ABSTRACT
We present the LGAAP computational pipeline, which was successfully used to assemble six genomes of the parasite subfamily Leishmaniinae to chromosome-scale completeness from a combination of long- and short-read sequencing data. LGAAP is open source, and we suggest that it may easily be ported for assembly of any genome of comparable size (∼35 Mb).
ANNOUNCEMENT
We developed an automated genome assembly and annotation pipeline, successfully applying it to six genomes in the parasite subfamily Leishmaniinae, namely, (i) Leishmania martiniquensis (MHOM/TH/2012/LSCM1, LV760), (ii) Leishmania orientalis (MHOM/TH/2014/LSCM4, LV768), (iii) Leishmania enriettii (MCAV/BR/2001/CUR178, LV763), (iv) Leishmania sp. Ghana (MHOM/GH/2012/GH5, LV757), (v) Leishmania sp. Namibia (MPRO/NA/1975/252, LV425), and (vi) Porcisia hertigi (MCOE/PA/1965/C119, LV43). This paper closes the “protocol gap” (1) for this project by making all methods fully available.
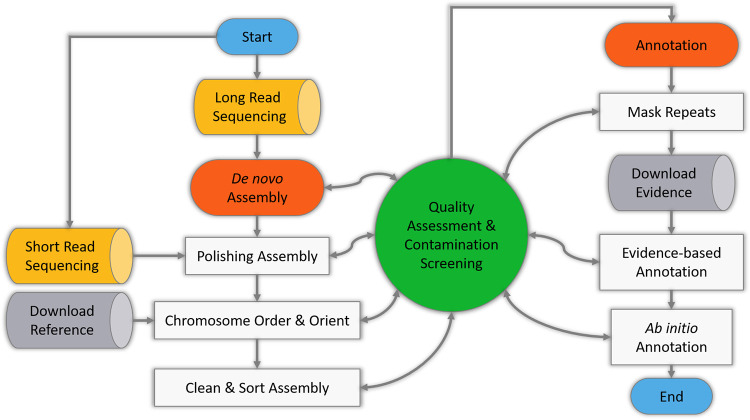
The pipeline was written and executed using the Snakemake (2) workflow management system and consists of a total of 314 computational steps, divided into 21 sequential processes in two main phases (Fig. 1). Genomic DNA was extracted from a previously developed culture system for L. orientalis axenic amastigotes (3) and sequenced using two standard technologies, i.e., short read (Illumina) and long read (Oxford Nanopore Technologies [ONT]).
FIG 1.
Graphical representation of the LGAAP protocol.
The first (assembly) phase of the pipeline comprises eight sequential processes, i.e., (i) long-read assembly using Flye (version 2.8.2) (4), (ii) mapping of short reads onto assemblies using Minimap2 (version 2.17) (5), (iii) creation of consensus sequences using SAMtools (version 1.11) (6), (iv) polishing of assemblies using Pilon (version 1.23) (7), (v) revision of consensus sequences using SAMtools, (vi) ordering and orientation of the chromosomes and breakage of any chimeric sequences using RaGOO (version 1.1) (8), (vii) sorting and removal of any duplicated scaffolds or contigs using Funannotate (version 1.5.3) (9), and (viii) generation of a quality report using QUAST (version 5.0.2) (10).
The second (annotation) phase of the pipeline comprises 13 sequential processes, i.e., (i) scanning of assemblies for vector contamination using BLAST+ (version 2.10.1) (11) against UniVec (12), (ii) masking of contaminants using BEDTools (version 2.30) (13), (iii) quality statistics preannotation using AGAT (version 0.6.0) (14), (iv) detection of repeats using RepeatModeler (15) running from Dfam TE Tools Container (version 1.3.1) (16), (v) classification of transposable elements using TEclass (16) running from a docker container (version 2.1.3b) (17), (vi) masking of identified complex repeats using RepeatMasker (version 4.1.2-p1) (18), (vii) downloading of protein and transcript evidence from TriTrypDB (release 47) (19), (viii) evidence-based annotation using MAKER2 (20) running from a docker container (version 2.31.10) (21), (ix) quality checking of annotation using GenomeTools (version 1.2.1) (22) and GAAS (version 1.2.0) (23), (x) ab initio annotation using AUGUSTUS (version 3.3.2) (24) within MAKER2, (xi) repeating of the ninth step, (xii) annotation assignments using BLAST+ against UniProt (25) and InterProScan (version 5.22-61.0) (26), and (xiii) finalization of the longest isoforms of each predicted protein using AGAT.
The final product of the analysis pipeline is five files per genome, i.e., the chromosome-scale assembly, proteins, and transcripts in FASTA format and two general feature format (GFF) files, one containing the coordinates of each feature and one with the longest isoforms. Testing on genomes longer than 35 Mb is a future optimization priority. Comparison of the performance of LGAAP with all 50 Leishmania genome assemblies in GenBank is shown in Table 1.
TABLE 1.
Assembly metrics for Leishmania genome assemblies deposited in GenBanka
| Organism | NCBI assembly no. | Strain | Sequencing technology(ies) | Assembly method | No. of scaffolds | Total length (bp) | N50 (bp) |
|---|---|---|---|---|---|---|---|
| L. aethiopica | GCA_003992445 | 209-622 | PacBio RS II | CANU | 118 | 33,648,436 | 763,733 |
| L. aethiopica | GCA_000444285 | L147 | Illumina | Allpaths-LG | 160 | 31,630,816 | 1,001,864 |
| L. amazonensis | GCA_003992505 | 210-660 | PacBio RS II | CANU | 92 | 33,504,997 | 850,106 |
| L. amazonensis | GCA_000438535 | NA | Roche 454, Illumina | Newbler, Velvet, Zorro | 2,627 | 29,029,348 | 22,901 |
| L. amazonensis | GCA_005317125 | UA301 | Illumina | SMALT | 34 | 32,156,470 | NA |
| L. arabica | GCA_000410695 | LEM1108 | Illumina | AllPaths-LG | 168 | 31,269,090 | 1,057,807 |
| L. braziliensis | GCA_003304975 | IOC-L 3564 | IonTorrent | SPAdes | 1,029 | 38,003,648 | 758,103 |
| L. braziliensis | GCA_000340355 | MHOM/BR/75/M2903 | Roche 454 | Newbler | 744 | 35,210,150 | 1,030,512 |
| L. braziliensis | GCA_000002845 | MHOM/BR/75/M2904 | Sanger | NA | 138 | 32,068,771 | 992,961 |
| L. braziliensis | GCA_900537975 | MHOM/BR/75/M2904 | PacBio, Illumina | NA | 35 | 32,301,632 | NA |
| L. chagasi | GCA_014466975 | MCER/BR/1981/M6445/Salvaterra | Illumina | SOAPdenovo | 36 | 31,924,566 | 1,043,794 |
| L. chagasi | GCA_014466935 | MHOM/HD/2017/M32502/Amapala | Illumina | SOAPdenovo | 36 | 31,924,975 | 1,043,719 |
| L. donovani | GCA_000470725 | BHU 1220 | Illumina | Bowtie | 36 | 32,414,853 | 1,024,085 |
| L. donovani | GCA_000227135 | BPK282A1 | Roche 454, Illumina | NA | 36 | 32,444,968 | 1,024,085 |
| L. donovani | GCA_003730175 | FDAARGOS_360 | PacBio, Illumina | CANU | 71 | 34,011,430 | 828,097 |
| L. donovani | GCA_003730215 | FDAARGOS_361 | PacBio, Illumina | CANU | 56 | 33,453,722 | 1,033,854 |
| L. donovani | GCA_900635355 | HU3 | Illumina | NA | 36 | 33,035,865 | NA |
| L. donovani | GCA_000283395 | Ld 2001 | SOLiDb | Velvet | 14,518 | 27,466,456 | 3,370 |
| L. donovani | GCA_000316305 | Ld 39 | SOLiD | Velvet | 16,323 | 23,683,296 | 1,772 |
| L. donovani | GCA_003719575 | LdCL | PacBio, Illumina | HGAP, Celera Assembler, CANU | 36 | 32,959,864 | NA |
| L. donovani | GCA_001989955 | MHOM/IN/1983/AG83 | Illumina | AllPaths, STLab-assembler | 36 | 32,148,377 | 1,015,993 |
| L. donovani | GCA_001989975 | MHOM/IN/1983/AG83 | Illumina | AllPaths | 36 | 32,196,393 | 1,029,368 |
| L. donovani | GCA_002243465 | Pasteur | PacBio | HGAP | 37 | 33,545,875 | 1,079,609 |
| L. enriettii | GCA_000410755 | LEM3045 | Illumina | AllPaths-LG | 495 | 30,761,861 | 868,233 |
| L. enriettii* | GCA_017916305* | MCAV/BR/2001/CUR178, LV763 | ONT, Illumina | LGAAP | 54 | 33,318,864 | 1,075,649 |
| L. gerbilli | GCA_000443025 | LEM452 | Illumina | AllPaths-LG | 492 | 31,398,648 | 379,527 |
| L. guyanensis | GCA_003664525 | 204-365 | PacBio RS II | CANU | 123 | 33,816,023 | 683,170 |
| L. infantum | GCA_003671315 | HUUFS14 | Illumina | ABySS | 2,507 | 32,578,914 | 29,848 |
| L. infantum | GCA_000002875 | JPCM5 | Sanger | NA | 76 | 32,122,061 | 1,043,848 |
| L. infantum | GCA_900500625 | JPCM5 | PacBio, Illumina | NA | 36 | 32,803,248 | NA |
| L. infantum | GCA_003020905 | TR01 | Illumina | Geneious | 36 | 32,009,138 | NA |
| L. lainsoni | GCA_003664395 | 216-34 | PacBio RS II | CANU | 137 | 34,152,029 | 638,860 |
| L. major | GCA_000002725 | Friedlin | Sanger | NA | 36 | 32,855,089 | NA |
| L. major | GCA_000331345 | LV39c5 | Roche 454 | Newbler | 849 | 32,327,517 | 978,401 |
| L. major | GCA_000250755 | SD 75.1 | Roche 454 | Newbler | 36 | 31,242,750 | 1,022,795 |
| L. martiniquensis | GCA_000409445 | LEM2494 | Illumina | AllPaths-LG | 251 | 30,813,970 | 873,628 |
| L. martiniquensis* | GCA_017916325* | MHOM/TH/2012/LSCM1, LV760 | ONT, Illumina | LGAAP | 42 | 32,413,670 | 1,046,741 |
| L. mexicana | GCA_003992435 | 215-49 | PacBio RS II | CANU | 55 | 32,057,209 | 825,953 |
| L. mexicana | GCA_000234665 | MHOM/GT/2001/U1103 | Sanger | NA | 588 | 32,108,741 | 1,044,075 |
| L. orientalis* | GCA_017916335* | MHOM/TH/2014/LSCM4, LV768 | ONT, Illumina | LGAAP | 98 | 34,194,276 | 1,120,138 |
| L. panamensis | GCA_000340495 | MHOM/COL/81/L13 | Illumina | SOAP denovo | 952 | 31,263,945 | 156,905 |
| L. panamensis | GCA_000755165 | MHOM/PA/94/PSC-1 | Roche 454, Illumina | Newbler, PAGIT | 35 | 30,688,794 | 1,043,456 |
| L. peruviana | GCA_001403695 | LEM-1537 | NA | NA | 37 | 33,890,200 | 1,047,715 |
| L. peruviana | GCA_001403675 | PAB-4377 | NA | NA | 37 | 32,907,781 | 1,015,393 |
| Leishmania sp. | GCA_000981925 | AIIMS/LM/SS/PKDL/LD-974 | Illumina | A5 assembly pipeline | 1,100 | 27,848,322 | 61,709 |
| Leishmania sp. Ghana* | GCA_017918215* | MHOM/GH/2012/GH5, LV757 | ONT, Illumina | LGAAP | 116 | 35,953,538 | 1,100,365 |
| Leishmania sp. Namibia* | GCA_017918225* | MPRO/NA/1975/252, LV425 | ONT, Illumina | LGAAP | 67 | 34,118,624 | 1,066,046 |
| L. tarentolae | GCA_009731335 | Parrot Tar II | PacBio RS II | HGAP | 179 | 35,416,496 | 663,019 |
| L. tarentolae | GCA_009770625 | Parrot Tar II | Roche 454 | Newbler | 7,227 | 31,556,583 | 7,432 |
| L. tropica | GCA_011316065 | ATCC 50129 | Illumina | CLC Genomics Workbench | 1,928 | 30,870,161 | 32,161 |
| L. tropica | GCA_014139745 | CDC216-162 | PacBio RS II, Illumina | Flye | 43 | 32,700,668 | 1,070,514 |
| L. tropica | GCA_000410715 | L590 | Illumina | AllPaths-LG | 448 | 32,989,014 | 303,214 |
| L. tropica | GCA_003067545 | MHOM/LB /2017/IK | Illumina | CLC NGS Cell | 9,499 | 32,139,927 | 13,854 |
| L. tropica | GCA_003352575 | MHOM/LB/2015/IK | Illumina | CLC NGS Cell | 17,013 | 32,280,712 | 7,721 |
| L. turanica | GCA_000441995 | LEM423 | Illumina | AllPaths-LG | 336 | 32,320,007 | 397,299 |
| Porcisia hertigi* | GCA_017918235* | MCOE/PA/1965/C119, LV43 | ONT, Illumina | LGAAP | 74 | 34,958,538 | 967,170 |
Asterisks indicate the six genomes assembled using LGAAP. NA, either not applicable to the technology used or not available from the GenBank record.
SOLiD, sequencing by oligonucleotide ligation and detection.
Data availability.
Genomes assembled using this protocol are available in the NCBI Assembly database with the following accession numbers: L. martiniquensis, GCA_017916325.1; L. orientalis, GCA_017916335.1; L. enriettii, GCA_017916305.1; Leishmania sp. Ghana, GCA_017918215.1; Leishmania sp. Namibia, GCA_017918225.1; and Porcisia hertigi, GCA_017918235.1. Raw sequencing data are available with the following NCBI BioProject accession numbers: L. martiniquensis, PRJNA691531; L. orientalis, PRJNA691532; L. enriettii, PRJNA691534; Leishmania sp. Ghana, PRJNA691536; Leishmania sp. Namibia, PRJNA689706; and Porcisia hertigi, PRJNA691541. The workflow is available at GitHub (https://github.com/hatimalmutairi/LGAAP) and Zenodo (https://doi.org/10.5281/zenodo.4663265).
ACKNOWLEDGMENT
This work is funded by a Ph.D. studentship grant to H.A. from the Saudi Arabian Ministry of Health.
Contributor Information
Derek Gatherer, Email: d.gatherer@lancaster.ac.uk.
Irene L. G. Newton, Indiana University, Bloomington
REFERENCES
- 1.Weller MG. 2021. The protocol gap. Methods Protoc 4:12. doi: 10.3390/mps4010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mölder F, Jablonski KP, Letcher B, Hall MB, Tomkins-Tinch CH, Sochat V, Forster J, Lee S, Twardziok SO, Kanitz A, Wilm A, Holtgrewe M, Rahmann S, Nahnsen S, Köster J. 2021. Sustainable data analysis with Snakemake. F1000Res 10:33. doi: 10.12688/f1000research.29032.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chanmol W, Jariyapan N, Somboon P, Bates MD, Bates PA. 2019. Axenic amastigote cultivation and in vitro development of Leishmania orientalis. Parasitol Res 118:1885–1897. doi: 10.1007/s00436-019-06311-z. [DOI] [PubMed] [Google Scholar]
- 4.Kolmogorov M, Yuan J, Lin Y, Pevzner PA. 2019. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol 37:540–546. doi: 10.1038/s41587-019-0072-8. [DOI] [PubMed] [Google Scholar]
- 5.Li H. 2016. Minimap and miniasm: fast mapping and de novo assembly for noisy long sequences. Bioinformatics 32:2103–2110. doi: 10.1093/bioinformatics/btw152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, Li H. 2021. Twelve years of SAMtools and BCFtools. Gigascience 10:giab008. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alonge M, Soyk S, Ramakrishnan S, Wang X, Goodwin S, Sedlazeck FJ, Lippman ZB, Schatz MC. 2019. RaGOO: fast and accurate reference-guided scaffolding of draft genomes. Genome Biol 20:224. doi: 10.1186/s13059-019-1829-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palmer J, Stajich J. 2019. nextgenusfs/funannotate: funannotate v1.5.3. Zenodo doi: 10.5281/zenodo.2604804. [DOI] [Google Scholar]
- 10.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.NCBI. The UniVec database. https://www.ncbi.nlm.nih.gov/tools/vecscreen/univec. Accessed 14 April 2021.
- 13.Quinlan AR. 2014. BEDTools: the Swiss-Army tool for genome feature analysis. Curr Protoc Bioinformatics 47:11.12.1–11.12.34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dainat J, Hereñú D, Pucholt P. 2020. NBISweden/AGAT: AGAT-v0.7.0. Zenodo doi: 10.5281/zenodo.5036996. [DOI] [Google Scholar]
- 15.Flynn JM, Hubley R, Goubert C, Rosen J, Clark AG, Feschotte C, Smit AF. 2020. RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci U S A 117:9451–9457. doi: 10.1073/pnas.1921046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abrusan G, Grundmann N, DeMester L, Makalowski W. 2009. TEclass: a tool for automated classification of unknown eukaryotic transposable elements. Bioinformatics 25:1329–1330. doi: 10.1093/bioinformatics/btp084. [DOI] [PubMed] [Google Scholar]
- 17.Almutairi H. 2021. hatimalmutairi/teclass-2.1.3b. https://hub.docker.com/r/hatimalmutairi/teclass-2.1.3b.
- 18.Smit A, Hubley R, Glusma G. 2021. RepeatMasker. http://www.repeatmasker.org.
- 19.Aslett M, Aurrecoechea C, Berriman M, Brestelli J, Brunk BP, Carrington M, Depledge DP, Fischer S, Gajria B, Gao X, Gardner MJ, Gingle A, Grant G, Harb OS, Heiges M, Hertz-Fowler C, Houston R, Innamorato F, Iodice J, Kissinger JC, Kraemer E, Li W, Logan FJ, Miller JA, Mitra S, Myler PJ, Nayak V, Pennington C, Phan I, Pinney DF, Ramasamy G, Rogers MB, Roos DS, Ross C, Sivam D, Smith DF, Srinivasamoorthy G, Stoeckert CJ, Jr, Subramanian S, Thibodeau R, Tivey A, Treatman C, Velarde G, Wang H. 2010. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res 38:D457–D462. doi: 10.1093/nar/gkp851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holt C, Yandell M. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12:491. doi: 10.1186/1471-2105-12-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Almutairi H. 2021. hatimalmutairi/lmgaap-maker. https://hub.docker.com/r/hatimalmutairi/lmgaap-maker.
- 22.Gremme G, Steinbiss S, Kurtz S. 2013. GenomeTools: a comprehensive software library for efficient processing of structured genome annotations. IEEE/ACM Trans Comput Biol Bioinform 10:645–656. doi: 10.1109/TCBB.2013.68. [DOI] [PubMed] [Google Scholar]
- 23.Genome Assembly and Annotation Service. 2021. Genome Assembly and Annotation Service code. https://github.com/NBISweden/GAAS.
- 24.Hoff KJ, Stanke M. 2019. Predicting genes in single genomes with AUGUSTUS. Curr Protoc Bioinformatics 65:e57. doi: 10.1002/cpbi.57. [DOI] [PubMed] [Google Scholar]
- 25.UniProt Consortium. 2021. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res 49:D480–D489. doi: 10.1093/nar/gkaa1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, McWilliam H, Maslen J, Mitchell A, Nuka G, Pesseat S, Quinn AF, Sangrador-Vegas A, Scheremetjew M, Yong SY, Lopez R, Hunter S. 2014. InterProScan 5: genome-scale protein function classification. Bioinformatics 30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Genomes assembled using this protocol are available in the NCBI Assembly database with the following accession numbers: L. martiniquensis, GCA_017916325.1; L. orientalis, GCA_017916335.1; L. enriettii, GCA_017916305.1; Leishmania sp. Ghana, GCA_017918215.1; Leishmania sp. Namibia, GCA_017918225.1; and Porcisia hertigi, GCA_017918235.1. Raw sequencing data are available with the following NCBI BioProject accession numbers: L. martiniquensis, PRJNA691531; L. orientalis, PRJNA691532; L. enriettii, PRJNA691534; Leishmania sp. Ghana, PRJNA691536; Leishmania sp. Namibia, PRJNA689706; and Porcisia hertigi, PRJNA691541. The workflow is available at GitHub (https://github.com/hatimalmutairi/LGAAP) and Zenodo (https://doi.org/10.5281/zenodo.4663265).