Abstract
miRNAs play critical roles during embryonic development and their dysregulation causes cancer1,2. Altered global miRNA abundance is observed in different tissues and tumors, implying precise control of miRNA dosage is important1,3,4, yet the underlying mechanism(s) remains unknown. Microprocessor, comprising one DROSHA and two DGCR8 proteins, is essential for miRNA biogenesis5–7. Here we identify a developmentally-regulated miRNA dosage control mechanism involving alternative transcription initiation (ATI) of DGCR8. ATI occurs downstream of a stem-loop in DGCR8 mRNA to bypass an autoregulatory feedback loop during mouse embryonic stem cell (ESC) differentiation. Deletion of the stem-loop causes imbalanced DGCR8:DROSHA protein stoichiometry that drives irreversible Microprocessor aggregation, reduced primary miRNA processing, decreased mature miRNA abundance, and widespread de-repression of lipid metabolic mRNA targets. While global miRNA dosage control is dispensable for mouse ESC exit from pluripotency, its dysregulation alters lipid metabolic pathways and interferes with embryonic development by disrupting germ layer specification in vitro and in vivo. Finally, this miRNA dosage control mechanism is conserved in humans. Our study uncovers a promoter switch that balances Microprocessor autoregulation and aggregation to precisely control global miRNA dosage and govern stem cell fate decisions during early embryonic development.
Microprocessor, a complex containing the ribonuclease DROSHA together with two subunits of the essential protein co-factor DGCR87–9, mediates the biogenesis of almost all mammalian microRNAs (miRNAs). Microprocessor specifically cleaves primary miRNAs (pri-miRNAs) to generate ~60–80 nucleotide (nt) hairpin-shaped precursor miRNAs (pre-miRNAs) that are further processed to ~22 nt duplexes by DICER1,2,6,7,10. Interestingly, two stem-loops (SLs) at the 5’-end of DGCR8 mRNA were reported to be cleaved by Microprocessor as part of a possible autoregulatory feedback regulation of DGCR8 expression11,12, however, the related molecular function and biological relevance remain unknown.
miRNAs play critical roles during development1,2,13,14, and global miRNA loss (DGCR8−/− or DICER−/−) caused an early embryonic lethal phenotype in mouse and failure of ESC differentiation15,16. Interestingly, altered global miRNA expression is widely observed in different tissues and tumors, suggesting the potential important roles of precise miRNA dosage control in development and tumorigenesis1,3,4,17,18. However, the underlying regulatory mechanism has remained unknown for over a decade.
A DGCR8 mRNA that escapes autoregulation
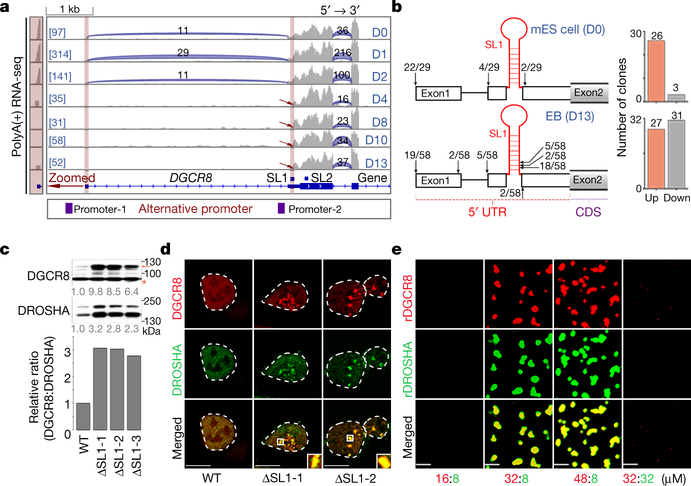
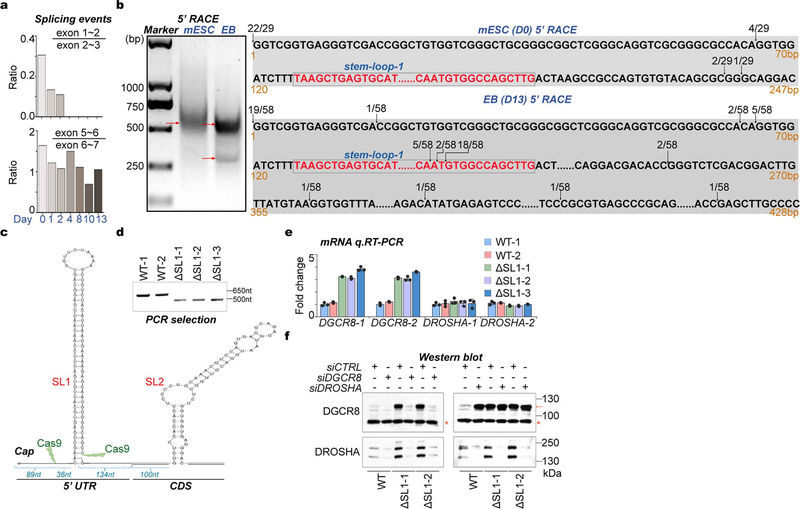
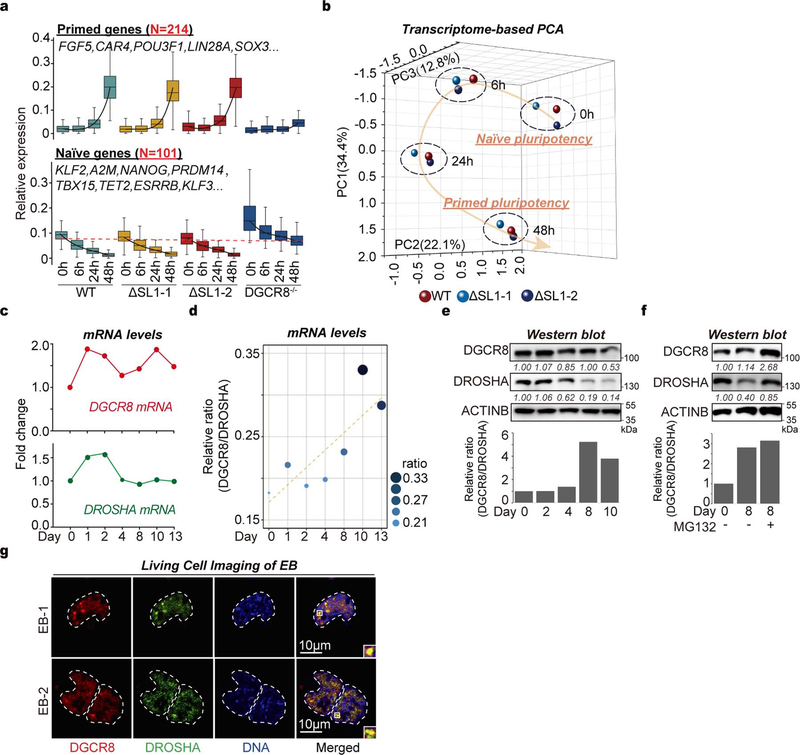
By analyzing RNA-seq data, we unexpectedly found a short isoform of DGCR8 mRNA without the canonical first exon that first appeared at 4 days of mouse ESC-to-embryoid body (EB) differentiation, which was supported by the reduced sequencing abundance of the annotated first exon and splicing events on the first intron. This finding matches well with two predicted alternative promoters (Fig. 1a, Extended Data Fig. 1a). To directly verify DGCR8 mRNA isoforms produced by this ATI event, we performed 5’ Rapid Amplification of cDNA End (5’RACE) assay. As expected, we found that in mouse ESCs the 5’-ends of DGCR8 mRNAs (22/29) were mainly as annotated, however in EB cells about half of the 5’-ends (31/58) were localized downstream of one SL structure (SL1) in Exon 2 (Fig. 1b, Extended Data Fig. 1b).
Fig. 1 |. ATI of DGCR8 links dynamic Microprocessor autoregulation to altered DGCR8: DROSHA stoichiometry and aggregation.
a, Sequencing reads mapping to 5’-end of DGCR8 mRNA from PolyA(+) RNA-seq on mESC to EB differentiation over a 13-day time course. Predicted alternative promoters are from UCSC dataset. Reads number of exon junctions (above the arcs) and sequencing abundance (in brackets) are indicated. b, 5’RACE for DGCR8 mRNA in mESC (Day 0) and EB (Day 13). Positions of 5’RACE ends and the number of colonies localized upstream and downstream of SL1 structure of annotated DGCR8 gene are shown in the histogram. c, Western blot of DGCR8 and DROSHA in WT and ΔSL1 mESCs. Arrow indicates the DGCR8 protein and star indicates unspecific band used as loading control. Density of the DROSHA and DGCR8 bands were normalized to the unspecific band to calculate the relative protein ratios. d, Images of tagged mCherry-DGCR8 and eGFP-DROSHA expressing in living WT and ΔSL1 mESCs transfected with corresponding plasmids. e, Images of endogenous mCherry-DGCR8 fusion protein in the WT and ΔSL1 reporter ESCs. f, Images of Microprocessor aggregates before and after treatment with 10% 1,6-hexanediol in living ΔSL1 cells. g, FRAP analysis of Microprocessor aggregates in living ΔSL1 cells. Normalized fluorescence intensity of DGCR8 and DROSHA are shown. Data are represented as mean +/− SD (N=6). See also Extended Data Fig. 2d. h, Images of puncta formation of rDGCR8 and rDROSHA mixed with different ratios are shown. i, j, Images of pre-formed Microprocessor aggregates (32μM rDGCR8: 8μM rDROSHA) followed by adding extra rDROSHA to achieve a 2:1 ratio or treatment with 10% 1,6-hexanediol. k, FRAP analysis of Microprocessor aggregates (32μM rDGCR8: 8μM rDROSHA). Normalized fluorescence intensity is shown. Data are represented as mean +/− SD (N=7).
Since SL structures at the 5’-end of DGCR8 mRNA were suggested to mediate Microprocessor autoregulation11,12, we speculated that ATI might impact the miRNA biogenesis pathway by bypassing this feedback control. To test this, we deleted the SL1 using CRISPR-Cas9 technology ( Extended Data Figs. 1c, 1d). q.RT-PCR analysis showed around 4-fold accumulation of DGCR8 mRNA in ΔSL1 cells compared with WT cells ( Extended Data Fig. 1e), indicating that SL1 can indeed mediate DGCR8 mRNA cleavage and instability. Western blot further revealed a dramatic accumulation of both DGCR8 and DROSHA proteins, however interestingly the accumulation of DGCR8 protein was ~3-fold greater than that of DROSHA protein in ΔSL1 cells resulting in an excess of DGCR8 protein (Fig. 1c). By transfecting siRNAs targeting either DROSHA or DGCR8 into WT and ΔSL1 cells, we verified the Microprocessor feedback regulation identified previously that DGCR8 can stabilize DROSHA protein and DROSHA mediates cleavage of DGCR8 mRNA11,12. Interestingly, we found that DROSHA knockdown in ΔSL1 cells had no effect on DGCR8 expression, supporting that DROSHA-mediated control of DGCR8 expression is entirely through SL1 in the 5’ UTR ( Extended Data Fig. 1f). Together, these results demonstrate that the 5’ UTR SL is responsible for autoregulation of the Microprocessor to maintain the precise DGCR8:DROSHA stoichiometry, which is governed by ATI of DGCR8 mRNA during ESC differentiation and early embryonic development.
Excess DGCR8 protein causes aggregation
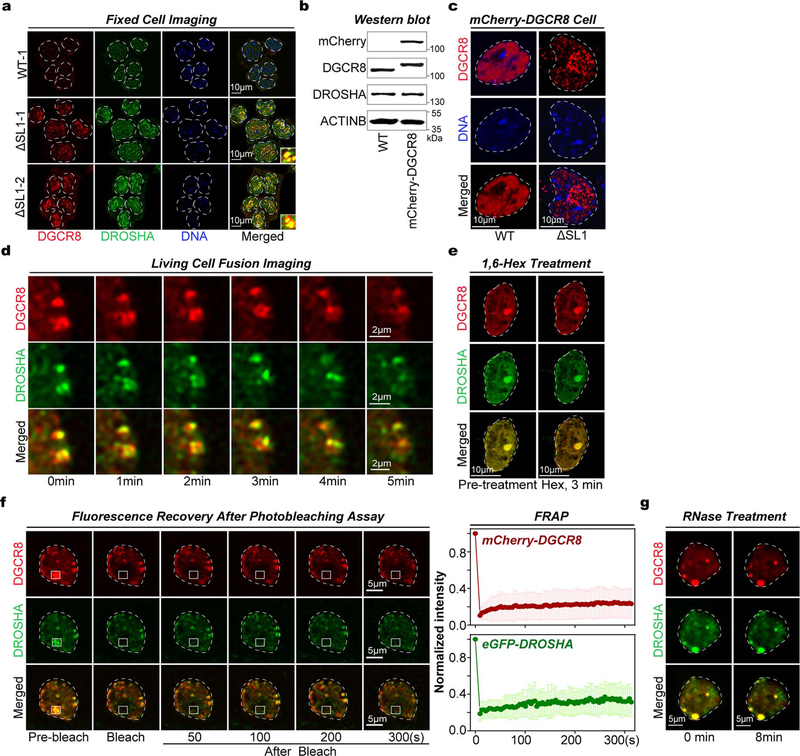
As several macromolecular complexes have been reported to undergo phase separation to influence various biochemical reactions19,20 and stoichiometry is important for biomolecule condensate formation21, we next explored the phase-separated potential of Microprocessor with imbalanced stoichiometry. Interestingly, in both fixed cells monitored by immunofluorescence and living cells transfected with two vectors expressing mCherry-conjugated DGCR8 and eGFP-conjugated DROSHA proteins respectively, we detected many obvious phase-separated puncta of Microprocessor (shown in yellow) in ΔSL1 but not in WT cells (Fig. 1d, Extended Data Fig. 2a). Furthermore, we generated a reporter ESC line expressing endogenous mCherry-DGCR8 fusion protein, in which we then subsequently deleted SL1 ( Extended Data Fig. 2b). As expected, we discovered SL1 knockout induces condensed assemblies of endogenous Microprocessor (Fig. 1e). Notably, the condensates of Microprocessor in ΔSL1 cells are not spherical in shape, and did not fuse with each other (Figs. 1d, 1e, Extended Data Figs. 2a, 2c), suggesting the Microprocessor puncta might not be in a mobile liquid state. We therefore treated the living ΔSL1 cells expressing mCherry-DGCR8 and eGFP-DROSHA proteins with 1,6-hexanediol for 3 minutes and did not detect the disruption of any condensates (Fig. 1f). Fluorescence recovery after photobleaching (FRAP) analysis showed that fluorescence signals of the Microprocessor assemblies did not recover (Fig. 1g, Extended Data Fig. 2d). All the above assays directly indicate that Microprocessor forms irreversible aggregates in ΔSL1 cells with imbalanced DGCR8:DROSHA stoichiometry.
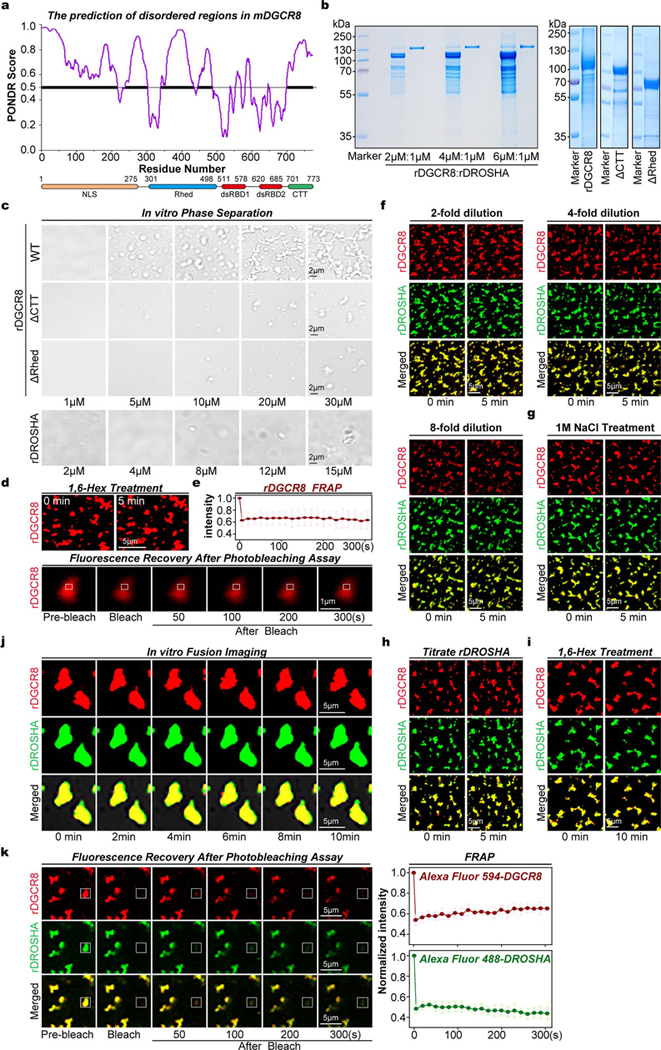
We next thought to further verify Microprocessor aggregation in vitro. Interestingly, several domains of DGCR8 are predicted to be disordered ( Extended Data Fig. 3a). WT recombinant DGCR8 (rDGCR8) proteins, as well as two mutant versions of rDGCR8 proteins lacking CTT or Rhed domains (ΔCTT and ΔRhed), were expressed and purified from bacteria. While recombinant DROSHA (rDROSHA) (amino acids 390–1374) protein was expressed and purified from insect cells ( Extended Data Fig. 3b). We found that WT rDGCR8 proteins alone can form phase separated droplets at physiological salt concentration, which transitioned to a gel-to-solid like state with increasing rDGCR8 concentration, but ΔCTT or ΔRhed proteins cannot even at 30 μM protein concentration, suggesting these two domains are indispensable for the phase separation of DGCR8 protein. While rDROSHA protein did not form visible puncta ( Extended Data Fig. 3c). Then we labeled rDGCR8 and rDROSHA proteins with Alexa 594 (red) and 488 (green), respectively. Since structural studies showed that two DGCR8 and one DROSHA protein comprise one Microprocessor 8,9, we mixed rDRCG8 and rDROSHA with 2:1 (16μM: 8μM) ratios, and no obvious phase separation was detected. However, in the mixtures with increased DGCR8:DROSHA ratios including 4:1 and 6:1, many obvious Microprocessor puncta appeared (Fig. 1h). Considering that 30 μM rDGCR8 alone can form irreversible condensates which were stable under the treatment with 1,6-hexanediol for 10 min and cannot recover in FRAP assay ( Extended Data Figs. 3d, 3e), we propose that DGCR8:DROSHA stoichiometry determines Microprocessor aggregation. Indeed, we found that the pre-formed Microprocessor aggregates in vitro were irreversible and steady under the treatments of adding extra rDROSHA (Fig. 1i), dilution ( Extended Data Fig. 3f), high salt concentration ( Extended Data Fig. 3g), 1,6-hexanediol (Fig. 1j), did not fuse even after 10 min ( Extended Data Fig. 3h), and cannot recover after photobleaching (FRAP) (Fig. 1k, Extended Data Fig. 3i). Finally, as the rDGCR8 and rDROSHA proteins used for the above phase separation assays were treated with RNase during purification ( Extended Data Fig. 3b) and RNase treatment did not disrupt Microprocessor aggregates in living cells ( Extended Data Fig. 2e), we conclude that RNA is not a significant component of DGCR8/DROSHA aggregates20. Based on the above in vitro and in vivo analysis, we conclude that a regulated DGCR8:DROSHA protein ratio is important to maintain soluble Microprocessor in the nucleoplasm, however imbalanced DGCR8:DROSHA stoichiometry, caused by disrupting the posttranscriptional autoregulation mechanism, directly leads to irreversible phase-separated aggregation of Microprocessor lacking the properties of fluidity and fusion and are in a gel-to-solid state and cannot exchange with the surrounding aqueous solution.
A miRNA dosage control mechanism
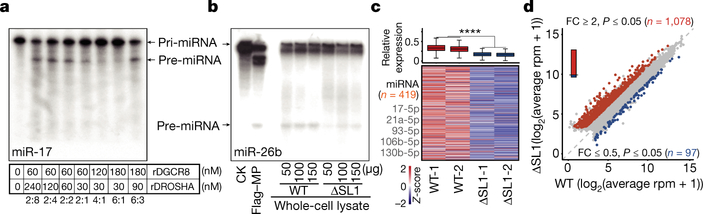
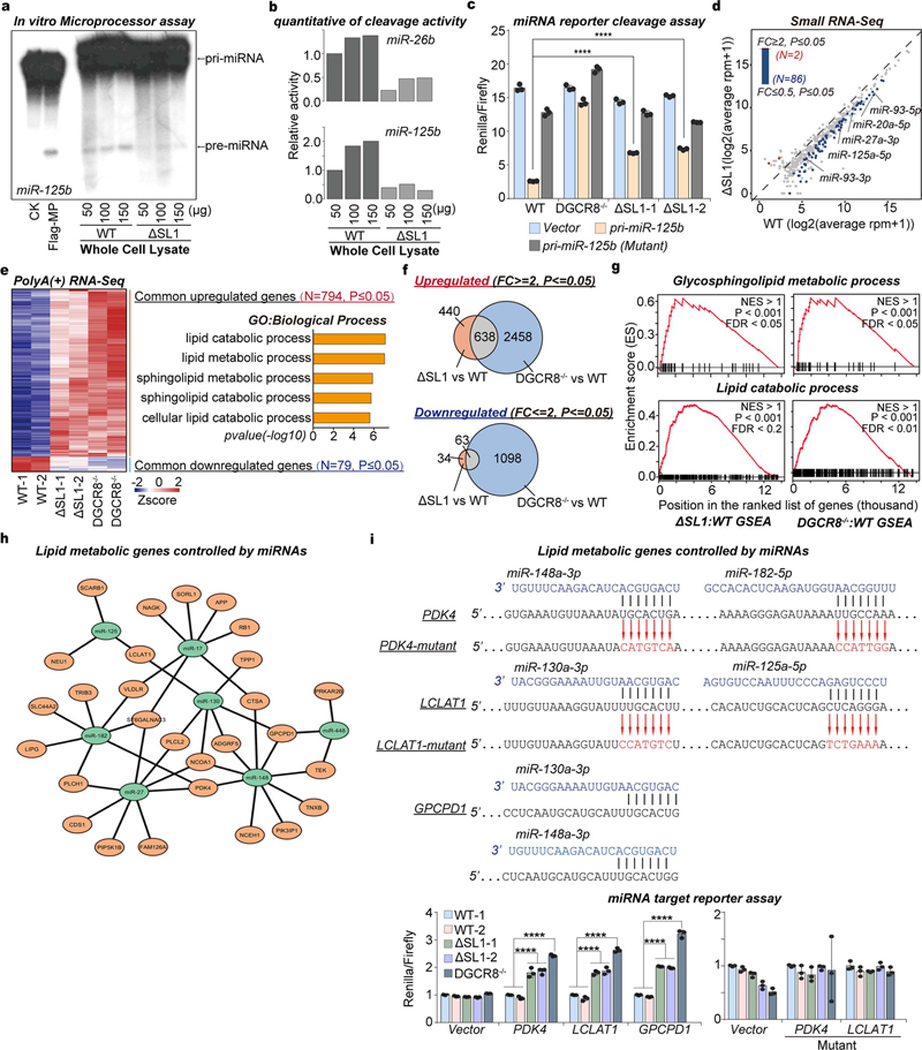
As aggregated condensation normally leads to functional inhibition of macromolecules22,23, we hypothesized that the irreversible aggregation of Microprocessor could inhibit miRNA biogenesis. To test this, we performed biochemical cleavage assays using in vitro transcribed pri-miR-17 as a substrate, which was incubated with mixtures of rDGCR8 and rDROSHA proteins with different ratios. We found that mixtures containing a consistent amount of rDGCR8 with increasing amounts of rDROSHA (2:2, 2:4, and 2:8) displayed similar cleavage efficiencies as measured by the relative levels of pre-miR-17 products. Interestingly however, an excess of rDGCR8 protein in these processing assays (4:1 and 6:1) repressed pri-miRNA cleavage efficiency and production of pre-miRNA (Fig. 2a). Thus, precise DGCR8:DROSHA stoichiometry determines the biochemical activity of Microprocessor in vitro.
Fig. 2 |. Microprocessor aggregation reduces the efficiency of miRNA processing and global miRNA dosage which leads to the de-repression of lipid metabolic genes.
a, Microprocessor in vitro cleavage assay of mouse pri-miR-17 using rDGCR8 and rDROSHA with the indicated ratios. b, Microprocessor in vitro cleavage assay of mouse pri-miR-26b using whole cell lysate from WT and ΔSL1 mESCs. Microprocessor purified by immunoprecipitation from Flag-DROSHA-293T cells was used as a control. c, Luciferase reporter in vivo cleavage assay of pri-miR-125b in WT, DGCR8−/−, and ΔSL1 mESCs. Data are represented as mean +/− SD (N=3). **p<0.01, Two-tailed Student’s t-test. d, Box plot and heatmap of the expression of global mature miRNAs in WT and ΔSL1 mESCs. The small RNA-seq data were normalized based on spike-in RNAs. Number of miRNAs analyzed is shown and representative miRNAs are listed. ****p<0.0001, Two-tailed Student’s t-test. e, Scatter plot of global mRNA expression in WT and ΔSL1 mESCs. Differentially expressed mRNAs are represented by colored circles, and number of up- and downregulated mRNAs is shown. FC: fold change, P: P-Value from Two-tailed Student’s t-test. f, Heatmap of the expression of common up- or downregulated genes in ΔSL1 and DGCR8−/− mESCs compared with WT mESCs. The enrichment of Gene Ontology (GO) terms and the number of genes in each group are shown.. DGCR8−/− samples are from two technical replicates. P: P-Value from Two-tailed Student’s t-test. g, Luciferase miRNA target reporter assay in WT, ΔSL1 and DGCR8−/− mESCs. Data are represented as mean +/− SD (N=3). **p<0.01, Two-tailed Student’s t-test.
Furthermore, we performed in vitro cleavage assays using pri-miR-26b or pri-miR-125b as the substrates and cell lysates prepared from WT and ΔSL1 cells. This revealed reduced pri-miRNA cleavage activity using cell lysate from ΔSL1 compared to WT cells (Fig. 2b, Extended Data Figs. 4a, 4b). Next we used dual luciferase reporter vectors containing WT or as a control a mutant version of pri-miR-125b lacking the pre-miRNA hairpin to measure relative Microprocessor cleavage activity in WT, DGCR8−/−, and ΔSL1 cells. Consistent with in vitro assays above, pri-miRNA cleavage activity was compromised in ΔSL1 cells compared with WT cells, and as a control, no pri-miRNA cleavage was detected in DGCR8 knockout cells (Fig. 2c). The above data suggests that aggregated assembly of Microprocessor represses efficient processing of pri-miRNA in vivo.
To measure the effects of Microprocessor aggregation on mature miRNA expression, we performed small RNA-seq on WT and ΔSL1 cells with insect RNAs spiked-in for normalization. This analysis revealed generally diminished, but not complete loss of global miRNA expression in ΔSL1 cells compared with WT cells (Fig. 2d, Extended Data Fig. 4c, Supplementary Table 1). In summary, our data indicates that the SL1 in the 5’ UTR of DGCR8 mRNA is required to maintain the precise DGCR8:DROSHA stoichiometry, which is indispensable to avoid aggregated assembly of Microprocessor to maintain miRNA processing efficiency and abundance in ESCs. We therefore refer to this ATI-mediated Microprocessor autoregulatory pathway as a ‘miRNA dosage control’ mechanism.
Molecular role of miRNA dosage control
To understand the molecular function of miRNA dosage control in mouse ESCs, we analyzed the transcriptomic changes caused by loss of miRNA dosage control. We found that reduced miRNA dosage led to increased expression of 1,078 genes, whereas only 97 genes were downregulated significantly in ΔSL1 cells compared with WT cells (Fig. 2e, Supplementary Table 2). We further compared effects of disruption of miRNA dosage control (ΔSL cells) with the complete loss of miRNAs (DGCR8−/− cells). This analysis revealed a large number of genes commonly upregulated in ΔSL cells and DGCR8−/− cells, which were interestingly enriched in “lipid metabolic process”, however few were commonly downregulated (Fig. 2f, Extended Data Figs. 4d, 4e, Supplementary Table 2). To experimentally verify lipid metabolic mRNA targets ( Extended Data Fig. 4f), we cloned the 3’ UTRs of three of the upregulated lipid metabolic genes, including PDK4, LCLAT1, and GPCPD1, into the dual luciferase reporter. These constructs were then transfected into WT, DGCR8−/−, and ΔSL1 cells and relative luciferase activity was measured. Increased luciferase expression was detected from these miRNA-sensors in both DGCR8−/− and ΔSL1 cells compared with WT cells, while mutation of the miRNA target sites erased the repressive effects of miRNAs (Fig. 2g, Extended Data Fig. 4g), strongly supporting that expression of these mRNAs is directly controlled by miRNAs. We therefore conclude that reduced miRNA dosage leads to the de-repression of lipid metabolic genes in ESCs.
Cellular role of miRNA dosage control
We next sought to understand the potential physiological relevance of miRNA dosage control during ESC differentiation. Since previous studies showed that global miRNA deficiency (DGCR8−/−) blocks ESC naïve-to-primed pluripotency transition and early embryonic development during embryo implantation in vivo14,16,24,25, we next tested whether miRNA dosage impacts this earliest stem cell transition using the epiblast-like cell (EpiLC) differentiation system26. Transcriptomic analysis showed as expected DGCR8−/− cells were unable to establish the primed pluripotent state after 48 hours differentiation with an inability to activate expression of primed genes as well as to silence expression of naïve genes. Strikingly, however, we found that ΔSL1 ESCs could readily exit the naïve state and establish primed pluripotency, behaving like WT ESCs in this assay (Fig. 3a, Extended Data Fig. 5a, Supplementary Table 3). Thus, unlike global miRNA loss, altered miRNA dosage control did not impact ESC exit from naïve pluripotency and transition to the primed pluripotent state.
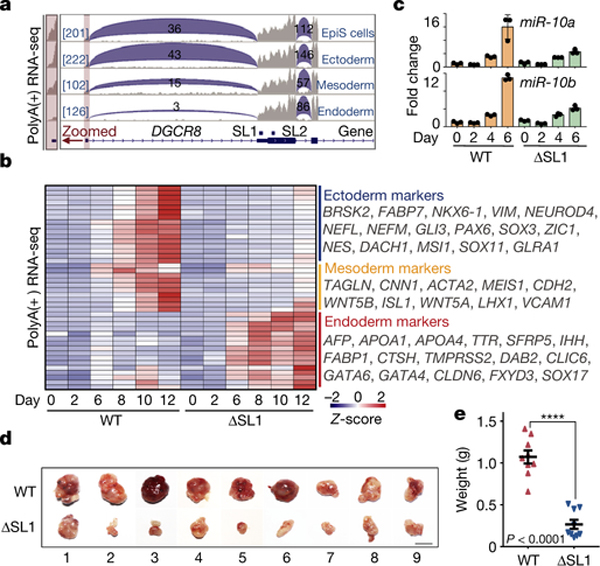
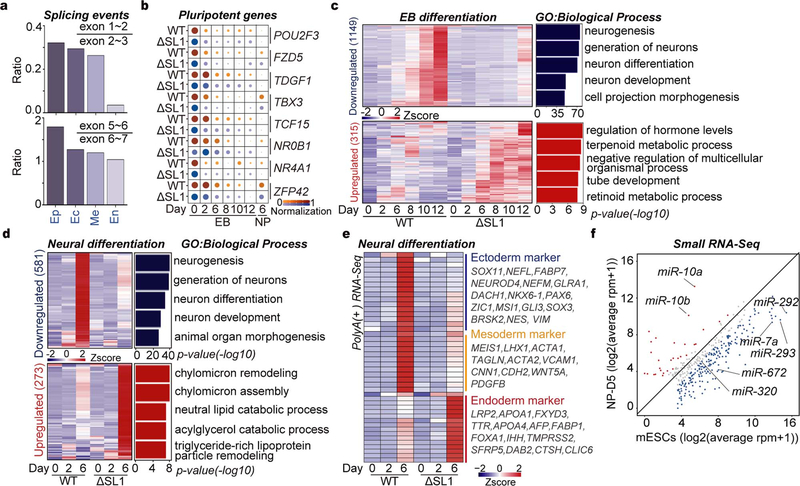
Fig. 3 |. ATI-mediated miRNA dosage control determines germ layer specification during ESC differentiation.
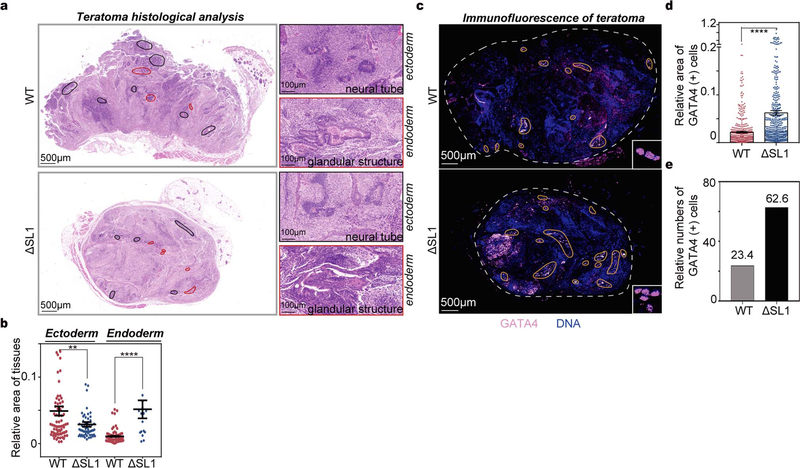
a, Box plots of the relative expression of naïve and primed genes during EpiLC differentiation of WT, ΔSL1 and DGCR8-\- mESCs. The number of naïve and primed genes is shown and representative markers are listed. b, Sequencing reads mapping to 5’-end of DGCR8 mRNA from PolyA(+) RNA-seq on EpiSC and three embryonic germ layers derived from mESCs. Regions containing the first exon are zoomed in. Reads number of exon junctions (above the arcs) and sequencing abundance (in brackets) are indicated. c, Heatmap of gene expression showing up- or downregulation in ΔSL1 cells compared with WT cells during mESC-to-neural progenitor differentiation. The enrichment of Gene Ontology (GO) terms and the number of genes in each group are shown. d, Heatmap of the expression of marker genes for three germ layers during EB differentiation. Representative genes are listed on the right. e, q.RT-PCR analysis of the expression of miR-10a and −10b during neural differentiation. Data were normalized to U2 snoRNA. Data are represented as mean +/− SD. f, Morphology of teratomas derived from WT and ΔSL1 mESCs in 9 donor mice. g, The weight of teratomas in f is shown. Data are represented as mean +/− SEM (N=9). p<0.0001, Two-tailed Student’s t-test. h, Comparison of the relative area of ectoderm and endoderm tissues between WT and ΔSL1 teratomas. The beeswarm plot showing relative area of ectoderm and endoderm tissues compared to the total area according to the HE staining. 16 (WT:8, ΔSL1:8) sections of 8 (WT:4, ΔSL1:4) teratomas from 4 mice are used for analysis. Data are represented as mean +/− SEM. **p<0.01, ****p<0.0001, Two-tailed Student’s t-test.
We then speculated that miRNA dosage control governed by ATI of DGCR8, which began to appear after 4 days of EB differentiation (Fig. 1a), might influence cell fate decisions at later stages of stem cell differentiation. Accordingly, we detected gradual increase of DGCR8:DROSHA stoichiometry at both mRNA and protein levels during EB differentiation (Extended Data Figs. 5b–d). While DGCR8 and DROSHA seemed to be degraded by the proteasome in later stage EBs ( Extended Data Fig. 5e). Finally, using a dual-reporter ESC line endogenously expressing both mCherry-DGCR8 and eGFP-DROSHA fusion proteins, we observed aggregated Microprocessor puncta in differentiated EB cells ( Extended Data Fig. 5f), which directly links ATI of DGCR8 to DGCR8:DROSHA stoichiometry and aggregation during ESC differentiation. Intriguingly, further data mining revealed the short isoform of DGCR8 caused by ATI was mainly detected in endoderm cells, but not the other two germ layers or EpiSCs (Fig. 3b, Extended Data Fig. 6a)27. We next performed ESC differentiation towards neural progenitors (ectoderm) or EBs. RNA-seq based transcriptomic comparison showed that although expression of typical pluripotency genes was obviously decreased in both WT and ΔSL1 cells, expression of 581 and 1,149 genes was highly increased in WT, but not ΔSL1 cells for neural and EB differentiation respectively (Fig. 3c, Extended Data Figs. 6b, 6c, Supplementary Table 4). By analyzing the expression of typical markers for the three germ layers, we discovered that the loss of miRNA dosage control caused by SL1 depletion significantly suppressed stem cell differentiation toward ectoderm and mesoderm layers, while instead enhanced endoderm development (Fig. 3d, Extended Data Fig. 6d, Supplementary Table 5). Interestingly q.RT-PCR analysis revealed the strong repression of miR-10a and −10b, both of which were particularly activated during neural differentiation ( Extended Data Fig. 6e), in ΔSL1 cells compared to WT cells (Fig. 3e). Therefore, although loss of miRNA dosage control does not impact ESC exit from pluripotency, it strongly interferes with ESC fate transition towards three germ layers in vitro.
To directly test this in mice, we injected WT and ΔSL1 ESCs into immune-deficient mice for teratoma formation assays. Interestingly, we observed the dramatic reduction in the size and weight of teratomas derived from ΔSL1 cells compared with those from WT cells, implying developmental deficiency due to the loss of miRNA dosage control (Figs. 3f, 3g). Both morphology and Immunofluorescence (using GATA4 antibody) analysis of endoderm cells of the teratoma sections showed that ΔSL1 cells generated relatively more endoderm tissues compared with WT cells (Fig. 3h, Extended Data Figs. 7a–d).
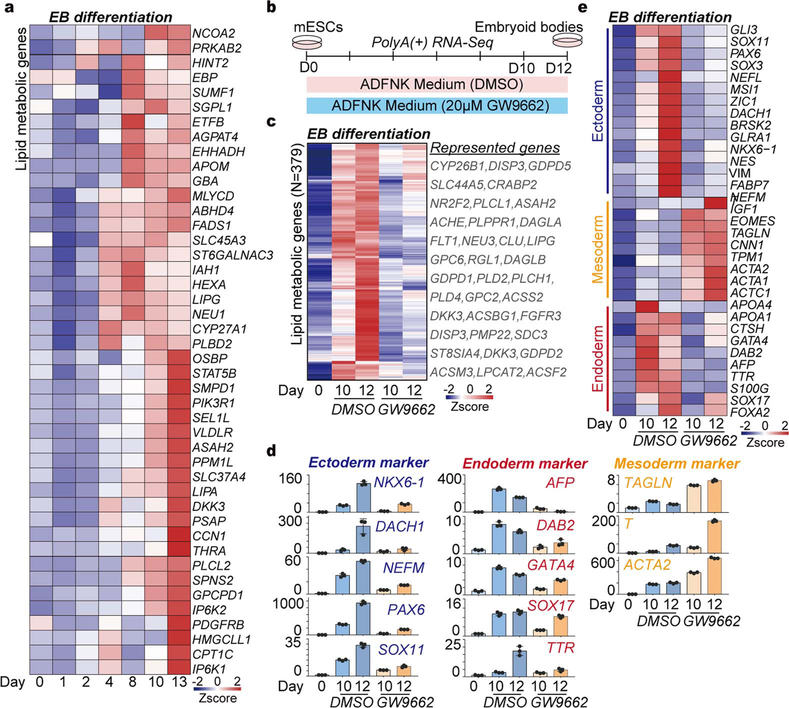
Since lipid metabolic genes were activated during EB differentiation, as well as in ESCs and differentiated EB cells with disrupted miRNA dosage control (Figs.2f, 3c, Extended Data Figs. 4e, 6c, 8a), we measured the effects of lipid metabolism on germ layer specification using a small molecule inhibitor of lipid metabolism (GW9662) to treat cells during EB differentiation ( Extended Data Fig. 8b). q.RT-PCR and RNA-seq analysis revealed that inhibition of lipid metabolism represses ESCs fate transition to endoderm, but enhances mesoderm development ( Extended Data Figs. 8c–8e, Supplementary Table 6), which is further supported by a recent study in human ESCs28.
Overall, our data provide strong evidence that distinct from global miRNA loss that inhibits ESCs exit from pluripotency, ATI-mediated miRNA dosage control is dispensable for the naïve-to-primed ESC transition but is a key mediator of germ layer specification, with cells expressing the shorter DGCR8 mRNA isoform or with ΔSL1 being strongly biased towards endoderm development, through modulating lipid metabolism.
miRNA dosage control conserved in humans
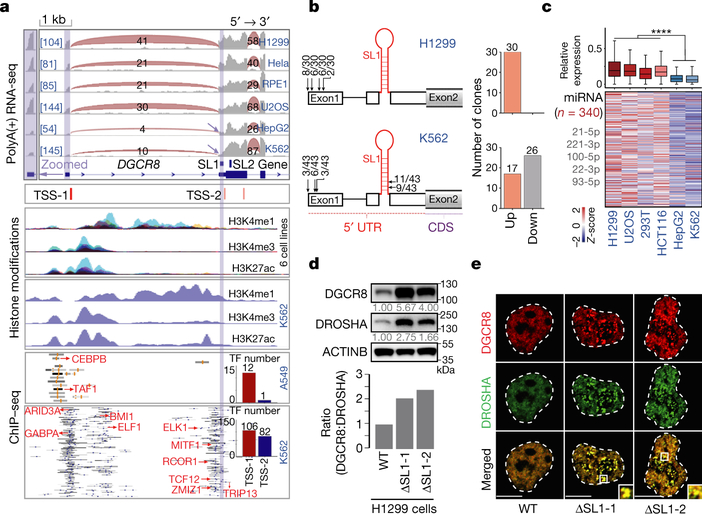
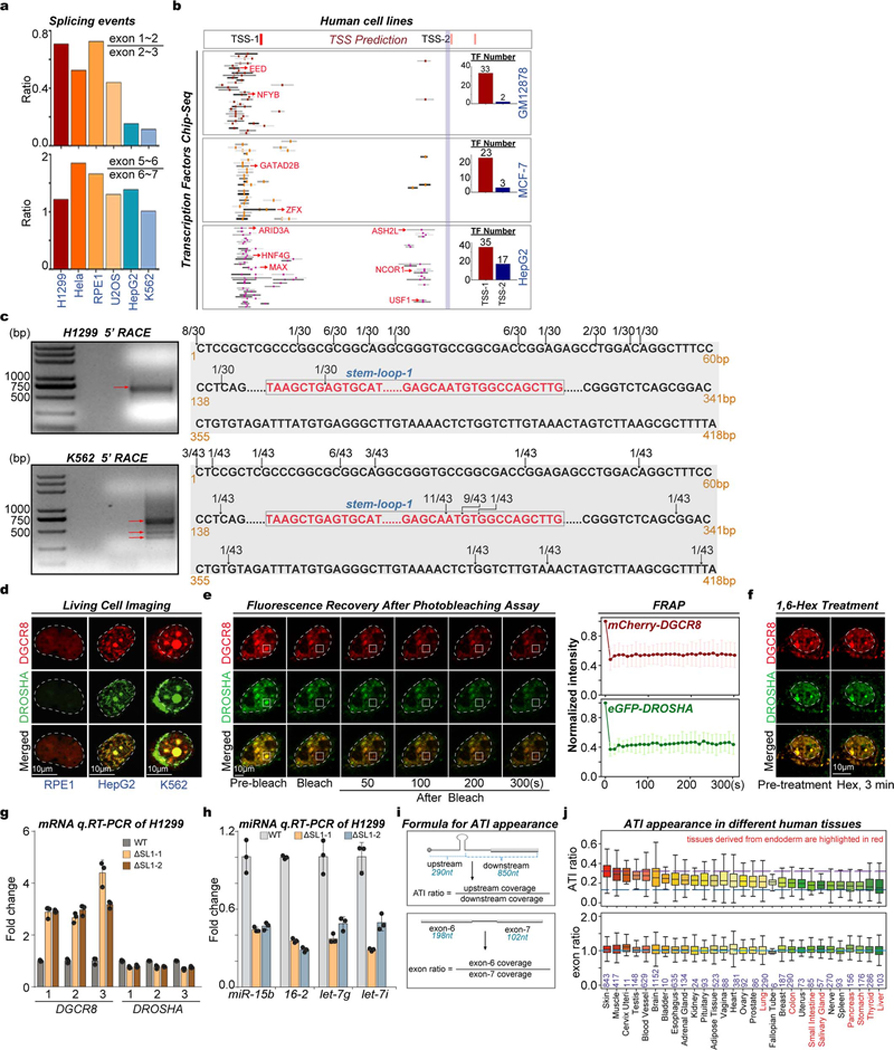
Since SL1-medaited Microprocessor autoregulation are conserved between mouse and human11,12, we further explored the above model in humans. Among 6 different human cell lines, we discovered obvious appearance of the short DGCR8 isoform particularly in hepatocellular cells (HepG2) and hematopoietic cells (K562), fitting well with one of the predicted ATIs close to SL1 structure at 5’ UTR of DGCR8 mRNA (Fig. 4a, Extended Data Fig. 9a). Further analysis of various ChIP-seq data for different histone modifications and hundreds of transcription factors (TFs) showed, particularly in K562 and HepG2 cells, there are two signal peaks for both H3K4me1 and H3K27Ac, corresponding to two distinct binding sites enriched with clusters of TF binding sites, at the 5’-end of DGCR8. Notably, the downstream site is in close proximity to SL1 (Fig. 4a, Extended Data Fig. 9b). 5’ RACE showed that most 5’ ends of DGCR8 mRNA were localized downstream of the SL1 structure in K562 cells, that was different from H1299 cells that expresses DGCR8 mRNA from the annotated 5’ end (Fig. 4b, Extended Data Fig. 9c). Similar as in mice, we detected obvious irreversible Microprocessor aggregates (Fig. 4c, Extended Data Figs. 9d, 9e), that resulted in obvious reduction of global miRNA dosage in living HepG2 and K562 cells compared with in U2OS, HCT116, and H1299 cells (Fig. 4d, Supplementary Table 7). To directly prove SL1 mediated miRNA dosage control mechanism in human cells, we deleted SL1 in H1299 cells, which as expected caused imbalanced DGCR8:DROSHA stoichiometry at both RNA and protein levels (Fig. 4e, Extended Data Fig. 9f) that led to Microprocessor phase-separated aggregation and decreased expression of mature miRNAs (Figs. 4f, 4g). Finally, by analyzing RNA-seq data from the GTEX dataset ( Extended Data Fig. 9h), we identified the potential DGCR8 ATI events in certain tissues, including liver, thyroid, stomach, and pancreas, which interestingly are commonly derived from embryonic endoderm (Fig. 4g). Therefore, miRNA dosage control mechanism is conserved in human cells and tissues.
Fig. 4 |. ATI-mediated miRNA dosage control mechanism is conserved in human cells and tissues.
a, Sequencing reads mapping to the 5’-end of the DGCR8 gene from RNA-seq and ChIP-seq for histone modifications and transcription factors (TFs) in several different cell lines. Transcription start sites (TSSs) predicted by SwitchGear Genomics from UCSC dataset are shown. CHIP-seq signals for various TFs at this region are shown, and representative TFs are listed in red. The number of TFs binding to the two distinct promoters is shown by Histograms. Reads number of exon junctions (above the arcs) and sequencing abundance (in brackets) are indicated. b, 5’RACE for DGCR8 mRNA in H1299 and K562 cells. Positions of 5’RACE ends and the number of colonies localized upstream and downstream of SL1 structure of annotated DGCR8 gene are shown. c, Imaging of tagged mCherry-DGCR8 and eGFP-DROSHA in living RPE1, HepG2, and K562 cells transfected with corresponding plasmids. d, Box plot and heatmap of the expression of global mature miRNAs in U2OS, H1299, HCT116, K562, and HepG2 cells. The small RNA-seq data were normalized based on spike-in RNAs. Number of miRNAs analyzed is shown and representative miRNAs are listed. ****p<0.0001, Two-tailed Student’s t-test. e, Western blot of DGCR8 and DROSHA in WT and ΔSL1 H1299 cells. Density of the DROSHA and DGCR8 bands were normalized to the ACTINB band to calculate the relative protein ratios. f, Imaging of tagged mCherry-DGCR8 and eGFP-DROSHA expressing in living WT and ΔSL1 H1299 cells transfected with corresponding plasmids. g, q.RT-PCR analysis of mature miRNA expression. Data were normalized to U2 snoRNA. Data are represented as mean +/− SD (n=2). h, Box plot shows the ATI of DGCR8 mRNA appearance in different human tissues based on RNA-seq from GTEX dataset. Tissues mainly derived from embryonic endoderm are highlighted in red font.
Discussion
As an indispensable layer of posttranscriptional gene regulation, miRNAs are critical during development and in cancer 1,2,14,24, and more evidence is emerging to support that precise global miRNA dosage control is important in these contexts1,3,4,17. However, a molecular explanation for this global suppression of miRNA expression has remained unknown. Here our study directly demonstrates that endogenous miRNA dosage control is mediated by a DGCR8 promoter switch that balances Microprocessor autoregulation and aggregation to control global miRNA expression that impacts stem cell fate decision during differentiation and germ layer specification.
We reveal that precise DRCR8:DROSHA ratio is indispensable to keep Microprocessor soluble in the nucleoplasm for efficient pri-miRNA cleavage, however, imbalanced DRCG8:DROSHA stoichiometry leads to the formation of irreversible aggregation and reduced miRNA dosage. A recent study discovered that in plants pri-miRNA processing complex (Dicing body) forms liquid-liquid phase separation for efficiently miRNA processing, that is quite different from our finding in animals29.
Methods
Cell Culture
All mouse and human cell lines were cultured at 37°C in a 90% (v/v) humidified atmosphere with 5% (v/v) CO2. Unless stated otherwise, all mouse embryonic stem cells (ESCs) were cultured in Serum/leukemia inhibitory factor (LIF) medium [DMEM (Gibco) with 1000 U/ml mLIF (Gemini), 15% (v/v) stem cell fetal bovine serum (FBS) (Gemini), 1X sodium pyruvate (Gibco), 1X non-essential amino acid (NEAA) (Gibco), 1X L-glutamine (Gibco), 50 μM 2-mercaptoethanol (ThermoFisher) and 1% penicillin-streptomycin (Gibco)] on 0.1% gelatin-coated dishes. To convert ESCs into ‘ground’ state for EpiLC differentiation, cells were cultured in 2i/LIF medium [1:1 DMEM/F12 (Gibco) and Neurobasal medium (Gibco) containing 1X N2 and B27 supplements (Gibco), 1 μM PD03259010 (Stemgent), 3 μM CHIR99021 (Stemgent), 1000 U/ml mLIF (Gemini), 1X Sodium Pyruvate (Gibco), 1X NEAA (Gibco), 1X L-glutamine (Gibco), 50 μM 2-mercaptoethanol (ThermoFisher) and 1% penicillin-streptomycin (Gibco)] on 0.1% gelatin-coated dishes without feeders for at least two weeks.
HEK293T cells expressing Flag-DROSHA (Flag-DROSHA-293T) were cultured in DMEM with 15% (v/v) FBS for Microprocessor purification. Other human cell lines (H1299, Hela, U2OS, RPE1 and HepG2) were cultured in DMEM (Gibco) with 15% (v/v) FBS (Gemini), 1X sodium pyruvate (Gibco), 1X L-glutamine (Gibco), 50 μM 2-mercaptoethanol (ThermoFisher) and 1% (v/v) penicillin-streptomycin (Gibco). K562 cells were cultured in RPMI 1640 (Gibco) with 10% (v/v) FBS (Gemini) and 1% penicillin-streptomycin (Gibco).
For insect cell culture, Sf21 cells were cultured in SIM SF medium (Sino Biological Inc) with 1‰ penicillin-streptomycin (Gibco) shaking at 100 rpm at 27°C. Drosophila S2 cells were cultured in Schneider’s Drosophila medium (Gibco) with 10% (v/v) FBS (Cellmax) and 1% penicillin-streptomycin (Gibco) at 25°C.
Construction of endogenously-tagged cell line
Homology mediated End Joining-based Strategy (HMEJ) based CRISPR/Cas9 system30 was used to generate endogenously-tagged DGCR8-mCherry and eGFP-DROSHA cell lines. Repair templates were cloned into a pBabe vector containing eGFP/mCherry, a (GGGGS)5 linker and 800 bp homology arms flanking the insert. mESCs were transfected with px330 vector and repair templates using Lipofectmine 3000. After 3 days, the eGFP+/mCherry+ cells were sorted and seeded at a low density. Single cell clones appeared within one week. About 100 clones were then picked up, digested and expanded. When passaged, some cells were used for genotyping, others were frozen.
Cell Sorting Assay
For screening of cell lines, the cells were digested into single cells by using 0.25% trypsin-EDTA and resuspended in DMEM medium. Cell suspension was then filtered through 40 μm cell strainers. BD FACSAria™ III cell sorter device was used to sort living eGFP+/mCherry+ cells.
Mouse ESC-to-EB Differentiation
The polyHEMA-coated plates were used for Embryoid Body (EB) differentiation. For coating the plates, 4 ml polyHEMA (Sigma) (20 mg/ml in 95% ethanol) was added into a 10 cm dish, and dried in cell culture hood for more than 4 h. 2×106 mESCs were plated into ADFNK differentiation medium [1:1 DMEM/F12 (Gibco) and Neurobasal medium (Gibco) with 10% knockout serum replacement (Gibco), 1X L-glutamine (Gibco), 50 μM 2-mercaptoethanol (ThermoFisher) and 1% (v/v) penicillin-streptomycin (Gibco)] on polyHEMA-coated dishes to induce EB formation for 13 days, and the samples were collected at the indicated time points.
To observe the phase separation in EBs, 2×106 dual-reporter mESCs (endogenously expressing both mCherry-DGCR8 and eGFP-DROSHA fusion proteins) were plated into ADFNK differentiation medium for 13 days. Then, EBs were collected and dissociated into single cell with 0.25% trypsin. Furthermore, these single cells were plated on Matrigel-coated glass bottom dishes, and on the following day the cells were subjected to confocal microscope imaging (see below).
To detect the effect of lipid metabolism on EB differentiation, 2×106 mESCs were plated into ADFNK differentiation medium with 20 μM GW9662 (a PPARγ inhibitor, dissolved in DMSO) on polyHEMA-coated dishes to induce EB formation for 12 days. As a control, 2×106 mESCs were plated into ADFNK differentiation medium with DMSO (10 μl DMSO per 10 ml medium) on polyHEMA-coated dishes to induce EB formation for 12 days. Samples were collected at the indicated time points.
For MG132 treat experiment, 2×106 mESCs were plated into ADFNK differentiation medium on polyHEMA-coated dishes to induce EB formation. The EBs were treated with 10 μM MG132 for 2 h on the 8th day to inhibit Microprocessor complex degradation. Then the treated and untreated EBs were collected for the next experiment.
EpiLC Differentiation
Fibronectin-coated dishes were firstly prepared for epiblast-like cell (EpiLC) differentiation. Briefly, 3 ml fibronectin (10 μg/ml in water) was added to 6 cm dishes, which were incubated at 37°C for 2–3 hours or at 4°C overnight. Subsequently, 5 × 105 WT, DGCR8−/−, or ΔSL1 mESCs cultured in 2i/LIF medium were re-plated into EpiLC differentiation medium [1:1 DMEM/F12 (Gibco) and Neurobasal medium (Gibco) containing 1X N2 and B27 supplements (Gibco), 1X Sodium Pyruvate (Gibco), 1X NEAA (Gibco), 1X L-glutamine (Gibco), 50 μM 2-mercaptoethanol (ThermoFisher), 12.5 ng/ml bFGF (Peprotech), 20ng/ml Activin A (Peprotech), and 1% knockout serum replacement (Gibco), and 1% penicillin-streptomycin (Gibco)] on fibronectin-coated plates as described previously26 for 2 days.
Neural Differentiation
The polyHEMA-coated plates were used for neural differentiation. For coating the plates, 4 ml polyHEMA (Sigma) (20 mg/ml in 95% ethanol) was added into 10cm dishes, and dried in cell culture hood for more than 4 h. To induce mESC differentiation into neural progenitor cells, 2×106 mESCs were plated into ADFNK differentiation medium [1:1 DMEM/F12 (Gibco) and Neurobasal medium (Gibco) with 10% knockout serum replacement (Gibco), 1X L-glutamine (Gibco), 50 μM 2-mercaptoethanol (ThermoFisher) and 1% (v/v) penicillin-streptomycin (Gibco)] on polyHEMA-coated dishes. Retinoic acid (1 μM, Sigma) was added to the ADFNK differentiation medium from the third day on. Recombinant Mouse Sonic Hedgehog (Shh) (R&D systems) was added at 10 ng/ml to the ADFNK differentiation medium from the fourth day on. Medium was replaced every day. ESCs were induced for six days, and the samples were collected at the indicated time points.
Cell Transfection
Lipofectamine 2000 was used for siRNA transfection according to the manufacturer’s instructions for three days unless stated otherwise. Briefly, siRNAs were incubated with Lipofectmine 2000 in 1 ml Opti-MEM™ I Reduced Serum Medium (Gibco) for 15 min at room temperature. Then Lipofectmine 2000 and siRNA mixtures were added into cell culture medium during splitting the cells. Medium was changed after 6 hours. siRNA sequences used were reported previously13. Lipofectamine 3000 and P3000 were used for plasmids transfection according to the manufactures’ instructions. The steps are similar with siRNA transfection.
Plasmids Construction
Primers used for CRISPR/Cas9 mutagenesis were designed (http://crispr.mit.edu/) and cloned into the PX330 vector. The cDNA of mouse pri-miR-26b was amplified by PCR and cloned into the EcoRI and XhoI sites of pcDNA3 vector (Invitrogen). The plasmid of mouse pri-miR-17 was previously reported13. The cDNA of 3’ untranslated regions (UTRs) of mouse PDK4, LCLAT1, and GPCPD1 was amplified and cloned into the XhoI and NotI sites of psiCHECK2 vector (Promega). As a control, the miRNA sites in the mouse PDK4, LCLAT1 UTRs were mutated. psiCHECK2 plasmids for pri-miR-125b and the mutant version were reported previously31. The full-length mouse DGCR8 fused with mCherry and full-length human DROSHA fused with GFP were cloned into the pCMV2 Vector. The cDNA of full-length mouse DGCR8 was also amplified by PCR and cloned into the SalI and NotI sites of the pETDuet-1 vector containing a His6-tag. Moreover, the Rhed or the CTT domain of DGCR8 was deleted using the Q5® Site-Directed Mutagenesis Kit. The cDNA human DROSHA spanning residues 390–1374 was amplified by PCR and cloned into the BamHI and NotI sites of the pFastBac-Dual vector carrying a GST-tag and a TEV cleavage site.
Immunofluorescence
For cell immunofluorescence, cells were cultured on glass bottom dishes (Cellvis). After washing the cells twice with PBS, the cells were fixed with 4% paraformaldehyde (BBI Life Science) at room temperature for 20 min. Following another two washing steps with PBS, the cells were permeabilized with PBS containing 0.1% Triton X-100 (Sigma) at room temperature for 15 min. After washing the cells twice with PBS, the cells were blocked with PBS containing 5% BSA at room temperature for 1 h. Subsequently, the cells were washed twice with PBS and incubated overnight at 4°C with the primary antibodies mouse monoclonal anti-DROSHA (Santa Cruz Biotechnology) and rabbit polyclonal anti-DGCR8 (Proteintech) diluted at 1:200 in 5% BSA. After washing three times with PBS, the cells were incubated for 2 h at room temperature with the secondary antibodies donkey anti-mouse IgG (Abcam, Alexa Fluor 488) and donkey anti-rabbit IgG (Abcam, Alexa Fluor 647), diluted at 1:500 in 5% BSA. Finally, the cells were washed three times with PBS and incubated with 1 μg/ml DAPI (Solarbio) at room temperature for 20 min. Following washing three times with PBS, the cells were subjected to confocal microscope imaging (see below).
For Paraffin section immunofluorescence, the sections were deparaffinize and rehydrate firstly, incubate sections twice in xylene 15 min, and dehydrate twice with pure ethanol for 5 min, followed by dehydrate in gradient ethanol of 85% and 75% for 5 min, and washed in distilled water for 10min. For antigen retrieval, immerse the slides in EDTA antigen buffer (pH 8.0) and maintain at a sub-boiling temperature for 8 min, standing for 8 min and followed by another sub-boiling temperature for 7 min. After washing 3 times with PBS, mark the objective tissue with liquid blocker pen, and the tissues were blocked with PBS containing 3% BSA at room temperature for 30 min. Subsequently, the tissues were washed twice with PBS and incubated overnight at 4°C with the primary antibodies mouse monoclonal anti-GATA4 (Santa Cruz Biotechnology) diluted at 1:100 in 3% BSA. After washing three times with PBS, the cells were incubated for 2 h at room temperature with the secondary antibodies donkey anti-mouse IgG (Abcam, Alexa Fluor 488) diluted at 1:500 in 5% BSA. Then, the cells were washed three times with PBS and incubated with 1 μg/ml DAPI (Solarbio) at room temperature for 20 min. After washing three times with PBS, add spontaneous fluorescence quenching reagent to incubate for 5 min, followed by washing in water for 10 min, then cover slip with anti-fade mounting medium.
Recombinant Proteins Expression and Purification
Recombinant full-length mouse DGCR8, ΔRhed-mDGCR8, and ΔCTT-mDGCR8 were expressed and purified from Escherichia coli BL21 Rosetta (DE3) cells. Bacteria containing the pETDuet-1-DGCR8 construct were cultured in LB media containing ampicillin (50 μg/ml) and chloramphenicol (25 μg/ml). In addition, 1 mM Isopropyl β-D-Thiogalactoside (IPTG) was added to induce DGCR8 expression and the bacteria were cultured at 16°C for 16 h. Subsequently, the bacteria were collected by centrifugation at 6000 rpm for 10 min and resuspended in lysis buffer [20 mM Tris-HCl pH 8.0, 500 mM NaCl, 10 mM imidazole, 1 μg/ml Benzonase Nuclease, 2 μg/ml RNaseA/T1 mix, 1 mM Dithiothreitol (DTT) and 1 mM phenylmethylsulfonyl fluoride (PMSF)]. Bacteria were lysed using a high-pressure homogenizer. Following centrifugation at 16,000 rpm at 4°C for 1 h, the supernatant was transferred to the Ni-IDA Agarose beads (Senhui Microsphere Technology), and rotated at 4°C for 1 h. Next, the mixture was washed with 10 volumes of wash buffer (20 mM Tris-HCl pH 8.0, 500 mM NaCl, 1mM DTT) containing 40 mM imidazole, and washed again with wash buffer containing 80 mM imidazole. Proteins were then eluted with elution buffer (20 mM Tris-HCl pH 8.0, 150 mM NaCl, 250 mM imidazole, 1 mM DTT).
The GST-tagged human DROSHA390−1374 was expressed and purified from Sf21 insect cells using a baculovirus expression system. Briefly, Sf21 cells were transfected with pFastBac-Dual-DROSHA plasmids using X-tremeGENE 9 DNA Transfection Reagent (Roche) according to the manufacturer’s instructions. Then, the cells were cultured at 27°C for four days. Supernatant containing baculoviruses was used to infect large scale of Sf21 cells. Subsequently, the Sf21 cells were cultured for 60 h and collected by centrifugation at 3000 rpm for 10 min. The cell pellet was resuspended in lysis buffer (PBS, 1% Triton X-100, 1 μg/ml Benzonase Nuclease, 2 μg/ml RNaseA/T1 mix, 1 mM DTT, 1 mM PMSF and protease inhibitor cocktail) and rotated at 4°C for 30 min. The lysates were cleared by centrifugation at 18,000 rpm at 4°C for 1 h. The supernatant was transferred to the GST Agarose beads (Senhui Microsphere Technology) and rotated at 4°C for 2 h. This mixture was washed three times with lysis buffer. The GST tag was cleaved and removed by TEV protease in the elution buffer (20 mM Tris pH 8.0, 150 mM NaCl, 1 mM DTT and 2 mM MgCl2), incubated overnight at 4°C.
Both proteins were further purified using anion exchange (Mono-Q) (GE Healthcare) according to the manufacturer’s instructions and eluted with a linear gradient of NaCl (0–1M) in 20 mM Tris-HCl pH 8.0 and 1 mM DTT. Fractions containing target proteins were pooled together and further purified by gel filtration using Superdex 200 column (GE Healthcare), which was equilibrated with 20 mM Tris-HCl pH 8.0, 150 mM NaCl and 1 mM DTT before use. Protein fractions were then collected using Amicon Ultra centrifugal filters (30K MWCO, Millipore) according to the manufacturer’s instructions.
Protein Labeling
The above rDGCR8 protein was labeled with Alexa Fluor 594 by using Alexa Fluor™ 594 NHS Ester (Succinimidyl Ester, ThermoFisher), and the rDROSHA protein was labeled with Alexa Fluor 488 by using Alexa Fluor™ 488 NHS Ester (Succinimidyl Ester, ThermoFisher) according to the manufacturer’s instructions. Briefly, recombinant proteins were incubated with the protein reaction buffer (20 mM HEPES pH 8.3, 150 mM NaCl and 1 mM DTT), and rotated at room temperature for 1 h. Free dye was removed by gel-filtration.
1,6-Hexanediol Treatment in vivo
The transfected cells were cultured for three days before imaging and 1,6-hexanediol treatment. As control, the transfected cells in 1 ml of complete cell culture medium were imaged using Nikon A1RSi+ Confocal microscope as described above. For the 1,6-hexanediol treatment, the medium was replaced with complete cell culture medium containing 10% 1,6-hexanediol (Sigma) and the cells were incubated at 37°C. After 3 min of incubation, the cells were imaged as described above.
In Vitro Phase Separation Assay
In vitro phase separation assay was performed in reaction buffer containing 20 mM Tris pH 8.0, 150 mM NaCl and 1 mM DTT, and the protein concentration was determined using Coomassie Blue staining (TIANGEN) according to the manufacturer’s instructions. Labeled rDGCR8 and rDROSHA proteins were mixed at indicated stoichiometry, and 10% (v/v) PEG-8000 (Sigma) was added to each reaction. The mixtures were added to glass bottom dishes (Cellvis), and imaging was performed using Nikon A1RSi+ Confocal microscope with 60× oil immersion objective.
Labeled rDGCR8 (32 μM) and rDROSHA (8 μM) proteins were mixed to form condensates. An equal volume of buffer was added to the pre-formed condensates. Immediately after mixing the condensates were imaged. After 5 min of incubation, the condensates were imaged again as described above. Then the condensates were diluted by adding buffer twice in the same way.
For 1,6-hexanediol treatment, the condensates were formed as described above, and then 1,6-hexanediol with a final concentration of 10% was added, and the condensates were imaged immediately after mixing. After 10 min of incubation, the condensates were imaged again as described above.
For NaCl treatment, the condensates were formed as described above, then NaCl with a final concentration of 1 M was added and the condensates were imaged immediately after mixing. After 5 min of incubation, the condensates were imaged again as described above.
For high concentration rDROSHA treatment, the condensates were formed as described above, then high concentration rDROSHA (The final concentration is 16 μM) was added and the condensates were imaged immediately after mixing. After 5 min of incubation, the condensates were imaged again as described above.
Fluorescence Recovery after Photobleaching (FRAP) Assay
For the cellular FRAP assay, the cells were transfected with pCMV2-mCherry-DGCR8 and pCMV2-eGFP-DROSHA, and cultured for three days. The assay was performed using Nikon A1RSi+ Confocal microscope with 100× oil immersion objective at 37°C in a live cell imaging chamber. The condensates were photobleached for 3 seconds using 488nm (for eGFP-DROSHA) and 561nm (for mCherry-DGCR8) laser with 100% laser power, and time-lapse images were collected for 5 min.
For the in vitro FRAP assay, the rDGCR8 and rDROSHA proteins were purified and mixed as described above. Images were collected using Nikon A1RSi+ Confocal microscope with 60× oil immersion objective. To make the initial bleaching be 50%~75% of the original signal, the condensates were photobleached for 0.1 seconds using 488 nm (for DROSHA) and 561 nm (for DGCR8) laser with 1% laser power, and time-lapse images were collected for 5 min with 15 seconds intervals. The fluorescence intensities of phase-separated puncta were acquired using Nikon NIS-Elements AR (Advanced Research) software. Fluorescence intensities of regions of interest (ROIs) were normalized to pre-bleached intensities of the ROIs. The images were analyzed using FIJI/ImageJ, the FRAP curves were analyzed using GraphPad Prism 7.
Single cell Injection with FluidFM BOT
The transfected ΔSL1 mESCs were cultured for three days before imaging and single cell injection. For injection, the cells were cultured in Serum/LIF medium with 10 mM HEPES, and imaged using Carl Zeiss LSM 710 NLO & DuoScan System. Single cell injection was performed by robotized FluidFM BOT platform (Cytosurge AG) which connects AFM to a microfluidics system. A rectangular, micro-channeled probe FluidFM nanosyringe (Cytosurge AG) with an aperture of 600 nm milled beside the pyramid apex of the tip and a nominal spring constant of 2.2 N/m was used for injection. The probe was filled with 1 mg/ml RNase A/T1 mix after coating with Sigmacote (Sigma Aldrich) to prevent fouling. The localization of the target cells in the gridded dish and the entire injection process was monitored by an iX83 inverted microscope (Olympus). By using injection workflow of the ARYA software, the probe was approached on top of the target cell with a set point of 140 mV at a velocity of 500 nm/s after choosing the middle of nucleus as the desired point of insertion. An over-pressure pulse of 5 mbar for 2 s was applied to deliver the solution into the cell nuclei when reaching the set point. The probe was then retracted to a height of 20 μm at a velocity of 1 μm/s. Two typical discontinuities in force-distance curves corresponding to indentation jumps of the membrane of the cell and nucleus reveal the injection into cell nuclei. The whole injection process was performed at 37°C with 5% CO2 incubation chamber. After injection, the cell culture dish was moved to the microscope immediately to image the target cell. Then the time-lapse images were collected for 8 min.
In Vitro Transcription and Microprocessor Cleavage Assay
A T7 primer and gene-specific primers were used to amplify pri-miR-26b sequences from pcDNA3-pri-miR-26b plasmid and pri-miR-17 sequences from pcDNA3-pri-miR-17–92 plasmid by PCR. PCR products were gel-purified and used as templates for in vitro transcription using the Riboprobe® Combination Systems (Promega) with 32P-UTP for radioactive labeling. Whole cell lysate was obtained from WT and ΔSL ESCs. Microprocessor was purified from Flag-DROSHA-293T cells, and recombinant proteins were purified as described above. In vitro Microprocessor cleavage assay was performed as reported previously7,13. Briefly, RNA was incubated with cell lysates, Microprocessor or recombinant proteins, and including 6.4 mM MgCl2, 1 μl RNasin (Progema) in 30 μl BC100 solution (20 mM Tris-HCl, pH 7.6, 10% glycerol and 100 mM KCl) at 37°C for 1 h. To separate the cleavage products, 10% or 15% TBE-Urea Polyacrylamide gel was used.
Luciferase Reporter Assay
For luciferase reporter assay, cells were transfected with related plasmid for three days, and washed twice with DPBS. TransDetect Double-Luciferase Reporter Assay Kit (TRANSGEN BIOTECH) was used to measure the Renilla and Firefly activity according to the manufacturer’s instructions.
Western Blot
Whole cell lysates were prepared as described above and the DGCR8, mCherry and DROSHA proteins were detected using the antibodies α-DGCR8 (Proteintech, 1:2000), mCherry (Qualityard, 1:5000) and α-DROSHA (Cell Signaling, 1:2000).
5’ RACE
5’ RACE was performed on 2 μg of total RNA using the SMARTer RACE 5’/3’ Kit (Clontech). 5’-CDS Primer A was used for reversed transcription and then cDNAs were diluted with Tricine-EDTA buffer. Two rounds of PCR were performed and the products were cloned into the pRACE vector. Different clones were selected for Sanger sequencing. Primers used for 5’ RACE are listed in Supplementary Table 8.
Teratoma Assay
WT and ΔSL1 cells were collected, and approximately 5 × 106 cells were injected subcutaneously into two sides of immunodeficient NPG mice. The teratomas were dissected when the size reached 1.8 cm3. Then the teratomas were rinsed with PBS, weighed and photographed. They were then embedded in paraffin and processed for hematoxylin and eosin (HE) staining and immunofluorescence. The paraffin sections for HE staining and immunofluorescence were scanned using Pannoramic DESK. Data analysis was performed using Caseviewer (3D HISTECH) software.
To compare the relative area of differentiated tissues between WT and ΔSL1 teratomas, each differentiated tissue (ectoderm tissues: neural tube structure composed of pseudostratified columnar epithelium and skin structure formed by stratified squamous epithelium; endoderm tissues: glandular structure formed by ciliated columnar epithelium or cubic epithelium) area relative to the total area was calculated based on the HE stainings. The number of animals, teratomas and sections for analysis is 4, 8 (WT:4, ΔSL1:4) and 16 (WT:8, ΔSL1:8), respectively.
The GATA4 (+) cells were counted according to the positive signal (co-localize with cells indicated by DAPI staining) of immunofluorescence in Extended Data Fig. 7b, and the following formula was used: relative number of GATA4 (+) cells= [GATA4 (+) cell numbers/total area]*100. For the comparison of the relative tissue area with GATA4 (+) expressing cells between WT and ΔSL1 teratomas, the positive signal area relative to the total area was determined according to immunofluorescence of GATA4 proteins. For the area statistics, the number of animals, teratomas and sections analyzed is 2, 4 and 7, respectively.
Total RNA Extraction and q. RT-PCR
TRIzol reagent (Invitrogen) was used to isolate total RNA according to the manufacturer’s instructions. For mRNA qRT-PCR, 4 μg of total RNA was treated with DNase (Promega) overnight at 37°C to remove genomic DNA. Superscript III Reverse Transcriptase (Invitrogen) and random primers were used to synthesize cDNA. SYBR Green Master Mix (ThermoFisher) was used to quantify the cDNA. For miRNA qRT-PCR, 1 μg of total RNA and the miScript II RT Kit (QIAGEN) were used to synthesize cDNA. Subsequently, miScript SYBR Green PCR Kit (QIAGEN) was used to quantify the cDNA. Primers used for q.RT-PCR are listed in Supplementary Table 8
Small RNA Libraries Construction and Bioinformatics Analysis
For neural differentiation, small RNA libraries preparation was performed as reported previously14. Briefly, 20 μg total RNA extracted from mESCs (Day 0) and neuronal progenitor cells (Day 5) were loaded into 15% TBE-Urea gel and 15–50 nt small RNAs were separated and purified, which were then used for library preparation using NEBNext® Small RNA Library Prep Set for Illumina kit according to the manufacturer’s instructions. High-throughput sequencing was then performed by NextSeq 500.
For bioinformatics analysis, after getting the high-throughput sequencing data, we used Perl script (remove.adaptor.pl) to realize three operations. Perl scripts were available from https://github.com/lyuxuehui/ATI-of-DGCR8. Firstly, if a raw sequence contains the adaptor sequence, the sequence from the adaptor sequence to the 3’ end was removed. The 5’ adaptor was removed after sequencing so that we only needed to deal with 3’ adaptor in this step. Secondly, after removing the adaptor, the sequence of 20–25 nt would be reserved, which covered the main length of mature miRNA. Thirdly, for there were many duplicates in our small RNA-seq data, we used PERL script to merge them. In the output FASTA file, the first line started with a “>” symbol, then the sequence and a “_” symbol, and the counts of that sequence.
Bowtie software32 (v1.0.0) was used to align the output FASTA file to mature miRNA sequences without any mismatches permitted. Align reads using bowtie with options: -v 0, other parameters were set to defaulted value. The reference sequences of mature miRNA were obtained from miRBase database33 (Release 22.1) and we lengthened the mature miRNA sequences by 3 nt both in the 3’ end and 5’ end according to the annotations of hairpin miRNA sequences.
After mapping to the mature miRNA reference, we used another Perl script (boat reads.pl) to extract the information of counts from the SAM file. Total reads numbers of 20–25 nt small RNAs were used for normalization (RPM: reads per million).
For different cell lines, we used spike-in method to compare the expression level of miRNA among these samples. The same number of cells for different cell lines were used to extract RNA, and the same amount of S2 cell RNA (equal to 10% of the RNA of the sample with the highest RNA) was added to each sample. Small RNA libraries preparation were performed as above. High-throughput sequencing was performed by Illumina HiSeq X Ten sequencing system.
For the bioinformatics analysis, most steps were performed as above. Adaptor sequences were removed and sequences were aligned to human/mouse and fly mature miRNA reference sequences using Bowtie software as mentioned before. Identical mature miRNAs in human/mouse and fly, such as miR-7–5p, miR-124–3p, miR-125–5p and miR-219–5p, were excluded for fur analysis, as these identical miRNAs made it difficult to determine the origin. In order to normalize the spike-in small RNA-seq data, miRNA counts were multiplied by a coefficient to make the expression level of total fly-specific miRNA equal in every cell line. Total miRNA expressions of the cell line with the highest total miRNA counts were set to 1,000,000 for normalization (RPM: reads per million). After normalization, miRNA expressed higher than 10 RPM in at least one cell line was used for further analysis. The following formula was used to calculate expression changes in WT mESCs and ΔSL1 mESCs: FC (fold change) = (Average rpmΔSL1+2) / (Average rpmWT+2).
PolyA(+) RNA-seq and Bioinformatics Analysis
For sample preparation, 3 μg total RNA and TruSeq Stranded mRNA Sample Prep Kits (Illumina) were used according to the manufacturer’s instructions. Library samples of neural and EB differentiation were subjected to Illumina HiSeq X Ten sequencing system. Other library samples were subjected to Illumina NextSeq 500 sequencing system.
For RNA-seq data analysis, the sequencing quality was evaluated using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and the adapters were removed by Trimmomatic34 (http://www.usadellab.org/cms/?page=trimmomatic). The data were aligned to mouse cDNA database (UCSC, NCBI37) using HiSat2 software35 (https://daehwankimlab.github.io/hisat2/). The read number of individual genes was counted using featureCounts software36 (http://subread.sourceforge.net/), and normalized to total reads of each library (RPM: reads per million). The following formula was used to calculate expressional changes: FC (fold change) = (RPMsample-a+2)/(RPMsample-b+2). The heatmaps were generated using R packages “pheatmap”. For box plot analysis, the following method was used: (gene expression in a sample) / (sum of gene expression in all sample). For normalizing the expression levels of the pluripotent genes, the following formula was used: (x-min(x))/(max(x)-min(x)). The Toppgene online analysis37 (https://toppgene.cchmc.org/) was used for Gene Ontology term enrichment analysis, and terms that had a P-value < 0.05 was defined as significantly enriched. The GSEA software38 (https://www.gseamsigdb.org/gsea/index.jsp) was used to perform Gene Set Enrichment Analysis (permutation = 1000, permutation type = gene_set, other parameters were default).
Parameters of the above software were as below:
Trimmomatic SE -threads 15 $file1 $file2 ILLUMINACLIP: TruSeq3-SE.fa:2:30:10:1:true LEADING:3 TRAILING:3 SLIDINGWINDOW:4:15 MINLEN:36.
Hisat2 -x $Reference.fa -p 10 -U $file1 -S $Sample.sam
FeatureCounts -T 10 -t exon -g gene_id -a $Annatation.gtf -o $sample.count $sample.sorted.bam
The previously published PolyA(+) RNA-seq data of mESC differentiation (GEO, GSE112334)14, and the previously published polyA(+) RNA-seq of EpiSC and cells of three germ layers (ArrayExpress, E-MTAB-4904)27, were aligned to mouse cDNA database (UCSC, NCBI37) using the Hisat2 software. The analysis was performed as described above. Integrative Genomics Viewer (IGV) software was used to visualize the genome.
Promoter Prediction
In human, histone modification (H3K4me1, H3K4me3, and H3K27Ac) levels in seven cell lines from ENCODE and the transcription factor binding sites from ENCODE 3 were compared. The predicted transcription start sites were obtained from SwitchGear Genomics (https://switchgeargenomics.com/). In mouse, the Alternative Promoter was shown in Fig. 1a. All data were analyzed in UCSC Genome Browser (Human GRch37/hg19, Mouse NCBI37/mm9).
Formula for ATI appearance
The ratio of average coverage depth between upstream and downstream of the first stem-loop of DGCR8 for each RNA-seq sample was called the “ATI ratio”. As a control, the ratio between the 6th and 7th exon of DGCR8 was called the “exon ratio”.
GTEx Data Analysis
For analysis in normal human tissues, we obtained 7,338 aligned GTEx RNA-seq samples (in bam-format) from RefLnc39. The upstream and downstream read coverage depth of the first stem-loop of DGCR8 was extracted by samtools (samtools depth -a -r). Next, the “ATI ratio” and the “exon ratio” were calculated.
Quantification and Statistical Analysis
Statistical analysis for gene expression was performed using Graphpad Prism. Significance was calculated with Student’s t-test and represented as mean +/− SD (The data analysis of Teratoma was used mean +/− SEM). P-values <0.05 were considered statistically significant.
In Fig. 2e and Extended Data Fig. 4c, differentially expressed genes or miRNAs were defined by P-values ≤ 0.05 and Fold change=(RPMsample-a+2)/(RPMsample-b+2) ≥2 (or ≤0.5).
Two-tailed Student’s t-test was used to calculate the P-values using the rpm of each gene as input data.
For q.RT-PCR or reporter assay (Fig. 2c), Two-tailed Student’s t-test was used to calculate the P-values, and data are represented as mean +/− SD.
For Fig. 2d and Fig. 4d, the rpm of all miRNA in heatmap of each sample were used to calculate the P-values by two-tailed Student’s t-test to dertermine the difference of the global miRNA dosage.
For teratoma assay including the weight (Fig. 3g), relative area of tissues (Fig. 3h), relative number of GATA4 (+) cells (Extended Data Fig. 7c), two-tailed Student’s t-test was used to calculate the P-values and data are represented as mean +/− SEM.
For GESA assay, the P-values and FDR were calculated by the GSEA software (https://www.gseamsigdb.org/gsea/index.jsp).
Extended Data
Extended Data Fig. 1 |. ATI of DGCR8 links dynamic Microprocessor autoregulation to altered DGCR8: DROSHA stoichiometry.
a, Ratio of splicing events covering different exons based on RNA-seq data during mESC-to-EB differentiation over a 13-day time course. See also Fig. 1a. b, Agarose gel analysis of 5’ RACE products (left) and summary of Sanger sequencing of 5’ RACE clones (right). PCR products outlined by red arrows were purified and further processed for cloning and Sanger sequencing. The numbers indicate the proportion of all sequenced clones that map to a particular nucleotide. Stem-loop-1 (SL1) sequence in 5’ UTR is highlighted in red font. See also Fig. 1b. c, Stem-loop structures (SL1 and SL2) in the 5’ UTR and CDS region of DGCR8 mRNA. Green arrowheads indicate CRISPR-Cas9 design for SL1 deletion. d, PCR analysis of genomic DNA for SL1 knockout (ΔSL1) in mESCs. e, q.RT-PCR analysis of DGCR8 and DROSHA mRNA expression in WT and ΔSL1 mESCs. Data were normalized to GAPDH, and error bars indicate SD (n=3). f, Western blot of DGCR8 and DROSHA proteins in WT and ΔSL1 mESCs transfected with corresponding siRNAs.
Extended Data Fig. 2 |. SL1 depletion drives irreversible Microprocessor aggregation in mouse ESCs.
a, Immunofluorescence (IF) followed by confocal imaging of DGCR8 and DROSHA proteins in WT and ΔSL1 mESCs. Fluorescence signals of DGCR8 and DROSHA are shown in red and green, respectively, and DAPI stain is shown in blue. The merged signals are also shown. b, Western blot of DGCR8 and DROSHA proteins in WT and reporter mESCs expressing endogenous mCherry-DGCR8 fusion protein. c, Representative time-lapse images of two proximate assemblies of Microprocessor in living ΔSL1 mESCs transfected with plasmids expressing tagged mCherry-DGCR8 and eGFP-DROHSA. d, Representative images of fluorescence recovery after photobleaching (FRAP) analysis of Microprocessor aggregates in living ΔSL1 mESCs transfected with plasmids expressing tagged mCherry-DGCR8 and eGFP-DROHSA. Targeted region is highlighted in a white box, and DGCR8 (red), DROSHA (green) and merged (yellow) signals are shown. See also Fig. 1g. e, Images of Microprocessor aggregates in ΔSL1 mESCs transfected with plasmids expressing tagged mCherry-DGCR8 and eGFP-DROSHA before and after microinjection with RNase.
Extended Data Fig. 3 |. Imbalanced DGCR8: DROSHA stoichiometry drives irreversible Microprocessor aggregation in vitro.
a, Top, prediction of disordered regions in DGCR8 protein by “PONDR” (http://pondr.com/). Bottom, schematic diagram showing the domains of DGCR8. b, Coomassie blue staining of purified rDGCR8 and rDROSHA proteins at different concentrations, as well as two mutant versions of rDGCR8 proteins with deletion of ΔCTT and ΔRhed domains respectively. c, Representative images of phase separation of rDGCR8, rDGCR8-ΔCTT, rDGCR8-ΔRhed and rDROSHA at different concentrations in physiological buffer. d, Representative images of the aggregates of labeled rDGCR8(30uM) before and after treatment with 10% 1,6-hexanediol for 5 min. e, Representative images of FRAP analysis of rDGCR8 puncta. Targeted region is highlighted in a white box. Normalized fluorescence intensity of rDGCR8 in FRAP analysis is represented as mean +/− SD (N=6). f, g, Representative confocal images of pre-formed Microprocessor aggregates (32μM rDGCR8: 8μM DROSHA) under the treatments of dilution and high salt (1M NaCl). rDGCR8 (red), rDROSHA (green) and merged (yellow) signals are shown. h, Representative time-lapse images of two proximate Microprocessor aggregates for the times indicated. rDGCR8 (red), rDROSHA (green) and merged (yellow) signals are shown. i, FRAP analysis of Microprocessor aggregates in vitro. Targeted droplet region is highlighted in a white box, and rDGCR8 (red), rDROSHA (green) and merged (yellow) signals are shown. See also Fig. 1k.
Extended Data Fig. 4 |. Microprocessor aggregation reduces the efficiency of pri-miRNA processing and global miRNA dosage which leads to the de-repression of lipid metabolic genes.
a, Microprocessor in vitro cleavage assay of mouse pri-miR-125b using whole cell lysate from WT and ΔSL1 mESCs. Microprocessor purified by immunoprecipitation from 293T-Flag-DROSHA cells was used as a control. b, Quantification of pri-miRNA cleavage activity calculated based on the density of pre-miRNA bonds in the assays shown in Fig. 2a and Extended Data Fig. 4a, respectively. c, Scatter plot of global miRNA expression based small RNA-seq data in WT and ΔSL1 mESCs. The small RNA-seq data were normalized based on spike-in RNAs. Differentially expressed miRNAs are represented by colored circles, and number of up- and downregulated miRNAs is shown. FC: fold change, P: P-Value from ****p<0.0001, Two-tailed Student’s t-test. d, Venn diagram of mRNAs with expression changes in ΔSL1 and DGCR8−/− cells compared with WT mESCs. Number of genes in each group is shown. FC: fold change. P: P-Value from Two-tailed Student’s t-test. e, GSEA analyses of lipid metabolic gene sets by comparing ΔSL1 and DGCR8−/− cells with WT mESCs. NES, normalized enrichment score. P-value, indicated the nominal p-value. FDR, false discovery rate. f, Network of miRNAs and lipid metabolic genes. g, miRNA target sites on PDK4, LCLAT1 and GPCPD1 mRNAs, and luciferase miRNA target reporter assay in WT, ΔSL1 and DGCR8−/− mESCs. Data are represented as mean +/− SD (n=3). **p<0.01, Student’s t-test. Mutations introduced into the miRNA target sites on PDK4 and LCLAT1, and the mutation sequences are shown in red font.
Extended Data Fig. 5 |. Imbalanced DGCR8: DROSHA stoichiometry during EB differentiation drives Microprocessor aggregation.
a, PCA of the WT and ΔSL1 cells by all expressed mRNAs during mESC-to-EpiLC differentiation. b, c, The relative expression and calculated ratio of DGCR8 and DROSHA mRNA based on RNA-seq data during EB differentiation over a 13-day time course. d, Western blot of DGCR8 and DROSHA at different time points of WT mESC-to-EB differentiation. Density of the DROSHA and DGCR8 bands were normalized to the ACTINB band to calculate the relative protein ratios. e, Western blot of DGCR8 and DROSHA in differentiated EB cells (Day 8) before and after treatment with a protease inhibitor MG132(10uM) for 2 hours. Density of the DROSHA and DGCR8 bands were normalized to the ACTINB band to calculate the relative protein ratios. f, Representative images of Microprocessor aggregates in differentiated EB cells (Day 8) derived from dual-reporter ESCs endogenously expressing both mCherry-DGCR8 and eGFP-DROSHA fusion proteins.
Extended Data Fig. 6 |. ATI-mediated miRNA dosage control determines germ layer specification during ESC differentiation in vitro.
a, Ratios of splicing events covering different exons based on RNA-seq data from various germ layer cells. Ep: EpiSC, Ec: Ectoderm, Me: Mesoderm, En: Endoderm. b, The relative expression of pluripotent genes in WT and ΔSL1 cells during neural and EB differentiation. NP: neural progenitor. c, Heatmap of the relative expression of up- or downregulated genes in ΔSL1 cells compared with WT cells during EB differentiation. The enrichment of Gene Ontology (GO) terms and the number of genes in each group are shown. d, Heatmap of the relative expression of marker genes for three germ layers during neural differentiation. Representative markers are listed on the right e, Scatter plot of global miRNA expression based small RNA-seq data in mESCs and neuronal progenitor cells (Day 5).
Extended Data Fig. 7 |. miRNA dosage control impacts germ layer specification during teratoma formation in mouse.
a, Histology of teratomas stained with hematoxylin and eosin (HE). Left: low power view of teratoma, differentiated areas are shown with different colors (ectoderm: marked with black; endoderm: marked with red). Right: representative images of ectoderm (neural tube) and endoderm (glandular structure) tissues. b, Immunofluorescence (IF) of GATA4 proteins (endoderm marker) in sections of teratomas derived from WT and ΔSL1 mESCs. Fluorescence signals of GATA4 are shown in pink, and DAPI stain is shown in blue. c, Comparison of the relative area of GATA4 (+) cells in teratomas derived from WT and ΔSL1 mESCs according to the immunofluorescence of 34 (WT:17, ΔSL1:17) sections of 18 (WT:9, ΔSL1:9) teratomas from 9 mice. d, The bar plot showing the GATA4 (+) cell numbers relative to the total area in teratomas derived from WT and ΔSL1 mESCs according to the staining in b.
Extended Data Fig. 8 |. Inhibition of lipid metabolism impacts germ layer specification during EB differentiation in vitro.
a, Heatmap of the relative expression of lipid metabolic genes, which were controlled by miRNAs dosage in mESCs, during mESC-to-EB differentiation over a 13-day time course. b, Protocol of mESC-to-EB differentiation under the treatment with lipid metabolic inhibitor GW9662. c, Heatmap of the relative expression of lipid metabolic genes suppressed by GW9662 in differentiated EB cells. Gene number and representative genes are listed. The lipid metabolic gene are from the GO terms “lipid metabolic process”. d, q.RT-PCR analysis of the relative expression of germ layer marker genes during EB differentiation under the treatment with GW9662. Data are normalized to GAPDH and represented as mean +/− SD (n=2). e, Heatmap of the relative expression of germ layer marker genes during mESC-to-EB differentiation under the treatment.
Extended Data Fig. 9 |. ATI-mediated miRNA dosage control mechisiam is conserved in human cells and tissues.
a, Ratios of splicing events covering different exons based on RNA-seq data in different human cell lines. b, CHIP-seq signals for various TFs at DGCR8 5’ UTR are shown in GM12878, MCF-7 and HepG2 cells, and representative TFs are listed in red. The number of TFs binding to the two distinct promoters is shown by Histograms. c, 5’RACE experiment for DGCR8 mRNA in H1299 and K562 cells. Agarose gel analysis of 5’ RACE products (left) and summary of Sanger sequencing of 5’ RACE clones (right). PCR products outlined by red arrows were purified and further processed for cloning and Sanger sequencing. The numbers indicate the proportion of all sequenced clones that map to a particular nucleotide. Stem-loop-1 (SL1) sequence in 5’ UTR is highlighted in red font. See also Fig. 4b. d, Fluorescence recovery after photobleaching (FRAP) analysis of Microprocessor aggregates in HepG2 cells transfected with plasmids expressing tagged mCherry-DGCR8 and eGFP-DROSHA. Targeted region is highlighted in a white box. DGCR8 (red), DROSHA (green) and merged (yellow) signals are shown. Normalized fluorescence intensity of DGCR8 and DROSHA in FRAP analysis. Data are presented as mean +/− SD (N=6). e, Representative images of Microprocessor aggregates in HepG2 cells transfected with plasmids expressing tagged mCherry-DGCR8 and eGFP-DROSHA before and after treatment with 10% 1,6-hexanediol for 3 min. f, q.RT-PCR analysis of DGCR8 and DROSHA mRNA expression in WT and ΔSL1 H1299 cells. Data are normalized to GAPDH and represented as mean +/− SD (n=2). g, Formula used to calculate the ATI ratio (representing ATI appearance) and exon ratio based on sequencing coverage of RNA-seq data.
Supplementary Material
Acknowledgements:
We thank Drs. Narry Kim (Seoul National University), Pilong Li and Yijun Qi (Tsinghua University), and Xiangdong Fu (University of California, San Diego) for helpful discussions. We thank Biopolymer Facility at Harvard Medical School for RNA-seq and small RNA-seq Illumina high-throughput sequencing. We thank the Core Facilities of the School of Life Sciences at Peking University, particularly Drs. Siying Qin, Chunyan Shan, Liqin Fu and Shiqiang Huang for technical helps with confocal imaging and radiolabeling assay. We thank the flow cytometry Core at National Center for Protein Sciences at Peking University, particularly Hongxia Lu and Huan Yang, for technical help. Some work on protein purification was performed in Ning Gao’s laboratory (Peking University), we thank them for assistance. We thank Junyu Xiao’s laboratory (Peking University) for providing the pFastBac-Dual vector and their assistance. We thank Dr. Feng Guo (University of California, Los Angeles) for providing the pFastBac-HTb-His6-DROSHA390−1374 vector.
Funding:
This work was supported by grants to P. D. from the Natural Science Foundation of China (32050214 and 32090012) and the National Key Research and Development Program of China (2019YFA0110000), and a grant to R.I.G. from the US National Institute of General Medical Sciences (NIGMS) (R01GM086386).
Footnotes
Data and materials availability: The RNA-seq and small RNA-seq data that support the finding of this study have been deposited in GEO with accession number GSE165017. Published PolyA(+) RNA-seq data of mESC differentiation is from Gene Expression Omnibus (GEO) database under accession number GSE11233414. The published polyA(+) RNA-seq of EpiSC and cells of the three germ layers reported in this paper is available on ArrayExpress with accession E-MTAB-490427.
References
- 1.Lin S & Gregory RI MicroRNA biogenesis pathways in cancer. Nat Rev Cancer 15, 321–333, doi: 10.1038/nrc3932 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartel DP Metazoan MicroRNAs. Cell 173, 20–51, doi: 10.1016/j.cell.2018.03.006 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu J et al. MicroRNA expression profiles classify human cancers. Nature 435, 834–838, doi: 10.1038/nature03702 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Landgraf P et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 129, 1401–1414, doi: 10.1016/j.cell.2007.04.040 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee Y et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 425, 415–419, doi: 10.1038/nature01957 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Denli AM, Tops BB, Plasterk RH, Ketting RF & Hannon GJ Processing of primary microRNAs by the Microprocessor complex. Nature 432, 231–235, doi: 10.1038/nature03049 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Gregory RI et al. The Microprocessor complex mediates the genesis of microRNAs. Nature 432, 235–240, doi: 10.1038/nature03120 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Kwon SC et al. Structure of Human DROSHA. Cell 164, 81–90, doi: 10.1016/j.cell.2015.12.019 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Partin AC et al. Cryo-EM Structures of Human Drosha and DGCR8 in Complex with Primary MicroRNA. Mol Cell 78, 411–422 e414, doi: 10.1016/j.molcel.2020.02.016 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ha M & Kim VN Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15, 509–524, doi: 10.1038/nrm3838 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Han J et al. Posttranscriptional crossregulation between Drosha and DGCR8. Cell 136, 75–84, doi: 10.1016/j.cell.2008.10.053 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Triboulet R, Chang HM, Lapierre RJ & Gregory RI Post-transcriptional control of DGCR8 expression by the Microprocessor. RNA 15, 1005–1011, doi: 10.1261/rna.1591709 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Du P, Wang L, Sliz P & Gregory RI A Biogenesis Step Upstream of Microprocessor Controls miR-17 approximately 92 Expression. Cell 162, 885–899, doi: 10.1016/j.cell.2015.07.008 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du P et al. An Intermediate Pluripotent State Controlled by MicroRNAs Is Required for the Naive-to-Primed Stem Cell Transition. Cell Stem Cell 22, 851–864 e855, doi: 10.1016/j.stem.2018.04.021 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernstein E et al. Dicer is essential for mouse development. Nat Genet 35, 215–217, doi: 10.1038/ng1253 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Medvid R, Melton C, Jaenisch R & Blelloch R DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet 39, 380–385, doi: 10.1038/ng1969 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang B et al. A dosage-dependent pleiotropic role of Dicer in prostate cancer growth and metastasis. Oncogene 33, 3099–3108, doi: 10.1038/onc.2013.281 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lambo S et al. The molecular landscape of ETMR at diagnosis and relapse. Nature 576, 274–280, doi: 10.1038/s41586-019-1815-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alberti S, Gladfelter A & Mittag T Considerations and Challenges in Studying Liquid-Liquid Phase Separation and Biomolecular Condensates. Cell 176, 419–434, doi: 10.1016/j.cell.2018.12.035 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maharana S et al. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 360, 918–921, doi: 10.1126/science.aar7366 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Case LB, Zhang X, Ditlev JA & Rosen MK Stoichiometry controls activity of phase-separated clusters of actin signaling proteins. Science 363, 1093–1097, doi: 10.1126/science.aau6313 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patel A et al. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell 162, 1066–1077, doi: 10.1016/j.cell.2015.07.047 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Lin Y, Protter DS, Rosen MK & Parker R Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol Cell 60, 208–219, doi: 10.1016/j.molcel.2015.08.018 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greve TS, Judson RL & Blelloch R microRNA control of mouse and human pluripotent stem cell behavior. Annu Rev Cell Dev Biol 29, 213–239, doi: 10.1146/annurev-cellbio-101512-122343 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gu KL et al. Pluripotency-associated miR-290/302 family of microRNAs promote the dismantling of naive pluripotency. Cell Res 26, 350–366, doi: 10.1038/cr.2016.2 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hayashi K, Ohta H, Kurimoto K, Aramaki S & Saitou M Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 146, 519–532, doi: 10.1016/j.cell.2011.06.052 (2011). [DOI] [PubMed] [Google Scholar]
- 27.Sladitschek HL & Neveu PA A gene regulatory network controls the balance between mesendoderm and ectoderm at pluripotency exit. Mol Syst Biol 15, e9043, doi: 10.15252/msb.20199043 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fang Y et al. Histone crotonylation promotes mesoendodermal commitment of human embryonic stem cells. Cell Stem Cell, doi: 10.1016/j.stem.2020.12.009 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie D et al. Phase separation of SERRATE drives dicing body assembly and promotes miRNA processing in Arabidopsis. Nat Cell Biol 23, 32–39, doi: 10.1038/s41556-020-00606-5 (2021). [DOI] [PubMed] [Google Scholar]
- 30.Yao X et al. CRISPR/Cas9-mediated Targeted Integration In Vivo Using a Homology-mediated End Joining-based Strategy. Journal of visualized experiments: JoVE, doi: 10.3791/56844 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mori M et al. Hippo signaling regulates microprocessor and links cell-density-dependent miRNA biogenesis to cancer. Cell 156, 893–906, doi: 10.1016/j.cell.2013.12.043 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Langmead B, Trapnell C, Pop M & Salzberg SL Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology 10, R25, doi: 10.1186/gb-2009-10-3-r25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Griffiths-Jones S, Saini HK, van Dongen S & Enright AJ miRBase: tools for microRNA genomics. Nucleic acids research 36, D154–158, doi: 10.1093/nar/gkm952 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolger AM, Lohse M & Usadel B Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England) 30, 2114–2120, doi: 10.1093/bioinformatics/btu170 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim D, Paggi JM, Park C, Bennett C & Salzberg SL Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature biotechnology 37, 907–915, doi: 10.1038/s41587-019-0201-4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liao Y, Smyth GK & Shi W featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics (Oxford, England) 30, 923–930, doi: 10.1093/bioinformatics/btt656 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Chen J, Bardes EE, Aronow BJ & Jegga AG ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic acids research 37, W305–311, doi: 10.1093/nar/gkp427 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 102, 15545–15550, doi: 10.1073/pnas.0506580102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang S et al. An expanded landscape of human long noncoding RNA. Nucleic acids research 47, 7842–7856, doi: 10.1093/nar/gkz621 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.