (XGy)z domain is only a few mass percent of the ELP, so that these diblock polymers are highly asymmetric amphiphiles, that would not, a priori, be expected to show self-assembly behavior. Surprisingly, these ELP peptide amphiphiles assembled into cylindrical micelles, with a cylinder length that could be tuned by varying the peptide sequence.[68,69] In an extension of this work, Bhattacharyya et al. used ELP-peptide amphiphiles as carriers for water-soluble chemotherapeutics.[70] They showed that various hydrophilic therapeutics could be conjugated within the core of the micelle, and that the ELP micelle improved the pharmacokinetics, biodistribution, and antitumor efficacy of the chemically conjugated—and sequestered—drug in vivo.
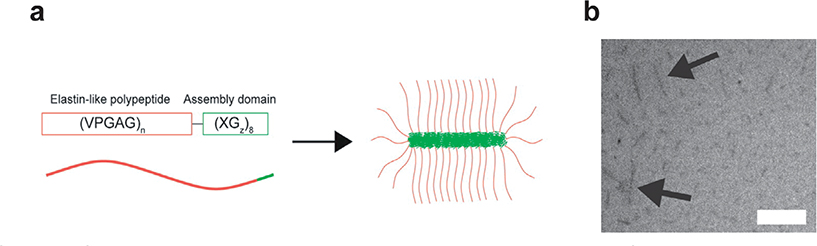
Figure 4.
Self-assembly of highly asymmetric ELPs with a short (XGz)8 assembly domain. a) Schematic of ELP asymmetric amphiphiles, where n is the number of pentameric repeats, X is an aromatic hydrophobic amino acid (Tyr, Phe, or Trp) responsible for driving self-assembly and z is the number glycine residues (G). b) Cryo-TEM micrographs of (VPGAG)160-(FGG)8 shows that this asymmetric amphiphile assembles into cylindrical micelles. Scale bar represents 200 nm. Reproduced with permission.[71] Copyright 2014, American Chemical Society.
More recently, MacKay and colleagues showed that fusing small proteins or peptides to hydrophilic ELP blocks can enable self-assembly into other nanoscale morphologies that include worm-like micelles and vesicles.[72,73] They showed that fusing an scFv with an ELP block results in self-assembly of a worm-like micelle with an scFv core and ELP corona. Although the scFv was located in the core, it remained active, likely because the corona was loosely packed.[73] When tested using a lymphoma xenograft model in vivo, these drug-loaded nanoworm structures outperformed their free monoclonal antibody counterpart (Rituximab). Another interesting example of ELP self-assembly is the formation of hollow spheres or vesicles. Pastuszka et al. demonstrated that fusing a small α-helical peptide L4F to an ELP forms vesicles with a radius of 49 nm and a 8 nm lamellae.[72] Self-assembly into a vesicle is driven by the amphipathic and self-association nature of L4F peptide.
The work described above used ELPs containing tens to hundreds of pentapeptide repeats. Shorter ELPs, with <10 pentapeptide repeats, are difficult to study due to their lack of phase behavior and the difficulty in synthesizing them recombinantly. Kiick and co-workers studied the self-assembly of short ELP sequences by using solid phase peptide synthesis and conjugating the ELP with hydrophilic, triple-helix-forming collagen-like peptides (CLPs).[74–77] A fusion of the ELP sequence (VPFGF)6 with the CLP motif (GPO)8 showed self-assembly of nanoparticles 50–200 nm in diameter, at temperatures ranging from 4 to 65 °C. These results were unexpected because the Tt of ELPs (and other thermoresponsive polymers) was thought to always increase when fused to a hydrophilic protein.[21,78–80] The pentameric repeat, (VPFGF)6, alone, was found to have a Tt of ≈37 °C in physiological buffer; fusing it to the hydrophilic (GPO)8 reduced the Tt dramatically, to less than 4 °C, which resulted in formation of nano-assemblies.[75] This large decrease in Tt was caused by the formation of a triple helix by the (GPO)8 collagen-like peptide, bringing three ELP domains together, raising the local ELP concentration that is ≈100-fold higher than that of ELP monomers in solution. This increased local crowding reduces the entropy necessary for ELP aggregation, thereby reducing the Tt. These nanostructures disassembled above 65 °C, the temperature at which the triple helix melts. Codon et al. subsequently demonstrated this ELP-crowding effect in atomistic and coarse-grained molecular dynamics simulations.[74] Self-assembly of ELP-CLP diblocks can also result in additional morphologies, including vesicles and platelets. Vesicle formation was achieved by using phenylalanine as a guest in the ELP motif, while platelet-like structures were formed when a more hydrophobic amino acid—tryptophan—was used as a guest residue.[77] Assembly into the platelet-like structures is thought to be driven by the stability afforded to the ELP layer from pi–pi stacking between the tryptophan indole side chains. TEM imaging experiments confirmed the predicted bilayer morphology with hydrophobic ELP chains forming the interior. The hydrophobic aromatic rings in the bilayer also allowed encapsulation of hydrophobic drugs within the bilayer, and hydrophilic drugs in the interior of the vesicle.[76] These nanostructures are highly stable, with a thermal stability up to 80 °C, making them useful for drug delivery applications.
Only a few studies have characterized multiblock ELP copolymers. MacEwan et al. studied the self-assembly of 11 multiblock ELP copolymers into higher-order structures as a function of temperature.[81] They maintained a constant overall amino acid composition and chain length while varying the size and distribution of the hydrophilic block (VPGSG) and hydrophobic block (VPGVG). The designs included alternating triblocks, alternating diblocks, and an architecture known as a gradient copolymer. They observed formation of micelles only when the number of contiguous hydrophobic pentapeptides was >35, which suggested that the hydrophobic block must be sufficiently long in order to drive self-assembly. The size and compactness of the micelles varied between the different block designs and were controlled by the block architecture (the gradient between the hydrophobic and hydrophilic blocks).[81] Martín et al. synthesized linear triblock ELP copolymers of the form “AEA,” where the A block was based on (VPGAG) and the E block was based on [(VPGVG)2-(VPGEG)-(VPGVG)2].[82] They showed that this triblock formed a hollow sphere or vesicle, observed initially as particles having a shape ratio (ρ, the ratio of the radius of gyration to the hydrodynamic radius) of 0.91, and confirmed by imaging the particles with TEM and AFM.
4. Self-Assembly of ELPs by Drug Conjugation
The clinical utility of many small molecule drugs is limited by their short half-life and nonspecific toxicity in vivo. Most small molecule therapeutics used in clinics are hydrophobic, which makes systemic administration inefficient. Packaging these drugs within hydrophilic macromolecular carriers can improve their solubility and efficacy. Moreover, packaging hydrophobic drugs within the core of soluble polymeric nanoparticles can also increase the accumulation of drugs in tumors via the enhanced permeation and retention effect that arises as a consequence of the leaky vasculature and poorly developed lymphatic drainage system present in the tumors.[83,84]
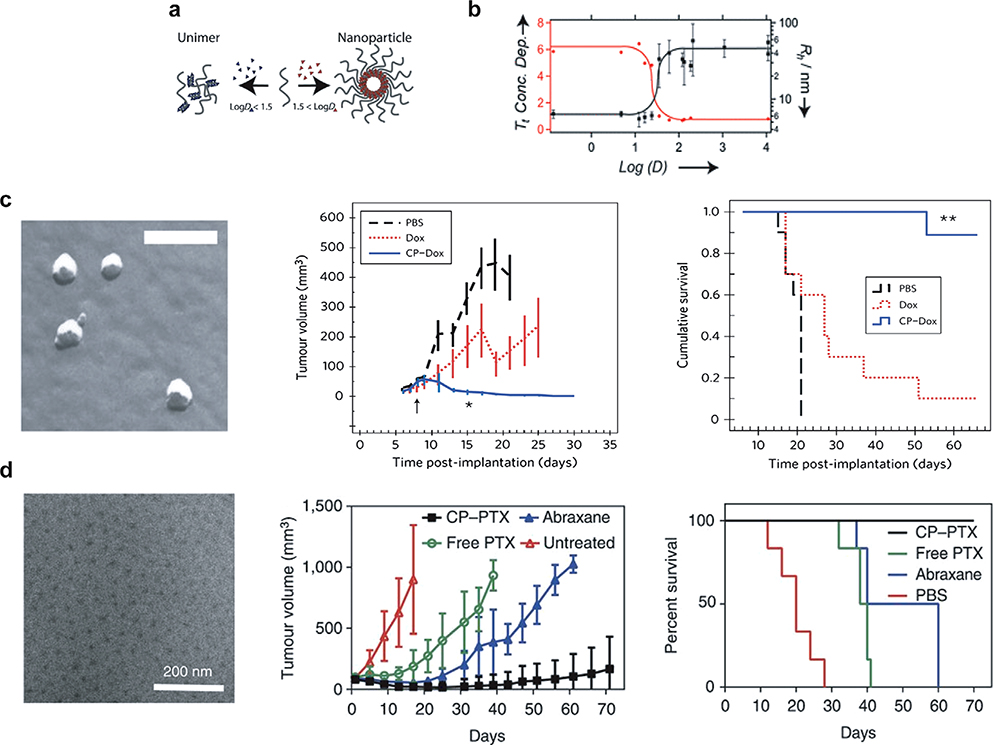
Many efforts have been made to improve the solubility and delivery of hydrophobic chemotherapeutics by using nanoscale formulations. Our group has demonstrated a simple method called attachment-directed assembly of micelles (ADAM) to create self-assembled ELP nanoparticles that sequester hydrophobic drugs within a micelle core.[85–88] ADAM enables the synthesis of ELP micelles by covalently attaching multiple copies of a small hydrophobic (CGG)8 domain to the C-terminus of a hydrophilic ELP.[37] This 1.6 kDa cysteine-rich domain provides eight sites for conjugating hydrophobic molecules with a diglycine spacer to minimize steric hindrance between conjugated drugs. Attachment of multiple copies of hydrophobic drugs to this ELP segment provides sufficient amphiphilicity to the ELP to trigger its self-assembly into near-monodisperse spherical micelles (Figure 5a,b). In an extensive study of 14 maleimide derivatives of small molecules spanning a range of hydrophobicity (measured using their octanol–water distribution coefficient, log D), McDaniel et al. found that a log D > 1.5 was necessary to trigger ELP self-assembly upon conjugation.[87] This work provides a basis for rationally designing micelle forming ELPs as drug carriers by predicting the propensity of a small molecule to trigger self-assembly of the ELP, based on the hydrophobicity of the drug.
Figure 5.
Self-assembly of ELP (labeled chimeric polypeptide (CP) in the figure panel) by drug conjugation. a) Attachment of three to six copies of small molecules with log D above 1.5 (shown in red) triggers self-assembly, whereas small molecules with log D less than 1.5 (shown in blue) does not trigger self-assembly. b) Hydrodynamic radius (Rh) (black curve) and transition temperature (Tt) (red curve) of ELP-small molecule conjugates as a function of log D. Below a log D of 1.5, conjugation of molecules does not trigger self-assembly, as the conjugates have an Rh of 6 nm, consistent with ELP unimers. As the log D of conjugate small molecules increase above 1.5, the ELP-conjugate self-assembles into micelles with an Rh of ≈30 nm. The concentration dependence of the Tt also decreases, indicating micelle formation. c) Conjugation of doxorubicin (Dox) to ELP triggers self-assembly into monodisperse micelles with an Rh of 22 nm as seen by freeze fracture scanning electron microscopy. In the C26 colon cancer model, ELP-Dox micelles (CP-Dox) at their MTD outperformed free Dox in reducing tumor growth rate and promoting survival. d) Conjugation of paclitaxel (PTX) to ELPs (labeled CP in the figure panel) drives self-assembly into mono-disperse spherical micelles as seen in the cryo-TEM micrographs. A single dose of ELP-PTX (CP-PTX) micelles outperformed both free PTX and Abraxane in a subcutaneous human prostate tumor model, PC3. All the mice in ELP-PTX group survived beyond 70 days (scale bar = 200 nm). (a-b) Reproduced with permission.[95] Copyright 2013, John Wiley and Sons. (c) Reproduced with permission.[96] Copyright 2015, Springer Nature. (d) Reproduced with permission.[97] Copyright 2009, Springer Nature.
Applying this methodology to clinically relevant small molecule therapeutics, we showed that conjugation of doxorubicin and paclitaxel to ELPs triggers self-assembly into monodisperse spherical micelles <100 nm in diameter (Figure 5c). Conjugation of doxorubicin or paclitaxel was mediated by a pH-labile hydrazone linker that enabled drug release in the acidic tumor microenvironment and endolysosomal compartments. In vivo, ELP-doxorubicin nanoparticles exhibited 14-fold greater drug accumulation in tumor than free drug.[85] A single dose of ELP-doxorubicin at its maximum tolerated dose (MTD) abolished subcutaneous C26 (colon carcinoma) tumors in mice (Figure 5c). Similarly, ELP-paclitaxel micelles showed favorable pharmacokinetics, with a sevenfold improvement in bioavailability versus free drug and a twofold improvement versus Abraxane, an FDA-approved taxane nanoformulation that is considered to be the gold standard for paclitaxel delivery (Figure 5d).[88] Notably, when mice with highly aggressive triple-negative breast cancer or prostate cancer were treated with a single dose of ELP-paclitaxel nanoparticles, their median survival was greater than 70 days, a significantly greater survival than when using Abraxane or free paclitaxel, consistent with the greater tumor growth inhibition achieved by treatment with ELP-paclitaxel than free drug or Abraxane.
Despite advances in cancer drug delivery using nanoparticles, upon systemic administration, most nanoparticles are taken up by macrophages in the liver and spleen and are cleared rapidly from the body. Rapid clearance leads to poor drug accumulation in the tumor, with a median of <1% of administered dose reaching the tumor. The other 99% is thought to accumulate in off-target organs or to be cleared by the liver and spleen.[89] A major advantage of the ELP drug delivery system is the ability to modify the micelle corona with stealth coatings that can increase drug circulation time. To that end, Banskota et al. showed that engineering the ELP repeat motif with two oppositely charged amino acids (a zwitterionic pair) can impart stealth behavior and increase the pharmacological efficacy of the carrier.[90] The optimal repeat unit of this class of stealth biopolymers, named zwitterionic polypeptides (ZIPPs), were identified by systematically varying the identity of the oppositely charged amino acids (X and Y) in the VPXYG repeat unit and the chain length to determine the sequence and chain length that optimizes the pharmacokinetics for intravenous and subcutaneous administration. A combination of lysine and glutamic acid in the ZIPP repeat unit was found to confer superior pharmacokinetics compared to an uncharged ELP of similar chain length and molecular weight. Similar to the parent ELPs that ZIPPs are derived from, ZIPPs also form self-assembled nanoparticles when hydrophobic molecules are conjugated to one end of the ZIPP chain, and drug-loaded ZIPP nanoparticles also exhibit stealth behavior and outperform ELP-drug nanoparticles in treating tumor xenografts in mice (unpublished).
In another approach to impart stealth properties to ELP nanoparticles, Yousefpour et al. developed a method to spontaneously coat the corona of ELP-doxorubicin nanoparticles with endogenous albumin in vivo.[91] This was achieved by fusing a ≈5 kDa protein—an albumin-binding domain (ABD)—to the N-terminus of a hydrophilic ELP which contains a (CGG)8 domain at the C-terminus for conjugation of doxorubicin or other hydrophobic drugs. Chemical conjugation of multiple doxorubicin molecules to one end of the ABD-ELP-triggered self-assembly into spherical micelles of ≈100 nm diameter. ABD-ELP-doxorubicin nanoparticles bound albumin with high-nanomolar affinity and had a significantly longer half-life and bioavailability in vivo than nonalbumin decorated ELP-doxorubicin nanoparticles. Moreover, the albumin coating lowered the uptake of nanoparticles by the liver and spleen, likely by preventing adsorption of serum proteins such as opsonins that help in phagocytosis of nanoparticles by macrophages.[92–94] The lower uptake in the liver and spleen resulted in higher accumulation in the tumor. The ability of these drug conjugation-triggered ELP micelles to outperform free drugs in tumor regression studies across multiple murine cancer models indicates that this ELP self-assembly platform is a robust approach to improving the efficacy of hydrophobic drugs.
5. Using Protein Order and Disorder in Hierarchical Assembly
A unique feature of ELP polymers that is useful for therapeutic applications is their LCST phase behavior, which allows stimulus-triggered assembly or disassembly.[43,98–100] This LCST behavior is intricately tied to the ELP molecular structure—or more accurately, the lack thereof.[101] Chilkoti—and others—have argued that ELPs represent a near-ideal elastomeric disordered protein,[102–104] and that the molecular malleability afforded by this disorder is a critical component of the thermodynamic origin of the polymer’s phase behavior.[101,105,106] Though critical for stimuli-responsive behavior, the structural disorder of ELPs limits the structural complexity available after aggregation. ELPs alone are largely constrained to the formation of micelles or microscale liquid-like coacervates with no internal architecture.[65,81,107] To expand on the nanostructures available to elastin-derived polymers, researchers have focused on introducing order into this otherwise disordered system.
Although the importance of protein disorder throughout the proteome (and its disruption of the well-established structure–function paradigm) is only now emerging,[108–116] the significance of molecular disorder has long been understood by researchers studying structural proteins such as silk, collagen, and tropoelastin. Such structural proteins derive their unique material properties from the synergistic combination of amorphous and structured regions.[117] Because ELPs provide a disordered stimuli-responsive template, they have been at the forefront of research to understand and exploit order–disorder interactions to create functional materials. Structured domains, ranging from short cell-binding domains to large globular proteins, have also been linked to, or encoded within ELPs to impart biological activity.[60,65,118–120] This article will not cover these efforts, as they are primarily concerned with adding biological activity while minimizing changes in architectural or biophysical properties. Rather, we will focus on the deliberate incorporation of ordered protein domains to affect the assembly of ELPs and other elastin-derived polymers.
5.1. Beta Structures
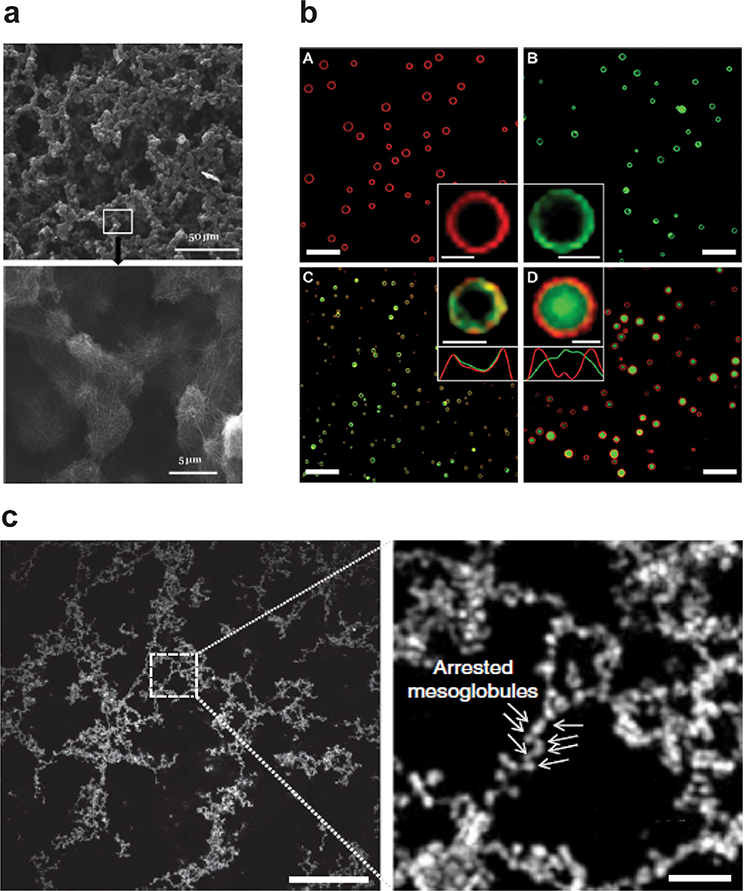
Silk has been studied intensively as a biomaterial due to its extensibility and strength.[113,115,116,121–126] This unique combination of material properties is derived from molecular interactions between amorphous protein chains and insoluble, tightly packed, β-sheet crystalline regions.[116] Efforts to functionalize silk for biomedical applications led to the development of silk-elastin-like polypeptides (SELPs), which combine tandem repeats of silk-like domains adopted from Bombyx mori silk heavy chains (GAGAGS) with ELP peptide blocks.[121,127] This work was pioneered in the mid-1990s by Ferrari[128] and Cappello[129] who, having recently developed new methods to create recombinant protein block copolymers,[130] sought to decrease the overall crystallinity of recombinant silk proteins. These first SELPs formed biocompatible, irreversible gels at body temperature with an increase in the number of silk blocks reducing the gelation time.[131] The first reversible SELPs were developed by Nagas et al. by substituting glutamine for a portion of the ELP guest residues, increasing the hydrophilicity of the polymer and making gelation sensitive to pH.[132] Though SELPS are known to create porous hydrogels, their self-assembly mechanisms have only recently been described. Xia et al. proposed that SELP aggregation proceeds via a two-step process where the structured silk domains promote the formation of micelles, which then serve as nucleation sites for gel formation above the polymer’s Tt.[133] Their work showed that increasing the ratio of β-sheet to disordered blocks reduces gel reversibility and leads to more fibrous assemblies. Zeng et al. showed that microscale porous hydrogels are derived from interconnected clusters of polymer nanofibers (Figure 6).[134] The stimuli-responsive behavior of SELPs and the diversity of structures they form, combined with post-processing techniques such as electrospinning or thin film deposition,[135,136] have made them useful for various applications in drug delivery (chemoembolics,[137] in situ enemas,[138] and adenovirus delivery[139]) as well as in tissue engineering,[140] and wound healing.[141]
Figure 6.
Incorporation of an ordered peptide domain into otherwise random ELP structure results in hierarchical self-assembly. a) Silk-elastin-like polypeptide self-assembles into nanofibers that form micrometer-sized clusters. The clustures are further organized into macroscopically continuous structures with defined microscale order as revealed by SEM images (scale bar 50 and 5 μm). b) Confocal micrographs show that incorporation of leucine zipper coiled coil into ELP backbone forms vesicles (scale bar 10 μm and in inset 1 μm). c) SEM analysis illustrates that at the mesoscale, a partially ordered polymer (POP) consisitng of an ELP and multiple copies of a periodically spaced oligoalanine helix forms a network of interconnected beads (scale bar 10 μm). Temperature-triggered phase separation of these POPs yields porous, physically crosslinked, viscoelastic networks. (a) Reproduced with permission.[156] Copyright 2014, AIP Publishing. (b) Reproduced with permission. [157] Copyright 2014, American Chemical Society. (c) Reproduced with permission [158] Copyright 2018, Springer Nature.
5.2. Alpha Helices
Unlike β-sheets, α-helices can introduce rigidity into a polymer via intramolecular interactions. This is similar to tacticity in synthetic polymers, where the organization of monomer chirality can be used to vary flexibility across the polymer backbone,[142] a strategy that has been used in the thermoresponsive protein PNIPAAM to tune its phase behavior.[143,144] α-helices can also be used to align amino acids via intermolecular interactions. The combination of both of these mechanisms is important in the formation of elastin fibers. Tropoelastin, the soluble precursor to elastin and the source of the ELP repeat, is composed primarily of alternating disordered hydrophobic domains and ordered polyalanine helical domains.[145] Initial coacervation is driven by tropoelastin’s disordered domains, followed by alignment of the more rigid helical domains, leading to crosslinking of the helical domains and assembly of fibers and networks.[106,146–149]
Building on this tropoelastin model, Roberts et al. recently developed a library of protein polymers for applications in tissue engineering and wound healing via the controlled introduction of helical polyalanine domains into an ELP backbone.[150] By systematically encoding helical domains similar to those of native elastin, they demonstrated that temperature-triggered phase separation of these partially ordered polymers (POPs) does not yield homogeneous coacervates, but instead yields porous, physically crosslinked, viscoelastic networks (Figure 6). By altering the composition and spatial distribution of the helical domains, mechanically stable protein networks could be created with controlled porosity, high surface-to-volume ratio, and fractal dimensions similar to native elastin networks. These physically crosslinked networks are formed by a unique α-helical domain swapping mechanism which provides them high kinetic and thermodynamic stability. Though the networks retain the thermal reversibility of disordered ELPs, the aggregation and dissolution temperatures can be controlled separately by adjusting the helical domains. The reversibility of phase separation could be tuned because the ordered components that drive thermal hysteresis are distinct from the disordered sequences that control the initial phase separation on heating.
Helical domains in ELPs can also be used to create hierarchical structures through sequence-designed biological interactions with high specificity and affinity. For example, dimerization of an α-helical leucine zipper has been used to drive ELP assembly. Leucine zippers are α-helical domains characterized by heptad repeat units that dimerize into coiled coils that are stabilized by intermolecular electrostatic and hydrophobic interactions. Leucine zippers have been used in biocompatible hydrogels,[151,152] and are particularly useful in protein polymers as they can be encoded into the protein backbone, eliminating the need for modification. Fernandez-Colino et al. created ELPs with paired leucine zippers at the N- and C-termini.[153] Above the Tt of the ELP, the synergistic effect of hydrophobic interactions of ELP and the dimerization domains resulted in the formation of an interconnected microporous network that could be used as a cell culture scaffold. Park and Champion used a single terminal leucine zipper to noncovalently bind globular fluorescent proteins to ELPs (Figure 6).[154] Unexpectedly, these constructs were able to self-assemble into microscale protein vesicles above the Tt of the ELP. The vesicles were stable for several days and could encapsulate a payload of polystyrene nanoparticles. The authors proposed that the rigid, rod-shaped conformation of the coiled coils, combined with the steric hindrance of the globular fluorescent proteins drove the formation of these vesicles.[155]
6. ELP-Hybrid Self-Assemblies
ELPs can be engineered with peptide domains to obtain higher-order structures. These ELPs are, in most cases, synthesized using recombinant DNA methods and using the 20 canonical amino acid sequences, which restricts their design space. Their recombinant expression can be precisely controlled because of the highly regulated transcription and translation processes that create monodisperse polypeptides. In contrast, synthetic polymers have a vast chemical repertoire to tune their chemical and physical properties, but control over their structure, stereochemistry, and dispersity is limited. Hybrid materials that combine the far larger chemical space accessible to synthetic polymers with the precision of recombinant polypeptides have been created by modifying ELPs with various biomolecules and synthetic molecules, including: lipids, unnatural amino acids, silica, hyaluronic acid, and synthetic polymers such as poly(ethylene glycol) (PEG), poly(acrylic acid) (PAA), polyethylenimine, poly(γ-benzyl-l-glutamate) (PBLG), poly(2-vinyl-4,4-dimethylazlactone), and polydimethylsiloxane. This has been achieving using recombinant DNA methods, chemical synthesis, and a combination of the two (Table 1).
Table 1.
Self-assembling behavior of ELP-hybrid materials.
| Synthetic motif | ELP sequence | Method of appendment | Self-assembling behavior | References |
|---|---|---|---|---|
| Palmitoyl chain | (VPGXG)3; X = A/I/V | Synthetic: Fmoc-chemistry | Nanofiber | [159] |
| Myristoyl chain | (VPGXG)40–120; X = A:V 9:1 | Recombinant: PTM | Spherical micelle | [160] |
| Peptide amphiphile | (VPGVG)40 | Recombinant: PTM | Dynamic: worm-like to nanofiber | [161] |
| Peptide amphiphile | [(VPGVG VPGEGVPGVG VPGVG)10-(VGIPG)60]2-[(VPGIG)10-AVTGRGDSPASS(VPGIG)10]2 | Physical mixing | Dynamic: membrane to 3D tube | [162] |
| Cholesterol | [G(V8/A2)GVP]10GKG] | Recombinant: PTM | Ellipsoid micelle | [161] |
| DSPE-PEG | (VPGVG)20 (VPGVG)128 |
Synthetic: NHS-chemistry | Liposomes | [163,164] |
| Stearyl | (VPGVG)2–6 | Synthetic: coupling chemistry | Liposomes | [165–167] |
| p-azidophenyl alanine | (VPGVG)20 & (VPGVG)20-(VPGSG)80 | Recombinant: UAA technology | Nano to microscale gel particle | [168] |
| PEG | (VPGXG)40–130; X = V:L:G 5:2:3 | Synthetic: SPAAC | Spherical micelle | [169] |
| PAA | (VPGVG)2 | Synthetic: Cu-catalyzed click reaction | Spherical aggregate | [170] |
| PBLG | [(VPGVG)(VPGMG)(VPGVG)2]10 | Synthetic: ROP | Spherical micelle, worm-like, vesicle | [171] |
| Silica | (VPGVG)60-(VPGXG)60; X = A:G 1:1 | Synthetic: Silification | Clustered sphere to discrete particle | [172] |
Self-assembly behavior depends on extrinsic parameters like temperature, concentration, solvent, self-assembling technique, etc. These parameters have been described in the text but not in this table.
6.1. ELP-Lipid Hybrid Materials
Adding only a single lipid moiety to an ELP can provide sufficient amphiphilicity to drive self-assembly. The first ELP-lipid hybrid that could independently self-assemble into a nanostructure was reported by Aluri et al. in 2012 (Figure 7a).[159] They used Fmoc chemistry to conjugate two palmitoyl chains to the N-terminal lysine of an ELP consisting of (VPGXG)3, where X is alanine, valine, or isoleucine. This ELP-palmitoyl hybrid was insoluble in water with valine or isoleucine as the guest residue, but was soluble with alanine as guest residue and underwent a phase transition to form a nanofiber at ≈56 °C at 1 mm concentration. Without the palmitoyl chains, the ELP sequence exhibited no phase separation below 100 °C. At 25 °C, CD spectra of the ELP alone showed a random coil structure, while spectra of ELP-palmitoyl showed significant proportion of both random coil and β-sheet conformation. As the solution temperature approaches the Tt, the proportion of β-type-1 turn increased. Interestingly, a highly disordered structure of ELP-palmitoyl (similar to native tripeptide) above its Tt led the author to suggest that the free energy of mixing during bulk phase separation may be partly compensated by the change in entropy from a highly ordered state (below the Tt) to a disordered state (above the Tt). The length of the ELP-palmitoyl nanofiber could be tuned by using dioleoylphosphatidylethanolamine (DOPE) as a capping agent without affecting the nanofiber width: increasing the concentration of DOPE reduced the fiber length. The aspect ratio of the nanofiber correlated with cell uptake, which increased with DOPE concentration. It has been proposed that nanoparticle–biomembrane interaction, cellular uptake, and intracellular trafficking of the nanoparticle invariably depend on the physicochemical properties of the nanoparticle such as shape, size, and surface chemistry.[173] The increased cellular uptake of ELP-palmitoyl nanostructure could be due to increased fusogenicity of the particle (doped with DOPE) to the phospholipid bilayer of the cells or due to the reduced size of the ELP-palmitoyl nanofiber caused by adding DOPE. The reduced size of the nanofiber may increase cellular internalization through nonphagocytic pathways like clathrin-mediated endocytosis.[159] The hydrophobic core of the fiber effectively solubilized paclitaxel, and the nanostructure showed cytotoxicity against multiple cancer cell lines.[159]
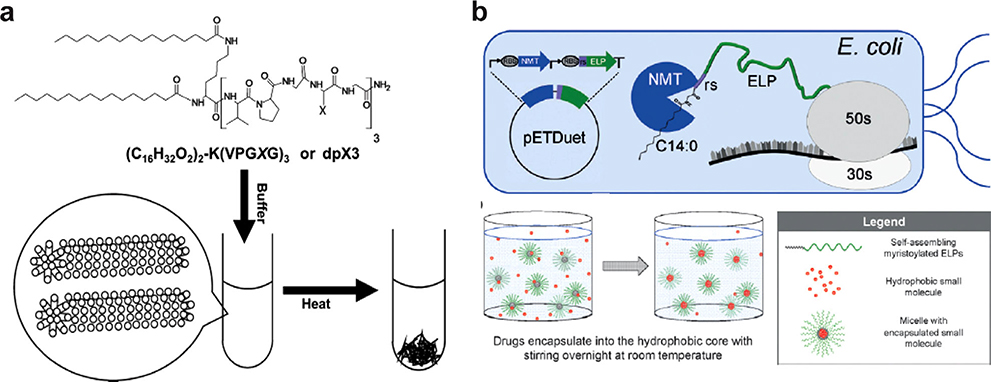
Figure 7.
Conjugating two C16 lipids to a short ELP oligomer leads to formation of cylindrical micelles with a) inverse phase transition behavior whereas attaching a single C14 lipid to a long ELP results in formation of b) monodisperse spherical micelles. The lipid chain can be appended through multi-step chemical synthesis in a reaction vessel (a) or recombinantly via a single step, one-pot post-translational modification in a reprogrammed bacterial cell (b). (a) Reproduced with permission.[159] Copyright 2012, American Chemical Society. (b) Reproduced with permission.[160] Copyright 2017, John Wiley and Sons.
The attachment of lipids to ELPs to create an amphiphile and drive self-assembly can also be achieved recombinantly by post-translational modification (PTM), a strategy evolved by nature to diversify the proteome beyond the 20 canonical amino acids. This approach was first used by Luginbuhl et al. through in situ enzymatic myristoylation of an ELP that contained a peptide substrate for myristoylation at the N-terminus (Figure 7b).[160] They did so by a single step, one-pot recombinant synthesis that used the transcription machinery of an E. coli reprogrammed to perform enzymatic lipidation within the cell. To do so, a 11 amino acid substrate peptide from the native myristoylated yeast protein Arf2[174] was fused to the N-terminus of a hydrophilic ELP consisting of (VPGXG)n, where X is 90% alanine and 10% valine and n = 40, 60, and 120. The ELP-peptide fusion was co-expressed with an N-myristoyltransferase in BL21(DE3) E. coli using a bicistronic expression vector. N-myristoyltransferase catalyzes the reaction between the N-terminal amine of the glycine and the activated thioester of myristoyl-CoA.[175] The addition of exogenous myristic acid during protein expression enabled in situ myristoylation of the ELP backbone. Myristoylation of the ELP introduced sufficient amphiphilicity to drive self-assembly, with myristic acid forming the core, and the hydrophilic ELP forming the corona. Interestingly, the larger 80- and 120-meric ELPs formed more stable spherical micelles, whereas the smaller 40-meric ELP formed a rod-like micelle which grew in hydrodynamic size over time. These lipid-ELP biopolymers differ from ELP diblocks and ELP-peptide fusions because the micellar core of lipid-ELPs contains lipids rather than only peptides, so it is significantly more dehydrated, allowing physical encapsulation of hydrophobic molecules in the core. Indeed, the ELP-myristoyl micelle encapsulates hydrophobic chemotherapeutics such as paclitaxel and doxorubicin more efficiently than the ELP diblock micelle.[160]
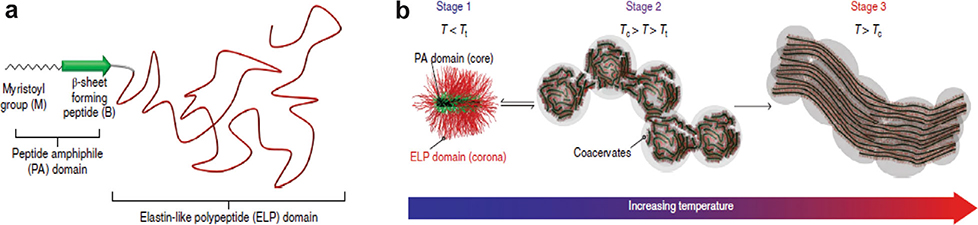
To induce the formation of additional structures by ELP-myristoyl, Mozhdehi et al. introduced a β-sheet-forming peptide with a myristoylation substrate at the ELP N-terminus to create a peptide amphiphile which they name a fatty-acid-modified elastin-like-polypeptide, or FAME (Figure 8a).[161] Upon myristoylation, the β-sheet-forming peptide acts as a peptide amphiphile (PA) that is known to exhibit an array of self-assembled structures with different morphologies.[176,177] They hypothesized that upon myristoylation, the peptide amphiphile would drive formation of hierarchical structures. This lipid-modified polypeptide formed a worm-like micelle below the ELP Tt, and above the Tt, underwent a phase transition to a liquid-like coacervate and formed spherical droplets. Above a critical temperature Tc (Tc > Tt), as the ELP corona was dehydrated, promoting noncovalent crosslinking in the core and irreversible formation of bundled fibers (Figure 8b). The attractive feature of FAMEs is that they retain both the temperature-triggered phase separation behavior of ELPs and hierarchical self-assembly of PAs. The ability of a FAME to transition from a solution to bundled fibers between room and body temperature make them attractive as injectable biomaterials for tissue regeneration and repair. In a similar example, Karla et al. co-assembled a peptide amphiphile containing a saturated alkyl chain and a short peptide sequence with an ELP in aqueous solution, resulting in dynamic interfacial assembly into a 3D tube above the ELP Tt.[162]
Figure 8.
a) There are three distinct building blocks in FAMEs: a myristoyl group, an ordered β-sheet-forming domain, and a disordered ELP domain. The myristoyl group and β-sheet-forming domain together forms a peptide amphiphile (PA). b) Below Tt (T < Tt), cylindrical micelles are formed due to the attractive forces of the PA core and the repulsive force of the hydrated ELP corona (stage 1). Above Tt (Tc > T > Tt) the dehydrated ELP domain undergoes an LCST phase transition into a liquid-like coacervate and forms spherical droplets (stage 2). Above Tc the ELP gets further dehydrated decreasing repulsion between the coronas that drive macroscale self-assembly (stage 3). Reproduced with permission.[178] Copyright 2018, Springer Nature.
PTM of ELPs is not limited to the ELP N-terminus or to incorporation of saturated alkyl chains. The Chilkoti group incorporated a cholesterol moiety at the C-terminus of an ELP fusion.[161] Previous work by Beachy and colleagues had demonstrated that PTM with cholesterol could be achieved by using proteins in the Hedgehog family.[179] In Hedgehog (Hh) proteins, an N-terminal signal processing domain (HhN) is fused to an autoprocessing C-terminal domain (HhC). HhC is homologous to intein-like proteins and contains a sterol-binding site that undergoes an N→S acyl shift upon binding cholesterol.[180] This intramolecular rearrangement leads to the formation of an active thioester intermediate that reacts with the 3β-hydroxyl of the bound cholesterol, resulting in modification of the signal processing domain with cholesterol and the release of HhC. To attach a cholesterol moiety recombinantly, Chilkoti and co-workers designed a fusion protein with three segments: ELP, HhC, and a histidine tag. The hydrophilic ELP replaces the N-terminal signaling domain of Hh and contains a linker peptide, (GGS)2, at the C-terminus to ensure minimal interference with the autoprocessing activity of the HhC. The HhC processes exogenous cholesterol to tether it to the fusion protein. The histidine tag allows visualization of the expressed protein by Western Blot and purification by affinity chromatography. The fusion peptide was expressed in BL21(DE3) E. coli and cholesterol was solubilized in the cell lysate by addition of a surfactant, leading to efficient lipidation of the ELP at the C-terminus and concomitant release of HhC-His.
These cholesterol-modified polypeptides (CHaMPs) exhibited programmable self-assembly, forming spherical micelles. A hydrophilic ELP was chosen to compensate for the attachment of cholesterol (log P > 7) so that the Tt would be physiologically relevant. To demonstrate the utility of CHaMPs, Mozhdehi et al. decorated ELP-HhC micelles with the peptide exendin-4, a peptide drug approved for the treatment of Type II diabetes.[161] The exendin-ELP-cholesterol conjugate (ExeELP-HhC) self-assembled into spherical micelles. Depending on the blood glucose level, exendin-4 binds glucagon-like peptide-1 receptor (GLP-1R) to increase the intracellular concentration of cyclic adenosine monophosphate (cAMP), which initiates a cascade of signaling leading to secretion of insulin. By using HEK293 cells that express GLP-1R, they demonstrated that the Exe-ELP-HhC micelle is a potent agonist of GLP-1R that outperforms monomeric Exe-ELP.
6.2. ELP-Liposomes
Liposomes—biomimetic spherical vesicles that enclose a watery interior—are composed of lipid bilayers. As drug carriers, liposomes exhibit extended plasma circulation, biodegradability, immunogenicity, and low toxicity in vivo.[181] Stimuli-responsive liposomes have been developed for drug release triggered by pH, ultrasound, light, magnetic fields, and temperature.[182–191] Recently, thermoresponsive ELP-liposomes have gained attention for their ability to allow release of drugs when exposed to mild hyperthermia.[163–167,192] Due to the hydrophobicity of the lipid bilayer, ELPs in ELP-liposomes have been modified with either a reactive group to decorate the hydrophilic head group of the liposomes that is exposed to aqueous solution,[163,164] or a lipid moiety to anchor the ELPs into the lipid bilayer.[165–167] The Tt of the ELPs has been tuned to ≈40 °C to match the temperature suitable for mild clinical hyperthermia of tissues. The hydrophilic head group of lipid amphiphiles such as DSPE-PEG-amine has been designed to accommodate an active ester functionality that can be reacted with the N-terminus of the ELP.[163,164] Such modification results in exposure of the ELP on the outer surface of liposomes, making the liposomes thermosensitive. Above their Tt, ELPs conjugated to the liposome surface desolvate and become hydrophobic, leading to aggregation of the liposomes. This aggregation distorts the liposome membrane, triggering release of cargo. Kyunga et al. showed that the ELP phase transition leads to enhanced cell uptake of ELP-liposomes compared to PEGylated liposomes at 42 °C, while at 37 °C both types of liposomes were internalized to the same extent.[163] Above Tt, the dehydrated ELPs on the liposome surface improved cell adhesion, presumably by creating a lower steric barrier to cell adhesion than PEGylated liposomes.[193,194]
Anchoring ELP into the liposome lipid bilayer requires adding a hydrophobic tail to the ELP. A saturated, 18C stearyl group is often added at the ELP N-terminus to append a hydrophobic tail. Kim and co-workers showed that by controlling the ELP length, ELP-stearyl concentration, and co-lipids, it is possible to create a liposome which releases doxorubicin under mild hyperthermia.[165–167] Under hyperthermic conditions, ELP-stearyl conjugates phase separate and the ELP undergoes a conformational change from random coil to β-turn that destabilizes the liposome membrane, leading to release of doxorubicin. Veneti et al. used a pH-sensitive ELP linker to decorate a liposome surface with the cRGD tumor cell targeting domain.[192] They showed that a pH-triggered conformational change of the ELP can activate RGD-mediated cancer cell targeting of liposomes when the pH changes from 7.4 to 6.
6.3. Unnatural Amino Acids in ELP Self-Assembly
Incorporation of noncanonical amino acids into ELPs expands the design space and allows additional functionality to the ELP and the option of bio-orthogonal conjugation of extrinsic moieties to the ELP. Different strategies have been used to introduce noncanonical amino acids into ELPs. Wu et al. used E. coli MRA30, which supports multi-site suppression for noncanonical amino acid incorporation to produce elastin-mimetic polymers with noncanonical amino acids at up to 22 positions.[195] Amiram et al. incorporated noncanonical amino acids in polypeptides by E. coli expression by developing aminoacyl-tRNA synthetases with tunable specificities for 14 noncanonical amino acids.[196] They synthesized ELPs with a VPGXG scaffold, where X is a noncanonical amino acid, and incorporated 30 noncanonical amino acids per ELP with yields of up to ≈50 mg L−1 in shaker flask culture. The site-specific incorporation of the noncanonical amino acids p-azidophenylalanine and p-acetylphenylalanine in these studies allow biorthogonal click chemistry and ketone-mediated conjugation to ELPs. Similarly, Teeuwen et al. replaced an ELP guest residue with azidohomoalanine and homoproparylglycine and showed that the resulting ELPs could be coupled through copper-mediated click chemistry to various biomolecules including probes, polymers, and enzymes.[197]
Incorporation of unnatural amino acids can affect ELP self-assembly. Kim et al. assessed the role of stereoelectronic effects on the conformational stability of elastin-mimetic polypeptides with the motif VPGVG.[198] They synthesized three different ELPs—that they named elastin-1, elastin-2, and elastin-3—by incorporating (2S)-proline, (2S,4S)-4-fluoroproline, and (2S,4R)-4-fluoroproline, respectively, into the elastin scaffold. They then conducted calorimetric and spectroscopic analyses of these protein polymers. Elastin-3 exhibited a lower Tt and a greater Type II β-turn population versus the parent ELP, while elastin2 demonstrated the opposite. The experimental data supported the notion that proline substitution alters the energetics of the β-turn conformation peptide self-assembly via stereoelectronic effects. These results were confirmed by density functional theory modeling of the turn types for the elastin-mimetic repeat. Incorporation of (2S, 4R)-fluoroproline stabilized Type II β-turn structures, while incorporation of (2S, 4S)-fluoroproline destabilized these β-turn structures when compared to unmodified ELP.
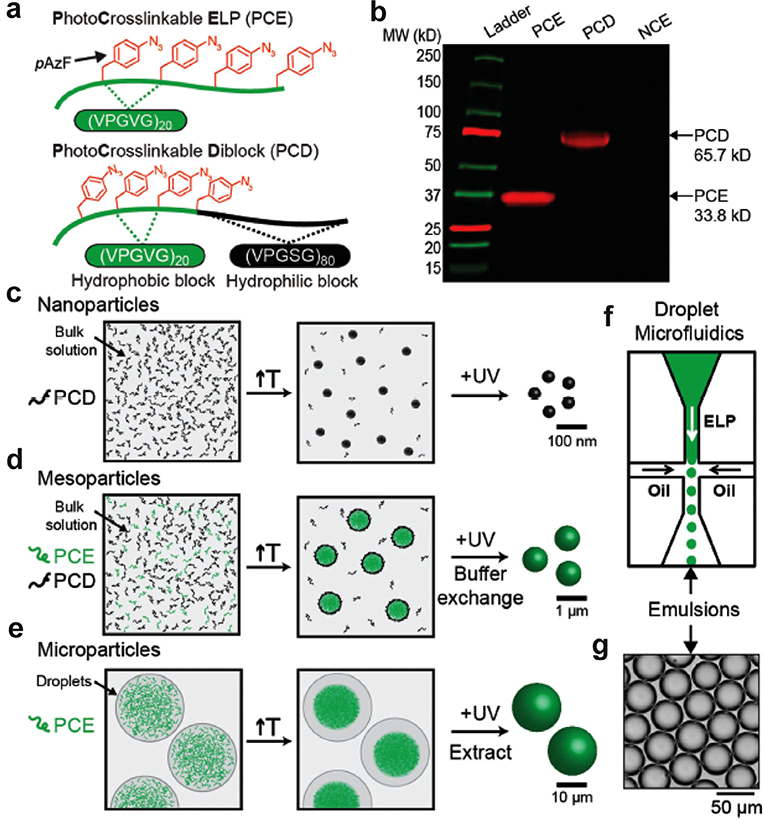
Costa et al. developed a bio-orthogonal crosslinking approach by genetically encoding a photoreactive unnatural amino acid, para-azidophenylalanine (pAzF), into an ELP at the X position of the VPGXG repeat (Figure 9).[168] They designed two pAzF-containing ELPs: i) a monoblock photocrosslinkable ELP (PCE) consisting of (VPGVG)80 with four regularly spaced pAzF residues, and ii) a self-assembling photocrosslinkable diblock (PCD) with an N-terminal hydrophobic ELP block containing four equispaced photocrosslinkable pAzF residues and a C-terminal hydrophilic ELP block that did not contain any pAzF residues. Following synthesis, crosslinked hydrogel particles were fabricated by tuning the temperature and by UV crosslinking with sizes that spanned multiple orders of magnitude from the nanoscale to microscale. At the nanoscale, a solution of the PCD formed spherical micelles above its CMT, and the micelles could be photocrosslinked by UV irradiation to form a nanogel. At the mesoscale, they created particles that ranges from several hundred nanometers to a micrometer by using a mixture of a PCE with PCD, where the PCD acts as a surfactant. By tuning the ratio of the PCD to PCE, the size of the coacervate of the PCE could be systematically tuned from several hundred nanometers to the micron size regime. UV irradiation was then used to photocrosslink the coacervate particles and the size of the particles was dictated by the ratio of the diblock ELP surfactant and PCE in solution. At the microscale level, a microfluidic droplet generator was used to generate microscale water-in-oil emulsion droplets with the photocrosslinkable ELP. Coacervates were formed by increasing the temperature above the Tt of the ELP, and the coacervates were then UV crosslinked to form microscopic gel particles. This work illustrates the incorporation of unnatural amino acids into ELP scaffolds as a strategy to create hydrogels with a range of particle sizes.
Figure 9.
Disordered ELPs containing photocrosslinkable unnatural amino acids form thermoresponsive gel particles with controllable sizes. a) Schematic representation of the two disordered ELPs—PCE and PCD. b) SDS-PAGE gel electrophoresis of PCE, PCD, and NCE (non-crosslinkable ELP, (VPGVG)80) stained with DBCO-Cy5. Strategy for generating: c) nanoparticles, d) mesoparticles, and e) microparticles. f) Chip-based microfluidic droplet generator yields g) monodisperse ELP containing water droplets. Reproduced with permission.[196] Copyright 2018, John Wiley and Sons.
6.4. Synthetic Polymer-ELP Hybrids: The Best of Both Worlds
PEG has been studied extensively as a hydrophilic polymer block that drives self-assembly of hydrophobic core-forming polymers and confers colloidal stability and stealth properties to the PEG decorated nanoparticle. Nanoparticle delivery vehicles have often been decorated with small (1–5 kDa) PEG chains to escape opsonization and prevent enzymatic degradation.[200,201] The use of PEG as the hydrophilic corona of ELP-micelles was first studied in 2014 by Hest and co-workers.[169] They modified the two free amine groups of an ELP with azide using a metal-free diazo transfer reaction, and modified the PEG backbone with a bicyclo-nonyne moiety. The ELP-PEG conjugate was synthesized by using a strain-promoted alkyne–azide cycloaddition (SPAAC). Conjugation of the hydrophilic PEG to the ELP lowered the Tt of the conjugate compared to the unmodified ELP. The ELP-PEG conjugate self-assembled into a spherical micelle in which the ELP formed the core and PEG formed the corona of the micelle. The micelle size and polydispersity depended on i) ELP length, ii) PEG molecular weight, and iii) the number of PEG chains per ELP. They showed that di-PEGylated ELPs formed monodisperse micelles with a lower polydispersity index than their mono-PEGylated counterpart. However, even with mono-PEGylated ELPs, higher molecular weight PEG formed micelles with lower polydispersity than lower molecular weight PEG. The data also showed that polydispersity increased with decreasing ELP chain length, possibly because of diminished hydrophobic interactions within the micellar core.[169]
Kiick and co-workers synthesized PAA containing elastin-mimetic copolymers by using copper-catalyzed azide–alkyne cycloaddition of poly(tert-butyl acrylate) diazide and a short elastin-like peptide X(VPGVG)2X (X: propargyl glycine).[170] These block polymers formed a polydisperse spherical aggregate upon slow titration of dimethyl sulfoxide (DMSO) solution with water. Interestingly, the block polymers did not contain a specific hydrophobic domain; instead, their self-assembly was driven by inter- and intramolecular hydrogen bonding. By using coarse-grained modeling in conjunction with replica-exchange molecular dynamics simulations, the authors showed that self-assembly was driven by intra- and intermolecular interactions of the polymer block. The interactions between ELP blocks did not significantly influence the shape, size, or conformational stability of the aggregates. The authors concluded that the PAA block is not ideal for use in a polymer-polypeptide hybrid in which protein domain interactions are intended to drive assembly.
ELPs have been also used as the hydrophilic domain in ELP-synthetic hybrid polymers. A study by Le Fer et al. showed that self-assembly and hierarchical nanostructure of these hybrid polymers depended not only on the ratio of hydrophilic and hydrophobic repeats in the block, but also on the methods used to induce self-assembly.[171] They synthesized a diblock polypeptide with a high control of polymerization by using the N-terminal group of a hydrophilic ELP as the macroinitiator for a ring-opening polymerization of γ-benzyl-l-glutamate N-carboxyanhydride (γ-BLG NCA). Using this approach, they synthesized a series of hybrid co-polypeptides with different PBLG block lengths (and thus different hydrophobic contents) with low dispersity. The amphiphilic hybrid diblocks were self-assembled using direct dialysis against water or by using a microfluidic chip system where the solvent polarity was controlled by changing the DMSO:water ratio. Diblocks with lower hydrophobic fractions (25–37%) formed spherical micelles irrespective of the self-assembly method. When the hydrophobic fraction was increased to 55%, the diblocks formed interconnected worm-like micelles. However, when the DMSO content was reduced, distorted aggregates were formed. The decrease in DMSO reduced the solubility of the hydrophobic block, likely reducing the chain mobility of the polymer to the extent that self-assembly and structural reorganization of the aggregates became too slow to establish a dynamic equilibrium.[171] The authors concluded that a diblock polymer with higher hydrophobic content became kinetically trapped during the experimental time scale to form a metastable, distorted aggregate. Direct dialysis against water resulted in a polydisperse spherical micelle, whereas the microfluidic chip produced a well-defined vesicular structure when the hydrophobic content was further increased to 70%.[171]
López and colleagues reported the synthesis of near-monodisperse, discrete hybrid ELP-silica particles by inducing a silification reaction on the micellar corona.[172,202,203] A silicon affinity domain from lysine-rich silaffin R5 peptide was fused with the hydrophilic N-terminus of a diblock ELP.[172] Formation of the micelle exposed the silaffin domain, resulting in a high density of positively charged residues at the corona. This facilitated mineralization of silica on the corona in presence of silicic acid. The silification reaction in the presence of phosphate anions (at 37 °C for 30 s) resulted in large, highly polydisperse, clustered silica spheres. The control diblock ELP, lacking the silaffin domain, did not induce mineralization. Interestingly, in the absence of the phosphate anion, the silification produced discrete, monodisperse ELP-silica particles. Cryo-TEM images showed solid particles packed in a near-hexagonal lattice.[172] Similar ELP-silica conjugates have been developed for drug delivery[202] and for functional modification of silica surfaces.[203]
7. Injectable ELP Drug Depots and Hydrogels
Precise control of the ELP Tt via sequence, size, concentration, and solvent allows the creation of hydrogels from soluble ELPs or ELP-hybrids via phase separation. ELP Tt can be designed to have a Tt below physiological temperature and can be injected in their soluble form at room temperature. The physiological temperature experienced by the injected ELP then triggers their phase separation into an insoluble coacervate in vivo. The Tt can be tuned to between room and physiological temperature by control of two sequence-specific variables: 1) hydrophobicity of the guest (X residue). ELPs that contain guest amino acids with moderately hydrophobic side chains (e.g., valine, isoleucine, and alanine) phase separate below 37 °C across a range of ELP MWs.[204–212] 2) MW: for a given ELP sequence the Tt can be further tuned by its MW as the Tt decreases with increasing MW. Simultaneous optimization of both variables is usually sufficient to design an ELP that will phase separate at a defined concentration in a narrow temperature range of interest. Chilkoti and co-workers have previously published number of papers that provide similar analytical models that relate ELP composition and MW to its Tt.[25,43,45] Optimization of the ELP MW for in vivo applications also needs to take account into the impact of MW on pharmacokinetics and tissue distribution.
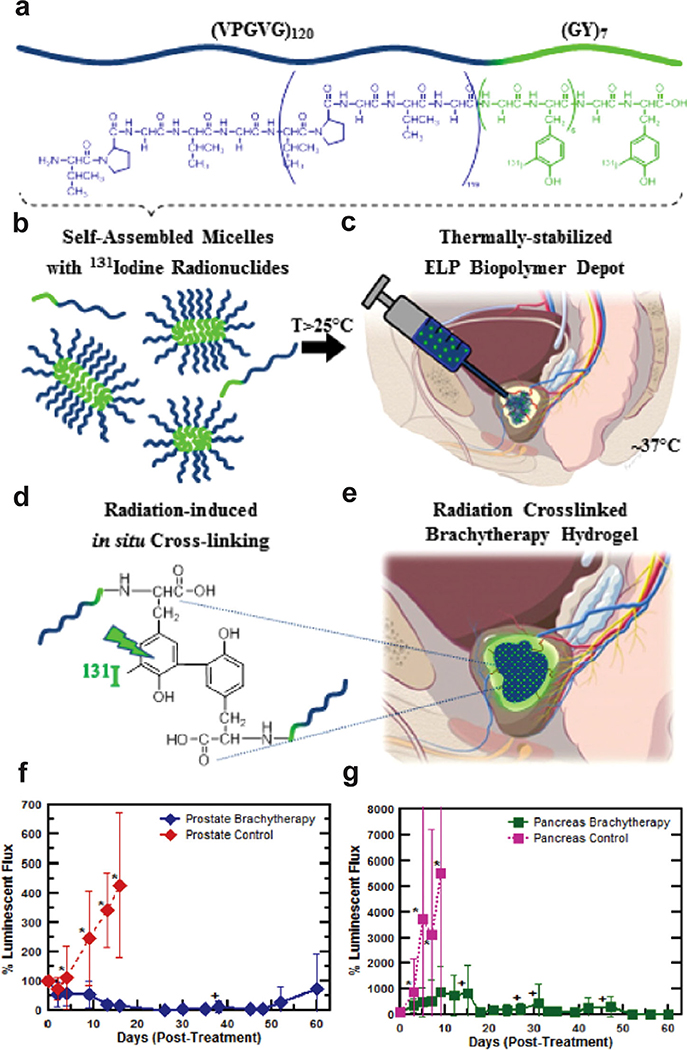
Chilkoti and co-workers have used the hydrogel-forming polymeric VPGVG repeat extensively to synthesize conjugates and fusion proteins for biomedical applications. (VPGVG)60, with its subphysiological Tt of ~28 °C, forms a viscous coacervate upon intratumoral (i.t.) injection.[207,208] In a tumor xenograft mouse model, radiolabeled ELPs containing (VPGVG)60 were found to be distributed across the tumor and was retained in the tumor for more than a week.[207] These ELPs exhibited low systemic toxicity even at high radionuclide doses and resulted in enhanced tumor regression and enhanced survival in tumor-bearing mice when compared to a soluble ELP radioconjugate. Liu et al. systematically analyzed the parameters that affect the physical properties of the hydrogel, including ELP molecular weight, concentration, composition, and attachment of a tyrosine-rich peptide for radiolabeling.[208] They found that a decrease in Tt in response to increased ELP concentration resulted in a higher tumor retention time. However, the Tt reached a plateau as the number of ELP repeats increased beyond 120. A systematic increase in the number of repeats of the hydrophobic, tyrosine-rich peptide (YG) increased the ELP’s viscosity and tumor accumulation. Attachment of the tyrosine-rich sequence also reduced Tt and resulted in a significant micelle population for (VPGVG)120-(YG)7, but only soluble unimers for the shorter YG repeats (VPGVG)120-(YG)or(VPGVG)120-(YG)4.Thisoptimized(VPGVG)120-(YG)7 sequence, which forms rod-shaped micelles, provides three advantages for cancer therapy: i) it can be labeled with radioiodine at the tyrosine; ii) it allows a large range of injection concentrations since Tt is independent of ELP concentration; and iii) the micelle-to-coacervate phase separation is less responsive to in vivo dilution than a unimer-to-coacervate transition. Upon i.t. administration, the radioiodine conjugate formed a seed-like hydrogel with prolonged intratumoral retention, potent antitumor efficacy, and degradation into nontoxic peptides. The radiation also induced irreversible crosslinking between tyrosine moieties that provided secondary stabilization of the coacervate (Figure 10).[209]
Figure 10.
a–e) Schematic of ELP depot formation for brachytherapy. f–g) Pre-clinical anti-tumor efficacy study of 131I-ELP brachytherapy for the treatment of f) orthotopic, human PC-3 M-luc-C6 prostate tumor xenografts and g) orthotopic BxPc3-luc2 pancreatic tumor in athymic, nude mice. Reproduced with permission.[209] Copyright 2016, Elsevier.
A similar strategy was used to deliver the type 2 diabetes drug, glucagon-like peptide 1 (GLP-1)[210,211] and fibroblast growth factor 21.[212] Unlike i.t. administration to a solid tumor in which the site of treatment is local, s.c. therapeutic depots for systemic treatment of diabetes require diffusion of the drug from the depot into the blood to reach specific organs and tissues to exert their therapeutic effect.[211] By adjusting the molecular weight of the ELP and the hydrophobicity of the guest residue, a GLP-1-ELP formulation was created that quickly formed an insoluble coacervate upon s.c. injection; showed sustained release from the coacervate depot with zero-order kinetics; exhibited excellent potency to control the blood glucose for up to 10 days following a single administration in three different mouse models of type 2 diabetes; and exhibited sustained drug release for up to 17 days in nonhuman primates.[211]
ELPs can also be physically or chemically crosslinked to create hydrogels. The first examples of ELP hydrogels were reported by Urry and co-workers who used radiation to crosslink ELPs into hydrogels.[213–215] The first studies on chemical crosslinking were carried by Conticello and co-workers who used the electrophilic crosslinker bis(sulfosuccinimidyl) suberate to covalently crosslink a lysine-rich ELP to form hydrogels in either phosphatebufferatpH8.5oranhydrousdimethylsulfoxide.They found that choice of solvent affected the crosslinking density and the resulting microstructure of the gel.[42,216] Trabbic-Carlson et al. also used lysine-based ELPs to form hydrogels by chemically crosslinking with tri-succinimidyl aminotriacetate in an organic solvent mixture of dimethylsulfoxide and dimethylformamide. The physical properties of the gel were found to be tunable by control of three parameters: ELP molecular weight, concentration, and lysine content.[24]
Because many applications of hydrogels involve the encapsulation of cells, radiation crosslinking is not a viable strategy. In order to create cytocompatible crosslinked hydrogels, Lim et al. synthesized ELPs with periodic Lys residues and crosslinked them using a nontoxic water-soluble organophosphorous crosslinker, β-[tris(hydroxymethyl)phosphino]propionic acid (THPP) that reacts with the amines of the lysine residues in the ELP to create trifunctional intra- or intermolecular crosslinks. The ELPs undergo gelation within minutes under physiological conditions, indicating that they are suitable for use as injectable biomaterials. These hydrogels are noncytotoxic and maintain cell survival.[217] Following up on these studies, Heilshorn and co-workers showed that hydrogel formation using an ELP composed of repeats of [(VPGIG)2(VPGKG)(VPGIG)2] requires stabilization through intermolecular crosslinking between two ELP strands and/or between an ELP strand and a secondary polymer.[218] Using a similar crosslinking strategy as Lim et al., they chemically crosslinked the ELP using a bifunctional disuccinimidyl suberate crosslinker and trifunctional β-[tris(hydroxymethyl) phosphino] propionic acid. These multifunctional amine-reactive crosslinkers reacted with the free amine groups of ELP lysine residues to form 3D hydrogels.
Another challenge in the design of hydrogels for biological applications is the need to reserve optical transparency so that encapsulated cells in the hydrogel can be imaged. Mathew et al. demonstrated the modification of the ELP sequence to create translucent ELP hydrogels.[219] Two peptide sequences, (XPGVG)50 and (XPAVG)50, where X = V:I 4:1, both had a Tt of 37 °C; however, (XPGVG)50 formed an ELP-rich coacervate while (XPAVG)50 formed a rigid hydrogel with >95% translucency for a 1 mm thick gel. Using temperature-dependent turbidimetry, rheology, and differential scanning calorimetry, they showed that gelation of (XPAVG)50 solutions occurred due to the interactions of hydrophobic chains and arrested phase separation above Tt at a threshold concentration of 15 wt%. Rheological measurements showed that an increase in ELP chain length or hydrophobic content of the guest residue (X = V:I 2:3) stiffens the hydrogel network.[219]
ELPs have also been modified with hydrazide to form hydrogels by allowing dynamic covalent condensation with aldehyde groups of oxidized hyaluronic acid.[220,221] The ELP-hyaluronic acid conjugates formed a stiff hydrogel at body temperature that has been applied to regenerating cartilage[220] and delivering stem cells.[221] The stiffness of the hydrogel can be tuned by varying temperature, ELP concentration, and the stoichiometry of the aldehyde and hydrazide groups. These hydrogels are attractive for regenerative cell delivery therapies because their shear-thinning nature makes them injectable without any need of predefined shape/mold, and the phase transition of their ELP domains at body temperature allows them to self-heal and form a potentially cell-adhesive scaffold with a long retention time. The ELP phase transition stabilizes the hydrogel and results in tenfold slower erosion than a hydrogel that is not thermally responsive.[221] To reduce the opacity of RGD-fused ELP hydrogels to make them suitable for light-based observation of encapsulated cells, Wang et al. synthesized a hydrogel network by crosslinking PEG bis(amine) and the ELP lysine via a Mannichtype condensation with tris(hydroxymethyl)-phosphine, a trifunctional crosslinker.[222] Grafting hydrophilic PEG onto the hydrophobic ELP backbone reduced the formation of hydrophobic aggregates within the hydrogel (even at 37 °C), resulting in greater optical transmittance.
ELPs with cysteine residues can also be chemically modified to yield novel hydrogels. Asai et al. demonstrated that ELPs with periodic cysteines can form crosslinked hydrogels based on the formation of intermolecular disulfide crosslinks.[223] Similarly, Xu et al. reported that cysteine containing ELPs can be mixed with hydrogen peroxide to form cysteine-ELP hydrogels with covalently crosslinked networks.[224] Because these ELPs are thermally responsive and form gels under physiological conditions, they are also useful for drug delivery and tissue engineering applications. Zhang et al. synthesized a highly elastic ELP hydrogel by including a pair of cysteine residues which formed disulfide bonds when exposed to UV light.[225]
Enzymatic crosslinking is an alternative to chemical crosslinking to create cytocompatible hydrogels. McHale et al. showed that ELPs containing glutamine and lysine residues can be crosslinked by the enzymatic activity of tissue transglutaminase in a biocompatible process. The resulting ELP hydrogels were used to encapsulate chondrocytes, which were then able to synthesize a cartilage matrix rich in sulfated glycosaminoglycans and Type II collagen. They also recorded an increase in mechanical integrity after the incubation with chondrocytes, suggesting that the ELP matrix had been restructured by the deposition of cartilage ECM components.[226]
8. Conclusions and Future Prospects
The studies described in this review highlight the versatility of ELP self-assembly systems. The ability to tune self-assembly of these nanoarchitectures by manipulating the ELP sequence and composition gives us the power to trigger phase separation and self-assembly using external stimuli such as temperature, pH, and light. Advances in genetic engineering, synthetic biology, and chemistry have enabled incorporation of structural motifs such as bioactive peptides, leucine zippers, β-sheet-forming domains, unnatural amino acids, lipids, and synthetic polymers into the ELP backbone to create self-assembled biofunctional nanostructures for a range of biomedical applications including drug delivery, regenerative medicine, and biosensing.
With their scalability and customizable structure and bioactivity, ELPs and ELP-hybrid nanostructures will continue to influence the understanding of self-assembly phenomena and the development of new biotechnologies. There are a few especially promising areas of research in ELP self-assembly. While substantial efforts have been made to understand extrinsic and intrinsic variables that influence the self-assembly of ELP block copolymers, this has not been done thoroughly for ELP-synthetic hybrid materials. Advances in molecular biology will push production of hybrid ELP material by recombinant DNA rather than chemical synthesis methods and will take advantage of PTM and via incorporation of unnatural amino acids. Selective and quantitative chemical synthesis reactions such as cycloaddition, click reactions, β-elimination, and substitution will continue to be useful for appending synthetic motifs. Harnessing new external stimuli to trigger self-assembly into new morphologies will continue to be important. De novo design of morphologies based on experimental results, theoretical calculation, and molecular simulations will improve the understanding of the structure–function relationships for self-assembled nanostructures and will enable the creation of new architectures. Although much of our present understanding of self-assembly comes from the study of static systems, the greatest challenges and opportunities lie in the study of dynamic systems. Substantial effort is needed to better understand dynamic self-assembling ELP-hybrid materials, particularly in terms of the interactions of ordered motifs with disordered ELP systems. This effort requires a systemic search for new peptide polymers that go beyond the commonly used VPGXG ELP pentapeptide repeat.[34,227]
Acknowledgements
S.S. and S.B. contributed equally to this work. The research described herein has been supported by the NIH through grants to A.C. from the National Institutes of Health (R21HL115410, R41HL123871, UG3CA211232, R21HL141028, R01-GM061232, R21-EB009904, R01-EB000188, R01-EB007205, R01-DK091789, R21-CA237705, MIRA-R35GM127042), the National Science Foundation (NSF-DMREF1729671), and by the Whitaker Foundation and the Coulter Foundation. S.B. would like to acknowledge the Pratt School of Engineering at Duke University for support from the Pratt-Gardner Fellowship.
Biography

SoumenSaha got his Ph.D. (2017) in chemical sciences from Academy of Scientific and Innovative Research, India. Working as an SPM fellow, his doctoral research focused on cationic lipid nanoparticles for targeted cancer chemo- and immuno-therapy. He is a postdoctoral associate in the Chilkoti lab where he works on genetically encoded peptide polymer for sustained drug release.

Samagya Banskota received her Ph.D. degree in biomedical engineering from Duke University. She previously obtained her B.S. degree in bioengineering from the Pennsylvania State University. Her doctoral work in the Chilkoti lab focused on the development of genetically engineered stealth biopolymers for drug delivery.

Ashutosh Chilkoti is the Alan L. Kaganov Professor and the chair of the Department of Biomedical Engineering at Duke University. His areas of research includegenetically encoded materials and biointerfacescience. He has worked extensively on developing elastin-like polypeptides for protein purification and drug delivery. He is the founder of five start-up companies.
Footnotes
Conflict of Interest
A.C. is an inventor of the ELP fusion technology, which has been licensed by Duke University to PhaseBio Pharmaceuticals, a company co-founded by A.C. A.C. serves as a paid scientific adviser to PhaseBio and has equity in the company.
Contributor Information
Soumen Saha, Department of Biomedical Engineering, Duke University, Durham, NC 27708, USA.
Samagya Banskota, Department of Biomedical Engineering, Duke University, Durham, NC 27708, USA.
Stefan Roberts, Department of Biomedical Engineering, Duke University, Durham, NC 27708, USA.
Nadia Kirmani, Department of Biology, Trinity College of Arts and Sciences, Duke University, Durham, NC 27708, USA.
Ashutosh Chilkoti, Department of Biomedical Engineering, Duke University, Durham, NC 27708, USA.
References
- [1].O’Leary LER, Fallas JA, Bakota EL, Kang MK, Hartgerink JD, Nat. Chem. 2011, 3, 821. [DOI] [PubMed] [Google Scholar]
- [2].García-Manrique P, Gutiérrez G, Blanco-López MC, Trends Biotechnol. 2018, 36, 10. [DOI] [PubMed] [Google Scholar]
- [3].Hubbell JA, Nat. Biotechnol. 1995, 13, 565. [Google Scholar]
- [4].Place ES, Evans ND, Stevens MM, Nat. Mater. 2009, 8, 457. [DOI] [PubMed] [Google Scholar]
- [5].Christman KL, Science 2019, 363, 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Macewan SR, Chilkoti A, J. Controlled Release 2014, 190, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Mastria E, Chilkoti A, MRS Bull. 2014, 39, 35. [Google Scholar]
- [8].Rodríguez-Cabello JC, Arias FJ, Rodrigo MA, Girotti A, Adv. Drug Delivery Rev. 2016, 97, 85. [DOI] [PubMed] [Google Scholar]
- [9].Swierczewska M, Han HS, Kim K, Park JH, Lee S, Adv. Drug Delivery Rev. 2016, 99, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Pelesko JA, Self Assembly. The Science of Things that Put Themselves Together. By John A. Pelesko, John Wiley & Sons, Ltd., Hoboken, NJ: 2007. [Google Scholar]
- [11].Whitesides GM, Mathias JP, Seto CT, Science 1991, 254, 1312. [DOI] [PubMed] [Google Scholar]
- [12].Dahman Y, Caruso G, Eleosida A, Hasnain ST, in Nanotechnology and Functional Materials for Engineers (Ed: Dahman Y), Elsevier, Amsterdam: 2017, pp. 207–228. [Google Scholar]
- [13].Zhang S, Nat. Biotechnol. 2003, 21, 1171. [DOI] [PubMed] [Google Scholar]
- [14].Whitesides GM, Science 2011, 2418, 2418. [Google Scholar]
- [15].Chang R, Zou Q, Xing R, Yan X, Adv. Ther. 2019, 1900048. [Google Scholar]
- [16].Mandal D, Nasrolahi Shirazi A, Parang K, Org. Biomol. Chem. 2014, 12, 3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Subramani K, Khraisat A, George A, Self-Assembly of Proteins and Peptides and their Applications in Bionanotechnology, 1st ed., Vol. 4, Elsevier Inc., Amsterdam: 2008. [Google Scholar]
- [18].Smits FCM, Buddingh BC, Van Eldijk MB, Van Hest JCM, Macromol. Biosci. 2015, 15, 36. [DOI] [PubMed] [Google Scholar]
- [19].Wright ER, Conticello VP, Adv. Drug Delivery Rev. 2002, 54, 1057. [DOI] [PubMed] [Google Scholar]
- [20].Hassouneh W, Christensen T, Chilkoti A, Curr. Protoc. Protein Sci. 2010, 1, ps0611s61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Chilkoti A, Dreher MR, Meyer DE, Adv. Drug Delivery Rev. 2002, 54, 1093. [DOI] [PubMed] [Google Scholar]
- [22].Meyer DE, Chilkoti A, Nat. Biotechnol. 1999, 17, 1112. [DOI] [PubMed] [Google Scholar]
- [23].Strzegowski LA, Martinez MB, Gowda DC, Urry DW, Tirrell DA, J. Am. Chem. Soc. 1994, 116, 813. [Google Scholar]
- [24].Trabbic-Carlson K, Setton LA, Chilkoti A, Biomacromolecules 2003, 4, 572. [DOI] [PubMed] [Google Scholar]
- [25].MacKay JA, Callahan DJ, FitzGerald KN, Chilkoti A, Biomacromolecules 2010, 11, 2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Urry DW, Hayes LC, Gowda DC, Harris CM, Harris RD, Biochem. Biophys. Res. Commun. 1992, 188, 611. [DOI] [PubMed] [Google Scholar]
- [27].Urry DW, Long MM, Cox BA, Ohnishi T, Mitchell LW, Jacobs M, Biochim. Biophys. Acta, Protein Struct. 1974, 371, 597. [DOI] [PubMed] [Google Scholar]
- [28].Ohgo K, Niemczura WP, Ashida J, Okonogi M, Asakura T, Kumashiro KK, Biomacromolecules 2006, 7, 3306. [DOI] [PubMed] [Google Scholar]
- [29].Isidro-Llobet A, Álvarez M, Albericio F, Chem. Rev. 2009, 109, 2455. [DOI] [PubMed] [Google Scholar]
- [30].Rodríguez-Cabello JC, Arias FJ, Rodrigo MA, Girotti A, Adv. Drug Delivery Rev. 2016, 97, 85. [DOI] [PubMed] [Google Scholar]
- [31].Sarangthem V, Cho EA, Yi A, Kim SK, Lee BH, Park RW, Sci. Rep. 2018, 8, 3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].White MJ, Fristensky BW, Thompson WF, Anal. Biochem. 1991, 199, 184. [DOI] [PubMed] [Google Scholar]
- [33].McDaniel JR, MacKay JA, Quiroz FG, Chilkoti A, Biomacromolecules 2010, 11, 944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Amiram M, Quiroz FG, Callahan DJ, Chilkoti A, Nat. Mater. 2011, 10, 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Quiroz FG, Chilkoti A, in ACS Symp. Series, (Eds: Lutz J-F, Meyer TY, Ouchi M, Sawamoto M), American Chemical Society, Washington, 2014, 1170, 15. [Google Scholar]
- [36].Mozhdehi D, Luginbuhl KM, Roberts S, Chilkoti A, in Sequence-Controlled Polymers (Ed: Lutz J-F), Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany: 2017, p. 91. [Google Scholar]
- [37].Roberts S, Costa S, Schaal J, Simon JR, Dzuricky M, Quiroz FG, Chilkoti A, in Comprehensive Biomaterials II, Vol. 2 (Eds: Healy K, Hutmacher DW, Grainger DW, Kirkpatrick CJ), Elsevier, Amsterdam; 2017, pp. 90–108. [Google Scholar]
- [38].Tang NC, Chilkoti A, Nat. Mater. 2016, 15, 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Kim JY, O’Malley S, Mulchandani A, Chen W, Anal. Chem. 2005, 77, 2318. [DOI] [PubMed] [Google Scholar]
- [40].Urry DW, Haynes B, Harris RD, Biochem. Biophys. Res. Commun. 1986, 141, 749. [DOI] [PubMed] [Google Scholar]
- [41].Urry DW, Trapane TL, Long MM, Prasad KU, J. Chem. Soc., Faraday Trans. 1 1983, 79, 853. [Google Scholar]
- [42].McMillan RA, Caran KL, Apkarian RP, Conticello VP, Macromolecules 1999, 32, 9067. [Google Scholar]
- [43].Meyer DE, Chilkoti A, Biomacromolecules 2004, 5, 846. [DOI] [PubMed] [Google Scholar]
- [44].Catherine C, Oh SJ, Lee KH, Min SE, Won JI, Yun H, Kim DM, Biotechnol. Bioprocess Eng. 2015, 20, 417. [Google Scholar]
- [45].McDaniel JR, Radford DC, Chilkoti A, Biomacromolecules 2013, 14, 2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].McDaniel JR, MacKay JA, Quiroz FG, Chilkoti A, Biomacromolecules 2010, 11, 944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lee TAT, Cooper A, Apkarian RP, Conticello VP, Adv. Mater. 2000, 12, 1105. [Google Scholar]
- [48].Rodríguez-Cabello JC, Reguera J, Girotti A, Arias FJ, Alonso M, in Advances in Polymer Science, Vol. 200 (Ed: Buchmeiser MR), Springer, Berlin: 2005, pp. 119–167. [Google Scholar]
- [49].Meyer DE, Chilkoti A, Biomacromolecules 2002, 3, 357. [DOI] [PubMed] [Google Scholar]
- [50].Dreher MR, Simnick AJ, Fischer K, Smith RJ, Patel A, Schmidt M, Chilkoti A, J. Am. Chem. Soc. 2008, 130, 687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Janib SM, Pastuszka MF, Aluri S, Folchman-Wagner Z, Hsueh PY, Shi P, Lin YA, Cui H, Mackay JA, Polym. Chem. 2014, 5, 1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hassouneh W, Zhulina EB, Chilkoti A, Rubinstein M, Macromolecules 2015, 48, 4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Massodi I, Bidwell GL, Raucher D, J. Controlled Release 2005, 108, 396. [DOI] [PubMed] [Google Scholar]
- [54].Bessa PC, Machado R, Nürnberger S, Dopler D, Banerjee A, Cunha AM, Rodríguez-Cabello JC, Redl H, van Griensven M, Reis RL, Casal M, J. Controlled Release 2010, 142, 312. [DOI] [PubMed] [Google Scholar]
- [55].García-Arévalo C, Bermejo-Martín JF, Rico L, Iglesias V, Martín L, Rodríguez-Cabello JC, Arias FJ, Mol. Pharmaceutics 2013, 10, 586. [DOI] [PubMed] [Google Scholar]
- [56].Simnick AJ, Valencia CA, Liu R, Chilkoti A, ACS Nano 2010, 4, 2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Macewan SR, Chilkoti A, Nano Lett. 2014, 14, 2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Costa SA, Mozhdehi D, Dzuricky MJ, Isaacs FJ, Brustad EM, Chilkoti A, Nano Lett. 2019, 19, 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bidwell GL, Davis AN, Raucher D, J. Controlled Release 2009, 135, 2. [DOI] [PubMed] [Google Scholar]
- [60].Hassouneh W, Fischer K, MacEwan SR, Branscheid R, Fu CL, Liu R, Schmidt M, Chilkoti A, Biomacromolecules 2012, 13, 1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].MacEwan SR, Chilkoti A, Nano Lett. 2012, 12, 3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Bidwell GL, Mol. Cancer Ther. 2005, 4, 1076. [DOI] [PubMed] [Google Scholar]
- [63].Simnick AJ, Amiram M, Liu W, Hanna G, Dewhirst MW, Kontos CD, Chilkoti A, J. Controlled Release 2011, 155, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Callahan DJ, Liu W, Li X, Dreher MR, Hassouneh W, Kim M, Marszalek P, Chilkoti A, Nano Lett. 2012, 12, 2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].MacEwan SR, Chilkoti A, Biopolymers 2010, 94, 60. [DOI] [PubMed] [Google Scholar]
- [66].McDaniel JR, Weitzhandler I, Prevost S, Vargo KB, Appavou MS, Hammer DA, Gradzielski M, Chilkoti A, Nano Lett. 2014, 14, 6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Urry DW, Chem. Phys. Lett. 2004, 399, 177. [Google Scholar]
- [68].Srinivas G, Discher DE, Klein ML, Nat. Mater. 2004, 3, 638. [DOI] [PubMed] [Google Scholar]
- [69].Vargo KB, Parthasarathy R, Hammer DA, Proc. Natl. Acad. Sci. USA 2012, 109, 11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Bhattacharyya J, Weitzhandler I, Ho SB, McDaniel JR, Li X, Tang L, Liu J, Dewhirst M, Chilkoti A, Adv. Funct. Mater. 2017, 27, 1605421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].McDaniel JR, Weitzhandler I, Prevost S, Vargo KB, Appavou MS, Hammer DA, Gradzielski M, Chilkoti A, Nano Lett. 2014, 14, 6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Pastuszka MK, Wang X, Lock LL, Janib SM, Cui H, Deleve LD, MacKay JA, J. Controlled Release 2014, 191, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Aluri SR, Shi P, Gustafson JA, Wang W, Lin YA, Cui H, Liu S, Conti PS, Li Z, Hu P, Epstein AL, Mackay JA, ACS Nano 2014, 8, 2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Condon JE, Martin TB, Jayaraman A, Soft Matter 2017, 13, 2907. [DOI] [PubMed] [Google Scholar]
- [75].Luo T, Kiick KL, J. Am. Chem. Soc. 2015, 137, 15362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Luo T, David MA, Dunshee LC, Scott RA, Urello MA, Price C, Kiick KL, Biomacromolecules 2017, 18, 2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Qin J, Luo T, Kiick KL, Biomacromolecules 2019, 20, 1514. [DOI] [PubMed] [Google Scholar]
- [78].Zhao C, Zhuang X, He C, Chen X, Jing X, Macromol. Rapid Commun. 2008, 29, 1810. [Google Scholar]
- [79].Isoda K, Kanayama N, Miyamoto D, Takarada T, Maeda M, React. Funct. Polym. 2011, 71, 367. [Google Scholar]
- [80].MacKay JA, Callahan DJ, FitzGerald KN, Chilkoti A, Biomacromolecules 2010, 11, 2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].MacEwan SR, Weitzhandler I, Hoffmann I, Genzer J, Gradzielski M, Chilkoti A, Biomacromolecules 2017, 18, 599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Martín L, Castro E, Ribeiro A, Alonso M, Rodríguez-Cabello JC, Biomacromolecules 2012, 13, 293. [DOI] [PubMed] [Google Scholar]
- [83].Maeda H, Bioconjugate Chem. 2010, 21, 797. [DOI] [PubMed] [Google Scholar]
- [84].Maeda H, Tsukigawa K, Fang J, Microcirculation 2016, 23, 173. [DOI] [PubMed] [Google Scholar]
- [85].Mackay JA, Chen M, Mcdaniel JR, Liu W, J A, Chilkoti A, Nat. Mater. 2009, 8, 2352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Bhattacharyya J, Ren XR, Mook RA, Wang J, Spasojevic I, Premont RT, Li X, Chilkoti A, Chen W, Nanoscale 2017, 9, 12709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].McDaniel JR, Bhattacharyya J, Vargo KB, Hassouneh W, Hammer DA, Chilkoti A, Angew. Chem., Int. Ed. 2013, 52, 1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Bhattacharyya J, Bellucci JJ, Weitzhandler I, McDaniel JR, Spasojevic I, Li X, Lin CC, Chi JTA, Chilkoti A, Nat. Commun. 2015, 6, 7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].MacEwan SR, Chilkoti A, Angew. Chem., Int. Ed. 2017, 56, 6712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Banskota S, Yousefpour P, Kirmani N, Li X, Chilkoti A, Biomaterials 2019, 192, 475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Yousefpour P, McDaniel JR, Prasad V, Ahn L, Li X, Subrahmanyan R, Weitzhandler I, Suter S, Chilkoti A, Nano Lett. 2018, 18, 7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].ichi Yokoe J, Sakuragi S, Yamamoto K, Teragaki T, ichi Ogawara K, Higaki K, Katayama N, Kai T, Sato M, Kimura T, Int. J. Pharm. 2008, 353, 28. [DOI] [PubMed] [Google Scholar]
- [93].Furumoto K, Yokoe JI, ichi Ogawara K, Amano S, Takaguchi M, Higaki K, Kai T, Kimura T, Int. J. Pharm. 2007, 329, 110. [DOI] [PubMed] [Google Scholar]
- [94].Ogawara KI, Furumoto K, Nagayama S, Minato K, Higaki K, Kai T, Kimura T, J. Controlled Release 2004, 100, 451. [DOI] [PubMed] [Google Scholar]
- [95].McDaniel JR, Bhattacharyya J, Vargo KB, Hassouneh W, Hammer DA, Chilkoti A, Angew. Chem., Int. Ed. 2013, 52, 1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Bhattacharyya J, Bellucci JJ, Weitzhandler I, McDaniel JR, Spasojevic I, Li X, Lin C-C, Chi J-TA, Chilkoti A, Nat. Commun. 2015, 6, 7939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Andrew MacKay J, Chen M, McDaniel JR, Liu W, Simnick AJ, Chilkoti A, Nat. Mater. 2009, 8, 993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Christensen T, Hassouneh W, Trabbic-Carlson K, Chilkoti A, Biomacromolecules 2013, 14, 1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].McDaniel JR, Radford DC, Chilkoti A, Biomacromolecules 2013, 14, 2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].MacKay JA, Callahan DJ, FitzGerald KN, Chilkoti A, Biomacromolecules 2010, 11, 2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Rauscher S, Pomes R, eLife 2017, 6, e26526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Urry DW, Hugel T, Seitz M, Gaub HE, Sheiba L, Dea J, Xu J, Parker T, Philos. Trans. R. Soc., B 2002, 357, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Roberts S, Dzuricky M, Chilkoti A, FEBS Lett. 2015, 589, 2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Fuxreiter M, Mol. BioSyst. 2012, 8, 168. [DOI] [PubMed] [Google Scholar]
- [105].Li NK, Garcia Quiroz F, Hall CK, Chilkoti A, Yingling YG, Biomacromolecules 2014, 15, 3522. [DOI] [PubMed] [Google Scholar]
- [106].Yeo GC, Keeley FW, Weiss AS, Adv. Colloid Interface Sci. 2011, 167, 94. [DOI] [PubMed] [Google Scholar]
- [107].Smits FC, Buddingh BC, van Eldijk MB, van Hest JC, Macromol. Biosci. 2015, 15, 36. [DOI] [PubMed] [Google Scholar]
- [108].van der Lee R, Buljan M, Lang B, Weatheritt RJ, Daughdrill GW, Dunker AK, Fuxreiter M, Gough J, Gsponer J, Jones DT, Kim PM, Kriwacki RW, Oldfield CJ, V Pappu R, Tompa P, Uversky VN, Wright PE, Babu MM, Chem. Rev. 2014, 114, 6589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Gosline J, Lillie M, Carrington E, Guerette P, Ortlepp C, Savage K, Philos. Trans. R. Soc., B 2002, 357, 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Pometun MS, Chekmenev EY, Wittebort RJ, J. Biol. Chem. 2004, 279, 7982. [DOI] [PubMed] [Google Scholar]
- [111].Coyne KJ, Qin XX, Waite JH, Science 1997, 277, 1830. [DOI] [PubMed] [Google Scholar]
- [112].Meyers MA, Chen P-Y, Lin AY-M, Seki Y, Prog. Mater. Sci. 2008, 53, 1. [Google Scholar]
- [113].Vollrath F, Porter D, Soft Matter 2006, 2, 377. [DOI] [PubMed] [Google Scholar]
- [114].Kim W, Conticello VP, Polym. Rev. 2007, 47, 93. [Google Scholar]
- [115].Keten S, Buehler MJ, Soc JR., Interface 2010, 7, 1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Keten S, Xu Z, Ihle B, Buehler MJ, Nat. Mater. 2010, 9, 359. [DOI] [PubMed] [Google Scholar]
- [117].Elvin CM, Carr AG, Huson MG, Maxwell JM, Pearson RD, Vuocolo T, Liyou NE, Wong DC, Merritt DJ, Dixon NE, Nature 2005, 437, 999. [DOI] [PubMed] [Google Scholar]
- [118].Cai L, Dinh CB, Heilshorn SC, Biomater. Sci. 2014, 2, 757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Amiram M, Luginbuhl KM, Li X, Feinglos MN, Chilkoti A, Proc. Natl. Acad. Sci. USA 2013, 110, 2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Liu JC, Heilshorn SC, Tirrell DA, Biomacromolecules 2004, 5, 497. [DOI] [PubMed] [Google Scholar]
- [121].Hardy JG, Scheibel TR, Biochem. Soc. Trans. 2009, 37, 677. [DOI] [PubMed] [Google Scholar]
- [122].Heidebrecht A, Eisoldt L, Diehl J, Schmidt A, Geffers M, Lang G, Scheibel T, Adv. Mater. 2015, 27, 2189. [DOI] [PubMed] [Google Scholar]
- [123].Lazaris A, Arcidiacono S, Huang Y, Zhou JF, Duguay F, Chretien N, Welsh EA, Soares JW, Karatzas CN, Science 2002, 295, 472. [DOI] [PubMed] [Google Scholar]
- [124].V Lewis R, Hinman M, Kothakota S, Fournier MJ, Protein Expression Purif. 1996, 7, 400. [DOI] [PubMed] [Google Scholar]
- [125].Romer L, Scheibel T, Prion 2008, 2, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Scheibel T, Microb. Cell Fact. 2004, 3, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Hu X, Wang X, Rnjak J, Weiss AS, Kaplan DL, Biomaterials 2010, 31, 8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Ferrari FA, Richardson C, Chambers J, Causey SC, Pollock TJ, Capello J, Crissman JW, US Patent No. 5243038A, 1993.
- [129].Cappello J, Handb. Biodegrad. Polym. 1998, 7, 387. [Google Scholar]
- [130].Cappello J, Crissman J, Dorman M, Mikolajczak M, Textor G, Marquet M, Ferrari F, Biotechnol. Prog. 1990, 6, 198. [DOI] [PubMed] [Google Scholar]
- [131].Cappello J, Crissman JW, Crissman M, Ferrari FA, Textor G, Wallis O, Whitledge JR, Zhou X, Burman D, Aukerman L, Stedronsky ER, J. Controlled Release 1998, 53, 105. [DOI] [PubMed] [Google Scholar]
- [132].Nagarsekar A, Crissman J, Crissman M, Ferrari F, Cappello J, Ghandehari H, J. Biomed. Mater. Res. 2002, 62, 195. [DOI] [PubMed] [Google Scholar]
- [133].Xia XX, Xu Q, Hu X, Qin G, Kaplan DL, Biomacromolecules 2011, 12, 3844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Zeng L, Teng W, Jiang L, Cappello J, Wu X, Appl. Phys. Lett. 2014, 104, 033702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Putzu M, Causa F, Parente M, Gonzalez de Torre I, Rodriguez-Cabello JC, Netti PA, Regener. Biomater. 2019, 6, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Chen L, Zhou M-L, Qian Z-G, Kaplan DL, Xia X-X, ACS Biomater. Sci. Eng. 2017, 3, 335. [DOI] [PubMed] [Google Scholar]
- [137].Poursaid A, Price R, Tiede A, Olson E, Huo E, McGill L, Ghandehari H, Cappello J, Biomaterials 2015, 57, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Jensen MM, Jia W, Isaacson KJ, Schults A, Cappello J, Prestwich GD, Oottamasathien S, Ghandehari H, J. Controlled Release 2017, 263, 46. [DOI] [PubMed] [Google Scholar]
- [139].Hatefi A, Cappello J, Ghandehari H, Pharm. Res. 2007, 24, 773. [DOI] [PubMed] [Google Scholar]
- [140].Qiu W, Huang Y, Teng W, Cohn CM, Cappello J, Wu X, Biomacromolecules 2010, 11, 3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Vasconcelos A, Gomes AC, Cavaco-Paulo A, Acta Biomater. 2012, 8, 3049. [DOI] [PubMed] [Google Scholar]
- [142].Lemieux E, Prudhomme RE, Forte R, Jerome R, Teyssie P, Macromolecules 1988, 21, 2148. [Google Scholar]
- [143].Ray B, Okamoto Y, Kamigaito M, Sawamoto M, Seno K, Kanaoka S, Aoshima S, Polym. J. 2005, 37, 234. [Google Scholar]
- [144].Nishi K, Hiroi T, Hashimoto K, Fujii K, Han Y-S, Kim T-H, Katsumoto Y, Shibayama M, Macromolecules 2013, 46, 6225. [Google Scholar]
- [145].Pepe A, Guerra D, Bochicchio B, Quaglino D, Gheduzzi D, Pasquali Ronchetti I, Tamburro AM, Matrix Biol. 2005, 24, 96. [DOI] [PubMed] [Google Scholar]
- [146].Clarke AW, Arnspang EC, Mithieux SM, Korkmaz E, Braet F, Weiss AS, Biochemistry 2006, 45, 9989. [DOI] [PubMed] [Google Scholar]
- [147].Muiznieks LD, Jensen SA, Weiss AS, Arch. Biochem. Biophys. 2003, 410, 317. [DOI] [PubMed] [Google Scholar]
- [148].Vrhovski B, Jensen S, Weiss AS, Eur. J. Biochem. 1997, 250, 92. [DOI] [PubMed] [Google Scholar]
- [149].Vrhovski B, Weiss AS, Eur. J. Biochem. 1998, 258, 1. [DOI] [PubMed] [Google Scholar]
- [150].Roberts S, Harmon TS, Schaal JL, Miao V, Li KJ, Hunt A, Wen Y, Oas TG, Collier JH, V Pappu R, Nat. Mater. 2018, 17, 1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Shen W, Lammertink RGH, Sakata JK, Kornfield JA, Tirrell DA, Macromolecules 2005, 38, 3909. [Google Scholar]
- [152].Huang CC, Ravindran S, Yin Z, George A, Biomaterials 2014, 35, 5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Fernandez-Colino A, Arias FJ, Alonso M, Rodriguez-Cabello JC, Biomacromolecules 2015, 16, 3389. [DOI] [PubMed] [Google Scholar]
- [154].Park WM, Champion JA, J. Am. Chem. Soc. 2014, 136, 17906. [DOI] [PubMed] [Google Scholar]
- [155].Jang Y, Choi WT, Heller WT, Ke Z, Wright ER, Champion JA, Small 2017, 13, 1700399. [DOI] [PubMed] [Google Scholar]
- [156].Zeng L, Teng W, Jiang L, Cappello J, Wu X, Appl. Phys. Lett. 2014, 104, 033702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Park WM, Champion JA, J. Am. Chem. Soc. 2014, 136, 17906. [DOI] [PubMed] [Google Scholar]
- [158].Roberts S, Harmon TS, Schaal JL, Miao V, (Jonathan) Li K, Hunt A, Wen Y, Oas TG, Collier JH, Pappu RV, Chilkoti A, Nat. Mater. 2018, 17, 1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Aluri S, Pastuszka MK, Moses AS, MacKay JA, Biomacromolecules 2012, 13, 2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Luginbuhl KM, Mozhdehi D, Dzuricky M, Yousefpour P, Huang FC, Mayne NR, Buehne KL, Chilkoti A, Angew. Chem., Int. Ed. 2017, 56, 13979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Mozhdehi D, Luginbuhl KM, Dzuricky M, Costa SA, Xiong S, Huang FC, Lewis MM, Zelenetz SR, Colby CD, Chilkoti A, J. Am. Chem. Soc. 2019, 141, 945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Inostroza-Brito KE, Collin E, Siton-Mendelson O, Smith KH, Monge-Marcet A, Ferreira DS, Rodríguez RP, Alonso M, Rodríguez-Cabello JC, Reis RL, Sagués F, Botto L, Bitton R, Azevedo HS, Mata A, Nat. Chem. 2015, 7, 897. [DOI] [PubMed] [Google Scholar]
- [163].Na K, Lee SA, Jung SH, Hyun J, Shin BC, Colloids Surf., B 2012, 91, 130. [DOI] [PubMed] [Google Scholar]
- [164].Choi H, Chu HS, Chung M, Kim B, Won JI, Biotechnol. Bioprocess Eng. 2016, 21, 620. [Google Scholar]
- [165].Park SM, Kim MS, Park SJ, Park ES, Choi KS, Kim YS, Kim HR, J. Controlled Release 2013, 170, 373. [DOI] [PubMed] [Google Scholar]
- [166].Park SM, Cha JM, Nam J, Kim MS, Park SJ, Park ES, Lee H, Kim HR, PLoS One 2014, 9, e103116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Kim MS, Lee DW, Park K, Park SJ, Choi EJ, Park ES, Kim HR, Colloids Surf., B 2014, 116, 17. [DOI] [PubMed] [Google Scholar]
- [168].Costa SA, Simon JR, Amiram M, Tang L, Zauscher S, Brustad EM, Isaacs FJ, Chilkoti A, Adv. Mater. 2018, 30, 1704878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Van Eldijk MB, Smits FCM, Vermue N, Debets MF, Schoffelen S, Van Hest JCM, Biomacromolecules 2014, 15, 2751. [DOI] [PubMed] [Google Scholar]
- [170].Paik BA, Blanco MA, Jia X, Roberts CJ, Kiick KL, Soft Matter 2015, 11, 1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Le Fer G, Portes D, Goudounet G, Guigner JM, Garanger E, Lecommandoux S, Org. Biomol. Chem. 2017, 15, 10095. [DOI] [PubMed] [Google Scholar]
- [172].Wei Han GPL, Sarah MacEwan, Ashutosh Chilkoti, Nanoscale 2015, 7, 12038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Nel AE, Mädler L, Velegol D, Xia T, Hoek EMV, Somasundaran P, Klaessig F, Castranova V, Thompson M, Nat. Mater. 2009, 8, 543. [DOI] [PubMed] [Google Scholar]
- [174].Duronio RJ, Jackson-Machelski E, Heuckeroth R0, Olinst P0, Devinet CS, Yonemotot W, Slicet LW, Taylort SS, Gordon JI, Proc. Natl. Acad. Sci. USA 1990, 87, 1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Farazi TA, Waksman G, Gordon JI, Biochemistry 2001, 40, 6335. [DOI] [PubMed] [Google Scholar]
- [176].Cui H, Webber MJ, Stupp SI, Biopolymers 2010, 94, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Hamley IW, Soft Matter 2011, 7, 4122. [Google Scholar]
- [178].Mozhdehi D, Luginbuhl KM, Simon JR, Dzuricky M, Berger R, Varol HS, Huang FC, Buehne KL, Mayne NR, Weitzhandler I, Bonn M, Parekh SH, Chilkoti A, Nat. Chem. 2018, 10, 496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Porter JA, Young KE, Beachy PA, Science 1996, 274, 255. [DOI] [PubMed] [Google Scholar]
- [180].Ciulla DA, Jorgensen MT, Giner JL, Callahan BP, J. Am. Chem. Soc. 2018, 140, 916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Torchilin VP, Nat. Rev. Drug Discovery 2005, 4, 145. [DOI] [PubMed] [Google Scholar]
- [182].Yatvin MB, Kreutz W, Horwitz BA, Shinitzky M, Science 1980, 210, 1253. [DOI] [PubMed] [Google Scholar]
- [183].Dromi S, Frenkel V, Luk A, Traughber B, Angstadt M, Bur M, Poff J, Xie J, Libutti SK, Li KCP, Wood BJ, Clin. Cancer Res. 2007, 13, 2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].De Smet M, Heijman E, Langereis S, Hijnen NM, Grüll H, Controlled Release J 2011, 150, 102. [DOI] [PubMed] [Google Scholar]
- [185].Miller CR, Clapp PJ, O’Brien DF, FEBS Lett. 2000, 467, 52. [DOI] [PubMed] [Google Scholar]
- [186].Pidgeon C, Hunt CA, Photochem. Photobiol. 1983, 37, 491. [Google Scholar]
- [187].Dandamudi S, Campbell RB, Biochim. Biophys. Acta, Biomembr. 2007, 1768, 427. [DOI] [PubMed] [Google Scholar]
- [188].Nakao R, Matuo Y, Mishima F, Taguchi T, Maenosono S, Nishijima S, Phys. C 2009, 469, 1840. [Google Scholar]
- [189].Lindner LH, Eichhorn ME, Eibl H, Nicole Teichert A, SchmittSody M, Issels RD, Dellian M, Clin. Cancer Res. 2005, 10, 2168. [DOI] [PubMed] [Google Scholar]
- [190].Paoli EE, Kruse DE, Seo JW, Zhang H, Kheirolomoom A, Watson KD, Chiu P, Stahlberg H, Ferrara KW, J. Controlled Release 2010, 143, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].de Smet M, Langereis S, van den Bosch S, Grüll H, J. Controlled Release 2010, 143, 120. [DOI] [PubMed] [Google Scholar]
- [192].Veneti E, Tu RS, Auguste DT, Bioconjugate Chem. 2016, 27, 1813. [DOI] [PubMed] [Google Scholar]
- [193].Biochips C, Na K, Jung J, Kim O, Lee J, Lee TG, Park YH, Hyun J, Synthesi 2008, 24, 4917. [DOI] [PubMed] [Google Scholar]
- [194].Raucher D, Chilkoti A, Cancer Res. 2001, 61, 7163. [PubMed] [Google Scholar]
- [195].Wu IL, Patterson MA, Carpenter Desai HE, Mehl RA, Giorgi G, Conticello VP, ChemBioChem 2013, 14, 968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Amiram M, Haimovich AD, Fan C, Wang YS, Aerni HR, Ntai I, Moonan DW, Ma NJ, Rovner AJ, Hong SH, Kelleher NL, Goodman AL, Jewett MC, Söll D, Rinehart J, Isaacs FJ, Nat. Biotechnol. 2015, 33, 1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Teeuwen RLM, Van Berkel SS, Van Dulmen THH, Schoffelen S, Meeuwissen SA, Zuilhof H, De Wolf FA, Van Hest JCM, Chem. Commun. 2009, 4022. [DOI] [PubMed] [Google Scholar]
- [198].Kim W, McMillan RA, Snyder JP, Conticello VP, J. Am. Chem. Soc. 2005, 127, 18121. [DOI] [PubMed] [Google Scholar]
- [199].Costa SA, Simon JR, Amiram M, Tang L, Zauscher S, Brustad EM, Isaacs FJ, Chilkoti A, Adv. Mater. 2018, 30, 1704878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Torchilin VP, Pharm. Res. 2007, 24, 1. [DOI] [PubMed] [Google Scholar]
- [201].Knop K, Hoogenboom R, Fischer D, Schubert US, Angew. Chem., Int. Ed. 2010, 49, 6288. [DOI] [PubMed] [Google Scholar]
- [202].Han W, Chilkoti A, López GP, Nanoscale 2017, 9, 6178. [DOI] [PubMed] [Google Scholar]
- [203].Li L, Li NK, Tu Q, Im O, Mo CK, Han W, Fuss WH, Carroll NJ, Chilkoti A, Yingling YG, Zauscher S, López GP, Biomacromolecules 2018, 19, 298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Sinclair SM, Bhattacharyya J, McDaniel JR, Gooden DM, Gopalaswamy R, Chilkoti A, Setton LA, J. Controlled Release 2013, 171, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Kimmerling KA, Furman BD, Mangiapani DS,Moverman MA, Sinclair SM, Huebner JL, Chilkoti A, Kraus VB, Setton LA, Guilak F, Olson SA, Eur. Cells Mater. 2015, 29, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Mukerji R, Schaal J, Li X, Bhattacharyya J, Asai D, Zalutsky MR, Chilkoti A, Liu W, Biomaterials 2016, 79, 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Liu W, MacKay JA, Dreher MR, Chen M, McDaniel JR, Simnick AJ, Callahan DJ, Zalutsky MR, Chilkoti A, J. Controlled Release 2010, 144, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Liu W, McDaniel J, Li X, Asai D, Quiroz FG, Schaal J, Park JS, Zalutsky M, Chilkoti A, Cancer Res. 2012, 72, 5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Schaal JL, Li X, Mastria E, Bhattacharyya J, Zalutsky MR, Chilkoti A, Liu W, J. Controlled Release 2016, 228, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Amiram M, Luginbuhl KM, Li X, Feinglos MN, Chilkoti A, J. Controlled Release 2013, 172, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Luginbuhl KM, Schaal JL, Umstead B, Mastria EM, Li X, Banskota S, Arnold S, Feinglos M, D’Alessio D, Chilkoti A, Nat. Biomed. Eng. 2017, 1, 0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Gilroy CA, Roberts S, Chilkoti A, J. Controlled Release 2018, 277, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Lee J, Macosko CW, Urry DW, Macromolecules 2001, 34, 5968. [Google Scholar]
- [214].Lee J, Macosko CW, Urry DW, Macromolecules 2001, 34, 4114. [Google Scholar]
- [215].Lee J, Macosko CW, Urry DW, J. Biomater. Sci., Polym. Ed. 2001, 12, 229. [DOI] [PubMed] [Google Scholar]
- [216].McMillan RA, Conticello VP, Macromolecules 2000, 33, 4809. [Google Scholar]
- [217].Lim DW, Nettles DL, Setton LA, Chilkoti A, Biomacromolecules 2007, 8, 1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Straley KS, Heilshorn SC, Adv. Mater. 2009, 21, 4148. [Google Scholar]
- [219].Glassman MJ, Olsen BD, Biomacromolecules 2015, 16, 3762. [DOI] [PubMed] [Google Scholar]
- [220].Zhu D, Wang H, Trinh P, Heilshorn SC, Yang F, 2017, 127, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Wang H, Zhu D, Paul A, Cai L, Enejder A, Yang F, Heilshorn SC, Adv. Funct. Mater. 2017, 27, 1605609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Wang H, Cai L, Paul A, Enejder A, Heilshorn SC, Biomacromolecules 2014, 15, 3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Asai D, Xu D, Liu W, Garcia Quiroz F, Callahan DJ, Zalutsky MR, Craig SL, Chilkoti A, Biomaterials 2012, 33, 5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Xu D, Asai D, Chilkoti A, Craig SL, Biomacromolecules 2012, 13, 2315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Zhang YN, Avery RK, Vallmajo-Martin Q, Assmann A, Vegh A, Memic A, Olsen BD, Annabi N, Khademhosseini A, Adv. Funct. Mater. 2015, 25, 4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].McHale MK, Setton LA, Chilkoti A, Tissue Eng. 2005, 11, 1768. [DOI] [PubMed] [Google Scholar]
- [227].Quiroz FG, Chilkoti A, Nat. Mater. 2015, 14, 1164. [DOI] [PMC free article] [PubMed] [Google Scholar]